Abstract
The genes involved in isoprene (2-methyl-1,3-butadiene) utilization in Rhodococcus sp. strain AD45 were cloned and characterized. Sequence analysis of an 8.5-kb DNA fragment showed the presence of 10 genes of which 2 encoded enzymes which were previously found to be involved in isoprene degradation: a glutathione S-transferase with activity towards 1,2-epoxy-2-methyl-3-butene (isoI) and a 1-hydroxy-2-glutathionyl-2-methyl-3-butene dehydrogenase (isoH). Furthermore, a gene encoding a second glutathione S-transferase was identified (isoJ). The isoJ gene was overexpressed in Escherichia coli and was found to have activity with 1-chloro-2,4-dinitrobenzene and 3,4-dichloro-1-nitrobenzene but not with 1,2-epoxy-2-methyl-3-butene. Downstream of isoJ, six genes (isoABCDEF) were found; these genes encoded a putative alkene monooxygenase that showed high similarity to components of the alkene monooxygenase from Xanthobacter sp. strain Py2 and other multicomponent monooxygenases. The deduced amino acid sequence encoded by an additional gene (isoG) showed significant similarity with that of α-methylacyl-coenzyme A racemase. The results are in agreement with a catabolic route for isoprene involving epoxidation by a monooxygenase, conjugation to glutathione, and oxidation of the hydroxyl group to a carboxylate. Metabolism may proceed by fatty acid oxidation after removal of glutathione by a still-unknown mechanism.
2-Methyl-1,3-butadiene (isoprene) is a volatile compound that is emitted in large quantities from plants, especially under thermal stress conditions. The annual global emission from plants is estimated to be 500 million tons, which is similar to the global emission of methane (35). In addition, isoprene emission from several bacteria has been detected (50). Isoprene plays an important role in atmospheric chemistry since it is involved in the generation of ozone and carbon monoxide (42). Despite this, little work on the microbial degradation of isoprene has been done. Cleveland and Yavitt (8) showed that microorganisms consume isoprene even when it is present at trace level concentrations and that soil microorganisms may provide a significant biological sink for atmospheric isoprene. Information about the physiology of isoprene degradation is scarce, however. van Ginkel et al. (44) have shown that degradation in a Nocardia sp. starts with a monooxygenase which oxidizes isoprene to 1,2-epoxy-2-methyl-3-butene and 1,2-3,4-diepoxybutane. Ewers and Knackmuss (13) reported the presence of a glutathione-dependent activity towards 1,2-epoxy-2-methyl-3-butene in cell extracts of an isoprene-utilizing Rhodococcus sp., but the enzyme catalyzing this reaction and the products formed were not further characterized. Moreover, no work on the genetics of isoprene metabolism has been published.
In our laboratory, Rhodococcus sp. strain AD45 was isolated for its capability to use isoprene as the sole source of carbon and energy. The pathway of isoprene degradation in strain AD45 starts with oxidation by a monooxygenase to yield 1,2-epoxy-2-methyl-3-butene (45). A glutathione S-transferase catalyzes the nucleophilic attack of glutathione at the sterically most-hindered carbon atom. The reaction product, 1-hydroxy-2-glutathionyl-2-methyl-3-butene (HGMB), is then oxidized in two consecutive steps to 2-glutathionyl-2-methyl-3-butenoic acid (GMBA) by a NAD+-dependent dehydrogenase that is highly specific for HGMB. The glutathione S-transferase and the dehydrogenase have been purified to homogeneity and characterized (46).
Strain AD45 is also capable of oxidizing chlorinated ethenes to the corresponding chlorinated epoxyethanes (45). These epoxides and their decomposition products cause the toxicity that is associated with the oxidative cometabolic degradation of chlorinated ethenes in other organisms (47). Since this toxicity is the main limiting factor for the application of monooxygenase-expressing organisms for dichloroethene removal, it is desirable to obtain insight in the pathways that lead to biological detoxification of these reactive transformation products. The glutathione S-transferase of strain AD45 converts cis-1,2-dichloroepoxyethane without the formation of toxic chlorinated reaction products and thus may effectively detoxify chlorinated ethene epoxides (45).
Currently, no information on the genetics of isoprene metabolism is available, and the metabolic pathway is still not completely known. In the present study we report the complete nucleotide sequence and polypeptide analysis of an 8,456-bp region encoding the putative isoprene monooxygenase, the glutathione S-transferase with activity towards epoxides, a second glutathione S-transferase, the HGMB-specific dehydrogenase, and a putative racemase. Furthermore, we have overexpressed both glutathione S-transferases, and data on their activities with 1,2-epoxy-2-methyl-3-butene, 1,2-dichloro-4-nitrobenzene (DCNB), and 1-chloro-2,4-dinitrobenzene (CDNB) are reported. The implications for the catabolic pathway of isoprene will be discussed.
MATERIALS AND METHODS
Bacterial strains and media.
Rhodococcus sp. strain AD45 was grown on isoprene in batch culture or continuous culture using a mineral medium supplemented with 20 mg of yeast extract liter−1 as described before (45) or maintained at 30°C on 0.8% nutrient broth agar plates. For the isolation of genomic DNA, the organism was cultivated in 0.8% nutrient broth medium. Escherichia coli strains HB101, JM101, and TOP10F′ (Invitrogen, Carlsbad, Calif.), which were used for cloning purposes, were grown at 37°C in Luria-Bertani medium. For plasmid selection the appropriate antibiotic was added at the following concentrations: 50 μg ml−1 for kanamycin, 50 μg ml−1 for ampicillin, and 12.5 μg ml−1 for tetracycline. E. coli BL21(DE3) grown at 17°C was used for high-level expression of both glutathione S-transferases of strain AD45.
DNA preparation and manipulation.
Genomic DNA of strain AD45 was isolated from an exponentially growing nutrient broth culture (100 ml) as described before (45).
Small-scale preparation of plasmid DNA (<10 kb) from 1.5-ml cultures of E. coli was performed using the High Pure plasmid isolation kit (Boehringer, Mannheim, Germany) according to the recommendations of the manufacturer. Small-scale isolation of DNA of large plasmids (>10 kb) was performed using the alkaline lysis method (31) with two modifications. The lysozyme step was omitted, and ethanol instead of isopropanol was used for DNA precipitation.
Large-scale preparation of plasmid DNA from 100-ml cultures of E. coli was performed using the Plasmid Midi kit (Qiagen, Hilden, Germany) according to the recommendations of the manufacturer. DNA of pLAFR3 used for the construction of the gene library was isolated by the alkaline lysis method and purified by ultracentrifugation using a CsCl gradient as described by Sambrook et al. (31).
PCR products were purified using a Qiaquick PCR purification kit (Qiagen). DNA restriction, ligations, and dephosphorylations were performed according to protocols described by Sambrook et al. (31).
CNBr cleavage and N-terminal sequencing.
Two milligrams of purified glutathione S-transferase (46) was digested with CNBr as described before (19). Peptides were separated by reversed-phase high-pressure liquid chromatography (19), and the N-terminal amino acid sequence of one of the peptides was determined as described before (46). The following sequence was obtained: RVGLDAFRARILDGFNGQG(HoSer). The carboxy-terminal residue was tentatively identified as a homoserine (HoSer), which is the expected product of methionine after CNBr cleavage.
PCR amplification and overexpression of the isoI and isoJ genes.
Degenerate primers for amplification of fragments of the glutathione S-transferase gene were designed on the basis of peptide sequences of the purified glutathione S-transferase (46). Forward primer PGT1, 5′-AAACCATGGT(T/A/C)ACZGTZTAXCGZTAXGTZCCZGCZTGG-3′ (X = C or T; Y = G or A; Z = G, A, C, or T), was based on the N-terminal amino acid sequence of the protein (MITVYGYVPAWGIPDISPYVTKVXNYXTFTGI). Reverse primer PGT3, 5′-AAACCATGGCATZCCXTGZCCYTTYAAZCCYTCZAG(A/G/T)AT-3′, was based on the N-terminal amino acid sequence of a peptide isolated after CNBr cleavage (see above). For overexpression of the isoI and isoJ genes the plasmids pGEFIsoI and pGEFIsoJ were constructed. In these plasmids the isoI and isoJ genes were cloned in the NcoI site behind the T7 promoter in the expression vector pGEF+ (32). The isoI gene was amplified with forward primer PFI, 5′-AACATACCATGGTCACCGTTTA-3′, and reverse primer PRI, 5′-ATGCTCCAAGATCCATGGTTTCGGACT-3′ (NcoI sites are underlined; the start codon is in boldface; substituted nucleotides are in italics). The isoJ gene was amplified with forward primer PFJ, 5′-ATCGTTCCATGGTTGACTTCTA-3′, and reverse primer PRJ, 5′-AAGAATCGGCCATGGCGCTCACTTTTGACA-3′. The isoABCDEF operon was amplified with forward primer PFM2, 5′-TGCTATTGAACAGGGACGATTGGTACGACA-3′, and reverse primer PRM2, 5′-GATGGGTCCTTCCGGGATTCGGGGGTAGTTC-3′. The genes were cloned behind the araBAD promoter in the expression vector pBAD-TOPO (Invitrogen), and protein expression was induced according to the recommendations of the manufacturer.
PCR amplification was performed as described by Innis and Gelfand (18). Each 50-μl reaction mixture contained 5 μl of 10× DNA polymerase buffer, 50 μM deoxynucleoside triphosphates, 1 to 2 U of GoldStar DNA polymerase (Eurogentec, Seraing, Belgium) or Pwo DNA polymerase (Boehringer Mannheim B.V., Almere, The Netherlands), 20 pmol of each primer (synthesized by Eurosequence BV, Groningen, The Netherlands), and 10 to 100 ng of template DNA. Amplification of the ISO1 fragment with PGT1 and PGT3 primers was performed by denaturation at 94°C for 5 min followed by “touchdown” PCR that consisted of 14 cycles of denaturation at 94°C for 1 min, hybridization starting at 60°C for 1 min (2°C decrease every second cycle to a touchdown at 48°C), and elongation at 72°C for 1 min (11). Amplification was completed by 25 additional cycles during which the hybridization temperature was 48°C. Amplification of the complete isoI and isoJ genes was performed by 30 cycles of denaturation at 94°C for 1 min, hybridization at 55°C for 1 min, and elongation at 72°C for 1 min.
A partial purification of IsoJ from E. coli BL21(DE3) was carried out by anion exchange chromatography as described by van Hylckama Vlieg et al. (46).
Preparation of the gene library.
Sixty micrograms of genomic DNA of strain AD45 was partially digested with Sau3A to yield an average fragment size of approximately 23 kb. The digest was dephosphorylated and ligated into the BamHI site of the pLAFR3 cosmid cloning vector (39). The ligation mixture was packaged with the DNA packaging kit (Boehringer, Mannheim, Germany) and transduced to E. coli HB101 according to the recommendations of the manufacturer.
Colonies of the gene library were tested for monooxygenase activity by the indole assay as described by O'Connor et al. (27) or by spraying colonies with 10 mM indole in diethylether. As a control, isoprene-grown colonies of Rhodococcus sp. strain AD45 were used.
DNA-DNA hybridization experiments.
For Southern blotting experiments, DNA was digested with the appropriate restriction enzyme and separated by agarose gel electrophoresis. The gel was incubated in 0.25 M HCl to nick the DNA before it was transferred to a nylon membrane (Boehringer) by the capillary flow method (31). The DNA was fixed to the membrane by illumination with UV radiation for 3 min.
For colony blotting, colonies grown overnight were transferred to a nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany). After the membrane was dried in air, it was incubated for 5 min in four consecutive steps on Whatman no. 1 filters that were saturated with 10% sodium dodecyl sulfate (SDS), 0.5 N NaOH with 1.5 M NaCl, 1 M Tris-HCl with 1.5 M NaCl (pH 7.5), or 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The DNA was fixed to the membrane by incubation at 80°C for 30 min.
DNA probes were prepared by incorporation of digoxigenin-dUTP with the DIG DNA-labeling kit (Boehringer). The membrane carrying the DNA was prehybridized by incubation at 68°C for 1 h with 5× SSC containing 0.1% N-lauroylsarcosine, 0.02% SDS, and 1% blocking agent (Boehringer). Hybridization was performed by overnight incubation at 68°C in this buffer to which 20 ng of denatured probe ml−1 was added, and subsequently the membrane was washed twice in 0.1× SSC containing 0.1% SDS at 68°C. Detection of the hybridized probe was done with anti-digoxigenin-alkaline phosphatase Fab fragments (Boehringer) following the manufacturer's recommendations.
DNA sequencing and analysis.
Cycle sequencing (26) was performed on double-stranded DNA using the Thermo Sequenase cycle sequencing kit (Amersham BV, Roosendaal, The Netherlands) with 7-deaza-dGTP and 5′Cy5 fluorescent primers. Sequence reactions were run on the ALF-Express automatic sequencing machine (Pharmacia, Uppsala, Sweden) at the BioMedical Technology Centre (Academic Hospital, Groningen, The Netherlands).
The BLAST program (3) was used to search DDBJ, EMBL, and GenBank databases for proteins that had sequence similarity. Multiple sequence alignments were made with ClustalW, version 1.7 (43) using a gap penalty of 6 and a gap opening penalty of 20. Other parameters were default, and, if necessary, minor adjustments were made manually. Pairwise similarities were calculated with the ALIGN program, which uses the BLOSSUM 50 matrix (17).
Enzyme assays.
Glutathione S-transferase activity with epoxides was determined with online gas chromatographic monitoring of substrate depletion as described previously (46). Enantiomers of 1,2-epoxy-2-methyl-3-butene were separated with chiral gas chromatography as described by Wistuba et al. (51). Separation of enantiomers of other chiral compounds was carried out as described before (22). Activities towards DCNB and CDNB were assayed at 30°C in a 50 mM Tris-HCl buffer containing 1 mM substrate (16). Activities were determined at pH 7.0 or 8.5 with CDNB and at pH 7.0 with DCNB since non-enzyme-catalyzed reaction rates were too high for an accurate assay of specific activities at pH 8.0. Substrate was added from a 20 mM stock solution in ethanol. The formation of glutathione conjugates was assayed by monitoring the increase of the absorbancy at 340 nm in the presence of 2 mM glutathione for DCNB or at 345 nm in the presence of 10 mM glutathione for CDNB. Activities were calculated using extinction coefficients of 8.5 and 9.6 mM−1 cm−1 for DCNB and CDNB conjugates, respectively (16). Activities were corrected for nonenzymatic reaction rates and for background glutathione S-transferase activity of E. coli using extracts of E. coli BL21(DE3)pGEF+.
Nucleotide sequence accession number.
The nucleotide sequence described in this article was deposited at the EMBL, DDBJ, and GenBank databases under accession no. AJ249207.
RESULTS AND DISCUSSION
Cloning of the genes involved in isoprene metabolism.
In order to clone a fragment of the glutathione S-transferase of Rhodococcus sp. strain AD45 (46) the degenerate primers PGT1 and PGT3 were designed on the basis of the N terminus and an internal peptide obtained after CNBr digestion. With these primers a 471-bp fragment, designated ISO1, was amplified from genomic DNA of strain AD45. The PCR product was cloned and sequenced, and comparison of the deduced peptide sequence with the N-terminal sequence of the protein and with the sequence of the isolated peptide confirmed that the PCR product represented part of the gene encoding IsoI.
A gene library of Rhodococcus sp. strain AD45 in the cosmid pLAFR3 was constructed. Approximately 6,000 independent clones were obtained when the library was transduced to E. coli HB101. An analysis of 28 clones showed that 12 contained plasmids with inserts. The library was screened for the presence of DNA that hybridized with the ISO1 probe by colony blotting. Out of 1,200 insert-containing recombinants tested, three clones that contained plasmids, designated pHL1, pHL8, and pHL9, that hybridized with the ISO1 probe were identified. The clones did not utilize isoprene as a growth substrate. In extracts of cells grown on Luria-Bertani medium no activity towards 1,2-epoxy-2-methyl-3-butene could be detected, indicating that the glutathione S-transferase was not functionally expressed. The gene library was also screened for the presence of monooxygenase activity by the indole assay, but no active clones were identified, even when plates were incubated overnight with 0.1 mM isoprene in the gas phase for induction.
Nucleotide sequence analysis.
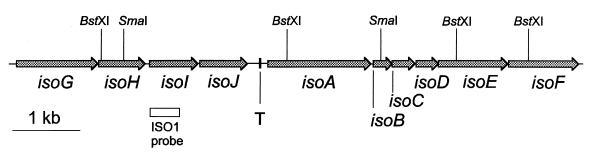
Starting from the ISO1 sequence, an 8,456-bp fragment was sequenced from plasmids pHL8 and pHL9 (Fig. 1). The overall G+C content of the sequenced fragment was 58.9%, which is low compared to the high G+C content typically found for rhodococci, 67 to 72% (14). A sequence similarity search with the BLAST program in various protein and nucleotide databases helped to identify 10 open reading frames (ORFs) that all encoded (putative) proteins that showed similarity with entries in the databases (Fig. 1; Table 1). All ORFs were encoded on the same strand and were preceded by plausible ribosome binding sites. One ORF (isoH) starts with a GTG codon, which frequently occurs in Rhodococcus mRNA translation (21). A G+C-rich region of almost perfect dyad symmetry (nucleotides 3685 to 3704), followed by an A+T-rich region, is located downstream of isoJ (Fig. 1). This structure is characteristic of a rho-independent transcription terminator (29).
FIG. 1.
Schematic representation of the 8,456-bp DNA region of Rhodococcus sp. strain AD45 carrying the isoABCDEFGHIJ ORFs and a putative rho-independent transcription terminator (T).
TABLE 1.
Genes and gene products of the iso locus and homologies to proteins in the databases
| ORF | Position in sequence (nucleotides) | Gene product
|
Similar gene product | Organism | Identity (%)a | Accession no. | |
|---|---|---|---|---|---|---|---|
| Calculated molecular mass (Da) | Identification | ||||||
| isoG | 57–1274 | 43,450 | Putative racemase | YfdE | Escherichia coli | 31.6 | AE000325 |
| Mcr | Mus musculus | 30.5 | AAB72146 | ||||
| Fct | Oxalobacter formigenes | 26.4 | AAC45298 | ||||
| CaiB | Escherichia coli | 24.6 | CAA47971 | ||||
| isoH | 1291–1971 | 24,033 | NAD-dependent HGMB-dehydrogenase | CsgA | Myxococcus xanthus | 33.8 | 1587000 |
| XpIII | Xanthobacter sp. strain Py2 | 24.2 | CAA56244 | ||||
| 2bhd | Streptomyces exfoliatus | 22.2 | 1611236A | ||||
| XpIV | Xanthobacter sp. strain Py2 | 22.0 | CAA56245 | ||||
| LigD | Sphingomonas paucimobilis SYK-6 | 18.6 | BAA02030 | ||||
| isoI | 2038–2754 | 27,096 | Glutathione S-transferase active with epoxides | Faxb | Drosophila melanogaster | 27.0 | AAB07806 |
| LigF | Sphingomonas paucimobilis SYK-6 | 24.3 | BAA02031 | ||||
| IsoJ | Rhodococcus sp. strain AD45 | 23.8 | AJ249207 | ||||
| ORF3 | Burkholderia cepacia AC1100 | 22.3 | AAC43335 | ||||
| PcpC | Flavobacterium sp. strain ATCC 39723 | 21.1 | Q03520 | ||||
| GSTB1-1 | Proteus mirabilis | 20.5 | P15214 | ||||
| ORF4 | Burkholderia cepacia AC1100 | 18.5 | AAC43336 | ||||
| isoJ | 2789–3490 | 26,342 | Glutathione S-transferase active with CDNB and DCNB | YFCG | Escherichia coli | 43.8 | P39100 |
| ORF3 | Burkholderia cepacia AC1100 | 41.9 | AAC43335 | ||||
| LigF | Sphingomonas paucimobilis SYK-6 | 27.9 | BAA02031 | ||||
| PcpC | Flavobacterium sp. strain ATCC 39723 | 24.5 | Q03520 | ||||
| GSTB1-1 | Proteus mirabilis | 24.3 | P15214 | ||||
| IsoI | Rhodococcus sp. strain AD45 | 23.8 | AJ249207 | ||||
| isoA | 3796–5340 | 58,461 | Oxygenase α-subunit | XamoA | Xanthobacter sp. strain Py2 | 67.9 | AJ012090 |
| TouA | Pseudomonas stutzeri OX1 | 49.0 | AJ005663 | ||||
| BmoA | Pseudomonas aeruginosa JI104 | 48.4 | D83068 | ||||
| TmoA | Pseudomonas mendocina KR1 | 46.6 | M65106 | ||||
| TbuA1 | Ralstonia pickettii PK01 | 46.6 | U04052 | ||||
| MemA | Methylococcus capsulatus (Bath) | 23.4 | P22869 | ||||
| AmoC | Rhodococcus rhodochrous B-276 | 23.3 | D37875 | ||||
| isoB | 5376–5660 | 10,331 | Oxygenase γ-subunit | XamoB | Xanthobacter sp. strain Py2 | 55.3 | AJ012090 |
| TouB | Pseudomonas stutzeri OX1 | 37.9 | AJ005663 | ||||
| TbuU | Ralstonia pickettii PK01 | 34.7 | U04052 | ||||
| TmoB | Pseudomonas mendocina KR1 | 32.6 | M65106 | ||||
| BmoB | Pseudomonas aeruginosa JI104 | 30.2 | D83068 | ||||
| isoC | 5653–5997 | 12,772 | Ferredoxin | BmoC | Pseudomonas aeruginosa JI104 | 44.4 | D83068 |
| XamoC | Xanthobacter sp. strain Py2 | 43.4 | AJ012090 | ||||
| TmoC | Pseudomonas mendocina KR1 | 41.2 | M65106 | ||||
| TbuB | Ralstonia pickettii PK01 | 40.2 | U04052 | ||||
| isoD | 6016–6348 | 12,380 | Effector or coupling protein | XamoD | Xanthobacter sp. strain Py2 | 51.8 | AJ012090 |
| TbuV | Ralstonia pickettii PK01b | 38.4 | U04052 | ||||
| TmoD | Pseudomonas mendocina KR1 | 35.5 | M65106 | ||||
| TouD | Pseudomonas stutzeri OX1 | 33.3 | AJ005663 | ||||
| isoE | 6345–7373 | 38,510 | Oxygenase β-subunit | XamoE | Xanthobacter sp. strain Py2 | 52.5 | AJ012090 |
| TbuA2 | Ralstonia pickettii PK01 | 40.2 | U04052 | ||||
| TouE | Pseudomonas stutzeri OX1 | 38.1 | AJ005663 | ||||
| TmoE | Pseudomonas mendocina KR1 | 34.5 | M65106 | ||||
| isoF | 7387–8424 | 37,322 | Reductase | XamoF | Xanthobacter sp. strain Py2 | 38.8 | AJ012090 |
| TbuC | Ralstonia pickettii PK01 | 36.5 | U04052 | ||||
| TouF | Pseudomonas stutzeri OX1 | 34.4 | AJ005663 | ||||
| AmoD | Rhodococcus rhodochrous B-276 | 32.7 | D37875 | ||||
| MemC | Methylococcus capsulatus (Bath) | 29.9 | P22869 | ||||
| TmoF | Pseudomonas mendocina KR1 | 29.1 | M65106 | ||||
The percentage of similarity was calculated by pairwise alignment using the program ALIGN with default parameterization.
Only residues 105 to 340 of the failed axon connection protein were included in calculation of sequence identity.
The ORF that encodes the glutathione S-transferase with activity for epoxides was designated isoI. The 471-bp ISO1 sequence is identical to that of a fragment of the isoI gene, and the molecular mass of the isoI-encoded deduced protein (Table 1) is in close agreement with the experimentally determined value of 27 kDa (46). Downstream of the isoI gene an ORF designated isoJ was found (Table 1).
Upstream of isoI an ORF designated isoH was found, and it was concluded that this ORF encodes the dehydrogenase with activity for HGMB, since the deduced amino acid sequence of residues 2 to 23 is identical to the N-terminal sequence of the dehydrogenase purified from strain AD45 (46). This indicates that in the mature protein the initiator methionine residue is cleaved off.
Upstream of isoH an ORF designated isoG was found. A few ATG codons were available as starting points of translation. The start codon that results in the deduced protein that aligns best with similar proteins and that is preceded by a plausible ribosome binding site is included in Table 1.
A gene cluster of six closely spaced ORFs was found downstream of isoJ. The ORFs were designated isoABCDEF on the basis of the high sequence similarity with the tmoABCDEF gene cluster encoding the toluene 4-monooxygenase of Pseudomonas mendocina KR1 and the recently published aamABCDEF alkene monooxygenase gene cluster of Xanthobacter sp. strain Py2 (52–55). The first 7 bp of isoC overlapped with the 3′ end of isoB, and the TGA stop codon of isoD overlapped with the presumed ATG start codon of isoE.
The flanking regions of the 8,456-bp fragment were further analyzed by single-strand sequencing. Upstream of isoG, sequencing a 912-bp fragment revealed the presence of an ORF similar to those encoding γ-glutamylcysteine synthetases, proteins that catalyze the first step in glutathione biosynthesis. Sequence analysis of a 1,892-bp fragment downstream of isoABCDEF showed the presence of an ORF similar to those encoding aldehyde dehydrogenases and an ORF similar to those encoding glutathione synthetases. Glutathione synthetases couple glycine to γ-glutamylcysteine yielding glutathione. Hence, on a 11,260-bp DNA fragment in strain AD45, the isoABCDEFGHIJ genes, a gene encoding a putative aldehyde dehydrogenase, and two genes required for glutathione biosynthesis are present.
Copy number of the isoI gene.
In order to determine the copy number of isoI in Rhodococcus sp. strain AD45, total DNA was digested with BamHI, BstXI, EcoRI, SacI, SalI, or SmaI, separated on an agarose gel, and hybridized with the ISO1 probe. Only one band hybridized with all six restriction enzymes that were tested. For two of the enzymes tested, BstXI and SmaI, the sites flank the ISO1 sequence in the 8,456-bp genomic fragment. With these enzymes hybridizing fragments of 3 and 3.5 kb, respectively, were observed, which is in agreement with the sequence data (Fig. 1). The largest hybridizing fragment, a 10-kb fragment, was observed when the DNA was digested with SacI. These data suggest that only one copy of the isoI gene is present in Rhodococcus sp. strain AD45, although the possibility that there are more copies of a large highly conserved DNA fragment (>10 kb) cannot be excluded.
Similarity of isoI and isoJ gene products with glutathione S-transferases.
A sequence similarity search with the isoI and isoJ gene products showed that both proteins are similar to each other and to other glutathione S-transferases (Table 1). The presence of two (putative) glutathione S-transferase genes has also been observed in the 2,4,5-trichlorophenoxyacetic acid-degrading Burkholderia cepacia strain AC1100 (9) and in Sphingomonas paucimobilis strain SYK-6, which grows on lignin (23, 24). Mutagenesis and complementation studies have shown that these genes are not essential for metabolism of 2,4,5-trichlorophenoxyacetic acid, and no physiological function could be assigned to these genes (9).
Similarities of IsoJ to glutathione S-transferases in the databases were much higher than similarities of IsoI to glutathione S-transferases (Table 1). Proteins IsoI and IsoJ both displayed the highest degree of sequence similarity to other glutathione S-transferases in a region located in the N-terminal domain. This domain is involved in glutathione binding (4, 10, 48). Typically, glutathione S-transferases contain one or more tyrosine residues at the N terminus. These residues are also present in IsoI and IsoJ. In some glutathione S-transferases the N-terminal tyrosine decreases the pKa of the glutathione thiol and thus enhances its nucleophilicity (4). In class theta glutathione S-transferases, to which many bacterial enzymes belong, this role is performed by a serine that is located in the N-terminal part of the protein, whereas the tyrosine residues are not essential for catalysis (6, 49). Mutagenesis studies may reveal whether serine residues also play a critical role in glutathione conjugation catalyzed by IsoI or IsoJ.
Two bacterial glutathione S-transferases, FosA (accession no. M85195) and FosB (accession no. Q03377), that convert epoxides have been described. The proteins are involved in bacterial resistance to the antibiotic fosfomycin (1,2-epoxypropylphosphonic acid); they catalyze the conjugation of glutathione to the secondary carbon atom in the epoxide ring (5). Despite the analogy of the reaction catalyzed, IsoI has little sequence similarity to FosA and FosB, which is in agreement with the observation that IsoI does not require metal ions for activity (46).
Sequence similarity of IsoH with SDRs.
A sequence similarity search with IsoH, which is the HGMB dehydrogenase, in various protein and DNA databases showed that this protein is similar to proteins of the short-chain dehydrogenase/reductase (SDR) family (20) including the XpIII and XpIV proteins of the epoxide carboxylase system of Xanthobacter sp. strain Py2 (2). Multiple sequence alignments with representative and well-studied members (accession no. P25529, 226912, and P00334) of this protein family showed that IsoH contains most of the highly conserved and functionally or structurally important residues (20, 41), which suggests that it has the same topology. The SDR protein family is a large group of enzymes that catalyze the oxidation or reduction of a broad range of substrates including primary alcohols, steroids, and sugars (20). They use NAD(H) or NADP(H) as a coenzyme, do not require metal ions for activity, are approximately 250 residues in length, and are active as monomers, dimers, or tetramers. Several proteins of this family have been subjected to detailed biochemical and structural studies.
The N-terminal parts of SDR proteins typically contain the Rossman fold for cofactor binding. It is characterized by a GlyXXXGlyXGly motif. In IsoH the first two residues are present as Gly-12 and Gly-15, whereas an alanine residue is located at position 17. SDR proteins contain three highly conserved residues, Ser, Tyr, and Lys, that play a critical role in catalysis (20, 41). These residues are also conserved in IsoH as Ser-132, Tyr-144, and Lys-147. The C-terminal domains of SDR proteins are involved in substrate binding. They are highly variable, which may reflect the adaptation to diverse substrates. Since IsoH catalyzes the oxidation of a glutathione conjugate, it may be expected that substrate binding involves specific interactions with the glutathione moiety of the substrate. Therefore, we looked for similarity with other glutathione binding enzymes, but no glutathione binding site could be identified.
Similarity of isoABCDEF gene products to monooxygenases.
The putative isoABCDEF gene products all have a high degree of similarity with peptides of the four-component monooxygenases which are involved in the oxidation of alkenes and aromatic compounds (Table 1). Moreover, the gene orders in the similar clusters are identical. For all gene products except IsoD the highest sequence identity was observed with the alkene monooxygenase of the propene utilizer Xanthobacter sp. strain Py2, whose sequence was recently published by Zhou et al. (55). In fact, many of the features of the isoprene monooxygenase gene cluster and the deduced proteins are very similar to those described for the alkene monooxygenase of strain Py2. Lower similarity was observed with polypeptides of three-component monooxygenases, such as methane monooxygenases of Methylococcus capsulatus (Bath) (38) and the alkene monooxygenase of Rhodococcus rhodochrous (previously Nocardia corallina) B-276 (30).
Detailed biochemical and biophysical analysis of toluene 4-monooxygenase of P. mendocina KR1 and alkene monooxygenase of Xanthobacter sp. strain Py2 (28, 37) has shown that these proteins consist of four dissociable components. This suggests the following model for isoprene monooxygenase of Rhodococcus sp. strain AD45. Oxidation of the substrate occurs at a di-iron (αβγ)2 oxygenase component consisting of the proteins encoded by isoA, isoB, and isoE. Electrons needed for O2 activation are provided by an NADH reductase, which is a di-iron disulfur flavoprotein encoded by the isoF gene. A Rieske-type ferredoxin, encoded by isoC, mediates electron transfer from the reductase to the oxygenase. The fourth component is a regulatory or effector protein, encoded by isoD, that may influence reaction rates or product distribution.
The isoprene monooxygenase of Rhodococcus sp. strain AD45 is induced in the presence of isoprene and catalyzes the oxidation of aliphatic and aromatic alkenes to the corresponding epoxides. Oxidation of indole by the isoprene monooxygenase in these cells resulted in the rapid formation of indigo. With several alkenes the enzyme produced predominantly one enantiomer of the corresponding epoxides such as (R)-1,2-epoxy-2-methyl-3-butene (enantiomeric excess = 95%), (R)-styrene oxide (87%), and (R)-p-chlorostyrene (59%), indicating that the enzyme may be useful for the biocatalytic production of optically active epoxides. Cells of strain AD45 grown on nutrient broth did not oxidize indole, which suggests that the isoprene monooxygenase is responsible for indigo formation.
The E. coli HB101 clones containing plasmid pHL8 or pHL9, both of which carry the isoABCDEF gene cluster, were not active with isoprene or indole (see above). From this we concluded that the isoprene monooxygenase is not functionally expressed in E. coli. Similarly, the alkene monooxygenase gene cluster of Xanthobacter sp. strain Py2 is also not expressed in E. coli (54, 55). When the plasmids pHL8 and pHL9 were transferred by triparental mating to Xanthobacter, Pseudomonas, or Burkholderia spp., the resulting transconjugants were not active with indole or isoprene, indicating that transcription or translation signals were also not recognized in these organisms. To further investigate the possibility of functional expression of the isoprene monooxygenase, the isoABCDEF gene cluster was cloned behind the araBAD promoter in the expression vector pBAD-TOPO. No active protein was produced, probably because one or several of the ribosome binding sites preceding the isoBCDEF, which significantly deviate from the E. coli consensus sequence, are poorly recognized in E. coli or because of improper protein folding. Complementation of mutants of strain AD45 defective in monooxygenase production may be necessary to obtain final proof that the isoABCDEF gene cluster encodes the isoprene monooxygenase.
Similarity searches with IsoG.
Database searches with the isoG-encoded protein showed that it is similar to racemases, carnitine dehydratases (12), and formyl-coenzyme A (CoA) transferases (Table 1). The functionally characterized protein to which IsoG is most similar is α-methylacyl-CoA racemase of Mus musculus. This enzyme catalyzes the racemization of an α-methyl group in α-methylacyl-CoA esters that thus become available for enzymes of the β-oxidation pathway, which may only accommodate one enantiomer (33, 34). Furthermore, IsoG is similar to carnitine dehydratase of E. coli and the formyl-CoA transferase of Oxalobacter formigenes (Table 1). The latter enzyme is involved in the detoxification of oxalic acid, a toxic byproduct of the metabolism of virtually all life forms. It catalyzes the transfer of the CoA moiety from formyl-CoA to oxalic acid, which is subsequently decarboxylated (36).
Overexpression and activity of IsoI and IsoJ.
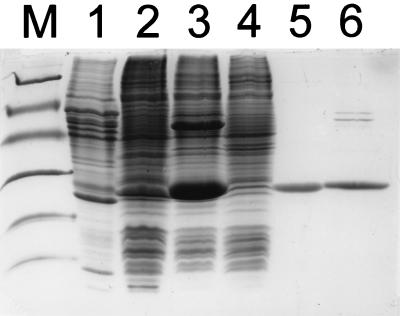
The genes isoI and isoJ were amplified from the cosmid clone pHL8 and placed under the control of a T7 promoter in the expression vector pGEF+ (32). Due to the introduction of the NcoI restriction site, the second residue, which is an isoleucine in both proteins, was replaced by a valine. The resulting plasmids were designated pGEFIsoI and pGEFIsoJ, respectively, and were introduced into E. coli BL21(DE3). IsoI and IsoJ were expressed in a soluble and active form at levels of 3 and 35% of the total soluble cellular protein content, respectively (Fig. 2). Microscopic analysis and SDS-polyacrylamide gel electrophoresis (PAGE) analysis of whole-cell lysates showed that inclusion bodies were formed in E. coli BL21(DE3)(pGEFIsoI) even when cells were grown at 17°C and when no isopropyl-β-d-thiogalactside was used to induce expression of the T7 RNA polymerase. The molecular masses of the recombinant proteins were estimated to be 27 and 26 kDa, respectively, as judged by SDS-PAGE (Fig. 2). The extracts with recombinant IsoI and IsoJ were analyzed for activity with 1,2-epoxy-2-methyl-3-butene and with the typical glutathione S-transferase substrates CDNB and DCNB.
FIG. 2.
SDS-PAGE of crude extracts of E. coli overexpressing IsoI or IsoJ, purified IsoI, and partially purified IsoJ. Lanes: M, marker proteins; 1, crude extract of Rhodococcus sp. strain AD45 grown on isoprene; 2, crude extract of E. coli BL21(DE3)(pGEFIsoI); 3, crude extract of E. coli BL21(DE3)(pGEFIsoJ); 4, crude extract of E. coli BL21(DE3); 5, IsoI purified from strain AD45 (46); 6, partially purified IsoJ from E. coli BL21(DE3)(pGEFIsoJ). Marker proteins: phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), soybean trypsin inhibitor (20 kDa), and α-lactalbumin (14 kDa).
A glutathione-dependent activity with CDNB of 9.0 nmol min−1 mg−1 was detected at pH 7.0 with recombinant IsoJ. Using DCNB as a substrate, activities were 6.4 and 0.20 nmol min−1 mg−1 at pH 7.0 and 8.5, respectively. IsoI purified from Rhodococcus sp. strain AD45 (46) and recombinant IsoI were not active with CDNB and DCNB. Hence, it was concluded that IsoJ has low but significant activity towards CDNB and DCNB whereas IsoI is not active with these typical glutathione S-transferase substrates. Optimal activity was observed at pH 6 to 7 (46), and a specific activity of 26 nmol min−1 mg−1 for CDNB at pH 7.0 was calculated assuming that recombinant IsoJ represented 35% of the total soluble protein (Fig. 2).
Recombinant IsoJ was not active with 1,2-epoxy-2-methyl-3-butene at pH 7.0 or 8.5. The highest specific activity with recombinant IsoI was observed at pH 8.5, and a specific activity of 6 to 10 μmol min−1 mg−1 at pH 8.5 was calculated assuming that recombinant IsoI represented 3 to 5% of the total soluble protein (Fig. 2). The six- to sevenfold-lower specific activity of recombinant IsoI compared to the activity of IsoI that was purified from strain AD45 (46) may be caused by partial misfolding or the Ile-2-to-Val-2 substitution that was introduced in the cloning procedure.
Pathway of isoprene degradation and comparison with the pathway of propene degradation.
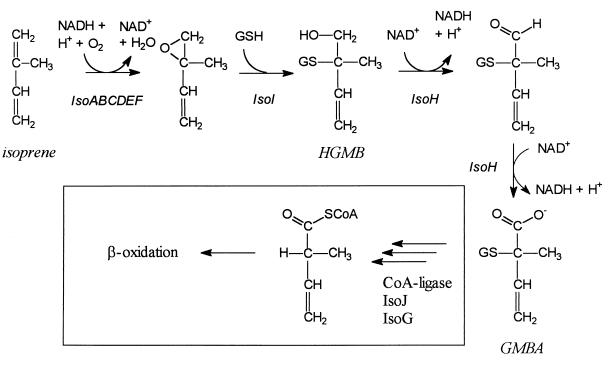
The data presented in this study are consistent with a pathway for isoprene degradation that starts with the oxidation of the methyl-substituted double bond by a monooxygenase, which might be encoded by isoABCDEF (Fig. 3). This reaction yields 1,2-epoxy-2-methyl-3-butene, which is a substrate for IsoI. This enzyme catalyzes the glutathione-dependent opening of the epoxide ring, yielding the stable glutathione conjugate HGMB. Two consecutive oxidation steps by IsoH in which the hydroxyl is oxidized to a carboxylate then result in the formation of GMBA.
FIG. 3.
Pathway of isoprene degradation in Rhodococcus sp. strain AD45. Box, proposed pathway for further GMBA metabolism (see text for details).
Propene metabolism in Xanthobacter sp. strain Py2 and R. rhodochrous strain B-276 also starts with oxidation by a monooxygenase yielding epoxypropane. Interestingly, the putative isoprene monooxygenase of Rhodococcus sp. strain AD45 is more similar to the alkene monooxygenase of Xanthobacter sp. strain Py2 than to the alkene monooxygenase of the phylogenetically more closely related propene utilizer R. rhodochrous B-276 (15, 30, 55). The latter is a three-component rather than a four-component monooxygenase. It consists of an (αβ)2 oxygenase that is encoded by the amoA and amoC genes, a reductase that directly transfers electrons to the oxygenase that is encoded by amoD, and an effector protein that is encoded by amoB (15, 30).
In both propene utilizers the conversion of epoxypropane proceeds with the carboxylation of epoxypropane via a three-step pathway involving four different proteins to yield acetoacetate (1, 2). A detailed study of these proteins for Xanthobacter sp. strain Py2 revealed that an epoxyalkane-coenzyme M (CoM) transferase catalyzes the conjugation of CoM to epoxypropane to yield 2-hydroxypropyl–CoM. Hence, 2-hydroxyethers are the primary products of epoxide conversion in isoprene and propene metabolism. However, the enzymes catalyzing the formation of these compounds, XpI encoded by orf1 in strain Py2 (CAA56241) (1, 2, 40) and IsoI in strain AD45, are not closely related. Furthermore, the next step in both pathways is the dehydrogenation of the 2-hydroxyethers by SDR proteins to the corresponding oxocompounds. Enzymes belonging to the SDR protein family, IsoH in strain AD45 and XpIII and XpIV in strain Py2, catalyze these reactions. Propene metabolism continues with the conversion of 2-ketopropyl–CoM to acetoacetate by an NADPH:2-ketopropyl–CoM oxidoreductase/carboxylase, whereas in isoprene metabolism the aldehyde in 1-oxo-2-glutathionyl-2-methyl-3-butene is oxidized to a carboxylate to yield GMBA.
It remains to be established how in strain AD45 the conversion of GMBA proceeds. Removal of the glutathione moiety is a key step. We did not detect glutathionyl removal, carboxylation, or any other activity with GMBA added to crude extracts of isoprene-grown cells of strain AD45 even when NADH or NADPH was added (46). Both in strains AD45 and Py2 (40) the genes encoding enzymes involved in epoxide conversion are clustered. The isoH gene, which encodes the HGMB dehydrogenase, is similar to the genes encoding XpIII and XpIV in Xanthobacter sp. strain Py2, whereas IsoI and the two gene products of the iso gene cluster, IsoG and IsoJ, for which no role in isoprene degradation has been established, do not have significant homology with genes of propene catabolism in strain Py2. This is in agreement with the fact that epoxide metabolism in strains AD45 and Py2 proceeds via different mechanisms. Interestingly, some glutathione S-transferases catalyze the elimination of glutathionyl groups in a reversed Michael reaction (7), but such a reaction may be unlikely with glutathione conjugates in isoprene metabolism since this would require the energetically unfavorable elimination of a proton from the unactivated methyl group. Furthermore, we did not observe conversion of GMBA with extracts of E. coli BL21(DE3)(pGEFIsoJ) (unpublished data), although active IsoJ enzyme was produced, as concluded from activity measurements with CDNB or DCNB as the substrate. Reductive removal with a second glutathione molecule, thus generating glutathione disulfide, seems more feasible, but with overproduced IsoJ or with extracts of isoprene-grown cells of strain AD45 no glutathionyl removal from GMBA in the presence of 5 mM glutathione was detected.
An alternative possibility is that IsoJ catalyzes the reductive glutathione removal from a downstream metabolite. This may be the CoA-thioester of GMBA, which could be formed by an unidentified ATP-dependent CoA ligase (Fig. 3). Such a pathway seems feasible since a reductive elimination of glutathione is greatly facilitated when the carboxyl group is replaced by a more-electron-withdrawing CoA-thioester. As described above, IsoG is similar to racemases that catalyze the racemization of an α-methyl group in methylacyl-CoA thioester. Hence, both diastereomers of the GMBA-CoA thioester would become available for further metabolism, which for α-methylacyl–CoA esters mainly occurs through the 2S-diastereomer (34). This supports the hypothesis that a CoA-thioester is an intermediate in the isoprene degradation pathway where IsoG could have a function in racemization. These reactions would result in the formation of 2-methyl-3-butenyl–CoA, which after reduction of the double bond could be further degraded via β-oxidation as in the isoleucine degradation pathway (25).
ACKNOWLEDGMENTS
The work of J.E.T.H.V. was financed by grant IOP91204 from the Dutch IOP Environmental Biotechnology program and grant ENV5-CT95-0086 from the EU Environment and Climate Programme.
Jaap Kingma is acknowledged for performing CNBr digestion and peptide isolation.
REFERENCES
- 1.Allen J R, Clark D D, Krum J G, Ensign S A. A role for coenzyme M (2-mercaptoethanesulfonic acid in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8447. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J R, Ensign S A. Two short-chain dehydrogenases confer stereoselectivity for enantiomers of epoxypropane in the multiprotein epoxide carboxylating systems of Xanthobacter strain Py2 and Nocardia corallina B276. Biochemistry. 1999;38:247–256. doi: 10.1021/bi982114h. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong R N. Structure, catalytic mechanism, and evolution of the glutathione S-transferases. Chem Res Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 5.Bernat A B, Laughlin L T, Armstrong R N. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase 1 and the extradiol dioxygenases. Biochemistry. 1997;36:3050–3055. doi: 10.1021/bi963172a. [DOI] [PubMed] [Google Scholar]
- 6.Board P G, Coggan M, Wilce M C J, Parker M W. Evidence for an essential serine residue in the active site of the theta class glutathione transferases. Biochem J. 1995;311:247–250. doi: 10.1042/bj3110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Armstrong R N. Stereoselective catalysis of a retro-Michael reaction by mu class glutathione transferases. Consequences for the internal distribution of products in the active site. Chem Res Toxicol. 1995;8:580–585. doi: 10.1021/tx00046a012. [DOI] [PubMed] [Google Scholar]
- 8.Cleveland C C, Yavitt J B. Microbial consumption of atmospheric isoprene in a temperate forest soil. Appl Environ Microbiol. 1998;64:172–177. doi: 10.1128/aem.64.1.172-177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daubaras D L, Hershberger C D, Kitano K, Chakrabarty A M. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1995;61:1279–1289. doi: 10.1128/aem.61.4.1279-1289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirr H, Reinemer P, Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur J Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 11.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touch-down” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichler K, Schunck W H, Kleber H P, Mandrand-Berthelot M A. Cloning, nucleotide sequence, and expression of the Escherichia coli gene encoding carnitine dehydratase. J Bacteriol. 1994;176:2970–2975. doi: 10.1128/jb.176.10.2970-2975.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewers J W C, Knackmuss H-J. Biodegradation of chloroethenes using isoprene as a co-substrate. In: Verachtert H, Verstraete W, editors. Proceedings of the International Symposium on Environmental Biotechnology. Oostende, Belgium: Royal Flemish Society of Engineers; 1991. pp. 77–83. [Google Scholar]
- 14.Finnerty W R. The biology and genetics of the genus Rhodococcus. Annu Rev Microbiol. 1992;46:193–218. doi: 10.1146/annurev.mi.46.100192.001205. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher S C, Cammack R, Dalton H. Alkene monooxygenase from Nocardia corallina B-276 is a member of the class of dinuclear iron proteins capable of stereospecific epoxygenation reactions. Eur J Biochem. 1997;247:635–641. doi: 10.1111/j.1432-1033.1997.00635.x. [DOI] [PubMed] [Google Scholar]
- 16.Habig W H, Pabst M J, Jakoby W B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 17.Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Innis M A, Gelfand D H. A guide to methods and amplification. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 3–11. [Google Scholar]
- 19.Jacobs M H, van den Wijngaard A J, Pentenga M, Janssen D B. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur J Biochem. 1991;202:1217–1222. doi: 10.1111/j.1432-1033.1991.tb16493.x. [DOI] [PubMed] [Google Scholar]
- 20.Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffrey J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6004–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 21.Larkin M J, deMot R, Kulakov L A, Nagy I. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek. 1998;74:133–153. doi: 10.1023/a:1001776500413. [DOI] [PubMed] [Google Scholar]
- 22.Lutje Spelberg J H, Rink R, Kellogg R, Janssen D B. Enantioselectivity of a recombinant epoxide hydrolase from Agrobacterium radiobacter. Tetrahedron Asymmetry. 1998;9:459–482. [Google Scholar]
- 23.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione S-transferase superfamily. FEBS Lett. 1993;323:135–140. doi: 10.1016/0014-5793(93)81465-c. [DOI] [PubMed] [Google Scholar]
- 25.Massey L K, Sokatch J R, Conrad R S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976;40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray V. Improved double-stranded DNA sequencing using the linear polymerase chain reaction. Nucleic Acids Res. 1989;17:8889. doi: 10.1093/nar/17.21.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connor K E, Dobson A D, Hartmans S. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl Environ Microbiol. 1997;63:4287–4291. doi: 10.1128/aem.63.11.4287-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pikus J D, Studs J M, Achim C, Kauffmann K E, Münck E, Steffan R J, McClay K, Fox G. Recombinant toluene-4-monooxygenase: catalytic and Mössbauer studies of the purified diiron and Rieske components of a four-protein complex. Biochemistry. 1996;35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 29.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 30.Saeki H, Furuhashi K. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J Ferment Bioeng. 1994;78:339–406. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schanstra J P, Rink R, Pries F, Janssen D B. Construction of an expression and site-directed mutagenesis system of haloalkane dehalogenase in Escherichia coli. Protein Expr Purif. 1993;4:479–489. doi: 10.1006/prep.1993.1063. [DOI] [PubMed] [Google Scholar]
- 33.Schmitz W, Fingerhut R, Conzelmann E. Purification and properties of α-methylacyl-CoA racemase from rat liver. Eur J Biochem. 1994;222:313–323. doi: 10.1111/j.1432-1033.1994.tb18870.x. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz W, Helander H M, Hiltunen J K, Conzelmann E. Molecular cloning of cDNA species for rat and mouse liver-methylacyl-CoA racemases. Biochem J. 1997;326:883–889. doi: 10.1042/bj3260883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharkey T D. Isoprene synthesis by plants and animals. Endeavour (Oxford) 1996;20:74–78. doi: 10.1016/0160-9327(96)10014-4. [DOI] [PubMed] [Google Scholar]
- 36.Sidhu H, Ogden S D, Lung H Y, Luttge B G, Baetz A L, Peck A B. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J Bacteriol. 1997;179:3378–3381. doi: 10.1128/jb.179.10.3378-3381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small F J, Ensign S A. Alkene monooxygenase from Xanthobacter strain Py2. Purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J Biol Chem. 1997;272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 38.Stainthorpe A C, Lees V, Salmond G P, Dalton H, Murrell J C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath) Gene. 1990;91:27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- 39.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swaving J, Weijers C A, van Ooyen A J, de Bont J A M. Complementation of Xanthobacter Py2 mutants defective in epoxyalkane degradation, and expression and nucleotide sequence of the complementing DNA fragment. Microbiology. 1995;141:477–484. doi: 10.1099/13500872-141-2-477. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. Crystal structures of the binary and ternary complexes of 7alpha-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 42.Thompson A M. The oxidizing capacity of earth's atmosphere; probable past and future changes. Science. 1992;256:1157–1165. doi: 10.1126/science.256.5060.1157. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ginkel C G, de Jong E, Tilanus J W R, de Bont J A M. Microbial oxidation of isoprene, a biogenic foliage volatile, and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol Ecol. 1987;45:275–279. [Google Scholar]
- 45.van Hylckama Vlieg J E T, Kingma J, van den Wijngaard A J, Janssen D B. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl Environ Microbiol. 1998;64:2800–2805. doi: 10.1128/aem.64.8.2800-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Hylckama Vlieg J E T, Kingma J, Kruizinga W, Janssen D B. Purification of a glutathione S-transferase and dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol. 1999;181:2094–2101. doi: 10.1128/jb.181.7.2094-2101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Hylckama Vlieg J E T, de Koning W, Janssen D B. Effect of chlorinated ethene conversion on viability and activity of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1997;63:4961–4964. doi: 10.1128/aem.63.12.4961-4964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vuilleumier S, Leisinger T. Protein engineering studies of dichloromethane dehalogenase/glutathione S-transferase from Methylophilus sp. strain DM11. Ser12 but not Tyr6 is required for enzyme activity. Eur J Biochem. 1996;239:410–417. doi: 10.1111/j.1432-1033.1996.0410u.x. [DOI] [PubMed] [Google Scholar]
- 50.Wagner W P, Nemecek M, Fall R. Three distinct phases of isoprene formation during growth and sporulation of Bacillus subtilis. J Bacteriol. 1999;181:4700–4703. doi: 10.1128/jb.181.15.4700-4703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wistuba D, Weigand K, Peter H. Stereoselectivity of in vitro isoprene metabolism. Chem Res Toxicol. 1994;7:336–343. doi: 10.1021/tx00039a010. [DOI] [PubMed] [Google Scholar]
- 52.Yen K M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yen K M, Karl M R. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1992;174:7253–7261. doi: 10.1128/jb.174.22.7253-7261.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou N-Y, Chan Kwo Chion C K N, Leak D J. Cloning and expression of the genes encoding the propene monooxygenase from Xanthobacter, Py2. Appl Microbiol Biotechnol. 1996;44:582–588. [Google Scholar]
- 55.Zhou N-Y, Chan Kwo Chion C K N, Leak D J. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl Environ Microbiol. 1999;65:1589–1595. doi: 10.1128/aem.65.4.1589-1595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]