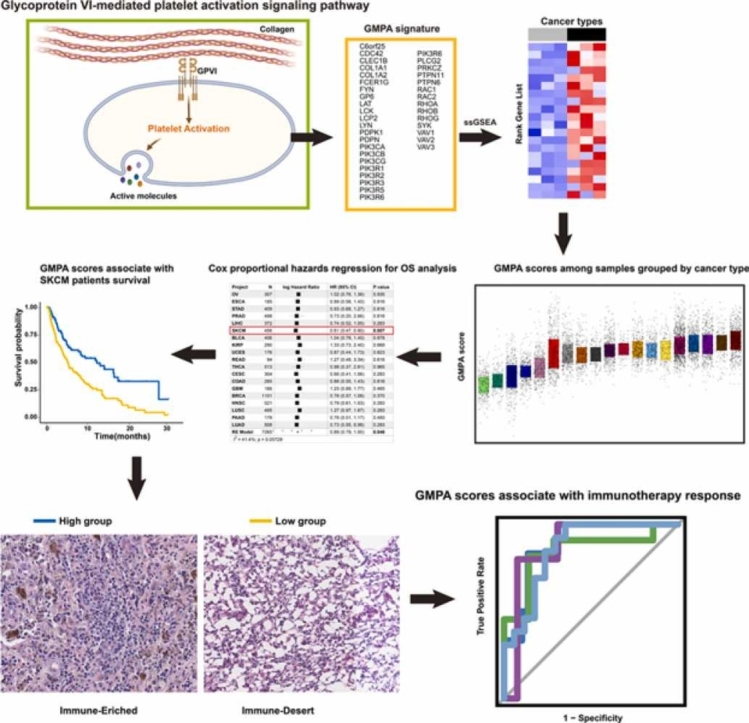
Abstract
Platelets play a vital role in cancer and immunity. However, few comprehensive studies have been conducted on the role of platelet-related signaling pathways in various cancers and their responses to immune checkpoint blockade (ICB) therapy. In the present study, we focused on the glycoprotein VI-mediated platelet activation (GMPA) signaling pathway and comprehensively evaluated its roles in 19 types of cancers listed in The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO). Cox regression and meta-analyses showed that for all 19 types of cancers, patients with high GMPA scores tended to have a good prognosis. Furthermore, the GMPA signature score could serve as an independent prognostic factor for patients with skin cutaneous melanoma (SKCM). The GMPA signature was linked to tumor immunity in all 19 types of cancers, and was correlated with SKCM tumor histology. Compared to other signature scores, the GMPA signature scores for on-treatment samples were more robust predictors of the response to anti-PD-1 blockade in metastatic melanoma. Moreover, the GMPA signature scores were significantly negatively correlated with EMMPRIN (CD147) and positively correlated with CD40LG expression at the transcriptomic level in most cancer patient samples from the TCGA cohort and on-treatment samples from anti-PD1 therapy cohorts. The results of this study provide an important theoretical basis for the use of GMPA signatures, as well as GPVI-EMMPRIN and GPVI-CD40LG pathways, to predict the responses of cancer patients to various types of ICB therapy.
Keywords: Glycoprotein VI-mediated platelet activation signaling pathway, Immune checkpoint blockade therapy, Platelets, EMMPRIN-GPVI, CD40L-GPVI
Graphical Abstract
1. Introduction
Platelets, a nucleated cells derived from megakaryocytes, regulate immunity and tumor metastasis [1], [2]. Platelet–tumor cell interactions are critical for metastatic dissemination during tumor progression [3]. However, platelet activation is not always harmful in cancer. Platelet-derived growth factor subunit B is formed during platelet activation; it helps maintain vascular barriers in tumors and inhibits tumor cell proliferation [4]. Platelet-derived microparticles infiltrate solid tumors and transfer platelet-derived miRNAs to the cells in solid tumors in vivo. In this manner, they downregulate tumor cell genes and inhibit solid tumor growth[5]. Platelets contain abundant soluble and cell-related immunoregulatory molecules that may promote or inhibit the immune response in different environments [6]. Platelets may also promote innate immunity by interacting with Triggering Receptor Expressed On Myeloid Cells 1(TREM1) receptors expressed in the bone marrow [7].
Glycoprotein VI (GPVI), a major platelet-activating collagen receptor, is crucial for mediating collagen-induced platelet activation and potentiating other platelet activation pathways [8]. Upon binding of a GPVI dimer with collagen, the affinity between the aggregated GPVI and collagen is increased and the proximity of GPVI-related signal molecules is enhanced. The cytoplasmic tail of GPVI connects with the Fc receptor of immune cells through a salt bridge γ chain (FCR γ). The ITAM motif in the FCR γ chain is exposed and phosphorylated by the Src kinases Lyn and Fyn. In turn, phosphorylated ITAM recruits and activates Syk, which assembles a signal complex composed of LAT and slp76 downstream. This complex eventually leads to effective platelet activation and diffusion [9]. Nevertheless, little is known regarding the roles of the GPVI-mediated platelet activation (GMPA) signaling pathway in pan-cancer immunity. In the present study, we performed a pan-cancer expression analysis of the GMPA signaling pathway in 19 types of cancers and explored the role of GMPA in immunotherapy across multiple cancer types, especially melanoma.
2. Methods
2.1. Patient characteristics
Normalized mRNA expression levels and clinical data of 19 types of cancers were downloaded from the Pan-Cancer Atlas (PanCanAtlas) (https://gdc.cancer.gov/about-data/publications/pancanatlas). Eight published datasets with RNA-Seq data for metastatic urothelial cancer, metastatic melanoma, renal cell carcinoma and gastric cancer were also collected. All of these cancers had been subjected to immune checkpoint blockade (ICB) therapy. The datasets from Lee et al., MGH, Abril-Rodriguez et al., and Gide et al.[10], [11], [12], [13]. included both pre-treatment and on-treatment samples. The datasets from Kim et al., IMvigor210, MGSP, and Ascierto et al. included only pre-treatment samples[14], [15], [16], [17]. Clinical responses to treatment were obtained for all eight cohorts based on the Response Evaluation Criteria in Solid Tumors (RECIST; https://recist.eortc.org). Details of the responses of the patients to the treatment are presented in Table S1.
2.2. GMPA signaling pathway score
A GMPA signaling pathway-related set of 35 genes was collected based on Reactome (R-HSA-114604) (https://reactome.org/PathwayBrowser/#/R-HSA-114604). The genes are listed in Table S2. The GMPA signaling pathway scores across all samples from The Cancer Genome Atlas (TCGA) and the ICB cohorts were calculated by the single-sample gene set enrichment analysis (ssGSEA) algorithm in the Gene Set Variation Analysis (GSVA) package in R (R Core Team, Vienna, Austria) [18]. For showing the relative expression levels of the 35 genes, normalized data + 1 were log2-transformed.
2.3. Histological examination of skin cutaneous melanoma (SKCM) samples.
Histological data were obtained for the skin cutaneous melanoma (SKCM) samples of Bagaev et al. (annotation.tsv) and the GSE8401 dataset [19], [20]. Sixty-five formalin-fixed, paraffin-embedded tissue slides were provided by the TCGA data portal (https://portal.gdc.cancer.gov/). Lymphocytic scores were graded with a semiquantitative scoring system (0−5) and described tumor inflammation.
2.4. Gene set enrichment analysis (GSEA)
GSEA was performed on hallmark gene sets in the GSVA package of R [18]. They represent well-defined biological states or processes based on GMPA signature scores for the top 30% vs. the bottom 30% of each type of cancer. Normalized enrichment scores> 1 and false discovery rates< 0.25 were considered statistically significant.
2.5. Immunity analysis
A list of immune-related genes was obtained from a recently published study and used to develop four conserved pan-cancer microenvironment subtypes [19]. Four TCGA conserved pan-cancer microenvironment subtypes, including (1) immune-enriched, fibrotic (IE/F), (2) immune-enriched, non-fibrotic (IE), (3) fibrotic (F), and (4) immune-depleted (D) were downloaded from the dataset of Bagaev et al. [19]. TCGA immune clusters C1–C6 were downloaded from Thorsson et al. [21]. Spearman’s correlation coefficients between the GMPA signature scores and the mRNA expression levels of the immune-related subtypes were calculated for each cancer type. T cell receptor Shannon indices and richness scores across patients with the 19 different types of cancer in TCGA were obtained from Bagaev et al. [19]. Immunophenoscores (IPSs) for CTLA4 and PDCD1 among the TCGA samples were obtained from Charoentong et al. [22].
2.6. Statistical analysis
Associations between the GMPA signaling scores and the expression level of immune-related gene were evaluated by Spearman’s correlation analysis. We used area under the receiver operating characteristic curves (AUROCs) to evaluate the predictive performance of the GMPA signature scores. The Survival and Survminer packages in R were used to determine the optimal GMPA score cutoff values for the high and low groups. Survival analyses were performed using Kaplan–Meier (K-M) estimates of survival probability and log rank tests. Cox regression analyses were performed using the survival and metafor package in R to calculate the hazard ratios (HR) based on the random-effects model [23]. P < 0.05 was considered statistically significant. All statistical analyses were performed and data were plotted with R v. 4.1.2. and GraphPad Prism v. 7.0 (https://www.graphpad.com/scientific-software/prism/). Graphical Abstract was partially made using Biorender (https://biorender.com/).
3. Results
3.1. Expression and alteration landscapes of GMPA signaling pathway-related genes in human cancers
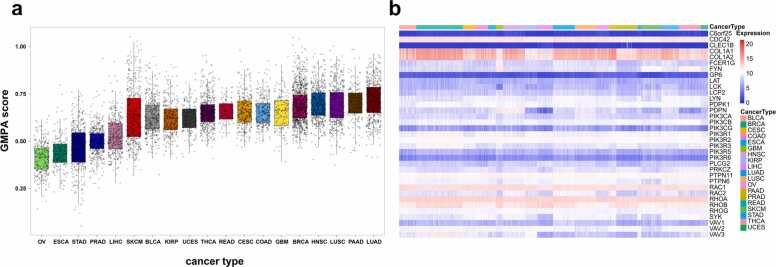
We used ssGSEA to calculate 35 GMPA-related genes and described their expression landscapes in 19 types of cancer in TCGA. They significantly differed in terms of GMPA score (ANOVA, P < 0.0001, Fig. 1a, Table S3). Lung adenocarcinoma (LUAD) and ovarian serous cystadenocarcinoma had the highest and lowest average GMPA scores, respectively. Analysis of the GMPA pathway-related gene expression profiles for the 19 cancer types showed that COL1A1, COL1A2, RHOA, RHOB, CDC42, and RAC1 were significantly upregulated in all cancers (Fig. 1b, Table S4). These findings indicate significant differences in the GPVI-mediated platelet activation levels among various types of cancer.
Fig. 1.
Expression landscapes of GMPA signaling pathway-related genes in 19 different types of cancer. (a) ssGSEA analysis scores of GMPA signatures among samples grouped by cancer type. (b) Heatmap showing expression levels of GMPA signature-related genes in 19 types of cancer listed in TCGA.
3.2. GMPA signaling pathway signature scores are significantly associated with survival in patients with SKCM
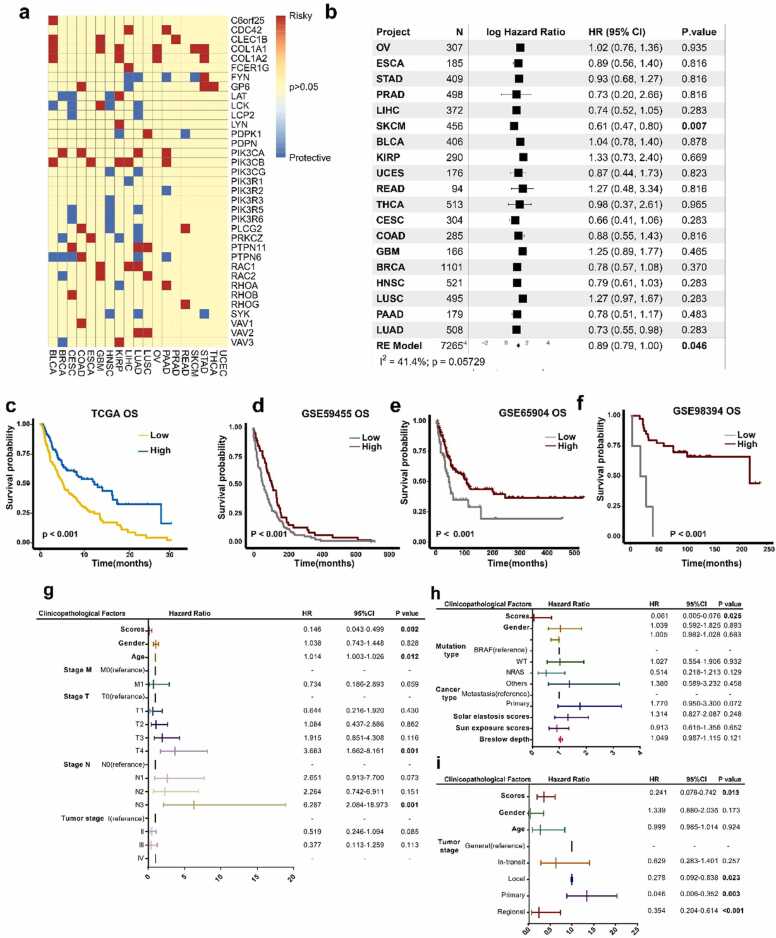
We then investigated the relationships among the GMPA signaling pathway-related gene expression levels, cancer prognosis, and cancer survival. To this end, we evaluated the prognostic performance of 35 genes via univariate survival analysis and Cox proportional hazards models based on their expression levels in the 19 types of cancer in TCGA. LUAD had the highest number of prognostic genes (13/35). By contrast, none of the 35 genes were significantly associated with overall survival (OS) in uterine corpus endometrial carcinoma (Fig. 2a). We performed a univariate Cox regression analysis based on the GMPA scores and observed that GMPA-related gene expression was only associated with OS in SKCM (hazard ratio [HR] = 0.61, 95% confidence interval [CI] = 0.47–0.80, P = 0.007, Fig. 2b, Table S5). A meta-analysis of the HR values for all cancer types showed that the GMPA scores were correlated with prognosis (HR = 0.89, 95% CI = 0.79–1.00, P = 0.046, Fig. 2b, Table S5).
Fig. 2.
Identification of GMPA signatures associated with patient survival. (a) Prognostic value of GMPA signature-related genes for OS in 19 types of cancer in TCGA. (b) (b) Forest plots depicting results of Cox proportional hazards regression for OS analysis using GMPA signature scores. A random effects model was used to calculate the pooled hazard ratio and the p-values. Forest plots showing loge hazard ratio (95% confidence interval). P-values were adjusted for multiple testing using the FDR method (significance threshold of 5%). I-square (I2) statistic test was used to evaluate the proportion of statistical heterogeneity. (c-f) K-M curves of OS by GMPA signature scores in TCGA (c), GSE59455 (d), GSE65904 (e), and GSE98394 (f) datasets. P values for survival analysis were calculated using log rank tests. (g-i) Multivariate regression analysis of OS in patients with SKCM from TCGA (g), GSE65904 (h), and GSE59455 datasets (i). P-values less than 0.05 are in bold in the Figure.
A K-M curve analysis showed that the subjects in the high-risk cohort had comparatively better OS (log-rank P < 0.001, Fig. 2c, Table S6), disease-specific survival (DSS) (log-rank P < 0.001, Fig. S1a), and disease-free survival (DFS) (log-rank P < 0.001, Fig. S1b) than those in the low-risk cohort among SKCM patients. We applied ssGSEA to calculate the GMPA signature scores for three GEO datasets. Figs. 2d–2 f show that according to the three GEO datasets, a low GMPA signature score was associated with a poor OS in SKCM (all log-rank P < 0.001, Table S6).
We then performed a multivariate Cox regression analysis to establish whether the expression of GMPA signature scores can serve as an independent prognostic factor for patients with SKCM. The GMPA signature scores were independent prognostic factors with HR = 0.146 (0.043, 0.499, P = 0.002) for OS after adjusting for the other factors in TCGA-SKCM (Fig. 2 g). These results were also validated for the GSE65904 and GSE59455 cohorts and demonstrated significant associations with high GMPA signature scores and better OS (all P < 0.05, Fig. 2 h and 2i) in SKCM. We used a Cox regression model to verify the prognostic value of the GMPA signature on the DSS and DFS of the patients in TCGA (P < 0.05, Fig. S1c) and the GSE65904 cohort (P < 0.05, Fig. S1d). The original data are listed in Table S6.
Further results revealed that GMPA signature scores were independent prognostic factors for OS after adjusting for tumor stage in head and neck squamous cell carcinoma (Fig. S2d), LUAD (Fig. S2g), and SKCM (Fig. S2k). Previous research has found that GPVI could serve as a novel adhesion receptor binding to EMMPRIN (CD147)[24]. We analyzed the correlation between CD147 and GMPA signature scores across 19 cancer types. Notably, we found that the GMPA signature scores were negatively correlated with the expression level of CD147 in most cancer types at the transcriptomic level (Fig. S3, Table S7).
3.3. GMPA scores are significantly correlated with immunity in 19 types of cancer
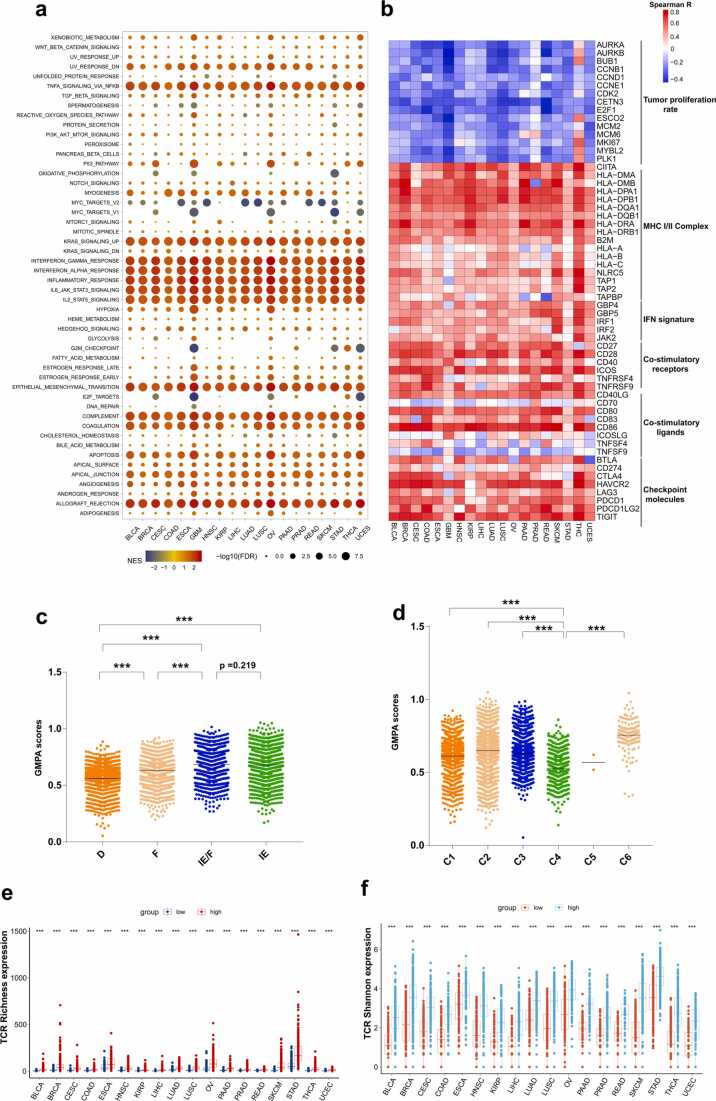
We performed GSEA to identify the differentially enriched hallmarks and pathways in the groups with low and high GMPA signature scores. We found that the interferon (IFN)-alpha, IFN-gamma, and inflammatory responses were markedly enriched in samples with high GMPA signature scores (Fig. 3a, Table S8). In the Kyoto Encyclopedia of Genes and Genomes (KEGG) database gene sets, an enrichment analysis based on GSEA indicated that the high-GMPA signature score group was associated with immunocyte function-related pathways such as antigen processing and presentation, T cell receptor signaling, and natural killer cell-mediated cytotoxicity (Fig. S4, Table S9).
Fig. 3.
Analyses of correlations among GMPA signature scores and the tumor immunity and pan-cancer microenvironment subtypes. (a) Significant alterations in the pathways of the high GMPA signature score group relative to the low-score group were assessed by GSEA. The sizes of points represent log10-transformed false discovery rates (FDR) value and the colors of points represent Normalized enrichment scores (NES). (b) Heatmap showing Spearman’s correlation coefficients between the GMPA signature scores and immunomodulator expression in the 19 types of cancer. (c-d) Distribution of GMPA signature scores in two pan-cancer microenvironment subtypes. P-values are based on the Mann–Whitney test. (e-f) Relative TCR richness and Shannon indices for high- and low GMPA signature scores. P-values are based on the Mann–Whitney test. (***P < 0.001). The horizontal line represents the median.
We found strong positive correlations between the GMPA signature scores and the immune gene lists such as the major histocompatibility (MHC) I/II complex signature, IFN signatures, and the co-stimulatory receptors and ligands (Fig. 3b, Table S10). Most cancer types presented with GPMA scores that were negatively correlated with tumor proliferation rate-related genes. Notably, Spearson's correlation analysis revealed a significant positive correlation between the GMPA signature scores and the expression level of CD40LG in 19 cancer types (Fig. 3b and Fig. S5, Table S7, all p < 0.001).
Then, we compared the expression levels of the classic checkpoint molecules BTLA, CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, and TIGIT between the groups with high and low GMPA scores (Fig. 3b, Table S10). Significant positive correlations were observed between the GMPA scores and the checkpoint molecules among all 19 types of cancer. The correlations among the GMPA scores and PDCD1LG2, PDCD1, HAVCR2, and TIGIT were particularly strong.
Transcriptomic analyses have confirmed that numerous cancer types are divided into four distinct tumor microenvironment (TME) subtypes. The IE cancers are characterized by high levels of immune cell infiltration, and the F and D subtypes demonstrated little or no leukocyte/lymphocyte infiltration. In addition, the D subtype had the highest percentage of malignant cells[19]. We explored the differences in the GMPA signature scores among the TME subtypes and found that IE/F and IE had higher median scores than the other subtypes and D had the lowest median score (Fig. 3c, Table S11).
Immunogenomic analyses of> 10,000 cancer patients across 33 different types of cancer identified six immune subtypes [21]. C4 (lymphocyte-depleted) had low lymphocytic infiltration and the lowest median GMPA signature score (Fig. 3d, Table S11). This finding was consistent with the results of the analysis of the four TME subtypes. The aforementioned results suggest that the GMPA scores may be closely related to the cancer immune status.
Clonality in peripheral blood PD-1 + CD8 + T cells may serve as a noninvasive predictor of patient response to ICB and survival outcomes in non-small cell lung cancer (NSCLC) [25], [26]. We therefore analyzed the T cell receptor (TCR) repertoires from the RNA-Seq data for 18 cancer types in TCGA. The values for the TCR repertoire variables were missing for glioblastoma multiforme (GBM). For all 18 types of cancer, patients with high GMPA signature scores tended to have relatively higher TCR richness and Shannon indices than those with low GMPA scores (Fig. 3e–f, Table S11).
The IPS is a superior predictor of patient response to anti-CTLA-4 and anti-PD-1 immunotherapy. A high IPS is associated with an immunogenic tumor [22]. We categorized 18 cancer patients as IPS-CTLA4-neg-PD1-neg, IPS-CTLA4-neg-PD1-pos, IPS-CTLA4-pos-PD1-neg, or IPS-CTLA4-pos-PD1-pos and compared the IPSs among the high and low GMPA score groups. Most IPS-CTLA4-neg-PD1-pos, IPS-CTLA4-pos-PD1-neg, and IPS-CTLA4-pos-PD1-pos cancer patients with high GMPA scores expressed relatively higher IPS values (Figs. S6a–d, Table S11). These results suggest that patients with high GMPA scores may be relatively more immunogenic and responsive to ICB therapy than those with low GMPA scores.
3.4. GMPA signature scores are significantly correlated with tumor histology in melanoma
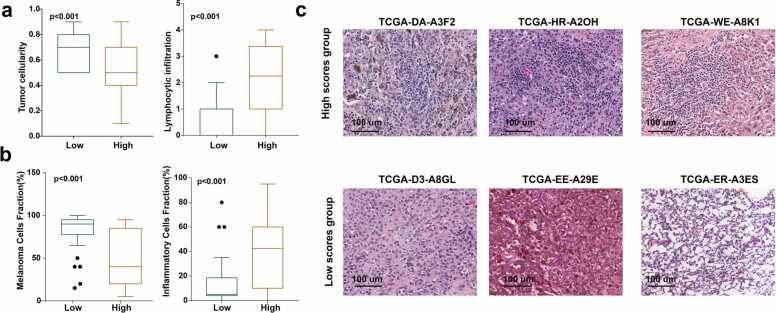
Given that GMPA signature scores were only associated with OS and were independent factors in melanoma prognosis (Figs. 2b–2i), we analyzed representative melanoma tumor specimens from TCGA-SKCM (n = 65) and histologically validated their GMPA scores. Once again, this analysis demonstrated that the low GMPA signature score group had a relatively higher malignant cell density and lower immunocyte infiltration (Fig. 4a, Table S12). This discovery was validated using an independent melanoma dataset (Fig. 4b, Table S13). Lymphocytes were highly abundant in the high GMPA score group, whereas the low GMPA score group displayed relatively greater stromal content, collagen formation, and tumor cell percentage (Fig. 4c).
Fig. 4.
Correlations among GMPA signature scores, tumor histology, and tumor immunity in melanoma. (a-b) Boxplots depicting relative malignant cell and lymphocyte % based on the GMPA signature scores in SKCM from TCGA (a) and the GSE8401 dataset (b). In all box plots, the upper whisker indicates the 75th percentile + 1.5 IQR; the lower whisker indicates the 25th percentile − 1.5 IQR. (c) Representative TCGA hematoxylin-eosin (H&E) histological images of high- and low GMPA signature scores in SKCM.
3.5. GMPA scores for pre-treatment samples
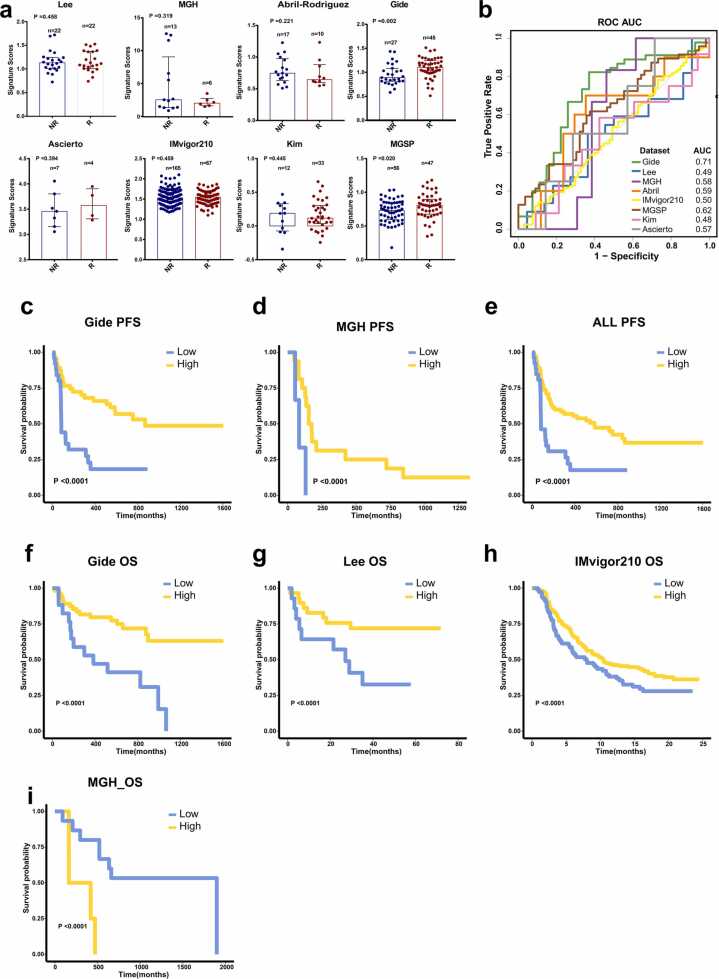
We performed ssGSEA based on the GMPA genes in pre-treatment samples from eight cohorts of patients with multiple cancer types treated with ICB therapy. The datasets of pre-treatment samples were derived from Lee et al., Gide et al., MGH, Abril-Rodriguez et al., Kim et al., IMvigor210, MGSP, and Ascierto et al. Using the Mann–Whitney U test, we found no significant differences between responders (R) and non-responders (NR) in any of the foregoing datasets except those of Gide et al. and MGSP (Fig. 5a). In these two datasets, the GMPA scores were significantly higher for R than NR (P < 0.05, Fig. 5a). We then used the receiver operator characteristic (ROC) to evaluate and quantify the predictive power of the GMPA scores. The areas under the curve (AUC) were 0.49, 0.71, 0.58, 0.59, 0.50, 0.62, 0.48, and 0.57 for the Lee et al., Gide et al., MGH, Abril-Rodriguez et al., IMvigor210, MGSP, Kim et al., and Ascierto et al. datasets, respectively (Fig. 5b).
Fig. 5.
GMPA signatures of pre-treatment samples. (a) Boxplot of GMPA signature scores for pre-treatment samples from Lee et al., Gide et al., MGH, Abril-Rodriguez et al., Kim et al., IMvigor210, MGSP, and Ascierto et al. datasets. R: responders; NR: non-responders. R: responders; NR: non-responders. In all box plots, the upper whisker indicates the 75th percentile + 1.5 IQR; the lower whisker indicates the 25th percentile − 1.5 IQR. P-values are based on the Mann–Whitney test. (b) ROC and AUC of GMPA signatures for pre-treatment samples from Lee et al., Gide et al., MGH, Abril-Rodriguez et al., Kim et al., IMvigor210, MGSP, and Ascierto et al. datasets. (c–e) K-M curves of PFS for on-treatment samples based on GMPA signature scores for Gide et al. (c) and MGH (d) cohortsand all pre-treatment samples (e). (f–i) K-M curves of OS for on-treatment samples based on GMPA signature scores in Gide et al. (f), Lee et al. (g), IMvigor210 (h), and MGH (i) cohorts.
We stratified the patients into high and low groups based on their optimal GMPA scores and performed K-M survival analyses to determine OS and progression-free survival (PFS). Patients with high GMPA scores had significantly improved PFS compared to those with low GMPA scores (Figs. 5c–5e). The K-M survival analyses of the samples from the Lee et al., Gide et al., and IMvigor210 datasets suggested that patients with high GMPA scores had prolonged OS (Figs. 5f–5 h). However, analysis of the MGH dataset revealed the opposite result (Fig. 5i). Thus, the predictive performance of the GMPA scores was generally poor across all pre-treatment samples from the various datasets.
3.6. GMPA scores for on-treatment samples
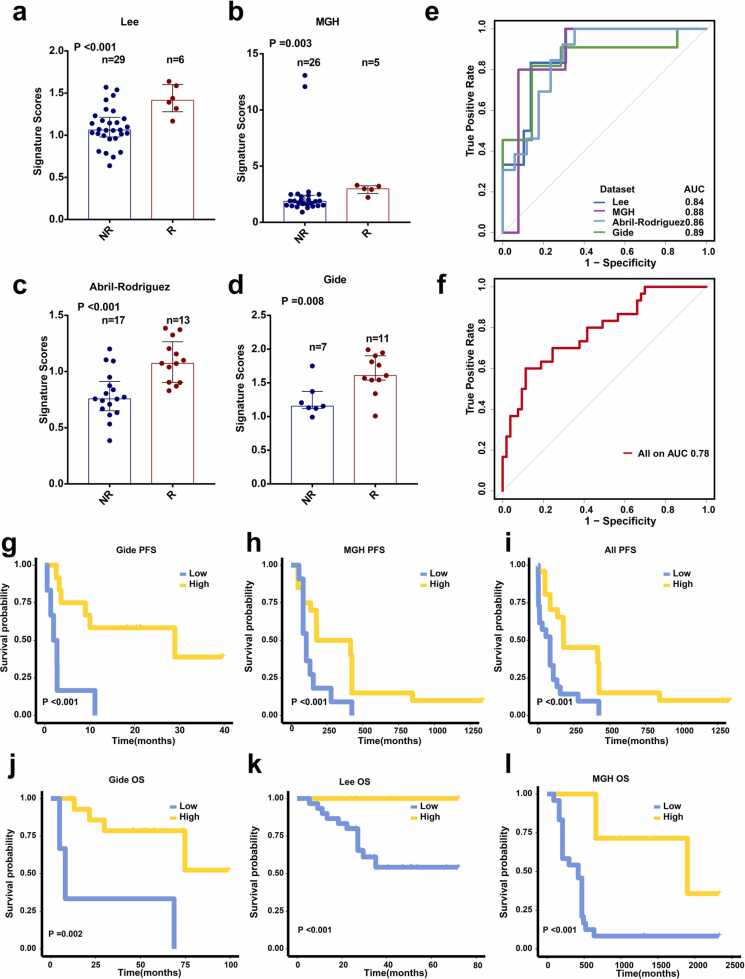
We investigated the predictive performance of GMPA signaling pathway signatures derived from on-treatment samples in metastatic melanoma. The ssGSEA scores were calculated for each patient among the on-treatment samples in the Lee et al., MGH, Abril-Rodriguez et al., and Gide et al. datasets. A Mann–Whitney U test demonstrated that the signature scores were significantly higher for R than NR in the Lee et al. (P < 0.001), MGH (P = 0.003), Abril-Rodriguez et al. (P < 0.001), and Gide et al. (P = 0.008) datasets (Figs. 6a–6d). The AUCs were 0.84, 0.88, 0.86, and 0.89 for the Lee et al., MGH, Abril-Rodriguez et al., and Gide et al. datasets, respectively (Fig. 6e). Combining the on-treatment samples in the Lee et al., MGH, Abril-Rodriguez et al., and Gide et al. datasets generated an AUC of 0.78 (Fig. 6 f). Patients with high GMPA scores had significantly improved PFS compared to those with low GMPA scores in the Gide et al., MGH, and combined datasets (Figs. 6g–6i). OS was also significantly longer for the patients with high GMPA scores than those with low GMPA scores in the Gide et al., Lee et al., and MGH datasets (Figs. 6j–6 l). Analysis of the results for the on-treatment samples disclosed that the GMPA scores were robust and could effectively predict the clinical responses and survival outcomes in melanoma patients undergoing anti-PD-1 therapy.
Fig. 6.
GMPA signatures for on-treatment samples. (a-d) Boxplot of GMPA signature scores for on-treatment samples in Lee et al. (a), MGH (b), Abril-Rodriguez et al. (c), and Gide et al. (d) cohorts. R: responders; NR: non-responders. In all box plots, the upper whisker indicates the 75th percentile + 1.5 IQR; the lower whisker indicates the 25th percentile − 1.5 IQR. P-values are based on the Mann–Whitney test. (e) ROC and AUC for GMPA signatures of on-treatment samples from Lee et al., MGH, Abril-Rodriguez et al., and Gide et al. cohort. (f) ROC and AUC for GMPA signatures of all on-treatment samples. (g-i) K-M curves for PFS of on-treatment samples based on GMPA signature scores of Gide et al. (g) and MGH (h) cohorts and all on-treatment samples (i). (j-l) K-M curves for OS of on-treatment samples based on GMPA signature scores in Gide et al. (j), Lee et al. (k), and MGH (l) cohorts.
Furthermore, Spearson’s correlation analysis revealed that GMPA signature scores were significantly negatively correlated with CD147 expression and positively correlated with CD40LG expression in on-treatment samples from four anti-PD1 therapy cohorts (Fig. S7 and Fig. S8), which is consistent with the results of the analysis of the TCGA cohort.
We also obtained information on BRAF mutation in the samples from the Abril-Rodriguez et al. database. A Mann–Whitney U test on the GMPA scores derived from the on-treatment samples with a BRAF mutation demonstrated that the signature scores were significantly higher for R than NR in the Abril-Rodriguez et al. dataset (Fig. S9a). Because the Mann–Whitney U test requires at least two values in each group, we were unable to calculate the P value for the BRAF wildtype (WT) subgroup (Fig. S9b). The AUCs were 1 and 0.67 for the patients with BRAF V600E and BRAF WT, respectively (Fig. S9c). These results indicate that the GMPA scores were robust and could effectively predict the clinical responses and survival outcomes in melanoma patients with a BRAF mutation undergoing anti-PD-1 therapy.
3.7. Comparison of GMPA and published predictive signatures
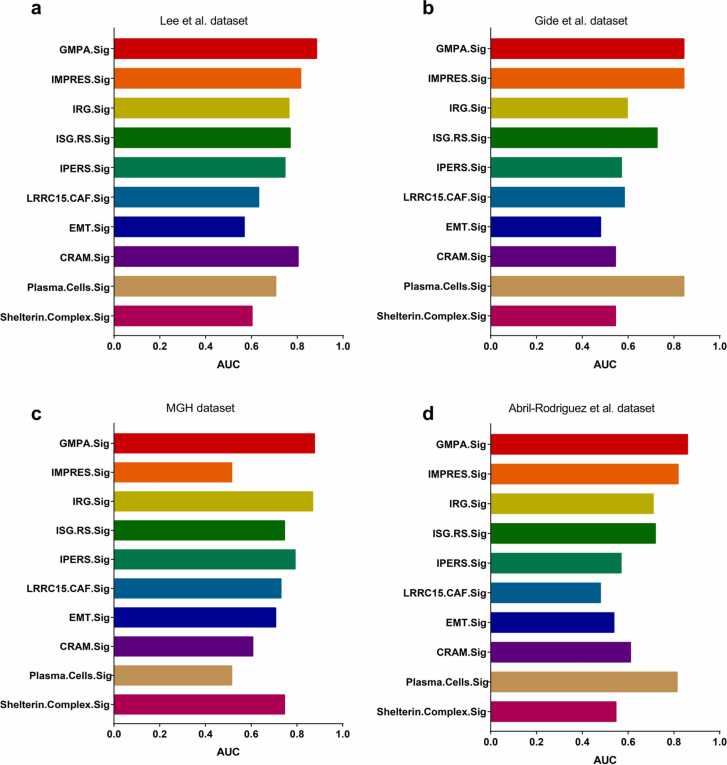
We then evaluated the predictive performance of the GMPA signatures derived from on-treatment specimens. To this end, we compared them against those of previously reported transcriptome-based predictive signatures including IMPRES.Sig [27], IRG.Sig [28], ISG.Sig [29], IPRES.Sig [30], [31], LRRC15. CAF.Sig [32], EMT.Sig [30], CRAM.Sig [33], Plasma.Cells.Sig [34], and Shelterin.Complex.Sig [35]. This analysis indicated that GMPA.Sig derived from on-treatment specimens most effectively predicted the responses to ICB therapy across all four datasets (Fig. 7).
Fig. 7.
Comparison of predictive performance of GMPA.Sig against other ICB response signatures. Barplots of AUCs for 10 ICB response signatures are shown for the Lee et al. (a), Gide et al. (b), MGH (c), and Abril-Rodriguez et al. (d) cohorts. Each cohort’s sample number were showed in the legend.
4. Discussion
The GMPA signaling pathway is one of the pathways involved with platelet activation, signaling, and aggregation [36]. In this study, we mined TCGA RNA data and performed a comprehensive analysis of the GMPA signaling pathways in> 7000 samples of 19 different types of cancer. We also assessed the predictive value of the GMPA signaling pathway in eight datasets of pre-treatment and on-treatment metastatic melanoma tumor samples subjected to anti-PD-1 therapy.
Platelets might play important roles in the adaptive immune response. As GMPA signaling is an essential factor in platelet activation, we systematically analyzed the correlations between the GMPA signaling scores and tumor immunity. The immune activation-related pathways were significantly upregulated in the high GMPA signature score group. For all 19 types of cancer, the GMPA signature scores were strongly correlated with the immune signatures of the MHCI/II class, IFN response, co-stimulatory receptors and ligands, and checkpoint molecules. For most cancer types, the GMPA scores were negatively correlated with the tumor proliferation rate signatures. This observation was consistent with a previous study reporting that platelet activation may inhibit tumor cell proliferation [5].
Previous studies have shown that matrix metalloproteinases (MMPs) are closely related to tumor immunity and that targeting MMPs significantly improves survival time by reducing the tumor burden and promoting anti-tumor immunity [37], [38]. EMMPRIN–EMMPRIN interactions cause an increase in the expression of MMPs [39]. In particular, GPVI has been identified as a novel adhesion receptor for EMMPRIN on platelets [24]. Thus, the binding of EMMPRIN to GPVI may maintain EMMPRIN in a conformation that suppresses the induction of MMP by EMMPRIN, thereby reducing tumor progression and improving the response of cancer to anti-PD1 therapies. In line with this hypothesis, EMMPRIN was shown to regulate the response to immunotherapy [40], [41]. Notably, correlation analysis revealed that GMPA signature scores are negatively correlated with the expression level of CD147 at the transcriptomic level in most TCGA cancer types and in on-treatment samples from four anti-PD1 therapy cohorts. These clues suggest that the GMPA signature and CD147 might have a direct or indirect regulatory relationship at the transcriptomic level and that CD147-GPVI signaling could be used to predict GPVI-associated signatures. This is an interesting hypothesis that needs to be further explored.
In metastatic melanoma patients, AdCD40L (adenovirus-based CD40LG gene therapy) intratumoral injections increase the T-effector/regulatory cell ratio and upregulate expression of the death receptor, which is correlated with prolonged survival [42]. CD40LG can activate the Caspase cascade and ultimately lead to CD40 + tumor cell apoptosis [43]. Because melanoma cells usually express CD40, they are suitable targets for CD40LG-mediated apoptosis [44], [45]. Notably, a previous study showed that GPVI-stimulated platelets could significantly enhance the secretion of CD40LG [46]. Using Spearman’s correlation analysis, we demonstrated that the GMPA signature scores are significantly positively correlated with the CD40LG expression level within TCGA and ICB cohorts. This finding is of particular interest because platelet activation results in the release of multiple bioactive factors, one of which is CD40LG, which was reported to be involved in improving the anti-tumor immune response mediated by T lymphocytes [47], [48]. GPVI-stimulated platelets increased the secretion of CD40LG, which kills CD40 + target melanoma cells. Moreover, the strong positive correlation between GPVI and CD40LG suggested that as the upstream mediator of the GPVI activation pathway, GPVI-CD40LG might be able to predict SKCM survival and the response to anti-PD-1 immunotherapy. These results could also explain why patients with high GMPA signature scores are associated with favorable prognoses in the anti-PD1 cohort.
Furthermore, we evaluated the ability of GMPA signature scores to predict the responses of various types of cancer to anti-PD-1 therapy and found that the AUCs for the GMPA signatures derived from the on-treatment samples were in the range of 0.84–0.89 and were more informative than the AUCs for the GMPA signatures derived from the pre-treatment samples. Importantly, GMPA signatures derived from on-treatment samples are more stable and robust than previously published signatures. Previous studies have shown that on-treatment tumor samples had a relatively better predictive ability than pre-treatment tumor samples in breast cancer patients being administered endocrine therapy [49], [50], [51]. Pathway signatures and functional gene expression signatures derived from on-treatment samples accurately predicted the responses of patients with metastatic melanoma to anti-PD-1 therapy [13], [52]. Overall, our results, which demonstrate that the GMPA signature scores for on-treatment samples are more robust predictors of the response to anti-PD-1 blockade in metastatic melanoma, agree well with the above phenomenon. To the best of our knowledge, the present study may be the first to assess the reliability of the gene signatures in the GMPA signaling pathway for predicting the responses of cancer patients to ICB therapy.
This study had several limitations. First, it was difficult to distinguish the hemostatic pathways from non-hemostatic pathways for almost all platelet receptors using ssGSEA from the Reactome database. Although the other pathway genes did not contribute to the GMPA signature scores, they may still cause potential biases in the results. Second, we lacked information on the BRAF inhibitor treatment that the patients may have received; thus, we could not evaluate the response to anti-PD1 in combination with BRAF inhibitors in melanoma cancer patients. Finally, all on-treatment samples from four ICB cohorts were administered only anti-PD-1/anti-PDL-1 rather than anti-CTLA4.
In this study, we conducted systematic analyses to evaluate the clinicopathological and prognostic value of GMPA signatures in various types of cancer. The data suggested that GMPA signatures may serve as independent prognostic factors for SKCM. Moreover, GMPA signature scores are closely related to tumor immunity in different types of cancer and histological types of SKCM. We also found that the GMPA scores derived from on-treatment tumor specimens were better predictors of responses to anti-PD-1 therapy than those derived from pre-treatment tumor specimens, and that GMPA signaling pathway scores are promising for the optimization of ICB in cancer immunotherapy. As the mediator of upstream signaling, the GPVI-EMMPRIN and GPVI-CD40LG model could predict GMPA signature scores favorably in metastatic melanoma and are worth developing as a new concept and approach for predicting SKCM survival and responses to anti-PD-1 immunotherapy.
Funding
YL is supported by Sun Yat-sen Universit Start-up Funding (201603). YW is supported by the National Natural Science Foundation of China (Grant No. 82100184), the Natural Science Foundation of Guangdong (Grant No. 2022A1515012521) and Beijing Xisike Clinical Oncology Research Foundation (Grant No. Y-Young20220281).
CRediT authorship contribution statement
Yang Liang and Yun Wang: Conceptualization, Visualization, Supervision, Writing – review & editing, Shuzhao Chen: Methodology, Software, Visualization, Data curation, Writing – original draft preparation, Writing – review & editing, Limei Zhang: Data curation, Writing – original draft preparation, Writing – review & editing, Lezong Zhang: Writing – original draft preparation, Qianqian Huang: Data curation.
Conflict of interest
There are no conflicts of interest.
Acknowledgements
The authors would like to acknowledge the contribution of all team members. We thank Xia Niu (Tangdu Hospital, Air Force Medical University) for generously sharing her experiences on TCGA hematoxylin-eosin histological images. We also thank Jianming Zeng (University of Macau) and all the members of his bioinformatics team and biotrainee for generously sharing their experiences and codes. We thank all authors who contributed valuable data and made them public.
Author contributions
S.C and L.Z collected data, analyzed data and wrote the manuscript. L.C wrote the manuscript. Q.H collected the data. Y.L and Y.W conceptualized the study and edited the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.04.002.
Contributor Information
Yun Wang, Email: wangyun@sysucc.org.cn.
Yang Liang, Email: liangyang@sysucc.org.cn.
Appendix A. Supplementary material
Supplementary material Supplementary Figure 1. Identification of GMPA signatures associated with patient survival. (a) GMPA signature score curve for DSS in SKCM patients from TCGA. (b) GMPA signature score curve for DFS in SKCM patients from the GSE65904 dataset. (c) Multivariate analysis including GMPA signature scores and clinical factors for DSS in SKCM patients from TCGA. (d) Multivariate analysis including GMPA signature scores and clinical factors for DFS in SKCM patients from the GSE65904 dataset.
.
Supplementary material Supplementary Figure 3. Spearman’s correlation coefficients between the GMPA signature scores and CD147 expression in the 19 types of cancer.
.
Supplementary material Supplementary Figure 4. Relationships among GMPA signatures and KEGG signaling pathways in cancer patients with high- and low-GMPA signature scores.
.
Supplementary material Supplementary Figure 5. Spearman’s correlation coefficients between the GMPA signature scores and CD40LG expression in the 19 types of cancer.
.
Supplementary material Supplementary Figure 6. Immunophenoscores (IPS) of CTLA4 and PDCD1 between high- and low-GMPA signature scores in 19 types of cancer from the TCGA database.
.
Supplementary material Supplementary Figure 7. Spearman’s correlation coefficients between the GMPA signature scores and CD147 expression in on-treatment samples from four anti-PD1 therapy cohorts of metastatic melanoma.
.
Supplementary material Supplementary Figure 8. Spearman’s correlation coefficients between the GMPA signature scores and CD40LG expression in on-treatment samples from four anti-PD1 therapy cohorts of metastatic melanoma.
.
Supplementary material Supplementary Figure 9. GMPA signatures for on-treatment samples with BRAF mutation and BRAF WT. (a, b) Boxplot of GMPA signature scores for on-treatment samples with BRAF V600E mutation (a) and BRAF WT (b) in the Abril-Rodriguez et al. dataset. R: responders; NR: non-responders. (c) ROC and AUC for GMPA signatures of on-treatment samples with BRAF V600E mutation and BRAF WT from Abril-Rodriguez et al. dataset.
.
Supplementary material
.
Supplementary material Supplementary Figure 2. Multivariate analysis including GMPA signature scores and tumor stage for OS in 19 types of cancer from the TCGA database.
.
References
- 1.Haemmerle M., et al. The platelet lifeline to cancer: challenges and opportunities. Cancer Cell. 2018;33(6):965–983. doi: 10.1016/j.ccell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., et al. Signaling during platelet adhesion and activation. Arterioscler Thromb Vasc Biol. 2010;30(12):2341–2349. doi: 10.1161/ATVBAHA.110.207522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsig L. The role of platelet activation in tumor metastasis. Expert Rev Anticancer Ther. 2008;8:1247–1255. doi: 10.1586/14737140.8.8.1247. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y., et al. Platelet-specific PDGFB ablation impairs tumor vessel integrity and promotes metastasis. Cancer Res. 2020;80(16):3345–3358. doi: 10.1158/0008-5472.CAN-19-3533. [DOI] [PubMed] [Google Scholar]
- 5.Michael J.V., et al. Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood. 2017;130(5):567–580. doi: 10.1182/blood-2016-11-751099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elzey B., et al. Platelet-derived CD 154 enables T-cell priming and protection against Listeria monocytogenes challenge. Blood. 2008;111:3684–3691. doi: 10.1182/blood-2007-05-091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapur R., Semple J.W. In: Molecular and Cellular Biology of Platelet Formation: Implications in Health and Disease. Schulze H., Italiano J., editors. Springer International Publishing; Cham: 2016. Platelet functions beyond hemostasis; pp. 221–237. [Google Scholar]
- 8.Bültmann A., et al. Impact of glycoprotein VI and platelet adhesion on atherosclerosis—A possible role of fibronectin. J Mol Cell Cardiol. 2010;49(3):532–542. doi: 10.1016/j.yjmcc.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Kato K., et al. The contribution of glycoprotein VI to stable platelet adhesion and thrombus formation illustrated by targeted gene deletion. Blood. 2003;102(5):1701–1707. doi: 10.1182/blood-2003-03-0717. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.H., et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat Commun. 2020;11(1):1897. doi: 10.1038/s41467-020-15726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abril-Rodriguez G., et al. PAK4 inhibition improves PD-1 blockade immunotherapy. Nature Cancer. 2020;1(1):46–58. doi: 10.1038/s43018-019-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gide T., et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell. 2019;35:238–255. doi: 10.1016/j.ccell.2019.01.003. e6. [DOI] [PubMed] [Google Scholar]
- 13.Du K., et al. Pathway signatures derived from on-treatment tumor specimens predict response to anti-PD1 blockade in metastatic melanoma. Nat Commun. 2021;12(1):6023. doi: 10.1038/s41467-021-26299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S.T., et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449–1458. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 15.Van Allen E.M., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ascierto M.L., et al. The intratumoral balance between metabolic and immunologic gene expression is associated with anti–PD-1 response in patients with renal cell carcinoma. Cancer Immunol Res. 2016;4(9):726–733. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariathasan S., et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagaev A., et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39 doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Xu L., et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6(5):760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charoentong, P., et al., Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. 2016. [DOI] [PubMed]
- 23.Viechtbauer W. Conducting meta-analyses in R with the metafor Package. J Stat Softw. 2010;36(3):1–48. [Google Scholar]
- 24.Seizer P., et al. EMMPRIN (CD147) is a novel receptor for platelet GPVI and mediates platelet rolling via GPVI-EMMPRIN interaction. Thromb Haemost. 2009;101(4):682–686. doi: 10.1160/th08-06-0368. [DOI] [PubMed] [Google Scholar]
- 25.Charles J., et al. T-cell receptor diversity as a prognostic biomarker in melanoma patients. Pigment Cell Melanoma Res. 2020;33(4):612–624. doi: 10.1111/pcmr.12866. [DOI] [PubMed] [Google Scholar]
- 26.Han J., et al. TCR repertoire diversity of peripheral PD-1+CD8+ T cells predicts clinical outcomes after immunotherapy in patients with non–small cell lung cancer. Cancer Immunol Res. 2020;8(1):146–154. doi: 10.1158/2326-6066.CIR-19-0398. [DOI] [PubMed] [Google Scholar]
- 27.Auslander N., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S., et al. Identification of a prognostic immune signature for cervical cancer to predict survival and response to immune checkpoint inhibitors. OncoImmunology. 2019;8 doi: 10.1080/2162402X.2019.1659094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell. 2019;178:933–948. doi: 10.1016/j.cell.2019.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J.C., et al. Gene signatures of tumor inflammation and epithelial-to-mesenchymal transition (EMT) predict responses to immune checkpoint blockade in lung cancer with high accuracy. Lung Cancer. 2020;139:1–8. doi: 10.1016/j.lungcan.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hugo W., et al. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez C.X., et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 2020;10(2):232–253. doi: 10.1158/2159-8290.CD-19-0644. [DOI] [PubMed] [Google Scholar]
- 33.Shukla S., et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell. 2018;173 doi: 10.1016/j.cell.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patil N.S., et al. Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell. 2022;40(3):289–300. doi: 10.1016/j.ccell.2022.02.002. e4. [DOI] [PubMed] [Google Scholar]
- 35.Luo Z., et al. Pan-cancer analyses reveal regulation and clinical outcome association of the shelterin complex in cancer. Brief Bioinforma. 2021;22 doi: 10.1093/bib/bbaa441. [DOI] [PubMed] [Google Scholar]
- 36.Jassal B., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y., et al. Small-molecule MMP2/MMP9 inhibitor SB-3CT modulates tumor immune surveillance by regulating PD-L1. Genome Med. 2020;12(1):83. doi: 10.1186/s13073-020-00780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foley C., et al. Matrix metalloprotease-1a promotes tumorigenesis and metastasis. J Biol Chem. 2012;287:24330–24338. doi: 10.1074/jbc.M112.356303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toole B. EMMPRIN (CD147), a cell surface regulator of matrix metalloproteinase production and function. Curr Top Dev Biol. 2003;54:371–389. doi: 10.1016/s0070-2153(03)54015-7. [DOI] [PubMed] [Google Scholar]
- 40.Vonderheide R.H. Prospect of targeting the CD40 pathway for cancer therapy. Clin Cancer Res. 2007;13(4):1083–1088. doi: 10.1158/1078-0432.CCR-06-1893. [DOI] [PubMed] [Google Scholar]
- 41.Zhu X., et al. CD147: a novel modulator of inflammatory and immune disorders. Curr Med Chem. 2013:21. doi: 10.2174/0929867321666131227163352. [DOI] [PubMed] [Google Scholar]
- 42.Schiza A., et al. Adenovirus-mediated CD40L gene transfer increases Teffector/Tregulatory cell ratio and upregulates death receptors in metastatic melanoma patients. J Transl Med. 2017;15(1):79. doi: 10.1186/s12967-017-1182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loskog A., Tötterman T. CD40L - a multipotent molecule for tumor therapy. Endocr Metab Immune Disord Drug Targets. 2007;7:23–28. doi: 10.2174/187153007780059432. [DOI] [PubMed] [Google Scholar]
- 44.Tong A., Stone M. Prospects for CD40-directed experimental therapy of human cancer. Cancer gene Ther. 2003;10:1–13. doi: 10.1038/sj.cgt.7700527. [DOI] [PubMed] [Google Scholar]
- 45.Oord J., et al. CD40 is a prognostic marker in primary cutaneous malignant melanoma. Am J Pathol. 1997;149:1953–1961. [PMC free article] [PubMed] [Google Scholar]
- 46.Cabeza N., et al. Surface expression of collagen receptor Fc receptor-γ/glycoprotein VI is enhanced on platelets in type 2 diabetes and mediates release of CD40 ligand and activation of endothelial cells. Diabetes. 2004;53(8):2117–2121. doi: 10.2337/diabetes.53.8.2117. [DOI] [PubMed] [Google Scholar]
- 47.Elzey B.D., et al. Platelet-mediated modulation of adaptive immunity: a communication link between innate and adaptive immune compartments. Immunity. 2003;19(1):9–19. doi: 10.1016/s1074-7613(03)00177-8. [DOI] [PubMed] [Google Scholar]
- 48.Iannacone M., et al. Platelets mediate cytotoxic T lymphocyte–induced liver damage. Nat Med. 2005;11(11):1167–1169. doi: 10.1038/nm1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain S., Russell S., Ware J. Platelet glycoprotein VI facilitates experimental lung metastasis in syngenic mouse models. J Thromb Haemost. 2009;7(10):1713–1717. doi: 10.1111/j.1538-7836.2009.03559.x. [DOI] [PubMed] [Google Scholar]
- 50.Turnbull A.K., et al. Accurate prediction and validation of response to endocrine therapy in breast cancer. J Clin Oncol. 2015;33(20):2270–2278. doi: 10.1200/JCO.2014.57.8963. [DOI] [PubMed] [Google Scholar]
- 51.Ellis M.J., et al. Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: results from the american college of surgeons oncology group Z1031 trial (Alliance) J Clin Oncol. 2017;35(10):1061–1069. doi: 10.1200/JCO.2016.69.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., et al. Functional gene expression signatures from on-treatment tumor specimens predict anti-PD1 blockade response in metastatic melanoma. Biomolecules. 2023:13. doi: 10.3390/biom13010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supplementary Figure 1. Identification of GMPA signatures associated with patient survival. (a) GMPA signature score curve for DSS in SKCM patients from TCGA. (b) GMPA signature score curve for DFS in SKCM patients from the GSE65904 dataset. (c) Multivariate analysis including GMPA signature scores and clinical factors for DSS in SKCM patients from TCGA. (d) Multivariate analysis including GMPA signature scores and clinical factors for DFS in SKCM patients from the GSE65904 dataset.
Supplementary material Supplementary Figure 3. Spearman’s correlation coefficients between the GMPA signature scores and CD147 expression in the 19 types of cancer.
Supplementary material Supplementary Figure 4. Relationships among GMPA signatures and KEGG signaling pathways in cancer patients with high- and low-GMPA signature scores.
Supplementary material Supplementary Figure 5. Spearman’s correlation coefficients between the GMPA signature scores and CD40LG expression in the 19 types of cancer.
Supplementary material Supplementary Figure 6. Immunophenoscores (IPS) of CTLA4 and PDCD1 between high- and low-GMPA signature scores in 19 types of cancer from the TCGA database.
Supplementary material Supplementary Figure 7. Spearman’s correlation coefficients between the GMPA signature scores and CD147 expression in on-treatment samples from four anti-PD1 therapy cohorts of metastatic melanoma.
Supplementary material Supplementary Figure 8. Spearman’s correlation coefficients between the GMPA signature scores and CD40LG expression in on-treatment samples from four anti-PD1 therapy cohorts of metastatic melanoma.
Supplementary material Supplementary Figure 9. GMPA signatures for on-treatment samples with BRAF mutation and BRAF WT. (a, b) Boxplot of GMPA signature scores for on-treatment samples with BRAF V600E mutation (a) and BRAF WT (b) in the Abril-Rodriguez et al. dataset. R: responders; NR: non-responders. (c) ROC and AUC for GMPA signatures of on-treatment samples with BRAF V600E mutation and BRAF WT from Abril-Rodriguez et al. dataset.
Supplementary material
Supplementary material Supplementary Figure 2. Multivariate analysis including GMPA signature scores and tumor stage for OS in 19 types of cancer from the TCGA database.