Highlights
-
•
Emission fluorescence is a versatile and powerful biophysical technique.
-
•
Changes in the nano-environment provoke alterations in the fluorophore behaviour.
-
•
Conformational changes and intermolecular contacts can be studied using fluorescence.
-
•
Fluorescent, photo-switchable, -convertible, large Stokes shifts proteins are known.
-
•
Fluorescence microscopy provides valuable live-cell investigations.
Keywords: fluorescence, protein-ligand, protein-protein, quenching, Stern-Volmer, fluorescent proteins, and fluorescence microscopy
Abstract
Emission fluorescence is one of the most versatile and powerful biophysical techniques used in several scientific subjects. It is extensively applied in the studies of proteins, their conformations, and intermolecular contacts, such as in protein-ligand and protein-protein interactions, allowing qualitative, quantitative, and structural data elucidation. This review, aimed to outline some of the most widely used fluorescence techniques in this area, illustrate their applications and display a few examples. At first, the data on the intrinsic fluorescence of proteins is disclosed, mainly on the tryptophan side chain. Predominantly, research to study protein conformational changes, protein interactions, and changes in intensities and shifts of the fluorescence emission maximums were discussed. Fluorescence anisotropy or fluorescence polarization is a measurement of the changing orientation of a molecule in space, concerning the time between the absorption and emission events. Absorption and emission indicate the spatial alignment of the molecule's dipoles relative to the electric vector of the electromagnetic wave of excitation and emitted light, respectively. In other words, if the fluorophore population is excited with vertically polarized light, the emitted light will retain some polarization based on how fast it rotates in solution. Therefore, fluorescence anisotropy can be successfully used in protein-protein interaction investigations. Then, green fluorescent proteins (GFPs), photo-transformable fluorescent proteins (FPs) such as photoswitchable and photoconvertible FPs, and those with Large Stokes Shift (LSS) are disclosed in more detail. FPs are potent tools for the study of biological systems. Their versatility and wide range of colours and properties allow many applications. Finally, the application of fluorescence in life sciences is exposed, especially the application of FPs in fluorescence microscopy techniques with super-resolution that enables precise in vivo photolabeling to monitor the movement and interactions of target proteins.
Graphical abstract
Emission fluorescence
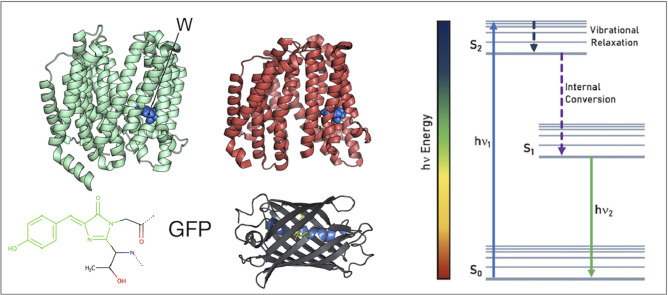
Fluorescence in protein chemistry and biophysics is a phenomenon by which a protein emits light when exposed to radiation such as ultraviolet [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. The absorbed radiation (invisible to the human eye) is transformed into light with a wavelength longer than the incident radiation [1], [2], [3]. Fluorescence usually occurs in real time, with the fluorescent light emission just after the protein is exposed to the primary energy source [2]. Fluorescence originates from electronic transitions from aromatic amino acids [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], such as tryptophan (Trp, W), phenylalanine (Phe, F), and tyrosine (Tyr, Y) that take electrons previously displaced by incident radiation to excited states in the protein structure again to ground states (Fig. 1). But fluorescence may also come from the in-site formed chromophores such as those with heteroaromatic (imidazolinone) ring typical for fluorescent proteins [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47]. The last phenomena are explored with success in fluorescence microscopy [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101]. Using fluorescence tools, chemical, and biological systems are being studied at a molecular level and in real time. The fluorescence images are successfully used in structure elucidation, but also for elucidation of interconnectivity and motion of target proteins.
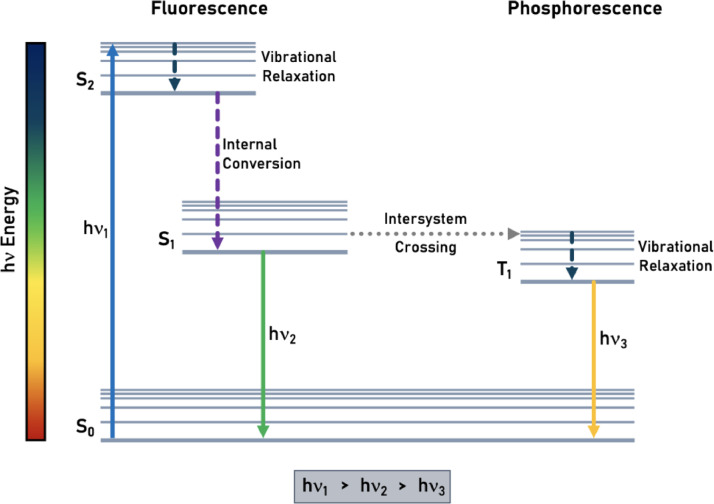
Fig. 1.
Jablonski diagram, showing the different stages of fluorescence and phosphorescence. Note that emission happens at the lowest excited state in each system, as described by Kasha's rule. The difference between and , for example, is explained by the occurrence of Stokes shift (i.e., the difference between excitation and emission spectra).
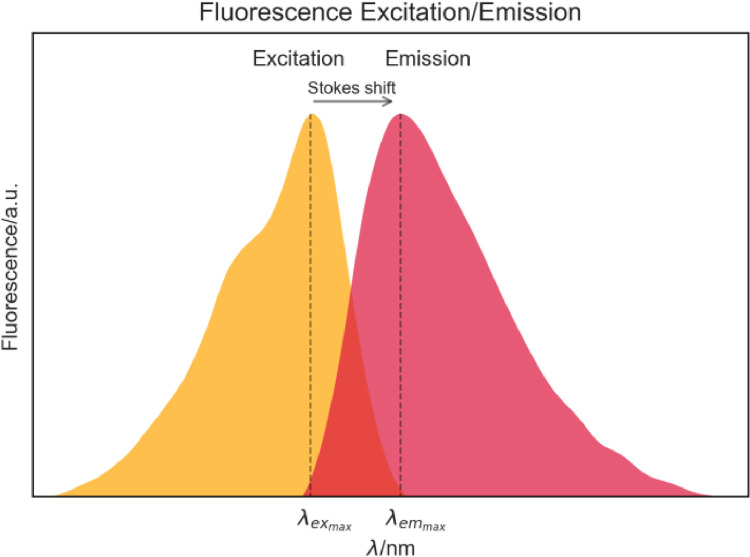
Our first topic accounts for aromatic acid residues in proteins [2,3]. The aromatic residues undergo π–π⁎ transitions under excitation with light of a particular wavelength and then release photons, emitting fluorescence. The protein fluorescence emission maximum is always observed at longer wavelengths than the excitation ( > ). This phenomenon occurs due to vibrational relaxation in the excited singlet state and is called Stokes Shift [17], as observed in Fig. 2. It is important to have in mind that emission fluorescence and a lifetime of the aromatic amino acids are very sensitive to changes in protein's secondary or tertiary structure, principally because of the relaxation mechanisms of their excited states and energy transfer to the surrounding environment [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]. The charge (π-electron) and energy transfer processes are very important for intrinsic protein properties, and fluorophores' intra- and inter-molecular interactions can drastically disturb the intrinsic properties. Therefore, protein structural features and interactions with other molecules can be successfully studied by applying fluorescence [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20].
Fig. 2.
Fluorescence excitation and emission spectra, and corresponding Stokes shift.
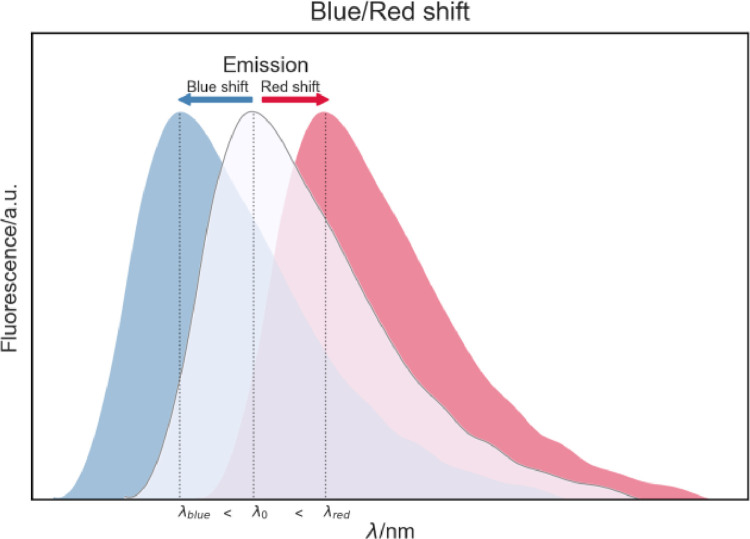
An intriguing aspect of protein fluorescence emission is the occurrence of changes in the maximum emission wavelength towards larger wavelengths (redshift) or smaller wavelengths (blueshift) [2,3]. These variations can occur, for instance, when a fluorophore-containing protein experiences conformational changes. For example, a protein with just one fluorophore, such as tryptophan, has a specific maximum fluorescence emission wavelength (). If this protein undergoes a conformational change that affects the surroundings of the fluorophore, its maximum emission wavelength can be shifted to higher or lower wavelengths. If the shifts to a higher value, a redshift is observed, and if the opposite happens, i.e., the shift is towards smaller wavelengths, then a blueshift is observed (Fig. 3). The protein fluorophore takes part in intermolecular interactions, such as hydrogen bonds, and hydrophobic and electrostatic interactions. Therefore, even a tiny protein structural change may affect the fluorophore nano-environment and cause a change in intermolecular interactions, observable in maximum emission wavelength change [20]. If the observed maximum emission wavelength suffered a blue shift, it is expected that fluorophore suddenly experienced a more hydrophobic nano-environment and might be engaged in stronger affinity interactions when compared to the protein's previous structure.
Fig. 3.
Fluorescence emission wavelength changes – redshift and blueshift.
Proteins exert broad biological functions, in which the spatial conformation can vary their function which can be influenced by the environment [5], [6], [7], [8]. One example is linked to how conformational changes can become essential for the binding of protein structures in the cells of the organism [12]. In the study of protein conformational changes, the fluorescence technique can be very informative and provide valuable answers on protein chemistry and biophysics through emission maximum shift phenomenon that can be combined with quenching/self-quenching.
The effect of protein conformational changes can be exemplified on cellular retinoic acid-binding protein I (CRABP I), as a function of various anions such as SO42−, HPO42− and Cl− employing fluorescence. In this study, a fixed wavelength of 295 nm for excitation was used to avoid interference with tyrosine amino acid residues, and emission wavelengths were observed between 305 and 450 nm. The wavelength of the emission and excitation slits was 5 nm. It was found that regardless of the anion used, the phenomenon of salt-enhanced fluorescence (SEF) occurs, in which there is an increase in intensity proportional to the salt concentration. This is because, due to the conformation change caused by the addition of salt, some amino acid residues such as arginine and asparagine which are responsible for the suppression of fluorescence, have their 3D structural features changed, leading to a fluorescence intensity enhancement.
Also, surrounding conditions, such as solution conditions and pH, influence the binding of the polyphenol quercetin to the wheat protein gliadin and can be studied using the fluorescence technique [23]. Fluorescence tests were performed from pH 2 to 9. The excitation range of the protein was kept fixed at 295 nm, and a scan was made for the emission wavelengths between 300 to 500 nm. The concentration of the protein was 5 μmol L−1 and the concentration of quercetin varied between 0, 1.5, 2.5, 5, 10, 15, 30, and 50 μmol L−1. Upon reaching 50 μmol L−1 of quercetin added to gliadin, the quenching phenomenon was observed having a decrease of 85% - 89% of the fluorescence at pH 2.0 - 9.0. Furthermore, at pH 3.0, 5.0, 7.0, and 9.0 a blue shift of around 10 nm occurred, which may be related to the displacement of W residues to a more hydrophobic nano-environment. Another observation was that in alkaline conditions (pH 7.0 - 9.0), there were more intense fluorescence peaks compared to the fluorescence in acidic conditions (pH 2.0 - 6.0), which may indicate that there was an increased exposure of -OH groups from the quencher that is polyphenol compound, and their deprotonation in higher pH values. The observed differences in intensity and shift in maximums of the target protein fluorescence emission strongly depended on the polarity of the fluorophores' nanoenvironments. For example, the pH change caused the charge of proteins from a negative at high pH to a positive value (low pH), therefore, provoking the protein to unfold and rearrange.
Conformational changes
Proteins may change their structure by undergoing conformational changes under various influences and events, such as after a covalent modification by phosphorylation, then during interactions with other molecules from their environment or because of the molecular recognition with other biomolecules, by binding to a drug, among others. The resulting conformational change can be monitored by applying intrinsic protein fluorescence, especially if fluorophores (W, Y, F) have changed their original locations and, consequently, their intermolecular interactions. Being very sensitive to the polarity of the local nano environment, protein fluorophores, such as W, show maximums of the emission light (λmax) in a wide range from ∼308 nm (azurin) to ∼355 nm (glucagon).
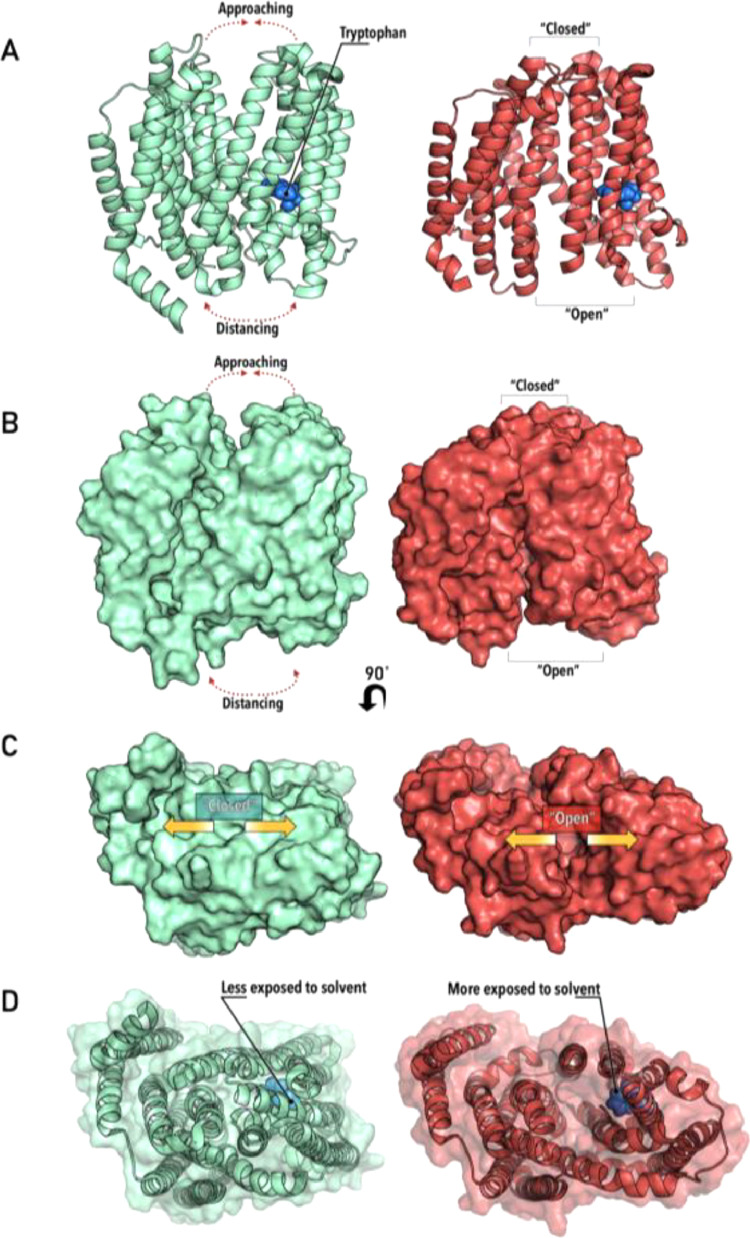
Conformational changes in a fluorophore-containing protein are observed by a phenomenon called blueshift/redshift. Fig. 4 illustrates a hypothetical protein P, based on PDBs 6NC6 and 6NC9, which passes through a conformational change, transitioning from an open/closed (PO/C) state to a closed/open one (PC/O). This protein has only one fluorophore in its primary structure (W).
Fig. 4.
(A) Conformations PO/C (green) and PC/O (red), showing the protein cartoon structure, “opening/closing” mechanism and fluorophore (tryptophan, in blue). (B) Surface representation of conformations PO/C (green) and PC/O (red) – note the opening/closing cavities. (C) 90˚ rotated surface representation of conformations PO/C (green) and PC/O (red). (D) Surface/cartoon representation of conformations PO/C (green) and PC/O (red), showing fluorophore (tryptophan) exposure to solvent.
In this example, two possible outcomes could be expected – if after the conformational conversion, the fluorophore is more “buried” inside the protein structure, and therefore less exposed to the solvent, a blueshift in is expected. If the opposite happens, i.e., the fluorophore is more exposed to the solvent, a redshift would be observed (Fig. 5).
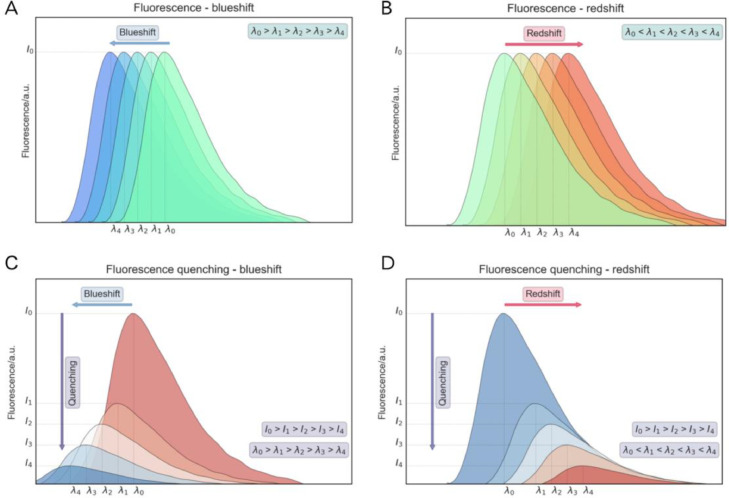
Fig. 5.
(A) shift towards smaller wavelengths – blueshift; (B) shift towards larger wavelengths – redshift. (C) Blueshift combined with fluorescence quenching; (D) redshift combined with fluorescence quenching.
Careful observation of the fluorophore in the protein, P structure in the conformation PC/O, suggests that it is more exposed – this behaviour is reinforced by the redshift in fluorescence emission, and can therefore be used as a robust indicator to monitor conformational change in this hypothetical example.
When a quencher is added to a protein, for example, it is common to observe not only quenching, but a combination of quenching and red/blueshift, as illustrated in Fig. 5C and D. Such behaviour provides interesting information on how the quenching molecules affect the protein conformation.
Fluorescence quenching
The fluorescence intensity of a fluorophore can be diminished by several conditions, denominated fluorescence quenching. This reduction of fluorescence intensity can occur by several processes, some via dynamic quenching, while others via static quenching.
A molecule that causes the reduction of fluorescence intensity of a fluorophore is called a quencher. In a dynamic quenching, a quencher collides with the fluorophore, deactivating it, and therefore, reducing the fluorescence intensity. This type of quenching, based on a collisional effect, can be described mathematically by the Stern-Volmer equation – it will be described in more detail later.
Another possibility for fluorescence quenching can take place when non-fluorescent complexes are formed, in what is called static quenching. For this, a quencher binds to the fluorophore, forming a complex and preventing the occurrence of fluorescence before any excitation/emission happens.
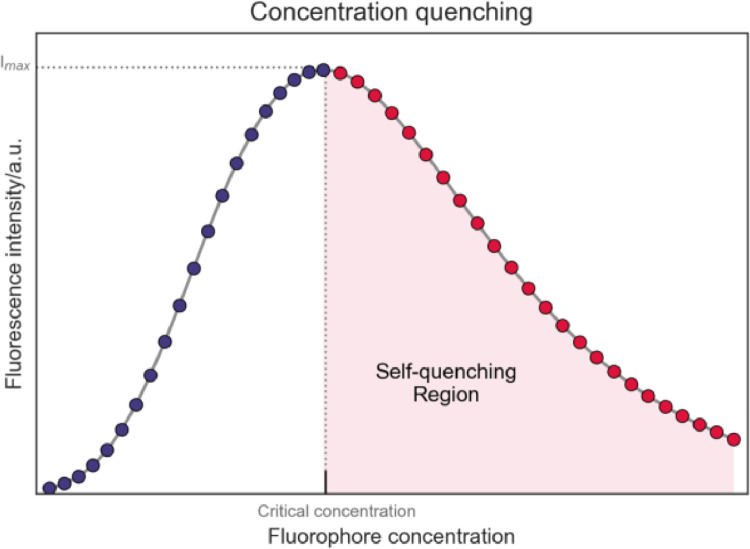
Temperature, viscosity, presence of other molecules, and even fluorophore concentration can result in fluorescence quenching [35,36]. Self-quenching is a phenomenon that occurs at high fluorophore concentrations – at low concentrations, the fluorescence signal is directly proportional to fluorophore concentration, until it reaches a critical concentration, with maximum fluorescence intensity (Fig. 6). Above this concentration, the opposite is observed, with fluorescence intensity being inversely proportional to fluorophore concentration. A proposed explanation for this behaviour lies in the collision between fluorophore molecules that cause the inner filter effect, which involves depletion of excitation light and re-absorption of emission light.
Fig. 6.
Self-quenching of a fluorophore. Fluorescence intensity increases with fluorophore concentration until reaching a critical concentration (purple markers); above this concentration, fluorescence starts to decrease (red markers) – self-quenching.
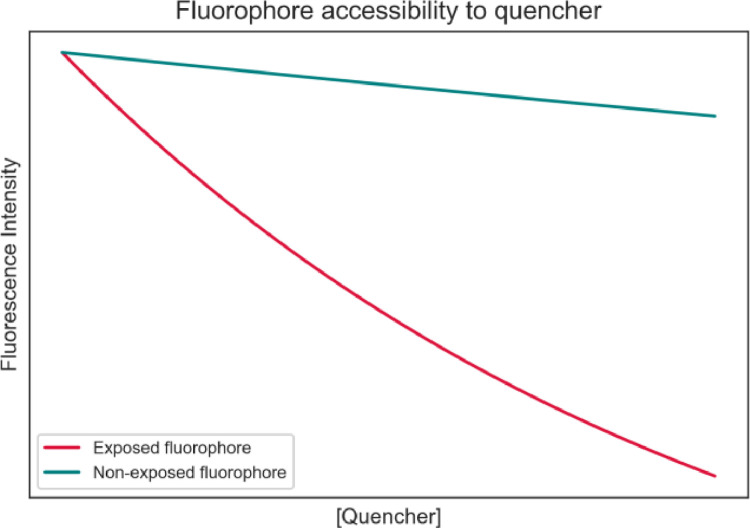
The utilization of a quencher can also provide information about fluorophore accessibility. More accessible fluorophores are more sensitive to quencher concentration and therefore display a more dramatic decrease in fluorescence intensity. Less accessible fluorophores, however, are less sensitive to quencher concentration, having modest or even non-existent decreases in fluorescence concentration. Fig. 7 illustrates such behavior.
Fig. 7.
Fluorophore accessibility can be assessed by quencher concentration. A more exposed (i.e., accessible fluorophore) shows a significant decrease in fluorescence intensity with the increase of quencher concentration (in red); a less exposed (i.e., less accessible) shows a modest decrease in fluorescence, with the increase of quencher concentration.
The Stern-Volmer equation and its implication in fluorescence studies
In the decade of 1920s, due to the optic resolution and improved electronics of instruments, the fluorescence studies of organic, inorganics, and biological compounds were limited to the excitation, emission, and lifetime values focusing on the steady-state and letting the dynamics of the fluorophore unconsidered. The 1943 Nobel Prize winners Otto Stern and Max Volmer proposed the, subsequently called, Stern and Volmer equation in the year 1919, used in fluorophore dynamics studies.
The Stern-Volmer equation can be described with varied degrees of complexity, depending on the desired application. The most common version of Eq. (1) is:
| (1) |
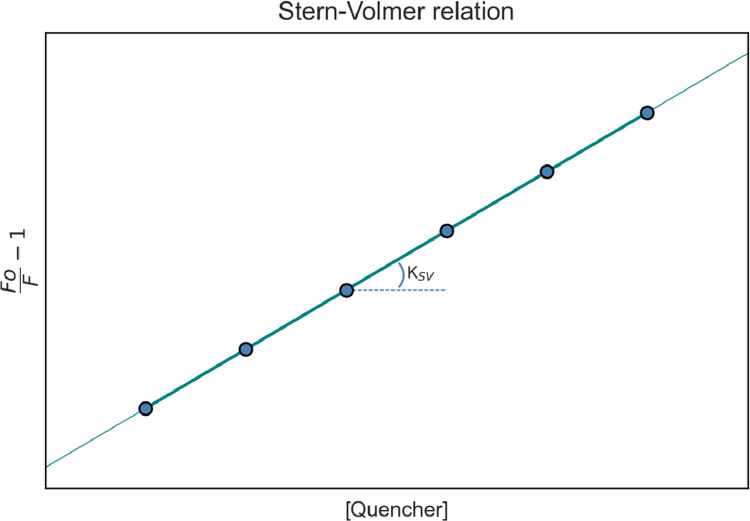
Where F0 and F are fluorescence intensities at starting point and after the addition of a known quencher concentration, respectively, is the Stern-Volmer quenching constant, is the bimolecular quenching constant, is the unquenched lifetime and is the quencher concentration. This form of the Stern-Volmer equation is very convenient for linear fluorescent systems, such as those with only one fluorophore (W), and can correlate to the graph shown in Fig. 8.
Fig. 8.
Stern-Volmer graph, for a single type of fluorophore – note the Stern-Volmer constant () as the curve slope.
However, for more complex systems, a more detailed equation is often required, and the deactivation of an excited molecular population or excited state of the protein must be considered.
Deactivation of an excited state is composed of important steps as follows: absorption, internal conversion, inter-system crossing, and fluorescence. Each step has a characteristic chemical equation that involves a binding constant and a rate expression which are shown in Table 1.
Table 1.
Steps involved in the excited molecular population or excited state of the protein deactivation. A is the fluorophore, Ka, KISC, KIC and Kf are the binding constants in the excitation, intercrossing system relaxation, internal conversion, fluorescence, and quenching steps. The quenching steps are considered only when a quencher is present, and it is symbolised as Q
| Step | Chemical equation | Rate |
|---|---|---|
| Absorption | ||
| Inter system crossing | ||
| Internal conversion | ||
| Fluorescence | ||
| Quenching* |
A fluorescence process has two possible scenarios: A quencher can be absent or present. The quantum efficiency in the first case can be calculated using the Eq. (2):
| (2) |
The fluorescence rate is shown in Table 1, as well as the absorption rate, the formation rate of the excited fluorophore is equal to the deactivation rate and this deactivation includes the intersystem crossing, internal conversion, and fluorescence steps when steady state is considered. Eq. 2 transforms to 4, and it represents the quantum efficiency of a system with the absence of a quencher.
| (3) |
| (4) |
The second case, when a quencher is added, the deactivation rate is given by the intersystem crossing, internal conversion, fluorescence, and quenching steps. Then, Eq. (2) is transformed to 6.
| (5) |
| (6) |
The ratio of quantum yield of bimolecular collisional quenching in absence and presence of a quencher Q is given by 7.
| (7) |
Associating the K values for each decay pathway, the fluorescence lifetime expression appears and (7) modifies to (9).
| (8) |
| (9) |
Where τ is the fluorescence lifetime and KDSV is the dynamic Stern-Volmer constant.
A third scenario can be studied and is when a complex AQ is formed, in this case, the KSSV value changes to a relation between the complex, the fluorophore, and the quencher concentration, given the static Stern-Volmer Eq. (10).
| (10) |
The Stern-Volmer equation [14,17,21] can be used in practical biochemistry as a tool to measure protein inhibition when the relationship between quencher concentration and is plotted. In this plot, the slope represents the KDSV or KSSV constants and those can be differentiated by the lifetime value influence.
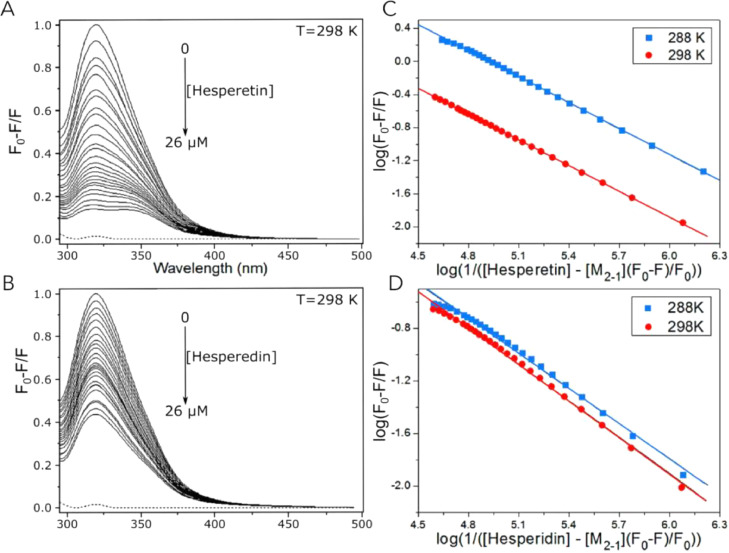
The interaction of the M2-1 protein present in human respiratory syncytial viruses and flavonoids as hesperidin and hesperetin was studied by Piva and collaborators [22] and it's an example of the Sterm-Volmer equation use. Variating the concentration of quencher from 0 to 26 μmol L−1 and using a protein concentration of 6 μmol L−1 was made a double log plot and with this information was possible to measure the binding constant (Kb) between the M2-1 protein and the flavonoid. Due to M2-1 and flavonoid interaction making a complex, the Kb value is obtained from Eq. (9) and it is related to protein and quencher concentrations. Fig. 9 shows the protein M2-1 emission spectra in the absence and presence of hesperetin (A) and hesperidin (B) and their respective double log graphs (C and D). This result shows that the presence of flavonoids can perturb the protein nanoenvironment, affecting its tertiary structure [22,23].
Fig. 9.
A and B show interactions between protein M2-1 and hesperetin and hesperidin, respectively, the concentration of quencher increase from 0 μmol L−1 to 26 μmol L−1, provoking a decrease in fluorescence. C and D correspond to M2-1 and hesperetin (C) and hesperidin (D) double log plots, in blue squares are shown data obtained at 288 K, and red circles show data at 298 K. Adapted from Piva [22], 2020.
Fluorescence anisotropy
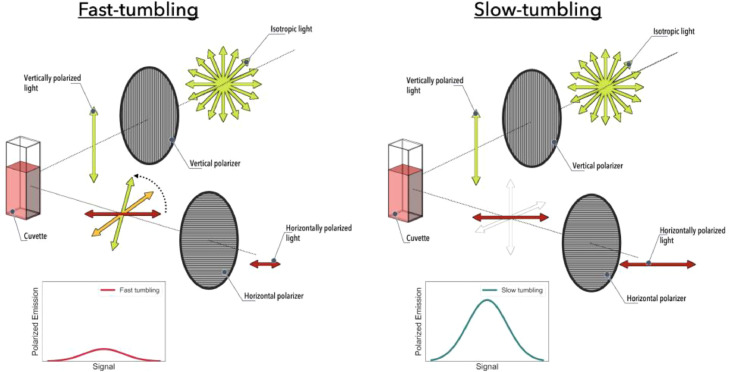
Fluorescence anisotropy is a powerful technique that is commonly used in protein-protein interaction investigations [24], [25], [26]. The technique uses changes in the tumbling time of a fluorescent protein when a ligand (or protein partner) binds to it. This is executed by the usage of perpendicularly oriented polarizers – often, isotropic light at the excitation wavelength is polarized vertically, which is in turn used to excite the fluorescent protein [25]. The fluorescent protein emits light (which is also polarized), which then passed through a horizontal polarizer, and then is detected. The fluorescent protein freely tumbles in solution, changing the angle of the emitted polarized light. A protein without any ligand/partner tends to tumble at a faster speed, while when a ligand/partner is bound, it tumbles at a slower speed. Since the polarizers are perpendicular to each other, in principle, if the fluorescent protein quickly tumbles, the emitted light reaches the second polarizer at an angle that will reduce its intensity (due to the polarizer orientation). If the fluorescent protein tumbles slowly, this change in angle is less pronounced, and therefore, the emitted light reaches the second polarizer at an angle in which the reduction in its signal is significantly smaller. Fig. 10, Fig. 11 illustrate the processes.
Fig. 10.
Schematics of fluorescence anisotropy experimental setup. Fast-tumbling system (left) – fluorophore rotation happens very quickly, resulting in a significant vertical component for the emitted polarised light, and therefore, after passing through the horizontal polarizer, a small signal of horizontally polarised light is recorded. Slow-tumbling system (right) – fluorophore rotation happens very slowly, resulting in minimal vertical component for the emitted polarised light, and therefore, after passing through the horizontal polarizer, a larger signal of horizontally polarised light is measured.
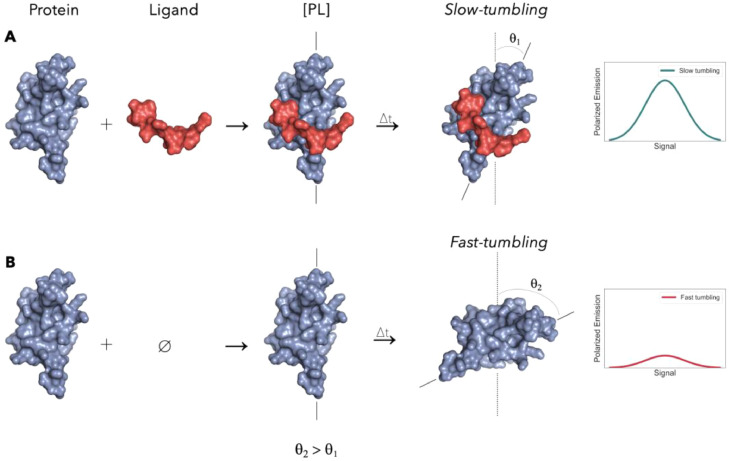
Fig. 11.
(A) Slow-tumbling system. After the ligand binds to the target protein, the complex tumbles at a slower speed due to the increase in size/mass. (B) Fast-tumbling system. Since there is no ligand, the free protein tumbles at a faster speed.
The anisotropy (r) then can be calculated by the variation of the emission intensity of light vertically and horizontally polarised, as indicated by the Eq. (11).
| (11) |
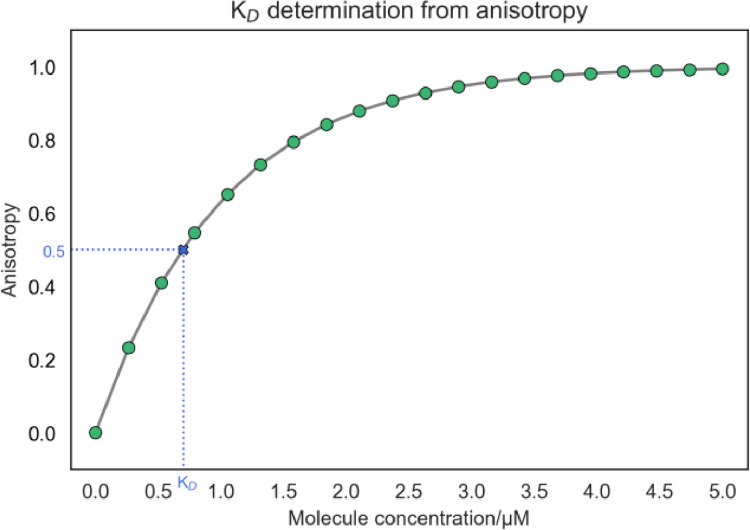
An example of the fluorescence anisotropy binding curve obtained from the titration of the ligand to the target protein is shown in Fig. 12. The shape of the graph points that the two species interacted, and an increase in the anisotropy signal upon the addition of the ligand reached a plateau suggesting that at the end of the titration, just the protein/ligand complex is present in the solution. Then, we can quantitatively describe the interaction between the ligand and the protein by fitting the data using a nonlinear least squares regression to a presumed binding model and obtain the corresponding dissociation equilibrium constant (KD). In this case, fitting to a one-binding site model appropriately described the experimental data.
Fig. 12.
Calculation of KD value using fluorescence anisotropy.
Fluorescence anisotropy can also use fluorescently labelled molecules, such as cysteine-containing client peptides [26], by exploring the change from low anisotropy, to an increased anisotropy provoked by binding to the client protein.
Changes in fluorescence anisotropy upon binding may also explore ligand fluorescence, in which the ligand, when free, shows a rapid rotational diffusion, and when illuminated with linearly polarized light, emits fluorescence that is depolarized because the emitting dipole rotates during the time interval between fluorescence excitation and emission. On the other side, when the ligand interacts with the receptor, the bound ligand shows slower rotation and fluorescence emission maintains linear polarization. The extent of polarization of a solution of fluorescent ligands and receptors is a weighted average of the emissions from the free and bound populations of ligands.
Time-resolved fluorescence
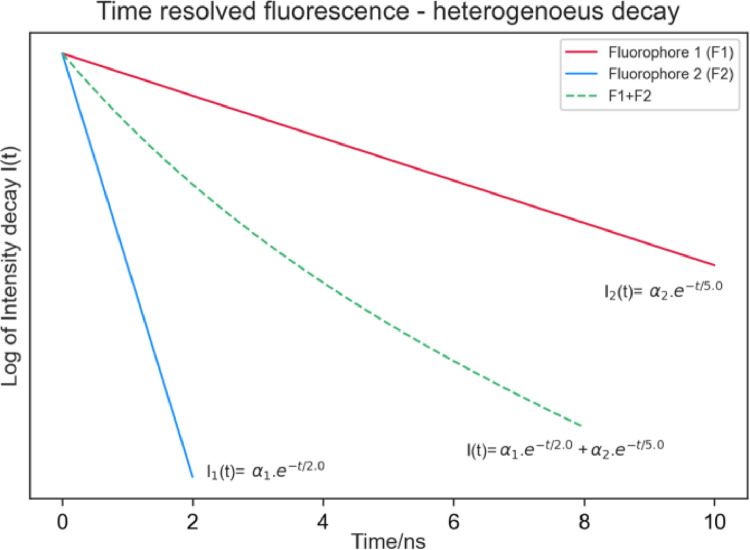
The technique of time-resolved fluorescence spectroscopy has wide application in the study of the structure and dynamics of biological macromolecules, such as proteins [46]. Through this study, it is possible to monitor the interactions and motions that occur in picosecond-nanosecond intervals. One type of time-resolved fluorescence spectroscopy is to investigate the decay of the total fluorescence intensity, which aims to determine the fluorescence lifetime of a fluorophore (Fig. 13). Another type of time-resolved fluorescence corresponds to the decay of polarisation anisotropy and is used to characterise molecular motions, for example, the rotational diffusion of a protein or nucleic acid [46]. The time-resolved fluorescence spectroscopy is one of the only tools with the ability to evaluate processes of reaction mechanisms that occur in picosecond-nanosecond duration, to gain more detailed insights into reactions [47].
Fig. 13.
Time-resolved fluorescence graph. Note that for a system with two distinct fluorophores (F1 and F2, red and blue lines, respectively), the observed decay is a combination of the decays of the two individual fluorophores (green dotted line).
An advantage of the time-resolved technique is that it does not depend on the excitation intensity or the duration of light exposure and is therefore extracted from the decay of fluorescence intensity [48].
In a protein-protein interaction assay, it is possible to apply the time-resolved fluorescence energy transfer technique, which combines a practice of time-resolved fluorimetry with Förster resonance energy transfer, to characterise the modes of action, efficacy, and binding affinities of ligands [49]. The time-resolved method for protein analysis is capable of quantifying concentrations of proteins of interest. To do this, the protein of interest or conformational state has a fluorescence decay signature capable of characterising a particular protein species. The method used for this quantification is based on a series of measurements with intensity in the range of 250-280 nm, in which the time of each measurement varies by at least 10 nanoseconds (ns) so that the fluorescence intensity decay allows the construction of a fluorescence decay curve [50].
The orientation of the fluorophore plays a role in both absorption and emission processes, due to their dipolar nature. A protein's three-dimensional (3D) orientation affects the way in which it interacts with the surrounding electromagnetic field. Protein fluorophores interact with the electromagnetic field primarily through their electric transition dipole moments.
Fluorescent Proteins - FPs
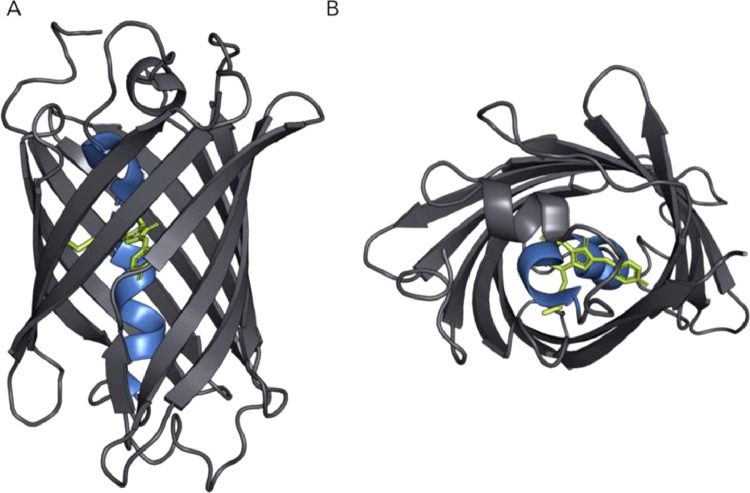
Fluorescent proteins have extensive use in science in a wide range of applications. For instance, they can be used as reporters for gene expression, as fusion tags, and to investigate protein-protein interactions [44]. The Green Fluorescent Protein (GFP) was the first fluorescent protein identified and further developed, and in 2008 Osamu Shimomura, Martin Chalfie, and Roger Tsien were awarded the Nobel Prize for this work. GFP (Fig. 14) was initially purified from the jellyfish Aequorea victoria and named avGFP. Structure-wise, it has a β-roll structure made of antiparallel β-sheets, and a central α-helix, which contains the chromophore, responsible for the protein's fluorescence [45].
Fig. 14.
(A) Frontal and (B) top views of a fluorescent protein, with the central α-helix in blue, and chromophore is shown in green.
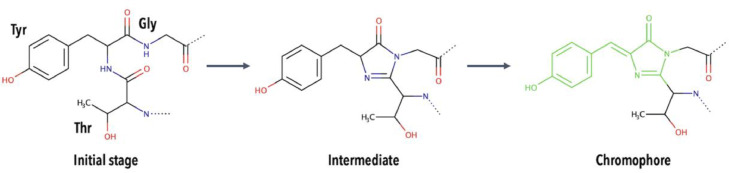
The GFP chromophore [31] is formed by a folding/cyclization/oxidation reaction involving three amino acid residues from the central α-helix. The amino acids involved in chromophore formation can vary but are often glycine (Gly, G), threonine (Thr, T), and tyrosine (Tyr, Y). The exact mechanism for chromophore formation is not completely elucidated, and there are several proposed mechanisms. However, it is believed that its formation can be split into three stages, as illustrated in Fig. 15, starting with the unmodified amino acids (G, T, Y) that pass through an intermediate state, and finally, convert into the chromophore [30], [31], [32], [33], [34].
Fig. 15.
Simplified illustration of the proposed mechanism for chromophore formation in green fluorescent proteins, such as avGFP. The chromophore is shown in green.
Fluorescent proteins (FPs), with dissimilar primary structures, can take different times to complete the chromophore formation, and this process is therefore called chromophore maturation time. The maturation time can vary greatly, taking a few minutes for some FPs, and several hours for others. This can significantly impact FP usage and, therefore, care must be taken when selecting FP, ensuring that it has an appropriate maturation time for the intended use.
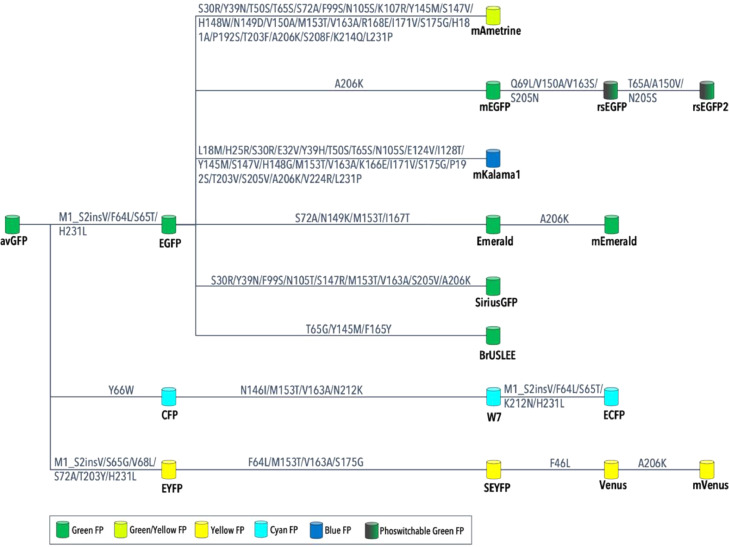
Aiming for the improvement of FP properties, avGFP was further engineered, undergoing through an extensive process of point mutations. These structural changes resulted in new FP lineages, with varied advancements in their properties, such as brightness, maturation time, thermal sensitivity, and even colour. Fig. 16 shows a few of the derivative FP lineages that originated from avGFP, and the corresponding point mutations [32], [33], [34], [35].
Fig. 16.
Example of FP lineages derived from avGFP, and the corresponding mutations that resulted in each individual FP.
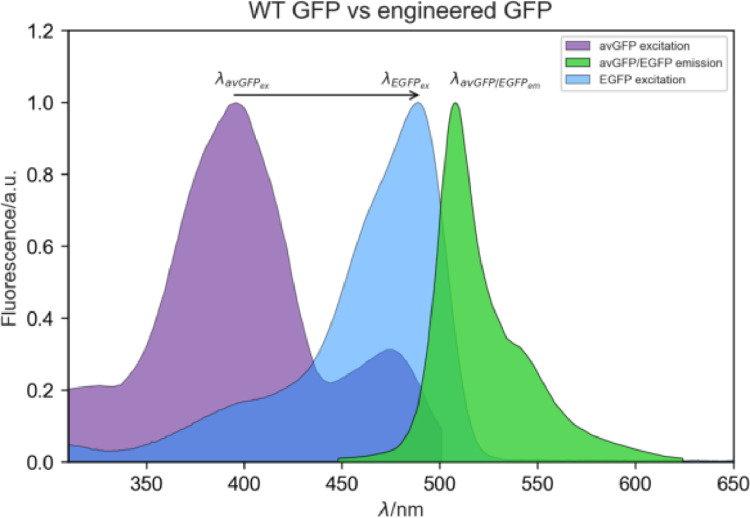
One interesting characteristic observed in the engineered FPs, when compared to avGFP, is the major change in excitation wavelength (). This change, of almost 100 nm, results in an increased fluorescence signal when compared to avGFP. Fig. 17 illustrates this change in .
Fig. 17.
Excitation and emission spectra of avGFP and the engineered EGFP. Note the extensive change in (∼95 nm), while remains unchanged.
Interestingly, some mutations are recurrent in several derivative FPs, even with different colours. For example, the A206K mutation was used in the development of several FPs, and it was used to ensure a monomeric state for these proteins [35].
Currently, there are around 857 FPs deposited in the FPbase [36], with an extensive range of different properties available, like colour, brightness, oligomerization, maturation time, stability, and even photoswitchable and photoconvertible FPs, among others [37].
Fluorescent proteins are powerful tools for the study of biological systems. Their versatility and wide range of colours and properties allow a myriad of applications. To further increase their capability, new types of FPs, with new properties, have been developed. In this section, we will describe three of them, and how they function: Large Stokes Shift (LSS), Photoswitchable and Photoconvertible FPs.
Photoswitchable FPs adopt a classic 11-strand beta-barrel FP structure that encloses an autocatalytically generated chromophore. The cis–trans isomerization of the chromophore methylene bridge between the two rings of the chromophore can account for the photoswitching mechanism. FPs can switch from on to off, with the chromophore at the cis conformer at rest. On the other side, FPs with off–on switching show the trans conformer in the resting state. The chromophore interacts with the surrounding residues, and those determine their resting states, for example, some strong hydrogen bonding interaction stabilises cis conformation enabling an on–off switch, while some FPs, because of stabilising the trans conformer of the chromophore show reversed switching direction. Another mechanism involves a reversible hydration/dehydration reaction on a carbon atom in the imidazolinone ring of the chromophore of Dreiklang. In this reaction, the surrounding residues take part in a reversible hydration/dehydration, and the chromophore π-electron system changes, which shifts absorption wavelengths to the blue. Thus, FPs can show a cis-trans isomerisation in Dronpa and trans-cis isomerisation in Padron; but can also undergo conformational changes in Dreiklang photoswitching where the chromophore suffers a reversible hydration/dehydration reaction on a carbon atom in the imidazolinone ring.
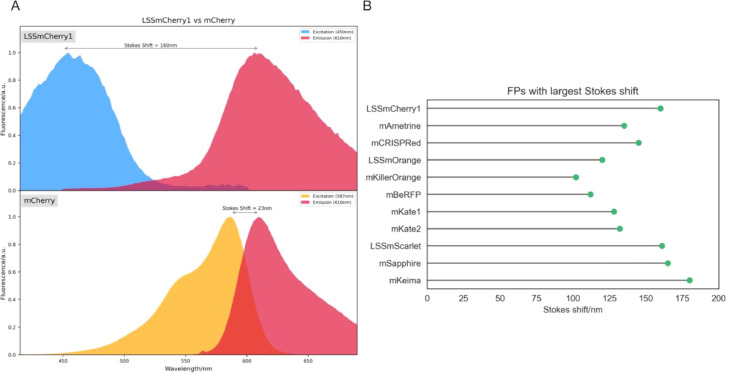
Fluorescent proteins have a difference between their maximum excitation and emission wavelengths. This difference is called Stokes shift, and usually, this difference is in the range of 10-20 nm. Sometimes a greater difference is beneficial for a specific application, or if the Stokes shift is too small, this can impair a measurement. For this reason, FPs with a large difference between their maximum excitation and emission wavelengths were developed, and these are LSS-FPs. There are LSS versions of several of the most “traditional” FPs, such as LSS-mCherry, LSS-mScarlet, aGFP, LSS-mKate2, among several others, as illustrated in Fig. 18.
Fig. 18.
(A) Fluorescence spectra of mCherry (bottom) and its large Stokes shift version, LSS-mCherry (top). Note that there is a dramatic difference (137 nm) in between mCherry and LSS-mCherry, while remains unchanged. (B) Chart of FPs with the largest Stokes shift (i.e., greater than 100 nm).
There are several applications for LSS-FPs, but an interesting application involves multi-colour fluorescence imaging [37]. LSS-FPs are very useful in such circumstances since they reduce the overlap of different emission wavelengths (from the usage of multiple FPs or fluorescent dyes).
Some LSS-FPs experience an increase in acidity that leads to chromophore deprotonation and excited state proton transfer if applicable, which red-shifts the emission peak leading to an LSS protein/fluorophore complex. Essential for selective photo-base activity is the intimate involvement of the target protein structure and sequence that enables excited state proton transfer.
Photoswitchable fluorescent proteins (FPs)
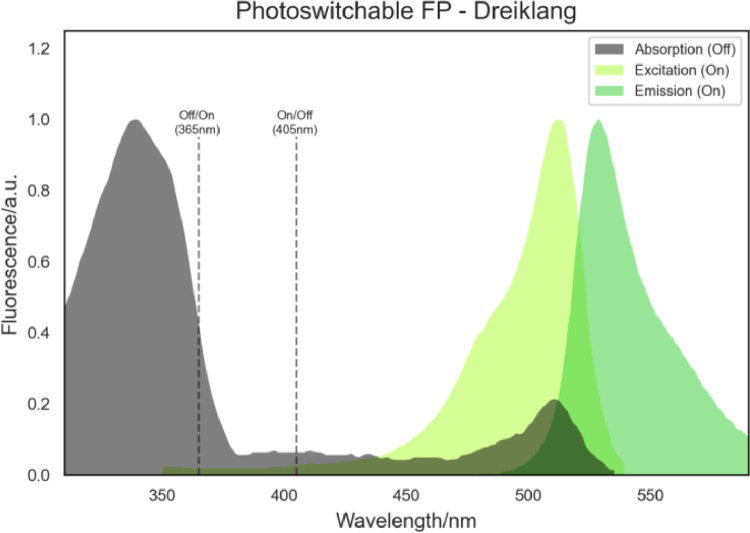
There are circumstances in which having an FP constantly emitting light is not convenient, or even being able to “switch off” an FP in certain circumstances, which can be very advantageous. Photoswitchable (Fig. 19) FPs were developed to have such properties, i.e., to be able to “switch on and off” an FP [40], [41], [42], [43], [44], [45].
Fig. 19.
Example of the spectra of a photoswitchable FP, Dreiklang. Note the off/on and on/off switch wavelengths.
These FPs are switched on () and off () by irradiating the FP with the light of a specific wavelength. Therefore, if a photoswitchable FP is irradiated with , it is converted to its off-state, and even if irradiated with its , no fluorescence emission takes place. However, if irradiated with , the FP is then converted to its on-state, and the normal / condition takes place (Fig. 19).
Photoconvertible fluorescent proteins (FPs)
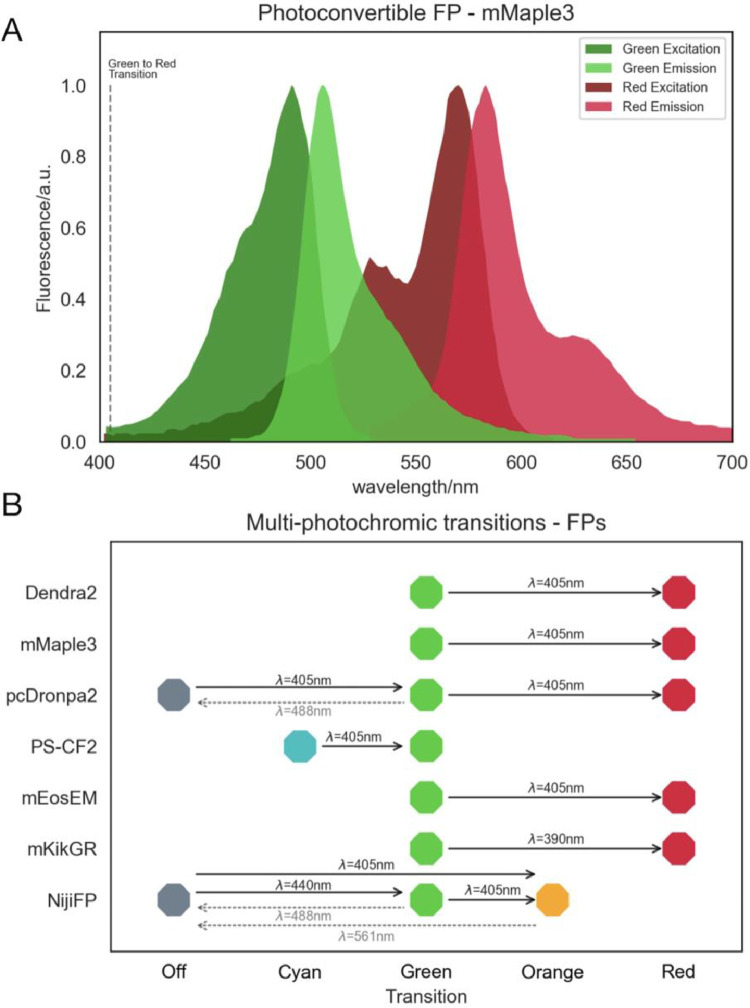
Photoconvertible FPs have a behaviour similar to photoswitchable ones. The main difference is that photoconvertible FPs can switch between different , and, therefore, emit different colours [57]. Some of these can not only switch between different colours but also between on and off states as exemplified in the illustrations shown in Fig. 20.
Fig. 20.
(A) Spectra of the photoconvertible FP mMaple3. Note the two and two , and the green-to-red transition switch wavelength. (B) Illustration of multi-photochromic transitions for photoconvertible FPs, and the corresponding switch wavelengths.
Applications
Protein fluorescence is a phenomenon where a protein chromophore interacts with light and absorbs it, remains in an excited state for a few nanoseconds, and then emits light with lower energy. It is a very sensitive spectroscopic technique that needs just a small amount of a chromophore, mostly in the pmol L−1 to nmol L−1 range. The most important protein chromophore is tryptophan (Trp, W) because of the quantum yield and emission maximum. It is very sensitive to the polarity of the nanoenvironment, pH, and intramolecular interactions, such as with ligands. Important protein fluorescence spectroscopy data include emission spectra obtained in different working conditions, where especially maximum wavelengths of the emission spectra and their intensities, then, lifetime (or decay) and anisotropy (or polarization) measurements are very useful for elucidating protein structure features, conformational changes and interactions within the studied protein, or with different ligands. Many aspects of protein structure and motion, rotation, then, protein interactions with different ligands, and mechanisms of such interactions can be revealed by analysing fluorescence data, such as changes in emission intensity, and maximums of the emitted light. Because it is non-destructive, protein fluorescence can also be monitored as a function of time, therefore, the dynamic nature of the signal can be used to study protein folding, dynamics, assembly, and interactions. The fluorescence emission lifetimes are in the nanosecond range, and commonly faster than most protein conformational transitions, therefore, can even enable the investigation of fast protein conformational changes [46], [47], [48], [49], [50].
Another interesting application of fluorescence spectroscopy came to the light with protein labelling through mutagenesis and chemical modifications [51], [52], [53], [54], [55], [56], and site-specific labelling with external fluorophores. Those fluorescent proteins and fluorescent probes can be used for many applications and are extremely important for in situ investigations of protein interactions, protein assemblies, and protein conformational and 3D studies [57], [58], [59].
The Nobel Laureates in Chemistry 2014, Eric Betzig, Stefan W. Hell, and William E. Moerner were the pioneers in the development of super-resolved fluorescence microscopy. The method of stimulated emission depletion (STED) microscopy explores two laser beams where one stimulates fluorescent molecules to glow, and another cancels out all fluorescence except for that in a nanometre-sized volume. Scanning over the sample, nanometre for nanometre, yields an image with a high resolution. The single-molecule microscopy relies upon the possibility to turn the fluorescence of individual molecules on and off. The same area is scanned multiple times, letting just a few interspersed molecules glow each time. Superimposing these images yields a dense super-image resolved at the nano-level.
Fluorescent microscopy makes use of fluorescent proteins (FPs) [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103] to label target proteins and monitor their localisation in cells, for example, and reveal information about the target protein's relationships with other molecules within cells. It has become an essential tool in biology and the biomedical sciences. The application of an array of fluorochromes has made it possible to identify cells and sub-microscopic cellular components with a high degree of specificity. Through the use of multiple fluorescence labelling, different probes can simultaneously identify several target molecules.
Additionally, there are fluorescence microscopy techniques designed for monitoring the single-molecule orientation that is based on modulation of the absorption/emission polarization(s), exploring the spatial dependence of absorption, or based on the spatial distributions of emitted fluorescence.
Conclusions and perspectives
The fluorescence of proteins is a process consisting of the excitation of a protein fluorophore by an incoming photon (UV range), which is the fastest event, followed by a vibrational relaxation of excited state electrons to the lowest energy level, and ended up by the emission of a longer wavelength photon that returns the molecule to the ground state. The entire molecular fluorescence lifetime takes around billionths of a second and forms the basis for the expansive fields of steady-state and time-resolved fluorescence spectroscopy and microscopy.
As fluorescence emission spectra and lifetimes are very sensitive to molecular interactions and surrounding conditions, therefore, fluorescence is a remarkably useful biophysical technique for the investigations of the structure, function, and interactions of proteins. The most explored protein intrinsic fluorophore is tryptophan. Fluorescence spectroscopy explores not only excitation and emission spectra, and emission maximums, but also quenching phenomena, and anisotropy i.e. polarization. Also, the fluorescence-based techniques provide diverse ways of probing the chemical environments and intermolecular interactions that can be successfully applied to understanding receptor-mediated signaling.
A very important discovery of fluorescent proteins revolutionised biophysics and microscopy and opened up advantageous monitoring of living cells. Many important fluorescence proteins' photophysical properties have been explored so far, and those showing photoconversion and photoswitching, as well as large Stokes shifts found their way to exiting investigations. Additionally, improvements have led to the application of super-resolved fluorescence microscopy to proteins' structure, function, and relationships investigations in living cells at nanometer resolution, up to 20 nm, with a single-molecule sensitivity. There are still many challenges to overcome. For example, fluorescence labelling changes the native cell conditions, so the control conditions must be very well-designed and defined. If the studied complexes or species are smaller than the current limit of super-resolution, critical dynamic processes, such as self-association and oligomerization, fundamental for the functioning of many proteins, cannot be studied. Some current and future studies introduce proposals for the application of new fluorescence proteins, such as red-emitting ones, for resolution and image improvements.
Finally, as in many other techniques, fluorescence experiments are quite rich in information that needs to be understood and interpreted in an unbiased manner, mostly because the data are the sum of populations of protein molecules in the sample, except when analysed data from a single‐molecule protein fluorescence experiment.
Author contributions
All authors took part in the formal analysis, writing, reviewing, and editing of the original draft. Dr. Rodrigues and Dr. Tasic cooperated in the conceptualization, writing, and final revision of the manuscript. Dr. Rodrigues designed the figures of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the funding agencies, especially, the Sao Paulo Research Foundation (FAPESP), Grant numbers # 2020/08615-8 and # 2023/02338-0, National Council for Scientific and Technological Development (CNPq), and the Coordination of Improvement of Higher Education Personnel (CAPES, Grant number 001). We also acknowledge National Institute for Bioanalytics (INCTBio) sponsored by FAPESP (Grant number # 2014/50867-3).
Data availability
This is a review article.
References
- 1.Albrecht C., Lakowicz J.R. Principles of fluorescence spectroscopy. Anal. Bioanal. Chem. 2008;390:1223–1224. [Google Scholar]
- 2.Lakowicz J.R. 2nd. Plenum; New York: 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 3.Meech S.R. Excited state reactions in fluorescent proteins. Chem. Soc. Rev. 2009;38:2922. doi: 10.1039/b820168b. [DOI] [PubMed] [Google Scholar]
- 4.Misra G., Ramachandran R. Hsp70-1 from Plasmodium falciparum: protein stability, domain analysis and chaperone activity. Biophys. Chem. 2009;142:55–64. doi: 10.1016/j.bpc.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Michalet X., Weiss S., Jäger M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem. Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M., Li Q., Wang R. Current experimental methods for characterizing protein–protein interactions. ChemMedChem. 2016;11:738–756. doi: 10.1002/cmdc.201500495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobiilka B.K., Gether U. Use of fluorescence spectroscopy to study conformational changes in the β2-adrenoceptor. Methods in Enzymology. 2002;343:170–182. doi: 10.1016/s0076-6879(02)43134-5. [DOI] [PubMed] [Google Scholar]
- 8.Sengupta I., Udgaonkar J. Monitoring site-specific conformational changes in real-time reveals a misfolding mechanism of the prion protein. eLife. 2019;8:e44698. doi: 10.7554/eLife.44698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misra G., Ramachandran R. Exploring the positional importance of aromatic residues and lysine in the interactions of peptides with the Plasmodium falciparum Hsp70-1. Biochim Biophys Acta (BBA)-Prot Proteom. 2010;1804:2146–2152. doi: 10.1016/j.bbapap.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Shang N., Meram C., Bandara N., Wu J. Protein and peptides for elderly health. Adv Protein Chem Struct Biol. 2018;112:265–308. doi: 10.1016/bs.apcsb.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Backman L. Protein Chemistry. 1st. De Gruyter; Berlin, Boston: 2020. [Google Scholar]
- 12.Orellana L. Large-scale conformational changes and protein function: Breaking the in silico barrier. Frontiers in Molecular Biosciences. 2019;6:117. doi: 10.3389/fmolb.2019.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millan S., Swain B.C., Tripathy U., Mishra P., Sahoo H. Effect of micro-environment on protein conformation studied by fluorescence-based techniques. J. Mol. Liq. 2020;320 [Google Scholar]
- 14.Wahba M.E.K., El-Enany N., Belal F. Application of the Stern–Volmer equation for studying the spectrofluorimetric quenching reaction of eosin with clindamycin hydrochloride in its pure form and pharmaceutical preparations. Anal. Methods. 2015;7:10445–10451. [Google Scholar]
- 15.Zhao H., Zastrow M.L. Transition metals induce quenching of monomeric near-infrared fluorescent proteins. Biochemistry. 2022;61:494–504. doi: 10.1021/acs.biochem.1c00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sillen A., Verheyden S., Delfosse L., Braem T., Robben J., Volckaert G., Engelborghs Y. Mechanism of fluorescence and conformational changes of the sarcoplasmic calcium binding protein of the sand worm Nereis diversicolor upon Ca2+ or Mg2+ binding. Biophys J. 2003;85:1882–1893. doi: 10.1016/S0006-3495(03)74616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehlen M.H. The centenary of the Stern-Volmer equation of fluorescence quenching: From the single line plot to the SV quenching map. Journal of Photochemistry and Photobiology C: Photochemistry Reviews. 2020;42 [Google Scholar]
- 18.Bizzarri A.R., Cannistraro S. Temperature modulation of the DBDp53 structure as monitored by static and time-resolved fluorescence combined with molecular dynamics simulations. The Journal of Physical Chemistry B. 2021;125:10166–10173. doi: 10.1021/acs.jpcb.1c05909. [DOI] [PubMed] [Google Scholar]
- 19.Mohan V., Sengupta B., Acharyya A., Yadav R., Das N., Sen P. Region-specific double denaturation of human serum albumin: Combined effects of temperature and GnHCl on structural and dynamical responses. ACS Omega. 2018;3:10406–10417. doi: 10.1021/acsomega.8b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellmann N., Schneider D. Hands on: Using tryptophan fluorescence spectroscopy to study protein structure. Methods Mol. Biol. 2019;1958:379–401. doi: 10.1007/978-1-4939-9161-7_20. [DOI] [PubMed] [Google Scholar]
- 21.Hamann S., Kiilgaard J.F., Litman T., et al. Measurement of cell volume changes by fluorescence self-quenching. Journal of Fluorescence. 2002;12:139–145. [Google Scholar]
- 22.Piva H.M.R., Sá J.M., et al. Insights into interactions of flavanones with target human respiratory syncytial virus M2-1 protein from STD-NMR, fluorescence spectroscopy, and computational simulations. International Journal of Molecular Sciences. 2020;21:2241. doi: 10.3390/ijms21062241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Tang Y., Yang Y., et al. Interaction between wheat gliadin and quercetin under different pH conditions analyzed by multi-spectroscopy methods. Spectrochim Acta A Mol Biomol Spectrosc. 2020;229 doi: 10.1016/j.saa.2019.117937. [DOI] [PubMed] [Google Scholar]
- 24.Gijsbers A., Nishigaki T., Sánchez-Puig N. N. Fluorescence anisotropy as a tool to study protein-protein Interactions. J. Vis. Exp. 2016;116:54640. doi: 10.3791/54640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi A., Taylor C. Analysis of protein-ligand interactions by fluorescence polarization. Nat Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sijbesma E., Somsen B.A., Miley G.P., Leijten-van de Gevel I.A., Brunsveld L., Arkin M.R., Ottmann C. Fluorescence anisotropy-based tethering for discovery of protein–protein interaction stabilizers. ACS Chem. Biol. 2020;15:3143–3148. doi: 10.1021/acschembio.0c00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besson B., Eun H., Kim S., Windisch M.P., Bourhy H., Grailhe R. Optimization of BRET saturation assays for robust and sensitive cytosolic protein–protein interaction studies. Sci. Rep. 2022;12:1–11. doi: 10.1038/s41598-022-12851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kremers G.J., Gilbert S.G., Cranfill P.J., Davidson M.W., Piston D.W. Fluorescent proteins at a glance. J. Cell Sci. 2011;124:2676. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craggs T.D. Green fluorescent protein: Structure, folding and chromophore maturation. Chem. Soc. Rev. 2009;38:2865–2875. doi: 10.1039/b903641p. [DOI] [PubMed] [Google Scholar]
- 30.Grigorenko B.L., Krylov A.I., Nemukhin A.V. Molecular modeling clarifies the mechanism of chromophore maturation in the Green Fluorescent Protein. J. Am. Chem. Soc. 2017;139:10239–10249. doi: 10.1021/jacs.7b00676. [DOI] [PubMed] [Google Scholar]
- 31.Reid B.G., Flynn G.C. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 32.Dedecker P., De Schryver F.C., Hofkens J. Fluorescent proteins: Shine on, you crazy diamond. J. Am. Chem. Soc. 2013;135:2387–2402. doi: 10.1021/ja309768d. [DOI] [PubMed] [Google Scholar]
- 33.Den-Dunnen J.T., Antonarakis E. Nomenclature for the description of human sequence variations. Hum. Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 34.Hirano M., Ando R., Shimozono S., et al. Author Correction: A highly photostable and bright green fluorescent protein. Nat. Biotechnol. 2022;40:1412. doi: 10.1038/s41587-022-01469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cormack B.P., Valdivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 36.Lambert T.J. FPbase: a community-editable fluorescent protein database. Nature Methods. 2019;16:277–278. doi: 10.1038/s41592-019-0352-8. [DOI] [PubMed] [Google Scholar]
- 37.Costantini L., Baloban M., Markwardt M., et al. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015;6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh M., Nath S., Hajra A., Sinha S. Fluorescence self-quenching of tetraphenylporphyrin in liquid medium. J. Lumin. 2013;141:87–92. [Google Scholar]
- 39.Lichtenthaler H.K. Multi-colour fluorescence imaging of photosynthetic activity and plant stress. Photosynthetica. 2021;59:364–380. [Google Scholar]
- 40.Berlin S., Carroll E., Newman Z., et al. Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat. Methods. 2015;12:852–858. doi: 10.1038/nmeth.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subach F., Patterson G., Manley S., et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson G.H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 43.Paez-Segala M., Sun M., Shtengel G., et al. Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat. Methods. 2015;12:215–218. doi: 10.1038/nmeth.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roebroek T., Vandenberg W., Sipieter F., et al. Simultaneous readout of multiple FRET pairs using photochromism. Nat. Commun. 2021;12:2005. doi: 10.1038/s41467-021-22043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scruggs A.W., Flores C.L., Wachter R., Woodbury N.W. Development and characterization of green fluorescent protein mutants with altered lifetimes. Biochemistry. 2005;44:13377–13384. doi: 10.1021/bi050550f. [DOI] [PubMed] [Google Scholar]
- 46.Millar D.P. Fluorescence studies of DNA and RNA structure and dynamics. Curr Opin Struct Biol. 1996;6:322–326. doi: 10.1016/s0959-440x(96)80050-9. [DOI] [PubMed] [Google Scholar]
- 47.Chukhutsina V.U., Holzwarth A.R., Croce R. Time-resolved fluorescence measurements on leaves: Principles and recent developments. Photosynth Res. 2019;140:355–369. doi: 10.1007/s11120-018-0607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandao M.P., de Carvalho dos Anjos V., Bell M.J.v. Time resolved fluorescence of cow and goat milk powder. Spectrochim Acta A Mol Biomol Spectrosc. 2017;171 doi: 10.1016/j.saa.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Lin W., Wang Y.M., Chai S.C., et al. SPA70 is a potent antagonist of human pregnane X receptor. Nat. Commun. 2017;8:741. doi: 10.1038/s41467-017-00780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hales J.E., Aoudjane S., Aeppli G., Dalby P.A. Proof-of-concept analytical instrument for label-free optical deconvolution of protein species in a mixture. J Chromatogr A. 2021;1641 doi: 10.1016/j.chroma.2021.461968. [DOI] [PubMed] [Google Scholar]
- 51.Poudel C., Mela I., Kaminski C.F. High-throughput, multi-parametric, and correlative fluorescence lifetime imaging. Methods Appl. Fluoresc. 2020;8 doi: 10.1088/2050-6120/ab7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghuraman H., Chattopadhyay A. Interaction of melittin with membrane cholesterol: A fluorescence approach. Biophys. J. 2004;87:2419–2432. doi: 10.1529/biophysj.104.043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernhard S., Goodman C.K., Norton E.G., et al. Time-dependent fluorescence spectroscopy to quantify complex binding interactions. ACS Omega. 2020;5:29017–29024. doi: 10.1021/acsomega.0c03416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunn S. Handbook of Neurochemistry and Molecular Neurobiology. Springer; New York, NY: 2007. Fluorescence Measurements of Receptor–Ligand Interactions. Lajtha, A., Baker, G., Dunn, S., Holt, A. [DOI] [Google Scholar]
- 55.Heine V., Dey C., Bojarová P., Křen V., Elling L. Methods of in vitro study of galectin-glycomaterial interaction. Biotechnology Advances. 2022;58 doi: 10.1016/j.biotechadv.2022.107928. [DOI] [PubMed] [Google Scholar]
- 56.Raghuraman H., Chatterjee S., Das A. Site-directed fluorescence approaches for dynamic structural biology of membrane peptides and proteins. Front. Mol. Biosci. 2019;6:96. doi: 10.3389/fmolb.2019.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toseland C.P. Fluorescent labeling and modification of proteins. J Chem Biol. 2013;6:85–95. doi: 10.1007/s12154-013-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimogawa M., Petersson E.J. New strategies for fluorescently labeling proteins in the study of amyloids. Current Opinion in Chemical Biology. 2021;64:57–66. doi: 10.1016/j.cbpa.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meera J., Patel G.Y., Lavesh B., et al. Site-specific fluorescence double-labeling of proteins and analysis of structural changes in solution by Fluorescence Resonance Energy Transfer (FRET) MethodsX. 2018;5:419–430. doi: 10.1016/j.mex.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao R., Wang N., Shen T., et al. High-fidelity imaging of amyloid-beta deposits with an ultrasensitive fluorescent probe facilitates the early diagnosis and treatment of Alzheimer's Disease. Theranostics. 2022;12:2549–2559. doi: 10.7150/thno.68743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y., Tsao K., Keillor J.W. Fluorogenic protein labelling: A review of photophysical quench mechanisms and principles of fluorogen design. Can. J. Chem. 2015;93:389–398. [Google Scholar]
- 62.Tsao K.K., Lee A.C., Racine K.É., Keillor J.W. Site-specific fluorogenic protein labelling agent for bioconjugation. Biomolecules. 2020;10:369. doi: 10.3390/biom10030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimm J.B., English B.P., Chen J., et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canty L., Hariharan S., Liu Q., Haney S.A., Andrews D.W. Peak emission wavelength and fluorescence lifetime are coupled in far-red, GFP-like fluorescent proteins. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0208075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becker W. Fluorescence lifetime imaging - techniques and applications. J Microsc. 2012;247:119–136. doi: 10.1111/j.1365-2818.2012.03618.x. [DOI] [PubMed] [Google Scholar]
- 66.Cao R., Wallrabe H., Periasamy A. Multiphoton FLIM imaging of NAD(P)H and FAD with one excitation wavelength. J. Biomed. Opt. 2020;25:1–16. doi: 10.1117/1.JBO.25.1.014510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D., Moon S., Hwang W., Kim D.Y. Use of nanosecond excitation pulses in fluorescence lifetime measurement via phasor analysis. Optics Express. 2022;30:14677. doi: 10.1364/OE.450761. [DOI] [PubMed] [Google Scholar]
- 68.Kage D., Hoffmann K., Hoffmann H., Schedler U., Resch-Genger U. Lifetime encoding in flow cytometry for bead-based sensing of biomolecular interaction. Sci. Rep. 2020;10:19477. doi: 10.1038/s41598-020-76150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nienhaus K., Nienhaus G.U. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014;43:1088–1106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 70.Bourgeois D., Adam V. Reversible photoswitching in fluorescent proteins: A mechanistic view. IUBMB Life. 2012;64:482–491. doi: 10.1002/iub.1023. [DOI] [PubMed] [Google Scholar]
- 71.Tang L., Fang C. Photoswitchable fluorescent proteins: Mechanisms on ultrafast timescales. Int. J. Mol. Sci. 2022;23:6459. doi: 10.3390/ijms23126459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henderson J.N., Remington S.J. The kindling fluorescent protein: A transient photoswitchable marker. Physiology. 2006;21:162–170. doi: 10.1152/physiol.00056.2005. [DOI] [PubMed] [Google Scholar]
- 73.Kim I.J., Xu Y., Nam K.H. Metal-induced fluorescence quenching of photoconvertible fluorescent protein DendFP. Molecules. 2022;27:2922. doi: 10.3390/molecules27092922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedersen M., Jamali S., Saha I., Daum R., Bendjennat M., Saffarian S. Correlative iPALM and SEM resolves virus cavity and Gag lattice defects in HIV virions. Eur. Biophys. J. 2019;48:15–23. doi: 10.1007/s00249-018-1324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S.E., Hwang K.Y., Nam K.H. Spectral and structural analysis of a red fluorescent protein from Acropora digitifera. Protein Sci. 2019;28:375–381. doi: 10.1002/pro.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nam K.H., Kwon O.Y., Sugiyama K., Lee W.H., Kim Y.K., Song H.K., Kim E.E., Park S.Y., Jeon H., Hwang K.Y. Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S. Biochem. Biophys. Res. Commun. 2007;354:962–967. doi: 10.1016/j.bbrc.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 77.Yang Y., Feng R.-r., Gai F. 4-Cyanotryptophan as a sensitive fluorescence probe of local electric field of proteins. The Journal of Physical Chemistry B. 2023;127:514–519. doi: 10.1021/acs.jpcb.2c07605. [DOI] [PubMed] [Google Scholar]
- 78.Oscar B.G., Liu W., Zhao Y., Tang L., Wang Y., Campbell R.E., Fang C. Excited-state structural dynamics of a dual-emission calmodulin-green fluorescent protein sensor for calcium ion imaging. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10191–10196. doi: 10.1073/pnas.1403712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang C., Frontiera R.R., Tran R., Mathies R.A. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature. 2009;462:200–204. doi: 10.1038/nature08527. [DOI] [PubMed] [Google Scholar]
- 80.Jones C.M., List N.H., Martínez T.J. Resolving the ultrafast dynamics of the anionic green fluorescent protein chromophore in water. Chem. Sci. 2021;12:11347–11363. doi: 10.1039/d1sc02508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boulanger S.A., Chen C., Tang L., Zhu L., Baleeva N.S., Myasnyanko I.N., Baranov M.S., Fang C. Shedding light on ultrafast ring-twisting pathways of halogenated GFP chromophores from the excited to ground state. Phys. Chem. Chem. Phys. 2021;23:14636–14648. doi: 10.1039/d1cp02140k. [DOI] [PubMed] [Google Scholar]
- 82.Cui G., Lan Z., Thiel W. Intramolecular hydrogen bonding plays a crucial role in the photophysics and photochemistry of the GFP chromophore. J. Am. Chem. Soc. 2012;134:1662–1672. doi: 10.1021/ja208496s. [DOI] [PubMed] [Google Scholar]
- 83.Baranov M.S., Lukyanov K.A., Borissova A.O., Shamir J., Kosenkov D., Slipchenko L.V., Tolbert L.M., Yampolsky I.V., Solntsev K.M. Conformationally locked chromophores as models of excited-state proton transfer in fluorescent proteins. J. Am. Chem. Soc. 2012;134:6025–6032. doi: 10.1021/ja3010144. [DOI] [PubMed] [Google Scholar]
- 84.Chen C., Fang C. Devising efficient red-shifting strategies for bioimaging: A generalizable donor-acceptor fluorophore prototype. Chem. Asian J. 2020;15:1514–1523. doi: 10.1002/asia.202000175. [DOI] [PubMed] [Google Scholar]
- 85.Sedgwick A.C., Wu L., Han H.-H., Bull S.D., He X.-P., James T.D., Sessler J.L., Tang B.Z., Tian H., Yoon J. Excited-state intramolecular proton-transfer (ESIPT) based fluorescence sensors and imaging agents. Chem. Soc. Rev. 2018;47:8842–8880. doi: 10.1039/c8cs00185e. [DOI] [PubMed] [Google Scholar]
- 86.Molina R.S., Qian Y., Wu J., Shen Y., Campbell R.E., Drobizhev M., Hughes T.E. Understanding the fluorescence change in red genetically encoded calcium ion indicators. Biophysical Journal. 2019;116:1873–1886. doi: 10.1016/j.bpj.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Acharya A., Bogdanov A.M., Grigorenko B.L., Bravaya K.B., Nemukhin A.V., Lukyanov K.A., Krylov A.I. Photoinduced chemistry in fluorescent proteins: Curse or blessing? Chem. Rev. 2017;117:758–795. doi: 10.1021/acs.chemrev.6b00238. [DOI] [PubMed] [Google Scholar]
- 88.Brejc K., Sixma T.K., Kitts P.A., Kain S.R., Tsien R.Y., Ormö M., Remington S.J. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKinney S.A., Murphy C.S., Hazelwood K.L., Davidson M.W., Looger L.L. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;2:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 91.Harroun S.G., Lauzon D., Ebert M.C.C.J.C., et al. Monitoring protein conformational changes using fluorescent nanoantennas. Nat Methods. 2022;19:71–80. doi: 10.1038/s41592-021-01355-5. [DOI] [PubMed] [Google Scholar]
- 92.Talukder P., Chen S., Roy B., Yakovchuk P., Spiering M.M., Alam M.P., Madathil M.M., Bhattacharya C., Benkovic S.J., Hecht S.M. Cyanotryptophans as novel fluorescent probes for studying protein conformational changes and DNA–protein interaction. Biochemistry. 2015;54:7457–7469. doi: 10.1021/acs.biochem.5b01085. [DOI] [PubMed] [Google Scholar]
- 93.Micikas R.J., Acharyya A., Smith A.B., Gai F. Synthesis and characterization of the fluorescence utility of two visible-light-absorbing tryptophan derivatives. Chemical Physics Letters. 2022;795 [Google Scholar]
- 94.Gu K., Xu Y., Li H., Guo Z., Zhu S., Zhu S., Shi P., James T.D., Tian H., Zhu W.-H. Real-time tracking and in vivo visualization of β-galactosidase activity in colorectal tumor with a ratiometric near-infrared fluorescent probe. Journal of the American Chemical Society. 2016;138:5334–5340. doi: 10.1021/jacs.6b01705. [DOI] [PubMed] [Google Scholar]
- 95.Lim J.-W., Petersen M., Bunz M., Simon C., Schindler M. Flow cytometry based-FRET: basics, novel developments and future perspectives. Cellular and Molecular Life. 2022;217 doi: 10.1007/s00018-022-04232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwak M.S., Rhee W.J., Lee Y.J., Kim H.S., Kim Y.H., Kwon M.K., Shin J.-S. Reactive oxygen species induce Cys106-mediated anti-parallel HMGB1 dimerization that protects against DNA damage. Redox Biology. 2021;40 doi: 10.1016/j.redox.2021.101858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee S., Kim J., Koh M. Recent advances in fluorescence imaging by genetically encoded non-canonical amino acids. Journal of Molecular Biology. 2022;434 doi: 10.1016/j.jmb.2021.167248. [DOI] [PubMed] [Google Scholar]
- 98.Markiewicz B.N., Mukherjee D., Troxler T., Gai F. Utility of 5-cyanotryptophan fluorescence as a sensitive probe of protein hydration. The Journal of Physical Chemistry B. 2016;120:936–944. doi: 10.1021/acs.jpcb.5b12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Acharyya A., Zhang W., Gai F. Tryptophan as a template for development of visible fluorescent amino acids. The Journal of Physical Chemistry B. 2021;125:5458–5465. doi: 10.1021/acs.jpcb.1c02321. [DOI] [PubMed] [Google Scholar]
- 100.Hebestreit M.-L., Lartian H., Henrichs C., Kühnemuth R., Meerts W.L., Schmitt M. Excited state dipole moments and lifetimes of 2-cyanoindole from rotationally resolved electronic Stark spectroscopy. Physical Chemistry Chemical Physics. 2021;23:10196–10204. doi: 10.1039/d1cp00097g. [DOI] [PubMed] [Google Scholar]
- 101.Micikas R.J., Ahmed I.A., Acharyya A., Smith A.B., Gai F. Tuning the electronic transition energy of indole via substitution: Application to identify tryptophan-based chromophores that absorb and emit visible light. Physical Chemistry Chemical Physics. 2021;23:6433–6437. doi: 10.1039/d0cp06710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heilemann M. Comprehensive Biophysics. Elsevier; 2012. Super-Resolution Microscopy; pp. 39–58. Editor(s): Edward H. Egelman. [Google Scholar]
- 103.K.Zhanghao X.Chen, Liu W., et al. Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy. Nat Commun. 2019;10:4694. doi: 10.1038/s41467-019-12681-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review article.