Abstract
Pseudomonas aeruginosa is a major cause of life-threatening acute infections and life-long lasting chronic infections. The characteristic biofilm mode of life in P. aeruginosa chronic infections severely limits the efficacy of antimicrobial therapies, as it leads to intrinsic tolerance, involving physical and physiological factors in addition to biofilm-specific genes that can confer a transient protection against antibiotics promoting the development of resistance. Indeed, a striking feature of this pathogen is the extraordinary capacity to develop resistance to nearly all available antibiotics through the selection of chromosomal mutations, evidenced by its outstanding and versatile mutational resistome. This threat is dramatically amplified in chronic infections, driven by the frequent emergence of mutator variants with enhanced spontaneous mutation rates. Thus, this mini review is focused on describing the complex interplay of antibiotic resistance mechanisms in P. aeruginosa biofilms, to provide potentially useful information for the design of effective therapeutic strategies.
Keywords: Pseudomonas aeruginosa, Biofilms, Antimicrobial resistance, Antimicrobial tolerance, Resistance mechanisms
1. Introduction
Biofilms, first described as microbial cells embedded in a self-produced extracellular polymeric matrix attached to either biotic or abiotic surfaces [1], are thought to stem from an adaptive social behaviour to survive in hostile environments [2]; including the human body during infection [3]. However, the definition of what a “biofilm” is has been adapted to advances in research to suit new findings. For instance, it has been found that some pathogenic biofilm aggregates can be formed without the need to attach to a surface, thus being able to grow within the mucus of individuals with cystic fibrosis (CF) and not in or on the lung tissue [4]. Due to the matrix protection, bacterial cells within a biofilm are significantly more recalcitrant to antibiotics and host immune defences than their planktonic counterparts, as shown by in vitro and in vivo evidence [[5], [6], [7], [8]]. This fact poses a health issue, as the inability to clear the bacteria is directly related to the chronicity of the infections [9,10]. It is estimated that up to 65–80% of all infections are associated with biofilm formation [11], with chronic infections encompassing a myriad of diseases. Besides chronic respiratory infections, such as those occurring in the lungs of people with CF [12], biofilm-related chronic infections include, among others, those related to indwelling medical devices, (e.g., catheters, prosthetic joints, and surgical implants), tissue infections, such as otitis media, rhinosinusitis, osteomyelitis, and chronic wounds.
1.1. Pseudomonas aeruginosa: a paradigmatic microorganism
Pseudomonas aeruginosa is an opportunistic pathogen that causes severe infections, particularly in health care settings, mostly affecting immunocompromised patients [13]. There is a growing prevalence of nosocomial infections caused by P. aeruginosa strains, which are associated with significantly increased morbidity and mortality [14]. Likewise, P. aeruginosa is the main driver of chronic respiratory infections in CF and other respiratory diseases [10]. One of the most striking features of P. aeruginosa, which by itself is intrinsically resistant to many antibiotics, is its capacity to develop resistance to nearly all available antibiotics, through mutations in chromosomal genes, as well as acquire resistance markers through horizontal transfer, which has led to the worldwide spreading of a few specific multidrug-resistant (MDR) and extensively drug-resistant (XDR) high-risk clones [15,16]. Indeed, the outstanding capacity of P. aeruginosa for developing antimicrobial resistance is for seen in the extraordinary versatility of is vast mutational resistome, not only dependent on the exposure to a specific antibiotic but conditioned by simultaneous or even previous exposures to other [17,18]. Recent studies have estimated 4.95 million deaths associated with antimicrobial resistance (AMR) in 2019, including 1.27 million deaths attributable to bacterial AMR, P. aeruginosa being the sixth pathogen on the list [19].
In addition to the threat that poses this pathogen regarding AMR, P. aeruginosa infections are also important to be related with biofilms, being an archetypal microorganism for their study.
1.2. Tolerance to antibiotics in Pseudomonas aeruginosa biofilms
Biofilm antibiotic tolerance mechanisms involve a wide range of environmental, physical, and physiological factors that, in principle, confer a transient protection; in opposition to conventional resistance mechanisms; meaning that individual cells remain susceptible to previously exposed antibiotics despite being able to survive to lethal doses during biofilm growth mode.
The presence of an extracellular matrix is probably one of the most characteristic factors involved in biofilm antibiotic tolerance. As it is well known, the matrix consists of exopolysaccharides, proteins, extracellular DNA (eDNA) and lipids [20,21], yet its exact composition will vary depending on the bacterial strain, the growth conditions, and the biofilm maturation stage [22]. Interactions with the matrix components, as anionic eDNA and alginate [[23], [24], [25]] can capture positively charged classes of antibiotics, such as colistin or tobramycin, and consequently reduce their activity [23,[26], [27], [28]], whereas neutral antibiotics, like ciprofloxacin, penetrate more easily into the biofilms [24]. In general, the transition through the matrix entails a delay in the antibiotic-bacteria contact and the exposition to lower concentrations, giving bacteria time to become tolerant [29,30]. In fact, regarding the pharmacokinetic (PK) and pharmacodynamic (PD) parameters of antibiotics, biofilms have been proposed as a third pharmacological compartment, in addition to the blood and the target tissue of the infection [31,32]. Accordingly, the existence of nutrients and oxygen gradients results in differential subcompartments [33] differentiating two main subpopulations; one metabolically active, frequently in the outer layers of the biofilm, and one less active (or even inactive), in the deeper ones [34,35]. This should be considered, since several antibiotics only target processes of growing bacteria (like replication, transcription, translation, or cell wall synthesis), while others are effective on metabolically inactive cells, to establish the basis for combination therapies [33,36,37].
The most extreme case of decreased metabolism, also found in the biofilm, is represented by the persisters. This is a special growth state that means less than 0.1% of the biofilm population, refractory to antibiotics, a kind of spore-like cell state activity that can become active after finishing the treatment [38,39].
The lack of oxygen inside the biofilm also plays an important role in tolerance to some antibiotics that work under aerobic conditions (e.g., beta-lactams, aminoglycosides, fluoroquinolones and tetracyclines) [40], being only effective against bacteria on the periphery [23,41]. Fortunately, polymyxins and other membrane-targeting compounds such as SDS, EDTA, and chlorhexidine, maintain their anti-biofilm activity [34,[42], [43], [44]] despite hypoxia.
Loss of antibiotic activity could also be partly explained by the deficiency of reactive oxygen species (ROS) production under anaerobic conditions. Induction of ROS by some bactericidal antibiotics is thought to contribute to their killing effect, as evidenced by the emergence of cytotoxic hydroxyl radicals (•OH) in P. aeruginosa biofilms treated with ciprofloxacin [45]. The antioxidant systems are also upregulated due to the stringent response in biofilms [46,47], participating in their tolerance to antibiotics. On the contrary, in response to different forms of stress (oxidative stress, nitrosative stress and membrane-damaging agents) the up-regulation of the efflux pumps, like MexXY-OprM, MexEF-OprN and MexCD-OprJ, can be triggered in P. aeruginosa as non-specific antibiotic tolerance mechanism [48,49].
Other than biofilm environment intrinsic factors leading to physiological tolerance, there is also an in vivo contribution of the host immune system mediated, for example, by the polymorphonuclear (PMNs) leukocytes action as it is demonstrated in the endotracheal mucus from people with CF infected by P. aeruginosa [50,51] where PMNs are known to consume oxygen and release eDNA that traps cationic antibiotics [[51], [52], [53]].
The differences between planktonic and biofilm modes of life are also related with differential gene expression even in the absence of antibiotics [[54], [55], [56], [57], [58]]. This is the case, for instance, of brlR, which is a Mer-like transcriptional activator [59] that stimulates the expression of the MexAB-oprM and MexEF-oprN efflux pumps [60], the production of the ABC (ATP-Binding Cassette) transporters, (like the ABC transporter PA1874-1877, 10 times more expressed in P. aeruginosa biofilms [59,61]), or alters the expression of genes encoding modification of lipopolysaccharide (LPS), membrane protein composition, or metabolism and energy generation [62]. Consequently, increased expression of brlR in biofilms lowered the susceptibility to hydrogen peroxide and five different classes of antibiotics by increasing the minimum inhibitory concentrations (MICs) up to 6-fold [63]. On the contrary, brlR represses phoPQ expression increasing susceptibility to colistin so; the reciprocal role of brlR enhancing colistin susceptibility while increasing resistance to other antibiotics, like tobramycin, provides the genetic basis for their use in combination [64]. The expression of other genes, like ndvB, coding a glycosyltransferase involved in the formation of cyclic glucans [65] that can sequester aminoglycoside antibiotics is also augmented in biofilms [64,66,67].
Besides the increased expression of specific genes on biofilms compared to planktonic, adaptive tolerance mechanisms induced by the presence of the antibiotic (especially favoured by sub-inhibitory concentrations [[68], [69], [70]]) results in an emerging transient tolerant phenotype that reverts to susceptibility once the molecules have disappeared. For example, the presence of colistin upregulates the two-component regulatory system pmr which, in turn, regulates arn genes, leading to a reduction of the negative charge of the LPS thus, protecting the biofilm surface against the cationic peptide colistin [34].
The induction of AmpC β-lactamase by the exposure to β-lactam antibiotics, as imipenem or ceftazidime, is probably the main adaptive tolerance mechanism in P. aeruginosa biofilms [71,72]. Bagge et al. demonstrated that the expression of the enzyme showed a special structural distribution characteristically concentrated at the periphery of the biofilms [29]. In addition, as seen in P. aeruginosa biofilms from people with CF, these β-lactamases are partially excreted by membrane vesicles [73], consistent with their extracellular location [71].
Hence, adaptive tolerance mechanisms operate in a similar way than persisters and an on/off model with long term consequences can be evidenced; if survivor cells are present once the treatment has stopped, the re-emergence of biofilms might happen.
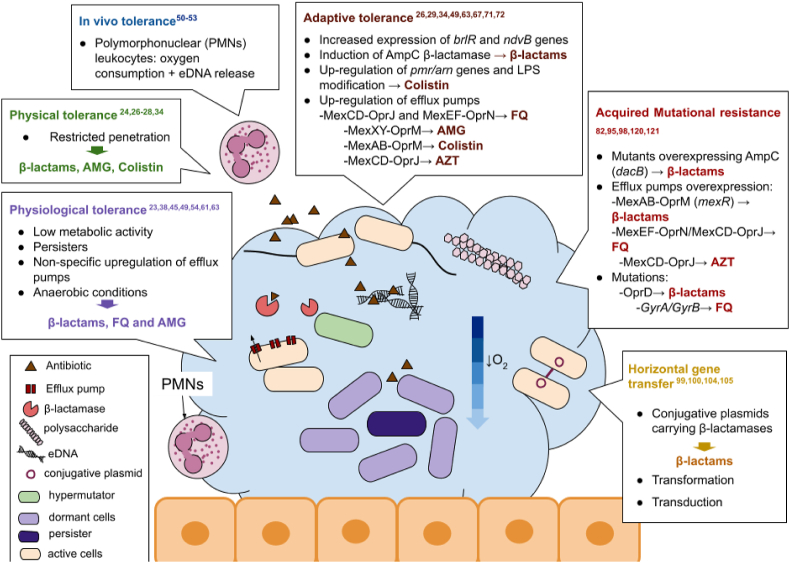
In summary, concerning recalcitrance of biofilms, there are different pathways that act synergistically with intrinsic tolerance providing a fertile ground that helps traditional antibiotic resistance mechanisms to develop [74]. Fig. 1 gives a general view of all the mechanisms of both, tolerance, and resistance, that will be discussed in the next section.
Fig. 1.
Mechanisms of antimicrobial tolerance and resistance found in biofilms and antibiotics affected by them. FQ: Fluoroquinolones; AMG: Aminoglycosides; AZT: Azitromycin; LPS: lipopolysaccharide. Figure adapted from Ciofu O et al., Nat rev, 2022 [62].
2. Mechanisms of conventional antibiotic resistance in biofilms
Whereas the previously reported tolerance mechanisms show clear impact on biofilm recalcitrance to antibiotics and have been broadly studied, the role that traditional resistance mechanisms play in the decreased susceptibility of biofilms towards antibiotics has been less examined.
Nonetheless, recent studies seem to point towards tolerance as a preceding stage that favours the establishment of antibiotic resistance mutations that could otherwise occur, but might not be selected for [74,75].
Furthermore, the tolerance conditions described above, particularly, the high density and proximity of cells, together with an accumulation of available genetic elements, make the biofilm a great environment for horizontal gene transfer [76,77].
2.1. Mutational resistance
As mentioned before, in biofilms under treatment, matrix restrictive penetration results in areas of lower antibiotic concentration that, together with nutrient limitation, can prompt stress responses and lead to mutation emergence [49,78]. In addition, heterogeneity of nutrient and oxygen availability, results in different ecological niches that ensure the formation of distinct subpopulations and, consequently, fixation of beneficial mutations is more easily enabled [78]. Typically, the balance between evolution and genetic change shapes the rate of mutation [74]. If that rate is augmented more than usual, a priori, premature death could happen due to the additive effect of deleterious mutations [79]. Even so, in certain situations, especially those involving stressful environments, increased mutation rate might be beneficial for the bacteria [80,81].
One characteristic of P. aeruginosa biofilms, particularly described in CF individuals with chronic respiratory infections, is the elevated prevalence of mutator (or hypermutable) strains, present in over one-third of the studied patients [82]. The high prevalence of mutators in this setting strongly contrasts with what has been documented for onset of chronic infections (10%) [83], environment (6%) [84], or acute infections (<1%) [85]. Mutators, bacteria with an increased spontaneous mutation rate, frequently emerge due to defects in DNA repair mechanisms, such as the mismatch repair system (MMR), or stress responses [[86], [87], [88]]. These mutators are especially relevant in the clinical setting as they are frequently associated with high antibiotic resistance rates [82,89]. Moreover, it has been demonstrated that mutagenesis is naturally increased in biofilms and that this condition favours development, adaptation, and diversification processes that could, ultimately, lead to the occurrence of resistance under antibiotic exposure [[90], [91], [92], [93], [94]].
Mutation-driven resistance has been shown to be relevant also for antibiotics showing potent activity against P. aeruginosa biofilms, but not against planktonically growing cells, such as azithromycin (AZM). Indeed, marked selection of resistant mutants was demonstrated to be linked to hyperproduction of the multidrug efflux pump MexCD-OprJ, associated with inactivation mutations in its negative regulator nfxB [95]. The emergence of such resistant mutants was dramatically enhanced in biofilms formed by hypermutable strains [95]. Hyperexpression of MexCD-OprJ, showed cross-resistance to other unrelated antipseudomonal agents as ciprofloxacin or cefepime but hypersusceptibility to others such as imipenem or tobramycin [95]. Therefore, this work was helpful in guiding the selection of appropriate antipseudomonal therapies in CF individuals under AZM maintenance treatment.
Selection and amplification of resistant mutants and, even the hypermutator strains themselves, has been demonstrated in P. aeruginosa biofilms treated with ciprofloxacin [70]. Results showed that mutational mechanisms were playing a major role in biofilm antibiotic resistance and that theoretically optimized PK/PD parameters failed to suppress resistance development, suggesting that the increased antibiotic tolerance driven by the special biofilm physiology and architecture probably raises the effective mutant prevention concentration (MPC), favouring gradual mutational resistance development, especially in mutator strains [70].
Differential acquisition of resistance mutations in P. aeruginosa planktonic and biofilm growth have been documented in some cases, such as aminoglycoside (tobramycin) resistance development seemingly linked to LPS biosynthesis genes (orfKHLN) and electron transport chain components (cyoAB) [96,97].
Other relevant mutations involved in biofilm antibiotic resistance are those connected to β-lactamase hyperexpression. In a study looking into mixed biofilm communities of wild-type PAO1 and mutants with hyperproduction of either, the AmpC β-lactamase (dacB knockout), or the MexAB-OprM efflux pump (mexR knockout), it was shown that, under treatment with cefepime PAO1ΔdacB resistant mutants were selected for and amplified [98]. Furthermore, both, PAO1ΔmexR and PAO1ΔdacB mutants, seemed to locate themselves in the outer biofilm layers surrounding the sensitive PAO1 subpopulation and exert a shielding effect, which did not happen during planktonic growth [98]. Thus, this study demonstrated that, in biofilms, mutants showing diverse resistance mechanisms such as β-lactamase hyperproduction protect the whole community, preserving wild-type susceptible populations from the effect of the antibiotics. Therefore, these findings represented a step forward to figure out antibiotic resistance dynamics in biofilms, as well as to understand the population biology of bacterial pathogens in chronic infections, where the coexistence of susceptible and resistant variants in dense communities is a hallmark.
Summing up, the biofilm microenvironment and population relationships and structure, together with its associated tolerance phenomenon, potentiate adaptive mutagenesis; all of which creates a breeding ground for the emergence of antibiotic resistance.
2.2. Horizontal gene transfer
Another traditional source of antimicrobial resistance is horizontal gene transfer, which can occur via conjugation, transformation or transduction, as it represents a major source of genetic variability.
For instance, it is known that conjugation in biofilms occurs at higher frequencies than in planktonic cultures [[99], [100], [101]]. Actually, biofilms could act as plasmid reserves maintaining and preserving MDR plasmids [102,103].
The biofilm architecture and the elevated bacterial density together facilitate the encounter of donor and recipient cells [77]. However, individual cells from within the diverse biofilm microenvironments were found to differ in their capacity to maintain incoming plasmids [100].
Thus, a high number of conjugation events does not guarantee effective gene transfer in all cases and could partially explain why some and not all cells acquire resistance mechanisms.
Similarly, transformation also seems to experience a boost within the biofilm domain [99]. The main hypothesis behind this is that the presence of eDNA in the matrix triggers a natural state of competence which in turn activates DNA release systems further promoting said state, that also contribute to the stabilisation of the biofilm matrix [77,99,104]. Whilst the state of competence might play an important part in biofilm matrix formation, it might also result in resistance gene acquisition as an after effect.
Lastly, along the same line as the case of transformation, given the role bacteriophages might play in biofilm matrix development and maintenance through the release of bacterial cytoplasmic components, the contribution of transduction to antibiotic resistance acquisition should not be ignored. On this note, phage mediated conversion of P. aeruginosa into a mucoid phenotype has already been reported and associated with poor outcomes for people with CF [105].
3. Final thoughts and future perspectives
Altogether, P. aeruginosa biofilms encompass an intricate environment conducive to antibiotic therapy failure; either through its natural tolerance and adaptive mechanisms or via an increased rate of conventional resistance events favourably selected.
So, improving knowledge regarding P. aeruginosa biofilm, research should be guided to find new strategies to overcome resistance, considering the multitude of factors that make the eradication of P. aeruginosa infections difficult. From a clinical perspective, the severity of chronic P. aeruginosa infections is having public health consequences, particularly affecting the most severely infected patients in our hospitals and people with CF.
After all the nuances about biofilms explained in this mini-review, it is obvious that, apart from antimicrobials, we need other compounds that help the action of our selected antibiotic, specifically targeting the biofilm mode of growth [106]. Starting with the numerous quorum sensing inhibitors that plays a key role in the regulation of P. aeruginosa biofilm formation but does not affect the viability of the bacteria. Other strategies to stand out would be the ones activating metabolic inactive cells by using SDS, EDTA and chlorhexidine, antibiotics like colistin or other compounds as organic acids or carbon sources [34,42,46,[107], [108], [109]]. More recently, the use of hyperbaric oxygen therapy (HBOT) metabolism-stimulating has been shown to redirect bacterial metabolism towards an antibiotic-susceptible phenotype [44,110,111]. Similarly, Oligo-G (alginate oligosaccharide), has been demonstrated to be able to inhibit biofilm formation and disrupt established biofilm matrix in vitro, as well as c-di-GMP modulators do [112,113]. Treatment with eDNAases [114]; use of efflux pumps inhibitors (EPIs), directed to the adaptive resistance [115]; use of antioxidants like N-acetylcysteine to target hypermutation, SOS, and oxidative stress [116]; bacteriophages [117]; photodynamic inactivation [118]; or even revert acidic environment to neutral conditions to prevent chronic P. aeruginosa infection and colonization [119], should be added to the list of possible therapies.
Beyond looking for new compounds or non-antibiotics strategies, it would be sensible to strive to improve therapeutic antibiotic strategies already available, as well as PK/PD parameters, to overwhelm mutation selection like combination of antibiotics [[120], [121], [122], [123], [124]], innovative sequential regimens [125,126] or improved drug-delivered for topical administration [127,128] recently demonstrated by ultrasound patches [129]. Progress in new alternative therapeutic approaches like formulation of antibiotics on nanoparticles [130,131], novel anti-biofilm compounds, CRISPR gene editing technologies and photodynamic therapies [[132], [133], [134]] is necessary. In addition, a determined attempt on transferring all these options into clinical practice for the treatment of biofilm-related infections will need to be done.
CRediT authorship contribution statement
María Fernández-Billón: Writing – original draft. Aina E. Llambías-Cabot: Writing – original draft. Elena Jordana-Lluch: Writing – review & editing. Antonio Oliver: Conceptualization, Writing – review & editing. María D. Macià: Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Aknowledgments
EJL holds a “Juan de la Cierva-Incorporación” Fellowship (IJC2019-038836-I) granted by The Ministry of Science and Innovation (Spain). AELC holds a CIBERINFEC (2322/2746) training contract. MDM is the scientific officer of the European Society for Clinical Microbiology and Infectious Disease Study Group for Biofilms (ESGB). Research of all authors is supported by the Spanish Network for Research in Infectious Diseases (REIPI RD16/0016) and Centro de Investigación Biomédica en Red en Enfermedades Infecciosas,Instituto de Salud Carlos III (CIBERINFEC).
Contributor Information
María Fernández-Billón, Email: maria.fernandez-billon@ssib.es.
Aina E. Llambías-Cabot, Email: ainabet1207@gmail.com.
Elena Jordana-Lluch, Email: elena.jordana@ssib.es.
Antonio Oliver, Email: antonio.oliver@ssib.es.
María D. Macià, Email: mariad.macia@ssib.es.
Data availability
No data was used for the research described in the article.
References
- 1.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Stoodley P., Sauer K., Davies D.G., Costerton J.W. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 3.Fux C.A., Costerton J.W., Stewart P.S., Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt T., Jensen P.Ø., Fiandaca M.J., Pedersen J., Hansen C.R., Andersen C.B., et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 5.Høiby N., Ciofu O., Johansen H.K., Song Z., Moser C., Jensen P.Ø., et al. The clinical impact of bacterial biofilms. Int J Oral Sci. 2011;3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hengzhuang W., Wu H., Ciofu O., Song Z., Høiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2011;55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hengzhuang W., Wu H., Ciofu O., Song Z., Høiby N. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob Agents Chemother. 2012;56:2683–2690. doi: 10.1128/AAC.06486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjarnsholt T., Jensen P.Ø., Burmølle M., Hentzer M., Haagensen J.A.J., Hougen H.P., et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 9.Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D’Argenio D.A., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver A., Mena A., Maciá M.D. Evol. Biol. Bact. Fungal pathog. John Wiley & Sons, Ltd; 2007. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections; pp. 433–444. [DOI] [Google Scholar]
- 11.Jamal M., Ahmad W., Andleeb S., Jalil F., Imran M., Nawaz M.A., et al. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81:7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Høiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., et al. ESCMID∗ guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21:S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Lyczak J.B., Cannon C.L., Pier G.B. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juan C., Peña C., Oliver A. Host and pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Dis. 2017;215:S44–S51. doi: 10.1093/infdis/jiw299. [DOI] [PubMed] [Google Scholar]
- 15.Potron A., Poirel L., Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Oliver A., Mulet X., López-Causapé C., Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updates. 2015;21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 17.López-Causapé C., Cabot G., Del Barrio-Tofiño E., Oliver A. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol. 2018;9:685. doi: 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López-Causapé C., Maruri-Aransolo A., Gomis-Font M.A., Penev I., Castillo M.G., Mulet X., et al. Cefiderocol resistance genomics in sequential chronic Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin Microbiol Infect. 2022;S1198–743X(22):586–589. doi: 10.1016/j.cmi.2022.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet Lond Engl. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colvin K.M., Irie Y., Tart C.S., Urbano R., Whitney J.C., Ryder C., et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14 doi: 10.1111/j.1462-2920.2011.02657.x. 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryder C., Byrd M., Wozniak D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann E.E., Wozniak D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walters M.C., Roe F., Bugnicourt A., Franklin M.J., Stewart P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng B.S., Zhang W., Harrison J.J., Quach T.P., Song J.L., Penterman J., et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jennings L.K., Dreifus J.E., Reichhardt C., Storek K.M., Secor P.R., Wozniak D.J., et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulcahy H., Charron-Mazenod L., Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao B., Christophersen L., Kolpen M., Jensen P.Ø., Sneppen K., Høiby N., et al. Diffusion retardation by binding of tobramycin in an alginate biofilm model. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang W.-C., Nilsson M., Jensen P.Ø., Høiby N., Nielsen T.E., Givskov M., et al. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagge N., Hentzer M., Andersen J.B., Ciofu O., Givskov M., Høiby N. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2004;48:1168–1174. doi: 10.1128/AAC.48.4.1168-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao B., Christophersen L., Thomsen K., Sønderholm M., Bjarnsholt T., Jensen P.Ø., et al. Antibiotic penetration and bacterial killing in a Pseudomonas aeruginosa biofilm model. J Antimicrob Chemother. 2015;70:2057–2063. doi: 10.1093/jac/dkv058. [DOI] [PubMed] [Google Scholar]
- 32.Christophersen L., Schwartz F.A., Lerche C.J., Svanekjær T., Kragh K.N., Laulund A.S., et al. In vivo demonstration of Pseudomonas aeruginosa biofilms as independent pharmacological microcompartments. J Cyst Fibros. 2020;19:996–1003. doi: 10.1016/j.jcf.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Stewart P.S., Zhang T., Xu R., Pitts B., Walters M.C., Roe F., et al. Reaction–diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. Npj Biofilms Microbiomes. 2016;2:1–8. doi: 10.1038/npjbiofilms.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pamp S.J., Gjermansen M., Johansen H.K., Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol. 2008;68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 35.Pamp S.J., Sternberg C., Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry. 2009;75A:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 36.Haagensen J., Verotta D., Huang L., Engel J., Spormann A.M., Yang K. Spatiotemporal pharmacodynamics of meropenem- and tobramycin-treated Pseudomonas aeruginosa biofilms. J Antimicrob Chemother. 2017;72:3357–3365. doi: 10.1093/jac/dkx288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrmann G., Yang L., Wu H., Song Z., Wang H., Høiby N., et al. Colistin-Tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis. 2010;202:1585–1592. doi: 10.1086/656788. [DOI] [PubMed] [Google Scholar]
- 38.Wilmaerts D., Windels E.M., Verstraeten N., Michiels J. General mechanisms leading to persister formation and awakening. Trends Genet. 2019;35:401–411. doi: 10.1016/j.tig.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Fauvart M., De Groote V.N., Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. 2011. [DOI] [PubMed] [Google Scholar]
- 40.Kohanski M.A., Dwyer D.J., Collins J.J. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borriello G., Werner E., Roe F., Kim A.M., Ehrlich G.D., Stewart P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang W.-C., Pamp S.J., Nilsson M., Givskov M., Tolker-Nielsen T. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol Med Microbiol. 2012;65:245–256. doi: 10.1111/j.1574-695X.2012.00929.x. [DOI] [PubMed] [Google Scholar]
- 43.Brochmann R.P., Toft A., Ciofu O., Briales A., Kolpen M., Hempel C., et al. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int J Antimicrob Agents. 2014;43:140–147. doi: 10.1016/j.ijantimicag.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Kolpen M., Appeldorff C.F., Brandt S., Mousavi N., Kragh K.N., Aydogan S., et al. Increased bactericidal activity of colistin on Pseudomonas aeruginosa biofilms in anaerobic conditions. Pathog Dis. 2015;74:ftv086. doi: 10.1093/femspd/ftv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen P.Ø., Briales A., Brochmann R.P., Wang H., Kragh K.N., Kolpen M., et al. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog Dis. 2014;70:440–443. doi: 10.1111/2049-632X.12120. [DOI] [PubMed] [Google Scholar]
- 46.Khakimova M., Ahlgren H.G., Harrison J.J., English A.M., Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol. 2013;195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martins D., McKay G., Sampathkumar G., Khakimova M., English A.M., Nguyen D. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115:9797–9802. doi: 10.1073/pnas.1804525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaible B., Schaffer K., Taylor C.T. Hypoxia, innate immunity and infection in the lung. Respir Physiol Neurobiol. 2010;174:235–243. doi: 10.1016/j.resp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Poole K. Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can J Microbiol. 2014;60:783–791. doi: 10.1139/cjm-2014-0666. [DOI] [PubMed] [Google Scholar]
- 50.Kolpen M., Kühl M., Bjarnsholt T., Moser C., Hansen C.R., Liengaard L., et al. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kragh K.N., Alhede M., Jensen P.Ø., Moser C., Scheike T., Jacobsen C.S., et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolpen M., Hansen C.R., Bjarnsholt T., Moser C., Christensen L.D., Gennip M van, et al. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax. 2010;65:57–62. doi: 10.1136/thx.2009.114512. [DOI] [PubMed] [Google Scholar]
- 53.Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K.C., et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteley M., Bangera M.G., Bumgarner R.E., Parsek M.R., Teitzel G.M., Lory S., et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 55.Waite R.D., Paccanaro A., Papakonstantinopoulou A., Hurst J.M., Saqi M., Littler E., et al. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genom. 2006;7:162. doi: 10.1186/1471-2164-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waite R.D., Papakonstantinopoulou A., Littler E., Curtis M.A. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kordes A., Preusse M., Willger S.D., Braubach P., Jonigk D., Haverich A., et al. Genetically diverse Pseudomonas aeruginosa populations display similar transcriptomic profiles in a cystic fibrosis explanted lung. Nat Commun. 2019;10:3397. doi: 10.1038/s41467-019-11414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi E., Falcone M., Molin S., Johansen H.K. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun. 2018;9:3459. doi: 10.1038/s41467-018-05944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poudyal B., Sauer K. The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with pa1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01981-17. e01981-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao J., Schurr M.J., Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol. 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Mah T.-F. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol. 2008;190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciofu O., Moser C., Jensen P.Ø., Høiby N. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022;20:621–635. doi: 10.1038/s41579-022-00682-4. [DOI] [PubMed] [Google Scholar]
- 63.Liao J., Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mah T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 65.Sadovskaya I., Vinogradov E., Li J., Hachani A., Kowalska K., Filloux A. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated β-(1→3)-glucans, which bind aminoglycosides. Glycobiology. 2010;20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 66.Mah T.-F., Pitts B., Pellock B., Walker G.C., Stewart P.S., O'Toole G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 67.Beaudoin T., Zhang L., Hinz A.J., Parr C.J., Mah T.-F. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J Bacteriol. 2012;194:3128–3136. doi: 10.1128/JB.06178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmed M.N., Porse A., Sommer M.O.A., Høiby N., Ciofu O. Evolution of antibiotic resistance in biofilm and planktonic Pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.00320-18. e00320-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed M.N., Abdelsamad A., Wassermann T., Porse A., Becker J., Sommer M.O.A., et al. The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance. Npj Biofilms Microbiomes. 2020;6:1–10. doi: 10.1038/s41522-020-00138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaborskyte G., Andersen J.B., Kragh K.N., Ciofu O. Real-time monitoring of nfxB mutant occurrence and dynamics in Pseudomonas aeruginosa biofilm exposed to subinhibitory concentrations of ciprofloxacin. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02292-16. e02292-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giwercman B., Meyer C., Lambert P.A., Reinert C., Høiby N. High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob Agents Chemother. 1992;36:71–76. doi: 10.1128/aac.36.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giwercman B., Jensen E.T., Høiby N., Kharazmi A., Costerton J.W. Induction of beta-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother. 1991;35:1008–1010. doi: 10.1128/aac.35.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciofu O., Beveridge T.J., Kadurugamuwa J., Walther-Rasmussen J., Høiby N. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 74.Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., Balaban N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 75.Santi I., Manfredi P., Maffei E., Egli A., Jenal U. Evolution of antibiotic tolerance shapes resistance development in chronic Pseudomonas aeruginosa infections. mBio. 2021;12 doi: 10.1128/mBio.03482-20. e03482-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 77.Molin S., Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 78.Coenye T., Bové M., Bjarnsholt T. Biofilm antimicrobial susceptibility through an experimental evolutionary lens. Npj Biofilms Microbiomes. 2022;8:82. doi: 10.1038/s41522-022-00346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radman M., Matic I., Taddei F. Evolution of evolvability. Ann N Y Acad Sci. 1999;870:146–155. doi: 10.1111/j.1749-6632.1999.tb08874.x. [DOI] [PubMed] [Google Scholar]
- 80.Taddei F., Radman M., Maynard-Smith J., Toupance B., Gouyon P.H., Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 81.Mao E.F., Lane L., Lee J., Miller J.H. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oliver A., Cantón R., Campo P., Baquero F., Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 83.Montanari S., Oliver A., Salerno P., Mena A., Bertoni G., Tümmler B., et al. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiol Read Engl. 2007;153:1445–1454. doi: 10.1099/mic.0.2006/003400-0. [DOI] [PubMed] [Google Scholar]
- 84.Kenna D.T., Doherty C.J., Foweraker J., Macaskill L., Barcus V.A., Govan J.R.W. Hypermutability in environmental Pseudomonas aeruginosa and in populations causing pulmonary infection in individuals with cystic fibrosis. Microbiology. 2007;153:1852–1859. doi: 10.1099/mic.0.2006/005082-0. [DOI] [PubMed] [Google Scholar]
- 85.Gutiérrez O., Juan C., Pérez J.L., Oliver A. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob Agents Chemother. 2004;48:3573–3575. doi: 10.1128/AAC.48.9.3573-3575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eliopoulos G.M., Blázquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 87.Blázquez J., Rodríguez-Beltrán J., Matic I. Antibiotic-induced genetic variation: how it arises and how it can Be prevented. Annu Rev Microbiol. 2018 doi: 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 88.Stewart P.S., Franklin M.J., Williamson K.S., Folsom J.P., Boegli L., James G.A. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2015;59:3838. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maciá M.D., Blanquer D., Togores B., Sauleda J., Pérez J.L., Oliver A. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother. 2005;49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woodford N., Ellington M.J. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2007;13:5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 91.Blázquez J., Oliver A., Gómez-Gómez J.-M. Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance? Curr Drug Targets. 2002;3:345–349. doi: 10.2174/1389450023347579. [DOI] [PubMed] [Google Scholar]
- 92.Webb J.S., Thompson L.S., James S., Charlton T., Tolker-Nielsen T., Koch B., et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Driffield K., Miller K., Bostock J.M., O'Neill A.J., Chopra I. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother. 2008;61:1053–1056. doi: 10.1093/jac/dkn044. [DOI] [PubMed] [Google Scholar]
- 94.Boles B.R., Singh P.K. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulet X., Maciá M.D., Mena A., Juan C., Pérez J.L., Oliver A. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob Agents Chemother. 2009;53:1552–1560. doi: 10.1128/AAC.01264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scribner M.R., Santos-Lopez A., Marshall C.W., Deitrick C., Cooper V.S. Parallel evolution of tobramycin resistance across species and environments. mBio. 2020;11 doi: 10.1128/mBio.00932-20. e00932-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Díez-Aguilar M., Morosini M.I., Tedim A.P., Rodríguez I., Aktaş Z., Cantón R. Antimicrobial activity of fosfomycin-tobramycin combination against Pseudomonas aeruginosa isolates assessed by time-kill assays and mutant prevention concentrations. Antimicrob Agents Chemother. 2015;59:6039–6045. doi: 10.1128/AAC.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rojo-Molinero E., Macià M.D., Oliver A. Social behavior of antibiotic resistant mutants within Pseudomonas aeruginosa biofilm communities. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madsen J.S., Burmølle M., Hansen L.H., Sørensen S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 100.Hausner M., Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65:3710–3713. doi: 10.1128/AEM.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abe K., Nomura N., Suzuki S. Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol Ecol. 2020;96 doi: 10.1093/femsec/fiaa031. fiaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Røder H.L., Trivedi U., Russel J., Kragh K.N., Herschend J., Thalsø-Madsen I., et al. Biofilms can act as plasmid reserves in the absence of plasmid specific selection. Npj Biofilms Microbiomes. 2021;7:1–6. doi: 10.1038/s41522-021-00249-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Metzger G.A., Ridenhour B.J., France M., Gliniewicz K., Millstein J., Settles M.L., et al. Biofilms preserve the transmissibility of a multi-drug resistance plasmid. Npj Biofilms Microbiomes. 2022;8:1–10. doi: 10.1038/s41522-022-00357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 105.Rice S.A., Tan C.H., Mikkelsen P.J., Kung V., Woo J., Tay M., et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yin R., Cheng J., Wang J., Li P., Lin J. Treatment of Pseudomonas aeruginosa infectious biofilms: challenges and strategies. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.955286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meylan S., Porter C.B.M., Yang J.H., Belenky P., Gutierrez A., Lobritz M.A., et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Donnert M., Elsheikh S., Arce-Rodriguez A., Pawar V., Braubach P., Jonigk D., et al. Targeting bioenergetics is key to counteracting the drug-tolerant state of biofilm-grown bacteria. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bao X., Bové M., Coenye T. Organic acids and their salts potentiate the activity of selected antibiotics against Pseudomonas aeruginosa biofilms grown in a synthetic cystic fibrosis sputum medium. Antimicrob Agents Chemother. 2022;66 doi: 10.1128/AAC.01875-21. e01875-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gade P.A.V., Olsen T.B., Jensen P.Ø., Kolpen M., Høiby N., Henneberg K.-Å., et al. Modelling of ciprofloxacin killing enhanced by hyperbaric oxygen treatment in Pseudomonas aeruginosa PAO1 biofilms. PLoS One. 2018;13 doi: 10.1371/journal.pone.0198909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lerche C.J., Christophersen L.J., Kolpen M., Nielsen P.R., Trøstrup H., Thomsen K., et al. Hyperbaric oxygen therapy augments tobramycin efficacy in experimental Staphylococcus aureus endocarditis. Int J Antimicrob Agents. 2017;50:406–412. doi: 10.1016/j.ijantimicag.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 112.Andersen J.B., Hultqvist L.D., Jansen C.U., Jakobsen T.H., Nilsson M., Rybtke M., et al. Identification of small molecules that interfere with c-di-GMP signaling and induce dispersal of Pseudomonas aeruginosa biofilms. Npj Biofilms Microbiomes. 2021;7:1–13. doi: 10.1038/s41522-021-00225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baker P., Hill P.J., Snarr B.D., Alnabelseya N., Pestrak M.J., Lee M.J., et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2 doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu S., Su T., Wu H., Liu S., Wang D., Zhao T., et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–1367. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kvist M., Hancock V., Klemm P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol. 2008;74:7376–7382. doi: 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Skov M., Pressler T., Lykkesfeldt J., Poulsen H.E., Jensen P.Ø., Johansen H.K., et al. The effect of short-term, high-dose oral N-acetylcysteine treatment on oxidative stress markers in cystic fibrosis patients with chronic P. aeruginosa infection — a pilot study. J Cyst Fibros. 2015;14:211–218. doi: 10.1016/j.jcf.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 117.Pires D.P., Oliveira H., Melo L.D.R., Sillankorva S., Azeredo J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100:2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 118.Pérez-Laguna V., García-Luque I., Ballesta S., Pérez-Artiaga L., Lampaya-Pérez V., Rezusta A., et al. Photodynamic therapy using methylene blue, combined or not with gentamicin, against Staphylococcus aureus and Pseudomonas aeruginosa. Photodiagnosis Photodyn Ther. 2020;31 doi: 10.1016/j.pdpdt.2020.101810. [DOI] [PubMed] [Google Scholar]
- 119.Lin Q., Pilewski J.M., Di Y.P. Acidic microenvironment determines antibiotic susceptibility and biofilm formation of Pseudomonas aeruginosa. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.747834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bilal H., Bergen P.J., Kim T.H., Chung S.E., Peleg A.Y., Oliver A., et al. Synergistic meropenem-tobramycin combination dosage regimens against clinical hypermutable Pseudomonas aeruginosa at simulated epithelial lining fluid concentrations in a dynamic biofilm model. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.01293-19. e01293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bilal H., Bergen P.J., Tait J.R., Wallis S.C., Peleg A.Y., Roberts J.A., et al. Clinically relevant epithelial lining fluid concentrations of meropenem with ciprofloxacin provide synergistic killing and resistance suppression of hypermutable Pseudomonas aeruginosa in a dynamic biofilm model. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.00469-20. e00469-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rodríguez-Rojas A., Couce A., Blázquez* J. Frequency of spontaneous resistance to fosfomycin combined with different antibiotics in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:4948–4949. doi: 10.1128/AAC.00415-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maciá M.D., Borrell N., Segura M., Gómez C., Pérez J.L., Oliver A. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:975–983. doi: 10.1128/AAC.50.3.975-983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Plasencia V., Borrell N., Maciá M.D., Moya B., Pérez J.L., Oliver A. Influence of high mutation rates on the mechanisms and dynamics of in vitro and in vivo resistance development to single or combined antipseudomonal agents. Antimicrob Agents Chemother. 2007;51:2574–2581. doi: 10.1128/AAC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rojo-Molinero E., Macià M.D., Rubio R., Moyà B., Cabot G., López-Causapé C., et al. Sequential treatment of biofilms with aztreonam and tobramycin is a novel strategy for combating Pseudomonas aeruginosa chronic respiratory infections. Antimicrob Agents Chemother. 2016;60:2912–2922. doi: 10.1128/AAC.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Müsken M., Pawar V., Schwebs T., Bähre H., Felgner S., Weiss S., et al. Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01635-18. e01635-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krajewski J., Bode-Böger S.M., Tröger U., Martens-Lobenhoffer J., Mulrooney T., Mittelstädt H., et al. Successful treatment of extensively drug-resistant Pseudomonas aeruginosa osteomyelitis using a colistin- and tobramycin-impregnated PMMA spacer. Int J Antimicrob Agents. 2014;44:363–366. doi: 10.1016/j.ijantimicag.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 128.Heijerman H., Westerman E., Conway S., Touw D. Inhaled medication and inhalation devices for lung disease in patients with cystic fibrosis: a European consensus. J Cyst Fibros. 2009;8:295–315. doi: 10.1016/j.jcf.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 129.Kvich L., Christensen M.H., Pierchala M.K., Astafiev K., Lou-Moeller R., Bjarnsholt T. The combination of low-frequency ultrasound and antibiotics improves the killing of in vitro Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Antibiotics. 2022;11:1494. doi: 10.3390/antibiotics11111494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pelgrift R.Y., Friedman A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev. 2013;65:1803–1815. doi: 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 131.Li X., Chen D., Xie S. Current progress and prospects of organic nanoparticles against bacterial biofilm. Adv Colloid Interface Sci. 2021;294 doi: 10.1016/j.cis.2021.102475. [DOI] [PubMed] [Google Scholar]
- 132.Beirão S., Fernandes S., Coelho J., Faustino M.A.F., Tomé J.P.C., Neves M.G.P.M.S., et al. Photodynamic inactivation of bacterial and yeast biofilms with a cationic porphyrin. Photochem Photobiol. 2014;90:1387–1396. doi: 10.1111/php.12331. [DOI] [PubMed] [Google Scholar]
- 133.Ghosh S., Lahiri D., Nag M., Sarkar T., Pati S., Edinur H.A., et al. Precision targeting of food biofilm-forming genes by microbial scissors: CRISPR-Cas as an effective modulator. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.964848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noirot-Gros M.-F., Forrester S., Malato G., Larsen P.E., Noirot P. CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens. Sci Rep. 2019;9 doi: 10.1038/s41598-019-52400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.