Abstract
The exquisite specificity, natural biological functions, and favorable development properties of antibodies make them highly effective agents as drugs. Monoclonal antibodies are particularly strong as inhibitors of systemically accessible targets where trough-level concentrations can sustain full target occupancy. Yet beyond this pharmacologic wheelhouse, antibodies perform suboptimally for targets of high abundance and those not easily accessible from circulation. Fundamentally, this restraint on broader application is due largely to the stoichiometric nature of their activity—one drug molecule is generally able to inhibit a maximum of two target molecules at a time. Enzymes in contrast are able to catalytically turnover multiple substrates, making them a natural sub-stoichiometric solution for targets of high abundance or in poorly accessible sites of action. However, enzymes have their own limitations as drugs, including, in particular, the polypharmacology and broad specificity often seen with native enzymes. In this study, we introduce antibody-guided proteolytic enzymes to enable selective sub-stoichiometric turnover of therapeutic targets. We demonstrate that antibody-mediated substrate targeting can enhance enzyme activity and specificity, with proof of concept for two challenging target proteins, amyloid-β and immunoglobulin G. This work advances a new biotherapeutic platform that combines the favorable properties of antibodies and proteolytic enzymes to more effectively suppress high-bar therapeutic targets.
Keywords: antibody engineering, amyloid, enzyme catalysis, immunoglobulin G (IgG), protein targeting, protein degradation, protease
Monoclonal antibodies are an immensely successful class of drugs that address major medical needs in a variety of therapeutic areas (1, 2). The widespread success of antibodies stems in part from their high specificity, capability for immune recruitment, long serum half-life, relatively low immunogenicity, and streamlined discovery methods. Despite these favorable features, an inherent limitation of antibodies is their general reliance on stoichiometric target binding to induce the desired therapeutic effect. This aspect of antibodies can impede their effective application to some targets of therapeutic interest, specifically those of high abundance and those for which there are barriers to site of action. The former category includes, for example, molecules such as immunoglobulins and complement proteins, which are of growing therapeutic interest due to their roles in autoimmune and inflammatory diseases (3, 4, 5). The latter category includes targets in the central nervous system (CNS), eye, and gastrointestinal tract. Indeed, the low exposure of systemic antibodies to the CNS (∼0.1%) (6, 7, 8) has demanded extraordinarily high doses of antibodies targeting pathogenic amyloid-β and tau proteins within the central nervous system, and may be a factor that has hindered their clinical success (7, 9, 10).
Enzymes are a class of catalytic proteins with a sub-stoichiometric mechanism of action. In contrast to antibodies, a single enzyme can react with many substrate molecules with a high catalytic rate and turnover, thus enabling low doses to maintain sufficient activity. Enzymes have been approved for the treatment of cancer, blood disorders, lysosomal storage disorders, and metabolic deficiencies, among many other conditions (11, 12, 13). However, several drawbacks limit more widespread application of this therapeutic class including short half-life, lack of tissue specificity, broad substrate specificity, and high immunogenicity when not of human origin.
In this work, we explore antibody-guided proteolytic enzymes as a means to achieve selective sub-stoichiometric turnover of therapeutic targets. We show that increased target engagement through antibody-antigen recognition can enhance the catalytic activity and specificity of genetically fused proteases, with proof of concept for two clinical stage yet difficult to target proteins, amyloid-β (Aβ) and immunoglobulin G (IgG). This approach can potentially be generalized to other targets of high abundance or within physiologic sites of low drug exposure, creating a unique format that can be used to treat unmet medical needs.
Results
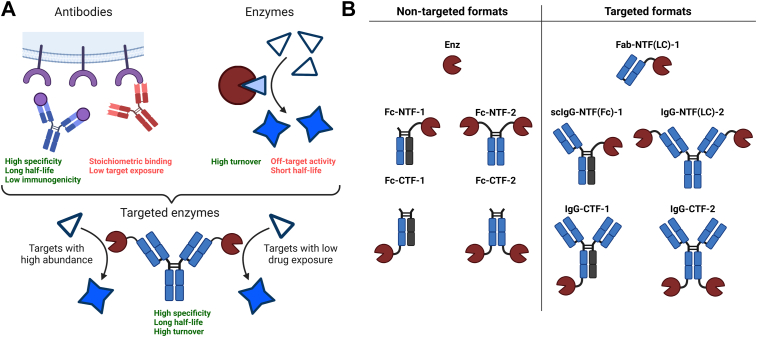
The complementary properties of antibodies and enzymes suggest that fusion of these two classes of proteins into a single molecular modality may have significant therapeutic potential (Fig. 1A). A so-called targeted catalyst could maintain the favorable properties from each class of molecule while mitigating the drawbacks. Ideally, an antibody-guided enzyme would have the specificity and long serum half-life of an antibody while demonstrating the high substrate turnover yet low dose requirements of an enzyme. For two distinct but challenging targets, Aβ and IgG, we engineered a series of formats that explored geometry and valency, including both N- and C-terminal fusions with either one or two enzymes per molecule (Fig. 1B). N-terminal enzymes were either fused to the antibody light chain (LC) or fragment crystallizable region (Fc), whereas C-terminal fusions were fused to the CH3 domain of the heavy chain (HC). Fc fusion versions lacking targeting arms or IgG formats targeting glycoprotein D of herpes simplex virus (gD) were constructed and tested as nontargeted controls.
Figure 1.
Schematic overview and naming convention of antibody-targeted proteases. A, the antibody-guided enzyme combines the favorable properties of antibodies and enzymes while mitigating the drawbacks of each. B, nontargeted and targeted formats for the antibody–enzyme fusion proteins used in this work. The naming convention is structured as follows. The first label refers to the molecule format (Fc: fragment crystallizable, Fab: fragment antigen binding, scIgG: monovalent single-chain Immunoglobulin G, IgG: immunoglobulin G). By definition, “Fab” is monovalent and “Fc” has no targeting arms. Enz refers to the free enzyme. The center label describes the fusion format as either NTF (N-terminal fusion) or CTF (C-terminal fusion). When necessary, further clarification of the fusion domain is specified within the parentheses. The third and final label denotes the number of proteases per molecule as either one or 2. LC, light chain.
Protease screen for Aβ cleavage activity
Numerous antibody-based drugs targeting Aβ have been investigated in clinical trials for the treatment of Alzheimer's Disease (AD) with thus far limited benefit to patients (14). Among the factors that have hindered clinical success of these drugs, a confounding element is the poor exposure of systemically administered antibody-based drugs to the CNS (∼0.1%) (6, 7, 8). We explored whether suppression of Aβ can be improved through antibody cotargeting of an enzyme to Aβ substrate.
Proteolytic degradation of Aβ is an important and natural elimination process to avoid pathogenic accumulation and as a consequence a diverse set of proteases have been found to play biological roles in endogenous Aβ elimination (15). This set varies in mechanism, Aβ substrate type, and subcellular localization. To select an optimal protease for our targeted catalyst approach, we screened a set of nine proteases previously implicated in Aβ degradation, including neprolysin (NEP), neprolysin-2 (NEP2), endothelin-converting enzyme 1 and 2 (ECE1 and ECE2), angiotensin-converting enzyme (ACE), insulin-degrading enzyme (IDE), matrix metalloproteinase 2 and 9 (MMP2 and MMP9), and matriptase (MTSP1).
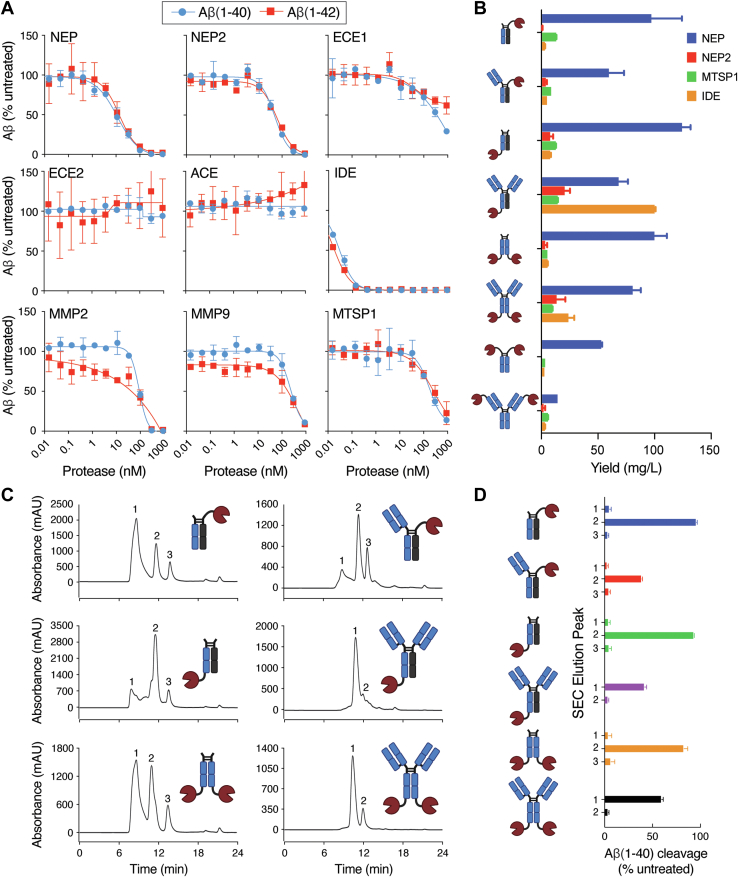
The activity of each protease was first tested on commercially available fluorogenic substrates (Fig. S1). All enzymes were active on control substrates with the exception of ECE2, which is known to have optimal activity at low pH (16). The activity of each enzyme was then determined on two isoforms of Aβ, 1 to 40 and 1 to 42, using an optimized ELISA-based assay that utilizes capture and detection antibodies specific for the N and C terminus of the Aβ peptides, respectively. The efficiency of Aβ cleavage was variable with a similar level of activity observed between the two isoforms (Fig. 2A). The most active enzymes against Aβ were IDE (EC50 ∼ 0.1 nM) and NEP (EC50 ∼ 12 nM), while ECE2 and ACE were completely inactive on both Aβ isoforms.
Figure 2.
Protease and fusion format screening for targeted degradation of Aβ. A, in vitro protease screening assay for cleavage of Aβ(1–40) (blue circles) and Aβ(1–42) (red squares). B, expression yields of Fc and IgG protease fusion formats. Four different proteases were expressed in the context of the eight formats shown in the icons. IgG fusions contained variable regions of the anti-Aβ antibody crenezumab, and all heavy chain constant regions were human IgG1. The bar graph shows the expression yields from duplicate 30 ml HEK293 expressions of each construct. C, purification of crenezumab NEP protease fusion formats. Each NEP fusion format was expressed in HEK293 cells and initially purified using a protein A resin. Size exclusion chromatography (SEC) coupled with sample fractionation was used for further purification. SEC chromatograms revealed the presence of multiple species with each sample containing 2 to 3 peaks. Nonreducing SDS-PAGE analysis to identify the peaks of similar antibody–protease fusions is shown in Fig. S7. D, the central fraction associated with each peak in the chromatograms from (C) was tested for Aβ(1–40) cleavage, and fractions of active peaks were pooled to obtain samples free of unwanted species. Error bars in (A, B, and D) represent standard error values with n = 2. Aβ, amyloid-β; ECE, endothelin-converting enzyme; Fc, fragment crystallizable; IgG, immunoglobulin G; IDE, insulin-degrading enzyme; MMP, matrix metalloproteinase; MTSP1, matriptase; NEP, neprolysin.
Protease targeting to Aβ substrate enhances catalytic potency
In order to identify the optimal geometry for our Aβ-degrading targeted protease fusions in terms of expression and activity, we explored a variety of molecules with differing proteases and fusion orientations to the anti-Aβ antibody, crenezumab, which is known to bind to Aβ monomers, oligomers, and fibrils with nanomolar affinity (17). Following purification using protein A resin, NEP fusions consistently showed the highest yield compared to the other proteases tested and were therefore chosen for further purification (Fig. 2B). For each NEP fusion format, protein A purified material was fractionated using size-exclusion chromatography (SEC) (Fig. 2C), and fractions from each of the major peaks were tested for Aβ cleavage activity to identify the fraction(s) of interest (Fig. 2D).
Fusion of two NEP copies to the N terminus of crenezumab [IgG-NTF(LC)-2] lacked sufficient expression for biochemical assays and was therefore abandoned. Sufficient material was obtained for the monovalent N-terminal Fc fusion [scIgG-NTF(Fc)-1] and both monovalent and bivalent C-terminal IgG fusions [IgG-CTF-1 and IgG-CTF-2]. Nontargeted NEP Fc fusion versions of each format that lacked the crenezumab fragment antigen-binding (Fab) were constructed and tested as controls.
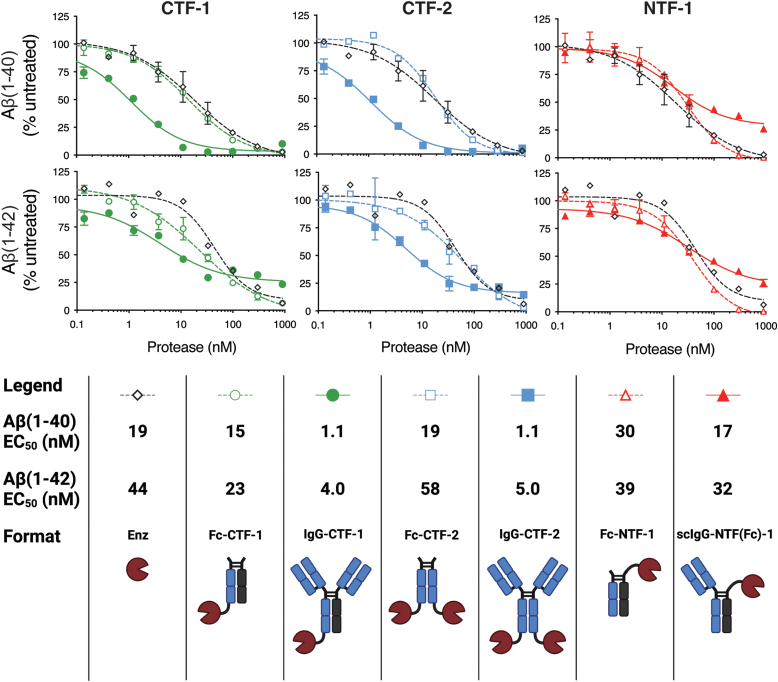
Enhancement in Aβ cleavage was observed with crenezumab-mediated targeting in the context of both C-terminal NEP fusions for both Aβ isoforms (Fig. 3, left and middle graphs), improving potency approximately 15-fold on Aβ(1–40) and 9-fold on Aβ(1–42) relative to their respective nontargeted controls (Fig. 3, legend). No difference in cleavage activity was observed between monovalent and bivalent C-terminal NEP fusions. The targeted monovalent N-terminal IgG fusion [scIgG-NTF(Fc)-1] did not enhance activity over the nontargeted constructs (Fig. 3, right graphs). Importantly, all Fc fusion controls showed comparable activity to unfused free enzyme, indicating that both N-terminal and C-terminal fusion does not impair enzyme activity. Together, these data suggest that the lack of enhancement for the N-terminal fusion [scIgG-NTF(Fc)-1] may be attributed to geometric and/or steric constraints of the fusion format, possibly exacerbated by the small substrate size of Aβ.
Figure 3.
Comparison of nontargeted and targeted proteolytic degradation of Aβ in different fusion formats. Antibody-targeting of C-terminal–fused NEP improves the EC50 value of Aβ(1–40) (top) and Aβ(1–42) (middle) by an average of 15-fold and 9-fold, respectively, compared to nontargeted controls, while the N-terminal fusion format shows no improved activity upon targeting. All targeted formats contain the variable domain of the anti-Aβ antibody crenezumab. Formats of the same enzyme and Fab valency and enzyme fusion site are compared in each plot along with enzyme alone. The key shows the EC50 values and visually depicts the targeted and nontargeted formats. Error bars represent standard error values with n = 2. Aβ, amyloid-β; CTF, C-terminal fusion; Fc, fragment crystallizable; IgG: immunoglobulin G; NEP, neprolysin; NTF, N-terminal fusion; scIgG, monovalent single-chain immunoglobulin G.
Faster off-rate and weaker Aβ affinity correlate with catalytic potency of enzyme fusions
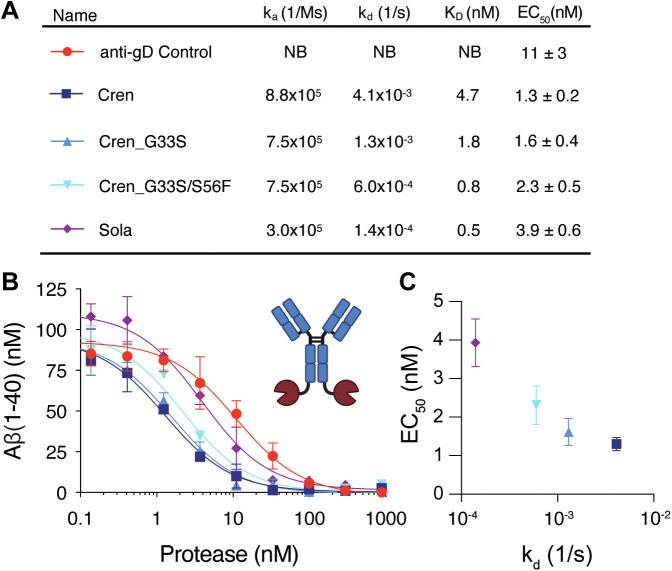
The anti-Aβ antibody, crenezumab, has both fast on and off rates (Figs. 4A and S2), which we hypothesized would facilitate rapid recycling of target antigen for cleavage by the fused protease. To investigate the dependence of targeted catalysis on antibody binding kinetics and affinity, we incorporated into the bivalent NEP IgG-CTF-2 format two variants of crenezumab G33S(HC) and G33S(HC)/S56F(LC) with slower off-rates and, therefore, stronger affinities. In addition, we also constructed the format with the variable region of another anti-Aβ antibody, solanezumab, which shares high sequence identity to crenezumab (93% ID in VH and 92% ID in VL) and binds a similar epitope but with higher affinity (18, 19, 20). Finally, a fusion containing a variable region targeting an unrelated antigen (gD) was produced as a nonbinding control. This resulted in a panel of five constructs of IgG-CTF-2 with varying off-rates and affinities yet similar on-rates (Figs. 4A and S2). Overall, higher Aβ cleavage activity correlated with faster off rate across the affinity variants with the crenezumab-NEP fusion (fastest off-rate) having a 3-fold lower EC50 than the solanezumab-NEP fusion (slowest off-rate) (Fig. 4, B and C). These data suggest that fast binding kinetics may be optimal, enabling the antibody to cycle through Aβ binding events for more effective enzymatic turnover. Overall, these results also illustrate the importance of tuning various activity parameters to optimize the engineered format.
Figure 4.
Effect of antibody epitope and off-rate on targeted enzyme activity. A, summary of kinetic parameters and EC50 values for the anti-Aβ antibody off-rate series. EC50's are presented as average and standard error from duplicates. B, faster anti-Aβ antibody off-rates lead to improved targeted Aβ proteolytic cleavage in the IgG-CTF-2 fusion format as shown in the in vitro Aβ(1–40) cleavage assay. Targeting of NEP with crenezumab (kd 4.1 × 10−3 1/s) leads to a 3-fold improvement in the EC50 value for Aβ(1–40) cleavage compared to targeting with solanezumab (1.4 × 10−4 1/s). C, correlation between the EC50 value for Aβ(1–40) cleavage and the off-rate for the anti-Aβ targeting antibody. Aβ, amyloid-β; CTF, C-terminal fusion; NEP, neprolysin.
Protease engineering for IgG cleavage
We further explored applicability of the antibody-guided protease platform to turnover a target of high abundance, serum IgG. Therapeutic IgG-suppression has been investigated for a wide range of autoimmune and inflammatory diseases with clinical success (21, 22, 23). Reduction of IgG levels is one of the putative mechanisms of action of intravenous immunoglobulin (24), which is approved for the treatment of immunothrombocytopenia, Guillain–Barré syndrome, Kawasaki’s disease, and chronic inflammatory demyelinating polyneuropathy. Furthermore, intravenous immunoglobulin is used off-label for a large number of chronic inflammatory diseases (25). More recently, biotherapeutics that block the neonatal Fc receptor (FcRn) have been advanced to directly suppress serum IgG (26). In patients, these agents demonstrate clear pharmacodynamic activity and efficacy in indications for which autoantibodies are disease drivers. The recent approval of efgartigimod for generalized myasthenia gravis in adults who test positive for the anti-acetylcholine receptor antibody validates IgG depletion as a therapeutic mechanism (21), with several anti-FcRn antibody–based medicines in clinical trials for generalized myasthenia gravis, immunothrombocytopenia, chronic inflammatory demyelinating polyneuropathy, and other indications (22, 23, 27). Despite this success, the high amount of FcRn and stoichiometric nature of blockade necessitates high doses and frequent dosing for anti-FcRn biotherapeutic agents yet still results in incomplete target suppression.
A more direct alternative to FcRn blockade is IgG depletion. Due to the extraordinarily high (∼10 mg/ml) concentration of IgG in human serum, catalytic turnover is essential. The IgG degrading enzyme IdeS is effective at depleting IgG in vitro and in vivo and has been tested clinically (28, 29, 30, 31). However, due to its bacterial origin, IdeS is highly immunogenic, hindering repeat dosing, and as a consequence, its therapeutic application is limited to acute treatment indications such as kidney transplantation. Several human enzymes with lower immunogenicity risk have been shown to cleave IgG (Fig. 5A) (32), but they are not IgG-selective and lack the efficiency needed to clear the high levels of substrate present in serum. We sought to use targeted catalysis as a means to enhance the cleavage efficiency and selectivity of human proteases to degrade serum IgG.
Figure 5.
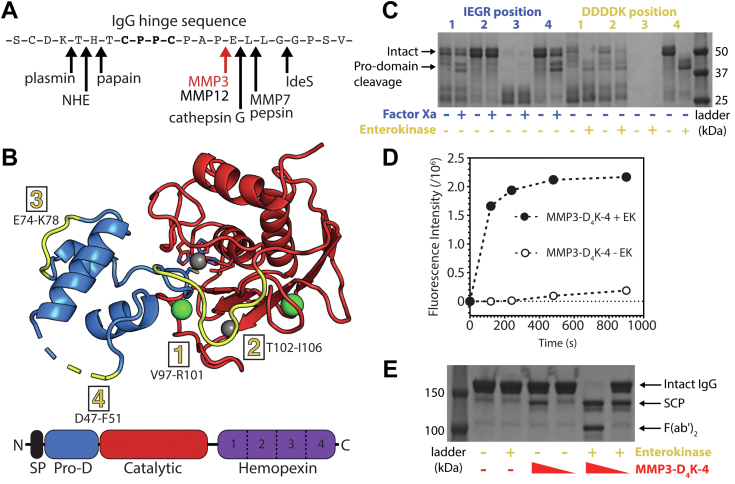
Engineering and characterization of matrix metalloproteinase 3 (MMP3) for targeted endogenous human IgG cleavage. A, human and bacterial proteases previously shown to cleave within the hinge of human IgG, with the MMP3 cleavage site indicated in red. B, structural representation of MMP3 (PDB ID: 1SLM (64)). The prodomain (Pro-D, blue) and catalytic domain (catalytic, red) are shown in cartoon representation, while the signaling peptide (SP, black) and the hemopexin domain (hemopexin, violet) are not present in the published crystal structure. Calcium atoms and zinc atoms are shown in green and gray, respectively. The catalytic zinc is coordinated by three histidines within the catalytic domain and one cysteine within the pro-domain (shown as sticks). The four substitution sites for the factor Xa (IEGR) and enterokinase (DDDDK) cleavage sequences are highlighted in yellow and listed with the corresponding MMP3 residues. C, SDS-PAGE gel depicting the eight protease cleavage site insertion variants before and after activation with their respective external protease (factor Xa in blue and enterokinase in yellow). D MMP3-D4K-4 represents the variant with an enterokinase cleavage site substituted within position four of the MMP3 prodomain. This variant efficiently cleaves a fluorescent MMP3 peptide substrate after the prodomain is removed with enterokinase (filled circles), while the intact form containing the prodomain minimally cleaves the substrate (open circles). E, SDS-PAGE gel showing the cleavage of human IgG by MMP3-D4K-4 either with or without the prodomain at different relative concentrations (10% and 1% w/w) after 24 h at 37 °C. Presence or absence of a component in the reaction is represented by + and −, respectively. MMP3-D4K-4 cleaves the lower hinge of intact human IgG in a sequential manner, first producing a single cleavage product (SCP, in which half of the Fc is lost upon denaturation), then producing F(ab’)2 and Fc (not shown) after the second cleavage. Enterokinase does not detectably cleave human IgG (lane 3). IgG, immunoglobulin G.
We selected three human proteases [matrix metalloproteinase 3 (MMP3), MMP7, and cathepsin G] based on their human origin and IgG cleavage properties (32) and screened them for expression (Fig. S3). For further development, we selected MMP3, a zinc matrix metalloproteinase expressed by a broad variety of cell types with promiscuous activity against matrix and bioactive substrates. The structure consists of an N-terminal signaling sequence, prodomain, catalytic domain, and hemopexin domain connected by a proline-rich linker (Fig. 5B) (33). Successful expression of MMP3 requires fusion to the inhibitory N-terminal prodomain, which is proteolytically removed to induce activity. While various approaches, including heat, addition of organo-mercury reagents, or partial proteolysis, have been reported to induce activation in vitro, each comes with significant risks to the structure and stability of a potential therapeutic.
To circumvent the problematic activation step (removal of inhibitory N-terminal prodomain), we designed MMP3 variants capable of prodomain cleavage by the highly specific proteases enterokinase (EK) and Factor Xa. In order to determine the optimal cleavage location to achieve full and selective MMP3 activity, we substituted the recognition sequences for EK or Xa within the prodomain of MMP3 at four unstructured locations to both allow for maximal EK or Xa protease accessibility and to minimize structural perturbation of the prodomain (Fig. 5B). SDS-PAGE analysis and MMP3 activity assays were performed on the eight variants with and without the addition of EK or Xa. The variant with the EK site insertion at location four showed the best combination of high stability with low catalytic activity in the native state (with prodomain) while yielding efficient removal of the prodomain in the presence of EK (Fig. 5, C and D). “Walking” the EK insertion site around position 4 in one residue increments did not show any significant improvements (Fig. S4). Therefore, the original variant with the EK site inserted at position 4, referred to as MMP3-D4K-4, was selected for further studies. At high concentrations and after EK cleavage of the prodomain, MMP3-D4K-4 is capable of cleaving the IgG hinge (Fig. 5E).
Engineering a nonself selective anti-IgG antibody for endogenous IgG targeting
To target MMP3 to IgG, we explored the use of rheumatoid factors (RFs), which are naturally occurring human autoantibodies that bind to IgG. We focused on a well-characterized RF, referred to as RF61, which was first isolated from a rheumatoid arthritis patient and binds to the antibody Fc region (34). Importantly, the crystal structure of RF61 in complex with Fc has been solved (35) and suggested the possibility for nonself selectivity engineering (below).
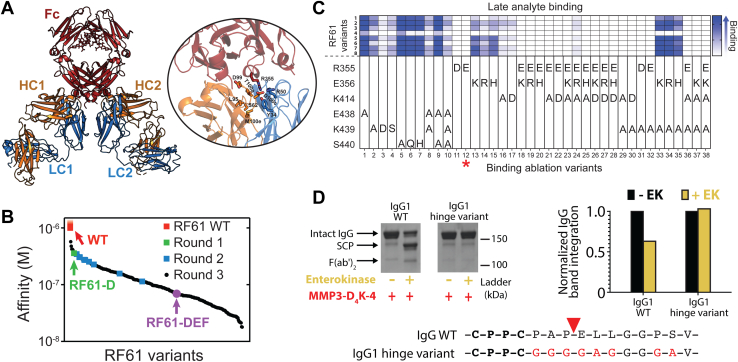
RF61 was initially identified as an IgM, which binds weakly to IgG1 Fc with a KD of approximately 600 nM (including avidity) (35). The crystal structure shows a stoichiometry of two RF61 Fabs per Fc with each Fab contacting residues from both CH3 domains (Fig. 6A) (35). To improve the affinity of RF61 for the IgG Fc, we performed saturation mutagenesis (excluding cysteine) at each residue of the complementarity-determining regions (CDRs) of both the HC (38 residues) and LC (31 residues), totaling 1242 single RF61 point mutants. While we were unable to detect binding of wildtype RF61 Fab to Fc in a monovalent surface plasmon resonance (SPR) binding format, screening of mutants using the same technique identified four promising mutations with significantly increased affinity all located in the LC (R50N, R50D, Y34F, and Y34N). A second round of screening was performed in which the R50N mutant LC was paired with the same HC single point mutant library (684 total variants). SPR screening revealed six HC mutations with improved binding upon combination with the R50N LC (S62P, L95N, D99H, T100aA, D100cE, and M100eF). For the third and final round, 320 variants with combinations of the selected LC and HC mutations were produced, ranging from one to eight mutations per variant. The three rounds of our saturation mutagenesis screen yielded RF61 variants spanning over two logs in affinity from >1 μM to ∼10 nM (Figs. 6B and S5).
Figure 6.
Development of an anti-human IgG antibody.A, structural representation of two RF61 Fabs (LC: blue, HC: orange) bound to human Fc (red) (PDB ID: 2J6E (35)). The inset highlights residues identified through saturation mutagenesis to be important for Fc binding. B, waterfall plot summarizing the affinities of RF61 variants to human IgG1 over three rounds of saturation mutagenesis and screening. The affinity of wildtype RF61 could not be determined using our monovalent SPR approach and can only be estimated as >1 μM. Affinities were measured via SPR on RF61 variants in a mouse IgG2a chimeric format against human IgG1 Fc (see Experimental procedures). C, quantification of binding between eight high affinity RF61 variants identified in round three of affinity maturation and 38 binding-ablation variants of human Fc. See Methods for specific RF61 variants in this experiment. Late analyte binding values from SPR sensograms representing binding of each RF61 variant to human Fc were used to evaluate the binding-ablation variants, with complete binding ablation shown in white and high binding levels shown in blue. The red asterisk highlights the Fc variant (R355E, variant 12) used in all RF61 IgG constructs to eliminate binding to self. D, mutation of the lower hinge sequence and N-terminal region of CH2 of human IgG1 effectively inhibits cleavage by MMP3-D4K-4. SDS-PAGE gel image (top left) and densitometric representation (top right) confirm resistance of the IgG1 hinge variant to proteolytic cleavage. EK, enterokinase; Fab, fragment antigen binding; Fc, fragment crystallizable; HC, heavy chain; IgG, immunoglobulin G; LC, light chain; MMP, matrix metalloproteinase; SPR, surface plasmon resonance.
Additional engineering of the RF61 IgG format was needed for an effective endogenous IgG targeting antibody. First, the Fc must be modified to avoid self-recognition. Using the crystal structure of RF61 bound to an IgG1 Fc as a guide (35), 38 Fc variants containing between one and four mutations were designed to ablate RF61 binding. SPR was used to screen the Fc variants against a panel of eight affinity-improved RF61 variants (Fig. 6C). The single mutant R355E showed exquisite ablation of RF61 binding, with minimal improvement from additional mutations, and was therefore selected. A second requisite is that the hinge must be resistant to proteolysis by MMP3 to avoid self-cleavage by the fused enzyme. We replaced the 10 residues following the hinge disulfides, effectively the lower hinge and N-terminal region of the CH2 domain, with a (G4A)2 linker to confer resistance to MMP3 cleavage (Fig. 6D). Altogether, the IgG-targeting antibody contains Fab arms with improved RF61 affinity, an Fc with the R355E mutation to avoid self-binding, and a mutated lower hinge to avoid self-cleavage by MMP3.
Antibody-enzyme fusion selectively enhances MMP3-mediated IgG degradation
We combined the two engineered modalities above into a single targeted protease. In a similar approach as Aβ, a variety of antibody-enzyme fusion formats were screened for expression, stability, affinity, and activity. Since C-terminal fusions of MMP3 to the antibody would result in separation of the proteolytic and targeting modalities upon the prodomain cleavage needed to activate MMP3, we focused our efforts on N-terminal fusions. Fusion of MMP3 and its prodomain to the N termini of the antibody HC, LC, or Fc yielded sufficient purified material. Activity screening against both fluorogenic peptide substrate and IgG substrate revealed the most promising IgG format as an MMP3 fusion to the LC N terminus via a (G4A)2 linker, referred to as IgG-NTF(LC)-2 (Figs. 7A and S6). Both IgG and Fab formats of the MMP3 LC N-terminal fusion were scaled up and purified for further study (Fig. S7).
Figure 7.
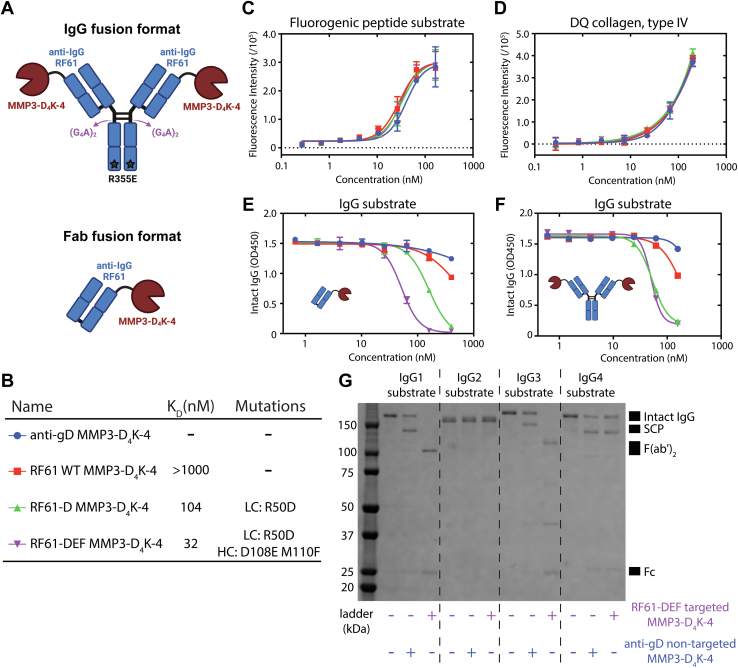
Engineering a targeted protease for human IgG cleavage. A, cartoon representations of the IgG-MMP3-D4K-4 and the Fab-MMP3-D4K-4 fusion protein formats. B, summary of affinity and relevant mutations for the anti-IgG antibody affinity series tested in (C–F). Affinity was measured for the anti-IgG Fabs of RF61 variants against human IgG1 Fc using SPR (Fig. S8). C–F, in vitro cleavage assays measuring proteolytic activity of an anti-IgG affinity series of antibody-MMP3-D4K-4 fusion proteins against a fluorogenic MMP3 peptide substrate (C), DQ collagen, type IV (D), and human IgG1 (E and F). The Fab-MMP3-D4K-4 fusion format was used in (C–E), while the IgG-MMP3-D4K-4 fusion format was used in (F). Cleavage of the substrates in (C and D) generates fluorescent signal through dequenching of fluorophores. Cleavage of IgG in (E and F) is measured with ELISA. Error bars represent standard error values with n = 2 (C and D) and n = 3 (E and F). G, dependence of targeted and nontargeted IgG cleavage on human IgG subtypes. MMP3 does not cleave IgG2, while RF61 does not bind IgG4 (Fig. S9). Fab, fragment antigen binding; Fc, fragment crystallizable; IgG, immunoglobulin G; MMP, matrix metalloproteinase; SPR, surface plasmon resonance.
In order to determine whether RF61-mediated IgG targeting of MMP3 could enhance its activity, we designed three antibody-MMP3 fusion proteins with varying affinity toward endogenous human IgG (Fig. 7B). Although we were unable to detect binding of wildtype RF61 to Fc using our SPR approach above, we nonetheless included it as a weak binder due to the reported binding affinity with avidity effects (35, 36). We also included two RF61 mutants from the saturation mutagenesis screen described above: RF61-D (104 nM) and RF61-DEF (32 nM) (Figs. 7B and S8). A control format with an anti-gD Fab was also produced as a true nonbinding control. Each affinity variant and control were produced with MMP3 fused to the LC of both a Fab and full-length IgG. Monovalent affinities for each targeting arm are summarized in Figure 7B. Nontargeted activity was first measured against two non-IgG substrates: a fluorogenic peptide substrate (Fig. 7C) and DQ collagen IV that represents an endogenous off-target substrate (Fig. 7D). As expected, the activity curves for each member of the affinity series overlap well, signifying nonselectivity of MMP3 for non-IgG substrate. To determine the effect of IgG targeting, Fab fusion (Fig. 7E) and IgG fusion (Fig. 7F) proteins were incubated at various concentrations with a human IgG1 substrate, and IgG cleavage was determined with an ELISA-based assay. EC50 values correlated remarkably well with IgG affinity of the targeting arm, with the Fab fusion but not IgG fusion format differentiating between the two higher affinity RF61 variants. The nontargeted anti-gD construct showed little to no cleavage at the maximum tested concentration. While a quantitative measure of enhancement over the nontargeted construct could not be determined due to the lack of IgG cleavage from the anti-gD control, the results suggest that antibody targeting provides at least one log enhancement of protease activity.
Interestingly, while the EC50 values match for the Fab and IgG fusion formats of the RF61-DEF affinity variant (Fig. 7, E and F, purple), the RF61-D IgG fusion had an EC50 3-fold lower than that of the Fab fusion. These data suggest that avidity in the IgG fusion may contribute to enhanced activity. Furthermore, the lack of enhanced activity upon increased affinity for the two tightest binding IgG fusion formats indicates that a maximum beneficial effect of IgG-targeting was achieved, and the activity of the MMP3-D4K-4 protease is the limiting factor. Accordingly, further improvement in targeted IgG cleavage may require protease engineering to improve catalytic rate.
Although IgG1 accounts for approximately two-thirds of all human IgG (37), there are three other human IgG subclasses present in serum: IgG2, IgG3, and IgG4. To assess the subtype dependence of our targeted protease, we measured the ability of targeted (RF61-DEF) and nontargeted (anti-gD) antibody-MMP3 fusions to cleave all four human IgG subtypes (Fig. 7G). IgG1 and IgG3 substrates showed similar enhanced targeting-dependent cleavage, with the RF61-DEF fusion fully cleaving IgG into Fc and F(ab’)2 fragments. In contrast, anti-gD control fusions were only capable of single hinge cleavage events for the same two subtypes. No cleavage of IgG2 was observed, which is consistent with the lack of an MMP3 cleavage site in its hinge sequence together with previous literature reports (38, 39). No difference between targeted and nontargeted cleavage was observed for the IgG4 substrate, which was anticipated based on the lack of RF61 binding to IgG4 (Fig. S9). While this IgG subclass cleavage profile may not be ideal from a therapeutic standpoint, altogether the results are consistent with the selectivity enhancement provided by targeted catalysis.
To assess the activity of the RF61-mediated IgG targeting of MMP3 in a more biologically relevant environment, we measured cleavage of IgG1 within human serum by both a targeted (RF61-DEF) and nontargeted (anti-gD) Fab fusion format (Fig. S10A). While the potency (EC50) of the targeted protease is reduced in the presence of serum compared to the results obtained with purified IgG1, there is still a clear enhancement in activity compared to the nontargeted control. Cleavage activity against a fluorogenic MMP3 peptide substrate under the same conditions suggests that endogenous protease inhibitors within human serum are not responsible for the reduced activity of these fusion constructs (Fig. S10B). The reduced activity could be due to a variety of other factors present in serum, including additional RF61 antigens (e.g., IgG2 and IgG3) that compete with binding to IgG1 and/or alternative MMP3 substrates (e.g., matrix and bioactive substrates including other immunoglobulin subtypes) that dilute IgG1 in the pool of potential MMP3 substrates. It may be possible to address some of these factors with further engineering of the formats, particularly the protease. Regardless, these results are a first and encouraging step toward exploring whether antibody-guided proteases can mediate target degradation in vivo, ultimately for therapeutic benefit.
Discussion
In this study, we combine the target-specificity of an antibody with the catalytic turnover of an enzyme to provide a new therapeutic approach for neutralization of difficult therapeutic targets, so-called higher hanging fruit (2). We report guided proteases for the degradation of Aβ, for which the site of action is not easily accessible from systemic circulation, and endogenous IgG, for which competitive inhibition is impractical due to its extremely high abundance. While the engineered proteases that cleave Aβ and IgG described here have the potential to offer advantages over existing agents in neurodegenerative and autoimmune diseases, there are undoubtedly numerous hurdles and risks to the development of these modalities. The present study is foremost an exploratory one, aimed principally at establishing proof-of-concept and providing a prototype for engineering an antibody-guided protease against a target of interest.
Previous work combining antibodies with enzymes has focused on the former's use as a delivery vehicle, with varied preclinical and limited clinical success (40). Antibody delivery of enzymes has been studied for three main applications. The first involves replacing the function of inactive native enzymes, termed enzyme replacement therapy, and has demonstrated clinical success for antibody-mediated delivery of enzymatic activity to the lysosome, cytosol, and brain (40, 41, 42, 43). The second, referred to as antibody-directed enzyme prodrug therapy, combines a tumor-targeting antibody–enzyme fusion with a systemically delivered inactive prodrug. Enzymatic activation of the prodrug locally at the tumor site is intended to minimize toxicity. Most antibody-directed enzyme prodrug therapy development has been preclinical, with minimal success in early clinical studies (40, 41, 44, 45). The third broad category of applications includes attempts to localize or direct enzymes to specific tissues, cell types, or subcellular locations. Antibodies that bind the human insulin receptor or transferrin receptor have been used to shuttle cargo, including enzymes, across the blood brain barrier to treat enzyme deficiencies (40, 46). Cancer therapies have been explored that combine a tumor-targeted antibody with cytotoxic enzymes, such as RNases and various proapoptotic enzymes (47, 48, 49). Intracellular targeting of alkaline phosphatase and catalase with an antinuclear antibody has also shown early preclinical success (50, 51).
Relative to these platforms, the distinction of our approach is the targeting of the enzymatic substrate. In effect, we impart some of the favorable selectivity of an antibody to the enzyme to improve catalytic activity and tune selectivity. Notably, for the anti-IgG, the selective enhancement in degradation of the targeted IgG substrate relative to an off-target, and potentially toxic substrate collagen, illustrates the promise of the approach. Such results suggest that, with appropriate optimization, antibody-guided catalysis may broaden the therapeutic window of proteases to enable their wider use as biotherapeutics. In turn and from the standpoint of the antibody, the protease brings sub-stoichiometric turnover to what otherwise would be a competitive inhibitor, enabling more complete suppression of tough targets.
The current study fills in some of the blueprint for discovering and optimizing a targeted catalyst. As the complexity of a molecular format increases, so too does the number of engineering parameters for optimization. In this study, we explored several such parameters, including enzyme selection, geometry, affinity, and for IgG some engineering solutions for the difficulty of targeting with a modality that is itself recognized by the antibody and enzyme. In this sense, IgG is not a straightforward pilot for the targeted catalysis approach, but nonetheless was chosen because it represents a sweet-spot application for the platform. All of the parameters we explored made a difference, and in the case of affinity, contrasting dependencies were observed for the two applications. Faster off-rates for anti-Aβ antibodies led to more potent Aβ degradation. A possible explanation is that faster off-rates may allow for more efficient substrate cycling and consequently greater substrate turnover. In contrast, for the anti-IgG antibodies, we generally found that increasing affinity resulted in improved IgG degradation activity. However, it is difficult to directly compare the anti-Aβ and anti-IgG results. The overall affinity of the anti-IgG antibodies (tens to hundreds of nM and greater) was not comparable to the affinity of the anti-Aβ antibodies (single digit nM to sub-nM). Overall, the reason(s) for the different affinity dependencies are likely complex and potentially difficult to interrogate, highlighting at this early stage the empirical nature of the approach and the benefit of engineering molecular parameters to tune performance and activity. Accordingly, while not explored in the present study, we would anticipate epitope and linker length to be additional and important elements for optimization.
While the in vitro results in the present work are promising, advancement of antibody-guided proteases as therapeutics requires further molecular optimization and substantial investigation of their in vivo pharmacology. Though it is evident that exposure from systemic administration would be favorable for serum IgG, exposure to the CNS may also be sufficient given the catalytic nature of the described molecules. Clinical results with anti-Aβ antibodies demonstrating plaque clearance in the brain, albeit slow, support the notion that systemically-administered biotherapeutics are able to elicit pharmacologic effect in the CNS (52, 53, 54, 55). Additionally, evolving technologies for transport and delivery of biotherapeutics into the brain (56, 57) may be investigated in the context of antibody-protease fusions to improve substrate accessibility and, consequently, activity. Beyond exposure, protease optimization is another area to be explored for advancement into drug development. Protein engineering approaches have been described that enable improvements in protease catalytic rate and selectivity and can even provide the means to alter cleavage sequence specificity if desired (58, 59, 60, 61). Protease optimization may mitigate the risk of potential pitfalls such as endogenous protease inhibitors, off-target substrate specificity, and poor stability. Regardless, our in vitro results with minimally engineered fusions provide an encouraging basis for exploring the pharmacokinetics and pharmacology of antibody-guided proteases in vivo.
While further study is needed, antibody-guided proteases offer the possibility for more complete suppression at lower doses than conventional biotherapeutic drugs, with broad therapeutic potential. The use of antibody targeting is an approach to addressing a subset of the hurdles to advancing enzymes as drugs. The natural mechanisms by which enzymes are regulated biologically, including for example selective expression, local activation, controlled expression of inhibitors, and colocalization via adaptor domains, are not in play for a systemically administered drug. Cotargeting may also be suited to enhance the pharmacologic performance of other catalytic modalities, including for example catalytic antibodies. In this frame, the present work represents a step toward advancing catalysts as biotherapeutics to expand the toolbox for drugging otherwise difficult-to-drug disease targets.
Experimental procedures
Molecular cloning
Gene fragments encoding all in-house–derived constructs with human codon optimization were synthesized and cloned into the pRK mammalian expression vector (Wuxi, Genewiz, Genscript). The pRK vector contains a cytomegalovirus enhancer and promotor to control gene expression, an N-terminal secretion signal (MGWSCIILFLVATATGVHS), a C-terminal simian virus 40 (SV40) PolyA sequence, and an ampicillin resistance gene for bacterial selection. For Aβ protease constructs, NEP (Y52-W750), NEP2 (R74-W770), IDE (M42-L1019), or MTSP1 (G596-V855) were fused to either Fc (D221-K447, EU numbering) or full-length human IgG1 via a GGGGS linker. For C-terminal protease fusions, the C-terminal lysine of the Fc was excluded. For IgG protease constructs, MMP3 (Y18-C477) was fused to the N terminus of either Fc (D221-K447, EU numbering), full-length human IgG1 HC, Fab HC (Q1-S113, Kabat numbering), or full-length human kappa LC via a (GGGGA)2 linker. For all monovalent protease fusions, knob-in-hole mutations were introduced into the Fc to enable heterodimerization (62). The gene for MMP3 only (Y18-C477) was synthesized with a C-terminal FLAG-tag for purification, and the EK (DDDDK) and factor Xa (IEGR) protease cleavage sites were inserted via site-directed mutagenesis using standard protocols (Qiagen, 210,513). Genes introducing Fc mutations for RF61 binding ablation (Fig. 6C) and an alternate hinge sequence for MMP3 resistance (Fig. 6D) were synthesized within a human IgG1 framework (Wuxi). Nucleotide sequences of the core constructs used in this work are provided in Text S1.
Protein expression and purification
Protein expression was performed by transfection of HEK293 cells with 30 μg DNA per 30 ml cell culture at a 1:1 HC:LC DNA ratio using standard protocols. Some formats only required transfection of a single DNA while others necessitated cotransfection of separate DNAs encoding, for example, HC and LCs or knob and hole constructs. Affinity chromatography was carried out using MabSelect SuRe resin (Cytiva, 17543803) for Fc-containing proteins, CaptureSelect CH1-XL resin (Thermo, 194346201L) for Fabs, and anti-Flag resin for MMP3-flag with elution using a buffer consisting of 50 mM sodium citrate at pH 3.0 and 150 mM NaCl. For most antibody-enzyme fusion proteins, analytical SEC revealed the presence of multiple species, likely representing a variety of cleavage products, mis-paired antibody subunits, and aggregates. To isolate the appropriate species within each sample, fractions were tested from each major peak within the SEC chromatogram for cleavage activity. Further SEC purification using a HiLoad 16/600 Superdex 200 column was required to isolate the desired monomeric species. Protein quality was determined by analytical SEC using a Waters xBridge BEH200A SEC 3.5 μm (7.8 × 300 mm) column (Waters, 176,003,596) and by SDS-PAGE. IgGs and Fabs were stored in buffer consisting of 20 mM histidine acetate and 150 mM NaCl at pH 5.5. MMP3 and all MMP3 fusion proteins were stored in 10 mM Hepes, 150 mM NaCl, and 10 mM CaCl2 at pH 7.5.
Aβ protease panel
NEP (R&D, 1182-ZNC-010), NEP2 (R&D, 2340-ZN-010), ECE-1 (R&D, 1784-ZN-010), ECE-2 (R&D, 1645-ZN-010), ACE (R&D, 929-ZN-010), IDE (R&D, 2496-ZN-010), MMP2 (R&D, 902-MP-010), MMP9 (R&D, 911-MP-010), and MTSP1 (R&D, 3946-SEB-010) were purchased commercially for initial Aβ cleavage activity screening. Activation of a 100 μg/ml solution of MMP2 or MMP9 was performed by incubation in 1 μM APMA (Sigma, A9563) for 1 or 24 h, respectively. Catalytic activity of each protease was confirmed using one of three control fluorogenic substrates: Mca-RPPGFSAFK(Dnp)-OH (R&D, ES005), Mca-PLGL-Dpa-AR-NH2 (R&D, ES001), or Boc-QAR-AMC (R&D, ES014). For this assay, a 3-fold dilution series starting at 100 nM (n = 2) was constructed for each protease in Aβ assay buffer: 50 mM Hepes pH 7.4, 150 mM NaCl, and 0.05% Brij-35, a nonionic surfactant known to enhance protein solubility (Sigma, B4184). Each dilution series was incubated with either 20 μM Mca-RPPGFSAFK(Dnp)-O, 60 μM Mca-PLGL-Dpa-AR-NH2, or 50 μM Boc-QAR-AMC in black 96-well plates (Corning, 3356) for 10 min. Fluorescence was measured on a Molecular Devices SpectraMax M2 microplate reader with 320 nm excitation and 460 nm emission for Mca-RPPGFSAFK(Dnp)-O and Mca-PLGL-Dpa-AR-NH2 or 380 nm excitation and 460 nm emission for Boc-QAR-AMC.
Aβ cleavage assay
Aβ(1–40) (Anaspec, AS-24236) and Aβ(1–42) (Anaspec, AS-20276) substrates were resuspended in 1% ammonium hydroxide (Anaspec, AS-61322) to a concentration of 1 mg/ml. The solution was sonicated twice for 30 s on ice, aliquoted, and stored at −80 °C. Prior to each assay, Aβ was thawed on ice, and a 200 nM working solution was made in Aβ assay buffer. A 3-fold dilution series of each protease or protease fusion was produced in Aβ assay buffer starting at 1.8 μM. Five microliter of protease dilution was added to 5 μl of Aβ and incubated at 37 °C for 1 h, and 10 μl of 20 μM 1,10-phanthroline (Sigma, 131377) was added to stop the reaction. Each sample was then diluted 10-fold using 180 μl of PBST (10 mM sodium phosphate pH 7.4, 150 mM NaCl, and 0.05% Tween-20), and the concentration of intact Aβ was assessed as previously described (63). Biotinylated capture antibody anti-Aβ(1–16) clone 6E10 (Biolegend, 803,009) was diluted to 1 μg/ml in PBST. Detection antibodies for Aβ(1–40) (in-house derived) and Aβ(1–42) (Thermo, 700254) were fluorescently labeled using an Alexa Fluor 647 antibody labeling kit (Thermo, A20186) and diluted to 25 nM in Rexxip F buffer (Gyros Protein Technologies, P0004825). A standard curve for Aβ(1–40) or Aβ(1–42) was constructed using a 3-fold, 12-point dilution series starting at 50 μM in PBST (n = 2). EC50 values were calculated in GraphPad Prism using a nonlinear dose–response model with variable slope and four parameters. All samples, including capture and detection antibodies, Aβ standards, and diluted protease reactions, were loaded into 96-well plates (Thermo, AB0800) and run on a Gyrolab xPand system using a 1000 nl CD (Gyros Protein Technologies, P0004253) and according to the manufacturer’s standard protocol for 3-step ELISA with two wash buffers: PBST and pH 11 wash buffer (Gyros Protein Technologies, P0020096). The Gyros software was used to measure Aβ concentration by fitting to the standard curve.
Aβ surface plasmon resonance
Solution affinity constants for anti-Aβ antibodies were assessed on a Biacore T200. Anti-Aβ fusions were diluted to 1 μg/ml in HBS-P+ (Cytiva, BR100671) and captured using a Series S protein A chip (Cytiva, 29,127,555). A 3-fold, 8-point dilution series of Aβ(1–28) (New England Peptide, 22,360) was constructed in HBS-P+ and injected for 5 min, followed by 5 min of dissociation. Affinity constants were obtained through kinetic fitting using the Biacore Evaluation Software (GE).
RF61 affinity maturation
Affinity maturation of RF61 was performed by mutating each residue within the HC and LC CDRs to the other 18 possible residues (excluding cysteine) with a 2-step PCR protocol using PrimeSTAR Max DNA polymerase (Takara, R045B) according to standard protocols, generating 18 single point mutants per CDR residue. For rounds 1 and 2, the HC template DNA contained only the VH and CH1 domains to produce recombinant Fab proteins. Fab protein variants were expressed via cotransfection of HC and LC DNAs at 1 ml scale in HEK293 cells and purified with CaptureSelect CH1-XL resin for affinity screening. For round 3, RF61 HC and LC variable domains were fused to mouse IgG2a constant domains to create chimeric full-length antibodies that do not bind to their own Fc domains, as RF61 does not bind to mouse IgG2a. Protein expression was performed as described above followed by purification using MabSelect SuRe resin.
RF61 surface plasmon resonance
RF61 affinity for human IgG was assessed with a Biacore 8K+ or T200. For rounds 1 and 2 of affinity maturation, the Fc domain of human IgG1 was captured on a Series S Protein A chip according to the manufacturer’s protocols. Serial dilutions of the RF61 Fab variants were prepared in HBS-P+ buffer. The dilutions were passed over the chip for 4 min, followed by a 5 min dissociation step. Variants were assessed using the response units at the point of late analyte binding normalized to Fc capture level. For round 3 of affinity maturation, RF61 chimeric antibody variants were captured on a Series S CM5 chip (Cytiva, 29104988) containing immobilized anti-mouse antibodies from a mouse antibody capture kit (Cytiva, 29215281). Serial dilutions of human IgG1 Fc in HBS-P+ were passed over the chip for 10 min, followed by a 6 min dissociation step. Affinity constants were obtained through kinetic fitting using the Biacore Evaluation Software (GE). To evaluate the Fc mutations for RF61 binding ablation, a selection of eight RF61 chimeric antibody variants spanning a range of binding strengths from round three of affinity maturation (R50D LC; R50N LC, L95N D99H T100aA HC; R50N LC, S62P L95N D99H T100aA D100cE M100eF HC; R50N Y34F LC, S62P D99H D100cE M100eF HC; R50D LC, S62P L95N T100aA D100cE M100eF HC; R50D Y34F LC, D100cE HC; R50D Y34F LC, S62P D99H M100eF HC; R50D Y34F LC, S62P L95N D100cE M100eF HC) were captured as described above. A single 1000 nM concentration of each binding ablation variant was passed over the chip for 10 min, followed by a 6 min dissociation step. The late analyte binding signal (RU) was normalized to antibody capture level to quantify binding, and the signal from the eight RF61 variants is presented in Figure 6C. To characterize affinity of the antibody-MMP3 fusion constructs, the IgG1 subtype of anti-HER2 antibody, 4D5, was captured on a Series S Protein L chip (Cytiva, 29205138) according to the manufacturer’s protocols. Serial dilutions of the RF61-MMP3 fusion constructs were prepared in HBS-P+ buffer. The dilutions were flowed over the chip for 3 min, followed by 8 min of dissociation. RF61 contains a lambda LC, so it does not bind to the Protein L chip. Affinity constants were determined as described above.
MMP3 activity assay
All MMP3-D4K-4 and MMP3-D4K-4 fusion protein samples were exchanged into cleavage buffer (10 mM Hepes, 150 mM NaCl, and 10 mM CaCl2 at pH 7.5). MMP3-D4K-4 was activated with 16 units of EK (NEB, P8070L) for every 25 μg protein through incubation at room temperature for 16 h. To inactivate the EK, 0.1 mg/ml soybean trypsin inhibitor (Sigma, 17075029) was added to the protein solution. Fifty microliter of 2.5 μM fluorogenic MMP3 peptide substrate (R&D Systems, ES002) or 50 μg/ml DQ-collagen-IV (Invitrogen, D12052) in cleavage buffer was combined with the desired concentration of activated MMP3-D4K-4 sample within a 96-well black flat-bottom plate (Corning, CLS3925), and the fluorescence signal was measured on a Molecular Devices SpectraMax M2 microplate reader (Molecular Devices) with 320/405 nm and 485/535 nm excitation/emission for the peptide substrate and DQ-collagen-IV, respectively. Concentration-dependent assays were performed in duplicate with 2.5-fold dilutions from 400 nM. Curve fits were generated in GraphPad Prism using a nonlinear dose–response model with variable slope and four parameters.
IgG cleavage assay
MMP3-D4K-4 fusion proteins were activated as described above with EK. Seven 2.5-fold dilutions of the activated fusion proteins were prepared in cleavage buffer starting at 800 nM (n = 3). Four microliter of each dilution was mixed with 4 μl of the antibody substrate (one-arm anti-gD IgG1 antibody at 800 nM). The cleavage reaction was incubated at 37 °C for 24 h. The extent of antibody cleavage was assessed via ELISA as follows. One hundred microliter of Affinipure goat anti-human Fc antibody (Jackson ImmunoResearch, 109–005–098) at 1.2 μg/ml was added to each well of a Maxisorp 96 well plate (Thermo, 44–2404–21). The plate was incubated at room temperature for 1 h and then washed 3 times with PBST. The wells were blocked with SuperBlock buffer (Thermo, 37515) for 1 h at room temperature followed by three washes with PBST. Each cleavage reaction was diluted to 100 ng/ml of the one-arm anti-gD substrate in PBST (400-fold), and then 100 μl of the dilutions were added to the blocked wells. The plate was incubated at room temperature for 1 h and then washed 5 times with PBST. One hundred microliter of a 1:40,000-fold dilution of a goat anti-human Fab HRP-conjugated antibody (Sigma, A0293) was added to each well and incubated for 1 h at room temperature. The wells were washed 5 times with PBST. One hundred microliter of TMB substrate (Thermo, N301) was added to each well, and the reaction proceeded for 15 min at room temperature before quenching with the stop solution (Thermo, N600). Absorbance was measured at 405 nm. Curve fits were generated in GraphPad Prism using a nonlinear dose–response model with variable slope and four parameters.
IgG and fluorogenic peptide substrate cleavage assays in human serum
MMP3-D4K-4 Fab fusion proteins were activated as described above with EK. Soybean trypsin inhibitor was not added to the fusion proteins in order to observe any effects from endogenous protease inhibitors in the serum. Eight 2-fold dilutions of the fusion proteins starting at 600 nM were prepared in the cleavage buffer described above (n = 3). The serum was prepared from a blood sample of a single human donor through centrifugation and collection of the supernatant. Before use, the serum was diluted 1:1 in cleavage buffer. 25 μl of serum was combined with 25 μl of the antibody dilution, resulting in a top fusion protein concentration of 300 nM. The cleavage reaction was incubated at 37 °C for 24 h. IgG1 cleavage was detected with the ELISA assay described above with two important modifications. First, the antibody used to coat the Maxisorp 96 well plate was a mouse anti-human IgG1 antibody at 1.0 μg/ml in PBS (Thermo, MH1015). This modification ensured that the ELISA would detect cleavage of only IgG1 rather than all subtypes of IgG in the serum. Second, the cleavage reactions were diluted 100-fold in PBST before addition to the Maxisorp plate. The optimal dilution factor was determined by performing the ELISA assay with a dilution series of the serum. The rest of the assay was performed as described above. For the fluorogenic peptide substrate cleavage assay, 25 μl of the cleavage reaction in serum was combined with 25 μl of the fluorogenic MMP3 peptide substrate (R&D Systems, ES002) at 5 μM in cleavage buffer, and the sample was added to wells of a 384-well black flat, clear bottom plate (Thermo, 242,764). The final top protease fusion concentration for the peptide cleavage assay was 150 nM with seven 2-fold dilutions. The plate was sealed and incubated at room temperature for 1 h before reading the fluorescence signal with a PerkinElmer Envision plate reader.
Data availability
All data are included in the manuscript and/or supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
All authors are current or former employees of Genentech, a member of the Roche Group, and are shareholders in Roche.
Acknowledgments
We thank Reina Bassil and Ben Chih for help with the Aβ cleavage assay, Kathryn Bewely and Farzam Farahi for help with protein purification, and Nick Agard for helpful discussions.
Author contributions
M. G. R., B. L., and G. A. L. conceptualization; G. R., B. L., and G. A. L. methodology; M. G. R., B. L., I. K., H. S. K., G. A. L. investigation; M. G. R., B. L., G. A. L. visualization; G. A. L. supervision; M. G. R., B. L., and G. A. L. writing-original draft.
Funding and additional information
This study was supported by internal Genentech funds, and the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
Supporting information
References
- 1.Mullard A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 2021;20:491–495. doi: 10.1038/d41573-021-00079-7. [DOI] [PubMed] [Google Scholar]
- 2.Carter P.J., Lazar G.A. Next generation antibody drugs: pursuit of the “high-hanging fruit. Nat. Rev. Drug Discov. 2018;17:197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- 3.Ricklin D., Lambris J.D. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J. Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thurman J.M., Yapa R. Complement therapeutics in autoimmune disease. Front. Immunol. 2019;10:672. doi: 10.3389/fimmu.2019.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkon K., Casali P. Nature and functions of autoantibodies. Nat. Clin. Pract. Rheum. 2008;4:491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Delva L., Weinreb P.H., Pepinsky R.B., Graham D., Veizaj E., et al. Monoclonal antibody exposure in rat and cynomolgus monkey cerebrospinal fluid following systemic administration. Fluids Barriers CNS. 2018;15:10. doi: 10.1186/s12987-018-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemere C.A. Immunotherapy for Alzheimer’s disease: hoops and hurdles. Mol. Neurodegener. 2013;8:36. doi: 10.1186/1750-1326-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y.J., Zhang Y., Kenrick M., Hoyte K., Luk W., Lu Y., et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- 9.Kwan A.T.H., Arfaie S., Therriault J., Rosa-Neto P., Gauthier S. Lessons learnt from the second generation of anti-amyloid monoclonal antibodies clinical trials. Dement Geriatr. Cogn. 2021;49:334–348. doi: 10.1159/000511506. [DOI] [PubMed] [Google Scholar]
- 10.Dyck C. H. van. Anti-amyloid-β monoclonal antibodies for Alzheimer’s disease: pitfalls and promise. Biol. Psychiat. 2018;83:311–319. doi: 10.1016/j.biopsych.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cioni P., Gabellieri E., Campanini B., Bettati S., Raboni S. Use of exogenous enzymes in human therapy: approved drugs and potential applications. Curr. Med. Chem. 2021;29:411–452. doi: 10.2174/0929867328666210713094722. [DOI] [PubMed] [Google Scholar]
- 12.Tandon S., Sharma A., Singh S., Sharma S., Sarma S.J. Therapeutic enzymes: discoveries, production and applications. J. Drug Deliv. Sci. Tec. 2021;63 [Google Scholar]
- 13.Fuente M. de la, Lombardero L., Gómez-González A., Solari C., Angulo-Barturen I., Acera A., et al. Enzyme therapy: current challenges and future perspectives. Int. J. Mol. Sci. 2021;22:9181. doi: 10.3390/ijms22179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Yang B., Ke J., Li W., Suen W.-C. Antibody-based drugs and approaches against amyloid-β species for Alzheimer’s disease immunotherapy. Drug Aging. 2016;33:685–697. doi: 10.1007/s40266-016-0406-x. [DOI] [PubMed] [Google Scholar]
- 15.Saido T., Leissring M.A. Proteolytic degradation of amyloid β-protein. Cold Spring Harb. Perspect. Med. 2012;2:a006379. doi: 10.1101/cshperspect.a006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mzhavia N., Pan H., Che F.-Y., Fricker L.D., Devi L.A. Characterization of Endothelin-converting Enzyme-2. Implication for a role in the nonclassical processing of regulatory peptides. J. Biol. Chem. 2003;278:14704–14711. doi: 10.1074/jbc.M211242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adolfsson O., Pihlgren M., Toni N., Varisco Y., Buccarello A.L., Antoniello K., et al. An effector-reduced anti-β-amyloid (Aβ) antibody with unique Aβ binding properties promotes neuroprotection and glial engulfment of Aβ. J. Neurosci. 2012;32:9677–9689. doi: 10.1523/JNEUROSCI.4742-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ultsch M., Li B., Maurer T., Mathieu M., Adolfsson O., Muhs A., et al. Structure of crenezumab complex with aβ shows loss of β-hairpin. Sci. Rep. 2016;6 doi: 10.1038/srep39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crespi G.A.N., Hermans S.J., Parker M.W., Miles L.A. Molecular basis for mid-region amyloid-β capture by leading Alzheimer’s disease immunotherapies. Sci. Rep. 2015;5:9649. doi: 10.1038/srep09649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J., Nussinov R., Ma B. Mechanisms of recognition of amyloid-β (Aβ) monomer, oligomer, and fibril by homologous antibodies. J. Biol. Chem. 2017;292:18325–18343. doi: 10.1074/jbc.M117.801514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard J.F., Bril V., Vu T., Karam C., Peric S., Margania T., et al. Safety, efficacy, and tolerability of efgartigimod in patients with generalised myasthenia gravis (ADAPT): a multicentre, randomised, placebo-controlled, phase 3 trial. Lancet Neurol. 2021;20:526–536. doi: 10.1016/S1474-4422(21)00159-9. [DOI] [PubMed] [Google Scholar]
- 22.Bril V., Benatar M., Andersen H., Vissing J., Brock M., Greve B., et al. Efficacy and safety of rozanolixizumab in moderate to severe generalized myasthenia gravis. Neurology. 2021;96:e853–e865. doi: 10.1212/WNL.0000000000011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling L.E., Hillson J.L., Tiessen R.G., Bosje T., Iersel M. P. van, Nix D.J., et al. M281, an anti-FcRn antibody: pharmacodynamics, pharmacokinetics, and safety across the full range of IgG reduction in a first-in-human study. Clin. Pharmacol. Ther. 2019;105:1031–1039. doi: 10.1002/cpt.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwab I., Nimmerjahn F. Intravenous immunoglobulin therapy: how does IgG modulate the immune system? Nat. Rev. Immunol. 2013;13:176–189. doi: 10.1038/nri3401. [DOI] [PubMed] [Google Scholar]
- 25.Perez E.E., Orange J.S., Bonilla F., Chinen J., Chinn I.K., Dorsey M., et al. Update on the use of immunoglobulin in human disease: a review of evidence. J. Allergy Clin. Immun. 2017;139:S1–S46. doi: 10.1016/j.jaci.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe G.I., Ward E.S., Haard H. de, Ulrichts P., Mozaffar T., Pasnoor M., et al. IgG regulation through FcRn blocking: a novel mechanism for the treatment of myasthenia gravis. J. Neurol. Sci. 2021;430 doi: 10.1016/j.jns.2021.118074. [DOI] [PubMed] [Google Scholar]
- 27.Robak T., Kaźmierczak M., Jarque I., Musteata V., Treliński J., Cooper N., et al. Phase 2 multiple-dose study of an FcRn inhibitor, rozanolixizumab, in patients with primary immune thrombocytopenia. Blood Adv. 2020;4:4136–4146. doi: 10.1182/bloodadvances.2020002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjellman C., Maldonado A.Q., Sjöholm K., Lonze B.E., Montgomery R.A., Runström A., et al. Outcomes at 3 years posttransplant in imlifidase-desensitized kidney transplant patients. Am. J. Transpl. 2021;21:3907–3918. doi: 10.1111/ajt.16754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorant T., Bengtsson M., Eich T., Eriksson B., Winstedt L., Järnum S., et al. Safety, immunogenicity, pharmacokinetics, and efficacy of degradation of anti-HLA antibodies by IdeS (imlifidase) in chronic kidney disease patients. Am. J. Transpl. 2018;18:2752–2762. doi: 10.1111/ajt.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonze B.E., Tatapudi V.S., Weldon E.P., Min E.S., Ali N.M., Deterville C.L., et al. IdeS (imlifidase): a novel agent that cleaves human IgG and permits successful kidney transplantation across high-strength donor-specific antibody. Ann. Surg. 2018;268:488–496. doi: 10.1097/SLA.0000000000002924. [DOI] [PubMed] [Google Scholar]
- 31.Jordan S.C., Legendre C., Desai N.M., Lorant T., Bengtsson M., Lonze B.E., et al. Imlifidase desensitization in crossmatch-positive, highly sensitized kidney transplant recipients: results of an international phase 2 trial (Highdes) Transplantation. 2021;105:1808–1817. doi: 10.1097/TP.0000000000003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan M.H., Petrone D., Nemeth J.F., Barnathan E., Björck L., Jordan R.E. Proteolysis of purified IgGs by human and bacterial enzymes in vitro and the detection of specific proteolytic fragments of endogenous IgG in rheumatoid synovial fluid. Mol. Immunol. 2008;45:1837–1846. doi: 10.1016/j.molimm.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 33.Sbardella D., Fasciglione G.F., Gioia M., Ciaccio C., Tundo G.R., Marini S., et al. Human matrix metalloproteinases: an ubiquitarian class of enzymes involved in several pathological processes. Mol. Aspects Med. 2012;33:119–208. doi: 10.1016/j.mam.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Harindranath N., Goldfarb I.S., Ikematsu H., Burastero S.E., Wilder R.L., Notkins A.L., et al. Complete sequence of the genes encoding the VH and VL regions of low- and high-affinity monoclonal lgM and lgA1 rheumatoid factors produced by CD5+ B cells from a rheumatoid arthritis patient. Int. Immunol. 1991;3:865–875. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duquerroy S., Stura E.A., Bressanelli S., Fabiane S.M., Vaney M.C., Beale D., et al. Crystal structure of a human autoimmune complex between IgM Rheumatoid factor RF61 and IgG1 Fc reveals a novel epitope and evidence for affinity maturation. J. Mol. Biol. 2007;368:1321–1331. doi: 10.1016/j.jmb.2007.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falkenburg W.J.J., Kempers A.C., Dekkers G., Heer P.O., Bentlage A.E.H., Vidarsson G., et al. Rheumatoid factors do not preferentially bind to ACPA-IgG or IgG with altered galactosylation. Rheumatology. 2017;56:2025–2030. doi: 10.1093/rheumatology/kex284. [DOI] [PubMed] [Google Scholar]
- 37.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brezski R.J., Oberholtzer A., Strake B., Jordan R.E. The in vitro resistance of IgG2 to proteolytic attack concurs with a comparative paucity of autoantibodies against peptide analogs of the IgG2 hinge. MAbs. 2011;3:558–567. doi: 10.4161/mabs.3.6.18119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gearing A.J.H., Thorpe S.J., Miller K., Mangan M., Varley P.G., Dudgeon T., et al. Selective cleavage of human IgG by the matrix metalloproteinases, matrilysin and stromelysin. Immunol. Lett. 2002;81:41–48. doi: 10.1016/s0165-2478(01)00333-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z., Austin G.L., Shaffer R., Armstrong D.D., Gentry M.S. Antibody-mediated enzyme therapeutics and applications in glycogen storage diseases. Trends Mol. Med. 2019;25:1094–1109. doi: 10.1016/j.molmed.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver A.B., Leonard E.K., Gould J.R., Spangler J.B. Engineered antibody fusion proteins for targeted disease therapy. Trends Pharmacol. Sci. 2021;42:1064–1081. doi: 10.1016/j.tips.2021.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi H., Sun T., Armstrong D., Borneman S., Yang C., Austin S., et al. Antibody-mediated enzyme replacement therapy targeting both lysosomal and cytoplasmic glycogen in Pompe disease. J. Mol. Med. 2017;95:513–521. doi: 10.1007/s00109-017-1505-9. [DOI] [PubMed] [Google Scholar]
- 43.Giugliani R., Giugliani L., Poswar F. de O., Donis K.C., Corte A.D., Schmidt M., et al. Neurocognitive and somatic stabilization in pediatric patients with severe Mucopolysaccharidosis type I after 52 weeks of intravenous brain-penetrating insulin receptor antibody-iduronidase fusion protein (valanafusp alpha): an open label phase 1-2 trial. Orphanet J. Rare Dis. 2018;13:110. doi: 10.1186/s13023-018-0849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S.K., Bagshawe K.D. Antibody directed enzyme prodrug therapy (ADEPT): trials and tribulations. Adv. Drug Deliver Rev. 2017;118:2–7. doi: 10.1016/j.addr.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Mayer A., Francis R.J., Sharma S.K., Tolner B., Springer C.J., Martin J., et al. A Phase I study of single administration of antibody-directed enzyme prodrug therapy with the recombinant anti–carcinoembryonic antigen antibody-enzyme fusion protein MFECP1 and a bis-iodo phenol mustard prodrug. Clin. Cancer Res. 2006;12:6509–6516. doi: 10.1158/1078-0432.CCR-06-0769. [DOI] [PubMed] [Google Scholar]
- 46.Boado R.J., Hui E.K.-W., Lu J.Z., Sumbria R.K., Pardridge W.M. Blood-brain barrier molecular trojan horse enables imaging of brain uptake of radioiodinated recombinant protein in the rhesus monkey. Bioconjug. Chem. 2013;24:1741–1749. doi: 10.1021/bc400319d. [DOI] [PubMed] [Google Scholar]
- 47.D’Avino C., Paciello R., Riccio G., Coppola C., Coppola M., Laccetti P., et al. Effects of a second-generation human anti-ErbB2 ImmunoRNase on trastuzumab-resistant tumors and cardiac cells. Protein Eng. Des. Sel. 2014;27:83–88. doi: 10.1093/protein/gzt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y.-M., Wang L.-F., Jia L.-T., Qiu X.-C., Zhao J., Yu C.-J., et al. A caspase-6 and anti-human epidermal growth factor receptor-2 (HER2) antibody chimeric molecule suppresses the growth of HER2-overexpressing tumors. J. Immunol. 2004;173:61–67. doi: 10.4049/jimmunol.173.1.61. [DOI] [PubMed] [Google Scholar]
- 49.Andrady C., Sharma S.K., Chester K.A. Antibody-enzyme fusion proteins for cancer therapy. Immunotherapy. 2011;3:193–211. doi: 10.2217/imt.10.90. [DOI] [PubMed] [Google Scholar]
- 50.Weisbart R.H., Baldwin R., Huh B., Zack D.J., Nishimura R. Novel protein transfection of primary rat cortical neurons using an antibody that penetrates living cells. J. Immunol. 2000;164:6020–6026. doi: 10.4049/jimmunol.164.11.6020. [DOI] [PubMed] [Google Scholar]
- 51.Zack D.J., Stempniak M., Wong A.L., Taylor C., Weisbart R.H. Mechanisms of cellular penetration and nuclear localization of an anti-double strand DNA autoantibody. J. Immunol. 1996;157:2082–2088. [PubMed] [Google Scholar]
- 52.Mintun M.A., Lo A.C., Evans C.D., Wessels A.M., Ardayfio P.A., Andersen S.W., et al. Donanemab in early Alzheimer’s disease. New Engl. J. Med. 2021;384:1691–1704. doi: 10.1056/NEJMoa2100708. [DOI] [PubMed] [Google Scholar]
- 53.Swanson C.J., Zhang Y., Dhadda S., Wang J., Kaplow J., Lai R.Y.K., et al. A randomized, double-blind, phase 2b proof-of-concept clinical trial in early Alzheimer’s disease with lecanemab, an anti-Aβ protofibril antibody. Alzheimer’s Res. Ther. 2021;13:80. doi: 10.1186/s13195-021-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein G., Delmar P., Voyle N., Rehal S., Hofmann C., Abi-Saab D., et al. Gantenerumab reduces amyloid-β plaques in patients with prodromal to moderate Alzheimer’s disease: a PET substudy interim analysis. Alzheimer’s Res. Ther. 2019;11:101. doi: 10.1186/s13195-019-0559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 56.Pulgar V.M. Transcytosis to cross the blood brain barrier, new advancements and challenges. Front. Neurosci. 2019;12:1019. doi: 10.3389/fnins.2018.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alajangi H.K., Kaur M., Sharma A., Rana S., Thakur S., Chatterjee M., et al. Blood–brain barrier: emerging trends on transport models and new-age strategies for therapeutics intervention against neurological disorders. Mol. Brain. 2022;15:49. doi: 10.1186/s13041-022-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denard C.A., Paresi C., Yaghi R., McGinnis N., Bennett Z., Yi L., et al. YESS 2.0, a tunable platform for enzyme evolution, yields highly active TEV protease variants. ACS Synth. Biol. 2021;10:63–71. doi: 10.1021/acssynbio.0c00452. [DOI] [PubMed] [Google Scholar]
- 59.Packer M.S., Rees H.A., Liu D.R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 2017;8:956. doi: 10.1038/s41467-017-01055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez M.I., Ting A.Y. Directed evolution improves the catalytic efficiency of TEV protease. Nat. Met. 2020;17:167–174. doi: 10.1038/s41592-019-0665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holstein J.M., Gylstorff C., Hollfelder F. Cell-free directed evolution of a protease in microdroplets at ultrahigh throughput. ACS Synth. Biol. 2021;10:252–257. doi: 10.1021/acssynbio.0c00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merchant A.M., Zhu Z., Yuan J.Q., Goddard A., Adams C.W., Presta L.G., et al. An efficient route to human bispecific IgG. Nat. Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 63.Bassil R., Shields K., Granger K., Zein I., Ng S., Chih B. Improved modeling of human AD with an automated culturing platform for iPSC neurons, astrocytes and microglia. Nat. Commun. 2021;12:5220. doi: 10.1038/s41467-021-25344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Becker J.W., Marcy A.I., Rokosz L.L., Axel M.G., Burbaum J.J., Fitzgerald P.M.D., et al. Stromelysin-1: three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci. 1995;4:1966–1976. doi: 10.1002/pro.5560041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or supporting information.