Abstract
Within the 5.9-kb DNA region between the tfdR and tfdK genes on the 2,4-dichlorophenoxyacetic acid (2,4-D) catabolic plasmid pJP4 from Ralstonia eutropha JMP134, we identified five open reading frames (ORFs) with significant homology to the genes for chlorocatechol and chlorophenol metabolism (tfdCDEF and tfdB) already present elsewhere on pJP4. The five ORFs were organized and assigned as follows: tfdDIICIIEIIFII and tfdBII (in short, the tfdII cluster), by analogy to tfdCDEF and tfdB (the tfdI cluster). Primer extension analysis of mRNA isolated from 2,4-D-grown R. eutropha JMP134 identified a single transcription start site in front of the first gene of the cluster, tfdDII, suggesting an operon-like organization for the tfdII genes. By expressing each ORF in Escherichia coli, we confirmed that tfdDII coded for a chloromuconate cycloisomerase, tfdCII coded for a chlorocatechol 1,2-dioxygenase, tfdEII coded for a dienelactone hydrolase, tfdFII coded for a maleylacetate reductase, and tfdBII coded for a chlorophenol hydroxylase. Dot blot hybridizations of mRNA isolated from R. eutropha JMP134 showed that both tfdI and tfdII genes are transcribed upon induction with 2,4-D. Thus, the functions encoded by the tfdII genes seem to be redundant with respect to those of the tfdI cluster. One reason why the tfdII genes do not disappear from plasmid pJP4 might be the necessity for keeping the regulatory genes for the 2,4-D pathway expression tfdR and tfdS.
Ralstonia eutropha JMP134(pJP4) was originally isolated in Australia from an unspecified soil sample by selection for its ability to use 2,4-dichlorophenoxyacetic acid (2,4-D) as sole carbon and energy source (6). The genes necessary for the metabolism of 2,4-D are located on a 22-kb DNA fragment of plasmid pJP4 (6) (Fig. 1). Among these, tfdA (33), tfdB, and tfdCDEF (8, 29) were the first genes to be identified. TfdA catalyzes the conversion of 2,4-D to 2,4-dichlorophenol (2,4-DCP) (13, 14), and TfdB catalyzes the conversion of 2,4-DCP to 3,5-dichlorocatechol (3,5-DCC) (12). The TfdCDEF enzymes catalyze the transformation of 3,5-DCC via 2,4-dichloromuconate (2,4-DCM) to 3-oxoadipate (12). Expression of the tfd pathway genes is regulated by the two identical LysR-type regulatory proteins, TfdR and TfdS (18, 19, 24, 27, 37). TfdT was long suspected to be a regulatory protein of the pathway as well, but it is actually a nonfunctional regulatory due to a C-terminal deletion caused by insertion of the insertion sequence (IS) element ISJP4 (24).
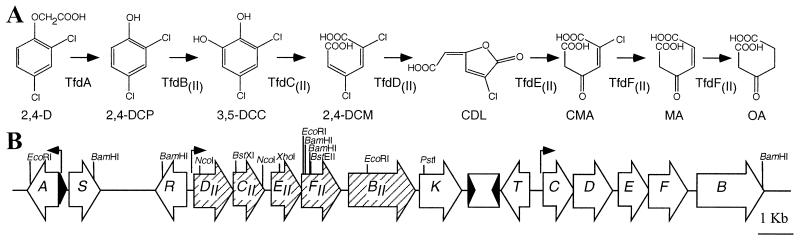
FIG. 1.
(A) Overview of the steps in 2,4-D degradation. Enzymes catalyzing the different conversion steps: TfdA, 2,4-D α-ketoglutarate dioxygenase; TfdB(II), chlorophenol hydroxylase; TfdC(II), chlorocatechol 1,2-dioxygenase; TfdD(II), chloromuconate cycloisomerase; TfdE(II), dienelactone hydrolase; TfdF(II), (chloro)maleylacetate reductase. (B) Organization of the tfd genes on plasmid pJP4. Arrows indicate the sizes and orientations of all tfd genes currently known. The solid line represents noncoding DNA regions of pJP4. The rectangle between tfdK and tfdT represents the IS element ISJP4; black triangles depict the inverted repeats (not to scale). Positions of promoters regulated by TfdR(S) are indicated above the gene structure. All sites of the restriction enzymes BamHI and EcoRI are indicated. Not all positions of the other depicted restriction enzymes are given. Abbreviations not given in the text: CDL, cis-DL; CMA, chloromaleylacetate; MA, maleylacetate; OA, 3-oxoadipate.
Plasmids for 2,4-D degradation such as pJP4 form a paradigm for the evolution of new catabolic pathways. The current hypothesis is that existing sets of genes from different organisms can be assembled into new structures, processes often catalyzed by mobile DNA elements (15). Indeed, mobile DNA elements associated with 2,4-D degradative genes are found in a number of plasmids such as pIJB1 and pJP4 (6, 36). In the case of plasmid pJP4, the IS element ISJP4 is thought to have been responsible for inserting a larger gene cassette containing (among others) the genes tfdR and tfdS (Fig. 1). Preliminary evidence had been obtained that another set of genes for chlorocatechol degradation might be present within this transposable element (17, 27), which was just recently confirmed (28), although this second cluster had not been previously detected by transposon mutagenesis studies (8). In addition, one novel function (tfdK) was discovered recently within this region (25). Located adjacent to ISPJ4, tfdK codes for an active transporter of 2,4-D at low-millimolar concentrations. This demonstrated that the 2,4-D pathway of R. eutropha JMP134 was even more complex than expected until then.
Here, we report the identification of a set of five genes in the 5.9-kb tfdR-tfdK intergenic region on plasmid pJP4. We provide evidence that this gene cluster, tfdII, is actively transcribed in its host R. eutropha JMP134 and that it encodes the enzymes for the complete conversion of 2,4-DCP to β-ketoadipate. Although similar in structure and function, tfdII is not simply a duplication of the tfdCDEF-tfdB (in short, tfdI) gene cluster. Its role in 2,4-D degradation is subtle, redundant, and imperative as well, all of which seems to be the consequence of the past insertion of the ISJP4-flanked mobile element.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. eutropha JMP134(pJP4) is able to use 2,4-D and 3-chlorobenzoate as sole carbon and energy source (6, 7). Escherichia coli DH5α (30) was used for routine cloning purposes. E. coli BL21(DE3)(pLysS) (34), which carries the T7 RNA polymerase gene under control of the lacUV5 promoter, was used for the T7-directed expression of pRSET6a-derived plasmids (31). E. coli cultures were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml). R. eutropha cultures were grown in nutrient broth (Biolife, Milan, Italy) or in Pseudomonas mineral medium (16) supplemented with 10 mM fructose. Induction experiments were carried out with R. eutropha JMP134(pJP4) cultivated in a 1.5-liter chemostat on 10 mM fructose at a dilution rate of 0.05 h−1. Induction of the 2,4-D pathway was achieved by addition of 2,4-D to the chemostat to a final concentration of 0.1 mM (23).
DNA manipulations, PCR, and sequence analysis.
Plasmid DNA isolations, transformations, and other DNA manipulations were carried out according to established procedures (30). Restriction enzymes and other DNA-modifying enzymes were obtained from Amersham Pharmacia Life Science (Cleveland, Ohio) or GIBCO/BRL Life Technologies Inc. (Gaithersburg, Md.) and used as specified by the manufacturer. Oligonucleotides for the PCR were obtained from Microsynth GmbH (Balgach, Switzerland). PCR mixtures contained 200 pmol of each primer per ml, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.05% (vol/vol) W-1, 2 mM MgCl2, 0.25 mM each deoxynucleotide, and 30 U of Taq DNA polymerase (Life Technologies). DNA sequencing was performed on double-stranded DNA templates with a Thermo Sequenase cycle sequencing kit with 7-deaza-dGTP (Amersham). For sequencing of the tfdII gene cluster, suitable overlapping subclones were generated for use as templates in sequencing reactions. Primers for sequencing were labeled with fluorescent dye IRD-800 or IRD-700 at the 5′ end and were purchased from MWG Biotech (Ebersberg, Germany). Fragments were separated on an automated DNA sequencer (model 4200 IR2; LI-COR Inc., Lincoln, Neb.). Sequence assembly and computer analysis of the DNA sequences were done with the DNASTAR software (DNASTAR Inc., Madison, Wis.).
RNA isolation and primer extension analysis.
RNA was isolated from chemostat-grown cultures of R. eutropha JMP134(pJP4), either under uninduced conditions (10 mM fructose) or induced with 0.1 mM 2,4-D, as described previously (2). DNase I-treated RNA samples were spotted on Hybond N+ membranes (Amersham) and hybridized with biotin-labeled antisense RNAs for each of the tfdII open reading frames (ORFs) as described elsewhere (23). Primer extension reactions were carried out as follows. One microgram of total RNA from induced R. eutropha was annealed with 1 pmol of IRD800-labeled primer (for tfdDII, 5′ CACGCTGCTCTGATGCTTGG 3′) in annealing buffer (Amersham) in a total volume of 5 μl. This amount was covered with one drop of mineral oil (Sigma), heated for 5 min at 68°C, and then cooled to 42°C. The reverse transcription reaction was started by addition of 3 μl of a mix containing avian myeloblastosis virus reaction buffer (Amersham), 6 U of avian myeloblastosis virus reverse transcriptase (Amersham), and 1.6 mM each deoxynucleotide. Reverse transcription reaction mixtures were incubated for 1 h at 42°C, then heated for 3 min at 95.5°C, and cooled on ice immediately. Samples of 1 μl from the reverse transcriptase reaction were mixed with 0.5 μl of formamide loading buffer and loaded onto a denaturing sequencing gel as described above. DNA sequencing reactions were prepared with the same IRD-labeled primer on double-stranded plasmid DNAs with the corresponding cloned pJP4 regions. Regions tested for cDNA synthesis on total RNA from induced R. eutropha cultures included tfdDII, tfdCII, tfdBII, tfdK, and tfdC.
Plasmids.
Plasmid pUC28 (3) was used as general cloning vector. pRSET6a (31) is a plasmid with a pBluescript (Stratagene, La Jolla, Calif.) backbone containing the specific expression elements of the pET3 vectors (34) and a newly designed multiple cloning site to facilitate cloning.
Translational fusions of each of the genes of the tfdII cluster individually were constructed by fusing the ATG triplet of the NdeI site located in the multiple cloning site downstream of the T7 promoter and ribosome binding site on pRSET6a to the start codon of the respective tfd gene. DNA fragments containing either the tfdCII, tfdDII, or tfdEII ORF were custom-amplified by PCR, thereby introducing an NdeI site at the start codon and a BamHI site downstream of the stop codon of each gene. The obtained PCR fragments were first cloned into the NdeI/BamHI site of pUC28 and sequenced to confirm their identity with the original nucleotide sequence. The fragments were then reisolated and cloned into pRSET6a cut with NdeI and BamHI. This resulted in plasmids pCBA199 (tfdCII), pCBA165 (tfdDII), pCBA202 (tfdEII). Note that the tfaDII ORF starts at the ATG codon at position 1610 (numbering according to U16782).
For cloning the tfdFII gene, first a 240-bp fragment was amplified by PCR from plasmid pCBA83IV. The PCR-derived fragment was digested with NdeI and BamHI and directly cloned to pRSET6a. A plasmid with the proper insert determined by sequencing was named pCBA179. To complete the ORF of tfdFII, a 1.3-kb EcoRI fragment of pCBA88 was cloned into the EcoRI site of pCBA179 to give rise to plasmid pCBA184. The tfdBII gene was cloned as follows. First, a 620-bp fragment was amplified from plasmid pCBA84IV, using primers 981203 (5′ CGA TAA GGA GAC CAT ATG AACG 3′; tfdBII sequence) and 931011 (5′ TGA GCG GAT AAC AAT TT 3′; pUC18 sequence), digested with NdeI-EcoRI, and ligated into pUC28 cut with the same enzymes. After transformation, this resulted in plasmid pCBA174. The insert of pCBA174 was sequenced to confirm its identity. In a three-point ligation, the NdeI-EcoRI fragment of pCBA174 and the EcoRI-PstI fragment of pCBA90 were ligated to pRSET6atfdD (U. Schell, Department of Microbiology, University of Stuttgart) cut with NdeI and PstI to give pCBA180 (Table 1).
TABLE 1.
Plasmids constructed in this work
| Plasmid | Description |
|---|---|
| pCBA83IV | pGEM-5Zf carrying the 870-bp NcoI-BamHI fragment of pJP4 containing tfdEII; Apr |
| pCBA84IV | pUC19 carrying the 1.3-kb BamHI-EcoRI fragment of pJP4 containing part of tfdFII and tfdBII; Apr |
| pCBA89 | 6.9-kb SacI fragment of pJP4 in pUC |
| pCBA90 | pUC19 carrying the 1.4-kb EcoRI-PstI fragment of pJP4 containing part of tfdBII and tfdK; Apr |
| pCBA122 | 1-kb SacII fragment of pCBA89; contains tfdCII |
| pCBA174 | pUC28 carrying a 550-bp NdeI-EcoRI fragment amplified by PCR from pCBA84IV; contains part of tfdBII, Apr |
| pCBA179 | pRSET6a carrying a 100-bp NdeI-BamHI fragment amplified by PCR from pCBA83IV; contains part of tfdFII; Apr |
| pCBA165 | pRSET6a carrying the 1.2-kb NdeI-BamHI fragment amplified from pCBA89; contains tfdDII; Apr |
| pCBA180 | pRSET6a carrying fragment NdeI-EcoRI from pCBA174 and fragment EcoRI-PstI from pCBA90; contains complete tfdBII; Apr |
| pCBA184 | pCBA179 with the 1.3-kb EcoRI fragment from pCBA88; contains tfdFII; Apr |
| pCBA192 | pUC28 carrying a 800-bp NdeI-BamHI fragment amplified from pCBA122; contains tfdCII; Apr |
| pCBA196 | pCBA165 with deletion of the NcoI site; contains frameshift mutation in tfdDII; Apr |
| pCBA197 | pCBA184 with deletion of the BstEII site; contains frameshift mutation in tfdFII; Apr |
| pCBA198 | pCBA180 with deletion of the EcoRI site; contains frameshift mutation in tfdBII; Apr |
| pCBA199 | pRSET6a carrying the 800 bp NdeI-BamHI fragment of pCBA192; contains tfdCII; Apr |
| pCBA200 | pCBA199 with deletion of the BstXI site; contains frameshift mutation in tfdCII; Apr |
| pCBA201 | pCBA202 with deletion of the XhoI site; contains frameshift mutation in tfdEII; Apr |
| pCBA202 | pRSET6a carrying a 700-bp NdeI-BamHI fragment amplified from pCBA89; contains tfdEII; Apr |
We then prepared frameshift mutations in each of the ORFs of the individually cloned tfdII genes. For this purpose, plasmids pCBA199 (tfdCII), pCBA165 (tfdDII), pCBA202 (tfdEII), pCBA184 (tfdFII), and pCBA180 (tfdBII) were each digested with a unique restriction site located in the respective tfdII gene, treated with Klenow DNA polymerase, and religated (Table 1). By analogy, the truncated genes were named tfdCIIΔ, tfdDIIΔ, tfdEIIΔ, tfdFIIΔ, and tfdBIIΔ, and the corresponding plasmids are pCBA200, pCBA196, pCBA201, pCBA197, and pCBA198 (Table 1).
Expression in E. coli.
E. coli BL21(DE3)(pLysS) strains harboring pRSET6a-derived plasmids were grown in 50 ml of LB at 37°C to an optical density at 546 nm of between 0.5 and 0.6. To achieve induction, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the medium at a final concentration of 0.4 mM, and strains were incubated at 30°C for an additional 2.5 h. Cells were then centrifuged for 15 min at 5,300 rpm (4°C), washed with 20 ml of washing buffer (containing 20 mM Tris-HCl [pH 7.5] supplemented with 1 mM MnSO4 in the case of E. coli expressing tfdDII), and resuspended in 1 ml of washing buffer. Samples of 50 μl were taken from these cell suspensions for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), which was performed by the method of Laemmli (22). Disruption of the remainder of the cell suspensions was performed by sonication (Branson Sonifier 450; SCAN AG, Basel, Switzerland). One-milliliter cell suspensions were sonicated (five pulses of 15 s each) on ice at an output of 30 to 40 W, with a 1-min pause between pulses. Subsequently, suspensions were centrifuged at 4°C for 30 min at 15,000 × g. The resulting supernatants, referred to as cell extracts, were used in enzyme assays. Protein concentrations in the cell extracts were determined as described by Bradford (5), using bovine serum albumin as a standard.
Enzyme assays.
All enzyme assays were performed by spectrophotometric methods in 0.5-ml quartz cuvettes at room temperature. Extinction coefficients were taken from Dorn and Knackmuss (9).
Chlorocatechol 1,2-dioxygenase activity was measured by determining product formation at 260 nm. Substrates tested included 3,5-DCC (ɛ2,4-DCM = 12,000 M−1 cm−1), 3-chlorocatechol (3-CC) (ɛ2-CM = 17,100 M−1 cm−1), or 4-CC (ɛ3-CM = 12,400 M−1 cm−1). Reaction mixtures contained 40 mM Tris-HCl (pH 7.5), 0.3 mM EDTA, and 0.1 mM substrate. The reaction was started by adding cell extract (0.01 to 0.2 mg of protein).
Chloromuconate cycloisomerase activity was measured by determining the disappearance rate of substrate at 260 nm. Substrates used were 2-chloromuconate (2-CM), 3-CM, or freshly made 2,4-DCM (see below). Reaction mixtures contained 30 mM Tris-HCl (pH 7.5), 1 mM MnSO4, and 0.1 mM substrate. Cycloisomerization of chloromuconates was assayed in the presence of an excess of dienelactone hydrolase to avoid accumulation of 4-carboxymethylene-but-2-en-4-olides. For the conversion of 2,4-DCM, an extinction coefficient of 5,800 M−1 cm−1 was used (21). The reaction was started by adding cell extract (0.01 to 0.2 mg of protein).
Dienelactone hydrolase activity was measured at 280 nm by determining the disappearance rate of substrate (cis-dienelactone [DL] [ɛDL = 17,000 M−1 cm−1] or trans-DL). Reaction mixtures contained 10 mM histidine-HCl (pH 6.5) and 0.1 mM substrate. The reaction was started by adding cell extract (0.01 to 0.2 mg of protein).
Maleylacetate reductase activity was measured by determining maleylacetate-dependent NADH oxidation at 340 nm. Reaction mixtures contained 50 mM Tris-HCl (pH 7), 0.4 mM NADH, and cell extract (0.01 to 0.2 mg of protein). After the unspecific oxidation rate of NADH (ɛNADH = 6,300 M−1 cm−1) was determined, the reaction was started by adding 0.4 mM freshly prepared maleylacetate (see below).
2,4-DCP dioxygenase was measured by determining 2,4-DCP-dependent NADPH oxidation at 340 nm. Reaction mixtures contained 60 mM phosphate buffer (pH 7.6), 0.03 mM flavin adenine dinucleotide, 0.3 mM NADPH, and cell extract (0.01 to 0.2 mg of protein). After the unspecific oxidation rate of NADPH (ɛNADPH = 6,300 M−1 cm−1) was determined, the reaction was started by adding 0.05 mM 2,4-DCP.
Chemicals.
3-CC was a kind gift of Barbara Jakobs, GFB, Braunschweig, Germany. 4-CC and 3,5-DCC were purchased from Promochem GmbH (Wesel, Germany). 3-CM, cis-DL, and trans-DL were a kind gift from Walter Reineke, Bergische Universität-Gesamthochschule Wuppertal, Wuppertal, Germany. 2,4-DCM was prepared by incubation of a solution of 1 mM 3,5-DCC in 30 mM Tris-HCl (pH 8) with cell extract of E. coli BL21(pCBA199) expressing tfdCII. The formation of 2,4-DCM was followed spectrophotometrically at 260 nm. After 20 min, the reaction mixture was centrifuged through a Centricon-10 (Amicon, Inc., Beverly, Mass.) filter at 5,000 × g. Maleylacetate was prepared by alkaline hydrolysis of cis-DL, as described elsewhere (11), by mixing 1 ml of 5 mM cis-DL with 7.5 μl of 2 N NaOH and incubating for 15 min at room temperature. 2,4-DCP was purchased from Fluka Chemie AG (Buchs, Switzerland).
Digital imaging.
Sequence images were exported as TIFF files. Autoradiographic films and protein gels were scanned on a laser densitometer (Molecular Dynamics, Sunnyvale, Calif.) and exported as TIFF files. All TIFF files were imported into Adobe Photoshop (version 4.0; Adobe Systems, Inc., Mountain View, Calif.), cropped to the appropriate size, enhanced whenever necessary for reproduction, saved as gray scale TIFF files, and placed into Adobe Illustrator (version 8.0) for text additions.
Nucleotide sequence accession number.
The sequence of the tfdII gene cluster was deposited in GenBank under accession number U16782.
RESULTS
Identification of a second gene cluster for chlorophenol and chlorocatechol metabolism on pJP4.
In the tfdR-tfdK intergenic region of plasmid pJP4, we located five ORFs with significant homology to genes for the metabolism of chlorinated phenols and catechols. The ORFs were arranged serially and in an orientation opposite that of the tfdR gene (Fig. 1). To signify their resemblance to genes from the tfdCDEFB cluster on pJP4, the ORFs were sequentially labeled tfdDII, tfdCII, tfdEII, tfdFII, and tfdBII. The percentages of amino acid identity between the predicted polypeptides from the tfdII genes and their counterparts from the tfdCDEFB genes varied substantially (Table 2). For example, TfdEII had only 15% predicted identical amino acids with TfdE, whereas TfdCII and TfdC shared 60% amino acid identity. The evolutionary relationships of the TfdII and TfdI gene products with other related proteins have been clearly pointed out elsewhere (10). These sequence comparisons indicated quite well that the tfdII genes were not simply a duplication of the tfdI cluster, or vice versa, but had a different evolutionary origin.
TABLE 2.
Features of the ORFs of the tfdI and tfdII gene clusters
| ORF | Ribosome binding site | bp between RBS and start codon | Start codon | Location on sequencea | Length of ORF (bp) | Predicted molecular mass (kDa) | Pairwise identity (%)b |
|---|---|---|---|---|---|---|---|
| tfdC | GGAGG | 4 | GTG | 337–1104 | 765 | 28.3 | |
| tfdCII | GAAGAG | 8 | ATG | 2856–3617 | 759 | 28.1 | 60 |
| tfdD | GGGGG | 7 | GTG | 1101–2213 | 1,110 | 39.7 | |
| tfdDIIc | −/GAAGCG | −/9 | ATG/GTG | 1610/1694–2659 | 1,047/963 | 36.9 | 35 |
| tfdE | GGAG | 7 | ATG | 2288–2992 | 702 | 25.4 | |
| tfdEII | GAAGG | 7 | ATG | 3639–4346 | 705 | 25.4 | 15 |
| tfdF | GAAGG | 7 | ATG | 2989–4053 | 1,062 | 37.9 | |
| tfdFII | GGGGG | 5 | GTG | 4348–5427 | 1,077 | 37.5 | 45 |
| tfdB | GGAGG | 5 | ATG | 4398–6194 | 1,794 | 65.4 | |
| tfdBII | GGAG | 5 | ATG | 5495–7255 | 1,758 | 64.4 | 62 |
This became evident also from a comparison of the gene organization of the two clusters. In the tfdII cluster, the tfdDII ORF preceded that of tfdCII, whereas the opposite was found in the tfdI cluster. Furthermore, no ORFs overlapped in the tfdI cluster, whereas two cases of translational coupling (i.e., tfdCD and tfdEF) occur in the tfdI cluster. In addition, the remainder of an ORF with unknown function exists between tfdD and tfdE which is similar to the tcbCDEF and clcABDE clusters but was absent in the tfdII cluster. The tfdII cluster showed highest percentages of identity to a set of tfd genes on plasmid pEST4011 of Pseudomonas putida and of Variovorax paradoxus (T. Valleys, L. Shengao, and K. Miyashita, unpublished data [DDBJ/EMBL/GenBank accession no. AB028643]). although smaller deletions or frameshift mutations must have occurred there. For example, the first 131 bp of the tfdDII ORF had 74% sequence identity to a region upstream of the tfdC gene of pEST4011, but no complete tfdD ORF exists. TfdCII had only 60% identical amino acids with TfdC, whereas it showed 83.5% identity with TfdC of pEST4011 (26). TfdEII carried 79.6% identical amino acid residues with the predicted polypeptide encoded by tfdE from V. paradoxus. Interestingly, the TfdE polypeptide predicted from tfdE on pEST4011 (26) had no significant similarity with TfdEII except for the first 22 amino acids, although the DNA sequence identity along the total 708-bp region in common was 77.4%. However, introducing a 2-bp frameshift into the tfdE ORF 66 bp downstream of the ATG start codon on pEST4011 would again result in a theoretical polypeptide with 79.6% similarity to TfdEII and 100% identity to TfdE of V. paradoxus. Finally, TfdBII carried 90.8% amino acid identity to TfdB of pEST4011.
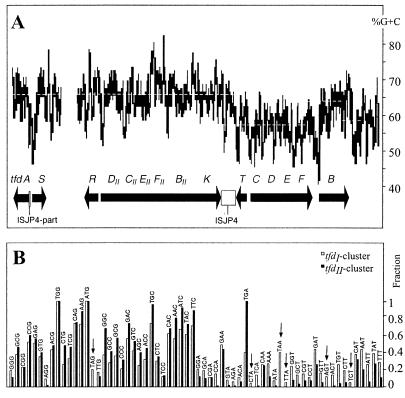
Codon usage among genes of the tfdII genes differed slightly from that of tfdI: the two lysine codons TTA and TCT, the two serine codons TCT and AGT, and the two stop codons TAG and TAA did not occur in any of the tfdII genes, whereas all possible codons occurred at least once in the tfdI genes (Fig. 2B). This difference in codon usage may have effects on relative translation efficiency of the tfdII or tfdI mRNAs, for example, when codon demand does not coincide with specific tRNA abundance in the cell (35). Both clusters tended to use the codons containing a G or C wobble base more abundantly than those with A or T (Fig. 2B). This bias was reflected in the high average G+C content of both gene clusters; for tfdCDEFB, however, the G+C content was remarkably lower (56%) than that for the tfdII cluster (66%) (Fig. 2A).
FIG. 2.
(A) G+C content of the tfd region on plasmid pJP4. The graph was created using the program Curve.It (ICGEB, Trieste, Italy) with a window of 100 bp. Dotted lines indicate the average G+C content for distinct fragments. (B) Codon usage of the tfdI and tfdII gene clusters. The black and white bars represent the fraction of usage of a certain codon among all possible codons for a certain amino acid. Addition of all fractions of all possible codons for a certain amino acid gives a total fraction of 1. The codons are ordered by G+C content and wobble base, rich G+C content (left) to poor G+C content (right).
Expression of each of the tfdII genes in E. coli.
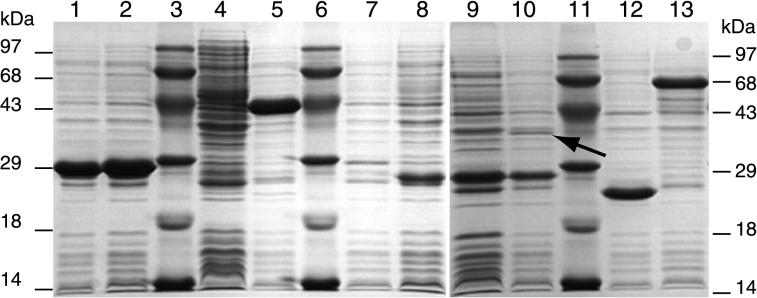
Cell extracts of E. coli BL21 cultures overexpressing each tfdII gene individually were analyzed by SDS-PAGE (Fig. 3) and assayed for enzyme activities from the chlorocatechol and chlorophenol oxidative pathway (Table 3). As negative controls, induced cultures of each of the E. coli strains containing the plasmids with mutated tfdII ORFs were used, along with E. coli BL21 without any pRSET plasmid. In total cell extract of E. coli BL21(pCBA165) expressing the tfdDII gene from its ATG start, we detected an overproduced polypeptide of about 42 kDa (Fig. 3, lane 5) which was absent in extracts of E. coli BL21(pCBA196) harboring a frameshift mutation in tfdDII (Fig. 3, lane 4). Chloromuconate cycloisomerase activity with 2,4-DCM and 3-CM as substrates was indeed detected in cell extracts of E. coli cultures harboring pCBA165 (Table 3). Significantly lower values were obtained when 2-CM was used as substrate. In cell extracts from cultures expressing the tfdDII frameshift mutant, however, no activity was found. Chlorocatechol 1,2-dioxygenase activity was clearly present in cell extracts from cultures expressing the tfdCII gene, with either 3,5-DCC, 3-CC, or 4-CC as substrate (Table 3). The predicted molecular mass of the polypeptide encoded by this ORF (28.1 kDa) fit well with the 27-kDa protein band determined by SDS-PAGE (Fig. 3, lane 2). No activity was detected in extracts of E. coli BL21 harboring the truncated gene tfdCIIΔ. The observed peptide of 27 kDa in cell extracts of E. coli expressing tfdCIIΔ is caused by the frameshift at the BstXI site, accidentally resulting in a peptide of the same size as TfdCII.
FIG. 3.
SDS-polyacrylamide gel of total cell extracts of IPTG-induced E. coli BL21(DE3)(pLysS) strains harboring pRSET6a-derived plasmids carrying one of the tfdII genes or its truncated derivative. Lanes: 1, pCBA200 (tfdCIIΔ); 2, pCBA199 (tfdCII); 3, molecular size marker; 4, pCBA196 (tfdDIIΔ); 5, pCBA165 (tfdDII); 6, molecular size marker; 7, pCBA201 (tfdEIIΔ); 8, pCBA202 (tfdEII); 9, pCBA197 (tfdFIIΔ); 10, pCBA184 (tfdFII); 11, molecular size marker; 12, pCBA198 (tfdBIIΔ); 13, pCBA180 (tfdBII). Positions of molecular masses are indicated on the left and right. The arrow in lane 10 points to the putative TfdFII protein. (Digital image was recorded as a TIFF file; background was enhanced for reproduction in Adobe Photoshop.)
TABLE 3.
Enzyme specific activities encoded by the tfdII genes measured in cell extracts of E. coli BL21
| Gene | Plasmid | Sp act (mU/mg of protein)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC 1,2-Db
|
CMC
|
DLH
|
MAR, MA | 2,4-DPH, 2,4-DCP | |||||||
| 3,5-DCC | 3-CC | 4-CC | 2,4-DCM | 2-CM | 3-CM | cis-DL | trans-DL | ||||
| tfdCII | pCBA199 | 103 | 0.8 × 103 | 0.6 × 103 | — | — | — | — | |||
| tfdCIIΔc | pCBA200 | 2 | 4 | 2 | — | — | — | — | |||
| tfdDII | pCBA165 | —d | 4.4 × 103 | 60 | 2 × 103 | — | — | — | |||
| tfdDIIΔ | pCBA196 | — | −3 | −1 | 6 | — | — | — | |||
| tfdEII | pCBA202 | — | — | 2.6 × 103 | 2 × 103 | — | — | ||||
| tfdEIIΔ | pCBA201 | — | — | 17 | 4 | — | — | ||||
| tfdFII | pCBA184 | — | — | — | 3.6 × 103 | — | |||||
| tfdFIIΔ | pCBA197 | — | — | — | 37 | — | |||||
| tfdBII | pCBA180 | — | — | — | — | 7 | |||||
| tfdBIIΔ | pCBA198 | — | — | — | — | 2 | |||||
| None | — | 1.1 | — | — | — | 1.7 | 1.1 | — | 31 | — | |
1 U = 1 μmol of substrate disappearance or product formation per min. Shown are representative values from one activity assay. Assays were performed at least in independent triplicates.
CC 1,2-D, chlorocatechol 1,2-dioxygenase; CMC, chloromuconate cycloisomerase; DLH, dienelactone hydrolase; MAR, maleylacetate reductase; 2,4-DPH, 2,4-dichlorophenol hydroxylase; DCC, dichlorocatechol; CC, chlorocatechol; DCM, dichloromuconate; CM, chloromuconate; DL, dienelactone; MA, maleylacetate; DCP, dichlorophenol
Frameshift in tfdCII.
—, not measured.
With cis- and trans-DL as substrates, dienelactone hydrolase activity was clearly detected in cell extracts from cultures expressing tfdEII. This activity coincided with the production of a 25-kDa protein (Fig. 3, lane 8). In contrast, we found no such polypeptide (Fig. 3, lane 7) and also no hydrolase activity with cells expressing tfdEIIΔ from plasmid pCBA201. Maleylacetate reductase activity was detected in cell extracts of E. coli BL21 harboring tfdFII by monitoring the maleylacetate-dependent oxidation of NADH (Table 3). In these cell extracts, SDS-PAGE revealed a protein with an apparent mass of about 37 kDa, which is close to the theoretically predicted mass of 37.5 kDa (Fig. 3, lane 10). This protein band was absent in total cell extracts of E. coli BL21(pCBA197), carrying a frameshift mutation in tfdFII (Fig. 3, lane 9). Both E. coli BL21(pCBA184) and E. coli (pCBA197) strongly produced a 27-kDa protein, which might be the result of a translational fusion protein starting at nucleotide position 3844 (numbering according to GenBank entry U16782) and continuing in pRSET6a. A slight background activity was found with cell extracts expressing a frameshift mutated tfdFII and in cell extracts from E. coli without any pRSET-type plasmid (Table 3). We suppose, therefore, that this low activity is due to native proteins from E. coli itself. Finally, the activity of TfdBII was determined in cell extracts of E. coli BL21 harboring tfdBII and compared to that in cell extracts of E. coli BL21 containing the frameshift mutated tfdBIIΔ. The activities measured for TfdBII were lower than those of the other TfdII enzymes but still higher than the activities measured in the negative control. The molecular mass of the TfdBII protein in cells harboring pCBA180 was as predicted (65 kDa) (Fig. 3, lane 13). In E. coli(pCBA198) containing a frameshift mutation in the tfdBII gene, we observed a protein of 24 kDa, which is the size of the generated truncated TfdBIIΔ protein (Fig. 3, lane 12). Based on these results, we conclude that the TfdII enzymes catalyzed the transformations expected from their similarities to the TfdI counterparts.
Expression of the tfdII genes in R. eutropha JMP134(pJP4).
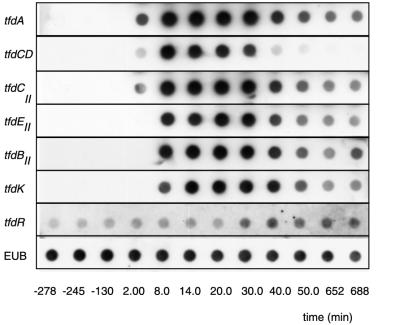
Since enzyme assays in extracts from R. eutropha JMP134 do not allow us to distinguish between activities from the tfdI cluster and the corresponding ones from the tfdII cluster, we relied on specific mRNA analysis to determine if transcription from the tfdII genes occurred. Induction of transcription was shown by hybridizing total RNA, isolated from a continuous culture at different time points after addition of 0.1 mM 2,4-D to the medium, with probes for the different tfdII genes. Immediately after addition of 2,4-D, levels of mRNA of the tfdCII gene increased rapidly, followed shortly by those of tfdEII, tfdBII, and tfdK (Fig. 4). Maximum levels of mRNA were reached 14 min after induction, after which there was a decrease in mRNA to a stable level that remained unchanged for over 10 h. We observed a similar pattern of gene induction for the tfdA and tfdCD genes (Fig. 4). Expression of tfdR, as well as that of the gene for 16S rRNA, remained essentially unchanged upon addition of 2,4-D (Fig. 4).
FIG. 4.
Dot blot hybridizations of total RNA isolated from a chemostat-grown culture of R. eutropha JMP134 before and after induction with 2,4-D. Antisense RNA probes are indicated on the left. Times at which the samples were taken from the chemostat are indicated at the bottom. 2,4-D was introduced into the chemostat at time zero. Panels for each hybridization were obtained in independent hybridization experiments; therefore, signal intensities cannot be directly compared between different probes. In addition, spot densities were not corrected for small differences among total RNA amounts spotted at each position. Note the distinct and immediate strong induction for all markers except tfdR and EUB (=16S rRNA). (Digital image was obtained by scanning of original autoradiograms; individual TIFF files were compiled in Adobe Photoshop.)
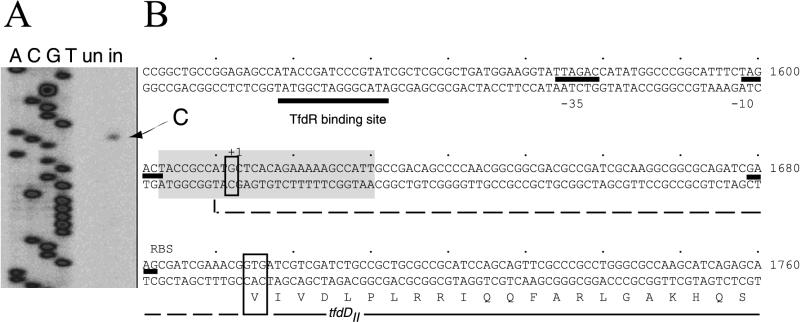
To map transcriptional start sites of the tfdII gene cluster, we performed primer extension analysis on total RNA that was isolated from chemostat-grown cells of R. eutropha JMP134(pJP4) under uninduced and induced (i.e., with 2,4-D) conditions. Using a primer positioned in the tfdDII gene, we were able to detect a specific cDNA transcript which was not observed from an uninduced culture (Fig. 5). The product identified a single transcription start site at a G residue at position −82 relative to the putative GTG or +3 relative to the postulated ATG translation start codon of tfdDII (Fig. 5). This makes the GTG a more likely candidate for the translation start codon of the tfdDII gene (Fig. 5). We found no primer extension products from primers positioned in the reading frames of any of the other genes of the tfdII cluster, including tfdK. These results suggest an operon-like organization for the genes tfdDII to tfdK. From the location of the transcriptional start site, we propose a TTAGAC/TAGACT promoter sequence for tfdDII (Fig. 5). Control primer extension reactions with a primer complementary to tfdC resulted in transcription starts at T and G (nucleotides 286 and 287), 4 nucleotides downstream of the −10 region proposed by Perkins (29) (not shown).
FIG. 5.
(A) Digital image from the gel region showing the size of the transcript synthesized from the tfdII mRNA and the sequence derived with the same primer. (Image recorded as a TIFF file on a LI-COR IR2 sequencer; background enhanced for reproduction purposes in Adobe Photoshop). The arrow points to the specific transcript observed under induced (in; with 2,4-D) conditions. un, uninduced. (B) Relevant part of the DNA sequence upstream of the tfdDII ORF. Translation of tfdDII is shown from the second possible start (Val at position 1694). The dotted line represents continuation of the ORF in the upstream direction. The first start codon (ATG at position 1612), however, was identified as the position of the transcription start (indicated with +1). Possible promoter elements and the TfdR binding site are indicated. The shaded region represents the sequence showed in the digital image.
DISCUSSION
Further exploration of the ISJP4-flanked transposable element on pJP4 led us to discover five ORFs, potentially encoding the metabolism of chlorocatechols and of chlorophenols. The ORFs were designated tfdDII, tfdCII, tfdEII, tfdFII, and tfdBII, by analogy to the tfdCDEFB genes on pJP4. We demonstrated by expressing each individual ORF in E. coli that the ORF designated tfdDII codes for a chloromuconate cycloisomerase, tfdCII codes for a chlorocatechol 1,2-dioxygenase, tfdEII codes for a dienelactone hydrolase, tfdFII codes for a maleylacetate reductase, and tfdBII codes for a chlorophenol hydroxylase. Together with the previously characterized tfd genes, this brings to eight the number of genes that are presently included in the transposable DNA: tfdS, tfdR, and tfdDIICIIEIIFIIBIIK. Substantial evidence leads us to believe that the tfdI and tfdII genes were acquired from different origins rather than evolved by duplication and divergence within one host. First, the actual percentages of identity among counterparts in the tfdI and tfdII clusters were rather low (15 to 62% at the amino acid level). Second, the G+C content of the tfdII genes is significantly higher (Fig. 2). Since tfdS and tfdR are fully identical, and perhaps themselves the result of a duplication event, we suppose that at one point a DNA fragment containing tfdR and tfdDIICIIEIIFIIBIIKII flanked by ISJP4 was mobilized into an ancestor pJP4 plasmid.
The results obtained with expression in E. coli showed that the tfdII genes can encode 2,4-DCP-metabolizing enzymes. Furthermore, we showed that the tfdII genes are transcribed in R. eutropha when cells are exposed to 2,4-D. Although we could not directly demonstrate that the tfdII genes are indeed translated into functional enzymes in R. eutropha JMP134, it seems rather unlikely that they would not be. First, all of the tfdII genes are transcribed and induced upon exposure of the cells to 2,4-D, and as strongly as the genes from the cluster tfdI (Fig. 5). Second, at least three gene products from the tfdII cluster are synthesized during growth on 2,4-D: TfdR, the regulatory protein of all the pathway genes; TfdK, a transporter protein for 2,4-D; and TfdFII, the maleylacetate reductase (19, 24, 25, 27, 32). Accidentally, the purified active enzyme catalyzing 2-chloromaleylacetate reduction in R. eutropha JMP134 turned out to be TfdFII, which was proven by NH2-terminal sequencing of the purified protein (32). Moreover, it was recently demonstrated that R. eutropha strains with a plasmid containing the tfdII gene cluster could actually grow on 3-chlorobenzoate (28). This makes it unlikely that the tfdII genes would not be translated in R. eutropha JMP134, although especially the tfdDII ORF has a very poor ribosome binding site. Strangely enough, the tfdII genes were not detected along with the tfdI genes in the original transposon mutagenesis studies performed by Don et al. (8). We can at least rule out that the tfdII cluster was inserted on pJP4 after their analysis, since the physical map of plasmid pJP4 as drawn by Don and Pemberton in 1985 (6) is identical to the current map determined from DNA sequencing data.
At this point, the question arises as to why the present-day configuration of the tfd genes with two sets of homologous genes is kept on plasmid pJP4 as it is. One answer is that at least some of the functions encoded within the tfdII cluster are favorable for growth on 2,4-D. Such a function might indeed be the chloromaleylacetate reductase TfdFII. TfdF transposon mutants grew poorly on 2,4-D but well on 3-chlorobenzoate (8), which suggests that not TfdF but TfdFII actually catalyzes the dechlorination of 2-chloromaleylacetate during growth on 2,4-D. Another function specific for the tfdII cluster is TfdK, a transporter protein which facilitates uptake of 2,4-D at low extracellular concentrations. However, TfdK does not seem to be indispensable for growth (25). A more important function, however, is carried by the regulatory protein TfdR (or its identical twin TfdS). Since TfdR is the transcriptional activator for tfdA expression and for both the tfdI and the tfdII genes (19, 24, 27), its loss would abolish 2,4-D pathway induction. Most likely, if the tfdII cluster were to become lost from pJP4, this would occur through recombination of homologous regions or activity of the ISJP4 element. Recombination between the right-end partial copy of ISJP4 (located between tfdS and tfdA) and ISJP4 (downstream of tfdK) would lead to loss of the regulatory genes. An alternative, perhaps more seldom, recombination between tfdT and tfdR would lead to loss of the tfdII cluster but could still restore the regulatory function. At least one plasmid with this type of recombination seems to exist, i.e., pMAB1. Restriction analysis of pMAB1 suggests identical tfdCDEF genes as on pJP4 but a recombination between tfdT and tfdR (21). This again points to the importance of maintaining proper regulation of the tfd pathway genes. Therefore, it seems that the current configuration is locked into a semistable state, due to the presence of the current regulatory genes within the tfdII cluster and the inactivated original regulatory gene (tfdT) lying in the tfdI cluster.
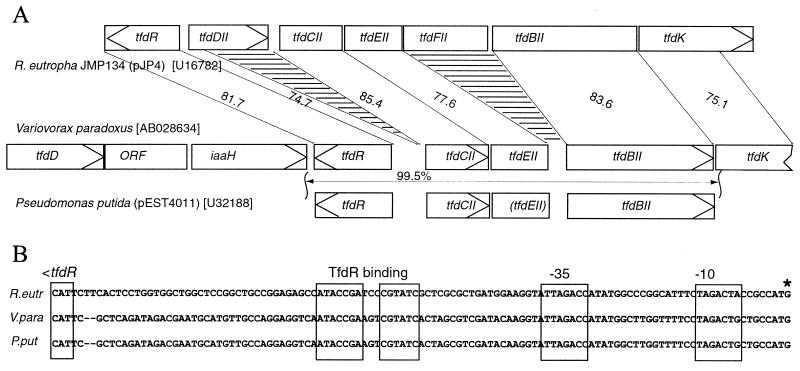
We can speculate a little on the genealogy of the tfdII cluster, since at least two other genetic systems for 2,4-D degradation are known to carry tfd-type genes with higher identities to the tfdII cluster of pJP4 than to the tfdI cluster (Fig. 6). One of these occurs on plasmid pEST4011, which is a derivative of a plasmid (pEST4002) originally isolated from P. putida strain EST4002 from 2,4-D-treated soils in Estonia (1). One region on this plasmid contains tfdII-type genes, although in a configuration, tfdR-tfdC-(tfdE)-tfdB, without tfdDII, tfdFII, or tfdK (20). Regulatable expression of chlorocatechol 1,2-dioxygenase and chlorophenol hydroxylase activities was demonstrated for this region of plasmid pEST4011 (26). The other system from V. paradoxus is basically identical to the tfd genes of pEST4011, although more sequence information is available on regions upstream of tfdR and downstream of tfdB (Valleys et al., unpublished). This indicated that a tfdK-like gene was downstream of tfdB and a tfdD-like gene was further upstream of tfdR (Fig. 6). Interestingly, in at least one region (between tfdR and tfdC), a deletion has occurred which removed part of the tfdDII ORF on pEST4011, leaving only 131 bp of the tfdDII ORF and the intergenic region between tfdR and tfdDII (Fig. 6A). This part, however, might be still important for a proper regulation of tfd gene expression, since it carries the TfdR binding site (Fig. 6B). At least in the V. paradoxus system, a complete tfdD gene copy exists, which, however, has a higher percentage sequence identity to tfdDI (64.2) than to tfdDII (55.2). Between tfdC and tfdB is a 708-bp sequence with 77.4% identity with tfdEII, but a frameshift hinders the production of a TfdEII-like polypeptide. This frameshift is not present in the V. paradoxus sequence. Curiously, no traces of the tfdFII gene can be found in the V. paradoxus or pEST4011 sequence. This finding suggests that this region of pEST4011 and of V. paradoxus was derived from a tfdII-like cluster and points to a wider distribution of the tfdII-type cluster among soil microorganisms rather than a single occurrence on pJP4.
FIG. 6.
(A) Comparison of the genetic organizations of the tfdII clusters of R. eutropha JMP134, V. paradoxus (Valleys et al., unpublished), and P. putida (pEST4011) (20). Connecting lines point to regions of high sequence identity among the different gene clusters. Percentages of sequence identity are given between the gene maps. The striped areas indicate regions deleted from the tfdII cluster of R. eutropha compared to the others. GenBank accession numbers are given in panel A. (B) Sequence alignment of the intergenic regions directly upstream of tfdR in the direction of tfdDII (for R. eutropha) or tfdC (for V. paradoxus and pEST4011). Boxed regions point to the conserved TfdR binding motif and to the −35 and −10 promoter sequences. The asterisk indicates the mapped transcription start site for the R. eutropha tfdII operon.
ACKNOWLEDGMENT
The work of C.M.L. was supported by grant 31-49222.96 from the Swiss National Science Foundation.
REFERENCES
- 1.Ausmees N R, Heinaru A L. New plasmids of herbicide 2,4-dichlorophenoxyacetic acid biodegradation. Genetika. 1990;26:770–772. [PubMed] [Google Scholar]
- 2.Baumann B, Snozzi M, Zehnder A J, van der Meer J R. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol. 1996;178:4367–4374. doi: 10.1128/jb.178.15.4367-4374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;130:151–152. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 4.Bhat M A, Tsuda M, Horiike K, Nozaki M, Vaidyanathan C S, Nakazawa T. Identification and characterization of a new plasmid carrying genes for degradation of 2,4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl Environ Microbiol. 1994;60:307–312. doi: 10.1128/aem.60.1.307-312.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the determination of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Don R H, Pemberton J M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid degradative plasmid pJP4. J Bacteriol. 1985;161:466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don R H, Weightman A J, Knackmuss H J, Timmis K N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134 (pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn E, Knackmuss H J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978;174:85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eulberg D, Kourbatova E M, Golovleva L A, Schlömann M. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J Bacteriol. 1998;180:1082–1094. doi: 10.1128/jb.180.5.1082-1094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans W C, Smith B S W, Fernley H N, Davies J I. Bacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem J. 1971;122:543–551. doi: 10.1042/bj1220543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farhana L, New P B. The 2,4-dichlorophenol hydroxylase of Alcaligenes eutrophus JMP134 is a homotetramer. Can J Microbiol. 1997;43:202–205. doi: 10.1139/m97-027. [DOI] [PubMed] [Google Scholar]
- 13.Fukumori F, Hausinger R P. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an alpha-ketoglutarate-dependent dioxygenase. J Bacteriol. 1993;175:2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumori F, Hausinger R P. Purification and characterization of 2,4-dichlorophenoxyacetate/alpha-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 15.Fulthorpe R R, McGowan C, Maltseva O V, Holben W E, Tiedje J M. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl Environ Microbiol. 1995;61:3274–3281. doi: 10.1128/aem.61.9.3274-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. [Google Scholar]
- 17.Ghosal D, You I-S. Nucleotide homology and organization of chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Mol Gen Genet. 1988;211:113–120. doi: 10.1007/BF00338401. [DOI] [PubMed] [Google Scholar]
- 18.Harker A R, Olsen R H, Seidler R J. Phenoxyacetic acid degradation by the 2,4-dichlorophenoxyacetic acid (TFD) pathway of plasmid pJP4: mapping and characterization of the TFD regulatory gene, tfdR. J Bacteriol. 1989;171:314–320. doi: 10.1128/jb.171.1.314-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaphammer B, Kukor J J, Olsen R H. Regulation of tfdCDEF by tfdR of the 2,4-dichlorophenoxyacetic acid degradation plasmid pJP4. J Bacteriol. 1990;172:2280–2286. doi: 10.1128/jb.172.5.2280-2286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koiv V, Marits R, Heinaru A. Sequence analysis of the 2,4 dichlorophenol hydroxylase gene tfdB and 3,5 dichlorocatechol 1,2 dioxygenase gene tfdC of 2,4 dichlorophenoxyacetic acid degrading plasmid pEST4011. Gene. 1996;174:293–297. doi: 10.1016/0378-1119(96)00043-1. [DOI] [PubMed] [Google Scholar]
- 21.Kuhm A E, Schlömann M, Knackmuss H-J, Pieper D H. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem J. 1990;266:877–883. [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Leveau J H J, König F, Füchslin H, Werlen C, van der Meer J R. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol Microbiol. 1999;33:396–406. doi: 10.1046/j.1365-2958.1999.01483.x. [DOI] [PubMed] [Google Scholar]
- 24.Leveau J H J, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134 (pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leveau J H J, Zehnder A J B, van der Meer J R. The tfdK gene product facilitates the uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134 (pJP4) J Bacteriol. 1997;180:2237–2243. doi: 10.1128/jb.180.8.2237-2243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maë A A, Marits R O, Ausmees N R, Kôiv V M, Heinaru A L. Characterization of a new 2,4-dichlorophenoxyacetic acid degrading plasmid pEST4011: physical map and localization of catabolic genes. J Gen Microbiol. 1993;139:3165–3160. [Google Scholar]
- 27.Matrubutham U, Harker A R. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alcaligenes eutrophus JMP134. J Bacteriol. 1994;176:2348–2353. doi: 10.1128/jb.176.8.2348-2353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Pantoja D, Guzman L, Manzano M, Pieper D H, Gonzalez B. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4) Appl Environ Microbiol. 2000;66:1602–1608. doi: 10.1128/aem.66.4.1602-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schoepfer R. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene. 1993;124:83–85. doi: 10.1016/0378-1119(93)90764-t. [DOI] [PubMed] [Google Scholar]
- 32.Seibert V, Stadler F K, Schlömann M. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4) J Bacteriol. 1993;175:6745–6754. doi: 10.1128/jb.175.21.6745-6754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Streber W R, Timmis K N, Zenk M H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Xia X. How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics. 1998;149:37–44. doi: 10.1093/genetics/149.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia X S, Aathithan S, Oswiecimska K, Smith A R W, Bruce I J. A novel plasmid pIJB1 possessing a putative 2,4-dichlorophenoxyacetic acid degradative transposon Tn5530 in Burkholderia cepacia strain 2a. Plasmid. 1998;39:154–159. doi: 10.1006/plas.1997.1332. [DOI] [PubMed] [Google Scholar]
- 37.You I S, Ghosal D. Genetic and molecular analysis of a regulatory region of the herbicide 2,4-dichlorophenoxyacetate catabolic plasmid pJP4. Mol Microbiol. 1995;16:321–331. doi: 10.1111/j.1365-2958.1995.tb02304.x. [DOI] [PubMed] [Google Scholar]