Abstract
Objective:
We assessed whether Prostate Health Index (phi) results improve prediction of grade reclassification for men on active surveillance.
Methods/Materials:
We identified men in Canary Prostate Active Surveillance Study with Grade Group (GG) 1 cancer. Outcome was grade reclassification to GG2+ cancer. We considered decision rules to maximize specificity with sensitivity set at 95%. We derived rules based on clinical data (R1) vs clinical data + phi (R3). We considered an “or”-logic rule combining clinical score and phi (R4), and a “two-step” rule using clinical data followed by risk stratification based on phi (R2). Rules were applied to a validation set, where values of R2 - R4 vs R1 for specificity and sensitivity were evaluated.
Results:
We included 1532 biopsies (n=610 discovery; n=922 validation) among 1142 men. Grade reclassification was seen in 27% of biopsies (23% discovery, 29% validation). Among discovery set, at 95% sensitivity, R2 yielded highest specificity at 27% vs 17% for R1. In validation set, R3 had best performance vs R1 with Δsensitivity = −4% and Δspecificity = +6%. There was slight improvement for R3 vs R1 for confirmatory biopsy (AUC 0.745 vs R1 0.724, ΔAUC = 0.021, 95%CI 0.002–0.041) but not for subsequent biopsies (ΔAUC = −0.012, 95%CI −0.031–0.006). R3 did not have better discrimination vs R1 among the biopsy cohort overall (ΔAUC = 0.007, 95%CI −0.007–0.020).
Conclusions:
Among active surveillance patients, using phi with clinical data modestly improved prediction of grade reclassification on confirmatory biopsy and did not improve prediction on subsequent biopsies.
Keywords: prostate cancer, active surveillance, biomarker
Introduction
Active surveillance is the preferred management strategy for men with low-risk prostate cancer.1 It is recommended that men on surveillance undergo prostate-specific antigen (PSA) tests and biopsies after their diagnosis.1 Repeat biopsies are invasive and burdensome but represent the only mechanism to identify grade reclassification and distinguish men at greater risk of disease progression. Some strategies—particularly those relying on annual biopsies—are likely too intense for patients. Ideally, repeat biopsies would be only recommended for men at greatest risk of harboring lethal cancer.
Risk calculators utilizing clinical information (e.g., patient age, PSA level) provide modest discrimination to predict those at risk for reclassification to Grade Group (GG) 2 or greater prostate cancer.2–4 It remains unclear whether prediction tools could be improved with information from biomarkers. For instance, a four-kallikrein panel in this setting gave mixed results, with some benefit with predicting higher-grade disease at confirmatory biopsy, but not subsequent biopsies.5 It is unknown whether other biomarkers, such as prostate health index (phi), would provide more robust improvement when added to existing calculators. Furthermore, it is unknown whether phi could outperform predictive capability of clinical data alone.
We evaluated how phi may improve prediction of which active surveillance patients would have higher-grade cancer on surveillance biopsies in combination with other clinical data, versus clinical data alone. We utilized data and serum from men in a multi-institutional, prospectively accrued active surveillance cohort (i.e., Canary Prostate Active Surveillance Study (PASS)) to address this question. The large sample of men permitted development and validation of decision rules that included phi results. We hypothesized that adding phi would increase discriminatory ability versus a validated predictive calculator. If confirmed, these results will help decrease burden of excessive surveillance biopsies among prostate cancer patients pursuing active surveillance.
Methods
Patients were identified from the Canary PASS, which is a cohort trial that has prospectively enrolled men with clinical stage T1-T2, GG1–2 prostate cancer eligible to pursue active surveillance (clinicaltrials.gov, NCT00756665).6 Participants are monitored with PSA testing and biopsies (12 months, 24 months, 48 months from diagnosis) and provide specimens for a biorepository every six months during the first 5 years on surveillance. We identified a preliminary cohort of men with GG1 cancer on biopsy. Our analysis was performed at biopsy-level, where men may have had multiple biopsies in the analysis.
Creation of Discovery/Validation Sets
We established a discovery set of biopsies where serum specimen was drawn prior to biopsy. We randomly selected biopsies to meet a pre-determined sample size based on power calculations. For validation, we used a two-stage process. First, we identified a unique set of biopsies (n=821) distinct from discovery set where sample size was again pre-determined by power calculations. We then supplemented this preliminary set with a purposefully sampled subset of biopsies (n=101) with our outcome of interest (i.e., grade reclassification) to ensure adequate precision for our validation. Details are described in the Appendix.
Prostate Health Index (phi)
Blood was collected in Serum Separator Tubes, allowed to clot for 45 min, centrifuged at 1600g-force, 4°C, and frozen at −70°C within 4 hours of collection. Frozen serum was stored until shipment on dry ice to the Johns Hopkins University EDRN Biomarker Reference Lab (Baltimore, MD) for analysis. Specimens were analyzed for PSA, free PSA (fPSA), and [−2]proPSA (p2PSA) on the Access 2 Immunoassay Analyzer (Beckman Coulter, Inc., Brea, CA, United States). The prostate health index (phi) was calculated as (p2PSA/fPSA) x PSA1/2. The analysis lab was blinded to all specimen and clinical information.
Outcome
Outcome was grade reclassification to GG2–5 prostate cancer on a biopsy. Biopsies were confirmatory (i.e., first biopsy after diagnosis) or subsequent.
Model Building and Decision Rule Derivation
To identify decision rules in our discovery cohort with clinical information, we fit a multivariable logistic regression model for predicting grade reclassification using factors validated in the most recent PASS risk calculator (i.e., clinical model).5 These included patient age, body mass index (BMI), PSA (log-transformed), and prostate volume (log-transformed) as continuous variables, confirmatory versus subsequent biopsy, having more than one prior negative biopsy, and a previous biopsy with more than 20% of cores on biopsy with cancer as dichotomous variables. We fit a multivariate model that included phi results plus clinical factors (clinical factors plus phi model). Generalized estimating equations were used to account for clustered nature of data for men with more than one biopsy.
We developed decision rules using clinical data alone or in combination with phi results in a variety of ways. Since rules are intended for clinical decision of avoiding an unnecessary biopsy, our objective was to identify rules with the highest observed specificity in the discovery cohort while keeping the false negative rate at or below 5% (i.e., at least 95% sensitivity). R1 was based on the risk scores from the clinical model. R2 was built in two steps. First, two thresholds were determined to divide patients into three risk groups using clinical data. Patients in the highest risk group would be recommended to undergo biopsy; patients with lowest risk would be recommended to forgo biopsy. For those with intermediate risk, a threshold for phi was identified for selecting higher-risk patients to undergo biopsy. R3 was derived based on the clinical factors plus phi in a single linear regression model. The final rule (R4) is a “or”-logic rule, having two thresholds (one based on the risk score from the clinical model, and one based on phi) where biopsy was recommended if risk exceeded either threshold. R2 required phi measurements only for the intermediate risk group. For R2 and R4, thresholds were determined via grid search for the highest specificity at 95% sensitivity (i.e., 5% false-negative rate).
Receiver Operating Characteristic (ROC) and Decision Curve Analyses (DCA)
We assessed absolute and incremental performance of each rule using the validation biopsy cohort. Rules were implemented using the same coefficients from models and thresholds from analyses performed on the discovery cohort. The sensitivity and specificity of the four rules (R1–R4) were determined using the validation cohort. These were compared to a base rule (R0), performing biopsy no matter what the clinical factors or PHI result (i.e., “biopsy all”), or R1. For R1 and R3, areas under the Receiver Operating Characteristic curve (AUC of ROC) were evaluated and compared using risk scores from multivariable models. As an exploratory analysis, we planned to validate rules in a stratified analysis by confirmatory and subsequent biopsies among the validation cohort. We performed a DCA to display net benefit of using R1 – R4 across a spectrum of probabilities of reclassification for all, confirmatory, and subsequent biopsies from the validation set. 95% confidence intervals of sensitivities, specificities, AUC, and their difference between rules were calculated via bootstrap resampling at the patient-level. Analyses were conducted using R v4.0.4. Participants consented to collection of research specimens under institutional regulatory board approval (IRB) at all participating study sites and this was approved by the central site IRB.
Results
We evaluated 1,532 biopsies among 1,142 men with GG1 prostate cancer. There were 610 biopsies in the discovery set and 922 biopsies in the validation set. The characteristics of the biopsy sets are in Table 1. Overall, 23.1% (141/610) and 29.4% (271/922) biopsies had grade reclassification in the discovery and validation sets, respectively (Supplemental Tables 1 and 2). Among the discovery set, grade reclassification was associated with age (66 vs 64 years, OR 1.03 per year, 95% CI 1.00 – 1.07) and PSA (4.9 vs 4.3 ng/mL, OR 1.62 per ng/mL, 95% CI 1.07 – 2.45). There were associations between reclassification and prostate volume (38.1 vs. 41.7 cc, OR 0.34, 0.20 – 058), prior biopsy with ≥20% positive cores (35% vs. 18%, OR 1.98, 95% CI 1.26 – 3.10), and 2+ prior negative biopsies (2% vs. 11%, OR 0.24, 95% CI 0.07 – 0.86). There was not a significant association between biopsy type (confirmatory vs surveillance; OR 1.20, 95% CI 0.79 – 1.84). For the validation set, biopsies with reclassification were more likely to be confirmatory (61% vs 46% no reclassification, p<0.001). There was a significant association between phi and grade reclassification in both sets (discovery: 44.9 vs 33.7, adjusted OR 2.09, 95% CI 1.24 – 3.53; validation: 40.8 vs 31.8, p<0.001). Results from calibration of multivariable model for the validation set are in Supplemental Figure 1.
Table 1:
Characteristics of discovery and validation cohorts.
| Discovery Cohort1 (n=610 biopsies) | Validation Cohort2 (n=922 biopsies) | |
|---|---|---|
| Age at Biopsy (median, IQR) | 64 (59 – 68) | 65 (60 – 69) |
| Body Mass Index (kg/m2) (median, IQR) | 27.2 (25.1 – 30.0) | 27.2 (25.0 – 30.6) |
| Prostate volume (cc) (median, IQR) | 40.4 (30.3 – 57.3) | 46.3 (32.5 – 62.2) |
| PSA (ng/mL) (median, IQR) | 4.5 (3.0 – 6.3) | 5.2 (3.6 – 7.3) |
| Biopsy Type (n, %) | ||
| Confirmatory Biopsy | 298 (49%) | 463 (50%) |
| Second Surveillance Biopsy | 218 (36%) | 285 (31%) |
| Third Surveillance Biopsy | 67 (11%) | 110 (12%) |
| Fourth Surveillance Biopsy | 18 (3%) | 42 (5%) |
| Fifth Surveillance Biopsy | 9 (1%) | 12 (1%) |
| Sixth Surveillance Biopsy | 0 | 7 (1%) |
| Seventh Surveillance Biopsy | 0 | 3 (<1%) |
| Prior Biopsy with 20% or more cores positive (n, %) | 132 (22%) | 240 (26%) |
| 2+ Prior Negative Biopsies (n, %) | 55 (9%) | 60 (7%) |
| Prostate Health Index (median, IQR) | 35.7 (27.4 – 49.6) | 34.0 (24.7 – 46.8) |
Comprised of 513 men.
Comprised of 629 men and includes 821 randomly sampled biopsies and 101 biopsies purposefully sampled for presence of GG2 disease.
The performance of the selected decision rules in the discovery and validation sets is in Table 2. In the discovery set, the two-step rule (R2) had the greatest specificity with sensitivity fixed at 95% (27% vs. 17% R1, 21% R3 and 25% R4). In the validation set, R1 and R3 performed similarly to counterparts in the discovery set: R1 yielded sensitivity of 97% (95%CI 95% - 99%) and specificity at 15% (95%CI 11 – 18%), and R3 had sensitivity of 93% (95% CI 90–96%) and specificity of 21% (95% CI 17–24%). Compared to R1, R3 had Δsensitivity = −4% (95% CI −6–2%) and Δspecificity = +6% (95% CI 4–8%), whereas R2 had decreased sensitivity (Δsensitivity = −7%, 95% CI −1 – −4%) and Δspecificity = +14% (95% CI 10–18%). Performance of the “or”-logic rule R4 was comparable to R3. When stratified by biopsy type, R3 had higher sensitivity (97%; 95% CI 95–99%) but lower specificity (12%; 8–16%) among confirmatory biopsies. Among subsequent biopsies, R3 had lower sensitivity (88%; 95% CI 81–93%) but higher specificity (28%; 95% CI 22–33%).
Table 2.
Performance of prediction rules among discovery and validation biopsy cohorts.
| Discovery (n=610) | Validation (n=922) | |||||||
|---|---|---|---|---|---|---|---|---|
| Rule | Specificity1 | Overall | Confirmatory Biopsy | Subsequent Biopsy | ||||
| Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | |||
| R1: clinical only | Biopsy if clinical score (CS)* > 0.083 | 17% | 97% (95–99%) | 15% (11–18%) | 99% (98–100%) | 7% (4–10%) | 93% (88–98%) | 22% (16–26%) |
| R2: clinical then PHI | Biopsy if (CS>0.185) or (CS>0.05 and phi>27.6) | 27% | 90% (86–93%) | 28% (24–33%) | 95% (91–98%) | 23% (19–28%) | 83% (76–90%) | 33% (27–39%) |
| R3: clinical + PHI(linear) | Biopsy if CS + phi score** > 0.092 | 21% | 93% (90–96%) | 21% (17–24%) | 97% (94–99%) | 12% (8–16%) | 88% (81–93%) | 28% (22–33%) |
| R4:clinical or PHI | Biopsy if (CS > 0.12) or (phi>39.0) | 25% | 93% (89–96%) | 22% (18–26%) | 95% (91–98%) | 16% (12–20%) | 90% (83–95%) | 27% (21–33%) |
Sensitivity fixed at 95% for performance assessment based on fitted model coefficients for discovery cohort.
CS = expit (-1.77 + 0.80*log PSA + 0.19*Initial Biopsy + 0.03*Age + 0.06*BMI + 0.75*Prior Cores ratio ≥ 0.2 – 1.57*Neg Bx ≥ 2 – 1.29*log prostate volume)
CS + phi = expit (-4.47 + 0.74*log phi + 0.48*log PSA + 0.18*Initial Biopsy + 0.033*Age + 0.05 * BMI + 0.68 * Prior Cores ratio ≥ 0.2 – 1.42 * Neg Bx ≥ 2 – 1.07 * log prostate volume)
AUC values and clinical consequences of applying different rules are displayed in Table 3. There was slight improvement for R3 vs R1 for men receiving confirmatory biopsy (AUC 0.745 vs R1 0.724, ΔAUC = 0.021, 95%CI 0.002–0.041). R3 did not have better discrimination vs R1 among the entire cohort (AUC 0.727 vs R1 0.720, ΔAUC = 0.007, 95%CI −0.007–0.020), nor for subsequent biopsies (AUC 0.681 vs R1 0.693, ΔAUC = −0.012, 95%CI −0.031–0.006). Due to R2 and R4 having multiple decision thresholds, we could not generate ROC curves. Applying R3 for every 1000 confirmatory biopsies would avoid 97 biopsies (95% CI 69–125) and miss 8 reclassifications (95% CI 2–16). Applying R1 for every 1000 subsequent biopsies would avoid 193 biopsies (95% CI 147–236) and miss 10 reclassifications (95% CI 3 – 18). Results from the DCA are in Figure 1. We also investigated potential heterogeneity in the influence of phi based on biopsy type (confirmatory vs surveillance). In a model with interaction term between phi and biopsy type, the estimate was not statistically significant (adjusted OR 0.81, 95% CI 0.33 – 2.01).
Table 3.
Clinical consequences for decision rules with and without phi.
| Rule | Overall | Confirmatory | Subsequent | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Biopsies avoided# (n, 95% CI) | Reclassifications Missed# (n, 95% CI) | AUC | Biopsies avoided (n, 95% CI) | Reclassifications Missed (n, 95% CI) | AUC | Biopsies avoided (n, 95% CI) | Reclassifications Missed (n, 95% CI) | AUC | |
| R1: clinical only | 123 (95 – 151) | 6 (2 – 11) | 0.720 | 51 (30 – 73) | 2 (0 – 5) | 0.724 | 193 (147 – 236) | 10 (3 – 18) | 0.693 |
| R2: clinical then PHI | 246 (210 – 282) | 21 (14 – 28) | * | 187 (149 – 225) | 14 (6 – 24) | * | 303 (252 – 356) | 26 (15 – 37) | * |
| R3: clinical + PHI (linear) |
177 (145 – 209) | 14 (8 – 20) | 0.727 | 97 (69 – 125) | 8 (2 – 16) | 0.745 | 255 (205 – 301) | 18 (10 – 28) | 0.681 |
| R4: clinical or PHI | 189 (158 – 222) | 15 (9 – 22) | * | 133 (102 – 163) | 14 (6 – 24) | * | 245 (195 – 297) | 16 (8 – 26) | * |
The number of biopsies avoided, and reclassifications missed from a population of 1000 biopsies with an event rate of 21%, 26% and 15% for the overall, confirmatory, and subsequent biopsies respectively, which are estimated from the cohort of 821 biopsies from the validation set.
Unable to calculate due to multiple thresholds in rule.
Figure 1.
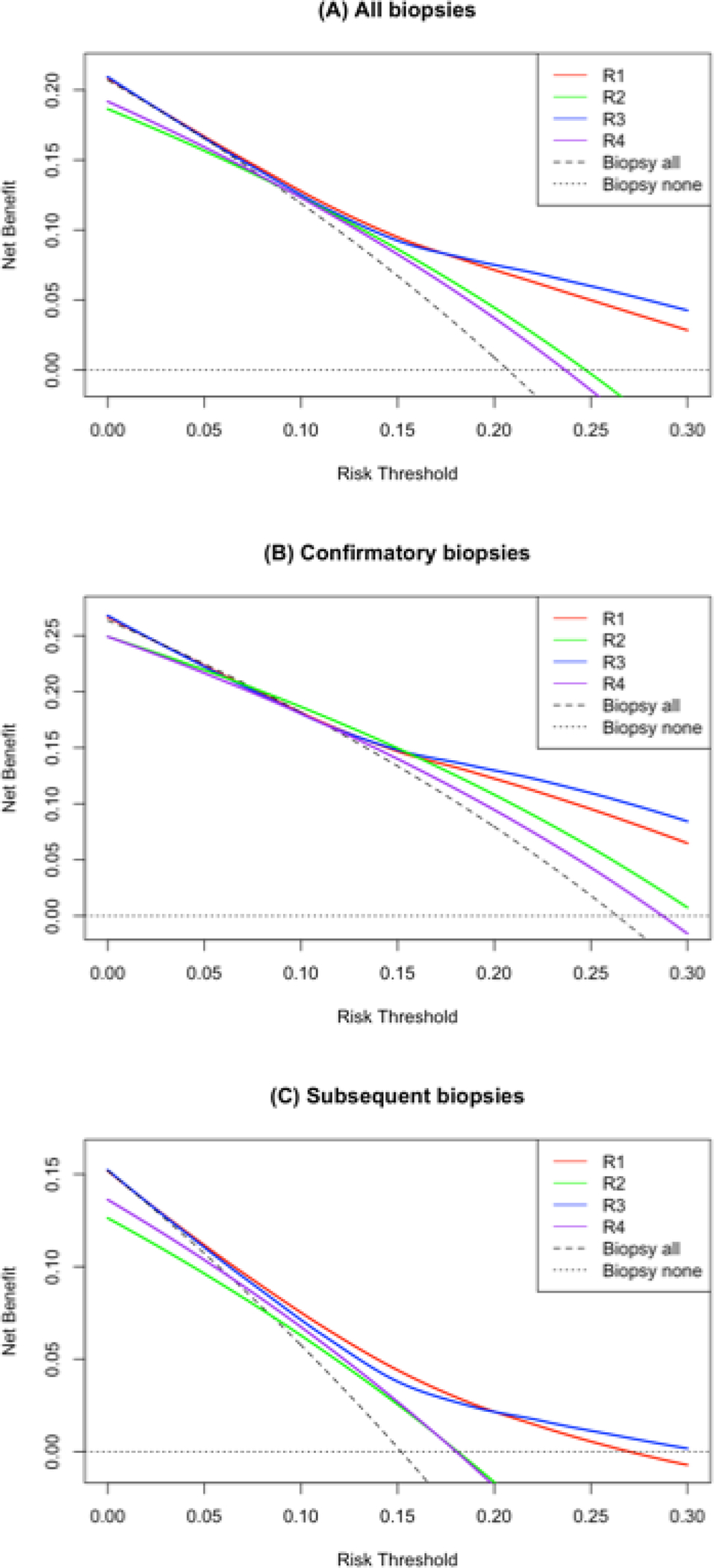
DCA from best performing prediction rule (R3. blue line) versus clinical-only rule (R1, red line). This figure demonstrates DCA for R3 (clinical data plus phi) compared to R1 (clinical data alone) for all biopsies (A), confirmatory biopsies (B), and subsequent biopsies (C). Of note this DCA was performed on 821 biopsies randomly sampled for the validation set and excluded 101 biopsies that were purposefully sampled for having GG2 on biopsy.
Discussion
In this analysis, we evaluated whether an established serum-based biomarker (phi) could improve the ability to discriminate whether a man with low-grade prostate cancer would be reclassified on a subsequent biopsy. There were two key findings from this analysis. First, addition of phi to clinical factors did not improve discrimination of grade reclassification overall. Second, we observed slightly improved discrimination with phi added to clinical factors among confirmatory biopsies, but not among subsequent biopsies.
Active surveillance for men with low-risk prostate cancer is the preferred approach by contemporary guidelines.1 This is reflected by expanding uptake of this strategy at a population-level in the U.S.; nearly half of men diagnosed with lower-risk tumors in 2015 were managed with active surveillance.7,8 The question emerging now is not whether to perform active surveillance for men with low-risk prostate cancer, but how to perform it. Guidelines are less clear on the optimal frequency of PSA tests and biopsies.9 Current biopsy protocols for surveillance might be too “intense”, exposing patients to increased anxiety, potential biopsy complications, and burdensome costs of care.10,11 Prior work using data from the PASS cohort showed that clinical factors can help predict the likelihood of grade reclassification on subsequent biopsies and perhaps help “de-intensify” surveillance by avoiding biopsies for some men.11
It is less clear whether biomarkers—beyond PSA—can supplement clinical factors to improve our ability to discriminate which men may or may not benefit from a repeat biopsy while on active surveillance. For instance, use of a four-kallikrein biomarker panel plus clinical factors improved ability to predict reclassification at confirmatory biopsy only slightly, and did not provide any additional benefit for men considering subsequent biopsies.5 The phi biomarker represents a promising candidate to augment biopsy decision-making for men on active surveillance. This biomarker helps predict the presence of significant prostate cancer for men considering initial prostate biopsy.12 Among institutional cohorts, phi has shown promise in predicting grade reclassification among active surveillance patients.13,14 However, among a large cohort of men across different practice sites, we did not observe a marked improvement in the ability to predict grade reclassification with the inclusion of phi in addition to other previously-validated clinical factors. Like the four-kallikrein panel, we did observe some improved discrimination for active surveillance patients undergoing their confirmatory biopsy after their prostate cancer diagnosis.
Our findings must be placed within the context of the limitations of this analysis. First, our primary outcome was grade reclassification, and there may be other critical factors (i.e., biopsy technique, ultrasound findings, etc.) left out of our models. However, the clinical factors that were included have been validated as important to predicting grade reclassification. Second, we retrospectively assessed phi among samples drawn months to years ago, and there may have been variation in the phi values over time. However, we ran sensitivity analyses assessing repeated measures of blinded duplicate samples and found very high correlation with re-testing. Finally, though grade reclassification is a clinically actionable endpoint, it may not always reflect the overall aggressiveness of the cancer based on sampling error and divergence between clinical risk and genomic risk.
Despite those limitations, we found phi may have a role in helping decide whether a man considering active surveillance needs a confirmatory biopsy. The clinical implications of finding a biomarker that can help decide whether to perform confirmatory biopsy are unclear. Among our cohort, grade reclassification was more common with confirmatory biopsy versus surveillance biopsy. Guidelines recommend the initial confirmatory biopsy most strongly for active surveillance patients, and efforts to “de-intensify” prostate biopsies focused on subsequent prostate biopsies. Notably, most biopsies among our cohort were not aided by MRI guidance, now being used more frequently during active surveillance.15 Among the PASS cohort, MRI findings did not improve discrimination for grade reclassification compared to clinical factors alone, but the presence of a suspicious PIRADS 5 lesion was associated with nearly a 3-fold increased odds for reclassification.16 A recent randomized trial showed that active surveillance patients randomized to an MRI to help guide their confirmatory biopsy had fewer grade reclassifications at 2 years and less frequent transition to treatment.17 It is unlikely that providers would recommend against a confirmatory biopsy with a favorable risk prediction based on biomarker results, in the face of an unfavorable MRI finding. With the modest observed effect, it is possible that incorporation of MRI findings into models could eliminate the impact phi has in predicting grade reclassification. It will be crucial to understand the interplay between serum and urine biomarkers and radiographic findings related to characterizing risk of grade reclassification for active surveillance patients, particularly for subsequent biopsies after the confirmatory biopsy.
Conclusions
Among active surveillance patients, using phi with clinical data only modestly improved ability to assess risk of grade reclassification on confirmatory biopsy versus clinical information alone. In addition, phi did not improve prediction of grade reclassification on subsequent biopsies.
Supplementary Material
Acknowledgments
Research support:
NIH/NCI Early Detection Research Network, U24 CA115102 (DWC).
PHI assay reagents were provided by Beckman Coulter, Inc
American Cancer Society MRSG 18-015-01-CPHPS (CPF)
NIH U01 CA224255 (DL)
NIH U01 CA113913 (YH, LN, MGS and DWL)
Abbreviation Key
- AUC
Area Under Curve
- GG
Grade Group
- IRB
Institutional Regulatory Board
- IQR
Interquartile range
- MRI
Magnetic resonance imaging
- OR
Odds ratio
- PASS
Prostate Active Surveillance Study
- phi
Prostate Health Index
- PSA
Prostate specific antigen
- ROC
Receiving Operating Characteristic
REFERENCES
- 1.Sanda MG, Cadeddu JA, Kirkby E, et al. : Clinically Localized Prostate Cancer: AUA/ASTRO/SUO Guideline. Part I: Risk Stratification, Shared Decision Making, and Care Options. J Urol 2018; 199: 683–690. [DOI] [PubMed] [Google Scholar]
- 2.Ankerst DP, Xia J, Thompson IM, et al. : Precision Medicine in Active Surveillance for Prostate Cancer: Development of the Canary–Early Detection Research Network Active Surveillance Biopsy Risk Calculator. Eur Urol 2015; 68: 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankerst DP, Straubinger J, Selig K, et al. : A Contemporary Prostate Biopsy Risk Calculator Based on Multiple Heterogeneous Cohorts. Eur Urol 2018; 74: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drost F-JH, Nieboer D, Morgan TM, et al. : Predicting Biopsy Outcomes During Active Surveillance for Prostate Cancer: External Validation of the Canary Prostate Active Surveillance Study Risk Calculators in Five Large Active Surveillance Cohorts. Eur Urol 2019; 76: 693–702. [DOI] [PubMed] [Google Scholar]
- 5.Lin DW, Newcomb LF, Brown MD, et al. : Evaluating the Four Kallikrein Panel of the 4Kscore for Prediction of High-grade Prostate Cancer in Men in the Canary Prostate Active Surveillance Study. Eur Urol 2017; 72: 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newcomb LF, Brooks JD, Carroll PR, et al. : Canary Prostate Active Surveillance Study: Design of a Multi-institutional Active Surveillance Cohort and Biorepository. Urology 2010; 75: 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahal BA, Butler S, Franco I, et al. : Use of Active Surveillance or Watchful Waiting for Low-Risk Prostate Cancer and Management Trends Across Risk Groups in the United States, 2010–2015. JAMA 2019; 321: 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Hall IJ, Filson C, et al. : Trends in the use of active surveillance and treatments in Medicare beneficiaries diagnosed with localized prostate cancer. Urol Oncol 2020. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruinsma SM, Bangma CH, Carroll PR, et al. : Active surveillance for prostate cancer: a narrative review of clinical guidelines. Nat Rev Urol 2016; 13: 151–167. [DOI] [PubMed] [Google Scholar]
- 10.Tomer A, Nieboer D, Roobol MJ, et al. : Personalised biopsy schedules based on risk of Gleason upgrading for patients with low-risk prostate cancer on active surveillance. BJU Int 2021; 127: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooperberg MR, Zheng Y, Faino AV, et al. : Tailoring Intensity of Active Surveillance for Low-Risk Prostate Cancer Based on Individualized Prediction of Risk Stability. JAMA Oncol 2020; 6: e203187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosoian JJ, Druskin SC, Andreas D, et al. : Prostate Health Index density improves detection of clinically significant prostate cancer. BJU Int 2017; 120: 793–798. [DOI] [PubMed] [Google Scholar]
- 13.Tosoian JJ, Loeb S, Feng Z, et al. : Association of [−2]proPSA with Biopsy Reclassification During Active Surveillance for Prostate Cancer. J Urol 2012; 188: 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwen ZR, Mamawala M, Tosoian JJ, et al. : Prostate Health Index and multiparametric magnetic resonance imaging to predict prostate cancer grade reclassification in active surveillance. BJU Int 2020; 126: 373–378. [DOI] [PubMed] [Google Scholar]
- 15.Fam MM, Yabes JG, Macleod LC, et al. : Increasing Utilization of Multiparametric Magnetic Resonance Imaging in Prostate Cancer Active Surveillance. Urology 2019; 130: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liss MA, Newcomb LF, Zheng Y, et al. : Magnetic Resonance Imaging for the Detection of High Grade Cancer in the Canary Prostate Active Surveillance Study. J Urol 2020; 204: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klotz L, Pond G, Loblaw A, et al. : Randomized Study of Systematic Biopsy Versus Magnetic Resonance Imaging and Targeted and Systematic Biopsy in Men on Active Surveillance (ASIST): 2-year Postbiopsy Follow-up. Eur Urol 2020; 77: 311–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


