Abstract
Fungal effectors play a pivotal role in suppressing the host defence system, and their evolution is highly dynamic. By comparative sequence analysis of plant‐pathogenic fungi and Magnaporthe oryzae, we identified the small secreted C2H2 zinc finger protein MoHTR3. MoHTR3 exhibited high conservation in M. oryzae strains but low conservation among other plant‐pathogenic fungi, suggesting an emerging evolutionary selection process. MoHTR3 is exclusively expressed in the biotrophic stage of fungal invasion, and the encoded protein localizes to the biotrophic interfacial complex (BIC) and the host cell nucleus. The signal peptide crucial for MoHTR3′ secretion to the BIC and the protein section required for its translocation to the nucleus were both identified by a functional protein domain study. The host‐nuclear localization of MoHTR3 suggests a function as a transcriptional modulator of host defence gene induction. After ΔMohtr3 infection, the expression of jasmonic acid‐ and ethylene‐associated genes was diminished in rice, in contrast to when the MoHTR3‐overexpressing strain (MoHTR3ox) was applied. The transcript levels of salicylic acid‐ and defence‐related genes were also affected after ΔMohtr3 and MoHTR3ox application. In pathogenicity assays, ΔMohtr3 was indistinguishable from the wild type. However, MoHTR3ox‐infected plants showed diminished lesion formation and hydrogen peroxide accumulation, accompanied by a decrease in susceptibility, suggesting that the MoHTR3‐induced manipulation of host cells affects host–pathogen interaction. MoHTR3 emphasizes the role of the host nucleus as a critical target for the pathogen‐driven manipulation of host defence mechanisms and underscores the ongoing evolution of rice blast's arms race.
Keywords: effector‐triggered immunity, jasmonic acid, Magnaporthe oryzae, nuclear effector, plant hormone, rice blast disease, signal peptide
The plant nucleus‐localized effector MoHTR3 is exclusively expressed in the biotrophic stage of fungal invasion, and it highlights the critical role of the host nucleus in the pathogen‐driven manipulation of plant defence mechanisms.
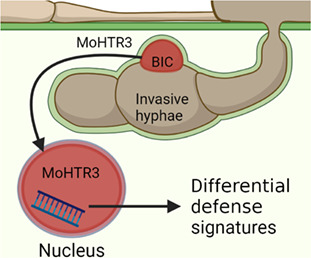
1. INTRODUCTION
Plants are under constant threat by pathogens, which can have diverse negative effects, including mortality or reduced yield. Magnaporthe oryzae is a hemibiotrophic fungus that invades living host plant tissue and takes up nutrients to maintain its pathogenic growth, eventually resulting in plant death (Talbot, 2003). Plants have a two‐layered immune system that defends against infection by numerous pathogens (Jones & Dangl, 2006). Membrane‐localized pattern recognition receptors perceive pathogen‐associated molecular patterns (PAMPs) such as flagellin22 or chitin, triggering the sequential activation of an underlying mitogen‐activated protein kinase cascade. PAMP‐triggered immunity activates a mild defence response, whereas cytosol‐localized receptors of the nucleotide‐binding site leucine‐rich repeat family sense pathogen virulence factors, or effectors, thereby triggering a strong, sustainable defence response. Effector‐triggered immunity is accompanied by programmed cell death, increased reactive oxygen species (ROS) accumulation, defence‐related gene expression, and hormonal changes (Chang et al., 2022; Tsuda & Katagiri, 2010). The biosynthesis and activation of defence‐related hormones and mitogen‐activated protein kinase signalling accompany both effector‐triggered immunity and PAMP‐triggered immunity (Peng et al., 2018). The plant defence hormones salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) coordinate the defence response according to the pathogen type (Li et al., 2019). SA‐mediated immunity mediates the defence against biotrophic and hemibiotrophic pathogens, whereas JA and ET synergistically manage the immune response after the perception of necrotrophic pathogens (Robert‐Seilaniantz et al., 2011; Völz, Kim, Mi, Mariappan, et al., 2019b).
Following infection, M. oryzae enlarges its invasive hyphal network to secrete effector proteins. Pathogen effectors can be classified into three groups according to their host subcellular localization as apoplastic, cytoplasmic, or nuclear effectors (Asai & Shirasu, 2015). Apoplastic effectors accumulate in an extra‐invasive hyphal membrane, whereas cytoplasmic effectors are secreted into the host cytosol and nuclear effectors target the host nucleus. Typically, effector proteins are composed of fewer than 300 amino acids and are classified as small secreted proteins (Kim et al., 2016). Cytoplasmic and nuclear effectors are secreted through the biotrophic interfacial complex (BIC) and then translocated to the plant cytoplasm. The subsequent spread of effectors among adjacent plant cells is facilitated by the plasmodesmata (Zhang & Xu, 2014).
Recent studies have reported that fungal effectors modulate the host internal environment to favour the pathogen (Figueroa et al., 2021; König et al., 2021; Tariqjaveed et al., 2021). For example, Avr2 of Cladosporium fulvum, which is required for plant basal defence, inhibits Cys proteases (van Esse et al., 2008), the Pleiades effector gene cluster of Ustilago maydis is involved in PAMP‐triggered ROS production (Navarrete et al., 2021), Mlp124478 of Melampsora larici‐populina functions in virulence activity (Ahmed et al., 2018), and CgEP1 of Colletotrichum graminicola is involved in the plant basal defence response and pathogenesis (Vargas et al., 2016). In M. oryzae, the effectors MoHTR1 and MoHTR2 contain a transcription factor domain and have features that are typical of small secreted proteins. Following secretion, these nuclear effectors bind to plant immunity‐associated genes such as OsMYB4, OsHPL2, and OsWRKY45 and compromise the plant defence system (Kim et al., 2020). Similarly, Mlp124478 and CgEP1 also have DNA‐binding functions (Ahmed et al., 2018; Vargas et al., 2016). Notably, Mlp124478 regulates the plant immune system‐related gene TGA1a after translocation to the nucleus. On the other hand, several effector studies using transgenic overexpression lines have revealed opposite effects on plant immunity (Bhadauria et al., 2013; Dagvadorj et al., 2017; Selin et al., 2016). Thus, our understanding of the plant‐beneficial behaviour patterns of these effector candidates is inconclusive.
In this study, we unravelled the function of MoHTR3 in pathogenicity during host plant infection. The closest relatives of MoHTR3 are the nuclear effectors MoHTR1 and MoHTR2 (Kim et al., 2020). We found MoHTR3 predominantly accumulated in the BIC and translocated to the host cell nuclei. MoHTR3 interferes with the expression of plant defence‐related genes that contribute to JA metabolism and immune adaptation. MoHTR3 overexpression interferes with M. oryzae virulence and results in reduced symptom development on the host plant, in accordance with previous results from MoHTR1‐ and MoHTR2‐expressing rice plants. Taken together, we found that MoHTR3 transcriptionally affects the plant defence system to promote host invasion.
2. RESULTS
2.1. Identification of the effector gene candidate MoHTR3
In a previous study of candidate effectors that determine the virulence of M. oryzae, we identified the effector gene candidate MoHTR3, which contains a putative Cys2‐His2 (C2H2) zinc finger domain that mediates DNA interaction (Kim et al., 2020). Protein sequence analysis suggested that MoHTR3 harbours a signal peptide (SP) at its N‐terminal region, a nuclear localization signal (NLS) in the middle region, and the C2H2 zinc finger domain at the C‐terminal region (Figure 1a). Sequence analysis revealed that the SP site resides within an intrinsically disordered region, whereas the NLS and the zinc finger domain are located within a region of higher conservation. Due to its short length, the protein MoHTR3 may act as a fungal effector after secretion into the host cell. Phylogenetic analysis revealed that MoHTR1, MoHTR2, and MoHTR3 possess a high degree of conservation within the MoHTR effector group (Figure 1b). Following protein sequence comparison, we found that C2H2 zinc finger domain‐containing proteins in M. oryzae have been subject to divergent evolution but appear to remain genetically close. The close relationship between these genes suggests an evolutionary specialization, with similar biological functions. To shed light on a putative function of MoHTR3, we analysed the phylogenetic distances of putative M. oryzae effectors that are considered to be C2H2 zinc finger domain‐containing transcription factors. We eliminated from the analysis disordered regions such as the SP sequence or protein regions with no predicted domain and considered only C2H2 zinc finger domains. The candidate genes were grouped into clades of particular dedicated biological functions, for example, virulence, germination, conidiation, and appressorium formation (Figures 1c and S1, Table S4). The clade containing MoHTR3 included ZFP3 and MoHTR1, whose encoded proteins function in conidial germination and virulence, suggesting a comparable function of MoHTR3 in pathogenesis. MoHTR1, MoHTR2, and MoHTR3 showed correlative protein structures presenting C2H2 zinc finger domains (Figures 1d and S6a). The secondary structure analysis suggests an effector‐related function of MoHTR3, which might be distinct from those of MoHTR1 and MoHTR2.
FIGURE 1.
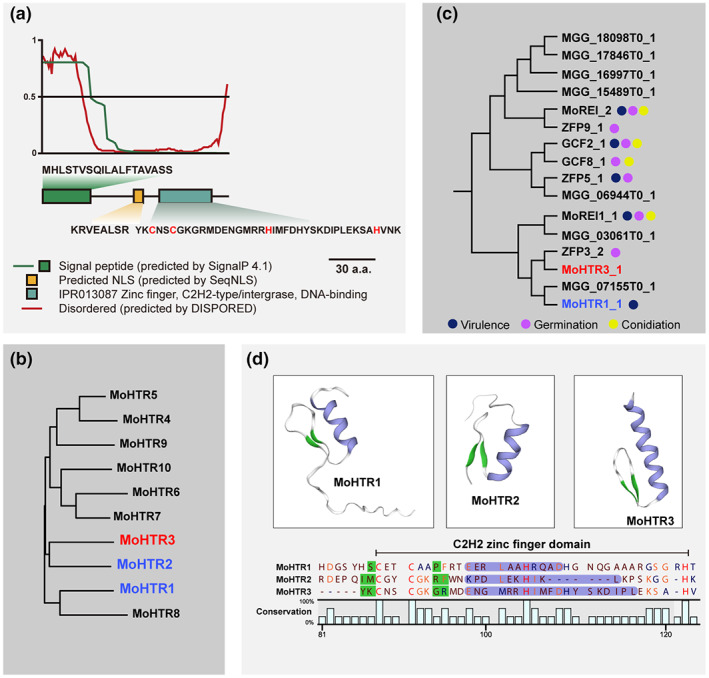
MoHTR3 harbours both a signal peptide (SP) and a nuclear localization signal (NLS). (a) Domain structural analysis of the entire amino acid sequence of MoHTR3. The distribution of putative domains, SP, and NLS is depicted. Disordered regions were predicted at a threshold of 0.5 and an NLS score cut‐off of 0.1. (b) Phylogenetic tree of the Cys2‐His2 (C2H2) zinc finger domain‐containing effectors in Magnaporthe oryzae. (c) A simplified phylogenetic tree generated by using genes encoding C2H2 zinc finger domain‐containing proteins in M. oryzae. A neighbour‐joining domain tree was generated using the single C2H2 zinc finger domains. Effector candidates and MoHTR3 are indicated in blue and red, respectively. (d) The models of secondary structures harbouring a C2H2 zinc finger domain of three MoHTRs were predicted via SWISS‐MODEL. Purple and green structures and boxes in alignments indicate α‐helices and β‐strands, respectively.
2.2. MoHTR3 is conserved in M. oryzae
We analysed the well‐annotated genomes of M. oryzae strains 70‐15 (from Oryza sativa), B71 (Triticum aestivum), Y34 (O. sativa), BR32 (T. aestivum), Guy11 (O. sativa), and MZ5‐1‐6 (Elusine coracana), with the outgroups Aspergillus nidulans and Fusarium graminearum (Chiapello et al., 2015; Farman et al., 2017; Inoue et al., 2017; Peng et al., 2019).
Interestingly, nuclear effector candidates of the C2H2 zinc finger family were only found in M. oryzae and could not be detected in the other fungi (Table S1). Furthermore, MoHTR3 was distributed in the genomes of differentially isolated M. oryzae strains including KJ201 (Table S2). The surrounding gene loci of MoHTR3's genetic position indicate strict conservation of MoHTR3 in the M. oryzae strains (Figure 2a). The M. oryzae strains KJ201, Y34, 70‐15, and Guy11 predominantly infect rice, whereas MZ5‐1‐6, BR32, and B71 infect other crops, for example, wheat and finger millet. Interestingly, the orthologous gene structures in each strain were identical in terms of the C2H2 zinc finger domain and SP (Figures 2b and S2). Among these strains, single amino acid changes in the sequences adjoining the C2H2 zinc finger domain indicate phylogenetic divergence during individual differentiation. The strains KJ201, Y34, 70‐15, and Guy11 showed sequence variation in front of the zinc finger domain in only one amino acid, at position 61. KJ201 and Y34 carry at this position an aspartate (D) residue, while 70‐15 and Guy11 harbour an asparagine (N) residue. However, MZ5‐1‐6, BR32, and B71 showed five amino acid variations, at positions 93, 99, 103, 105, and 117, compared with KJ201. Interestingly, these amino acid changes are highly conserved among MZ5‐1‐6, BR32, and B71, suggesting an evolutionary selection for this amino acid sequence. Four of these amino acid exchanges reside within the zinc finger domain. Zinc finger domains enable protein–DNA interaction and variations in the position and properties of particular amino acids interfere with the DNA‐binding ability of the zinc finger domain and eventually the activation of downstream target gene expression (Hirano et al., 2017; Völz, Kim, Mi, Rawat, et al., 2019a).
FIGURE 2.
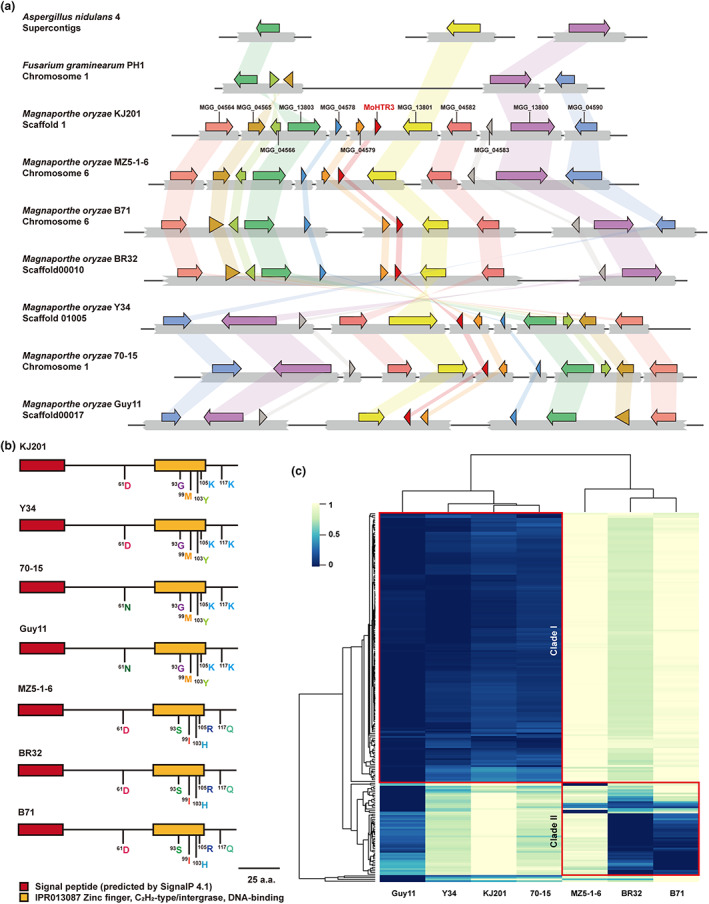
MoHTR3 is a Magnaporthe oryzae‐specific effector gene. (a) Comparative analysis of the genomic region containing MoHTR3 in various fungal strains. (b) Gene structure comparison of MoHTR3's orthologues in seven strains. (c) Genome distances analysed using GGDC.
The genome sequence similarity of all tested strains was about 90%; however, their genomic distances were distinct. Each of the four strains comprised independent clusters, with KJ201, Guy11, Y34, and 70‐15 grouped in Clade I and the other strains closely related within Clade II (Figure 2c). These results suggest that the detected divergence was unrelated to MoHTR3. Four strains isolated from O. sativa showed a close genetic relationship. Comparative analysis among the strains suggests that MoHTR3 plays a host‐specialized role in M. oryzae.
2.3. MoHTR3 localizes to the BIC and host cell nucleus
The effector candidate gene MoHTR3 was predominantly expressed in invasive hyphae at 36 h postinoculation (hpi) (Figure 3a). The expression patterns of MoHTR3 suggest that it may act as an effector at the biotrophic stage. Therefore, we analysed the localization of MoHTR3 within the invasive hyphae. Fluorescent protein‐tagged transformants expressing mRFP1 at the C‐terminus of MoHTR3 were used for infection assays. MoHTR3:mRFP1 driven by the native MoHTR3pro promoter showed dense accumulation in the BIC (Figure 3b). To gain more insight into MoHTR3:mRFP1 localization and to overcome observation constraints caused by the low protein abundance, in subsequent experiments MoHTR3:mRFP1 expression was driven by the PWL2pro promoter. The transcriptional activity of PWL2pro was determined to be about eightfold higher than that of MoHTR3pro in the transcriptome profiling of rice blast genes during infection (Figure S6b) (Jeon et al., 2020). MoHTR3:mRFP1 driven by PWL2pro showed high expression during the biotrophic stage, and we detected MoHTR3:mRFP1 fluorescence signals in the BIC and in the nuclei of infected and adjacent cells (Figure 3c). To confirm the localization of MoHTR3 in the BIC and nucleus, we used PWL2:eGFP:NLS, driven by PWL2pro (Khang et al., 2010). We found that MoHTR3:mRFP1 colocalized with PWL2:GFP in the BIC and the nucleus of infected plant cells (Figure 3c). The translocation of MoHTR3 from the BIC into nuclei implies that it may affect the expression of defence‐related genes.
FIGURE 3.
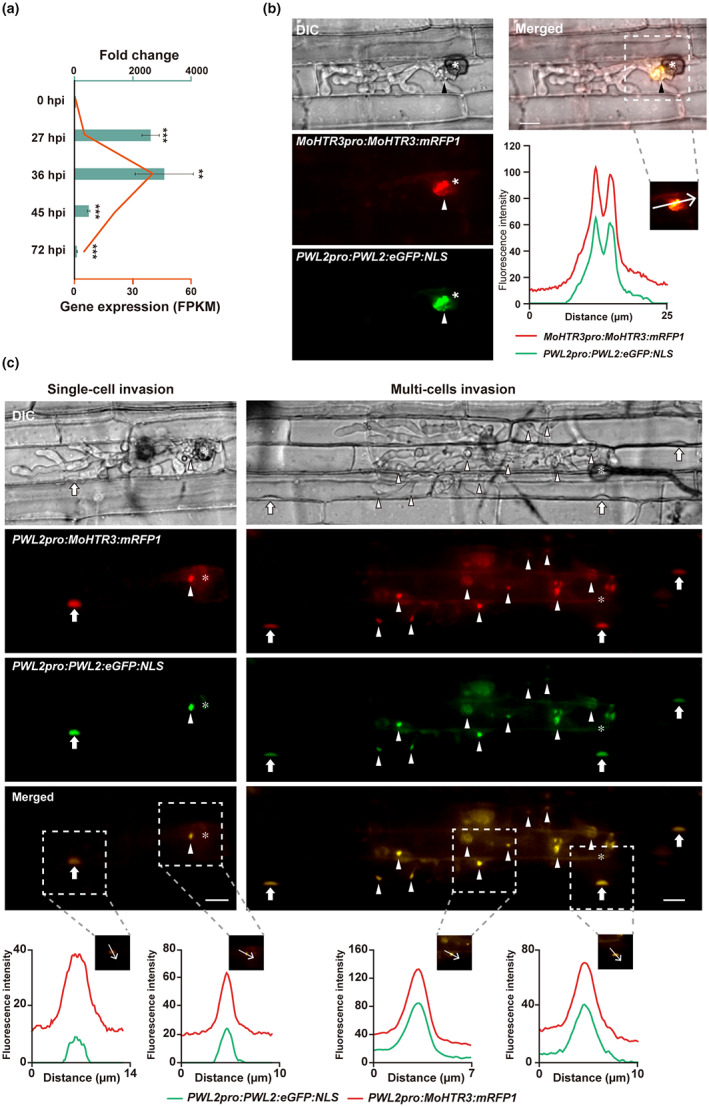
MoHTR3 is expressed in the biotrophic stage and accumulates in the biotrophic interfacial complex (BIC) and plant nuclei. (a) In planta expression of MoHTR3 at different time points after infection by reverse transcription‐quantitative PCR (RT‐qPCR). RNA sequencing‐based expression data (Jeon et al., 2020) were reanalysed on MoHTR3 transcript abundance (fragments per kb of transcript per million mapped reads [FPKM]). (b) To observe the location of MoHTR3, we used an mRFP1‐tagged strain under the control of the native promoter MoHTR3pro. PWL2pro:PWL2:eGFP:NLS was codetected after sheath inoculation at 36 h postinoculation (hpi). (c) Translocation and nuclear accumulation of MoHTR3, driven by the PWL2pro promoter, producing high protein abundance. MoHTR3:mRFP1 colocalizes with PWL2:eGFP:NLS in the host plant. Arrowhead, BIC; white arrow, plant nucleus; asterisk, infection site; DIC, differential interference contrast microscopy. The area of fluorescence intensity determination is indicated within the merged image. Scale bars, 10 μm.
2.4. The MoHTR3′ SP domain is required for the localization at the BIC
We assumed a localization peptide resided within the putative SP at the N‐terminus of MoHTR3, which might be required for targeting MoHTR3 to the BIC. Thus, we generated an M. oryzae mutant strain that expresses the MoHTR3pro:MoHTR3 ΔSP:mRFP1transgene, resulting in MoHTR3ΔSP, without the SP domain, driven by the endogenous promoter. Rice sheath cells were inoculated with this strain. We detected a fluorescence signal in the cytoplasm of invasive hyphae but we could not capture a signal within the BIC (Figures 4a,b and S3b,c, Movies S1 and S2). A determination of the fluorescence intensity confirmed the cytoplasmic localization of MoHTR3ΔSP (Figure 4b). These results reveal an inherent localization peptide within the SP domain that is necessary for MoHTR3's localization in the BIC.
FIGURE 4.
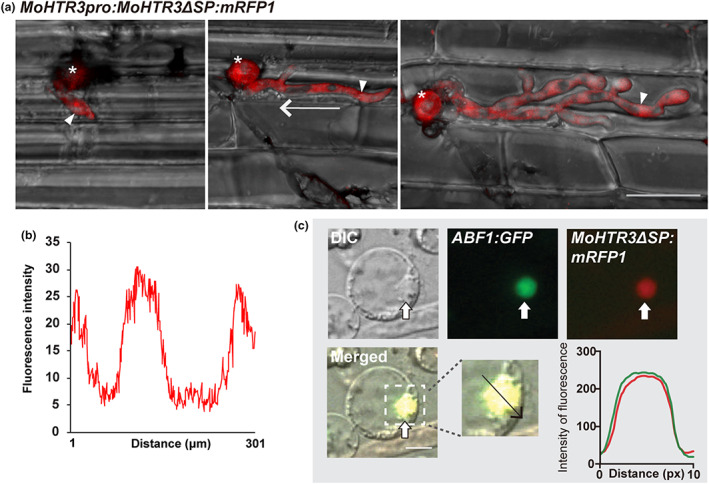
MoHTR3 localization to the biotrophic interfacial complex (BIC) depends on the signal peptide (SP) domain. (a, b) Representative confocal laser‐scanning microscopy images of the localization of MoHTR3ΔSP tagged with mRFP1 at different developmental stages until 36 h postinoculation. Depicted are 3D maximum projections of z‐stacks, with a focal plane distance of 1 μm within a total range of about 20 μm. Animated 3D projections are provided in Movies S1 and S2 (MoHTR3:mRFP1 control in Figure S3c and Movie S1). White arrowhead, invasive hypha; asterisk, infection site; white arrow indicates the area of fluorescence intensity measurement in (b). (c) MoHTR3 without the signal peptide domain (ΔSP) was localized in rice protoplasts. MoHTR3ΔSP colocalizes with the nuclear marker ABF1:GFP. White arrow, plant nucleus; the black arrow indicates the area of fluorescence intensity determination; DIC, differential interference microscopy. Scale bars, 20 μm (a) and 5 μm (c).
2.5. Nuclear localization of MoHTR3 in rice protoplasts
To further elucidate the mechanisms underlying MoHTR3 localization, we focused on the function of the SP at the N‐terminus (Figure 1a). We removed the SP sequence at the N‐terminus (MoHTR3ΔSP) to analyse its subcellular localization and expressed MoHTR3ΔSP driven by the CaMV 35S promoter in rice protoplasts. MoHTR3ΔSP tagged with mRFP1 was detected in the nuclei, and the fluorescence signals fully overlapped with the nuclear localization control ABF1:GFP (Figure 4c). This outcome shows that the NLS does not reside within the SP domain, but can be found in the remaining peptide sequence of MoHTR3. Furthermore, MoHTR3ΔSP linked to the NLS from simian virus 40 large T antigen was also detected in the nuclei (Figure S3). Together, these results indicate that the SP domain is unrelated to MoHTR3's nuclear localization and an NLS is predicted to be located towards the C‐terminus of MoHTR3.
2.6. MoHTR3 is involved in the generation of disease lesions
Functional characterization of MoHTR3 was performed using the knockout strain ΔMohtr3, a complementation strain, and the two overexpression strains MoHTR3ox1 and MoHTR3ox2, which express MoHTR3 under the control of the ubiquitously active EF1a promoter from Fusarium verticillioides (Figure S4). We inoculated the conidial suspension on rice leaves. At 5 days postinoculation into rice leaves, lesion formation in ΔMohtr3 and the complementation line were indistinguishable from that of the wild type (WT). However, MoHTR3ox strains showed reduced virulence (Figure 5a,b). On the WT‐infected leaves, disease lesions with dark margins were generated; however, infection with MoHTR3ox strains was weaker and resulted in small disease lesions, reflecting delayed invasive growth in rice sheaths. Thus, we analysed the infection process in WT and overexpression strains MoHTR3ox1 and MoHTR3ox2 at 36 hpi. We found that the invasive growth in the first and second host cell was delayed in MoHTR3ox1 and MoHTR3ox2, in accordance with the reduced lesion formation (Figure S5). These results are intriguing and correspond to the reduced susceptibility of rice plants that constitutively express the MoHTR3 relatives MoHTR1 and MoHTR2.
FIGURE 5.
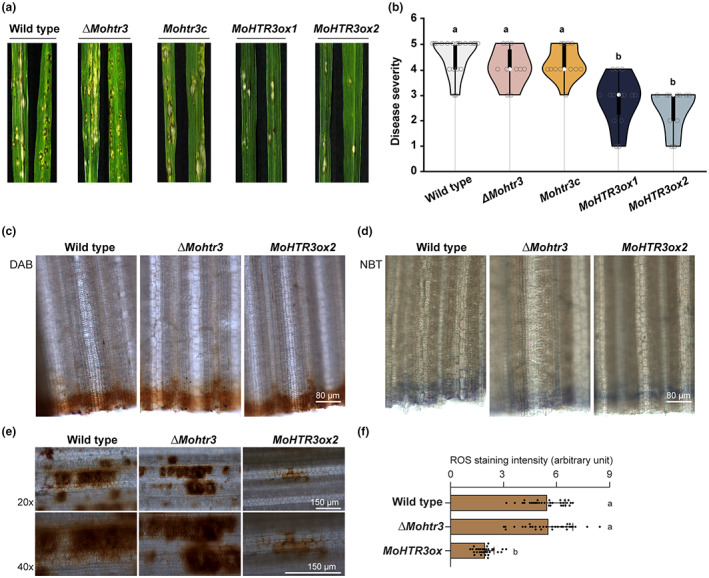
Pathogenicity study of knockout strain ΔMohtr3 and overexpression strains MoHTR3ox. (a) Disease symptoms on rice leaves at 5 days postinoculation following conidial spraying of Magnaporthe oryzae (WT), ΔMohtr3, complemented Mohtr3c, and MoHTR3ox strains. (b) Quantified data visualization was performed using the BoxPlotR web tool. (c, e) 3,3′‐Diaminobenzidine (DAB) staining was performed to detect hydrogen peroxide (H2O2). (d) Nitroblue tetrazolium (NBT) staining was performed to detect superoxide. The staining at the bottom (c, d) is due to the cut edge of the leaf and indicates consistent staining conditions. (f) Quantified DAB staining intensity at the site of infection. Letters over bars indicate significant difference as determined using one‐way analysis of variance (p < 0.05). All experiments were performed with three biological replicates. ROS, reactive oxygen species.
We conclude that MoHTR3 expressed in rice plants might prime the plant immune system, thereby interfering with the virulence of M. oryzae. Therefore, we assume that MoHTR3 targets the plant defence system by transcriptional reprogramming of immunity‐associated genes.
2.7. Different hydrogen peroxide accumulation after MoHTR3ox infection
The accumulation of ROS is one of the first defence reactions in response to a pathogen attack and determines the degree of susceptibility of a host plant (Apel & Hirt, 2004; Lee et al., 2021). Thus, to characterize the higher resistance phenotype of rice leaves after MoHTR3ox infection, we analysed ROS homoeostasis after WT and ΔMohtr3 infection. We carried out 3,3′‐diaminobenzidine (DAB) staining to detect the ROS hydrogen peroxide (H2O2) in plant cells. H2O2 is produced as a by‐product of respiration, and can also be released upon biotic stress. When DAB is added to a sample of plant cells, it reacts with H2O2 to form a brown precipitate that can be observed under a microscope. To detect the ROS superoxide (O2−) in plant cells, we conducted nitroblue tetrazolium (NBT) staining. When NBT is added to a sample of plant cells, it is reduced by the enzyme NADH‐dependent succinate dehydrogenase, which is present in the mitochondria of active plant cells. The reduced NBT forms a blue‐black precipitate, which can be detected under a microscope. Following MoHTR3ox application, the DAB and NBT staining intensity of infected leaves was indistinguishable from that of leaves infected with WT and the ΔMohtr3 strain (Figure 5c,d). This result suggests that systemic ROS homeostasis in rice leaves is not affected by MoHTR3 overexpression. However, we found that the local H2O2 accumulation at the infection site differed after MoHTR3ox or WT infection (Figure 5e,f). Following MoHTR3ox administration, the DAB staining intensity at the infection sites on rice leaves was significantly lower than after WT or ΔMohtr3 infection, suggesting that MoHTR3ox interferes with H2O2 production.
2.8. MoHTR3 modulates plant hormonal synthesis pathways
During M. oryzae–O. sativa interactions, genes related to plant immune responses are highly expressed (Jeon et al., 2020). These genes, which include basal defence response genes, plant hormone‐related genes, and hypersensitive response (HR)‐related genes, showed increased expression in the biotrophic stage (Figure 6a). To determine the function of MoHTR3 in planta, we examined the expression changes of plant defence‐related genes after inoculation of WT, the knockout strain, and two overexpression strains at 36 hpi. We found that specific sets of plant defence‐associated genes were differentially expressed after infection (Figure 6b–h). The JA biosynthesis genes AOS2 (encoding allene oxide synthase 2) (Haga & Iino, 2004), LOX2 (encoding lipoxygenase 2) (Wang et al., 2008; Zhang et al., 2018), and OPR7 (encoding 12‐oxophytodienoate reductase 7) (Tani et al., 2008) were down‐regulated in ΔMohtr3‐infected rice but up‐regulated in MoHTR3ox‐infected rice. These results indicate that MoHTR3 promotes the expression of genes that contribute to JA metabolism. Interestingly, differential gene expression was accompanied by substantial transcriptional changes of the JA signalling pathway (Figure 6b). JAMYB (encoding a MYB transcription factor involved in the JA signalling pathway) (Yokotani et al., 2013) and JMT1 (jasmonic acid carboxy methyltransferase 1) (Qi et al., 2016) showed high expression after MoHTR3ox infection and low expression after ΔMohtr3 infection. POX22.3 (peroxidase 22.3) (Chittoor et al., 1997) showed high expression during deletion mutant infection. EDS1 (enhanced disease susceptibility 1) (Xie et al., 2011), NPR1 (nonexpressor of pathogenesis‐related genes 1) (Chern et al., 2005), and PAL (phenylalanine ammonia‐lyase) (Tonnessen et al., 2015), which contribute to SA signalling (Figure 6e), APO1 (aberrant panicle organization 1) (Ikeda et al., 2005) and P450 (cytochrome P450) (Tanabe et al., 2005), which are involved in the P450‐related response (Figure 6c), the HR‐related gene HSR203J (HR‐related protein 203J) (Huang et al., 2007) (Figure 6g), and ACS1 (aminocyclopropane‐1‐carboxylate synthesis 1) (Zarembinski & Theologis, 1997) and EBP89 (ethylene‐responsive element binding protein transcription factor) (Shen & Wang, 2004), which contribute to ET metabolism/signalling, all showed significant expression changes following inoculation with MoHTR3ox or ΔMohtr3 (Figure 6f). The defence markers PR1a, PR1b, PR4, and PR5 showed reduced expression after ΔMohtr3 infection, indicating that MoHTR3 functions in fungal virulence (Figure 6d). After infection with either the deletion strain or an overexpression strain, PR3 expression was increased. Together, these data suggest that MoHTR3 regulates various genes contributing to defence‐associated pathways to modulate the plant immune system. We previously showed that the closest homologues of MoHTR3, MoHTR1 and MoHTR2, bind to specific effector‐binding elements in the promoter regions of their target genes, OsMYB4 and OsWRKY45. Ectopic expression of MoHTR1 and MoHTR2 in transgenic rice lines represses the expression of these genes involved in plant defence. We questioned whether MoHTR3 might act redundantly with MoHTR1 and MoHTR2 and affect the expression of OsMYB4 and OsWRKY45. To investigate this, we analysed the expression of OsMYB4 and OsWRKY45 after infection with WT and MoHTR3ox. As depicted in Figure S3d, we could not detect differences in the expression of OsMYB4 and OsWRKY45 following infection with WT and MoHTR3ox, suggesting a function of MoHTR3 that is distinguishable from that of MoHTR1 and MoHTR2.
FIGURE 6.
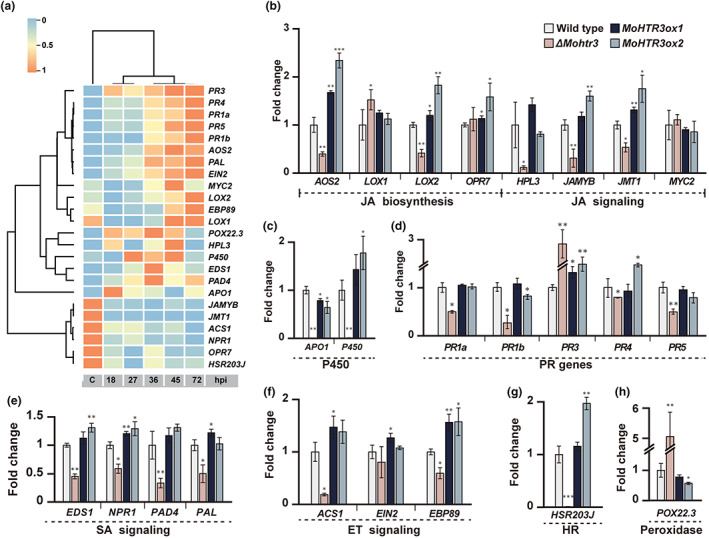
MoHTR3 affects the expression of plant defence‐associated genes. (a) Expression of rice defence‐related genes during the interaction between Magnaporthe oryzae and rice (Jeon et al., 2020). The expression of hypersensitive response (HR)‐related, peroxidase response‐related, P450 defence‐related, pathogenesis‐related (PR), ethylene (ET) signalling‐related, jasmonic acid (JA) signalling‐related, and salicylic acid (SA) signalling‐related genes is indicated. Three biological replicates were performed. (b) JA synthesis‐related genes. (c) P450 defence‐related genes. (d) PR genes. (e) SA signalling‐related genes. (f) ET signalling‐related genes. (g) HR‐related gene. (h) Peroxidase gene. Significance (b–h) was determined by the t test. Error bars indicate standard deviation (SD). Expression levels were calculated using the 2−ΔΔCt method, and β‐tubulin was used for normalization. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001).
3. DISCUSSION
Despite the economic importance of fungal plant pathogens, information about their nuclear effectors remains limited. Genetic studies of many fungal pathogens are challenging due to their diverse lifestyles, but various model systems have facilitated the discovery of fungal nuclear effectors (O'Connell et al., 2012; Völz et al., 2020). Here we show that the virulence factor MoHTR3 of M. oryzae functions as a host nucleus‐targeted effector that regulates the expression of specific subsets of defence‐associated genes involved in JA, SA, and ET metabolism and signalling. JA/ET are known to act antagonistically to SA in the coordination of defence against pathogens with different lifestyles (Koornneef & Pieterse, 2008; Pieterse et al., 2012).
We found that JA‐related genes were down‐regulated after infection with ΔMohtr3, whereas MoHTR3ox predominantly induced the expression of JA‐associated genes. In a previous study (Jeon et al., 2020), JA, ET, and SA signalling‐related genes showed elevated transcript levels during early M. oryzae infection in the biotrophic stage. We found that the JA metabolism‐related genes AOS2, LOX2, and JAMYB were up‐regulated during early MoHTR3ox infection, in accordance with the SA signalling genes EDS1 and PAL and the SA receptor gene NPR1. The ET biosynthesis gene ACS1 and the ET signalling gene EIN2 also showed higher transcript levels. This result suggests a multilayered control of hormonal pathways by MoHTR3. The transcriptional profiles of ΔMohtr3 and MoHTR3ox suggest redundant functions of MoHTR3 with other M. oryzae effectors that interfere with the plant's hormonal metabolism and defence response, in favour of plant colonization. We suggest that MoHTR3 misleads the plant immune system by promoting the JA‐mediated defence response, which tackles necrotrophic pathogen invasion, thereby counteracting defence strategies against hemibiotrophs such as M. oryzae.
Small, secreted effectors that influence the plant's hormonal system have been reported in various pathogens (Khan et al., 2018). For example, Cmu1, Pit, and Tin2 in U. maydis are secreted and influence host immune strategies, accompanied by changing plant hormone levels (Brefort et al., 2014; Djamei et al., 2011; Doehlemann et al., 2011). Botrytis cinerea promotes the accumulation of host‐derived indole‐3‐acetyl‐aspartate, and its secreted exopolysaccharide affects JA‐mediated signalling (El Oirdi et al., 2011; Prins et al., 2000). Sclerotinia sclerotiorum modulates the host SA concentration through a fungal effector protein (Penn & Daniel, 2013).
MoHTR3 can only be found in a subset of M. oryzae strains, showing that the arms race between pathogen and host continuously creates new factors to challenge the tightly interlocked host–pathogen interplay. Effector proteins in M. oryzae have been reported as pivotal factors of hemibiotrophic fungi that exhibit both biotrophy and necrotrophy (Stergiopoulos & de Wit, 2009). Rather than producing toxic secondary materials that kill its host, this plant‐pathogenic fungus fills the host cell, takes up all its nutrients, and infects the entire host plant (Horbach et al., 2011). For colonization of the host, the biotrophic stage, which starts right after the infection, is of paramount importance (Chaudhari et al., 2014; Koeck et al., 2011). After that, M. oryzae changes to a necrotrophic lifestyle accompanied by the generation of disease lesions on rice leaves (Wang et al., 2016). Necrotrophic pathogens produce analogues of plant hormones to influence plant metabolism for the benefit of the pathogens (Mengiste, 2012; Sharon et al., 2007). MoHTR3ox‐infected plants showed reduced lesion formation, H2O2 accumulation, and susceptibility, which might be caused by the MoHTR3‐coordinated modulation of plant hormone‐related gene expression. The invasive growth of MoHTR3ox strains was delayed at 36 hpi (Figure S5). This finding suggests that the colonization, after effector secretion in MoHTRox strains, is affected according to enhanced expression of plant defence genes. Furthermore, the amount of MoHTR3 might be increased in MoHTR3ox strains (Figure S4b) and consequently unbalanced, which seems to interfere with a properly fine‐tuned induction of plant defence gene expression, which results in the gain‐of‐function phenotype. We assume that MoHTR3 shapes the plant defence hormone repertoire and host colonization of M. oryzae. These noncanonical functions of MoHTR3 in the host plant might pave the way to discover the mechanism of direct/indirect interactions regulated by pathogenic effector proteins.
Effector‐encoding genes generally show high expression during the interaction with the host (Stergiopoulos & de Wit, 2009; Zhang & Xu, 2014). Avr effectors, MoHTRs, and other effector candidates in M. oryzae are differentially expressed in the biotrophic stage (Kim et al., 2019, 2020). The manipulation of nuclear processes by plant‐interacting fungi is a prevalent mechanism for their survival. Recently, two M. oryzae effectors (MoHTR1 and MoHTR2) were found to be translocated into the nuclei of infected rice cells, increasing their susceptibility to hemibiotrophic pathogens. These effectors reprogrammed clusters of immunity‐associated genes and down‐regulated the expression of OsMYB4 and OsWRKY45. These effectors have variable effects on rice immunity, depending on the lifestyle of the pathogen. MoHTR3 was highly expressed in the biotrophic stage and was identified as the C2H2 zinc finger‐containing effector candidate most closely related to MoHTR1 and MoHTR2 (Kim et al., 2020). Structural domains are highly similar among these three genes, which all contain a C‐terminal regulator domain, SP, and NLS (Figure S6a). The close relationship of MoHTR3 with MoHTR1 and MoHTR2 suggests a DNA‐binding ability of MoHTR3 and overlapping functions with MoHTR1 and MoHTR2.
In this concern, studies in Phytophthora sojae showed that the two effector proteins PsCRN161 and PsCRN115 enhance the plant defence response against the pathogen and tolerance to abiotic stress by up‐regulation of defence‐related genes (Rajput et al., 2015; Zhang et al., 2015). The M. oryzae effector MoSM1 belongs to the cerato‐platanin family and enhances plant immunity against different rice pathogens. In MoSM1‐overexpressing transgenic rice, the development of rice blast disease and bacterial blight disease is compromised, leading to hormonal changes in rice (Hong et al., 2017). MoSDT1 overexpression studies revealed that the effector protein improves rice blast disease resistance by modulating the expression of the host gene OsBsr‐d1, thereby inducing an immune response in rice (Wang, Li, et al., 2019a).
In general, small secreted proteins such as those nuclear effectors can be traced back to gene birth in the pathogen's genome (Stergiopoulos & de Wit, 2009). They frequently show a species‐specific distribution in their kingdom and evolved individually (Stergiopoulos et al., 2012). Likewise, these hallmarks imply that unconserved effectors, including MoHTR3, are adapted to differentiated pathogenesis among individual plant‐pathogenic fungi.
An in silico study suggested that MoHTR3 possesses a native NLS in the middle of the amino acid sequence. MoHTR3 was shown to localize to the BIC and nuclei of infected plant cells. An effector with the potential to reorchestrate the plant immune system should be able to move into the plant cell. Biotrophy‐associated secreted (BAS) proteins are secreted into the BIC; in particular, BAS4 has been shown to localize to the extra‐invasive hyphal membrane (Mosquera et al., 2009). BAS4 has been reported to contribute to plant immune responses during the biotrophic and necrotrophic stages by causing plant cell death (Wang, Liu, et al., 2019b). Indeed, a secreted protein that cannot be found in the plant cytoplasm may affect interactions between the plant and pathogen. The previously reported plant nucleus‐localized effector proteins PWL2 (Khang et al., 2010), MoHTR1, and MoHTR2 (Kim et al., 2020) in M. oryzae, See1 in U. maydis (Redkar et al., 2015), PstGSRE1 in Puccinia striiformis f. sp. tritici (Qi et al., 2019), and CgEP1 in C. graminicola (Vargas et al., 2016) have also been considered as cytoplasmic effectors modulating pathogenesis‐related processes. Likewise, cytoplasmic effectors are often translocated into plant nuclei during the reprogramming of plant immune responses (Giraldo & Valent, 2013).
In conclusion, we showed that the C2H2 zinc finger domain‐containing effector MoHTR3 is translocated into the plant nuclei after secretion into the BIC. During biotrophy, secreted MoHTR3 alters the expression of plant defence‐related genes with a consequential pathogenic effect. Together, these results demonstrate that MoHTR3 encodes a nuclear effector that modulates plant defence hormone pathways at various levels, benefitting the fungal invader.
4. EXPERIMENTAL PROCEDURES
4.1. Identification of MoHTR3 and culture conditions
We used a strain isolated from infected rice, M. oryzae KJ201. It was obtained from the Center for Fungal Genetic Resources (http://knrrb.knrrc.or.kr) at Seoul National University, Seoul, South Korea. MoHTR3's secretion ability and the domains it contains were introduced as previously described by using the information archived in the Fungal Secretome Database (http://fsd.snu.ac.kr) and the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr), respectively (Choi et al., 2010; Park et al., 2008).
To induce production of conidia, all strains were cultured on V8 juice agar (80 mL V8 juice, 310 μL 10 M NaOH, and 15 g agar per L) at 25°C under continuous light. Mycelial growth was measured after growing on modified complete agar medium as previously described (Talbot et al., 1997). Genomic DNA and RNA were prepared from vegetative mycelia cultured in liquid CM (6 g yeast extract, 6 g casamino acids, and 10 g sucrose per L) for 4 days at 25°C on a shaking incubator at 200 rpm.
4.2. Mutant strain generation using fungal protoplasts
MoHTR3 knockout mutant and overexpression strains were generated via homologous recombination using amplified fragments harbouring the hygromycin B phosphotransferase gene cassette derived from pBCATPH (Chung et al., 2013) as the selection marker. The individual fragments were constructed using a double‐joint PCR method (Yu et al., 2004). The final constructs were introduced into KJ201 protoplasts as previously reported (Bolton & Thomma, 2012). Transformants were regenerated on TB3 agar medium (20% sucrose, 1% glucose, 0.3% yeast extract, 0.3% casamino acids, and 0.8% agar) supplemented with hygromycin B (200 ppm). For the selection of mutants, we used a two‐step DNA‐based screening system, PCR, and Southern blot. Genomic DNA of each mutant was analysed by PCR in a C1000 thermal cycler (Bio‐Rad). Each PCR was performed in a reaction volume of 10 μL, composed of 1 μL of qRTF and qRTR primers for each gene (100 nM for each primer), 5 μL of 2× PCR Master mix solution (i‐StarMAX II), dNTPs, PCR buffer, i‐StarMAX DNA polymerase, and loading dye (iNtRON Biotechnology). In the second step, Southern hybridization was performed following the standard procedure (Sambrook & Russell, 2001). A 5′‐ or 3′‐flanking region of each mutagenized gene was used as a probe. Probe labelling with 32P was performed using the Rediprime II Random Prime Labeling System kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Complemented strains were generated by co‐transforming each gene construct, including a native promoter and an open reading frame region, and pII99, a vector containing the geneticin resistance gene as a selection marker. Each gene construct consisted of the open reading frame and its 5′‐ and 3′‐flanking regions. The resulting transformants were selected using TB3 agar medium supplemented with geneticin (800 ppm). All primers used in this study are listed in Table S2. All strains produced in this study were deposited into the Center for Fungal Genetic Resources database.
4.3. Rice protoplast generation and transfection
The rice protoplasts were extracted from both leaves and stems of 2‐week‐old etiolated rice seedlings incubated in 1/2× MS agar (2.2 g Murashige–Skoog medium, 10 g sucrose, and 8 g agar per L) using a previously described method (Kim et al., 2020).
The polyethylene glycol‐mediated transfection method was used in this study. The transfected rice protoplasts were incubated for at least 18 h at 25°C. To elucidate the localization of each protein, the promoter regions were replaced with the CaMV 35S promoter and fluorescent proteins (eGFP or mRFP1) were fused to their C‐terminal regions.
4.4. Rice infection assays
A conidial solution was used to infect rice leaves and sheaths. Harvested conidia (105 conidia/mL) suspended in 250 ppm Tween 20 were sprayed onto leaves of Oryza sativa ‘Nakdongbyeo’, a cultivar susceptible to KJ201, at the four‐leaf stage. The seedlings were incubated in a dew chamber at 25°C for 24 h in darkness and subsequently moved to a growth chamber at 28°C and 80% humidity with a photoperiod of 16 h light (Valent & Chumley, 1991). Five days after inoculation, each blast lesion was collected and evaluated using a previously used disease scoring system (Valent et al., 1991). For visualization of the pathogenicity data, we used the web‐based tool BoxPlotR (http://shiny.chemgrid.org/boxplotr/).
4.5. Gene expression analysis
Total RNA was extracted from frozen mycelia and rice leaves infected with the deletion mutant, overexpression strains, and WT M. oryzae using the Easy‐Spin total RNA extraction kit (iNtRON Biotechnology) according to the manufacturer's instructions. A total of 5 μg of RNA was reverse transcribed using oligo(dT) primers and an ImProm‐II reverse transcription system (Promega). Quantitative real‐time PCR (qPCR) was performed in a reaction volume of 10 μL, including 2 μL of cDNA template (12.5 ng/μL), 3 μL of primer pair (100 nM for each primer), and 5 μL of 2× Rotor‐Gene SYBR Green PCR Master Mix (Qiagen) in a Rotor‐Gene Q2plex (Qiagen). The cycling conditions were as follows: 3 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Relative gene expression levels were calculated via the 2−ΔΔCt method using the β‐tubulin gene for normalization (ΔΔCt = (Cttarget gene − Ct β‐tubulin )treated − (Cttarget gene − Ct β‐tubulin )control). The primers used for reverse transcription‐qPCR are listed in Table S3.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
Supporting information
Figure S1 Phylogenetic analysis of genes encoding C2H2 zinc finger domain‐containing proteins in Magnaporthe oryzae. A neighbour‐joining circular domain tree was generated based on the C2H2 zinc finger domain. Effector candidates and MoHTR3 are indicated in blue and red characters, respectively.
Figure S2 Alignment of MoHTR3 orthologues in seven Magnaporthe oryzae strains. Sequence alignment of seven MoHTR3 orthologues using ClustalW. Asterisks indicate single amino acid differences.
Figure S3 (a) MoHTR3 without signal peptide (SP) but with exogenous simian virus40 large T antigen nuclear localization signal (NLS) is localized in the nuclei in rice protoplasts. Nuclei in the rice protoplasts are indicated with ABF1:GFP. ImageJ was used to measure the fluorescence intensity. Scale bar indicates 5 μm. (b) The localization of MoHTR3ΔSP tagged with mRFP1 was observed at 36 h postinoculation by an epifluorescence microscope. Scale bar indicates 10 µm. (a, b) White arrow, plant; asterisk, infection site. The measured area of fluorescence intensity is shown in the merged image. (c) Expression of OsMYB4 and OsWRKY45 following MoHTR3ox infection.
Figure S4 Generation of fungal mutant strains. (a) MoHTR3 was replaced with a hygromycin B phosphotransferase resistance (HPH) cassette in the Magnaporthe oryzae genome using homologous recombination. (b) The MoHTR3 promoter was replaced. Southern blot analysis was conducted to prove there is a single copy in the promoter region of MoHTR3. Expression levels were measured with three biological replicates. Significance is indicated with asterisks (t test) (p < 0.05).
Figure S5 Invasive growth of MoHTR3ox strains in rice sheath cells. Infection severity was quantified by rating the perception of infection progression. Representative microscopic images are shown. Scale bar = 10 μm.
Figure S6 (a) Protein structure comparison among MoHTR3, MoHTR1, and MoHTR2. Full amino acid sequences of these three effector proteins were analysed. The distribution of the putative domain, signal peptide, and nuclear localization signal (NLS) is shown. (b) FPKM values of the Magnaporthe oryzae genes PWL2, MoHTR1, MoHTR2, and MoHTR3 at different time points during infection at the biotrophic stage (Jeon et al., 2020).
Movie S1 Animated 3D projections of confocal laser‐scanning microscopy z‐stacks of MoHTR3:mRFP1.
Movie S2 Animated 3D projections of MoHTR3ΔSP:mRFP1 localization in the invasive hyphae.
Table S1 MoHTRs are Magnaporthe oryzae‐specific effectors.
Table S2 MoHTR3 is conserved in Magnaporthe oryzae strains.
Table S3 Primers used in this study.
Table S4 Previously studied C2H2 zinc finger transcription factor genes.
ACKNOWLEDGEMENTS
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Ministry of Science and ICT (MSIT) (2020R1A2B5B03096402, 2018R1A5A1023599, and 2021M3H9A1096935). S.L. is grateful for a graduate fellowship from the Brain Korea 21 Plus Program.
Lee, S. , Völz, R. , Lim, Y.‐J. , Harris, W. , Kim, S. & Lee, Y.‐H. (2023) The nuclear effector MoHTR3 of Magnaporthe oryzae modulates host defence signalling in the biotrophic stage of rice infection. Molecular Plant Pathology, 24, 602–615. Available from: 10.1111/mpp.13326
Sehee Lee and Ronny Völz equally contributed to this study.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and in its Supporting Information online at the publisher's website.
REFERENCES
- Ahmed, M.B. , DOS Santos, K.C.G. , Sanchez, I.B. , Petre, B. , Lorrain, C. , Plourde, M.B. et al. (2018) A rust fungal effector binds plant DNA and modulates transcription. Scientific Reports, 8, 14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K. & Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology, 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Asai, S. & Shirasu, K. (2015) Plant cells under siege: plant immune system versus pathogen effectors. Current Opinion in Plant Biology, 28, 1–8. [DOI] [PubMed] [Google Scholar]
- Bhadauria, V. , Banniza, S. , Vandenberg, A. , Selvaraj, G. & Wei, Y. (2013) Overexpression of a novel biotrophy‐specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryotic Cell, 12, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.D. & Thomma, B.P. (2012) Plant fungal pathogens methods and protocols. In: Bolton M.D. (Ed.) Methods in molecular biology, Vol. 835. Totowa, NJ: Humana. [Google Scholar]
- Brefort, T. , Tanaka, S. , Neidig, N. , Doehlemann, G. , Vincon, V. & Kahmann, R. (2014) Characterization of the largest effector gene cluster of Ustilago maydis . PLoS Pathogens, 10, e1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M. , Chen, H. , Liu, F. & Fu, Z.Q. (2022) PTI and ETI: convergent pathways with diverse elicitors. Trends in Plant Science, 27, 113–115. [DOI] [PubMed] [Google Scholar]
- Chaudhari, P. , Ahmed, B. , Joly, D.L. & Germain, H. (2014) Effector biology during biotrophic invasion of plant cells. Virulence, 5, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, M. , Fitzgerald, H.A. , Canlas, P.E. , Navarre, D.A. & Ronald, P.C. (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Molecular Plant‐Microbe Interactions, 18, 511–520. [DOI] [PubMed] [Google Scholar]
- Chiapello, H. , Mallet, L. , Guerin, C. , Aguileta, G. , Amselem, J. , Kroj, T. et al. (2015) Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biology and Evolution, 7, 2896–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittoor, J.M. , Leach, J.E. & White, F.F. (1997) Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae . Molecular Plant‐Microbe Interactions, 10, 861–871. [DOI] [PubMed] [Google Scholar]
- Choi, J. , Park, J. , Kim, D. , Jung, K. , Kang, S. & Lee, Y.‐H. (2010) Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics, 11, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H. , Choi, J. , Park, S.‐Y. , Jeon, J. & Lee, Y.‐H. (2013) Two conidiation‐related Zn (II) 2Cys6 transcription factor genes in the rice blast fungus. Fungal Genetics and Biology, 61, 133–141. [DOI] [PubMed] [Google Scholar]
- Dagvadorj, B. , Ozketen, A.C. , Andac, A. , Duggan, C. , Bozkurt, T.O. & Akkaya, M.S. (2017) A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Scientific Reports, 7, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. , Ghosh, A. , Vincon, V. , Kahnt, J. et al. (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Reissmann, S. , Aßmann, D. , Fleckenstein, M. & Kahmann, R. (2011) Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis‐induced tumour formation. Molecular Microbiology, 81, 751–766. [DOI] [PubMed] [Google Scholar]
- EL Oirdi, M. , EL Rahman, T.A. , Rigano, L. , EL Hadrami, A. , Rodriguez, M.C. , Daayf, F. et al. (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. The Plant Cell, 23, 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, M. , Peterson, G. , Chen, L. , Starnes, J. , Valent, B. , Bachi, P. et al. (2017) The Lolium pathotype of Magnaporthe oryzae recovered from a single blasted wheat plant in the United States. Plant Disease, 101, 684–692. [DOI] [PubMed] [Google Scholar]
- Figueroa, M. , Ortiz, D. & Henningsen, E.C. (2021) Tactics of host manipulation by intracellular effectors from plant pathogenic fungi. Current Opinion in Plant Biology, 62, 102054. [DOI] [PubMed] [Google Scholar]
- Giraldo, M.C. & Valent, B. (2013) Filamentous plant pathogen effectors in action. Nature Reviews Microbiology, 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Haga, K. & Iino, M. (2004) Phytochrome‐mediated transcriptional up‐regulation of ALLENE OXIDE SYNTHASE in rice seedlings. Plant & Cell Physiology, 45, 119–128. [DOI] [PubMed] [Google Scholar]
- Hirano, Y. , Nakagawa, M. , Suyama, T. , Murase, K. , Shirakawa, M. , Takayama, S. et al. (2017) Structure of the SHR–SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nature Plants, 3, 17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y. , Yang, Y. , Zhang, H. , Huang, L. , Li, D. & Song, F. (2017) Overexpression of MoSM1, encoding for an immunity‐inducing protein from Magnaporthe oryzae, in rice confers broad‐spectrum resistance against fungal and bacterial diseases. Scientific Reports, 7, 41037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbach, R. , Navarro‐Quesada, A.R. , Knogge, W. & Deising, H.B. (2011) When and how to kill a plant cell: infection strategies of plant pathogenic fungi. Journal of Plant Physiology, 168, 51–62. [DOI] [PubMed] [Google Scholar]
- Huang, L. , Sun, Q. , Qin, F. , Li, C. , Zhao, Y. & Zhou, D.‐X. (2007) Down‐regulation of a SILENT INFORMATION REGULATOR2‐related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiology, 144, 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K. , Nagasawa, N. & Nagato, Y. (2005) ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Developmental Biology, 282, 349–360. [DOI] [PubMed] [Google Scholar]
- Inoue, Y. , Vy, T.T. , Yoshida, K. , Asano, H. , Mitsuoka, C. , Asuke, S. et al. (2017) Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science, 357, 80–83. [DOI] [PubMed] [Google Scholar]
- Jeon, J. , Lee, G.‐W. , Kim, K.‐T. , Park, S.‐Y. , Kim, S. , Kwon, S. et al. (2020) Transcriptome profiling of the rice blast fungus Magnaporthe oryzae and its host Oryza sativa during infection. Molecular Plant‐Microbe Interactions, 33, 141–144. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Khan, M. , Seto, D. , Subramaniam, R. & Desveaux, D. (2018) Oh, the places they'll go! A survey of phytopathogen effectors and their host targets. The Plant Journal, 93, 651–663. [DOI] [PubMed] [Google Scholar]
- Khang, C.H. , Berruyer, R. , Giraldo, M.C. , Kankanala, P. , Park, S.‐Y. , Czymmek, K. et al. (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell‐to‐cell movement. The Plant Cell, 22, 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.‐T. , Jeon, J. , Choi, J. , Cheong, K. , Song, H. , Choi, G. et al. (2016) Kingdom‐wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Frontiers in Plant Science, 7, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.‐T. , Ko, J. , Song, H. , Choi, G. , Kim, H. , Jeon, J. et al. (2019) Evolution of the genes encoding effector candidates within multiple pathotypes of Magnaporthe oryzae . Frontiers in Microbiology, 10, 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kim, C.‐Y. , Park, S.‐Y. , Kim, K.‐T. , Jeon, J. , Chung, H. et al. (2020) Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nature Communications, 11, 5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck, M. , Hardham, A.R. & Dodds, P.N. (2011) The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cellular Microbiology, 13, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König, A. , Müller, R. , Mogavero, S. & Hube, B. (2021) Fungal factors involved in host immune evasion, modulation and exploitation during infection. Cellular Microbiology, 23, e13272. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. & Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiology, 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Volz, R. , Song, H. , Harris, W. & Lee, Y.H. (2021) Characterization of the MYB genes reveals insights into their evolutionary conservation, structural diversity, and functional roles in Magnaporthe oryzae . Frontiers in Microbiology, 12, 721530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Han, X. , Feng, D. , Yuan, D.Y. & Huang, L.J. (2019) Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? International Journal of Molecular Sciences, 20, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste, T. (2012) Plant immunity to necrotrophs. Annual Review of Phytopathology, 50, 267–294. [DOI] [PubMed] [Google Scholar]
- Mosquera, G. , Giraldo, M.C. , Khang, C.H. , Coughlan, S. & Valent, B. (2009) Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1‐4 as biotrophy‐associated secreted proteins in rice blast disease. The Plant Cell, 21, 1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete, F. , Grujic, N. , Stirnberg, A. , Saado, I. , Aleksza, D. , Gallei, M. et al. (2021) The Pleiades are a cluster of fungal effectors that inhibit host defenses. PLoS Pathogens, 17, e1009641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G. , Kleemann, J. , Torres, M.F. et al. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics, 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Park, J. , Jang, S. , Kim, S. , Kong, S. , Choi, J. et al. (2008) FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics, 24, 1024–1025. [DOI] [PubMed] [Google Scholar]
- Peng, Y. , Van Wersch, R. & Zhang, Y. (2018) Convergent and divergent signaling in PAMP‐triggered immunity and effector‐triggered immunity. Molecular Plant‐Microbe Interactions, 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Peng, Z. , Oliveira‐Garcia, E. , Lin, G. , Hu, Y. , Dalby, M. , Migeon, P. et al. (2019) Effector gene reshuffling involves dispensable mini‐chromosomes in the wheat blast fungus. PLoS Genetics, 15, e1008272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn, C.D. & Daniel, S.L. (2013) Salicylate degradation by the fungal plant pathogen Sclerotinia sclerotiorum . Current Microbiology, 67, 218–225. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van Der Does, D. , Zamioudis, C. , Leon‐Reyes, A. & Van Wees, S.C. (2012) Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology, 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Prins, T.W. , Tudzynski, P. , v Von Tiedemann, A. , Tudzynski, B. , Ten Have, A. , Hansen, M.E. et al. (2000) Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In: Kronstad, J.W. (Ed.) Fungal pathology. Dordrecht: Springer, pp. 33–64. [Google Scholar]
- Qi, J. , Li, J. , Han, X. , Li, R. , Wu, J. , Yu, H. et al. (2016) Jasmonic acid carboxyl methyltransferase regulates development and herbivory‐induced defense response in rice. Journal of Integrative Plant Biology, 58, 564–576. [DOI] [PubMed] [Google Scholar]
- Qi, T. , Guo, J. , Liu, P. , He, F. , Wan, C. , Islam, M.A. et al. (2019) Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS‐promoting transcription factor TaLOL2 to defeat ROS‐induced defense in wheat. Molecular Plant, 12, 1624–1638. [DOI] [PubMed] [Google Scholar]
- Rajput, N.A. , Zhang, M. , Shen, D. , Liu, T. , Zhang, Q. , Ru, Y. et al. (2015) Overexpression of a Phytophthora cytoplasmic CRN effector confers resistance to disease, salinity and drought in Nicotiana benthamiana . Plant & Cell Physiology, 56, 2423–2435. [DOI] [PubMed] [Google Scholar]
- Redkar, A. , Hoser, R. , Schilling, L. , Zechmann, B. , Krzymowska, M. , Walbot, V. et al. (2015) A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. The Plant Cell, 27, 1332–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. & Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annual Review of Phytopathology, 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D.W. (2001) Molecular cloning: A laboratory manual. Cold Spring Harbor: CSHL Press. [Google Scholar]
- Selin, C. , d De Kievit, T.R. , Belmonte, M.F. & Fernando, W. (2016) Elucidating the role of effectors in plant–fungal interactions: progress and challenges. Frontiers in Microbiology, 7, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, A. , Elad, Y. , Barakat, R. & Tudzynski, P. (2007) Phytohormones in Botrytis–plant interactions. In: Elad, Y. , Williamson, B. , Tudzynski, P. & Delen, N. (Eds.) Botrytis: biology, pathology and control. Dordrecht: Springer, pp. 163–179. [Google Scholar]
- Shen, H. & Wang, Z.‐Y. (2004) Expressional analysis of an EREBP transcription factor gene OsEBP‐89 in rice. Acta Biochimica et Biophysica Sinica, 36, 21–26. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. & d De Wit, P.J. (2009) Fungal effector proteins. Annual Review of Phytopathology, 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Kourmpetis, Y.A.I. , Slot, J.C. , Bakker, F.T. , de DE Wit, P.J.G.M. & Rokas, A. (2012) In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Molecular Biology and Evolution, 29, 3371–3384. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J. (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea . Annual Review of Microbiology, 57, 177–202. [DOI] [PubMed] [Google Scholar]
- Talbot, N. , McCafferty, H. , Ma, M. , Moore, K. & Hamer, J. (1997) Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiological and Molecular Plant Pathology, 50, 179–195. [Google Scholar]
- Tanabe, S. , Ashikari, M. , Fujioka, S. , Takatsuto, S. , Yoshida, S. , Yano, M. et al. (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. The Plant Cell, 17, 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, T. , Sobajima, H. , Okada, K. , Chujo, T. , Arimura, S.‐I. , Tsutsumi, N. et al. (2008) Identification of the OsOPR7 gene encoding 12‐oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta, 227, 517–526. [DOI] [PubMed] [Google Scholar]
- Tariqjaveed, M. , Mateen, A. , Wang, S. , Qiu, S. , Zheng, X. , Zhang, J. et al. (2021) Versatile effectors of phytopathogenic fungi target host immunity. Journal of Integrative Plant Biology, 63, 1856–1873. [DOI] [PubMed] [Google Scholar]
- Tonnessen, B.W. , Manosalva, P. , Lang, J.M. , Baraoidan, M. , Bordeos, A. , Mauleon, R. et al. (2015) Rice phenylalanine ammonia‐lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Molecular Biology, 87, 273–286. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. & Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Current Opinion in Plant Biology, 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Valent, B. & Chumley, F.G. (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea . Annual Review of Phytopathology, 29, 443–467. [DOI] [PubMed] [Google Scholar]
- Valent, B. , Farrall, L. & Chumley, F.G. (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics, 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H.P. , Van't Klooster, J.W. , Bolton, M.D. , Yadeta, K.A. , van VAN Baarlen, P. , Boeren, S. et al. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. The Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W.A. , Sanz‐Martín, J.M. , Rech, G.E. , Armijos‐Jaramillo, V.D. , Rivera, L.P. , Echeverria, M.M. et al. (2016) A fungal effector with host nuclear localization and DNA‐binding properties is required for maize anthracnose development. Molecular Plant‐Microbe Interactions, 29, 83–95. [DOI] [PubMed] [Google Scholar]
- Völz, R. , Kim, S.‐K. , Mi, J. , Rawat, A.A. , Veluchamy, A. , Mariappan, K.G. et al. (2019) INDETERMINATE‐DOMAIN 4 (IDD4) coordinates immune responses with plant‐growth in Arabidopsis thaliana . PLoS Pathogens, 15, e1007499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völz, R. , Kim, S.K. , Mi, J. , Mariappan, K.G. , Siodmak, A. , AL‐Babili, S. et al. (2019) A chimeric IDD4 repressor constitutively induces immunity in Arabidopsis via the modulation of salicylic acid and jasmonic acid homeostasis. Plant & Cell Physiology, 60, 1536–1555. [DOI] [PubMed] [Google Scholar]
- Völz, R. , Park, J.Y. , Kim, S. , Park, S.Y. , Harris, W. , Chung, H. et al. (2020) The rice/maize pathogen Cochliobolus spp. infect and reproduce on Arabidopsis revealing differences in defensive phytohormone function between monocots and dicots. The Plant Journal, 103, 412–429. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Shen, W. , Liu, L. , Jiang, L. , Liu, Y. , Su, N. et al. (2008) A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Molecular Biology, 66, 401–414. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Ning, Y. , Shi, X. , He, F. , Zhang, C. , Fan, J. et al. (2016) Immunity to rice blast disease by suppression of effector‐triggered necrosis. Current Biology, 26, 2399–2411. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Li, C. , Duan, G. , Wang, Y. , Zhang, Y. & Yang, J. (2019) Overexpression of Magnaporthe oryzae systemic defense trigger 1 (MoSDT1) confers improved rice blast resistance in rice. International Journal of Molecular Sciences, 20, 4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Liu, Y. , Liu, L. , Wang, Y. , Yan, J. , Wang, C. et al. (2019) The biotrophy‐associated secreted protein 4 (BAS4) participates in the transition of Magnaporthe oryzae from the biotrophic to the necrotrophic phase. Saudi Journal of Biological Sciences, 26, 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X.‐Z. , Xue, Y.‐J. , Zhou, J.‐J. , Zhang, B. , Chang, H. & Takano, M. (2011) Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea . Molecular Plant, 4, 688–696. [DOI] [PubMed] [Google Scholar]
- Yokotani, N. , Ichikawa, T. , Kondou, Y. , Iwabuchi, M. , Matsui, M. , Hirochika, H. et al. (2013) Role of the rice transcription factor JAmyb in abiotic stress response. Journal of Plant Research, 126, 131–139. [DOI] [PubMed] [Google Scholar]
- Yu, J.‐H. , Hamari, Z. , Han, K.‐H. , Seo, J.‐A. , Reyes‐Domínguez, Y. & Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genetics and Biology, 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Zarembinski, T.I. & Theologis, A. (1997) Expression characteristics of OS‐ACS1 and OS‐ACS2, two members of the 1‐aminocyclopropane‐1‐carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Molecular Biology, 33, 71–77. [DOI] [PubMed] [Google Scholar]
- Zhang, S. & Xu, J.‐R. (2014) Effectors and effector delivery in Magnaporthe oryzae . PLoS Pathogens, 10, e1003826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Rajput, N.A. , Shen, D. , Sun, P. , Zeng, W. , Liu, T. et al. (2015) A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana . Scientific Reports, 5, 10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Bao, Y. , Shan, D. , Wang, Z. , Song, X. , Wang, Z. et al. (2018) Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiology, 177, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analysis of genes encoding C2H2 zinc finger domain‐containing proteins in Magnaporthe oryzae. A neighbour‐joining circular domain tree was generated based on the C2H2 zinc finger domain. Effector candidates and MoHTR3 are indicated in blue and red characters, respectively.
Figure S2 Alignment of MoHTR3 orthologues in seven Magnaporthe oryzae strains. Sequence alignment of seven MoHTR3 orthologues using ClustalW. Asterisks indicate single amino acid differences.
Figure S3 (a) MoHTR3 without signal peptide (SP) but with exogenous simian virus40 large T antigen nuclear localization signal (NLS) is localized in the nuclei in rice protoplasts. Nuclei in the rice protoplasts are indicated with ABF1:GFP. ImageJ was used to measure the fluorescence intensity. Scale bar indicates 5 μm. (b) The localization of MoHTR3ΔSP tagged with mRFP1 was observed at 36 h postinoculation by an epifluorescence microscope. Scale bar indicates 10 µm. (a, b) White arrow, plant; asterisk, infection site. The measured area of fluorescence intensity is shown in the merged image. (c) Expression of OsMYB4 and OsWRKY45 following MoHTR3ox infection.
Figure S4 Generation of fungal mutant strains. (a) MoHTR3 was replaced with a hygromycin B phosphotransferase resistance (HPH) cassette in the Magnaporthe oryzae genome using homologous recombination. (b) The MoHTR3 promoter was replaced. Southern blot analysis was conducted to prove there is a single copy in the promoter region of MoHTR3. Expression levels were measured with three biological replicates. Significance is indicated with asterisks (t test) (p < 0.05).
Figure S5 Invasive growth of MoHTR3ox strains in rice sheath cells. Infection severity was quantified by rating the perception of infection progression. Representative microscopic images are shown. Scale bar = 10 μm.
Figure S6 (a) Protein structure comparison among MoHTR3, MoHTR1, and MoHTR2. Full amino acid sequences of these three effector proteins were analysed. The distribution of the putative domain, signal peptide, and nuclear localization signal (NLS) is shown. (b) FPKM values of the Magnaporthe oryzae genes PWL2, MoHTR1, MoHTR2, and MoHTR3 at different time points during infection at the biotrophic stage (Jeon et al., 2020).
Movie S1 Animated 3D projections of confocal laser‐scanning microscopy z‐stacks of MoHTR3:mRFP1.
Movie S2 Animated 3D projections of MoHTR3ΔSP:mRFP1 localization in the invasive hyphae.
Table S1 MoHTRs are Magnaporthe oryzae‐specific effectors.
Table S2 MoHTR3 is conserved in Magnaporthe oryzae strains.
Table S3 Primers used in this study.
Table S4 Previously studied C2H2 zinc finger transcription factor genes.
Data Availability Statement
All relevant data can be found within the manuscript and in its Supporting Information online at the publisher's website.


