Abstract
Taxonomy: Cotton leafroll dwarf virus (CLRDV) is a member of the genus Polerovirus, family Solemoviridae.
Geographical Distribution: CLRDV is present in most cotton‐producing regions worldwide, prominently in North and South America.
Physical Properties: The virion is a nonenveloped icosahedron with T = 3 icosahedral lattice symmetry that has a diameter of 26–34 nm and comprises 180 molecules of the capsid protein. The CsCl buoyant density of the virion is 1.39–1.42 g/cm3 and S20w is 115–127S.
Genome: CLRDV shares genomic features with other poleroviruses; its genome consists of monopartite, single‐stranded, positive‐sense RNA, is approximately 5.7–5.8 kb in length, and is composed of seven open reading frames (ORFs) with an intergenic region between ORF2 and ORF3a.
Transmission: CLRDV is transmitted efficiently by the cotton aphid (Aphis gossypii Glover) in a circulative and nonpropagative manner.
Host: CLRDV has a limited host range. Cotton is the primary host, and it has also been detected in different weeds in and around commercial cotton fields in Georgia, USA.
Symptoms: Cotton plants infected early in the growth stage exhibit reddening or bronzing of foliage, maroon stems and petioles, and drooping. Plants infected in later growth stages exhibit intense green foliage with leaf rugosity, moderate to severe stunting, shortened internodes, and increased boll shedding/abortion, resulting in poor boll retention. These symptoms are variable and are probably influenced by the time of infection, plant growth stage, varieties, soil health, and geographical location. CLRDV is also often detected in symptomless plants.
Control: Vector management with the application of chemical insecticides is ineffective. Some host plant varieties grown in South America are resistant, but all varieties grown in the United States are susceptible. Integrated disease management strategies, including weed management and removal of volunteer stalks, could reduce the abundance of virus inoculum in the field.
Keywords: cotton blue disease, cotton leafroll dwarf disease, cotton leafroll dwarf virus, Gossypium hirsutum, Polerovirus
This review provides insight into cotton leafroll dwarf disease, which is caused by cotton leafroll dwarf virus, including its host range, evolution, diagnosis, tissue tropism, yield loss, and management.
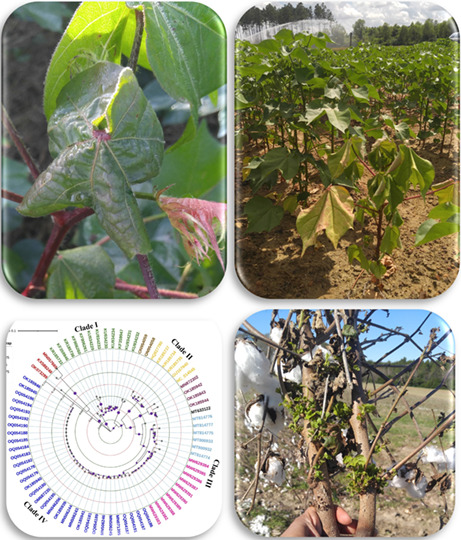
1. INTRODUCTION
Cotton (Gossypium hirsutum) is a major cash crop in many parts of the world, including the United States. In 2021, 4.1 million hectares of cotton were harvested in the United States, valued at approximately $7.4 billion (USDA, 2021). Cotton blue disease (CBD) is a viral disease capable of causing significant losses in the cotton industry. CBD was first described in the Central African Republic in 1949 and has since then been reported from several regions in Africa, Asia, and the Americas (Cauquil, 1977). However, the nature of the causal agent of CBD was not known until 2005, when the entire capsid gene and a partial RNA‐dependent RNA polymerase gene revealed its association with a virus belonging to the genus Polerovirus, family Luteoviridae, and named it cotton leafroll dwarf virus (CLRDV) (Corrêa et al., 2005). Distéfano et al. (2010) sequenced the complete genome of a CLRDV isolate from Argentina. Recently, ICTV reclassified CLRDV as a member of the family Solemoviridae (Sõmera et al., 2021). In 2006, a less aggressive resistance‐breaking genotype of CLRDV was observed in Brazil on cotton varieties known to be resistant against CBD. This new disease was referred to as “atypical” CBD (Agrofoglio et al., 2017; da Silva et al., 2015).
In the United States, CLRDV was first reported in Alabama from cotton in 2019. Plants infected with CLRDV showed symptoms including intense dark green to bluish foliage, reddening of stems and petioles, curling and drooping of leaves, internodal shortening, and moderate to severe stunting. The genome sequences from Alabama and Georgia isolates were characterized. The isolates present in the United States differ from those causing CBD in South America and other regions. The disease caused by CLRDV in the United States is hence referred to as cotton leafroll dwarf disease (CLRDD; Brown et al., 2019). It has been observed that cotton plants infected in younger stages could suffer complete yield loss, whereas the losses decrease when plants are infected at the mature stages of plant development (Parkash et al., 2021). Although the incidence of the virus in some commercial cotton fields in Georgia and Alabama is 80%–100%, no significant yield losses have been reported (Mahas et al., 2022).
2. GEOGRAPHICAL DISTRIBUTION
CBD was first reported around 1949 in the Central African Republic. The disease spread towards Benin (previously known as Dahomey), Chad, Cameroon, Congo, and Ivory Coast (Cauquil, 1977; Cauquil & Follin, 1983; Cauquil & Vaissayre, 1971). Later, CBD was introduced in Brazil and Argentina (Corrêa et al., 2005; Costa & Carvalho, 1962) and Asian countries including India (Mukherjee et al., 2012), Thailand (Sharman et al., 2015), and Timor‐Leste (Ray et al., 2016; Figure 1a). More recently, CLRDV was reported in China (Feng et al., 2017), South Korea (Igori et al., 2022), and Uzbekistan (Kumari et al., 2020; Figure 1a). In North America, symptoms associated with CBD were first observed in the autumn of 2017 on the Gulf Coast of Alabama, and the identity of CLRDV was confirmed in 2019 (Avelar et al., 2019). Currently, CLRDV is widespread in all major cotton‐growing regions in the United States (Aboughanem‐Sabanadzovic et al., 2019; Alabi et al., 2020; Ali & Mokhtari, 2020; Ali et al., 2020; Avelar et al., 2019; Faske et al., 2020; Ferguson & Ali, 2022; Iriarte et al., 2020; Price et al., 2020; Tabassum et al., 2019; Thiessen et al., 2020; Wang et al., 2020).
FIGURE 1.
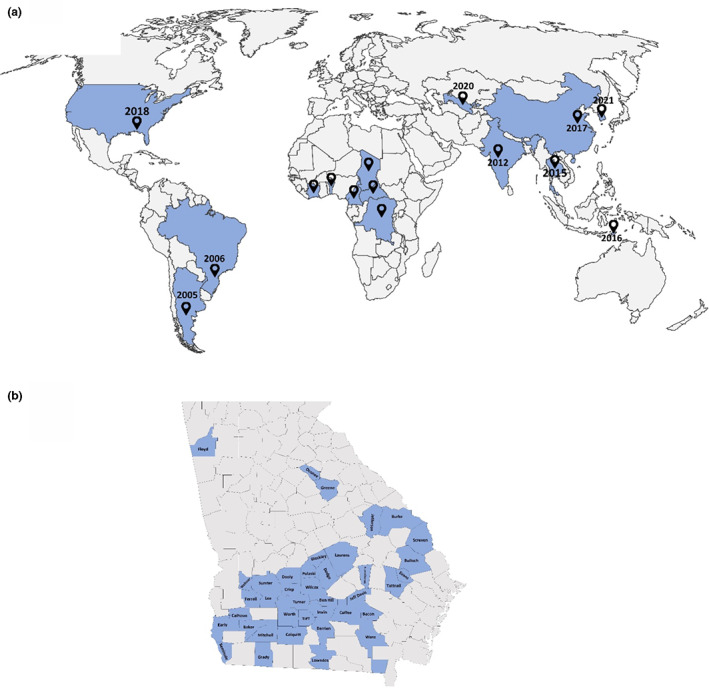
(a) Worldwide distribution of cotton blue disease and cotton leafroll dwarf disease. (b) Counties where cotton leafroll dwarf virus (CLRDV) was detected on cotton fields in Georgia, USA, are highlighted in blue. Pointers in (a) indicate where CLRDV was discovered, and the year of the first report in the respective country. For the African continent, virus discovery dates are not known; hence, only disease presence is shown.
The incidence of CLRDV infection in different states in the United States as estimated based on symptomatology ranges from 2% to 100% (Aboughanem‐Sabanadzovic et al., 2019; Alabi et al., 2020; Ali & Mokhtari, 2020; Avelar et al., 2019, 2020). In Georgia, a disease incidence of less than 10% was recorded, but CLRDV was detected in many plant tissues with and without symptoms in commercial fields and research plots (Figure 1b; Tabassum et al., 2020, 2021).
3. SYMPTOMATOLOGY
Symptoms including leaf rolling, crumpling, and a bushy top were reported in the province of Chaco in north‐west Argentina (Agrofoglio et al., 2017). Plants infected in the mature stages had normal morphology in the lower plant parts (stem and internodes) but a bushy phenotype in the top, with deformed apical leaves (Agrofoglio et al., 2017). Symptomless CLRDV infections in cotton are very common in the United States (Bag et al., 2021; Tabassum et al., 2020, 2021). In addition, CLRDV‐induced symptoms are highly variable at different locations in the United States. On the Gulf Coast of Alabama, symptoms such as leaf deformation with bluish‐green discolouration, vein clearing, leaf curling and rolling, and dwarf, stacked internodes were observed during the autumn of 2017 (Avelar et al., 2019). In Georgia, CLRDV‐infected cotton plants exhibit a multitude of symptoms depending on the plant growth stage (Bag et al., 2021; Figure 2a,b). Plants infected early (within 50 days after emergence) exhibit symptoms such as reddening of leaves, petioles, and stems, puckering, crinkling, deformation of the leaf lamina, wilting, and downward leaf drooping with V‐shaped lamina folding (Figure 2c–f). Infected plants often wilt and some may recoverbut do not produce any bolls as they mature (Parkash et al., 2021). Plants infected later in the season at maturity exhibit more turgidity with a leathery texture, crinkling, and square/leaf dropping along with wilting (Figure 2g–j). Often, these plants have more vegetative growth in the upper branches of the stem, exhibiting stacked internodes and ceasing reproductive development (Figure 2k,l). Terminal whips or accentuated verticality and a bushy phenotype are also observed, indicating infection during the early flowering or boll setting stage that leads to noticeable yield loss (Figure 2m,n). Other structural abnormalities such as parrot beak fruits (Figure 2o) with reduced seeds are also observed in infected plants. Some of these symptoms are observed across Georgia irrespective of the variety. The development of symptoms is influenced by plant age during infection, plant vigour, environmental conditions, and soil health. In plants infected at early stages exhibiting reddening and wilting of foliage (Figure 2p), it has also been observed that the symptoms diminish and the plants reappear green but remain dwarf without any harvestable bolls (Figure 2q,r).
FIGURE 2.

Symptoms observed on cotton plants infected with cotton leafroll dwarf virus (CLRDV) grown in commercial cotton fields in Georgia, USA. (a) Leaf deformation with bluish‐green discolouration. (b) Vein clearing, leaf curling, rolling, and deformation. (c) Reddening of leaf, petioles, and stem. (d) Puckering and crinkling. (e) Wilting and downward leaf drooping (V‐shaped lamina folding). (f) Deformation of the leaf lamina. (g) Leaf turgidity with a leathery texture. (h) Crinkling. (i) Square/leaf dropping. (j) Wilting. (k,l) Vegetative growth in the upper branches of the stem and stacked internodes and ceased reproductive growth. (m) Bushy phenotype. (n) Terminal whips or accentuated verticality. (o) Parrot beak fruits. (p) Plant infected in the early stage. (q,r) Symptoms diminish in early infected plants but plants remain dwarf without any harvestable bolls. (s,t) Volunteer cotton stalks.
Symptoms described for CLRDD are strikingly similar to the symptoms of bronze wilt (Parkash et al., 2021), another wilting disease with unknown causes. Bronze wilt, synonymous with “copper top, sudden wilt, and phloem wilt",was first identified in Mississippi and Louisiana in 1995, causing significant yield loss (Bell et al., 2002). Outbreaks were later reported in the late 1990s from Missouri without much yield loss (Phipps, 2000). But in 1998, Georgia recorded about $25 million in losses due to severe disease pressure during a long dry spell with temperatures above 35°C (Brown, 2000; McGraw, 2000). Short‐season varieties of Upland and Pima cotton have been reported to be more susceptible to bronze wilt, especially if the pedigree involved crosses with Tamcot SP‐37 or its progeny Miscot T8‐27 (Bell et al., 2002). In other countries, symptoms such as red leaves, red wilt, and anthocyanosis are associated with bronze wilt and are considered a physiological disorder induced by numerous abiotic factors (Gade et al., 2013). Studies that were conducted to understand the aetiology of bronze wilt and its causal biotic or abiotic factors were inconclusive due to the inconsistency of the disease symptoms and severity (Padgett et al., 2004). The combined role of biotic (virus) and abiotic factors in causing bronze wilt disease has never been evaluated. Due to the similarity of CLRDD and bronze wilt symptoms, the cause of symptoms observed in cotton in Georgia and the surroundings is still under discussion.
4. PHYSIOLOGY OF CLRDV‐INFECTED COTTON
Because CLRDV is a relatively new cotton pathogen in the United States, minimal research has been conducted on the physiological response of cotton plants to CLRDV infection. However, leaf reddening in response to other stresses is a result of anthocyanin production, which is speculated to attenuate solar radiation or to reduce reactive oxygen species levels (Close & Beadle, 2003). Similar to plants exposed to other wilt‐inducing stresses like drought (Chastain et al., 2014, 2016), recent research has shown that plants with CLRDV symptoms significantly reduce stomatal conductance and the net photosynthetic rate under field conditions (Parkash et al., 2021). Even in the earliest stages of the disease, when severe wilting is not yet observable, stomatal conductance and the net photosynthetic rate were reduced by 94% and 84%, respectively. Because declines in conductance limit transpirational cooling, leaf temperature also significantly increased by 0.5–3.8°C at advanced stages of the disease (Parkash et al., 2021). Using stomatal conductance as a reference indicator of stress, Medrano et al. (2002) and Parkash et al. (2021) were also able to show that the electron transport rate through photosystem II was less sensitive to CLRDV‐induced stress than carbon assimilation. The possibility that these differences in sensitivity lead to oxidative stress and subsequent increases in anthocyanin production should be explored further.
5. HOST RANGE
Crops and weeds are vital components in virus disease epidemiology and spread into new geographical locations. While cotton is the primary host of CLRDV, weeds and other crops that serve as hosts for this virus can influence its spread and aid in reoccurrence in agricultural landscapes. In Georgia, where the winters are mild, CLRDV was detected in a large number of overwintering cotton stalks and regrowth from these stalks (Figure 2s,t; Sedhain et al., 2021). CLRDV was also detected in 23 weed species growing near cotton fields in Georgia (Sedhain et al., 2021). In greenhouse experiments, CLRDV was detected by reverse transcription (RT)‐PCR following aphid‐mediated transmission in hibiscus (Hibiscus acetosella), okra (Abelmoschus esculentus), Nicotiana benthamiana, Palmer amaranth (Amaranth palmeri), and prickly sida (Sida spinosa) (Pandey et al., 2022). Aphids feeding on CLRDV‐infected malvaceous hosts including hibiscus, prickly sida, and okra were able to acquire CLRDV and back‐transmitted it to noninfected cotton seedlings (Pandey et al., 2022). These volunteer cotton stalks and weed hosts could potentially act as a “green bridge” and as a source of primary inoculum for the next growing season (Bag et al., 2021; Sedhain et al., 2021).
In addition to cotton, CLRDV also infects a number of crop species worldwide. For example, chickpea (Cicer arietinum) was shown to be a host for CLRDV (Kumari et al., 2020). Symptoms induced by CLRDV on chickpea are similar to those observed on cotton, including chlorosis, stunting, necrosis, yellowing, and reddening (Kumari et al., 2020). In addition, CLRDV capsid protein gene sequences share 89.4% to 100% homology with chickpea stunt disease‐associated virus (CpSDaV) isolates from India (Mukherjee et al., 2016). The virus was also transmitted from cotton to chickpea, suggesting that CLRDV and CpSDaV are strains of the same virus (Mukherjee et al., 2016). Natural infection of CLRDV has also been observed on Hibiscus syriacus exhibiting vein‐clearing symptoms in South Korea upon mixed infection with other viruses (Igori et al., 2022). CLRDV was reported infecting cacao (Theobroma cacao) trees in Bahia state in north‐eastern Brazil (Ramos‐Sobrinho et al., 2022), suggesting its movement to different cultivated crop species.
6. MORPHOLOGY AND GENOME ORGANIZATION
As a primary confirmation of Koch's postulates, CLRDV was detected in infected plants using electron microscopy. Virion particles were partially purified from young bark tissues from cotton plants with CLRDV symptoms (Figure 2c,d). The virion preparation was examined under a transmission electron microscope as described by Al Rwahnih et al. (2021). A minimal number of spherical virion particles with a diameter of 25–35 nm were observed (Figure 3a), suggesting the titre of the virus particles in the sample was low. RT‐PCR further confirmed the presence of CLRDV in the same bark tissues following the protocol described in Tabassum et al. (2020). Previously Takimoto et al. (2009) conducted anatomical studies on CLRDV‐infected cotton petioles and midrib tissues that exhibited more callose depositions and inclusion bodies in phloem cells (Takimoto et al., 2009).
FIGURE 3.
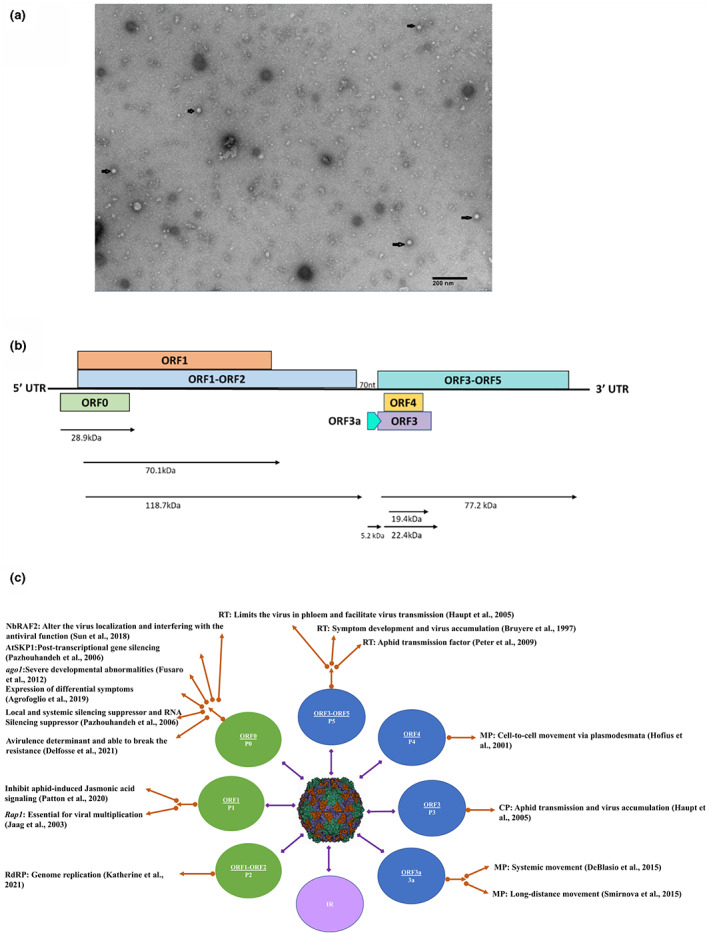
(a) Transmission electron micrograph of virion particles of cotton leafroll dwarf virus (CLRDV) from bark tissues of an infected cotton plant with symptoms. The presence of CLRDV in these bark tissues was further confirmed by reverse transcription‐PCR. (b) Schematic representation of the genome of CLRDV and open reading frames (ORFs). (c) Functional overview of proteins associated with CLRDV, including a schematic representation and their functions. An artificially coloured cryo‐electron microscopy image of rice yellow mottle virus (family Solemoviridae) is shown in the centre (ICTV; https://ictv.global/report/chapter/solemoviridae/solemoviridae and Opalka et al., 2000). The functional role played by each viral protein (shown in green, blue, and lavender‐coloured circles) encoded by CLRDV and other members of the genus Polerovirus are depicted at the periphery corresponding to their functional characterization in that virus species.
The CLRDV genome organization resembles those of other poleroviruses (Avelar et al., 2020; Corrêa et al., 2005; Distéfano et al., 2010; Tabassum et al., 2020, 2021). The virus genome comprises seven open reading frames (ORFs) with an intergenic region between ORF2 and ORF3a (Figure 3b). ORF0 encodes the P0 protein (28.9 kDa), a silencing suppressor (Delfosse et al., 2014). ORF1 encodes the P1 protein (70.1 kDa), predicted to be expressed through leaky scanning. ORF1–2 encodes a fused protein P1–P2 replication‐related protein of 118.7 kDa through ribosomal frameshift. ORF3–5 are expressed through subgenomic RNAs. ORF3 encodes the P3 (capsid) protein (22.4 kDa), ORF4 encodes the P4 (movement) protein (19.4 kDa), and P5 is translated through an in‐frame read through the P3 stop codon. The P3–P5 protein (77.2 kDa) is essential for aphid transmission and virus accumulation in plants (Agrofoglio et al., 2017; Distéfano et al., 2010; Silva et al., 2008; Figure 3b).
ORF0 is a highly variable genomic region of CLRDV (Cascardo et al., 2015; Delfosse et al., 2014); the P0 protein is the most diverse protein. The P0 protein contains a conserved F‐box (LPxx[L/I]) domain located near the N‐terminus and a conserved sequence ([K/R]IYGEDGXXXFWR) at the C‐terminus (Delfosse et al., 2021) considered essential for viral silencing suppressor activity (Pazhouhandeh et al., 2006; Zhuo et al., 2014). Silencing suppression activity varies in different species and can be systemic or local, and a few proteins have null activity (Delfosse et al., 2021). In isolates collected from the United States, five unique amino acid substitutions were present; however, their role in disease development and expression is unknown. The amino acid substitution from isoleucine (I) to valine (V) at the 72nd position (I72V), in the F‐box domain, differentiates between the typical and atypical genotypes of CLRDV (Delfosse et al., 2014). The significance of this substitution in symptom development and virulence is not yet determined. The sequenced isolates from Georgia and Alabama resemble the South American resistance‐breaking atypical genotypes with valine (V) at the 72nd position, whereas isolates from Texas have both typical (I) and atypical (V) residues at the 72nd position (Tabassum et al., 2020, 2021).
7. DIVERSITY AND EVOLUTION OF CLRDV
Since the first report of CLRDV in Alabama, isolates from different host species and geographical locations in the United States were sequenced and compared with the isolates of other parts of the world to further understand the diversity and evolution of CLRDV (Avelar et al., 2020; Ramos‐Sobrinho et al., 2021; Tabassum et al., 2020, 2021). A study by Tabassum et al. (2021) revealed that the P0 proteins of CLRDV isolates from Texas and Alabama were >90% identical to those of the CLRDV isolates from Georgia. However, Georgia isolates have >10% divergence in their amino acid sequences compared to other South American CLRDV sequences. These data might be helpful in further understanding the diversity, degree of genetic differentiation, and population dynamics of CLRDV. A recent study suggests that eight newly determined full‐length genome sequences of CLRDV isolated from cotton plants in Alabama, Florida, and Texas were phylogenetically related with a CLRDV genotype from Alabama (Ramos‐Sobrinho et al., 2021). Furthermore, these isolates have been shown to cluster into a monophyletic group among the CLRDV populations in North America. These data further suggest that horizontal gene flow may have occurred between CLRDV isolates during the course of evolution.
A more in‐depth analysis of ORF0 of various CLRDV isolates was performed with the sequence information available as of 2023‐01‐10 (n = 76) using MEGA‐XI and iTOL (Table S1; Letunic & Bork, 2021; Tamura et al., 2021). CLRDV isolates were separated into four major clades mostly based on geographical location (Figure 4a). The Asian clade consisted of three CLRDV isolates, of which two infect the family Malvaceae (H. syriacus and Malvaviscus arboreus) in South Korea and one was reported from soybean aphid (Aphis glycines) in China. The CLRDV P0 sequences (n = 20) from South American cotton isolates diverged into three separate clades: (i) one clade containing “typical” CLRDV genotypes (mostly from Brazil), (ii) one clade containing “atypical” CLRDV genotypes (mostly from Argentina; da Silva et al., 2015), and (iii) one clade containing two “atypical” isolates from cacao (T. cacao) trees in Brazil (Ramos‐Sobrinho et al., 2022). The most diverse and most extensive clade consisted of 53 sequences from North America. Among the CLRDV isolates reported from the United States, isolates associated with cotton (n = 43) and weed (n = 10) species were segregated into two distinct clusters. Such segregation suggests genetic recombination, which might help the viruses to broaden their host range and foster successful establishment in the ecosystem (Vassilakos et al., 2016).
FIGURE 4.
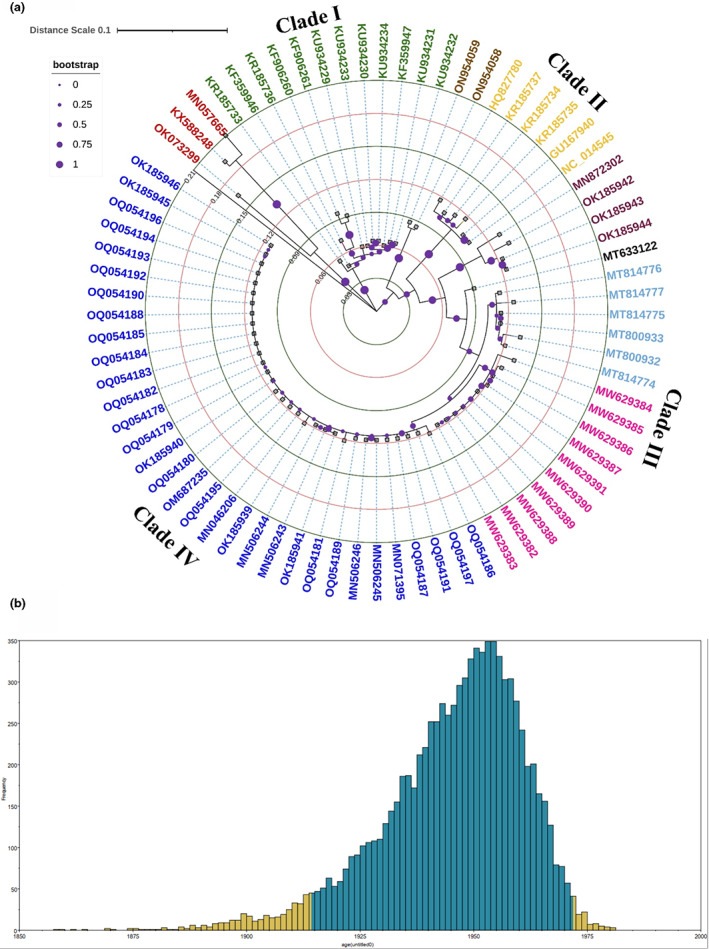
(a) The evolutionary history was inferred by using the maximum‐likelihood method and a JTT matrix‐based model with 1000 bootstrap replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 76 nucleotide sequences. All positions with less than 95% site coverage were eliminated. The blue colour indicates cotton leafroll dwarf virus P0 (CLRDV‐P0) sequences obtained from cotton plants in Alabama, Florida, Georgia, Oklahoma, and Texas, in 2018–2022. The light pink colour indicates CLRDV‐P0 sequences from weed species in Georgia. The CLRDV‐P0 sequences obtained from cotton in Georgia/Alabama in 2018 are represented in teal. The typical CLRDV‐P0 sequences from Brazil are shown in mustard yellow, whereas atypical CLRDV‐P0 sequences from cotton are shown in green. Four sequences from Texas forming a subclade are shown in purple. Two sequences (accessions ON954058 and ON954059) shown in golden brown were isolated from cacao (Theobroma cacao) trees in Brazil. Three CLRDVs (accessions UID85580, QLJ58259, and ARU09826), isolated from Hibiscus sp., Malvaviscus sp., and aphid species, are shown in red. The phylogenetic tree was visualized using iTOL v. 4 (Letunic & Bork, 2021). (b) Graph representing the evolution of CLRDV. The virus epidemic may have started around 1945. Analysis was conducted in BEAST v. 1.10.4 and the results were further visualized using Tracer v. 1.7.1.
The same set of sequences was also used to study the amino acid substitution rates and evolutionary pattern of CLRDV using BEAST v. 1.10.4 and Tracer v. 1.7.1 (Rambaut et al., 2018; Suchard et al., 2018). BEAST analysis showed that the amino acid substitution rate of P0 protein from different hosts combined was 2.194 × 10−3 substitutions per site per year. However, when P0 from only weeds was considered, the amino acid substitution rate was higher (6.313 × 10−10 substitutions per site per year) (Table 1). Genome‐wide mutations and recombination in viruses allow them to jump from one host to another (Calvo et al., 2014). The higher amino acid substitution rate in cultivated and noncultivated host species emphasizes the potential threat to cotton cultivation and other crops in the future.
TABLE 1.
Analysis of the rate of mutation and the age (root) of the CLRDV P0 protein using the BEAST tool (v. 1.10.4).
| Sequence source | Mean rate | 95% HPD interval | S | Variation | ESS/no. of samples | Age (root) | 95% HPD interval |
|---|---|---|---|---|---|---|---|
| All 76 sequences | 2.194 × 10−3 | 1.293 × 10−3 to 3.081 × 10−3 | 0.1015 | 0.0103 | 8,871/9,001 | 1945 | 1914 to 1971 |
| Only weed species | 6.313 × 10−10 | 1.266 × 10−23 to 1.398 × 10−10 | 1.129 × 10−12 | 1.275 × 10−22 | 566/9,001 | – | – |
Abbreviations: ESS, effective sample size, that is, the number of independent samples or the chain length; HPD, highest posterior density; Stdev, standard deviation of the mean.
Based on the CLRDV ORF0 sequences available in GenBank from 2005 to 2022, BEAST analysis estimated the age (root) of the CLRDV population in the year 1945 during the period 1938–1962 when CLRDV symptoms were first observed and documented in the Central African Republic (Cauquil, 1977; Cauquil & Follin, 1983; Costa & Carvalho, 1962). However, the 95% highest posterior density credible interval spans from 1914 to 1971 (Bryant et al., 2007; Figure 4b). These data provide insight into virus transmission among cotton and other species and suggest that CLRDV was probably introduced in South America from the African continent (as a CBD).
8. INSECT VECTOR AND TRANSMISSION
The cotton or melon aphid Aphis gossypii is a major agricultural pest found worldwide in over 170 countries (CABI Compendium, 2021). The earliest known investigations of CLRDV transmission were conducted in Africa in the early 1970s when CLRDV was commonly known as CBD. Aphids collected from cotton plants with symptoms in the field were able to transmit the virus to cotton (Cauquil & Vaissayre, 1971). Studies conducted in Brazil determined that susceptible cultivars required fewer viruliferous aphids to develop symptoms while more resistant cultivars required a minimum of 10 aphids to develop symptoms (Takimoto, 2003). A. gossypii transmits CLRDV to cotton plants in a circulative and nonpropagative manner (Michelotto & Busoli, 2003). Both winged and wingless morphs can transmit the virus after a 48‐h inoculation access period for up to 12 days (Michelotto & Busoli, 2006, 2009).
A. gossypii is an annual pest of cotton in the south‐eastern United States. Seasonal dynamics of cotton aphid populations and their potential role in the spread of CLRDV were investigated in field plots in Georgia and Alabama. A higher incidence of A. gossypii was observed during June and July. A. gossypii was detected in all test plots irrespective of the intensity of insecticide use surveyed in 2019. CLRDV was detected in 60% to 100% of the samples tested (Mahas et al., 2022), suggesting a correlation between vector and disease spread in the region.
Soybean aphids (A. glycines) collected in 2017 in China were found to harbour CLRDV, although cotton is a nonpreferred host of this aphid (Feng et al., 2017). Aphis craccivora and the green peach aphid Myzus persicae transmit CpSDaV (Horn et al., 1995), which is suspected to be another strain of CLRDV (Mukherjee et al., 2016). These findings indicate that more than one aphid species could transmit CLRDV and might contribute to the introduction and spread of the virus to different hosts and geographical regions. In the United States, A. craccivora, Aphis fabae, Macrosiphum euphorbiae, Protaphis middletonii, and Rhopalosiphum rufiabdominale are known to colonize cotton (Blackman & Eastop, 2000; Mahas et al., 2022). However, transmission experiments with these aphids need to be conducted to confirm their ability to transmit CLRDV. Due to the presence of sweet potato whitefly (Bemisia tabaci) on cotton in the south‐eastern United States, its ability to transmit CLRDV on cotton was tested. B. tabaci could neither acquire nor transmit the virus, suggesting A. gossypii is the only efficient vector transmitting CLRDV onto cotton in the United States (Heilsnis et al., 2022).
9. DIAGNOSIS
The symptoms of CLRDV‐infected cotton plants are complex and depend on the plant growth stage at the time of infection, the variety, and the geographical location, making it difficult to diagnose solely on the basis of symptomology. Serological tests performed on CBD samples from north‐eastern Argentina suggested its relationship to barley yellow dwarf virus serotypes RPV and PAV members of the family Luteoviridae (Lenardon, 1994). Recently, antibodies raised against CLRDV capsid protein antigens were used in double antibody sandwich‐ELISA to detect CLRDV in Commelina sp. weed samples (Hoffmann et al., 2022).
After the CLRDV genome was sequenced, PCR‐based methods were developed for detection. Initially, degenerate primers targeting ORF0 and ORF3 were used to detect the virus (Corrêa et al., 2005). Later, specific primers targeting ORF3 were developed for detection assays (Silva et al., 2008). With the emergence of CLRDV in the United States, PCR assays using primers designed specifically targeting the US isolates were developed (Tabassum et al., 2020). Another set of degenerate primers targeting ORF3 were designed to detect several polero‐ and luteoviruses from Argentina, Brazil, and India, including CLRDV isolates (Sharman et al., 2015). SYBR Green‐based RT‐quantitative PCR (RT‐qPCR) assays targeting ORF3 were developed for more precise and quick detection (Pandey et al., 2022).
High‐throughput sequencing plays a significant role in the early detection of CLRDV on plants with symptoms in commercial fields in Alabama, Florida, and Texas (Avelar et al., 2019, 2020; Ramos‐Sobrinho et al., 2021). CLRDV was found as coinfections with other viruses in cotton plants exhibiting leaf roll and vein yellowing symptoms (Yang et al., 2021) in China and on H. syriacus exhibiting vein clearing symptoms (Igori et al., 2022) in South Korea.
10. COTTON LEAFROLL DWARF VIRUS TISSUE TROPISM
CLRDV is phloem‐limited, and during initial studies conducted in Georgia, detection in whole leaf tissues gave inconsistent results due to the low virus titre (Figure 3a). To determine the best tissue to be sampled for accurate detection of CLRDV in cotton, virus titre was estimated in different tissues from plants exhibiting symptoms associated with CLRDV. The copy number of the virus in samples was estimated by the method described by Kavalappara et al. (2022). A partial capsid protein gene amplified and cloned in a Topo‐TA PCR cloning vector (Invitrogen) was used as standard. Six 10‐fold serial dilutions of the linearized plasmids were prepared and a standard curve was made. The number of copies of standard was calculated based on the spectrophotometric determination of the plasmid DNA concentration using a previously reported formula (Rotenberg et al., 2009). In general, CLRDV titre was higher in the upper parts of infected cotton plants than in the lower parts. CLRDV titre was highest in the main stem, followed by the branches and petioles. Fruits carried the lowest CLRDV titre among various tissues tested (Table 2). Thus, young phloem tissues in the upper canopy would be the ideal candidate for sample collection for accurate and efficient detection.
TABLE 2.
Estimated copy number of cotton leafroll dwarf virus (CLRDV) in different parts of infected cotton plants.
| Part of the plant | Estimated copy number per ng of total RNA | |
|---|---|---|
| Underground | Roots | 1.78 × 105 |
| Lower plant parts | Main stem | 5.01 × 105 |
| Branches | 4.47 × 105 | |
| Petioles | 2.63 × 103 | |
| Fruit bracts and skin | 2.00 × 103 | |
| Upper plant parts | Main stem | 8.91 × 105 |
| Branches | 6.15 × 106 | |
| Petioles | 1.58 × 105 | |
| Fruit bracts and skin | 1.26 × 104 | |
Note: Values are averages from eight CLRDV‐infected plants as determined by reverse transcription‐PCR. Samples from each plant were run in triplicate.
11. ECONOMIC IMPORTANCE
In Argentina and Brazil, yield losses of up to 80% were reported due to CLRDV infection in susceptible varieties (Distéfano et al., 2010; Silva et al., 2008). Yield reductions of up to 99% due to a decrease in boll number and boll mass resulting from a lower number of seeds per boll have been recorded on individual plants (Parkash et al., 2021). However, overall yield losses are variable in the United States (Avelar et al., 2019; Mahas et al., 2022). Many commercial cotton fields in the south‐eastern United States, where CLRDV was first detected in 2019, have met their production goals (authors' personal observations). It is unknown whether variation in disease and yield loss among locations is due to varietal, environmental, crop age, or vector‐related factors. Most of the cotton varieties cultivated in the United States are tolerant to CLRDV; the virus may be present, but the host plants do not exhibit any symptoms. Furthermore, only a small number of commercial cotton varieties exhibit disease symptoms such as reddening of leaves, wilting, and downward leaf drooping with V‐shaped lamina when the plant is infected by the virus early in the season. Consequently, CLRDD seems to be a minor concern at present. However, given the previous virus outbreaks and the yield losses in South American countries, monitoring of the evolving virus species/strains and its epidemics in commercial cotton varieties may be a good preventive measure. A thorough study of virus–host interactions will help in developing management strategies for future outbreaks.
12. DISEASE MANAGEMENT
Vector management is one of the major components of integrative pest management strategies for virus management. However, aggressive aphid sprays (weekly application) during 2019–2020 did not reduce the occurrence of CLRDV in an experimental plot in Georgia. It was also observed that insecticide application did not prevent aphid colonization or decrease CLRDV incidence in both Alabama and Georgia. Moreover, it could adversely impact other beneficial insects as well as elevate the risk of insects developing insecticide resistance. In addition, adjusting the planting date was also found to be ineffective in reducing the incidence of CLRDD (Mahas et al., 2022).
CBD was effectively mitigated in South America by deploying varieties possessing a single dominant resistance gene, Rghv1 (Pupim Junior et al., 2008). Morello et al. (2010) developed BRS‐293, a midseason high‐yielding cultivar that is moderately resistant to typical and atypical CLRDV but was found to be susceptible to the prevalent CLRDV in the south‐eastern United States (Brown et al., 2019). While assessing resistant populations obtained from cotton cultivar Delta Opal, possessing a single dominant resistance gene, Cbd, Fang et al. (2010) identified two tightly linked simple‐sequence repeat markers associated with the Cbd gene, localized on chromosome 10. However, genetic analyses have indicated that varieties carrying the Cbdgene were susceptible to CLRDV (Brown et al., 2019), suggesting that breeding for resistance would necessitate screening to identify new sources of resistance.
13. CONCLUSION
CLRDD is an emerging viral disease in cotton‐producing regions in the United States. Several weed species and overwintering cotton stalks might act as a potential green bridge and source of virus inoculum. Early and late season symptoms are yield‐limiting in the infected plant but not detrimental over a large acreage due to low disease incidence since its first report in 2018. Earlier recorded yield losses, the increase in host range, and its worldwide distribution are still concerning. At present, CLRDD may not be yield‐limiting in the United States, but it can cause significant yield loss in individual infected plants, as observed in commercial fields and experimental plots in Georgia (Parkash et al., 2021) and in other cotton‐producing countries. Basic research on virus–host interactions using a reverse genetic approach will be advantageous to devise appropriate management strategies and breeding approaches for future crops.
CLRDV is phloem‐limited, but little is known about its interactions with phloem proteins and interference with host phloem activities (Ψw, Ψp, Ψs, phloem loading, and unloading), highlighting that further research is needed. Systemic infection with plant viruses can affect the phloem tissue and its translatome (Collum & Culver, 2017). CBD viruses trigger severe symptoms and yield losses to which resistant germplasm was developed, which was soon overcome by the resistance‐breaking atypical strain causing mild symptoms. Reasons for the varied host response to different virus strains are not well known, and could include abiotic factors, virus–host interactions, or a combination of both, resulting in leaf drooping, wilting, reddening, stunting, and stacking internode symptoms. The CLRDV P0 protein is a silencing suppressor; it contains a highly variable region with five amino acid substitutions that are uniquely present in the isolates collected in Georgia, USA. The effects of these amino acid substitutions in P0 on symptom development and virulence need further investigation. The number of aphid vectors is very high from early June to mid‐July in Georgia. Aggressive sprays (once weekly) reduced the aphid populations but did not inhibit the occurrence of CLRDV and negatively influenced beneficial insects. The soilborne fungus Neozygites fresenii acts as a biological (natural) control and collapses the aphid population in the region, reducing the dependence on chemical control. The development and deployment of resistant varieties remain the most efficacious strategy for CLRDD management. Further investigations of the disease symptomatology, virus interactions with crop and noncrop hosts, vectors, the virus population, and cultural practices to develop and implement integrated disease management strategies are necessary.
CONFLICT OF INTEREST STATEMENT
The authors read the manuscript and declare no conflict of interest.
Supporting information
Table S1
ACKNOWLEDGEMENTS
This work was supported by the Georgia Cotton Commission, Cotton Incorporated, USDA‐HATCH (#1020319), USDA‐NIFA‐CPPM (2019‐70006‐30441), and USDA‐NIFA‐CARE (2021‐67028‐34097). The UGA‐Cotton team acknowledges the support provided by UGA‐ANR agents, growers, consultants, and the other stakeholders. S.B. is thankful to late Dr Robert “Bob” Nichols for his support during the project. The authors also acknowledge the constructive comments and edits by Saritha R.K. and anonymous reviewers. The funders had no role in the study design, data collection and analysis, publishing decision, or manuscript preparation.
Edula, S.R. , Bag, S. , Milner, H. , Kumar, M. , Suassuna, N.D. , Chee, P.W. et al. (2023) Cotton leafroll dwarf disease: An enigmatic viral disease in cotton. Molecular Plant Pathology, 24, 513–526. Available from: 10.1111/mpp.13335
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created.
REFERENCES
- Aboughanem‐Sabanadzovic, N. , Aboughanem‐Sabanadzovic, N. , Allen, T.W. , Wilkerson, T.H. , Conner, K.N. , Sikora, E.J. et al. (2019) First report of cotton leafroll dwarf virus in upland cotton (Gossypium hirsutum) in Mississippi. Plant Disease, 103, 1798. [Google Scholar]
- Agrofoglio, Y.C. , Delfosse, V.C. , Casse, M.F. , Hopp, H.E. , Kresic, I.B. & Distéfano, A.J. (2017) Identification of a new cotton disease caused by an atypical cotton leafroll dwarf virus in Argentina. Phytopathology, 107, 369–376. [DOI] [PubMed] [Google Scholar]
- Al Rwahnih, M. , Alabi, O.J. , Hwang, M.S. , Tian, T. , Mollov, D. & Golino, D. (2021) Characterization of a new nepovirus infecting grapevine. Plant Disease, 105, 1432–1439. [DOI] [PubMed] [Google Scholar]
- Alabi, O.J. , Isakeit, T. , Vaughn, R. , Stelly, D. , Conner, K.N. , Gaytán, B.C. et al. (2020) First report of cotton leafroll dwarf virus infecting upland cotton (Gossypium hirsutum) in Texas. Plant Disease, 104, 998. [Google Scholar]
- Ali, A. & Mokhtari, S. (2020) First report of cotton leafroll dwarf virus infecting cotton (Gossypium hirsutum) in Kansas. Plant Disease, 104, 1880. [Google Scholar]
- Ali, A. , Mokhtari, S. & Ferguson, C. (2020) First report of cotton leafroll dwarf virus from cotton (Gossypium hirsutum) in Oklahoma. Plant Disease, 104, 2531. [Google Scholar]
- Avelar, S. , Ramos‐Sobrinho, R. , Conner, K. , Nichols, R.L. , Lawrence, K. & Brown, J.K. (2020) Characterization of the complete genome and P0 protein for a previously unreported genotype of cotton leafroll dwarf virus, an introduced polerovirus in the United States. Plant Disease, 104, 780–786. [DOI] [PubMed] [Google Scholar]
- Avelar, S. , Schrimsher, D.W. , Lawrence, K. & Brown, J.K. (2019) First report of cotton leafroll dwarf virus associated with cotton blue disease symptoms in Alabama. Plant Disease, 103, 592. [Google Scholar]
- Bag, S. , Roberts, P.M. & Kemerait, R.C. (2021) Cotton leafroll dwarf virus: an emerging virus disease on cotton in the US. Crops and Soils, 54, 18–22. [Google Scholar]
- Bell, A.A. , Nichols, R.L. & Lemon, R. (2002) Bronze wilt of cotton, Texas Cooperative Extension . Available from: http://cotton.tamu.edu/Nematodes/bronzewilt.pdf [Accessed 18th March 2023]
- Blackman, R.L. & Eastop, V.F. (2000) Aphids on the world's crops: an identification and information guide. John Wiley & Sons. [Google Scholar]
- Brown, S. , Conner, K. , Hagan, A. , Jacobson, A. , Koebernick, J. , Lawrence, K. et al. (2019) Report of a research review and planning meeting on cotton leafroll dwarf virus, Cotton Incorporated . Available from: https://www.cottoninc.com/cotton‐production/ag‐research/plant‐pathology/cotton‐leafroll‐dwarf‐virus‐research/ [Accessed 18th March 2023]
- Brown, S.M. (2000) Bronze wilt field symptoms in Georgia. In: Dugger, C.P. & Richter, D.A. (Eds.) Proceedings of the 2000 Beltwide cotton conferences, 4–8 January 2000. Memphis, TN: National Cotton Council, p. 151. [Google Scholar]
- Bruyère, A. , Brault, V. , Ziegler‐Graff, V. , Simonis, M.T. , v Van den Heuvel, J. , Richards, K. et al. (1997) Effects of mutations in the beet western yellows virus readthrough protein on its expression and packaging and on virus accumulation, symptoms, and aphid transmission. Virology, 230, 323–334. [DOI] [PubMed] [Google Scholar]
- Bryant, J.E. , Holmes, E.C. & Barrett, A.D.T. (2007) Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathogens, 3, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABI Compendium . (2021) Aphis gossypii (cotton aphid) . Available from: 10.1079/cabicompendium.6204. [Accessed 29th December 2022]. [DOI]
- Calvo, M. , Malinowski, T. & García, J.A. (2014) Single amino acid changes in the 6K1‐CI region can promote the alternative adaptation of Prunus and Nicotiana propagated plum pox virus C isolates to either host. Molecular Plant‐Microbe Interactions, 27, 136–149. [DOI] [PubMed] [Google Scholar]
- Cascardo, R.S. , Arantes, I.L.G. , Silva, T.F. , Sachetto‐Martins, G. , Vaslin, M.F.S. & Corrêa, R.L. (2015) Function and diversity of P0 proteins among cotton leafroll dwarf virus isolates. Virology Journal, 12, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauquil, J. (1977) Etudes sur une maladie d'origine virale du cotonnier, la maladie bleue. Coton et Fibres Tropicales, 32, 259–278. [Google Scholar]
- Cauquil, J. & Follin, J.C. (1983) Presumed virus and mycoplasma‐like organism diseases in sub‐Saharan Africa and in the rest of the world. Cotton et Fibres Tropicales, 38, 293–317. [Google Scholar]
- Cauquil, J. & Vaissayre, M. (1971) La "maladie bleue" du cotonnier en Afrique: transmission de cotonnier à cotonnier par Aphis gossypii Glover. Coton et Fibres Tropicales, 26, 463–466. [Google Scholar]
- Chastain, D.R. , Snider, J.L. , Choinski, J.S. , Collins, G.D. , Perry, C.D. , Whitaker, J. et al. (2016) Leaf ontogeny strongly influences photosynthetic tolerance to drought and high temperature in Gossypium hirsutum . Journal of Plant Physiology, 199, 18–28. [DOI] [PubMed] [Google Scholar]
- Chastain, D.R. , Snider, J.L. , Collins, G.D. , Perry, C.D. , Whitaker, J. & Byrd, S.A. (2014) Water deficit in field‐grown Gossypium hirsutum primarily limits net photosynthesis by decreasing stomatal conductance, increasing photorespiration, and increasing the ratio of dark respiration to gross photosynthesis. Journal of Plant Physiology, 171, 1576–1585. [DOI] [PubMed] [Google Scholar]
- Close, D.C. & Beadle, C.L. (2003) The ecophysiology of foliar anthocyanin. The Botanical Review, 69, 149–161. [Google Scholar]
- Collum, T.D. & Culver, J.N. (2017) Tobacco mosaic virus infection disproportionately impacts phloem associated translatomes in Arabidopsis thaliana and Nicotiana benthamiana . Virology, 510, 76–89. [DOI] [PubMed] [Google Scholar]
- Corrêa, R.L. , Silva, T.F. , Simões‐Araújo, J.L. , Barroso, P.A. , Vidal, M.S. & Vaslin, M.F. (2005) Molecular characterization of a virus from the family Luteoviridae associated with cotton blue disease. Archives of Virology, 150, 1357–1367. [DOI] [PubMed] [Google Scholar]
- Costa, A.S. & Carvalho, A. (1962) Virus diseases of the cotton plant in the state of São Paulo. Bragantia, 21, 45–62. [Google Scholar]
- da Silva, A.K. , Romanel, E. , Silva Tda, F. , Castilhos, Y. , Schrago, C.G. , Galbieri, R. et al. (2015) Complete genome sequences of two new virus isolates associated with cotton blue disease resistance breaking in Brazil. Archives of Virology, 160, 1371–1374. [DOI] [PubMed] [Google Scholar]
- DeBlasio, S.L. , Johnson, R. , Mahoney, J. , Karasev, A. , Gray, S.M. , MacCoss, M.J. et al. (2015) Insights into the polerovirus–plant interactome revealed by coimmunoprecipitation and mass spectrometry. Molecular Plant‐Microbe Interactions, 28, 467–481. [DOI] [PubMed] [Google Scholar]
- Delfosse, V.C. , Agrofoglio, Y.C. , Casse, M.F. , Kresic, I.B. , Hopp, H.E. , Ziegler‐Graff, V. et al. (2014) The P0 protein encoded by cotton leafroll dwarf virus inhibits local but not systemic RNA silencing. Virus Research, 180, 70–75. [DOI] [PubMed] [Google Scholar]
- Delfosse, V.C. , Barrios Barón, M.P. & Distéfano, A.J. (2021) What we know about poleroviruses: advances in understanding the functions of polerovirus proteins. Plant Pathology, 70, 1047–1061. [Google Scholar]
- Distéfano, A.J. , Bonacic Kresic, I. & Hopp, H.E. (2010) The complete genome sequence of a virus associated with cotton blue disease, cotton leafroll dwarf virus, confirms that it is a new member of the genus Polerovirus . Archives of Virology, 155, 1849–1854. [DOI] [PubMed] [Google Scholar]
- Fang, D.D. , Xiao, J. , Canci, P.C. & Cantrell, R.G. (2010) A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theoretical and Applied Genetics, 120, 943–953. [DOI] [PubMed] [Google Scholar]
- Faske, T.R. , Stainton, D. , Aboughanem‐Sabanadzovic, N. & Allen, T.W. (2020) First report of cotton leafroll dwarf virus from upland cotton (Gossypium hirsutum) in Arkansas. Plant Disease, 104, 2742. [Google Scholar]
- Feng, Y. , Krueger, E.N. , Liu, S. , Dorman, K. , Bonning, B.C. & Miller, W.A. (2017) Discovery of known and novel viral genomes in soybean aphid by deep sequencing. Phytobiomes Journal, 1, 36–45. [Google Scholar]
- Ferguson, C. & Ali, A. (2022) Complete genome sequence of cotton leafroll dwarf virus infecting cotton in Oklahoma, USA. Microbiology Resource Announcements, 11, e0014722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro, A.F. , Correa, R.L. , Nakasugi, K. , Jackson, C. , Kawchuk, L. , Vaslin, M.F. et al. (2012) The enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology, 426, 178–187. [DOI] [PubMed] [Google Scholar]
- Gade, R.M. , Tanwar, R.K. , Jeyakumar, P. & Kanwar, V. (2013) Leaf reddening and its management in cotton . ICAR ‐ National Centre for Integrated Pest Management, New Delhi, Report number: 30.
- Haupt, S. , Stroganova, T. , Ryabov, E. , Kim, S.H. , Fraser, G. , Duncan, G. et al. (2005) Nucleolar localization of potato leafroll virus capsid proteins. Journal of General Virology, 86, 2891–2896. [DOI] [PubMed] [Google Scholar]
- Heilsnis, B. , McLaughlin, A. , Conner, K. , Koebernick, J. & Jacobson, A.L. (2022) Vector competency of Aphis gossypii and Bemisia tabaci to transmit cotton leafroll dwarf virus. Journal of Cotton Science, 26, 23–30. [Google Scholar]
- Hoffmann, L.V. , Branquinho, A.A. , Barroso, P.A.V. & Vaslin, M.F.S. (2022) Antibodies for the coat protein of cotton leafroll dwarf virus detect Commelina sp. as an intermediary host for cotton blue disease. Frontiers in Plant Science, 13, 814119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D. , Herbers, K. , Melzer, M. , Omid, A. , Tacke, E. , Wolf, S. et al. (2001) Evidence for expression level‐dependent modulation of carbohydrate status and viral resistance by the potato leafroll virus movement protein in transgenic tobacco plants. The Plant Journal, 28, 529–543. [DOI] [PubMed] [Google Scholar]
- Horn, N.M. , Reddy, S.V. , van den Heuvel, J.F.J.M. & Reddy, D.V.R. (1995) Survey of chickpea (Cicer arietinum L.) for chickpea stunt disease and associated viruses in India and Pakistan. Plant Disease, 80, 286–290. [Google Scholar]
- Igori, D. , Shin, A.‐Y. , Kim, S.E. , Kwon, S.‐Y. & Moon, J.S. (2022) First report of cotton leafroll dwarf virus infecting Hibiscus syriacus in South Korea. Plant Disease, 106, 3003. [DOI] [PubMed] [Google Scholar]
- Iriarte, F.B. , Dey, K.K. , Small, I.M. , Conner, K.N. , O'Brien, G.K. , Johnson, L. et al. (2020) First report of cotton leafroll dwarf virus in Florida. Plant Disease, 104, 2744–2020. [Google Scholar]
- Jaag, H.M. , Kawchuk, L. , Rohde, W. , Fischer, R. , Emans, N. & Prüfer, D. (2003) An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication‐associated protein. Proceedings of the National Academy of Sciences of the United States of America, 100, 8939–8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalappara, S.R. , Riley, D.G. , Cremonez, P.S.G. , Perier, J.D. & Bag, S. (2022) Wild radish (Raphanus raphanistrum L.) is a potential reservoir host of cucurbit chlorotic yellows virus. Viruses, 14, 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, S.G. , Sharman, M. , Moukahel, A. , Ziyaev, Z. & Ahmed, S. (2020) First report of cotton leafroll dwarf virus affecting chickpea (Cicer arietinum) in Uzbekistan. Plant Disease, 104, 2532. [DOI] [PubMed] [Google Scholar]
- Latourrette, K. , Holste, N.M. & Garcia‐Ruiz, H. (2021) Polerovirus genomic variation. Virus Evolution, 7, veab1021–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardon, S. (1994) Caracterización del agente causal de la enfermedad azul del algodón en el área centro chaqueña . Ing. Agr. Bonacic Ivan ‐ Grupo Protección Vegetal, Informe del PIC 320, 536.
- Letunic, I. & Bork, P. (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 49, 293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahas, J.W. , Hamilton, F.B. , Roberts, P.M. , Ray, C.H. , Miller, G.L. , Sharman, M. et al. (2022) Investigating the effects of planting date and Aphis gossypii management on reducing the final incidence of cotton leafroll dwarf virus. Crop Protection, 158, 106005. [Google Scholar]
- McGraw, L. (2000) The cause of bronze wilt of cotton. Agricultural Research Report Number11 . Available from: https://agresearchmag.ars.usda.gov/ar/archive/2000/nov/wilt1100.pdf. [Accessed on 31st December 2022]
- Medrano, H. , Escalona, J.M. , Bota, J. , Gulías, J. & Flexas, J. (2002) Regulation of photosynthesis of C3 plants in response to progressive drought: stomatal conductance as a reference parameter. Annals of Botany, 89, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotto, M.D. & Busoli, A.C. (2003) Eficiência de ninfas e adultos de Aphis gossypii Gloverna transmissão do vírus do mosaico das nervuras do algodoeiro. Bragantia, 62, 255–259. [Google Scholar]
- Michelotto, M.D. & Busoli, A.C. (2006) Efeito da época de inoculação do vírus do mosaico das nervuras por Aphis gossypii Glover (Hemiptera: Aphididae) no desenvolvimento e na produção do algodoeiro. Neotropical Entomology, 35, 251–256. [DOI] [PubMed] [Google Scholar]
- Michelotto, M.D. & Busoli, A.C. (2009) Biologia de Aphis gossypii em plantas infectadas pelo vírus do mosaico das nervuras do algodoeiro. Bragantia, 68, 1017–1024. [Google Scholar]
- Morello, C.D.L. , Nelson, D.S. , Farias, F.J.C. , Lamas, F.M. , Pedrosa, M.B. , Ribeiro, J.L. et al. (2010) BRS 293: a midseason high‐yielding upland cotton cultivar for Brazilian savanna. Crop Breeding and Applied Biotechnology, 10, 180–182. [Google Scholar]
- Mukherjee, A.K. , Chahande, P.R. , Meshram, M.K. & Kranthi, K.R. (2012) First report of Polerovirus of the family Luteoviridae infecting cotton in India. New Disease Reports, 25, 22. [Google Scholar]
- Mukherjee, A.K. , Mukherjee, P.K. & Kranthi, S. (2016) Genetic similarity between cotton leafroll dwarf virus and chickpea stunt disease associated virus in India. Plant Pathology Journal, 32, 580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka, N. , Tihova, M. , Brugidou, C. , Kumar, A. , Beachy, R.N. , Fauquet, C.M. et al. (2000) Structure of native and expanded Sobemoviruses by electron cryo‐microscopy and image reconstruction. Journal of Molecular Biology, 303, 197–211. [DOI] [PubMed] [Google Scholar]
- Padgett, G.B. , Colyer, P.D. & Whitam, H.K. (2004) Bronze wilt in Louisiana cotton. Louisiana Agriculture, 47, 24–25. [Google Scholar]
- Pandey, S. , Bag, S. , Roberts, P. , Conner, K. , Balkcom, K.S. , Price, A.J. et al. (2022) Prospective alternate hosts of an emerging polerovirus in cotton landscapes in the southeastern United States. Viruses, 14, 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash, V. , Sharma, D.B. , Snider, J. , Bag, S. , Roberts, P. , Tabassum, A. et al. (2021) Effect of cotton leafroll dwarf virus on physiological processes and yield of individual cotton plants. Frontiers in Plant Science, 12, 734386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, M.F. , Bak, A. , Sayre, J.M. , Heck, M.L. & Casteel, C.L. (2020) A polerovirus, potato leafroll virus alters plant–vector interactions using three viral proteins. Plant, Cell and Environment, 43, 387–399. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Marrocco, K. , Lechner, E. , Berry, B. , Brault, V. et al. (2006) F‐box‐like domain in the polerovirus protein P0 is required for silencing suppressor function. Proceedings of the National Academy of Sciences of the United States of America, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, K.A. , Gildow, F. , Palukaitis, P. & Gray, S.M. (2009) The C terminus of the polerovirus P5 readthrough domain limits virus infection to the phloem. Journal of Virology, 83, 5419–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps, B.J. (2000) Cotton variety response to bronze wilt in Missouri and northern Tennessee. In: Dugger, C.P. & Richter, D.A. (Eds.) Proceedings of the 2000 Beltwide cotton conferences, 4–8 January 2000. Memphis, TN: National Cotton Council, pp. 152–154. [Google Scholar]
- Price, T. , Valverde, R. , Singh, R. , Davis, J. , Brown, S. & Jones, H. (2020) First report of cotton leafroll dwarf virus in Louisiana. Plant Health Progress, 21, 142–143. [Google Scholar]
- Pupim Junior, O. , Schuster, I. , Pinto, R.B. , Pires, E. , Belot, J.L. , Silvie, P. et al. (2008) Herança da resistência do algodoeiro à doença‐azul. Pesquisa Agropecuária Brasileira, 43, 661–665. [Google Scholar]
- Rambaut, A. , Drummond, A.J. , Xie, D. , Baele, G. & Suchard, M.A. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos‐Sobrinho, R. , Adegbola, R.O. , Lawrence, K. , Schrimsher, D.W. , Isakeit, T. , Alabi, O.J. et al. (2021) Cotton leafroll dwarf virus US genomes comprise divergent subpopulations and harbor extensive variability. Viruses, 13, 2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos‐Sobrinho, R. , Ferro, M.M.M. , de Lima, G.S.A. & Nagata, T. (2022) First report of cotton leafroll dwarf virus infecting cacao (Theobroma cacao L.) trees in Brazil. Plant Disease. Available from: 10.1094/PDIS-07-22-1570-PDN [DOI] [PubMed] [Google Scholar]
- Ray, J.D. , Sharman, M. , Quintao, V. , Rossel, B. , Westaway, J. & Gambley, C. (2016) Cotton leafroll dwarf virus detected in Timor‐Leste. Australasian Plant Disease Notes, 11, 29. [Google Scholar]
- Rotenberg, D. , Krishna Kumar, N.K. , Ullman, D.E. , Montero‐Astúa, M. , Willis, D.K. , German, T.L. et al. (2009) Variation in tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathology, 99, 404–410. [DOI] [PubMed] [Google Scholar]
- Sedhain, N.P. , Bag, S. , Morgan, K. , Carter, R. , Triana, P. , Whitaker, J. et al. (2021) Natural host range, incidence on overwintering cotton and diversity of cotton leafroll dwarf virus in Georgia USA. Crop Protection, 144, 105604. [Google Scholar]
- Sharman, M. , Lapbanjob, S. , Sebunruang, P. , Belot, J.L. , Galbieri, R. , Giband, M. et al. (2015) First report of cotton leafroll dwarf virus in Thailand using a species‐specific PCR validated with isolates from Brazil. Australasian Plant Disease Notes, 10, 24. [Google Scholar]
- Silva, T.F. , Correa, R.L. , Castilho, Y. , Silvie, P. , Belot, J.L. & Vaslin, M.F.S. (2008) Widespread distribution and a new recombinant species of Brazilian virus associated with cotton blue disease. Virology Journal, 5, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E. , Firth, A.E. , Miller, W.A. , Scheidecker, D. , Brault, V. , Reinbold, C. et al. (2015) Discovery of a small non‐AUG‐initiated ORF in poleroviruses and luteoviruses that is required for long‐distance movement. PLoS Pathogens, 11, e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sõmera, M. , Fargette, D. , Hébrard, E. , Sarmiento, C. & ICTV Report Consortium . (2021) ICTV virus taxonomy profile: Solemoviridae 2021. Journal of General Virology, 102, 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard, M.A. , Lemey, P. , Baele, G. , Ayres, D.L. , Drummond, A.J. & Rambaut, A. (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4, vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Li, Y.Y. , Wang, Y. , Zhao, H.H. , Zhao, T.Y. , Zhang, Z.Y. et al. (2018) Brassica yellows virus P0 protein impairs the antiviral activity of NbRAF2 in Nicotiana benthamiana . Journal of Experimental Botany, 69, 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum, A. , Bag, S. , Roberts, P. , Suassuna, N. , Chee, P. , Whitaker, J.R. et al. (2019) First report of cotton leafroll dwarf virus infecting cotton in Georgia, U.S.A. Plant Disease, 103, 1803. [Google Scholar]
- Tabassum, A. , Bag, S. , Suassuna, N.D. , Conner, K.N. , Chee, P. , Kemerait, R.C. et al. (2021) Genome analysis of cotton leafroll dwarf virus reveals variability in the silencing suppressor protein, genotypes, and genomic recombinants in the USA. PLoS One, 16, e0252523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum, A. , Roberts, P.M. & Bag, S. (2020) Genome sequence of cotton leafroll dwarf virus infecting cotton in Georgia, USA. Microbiology Resource Announcements, 9, e00812–e00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, J.K. (2003) Estudo da relação vetor‐patógeno‐hospedeiro para a doença azul do algodoeiro . MSc Thesis, Sao Paulo, Instituto Agronômico de Campinas.
- Takimoto, J.K. , Benetti Queiroz‐voltan, R. & Alberto, J. (2009) Alterações anatômicas em algodoeiro infectado pelo vírus da doença azul. Bragantia, 68, 109–116. [Google Scholar]
- Tamura, K. , Stecher, G. & Kumar, S. (2021) MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38, 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiessen, L.D. , Schappe, T. , Zaccaron, M. , Conner, K. , Koebernick, J. , Jacobson, A. et al. (2020) First report of cotton leafroll dwarf virus in cotton plants affected by cotton leafroll dwarf disease in North Carolina. Plant Disease, 104, 3275. [Google Scholar]
- USDA . (2021) NASSQuickstat, a comprehensive tool for accessing agricultural data published by National Agricultural Statistics Service . Available from: https://quickstats.nass.usda.gov [Accessed 28th December 2022].
- Vassilakos, N. , Simon, V. , Tzima, A. , Johansen, E. & Moury, B. (2016) Genetic determinism and evolutionary reconstruction of a host jump in a plant virus. Molecular Biology and Evolution, 33, 541–553. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Greene, J. , Mueller, J. , Conner, K. & Jacobson, A. (2020) First report of cotton leafroll dwarf virus in cotton fields of South Carolina. Plant Disease, 104, 2532. [Google Scholar]
- Yang, X. , Du, M. , Li, S. & Zhou, X. (2021) Coinfection of cotton plants with watermelon mosaic virus and a novel polerovirus in China. Viruses, 13, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, T. , Li, Y.Y. , Xiang, H.Y. , Wu, Z.Y. , Wang, X.B. , Wang, Y. et al. (2014) Amino acid sequence motifs essential for P0‐mediated suppression of RNA silencing in an isolate of potato leafroll virus from Inner Mongolia. Molecular Plant‐Microbe Interactions, 27, 515–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
Data sharing is not applicable to this article as no new data were created.


