Abstract
Broadly neutralizing antibodies against SARS-CoV-2 variants are sought to curb COVID-19 infections. Here we produced and characterized a set of mouse monoclonal antibodies (mAbs) specific for the ancestral SARS-CoV-2 receptor binding domain (RBD). Two of them, 17A7 and 17B10, were highly potent in microneutralization assay with 50% inhibitory concentration (IC50) ≤ 135 ng/ml against infectious SARS-CoV-2 variants, including G614, Alpha, Beta, Gamma, Delta, Epsilon, Zeta, Kappa, Lambda, B.1.1.298, B.1.222, B.1.5 and R.1. Both mAbs (especially 17A7) also exhibited strong in vivo efficacy in protecting K18-hACE2 transgenic mice from the lethal infection with G614, Alpha, Beta, Gamma and Delta viruses. Structural analysis indicated that 17A7 and 17B10 target the tip of the receptor binding motif (RBM) in the RBD-up conformation. A third RBD-reactive mAb (3A6) although escaped by Beta and Gamma, was highly effective in cross-neutralizing Delta and Omicron BA.1 variants in vitro and in vivo. In competition experiments, antibodies targeting epitopes similar to these 3 mAbs were rarely enriched in human COVID-19 convalescent sera or post-vaccination sera. These results are helpful to inform new antibody/vaccine design and these mAbs can be useful tools for characterizing SARS-CoV-2 variants and elicited antibody responses.
Keywords: SARS-CoV-2, receptor binding domain (RBD), RBD-reactive antibody, neutralizing antibody, cross-neutralization, structural analysis
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the deadliest disease outbreaks in modern history. As of December 2022, >600 million confirmed COVID-19 infections including >6.6 million deaths worldwide have been reported to the World Health Organization (WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data).
SARS-CoV-2 infection of human cells is initiated by its spike glycoprotein. The spike is a trimeric protein consisting of three copies of each S1 subunit and S2 subunit1. The S1 subunit contains the receptor binding domain (RBD) that is involved in direct binding to host cell receptor angiotensin-converting enzyme 2 (ACE2) for viral entry2,3. The S2 subunit is the fusion machinery that mediates fusion between the viral membrane and the host cell membrane4. Similar to SARS-CoV and MERS-CoV, the SARS-CoV-2 RBD also adopts two states of conformation — “up” and “down” positions1,4,5. When RBD is in the “down” conformation, the ACE2 binding site is partially buried and is inaccessible to the receptor and some neutralizing antibodies; when the RBD conformation changes to an “up” position, the ACE2 binding site is fully exposed for the receptor engagement and cell attachment1,4. The spike is also the major surface antigen that induces strong host immune responses6–9. Thus, spike is the primary target for vaccine design and therapeutic antibody development.
Most SARS-CoV-2 neutralizing antibodies reported in the literature directly target RBD and are classified into several groups according to their epitope specificities10–13. Class 1/2 antibodies described by Barnes et al.10, RBD-1/2 antibodies by Hastie et al.11, or RBS-A/B antibodies by Yuan et al.12,13 essentially refer to the same type of neutralizing antibodies that only bind RBDs in the “up” conformation and directly compete with ACE2 for binding. Representative antibodies in this group include Etesevimab (LY-CoV16 or CB6), Casirivimab (REGN10933), AZD8895, COVA2-04, CoVIC-259, etc.12–14. RBD-3 antibodies exemplified by CoVIC-080 and EMD-24346 also bind the center of ACE2 binding site in the RBD-up conformation toward the mesa of the receptor binding motif (RBM)11. RBD-4/5 or RBS-C/D or Class 3 antibodies bind the outer face of RBD in either “up” or “down” conformation10–14. Antibodies in this group include CoVIC-215, CoVIC-096, P2B-2F6, REGN10987, C135, C110, etc.10–12. Antibodies from the remaining structural groups exemplified by CR3022 bind a conserved cryptic epitope on the inner face of RBD away from the ACE2 binding site10–14.
Since the beginning of the COVID-19 pandemic, SARS-CoV-2 has evolved rapidly into many genetic lineages with different antigenic properties (https://nextstrain.org/), five of them (Alpha, Beta, Gamma, Delta and Omicron) have been named as variants of concern (VOCs) by the World Health Organization. Mutations acquired by VOCs are heavily concentrated in the RBD region, enabling them to escape recognition by antibodies11,12,15–21. VOC mutations also confer resistance to neutralization by convalescent sera and vaccine-elicited polyclonal antibodies likely contributing to reinfections and breakthrough infections20,22–26. Thus, highly potent and broadly neutralizing antibodies are desired to cope with the continuous emergence of novel SARS-CoV-2 variants.
In this study, we characterized a set of mouse RBD-reactive mAbs and identified two mAbs — 17A7 and 17B10 that broadly neutralize infectious SARS-CoV-2 from ancestral strains to Delta variants. A third RBD-reactive mAb — 3A6, sensitive to E484K and N501Y substitutions in the ACE2 binding site, is highly potent at neutralizing Delta and Omicron BA.1 variants in vitro and in vivo. These mAbs should be useful reagents for characterization of emerging variants and related vaccines.
Results
Binding characteristics of RBD-reactive mAbs
Five mAbs — 17A7, 17B10, 3A6, 2G5 and 20B5 (Supplementary Table S1) were selected from a pool of hybridoma clones derived from mice immunized with the RBD of the original Wuhan-Hu-1 (D614) strain. All mAbs showed high binding affinity to ancestral RBD as measured by Bio-layer Interferometry (BLI) (Figure 1A). They also exhibited high neutralization potency, with 50% inhibitory concentration (IC50) ≤ 198 ng/ml against pseudovirus bearing D614 spike (Figure 1B). The BLI epitope binning assay showed that 17B10 competed with the other 4 mAbs for binding to immobilized RBD but could not block the binding of a purified spike-specific rabbit polyclonal antibody (Figure 2A), suggesting that these 5 mAbs target similar epitopes on RBD. On the other hand, these 5 mAbs exhibited different ELISA binding affinity toward immobilized RBD variants (Figure 2B–2F). 17A7 had larger ELISA Kd against RBD from the original Wuhan-Hu-1 (D614) and variants of Alpha, Beta and Gamma than the other 4 mAbs (Figure 2B–2E). 2G5 had the smallest ELISA Kd against the original RBD and Alpha RBD (Figure 2B and 2C). mAb 3A6 exhibited minimal binding to immobilized Beta RBD (Figure 2D) and a much higher Kd against Delta RBD than the other mAbs (Figure 2F). These results suggest that point mutations in RBD variants affect the binding characteristics of these RBD-reactive mAbs.
Figure 1.
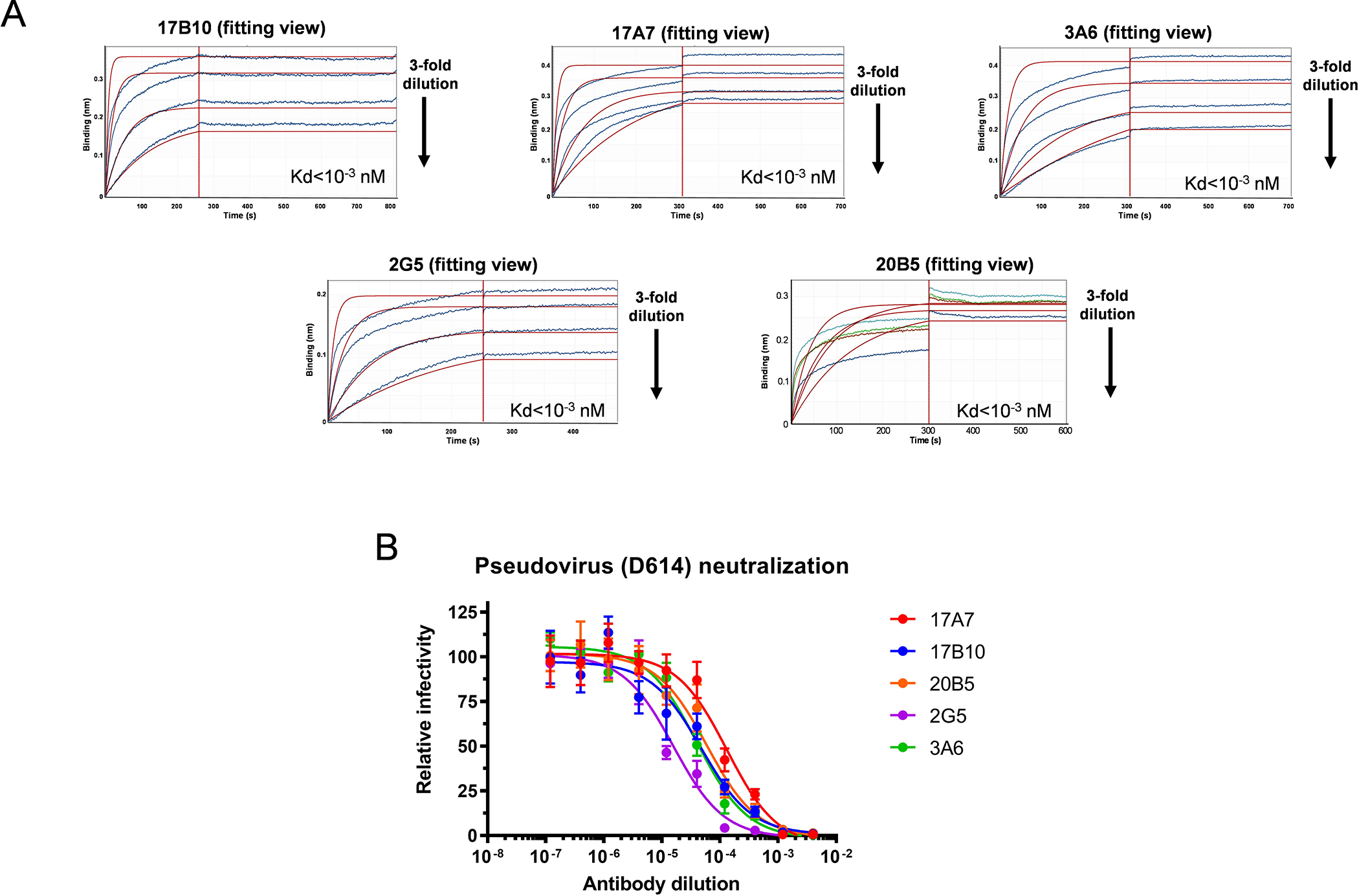
Mouse mAb clones with high affinity to the RBD of the original Wuhan-Hu-1 strain. Mouse hybridomas were generated using the original RBD as the immunogen. Five mAb clones (17B10, 17A7, 2G5, 3A6 and 20B5) were selected and evaluated for RBD binding by bio-layer interferometry (BLI). (A) Representative BLI sensorgrams of individual mAbs binding to immobilized RBD. All five mAbs were 3-fold diluted from the initial concentration of 15 μg/mL and collected data were applied to a built-in 1:2 global curve-fitting model with Kd values shown. (B) Pseudovirus-based neutralization against SARS-CoV-2 spike (D614).
Figure 2.
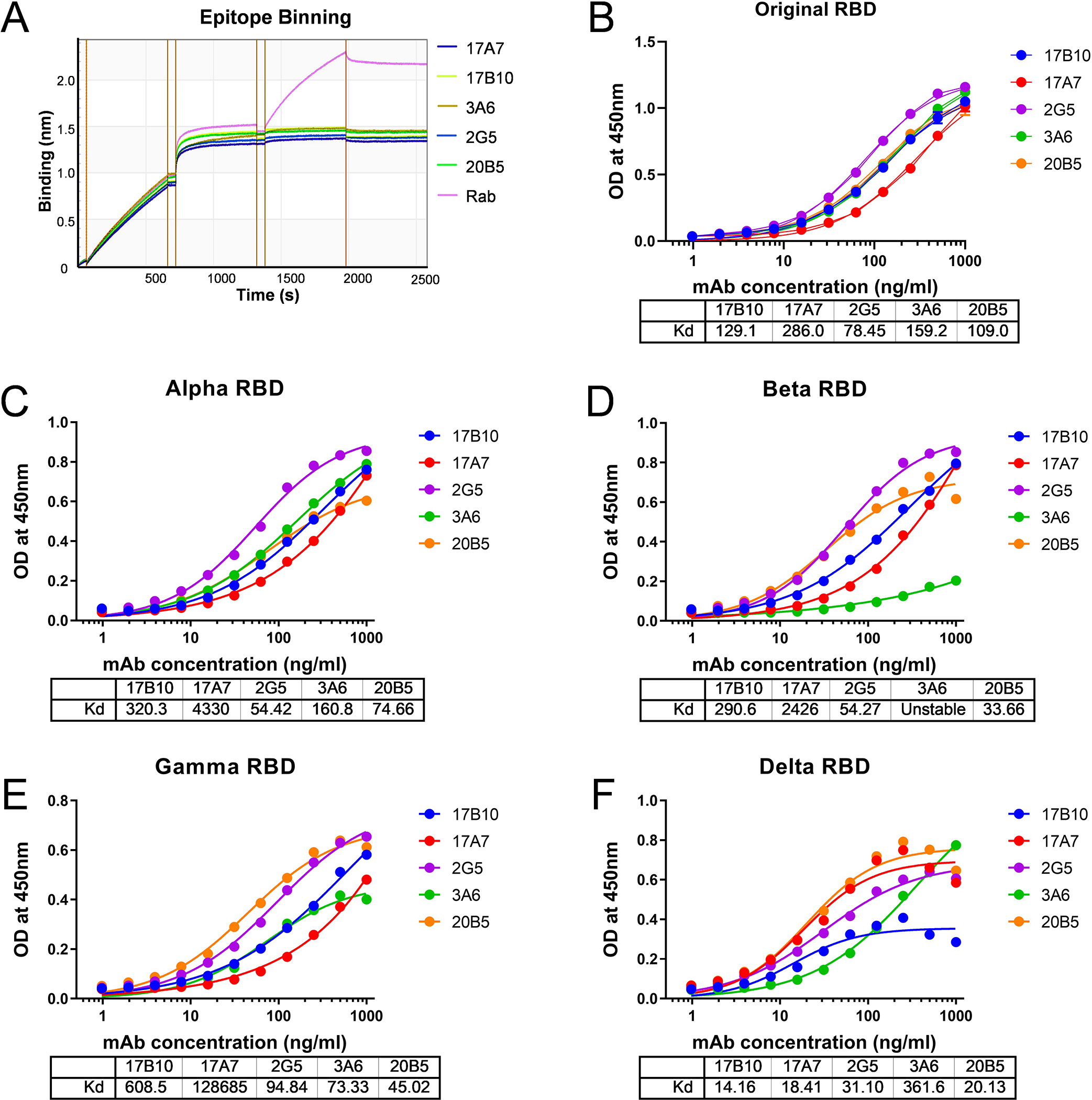
Distinct binding profiles to RBD variants. (A) Epitope binning of 17B10 mAb with competitive mAbs (17A7, 2G5, 3A6 or 20B5) for RBD binding. Spike-specific rabbit polyclonal Ab (Rab) was used as control. (B-F) ELISA binding to different RBD variants: (B) original RBD; (C) Alpha RBD; D) Beta RBD; (E) Gamma RBD; (F) Delta RBD. OD at 450 nm (mean± SD, n= 4 replicates per mAb dilution) were plotted by nonlinear regression curve fit (Specific binding with Hill slope) with individual Kd values shown.
Distinct neutralization profiles of RBD-reactive mAbs against live SARS-CoV-2 clinical isolates
When tested by live virus-based microneutralization (MN) assay, all 5 mouse mAbs were highly effective in inhibiting the original D614 and G614 (B.1.3) viruses, as well as variants that emerged before the Delta wave, including Alpha (B.1.1.7), B.1.1.298, B.1.222, Epsilon (B.1.429), B.1.5 and Lambda (C.37) viruses (Figure 3A–3H) with MN IC50 ranging from 0.9 ng/ml to 135 ng/ml (Table 1). In comparison, two well characterized human RBD-reactive mAbs COVA2–3910,27 and C63C76,28 were significantly less effective in neutralizing live D614 with very limited cross-reactivity toward other variants (Table 1). Among 5 mouse mAbs, 17A7 was the most potent with the lowest MN IC50 (≤ 29 ng/ml) against nearly all SARS-CoV-2 variants prior to the Delta (Figure 3A–3M, Table 1). 17B10 ranked second in neutralizing activity against pre-Delta variants, with MN IC50 ranging from 1.3 ng/ml to 60 ng/ml (Table 1). 3A6 was the only mAb that was unable to neutralize the Beta (B.1.351) variant and had significantly higher IC50 (392–5527 ng/ml) against R.1, Gamma (P.1) and Zeta (P.2) variants than the other mAbs (Figure 3I–3L, Table 1). Interestingly, 3A6 was highly effective in inhibiting the Delta variant with a significantly lower IC50 of 7.3 ng/ml compared to 17A7 (125 ng/ml), 17B10 (136 ng/ml), 20B5 (70 ng/ml) and 2G5 (888 ng/ml) (Figure 3N, Table 1). These results indicate that these RBD-reactive mAbs, especially 17A7 and 17B10 have high neutralizing activities against a wide range of SARS-CoV-2 clinical isolates from the original D614 strain to the Delta variant.
Figure 3.
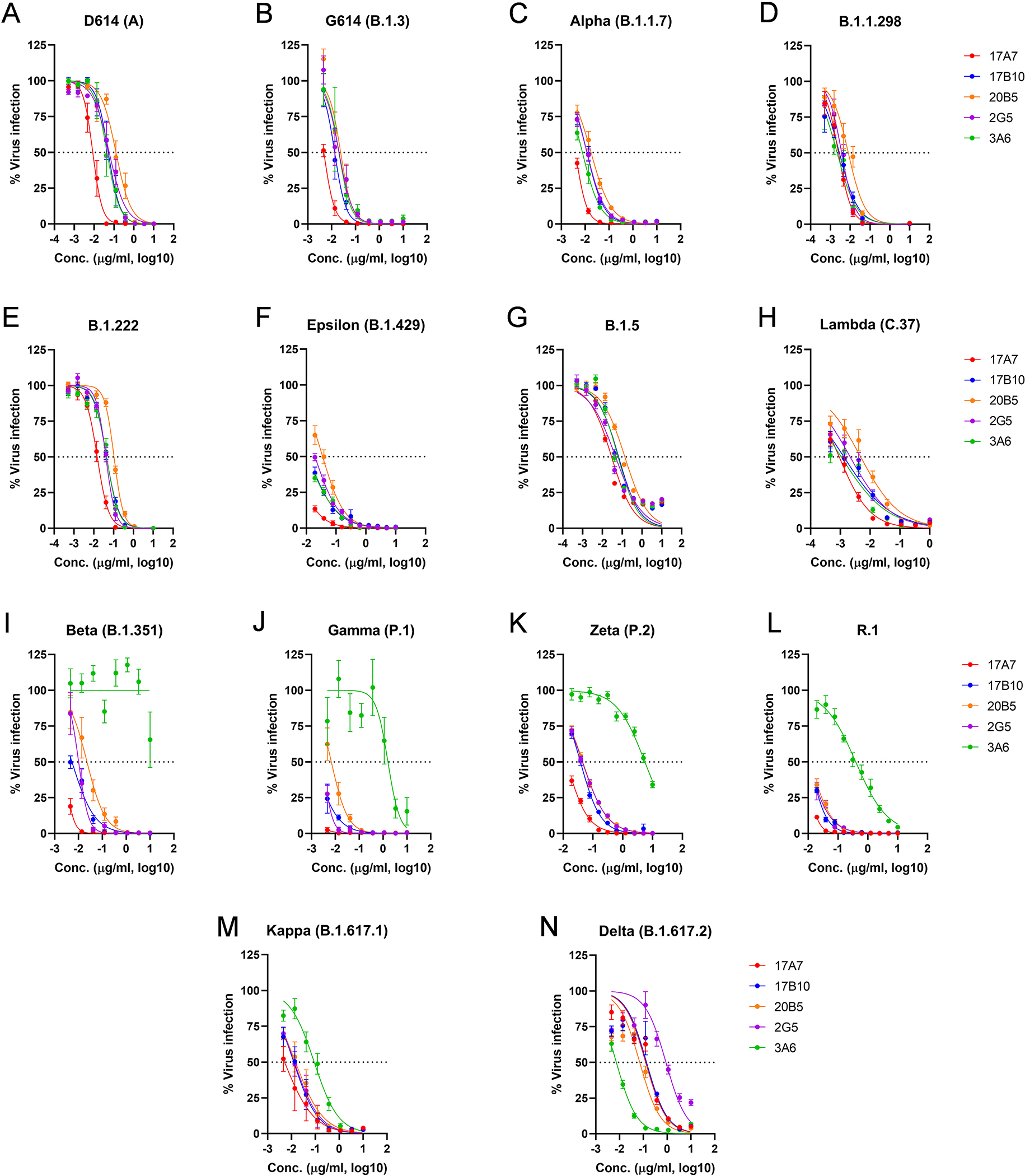
RBD-reactive mAbs cross-neutralizing multiple SARS-CoV-2 variants. Five mAb clones (17B10, 17A7, 2G5, 3A6 and 20B5) were assessed for neutralizing potential against live infectious SARS-CoV-2 strains, including (A) the ancestral D614 (A), (B) G614(B.1.3), (C) Alpha (B.1.1.7), (D) B.1.1.298, (E) B.1.222, (F) Epsilon (B.1.429), (G) B.1.5, (H_ Lambda (C.37), (I) Beta (B.1.351), (J) Gamma (P.1), (K) Zeta (P.2), (L) R.1, (M) Kappa (B.1.617.1) and (N) Delta (B.1.617.2). The % virus infection verse the wells that contained virus only were plotted by nonlinear regression. Data are expressed as mean± SD, n= 8 replicates per mAb concentration. Estimated IC50 values of individual mAbs are summarized in Table 1.
Table 1.
Microneutralization IC50 of mAbs against wild type live SARS-CoV-2 isolates
| Virus & Lineage | Microneutralization IC50 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| 2G5* | 3A6* | 17A7* | 17B10* | 20B5* | Cova2–39# | C63C7# | ||
| WA/1 (D614) (A) | 0.0570 | 0.0443 | 0.0085 | 0.0512 | 0.1255 | 0.150 | 3.657 | |
| NY (G614) (B.1.3) | 0.0204 | 0.0201 | 0.0047 | 0.0134 | 0.0238 | >20 | >20 | |
| Alpha (B.1.1.7) | 0.0118 | 0.0076 | 0.0039 | 0.0117 | 0.0175 | >20 | >20 | |
| B.1.1.298 | 0.0037 | 0.0022 | 0.0025 | 0.0028 | 0.0080 | n.d. | n.d. | |
| B.1.222 | 0.0407 | 0.0441 | 0.0149 | 0.0440 | 0.1049 | |||
| Epsilon (B.1.429) | 0.0195 | 0.0105 | 0.0038 | 0.0116 | 0.0354 | |||
| B.1.5 | 0.0462 | 0.0612 | 0.0291 | 0.0656 | 0.1352 | |||
| Lambda (C.37) | 0.0021 | 0.0011 | 0.0009 | 0.0013 | 0.0048 | |||
| Beta (B.1.351) | 0.0101 | >20 | 0.0025 | 0.0053 | 0.0226 | >20 | >20 | |
| Gamma (P.1) | 0.0030 | 1.5959 | 0.0002 | 0.0014 | 0.0067 | >10 | 0.297 | |
| Zeta (P.2) | 0.0444 | 5.5271 | 0.0130 | 0.0380 | 0.0457 | n.d. | n.d. | |
| R.1 | 0.0100 | 0.3920 | 0.0086 | 0.0114 | 0.0118 | |||
| Kappa (B.1.617.1) | 0.0132 | 0.0891 | 0.0051 | 0.0124 | 0.0146 | |||
| Delta (B.1.617.2) | 0.8881 | 0.0073 | 0.1249 | 0.1358 | 0.0701 | 0.330 | >20 | |
| Omicron (B.1.1.529) | BA.1 | >20 | 10.4954 | >20 | >20 | >20 | >20 | >20 |
| BA.1.1 | 15.4882 | |||||||
| BA.2 | >20 | |||||||
| BA.4 | ||||||||
| BA.5 | ||||||||
All five mouse RBD-reactive mAbs were tested against wild type live infectious SARS-CoV-2 isolates by microneutralization assay (n=8 replicates/mAb/dilution). The neutralization potency (IC50) of individual mAbs was estimated by nonlinear regression using GraphPad Prism (Version 9.3.1). The color scales indicate IC50 values <0.01 μg/ml (lavender), 0.01–0.1 μg/ml (turquoise), 0.1–1 μg/ml (green), 1–20 μg/ml (gold) and >20 μg/ml (white).
Two human RBD-reactive mAbs Cova2–39 and C63C7 were included as the references (n=4 replicates/mAb/dilution). n.d.: not determined.
In vivo protection of RBD-reactive mAbs against live SARS-CoV-2 clinical isolates
Passive antibody transfer and challenge experiments were then conducted to determine the in vivo protective efficacy of selected mAbs in SARS-CoV-2-susceptible K18-hACE2 mice (Figure 4A). 17A7 and 17B10 were selected because they were the most potent mAbs with broad in vitro neutralization spectra (Table 1). Naive K18-hACE2 mice were given 17A7 or 17B10 intraperitoneally, at 3–6 mg/kg body weight (BW), and subsequently challenged intranasally with a pre-determined lethal dose of G614 (103 TCID50/mouse, Figure 4B), Alpha (102 TCID50/mouse, Figure 4C), Beta (102 TCID50/mouse Figure 4D) or Gamma (102 TCID50/mouse, Figure 4E)25,29,30. Control mice that received an irrelevant isotype control showed widespread BW loss and all succumbed to death within 8 days of challenge (Figure 4B–4E). In contrast, administration of 17A7 or 17B10 at a dose of 3 mg/kg or higher protected recipient mice from the lethal infection of G614, Alpha, Beta or Gamma viruses (Figure 4B–4E). The in vivo efficacy results were consistent with in vitro neutralization activities of the mAbs. When recipient mice were challenged with a lethal dose of Delta variant at 102 TCID50/mouse, administration of 17A7 and 17B10 at the dose of 6 mg/kg was unable to prevent infection (Figure 4F). Only 75% (17A7) and 25% (17B10) of mice survived after suffering significant morbidity (Figure 4F). In comparison, mice that received 3A6 at the same dose were fully protected with no or minimal morbidity (Figure 4F). 3A6 administration also resulted in significantly reduced viral titers in lungs (~2 log) and brains (~1 log) as compared to control mice after the Delta challenge (Supplementary Figure S1A). 17A7 at the same dose also significantly reduced pulmonary viral titers in Delta-challenged mice, but the protective effect was marginal nor extended to extrapulmonary organs (Supplementary Figure S1A).
Figure 4.
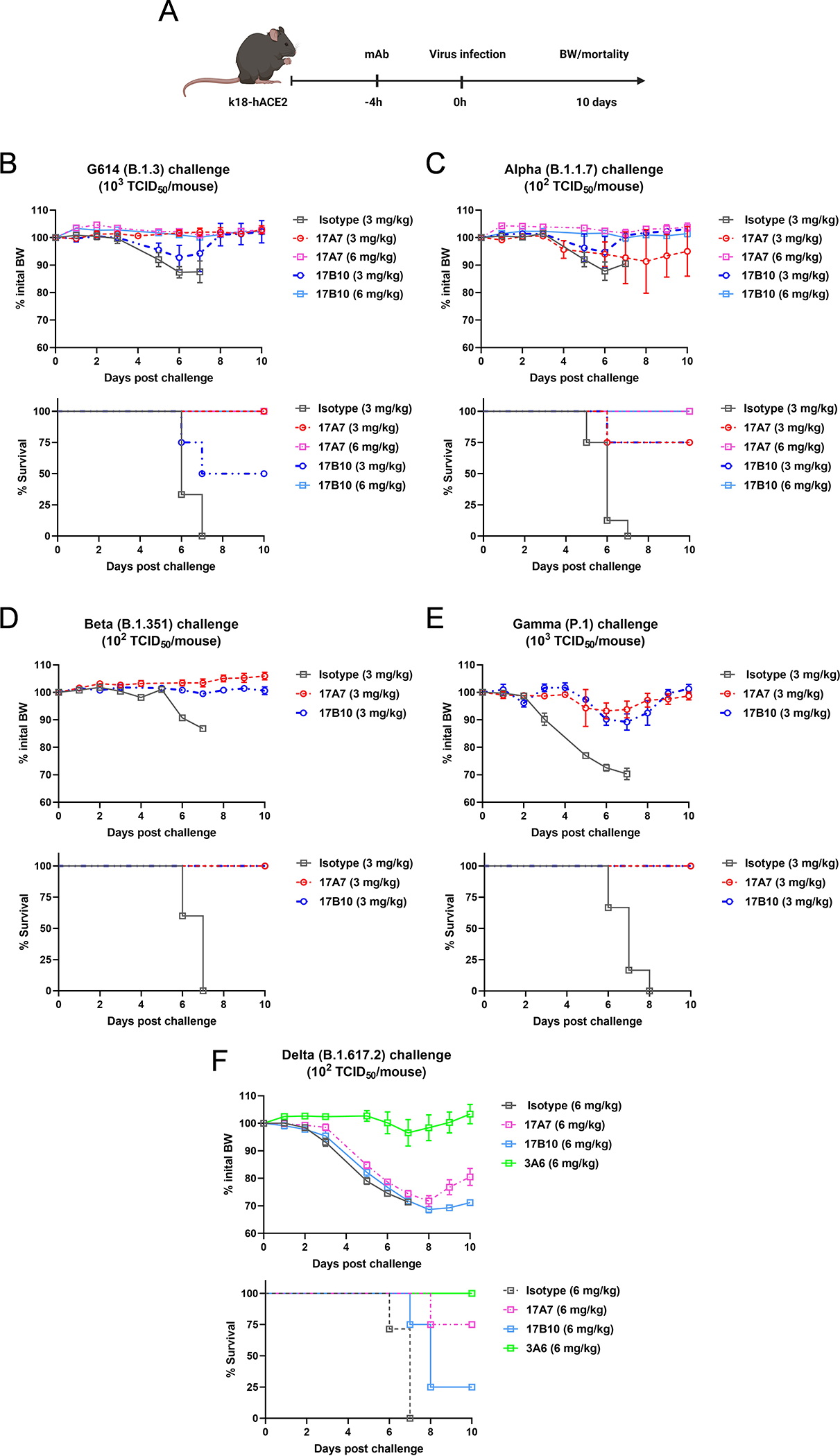
Efficacy of selected RBD mAbs in protecting K18-hACE2 mice against lethal challenges of SARS-CoV-2 and variants. Naïve K18-hACE2 mice were injected intraperitoneally with RBD-reactive 17A7, 17B10 or 3A6mAbs or a nonspecific mouse isotype control. Recipient mice were then challenged intranasally with a pre-determined lethal dose of live infectious SARS-CoV-2. (A) Schematic timelines of the passive transfer and challenge in K18-hACE2 mice. Body weight (BW) drop and mortality of infected mice were monitored for up to 10 days after a lethal challenge of (B) G614 (B.1.3), (C) Alpha (B.1.1.7), (D) Beta (B.1.351), (E) Gamma (P.1) or (F) Delta (B.1.617.2). % BW drops (mean ± s.e.m. of 4–6 mice/group) and % cumulative survivals (n=4–6 mice/group) are shown.
During the characterization of these RBD-reactive mAbs, the Omicron variants emerged and quickly evolved into multiple sub-lineages. Four of the RBD-reactive mAbs (17A7, 17B10, 2G5 and 20B5) completely lost binding to Omicron RBD by ELISA (Figure 5A) and were unable to neutralize live Omicron variants including BA.1, BA.1.1, BA.2, BA.4 and BA.5 in vitro (Figure 5B–5F). Although 3A6 remained reactive to Omicron BA.1 RBD by ELISA (Figure 5A), it lost the ability to recognize and bind to other Omicron variants. Additionally, 3A6 was the only mAb capable of neutralizing live Omicron BA.1 and BA.1.1 variants in vitro (Figure 5B–5D), but the potency was drastically reduced with MN IC50 values >10,000 ng/ml (Table 1). In the K18-hACE2 passive transfer and challenge model, mice injected with 3A6 at 6 mg/kg only had marginally reduced viral titers in lungs and hearts as compared to control mice after Omicron BA.1 challenge (Supplementary Figure S1B). Like the other mAbs, 3A6 completely lost in vitro neutralizing activity against antigenically further-evolved Omicron BA.4 and BA.5 subvariants (Figure 5E and 5G). Taken together, the in vivo protective effects of 17A7, 17B10 and 3A6 are consistent with their in vitro neutralization capacities against live SARS-CoV-2 clinical isolates.
Figure 5.
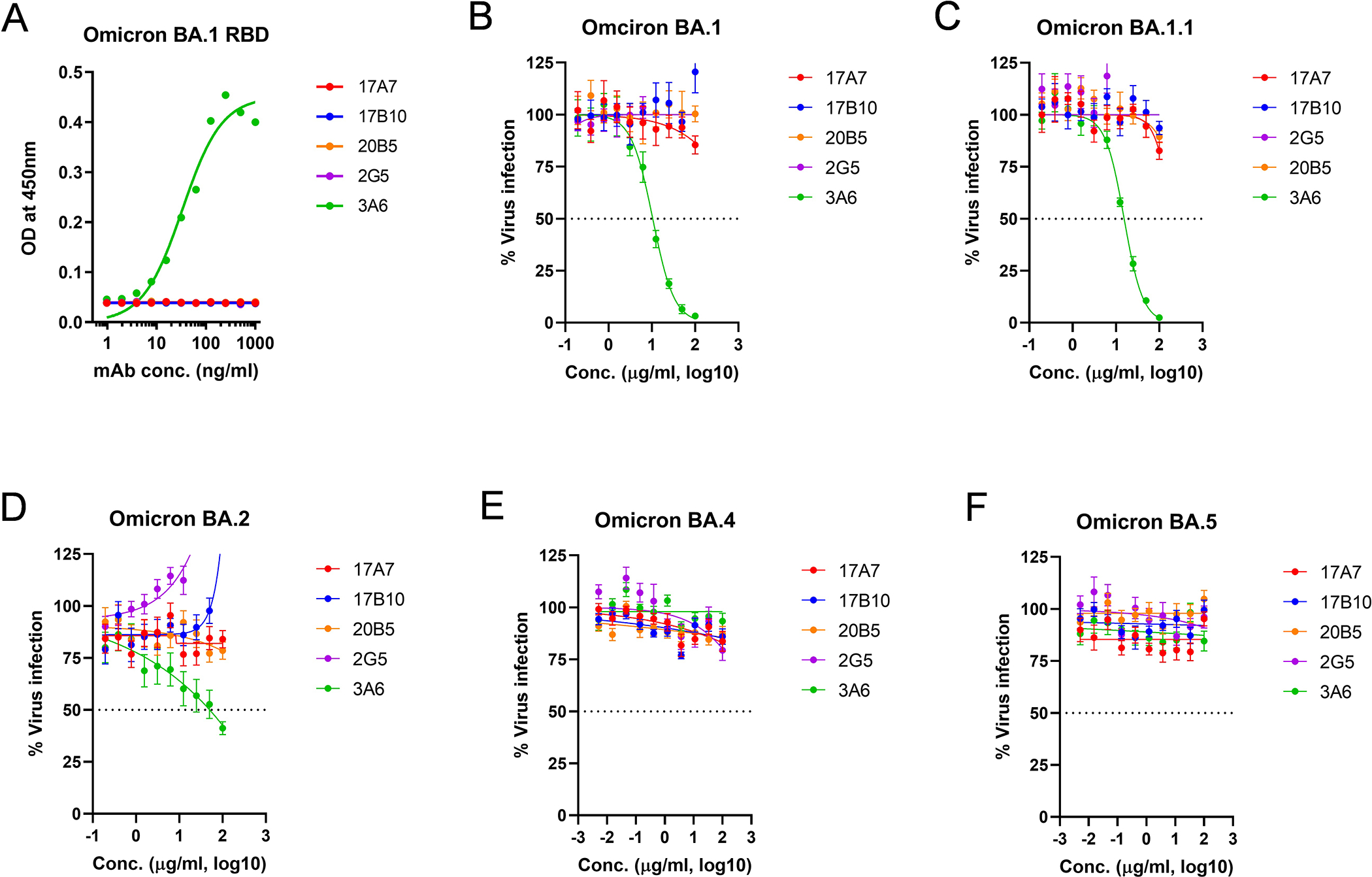
ELISA binding and cross-neutralization of RBD-reactive mAbs against Omicron subvariants. (A) Omicron BA.1 RBD ELISA. The OD at 450 nm (mean± SD, n= 4 replicates per mAb dilution) was plotted by nonlinear regression curve fit (Specific binding with Hill slope). Microneutralization against live infectious Omicron subvariants including (B) Omicron BA.1, (C) Omicron BA.1.1, (D) Omicron BA.2, (E) Omicron BA.4 and (F) Omicron BA.5. The % virus infection verse the wells that contained virus only (mean± SD, n= 8 replicates per mAb concentration) were plotted by 4-parameter logistic curve fitting. Estimated IC50 values of individual mAbs are summarized in Table 1.
Competition of RBD-reactive mAbs with human COVID-19 polyclonal sera
Competitive ELISAs were set up to assess if human COVID-19 polyclonal sera elicited by infection or vaccination could compete with 17A7, 17B10 or 3A6 for RBD binding (Figure 6A). Human COVID-19 convalescent sera elicited during the first wave of the pandemic showed no competition with any of the 3 mAbs tested (Figure 6B–6D). Human COVID-19 mRNA post-vaccination sera also showed no or minimal competition with 17A7 or 17B10 for ancestral RBD binding (Figure 6B and 6C). Although the polyclonal sera elicited by 2-dose COVID-19 mRNA vaccines marginally competed with 3A6 for the original RBD binding, the competitive ability was slightly enhanced after a vaccine booster dose (Figure 6D). These results suggest that human antibodies recognizing similar epitopes as 17A7, 17B10 or 3A6 are not commonly induced by SARS-CoV-2 antigen exposure (infection or vaccination).
Figure 6.
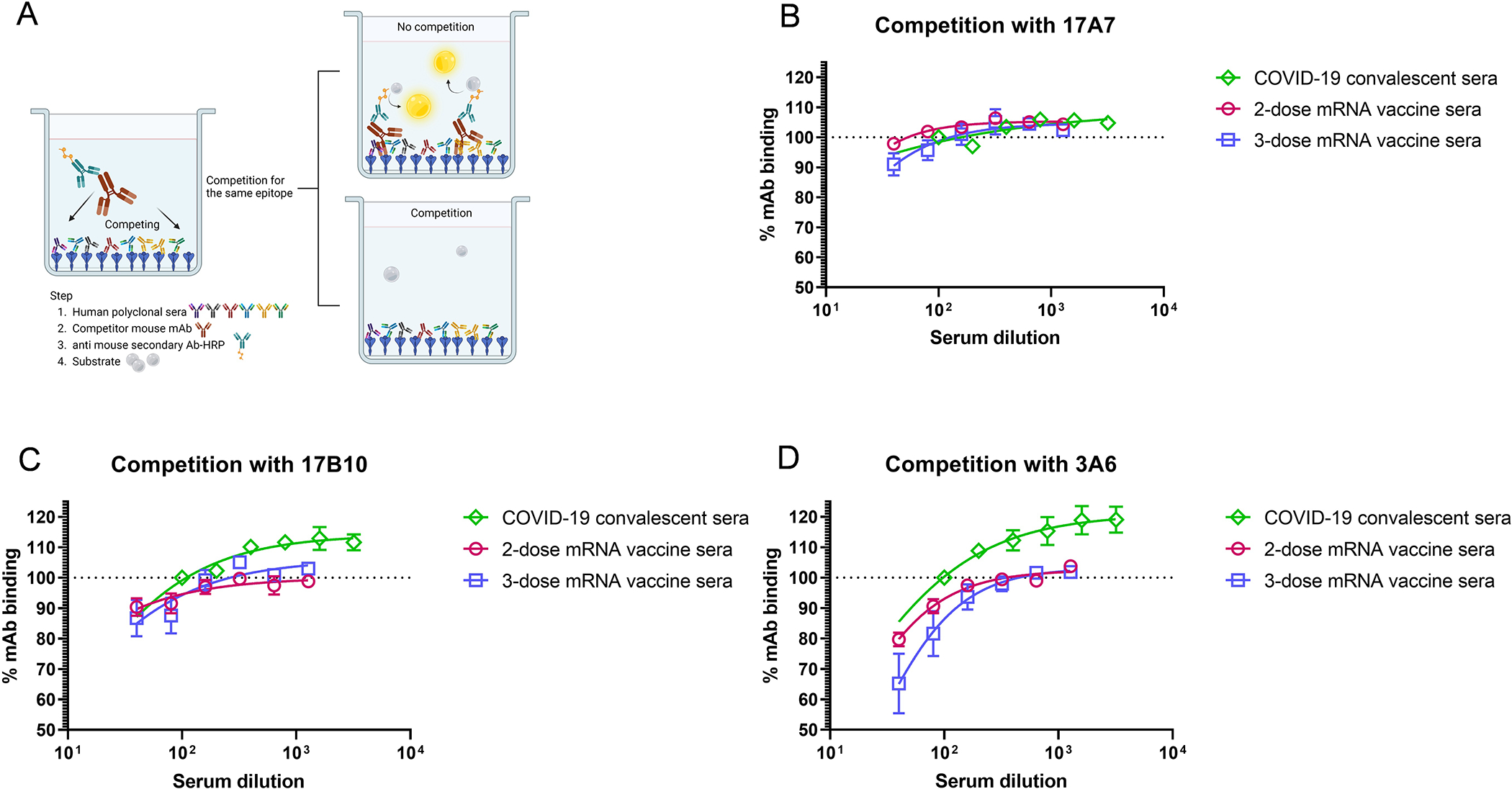
Competition of 17A7, 17B10 and 3A6 mAbs with human COVID-19 convalescent sera or mRNA post-vaccination sera for binding to immobilized original RBD. (A) Schematic illustration of competition between mouse mAbs and human COVID-19 specific polyclonal sera. The % of residual (B) 17A7, (C) 17B10 or (D) 3A6 after competing with human COVID-19 convalescent sera or post-vaccination sera as compared to the wells that contained only mAbs were plotted by nonlinear regression. Data are expressed as mean ± s.e.m (n = 6–8 sera/group).
Structural analysis of cross-neutralizing 17B10, 17A7 and 3A6 mAbs
Cryo-electron microscopy (Cryo-EM) was performed to determine the structure of the complex of 17B10 bound to the full-length spike (G614) trimer (Supplementary Figure S2 and S3). Multiple rounds of 3D classification and refinement led to a major class of one copy of 17B10 Fab in complex with the one-RBD-up spike trimer (Supplementary Figure S2). Further rounds of refinement produced an overall map at 2.8Å resolution and a local map focusing on the Fab-RBD interface at 3.6Å resolution. As shown in Figure 7A, both the CDR H1–3 (Ser52, Asn71, Ser76, Arg119, Tyr120 and Ser477) and CDR L1–3 (Tyr111, Ser113 and Tyr114) of 17B10 make direct contacts with the RBD. On the RBD side, the binding surface is formed primarily by residues 455–458 and residues 473–493, particularly with residues 458, 477, 478, 486, 487 and 489 as the key contacting sites (Figure 7D).
Figure 7.
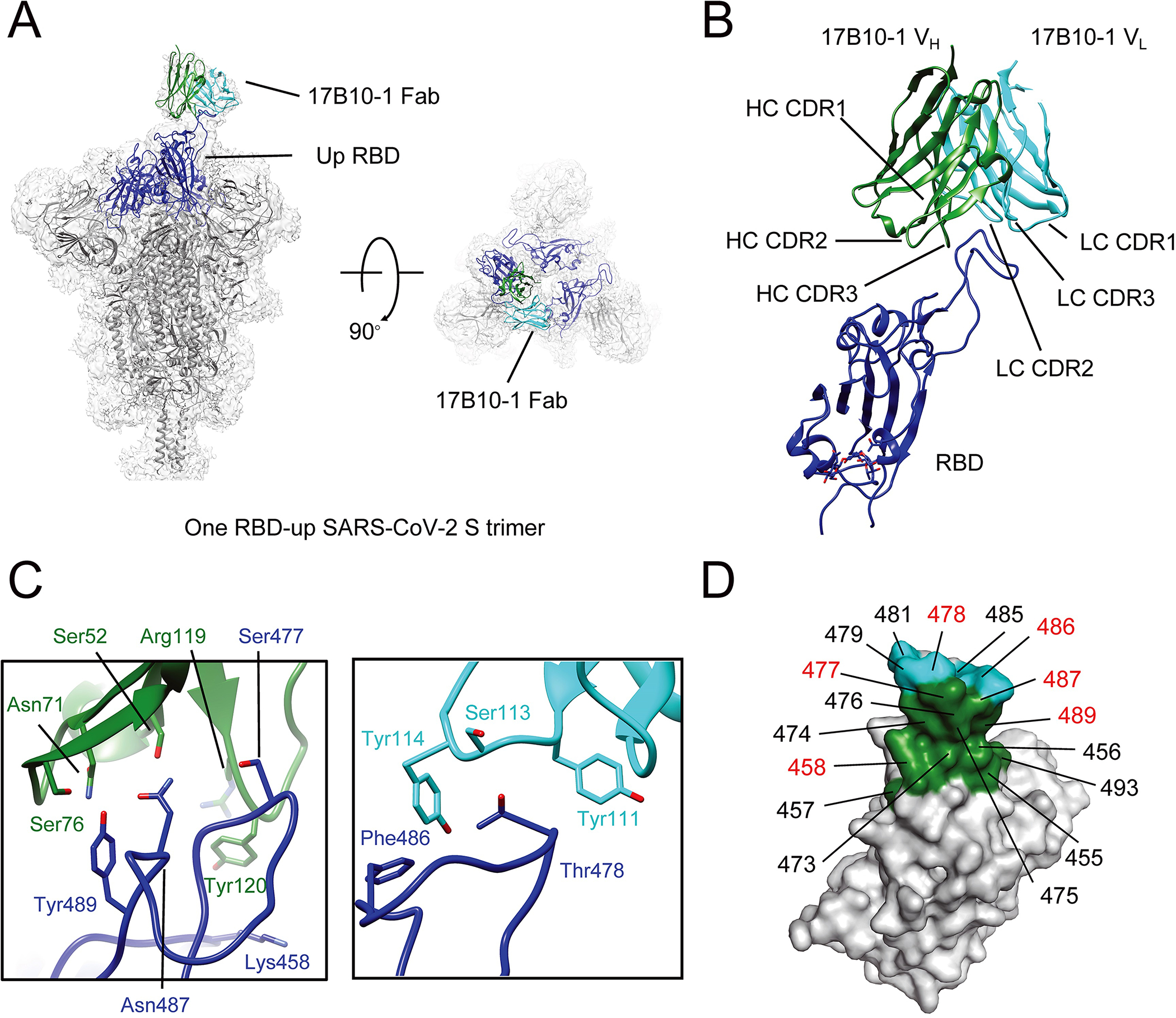
Cryo-EM structures of 17B10 Fab in complex of a full-length SARS-CoV-2 Spiks (G614) trimer. (A) One 17B10–1 Fab bound to one spike trimer in the one-RBD-up (2.8Å) conformation resolved by Cryo-EM. The EM density is colored in gray, and the structures are represented in ribbon diagram with the RBD in blue and the rest in dark gray. The heavy chain of 17B10–1 is shown in green and the light chain in cyan. (B) Close-up view of the interactions between 17B10–1 Fab and the RBD of the S trimer in the one-RBD-up conformation. (C) Zoom-in views of the binding interface between the heavy (green) or light chain (cyan) of 17B10–1 and the tip of one RBD-up conformation. (D) The footprints on the RBD interacting with the heavy chain (green) or light chain (cyan) of 17B10–1 with the major contacting residues highlighted in red.
To compare the binding mode of 17B10 with that of between 17A7 or 3A6, we used negative strain EM and reconstructed the 3D maps of the spike (G614) trimer in complex with 17A7 or 3A6 at a resolution of 11Å and 13Å, respectively (Figure 8). Like 17B10, 17A7 is an RBD-2 (or RBS-B or Class 1) mAb but binds all 3 RBDs in the up conformation in one spike trimer (Figure 8A and 8C, Supplementary Table S1). 3A6 also binds the RBD-up conformation with 3 copies of Fabs per S trimer (Figure 8A), but it has a longer CDR-H3 than 17A7 or 17B10 and may reach further down beyond the tip of the RBM (Supplementary Table S1 and Figure 8D). Many mutations in the RBD region acquired by Omicron subvariants reside in the binding footprints of 17A7, 17B10 and 3A6 (Figure 8E and 8F), possibly accounting for the loss in neutralization by these mAbs.
Figure 8.
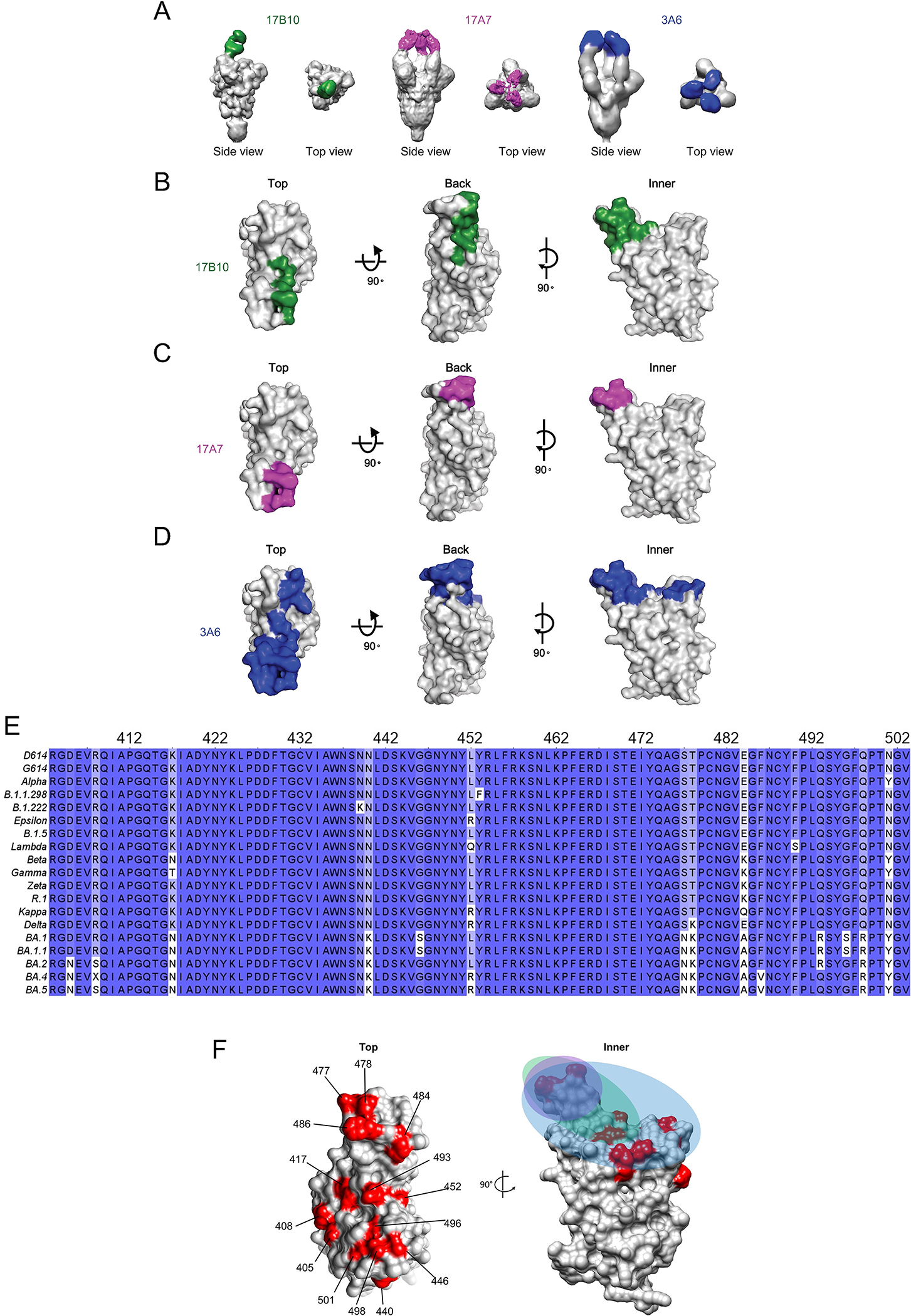
17B10, 17A7 and 3A6 Fab bound to different regions of RBD. The 3D map of 17B10-Spike complex was produced by low-pass filter 17B10-S complex cryo-EM map to 10Å. The 3D maps of 17A7-S and 3A6-S complexes were produced by negative stain EM with a resolution at 11Å and 13Å, respectively. (A) Top view and side view of 3D reconstruction of Fab-spike (gray) complexes. (B-D) Binding footprints of Fabs of three neutralizing antibodies, 17B10 (B), 17A7(C) and 3A6 (D) shading on the RBD in surface representation. The RBD surface (gray) was represented in three views (top, back and inner). The Fabs are colored as 17B10: green; 17A7: magenta; 3A6: blue. (E) Sequence alignment showing multiple mutations in the RBD region (aa 403–503) among SARS-CoV-2 variants. (F) The positions on the RBD being mutated in Omicron subvariants highlighted in red with the colored footprints of 17B10 (green), 17A7 (magenta) and 3A6 (blue).
Discussion
In this study, we show that two RBD-reactive mAbs — 17A7 and 17B10, are highly efficient in neutralizing infectious SARS-CoV-2 with the MN IC50 value ranging from 0.2–136 ng/ml against all SARS-CoV-2 variants that emerged prior to Omicron. 17A7 is a better neutralizer than 17B10 probably because it has 3 copies of Fabs that fully occupy all 3 RBDs of the spike trimer in contrast to only 1 copy of 17B10 Fab engaging in binding. Antibodies in the group of RBD-1/2 (or Class 1/2 or RBS-A/B) are primarily RBM-directed and strongly compete for ACE2 binding, thus generally more potent than antibodies targeting non-RBM-directed epitopes6,10,11,13,14,31. On the other hand, they are also very sensitive to mutations acquired in the RBM and are limited in the breadth of cross-reactivity10,11,15,17,31,32. S477N, T478K and F486V substitutions in Omicron subvariants are within the binding footprints of 17B10 and 17A7, resulting in complete loss of neutralizing activity against these variants. While T478K affects Class 2–4 mAbs more, S477N reduces the reactivity of all four classes of RBD mAb by 1.5- to 4.4-fold20. F486V severely affects Class 2 mAbs and reduce the potency of REGN10933, COV2–2196 and LYCoV555 by 272- to 10,000-fold against BA.4/BA.518.
In addition to 17A7 and 17B10, another RBD-reactive mAb — 3A6 is highly potent at neutralizing Delta but less effective against Gamma and Zeta, and unable to inhibit Beta. E484K and N501Y substitutions are shared among Beta, Gamma and Zeta. E484K resides in the tip of the RBM, while N501Y is on the other end of the RBM. The E484K/N501Y mutations improve virus fitness but have no impact on ACE2 binding affinity33. E484K/N501Y also confer resistance to RBD-3 antibodies, but only some RBD-2 or Class 2 antibodies are sensitive to E484K mutation10–12,14. RBS-C antibodies targeting the outer face of RBD are also affected by E484K which abolishes the interaction between Glutamate484 in the RBD and an arginine in the CDR-H312. Omicron subvariants possess E484A and show strong evasion of Class 2 mAbs34. Yet 3A6 retained binding to BA.1 RBD and was able to neutralize infectious BA.1 and BA.1.1 in vitro and BA.1 in vivo. This suggests that E484K has more detrimental than E484A to 3A6 neutralizing activity. Compared to the other RBD-reactive mouse mAbs, 3A6 has a longer CDR-H3 that can reach further toward the mesa of RBD, which could contribute to its high potency at neutralizing Delta variant (IC50: 7.3 ng/ml).
The competition with human convalescent or post-vaccination sera indicates that antibodies targeting similar epitopes as 17A7, 17B10 or 3A6 are rarely induced after COVID-19 infection or vaccination. It has been shown that the polyclonal response elicited by early COVID-19 infection consists of an abundance of Class 2 and Class 3 antibodies directed toward the RBD17,21. The RBM tip is constantly exposed to antibodies regardless of RBD in up or down conformation, which places those key residues (e.g., S477, T478, E484 and F486) in this region under high selection pressure for mutation. Omicron subvariants have acquired various mutations in spike including S477N, T478K, E484A and F486V in the RBM and confer greater immune evasion than prior VOCs18,20,32,34. Variants carrying ≥1 key mutation in the hotspots of RBD (i.e. L452R/Q, E484K/Q, F490S etc.) are more likely to cause SARS-CoV-2 reinfection and vaccine breakthrough infections due to enhanced neutralization resistance19–26.
Taken together, our results demonstrate that several RBD-reactive mAbs are highly potent at cross-neutralizing a wide range of live infectious SARS-CoV-2 variants. Although no mAbs extend the cross-neutralization to the latest Omicron subvariants, they can serve useful tools for characterizing SARS-CoV-2 variants and related vaccines and for profiling vaccination/infection elicited antibody responses. The findings obtained here may still provide useful information to inform future target-directed antibody/vaccine design, for instance, antibodies/vaccines targeting conformational epitopes involving key residues (e.g., 477, 478, 484 and 486 in the RBM tip) would be highly desired.
Methods
RBD-specific mAbs
SARS-CoV-2 RBD antigen was prepared in a baculovirus expression system (GenScript, Piscataway, New Jersey) using a synthetic DNA sequence encoding the RBD (residues 319–533) of Wuhan-Hu-1 strain (GenBank: NC_045512.2) with a 6xHis-tag. Purified RBD was used as the immunogen to generate mouse hybridoma according to the standard protocol (GenScript). Five hybridoma clones (17B10, 17A7, 3A6, 2G5 and 20B5) that secreted RBD-binding IgG in high titers were selected, sub-cloned, and expanded. The 5 mAbs were sequenced and subtyped as mouse IgG1 (Supplementary Table S1). Affinity purified mAbs were aliquoted and stored at −20°C until use. Two human RBD-reactive mAbs — C63C76 and Cova2–3927 were kindly provided by Dr. Duane Wesemann at Brigham and Women’s Hospital and Drs. Johnason Yewdell and Ivan Kosik at NIAID/NIH, respectively.
RBD-specific IgG ELISA
The following SEC-MALS verified His-tagged RBD variant proteins were purchased from Acrobiosystems (Newark, Delaware), including (1) Alpha RBD bearing N501Y (#SPD-C52Hn), (2) Beta RBD bearing K417N, E484K, and N501Y (#SPD-C52Hp), (3) Gamma RBD bearing K417T, E484K and N501Y (#SPD-C52Hr), (4) Delta RBD bearing L452R and T478K (#SPD-C52Hh), and (5) Omicron BA.1 RBD bearing G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H (#SPD-C522e). Ninety-six-well microtiter plates were pre-coated with 1 μg/ml of RBD variant protein. Serially diluted mouse mAbs were then added and were incubated at room temperature for 1h. Bound IgG was detected using peroxidase-conjugated human serum cross-absorbed goat anti-mouse IgG (H+L) (Seracare #5220-0341, 1:2000) followed by 1-Step™ Ultra TMB-ELISA substrate (ThermoFisher # 34028). Optical density (OD) at 450 nm was measured using a Victor V multilabel reader (PerkinElmer) equipped with Wallac 1420 Workstation (Version 3 Revision 4) and were plotted against serum dilutions by nonlinear regression using GraphPad Prism 9.3.1.
In competitive ELISA, deidentified post-vaccination human sera obtained after 2nd or 3rd dose of COVID-19 mRNA vaccines or convalescent sera from the first wave of the pandemic were serially diluted and incubated in RBD pre-coated 96-well microtiter plates at room temperature for 1 h. After thoroughly washing, mouse mAbs were then added and were incubate for another 1 h. After thoroughly washing, bound mAbs were detected using peroxidase-conjugated human serum cross-absorbed goat anti-mouse IgG (H+L) (Seracare #5220–0341, 1:2000) as described above. The % of residual mAbs as compared to the wells that contained only mAbs were plotted against serum dilutions by nonlinear regression using GraphPad Prism 9.3.1.
Bio-layer interferometry (BLI)
RBD-mAb binding was assessed using an Octet® RED96 system (ForteBio) with integrated data acquisition software 10.0. Streptavidin (SA) biosensors (Sartorius #18-5019) were saturated with biotinylated original RBD recombinant protein (Acrobiosystems #SPD-C82E9) in 1X Kinetics Buffer (Sartorius #18-1105). The RBD loaded SA biosensors were then dipped in 3-fold serially diluted mAbs with the initial concentration of 15 μg/ml in 1X Kinetics Buffer for up to 300 s for association, followed by immersion in 1X Kinetics Buffer alone for another 300–600 s for dissociation. In epitope binning experiments, the RBD loaded SA biosensors were dipped in 17B10 mAb (50 μg/ml in 1X Kinetics Buffer) for 600 s. After briefly washing in 1X Kinetics buffer for 60 s, the loaded biosensors were dipped in competing mAbs (50 μg/ml in 1X Kinetics Buffer) for another 600 s. Any binding shift indicates competing mAb targets a different epitope from 17B10. A spike-specific rabbit polyclonal Ab29 was used as the positive control. All steps were conducted at 26°C with constant shaking at 1,000 RPM. Collected data were analyzed using a built-in 1:2 global curve-fitting model and the Kd values were calculated using ForteBio Data Analysis software 10.0.
Pseudovirus-based neutralization assay
Serially diluted mAbs were incubated with SARS-CoV-2 pseudovirus particles bearing the original D614 spike (Codex #CB-97100-146) at 37°C for 1 h and were then incubated with HEK293-ACE2 target cells (7.5×103 cells/well, Codex #CB-97100-220) in Poly-D-lysine coated 384-well plates for 2 days. Pseudovirus infected HEK293-ACE2 were detected for luminescence using Codex’s Luciferase assay reagent (Codex #CB-80552-010) in a Biotek’s Synergy 2 Plate Reader. IC50 values were calculated based on the Sigmoidal dose-response curve fitting using GraphPad Prism. Wells containing cells only or cells plus pseudovirus without mAbs served as negative and positive controls, respectively.
Viruses
The seed viruses of SARS-CoV-2 clinical isolates were obtained through BEI Resources (Manassas, VA) or Centers for Disease Control and Prevention (Atlanta, GA). Seed viruses were amplified once in 90% confluent Vero E6 (ATCC CRL-1586) or Vero E6-TMPRSS2 (BPS Bioscience #78081) in DMEM supplemented with 3% FBS at 37 °C, 5% CO2 for 68–72 hours until >70% cells showed cytopathic effect. Amplified viruses were clarified, aliquoted and stored in a secured −80 °C freezer until use. Virus titers were determined using an ELISA-based 50% tissue culture infectious dose (TCID50) method29. The following lineages of wild type SARS-CoV-2 viruses were used in the study, including (1) lineage A (D614), (2) lineage B.1.3 (G614), (3) lineage B.1.1.7 (Alpha), (4) lineage B.1.351 (Beta), (5) lineage P.1 (Gamma), (6) lineage B.1.617.2 (Delta), (7) lineage B.1.429 (Epsilon), (8) lineage P.2 (Zeta), (9) lineage B.1.617.1 (Kappa), (10) lineage C.37 (Lambda), (11) lineage R.1, and (12) lineage B.1.1.529 (Omicron BA.1,BA.1.1, BA.2, BA.4 and BA.5). All experiments involving infectious SARS-CoV-2 viruses were performed in an FDA Animal Biosafety Level-3 (ABSL-3) laboratory equipped with advanced access control devices and by trained personnel equipped with powered air-purifying respirators.
Live virus-based microneutralization (MN) assay
Serially diluted mouse mAbs were incubated with 100 TCID50/well of live virus (1:1, v/v) at room temperature for 1 h. The virus-mAb mixtures were then incubated with Vero E6 cells pre-seeded in 96-well tissue culture plates at 37°C, 5% CO2 for 2 days29. Virus-infected cells were detected using human IgG1 specific for SARS-CoV-2 nucleocapsid (Acrobiosystems #NUN-S41, 0.2 μg/ml) followed by peroxidase-conjugated goat anti-human IgG (H+L) secondary antibody (Seracare #5220–0330, 1:2000) or rabbit anti-nucleocapsid monoclonal antibody (SinoBiol #40143-R001, 1:6000) followed by peroxidase-conjugated goat anti-rabbit secondary antibody (SeraCare #5220–0336, 1:2000). The % virus infection as compared to the wells that contained virus only were plotted by nonlinear regression to determine IC50 of individual mAbs for each virus using GraphPad Prism (Version 9.3.1).
Passive transfer and challenge
Hemizygous B6.Cg-Tg(K18-ACE2)2Prlmn/J (K18-hACE2) transgenic mice (JAX Stock No. 034860)35 were bred at FDA White Oak Vivarium. All K18-hACE2 mice were genotyping-confirmed (Transnetyx) and were used at 8–12 weeks old for passive transfer and challenge in the ABSL3 lab. Age-matched K18-hACE2 adult mice (male vs female at approximately 1:1 ratio) were randomly grouped and were injected intraperitoneally with 0.1 ml/mouse of 17B10, 17A7 or 3A6 in a dose of 3 or 6 mg/kg. K18-hACE2 mice receiving a nonspecific mouse IgG1 isotype served as controls. Two to four hours after passive transfer, recipient mice were challenged intranasally with a lethal dose of wild type SARS-CoV-2 and variants, including 103 TCID50/mouse of NY (G614) or Gamma variant, or 102 TCID50/mouse of Alpha, Beta or Delta variant, or 5X103 TCID50/mouse of Omicron BA.1. Infected mice were monitored daily for body weight (BW), cutaneous temperature using a rodent infrared thermometer (Braintree Scientific) and mortality for 10 days post infection (dpi). Mice becoming moribund or reaching humane endpoints (e.g., severe hypothermia and/or 30% BW loss) were immediately euthanized. All procedures were performed according to the animal study protocols approved by the FDA White Oak Animal Program Animal Care and Use Committee.
Viral load in mouse tissues
Mouse tissues (lung, brain and heart) were harvested on 3 dpi and were homogenized in sterile PBS (pH7.2) (10%, v/w) using a Fisherbrand™ Bead Mill 24 Homogenizer (FisherScientific) followed by total RNA extraction using RNeasy Plus Mini Kit (Qiagen #74136). Extracted RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific #4368813). SARS-CoV-2 nucleocapsid (N) gene in individual mouse organs was determined using QuantiNova SYBR Green PCR kit (Qiagen #208052) in combination of 500 nM of 2019-nCoV RUO Kit (Integrated DNA Technologies #10006713). The cycling program was performed in Stratagene MX3000p qPCR system (Agilent) as follows: 95 °C for 120 s, 95 °C for 5 s (50 cycles) and 60 °C for 18 s29. Threshold cycle (Ct) values were determined using MxPro qPCR software (Agilent). A standard curve was constructed using serially diluted pCC1-CoV2-F7 plasmid expressing SARS-CoV-2 N 36 and was used to interpolate the number of SARS-CoV-2 N gene copies in mouse tissues. A value of 1 was assigned if gene copies were below the detection limits.
Cryo-EM sample preparation and data collection
To prepare cryo EM grids, 4 mg/ml full-length G614 variant spike trimer37 in DDM and 5 mg/ml 17B10–1 Fab (1:3 S protein trimer: Fab ratio) were first incubated at 4°C for 30mins. Four μl of the mixture was then applied to a 1.2/1.3 Quantifoil gold grid (Quantifoil Micro Tools GmbH), which had been glow discharged with a PELCO easiGlow™ Glow Discharge Cleaning system (Ted Pella, Inc.) for 60 s at 15 mA. Grids were immediately plunge-frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific), and excess protein was blotted away by using grade 595 filter paper (Ted Pella, Inc.) with a blotting time of 4 s, a blotting force of −12 at 4°C with 100% humidity. The grids were first screened for ice thickness and particle distribution. Selected grids were used to acquire images by a Titan Krios transmission electron microscope (ThermoFisher Scientific) operated at 300 keV and equipped with a BioQuantum GIF/K3 direct electron detector. Automated data collection was carried out using SerialEM38 version 3.8.6 at a nominal magnification of 105,000× and the K3 detector in counting mode (calibrated pixel size, 0.825 Å) at an exposure rate of 27.448 electrons per pixel per second. Each movie added a total accumulated electron exposure of ~54.442 e-/Å2, fractionated in 52 frames. Data sets were acquired using a defocus range of 0.8–2.2 μm.
Image processing and 3D reconstructions
All data processing was carried out using cryoSPARC39 v.3.3.1. Drift correction for cryo-EM images was performed using patch mode, and contrast transfer function (CTF) was estimated by patch mode. Motion corrected sums with dose-weighting were used for all other image processing. A template was first produced by manual pick and template picking was performed for particle picking. There were 3,124,974 particles extracted from 18,927 images (box size 672Å, downsizing to 128Å). Particles were subjected to two rounds of 2D classification yielding 2,235,387 good particles. A low-resolution negative-stain reconstruction of the Wuhan-Hu-1 sample was low-pass filtered to 40Å resolution and used as an initial model. The good particles from 2D classification were used for three rounds of heterogeneous classification with six copies of initial model as the reference in C1 symmetry and three major classes (26.1% in total) with clear structural features were re-extracted to smaller box size (480Å) and subjected to another two rounds of heterogeneous refinement with six copies of initial model as the reference in C1 symmetry. Three major classes were produced, which all exhibit the one-RBD-up spike protein trimer binding with one copy of 17B10–1 fab antibody. These three classes were then subjected to one round of non-uniform refinement in C1 symmetry yielding a map at 3.0Å resolution from 439,265 particles. The overall resolution was further improved with local CTF refinement resulting in a map with C1 symmetry at 2.8Å. To improve the resolution at the interface between the RBD and 17B10–1 Fab, local refinement was performed with a soft mask covering a single RBD and the bound 17B10–1 Fab, resulting in a map at 3.6Å resolution (Supplementary Figure S2). The best density maps were used for model building. All resolutions were reported from the gold-standard Fourier shell correlation (FSC) using the 0.143 criterion. Density maps were corrected from the modulation transfer function of the K3 detector and sharpened by applying a temperature factor that was estimated using Sharpening Tools in cryoSPARC39. Local resolution was also determined using cryoSPARC39 (Supplementary Figure S3).
Model building
The initial templates for spike protein model building used the G614 spike trimer structures (PDB ID: 7KRR)37. The 17B10–1 antibody structure was firstly predicted by AlphaFold240 implementation in the ColabFold41 notebooks running on Google Colaboratory. Default settings were used with Amber relaxation; the sequence was entered in tandem and the best model was chosen based on pLDDT (predicted Local Distance Difference Test) score. The predicted model was then used for further modeling. Several rounds of manual building were performed in Coot. The model was then refined in Phenix42 against the 2.8Å (one-RBD-up) cryo-EM map of the G614 spike-17B10Fab complex, and the 3.6Å cryo-EM map of the RBD-17B10Fab complex. Iterative refinement was performed in both Phenix42 (real space refinement) and ISOLDE43. The Phenix42 refinement strategy included minimization_global, local_grid_search, and adp, with rotamer, Ramachandran, and reference-model restraints, using 7KRR as the reference model. The refinement statistics are summarized in Supplementary Table S2. Structural biology applications used in this project were compiled and configured by SBGrid44.
Statistical analysis
GraphPad Prism 9.3.1 was used to conduct one-way nonparametric ANOVA or two-way mixed ANOVA, or Mann-Whitney test, or Log-rank (Mantel-Cox) survival test. Viral loads were log transformed before statistical analysis. A p value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by FDA/CBER intramural SARS-CoV-2 pandemic fund (to H.X. and J.P.W.) and by COVID-19 Awards by MassCPR (to B.C.). The following SARS-CoV-2 clinical isolates (i.e., D614, G614, Alpha, Beta, Gamma, Epsilon, Zeta, R.1 and Omicron BA.1, BA.2, BA.4 and BA.5) were obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2. The authors sincerely thank Dr. Duane Wesemann at Brigham and Women’s Hospital and Drs. Jonathan Yewdell and Ivan Kosik at NIAID/NIH for providing human RBD-reactive mAb C63C7 and Cova2-39, respectively. The authors sincerely appreciate the support of FDA/CBER Biosafety team and White Oak Vivarium staff in this study. The findings and conclusions in this report are those of the authors and do not necessarily represent the views and official policies of the CDC and US government.
Footnotes
CONFLICT OF INTEREST DISCLOUSRE
The authors declare no competing interests.
DATA AVAILABILITY
Source data are provided with this paper. The data for cryo-EM structures have been deposited in the PDB (ID: 8GJM & 8GJN) and in the Electron Microscopy Databank (ID: 40094 & 40095), respectively. The unique materials/reagents used in this study are available from the corresponding authors upon reasonable request and by Material Transfer Agreement.
REFERENCES
- 1.Walls AC et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292 e286 (2020). 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 181, 894–904 e899 (2020). 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang J et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan Y et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature communications 8, 15092 (2017). 10.1038/ncomms15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong P et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Cell 184, 4969–4980 e4915 (2021). 10.1016/j.cell.2021.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan S et al. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife 10 (2021). 10.7554/eLife.68563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577 (2020). 10.1038/s41586-020-2599-8 [DOI] [PubMed] [Google Scholar]
- 9.Keeton R et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 603, 488–492 (2022). 10.1038/s41586-022-04460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes CO et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie KM et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science 374, 472–478 (2021). 10.1126/science.abh2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan M et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 373, 818–823 (2021). 10.1126/science.abh1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M, Liu H, Wu NC & Wilson IA Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem Biophys Res Commun 538, 192–203 (2021). 10.1016/j.bbrc.2020.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strohl WR et al. Passive Immunotherapy Against SARS-CoV-2: From Plasma-Based Therapy to Single Potent Antibodies in the Race to Stay Ahead of the Variants. BioDrugs 36, 231–323 (2022). 10.1007/s40259-022-00529-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaney AJ et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29, 44–57 e49 (2021). 10.1016/j.chom.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greaney AJ et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476 e466 (2021). 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaney AJ et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nature communications 12, 4196 (2021). 10.1038/s41467-021-24435-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature (2022). 10.1038/s41586-022-05053-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). 10.1038/s41586-021-03398-2 [DOI] [PubMed] [Google Scholar]
- 20.Liu L et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022). 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
- 21.Changrob S et al. Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike. mBio 12, e0297521 (2021). 10.1128/mBio.02975-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulliam JRC et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 376, eabn4947 (2022). 10.1126/science.abn4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen RE et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27, 717–726 (2021). 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Servellita V et al. Predominance of antibody-resistant SARS-CoV-2 variants in vaccine breakthrough cases from the San Francisco Bay Area, California. Nat Microbiol 7, 277–288 (2022). 10.1038/s41564-021-01041-4 [DOI] [PubMed] [Google Scholar]
- 25.Kwon HJ et al. Enhanced virulence and waning vaccine-elicited antibodies account for breakthrough infections caused by SARS-CoV-2 delta and beyond. iScience 25, 105507 (2022). 10.1016/j.isci.2022.105507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LL et al. Omicron variant susceptibility to neutralizing antibodies induced in children by natural SARS-CoV-2 infection or COVID-19 vaccine. Emerg Microbes Infect 11, 543–547 (2022). 10.1080/22221751.2022.2035195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouwer PJM et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643-+ (2020). 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science 374, 1353–1360 (2021). 10.1126/science.abl9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radvak P et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nature communications 12, 6559 (2021). 10.1038/s41467-021-26803-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong J et al. Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants. Proc Natl Acad Sci U S A 119, e2201433119 (2022). 10.1073/pnas.2201433119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- 32.Yuan M et al. A broad and potent neutralization epitope in SARS-related coronaviruses. Proc Natl Acad Sci U S A 119, e2205784119 (2022). 10.1073/pnas.2205784119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshpande A, Harris BD, Martinez-Sobrido L, Kobie JJ & Walter MR Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern. Front Immunol 12, 691715 (2021). 10.3389/fimmu.2021.691715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCallum M et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375, 864–868 (2022). 10.1126/science.abn8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCray PB Jr. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81, 813–821 (2007). 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie X et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 e843 (2020). 10.1016/j.chom.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372, 525–530 (2021). 10.1126/science.abf2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 39.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 40.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirdita M et al. ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682 (2022). 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croll TI ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530 (2018). 10.1107/S2059798318002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morin A et al. Collaboration gets the most out of software. Elife 2, e01456 (2013). 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. The data for cryo-EM structures have been deposited in the PDB (ID: 8GJM & 8GJN) and in the Electron Microscopy Databank (ID: 40094 & 40095), respectively. The unique materials/reagents used in this study are available from the corresponding authors upon reasonable request and by Material Transfer Agreement.


