Abstract
We recently identified Escherichia coli RNA polymerase (RNAP) mutants (RNAP β′ Δ215–220 and β RH454) that form extremely unstable complexes with rRNA P1 (rrn P1) core promoters. The mutant RNAPs reduce transcription and alter growth rate-dependent regulation of rrn P1 core promoters, because the mutant RNAPs require higher concentrations of the initiating nucleoside triphosphate (NTP) for efficient transcription from these promoters than are present in vivo. Nevertheless, the mutants grow almost as well as wild-type cells, suggesting that rRNA synthesis is not greatly perturbed. We report here that the rrn transcription factor FIS activates the mutant RNAPs more strongly than wild-type RNAP, thereby compensating for the altered properties of the mutant RNAPs. FIS activates the mutant RNAPs, at least in part, by reducing the apparent KATP for the initiating NTP. This and other results suggest that FIS affects a step in transcription initiation after closed-complex formation in addition to its stimulatory effect on initial RNAP binding. FIS and NTP levels increase with growth rate, suggesting that changing FIS concentrations, in conjunction with changing NTP concentrations, are responsible for growth rate-dependent regulation of rrn P1 transcription in the mutant strains. These results provide a dramatic demonstration of the interplay between regulatory mechanisms in rRNA transcription.
rRNA transcription is the rate-limiting step in ribosome synthesis and is subject to precise control by multiple regulatory systems (11, 19, 23). Since ribosome biosynthesis is an energetically expensive process, it is coupled to the cell's nutritional status by being regulated in proportion to the cell's growth rate (growth rate-dependent control).
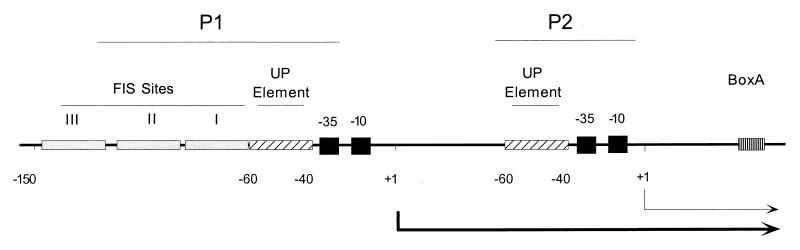
Multiple mechanisms contribute to rRNA transcription initiation. The seven rRNA operons are transcribed from tandem promoters, P1 and P2, spaced about 120 bp apart (20). The P1 promoters are the targets of most of the known regulatory signals affecting rRNA transcription initiation and are responsible for growth rate-dependent regulation (17, 40). The best studied of the rrn P1 promoters, rrnB P1 (Fig. 1), consists of core promoter elements 10 and 35 bp upstream of the transcription start site, recognized by the sigma subunit of RNA polymerase (RNAP), and an UP element upstream of the −35 hexamer, recognized by the α subunit of RNAP (14, 35). In addition, there are three binding sites for the transcription factor FIS, centered at positions −71, −102, and −143 upstream of the rrnB P1 start site (37).
FIG. 1.
The rrnB regulatory region, including the P1 and P2 promoters. FIS binding sites, UP elements, −10 and −35 elements, transcription start sites (+1), and the Nus factor binding site (BoxA) are indicated.
rrnB P1, rrnD P1, and perhaps all rrn P1 complexes with RNAP are unusually unstable. The stabilities of these promoter complexes are increased in vitro by the binding of the initiating nucleoside triphosphate (NTP) (GTP for rrnD P1 and ATP for the other six rrn P1 promoters), whose concentration increases with growth rate in vivo (15). We have suggested, therefore, that there is a kinetic competition between dissociation of the rrn P1 open complex and transcription initiation which is dependent on the concentration of the initiating NTP, leading to growth rate-dependent control of rRNA transcription (the NTP-sensing model) (7, 15). The core promoter (i.e., from about position −40 to the transcription start site) is sufficient for growth rate-dependent control of rrnB P1 and rrnD P1 transcription (6, 7).
We recently characterized two mutations, rpoCΔ215–220 and rpoBRH454 (in the genes for the β′ and β subunits of RNAP, respectively) that strongly reduce rrn P1 core promoter activity in vivo (7). The purified mutant RNAPs form less stable complexes with rrn P1 core promoters than wild-type RNAP and as a result require even higher levels of the initiating NTP than wild-type RNAP for efficient transcription in vitro. The mutant RNAPs respond to changes in the concentration of the initiating NTP in vitro, but NTP levels in cells apparently are never high enough for rrn P1 core promoters to reach normal activity in the mutants (7). Nevertheless, these mutations are not lethal, and mutant cells grow nearly as well as wild-type cells, despite the defects in rrn P1 core promoter-RNAP interactions. The transcriptional defects in these mutants were originally characterized using rrnB P1 and rrnD P1 promoter-lacZ fusions lacking the FIS binding sites normally present in rrn P1 promoters, and we speculated that FIS might compensate for the defects of the mutant RNAPs in vivo (7, 15).
FIS activates transcription from rrn P1 promoters (28, 37, 39, 45). At rrnB P1, where FIS increases transcription about fivefold (8, 37), most activation is attributable to site I, where FIS binds and interacts with the RNAP α subunit through surface-exposed patches on the two proteins (9, 16). The FIS concentration in vivo varies with growth phase and growth rate (5, 29, 30, 31), and occupancy of rrnB P1 FIS sites varies with FIS expression (3). However, neither the fis gene nor FIS sites are required for growth rate-dependent control of the rrnB P1 promoter (6, 37). On the other hand, FIS is absolutely required for growth rate-dependent control of several other promoters (e.g., some tRNA promoters [13] and the promoter of the 4.5S RNA gene [12]), and FIS is responsible for a major component (but not all) of the growth rate-dependent regulation observed for the leuV promoter (32, 36).
In this study, we have investigated the effects of FIS on rrnB P1 transcription by the mutant RNAPs β′ Δ215–220 and β RH454 in vivo and in vitro. We conclude that the mutant strains grow nearly normally in spite of the altered properties of their transcription initiation complexes, because FIS provides almost wild-type activity and regulation to rrn P1 promoters. Our results suggest that rrn P1 promoters integrate multiple signals, including changing NTP levels and changing FIS levels, in order to regulate rRNA transcription initiation. Furthermore, our results illustrate how regulation of ribosome synthesis remains qualitatively unchanged in the face of substantial changes to the system components; i.e., the system is robust.
MATERIALS AND METHODS
Bacterial strains and phages.
The strains used in this work are listed in Table 1. RNAP mutations and the fis::kan allele were moved between strains by bacteriophage P1 transduction (25). λ phage monolysogens were constructed essentially as described previously (17).
TABLE 1.
Bacterial strains
| Strain | Genotype | Reference(s) or source |
|---|---|---|
| MG1655 | Wild type | 4 |
| MGΔZB | MG1655 lacΔ145 | 42 |
| CAG4000 | MG1655 ΔlacX74 | D. J. Jin and C. Gross |
| RJ1617 | MC1000 fis::kan767 | 22 |
| CAG18500 | MG1655 thi-39::Tn10 | 42 |
| RLG3381 | MG1655 thi-39::Tn10 rpoCΔ215–220 | 7 |
| RLG1829 | MG1655 λ rrnB P1 (−88 to +50)-lacZ | 3, 17 |
| RLG1785 | MGΔZB λ rrnB P1 (−154 to +50)-lacZ | 8, 17 |
| RLG1679 | CAG4000 λ rrnB P1 (−88 to −37)-lac (−36 to +57)–lacZ | 3, 16 |
| RLG1680 | CAG4000 λ rrnB P1 (−88 to −37 Δ-72)-lac (−36 to +57)–lacZ | 16, 34 |
| RLG3950 | CAG18500 λ rrnB P1 (−61 to +50)-lacZ | 7, 37 |
| RLG3993 | RLG1829 thi-39::Tn10 | This work |
| RLG3398 | CAG18500 λ rrnB P1 (−154 to +50)-lacZ | 17; this work |
| RLG3951 | RLG3381 λ rrnB P1 (−61 to +50)-lacZ | 7, 37 |
| RLG3994 | RLG1829 thi-39::Tn10 rpoC Δ215–220 | This work |
| RLG3399 | RLG3381 λ rrnB P1 (−154 to +50)-lacZ | 17; This work |
| RLG4076 | RLG1784 thi-39::Tn10 rpoBRH454 | 7 |
| RLG4329 | RLG1785 thi-39::Tn10 rpoBRH454 | This work |
| RLG3974 | RLG3398 fis::kan-767 | This work |
| RLG3971 | RLG3399 fis::kan-767 | This work |
| RLG3979 | RLG1680 thi-39::Tn10 | This work |
| RLG3977 | RLG1679 thi-39::Tn10 | This work |
| RLG3980 | RLG1680 thi-39::Tn10 rpoCΔ215–220 | This work |
| RLG3978 | RLG1679 thi-39::Tn10 rpoCΔ215–220 | This work |
| RLG1784 | MGΔZB λ rrnB P1 (−61 to +50)-lacZ | 9, 17 |
| RLG3982 | RLG1784 thi-39::Tn10 rpoCΔ215–220 | 7 |
β-Galactosidase assays.
Cultures were grown at 30°C, and growth rates were varied using the media described previously (6, 7). For assays of rrnB P1 derivatives, strains were streaked from single colonies on plates containing media that supported growth rates lower than or equivalent to those supported by the media used in the experiment. Cells were scraped from the plate and diluted in the appropriate media, and after three to four generations of growth, mid-log-phase cultures were harvested, washed, sonicated, and assayed for β-galactosidase as described previously (7). All experiments were performed at least twice and on different days, and errors were less than 10% of the mean values.
In vitro transcription.
Multiple-round transcription reactions were performed at 22°C as described previously (7), using a 0.2 nM concentration of a supercoiled plasmid (pRLG597) (37) containing an rrnB P1 promoter (positions −154 to +50), making a 220-nucleotide transcript terminated by rrnB T1T2 terminators. The transcription buffer contained 115 mM NaCl (for Fig. 2), 100 mM NaCl (for Fig. 3A and B), or 130 mM NaCl (for Fig. 3C and D); 40 mM Tris-acetate (pH 7.9); 10 mM MgCl2; 1 mM dithiothreitol; 100 μg of bovine serum albumin per ml; 200 μM GTP; 200 μM UTP; 10 μM [α-32P]CTP (5 μCi); and the ATP concentrations indicated in the figure legends. Purified FIS protein (75 nM) was preincubated with the template for 16 min before the reactions were initiated by addition of wild-type or β′ Δ215–220 RNAP to 0.8 nM. The activities of the wild-type and β′ Δ215–220 RNAPs were similar on the lacUV5 promoter (data not shown). Reactions were allowed to proceed for 16 min before transcription was stopped by the addition of loading solution (35), and electrophoresis, phosphorimaging, and quantitation were as described previously (7). Fits to data points shown in Fig. 3 were made using SigmaPlot (Jandel Scientific).
FIG. 2.
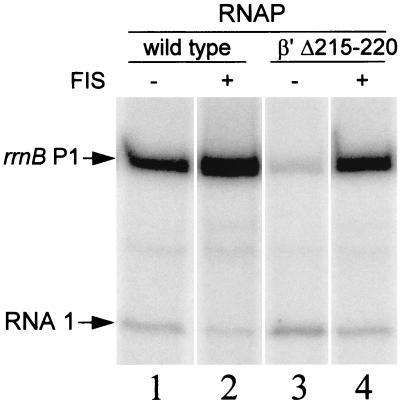
Activation of rrnB P1 transcription by FIS in vitro. The supercoiled template contained rrnB P1 positions −154 to +50. The reaction mixtures contained 200 μM ATP and wild-type RNAP (lanes 1 and 2) or β′ Δ215–220 RNAP (lanes 3 and 4) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of FIS. The transcripts derived from the rrnB P1 promoter and from the vector-encoded RNA I promoter are indicated. Since the reaction conditions were identical in each lane and the wild-type and mutant RNAPs had similar activities on a non-FIS-activated promoter (see Materials and Methods), activation by FIS was calculated directly from the relative amounts of rrnB P1 transcripts. Only one gel is shown, but the experiment was performed multiple times with similar results.
FIG. 3.
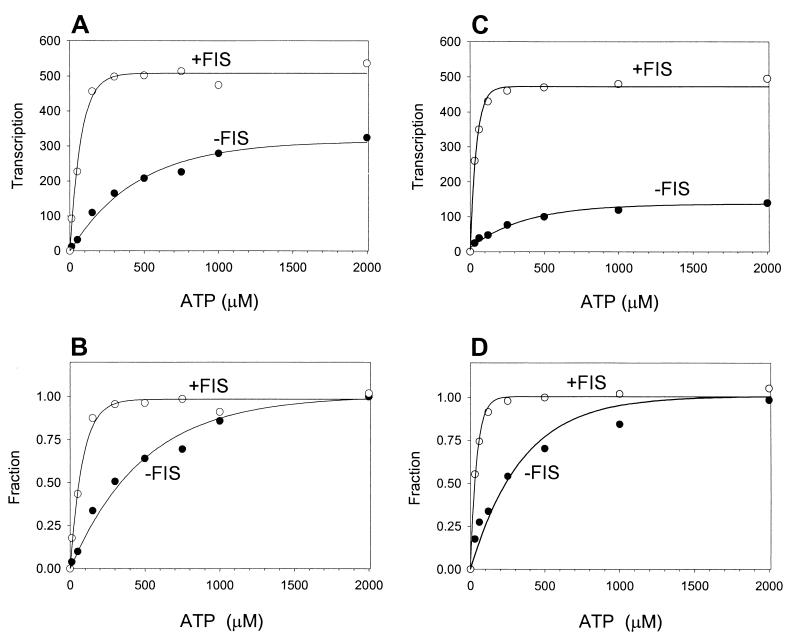
Effect of FIS on initiating NTP levels required for rrnB P1 transcription. (A) Transcription by the β′ Δ215–220 mutant RNAP at different ATP concentrations. In vitro transcription was performed as described in Materials and Methods with 100 mM NaCl, using supercoiled plasmid templates containing the rrnB P1 (−154 to +50) promoter in the absence or presence of FIS. (B) Results from panel A normalized to those obtained with 2 mM ATP. The graphed data represent averages from two experiments. The apparent KATPs in the absence and presence of FIS are about 330 and 60 μM, respectively. (C) Transcription by the wild-type RNAP at different ATP concentrations. In vitro transcription was performed as described in Materials and Methods with 130 mM NaCl, using supercoiled plasmid templates containing the rrnB P1 (−154 to +50) promoter in the absence or presence of FIS. (D) Results from panel A normalized to those obtained with 2 mM ATP. The graphed data represent averages from two experiments. The apparent KATPs in the absence and presence of FIS are about 240 and 30 μM, respectively.
We determined the apparent KATPs for transcription by mutant and wild-type RNAPs in the presence and absence of FIS using solution conditions that differed in NaCl concentration (see Results and Discussion); no single NaCl concentration was found where accurate determinations could be obtained for both enzymes. At 130 mM NaCl, where the apparent KATP for transcription by the wild-type RNAP could be quantified reliably, transcription by the mutant RNAP in the absence of FIS was too inefficient at lower ATP concentrations for accurate determination of an apparent KATP. Likewise, at 100 mM NaCl, where the apparent KATP for transcription by the mutant RNAP could be quantified reliably, transcription by the wild-type RNAP in the presence of FIS was too efficient even at lower ATP concentrations for accurate determination of an apparent KATP (data not shown).
Western analysis of FIS levels.
Western analysis was performed essentially as described previously (3). RLG1784 and RLG3982 (Table 1) were plated on a medium supporting the lowest growth rate, and colonies were suspended in appropriate media to an A600 of 0.025 to 0.030. As described previously (3), duplicate cultures were grown for three to four generations at different growth rates; 1-ml aliquots were pelleted, resuspended, and boiled for 5 min; equivalent A600 units of lysate were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the separated proteins were transferred to nitrocellulose membranes and probed with polyclonal anti-FIS antibody (a generous gift from R. Johnson, UCLA). Bound antibody was detected by enhanced chemiluminescence (Amersham), and bands were visualized by exposure to X-ray film and quantified using optical scanning and ImageQuant software (Molecular Dynamics). Purified FIS protein standards were used to calibrate the amounts of FIS detected and to ensure that samples were within the linear detection range.
RESULTS
Upstream DNA sequences restore rrnB P1 transcription activity in RNAP mutant strains.
Transcription of rrn P1 promoter-lacZ fusions lacking FIS binding sites is severely reduced in rpoCΔ215–220 and rpoBRH454 cells relative to wild-type cells (7, 15). However, the mutant strains grow reasonably well (doubling times are 80 to 90% of that of the wild type), suggesting that rRNA synthesis is not strongly perturbed. Since rrn P1 promoters in their natural context are activated by FIS (37), we tested whether the presence of their normal FIS sites would rescue the defects in transcription exhibited by rrnB P1 core promoter-lacZ fusions in the mutant strains.
We compared the activities of rrnB P1-lacZ fusions without FIS sites (positions −61 to +50), with only the proximal FIS site (−88 to +50), and with all three FIS sites (−154 to +50) in the wild-type and mutant strains (Table 2). The proximal FIS site increased transcription 3.8-fold, and three FIS sites increased transcription 4.7-fold in the wild-type strain, consistent with previous observations (8). However, the effect of the FIS sites was much greater in the mutant strains: e.g., in the rpoCΔ215–220 mutant, the proximal FIS site increased transcription 8.5-fold, and three FIS sites increased transcription 14-fold, restoring almost full rrnB P1 promoter activity. Thus, the defect in transcription caused by the mutant RNAPs is much greater for rrnB P1 promoters lacking FIS sites than for rrnB P1 promoters containing FIS sites.
TABLE 2.
Activation by FIS in RNAP mutant strain backgrounds
| RNAP allele | Growth rate (doublings/h) |
rrnB P1 activity (Miller units)a with promoter endpoints:
|
||
|---|---|---|---|---|
| −61 to +50 | −88 to +50 | −154 to +50 | ||
| Wild type | 1.29 | 2,297 (1.0) | 8,811 (3.8) | 10,822 (4.7) |
| rpoCΔ215–220 | 1.14 | 608 (1.0) | 5,152 (8.5) | 8,927 (14) |
| rpoBRH454 | 1.05 | 716 (1.0) | Not done | 7,036 (9.8) |
Results are means from two or three experiments; the variation was less than 10%. The promoter activity relative to −61 to +50 in each strain background is given in parentheses.
FIS is responsible for the effect of upstream sequences on rrnB P1 promoter activity in the rpoCΔ215–220 mutant.
In the previous section, we showed that upstream DNA sequences containing FIS sites compensated for the negative effect of the rpoB and rpoC mutations on rrnB P1 transcription. Consistent with the conclusion that activation by FIS was responsible for this effect of the upstream sequences in the RNAP mutant strains, transcription of an rrnB P1 promoter containing all three FIS sites dropped by about 60% in a fis::kan rpoCΔ215–220 double mutant compared to the rpoCΔ215–220 single mutant (Table 3). The requirement for FIS for the effect of the upstream sequence was further confirmed by measuring expression from an rrnB P1-lacZ fusion containing a single-base-pair deletion in FIS site I that eliminates FIS binding (−88 Δ−72 to +50) (37). This upstream sequence did not stimulate rrnB P1 transcription in the RNAP mutant strain (data not shown).
TABLE 3.
Effect of FIS and rpoCΔ215–220 on activity of rrnB P1 (−154 to +50)
| RNAP allele | fis allele | Growth rate (doublings/h) | Activity (Miller units)a | Relative activityb |
|---|---|---|---|---|
| rpoCΔ215–220 | Wild type | 1.14 | 8,927 | 1.00 |
| fis::kan | 1.06 | 3,736 | 0.42 | |
| Wild type | Wild type | 1.29 | 10,822 | 1.00 |
| fis::kan | 1.25 | 11,424 | 1.06 |
Results are means from two experiments; the variation was less than 10%.
Fraction of activity in the background containing the wild-type fis gene for each RNAP allele.
In contrast to the reduced transcription from rrnB P1 observed in the fis::kan rpoCΔ215–220 double mutant strain, deletion of the fis gene did not reduce rrnB P1 transcription substantially in the wild-type strain (Table 3), consistent with our previous reports (34, 37). This apparent paradox results from an increase in rrnB P1 core promoter activity in the fis::kan mutant (34, 37). We have attributed this increase in rrnB P1 core promoter activity to the homeostatic nature of the regulatory system(s) controlling rrn P1 transcription; i.e., rRNA transcription is feedback regulated such that disruptions that reduce ribosome synthesis increase rrn P1 core promoter activity (19, 21). In the RNAP mutant strain, this feedback response was not able to increase transcription from the rrnB P1 core promoter enough to compensate fully for the loss of the fis gene (see Discussion).
Although rrnB P1 promoter activity decreases by about 60% in the fis::kan rpoCΔ215–220 double mutant compared to the rpoC single mutant, the double mutant grows only about 10% slower than the rpoCΔ215–220 strain; i.e., the small growth defect of the rpoC mutant strain is exacerbated only slightly by the fis mutation (Table 3). To account for the double mutant strain's relative vigor, other mechanisms must increase rRNA transcription to compensate for the reduced activity of rrn P1 promoters (see Discussion).
Activation of β′ Δ215–220 RNAP by FIS in vitro.
The presence of FIS binding sites results in high rrnB P1 promoter activity in the rpoCΔ215–220 strain. To further confirm that the increased activation by FIS in vivo was direct, we examined rrnB P1 transcription in vitro in the presence of purified RNAP and FIS. Under these conditions (see Materials and Methods), FIS increased transcription by the wild-type RNAP about 2.0-fold (Fig. 2, lanes 1 and 2), while it increased transcription by the β′ Δ215–220 RNAP about 12-fold (Fig. 2, lanes 3 and 4), resulting in similar overall promoter activity with the two enzymes (Fig. 2, lanes 2 and 4). Thus, as predicted from the results obtained in vivo, FIS directly compensates for the defect of the mutant RNAP by activating the mutant enzyme to a greater extent than the wild-type enzyme.
Activation of transcription from the lac core promoter by FIS in the rpoCΔ215–220 mutant.
Although the mutant RNAPs formed less stable open complexes than wild-type RNAP at all promoters tested (7, 18; M. M. Barker, T. Gaal, and R. L. Gourse, unpublished data), they reduced transcription of only those promoters that formed intrinsically unstable open complexes with wild-type RNAP (e.g., rrnB P1). We predicted that the increased extent of activation by FIS observed in the mutant strains would be limited to promoters with kinetic properties similar to those of rrnB P1.
To test this hypothesis, we measured transcription from a hybrid rrnB-lac promoter which was shown previously to be activated by FIS in a wild-type strain (3). We compared the activity of the hybrid promoter (which contains FIS site I and the UP element from rrnB P1 fused to the lac core promoter) to that of an identical promoter with a Δ-72 FIS site (which eliminates FIS binding) (37) in the rpoCΔ215–220 and wild-type strains. FIS activated the rrnB-lac hybrid promoter to approximately the same extent in both the wild-type and the mutant strains (3.9- versus 3.0-fold) (Table 4), in contrast to its differential effect on the rrnB P1 promoter in the same two strains (Table 2). This result is consistent with the hypothesis that the differential effect of FIS in the wild-type strain versus the rpoC mutant strain is a function of a kinetic property of the promoter interaction with RNAP, presumably the intrinsic instability of the open complex.
TABLE 4.
Effect of rpoCΔ215–220 on activation of the lac core promoter by fis
| RNAP allele | Promoter | FIS site | β-Galactosidase activitya | Activation ratio |
|---|---|---|---|---|
| Wild type | rrnB (Δ-72)-lac | − | 1,878 | 1.0 |
| rrnB-lac | + | 7,359 | 3.9 | |
| rpoCΔ215–220 | rrnB (Δ-72)-lac | − | 2,560 | 1.0 |
| rrnB-lac | + | 7,614 | 3.0 |
Results (in Miller Units) are means from two experiments; the variation was less than 5%.
FIS reduces the concentration of the initiating NTP needed for rrnB P1 transcription in vitro.
We next attempted to determine how FIS is able to activate rrnB P1 transcription by the mutant RNAPs to a greater extent than by the wild-type RNAP (Table 2; Fig. 2). Transcription initiation at rrnB P1 (and at other promoters) involves a series of steps in which RNAP first forms a closed complex that then isomerizes through a series of intermediates to an open complex capable of transcription (34). We showed previously that FIS increases closed-complex formation (8). However, closed-complex formation at rrnB P1 is similar with mutant and wild-type RNAPs in the absence of FIS (7), suggesting that this might not be the step responsible for the differential effect of FIS on the mutant RNAPs. On the other hand, the mutant and wild-type RNAPs do differ in the stability of the open complexes they form at rrnB P1, which results in an increase in the required initiating NTP concentration for transcription with the mutant enzymes (7, 15). We therefore tested whether FIS might reduce the apparent KNTP for the initiating nucleotide in the mutants.
We measured the effect of FIS on transcription by β′ Δ215–220 RNAP at different ATP concentrations, using conditions that resulted in enough transcription in the absence of FIS for accurate measurement even at low ATP concentrations (Fig. 3A). FIS increased transcription by β′ Δ215–220 RNAP at all ATP concentrations under these conditions but did so the most at low ATP concentrations (Fig. 3A). We expressed transcription as a fraction of that obtained at a saturating ATP concentration (2 mM) in order to calculate apparent KATP values for transcription by the mutant RNAP in the presence and absence of FIS (Fig. 3B). FIS greatly reduced the apparent KATP needed for transcription initiation (from about 330 μM in the absence of FIS to about 60 μM in the presence of FIS). We conclude that FIS can affect both closed-complex formation (8) and a later step in transcription initiation. FIS thereby compensates for the decreased transcription exhibited by the mutant RNAP on rrn P1 promoters in vitro in part by reducing the concentration requirement for the initiating NTP.
We also determined the effect of FIS on the apparent KATP for transcription by the wild-type RNAP (Fig. 3C and D). FIS also greatly reduced the apparent KATP for transcription by wild-type RNAP (from about 240 μM in the absence of FIS to about 30 μM in the presence of FIS). We conclude that FIS can facilitate transcription by both the wild-type and the mutant RNAPs in vitro by reducing the apparent KATP.
We emphasize that the salt concentrations in the buffers used for transcription with the two RNAPs were not identical in these experiments (see Materials and Methods), nor do we presume that these solution conditions are similar to those present in growing cells. Thus, the apparent KATPs for transcription by the two enzymes should not be compared directly, nor should they be considered the absolute KATPs for transcription initiation in vivo (see Discussion).
FIS is responsible for normal growth rate-dependent control of rrnB P1 transcription in rpoCΔ215–220 and rpoBRH454 mutants.
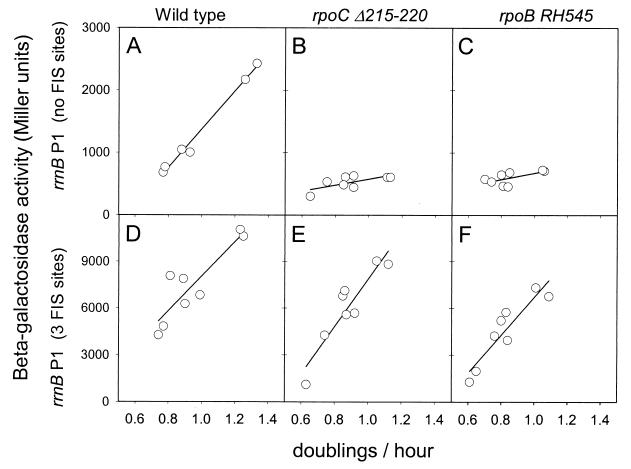
We previously established that transcription of rrnB P1 promoters lacking FIS sites is growth rate dependent in wild-type or in fis::kan strains (6, 7, 15, 17, 37) but that growth rate-dependent regulation is substantially reduced in the rpoB and rpoC mutant strains (7, 15). We proposed that the mechanism responsible for this regulation involves, at least in part, rrn P1 sensing of the initiating NTP concentration in vivo, consistent with the altered NTP-sensing properties of complexes containing the mutant RNAPs observed in vitro (7, 15). In the experiments shown in Fig. 4, we compared growth rate-dependent regulation of rrnB P1 promoters lacking FIS sites with that of rrnB P1 promoters containing FIS sites in the RNAP mutant strains. The presence of FIS sites not only increased rrnB P1 promoter activity at high growth rates (as shown in Table 2) but also resulted in nearly normal growth rate-dependent regulation of rrnB P1 (Fig. 4D to F). We conclude that FIS is an essential contributor to regulation of rrn P1 promoters in the RNAP mutant strains, although it is not essential for this purpose in wild-type cells.
FIG. 4.
Effect of FIS on growth rate-dependent regulation of rrnB P1 transcription in wild-type (A and D), rpoCΔ215–220 (B and E), and rpoBRH454 (C and F) strains. (A to C) Transcription from the rrnB P1 promoter without FIS sites (−61 to +50). (D to F) Transcription from the rrnB P1 promoter containing three FIS sites (−154 to +50). Promoter activities were determined from β-galactosidase activities of promoter-lacZ fusions. Growth rates of cultures were varied as described previously (7).
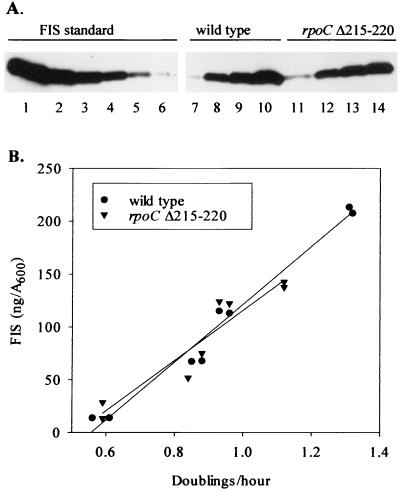
Previous studies from our lab and others have demonstrated that the FIS concentration and the level of FIS-dependent activation of the rrnB P1 promoter vary with growth conditions in wild-type cells (3, 5, 29, 33, 44). To determine whether changing FIS levels could contribute to growth rate-dependent regulation of rrn P1 transcription in the RNAP mutant strains, we examined FIS levels in cells grown in different media by using Western blot analysis with an anti-FIS antibody (Fig. 5). We found that FIS levels increased with growth rate similarly in the wild-type and rpoCΔ215–220 mutant strains; i.e., the rpoC mutation did not alter the expression of FIS. Thus, differential FIS-dependent activation with growth rate could contribute to the ability of FIS to restore growth rate-dependent regulation to rrn P1 promoters in the RNAP mutant strains.
FIG. 5.
Growth rate-dependent variation in FIS levels in wild-type and rpoCΔ215–220 strains. (A) Western blot with anti-FIS antibody. Lanes 1 to 6, 40, 20, 10, 5, 2.5, and 1.25 ng of purified FIS protein, respectively. Lanes 7 to 10, protein extracts from the wild-type strain grown at 0.56, 0.85, 0.93, and 1.32 doublings/h, respectively. Lanes 11 to 14, protein extracts from the rpoCΔ215–220 strain grown at 0.59, 0.88, 0.96, and 1.12 doublings/h, respectively. Aliquots of lysates representing equivalent numbers of cells (as determined from the optical density) were loaded in each lane. (B) Amounts of FIS as a function of growth rate. Quantitation is illustrated for two independent experiments, including the one pictured in panel A, lanes 7 to 14, using the purified standards of FIS protein in lanes 1 to 6 for calibration.
We showed above that FIS activates rrnB P1 transcription by the β′ mutant RNAP in vitro in part by reducing the apparent ATP concentration required for transcription initiation, and we suggested that this brings the initiating KNTP into a range sufficient for transcription by the mutant RNAP in vivo. Assuming that ATP and GTP concentrations change in the RNAP mutant strains as they do in the wild-type strain (15), we conclude that changing initiating NTP concentrations, in addition to changing FIS concentrations, likely contribute to growth rate-dependent regulation of rrn P1 transcription in the mutant strains.
DISCUSSION
Rescue of mutant RNAP function by FIS.
We found previously that the intrinsic instability of rrn P1 core promoter complexes is responsible for their regulation with changing initiating NTP concentrations and for their sensitivity to the destabilizing effects of mutant RNAPs (7, 15). In the work presented here, we propose that this intrinsic instability, exacerbated by the mutant RNAPs, leads to the increased extent of activation by FIS in the mutant strains. The increased activation by FIS accounts for the almost normal rrn P1 transcription and growth rate-dependent regulation observed in the mutants.
In theory, FIS could increase rrn P1 transcription with a mutant RNAP by increasing open-complex formation (KBkf), by decreasing open-complex dissociation, by decreasing the NTP concentration required for transcription initiation, or by some combination of these effects. We have shown previously that FIS activates transcription of rrnB P1 by wild-type RNAP in part by increasing the equilibrium constant for closed-complex formation (i.e., by increasing KB) (8) through direct contacts with the C-terminal domain of the RNAP α subunit (9). However, since the mutant RNAPs and the wild-type RNAP had similar equilibrium binding constants for closed-complex formation in the absence of FIS (7) and since the mutations are unlikely to affect the interaction between FIS and the C-terminal domain of the RNAP α subunit directly, we suggest that the differential effect of FIS on the mutant versus wild-type RNAPs is not likely to occur at this initial RNAP binding step.
Rather, the differential effect of FIS on the rrn P1 complex containing the mutant RNAP is most likely attributable to an effect on a step after initial closed-complex formation. Since FIS reduces the apparent KNTP for binding of the initiating ribonucleotide, which occurs in the open complex, we propose that FIS stabilizes an intermediate concurrent with or after strand opening, in addition to its effect on closed-complex formation described above. This proposal is consistent with the larger effect of FIS on the short-lived open complexes formed by the mutant RNAPs than on complexes containing the wild-type RNAP and with the fact that increasing the RNAP concentration did not alter the NTP concentration requirement for rrnB P1 transcription in vitro (T. Gaal, W. Ross, and R. L. Gourse, unpublished data). A role for FIS in kinetic steps after closed complex formation has been proposed previously for other promoters (rrnD P1 [39] and tyrT [26]). The effect of FIS on rrnB P1 that we have described here and that proposed for FIS on rrnD P1 and tyrT could have a similar mechanistic basis. However, our results do not rule out the possibility that FIS could facilitate still other steps in the transcription mechanism. In addition, we note that some effects of FIS differ from those resulting from UP element-α interactions, since the latter did not alter the ATP concentration requirement for rrnB P1 transcription (Gaal et al., unpublished data).
In the mutant strains, but not in the wild type (6, 37) FIS is essential for efficient transcription of rrn P1 promoters and for their regulation with growth rate. We ascribe this role in regulation in part to changing FIS concentrations with growth rate (Fig. 5) and thus to differential occupancy of the rrn P1 FIS sites. In addition, since FIS brings the apparent KNTP of the mutant RNAP into the range where changes in NTP concentrations would most likely affect rrn P1 transcription, rrn P1 regulation in the mutant strains also would be accomplished in part through the changes in NTP concentrations that accompany changes in growth rate.
We do not propose that the apparent KATPs derived from the in vitro transcription experiments reported here are the absolute binding constants in vivo; the apparent KATPs derived from these experiments are far below the millimolar ATP concentrations present in vivo (reference 15 and references therein). However, the ATP concentration needed for rrnB P1 transcription initiation in vitro is extremely sensitive to cation concentration and to template supercoiling (15), both of which greatly affect the lifetime of the open complex, which is crucial in determining the KNTP (15). In fact, maximal rrnB P1 transcription in vitro requires initiating NTP concentrations that are in the millimolar range when the reactions are performed with high salt concentrations and/or on linear templates. The relevance of the relative KATPs determined for rrn P1 transcription in vitro to transcription in vivo is supported strongly by the effects of artificial manipulation of ATP and GTP concentrations in vitro and in vivo (15) and by the correlation between the behaviors of the mutant RNAPs in vitro and in vivo (7, 15).
Role of FIS in growth rate-dependent control in wild-type strains.
We have shown previously that deletion of the rrnB P1 FIS sites or disruption of the fis gene has little effect on growth rate-dependent regulation of rrnB P1 transcription in wild-type strains (6, 37). Furthermore, the high occupancy of rrn P1 promoters with wild-type RNAP when NTP levels are maximal limits the potential for stimulation by FIS at high growth rates (Table 2) (J. A. Appleman, T. Gaal, M. S. Bartlett, W. Ross, and R. L. Gourse, unpublished data). Although seemingly paradoxical, the ability of FIS to rescue regulation of rrn P1 promoters in the mutant strains and yet to be dispensible for this purpose in wild-type strains is consistent with the results described above: the kinetic characteristics of the rrnB P1 complex are different in wild-type and RNAP mutant strains. FIS can thus affect the mutant RNAPs more than the wild-type RNAP and can confer growth rate-dependent regulation on rrn P1 transcription in the mutant strains. Nevertheless, the changing FIS concentrations that occur with growth rate in both wild-type and mutant strains and the effect of FIS on the apparent KATPs of both the wild-type and mutant initiation complexes suggest that FIS could potentially contribute to the regulation of rrnB P1 transcription under some conditions in wild-type strains, in conjunction with other regulatory mechanisms.
The data presented here also reinforce the previously recognized importance of FIS in growth rate-dependent control of other promoters. Some promoters are likely to owe their growth rate-dependent regulation almost entirely to changing FIS levels. Candidates for such promoters would be some tRNA promoters (13, 27) and the promoter for the 4.5S RNA (12), all of whose regulation is almost completely lost when FIS sites are deleted or in strains lacking the fis gene. Growth rate-dependent control of some other promoters may be attributable to the combined effects of changing FIS levels and changing NTP concentrations (or to the effects of other mechanisms). Candidates for such promoters would be those tRNAs whose growth rate-dependent regulation is aberrent, but not completely lost, in the absence of FIS sites or in strains lacking the fis gene (13, 27, 32, 36, 38). Finally, growth rate-dependent control of some promoters is likely to be independent of FIS. Candidates for such promoters would be tRNAs whose regulation is completely unaffected in fis::kan strains or which contain no FIS binding sites (13, 27).
Growth rate-dependent control of FIS levels.
The control of FIS expression has been studied extensively, and it appears to be complex. The promoter region of the fis operon contains binding sites for known regulatory proteins (integration host factor, FIS, and cyclic AMP receptor protein), and at least some of these clearly affect expression (5, 33, 44). In addition, the FIS promoter is sensitive to ppGpp levels; i.e., it displays a stringent response dependent on the relA gene product (30), but it is still subject to growth phase regulation in the complete absence of ppGpp (5).
Since changing FIS levels most likely contribute to growth rate-dependent regulation of rrn P1 transcription in the RNAP mutants, we investigated the expression of FIS promoter-lacZ fusions in the wild-type and RNAP mutant strains (M. S. Bartlett and R. L. Gourse, unpublished data). We found that transcription from the FIS core promoter (positions −36 to +7) was growth rate dependent and was unaffected by the rpoB and rpoC mutations. This suggests that the mechanism responsible for growth rate-dependent regulation of the FIS promoter differs from that for rrn P1 promoters. Further studies will be required to understand whether growth rate-dependent regulation of FIS expression is determined primarily at the transcription level and, if so, whether transcription of the fis gene is affected by changing NTP concentrations.
rRNA transcription regulation is robust.
Our results emphasize the resiliency of bacterial cells to the effects of mutation; i.e., like the bacteriophage lambda genetic switch (24), the rRNA transcription system is extremely robust. We have reported previously that although FIS activates rRNA transcription (as determined from the effects of FIS binding site mutations in vivo and in vitro), deletion of the fis gene does not reduce rRNA synthesis (34, 37). This apparent contradiction can be explained by an observed increase in rrn P1 core promoter function in fis::kan strains, an increase attributable to a feedback mechanism(s) that compensates for loss of FIS-dependent activation. We emphasize that the interpretation of effects of FIS binding site mutations is straightforward, because FIS binding site mutations reduce transcription of only the reporter constructs in which they are located and, unlike fis gene mutations, the site mutations do not have pleiotropic effects on cell metabolism that complicate interpretation of transcriptional outputs.
We report here that not only does the cell compensate for the loss of FIS-dependent activation caused by fis gene mutations, but conversely FIS compensates for altered NTP sensing at rrn P1 core promoters caused by rpoB or rpoC mutations. Thus, FIS and NTPs can each regulate rrn P1 transcription independently, but changes in one can alter effects resulting from the other.
The mechanism(s) responsible for feedback regulation of rRNA transcription has not been identified. It is possible that multiple mechanisms could contribute to this homeostatic regulation, with different mechanisms responding to particular stimuli. We have proposed previously that adjustments in cellular NTP levels could provide one such mechanism for feedback control of rRNA transcription (15). We are currently investigating whether changes in cellular NTP levels could account for the increase in rrn P1 core promoter activity observed in the fis::kan strain. However, since FIS apparently plays a role (direct or indirect) in additional cell functions that affect rRNA transcription (e.g., see references 41 and 43), identifying the specific mechanism(s) responsible for the feedback response of rrn P1 promoters that results from the loss of the fis gene presents a complex challenge for the future.
Since double mutants containing deletions of the fis gene and rpoCΔ215–220 have rrn P1 promoter activity reduced by 60% yet display growth defects only slightly greater than cells mutated in either single gene (Table 3), another regulatory mechanism must prevent rRNA underproduction under these circumstances. Furthermore, although rrnB P1 transcription was reduced in the fis::kan rpoCΔ215–220 double mutant, we note that this 60% reduction in transcription did not result in transcription as low as that of an rrnB P1 promoter lacking FIS sites in the RNAP mutant strain. Thus, an unidentified mechanism may also be responsible for partial derepression of the rrnB P1 promoter in the double mutant. Our results illustrate how potential components of the rRNA transcription machinery can be unmasked when other mechanisms contributing to transcription are eliminated.
In summary, our present view is that regulation of rRNA transcription is affected by multiple trans-acting factors that regulate rrn P1 promoter activity, e.g., FIS, NTPs, and ppGpp (10, 15, 18). However, regulatory roles for additional cis- and trans-acting factors cannot be excluded. For example, the rrn P2 promoters likely play a crucial role in upshifts (40; Appleman et al., unpublished data). Nus factors are required to prevent premature rRNA transcription termination (11), and it has been reported that the histone-like protein H-NS affects rRNA promoter activity during the transition to stationary phase (1, 2). The results reported here provide a dramatic example of the interplay between some of these regulatory factors during rRNA transcription.
ACKNOWLEDGMENTS
We thank Melanie Barker and other members of our laboratory for helpful comments on the manuscript and Alex Appleman for suggesting the title.
This work was supported by National Institutes of Health grant GM37048 to R.L.G.
REFERENCES
- 1.Afflerbach H, Schroder O, Wagner R. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 1998;28:641–653. doi: 10.1046/j.1365-2958.1998.00829.x. [DOI] [PubMed] [Google Scholar]
- 2.Afflerbach H, Schroder O, Wagner R. Conformational changes of the upstream DNA mediated by H-NS and FIS regulate E. coli rrnB P1 promoter activity. J Mol Biol. 1999;286:339–353. doi: 10.1006/jmbi.1998.2494. [DOI] [PubMed] [Google Scholar]
- 3.Appleman J A, Ross W, Salomon J, Gourse R L. Activation of Escherichia coli rRNA transcription by FIS during a growth cycle. J Bacteriol. 1998;180:1525–1532. doi: 10.1128/jb.180.6.1525-1532.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann B J. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 5.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 8.Bokal A J, Ross W, Gourse R L. The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol. 1995;245:197–207. doi: 10.1006/jmbi.1994.0016. [DOI] [PubMed] [Google Scholar]
- 9.Bokal A J, Ross W, Gaal T, Johnson R C, Gourse R L. Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J. 1997;16:154–162. doi: 10.1093/emboj/16.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 11.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Kirsebom L A, Nilsson L. Growth rate regulation of 4.5 S RNA and M1 RNA, the catalytic subunit of Escherichia coli RNase P. J Mol Biol. 1996;261:303–308. doi: 10.1006/jmbi.1996.0461. [DOI] [PubMed] [Google Scholar]
- 13.Emilsson V, Nilsson L. Factor for inversion stimulation-dependent growth rate regulation of serine and threonine tRNA species. J Biol Chem. 1995;270:16610–16614. doi: 10.1074/jbc.270.28.16610. [DOI] [PubMed] [Google Scholar]
- 14.Estrem S T, Ross W, Gaal T, Chen Z W S, Niu W, Ebright R H, Gourse R L. Bacterial promoter architecture: subsite structure of UP elements and interactions with the C-terminal domain of the RNA polymerase alpha subunit. Genes Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 16.Gosink K K, Gaal T, Bokal IV A J, Gourse R L. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 18.Gourse R L, Gaal T, Aiyar S E, Barker M M, Estrem S T, Hirvonen C A, Ross W. Strength and regulation without transcription factors: lessons from bacterial rRNA promoters. Cold Spring Harbor Symp Quant Biol. 1998;63:131–139. doi: 10.1101/sqb.1998.63.131. [DOI] [PubMed] [Google Scholar]
- 19.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 20.Jinks-Robertson S, Nomura M. Ribosomes and tRNA. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1358–1385. [Google Scholar]
- 21.Jinks-Robertson S, Gourse R L, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R C, Ball C A, Pfeffer D, Simon M L. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1417–1431. [Google Scholar]
- 24.Little J W, Shepley D P, West D W. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 26.Muskhelishvili G, Buckle M, Heumann H, Kahmann R, Travers A A. FIS activates sequential steps during transcription initiation at a stable RNA promoter. EMBO J. 1997;16:3655–3665. doi: 10.1093/emboj/16.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson L, Emilsson V. Factor for inversion stimulation-dependent growth rate regulation of individual tRNA species in Escherichia coli. J Biol Chem. 1994;269:9460–9465. [PubMed] [Google Scholar]
- 28.Nilsson L, Vanet A, Vijgenboom E, Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990;9:727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson L, Verbeek H, Vijgenboom E, van Drunen C, Vanet A, Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992;174:921–929. doi: 10.1128/jb.174.3.921-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninnemann O, Koch C, Kahmann R. The E. coli Fis promoter is subject to stringent control and autoregulation. EMBO J. 1992;11:1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osuna R, Lienau D, Hughes K T, Johnson R C. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokholok D K, Redlak M, Turnbough C L, Jr, Dylla S, Holmes W M. Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli. J Bacteriol. 1999;181:5771–5782. doi: 10.1128/jb.181.18.5771-5782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratt T S, Steiner T, Feldman L S, Walker K A, Osuna R. Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J Bacteriol. 1997;179:6367–6377. doi: 10.1128/jb.179.20.6367-6377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 35.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 36.Ross W, Salomon J, Holmes W M, Gourse R L. Activation of Escherichia coli leuV transcription by FIS. J Bacteriol. 1999;181:3864–3868. doi: 10.1128/jb.181.12.3864-3868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross W, Thompson J F, Newlands J T, Gourse R L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowley K B, Elford R M, Roberts I, Holmes W M. In vivo regulatory responses of four Escherichia coli operons which encode leucyl-tRNAs. J Bacteriol. 1993;175:1309–1315. doi: 10.1128/jb.175.5.1309-1315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sander P, Langert W, Mueller K. Mechanisms of upstream activation of the rrnD promoter P1 of Escherichia coli. J Biol Chem. 1993;268:16907–16916. [PubMed] [Google Scholar]
- 40.Sarmientos P, Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc Natl Acad Sci USA. 1983;80:7010–7013. doi: 10.1073/pnas.80.22.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 42.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wackwitz B, Bongaerts J, Goodman S D, Unden G. Growth phase-dependent regulation of nuoA-N expression in Escherichia coli K-12 by the Fis protein: upstream binding sites and bioenergetic significance. Mol Gen Genet. 1999;262:876–883. doi: 10.1007/s004380051153. [DOI] [PubMed] [Google Scholar]
- 44.Walker K A, Atkins C L, Osuna R. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol. 1999;181:1269–1280. doi: 10.1128/jb.181.4.1269-1280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zacharias M, Goringer H U, Wagner R. Analysis of the Fis-dependent and Fis-independent transcription activation mechanisms of the Escherichia coli ribosomal RNA P1 promoter. Biochemistry. 1992;31:2621–2628. doi: 10.1021/bi00124a024. [DOI] [PubMed] [Google Scholar]