Abstract
Cells of Escherichia coli growing on sugars that result in catabolite repression or amino acids that feed into glycolysis undergo a metabolic switch associated with the production and utilization of acetate. As they divide exponentially, these cells excrete acetate via the phosphotransacetylase-acetate kinase pathway. As they begin the transition to stationary phase, they instead resorb acetate, activate it to acetyl coenzyme A (acetyl-CoA) by means of the enzyme acetyl-CoA synthetase (Acs) and utilize it to generate energy and biosynthetic components via the tricarboxylic acid cycle and the glyoxylate shunt, respectively. Here, we present evidence that this switch occurs primarily through the induction of acs and that the timing and magnitude of this induction depend, in part, on the direct action of the carbon regulator cyclic AMP receptor protein (CRP) and the oxygen regulator FNR. It also depends, probably indirectly, upon the glyoxylate shunt repressor IclR, its activator FadR, and many enzymes involved in acetate metabolism. On the basis of these results, we propose that cells induce acs, and thus their ability to assimilate acetate, in response to rising cyclic AMP levels, falling oxygen partial pressure, and the flux of carbon through acetate-associated pathways.
Cells of Escherichia coli undergo a metabolic switch associated with the production and utilization of acetate (19, 30). During exponential growth on a mixture of amino acids such as tryptone broth, cells consume first l-serine and then l-aspartate in a strictly preferential order. Simultaneously, they produce and excrete acetate. Once they have consumed both the serine and aspartate, these cells resorb and utilize acetate instead of excreting it. This acetate-associated metabolic switch occurs just as the cells begin to decelerate growth, i.e., just as they begin the transition to stationary phase (30).
Acetate production depends on one acetate activation pathway, while under these growth conditions, utilization requires a second (Fig. 1A). The first pathway, catalyzed by the enzymes acetate kinase (AckA; ATP:acetate phosphotransferase; EC 2.7.2.1) and phosphotransacetylase (Pta; acetyl coenzyme A [acetyl-CoA]:Pi acetyltransferase; EC 2.3.1.8) proceeds through an unstable, high-energy, acetyl phosphate (acetyl-P) intermediate (34). Cells use this low-affinity pathway to activate large concentrations of acetate (4, 21). The second pathway, catalyzed by the enzyme acetyl-CoA synthetase (Acs; acetate:CoA ligase [AMP forming]; EC 6.2.1.1) proceeds through an enzyme-bound acetyladenylate (acetyl-AMP) intermediate (2). Cells use this high-affinity pathway to scavenge for small concentrations of acetate (4, 21).
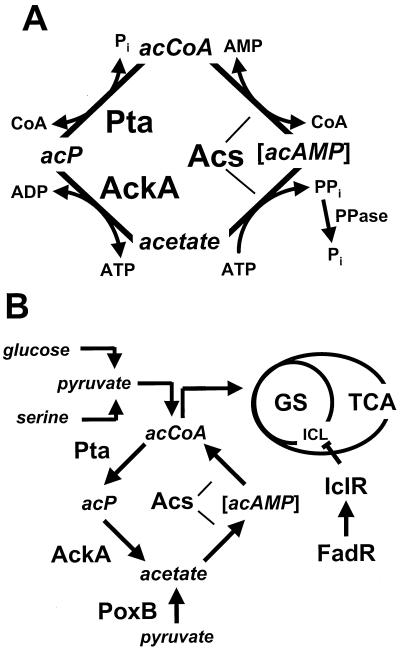
FIG. 1.
(A) Pathways of acetate activation in E. coli, acAMP, acetyl-AMP; acCoA, acetyl-CoA; AckA, acetate kinase; acP, acetyl P; Acs, acetyl-CoA synthetase; CoA, coenzyme A; Pi, inorganic phosphate; PPi, pyrophosphate; PPase, pyrophosphatase; Pta, phosphotransacetylase. (B) Carbon flux through Pta-AckA, Acs, and associated pathways during growth in TB or on glucose. GS, glyoxylate shunt; TCA, tricarboxylic acid cycle; PoxB, pyruvate oxidase; ICL, aceA gene product isocitrate lyase; IclR, repressor of the glyoxylate shunt operon aceBAK; FadR, regulator of fatty acid metabolism that also activates iclR.
In vivo, the Acs pathway is irreversible due to intracellular pyrophosphatases that remove pyrophosphate, a critical pathway intermediate. This pathway, therefore, functions only anabolically. In contrast, the Pta-AckA pathway is completely reversible. As such, it plays a critical catabolic role during both mixed acid fermentation and aerobic growth on excess glucose or other glycolytic intermediates (4). Under conditions that result in mixed acid fermentation, acetyl-CoA cannot enter the tricarboxylic acid (TCA) cycle. Thus, the cells convey acetyl-CoA through the Pta-AckA pathway, producing and excreting acetate while generating ATP (10). Similarly, under aerobic conditions, when the carbon flux into cells exceeds the amphibolic capacity of the central metabolic pathways, e.g., the TCA cycle, cells adjust by moving acetyl-CoA through the Pta-AckA pathway, again excreting acetate and generating ATP. As a consequence, such cells also accumulate the intermediate of this pathway, acetyl-P (31). Later, as they begin the transition to stationary phase, cells undergo the metabolic switch. Instead of excreting acetate, they resorb it, activate it to acetyl-CoA by means of Acs (21), and utilize it to generate energy and biosynthetic components via the TCA cycle and the glyoxylate shunt, respectively (4) (Fig. 1B). Simultaneously, the levels of acetyl-P decline (31).
Evidence exists that this metabolic switch can play a significant role in the regulation of certain two-component signal transduction pathways (reviewed in references 25 and 46). By serving as a phosphodonor for the autophosphorylation of the two-component response regulator, OmpR, acetyl-P functions in the control of flagellar synthesis (31, 37), cell division (29), and the expression of outer membrane porins (11). Acetyl-P also seems to play a critical role protecting cells against carbon starvation (27), presumably through some as yet unidentified response regulator. Furthermore, acetylation by the enzyme Acs can activate the chemotaxis response regulator, CheY (1, 32, 47), although the physiological relevance of this modification remains unclear.
In our attempt to identify the signals that trigger this acetate-associated metabolic switch and to dissect its underlying mechanisms, we focused on the enzyme Acs. We did so because the activity of this enzyme, strictly required for acetate assimilation (21), had been shown previously to vary as much as 22-fold depending on the nature of the available carbon source (4). In contrast, the levels and activities of the enzymes AckA and Pta and the expression of their respective genes, ackA and pta, vary no more than 2- to 10-fold (4, 22, 27, 43). By performing acs::lacZ reporter, Northern, and immunoblot analyses, we have learned that cells control Acs activity to a large degree by regulating the induction of its gene acs in response to both the phase of growth and the nature of the carbon source. We also have found that the timing and/or magnitude of this induction depends, in part, on the carbon regulator cyclic AMP (cAMP) receptor protein (CRP), the oxygen regulator FNR, the glyoxylate shunt repressor IclR and its activator FadR, and several enzymes involved in acetate metabolism. On the basis of these results, we propose that cells induce acs transcription, and thus the ability to assimilate acetate, in response to rising cAMP levels, falling oxygen partial pressure, and the flux of carbon through pathways associated with acetate metabolism.
MATERIALS AND METHODS
Chemicals.
Enzymes and substrates were obtained from Sigma Chemical Company (St. Louis, Mo.) or Promega (Madison, Wis.). Radiolabeled materials were from Amersham (Arlington Heights, Ill.), and Y-PER was obtained from Pierce Biochemicals (Rockford, Ill.).
Bacterial strains, plasmids, bacteriophage, and alleles.
All strains used in this study were derivatives of E. coli K-12 and are listed in Table 1 along with plasmids and phage.
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference(s) |
|---|---|---|
| AJW647 | CP750 proC::Tn5-132 | (P1)CP942→CP750 |
| AJW678 | AJW647 proC+ ΔlacX74 = thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136 ΔlacX74 | (P1)BW13711→AJW647 |
| AJW1074 | CP875 fadR::Km | (P1)RS3040→CP875 |
| AJW1075 | CP875 iclR::Km | (P1)ERL5R→CP875 |
| AJW1292 | CP875 crp::Tn5 | (P1)IT1133→CP875 |
| AJW1706 | AJW678 aceA1 zja::Tn10 | (P1)SM6009→AJW678 |
| AJW1727 | AJW678 fadR::Km | (P1)AJW1074→AJW678 |
| AJW1728 | AJW678 iclR::Km | (P1)AJW1075→AJW678 |
| AJW1729 | AJW678 poxB::Km | (P1)YYC877→AJW678 |
| AJW1730 | AJW678 fnr::Tn10 | (P1)M182fnr→AJW678 |
| AJW1786 | AJW678 λCB7 | λCB7 lysogeny of AJW678 |
| AJW1794 | AJW678 iclR::Km λCB7 | λCB7 lysogeny of AJW1728 |
| AJW1807 | AJW678 fadR::Km λCB7 | λCB7 lysogeny of AJW1727 |
| AJW1818 | AJW678 poxB::Km λCB7 | λCB7 lysogeny of AJW1729 |
| AJW1868 | AJW678 Δ(ackA pta hisJ hisP dhu) λCB7 | (P1)CP911→AJW1786 |
| AJW1876 | AJW678 aceA1 zja::Tn10 λCB7 | λCB7 lysogeny of AJW1706 |
| AJW1884 | AJW678 crp::Tn5 λCB7 | (P1)AJW1292→AJW1786 |
| AJW1938 | AJW678 fnr::Tn10 λCB7 | λCB7 lysogeny of AJW1730 |
| AJW1939 | AJW678 ackA::Km | This study |
| AJW1940 | AJW678 ackA::Km λCB7 | λCB7 lysogeny of AJW1939 |
| BW13711 | ΔlacX74 | B. Wanner |
| CP750 | thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136 | 30 |
| CP875 | thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136 ΔlacX74 λlacY | 30 |
| CP911 | CP875 Δ(ackA pta hisJ hisP dhu) | 30 |
| CP942 | proC::Tn5-132 | C. Park |
| DH5α | lacZΔM15 recA | Bethesda Research Laboratories |
| ERL5R | iclR::Km | 14, 33 |
| IT1133 | W3110 crp::Tn5 | H. Aiba |
| M182fnr | fnr::Tn10 | 17 |
| P90C | ara Δ(pro-lac) thi | 41 |
| RS3040 | fadR::Tn10 | 40 |
| SM6009 | aceA1 zja::Tn10 | 23 |
| YYC877 | KL333 poxB::Km | 7 |
| pAA121/pnrf53 | E. coli nrfA-acs intergenic region fragment carrying nucleotides −71 to −411 and −209 to +131 relative to the acs and nrfA promoters, respectively | 18, 45 |
| pCB26 | +130 nrfA to the 3′ end of acs open reading frame subcloned into pGEM-T | This study |
| pGEM-T | General cloning vector for PCR-amplified products | Promega |
| pMAK705 | rep(Ts) cam | 15 |
| pQE60 | fnrDA154-his6 | Wing et al., submitted |
| pREP4 | lacI+(Con) | Qiagen |
| pRS415 | bla′ lacZ+ (transcriptional fusion vector) | 41 |
| λCB7 | bla′ acs::lacZ imm434ind (acs transcriptional fusion) | This study |
| λRS88 | bla′ lacZ imm434ind (transcriptional fusion vector) | 41 |
The acs::lacZ transcriptional (operon) fusion λCB7 has been described previously (20). It was constructed by subcloning the nrfA-acs intergenic region into the multicopy vector pRS415 followed by recombination into the single-copy vector λRS88 using strain P90C (41). DH5α was used for constructing and propagating plasmids. Single lysogens of strain AJW678 (Δlac Ace+) were constructed and verified as described previously (41). Generalized transduction was performed using phage P1kc (39).
The ackA::Km allele was introduced into the AJW678 chromosome by means of homologous recombination using the temperature-sensitive suicide vector pMAK705 (15). The resultant ackA recombinants were verified by their poor ability to grow on 25 mM acetate as the sole carbon source (21) and their lack of motility due to an inability to form flagella at 35°C (31).
Media and growth conditions.
Cells were grown at 37°C in tryptone broth (TB; 1% [wt/vol] tryptone, 0.5% [wt/vol] sodium chloride) or in minimal salts medium (M63 [26]) containing either d-glucose (11 mM) or acetate (10 mM). The optical density at 590 nm (OD590) was monitored. For experiments involving a shift from one carbon source to another, cells were grown until the culture reached transition phase (defined as the point at which cells begin to grow at a lower rate), washed, and diluted 1:10 in fresh prewarmed M63 supplemented with acetate or glucose, further incubated as specified, harvested, washed, and resuspended in fresh prewarmed M63 supplemented with glucose or acetate, respectively.
Promoter activity assays.
β-Galactosidase activity was determined quantitatively using the Y-PER β-galactosidase assay kit from Pierce Biochemical. Each value is the mean ± standard error of the mean (SEM) of three independent measurements. Each experiment was repeated two to five times.
Overexpression and purification of CRP and FNR DA154.
CRP was overexpressed and purified as described previously (12). Purified FNR DA154 was generously provided by Helen Wing (University of Birmingham). FNR DA154, a constitutively active mutant of FNR that dimerizes stably in the presence of oxygen (48), was purified as a His6-tagged protein from strain M15 by a new method described by Wing et al. (H. Wing, J. Green, J. Guest, and S. Busby, submitted for publication). Strain M15 (Qiagen) carries plasmid pREP4 (derived from pACYC) that encodes constitutively expressed LacI (Qiagen). M15 cells were transformed with pQE60, encoding FNR DA154 His6 tagged at its C terminus. Transformants were grown at 37°C in 100 ml of L broth with appropriate antibiotics until cultures reached an OD600 of 0.5 to 0.6. Overexpression of the His6-tagged FNR DA154 protein was induced by the addition of 0.1 M isopropyl-β-d-thiogalactopyranoside for 1 h. Cells were harvested, and pellets were sonicated in 10 ml of lysis buffer at 4°C (1 mg of lysozyme per ml, 50 mM NaH2PO4-Na2HPO4 [pH 8.0], 750 mM NaNO3, 10 mM imidazole, 10 mM benzamidine). Sonicates were centrifuged at 10,000 × g and the amount of His6-tagged FNR DA154 was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Supernatants were applied to nickel-nitrilotriacetic acid agarose (Qiagen) columns at 4°C so that the binding capacity of the agarose (5 to 10 mg/ml) was exceeded (typical column volumes were 0.75 to 1.0 ml). Columns then were washed with 50 column volumes of wash buffer (50 mM NaH2PO4-Na2HPO4 [pH 8.0], 750 mM NaNO3, 20 mM imidazole), and FNR DA154 was eluted with elution buffer (50 mM NaH2PO4-Na2HPO4 [pH 8.0], 750 mM NaNO3, 250 mM imidazole). Protein was stored at 4°C in elution buffer and remained stable for up to 6 months.
EMSA.
Electrophoretic mobility shift assays (EMSA) using purified CRP were performed as described previously (35). EMSA using purified His6-tagged FNR DA154 protein were carried out essentially as detailed by Ziegelhoffer and Kiley (48). Purified nrfA-acs intergenic region fragments were end labeled with [γ-32P]ATP, and 2.5 to 0.5 ng of each fragment was incubated with various amounts of purified FNR DA154. The reaction buffer contained 10 mM potassium phosphate (pH 7.5), 100 mM potassium glutamate, 1 mM EDTA, 50 μM dithiothreitol, 5% glycerol, and 25 μg of herring sperm DNA per ml. The final reaction volume was 10 μl. After incubation at 37°C for 10 min, samples were run in 0.25× Tris-borate-EDTA on a 6% polyacrylamide gel (12 V/cm) containing 2% glycerol and analyzed by autoradiography.
RESULTS
Regulation of acs transcription.
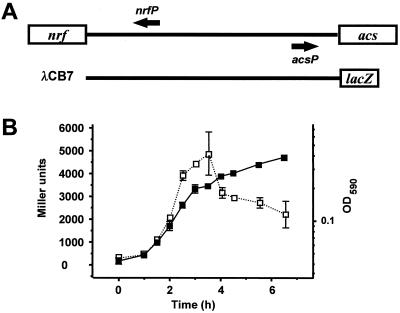
To determine whether cells regulate acs primarily at the level of transcription, we used the single-copy acs::lacZ transcriptional fusion carried by phage λCB7 (Fig. 2A). With λCB7, we lysogenized cells wild type for acetate metabolism but deleted for the lac locus (strain AJW678) and monitored the growth and β-galactosidase activity of the resultant lysogen (strain AJW1786 [Fig. 2B]). Immediately following resuspension of the overnight inoculum into fresh TB, acs transcription was low (∼200 Miller units [MU]). Within about 1 h, transcription began to rise, continued to increase throughout exponential growth, reached a maximum (∼5,000 MU) during transition, and decreased substantially as the culture approached stationary phase. Northern hybridization studies yielded similar results, while immunoblot analyses showed that Acs protein levels parallel transcription (data not shown).
FIG. 2.
(A) Schematic representation of the nrfA-acs intergenic region and the acs::lacZ operon fusion carried by λCB7. (B) OD590 (closed squares) and β-galactosidase activity in Miller units (open squares) for cells of strain AJW1786 (a λCB7 lysogen of the wild-type strain AJW678) grown at 37°C in TB. The SEM is shown only when it exceeded the size of the symbol.
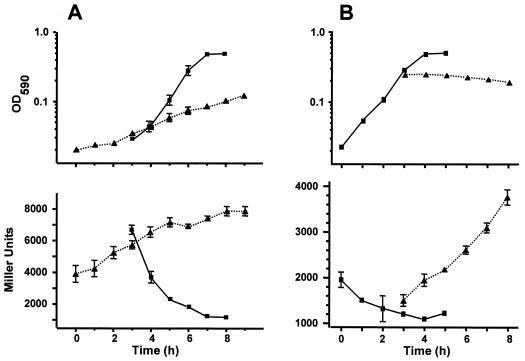
To investigate the transcriptional response to specific carbon sources, we grew cells of the acs::lacZ fusion strain (AJW1786) in M63 minimal medium supplemented with acetate or glucose and monitored their β-galactosidase activity. Cells grown on acetate as the sole carbon source (Fig. 3A) grew slowly (tD ∼210 min) and yielded high activity (∼8,000 MU). In contrast, those grown on glucose (Fig. 3B) grew rapidly (time of doubling [tD] of ∼50 min) and produced low activity (∼1,000 MU). Whereas cells grown initially on acetate and then exposed to glucose quickly shut off acs transcription (Fig. 3A), those grown first on glucose quickly induced acs transcription when shifted to acetate (Fig. 3B). Both Northern hybridization and immunoblot analyses yielded similar results (data not shown).
FIG. 3.
OD590 (top) and β-galactosidase activity in Miller units (bottom) for cells of the wild-type strain AJW1786 subjected to an acetate-to-glucose nutritional shift (A) or to a glucose-to-acetate nutritional shift (B). Cells were grown at 37°C in M63 supplemented either with 10 mM acetate (triangles) or with 11 mM glucose (squares). After 3 h of incubation, each culture was split. One half retained the original medium composition, while the other half was washed and resuspended in fresh, prewarmed medium supplemented with the other carbon source. The SEM is shown only when it exceeded the size of the symbol.
Involvement of CRP.
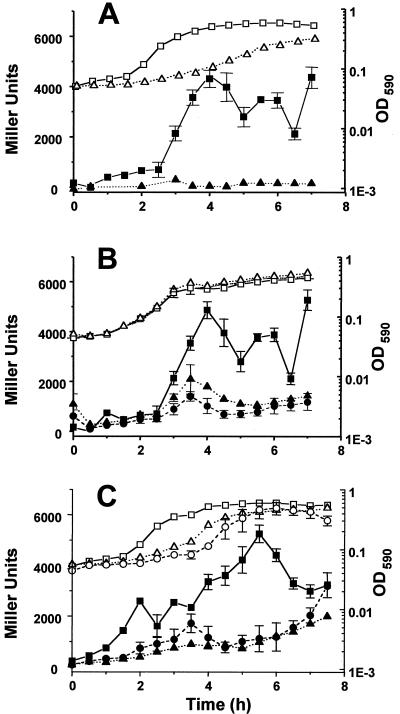
To determine whether the acetate-dependent increase in acs transcription depends on CRP, we constructed a crp derivative of the λCB7 lysogen AJW1786, grew the resultant strain and its parent in TB, and monitored their growth and β-galactosidase activity (Fig. 4A and 5). Whereas wild-type cells induced acs transcription reproducibly to about 5,000 MU, those that lacked CRP exhibited activity that was barely detectable. We observed similar results at the protein level. In contrast to wild-type cells, those lacking CRP did not induce Acs protein synthesis when shifted from glucose to acetate (data not shown).
FIG. 4.
OD590 (open symbols) and β-galactosidase activity in Miller units (closed symbols) for the wild-type strain AJW1786 and derivatives grown at 37°C in TB. (A) Strain AJW1786 (squares) and derivative lacking CRP (AJW1884; triangles); (B) strain AJW1786 (squares) and derivatives lacking IclR (AJW1794; circles) and FadR (AJW1807; triangles); (C) strain AJW1786 (squares) and derivatives lacking AckA (AJW1940; circles) and both AckA and Pta (AJW1868; triangles). The SEM is shown only when it exceeded the size of the symbol.
FIG. 5.
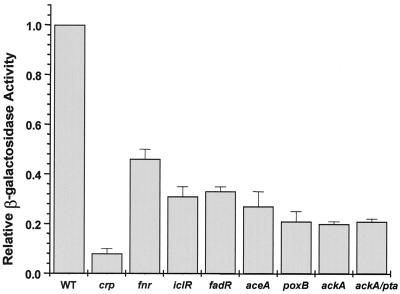
β-Galactosidase activity monitored at transition phase during growth in TB of the wild-type strain AJW1786 or isogenic derivatives lacking CRP (AJW1884), FNR (AJW1938), IclR (AJW1794), FadR (AJW1807), AceA (AJW1876), PoxB (AJW1818), AckA (AJW1940), or AckA and Pta (AJW1868). Activity is expressed as a percentage of the mean activity of the wild-type strain AJW1786 (4,993 ± 179; n = 7). Each value represents the mean ± SEM of at least three independent measurements.
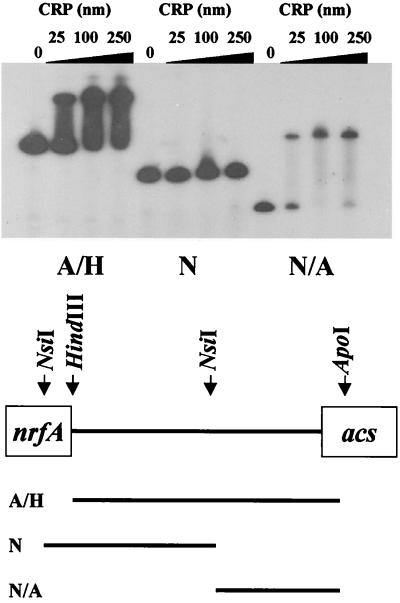
To determine whether CRP acts directly to facilitate acs transcription initiation, we performed EMSA (Fig. 6). We incubated fragments corresponding to the entire acs-nrfA intergenic region, its 5′ portion, or its 3′ portion with increasing concentrations of purified CRP protein in the presence of cAMP (0.2 mM). On the basis of these assays, we conclude that CRP binds to a single site located in the 3′ portion of the acs-nrfA intergenic region proximal to the putative acs promoter.
FIG. 6.
Autoradiogram of EMSA, analyzing the binding of purified CRP to end-labeled fragments of the acs-nrfA intergenic region. The fragments are depicted in the schematic below. These fragments were generated by NsiI, NsiI/ApoI, and ApoI/HindIII digestions of pCB26. A/H, entire intergenic region; N, 5′ portion; A/N, 3′ portion.
Involvement of FNR.
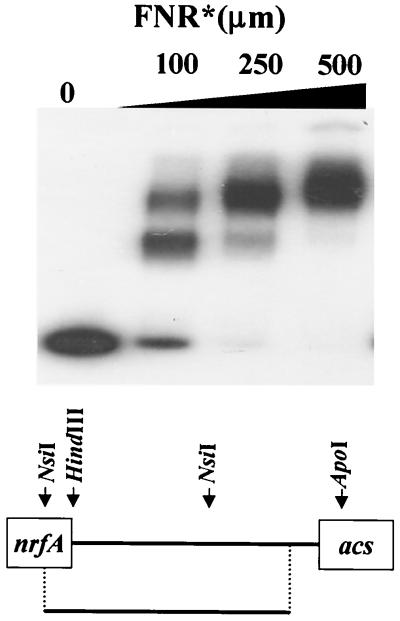
An FNR binding site, identified genetically and shown previously to be required for the anaerobic induction of the nrfA promoter, resides within the putative acs-nrfA intergenic region included in the acs::lacZ fusion carried by λCB7 (45). To determine whether Fnr actually binds this site, we performed an EMSA. We incubated a fragment of the acs-nrfA intergenic region with increasing amounts of purified FNR DA154 (FNR*). Surprisingly, we observed two FNR-dependent shifts, suggesting that FNR binds a second, previously unknown, site within this region. To determine whether the binding of FNR to either of these sites exerts any influence on acs transcription, we tested an fnr derivative of AJW1786 during growth in TB and found that this mutant transcribed acs at about half the level achieved by its wild-type parent (Fig. 7). Other evidence supports the hypothesis that FNR affects acs transcription through its ability to bind to this second FNR site and that this site is located 3′ of the intergenic NsiI site, i.e., proximal to acs: (i) a mutation in the nrf-proximal FNR site (p46A; a GC-to-AT mutation at positions −46 and −234 relative to the nrf and putative acs transcription initiation sites, respectively) that completely eliminates nrfA transcription (45) had no effect on acs transcription during growth on TB (data not shown) and (ii) an acs::lacZ fusion that does not include the 5′ region behaved transcriptionally in an FNR-dependent manner (data not shown). To date, we have not identified this second FNR binding site; however, several potential sites are located within the acs-proximal region.
FIG. 7.
Autoradiogram of EMSA, analyzing the binding of purified FNR DA154 (FNR*) to an end-labeled fragment of the acs-nrfA intergenic region. The fragment extends from +130 to −209 relative to nrfA and was generated by an EcoRI/HindIII digest of pAA121/nrf53 as depicted in the schematic below.
Involvement of the glyoxylate shunt.
To determine whether carbon flux through the glyoxylate shunt influences acs transcription, we tested iclR, fadR, and aceA derivatives of AJW1786. Intriguingly, null mutations (in iclR or fadR) that cause the glyoxylate shunt to operate constitutively yielded results similar to that of a null mutation (aceA) that incapacitates the shunt by eliminating synthesis of the first shunt enzyme, isocitrate lyase (23) (Fig. 4B and 5). Although all three strains exhibited a pattern of acs transcription whose timing resembled that of their wild-type parent, their peak expression reached less than 40% that of the parent. Surprisingly, poxB mutant cells lacking pyruvate oxidase, the enzyme that oxidizes pyruvate directly to acetate (7), exhibited very similar behavior (Fig. 5).
Involvement of acetate production.
To determine whether the ability to produce acetate influences acs transcription, we monitored ackA and ackA pta derivatives of AJW1786 (Fig. 4C and 5). Cells lacking only AckA or both AckA and Pta exhibited reduced acs transcription, a result consistent with immunoblot analyses that showed decreased steady-state levels of Acs protein in these mutants (data not shown). The exogenous addition of acetate to ackA pta mutant cells had no effect on the timing or levels of acs transcription (data not shown).
DISCUSSION
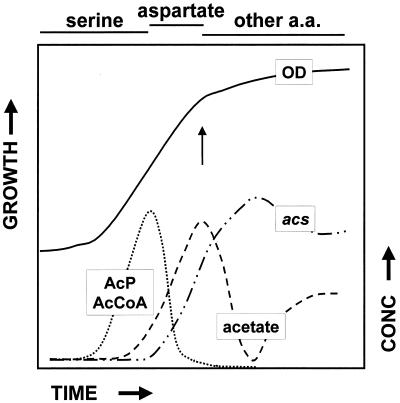
Observations made during these studies permit us to define the timing of acs induction (Fig. 8). During growth in TB, induction occurs at or near the conclusion of serine consumption and the beginning of aspartate consumption. This corresponds approximately to the time at which acetyl-CoA and acetyl-P pools reach their maximum (31). With the induction of Acs, both acetyl-CoA and acetyl-P pools decrease. The increasing presence of Acs likely contributes to this decrease by siphoning acetate, ATP, and CoA, the products of the Pta-AckA pathway. As long as acetyl-CoA remains in excess, such siphoning should continue to favor the generation of acetate and ATP. Indeed, the concentration of extracellular acetate continues to rise until the cells exhaust the supply of aspartate (31). At this time, the net acetate flux reaches zero (31) and the culture completes the metabolic switch. The extracellular acetate concentration starts to fall, the growth rate begins to decrease, and the culture commences its transition to stationary phase (30).
FIG. 8.
Schematic showing the relative timing of events during growth of wild-type E. coli cells at 37°C in TB. The arrow highlights the timing of the acetate-associated metabolic switch. Lines at the top delineate the duration of amino acid (a.a.) consumption. AcP, acetyl-P; AcCoA, acetyl-CoA; OD, optical density; acs, β-galactosidase activity observed from acs::lacZ fusion carried by λCB7; acetate, extracellular acetate concentration.
On the basis of reporter, Northern, and immunoblot analyses, we conclude that wild-type E. coli cells regulate this metabolic switch by inducing acs. This regulation occurs, in part, through the actions of the transcriptional regulators CRP, FNR, IclR, and FadR. Several acetate metabolic enzymes, including Pta, AckA, PoxB, and AceA, also contribute.
Evidence exists supporting direct roles for both CRP and FNR in regulating acs transcription initiation. A sequence that bears significant similarity to the CRP consensus binding site (5, 9) resides about 70 bp upstream of the proposed acs promoter (3, 44). EMSA and preliminary reporter analyses using selected CRP mutants support the relevance of this site (C. M. Beatty, D. Browning, S. Busby, and A. J. Wolfe, unpublished data). Thus, it seems likely that CRP acts directly to activate acs transcription and that rising cAMP levels help to trigger that transcription. EMSA data clearly support the existence of two FNR sites within the acs-nrfA intergenic region. One site has been identified previously by genetic means as required for the activation of the divergently transcribed nrfA promoter (45). We do not believe, however, that this site directly affects the transcription of acs. Instead, reporter analyses using mutant and deletion variants of the acs::lacZ fusion implicate a second site, which must reside considerably closer to the proposed acs promoter (Beatty et al., unpublished). Thus, it seems likely that FNR also acts directly to activate acs transcription and that falling oxygen partial pressure signals that transcription. If so, then acs would fall into the category of promoters controlled by tandem dimers of CRP or FNR or both (36).
Others previously reported the involvement of the glyoxylate shunt repressor IclR in acs transcription (38). We verified this report, showing that IclR affects the level but not the timing of acs promoter activity. We extended this observation to FadR, which activates iclR transcription directly (13) in addition to its role as the regulator of fatty acid metabolism genes (8). During growth in TB, which contains no fatty acids, FadR probably operates through IclR. Since we can find no sequences within the acs-nrfA intergenic region that closely resemble consensus binding sites for either IclR (28) or FadR (8), we believe it most likely that IclR operates upon acs transcription indirectly through its ability to control the synthesis of the glyoxylate shunt enzymes (Fig. 1B). Of course, we cannot rule out a direct effect until we test the ability of either protein to bind to this region. In fact, FadR binds to several sites that bear little resemblance to the reported consensus binding site (42). Because Acs produces the acetyl-CoA that functions as the glyoxylate shunt substrate, it seems reasonable that cells would coordinate the synthesis of Acs and the shunt enzymes. The fact that cells lacking the first shunt enzyme isocitrate lyase (aceA) displayed an acs transcription pattern almost indistinguishable from that of iclR and fadR mutants suggests that the feedback mechanism to acs senses both high and low glyoxylate shunt activity. Curiously, a poxB mutant that can no longer oxidize pyruvate directly to acetate transcribed acs in a manner strongly resembling that exhibited by iclR, fadR, and aceA mutants. This observation suggests that the mechanism that feeds back to acs senses some change in carbon flux.
We do not believe that either acetyl-P or acetyl-CoA functions as that feedback signal, despite the fact that both pools peak about the time that cells induce acs (31). We rule out acetyl-P because mutants lacking either AckA alone or both Pta and AckA behaved similarly with respect to acs transcription. Since the former accumulates acetyl-P and the latter fails to synthesize it at all, this observation argues strongly against any regulatory role for this intermediate of the Pta-AckA pathway. We also rule out acetyl-CoA because mutants lacking FadR display a pattern of acs transcription significantly different from that exhibited by wild-type cells. Since both cell types maintain acetyl-CoA pools at very similar levels (16), this observation argues that acetyl-CoA also cannot function in a regulatory capacity.
We are less certain concerning the regulatory role of acetate. Several observations support the hypothesis that acetate participates in the induction of acs. First, acs transcription correlates with extracellular acetate concentration. Second, cells inoculated into defined medium supplemented with acetate as the sole carbon source induce acs transcription rapidly. Third, the FadR-deficient mutant, which transcribes acs at reduced levels, utilizes acetate about five times faster than its wild-type parent (24). Such rapid utilization of acetate should keep the extracellular acetate pool low. If acetate functions as an inducing signal, however, then we must explain its failure to improve acs transcription by ackA pta cells that cannot excrete their own acetate. Such cells compensate for their inability to produce acetate by excreting nonacetate fermentation by-products, e.g., succinate and lactate (6). Perhaps one of these alternative products inhibits the response to acetate. If so, then this inhibitor does not affect the acetate-independent component of acs transcription. Alternatively, acetate itself may not signal acs induction.
Overall, the observations reported here suggest that the mechanisms used by E. coli cells to regulate acs expression are varied and complex. These seemingly include direct interactions by CRP and FNR to activate transcription initiation and indirect effects by acetate metabolic enzymes and transcription factors that control carbon flux. If so, then acs induction responds, in part, to rising cAMP levels, falling oxygen partial pressure, and changes in carbon flux. Such complexity should not be too surprising in light of the pivotal nature of this acetate-associated metabolic switch.
ACKNOWLEDGMENTS
S. Kumari and C. M. Beatty contributed equally to this work.
We thank C. Park, B. Wanner, D. LaPorte, S. Maloy, J. Cronan, Jr., R. W. Simons, and H. Aiba for strains; R. W. Simons for plasmids and phage; H. Wing for purified FNR DA154; N. J. Savery for purified CRP; G. S. Lloyd for help performing the CRP gel shifts; and H. M. Wols for construction of strain AJW1884. We also thank J. Foster, K. Visick, A. Driks, A. Stöver, and F. Catalano for thoughtful discussions and/or critical reading of the manuscript and D. Lewicki for assistance with graphics.
This work was supported by grant MCB-9630647 from the National Science Foundation.
REFERENCES
- 1.Barak R, Abouhamad W N, Eisenbach M. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli. J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg P. Acyl adenylates: an enzymatic mechanism of acetate activation. J Biol Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- 3.Blattner F R, Burland V, Plunkett G D, Sofia H J, Daniels D L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown T D K, Jones-Mortimer M C, Kornberg H L. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 5.Busby S J W. Positive regulation in gene expression. Symp Soc Gen Microbiol. 1986;39:51–77. [Google Scholar]
- 6.Chang D-E, Shin S, Rhee J-S, Pan J-G. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl-CoA flux for the growth and survival. J Bacteriol. 1999;181:6656–6663. doi: 10.1128/jb.181.21.6656-6663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y Y, Wang A Y, Cronan J E., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 8.Cronan J E, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expendiency. Mol Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- 9.Ebright R H, Cossart P, Gicquel-Sanzey B, Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein in E. coli. Nature. 1984;311:232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- 10.el-Mansi E M, Holms W H. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol. 1989;135:2875–2883. doi: 10.1099/00221287-135-11-2875. [DOI] [PubMed] [Google Scholar]
- 11.Forst S, Gelgado J, Rampersaud A, Inouye M. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J Bacteriol. 1990;172:3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosaini L R, Brown A M, Sturtevant J M. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988;27:5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- 13.Gui L, Sunnarborg A, LaPorte D C. Regulated expression of a repressor protein: FadR activates iclR. J Bacteriol. 1996;178:4704–4709. doi: 10.1128/jb.178.15.4704-4709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gui L, Sunnarborg A, Pan B, LaPorte D C. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J Bacteriol. 1996;178:321–324. doi: 10.1128/jb.178.1.321-324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackowski S, Rock C O. Consequences of reduced intracellular coenzyme A content in Escherichia coli. J Bacteriol. 1986;166:866–871. doi: 10.1128/jb.166.3.866-871.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaraman P S, Gaston K L, Cole J A, Busby S J. The nirB promoter of Escherichia coli: location of nucleotide sequences essential for regulation by oxygen, the FNR protein and nitrite. Mol Microbiol. 1988;2:527–530. doi: 10.1111/j.1365-2958.1988.tb00059.x. [DOI] [PubMed] [Google Scholar]
- 18.Kelsall A, Evans C, Busby S. A plasmid vector that allows fusion of the Escherichia coli galactokinase gene to the translation startpoint of other genes. FEBS Lett. 1985;180:155–159. [Google Scholar]
- 19.Kleman G L, Strohl W R. Acetate metabolism by Escherichia coli in high-cell-density fermentation. Appl Environ Microbiol. 1994;60:3952–3958. doi: 10.1128/aem.60.11.3952-3958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumari S, Simel E, Wolfe A J. ς70 is the principal sigma factor responsible for the transcription of acs, which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:551–554. doi: 10.1128/jb.182.2.551-554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumari S, Tishel R, Eisenbach M, Wolfe A J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwan H S, Chui H W, Wong K K. ack::Mu d1-8 (Aprlac) operon fusions of Salmonella typhimurium LT-2. Mol Gen Genet. 1988;211:183–185. doi: 10.1007/BF00338411. [DOI] [PubMed] [Google Scholar]
- 23.Maloy S R, Nunn W D. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol. 1982;149:173–180. doi: 10.1128/jb.149.1.173-180.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maloy S R, Nunn W D. Role of gene fadR in Escherichia coli acetate metabolism. J Bacteriol. 1981;148:83–90. doi: 10.1128/jb.148.1.83-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleary W R, Stock J B, Ninfa A J. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 27.Nystrom T. The glucose-starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol Microbiol. 1994;12:833–843. doi: 10.1111/j.1365-2958.1994.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 28.Pan B, Unnikrishnan I, LaPorte D C. The binding site of the IclR repressor protein overlaps the promoter of aceBAK. J Bacteriol. 1996;178:3982–3984. doi: 10.1128/jb.178.13.3982-3984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruss B M. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch Microbiol. 1998;170:141–146. doi: 10.1007/s002030050626. [DOI] [PubMed] [Google Scholar]
- 30.Pruss B M, Nelms J M, Park C, Wolfe A J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan R, Schuster M, Bourret R B. Acetylation of Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA. 1998;95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resnick E, LaPorte D C. Introduction of single copy sequences into the chromosome of Escherichia coli: application to gene and operon fusions. Gene. 1991;107:19–25. doi: 10.1016/0378-1119(91)90292-j. [DOI] [PubMed] [Google Scholar]
- 34.Rose I A, Grunberg-Manago M, Korey S R, Ochoa S. Enzymatic phosphorylation of acetate. J Biol Chem. 1954;211:737–756. [PubMed] [Google Scholar]
- 35.Savery N J, Lloyd G S, Kainz M, Gaal T, Ross W, Ebright R H, Gourse R L, Busby S J. Transcription activation at class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott S, Busby S, Beacham I. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- 37.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin S, Song S G, Lee D S, Pan J G, Park C. Involvement of iclR and rpoS in the induction of acs, the gene for acetyl coenzyme A synthetase of Escherichia coli K-12. FEMS Microbiol Lett. 1997;146:103–108. doi: 10.1111/j.1574-6968.1997.tb10178.x. [DOI] [PubMed] [Google Scholar]
- 39.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 40.Simons R W, Egan P A, Chute H T, Nunn W D. The regulation of fatty acid degradation in Escherichia coli. J Bacteriol. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 42.Subrahmanyam S, Cronan J. Isolation from genomic DNA of sequences binding specific regulatory proteins by the acceleration of protein electrophoretic mobility upon DNA binding. Gene. 1999;226:263–271. doi: 10.1016/s0378-1119(98)00548-4. [DOI] [PubMed] [Google Scholar]
- 43.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treves D S, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- 45.Tyson K L, Cole J A, Busby S J. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol Microbiol. 1994;13:1045–1055. doi: 10.1111/j.1365-2958.1994.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 46.Wanner B L. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetylphosphate. In: Hoch J A, Silhavy T J, editors. Two component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 203–221. [Google Scholar]
- 47.Wolfe A J, Conley M P, Berg H C. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci USA. 1988;85:6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziegelhoffer E C, Kiley P J. In vitro analysis of a constitutively active mutant form of the Escherichia coli global transcription factor FNR. J Mol Biol. 1995;245:351–361. doi: 10.1006/jmbi.1994.0029. [DOI] [PubMed] [Google Scholar]