Abstract
Since the second-half of the twentieth century, intracranial electroencephalography (iEEG), including both electrocorticography (ECoG) and stereo-electroencephalography (sEEG), has provided an intimate view into the human brain. At the interface between fundamental research and the clinic, iEEG provides both high temporal resolution and high spatial specificity but comes with constraints, such as the individual’s tailored sparsity of electrode sampling. Over the years, researchers in neuroscience developed their practices to make the most of the iEEG approach. Here we offer a critical review of iEEG research practices in a didactic framework for newcomers, as well addressing issues encountered by proficient researchers. The scope is threefold: (i) review common practices in iEEG research, (ii) suggest potential guidelines for working with iEEG data and answer frequently asked questions based on the most widespread practices, and (iii) based on current neurophysiological knowledge and methodologies, pave the way to good practice standards in iEEG research. The organization of this paper follows the steps of iEEG data processing. The first section contextualizes iEEG data collection. The second section focuses on localization of intracranial electrodes. The third section highlights the main pre-processing steps. The fourth section presents iEEG signal analysis methods. The fifth section discusses statistical approaches. The sixth section draws some unique perspectives on iEEG research. Finally, to ensure a consistent nomenclature throughout the manuscript and to align with other guidelines, e.g., Brain Imaging Data Structure (BIDS) and the OHBM Committee on Best Practices in Data Analysis and Sharing (COBIDAS), we provide a glossary to disambiguate terms related to iEEG research.
Keywords: Intracranial recording in humans, Stereotactic electroencephalography, sEEG, Electrocorticogram, EcoG, Good research practice
1 -. Data collection: the patient being a participant
1.0. Introduction
Treatment of neurological disease and some clinical circumstances may call for the transient implantation of electrodes in direct contact with the human brain (see Figure 1): the precise targeting of deep-brain stimulation (e.g., in movement disorders: (Benabid et al., 1987; Kringelbach et al., 2007), depression (Mayberg et al., 2005), obsessive-compulsive disorder (Nuttin et al., 1999) or dystonia (Vidailhet et al., 2005; Vidailhet and Pollak, 2005)), intraoperative mapping of cortical function during awake surgery (e.g., for tumor resection1 (DeAngelis, 2001; Sanai et al., 2008)), and the evaluation of potentially surgically remediable epilepsy using intracranial electroencephalography (iEEG) (Cardinale et al., 2016; Jobst et al., 2020; Krucoff et al., 2017). Implanted electrodes can be composed of electrode arrays placed on the brain’s surface (electrocorticography, ECoG) or on probes that penetrate into deep brain structures for recording stereotactic EEG (sEEG) or for deep-brain stimulation (DBS electrodes). The present article focuses on iEEG research carried out within the context of invasive monitoring for the treatment of drug-resistant epilepsy (Engel et al., 2005; Jacobs and Kahana, 2010; Lachaux et al., 2012; Mukamel and Fried, 2012). Surgeons can also use iEEG during intraoperative procedures which has significant advantages, such as the possibility to reposition the electrode array or the use of cooling probes. However, iEEG research conducted during acute application falls outside of this review’s scope. The opportunity to work with epilepsy patients presents several characteristics that favor cognitive neuroscience research: (i) the electrodes typically cover multiple cortical regions and subcortical structures in several lobes, sometimes bilaterally; (ii) electrodes remain implanted for several days to weeks, allowing time for research; and (iii) candidates for epilepsy surgery often have near-normal or normal neurological and cognitive functioning, which can enable comparisons to the healthy human brain. This section introduces iEEG data collection for research purposes. We discuss the clinical context, describe how research projects can be introduced within that context and finally, we address technical considerations.
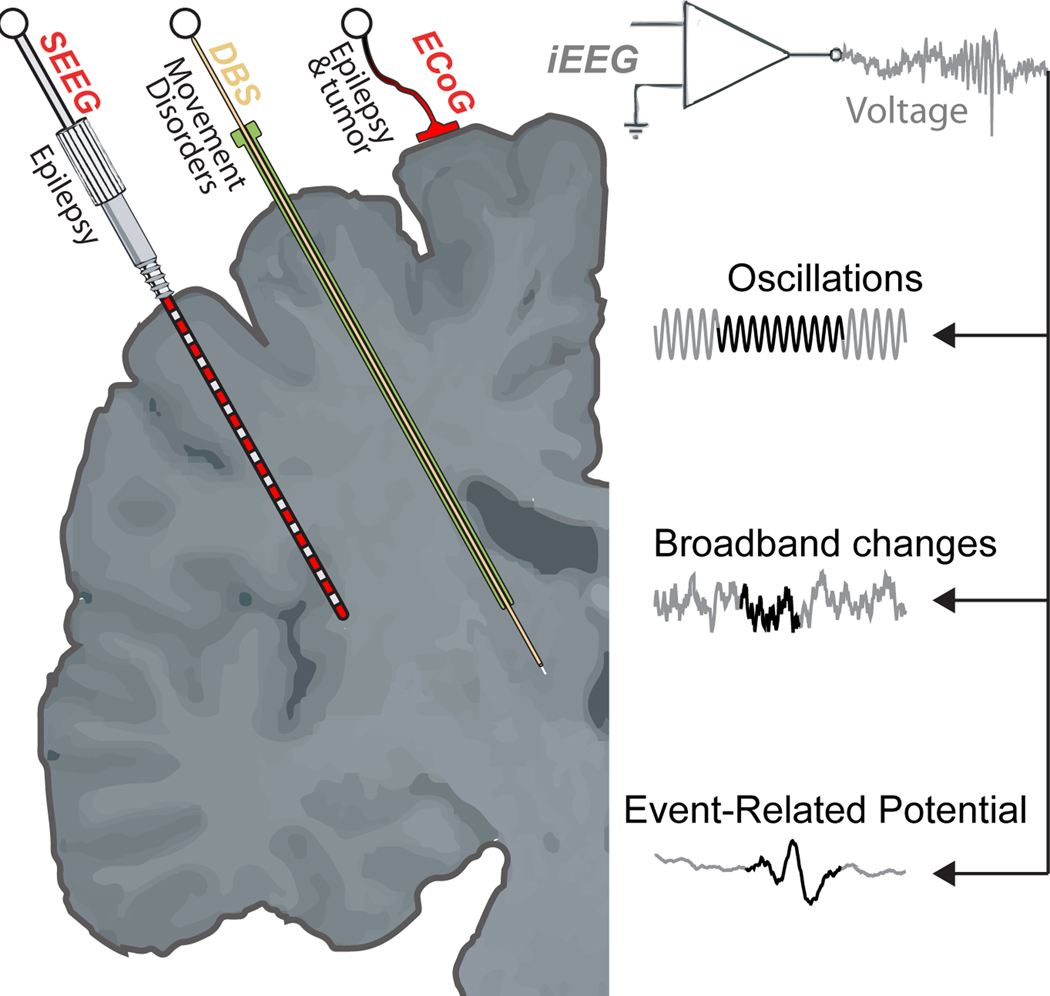
Figure 1.
Intracranial EEG recordings and basic signal features.
iEEG can be measured using three different recording strategies: stereotactic EEG (sEEG), Electrocorticogram (ECoG), and deep-brain stimulation (DBS). Signal features are generally characterized as oscillations, broadband changes, and event-related potentials.
1.1. Clinical context
1.1.1. The presurgical evaluation
In patients suffering from severe focal epilepsy that is pharmacologically intractable or that is not adequately controlled by anticonvulsant medications, epilepsy surgery represents a curative therapy, potentially providing patients with profound improvements in their quality of life (Chauvel et al., 2019; Engel et al., 2005; Englot et al., 2013; Kovac et al., 2017). In most cases, epilepsy surgery aims to resect the brain tissue generating the seizures (the epileptogenic zone), with the goal of suppressing their occurrence completely without causing significant neurological or cognitive deficits. Because epilepsy surgery is irreversible, it requires careful individualized presurgical assessment after review of the patient’s medical history, medications, and neurological examination. Typically, the multidisciplinary assessment comprises long-term monitoring of video-scalp EEG recordings of the patient’s habitual seizures (i.e., seizure semiology), structural magnetic resonance imaging (MRI) scans of the brain, and neuropsychological assessment (Baxendale et al., 2019; Bernasconi et al., 2019; Rosenow et al., 2016). Additional examinations may include positron-emission tomography (PET), ictal single photon-emission computed tomography (SPECT), surface electroencephalography (EEG) and/or magnetoencephalographic (MEG) source imaging, or an intracarotid anesthetic procedure (Lascano et al., 2016). At the conclusion of this non-invasive assessment, enough converging evidence may have been gathered to offer the patient a surgical option. In other cases, however, additional information must be collected to localize more precisely the putative epileptogenic zone or to ensure that the resective surgery will not impair normal cerebral functions. In such cases, an invasive evaluation with iEEG electrodes may be indicated.
1.1.2. Surgical implantation
Electrode implantation consists of a surgical procedure performed by the neurosurgeon in the operating room under general anesthesia. The pre-surgical evaluation helps surgeons determine the electrode placement, which typically targets the putative epileptogenic zone or zones. Invasive monitoring allows for the evaluation of multiple hypotheses regarding the cortical regions involved in seizure onset. Additionally, the cognitive functions of the implanted regions can be assessed by electrical stimulation mapping (see section 1.1.4.1). The implanted areas (i.e., spatial coverage) can be extensive, depending on estimated epileptic focus locations and seizure propagation. These electrodes may, or may not, record from epileptic tissue and are therefore especially relevant for fundamental research. Nonetheless, for ethical reasons, the electrode implantation must only be determined by clinical needs and not by research interests. Two types of implantation strategies are used, depending on the approach of the clinical team (see Figure 1).
Electrocorticography (ECoG) was first introduced by Penfield and colleagues in the mid 1950s at the Montreal Neurological Institute (Penfield et al., 1954) and was historically commonly used outside of Europe. It consists of one dimensional (1D) (strips) or 2D electrode arrays (grids) embedded in flexible silicone sheets placed onto the cortical surface just below the dura mater (outer cranial membrane). Regularly spaced contacts provide 1D or 2D spatial coverage (see section 1.3.3), typically extending over centimeters to cover large cortical areas (i.e., gyri), but are less sensitive to neural activity generated within sulci and do not access deep cortical structures (e.g., hippocampus or insular cortex). Surgical implantation involves a craniotomy to position the larger cortical grid(s) and/or a combination of associated strips. In some centers, 1D strip electrodes are inserted through burr holes, obviating the need for a large craniotomy. The implants are often, but not always, sutured in place to ensure that the electrodes do not move throughout the duration of the clinical monitoring period (see section 2.1.4).
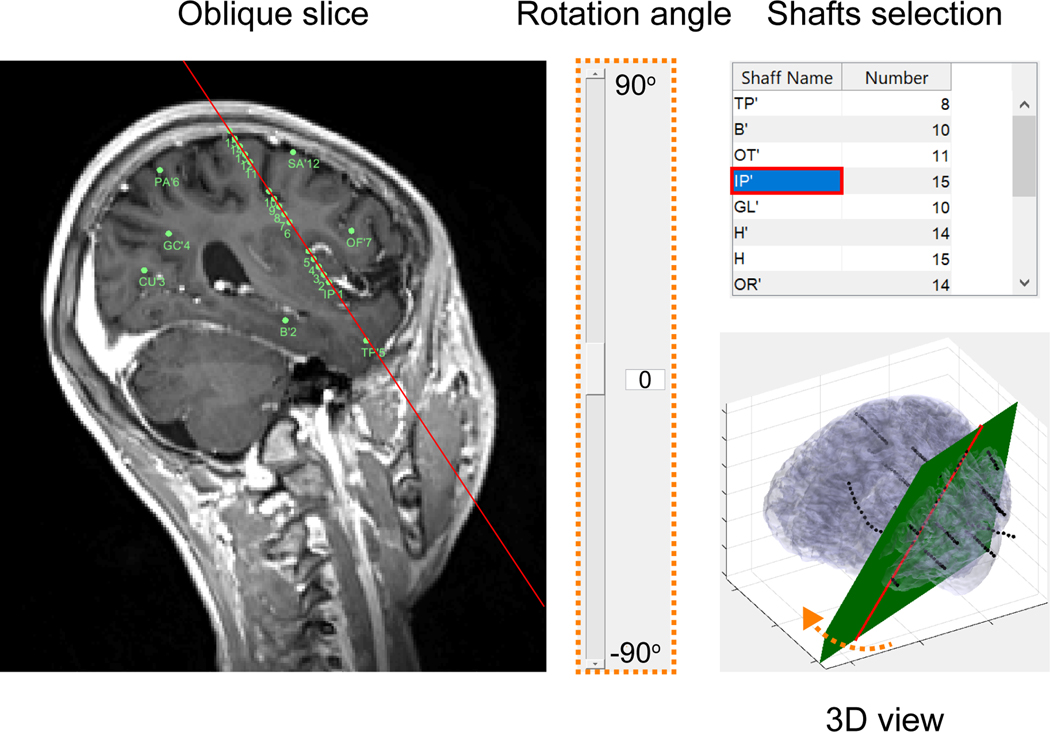
An alternative approach, the stereotactic EEG (sEEG), was developed a few years later in Paris by Bancaud and Talairach (Bancaud, J., 1965). The term ‘stereotactic’ refers to the way the electrodes are positioned: using a 3D stereotactic frame, which allows electrode placement in specific targeted deep brain structures. Targeted brain regions were formerly referenced relative to a set of idiosyncratic alphanumeric coordinates in a 3D standardized Talairach atlas (Talairach and Tournoux, 1988) that was based on a single post-mortem brain. Today, stereotactic placement of electrodes is guided by the individual patient’s structural imaging data - often a combination of MRI and X-ray computerized tomography (CT). sEEG uses needle-like semi-rigid shafts (often called “leads”), generally with regularly-spaced electrode contacts (Gonzalez-Martinez et al., 2013), but some manufacturers offer shafts with larger spacing in the middle of the shaft (e.g., where it traverses the white matter, see section 1.3.3). The shafts are inserted through small burr holes (typically a few millimeters in diameter) in the patient’s skull. The shafts can penetrate the brain with different approaches: laterally/orthogonally to reach the inter-hemispheric plane, obliquely for orbital or insular implantation, longitudinally/parietally to span the entire long axis of the hippocampus via a posterior implantation. The shafts are held in place with skull bolts (see Figure 1), ensuring that the electrodes remain at the same position for the duration of clinical monitoring. A typical sEEG implantation involves between eight and twenty shafts, with five to twenty electrode contacts along each shaft.
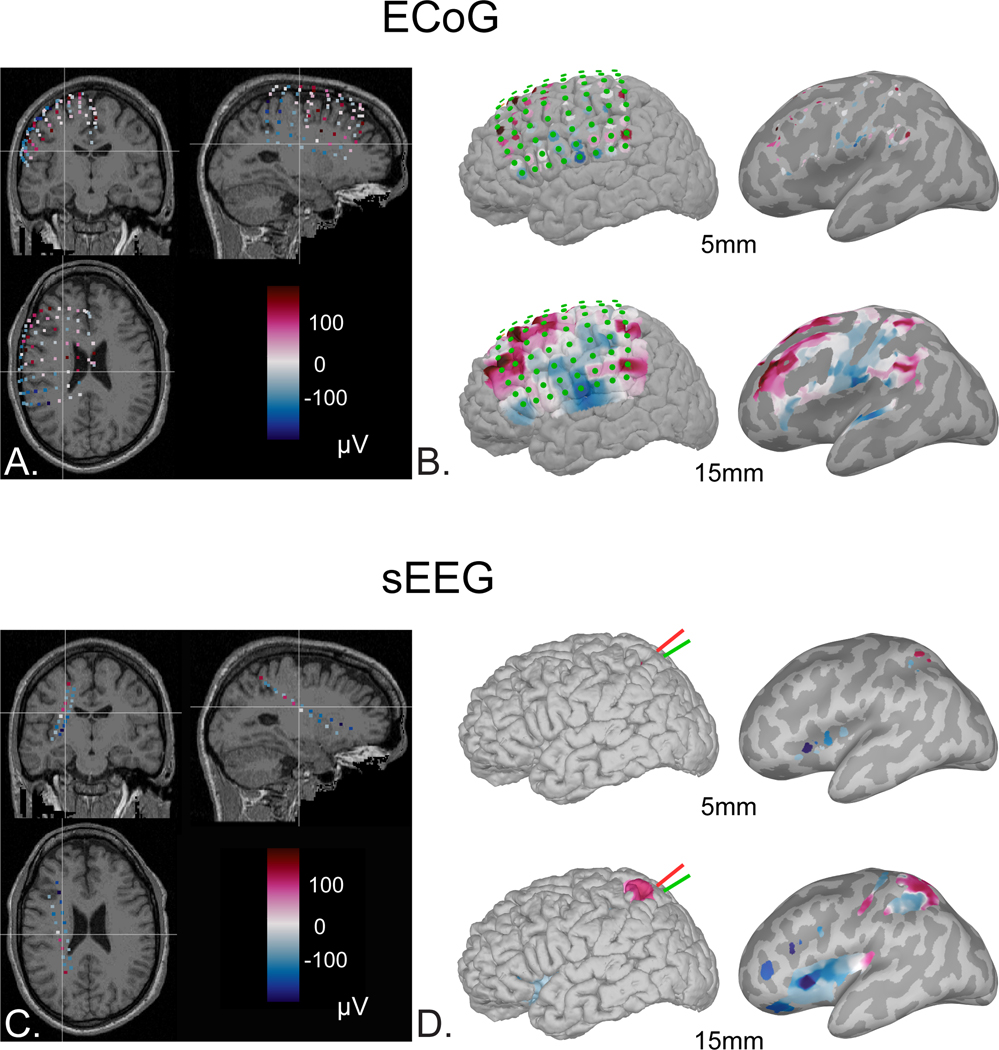
The main advantage of sEEG over ECoG is the access to activity in deep cortical structures and sulci, which, however, comes at the cost of the detailed 2D spatio-temporal mapping that ECoG provides (Minotti et al., 2018). That said, sEEG electrode contacts traverse both gray and white matter, which needs to be considered during signal analysis and data interpretation (see sections 2.1.4 and 3.1). Nowadays, many epilepsy surgery centers in Europe and North America are experienced with both ECoG and sEEG and select the most appropriate technique for each patient. Some centers use a combination of both ECoG and sEEG intracranial implantation in the same patient (Kim et al., 2011; Surbeck et al., 2011).
1.1.3. Clinical monitoring
Following electrode implantation, the patient remains under continuous medical observation in the epilepsy monitoring unit (EMU); typically for a period of several days to two weeks depending on the results of the evaluation and the patient’s clinical state. In the EMU, iEEG signals are continuously recorded together with the audio and video of the patient. The main objective is to let the patient’s habitual seizures occur spontaneously under careful withdrawal of anticonvulsant medication, and to identify, in relation to seizure semiology (e.g., overt behavioral manifestation), the iEEG sites with the earliest abnormal electrophysiological activity (i.e., the “seizure onset zone” from which seizures originate and spread to other brain regions). Temporal synchronization between recorded audio, video and intracranial EEG is crucial for accurate seizure characterization.
1.1.4. Functional exploration / mapping
1.1.4.1. Electrical stimulation
During the patient’s stay in the EMU, clinicians generally deliver intracranial electrical stimulations (iES) through iEEG electrodes (extraoperative ES mapping). It consists in delivering low-amplitude current electrical stimulations between pairs of contacts and is also known as direct cortical stimulation, electrical stimulation mapping, cortical stimulation mapping, or as electrocortical stimulation (Borchers et al., 2012; Cuello Oderiz et al., 2019; Kovac et al., 2016; Trébuchon and Chauvel, 2016). Information collected during iES is essential for surgical planning to potentially avoid resecting brain regions supporting critical functions, as the success of the epilepsy surgery depends not only on the resection of the epileptogenic zone but also on the absence of any consequent neurologic postoperative deficits (e.g., motor, sensory, linguistic or cognitive impairments (Corley et al., 2017; Kanner, 2016)). By means of iES, the functional brain regions or networks covered by the electrodes, are characterized based on a combination of observed behavior (e.g., motor movements when stimulating the motor cortex (Penfield and Boldrey, 1937), speech arrest when stimulating language areas (Ojemann and Whitaker, 1978)) or based on descriptions of experiential manifestations by the patient themselves, such as hallucinations (Aminoff et al., 2016; Mégevand et al., 2014; Penfield and Perot, 1963) or emotional reaction (Caruana et al., 2018, 2016; Fried et al., 1998). When the iES mapping reveals so-called eloquent tissue close to the epileptogenic focus, surgery can proceed with additional acute iES mapping in the operative room. In that case, during the surgical procedure, the depth of general anesthesia is decreased so that the patient can be mapped awake on the operating table (with local anesthesia of the surgical wound) to extend the resection margin to its maximum without creating post-surgical deficits.
iES can be used to identify sites where iES elicit after-discharges and/or symptoms similar to what the patient experiences at the beginning of their seizures. However, these evoked seizures may not match the patients’ naturally occuring seizures; and some neurologists prefer not to do stimulation until monitoring is complete and the patient is back on their anti-epileptic medications.
From a research perspective, iES can produce experiential phenomena that provide excellent functional localization information (Kern et al., 2019). These must be carefully documented (see section 1.3.8), because they can interfere with other test stimuli. For instance, the position of an illusory visual percept evoked by the iES can be pointed at, or mapped, using a white type of 'dartboard'. The patient can have this in front of them during the stimulation and can indicate the region in space where they experience illusory percepts; this can also helps to determine if the illusory percept moves (e.g., by measuring the eccentricity relative to the fovea or its position on an imaginary clock).
Yet, iES has limitations in identifying functional networks: stimulation cannot be repeated ad infinitum due to time constraints, patient overload and progressive loss of response specificity (e.g., behavioral fatigue and risk of unspecific after-discharges, (van ’t Klooster et al., 2011)). Furthermore, the effect on most cognitive functions can only be observed if stimulation produces an obvious change in the patient’s behavior, or if that function is being performed by the patient at the time of the stimulation (e.g., speech production, access to episodic memory, mental imagery, mental calculation). Many stimulation sites often lead to undetected subtle or no effects that provide no relevant functional information (Mazzola et al., 2019; Murphey et al., 2009).
1.1.4.2. Task-related exploration
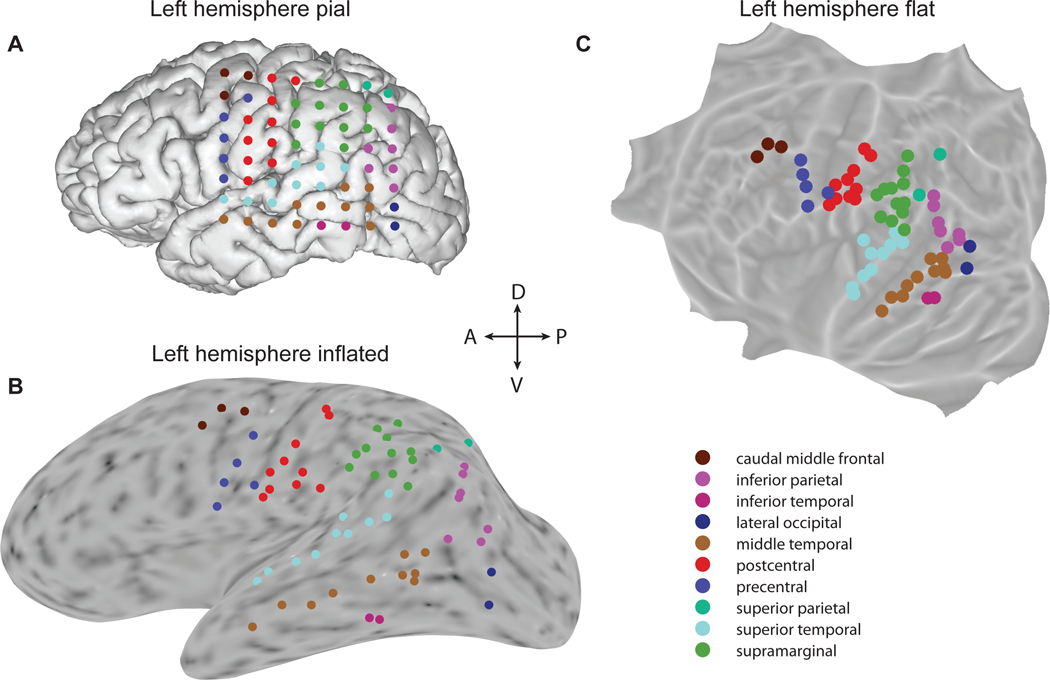
A complementary strategy to iES is to record evoked iEEG activity during a battery of short tasks engaging critical cognitive functions (i.e., “localizers” assessing sensory/motor processes, language, memory, attention, etc.). Changes in the activity of the neuronal populations supporting those functions is locally recorded by iEEG electrodes sited nearby, which provides additional information for producing functional maps of the patient’s brain. Some centers have implemented a systematic analysis pipeline to provide the clinical team with a detailed report of all significant iEEG task-induced responses within a few hours (Cheung and Chang, 2012; Lachaux et al., 2007b; Miller et al., 2007a; Schalk et al., 2008). Such localizer-based functional mapping procedures can be performed in a short period and be planned early during the hospital stay to inform clinicians and guide subsequent iES. Since electrode implantation is patient specific, and given that functional responses cannot be predicted based on structural anatomy alone, such reports are also valuable for research (Figure 2).
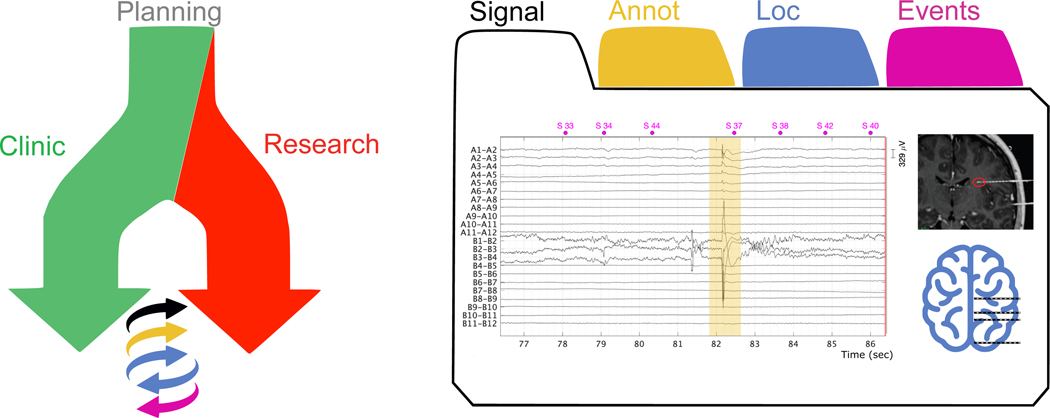
Figure 2.
Schematic representation of the interaction between clinic and research.
Left panel: starting from surgical planning, with involvement over time of the clinical and research procedure. The clinical team provides the research team with the iEEG data, imaging data, and clinical annotations (e.g., epileptic activity). In return, the research team(s) can provide the precise localization of the electrodes (Loc), and activation tasks with associated event-related results regarding brain functions (Events).
Right panel: illustration of the different types of data in an iEEG study: iEEG (Signal) and imaging data, clinical annotations (Annot), electrode locations (Loc) and (responses to) task-related events (Event).
1.1.5. Challenges, recommendations and reporting advice
Many factors may preclude a patient from being a suitable candidate for a given study. For example, the cognitive abilities of the patient may prevent them from completing demanding cognitive tasks, electrode placement may not be deemed relevant for the functions being studied, and/or persistent epileptogenic activity between seizures may hinder recording of artifact-free iEEG. Prospective recruitment of patients for research studies allows avoiding sampling biases, i.e., the selection of participants on a priori criteria which will limit the generalization of the results (see section 5.1). We recommend that researchers report inclusion and exclusion criteria in their publications.
1.2. Research and experimental aspects
A major technical challenge for iEEG research is to adapt research questions to the clinical environment, controlling sources of variability while maintaining high-quality iEEG signals. This section discusses considerations for running experiments in the EMU and addresses some ethical issues (see also (Chiong et al., 2018; Feinsinger et al., 2022)).
1.2.1. Interactions between researchers, clinical staff and the patient
Researchers should seek to minimize interference with normal clinical procedures. Should a seizure occur, it is essential that the experiment immediately cease and that research equipment does not hamper clinical intervention, both for the patient’s safety and to not obstruct the video recording of the behavioral characteristics of the seizure. Therefore, it is beneficial to use mobile, battery-powered setups that can be quickly moved without data loss. Should the participant become uncomfortable during the experimental recording, this should be respected: the experiment should be stopped. Then, only if the participant is willing, the experiment could be resumed or restarted at a later time. Intracranial implants can cause headaches and nausea, making it difficult or impossible for patients to concentrate and perform demanding tasks. Researchers must ensure that experiments do not cause stress, anxiety or discomfort, or take time away from visitors. The researcher may want to schedule the experimental recordings together with the participant. Giving autonomy to the patient benefits the relationship with the participant and his/her experience regarding research participation. Importantly, when scheduling experimental recordings, researchers should also liaise with clinical staff and make sure that they are not testing the patient at times when clinical staff need to interact with the patient, or when other clinical tests have been scheduled. When interacting with patients, three key perspectives must be taken into account:
From an ethical perspective, it is essential to inform the patient that his/her medical care will not be influenced by, or depend on, his/her participation (or lack of participation) in experimental research. It is also important that the patient understands that there is no direct benefit to them and that the research might not necessarily be directly clinical in nature. When the clinical staff is part of the research team, this can pose a particular challenge. Even if the clinician explicitly explains what is clinical necessity and what is voluntary research, for the patient these lines can remain blurry. For this reason, it can be advisable to not have the patient’s own clinical staff (neurologist or neurosurgeon) ask for informed consent/perform experiments.
From a human perspective, while the patient is aware that there is no direct benefit for him/her, curiosity often brings the patient to ask questions about the research. Here, our experience is that a sense of partnership can be established between researchers and the patient, who can enjoy the time spent in that ephemeral relationship at both a personal and intellectual level (e.g., knowledge sharing). A practical consequence is that the scheduled time-slot for the experimental interaction should also leave time for all important social interactions.
Finally, from a medical perspective, it is important to keep in mind that during this period the patient attempts to follow and understand the evolution of his/her clinical assessment. It is not appropriate for researchers to provide the patient with information concerning their medical assessment.
It can be a good idea to use gamification and story-telling to make experimental paradigms more enjoyable. Researchers should keep in mind that experimental conditions are unlikely to reach the quality of a proper laboratory setting due to many factors such as lighting conditions, electrical noise, distracting sounds, patient fatigue and fluctuating motivation. In that regard, it should never be forgotten that the iEEG research participant is a patient first.
1.2.2. Recording environment
Some centers have a dedicated experimental room to perform research in a controlled environment. While the additional time required for the patient transfer may reduce the total amount of time participating in the actual research paradigms, the advantage is the better standardization of experimental settings between patients. Such a controlled environment allows performing finer psychophysical manipulations,increasing the signal to noise ratio when it is possible to use a better electrical isolation (e.g., Faraday cage) or a high-quality research-oriented data acquisition system, reducing the risk of distractors during the experiment and permitting to record without other patient(s) in the same room. Yet, the transfer from the patienťs room to the dedicated experimental room can imply interrupting the clinical iEEG recording and the risk of missing seizures during the transfer which would have a negative clinical impact. We therefore recommend being extremely vigilant, that is, (i) to equip the experiment room with a microphone and a video camera to record potential seizure semiology, (ii) to start the recording as soon as possible and to continuously record from the participant in the experiment room in case a seizure occurs while the participant is not performing an experiment, (iii) to minimize the transfer time, (iv) a neurologist and/or a nurse should stay with the patient, (v) to ask the clinicians about the degree of patient medication (i.e., whether a reduction of anticonvulsant medication increases the probability of seizure), (vi) to use a wheel-chair or transport bed to reduce the risk of a fall should a seizure occur.
Other centers are equipped with a specific patient room that is optimized for both clinical observation and research with additional electromagnetic isolation. In this case, the EMU can be complemented with a research controlled setting, including, for example, conduits for cables allowing two-way communication between patients and research setting rooms. Hence, once arriving from the operating room the patient remains there for all investigations throughout the monitoring period (Billig et al., 2019; Lehongre et al., 2022).
1.2.3. Research projects in a clinical setting
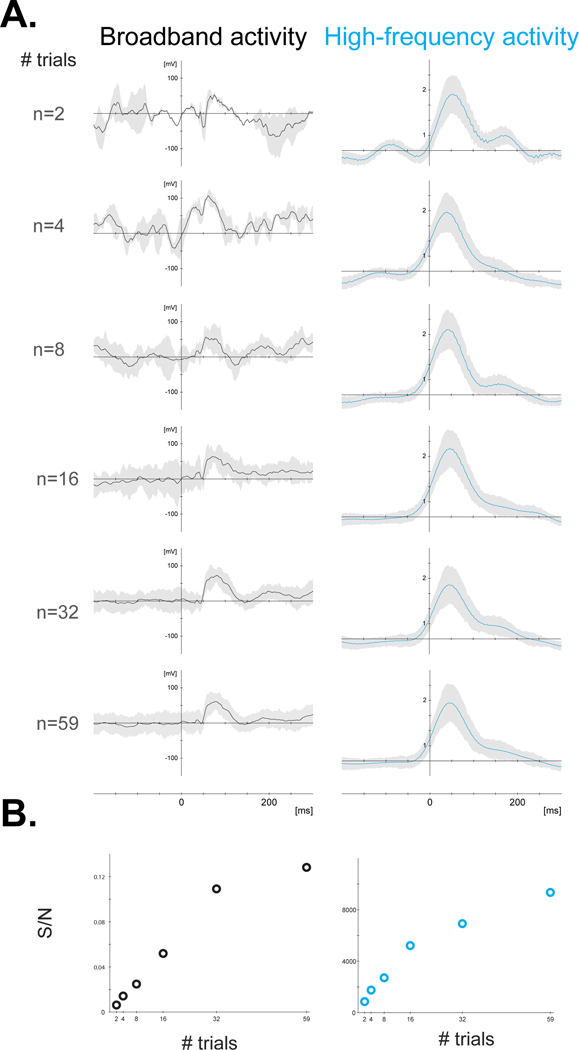
An iEEG experiment might be interrupted at any time due to clinical events (e.g., a seizure); therefore, experiments organized in short blocks ensure that data analysis remains possible, even if some data are missing. That said, when designing the experiment, the minimum number of ‘trials’ in each condition to be analyzed should be considered. Whenever possible, experiments should be designed to linearly increase the amount of useful data as time passes. All blocks should include all conditions, so later blocks will simply add more trials per condition and increase statistical power. Interictal epileptiform discharges can contaminate iEEG signals and their prevalence and frequency can vary from day to day. A check of the iEEG signal on the clinical system before commencing the experimental set-up may aid in a decision to go ahead with or postpone a recording session. It is generally preferable to not spread the recording of a single paradigm over multiple days, as the quality of the signal (Sillay et al., 2013), the medications taken by the patient, and their behavioral state may vary over time.
Since a patient can only participate in a limited number of experiments, it is important that those experiments are chosen and prioritized wisely. This inevitably leads to some selection bias, for instance with more patients recorded in the left hemisphere being included in language studies. Some centers send the results of their localizers (see section 1.1.4.2) to all research teams with ongoing projects and then prioritize incoming research requests according to protocol duration, particular requests to access certain brain regions, as well as the team’s or project’s history (e.g., how many patients have been recorded so far, and results obtained).
As multiple research teams often compete for a few time slots with the patient, it is always a good idea to keep an open register of all experiments performed and timing of recording sessions, to create a feeling of equity and transparency. This can be facilitated by organizing a regular schedule of meetings between clinicians and researchers to discuss new protocols and present results obtained from ongoing protocols from previous patients. These meetings provide an excellent opportunity to create bridges between clinical and fundamental science, and to increase the global understanding of a given patient’s brain, by combining all observations made by independent research groups for that same patient, in relation to his/her clinical records. Importantly, should some technical problem develop with the iEEG recordings themselves due to changes in software/hardware, regular team meetings can provide an excellent troubleshooting forum and communication to all about how to deal with any encountered technical issues.
1.2.4. Research procedure
The initial time spent by researchers with the patient is devoted to a careful explanation of the experimental procedure and making sure the patient has given formal written informed consent. When possible, we recommend discussing participation in research protocols with the patient prior to the implantation, and if possible asking for consent at that time. Thus, the patient can anticipate this dimension of his/her stay at the clinic and is less burdened during this period. Yet, it must be clear for the patient that the length of his/her stay is solely determined on a clinical basis.
Subsequently, careful installation of the necessary equipment (e.g. computer screens, audio outputs, response devices, eye-tracking device) and their connection/synchronization to the iEEG data recording system is required. We strongly recommend carefully performing all the needed tests (e.g., synchronization between presentation device and acquisition system) during a “dry run” with no patient (i.e., prior to performing the acquisition with the patient, see section 1.3.1).
When the EMU is not directly equipped for research (see section 1.2.2), the experimental equipment might be housed on a trolley/cart that can be moved from one room to another. This can facilitate easy removal of equipment from the field of view of the video camera to record the behavioral characteristics of a seizure, as well as allowing clinical staff access to the patient at any time (see section 1.2.1).
In some centers, a dedicated member of the staff is in charge of performing the experiments, assisted by the researcher. If the person in charge is part of the clinic, he/she should make it clear to the patient that at that time he/she is ‘wearing a research hat’ (i.e., the patient should feel free to say ‘no’ to a research request, see section 1.2.1). The obvious advantage of having the same person doing all the recording is that installation of experimental equipment is fast, efficient and reproducible, and that the interaction with the patient and the clinical team is optimal. The dedicated member of the staff is also in a good position to survey and provide information on the patient (e.g., handedness, age, moment of last seizure, medications, and technical information about the electrodes and recording setup, see section 1.3.8).
Because the interaction with the patient is usually attention-demanding, we recommend that experimenters prepare and use a checklist to keep track of each step in the experimental procedure. Such documentation reduces the cognitive load on the researcher, leads to more rigorous organization, can provide relevant meta-data, and makes data collection consistent. Working in teams of two researchers is also useful - one person can monitor the iEEG display continually (in case of seizure activity), while the other monitors the task computer and the patient’s performance of the activation task.
1.2.5. Challenges, recommendations and reporting advice
The clinical and research team should closely collaborate in organizing experimental recording sessions.
Written informed consent must be obtained from the patient for all research procedures being undertaken as well as for the clinical data that is used for the research (e.g., neuroimaging data). Make sure that a duly authorized professional collects this consent prior to testing (this is generally stipulated in the consent form and approved by the appropriate Ethical Review Committee).
Any person interacting with the patient during intracranial evaluation must behave and present a professional image at all times. This is important as it might influence the patient’s perception of his/her medical care.
Script the explanations/information given to the patient, use appropriate terminology, e.g., avoid using personalized phrasings like “test your abilities” but rather use terms such as “investigate human brain functions”. Be explicit with the purpose of fundamental research (e.g., “this research is not designed to help you, but the knowledge obtained may help future patients”).
Use template “recording sheets” to collect and document information relevant for subsequent analyzing and reporting (e.g., the time of the last seizure, the level of medication, handedness, reference/ground electrodes, comments on the recording quality and patient state). These records can be made on paper or digitally, but ultimately all data should be archived in digital format.
1.3. Signal monitoring, recording and supplementary data
1.3.1. Experimental acquisition setup
Most iEEG acquisition and visualization systems are optimized for clinical requirements and may not include some of the features desired for experimental research (e.g., the ability to input triggers, perform online averaging, or allow other online analysis for closed-loop experiments). Some acquisition systems allow starting a new recording with parameters for an experimental session (e.g., increased sampling rate) without interfering with the ongoing clinical recording. In other cases, a research amplifier may record data in parallel to the clinical system. Both good quality recordings and the local regulatory constraints for patient safety are the responsibility of a trained clinical engineer. We recommend coordinating with the clinical staff prior to taking any action that may alter the parameters of the long-term continuous clinical iEEG/video recordings. Additionally, when recording at the bedside with specialized research equipment that interacts with the clinical setup, it is important for patient safety that this equipment is approved for use in the clinical setting (which depends on local and national regulations), is battery operated, and does not lead to any discomfort to the patient (e.g. goggles or headphones exerting pressure on the bandaged head).
Practical considerations for stimulus presentation include what stimulus presentation software the researcher should use, how reliable the hardware and software of the recording environment are, and how well these serve the purposes of the research goal. Selection of stimulus presentation software may be constrained by the researcher’s ability to install new software on the available EMU equipment, or to synchronize their own equipment to the EMU set-up. The researchers should also be familiar with the hardware connection between the stimulus presentation computer and the recording device (e.g., whether a parallel port or other types of connections are supported). If the researcher can choose the stimulus presentation software, we recommend one that optimizes precise timing for the stimulus type being used in their study. A comparison between common stimulus delivery programs is available (Bridges et al., 2020).
Any electrophysiological recording that requires synchronization with a behavioral task needs a stimulus trigger channel that allows synchronization of event timing in the experimental software with electrophysiological events in the iEEG recordings. The trigger channel is the critical piece that renders our data useful - placing timestamps in the data file for specific task events. Given the idiosyncrasies of the iEEG recording environment, and the high value of such rare data, it is important that every trial be accurately accounted for. We recommend embedding redundant information in the data collection strategy, such as additional channels which directly record stimulus presentation, such as a photodiode attached to the computer screen, a microphone or direct audio recording from the amplifier (see section 1.3.4). These allow for a ‘ground-truth’ timing-reference in case the trigger channel does not record events, or does so with variable timing. Another source of redundant information could be the behavioral or task-related files generated by stimulus presentation scripts. While these may not as easily provide timing-related information, they can help the researcher to ensure a match between the number and types of events that occurred over the course of the experiment with the information embedded in the trigger channel. As such, a complete and verbose behavioral recording is desirable for any cognitive task.
Restrictions around the hardware and software permitted in the clinical environment (for reasons of clinical certification and patient safety) can introduce unforeseen sources of variability when running cognitive and behavioral experiments. In the ‘dry run’ that researchers should do before recording with an actual patient, they should aim to capture such variability in their experimental set-up, such as latency or jitter in stimulus presentation (see section 1.4). However, discrepancies between the timing of the trigger channel and other channels may be tolerable so long as the latency is consistent (i.e., correctable constant lag). An additional benefit of this dry run is to more accurately estimate the duration required for equipment set-up, providing realistic expectations of the amount of data the researcher can acquire in a session with the patient, while minimizing the appearance of uncertainty or unpreparedness that might make the patient uncomfortable. Accurately estimating the time required for set-up also allows for better communication with clinical staff and fellow research teams. Dry runs should be repeated if any hardware or software on either the clinical or the research side has been changed or updated.
1.3.2. Recording reference and ground
As with scalp EEG, recording iEEG signals relies upon low-noise differential amplifiers typically housed in a “headbox” located close to the patient, in combination with another amplifier component (the ADC converter and/or the interface and synchronization system) located closer to the data acquisition computer. Each differential amplifier channel takes a pair of electrodes (the electrode of interest and the reference, which is usually shared among all channels), amplifies the potential difference and converts it to a digital representation that can be visualized and stored on a computer.
iEEG signals typically range from 0.05 to 2 millivolts, about 10 to 100 times larger than scalp EEG signals. Besides the electrodes of interest that are placed at, or close to, the tissue of interest, biopotential recording systems also require a reference electrode (REF) and usually include a ground electrode (GND) that serves to suppress noise. As an alternative to the ground electrode, biopotential systems can also include a common sense (CMS) and a so-called driven right leg (DRL) electrode.
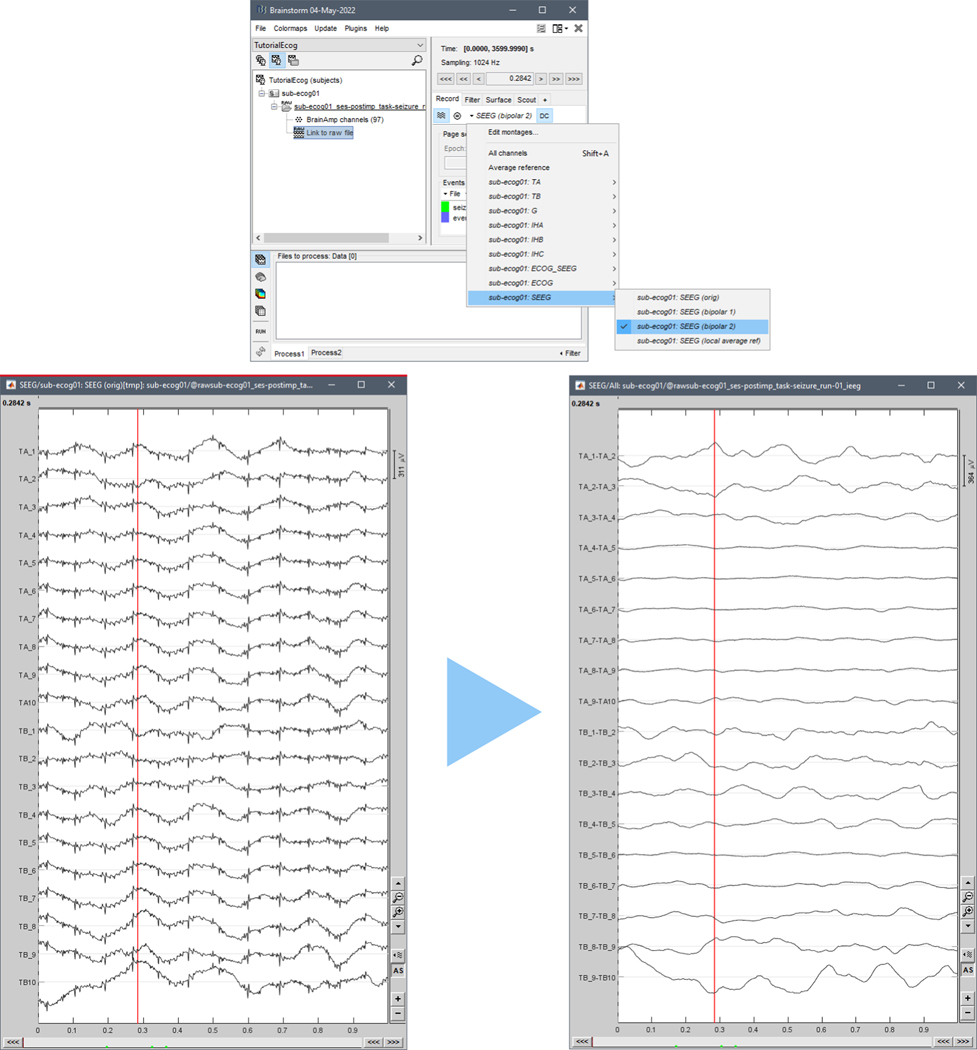
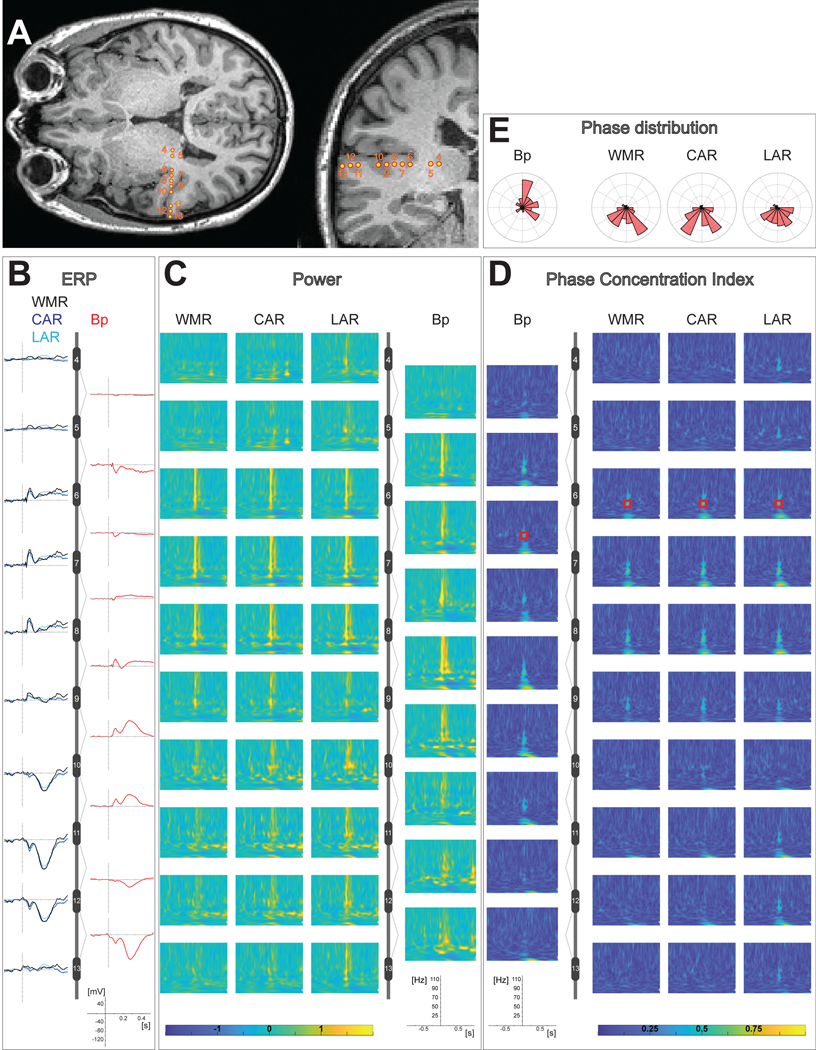
Electrical potentials by definition quantify a voltage difference. Thus, the iEEG signal reflects the voltage differences between a pair of electrode contacts, typically the electrode of interest relative to a reference electrode. It is important to distinguish the ‘online’ reference that is used during data acquisition itself, from the reference selected in post-hoc digital re-referencing for offline data reviewing, processing and/or analysis (see section 3.3). It is important to document what, and where, the online reference was and to share this with others that might analyze the data (e.g., scalp reference electrode at Cz, or sEEG shaft B contact 6, see section 1.3.8).
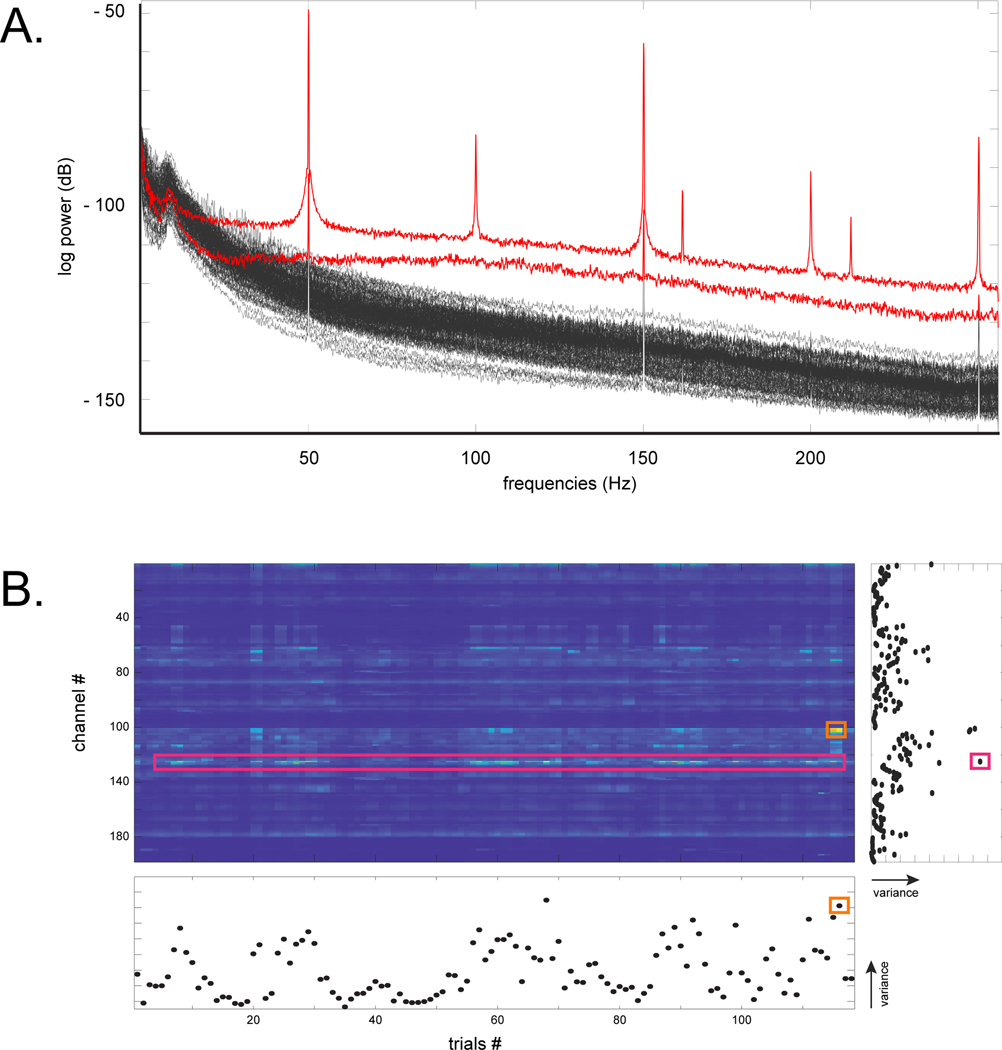
Differential physiological amplifiers use the GND electrode to reduce the effect of the common mode voltages present on both the electrode of interest and the reference electrode. This reduces common-mode interference, for example due to the 50/60 Hz power line and due to other non-physiological sources of noise (Scheer et al., 2006) (see sections 3.2 and 3.3).
At the electrode-tissue interface the displacement of electric charge in the electrode (a metal) consists of free electrons, whereas the displacement of electric charge in the tissue (an electrolyte) consists of ions. The electrochemical reaction at the electrode-electrolyte interface not only results in a specific electrode impedance, but also in an electrode potential. Different metals have different surface polarization potentials that can introduce offset potentials to the amplified and recorded potential difference between an electrode and the reference (Lee Stephen, n.d.). The impedance at the electrode-tissue interface (and thereby the electrode potential) fluctuates over time (about few minutes in case of scalp EEG, to days in the case of iEEG (Sillay et al., 2013)).
Besides being the source of slowly changing offset potentials, the electrode-tissue interface can also introduce broadband noise and affect the signal depending on the electrode impedance (Huigen et al., 2002). This also pertains to the impedance of the REF and GND, which are sometimes placed on the scalp or located in the skin, bone, meninges or white matter. Additionally, some metals can introduce filtering effects into measured signals, therefore the material for these electrodes needs to be carefully considered (Hari and Puce, 2017). When the researcher can choose these electrodes, he/she may consider the various possibilities that we have described here. However, in practice, the choice of the REF and GND is usually made by clinicians when the patient arrives in the EMU and usually maintained during the entire monitoring time. The criteria for a good REF/GND include: the patient’s safety and comfort, the complexity of the set-up/placement, the ease of maintenance and the ability to maintain a low impedance for a long time.
The two following subsections describe various options for choosing the recording reference and ground, ordered from the least to the most invasive. There is not a single best solution, but there are multiple reasonable solutions that depend on the electrode implantation schemes that impose constraints on where the REF and GND can be placed - largely due to the presence of sterile dressings that can cover large sections of the head. Despite this variation, considering signal quality and the influence of the REF and GND can have on planned analysis should be considered (e.g., for common pick-up on the measure of coherence, see (Zaveri et al., 2000), and see sections 3.3, 4.2.3.7 and 4.2.3.8).
1.3.2.1. External REF/GND
When opting for an external REF or GND, an electrode on the skin (e.g., tip of the nose, mastoids, clavicle) or scalp surface can be used to achieve a low impedance, although they require regular maintenance. The electrode paste or gel and tape used for their placement may not always be compatible with the sterile dressings covering the craniotomy and/or burr holes post-implantation.
These issues can be avoided with a subdermal needle electrode, although this is already more invasive (Vulliemoz et al., 2010). Alternatively, as part of the surgical procedure, an electrode such as a skull screw, or a cranial electrode (i.e., sEEG electrode localized in the skull), or an inverted subdural strip electrode, facing upward to the inner side of the skull, may serve as an indwelling reference.
The main disadvantage of an external reference is that it is more prone to pick up external noise (i.e., ambient 50/60Hz), some artifactual muscle activity (including blinks and eye or head movements) and/or global brain activity being passively volume conducted through the tissues. That said, invasive and semi-invasive reference electrodes can still pick-up volume-conducted artifactual activity such as eye blinks and movements or cardiac activity (see section 3). Importantly, as noted earlier, different material for electrodes should be avoided, as they would result in different half-cell potentials and thereby introduce unstable DC offset potentials which deteriorate the signal quality.
1.3.2.2. Internal REF/GND
An alternative solution for the REF and GND consists of using a pair of iEEG electrodes, e.g., adjacent electrodes in the white matter. This reduces the risk of using different electrode materials which inadvertently could introduce DC offset potentials. In this case the REF (like any other electrode) is sensitive to fluctuations of the bioelectrical activity generated in its vicinity, therefore it is advisable to choose an electrode site which picks up as little neural activity or bioelectrical artifacts as possible, as this would be introduced back into the full montage. An additional advantage with internal REF electrodes is that its impedance is likely to be comparable to the contacts of interest that form the other input to the differential physiological amplifier: impedance mismatches between pairs of contacts that provide the inputs to differential amplifiers can impair the ability of the amplifier to reject common mode signals.
1.3.3. Electrode characteristics
The materials used for ECoG and sEEG electrodes are in most cases platinum/iridium because of its biocompatibility and suitability for long-term implantation in human tissue. However, sEEG and ECoG electrodes vary substantially in terms of their geometry and many manufacturers custom-design probes on request (see section 1.1.1). Here we describe the most used macro-scale electrodes for both techniques.
ECoG subdural electrodes are flat discs. The diameter of the exposed surface varies from 1 to 4.5 mm and the surface from 5.3 to 28.2 mm2. The distance between individual electrodes within a grid or strip is typically 3 to 10 mm. Typically, grid arrays consist of 4x4, 4x5, 8x8 or 10x10 electrodes, and strips can include various electrode configurations e.g. 1x4, 1x6,1x8, 2x10 (Boatman et al. 2010). Strips can be linear arrays or form ‘L’ or ‘T’ shapes. Beneath an ECoG electrode with 2.3 mm diameter, given an estimate of about 100,000 neurons under 1 mm2 cortex, the electrode contact covers about 5 x 105 neurons (Miller et al., 2009). Estimates from macaque V1 suggest that each electrode measures activity from an area of about 3 mm in diameter (Dubey and Ray, 2019). Nonetheless, local field potentials also reflect volume-conducted activity from distant sources (Kajikawa and Schroeder, 2011).
sEEG depth shafts are cylinder-shaped with 5 to 20 contacts. For each contact the exposed surface length is around 2 mm with diameters ranging from 0.8 to 2 mm, leading to a surface area of 3.5 to 50 mm2. Various options exist for the spacing of the centers of adjacent electrodes, but distances typically range from 2.5 to 10 mm. Each sEEG electrode explores a small volume of brain tissue estimated to be a sphere of approximately 5 mm radius surrounding the electrode (von Ellenrieder et al., 2012).
ECoG electrodes measure cortical activity differently compared to sEEG electrodes. Their subdural position covers a large cortical surface area (i.e., up to tens of cm2) and captures the origin of grey matter signals orthogonal to the cortical surface; as compared to sEEG electrodes, which sample, from different spatial orientations, depths, and discrete brain volumes (i.e., a few cm3), by straddling different tissues and potentially different cortical layers. Finally, it is worth noting that the ability to pick up iEEG signals does not only depend on electrode properties, but also on the characteristics of the neural source itself: its spatial extent, its location and orientation relative to the electrode contact and the degree of synchronization among the active neural population (see (Cosandier-Rimélé et al., 2008; Ramantani et al., 2016) for a thorough investigation of the neurophysiological models of sources in sEEG and scalp EEG).
There are other specialized electrodes that we do not consider further here, for example microelectrodes specifically designed for the recording of activity of individual neurons or shafts with very closely spaced small electrodes to record the laminar potential distribution ((Kellis et al., 2016; Tóth et al., 2016) and see section 6.4). Their use requires recordings with much higher sampling rates and filter settings than regular ECoG and sEEG.
1.3.4. Supplementary channels and behavior
Along with iEEG signals, additional physiological signals may be recorded for clinic and/or research and may help identify artifacts (see sections 1.3.5 and 3.2). Electrocardiographic activity (i.e., ECG/EKG) is almost systematically recorded in the clinical environment, not only to monitor the patienťs cardiac activity, but also to allow distinguishing ECG/EKG artifacts from rhythmic interictal epileptiform discharges in the EEG. ECG/EKG can also be used to guide iEEG signal analysis (Kim et al., 2019; Park et al., 2018). Electrodermal activity (EDA, also referred to as galvanic skin response (GSR), psychogalvanic reflex (PGR), skin conductance response (SCR), or sympathetic skin response (SSR)) a marker for sympathetic skin activity, can be used for seizure monitoring, detection and prediction (Vieluf et al., 2021) as well as for studying cognitive processes (e.g., emotional processing, see for an example (Chen et al., 2021; D’Hondt et al., 2010)). For research purposes, electro-oculogram (i.e., the horizontal (HEOG) and vertical (VEOG) eye movements) or eye-tracking (i.e. eye gaze and/or a measure of pupillometry) can be recorded to monitor gross and fine eye movements, respectively (Ball et al., 2009; Cimbalnik et al., 2022; Golan et al., 2017, 2016; Jerbi et al., 2009a; Katz et al., 2020; Kern et al., 2021; Kovach, 2011; Lachaux et al., 2006; Podvalny et al., 2017). Similarly, surface electromyography (EMG) can be recorded to probe muscular activity (i.e.: voluntary and involuntary movement, (Talakoub et al., 2017)). Last, natural breathing can be recorded, for instance to investigate the influence of the respiratory cycle on the activity of specific brain areas and on cognitive functions (Herrero et al., 2018; Zelano et al., 2016), however, this requires specialized equipment that may require a lot of adjustment.
In most iEEG research paradigms, the participant receives instructions and performs a specific task. Therefore, iEEG data is often complemented by behavioral recordings which can be either discontinuous events (e.g., button presses) or a continuous data stream, e.g., from a microphone for speech tasks (Bouchard et al., 2013; Chartier et al., 2018; Hamilton et al., 2021, 2018; Mesgarani et al., 2014); eye tracking (Golan et al., 2017, 2016; Podvalny et al., 2017); or hand movements recorded with a dataglove (Miller et al., 2012). To relate the iEEG data to behavioral measure(s), synchronization between the two recordings is critical (see sections 1.2.4 and 1.3.1). For discontinuous events, corresponding time-points are either marked as triggers in a dedicated file accompanying the iEEG data (e.g., .vmrk in BrainVision Analyzer format one of the iEEG BIDS formats, see section 1.3.8), or events are tagged in a dedicated trigger channel (i.e., as binary pulses, numbers or strings). For some continuous streams a different sampling rate may be used (e.g., for audio as analog inputs, at least 16 kHz is needed, whereas the typical iEEG is sampled at about 1 kHz). Some systems allow implicit synchronization for recording iEEG along with other signals at different rates, whereas in other cases explicit synchronization between multiple systems is needed. The acquisition of continuous signals in parallel with the iEEG recording presents many advantages and moves forward to naturalistic neuroscience (see section 6.5). As mentioned in section 1.3.1, the continuous recording of supplementary channels can be used as ‘ground-truth’ in the timeline of iEEG data. Moreover, recording continuous signals allows for higher sensitivity in some research fields, like in the investigation of natural speech production where the specific spectrotemporal characteristics of the recorded audio stream can be analyzed and compared with neural activity (Ozker et al., 2022), which is not possible when only speech onset is marked as a discrete trigger-event.
Just like clinicians annotate the signals using the clinical monitoring system (e.g., to indicate seizure onset and/or associated behavior), researchers can document iEEG data to drive analyses (e.g., manual screening for real-life conversation or other activities, see for example (Glanz et al., 2018; Mercier et al., 2017), Figure 2). To that aim, the Hierarchical Event Descriptor tag (or HED schema2) offers a standard to annotate brain imaging data. It has been adopted as part of BIDS (see (Robbins et al., 2021) and section 1.3.8). Supplementary channels and annotations add value to iEEG data: they guide preprocessing (e.g., to identify artifacts; see section 3) and to integrate behavioral correlates in signal analyses (see sections 4.1.3 and 4.2.4 for common practices to relate behavior and iEEG signal through data visualization).
1.3.5. Monitoring artifacts
The term “artifact” refers to signal features that are not relevant or undesired for the research purpose, in iEEG these would be signals that do not originate from the brain or pathological neural activity. The signals not originating from the brain can further be divided into two: physiological and non-physiological artifacts. Pathological epileptiform activity during seizures or between seizures is discussed in section 3.2.1. If artifacts are identified during monitoring, the researcher can annotate them and possibly try to avoid or reduce them.
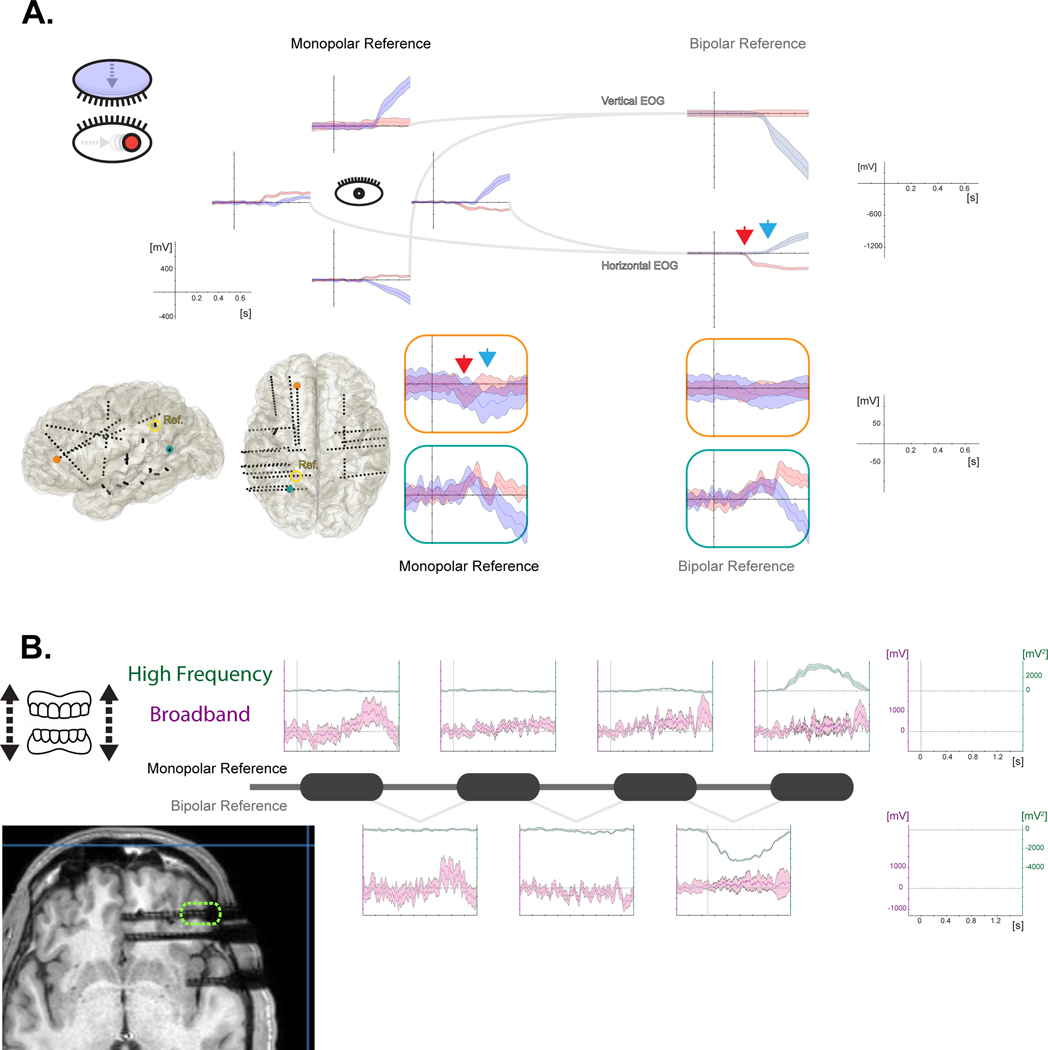
Physiological artifacts are generated by the patient, from sources other than the brain. The most common are eye blinks, eye movements (i.e., saccades or micro-saccades measured with EOG and/or eye-tracking), cardiac activity (EKG/ECG), and muscle activity (EMG) (Ball et al., 2009; Jerbi et al., 2009a; Kern et al., 2013, 2021; Kovach, 2011; Melloni et al., 2009; Mosher et al., 2020; Otsubo et al., 2008). Physiological artifacts are a major source of contamination in non-invasive neurophysiological techniques (e.g., MEG, EEG). While their presence in the iEEG signal is less evident to the naked eye, they can mix with the signal of interest spatially, temporally, and/or spectrally (see section 3.2.2). In addition, some unwanted brain responses can occur from experimental events (e.g., the patient hears the sound of their own button press when giving a behavioral response).
Non-physiological artifacts arise from a potentially large number of external elements including electrical stimulation artifacts, other electrical equipment (e.g., patient monitoring equipment, intravenous pumps, etc.), electrical motors (e.g., to incline the patienťs bed), electronic devices (e.g., screen, speakers, cell phones), and sway in improperly anchored electrode cables. Other sources of interference can produce low-frequency patterns, punctate high-frequency artifacts and continuous power line interference (50/60 Hz and harmonics, see section 3.2).
1.3.6. Combining intracranial and non-invasive electrophysio- logical recordings
iEEG has a high signal-to-noise ratio (SNR) and a high spatial specificity: it directly records brain signals and is mainly focused on activity originating from summated post-synaptic potentials from populations of neurons in the immediate vicinity of the electrode. Yet, iEEG is spatially sparse because the clinical exploration targets only a specific subset of brain regions.
MEG and EEG also record summated neurophysiological activity, but with sensors covering the whole scalp, therefore offering a more complete spatial coverage of the whole brain. However, these measurements are made outside of the brain and skull and have significantly reduced SNR relative to iEEG. Additionally, localizing and reconstructing the time course of activity in the brain requires source modeling and solving the inverse problem as measurements are not taken directly from the source locations.
Simultaneous invasive and non-invasive electrophysiological recordings can combine the qualities of each technique to describe as accurately (high SNR, high spatial specificity) and as comprehensively (whole brain) possible brain processes. For instance, a limited set of fronto-central EEG electrodes can help to identify sleep stages during iEEG. Since the development of high density EEG and MEG (M/EEG), there is increasing interest to investigate the relationship between surface signals and the spatiotemporal configuration of underlying brain sources. In this context, iEEG can only provide some “ground truth” for characterizing sources in brain areas where electrodes are implanted. Cosandier-Rimélé et al. (Cosandier-Rimélé et al., 2008) modeled the impact of several factors (e.g., distance to sources, skull conductivity, source area, source synchrony, background activity) on the observability of simultaneous rapid discharges in sEEG and scalp EEG signals. Empirical studies have tested ability for non-invasive techniques to detect activity originating in deep structures (Fahimi Hnazaee et al., 2020; Koessler et al., 2015; Pizzo et al., 2019; Seeber et al., 2019), to determine the lateralization of seizure onset (Sammaritano et al., 1987) and provided insights into the quality of source reconstruction (Mikulan et al., 2021, 2020). For cognitive research, the joint analysis of depth and surface signal evoked by individual stimuli can contribute to understanding the neural activity linked to cognitive processes (Dalal et al., 2009; Dubarry et al., 2014). Simultaneous recordings can also provide additional information relating to tissue that is actively generating the patient’s seizures (Gavaret et al., 2016; Kakisaka et al., 2012; Santiuste et al., 2008). For instance, MEG data time-locked to iEEG interictal epileptiform discharges revealed an additional generator in a region not covered by the iEEG implants (Gavaret et al., 2016).
While these studies provide unique perspectives on brain activity, simultaneous recordings are difficult to implement. There are infection control and pain-related issues due to the craniotomy and burr hole sites. It can be difficult to place non-sterile scalp EEG electrodes on patients with extensive intracerebral electrode implantation as they might contact iEEG probes (i.e., cables, screws). Scalp EEG electrode type can affect signal quality and practicality (e.g., transcutaneous electrodes can be more stable, while adhesive electrodes are easier to place). Scalp EEG recording should begin shortly after these electrodes are placed by the clinical team as the signals will be optimal at this time. Furthermore, when interpreting scalp EEG signals, changes in electric field propagation caused by the implants themselves, and discontinuities in the skull due to craniotomies and associated burr holes must be considered (Dalal et al., 2009; Kirchberger et al., 1998; Oostenveld and Oostendorp, 2002; Voytek et al., 2010).
Recording MEG simultaneously with iEEG presents physical challenges, e.g., transporting the implanted patient to the MEG laboratory, introducing iEEG hardware inside the shielded MEG environment, and getting the patient’s head successfully into the rigid MEG helmet. The amount of time the patient spends outside the EMU must be kept to a minimum, so the recording session must proceed efficiently (see section 1.2.2). Unfortunately, the quality of MEG signals can be severely compromised by the iEEG hardware. Because of these major technical challenges, it is possible, but in practice rare to acquire data in the three modalities (iEEG, EEG and MEG) simultaneously (Dubarry et al., 2014).
In summary, recording non-invasive and intracranial neural signals simultaneously can provide a more complete perspective for integrating and evaluating brain activity recorded by different methods. This conveys potentially important benefits for improving methods, and for interpreting clinical and cognitive questions that are addressed by these same approaches when used in isolation. However, concurrent neurophysiological recordings are technically and logistically challenging, from patient consent (explaining procedures), to data acquisition, analysis and interpretation. Performing such recordings should only be attempted when the research hypothesis clearly motivates this and there is strong support from the clinical staff.
1.3.7. Imaging data
As part of the clinical procedure, imaging data is routinely acquired and is available for research purposes if the patient gives their consent (Figure 2). Notably, pre-implantation structural MRI and post-implantation CT or MRI scans are critical for localizing and categorizing electrode contacts (e.g gray vs white matter, specific sulci/gyri) (see section 2), as well as for reporting and sharing data (see section 1.3.8). Sometimes additional complementary neuroimaging investigations may exist (e.g. fMRI, MRI scans with different imaging sequences, PET or SPECT scans), and these might also be informative for the research data analysis. Importantly, some patients have had previous brain resections or have brain lesions that may, or may not, be epileptogenic. This has important implications for matching individual to template brains (see section 2.3.2), and interpreting and modeling neurophysiological brain activity. Some centers also use fMRI studies with localizer tasks prior to implantation to perform a non-invasive brain mapping that can complement iEEG investigations (see sections 1.1.4).
Functional MRI and iEEG signals have been combined in many studies (for review see (Ojemann et al., 2013)). Most of these studies performed separate iEEG and pre- or postoperative fMRI recordings using the same task in the same individuals, and a few carried out simultaneous recordings. Several approaches have been used to compare separate recordings in the same individuals, including comparisons of the spatial distribution and comparisons of activity levels within electrodes across task conditions or time. Temporal approaches typically extract different iEEG measurements that are then convolved with a hemodynamic response function to predict the BOLD signal (Haufe et al., 2018; Mukamel et al., 2005). Initial spatial comparisons showed overlap between the sEEG high frequency and fMRI signal changes (Lachaux et al., 2007a; Nir et al., 2007). More complex relationships between these signals were revealed later (Hermes et al., 2012), where ECoG high frequency power increases and low frequency power decreases explain complementary variance in BOLD increases (see also (Haufe et al., 2018; Hermes et al., 2017)). One methodological consideration when integrating separately recorded fMRI and iEEG data is how to match voxels to iEEG electrodes (see section 2.1.4). A typical approach is to calculate a (weighted) average of the BOLD activity in a small region of gray matter cortex of about 3–8 mm around the electrode (excluding non-gray matter voxels), as correlations drop across larger regions (Hermes et al., 2012; Piantoni et al., 2021). When integrating iEEG with fMRI data, there may be serious fMRI signal dropout in anterior ventral temporal and orbitofrontal areas. These are often covered with iEEG electrodes (Ojemann et al., 1997; Shum et al., 2013), so there may be an absence of complementary information from both imaging modalities.
Simultaneous fMRI-iEEG studies have reported spatial correspondence in the trial-by-trial correlations between the BOLD signal increases and ECoG high frequency power increases and low frequency power decreases (Murta et al., 2017), and have also used simultaneous recordings to explore the effects of electrical stimulation on the BOLD signal (Oya et al., 2017). While scientifically interesting, simultaneous fMRI-iEEG recording requires specific authorization as it presents additional challenges with respect to fMRI compatibility with iEEG electrodes and recording devices (e.g., cables, amplifiers) and general patient comfort and safety. Surgical dressings themselves can be bulky and may impose space constraints for some MRI head coils - not to mention painful pressure points for the patient at the surgical site.
1.3.8. Reporting and sharing
An iEEG dataset consists of complex and often heterogeneous data, including information from the clinical team, pre- and post-operative neuroimaging data, results of neuropsychological assessments, electrical cortical mapping, assessments of patient participation in the experimental tasks, the actual neurophysiological and behavioral data recorded during the experimental tasks and associated annotations (Figure 2).
The Brain Imaging Data Structure (BIDS) provides a template for organizing neuroimaging data of various types in a standardized directory, filename and file structure that now covers multiple imaging modalities (for MRI, iEEG, MEG and EEG, see respectively (Gorgolewski et al., 2016; Holdgraf et al., 2019; Niso et al., 2018; Pernet et al., 2019)), with work continuing to incorporate additional modalities such as single unit activity. An iEEG dataset includes anatomical imaging data, location of intracranial electrodes, iEEG data in relation to the events in the experimental task, and behavioral measures (see Figure 2). As iEEG is acquired in clinical rather than lab settings, this introduces sources of variability across acquisition sessions and medical centers, so special attention is needed to document metadata. Minimally, those metadata shall document the experimental environment (e.g., hardware, software, equipment wiring diagram), information concerning additional equipment, participant behavioral status, and the experimental protocol and/or deviations from the protocol due to unforeseeable reasons such as medical interventions during testing. Specific pre-processing is required for the raw dataset both from the clinical and research perspective (e.g., determining electrode locations from imaging data, identifying channels with epileptiform activity or non-functioning contacts), and these must be documented.
We recommend a research data management plan that not only considers the iEEG data but also supplementary data. The latter includes important information that changes at a slower rate than the main data of interest such as medication and clinical state of the patient, research questionnaires, and version control where time-stamped (pre)processing steps are documented in a clear readable workflow. This is important for the easy identification and tracking of data and its derivatives. This ensures that future researchers (from the same or another lab) will see what has already happened to the data, without having to repeat any steps. This will improve efficiency, quality and reproducibility of science. For an example of iEEG datasets in the BIDS format, see 3.
While a standardized format offers the most universal application of data, some researchers share valuable data in a lab-specific manner. In that case, the interoperable (I) and reusable (R) criteria of FAIR4 should be considered (Wilkinson et al., 2016). Specifically, i) the format of physiological and behavioral data should be sufficiently simple, and variables corresponding to each measurement should be explicitly described in companion documents, ii) co-registered anatomy in standardized coordinates should be integrated into a provided workflow, and allow plots to be easily made from analyses of time series data, and iii) a clearly written workflow and all code (fully and extensively commented) to reproduce every element of published work should be provided. For examples of this approach, see recent libraries of publicly-available, open-source, ECoG data and code 5, (Miller, 2019), or multimodal iEEG-fMRI dataset (Berezutskaya et al., 2022).
If the patient data is to be shared publicly according to Open Science principles, additional constraints will apply. For example, to support a paper with primary research findings, or alternatively the form of a data descriptor paper, it is crucial to consider the legal (e.g., GDPR or HIPAA) and ethical considerations regarding patient privacy. Anatomical imaging data that are shared with the published dataset must be defaced or skull-stripped to remove facial features, but that does not guarantee that it is impossible for the participant to be re-identified (Abramian and Eklund, 2019; Schwarz et al., 2021). The same considerations must be applied to appropriately handle any supplementary data that is identifiable, such as audio and video recordings, but also clinical and demographic data that is represented in simple tabular format. Furthermore, the specific configuration of implanted electrodes and the clinical information itself (e.g. brain lesions) can contribute to the risk of potential reidentification of the individual (Rocher et al., 2019). However, the scientific utility of the shared data can be compromised by de-identification approaches which also remove the critical features that are required for data interpretation and reuse (on behalf of the MAGNIMS Study Group and Alzheimer’s Disease Neuroimaging Initiative et al., 2020). To balance the value of the shared data with legal and ethical responsibilities to the patient, we recommend sharing the data under a data use agreement that permits bona fide research, but ensures that the patient’s interests are not harmed, more specifically that the identity of the patient is protected (Bannier et al., 2021; Eke et al., 2021; Jwa and Poldrack, 2021). Since sharing anonymized iEEG data is challenging, new techniques may have to be developed and the awareness for existing deidentification strategies should be increased (e.g., (Meurers et al., 2021; Prasser et al., 2014; Vinding and Oostenveld, 2022), and see section 6.2).
1.3.9. Challenges, recommendations and reporting advice
Sharing iEEG data, or any other data from human research participants, requires written informed consent that explicitly specifies which data can be shared (e.g., iEEG signal, neuroimaging data, video), to which audience (e.g., other institute(s), public space) and what measures are undertaken to warrant data anonymization or de-identification to protect participant privacy ((Bannier et al., 2021), see also 6). In addition, one needs to consider the safety of the tools used to share the data, as well as the necessity of setting up a data sharing agreement.
Visualization of the raw signals and their power spectra permit the evaluation of data quality (if permitted by the acquisition software, the sanity check of power spectra should be performed during the actual recording, otherwise it must be performed offline, see section 3.2). When the signal from all electrodes is noisy (e.g., presence of 50/60 Hz and its harmonics) it is possible that the noise originates from the REF and/or the GND electrodes or cable; in that case these must be changed independently to identify the source of noise (see sections 1.3.2 and 3.3). If noise only contaminates a single ECoG grid/strip or sEEG shaft), its cabling should be verified (i.e., cables, adaptors and plugs).
Whenever possible, the reference electrode and ground electrode should be of the same material as the other electrodes to reduce the effect of half-cell electrode potentials (section 1.3.2).
As researchers operating in the clinical environment have inherently less control over the software and hardware, it is important to preemptively itemize the tools available for research in the EMU and to accommodate any deficiencies with redundancy in experimental design and recording scheme (section 1.3.1).
We recommend recording physiological artifact samples to ensure that some prototypical patterns are available when off-line reviewing and cleaning the data (see sections 1.3.5 and 3.2). These artifact templates are helpful to compare with those overlapping the signal during the experiment itself, especially when artifacts are suspected to be present in the data in line with the task (e.g., jaw, lip and tongue movements for language production).
As well as recording artifact templates, during data collection it is useful to keep track of any unexpected environmental or patient-related artifacts during the task (e.g., aura or prodrome prior to a seizure, seizure, sneezing, yawning, coughing, environmental noise, see (Mosher and Funke, 2020) in the context of MEG recordings). We recommend using annotation plug-ins directly in the acquisition software, or when that is not possible, to write notes in an experimental notebook or lab diary with times of occurrence (see section 1.3.8). Later, these annotations will allow the detection of artifacts in the recording and may provide insight into detecting ones that were not annotated.
Considering experimental designs, different conditions might result in different amounts of artifacts, for example 50/60 Hz line noise when touching a button-box/computer keyboard in one condition and not the other, or when one condition results in more participant movements. An experimental difference in artifacts should be considered as a potential confound for subsequent analysis (see section 3.2).
2 -. Electrode localization and anatomy
2.0. Introduction
Intracranial electrodes are in close contact with the brain tissues from which electrical activity is generated (see Figure 1). For ECoG implants, neural activity passively propagates through volume conduction across several layers of the cerebral cortex, pia mater, cerebrospinal fluid and arachnoid mater before reaching the electrodes. For sEEG implants, neural activity passively propagates through gray matter, white matter and cerebrospinal fluid (see section 1.1.1 and 1.3.3). This confers to iEEG both a high SNR and a high spatial specificity. The iEEG spatial specificity is in the range of millimeters, which cannot be matched with surface EEG or MEG recordings (Buzsáki et al., 2012). To make the most of the high iEEG spatial specificity, one needs to localize the electrodes on/in the brain and to identify the anatomical structures the electrodes are implanted in or in contact with. Electrode localization therefore more strongly depends on imaging data and anatomical processing than is common for M/EEG studies. We discuss how to localize the implanted electrodes on/in the individual patient’s brain in section 2.1 below.
The sparse coverage of iEEG results in data that is not sampled from the whole brain. The scattered electrode placement is participant dependent, as dictated by individual clinical needs. While typical M/EEG analysis relies on averaging and comparing recordings across multiple participants with standardized sensor positions, iEEG recordings are idiosyncratic and difficult to compare across patients. For instance, sEEG electrodes can sit in sulci and it can be difficult to gauge which side of sulcus is the source of the activity, and differences in cortical folding patterns can make direct comparisons between participants even more complex. Two main approaches are used to collate and interpret iEEG data at the group level: using a normalized space or delineated parcellations (see section 5). In section 2.2, we discuss how co-registering and normalizing the individual patienťs MRI (with corresponding electrode positions) permits work in a standard brain template space. In section 2.3, we discuss the procedure of matching anatomical or functional atlas labels from a template brain to an individual's brain. This procedure permits electrodes to be grouped with respect to the delineated feature in the individual’s anatomy.
There is no clear consensus in the iEEG research community about performing analyses either in normalized space or in native space. Yet, based on the literature and specialized toolboxes (see 7 for an updated list), a common workflow emerges (Figure 3). Here, we recommend a sequence of steps to obtain all information that may potentially be required in subsequent analyses of iEEG data. We assert that iEEG electrode localization is critical to fully utilize the high spatial specificity of iEEG, but also to acknowledge its limits and imperfections.
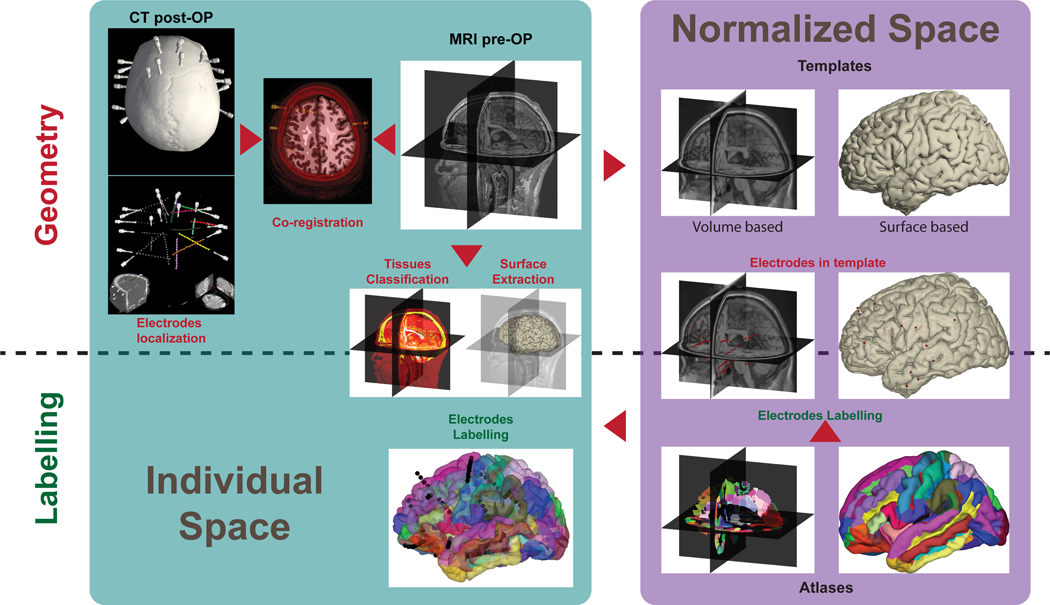
Figure 3.
Canonical framework of iEEG electrode localization using illustrations utilized as a sanity-check for the outputs of each step of the processing pipeline.
2.1. Individual space
The clinical procedure of electrode implantation involves a patient-specific surgery (see section 1.1). For sEEG, the positions (3D coordinates) of the sEEG shafts trajectories are pre-specified in the patient’s structural MRI and/or CT to guide stereotactic intervention by the neurosurgeon when targeting the region of interest while avoiding blood vessels. All information is uploaded to the neuro-navigation software to guide the planned stereotactic trajectory (Bakr et al., 2021; Brandmeir et al., 2018). However, during surgery the neurosurgeon may proceed differently than planned depending on the quality of perioperative recordings, the medical condition of the patient, existing vasculature or technical reasons. Surgical planning is usually not precise enough to properly estimate the final anatomical locations of the sEEG electrodes on the sole basis of the pre-surgical documentation, hence the complementary post-op imaging. With ECoG, the use of a neuro-navigator could provide decent approximates with a few millimeters error (Gupta et al., 2014; O’Shea et al., 2006).
To obtain accurate 3D coordinates and corresponding anatomical information (e.g., tissue types or brain regions), when available we recommend using a pre-implantation high-resolution whole brain T1-weighted MR and post-implantation imaging exams (if only one is available CT is generally preferable to post-implant structural MRI, see section 2.1.1 and 2.1.4). A three step procedure is then followed, which we describe in this section: (i) localization of each electrode contact from post-implantation anatomical images, (ii) coregistration of the post- and pre-operative images and (iii) segmentation of the pre-implantation MRI to obtain tissue classification. The order of execution of these steps does not critically affect the outcome, although reslicing of imaging data as an intermediate step may affect electrode localization (see section 2.1.2).
As the diagnostic neuroimaging protocols of epilepsy surgery programs may not include an MRI and/or CT scan, we indicate in the following some alternative, but non-preferred, options for localizing the electrodes.
2.1.1. Electrode localization
The 3D coordinates of each electrode centroid are determined on the post-implantation imaging data by pinpointing the center of mass of the electrode artifact visible in the imaging data (Blenkmann, 2017; Branco et al., 2018b; Deman et al., 2018; Groppe et al., 2017; Hamilton et al., 2017; Hermes et al., 2010; LaPlante et al., 2017; Narizzano et al., 2017; Sebastiano et al., 2006; Stolk, 2018; Tao et al., 2009). Post-implant CT-scans offer high-resolution images with excellent contrast between soft tissue and the electrodes and skull. Therefore, to achieve a high accuracy in the electrode localization, the CT-scan provides clear unambiguous data with a straightforward procedure (see Figure 3).
When post-implantation CTs are not available, alternative approaches can be used to localize the electrodes: post-implant MRI (Kovalev et al., 2005; Morris et al., 2004; Yang et al., 2012), intra-operative photographs taken during the surgery (Mahvash et al., 2007; Tao et al., 2009; Wellmer et al., 2002) or two orthogonal skull X-rays (Miller et al., 2007b; Winkler et al., 2000). However, these approaches lead to lower spatial precision: post-implant MRI shows large blurred artifacts around the electrode contacts due to their metallic structure, while photograph or skull X-rays imply some approximation in the reconstruction of the third dimension, and photography only applies to ECoG electrodes that are visible through the craniotomy and require to visually/manually relate them to underlying anatomical landmarks (e.g., gyri/sulci, blood vessels etc.). Usually, more than one of these approaches are used - taking advantage of their respective strengths and correcting for potential tissue deformations (see also section 2.1.4). For instance, Dalal and colleagues (Dalal et al., 2008) proposed a multimodal approach for ECoG, where: (i) perioperative photographs are used to localize the electrodes visible during the craniotomy, (ii) the coordinates are transferred to the pre-operative MRI rendering, (iii) a 3D-2D projective transform on the X-ray radiographs is used to back project to the cortical surface the location of the occluded electrodes. Two recent ECoG studies further propose defining anchor points on perioperative photographs and then using them to constrain semi-automated electrode localisation from a post-operative CT (Trotta et al., 2018) or post-operative MRI (Pieters et al., 2013).
2.1.2. Coregistration
To link electrode coordinates to neuroanatomy, the post-implant image (CT or MRI) is coregistered with a pre-implant MRI that serves as the reference. This registration is performed using a rigid transformation (i.e., rotation-translation) estimated mostly from the skull shape (Friston, 2007; Jenkinson et al., 2012). If electrodes were localized prior to coregistration with the pre-implant MRI, the same transformation must be applied to the 3D electrode positions as to the post-implant image.
Following coregistration, the registered image (CT or MRI) can optionally be resliced/resampled onto the reference image so that the two volumes match voxel-by-voxel. This is useful for quality control: overlaying the two images in the same view allows a visual check of whether coregistration was successful (see Figure 3). However, interpolation on a different voxel grid spacing does degrade image quality and spatial resolution of the post-implant image. CT scan resolution is typically higher than that of the pre-implant MR. Hence, we recommend performing localization of iEEG contacts on the post-implant (CT or MRI) images prior to reslicing. Once electrode coordinates are projected in the pre-implant MRI, electrodes can be visualized on/in the patienťs brain by scrolling through MRI slices.
2.1.3. Segmentation
The pre-implant whole head MRI can be processed to quantify and reconstruct anatomical features of the brain (see Figure 3). Voxel-based morphometry methods provide tissue classification (skin, skull, blood vessels, CSF, gray matter, white matter, lesions). As an imaging voxel is not a biologically meaningful unit, other methods have been developed such as surface-based morphometry, which makes use of segmentation boundaries into parametric surface meshes representing the separation between tissues in 3D (skull shape, pial envelope, gray-white interface) and providing related measures (e.g., cortical thickness, amount of CSF). T1-weighted MRI scans offer the optimal contrast between the tissues to be segmented, and are therefore recommended as the starting point for most segmentation pipelines (e.g., FreeSurfer, CAT, BrainSuite, BrainVISA, SimNIBS; (Fischl, 2012; Gaser et al., n.d.; Rivière et al., 2009; Shattuck and Leahy, 2002; Thielscher et al., 2015)). Some tools allow including additional T2 or FLAIR images to improve quality of the pial surface estimation (see for instance FreeSurfer, SimNIBS or SPM; (Lindig et al., 2018)). Specialized sequences for higher fidelity segmentation and tissue type identification such as the Fast Gray Matter Acquisition T1 Inversion Recovery (FGATIR see (Sudhyadhom et al., 2009)) should be used where possible as adjuncts to assist in segmentation.
Some surgical procedures require additional MR angiography scans to guide the implantation and avoid vessels: a contrast agent (e.g., Gadolinium, see recommendation from (Bernasconi et al., 2019)) is injected intravenously and results in a hypersignal in T1-weighted images around blood vessels and lesions, and possibly lower contrast between tissues in the rest of the image (Hannoun et al., 2018; Maekawa et al., 2019; Vakharia et al., 2018). These effects may affect the quality of segmentation pipelines (e.g., making gray and white matter difficult to separate, or meningeal vessels classified as gray matter), therefore the use of such images is not recommended as input for the segmentation pipeline. If the use of a contrast agent is requested, it is recommended to perform a T1-weighted scan before and after the infusion (see recommendations from the International League Against Epilepsy: (Bernasconi et al., 2019)), and only the former should be used for brain segmentation and then for iEEG electrode labeling.
The segmentation can be used to provide multiple labels or tissue metrics for each electrode (e.g., Tissue Proximal Density, see (Mercier et al., 2017) and see Figure 4), which can help identify which channels to include/exclude in analysis and visualization (e.g. some contacts in SEEG electrodes might not be in the brain, see section 2.1.4). The geometrical surface meshes can be used for projecting ECoG electrodes on the folded cortical surface or the inside of the skull, for visualization purposes, and/or for brain shift correction (see sections 2.1.4 and 3.1). The surface meshes and volumetric parcellations can also be used to compute volume conduction models of the head for modeling intracranial electrical currents (Medani et al., 2020).
Figure 4.
Examples of differences in signals related to the surrounding tissue.
A: ECoG signal as a function of electrode location.
The left panel shows the location of the ECoG grid on the individual's brain. Three electrodes are highlighted (but not at scale): over a blood vessel and sulcus (in yellow), on the bank of a gyrus and close to a blood vessel and sulcus (in orange), and over a gyrus (in red). The insert shows the angiogram (vessels) projected on the cortical surface. The right panel shows 30 sec of the high-frequency band signal (frequency analysis using Morlet wavelets, high-frequency band power extracted in 1-Hz bins and averaged over 45–120 Hz with 1-second smoothing) for the three electrodes (same amplitude scale).
B: sEEG signal as a function of electrode location.
The left panel shows the location of three sEEG electrodes (not at scale): one in the skull (in purple), one at the interface of the soft tissues (in pink), and one in the gray matter (in red). The right panel shows 2 sec of the raw signal for the three electrodes (same amplitude scale).
2.1.4. Challenges, recommendations and reporting advice
We recommend visual inspection of the output of each processing step as a sanity check (e.g., Figure 3). This verification can save a lot of time rather than trying to figure out a posteriori at which step there was a failure in the pipeline (e.g., when left and right axes are flipped).
Fiducial markers are reference points placed on the skin before performing brain imaging. Generally they are placed at the nasion, inion and/or pre-auricular points. Their use is recommended as these points help to reference and align different images. An additional fiducial, placed on one side of the head (e.g., the forehead), can prevent right/left confusion.
The 3D image acquisition can be performed with non-isotropic voxels and therefore to less detailed images along the axis with the coarser resolution; this results in more blurred localization accuracy of the electrode in that direction. When possible, we recommend using isotropic scans to avoid discrepancy between the three dimensions, so electrode localization is not impacted by the orientation of the electrode with respect to that of the scan.
Additional image data processing such as filtering or smoothing may introduce some approximation in the localization of iEEG electrodes. Only the center of mass of the iEEG electrode is localized, then the electrode is matched to a given voxel, which in most cases is smaller than the electrode itself (the size of an electrode contact is about a few millimeters, typically larger than the voxel size generally about 1 mm). As a consequence, electrode location must be considered as an estimation (Ken et al., 2007; Sebastiano et al., 2006) and see for a method review (Pieters et al., 2013)).
We recommend localizing the center of mass of each electrode contact on the original volume (e.g., on post-implantation CT-scan) prior to any further imaging processing, notably coregistration, which can blur the contact shape and decrease spatial specificity.
Electrode localization can be performed manually or using a semi-automated or fully automated algorithm (for sEEG see (Arnulfo et al., 2015; Granados et al., 2018), for ECoG see (Branco et al., 2018b; Centracchio et al., 2021; Hunter et al., 2005; O’Shea et al., 2006; Sebastiano et al., 2006; Yang et al., 2012).
For sEEG, some semi-automatic procedures identify two points along the electrode shaft (e.g., the proximal tip and the distal entry point of the shaft in the skull) and interpolate the coordinates of other contacts based on the description of the electrode’s geometry (i.e., inter-electrode spacing, electrode size, etc.). This approach assumes that the contacts are on a line, however the shaft can bend during implantation due to the resistance of the tissues (e.g., Figure 6, 3D view panel). Consequently, electrodes along a shaft may deviate from a straight trajectory (Cardinale et al., 2012). To improve on this issue, some tools fit a spline to three or more points along the electrode shaft and distribute the electrodes along the spline (e.g., 8). More automated procedures based on CT-scan segmentation were specifically developed to take into consideration the bending of electrode shafts (Arnulfo et al., 2015; Granados et al., 2018).
Figure 6.
Oblique slice aligned with sEEG shaft.
Graphical interface allowing the display of MRI slices aligned with a selected sEEG shaft highlighted in blue in the list. In the oblique slice sEEG contacts are represented as green dots. In the 3D view the plane corresponding with the shaft axis is depicted in green with a transparent brain, sEEG contacts are here represented as black spheres.
ECoG implants push the brain inward due to the thickness of the implant and because of a general reaction to the surgical intervention (e.g., CSF leakage due to dura opening, soft tissue swelling). These non-uniform and unpredictable brain shifts range up to 24 mm for cortical displacement and to 3 mm for deeper structures, ventricles and the midline (Dalal et al., 2008; Etame et al., 2011; Hartkens et al., 2003; Hastreiter et al., 2004; Hill et al., 1998; Roberts et al., 1998; Studholme et al., 2001; Tao et al., 2009). This deformation requires some correction of the 3D reconstruction by projecting ECoG electrodes at the surface of the brain using segmentation outputs (see below) or on the brain convex hull or the inner skull surface (see 9).
We recommend correcting for brain shifts. Several effective automatic methods have been developed. In Hermes et al. (Hermes et al., 2010), ECoG electrodes are orthogonally projected from the local vector norm of the grid to the point on the cortical surface. Yang et al. (Yang et al., 2012) use the inverse of the gnomonic projection to “fold” the grid onto the smoothed pial surface. Finally, in Dykstra et al. (Dykstra et al., 2012), the procedure consists of two steps: first the initial coordinate estimates are projected to the dura smoothed surface via an energy-minimization algorithm (constrained by a minimal local displacement relative to each electrode original position, and by a minimal global deformation of electrodes configuration), then the new coordinates are projected to the closest vertex of the pial surface (e.i., euclidean distance). All these methods require the use of the MRI segmentation output (i.e., cortical mesh) and they must be applied in the individual space. The pre versus post brain shift corrected distance can be used to visually check the quality of the method (see Figure 2 in (Groppe et al., 2017)).
The distal ends of sEEG shafts are screwed in place on the skull and can remain in a fixed position, but for ECoG electrodes some shift can occur due to blood and cerebrospinal fluid accumulation-pressure and soft tissue swelling, especially when they are loosely sutured in place. Although the position of the electrodes is only determined once from the imaging data, such a displacement over time has consequences for the actual location from where the signal is recorded (LaViolette et al., 2011; Wellmer et al., 2002).
With sEEG, while the interest is typically in gray matter, the shafts go through several tissues and consequently some sEEG electrodes can be located at the interface with, or in the skull, soft tissues (e.g., skin, dura, muscles), cerebro-spinal fluid, gray matter or white matter (see Figure 4).
We recommend using the brain mask computed from the volume segmentation to exclude the electrodes that are outside the brain from the analysis (see section 3.1).
Anatomical labels (i.e., tissue maps) are generated by the segmentation pipelines and can be binary or probabilistic (see section 2.1.3). In the case of binary masks, each voxel of the image is associated with a single label. The class of the voxel with the centroid of an iEEG electrode can be associated with the corresponding tissue label. For tissue probability maps (Ashburner and Friston, 2005; Lorio et al., 2016), the intensity of the voxel indicates a cumulative probability of belonging to each of the tissues (e.g., 0% skull, 10% soft tissues, 80% gray, 10% white matter) and reflects an uncertainty on the classification of each voxel.
For sEEG, because of the uncertainty on the 3D locations described above, we recommend taking a region around the contact centroid (e.g., 3x3x3mm, with respect to the size of the electrode that is no more than about 2mm wide) and counting the number of voxels of each label in this neighborhood volume (probabilistic approach in space, for an example of implementation see the Proximal Tissue Density index introduced in (Mercier et al., 2017)). Tissue probability maps can be combined with the uncertainty of a contact location by averaging the label probability across voxels around the centroid.
For ECoG, grids and strips generally cover multiple gyri and some electrodes may face blood vessels (see Figure 4 and (Bleichner et al., 2011; Wang et al., 2016)). In this case, the tissue map provides information about the electrode vicinity and allows to compute, for each electrode, the distance to the leptomeningeal surface.
Along the localization pipeline, imaging data can be registered to different coordinate spaces (relative to the scanner origin, relative to external head or internal brain anatomical landmarks, etc.). It is important to know which referential space is used at each processing step, because 3D coordinates are valid only in one given spatial reference.
It is crucial to document clearly in which coordinate system the spatial data and the position of electrodes are reported. The iEEG-BIDS specification (Holdgraf et al., 2019) provides guidelines to document the iEEG coordinates in a reproducible way (see 10).
2.2. Normalized space
2.2.1. Background
The first goal of registering the patient’s brain to a template is to project 3D electrode coordinates from individual space to a normalized space. Because of the anatomical variations between individuals, an accurate registration to a common brain template space requires a non-linear method. Two families of approaches exist to perform this registration. Volume-based pipelines (e.g., SPM12, FSL, Lead-DBS; (Friston, 2007; Horn and Kühn, 2015; Jenkinson et al., 2012)) compute volume deformation fields and volume parcellations from the individual MRI to the template space based on tissue probability maps from the segmentation. Surface-based pipelines (e.g., FreeSurfer, CAT, BrainVISA, BrainSuite; (Fischl, 2012; Gaser et al., n.d.; Rivière et al., 2009; Shattuck and Leahy, 2002)) reconstruct the cortical surface of the individual brain, which is then parametrized and registered to the brain template surfaces based on topological properties (e.g., curvature, sulcal depth, sulci tracking, etc.). This gives access to surface-based parcellations of the individual cortex, and is further used to compute volume deformation fields and volume parcellations.
A critical distinction must be made between these pipelines and their outputs. A volume-based pipeline can produce a normalized individual cortical surface mesh, even though the volume-based registration procedure does not rely on this surface. Conversely, a surface-based pipeline can, as an intermediate step, produce the individual brain as a volume in the normalized space. Some surface-based pipelines use the output of a volume-based pipeline (e.g., 11). The estimation of the "mapping quality" depends on the target(s): while volume-based mapping operates well for subcortical regions, surface-based mapping is more optimal for the cortex (Desai et al., 2005; Klein et al., 2010; Langers, 2014). We recommend the surface-based pipeline to label ECoG electrodes because it gives better individual-space cortical alignment (and parcellation) than the volume-based pipeline. We recommend volume-based pipeline output to label sEEG electrodes, as it operates on the full brain volume including subcortical regions, (see section 5.3.2.1).
Practically, volume and surface outputs differ in the way the spatial dimension is organized: respectively using a 3D matrix of voxels or using a sheet/mesh made of numerous vertices. Once 3D electrode coordinates are obtained for a given normalized space, it is possible to collate electrodes across patients on the basis of their normalized coordinates, to relate them to findings from other studies, and to report results relative to the normalized space (section 5.3.1). More broadly, once registered in a normalized space any individual data (e.g., MRIs, PET as with the iELVis toolbox (Groppe et al., 2017)) can be projected on the corresponding brain template to perform group-level analyses or comparisons with prior studies or databases.
The second goal of registering the patient’s brain to a template is to make use of various anatomical or functional information from atlas(es) matched to this template. While defined in space relative to a template, an atlas should not be equated with a brain template, rather an atlas is the projection of information on a template. Similar to an atlas of the world being defined by features over space (e.g., elevation, climate, social, geopolitical) projected on a given map, a brain atlas consists of labeled brain parcels with anatomical or functional information. Following this analogy, information from a brain atlas is projected to a spatial template brain. When working with data from multiple patients, a strategy can be to select and group electrodes from the different patients that fall within the same atlas parcel (see section 5.3.2.1).
2.2.2. Challenges, recommendations and reporting advice
Interpreting data in standardized space should be performed with care. Even within homologous brain areas across individuals, the representation for particular brain functions can be highly variable and be a product of that individual’s life experience, e.g., the remapping of auditory processing to the visual cortex in the blind (Klinge et al., 2010).
Each processing pipeline registers an individual brain to its own template (eg. fsaverage for FreeSurfer, USCBrain for BrainSuite, MNI152 for SPM12 and FSL, Colin27 for BioimageSuite), which are not always exactly compatible with each other (e.g., mapping between MNI volumetric and FreeSurfer surface coordinate systems see (Wu et al., 2018)). Over the past 20 years, the most widely used have been the successive MNI brain templates (MNI305, Colin27, MNI152, MNI452, see 12), jointly referred to as “MNI space” (Mazziotta et al., 2001, 1995). The deformation fields obtained can subsequently be applied to the iEEG electrode coordinates to map their location to the MNI standard space. Normalizing to the MNI space is a prerequisite for most group analysis approaches, yet it should be remembered that the MNI152 template is blurred, leading to a loss in spatial specificity. Various open-source software packages can perform the normalization to MNI space within a few minutes (SPM12, CAT, FSL, ANTs or Lead-DBS).
Reporting 3D MNI coordinates implies that the reader can understand them. However, the term “MNI space” refers to various spaces that are not exactly equivalent (Brett et al., 2002). Although the few millimeter difference between Colin27 and MNI152 may not dramatically impact fMRI or EEG source imaging results, they are concerning for iEEG and critical for DBS. See the iEEG-BIDS specification and Lead-DBS website for a thorough review of the different “MNI spaces” (see 13 and 14). Since the reference space and the transformations estimated are specific to each program, the exact reference template space (e.g., MNI152NLin2009Asym) should be mentioned, together with the software used for the normalization of electrode locations to the MNI space.
Brain lesions pose a challenge for registration algorithms that would try to match the locally abnormal imaging intensity to the template. As epileptic activity can originate from lesioned tissue, it is actually common to have electrodes surrounding lesions. The position of these electrodes in the template is therefore more susceptible to imprecision. We recommend being particularly vigilant regarding electrode position in a normalized space for patients with a lesion, and we advise to visually assess the quality of the registration procedure and the respective electrode positions around the brain lesion. When possible, cross-checking with a neurologist or a neurosurgeon is a good practice. To better handle brain lesions, some segmentation and registration algorithms can use multiple imaging contrasts (T1, FLAIR, T2, see section 2.1.3). Additionally, some dedicated toolbox extensions (Griffanti et al., 2016; Griffis et al., 2021; Guo et al., 2019; Schmidt et al., 2012; Schmidt, Paul, 2017), e.g. for FSL, FreeSurfer or SPM, implement methods such as lesion growth, machine learning, and Bayesian approaches to define lesion masks and improve registration (Cui et al., 2019; Despotović et al., 2015; Selvaganesan et al., 2019).
2.3. Transformations back to individual space
2.3.1. Background
Most MRI segmentation pipelines include the registration of the individual’s imaging data to a common template space and map it onto one or multiple atlases. This processing step is generally performed automatically. Even when working with data from only a single patient, establishing this mapping allows the transfer of useful atlas information to the individual level, i.e. reverse mapping (see Figure 3 and (Hamilton et al., 2017) Figure 1). Cortical or subcortical parcellations, and information associated with these parcels, can be used in the delineation of the individual brain volume or surface (see section 2.2.1). Consequently, iEEG electrodes can be automatically associated with various anatomical or functional labels, making the analysis, interpretation and reporting of an iEEG study easier (Taylor et al., 2021).
2.3.2. Challenges, recommendations and reporting advice
There are inherent approximations in the normalization process, such as anatomical data smoothing, cortical deformation, in addition to the approximation introduced by electrode location estimation (see section 2.1.4, e.g., the non-rigid deformation changes the distances between points; in sEEG whether an electrode is deep in the sulcus or more superficial; in ECoG whether an electrode facing a sulcus is associated with either of the two gyri).
To take these spatial approximations into account (i.e., inherent to volume reconstruction, interpolation artifacts, segmentation thresholds, implant deformation, for a discussion see (Brett et al., 2002)), we recommend adopting a probabilistic approach when defining brain regions in which iEEG electrodes are located (like in some atlas, see for instance (Amunts et al., 2020)). This probabilistic approach in space consists of taking into account the voxels surrounding the electrode centroid. The size of this volume should both consider the contact size (2 to 2.4 mm, see sections 1.3.3) and the passive spread of the intracranial signal that is about 6 to 10 mm (for sEEG see (Lachaux et al., 2012; Mercier et al., 2017), for ECoG see (Kellis et al., 2016), for both see (McCarty et al., 2022)). Considering the electrode contact size and signal spread, we suggest using a sphere of a certain size centered on the electrode centroid rather than considering the signal to originate from an infinitely small point (see 15). The probabilistic approach leads to a number of voxels around the centroid that may contribute to the recorded activity (Piantoni et al., 2021).
2.4. Data visualization
For spatial iEEG visualization, it is important to make the distinction between the projection of an electrode to the anatomy (i.e., the cortical sheet or the surface that outlines a subcortical structure) and the interpolation of the functional values on the anatomy (i.e., on a surface or in a volume). The projection is more qualitative; it relates to the anatomy, that is electrode visualization and/or electrode labeling. The interpolation is more quantitative; it is the spatial extent from the electrode to which the data is color-coded, which should take into account passive signal spread (see section 2.3.2).
- Different strategies exist for anatomical visualization of the position of iEEG electrodes, each with pros and cons:
- For ECoG, as electrodes are visible on the brain surface, three dimensional brain display is convenient. When not all ECoG electrodes are visible from a single point of view, the surface can be made transparent, the electrodes can be shown on the inflated pial surface or can be shown on an “flattened” or “unfolded” cortical sheet (see Figure 5). However, the rendering of an inflated or unfolded surface loses the relative distance between ECoG electrodes, alike between brain regions.
- In the case of sEEG, as the shafts pass through different tissues and often target deep structure(s), the 3D brain display needs to be transparent to see all electrodes (see 3D view in Figure 6). Visualizing sEEG electrodes on an inflated or an unfolded surface, the electrodes would have to be projected to the cortical mesh, which implies a non-trivial projection that must be explicitly reported; furthermore, deep electrodes should not be projected to the superficial surface. Moreover, as with ECoG, the rendering of sEEG electrodes projected on an inflated or unfolded surface loses the relative distance between electrodes, and with the anatomical tissue.
Many clinicians and researchers are able to identify brain structures in canonical axial, sagittal, and coronal views, but less easily along the shaft trajectory or in an inflated brain. When localization focuses on a specific sEEG shaft, it is convenient to depict a slice parallel to the electrode shaft. Some software offers this display by representing an arbitrary slice going through the shaft, plus a slider to rotate around the axis of the shaft (see Figure 6 from the Gardel software (Medina Villalon et al., 2018), see also IntrAnat 16 ).
The projection of an sEEG electrode that is located in a sulcus or an ECoG electrode that overlies a sulcus (and thus faces the gray matter on both sides), to a single vertex of the triangulated cortical surface (and thereby a single sulcal wall) is misleading. This anchored location should not be used for the interpolation and display of functional data.
Figure 5.
Brain depiction and electrode visualization
ECoG electrodes are color-coded according to the anatomical label that corresponds to their location. Electrodes are shown on (A) the original pial surface of the participant’s brain, where the grid structure may be best appreciated, (B) the inflated cortical surface and (C) the flattened cortical surface (in this case based on the projection onto the fsaverage brain). The inflated and flattened surfaces are colored according to surface curvature calculated in FreeSurfer, where darker gray indicates the sulci, and lighter gray indicates gyri. The best choice of representation (pial, inflated or flat) depends on the coverage of electrodes as well as which areas are to be compared.
Electrophysiological results of the iEEG analysis can be represented on the patient brain: any other measure/statistic (e.g., ERPs or spectral power, see section 3) can be interpolated on the cortical surface or on a slice of the MRI (usually based on Euclidean distance, see Figure 7).
Figure 7.
Electrode display and data interpolation.
For ECoG (Panel A) and sEEG (Panel C), the amplitude of the iEEG signal is represented at their corresponding 3D locations, then projected orthogonally along the three axes (as in a glass brain view), and depicted on MRI slices.
In panels B (ECoG) and D (sEEG), the amplitude of the signals is interpolated on a 3D brain (left column) and on an inflated brain (right column, with light gray indicating gyri and dark gray indicating sulci). Signal interpolation was done using a sphere of 5mm for the upper row and 15 mm for the lower row. In panel B the ECoG electrodes are indicated in green; in panel D two sEEG shafts are indicated in red and green. While activity is identical in the upper row and in the lower row, the extent of the interpolation changes the rendering.
Electrode projection and data interpolation are mutually exclusive operations; the interpolation of functional data on the anatomy should be performed using the original (non-projected) electrode location.
Due to the sparse spatial sampling of the iEEG, only a limited number of voxels or triangles (i.e., volume based or surface based) would be represented with a color-coded value, whereas fMRI and MEG/EEG source reconstruction typically result in whole brain coverage. The resulting iEEG images can be misleading if there is no distinction between a voxel or triangle with no data versus one with a value below the display threshold. We therefore recommend representing the parts of the brain where no data was interpolated with a different color than the parts where data was interpolated, but considered not significant (see panels B and D Figure 7).
The extent of this interpolation should be documented and should ideally reflect passive signal spread (i.e., about 6 mm see section 2.3.2). For instance, see panels B and D in Figure 7 and in (Stolk et al., 2018) Figure 6 of pt.51.
The interpretation of the interpolation can be ambiguous. For example, if the data at two electrodes are the same, they can be interpreted as activity originating from the in-between area, but it might also be that both electrodes pick up activity that is not seen by the other. To take into account the potential interplay between activity observed by different electrodes, we anticipate that future developments in electromagnetic modeling will provide better visualization solutions with estimates of intracranial currents using Finite Element Method (FEM) (Medani et al., 2021, 2020). The combination of better inverse methods, forward models and more detailed source models (see for example (Cosandier-Rimélé et al., 2008)) will enable source localization from iEEG, and reconstructed activity will for example suffer less from ambiguous representations on both sides of a sulcus.
3. - Preprocessing
3.0. Introduction
Preprocessing aims to provide better exploitable signals for subsequent analysis that targets the experimental question or explores a phenomenon of interest. Hence we define preprocessing as mostly neutral with respect to the research question, whereas analysis is specific to the research question. The preprocessing stage also provides an opportunity to familiarize oneself with the data, especially in case the researcher was not present during data acquisition. In this section, we specifically address three preprocessing operations that are commonly used in iEEG studies, and explain how these steps increase the specificity (e.g., local activity) and the selectivity of the signal (e.g., brain signal).
The first step consists of identifying channels (e.g., the signals measured between an electrode of interest and a reference electrode) that are expected to show meaningful neural signals (i.e., either electrodes in contact with the leptomeningeal surface for ECoG and/or the gray matter for sEEG). Specifically in the case of sEEG, some electrodes might be located outside the brain or in the white matter.
The second step aims at identifying channels and/or time segments that include epileptiform activity or that are contaminated by physiological and non-physiological artifacts. Although epileptiform activity is the prime reason for doing the clinical recordings and is the subject of research on the epileptiform features of iEEG signal, it is to be avoided for cognitive neuroscientific research questions for the sake of generalization to healthy brains.
The third step consists of re-referencing the signals. This operation focuses the signal on neural activity generated in the close vicinity of the electrode; and thus derives signals that are less affected by distant activity that can be observed through passive volume conduction. Re-referencing can also help to reduce common sources of noise.
The outcomes of these preprocessing steps consist of (1) tables or arrays indicating which electrodes to keep for the analysis, (2) tables with the timestamps of artifacts, and (3) montage(s) for re-referencing. Although these steps do not modify the raw data, they should be documented as they provide valuable information for sharing and reuse. Subsequently, the preprocessing completes with epoching, demeaning and filtering.
3.1. Electrode selection based on anatomy
3.1.1. Background
The properties of the measured signal depend on the tissue in/on which an electrode is located (see Figure 4). In iEEG, the differences in signal amplitude between channels are not only caused by brain activity elicited by an experimental task/protocol. Signal amplitude also depends on the distance between the electrode and the gray matter (i.e., geometry) and the properties of surrounding tissue(s) (i.e., tissue type). Some analyses, such as signal feature classification (e.g., using Receiver Operating Characteristic), are not necessarily affected by a weakened signal that is recorded centimeters away from the source (Torres Valderrama et al., 2010). However, weak signals are a concern when it comes to determining the location of the iEEG signal source.
With sEEG, while the gray matter is the main target, the electrode shafts inevitably go through different types of tissue and the majority of sEEG electrodes are surrounded by several tissue types. Some tissues, like the skull, are poorly conductive and passively spread the activity over long distances. Other tissues, such as the white matter, show a more complex profile with a mixture of passive volume conduction and active propagation (Mercier et al., 2017). The identification of the tissue surrounding sEEG electrodes requires a precise localization relative to the individual’s brain anatomy. Subsequent segmentation algorithms provide tissue classification and/or tissue probability maps (i.e., type/amount of tissue at each voxel). Knowing in which voxels(s) the electrode contact stands allows an easily automatized approach (see section 2.1.3 and 2.1.4). Not only sEEG electrodes located outside of the brain (e.g., in the skull or in soft tissues) can be identified and disregarded from further analysis; tissue classification and/or tissue probability map can also provide a quantification of the amount of gray matter surrounding electrodes in the brain and/or the distance to the nearest gray matter (see section 2.1).
With ECoG, it frequently occurs that a grid or strip extends over several gyri, leading to electrodes positioned on top of a blood vessel or over a sulcus, which are known to impact signal characteristics (see Figure 4). Specifically, signals recorded from electrodes on blood vessels have an attenuation of absolute power spectral density, especially in the high-frequency band (30–70Hz in (Bleichner et al., 2011) and 45–120 Hz in (Branco et al., 2018c)). Besides affecting the spectral characteristic of the signal, blood vessels increase the distance between electrode(s) and neural source(s), thereby being sensitive for activity from a larger area, as if it is a virtually larger electrode. Last, pulsation and smooth muscle activity of arteries can directly contribute to low frequency signals. Intraoperative photos (Bleichner et al., 2011; Miller et al., 2009; Wang et al., 2016), as well as imaging data (e.g., magnetic resonance angiography (Branco et al., 2018c)), can be used to determine which ECoG electrodes are in direct contact with the leptomeningeal surface. The tissue classification from the brain segmentation can also be used to estimate the distance to the cortical gray matter surface and thus to identify electrodes that are possibly placed over a vessel or a sulcus (see section 2.1.2).
3.1.2. Challenges, recommendations and reporting advice
When electrodes are selected based on anatomical considerations, we recommend reporting how the selection was performed. For instance, what is the tissue classification method, and what are the thresholds applied to electrode selection, such as the distance to the gray matter, or the fraction of gray matter voxels in the electrode surroundings.
3. 2. Data selection based on the signal
3.2.1. Epileptic activity
As the clinical motivation is to localize and characterize the epileptogenic network, some electrodes are prone to record epileptiform activity. This activity is not limited to seizures; interictal (between seizure) epileptiform discharges can also cause unwanted artifacts which can sometimes propagate widely. Epileptic activity is often not relevant for the cognitive processes under study, nonetheless this pathological activity can add to, or interact with, the physiological activity that is of interest (e.g., interictal epileptiform discharges can transiently impair cognitive function (Boly et al., 2017; Henin et al., 2020; Kleen et al., 2013; Leeman-Markowski et al., 2021)). Hence it cannot be excluded that epileptic activity reduces the ability to detect the relevant cognitive activity, especially if the epileptic activity is time-related to the event of interest (e.g., stimulus onset, behavioral response, time period of interest). Furthermore, epileptic activity may cause false positive activity, if the investigated cognitive function or the region of interest is linked to the individual characteristics of the epilepsy.
The information gathered by the clinical team provides the basis to reject sites that belong to the epileptogenic network, to identify interictal activity and thus to alleviate some of the uncertainty related to epileptiform activity. This knowledge should be shared by the clinical team and used by the researchers to shed light on the brain sites showing epileptic activity and on the cognitive processes that might be impaired (see section 1.3.8 and Figure 2).
3.2.2. Transient artifacts
Although in the past it was believed that iEEG is immune to the major movement-related artifacts that contaminate scalp EEG, this view has been challenged with several studies showing iEEG being contaminated by eye movements (Jerbi et al., 2009a; Katz et al., 2020; Kern et al., 2021; Kovach, 2011), blinks (Ball et al., 2009) and movements of cranio-facial muscles (Otsubo et al., 2008). These artifacts overlap within frequency ranges for which task-related physiological signals are analyzed and reported (see Figure 8). Despite re-referencing or more advanced spatial filters such as ICA to reduce the passive volume conduction impact of these artifacts, some residual still affect frontal regions as well as the ventral, medial and lateral parts of the temporal lobe (Katz et al., 2020; Kovach, 2011).
Figure 8.
Examples of artifacts due to eye and mouth movements.
A. Ocular artifacts in sEEG. The EOG and iEEG were recorded while the participant was asked to either blink or move his eyes leftward (upper left panel, color-coded in blue and red respectively). EOG single-trial averaging (n=16) shows time-locked muscular-related activity on cardinal EOGs (upper middle panel); and on vertical and horizontal EOGs obtained through bipolar referencing (upper right panel). Intracranial EEG was recorded from 252 electrodes relative to a reference located in the white matter (yellow circle on the lower left panel). Broadband Event Related Potentials computed with both a monopolar reference (WMR) and a bipolar reference (Bp) are depicted for a frontal and a posterior electrode. For the frontal sEEG electrode, the artifact time-locked to muscular activity recorded with EOG is visible with the monopolar reference and is reduced when the bipolar montage reference is applied. The posterior electrode shows activity that is not reduced by the bipolar referencing and that is delayed with regards to EOG activity. The frontal electrode shows muscular activity that has passively diffused from the eye muscles, while the posterior electrode shows local neural activity that is consecutive to the eyes movement (e.g. change in visual input or corollary signal).
B. Oral artifacts in sEEG. The participant was asked to clench his teeth ten times while sEEG was recorded. Broadband and high-frequency activity recorded from frontal electrodes reveal artifacts time-locked to jaw-contraction (color-coded in pink and in green respectively).
In addition, non-physiological artifacts can originate from stimulus delivery equipment, loose moving cables, or other electrical devices (see section 1.3.5). This contamination impacts not only data quality, but may bias the outcomes of certain analyses. A striking example of this was observed in speech decoding research, where some signal components initially thought to be speech-related turned out to originate from the mechanical action of the sound on the recording chain (Roussel et al., 2020). Similarly, using ECoG and DBS leads, it was reported that speech production itself induced high-frequency artifacts through mechanical vibration (Bush et al., 2021). More mundane but along a similar line, the auditory response to the audible click of a response-button could be mistakenly interpreted as evidence that the auditory cortex participates in task-related processes.
The operations of artifact detection and rejection benefit from knowing what artifact patterns look like. For instance, saccade-related artifacts occur as short, massive power increases above 50 Hz in the immediate vicinity of oculomotor muscles in the medial temporal lobe, coincident with eye movements (see Figure 8). Collecting and annotating a set of “artifact templates” can be used as training support for researchers that are new to iEEG, whereas for already skilled researchers they also provide documentation about which electrodes are susceptible to physiological artifacts (see sections 1.3.4, 1.3.5 and 1.3.9). Last, the spatio-temporal characteristics of “artifact templates” can be used in post-hoc control analyses (see below).
Some artifacts are more likely to co-occur with certain aspects of cognitive tasks, and are thus correlated with task-related neural activity and/or behavior (e.g., frowning during a difficult task, systematic blinking at the offset of visual stimuli, or muscle tension due to anxiety). Due to that correlation, the cleaning procedure can remove a mixture of artifactual and relevant data, and obscure the true relationship between physiology and behavior. A compelling example for this occurred during the detection of saccade-related artifacts in the anterior temporal lobe (Jerbi et al., 2009a). The hypothesis was that a “go-signal” in high-level visual areas would trigger the subsequent saccade when the local visual analysis was complete. The hypothesized signal was a burst of high-frequency energy in the anterior temporal lobe time-locked to saccades; however these turned out to have the same characteristics as saccade-related artifacts. This was subsequently verified from ‘artifact templates’ and EOG. Other examples report blink and saccade related suppression effects occurring in visual regions (Golan et al., 2016; Katz et al., 2020) and mediating visual processing during natural viewing (Kern et al., 2021), or pursuit related activity in occipital, frontal and parietal regions (Bastin et al., 2012).
In some clinical settings, electrical stimulation is performed during clinical testing, or during single pulse electrical stimulation to map networks (see section 6.6). The analysis of iEEG data from such settings requires some understanding of how these artifacts depend on recording and amplifier settings. Most amplifiers require cortical electrodes that are stimulated to be disconnected from recordings and do not record data. On the other electrodes that are recorded, the stimulation typically causes immediate artifacts that can saturate the signal; the duration of these effects should be carefully assessed (Miller et al., 2019) and/or can be modeled to correct the signal (Trebaul et al., 2016). Electrical stimulation can also result in slower offsets in electrodes close to the stimulated electrodes (Prime et al., 2020), where the signal then exponentially returns back to baseline. Besides, during electrical stimulation in the brain, electrical artifacts can also occur when performing somatosensory evoked response studies with an electrical stimulator located on the limbs, torso or face.
3.2.3. Data selection in practice
The primary aim of artifact detection and rejection is to obtain data that is as clean as possible. That said, statistical power can be jeopardized if an overly large proportion of data is discarded. Artifacts can have confounding effects on the analysis (e.g., increased heart rate or eye blink frequency in one condition than another, see section 3.2.2). Spending too much time in manual artifact-screening can be not as effective as re-referencing and/or getting more data, especially as some artifacts (i.e., confounds) may result in false positives, while other artifacts (e.g., uncorrelated interictal epileptiform discharges) are less likely to cause false positives but mostly decrease the sensitivity. An efficient artifact detection procedure requires training: it must be guided by information about the data and should be well-balanced in time for efficacy. The optimal data-cleaning strategy differs between studies as it depends on the research question (e.g., the time window of interest or electrodes of interest) and on the methods that are used in the analysis (e.g. frequency analysis).
Considering the experimental events (i.e., triggers) while reviewing the recordings can uncover relations between artifacts and the neural signals of interest for the study. The iEEG signal to noise ratio is so high that many response components can be seen in raw data at the single trial level (See Figure 11). Artifact detection is facilitated when individualized ‘artifact templates’ are available, as they identify susceptible electrodes (see sections 1.3.4, 1.3.5 and 1.3.9).
Figure 11.
Signal-to-noise ratio (SNR) as a function of the number of trials.
Activity was recorded in the primary auditory cortex of a participant while listening to pure tones (1000 Hz).
A. Activity is depicted for different numbers of averaged trials. The broadband ERP is depicted on the left, the magnitude envelope of high-frequency activity is shown on the right.
B. SNR varies with the number of trials used for averaging. SNR was computed using the maximum amplitude of the activity divided by the variance over the baseline (S/N).
Many research teams start the detection procedure by a quick visual inspection of the data, which provides a first understanding of the type and shape of artifacts and immediately reveals “bad channels”, either being flat (the signal shows no variance) or showing substantial amounts of noise or persistent epileptogenic activity. To complete this first examination, we recommend a systematic rapid evaluation by looking at the power spectral density (PSD; +/- its variance) of all channels over the entire recording (see Figure 9 panel A).
Figure 9.
Interfaces examples for artifact detection
A. The Power Spectrum Density (PSD) of all channels can help to identify artifacted channels with artifacts (in red).
B. A display per channel and per trial can be used to identify short-lived artifacts. The heatmap represents for every trial (along the horizontal axis) and every channel (along the vertical axis) a metric (here the variance) which is color-coded from blue to yellow. The panels to the right and below the heatmap represent the maximum variance over trials and over channels, respectively. A channel with high variance over most trials is highlighted by the magenta rectangle. A trial with high variance over channels is highlighted by the orange rectangle.
Semi-automated artifact detection procedures have been proposed, taking statistical signal metrics into account (e.g., normalized amplitude, variance, kurtosis or the correlation coefficient between neighboring electrodes, see for instance (Tuyisenge et al., 2018)). Often, multiple passes are carried out, such as on the broadband signal and on high-frequency activity (typically above 50 Hz), to accommodate the 1/f law and thus to avoid biasing artifact identification toward the lowest frequencies (i.e., to facilitate artifact identification in the higher frequency range usually concealed due to its lower amplitude, see section 4.2.3.4). No consensus exists currently within the iEEG community regarding an optimal semi-automated procedure. Nevertheless, the lack of objective procedures should not be taken as a hurdle to improve traceability: describing the (automatic or manual) methods and thresholds are part of the good practices.
Some researchers carefully check the result of (semi-) automatic procedures “by hand” to adjust rejection thresholds to individual SNR and keep a sufficient number of trials for the analysis (see section 4.1.2). In the ideal case, experiments should yield a great number of trials, but the time a patient participates in a single experiment is obviously limited and the number of trials might be small. Therefore experimental designs with many conditions and statistical contrasts should be avoided to retain sufficient statistical power (see also section 1.2.3). A recent meta-study by Meisler and colleagues (Meisler, 2019), evaluated different approaches to eliminate artifacts in iEEG: automated artifact rejection based on statistical properties of times series (standard deviation or kurtosis) and manual artifact rejection based on expert annotation. This study showed that with automatic thresholding, statistical power did not increase with more liberal criteria. In contrast, conservative approach reduced statistical power as much as with manual exclusion (i.e., based on experts' notes). Critically, bipolar referencing (BPR, see section 3.3) outperformed common average referencing (CAR) in increasing statistical power by attenuating noise for the automated and manual approaches. Overall, to increase statistical power, Meisler and colleagues advised collecting more data and the use of bipolar or local reference schemes, rather than spending a lot of time detecting artifacts. Thorough artifact scrutiny (e.g., performed manually, trial-by-trial) appears to be less time efficient than quick and semi-automatic approaches. In addition to the lack of real improvement of statistical power, it can be tedious, time-consuming and visual detection criteria can be subjective and unstable over time. Yet, manual cleaning permits the researcher to get a better feel for the signal. If detailed visual inspection is performed, it must be documented and traceable, similar to semi-automated approaches.
3.2.4. Challenges, recommendations and reporting advice
In M/EEG the channels usually show the underlying sources of interest with a similar order of magnitude and all channels are therefore processed in the same way. In iEEG, the sensitivity to the signal of interest can vary a lot across channels. Researchers familiar with M/EEG artifact rejection should therefore be aware of several peculiarities of iEEG data. In M/EEG, sensors are (i) uniformly distributed at systematic positions, (ii) centimeters away from the neural sources; with (iii) similar signal-to-noise ratio; and provide (iv) smooth topographical activity due to the passive volume conduction that causes spatial blurring. Conversely, iEEG electrodes (i) are scattered over more or less distributed brain areas, (ii) are directly in contact with the brain, sometimes right at the level of the neural sources, with a signal that is orders of magnitude stronger than in EEG; (iii) have a variable signal-to-noise ratio due to conductance differences between electrodes (especially as sEEG electrodes might be located in different types of tissues); (iv) provide signals that are rather independent from each other, and the sparsity in iEEG hinders topographic analysis or interpolation of noisy/bad channels. Such specificities therefore lead many iEEG researchers to homogenize or standardize (e.g., by z-scoring) the signals across electrodes prior to performing artifact detection to compare the normalized signals between electrodes.
When only few trials are available and only a few electrodes are relevant for the research question, one may consider preprocessing iEEG channels separately, without comparing the raw recordings across electrodes. Tailored signal selection/rejection is a convenient strategy when the effect of artifacts are limited to specific electrodes, since a global rejection would discard a data segment from all channels, whereas it is only “bad” for one channel. Such tailored individual rejection retains more “good” trials for each channel. Of course, this rejection strategy is not appropriate for multivariate analysis where distributed signals across channels are estimated.
If the online reference used during the recording was noisy, we recommend that the data is re-referenced to a common reference prior to inspecting the data for artifacts (see section 3.3 and Figure 10).
Figure 10.
Interface showing reference switch.
Illustration of displays with two different montages (unipolar reference on the left and bipolar reference on the right). The montages are selected graphically in a dedicated menu (here in Brainstorm). When switching from the unipolar to the bipolar montage, the low- and high-frequency artifacts shared over all channels are reduced.
3.2.5. Data visualization
The power-spectral density of the continuous data is useful to immediately detect artifacted channels and thereby electrode(s), as their power spectra often stand out from the non-artifacted electrodes (see Figure 9 panel A, as implemented in Brainstorm (Tadel et al., 2011)). Computed trial-by-trial (or epoch-by-epoch), it permits to additionally estimate variance across time and thus determine if the artifact is transient or sustained.
The complexity of rejecting artifact-contaminated data resides in finding a compromise between excluding epochs with transient spread of epileptic activity or other artifacts across artifacted channel(s) and excluding channel(s) with frequent epileptic activity or other artifacts. Some techniques/recommendations and clever data visualization can considerably accelerate the visual inspection step, for instance for assessing the number of epochs that would need to be rejected due to repetitive artifacts occurring on the same electrode. A convenient approach is to create heat-maps that show for each channel a metric of the signal (e.g., z-score of signal amplitude) as a function of time (trial/epoch on x-axis) and electrodes (y-axis) (see Figure 9 panel B, as implemented in the ft_rejectvisual function used with the summary method in FieldTrip (Oostenveld et al., 2011); see also (Tuyisenge et al., 2018) for alternative approaches). Maps can be created for broadband as well as frequency-specific signals (the effect of muscular or epileptiform activity is more visible above 50 Hz) and all channels can be visually inspected within a few minutes, to quickly identify those that should be analyzed further.
More generally, reviewing artifacts requires some training. At the beginning it implies finding the adequate display for data visualization, which is often subjective. Once the visualization strategy is defined (e.g., scaling, filters, layout), the review process is facilitated and accelerated.
3.3. Re-referencing
3.3.0. Introduction
During the acquisition, the online reference is chosen to obtain clean signals with minimal contamination by artifacts or noise (see section 1.3.2). After the recording, the signal can be re-referenced to an offline reference to improve the spatial interpretation of the signal. Typically, re-referencing is a subtraction that can be implemented as a linear combination of channels: depending on the reference scheme, some signal features are lowered (e.g., far field or distant activity) while other features are heightened (e.g., local activity). Therefore, the optimal reference scheme depends on the researcher’s definition of noise and their assumptions on the signal of interest.
The iEEG signal reflects a mixture of activity that originates close to the electrode of interest (see section 1.3.3), activity from sources further away that passively spreads by volume conduction (see section 2.3.2) and activity that is mostly captured by the reference and thereby also affects the potential difference (see section 1.3.2). Offline re-referencing permits: (i) to reduce common mode potentials with the original reference (line noise at 50Hz or 60Hz), (ii) to diminish the contribution of distant sources through passive volume conduction, (iii) to lower the correlation between channels and thus increase the SNR of some components of the signal (e.g., local features, see Figure 10). It is critical to know the original recording reference, as it might be the cause for distant activity appearing in the signals. In cases where a distributed signal is relevant to the research question, for example slow waves (4–7 Hz), the investigation does not benefit from a local reference scheme (Zaveri et al., 2006). A local reference scheme is more appropriate for analyzing focal high-frequency activity (e.g., bipolar activity acts as a high-pass filter because low-frequency signals tend to be more widespread). Each reference scheme has its own strengths and weaknesses, which explains the absence of a general consensus. Comparing results obtained with different referencing schemes helps to interpret the spatial origin of the analyzed signal (see Figure 12). Below, we present and discuss various alternatives for re-referencing.
Figure 12.
Influence of the reference montage on iEEG signal analysis.
A. Location of sEEG electrodes in the brain of the participant (axial slice on the left, coronal slice on the right).
B. ERP computed with different reference montages, respectively in black for monopolar white matter reference (WMR), dark blue for common average reference (CAR), light blue for local average reference (LAR, using all electrodes of the sEEG shaft), and red for bipolar reference (Bp). A schematic of the sEEG shaft is depicted to illustrate the spatial organization of the activity and especially the phase reversal revealed by the bipolar montage (e.g., at the level of channel #11).
C. Power from time-frequency analysis computed with different reference montages, respectively from left to right: WMR, CAR, LAR and Bp. Both CAR and LAR introduce a strong correlation of high-frequency activity over channels, absent in the WMR.
D. Phase Concentration Index from time-frequency analysis computed with different reference montages, respectively from left to right: Bp, WMR, CAR and LAR. Both CAR and LAR introduce a strong correlation of high-frequency activity over channels, absent in the WMR. Note the presence of induced activity, that is only visible in the power depiction (part C), and of evoked activity that is visible in both the depictions of power and Phase Concentration Index (e.g., see channel 8–9 with Bp montage).
E. Phase angle distribution represented for different reference montages, respectively from left to right: Bp, WMR, CAR and LAR. The data corresponds to the red square indicated in the time-frequency plane of channel 6. While the absolute angle measure varies with the reference montage, the relative measure (i.e., PCI) is consistent across reference montages.
3.3.1. Background
3.3.1.1. External reference
As discussed in Section 1.3.2, an external electrode can be utilized as an online reference during the acquisition. Keeping or applying an external electrode for data analysis or for offline re-referencing is less optimal, as it mixes the iEEG signal with external noise and global brain activity passively volume-conducted through the tissues. The resulting signals may thus comprise a combination of these components, leading to a feature common to all channels. Unless the subsequent analysis method permits removing this common undefined component (e.g., based on spatial filtering), we recommend re-referencing the signal to one or several internal electrodes.
3.3.1.2. Internal reference
When using an internal reference, at least four types of reference schemes can be considered (see Figure 12). Note that while some reference schemes are identical for all electrodes, other reference schemes are specific to each electrode (i.e., the reference is not the same for all electrodes).
Common Average Reference method (CAR): the mean signal over all artifact-free electrodes is computed and subtracted from every electrode. The rationale is that the noise which affects all electrodes can be approximated by the mean. The subtraction of the mean from all electrodes adds a (negative) common component to all channels and thereby can increase the correlation between channels, which is to be considered in certain analyses (e.g. connectivity analysis). The CAR makes spatially focal features less salient compared to widely distributed signal features. Last, the CAR can be spatially biased by the distribution of electrodes (e.g., in the case where the density of electrodes is higher over the temporal cortex as compared to the other cortical lobes, it leads to an over representation of activity of the temporal region).
The Local Average Reference method (LAR) is similar to the CAR, except that it consists in subtracting the mean over a subset of artifact-free electrodes from each of the electrodes of that same subset. The subset of electrodes is generally based on the type of implant, for instance a single ECoG grid or strip, or a sEEG shaft. Here, and as compared to the CAR, the assumption of having equal representation of the noise across electrodes of the subset is more likely due to the spatial proximity of the electrode subset (both in the brain and at the level of the recording device, e.g., cable sheath). Still, the LAR results in correlation between channels of the same subset due to the shared reference, and can inject high focal activity from one electrode into the entire subset.
BiPolar Reference method (BPR): each electrode is referenced to one of its nearest neighbors (one site only). BPR yields an estimate of the first-order spatial derivative. This is a popular choice in sEEG: the signals recorded along an electrode shaft are thus replaced by the subtraction of two spatially consecutive signals along the 1-D linear array, leading to a virtual channel shaft with one less channel than the real electrode shaft. In the absence of artifacted electrodes, the BPR is more systematized across all electrodes than with the Local Composite Reference (LCR, see next paragraph). The BPR gives the compound signal of the activity close to both electrodes and the volume-conducted activity of the tissue in between. The result of the BPR is often visualized as a virtual channel located at the mid-point between the two original electrodes (see section 3.2.2). The BPR reduces signal features shared between neighboring electrodes and neural response components with a wide spatial distribution.
Local Composite Reference method (LCR): each electrode is referenced to the average of its nearest neighbors (excluding artifacted electrodes). The LCR is an estimate of the second-order spatial derivative (i.e., Laplacian), where a stronger weight is assigned to local signal features compared to features shared between electrodes. For a 2-D ECoG grid for instance, the average that is subtracted is computed over n=4 neighboring electrodes for an electrode that lies inside the grid, n=3 if it lies on the border and n=2 if it lies on the corner. For narrow ECoG strips (i.e., 1-D) or sEEG, the mean signal is computed from the two neighboring electrodes. A disadvantage of the LCR is that for electrodes at the edge it has to be computed with a different number of neighbors, or that it has to be skipped for electrodes at the edge. Moreover, the LCR can spread highly focal activity from one electrode to its nearest neighbors that do not originally record that activity (e.g., neighboring electrodes located on different sides of a sulcus). Last, LCR reduces signal features that are local but shared between neighboring electrodes and neural response components with a wide spatial distribution.
Specific considerations about sEEG and white matter: As sEEG electrodes may be located in white matter, clinicians have commonly used such signals as a reference (online or offline), based on the observation that the signals recorded from the white matter typically show a lower variance, presumably because it does not correspond to actual neural activity. While it is true that neural activity is not directly generated in the white matter, activity generated in the distant gray matter actively spreads along fiber tracts. Therefore, the notion that white matter electrodes do not record neural activity is only an assumption and must be assessed (e.g., by computing the variance of the broadband signal as well as of the low amplitude high-frequency activity, see (Mercier et al., 2017) and (Uher et al., n.d.)). Further, the conclusion that a channel lies in the white matter is not always straightforward as there is some uncertainty in the precise location of an electrode (see sections 2.1.4 and 2.3.2). Last, a white matter electrode can be surrounded by gray matter, and thus records neural activity that has passively spread from it ((Mercier et al., 2017; Rizzi et al., 2021) and see section 6.8). There are two main types of white matter montage:
White matter reference (WMR or average white matter reference, AWMR) consists in subtracting from each channel the activity recorded at a chosen iEEG electrode located in the white matter (or the mean activity across all artifact-free electrodes located in the white matter).
Proximal white matter reference (PWMR): each iEEG site in the gray matter is re-referenced to the closest site in the white matter (Arnulfo et al., 2015).
Recently a low variance average reference montage was proposed by Uher and colleagues (Uher et al., n.d.). They reported that electrodes in the white matter can record signals with high variance and that such electrodes are thus not necessarily electrically neutral (as shown in (Mercier et al., 2017)). They proposed instead to use a virtual channel as reference, corresponding to the average signal of all channels with a low variance signal.
In iEEG research, less commonly used methods can serve the same purpose as re-referencing by using spatial filtering of the data to enhance signal-to-noise ratio (for a discussion see (Michael X Cohen, 2017)), for instance ICA (Michelmann et al., 2018; Whitmer et al., n.d.), PCA (Alexander et al., 2019), or Spatio-Spectral Decomposition (Schaworonkow and Voytek, 2021).
3.3.2. Challenges, recommendations and reporting advice
We recommend first processing the artifacts and then re-referencing the data, unless the online reference used during the recording was itself noisy, which complicates artifact detection.
While a local reference provides the highest human readability of local features of the iEEG signal, it can artificially mix activity from/to the neighboring electrodes. For instance when a non-local reference is used in ECoG, activity projected on the grid can reveal gyri and sulci, but when the local reference montage is applied it may shift some activity that jumps over a sulcus. It follows that in order to verify the genuine origin of a given activity, it is always informative to systematically compare the pattern of activity obtained using a low-variance reference scheme to the activity pattern obtained with a local reference montage. Finally, the use of a local reference montage may introduce artificial correlations between neighboring channels, which in turn may bias some subsequent analysis (e.g., connectivity) or statistical comparisons or corrections (e.g., dependent versus independent variables).
Reference schemes should be applied only as lightweight (“on the fly”) linear transformations, especially when doing the first pass of data reviewing. They can be used as a fully reversible montage, as long as data is processed linearly. Most software and toolboxes permit applying a reference scheme to the data almost instantaneously. The montage is generally provided as a matrix indicating for each electrode the weights specific to the reference scheme. Also, when it comes to sharing iEEG data, it is preferable to share the original signals plus the montages used to visualize and/or process them.
After applying a reference scheme, the output leads to virtual channels that do not necessarily spatially match the physical electrode used for acquisition. To obtain the coordinates of iEEG channels when a BPR is used, we recommend computing the position of the "virtual sensor" in between the two real sensors. This is done in the individual space first (see section 2.1): for each channel the coordinates are obtained by averaging the coordinates of the two original electrodes used from the bipolar montage. These channel coordinates shall then be transferred in the normalized space by applying the non-linear transform obtained from the non-linear registration of individual space to the normalized space (see section 2.2). As for spatial specificity, the signal of the virtual channels can be misleading for the neighboring electrodes along the shaft that sample from distinct functional areas (i.e., the functional specificity is then mixed in the virtual channel).
Ideally, bipolar referencing should be standardized to ease the comparisons between participants and between studies. In sEEG, electrode numbering is always performed from the most mesial contact to the most lateral, but there is a lack of common strategy when performing signal subtraction between two adjacent sEEG electrodes. We here propose to follow the same scheme used by most clinical teams, that is, to compute the bipolar referencing from the most inner electrode to the most external one (e.g., c1-c2, c2-c3, etc.; this is the by-default-approach followed in both the Brainstorm and the FieldTrip toolboxes, (Oostenveld et al., 2011; Tadel et al., 2011)). In ECoG, electrode numbering is not standardized with respect to brain anatomy; it is generally defined on the position of the cables on the grid or shaft. A common strategy for bipolar referencing is therefore not possible, yet in the case of ECoG grids the bipolar referencing should take place along all directions of the grid (i.e., vertical, horizontal and diagonal).
3.3.3. Data visualization
The choice of a reference scheme is not straightforward, as it builds on information relative to signal quality and electrode location, as well as on expertise. It is effective to be able to go back and forth between different reference schemes (e.g., using keyboard shortcuts as in the Brainstorm toolbox as illustrated in Figure 10 (Tadel et al., 2011), or using a drop-down menu as in the Anywave free software see Figure 5 in (Colombet et al., 2015)) while visualizing the data, especially if anatomical information is combined (e.g., to identify an electrode at the interface with the skull or over a blood vessel).
3.4. Additional preprocessing
Several steps are often associated with preprocessing, like epoching, demeaning-detrending, filtering, especially when data is processed with software that groups these steps together. As some of these processing steps limit the analyses that can or cannot be performed subsequently, they are not extensively developed in this section. In addition, they are identically applied in M/EEG and in iEEG. Therefore we only briefly introduce them and indicate some references for more in-depth consideration.
Epoching consists of segmenting the recorded signal into periods of interest, either in trials containing at least one experimental event (event-related paradigm), or in segments of a continuous recording (block paradigm). Epochs are defined in conjunction with the research question(s) and are then generally processed as repeated measures. Excluding trials due to artifacts should be done blind to the condition to avoid selection biases.
Demeaning (a.k.a DC-offset correction) corresponds to centering the signal around zero. Detrending consists of correcting for a linear drift. The signal features that we aim to remove with demeaning or detrending generally originate from the acquisition equipment and are not interesting. These corrections can be applied on an entire dataset or on single epochs depending on the planned analysis. For long trials, a high-pass filter at low frequency could be prefered (e.g., at 0.01 Hz). They facilitate signal visualization such that the average signal value during a predefined baseline epoch is zero.
Frequency filtering refers to the removal of specific spectral components, such as the ambient electrical noise (i.e., 50Hz or 60 Hz notch, plus harmonics). It can also be used to remove low-frequency components if the neural activity of interest is in higher frequencies, or to remove high-frequency components when the interest is in lower frequencies (e.g., Event Related Potentials). As such, filtering parameters depend on the brain dynamics that are being studied. Recommended filtering parameters are the same in iEEG and M/EEG and can impact signal measures (e.g., latency, amplitude). We redirect the reader to more specific references on filtering in human electrophysiological data (de Cheveigné, 2018; de Cheveigné and Nelken, 2019; Vanrullen, 2011; Widmann et al., 2015) and the subsequent commentaries from (Rousselet, 2012; Widmann and Schröger, 2012)). Filter parameters (such as filter type, order, frequency parameters, and direction) should always be reported.
4 -. Signal analysis
4.0. Introduction
Many aspects of iEEG signal analysis do not differ that much from M/EEG or other continuously sampled electrophysiological data such as LFPs in animal research (Pesaran et al., 2018). A number of the methods described below were initially developed and applied in M/EEG research; however, their application to intracranial data benefits from the high spatial specificity and SNR of iEEG. Therefore, we provide a didactic overview of the most analysis methods, their neurophysiological significance and some specific recommendations for iEEG. We will particularly focus on the iEEG analysis steps that differ from M/EEG.
The analysis methods listed here are not to be taken as a catalog of approaches to undertake sequentially until a potentially interesting pattern emerges. Rather, good research practices imply that the choice of a method is driven by the research question that is elaborated beforehand. This stands not only for hypothesis-driven research but also for exploratory research (see section 5), as in both cases the analysis method is inscribed in a more theoretical framework of neurophysiology of brain function which leads to the interpretation of the results. Last, to perform an analysis (e.g., frequency, time-frequency, connectivity), the wide range of measures (e.g., wavelet, coherence) and their algorithmic complexities, requires a motivated choice to prevent getting lost in the garden of forking paths (Gelman and Loken, 2014). The selection of a given measure must be based on a solid understanding of that measure, its strengths and its limitations.
First, we introduce Event Related Potentials (ERPs), which, by definition, implies that there is evoked activity that is elicited by experimental events. The ERPs computed from the iEEG signal are considered to reflect mainly post-synaptic synchronized activity of pyramidal neurons (Buzsaki et al 2012) due to coincident incoming inputs to a substantial population of neurons in the vicinity of the iEEG recording site.
Second, we present frequency analysis. It can be used to quantify neural activity in the absence of events, for example in spontaneous activity. When computed in a time-resolved fashion, frequency analysis can also be used to quantify neuronal activity related to but not necessarily precisely time-locked to events. One of the main interests in frequency analysis is to distinguish or focus on activity in specific frequency band(s), either reflecting wide modulatory inputs or more local neural processes.
Third, we present connectivity measures that express the relationship between signals recorded at spatially distant sites. Connectivity estimates provide information about long range communication between iEEG channels (i.e., nodes in brain-wide networks) and are essentially performed in the time-frequency domain.
Last, we introduce cross-frequency coupling (CFC) which characterizes the link between signal components occurring in different frequency bands. CFC can be used to investigate the local architecture (i.e., within the signal of one recording site), or at the network level (i.e., between signals recorded from distant sites).
4.1. Event related potentials
4.1.1. Background
Computing ERPs consists of averaging the brain's response over multiple trials that are defined by a specific type of experimental event (e.g., stimulus onset, motor response). Neural activity that is precisely time-locked and phase consistent over trials remains in the average, whereas activity that is uncorrelated over trials (the “noise”) tends to cancel out (see Figure 11). Note that activity that is elicited by events but whose timing is variable will not be seen by the averaging procedure (see section 4.2.1). The calculation of ERPs for intracranial data does not differ from scalp EEG, or event-related fields (ERFs) obtained from MEG. Good practices and reporting guidelines in ERP/ERF research are thoroughly documented elsewhere (Gross et al., 2013; Keil et al., n.d.; Pernet et al., 2020; Picton et al., 2000) and are not repeated here. In the following we will mention some critical aspects that are specific to iEEG research.
4.1.2. Challenges, recommendations and reporting advice
A recurring question pertains to the number of trials needed to obtain an ERP with a sufficient SNR. When designing the experimental protocol, it is of strategic importance to minimize the experimental duration for patients who are susceptible to seizures and who donate their time and energy at a challenging time for them (see section 1.2.3). Because of the high SNR of iEEG, especially in functionally specialized regions, the brain responses of interest are often visible in a single trial (see Figure 11). This high SNR makes it easier, as compared to M/EEG, to relate responses to single events to behavioral data (e.g., response times, see section 4.1.3). That said, patients may have frequent epileptiform abnormalities which can limit the number of artifact-free trials for task-related stimuli. We note that it is also important to emphasize that all of the preprocessing parameters—low-pass filter setting, reference scheme, baseline period, and artifact removal approach—interact to influence the final estimate of ERP amplitude (see section3.4 and (Clayson et al., 2021)).
Figure 11 shows ERPs recorded in the primary auditory cortex (AudCx) in response to pure tones. As expected, the SNR increases proportionally to the square root of the number of trials. The number of trials required to obtain a robust ERP depends on many factors, but the examples in Figure 11 provide a lower estimate of the minimum number of trials (for comparison with surface EEG, see (Boudewyn et al., 2018), for MEG see (Chaumon et al., 2021)). Following a procedure similar to the one used in Figure 11, it is possible to simulate the minimal number of trials needed to reach a stable estimate of a metric (e.g., ERP): (1) a given subset of trials is randomly selected (e.g., 10 out of the overall number of 20 trials); (2) the metric of interest is computed (e.g., ERP, magnitude at a frequency of interest); (3) the variance of the metric is calculated across many drawings of the chosen subset of trials (e.g., 10 trials); (4) the variance is compared for different subset sizes (e.g., n=3 to n=19); (5) the minimal number of trials corresponds to the subset size for which the variance across drawings does not decrease further as compared to the next-larger subset size (see for an illustration (Oehrn et al., 2015)).
In scalp EEG, the recommended nomenclature of ERPs consists of the letter P or N indicating a positive or negative deflection, combined with its nominal latency in milliseconds (e.g., N170, P300). Some early investigators have referred to event-related components by successive deflections in the EEG waveform (e.g., P1, N1, P2, N2, etc), however, this system of nomenclature is not generally recommended (Pernet et al., 2020). In iEEG, the polarity depends on the location of the electrode of interest and its reference relative to the active neural source, or the reference scheme. Using a bipolar reference scheme in sEEG focuses the sensitivity of the corresponding channel to the activity between two neighboring electrodes (see section 3.3). In that case, each electrode picks up another pole of the source and the polarity of the ERP is opposite at the two electrodes which comprise the bipolar channel (see phase reversal example in Figure 12). Consequently, the polarity is arbitrary, as it depends on whether the bipolar channel is computed as E1-E2 or E2-E1. Hence, the polarity of an intracranial ERP cannot be interpreted as-is, but should be put into perspective when compared with the classic EEG nomenclature (e.g., the “N170”, for a negative deflection at 170 ms, can show up as a positive deflection for an iEEG electrode in the fusiform gyrus (Allison et al., 1994; Puce et al., 1999); the “P3”, for a positive deflection at 300 ms can show up as a negative deflection for an iEEG electrode (Smith et al., 1990)).
To appreciate the relativity of ERP polarity in iEEG, it is useful to compare ERPs obtained with a local reference (e.g., bipolar) to the ones obtained with a common monopolar reference. Applying a systematic strategy when it comes to computing a bipolar reference scheme helps to compare ERP between individuals (for instance, by always subtracting the most internal electrode in each pair, as routinely done in clinic, see section 3.3.2). Anatomy also brings complementary information about polarity, especially in the case of sEEG where it indicates on which side of the gray matter an electrode is located (outermost or innermost layer) and at which distance. Last, in some cases the ERP polarity can be flipped to match the polarity of a known similar component recorded from the surface (see N2-P3 example in (Baudena et al., 1995; Halgren et al., 1995a, 1995b)).
DC offset removal, sometimes also referred to as baseline correction, is a generalized practice to center the ERP around zero, by subtracting the mean activity of the baseline from the entire epoch of interest (e.g. using pre-stimulus period). As for scalp EEG, we recommend including an integer number of cycles of the prevalent frequency (e.g., with a predominant frequency at 10 Hz, the length of the baseline period would be an integer multiple times 100 ms). When possible, the baseline should be chosen to be free of any cognitive process of interest for the study (see the last paragraph of section 4.2.3.3). If a prestimulus effect is suspected (e.g., due to attention fluctuation), averaging across the baseline period of all trials and then subtracting this trial-averaged baseline may weaken spurious effects. That is, it avoids prestimulus effects in a given trial being subtracted from the post-stimulus period of that same trial, which leads to ‘finding’ the effect during the post-stimulus period while it is actually in the prestimulus period. Alternatively, when the timing of such processes are unknown, or possibly encompasses the full trial length, the baseline might include the entire epoch or might be extracted from another part of the recordings, thereby avoiding a bias towards the activity at specific latencies.
When channels are compared (e.g., statistics between participants, see section 5), and as SNR can highly vary across iEEG electrodes (see section 3.2.4), a normalization within each channel might be performed before comparing channels (e.g., computing a z-score per channel by removing the mean and dividing by the baseline variance).
The high SNR of iEEG makes it a sound method to analyze the relationships between a continuous stimulus (or stimuli presented in very short succession causing the ERP to overlap) and the corresponding continuous brain response (for instance, cross-correlation analyses (Mégevand et al., 2020) or spectro-temporal response functions (Zion Golumbic et al., 2013)). Analyses like the General Linear Model are a means to address potential bias introduced by signal drifts when the baseline period is included in the statistical analysis as a covariate (Alday, 2019).
4.1.3. Data visualization
Two conventions have co-existed for plotting human electrophysiological data: in most recent publications the positive voltage points upward, but there is another (older) convention, where the positive voltage points downward (Hari and Puce, 2017). We recommend that positive up be used, consistent with other recommendations (Hari and Puce, 2017; Pernet et al., 2020).
Comparing ERPs obtained with different reference schemes helps to understand their anatomical origin. While local and bipolar reference schemes increase salience of local activity across electrodes (e.g., by revealing polarity reversal), a monopolar low-variance montage provides a broader picture including the attenuation of the ERP with distance to the source and passive volume conduction (see Figure 12 panel B or (Bidet-Caulet et al., 2007) Figure 4).
Systematic depiction of variance estimate (e.g., mean absolute deviation) of an ERP is advised to instantly inform about single-trial distribution and/or to infer difference between conditions (see section 5.4).
The signal over multiple trials can also be represented using a two dimensional heat-map, where color-coded lines (time: x-axis) are stacked (trial: y-axis). These ERP-images (Delorme et al., 2015) show the consistency over trials (i.e. variance) and can reveal specific temporal organization patterns when they are sorted to a given feature (e.g., the response times, in (Flinker et al., 2015) Figure 3).
4.2. Frequency and time-frequency analysis
Most cognitive processes trigger a rich repertoire of frequency-specific neural patterns whose signatures range from well-defined and quasi-periodic patterns (e.g., alpha rhythms around 10 Hz) to responses extending over wider, yet limited, frequency ranges (e.g., in the high-frequency activity, above 50 Hz). The orchestration of neural activity in different frequency bands permits to organize local assemblies of neurons and to coordinate interactions between distant sites (Buzsáki et al., 2012; Varela et al., 2001). iEEG is an ideal technique to study this dynamical architecture at the millimeter resolution, though care needs to be taken to ensure that oscillations are truly present when performing time-frequency analyses for this purpose (Donoghue et al., 2021; Lopes da Silva, 2013). Readers should be aware that different methods and nomenclatures are common in literature for describing the spectral changes related to an event, including for example Event-Related Band Power analysis (ERBP), Event-Related Spectral Perturbations (ERSP), Event Related Synchronization and Desynchronization (ERS and ERD).
4.2.1. Background
A theoretical neuroscience foundation helps to make physiologically informed methodological choices that are next expressed in directing proper signal computation. Depending on the aim of the study, frequency analysis can be performed on block-design epochs or on event-related epochs.
When a paradigm is block-design, the continuous data is epoched on the basis of long blocks corresponding to experimental conditions. Then the frequency analysis is generally applied on these single epochs to get repetitions for statistical assessment. Last, single epoch frequency spectra are averaged to obtain the magnitude-power spectra representation of each block/condition.
When the precise timing of stimuli or cognitive events is known, the frequency analysis can be made time-resolved with respect to the events (i.e., event-related). To assess periodic activity in different frequency bands, the time-frequency decomposition can be performed either on the ERP (i.e., after trial averaging) or on the single trial (i.e., before averaging across trials). Applied to the ERP, the time-frequency transform can only show evoked activity that is phase-consistent across trials and precisely time-locked to the event (events used to align the single trials before computing the ERP). Applied at the single trial level, the time-frequency transform reveals in addition the induced activity, the timing and phase of which vary across trials (Tallon-Baudry and Bertrand, 1999). The difference between evoked and induced activity tends to be more pronounced in high frequencies as the faster an oscillation is, the less likely it is for the phase of the oscillation to be time-locked across observations. For instance, if a stimulus elicits a strong and systematic oscillatory response at 40 Hz in the iEEG signal, but the phase of that oscillation jitters across trials, averaging the signals across trials will cancel that component. In contrast, averaging the time-frequency magnitude across trials will reveal an increase in magnitude after the stimulus around 40 Hz (the induced response).
4.2.2. Methods and neurophysiological interpretation
For block-design data for which precise timing of events of interest and related activity is unknown, the frequency analysis is commonly conducted using Fast Fourier Transforms (FFT). When the timing of events is known and time-frequency analysis is performed, a variety of methods are available, among others sliding-window FFT, the combination of band-pass filtering and Hilbert transform, wavelet analysis, multi-tapering, or matching pursuit analysis, or the demodulated band transform (Bénar et al., 2009; Kovach and Gander, 2016; Mitra and Pesaran, 1999; Tallon-Baudry et al., 1997). Some time-frequency analyses might be more appropriate for specific questions regarding specific types of neural responses (e.g., the precise frequency extent of a short broadband response, the number of cycles of well-defined sustained oscillation). For a particular situation, choosing “the” most appropriate method requires good understanding of the method and what aspects of the signal it captures best. In most cases, the different methods lead to similar results (Bruns, 2004; Le Van Quyen et al., 2001), especially in iEEG, thanks to the high SNR. A comprehensive presentation of the different methods available is beyond the scope of this article, and we encourage the interested reader to consult publications dedicated to time-frequency analysis of electrophysiological signal (Cohen, 2014; Gross, 2014; Herrmann et al., 2014; Roach and Mathalon, 2008; Wacker and Witte, 2013) and to related guidelines (Gross et al., 2013; Keil et al., n.d.; Pernet et al., 2020). Here, we introduce some key concepts for understanding different neurophysiological scenarios that are characterized more easily with iEEG (due to its spatial specificity) than with scalp EEG which is blurred by volume conduction.
Briefly, any frequency transform decomposes the original signal into a set of sinusoidal functions that are often taken to imply oscillatory signals. For each frequency (and each time-point in the case of time-frequency analysis), the output is a complex number from which two elemental measures are derived: the magnitude (sometimes referred as amplitude, we here prefer the term magnitude which implies an unsigned number that is the norm of a vector) and the phase (the shift of the oscillation along the time-axis). For a given data-point, the depiction of the complex number in the polar coordinate system provides an intuitive view: the magnitude is the norm of the vector, the phase is its angle from the origin (abscissa axis):
The magnitude represents the norm of the vector, i.e., the absolute value, whereas power refers to the norm squared, hence magnitude and power have a monotonic but non-linear relationship. The magnitude or power averaged across trials with respect to an event will show a modulation due to an induced (if it is jittered in time across trials) or evoked (if it occurs in-phase across trials) effect.
The phase indicates where the signal stands in its cycle with respect to its oscillatory estimate. Phase averaging across trials is generally performed by taking the norm of the normalized complex-vectors averaged across trials (i.e., magnitude is set at 1). In particular, the phase consistency across trials indicates if the phase of ongoing oscillations is reset by the same event across trials. This index is normed between 0 and 1, follows a right skew distribution and is generally named: Inter-trial coherence (ITC), Phase-Locking-Factor (PLF) or inter-trial phase clustering (ITPC).
Thus, the use of time-frequency analysis allows to draw four possible analytic scenarios following the occurrence of an event (Makeig et al., 2004): (a) a change in magnitude co-occurs with a phase reset; (b) a change in magnitude but no phase-reset (a.k.a. pure induced activity); (c) no change in magnitude but a phase-reset (a.k.a. pure phase-reset); (d) no change in magnitude and no phase-reset. The link between ERP, power modulation and phase resetting is still an ongoing debate (Klimesch et al., 2007; Lopes da Silva, 2006; Makeig et al., 2004; Sauseng et al., 2007; Shah et al., 2004) and some measures have been elaborated to investigate these questions (such as the phase-preservation index which evaluates the pre-to-post event phase relationship, see (Mazaheri and Jensen, 2006)). Critically, these scenarios are hard to dissociate using surface recordings because the passive spread of the signal mixes magnitude change and phase modulation across electrodes. The spatial specificity and SNR of iEEG permits a finer characterization of magnitude and phase modulation(s) caused by an event. For example, (Mégevand et al., 2020; Mercier et al., 2013) show pure phase reset as a mechanism for cross-modal modulation in the sensory cortex.
4.2.3. Challenges, recommendations and reporting advice
4.2.3.1. Distinguishing oscillations from activity in a canonical band
Time-frequency analyses allow activity at neighboring frequencies to be visualized and thereby are informative for distinguishing between general spectral magnitude or power changes in fixed canonical EEG bands versus the presence of specific oscillations (i.e. narrow band activity of a periodic, rhythmic nature) (Donoghue et al., 2021; Lopes da Silva, 2013). There is a growing literature that posits that some oscillatory phenomena are non-stationary, infrequent, and bursty (Jones, 2016) and that non-oscillatory periods are dominated by what is referred to as “aperiodic” activity (Donoghue et al., 2020; Gerster et al., 2022) that is itself dynamically modulated by task states (Podvalny et al., 2017), is likely physiologically relevant (Gao et al., 2017) and can be related to behavior (Brookshire, 2021; Wolpert and Tallon-Baudry, 2021).
4.2.3.2. Examining magnitude change and phase modulation
Time-frequency decomposition provides both magnitude and phase as a function of time and frequency. To dissociate induced responses from evoked responses (i.e., same phase across trials), we recommend considering both in relation to each other by visualizing them side by side. Beyond a more complete characterization of the brain response, grasping the bigger picture permits to reveal mathematically or physiologically questionable scenarios. For instance, some baselining choices (e.g., time-period method) can result in a decrease in magnitude accompanied by a phase-reset at the same frequency. Such a result is difficult to interpret from a physiological point of view, as it would mean that, across trials, the magnitude of local neural activity decreases and at the same time becomes more synchronized. Also, the potential consequences of the experimental design on the baseline activity should be considered when assuming the baseline to be “neutral” relative to the active period of interest (see also the last paragraph of the next bullet point). More specifically, some unlikely scenarios can be due to the reference scheme. In the example above, the decrease in power and the phase-reset at the same frequency can originate from different channels, with their signal being mixed by the use of a specific reference scheme. This example underlies the interest of comparing results obtained with different reference montages.
4.2.3.3. Baselining signal magnitude
It is common to investigate frequency-specific increases or decreases in magnitude related to a given cognitive process. Measuring modulation over time usually involves the comparison of time-frequency magnitude or power when the process is active versus when it is not active, the latter being referred to as the “baseline” period. Once a proper baseline period has been defined, a number of approaches are commonly used, although there is no consensus. While baselining signal magnitude is not specific to iEEG, it is a cornerstone of time-frequency analysis that should be minutely considered. Various methods commonly used in such analysis are described here (see Table 1 and Figures 12 and 13).
Table 1.
Baselining methods used in time-frequency analysis. A can be read as “active” and stands for the time-frequency resolved magnitude or power in the period of interest. B stands for the baseline period. An overscore indicates taking the mean over the time in the baseline; the standard-deviation σ is also computed over the time in the baseline.
| Absolute baseline | Z-scoring baseline | Relative baseline | Log-transform baseline | Absolute-relative baseline | Normed baseline |
|---|---|---|---|---|---|
Figure 13.
Baseline correction and time frequency representation.
The same time-frequency representation of power is depicted with different baselining methods computed from −1 sec to −0.5 s. Within a frequency range (orange) and two time ranges of interest (purple and red), the mean and the mean-absolute-deviation are depicted at the bottom and on the side. The choice of the baseline method enhances/reduces the visibility of particular features in the data (for instance the 1/f effect, see sections 4.2.3.3 and 4.2.3.4.1).
Absolute baseline
This method consists in subtracting for each frequency the mean magnitude over the baseline from the period of interest; the result is therefore expressed in the original units (e.g., μV2). This approach assumes that the task-induced magnitude or power adds linearly to the baseline power and equally across frequencies. While this is not a problem per se, it should be kept in mind that, due to the 1/f fall-off, some task-induced absolute magnitude increases can be stronger at low-frequencies (see Figure 13). Notice that this approach leads to different outcomes when applied on magnitude or on power because of their nonlinear relationship (e.g., when the power doubles the magnitude increases by one unit, making change in higher frequencies more visible).
Z-scoring baseline
Here the absolute baseline is further divided by the standard deviation of the magnitude/power over the baseline period. The result corresponds to Z-values that can be conveniently interpreted as standard scores. Note here that the result depends on baseline variance (SNR) which might not be equal across frequencies. Thus, what can be seen as an equal increase in power at two frequencies can be due to a real increase with respect to baseline at one frequency, while being due to a lower variance in the baseline at the other frequency without genuine increase with respect to the baseline. Only in the case where the variance is proportional across frequencies, the z-scoring baseline compensates for the 1/f frequency fall-off (see section 4.2.3.4).
Relative baseline
This approach relies on a gain model, where the period of interest is divided by the baseline mean and the result is expressed in percentage. With this method, event-related increase and decrease with respect to baseline are not treated equally: after baselining data-values are positively skewed: while task-induced decrease range from 0 to 1 (underestimation), task-induced increase range from 1 to +infinite (overestimation).
Log-transform baseline
Another way to correct for the 1/f fall-off is to express the relative magnitude or power compared to the baseline in decibel (dB) using a log-transform. As for the previous approaches, the log-transform does not lead to the same outcomes when applied on magnitude or on power (due to their nonlinear relationship, see above).
Absolute-relative baseline (relative change)
This method involves first subtracting the mean baseline activity and then dividing by the mean baseline activity. This approach has the advantage of centering the magnitude before expressing it in percentage, which avoids the asymmetry introduced when the signal is not centered first (as with the relative baseline).
Normed baseline
This method involves first subtracting the mean baseline activity and then dividing by the sum of the magnitude or power at each data point and the mean baseline activity over the baseline period. This approach is similar to the absolute-relative baseline.
A remaining question is whether the baseline correction should be performed at the single trial level and/or on the average. That issue has been assessed in two studies, which showed that both should be combined to minimize the contribution of outliers (Grandchamp and Delorme, 2011; Hu et al., 2014).
When choosing the baseline period, the temporal leakage of the time-frequency decomposition has to be kept in mind, otherwise the risk is that post-onset activity contributes to the baseline estimate over the pre-onset period. Due to the length of the window used to estimate the time-frequency magnitude or power, the estimated power immediately before a stimulus can be affected by oscillatory processes occurring after that stimulus. To avoid any overlap between the baseline period and the window of interest, we recommend using a baseline period that ends before any post-onset leakage (e.g., three cycles at 8 Hz = 375 ms, activity at time 0 ms backward leaks to –187.5 ms, therefore the baseline should be taken before).
The notion of “baseline correcting” or “normalization” has different meanings depending on the background of the researcher and/or the experimental paradigm. Here, we would adopt the notion that the baseline can initially be considered as any other condition. A baseline may be seen as ‘neutral’ with regard to an ‘active’ condition, but not with regard to the state of the brain. The brain is never fully switched off in the baseline period: when the experimenter presents a stimulus, this adds input to an already active system and the experimenter measures the subsequent interplay between the ongoing activity and that event. The baseline state of the brain can therefore influence the processing of an incoming stimulus, memory performance, the making of a decision, the construct of a prediction or a motor response (Benedetto et al., 2020; Fiebelkorn and Kastner, 2019; Makeig et al., 2004; Mayer et al., 2016; Norman et al., 2017; Schroeder et al., 2010; VanRullen, 2016). It follows that the relationship between the response to an event and the baseline preceding it, is not as symmetric as an ‘active’ versus a ‘passive’ period. We recommend bearing in mind this symmetry (i.e., the baseline being another condition) or asymmetry (i.e., the baseline having a causal effect on the processing of an event), especially as most of the approaches described above are asymmetric, except the absolute baseline and the log-transform baseline. That is, inverting the baseline-condition and the condition-of-interest is in most cases not equivalent to flipping the colormap of the time-frequency representation.
4.2.3.4. 1/f drop-off and broadband analysis
The power spectral density of neural signals is roughly inversely proportional to the frequency and follows a 1/f decrease (see (Donoghue et al., 2020; He et al., 2010; Miller et al., 2009; Milstein et al., 2009; Samaha and Cohen, 2022; Zhigalov et al., 2017, 2015). As different frequency bands contain signatures of different neural processes, some analyses focus on a single or a limited number of discrete frequency bands, rather than the entire spectrum. The choice can be driven by canonical frequencies from the literature, or be data-driven (e.g., by identifying individual frequencies where the power peaks or where the shape exceeds the 1/f spectrum). For instance, high-frequency activity (e.g., 50 Hz to 150 Hz) is often considered as a proxy of population-level local spiking activity (Le Van Quyen et al., 2010; Mukamel et al., 2005; Ray et al., 2008). Yet, broadband high-frequency activity and multi-unit activity index different features across layers (Leszczyński et al., 2020), furthermore broadband high-frequency can overlap with narrow-band oscillations (Arnulfo et al., 2020; Hermes et al., 2015). In the example of high-frequency activity, the result of an analysis conducted over the whole high frequencies range at once, is dominated by the lowest frequencies. Disentangling broadband, 1/f, signal changes can be difficult. The phenomenon can be obscured at high frequencies by the noise floor introduced by the amplifier system, and at low frequencies by coexistent oscillations (Miller et al., 2014).
When analyzing a wide range of frequencies, we recommend giving a similar weight to high and low-frequencies e.g., by normalizing each frequency before processing or averaging them or by “whitening” the PSD (as an example see (Groppe et al., 2013; Roehri et al., 2021; Vidal et al., 2010). Note that simply filtering the signal to perform a broadband analysis is not optimal; the characteristic of the signal is better described with a dedicated (time-)frequency analysis. Last, when a broadband analysis is conducted, we recommend further analyzing neighboring frequencies to assess the frequency specificity of the studied neural process. As a complement to methods that correct or adjust for the 1/f drop-off, there are several emerging approaches for explicitly modeling and parameterizing it. Several of these approaches—such as BOSC (Hughes et al., 2012), eBOSC (Kosciessa et al., 2020), and IRASA (Wen and Liu, 2016)—focus on modeling the 1/f drop-off in order to separate it from oscillations in order to better analyze oscillatory activity. Other approaches, such as spectral parameterization (Donoghue et al., 2020) conceive of the 1/f-like activity as a physiologically relevant signal that should be explicitly parameterized into its total broadband power, spectral exponent or slope, and a “knee” frequency, the latter of which describes the point where the 1/f drop-off occurs (Miller et al., 2009). This approach defines the 1/f-like activity as an aperiodic signal that arises from the total postsynaptic and transmembrane currents that underlie the local field potential and iEEG signal (Buzsáki et al., 2012). In this view, the total broadband power is related to total synaptic inputs, as highlighted above, while the power spectral exponent and knee arise as a natural consequence of the fact that the local currents exponentially rise and decay in time (Manning et al., 2009; Miller, 2010). This double-exponential in the time domain manifests as a knee and exponential decay in the frequency domain; as the relative contributions of e.g., excitatory and inhibitory currents into a region change, so too does the knee frequency and spectral exponent (Gao et al., 2020, 2017).
4.2.3.5. Phase across frequencies
The phase of a wide frequency band has little physiological meaning, as it mixes-up phases from oscillations at different frequencies. Thus, we recommend avoiding averaging phase across frequencies.
4.2.3.6. Phase quantification
Even if a phase estimate relies on good SNR, like with iEEG, the quantification of phase consistency across trials, or across time, is biased by the number of samples (i.e., the consistency is overestimated for too few trials). Sample size bias is inherent to the computation of any metric reflecting the magnitude of a vector like the Phase Concentration Index (Vinck et al., 2010). Such a metric always has non-zero positive values and is greater for a low number of observations. To circumvent this issue, an option is to compute every possible phase angle difference between all the trials, which leads to a distribution of pairwise circular differences next evaluated for non-uniformity around the circle (as proposed in (Gulbinaite et al., 2017) from the Pairwise Phase Consistency (Vinck et al., 2010)). Practically, if the phase angles of all single trials are similar, the distribution of every combination of paired-trial phase differences should not be random but rather tends towards zero degree. Thus, this combinatorial approach is making the most of single trials to be less subjected to small sample bias. When comparing the phase consistency across trials between conditions, equating the number of trials for the two conditions is a more simple way to elude the sample size bias. Nonetheless this must be a prerequisite when comparing conditions.
4.2.3.7. Influence of the reference on phase angle estimate
An ambiguity in the phase estimate comes from the reference as the phase angle of a signal is defined relative to the signal at the reference (i.e., 0 degree, see (Shirhatti et al., 2016)). With EEG, the use of the same reference montage across participants favors the correspondence of the phase angle estimate within the population. With iEEG, even if the same reference montage is used across participants, the peculiarity of electrode locations makes the phase angle hardly comparable between participants. Moreover, at the individual level, the phase angle estimate changes when the reference montage is changed (see comment on ERP phase reversal in section 4.1.2, and Figure 12). Changing the reference montage shifts the angle of the phase estimate, and can introduce some noise in the estimate (e.g., if the signal at the reference is artifactual or modulated by an experimental factor). Thus, as the angle is arbitrary, analysis should focus on phase angle relationships across trials or across time. We also recommend performing the time-frequency decomposition for phase analysis twice: once with a local reference scheme and once with a monopolar reference, to assert if the electrode(s) used as reference is/are ‘neutral’ phase wise or, in contrast, if it/they show(s) some phase angle consistency (see panel E in Figure 12).
4.2.3.8. The reference and the estimate of signal magnitude/power
Signal magnitude is impacted by the reference montage, as it represents the magnitude difference between a given electrode and the reference electrode(s) in the frequency domain (Shirhatti et al., 2016). Also, it is critical to make sure that the signal(s) used as reference is neither artifacted or modulated by experimental factor(s), which would then be mixed with the signal(s) of interest. We recommend looking at the same dataset with different reference montages to verify that the electrode(s) used as reference do(es) not contaminate the signal, enabling one to understand the spatial features of the referenced signal(s) (see Figure 12).
4.2.4. Data visualization
When plotting color-coded data, such as time-frequency responses, the choice of the colormap can lead to misleading effects. A colormap with a non-isoluminance/hue gradient (e.g., jet/rainbow colormap) can introduce illusory boundaries that are not present when the same data is plotted using a black and white colormap (Cooper et al., 2021). We recommend choosing an isoluminant color map (e.g., ‘batlow’) to avoid result misinterpretation (for examples and an in-depth discussion see (Crameri et al., 2020)).
When visually comparing a time-frequency response across frequencies, it is necessary to correct for the 1/f fall-off; otherwise the greatest modulations measured in the low frequencies bury the more modest modulations measured in the higher frequencies (see section 4.2.3.4). Frequency specific re-scaling requires baseline correction and we recommend the absolute relative or z-score approach to avoid a bias in the visualization (see previous section and Figure 13).
The visualization of a time-frequency response that is averaged over multiple trials does not provide variance estimates across the trials. As all display dimensions are already used (i.e., x-axis for time, y-axis for frequencies and color for the magnitude or power or phase consistency), one solution is to plot the mean magnitude or power (or phase consistency) and its variance, computed per frequency over time, on the side of the time-frequency map, for the baseline and for the active period (see Figure 13). When focusing on a specific frequency band, a 2D heat map depicts inter-trial variance over time either for the magnitude or the phase, like an ERP-image (see section 4.1.3). That is, trials are stacked above each other and eventually sorted by a variable of interest (e.g., a behavioral variable, see Figure 10 in Dubarry et al., 2022, or (Ossandón et al., 2012)). It supports a better understanding of single trial dynamics, for instance by revealing phasic or sustained response.
4.3. Connectivity analysis
4.3.0. Introduction
Following the connectome perspective, which aims at defining the structural connectivity underlying brain networks (Sporns et al., 2005), the dynome perspective proposes to investigate the dynamic interactions at play in brain networks (Kopell et al., 2014). This perspective posits a particular emphasis on neural oscillations and on relationships between the fine temporal structures of distributed neural activity (Sadaghiani et al., 2022; Siegel et al., 2012). Despite the limited and non-systematic coverage of iEEG, its high spatial and temporal resolution make it very suited to study such interactions. A large variety of connectivity measures have been proposed to detect long-range neural interactions, which are typically computed as bi-variate measures between pairs of iEEG channels. Connectivity measures can be split into two main classes: functional connectivity measures that are correlative and directed/effective connectivity measures that model causal relationships. Within each class, different measures applied to the same data can yield very different connectivity patterns (Strahnen et al., 2021; Wang et al., 2014). It is therefore important to understand which aspect of neural coupling is quantified by each measure (e.g., coupling between magnitude or phases). The coupling mechanisms considered are so diverse that so far no single connectivity measure has emerged as the tool of choice. A comprehensive review of different connectivity measures can be found in a number of dedicated didactic reviews (Bastos and Schoffelen, 2015; Greenblatt et al., 2012; O’Neill et al., 2018). The following section primarily introduces the main approaches used so far and some of the improvements to them that aim to resolve specific issues.
4.3.1. Functional connectivity
4.3.1.1. Background
One of the most intuitive connectivity measures is the correlation between amplitude envelopes of two signals (Bruns et al., 2000). This assesses the temporal correspondence in the magnitude fluctuations in a given frequency band in two electrodes. For instance, high-frequency activity (HFA) between 50 and 150 Hz is thought to represent a proxy of population-level spiking activity for neurons ((Buzsáki et al., 2012) and see section 4.2.3.4); therefore one can assume that two distant neural populations cooperating to process information should have HFA fluctuations that are correlated in time (Fries, 2005; Vidal et al., 2012).
As the excitability of neurons is modulated by sub-threshold fluctuations in cell-membrane potentials, the phase of ongoing oscillations can contribute to the exchange of information between distant groups of neurons (Fries, 2015; Varela et al., 2001). Rather than looking at magnitude (amplitude or power) correlations, the Phase-Locking Value (PLV) can be used to estimate phase synchronization. The PLV quantifies across trials or time the phase angle difference between two signals for a given latency and frequency window. When computed across trials, PLV measures if the phase difference between two signals is consistent across trials with respect to the event that defines the trials (i.e, the output is a mean across trials per time point). When the PLV is computed over time, the metric assesses the stability over time of the phase difference between the two signals (i.e., the output is a mean over time per trial). PLV values range between 0 (for totally random) and 1 (for perfect phase-locking). It is the bivariate homologue of the univariate index of phase consistency (see section 4.2) (Lachaux et al., 1999; Tass et al., 1998).
The dichotomy between amplitude and phase illustrates the intricacy of connectivity analysis and the relevance to inscribe any investigation in a physiologically grounded theoretical framework supporting communication in brain networks. Amplitude coupling and phase coupling lead to different but nevertheless non-exclusive measures. Distant coupling mechanisms might cause oscillations to rise and decay at the same time in two neural populations, with or without precise phase relationships. This illustrates that the application of different connectivity measures can provide a more comprehensive view of the coupling mechanisms within brain networks (Engel et al., 2013).
Coherency and coherence provide a mixed measure of amplitude and phase correlations. Coherency and coherence are based on the standardized cross-spectrum of two complex signals across time or trials. When expressed as a complex number, it is usually named coherency; taking the magnitude of coherency leads to the coherence coefficient. Ranging from 0 to 1, it quantifies the correlation in the frequency domain, taking both amplitude and phase consistency into account. Thus, coherence appears similar to PLV modulo signal magnitude: coherence considers both amplitude and phase consistency, while PLV is only sensitive to phase consistency. However, as any phase estimate depends on the amplitudes of the sine and cosine components, the PLV does depend on signal magnitude and SNR. Given a certain amount of noise, signals with a larger magnitude will result in a better phase estimate, therefore higher PLV values (Bastos and Schoffelen, 2015). Weighting events by their respective magnitudes can obscure possibly meaningful low amplitude events containing weak but genuine phase synchronization (see modeling study by (Hurtado et al., 2004)).
When getting started with connectivity analysis we recommend using either magnitude correlation, as it gives a straightforward and intuitive measure especially when analyzing HFA activity, or coherence for broader analysis scopes and when exploratory analysis is conducted. Once these initial measures have resulted in more explicit expectations or hypotheses of connectivity, other measures can be used to further characterize the underlying magnitude or phase related physiological mechanisms and address potential biases.
4.3.1.2. Advanced methods to work out potential biases
There are various aspects of the data and of the analysis of underlying activity that can cause spurious or biased estimates of basic connectivity measures. Additional methods have been developed to address these issues.
Sample size bias
Functional connectivity estimates (just like the phase consistency measure, see section 4.2.2) that are based on the magnitude of an averaged complex vector are dependent on the sample size even with high SNR (i.e., the number of vectors/trials that are averaged). The Pairwise Phase Consistency (PPC) index addresses this issue by computing the distribution of all pairwise phase angles differences, and tests for non-uniformity (Vinck et al., 2010). This measure does not test for consistency over all the trials (like the PLV) but across each possible pair of trials. By exploiting the combinatorial data over trials, PPC is more sensitive to true positives, less sensitive to false negatives and turns out to be equivalent to the squared PLV. PPC is therefore recommended to control for sample size bias. When the research question is to compare connectivity estimates across conditions, the sample size bias can also be addressed by stratifying the data (i.e., to equate the number of trials in the conditions). With this approach, if the number of trials is low, the estimate (i.e., the mean vector) for the two conditions should be equally inflated.
The common input problem
One common criticism is that most connectivity measures fail to distinguish direct and indirect interactions between neural populations (i.e., two populations driven by a common input would also be highly correlated if they are not directly connected). The partial coherence was proposed to remove the contribution of putative common inputs (Rosenberg et al., 1998). However, partial coherence requires data from potential common inputs. In the case of spatially sparsely sampled iEEG, those common sources may lie far from any electrodes. Yet, it can be used in some instances to estimate whether an increase in connectivity is due to a common artifact picked up by several electrodes. There the use of a ‘neutral’ electrode (i.e., with respect to the studied cognitive process) capturing the common artifact permits testing if the connectivity measure between two ‘active’ electrodes is due to the common artifact.
The passive volume conduction problem
Another source of spurious correlation between signals is volume conduction when two recording electrodes capture, with no delay, a common biophysical activity. Compared to M/EEG, iEEG is less prone to such effects due to the focal specificity of the implanted electrodes, especially when a local reference montage is used (see section 3.3). It is worth mentioning that several connectivity measures have been designed to reduce the effect of volume conduction. These are based on the notion that a physiological coupling mechanism between distant sources should impose a delay in information transmission, and therefore a time lag between correlated signals, whereas volume conduction is instantaneous. One approach is to only consider the imaginary part of Coherency (Nolte et al., 2004) and discard contributions along the real axis; this removes zero (in-phase) and anti-phase contributions to the signal. Similarly, the Phase Lag Index (PLI) measures the asymmetry, relative to the real axis, of the distribution of phase angles differences (Stam et al., 2007). PLI was further extended as the Weighted PLI (Cohen, 2015; Vinck et al., 2011), where the sign of the angle difference is scaled by the magnitude of the imaginary component (i.e., the distance from the real axis,). A similar approach was developed for power envelopes by orthogonalizing signals, that is to eliminate instantaneous correlation (Brookes et al., 2010; Hipp et al., 2012). In practice, these connectivity measures are recommended to characterize the immediacy/delay of connectivity (i.e., phase synchronization or magnitude correlation). If the connectivity result is replicated with these methods, the finding is strengthened and the lag can be reported (see next paragraph). Otherwise, the immediacy of the connectivity must be reported and/or tests should be performed to find the origin of a common source (e.g., common reference or artifact, see previous paragraph and section 4.3.3).
The temporal precedence problem
When considering two sinusoidal signals with a relative time shift between them, it is non-trivial to determine which of two signals leads or lags. If the two signals with a constant time delay are not merely constant oscillations but are represented in a broader frequency range, the phase difference between the two signals will increase linearly with the frequency of the oscillations. Building upon this, the Phase Slope Index quantifies the phase difference over a range of frequencies and provides an interpretation of temporal delays (Nolte et al., 2008). As for the correlation between amplitude envelopes, a straightforward extension is the cross-correlation measure, which further estimates the lag between the two correlated signals (Adhikari et al., 2010). We recommend these approaches to examine connectivity time-delay. Moreover, they purposefully back-up data visualization such as rose plots in the case of phase synchronization or cross-correlogram in the case of magnitude cross-correlation. While there are promising decomposition techniques aimed at disentangling overlapping oscillations (Nikulin et al., 2011) that have recently been adapted for iEEG (Schaworonkow and Voytek, 2021), great care still needs to be taken when interpreting such results.
4.3.2. Directed connectivity
Given that there is a correlation between the iEEG signals at distant sites, one can proceed to determine whether one of the neural sources drives or causes the other. A time difference in functional connectivity by itself is not sufficient to conclude statistical causation and is often mistakenly interpreted as a causal relationship (Friston, 2011). While an exhaustive review of directed/effective connectivity methods is beyond the scope of this paper, we mention the most common approaches used in electrophysiology. Model-based directed connectivity includes measures of Wiener-Granger causality (Bressler and Seth, 2011; Oya et al., 2007) or directed transfer function (Kamiński and Blinowska, 1991; Korzeniewska et al., 2008). Model-free methods derived from information theory (e.g., Mutual Information (Ince et al., 2017; Kraskov et al., 2004) include transfer entropy (Schreiber, 2000), Phase transfer entropy (Lobier et al., 2014) or Feature-specific Information transfer (Bím et al., 2019)). As the common input problem also applies to causal measures, partial approaches were developed to disambiguate the role of auxiliary node(s) in the assessment of the causal link between the two nodes of interest. These methodologies include the partial directed coherence (Baccalá and Sameshima, 2001), partial/conditional Granger causality (Ding et al., 2006; Guo et al., 2008; Wen et al., 2013), or the partial information decomposition (Wibral et al., 2017). While these methods have been less exploited in iEEG research due to the sparse spatial sampling and only partial coverage of the brain, multivariate autoregressive models include and extend the bivariate model, thus allowing causal inference at the full network scale (Kuś et al., 2004). Compared to functional connectivity analysis, effective connectivity analysis leads to more complete models of the large-scale cognitive networks, with detailed causal relationships and information spreads between brain areas.
We advise iEEG researchers to test the validity of the methods on simulated data prior to their application on acquired datasets. We also recommend comparing the differences between methods’ outputs and interpret the results in light of the literature related to the structural organization of the network of interest (including its hierarchical organization). Especially, the benefit remains to be explicit for hypothesis driven research and sometimes more risky for exploratory research.
4.3.3. Challenges, recommendations and reporting advice
The referencing scheme has a major impact on connectivity measures. A common reference adds a common component to all signals and therefore can artificially increase connectivity measures (Bastos and Schoffelen, 2015; Fein et al., 1988; Guevara et al., 2005). This also applies when connectivity is computed between two channels that share the same reference, for example with a bipolar reference scheme for three sEEG electrodes resulting in two neighboring channels. We recommend that each channel included in the connectivity estimation should be referenced to a different reference. When this is not possible, a less ideal solution is to contrast connectivity measures obtained in two different experimental conditions (see for example (Fell et al., 2001; Sehatpour et al., 2008)), or before and after an event of interest. In that case, the artificial inflation of connectivity is included in the comparison. Note that this solution assumes that the common component increases the connectivity measure similarly in the two conditions, or before and after the event of interest. As this assumption cannot be tested, results should be interpreted with caution and this potential pitfall of the analysis should be stated in the reporting.
Stationarity is one of the assumptions underlying the computation of connectivity over time or trials. When it is plausible that some jitter introduces variability, it can be critical to temporally re-align the data to the feature at the origin of the jitter. For instance, if an increase of phase synchronization is due to motor preparation, aligning the trials to the response times makes the phase synchronization visible, which otherwise may be absent if the trials are aligned to the stimulus onset (e.g., (Mercier et al., 2015)).
Both magnitude-based and phase-based connectivity measures are affected by SNR. In practice, the connectivity measure between two signals decreases when one of them is contaminated by strong noise, but increases if both signals are contaminated by the same noise source. Typically, we recommend comparing connectivity measures between conditions with similar SNR. One way to estimate SNR is to compute a variance estimate for each signal (see section 4.1.2).
Similarly, connectivity measures are dependent on the number of trials used in the computation. For instance, if there are fewer trials in one condition, the connectivity estimate is likely more biased as compared to another condition with more trials. Similar to phase quantification (see section 4.2.3.6), here we recommend using the same amount of trials to compute the connectivity measure for two conditions that are to be compared. This is easily done by randomly drawing a subsection of trials, based on the lowest number of trials between conditions.
Removing instantaneous interaction to circumvent passive volume conduction discards genuine in-phase or anti-phase interactions (i.e., at an angle of 0 or 180 degrees). To assess instantaneous interaction some controls exist, such as comparing between conditions (which assumes that passive volume conduction is identical between conditions), cross frequency comparisons as passive volume conduction is not entirely frequency specific (i.e., if an effect is not frequency specific it can be due to broadband passive volume conduction), or the use of a partial connectivity measure.
When applying directed connectivity measure, special consideration should be taken regarding (pre)processing steps (e.g., artifact removal, re-referencing or filtering) as they can lead to causal artifacts by introducing temporal correlation or disrupting time series (see for further details (Florin et al., 2010; Seth, 2010)).
Some causal inference often hypothesizes ideal unidirectional interactions while many network connections are bidirectional. Also, directed connectivity measurement should be performed in both directions to be able to assess cross-talk between signals.
4.3.4. Data visualization
The result of a complete bivariate connectivity analysis is a graph, in the sense of a mathematical structure used to model pairwise relations (i.e. graph theory approach). With bivariate connectivity measure, such a graph quantifies the strength of the connections (“edges”) between iEEG channels (“nodes”) used to compute the connectivity measure. Where functional connectivity provides only a global measure (i.e., non-directional) for each pair of nodes, effective connectivity also provides a directionality for those connections. Graph theoretical measures can subsequently be used to characterize properties of brain interaction architecture at the network level (Bassett and Sporns, 2017; Bullmore and Sporns, 2012; Sporns and Betzel, 2016). From a visualization perspective, rather than depicting connectivity measure results as “a spaghetti plot”, graph theory is an efficient way to sum up connectivity results. For instance, graph theory measures allow defining local/central hubs, path redundancies or time-dependent organization. One obvious limitation of iEEG is the sparse spatial sampling in single individuals. A global understanding of brain-level networks can therefore only be achieved by combining the connectivity measure graphs over a large number of participants (see as an example (Betzel et al., 2019)). Yet, another constraint relates to the localization of the electrodes. Either, because the granularity of some parcel(s) may exceed the spatial specificity of iEEG, (i.e., channels with different response profiles are projected and grouped into a wide parcel); or because some electrodes can be surrounded by different tissues which obscure spatial specificity (e.g. white matter). As such, limitations should be acknowledged when drawing a network-graph and when computing some graph metrics.
4.3.5. Statistical consideration
Considering the rich repertoire of methods to compute functional and effective connectivity, one obvious pitfall is to “try them all” until one provides “interesting” results that are consistent with the ongoing working hypothesis. Good research practices dictate to also report all negative results, as (i) the multiplication of attempts was part of the performed research, (ii) negative results provide insights about the type of connectivity measure supporting the studied cognitive process, and (iii) repetition of assessments has to be taken into account in the statistics (i.e multiple comparison problem).
4.4. Cross-frequency coupling analysis
4.4.1. Background
Because iEEG offers both high temporal and spatial specificity, advanced iEEG analysis faces a multiscale challenge that is to integrate neural activity across different temporal and/or spatial scales (Engel et al., 2013). For instance, many iEEG studies analyze and report separately task-related effects in different frequency bands (theta, alpha, beta, gamma), which might (mis)leadingly indicate that the different frequency components of iEEG signals are categorical and functionally independent from each other. Cross-frequency coupling (CFC) was developed to assess relationships between frequency bands, either locally or over some distance (i.e., CFC computed between electrodes). It offers a methodological framework that can bridge local computation and long-range functional connectivity. To assess the local hierarchy across frequency bands or the efficacy of communication on the modulation of local neural activity (Hyafil et al., 2015), four main CFC sketches were proposed (Jensen and Colgin, 2007): power to power, phase to phase (Palva et al., Siegel et al), phase to frequency (Colgin et al., 2009) and phase to amplitude (Canolty et al., 2006). Among those, Phase Amplitude Coupling (PAC) has attracted attention due to a plausible physiological mechanism with the phase of low frequencies reflecting neural excitability and the magnitude or power of high frequencies reflecting local neural activity (Canolty and Knight, 2010; Jacobs et al., 2007; Rutishauser et al., 2010). PAC has been proposed to represent a hierarchy across oscillatory activity (Lakatos et al., 2005), leading to compelling neuronal computation such as phase coding (Lisman, 2005). In this framework, the dynamic of interaction between frequencies represents how oscillations emerge from the intrinsic local activity or are imposed by another network. Altogether CFC can be computed at an electrode as well as between electrodes, making CFC a connectivity measure integrating time and space. Several methods exist to compute CFC that have been reviewed and compared in instructive publications (Canolty et al., 2012; Michael X. Cohen, 2017; Dvorak and Fenton, 2014; Miller et al., 2012; Penny et al., 2008; Tort et al., 2010).
4.4.2. Challenges, recommendations and reporting advice
The estimation and correct interpretation of cross-frequency coupling faces multiple methodological challenges and requires thorough controls in the analysis (Aru et al., 2015). For instance, nonstationarities in the signal, due to spectral correlations (i.e., harmonics), introduce spurious cross-frequencies coupling (Lozano-Soldevilla et al., 2016). This can be due to the non-sinusoidal morphology of the periodic signals due to sharp signal edges or asymmetrical waveforms, or due to filtering (Kramer and Eden, 2013; Scheffer-Teixeira and Tort, 2016). There is growing evidence that these non-sinusoidal morphologies are physiological (Cole and Voytek, 2017; Sherman et al., 2016) and are altered in disease states, such as Parkinson’s disease, which can manifest as apparent CFC (Cole et al., 2017). Because of this, it is recommended that CFC analyses be complemented by time-domain approaches that can quantify these non-sinusoidal morphologies (Cole and Voytek, 2019) and to help better identify multi-rhythm CFC from apparent CFC (Vaz et al., 2017).
5 -. Statistics
5.0. Introduction
When interpreting iEEG signal analysis and/or testing the significance of a research hypothesis, most statistical methods are shared with M/EEG. However, iEEG is remarkable for its (i) high SNR and (ii) sparse and variable spatial sampling across participants, which leads to a number of particularities in the statistical approach that we address in this chapter. First, the combination of the high SNR and high spatial specificity substantiate iEEG observation(s) established in a single participant and value the publication of single case studies (as compared with single case M/EEG study conducted in a healthy participant). Second, when a group of participants is considered to generalize results, the sparse and variable sampling requires defining some equivalence between recording sites, which may or not be defined a priori (i.e., hypothesis driven region of interest (ROI)). This equivalence relies on the electrode localization and related anatomical/spatial normalization processes (see section 2).
No consensus has emerged within the iEEG community regarding a standard statistical procedure, yet from the various practices described in the literature, two issues stand out that we will address: the type of research conducted (hypothesis driven vs. exploratory) and the number of participants. In the following, prior to moving into the specific aspects of iEEG, we first provide a general reflection on how the type of research relates to the particularities of statistics in iEEG. Second, from the literature we review the spectrum of approaches, we posit the challenge of generalizing findings from one or multiple iEEG participants to the population, and stress the importance of relating group analysis to individual analysis. Finally, we present concrete ways to combine electrodes over multiple participants.
5.1. Exploratory versus hypothesis-driven research
Dictated by the clinical dimension of iEEG (epilepsy prevalence, surgical planning, selection of participants...), the sparse spatial sampling and multiple comparison problem often motivates researchers to define ROIs, as is typical in fMRI research. Only electrodes placed in the chosen area(s) are investigated and participants without electrodes in these ROIs are excluded. Conversely, other investigations are conducted on the whole brain without any prior regions of interest; in these cases all electrodes available in all participants are investigated. These practices reflect two general scientific research approaches: hypothesis-driven and exploratory (Figure 14). In this section, after elaborating on these concepts, we put them in perspective within the framework of iEEG research and related statistical considerations.
Figure 14.
Distinction and interaction between prediction, i.e., hypothesis based confirmatory research, and postdiction or exploratory research.
5.1.1. The types of research conducted
Scientific research is based on a fundamental dissociation between exploratory research (a.k.a postdiction) and hypothesis-driven research (a.k.a. prediction). Discoveries stem from data-driven exploratory research that makes few, if any, prior assumptions. The investigation aims to make observations, build a model from these observations, and generate new questions and hypotheses. Next, the hypotheses or models have to be empirically tested according to a dedicated protocol. Here comes the hypothesis-driven approach, where the investigation is planned prior to observing a newly acquired dataset. The hypothesis/model that was stated beforehand is validated (or not) based on the data, and can be accepted or (partially) rejected and modified (see Figure 14).
It is often the case that during a hypothesis-driven study, some exploration also happens, as the researcher might notice an interesting pattern that warrants investigation in a follow-up study. It is generally admitted that scientific knowledge builds on this recursive loop between prediction and postdiction (Nosek et al., 2018), but the distinction is not always explicitly made. The need for a critical distinction has recently attracted attention in the field of cognitive neuroscience, as mixing the two types of research lies at the roots of circular reasoning (Kriegeskorte et al., 2009), post-hoc theorizing (Kerr, 1998) and other cognitive biases that impact good research practices, reproducibility and scientific credibility.
Acknowledging the distinction between exploratory and hypothesis-driven research is important at the onset of any research project:
Within the multidisciplinary iEEG research team that contributes different perspectives to a project: When a research proposal is discussed, the research type (exploratory or hypothesis-driven) should be explicitly stated. “Work in progress” meetings are propitious to verify that the project remains aligned with the initial plan, or to identify that an initially hypothesis-driven project turns into or leads to an exploratory project.
Within the framework of preregistered study: In recent years, with the aim to tackle the replication crisis and to recognize the foundation of hypothesis-driven research, registered reports were implemented to distinguish a priori planned analysis from unplanned or post-hoc analyses. Among others fostered by the Open-Science Framework (see 17), the principle relies on depositing or pre-registering a manuscript that details the hypotheses, methods, and planned analysis of a study prior to it being conducted. Several cognitive neuroscience journals have implemented the format of preregistered articles: after review and in principle acceptance, the study protocol is registered as a manuscript and data can be collected; following analysis, the report with the results is submitted to the same journal and is subjected to a reviewing process that does not focus on the outcomes but only on it adhering to the preregistration (for a guideline see (Paul et al., 2021)). This style of work is particularly suited for hypothesis-driven iEEG research, considering the time it can take to record data from a sufficiently large group of patients.
Because of the sparse spatial sampling combined with the high specificity and the high sensitivity, iEEG data is more prone to be explored at the individual participant level (c.f., the seminal work by (Penfield and Rasmussen, 1950)). Even if only a few electrodes are localized consistently in a handful of iEEG participants, their investigation can nevertheless result in serendipitous findings. In contrast to iEEG, M/EEG data from a single research participant often has a SNR that is too low to draw clear inferences and consequently M/EEG data is commonly acquired for a group of participants in well-controlled hypothesis-driven research.
At the larger scale, science advances through the alternation of exploratory and hypothesis-driven research; something that can be exploited within a single study using a cross-validation procedure that dissociates the two research types. Using a first (pilot) dataset that is already acquired, its exploratory analysis provides a hypothesis that is subsequently tested on a second dataset that remains to be recorded, or that is recorded but not yet analyzed (here the pilot dataset is not to be included in the subsequent analysis). Analyzing pilot data prior to the acquisition of the remaining dataset allows some time to get a similar electrode placement for the remaining dataset. As such, the reported choice of predefined ROI prevents circular analysis. The second dataset validates the hypothesis generated by the exploration of the first dataset. This procedure can be adapted when the dataset for an entire group is already available by simply splitting participants into an exploratory and validation group. Then, data from the validation cohort is held out from the initial analysis (performed on the exploratory cohort) and is used solely to validate the hypothesis/model.
This dichotomy, or perspective, is fundamental considering the general context of iEEG research, which is prone to selection bias (see sections 1.1.5 and 1.2.3). The limited recording time with the participant and the richness of the data often motivate the use of anatomical and/or functional criteria to select participants and/or electrodes. Typically, this choice can stem from the anatomical location of the electrodes and/or the working hypothesis guided by independent data collected from the same patients (e.g., fMRI performed prior to the implantation) or by the literature (e.g., (Lachaux et al., 2006) where ROIs are defined based on independent MRI studies). The decision to record a given protocol in a specific patient can also be motivated by the functional response to a ‘localizer’ (see section 1.1.4). In the case of complex analysis leading to a large amount of data (e.g., directed connectivity), a subset of electrodes can be selected on the basis of their signal features determined beforehand (e.g., by contrasting conditions, by using parametric regression or by using an informed spatial filter). In any case, it is a good research practice to document the choice of ROI to avoid a selection bias in the interpretation of the results. Conversely, if a protocol is run after checking that there was some response to the paradigm, the absence of acknowledgment in the recruitment would lead to circular reasoning: results are seen because it was known that they were there.
5.1.2. Descriptive statistics and inferential statistics
Although not systematic in practice, the two types of research are commonly associated with different statistical approaches: exploratory research mainly relies on descriptive statistics, and hypothesis-driven research requires inferential statistics. Descriptive statistics summarize quantitative measures of the data, such as the central tendency (e.g., mean or median) along with measures of dispersion (e.g., variance, median absolute deviation, quantiles). As in M/EEG, the descriptive metrics are valuable to data visualization, whereas inferential analysis is commonly based on null-hypothesis significance testing (Pernet, 2016) or decision-making, where the p-value cannot exist without a hypothesis. Some hypotheses are phrased prior to looking at the data, some after looking at the data. In the latter case, the researcher is not controlling the potential errors in the inferential decision.
Although the focus is often on the resulting p-value and the binary decision of significance, it is a good research practice to further provide the magnitude of the effect (i.e., effect size) and its precision (i.e., confidence interval) (Groppe, 2017). These measures are complementary to significance testing, as they set the weight on a quantitative dimension of the inference: how large (effect size) and how certain (interval estimate) it is. In addition to bringing a different view to interpreting the data, they permit better inference: by avoiding over confident claims from inadequate samples and improving comparisons of results (Calin-Jageman and Cumming, 2019). Last, the effect size estimate grounds cumulative evidence and permits meta-analysis. For these reasons, computing and reporting effect size and confidence intervals are now being adopted in scientific journals (Bernard, 2019).
5.1.3. Challenges, recommendations and reporting advice
We recommend documenting the progress of the iEEG project to keep track of its evolution and track the changes from the original plan. Starting with the rationale of the conducted research and the anticipated analysis/result; then the recording of participants and eventual selection; the analysis conducted with the unanticipated adaptation such as new ROIs; all the results found, even the unexpected ones. This strategy helps to distinguish when some part of the project departs (e.g., from hypothesis-driven to exploratory analysis) or when some new information comes into play and influences decisions (e.g., feed-back from a colleague regarding another experiment run on the same participants).
Papers reporting the results of iEEG studies should explicitly state in their introduction which a priori hypotheses will be tested (if any), whether post-hoc hypotheses will be tested, and whether results from exploratory analysis will be reported.
In ROI studies, sites outside the ROI might implicitly be considered as not showing the effect under investigation. Whether this conclusion can be drawn depends on whether those other regions were actually tested. To avoid an incorrect interpretation of the reported results, we recommend explicitly stating whether sites not included in the ROIs were tested and, in the case they were tested, to report the corresponding results (i.e., the non-ROI sites showing and not showing the effect of interest). While it could be argued that in hypothesis-driven ROI studies, sites outside the ROI relate to exploratory analysis, the use of a "negative-control" ROI allows showing that the effect is ROI-selective. To that aim, we recommend systematically selecting a "negative-control" ROI to verify the spatial specificity of the result(s) (i.e., by testing for a ROI x condition interaction).
Assessing a statistical difference between conditions must be done directly on the data from the two conditions, without corrections or normalizations that would reduce sensitivity or that would cause effects to appear differently. For instance, baseline correcting time-frequency analyses can hide differences present in the baseline window and make them appear in the post-stimulus window.
Assumption of statistical tests should be tested and reported (for guidance see (Wilcox and Rousselet, 2018)). For instance, parametric tests require verifying the type of distribution; in many cases they are not suitable for phase analysis (e.g., phase consistency across trials, due to the asymmetry and the bounds of the measure, see section 4.2.2 and 4.2.3).
When an iEEG study is published, we recommend sharing analysis code and making data available (conditional on the participants’ consent, see section 1.3.8). The reuse of data is valuable for replication purposes and for extending previously reported findings. It should be acknowledged that data sharing might come with some hidden risks: the multiplication of statistical tests and double dipping / circular reasoning. When the same data is re-analyzed, some tests are repeated without correction for multiple comparisons and with an inflated risk of false positives. Knowing what to look for can influence analysis processes in order to find again the initially postulated effect. To reduce these risks, we recommend in follow-up studies to take previous analyses into account.
5.2. The spectrum of practices
The literature of iEEG research describes a wide spectrum of practices, ranging from single case studies to investigations that include several tens of participants. Consequently, there are a myriad of statistical approaches. The scientific aims of a particular study but also the practical opportunities influence which path is chosen: either strongly conservative or more liberal. For instance, a clinically oriented study in a single patient going for surgical brain resection necessitates other risk/benefit considerations than a study that aims to generalize a correlation between cognition and brain activity to the healthy population. iEEG research practices can be considered from the perspective of quality (e.g., a single case is scrutinized or an investigation of a group with a very specific focus) or quantity (e.g., a big-data approach) (see (Tibon et al., 2022) for further discussion on small versus large scale studies and its implications for the interpretation of research findings).
From the perspective of an ideal neuroscientific or cognitive experimental design, the researcher would like to have a large group of participants (i.e., “N=many”) with largely consistent recordings, to be able to draw inferences that generalize to the population rather than being limited to the small sample. However, as the researcher is dealing with patients, it can also happen that recordings are only available in a single patient; these findings can still be shared as a single case (i.e., “N=1”) report. Finally, it can happen due to limited participant/patient availability, clinical or other experimental constraints that the sample size is larger than one, but not large enough to consider it a proper group study. In the following, we summarize the spectrum of practices (ordered by the number of participants) and underline their strengths and weaknesses.
5.2.1. Single case studies
The format of single case reports is historically linked to clinical practice, where the singularity of a medical case is reported to the community, pending replication. Single case reports in iEEG permit to provide, and/or to take into account, numerous peculiarities of the individual beyond its status of patient. It can be motivated by the rareness of electrodes placement and/or electrode type (e.g., prototype used in (Saleh et al., 2010)), the rareness of the patient’s specific skill set (e.g., a case study in a musician with iEEG electrodes (Martin et al., 2018)), the richness of the data available for the same participant (e.g., longitudinal study, PET, fMRI, electro-cortical stimulation, e.g., (Bruns and Eckhorn, 2004; Mégevand et al., 2014; Schippers et al., 2021)) or the wish to focus on the applicability/relevance of a series of methods applied on the human brain (Brovelli et al., 2005; Moses et al., 2021).
From the perspective of cognitive neuroscience studies, the analysis is performed on multiple trials and eventually validated through a test-retest procedure across two distinct datasets (for a discussion on inter-individual variability see (Jeffery et al., 2018)). While supported at the individual level by iEEG sensitivity (i.e., SNR), the main limitation of single case studies is the difficulty to generalize the result to the population and the significance of a single case study is reached when converging evidence from multiple patients (with various etiologies or foci) assure some generalisability. Yet, a thorough investigation of a single case permits to reach a high level of qualitative description, potentially leading to a “black swan” effect: an observation made for the first time can lead to a breakthrough. Therefore, a single case study must be well documented (i.e., including with meta-data), to permit both replication and the re-use of the data in future meta-analyses (see recommandation in section 1.3.8).
5.2.2. Multiple cases studies
To generalize findings from an individual and thus establish common features at the population level, the strategy of single case studies can be repeated over multiple participants across which the same effect(s) is (are) tested. Equivalence between participants’ recording sites is illustrated in every individual brain and/or determined in reference to a normalized space (e.g., MNI) or an atlas (see sections 2 and 5.3) and possibly supplemented with some recorded functional response (see sections 1.1.4 and 5.3.3). There are two statistical approaches commonly used: either the analysis is performed and reported at the level of single participants (i.e., trials being the repeated measures) and the results corroborated, or the trials are pooled across participants to create a ‘meta’ participant.
The first approach resembles most non-human primate (NHP) studies on old world monkeys, where often two individuals are used to cross-validate results from data recorded over many sessions within the same region in each individual. With human iEEG the number of participants tends to be larger, but with more variation in the placement of electrodes. The large number of sessions in NHP studies provide strong statistical power to individual data analyses. With human iEEG, time constraints do not allow recording a very large amount of data and consequently the inferential or descriptive meta-analyses performed over the individual results can differ from that of a NHP study. An option is to combine p-values across participants (Fisher, 1970) (Demerath, 1949) (Darlington and Hayes, 2000; Demerath, 1949; Fisher, 1970; Whitlock, 2005). The different methods have been compared (Bailey and Gribskov, 1998; Heard and Rubin-Delanchy, 2018; Loughin, 2004; Manolov and Solanas, 2012; Winkler et al., 2016; Won et al., 2009) to highlight their characteristics (e.g., sensitivity, more or less conservative) and the prerequisites of their use (e.g., independence between the combined p-values). This approach for multiple cases study presents the advantage of describing individuals as well as assessing the representativity of the findings across the participants.
In the second approach (i.e., the ‘meta’ participant), the trials from all participants are pooled and treated as if they were recorded from one participant. The statistic is similar to fixed-effect analysis, where the true effect size is assumed to be the same across participants and the only source of variation is presumed to be the measurement error. As any source of variability across participants (e.g., the pattern of brain response, the electrode localization, the number of trials per participant) would have a critical impact on the results, this ‘meta’ participant approach would require assessing the homogeneity across participants by looking at individual responses and at individual electrode locations before pooling all the data together. Critically, with this approach it follows that the inference is restricted to that population of participants. Therefore, this approach is no longer recommended in iEEG studies because it mixes intra-individual and inter-individual variance Null hypothesis significance testing can be performed within individual participants and then the prevalence of that effect in the population can be inferred from the proportion of participants showing an effect, see the Bayesian method proposed by (Ince et al., 2021).
5.2.3. Group studies
In a group study, statistical analysis is performed at the population level like in M/EEG studies. The only difference relates to the set of channels included in the analysis. Because electrode positions vary between participants, only electrodes with equivalent positions between participants shall be included in the analysis. Equivalence between electrodes relies on individuals’ brain registration to a normalized space (which then might be used for labeling in the individual space, see section 2 and 5.4). Then electrode coordinates or brain parcels are used to aggregate electrodes and participants are considered as independent measures. There, the statistic is similar to random-effect analysis, the true effect size is assumed to vary across participants, each representing a random sample of effect sizes, and the sources of variation are measurement error and response magnitude over participants. It follows that the inference is not restricted to that population.
Group studies require a commitment to the assumed spatial scale at which electrodes can be grouped, e.g., an anatomically defined ROI, which almost inevitably sacrifices spatial specificity that is available on the single participant level (see below). They critically rely on the procedure used to aggregate electrodes across participants. Hence, group studies may be considered less detailed as compared to the multiple cases approach. Here, we therefore also recommend providing quantitative estimates to assess the similarity/variability across participants within the assumed spatial scale (e.g. as in Figure 15 from the free software HiBoP18, also available as an online platform19, or in the MIA software (Dubarry et al. 2022)). Other approaches support both fixed- and random-effect models to take into account inter-individual variability (e.g. Frites20 toolbox (Combrisson et al., 2021), see also section 6.9.1)).
Figure 15.
Combining group-level and patient-level anatomical analysis.
Group-level functional activations show HFA response components at specific latencies. This allows the definition of a Region of Interest (ROI) for further investigation. Panel A shows a strong group-level (n=67) HFA response in the basal temporal cortex 400 ms after the display of a written word during a reading task contrasting attended vs. ignored words. Panel B shows a group representation of all iEEG sites in the ROI in the left temporal lobe (pink sphere), across all patients. Subsequently, each site can be visualized onto a 3D representation of the corresponding individual brain (panel C), to understand how the strength of the response depends on its precise localization relative to specific individual anatomical landmarks, such as the lingual gyrus. Repeating the same procedure for all subjects and sites within the ROI leads to a detailed anatomical characterization of iEEG sites with a homogenous functional response (visualizations and analysis were made with HiBoP).
5.2.4. Insights about the necessary number of participants
In cognitive neuroscience, a good practice consists in establishing a priori how many participants should be included in a study to support an exploratory study or to obtain a specific statistical power in a hypothesis-driven study. Similarly to M/EEG studies, a power analysis (and forthcoming result robustness) is more difficult when there are many different analytical measures, each coming with different effect sizes that may vary between frequencies, regions, etc., which becomes more complicated if several of these measures are combined. In iEEG, the complexity of the answer stems from the association of four aspects of iEEG: the SNR, the spatial specificity, the spatial sparsity of the sampling that is variable across patients and the possible pathological physiology.
Concretely, iEEG is so spatially specific that it is not rare to observe a task-induced response pattern on one recording site that is absent at a neighboring site just a few millimeters away. Considering the high SNR, the effect on that electrode can outclass the criteria for significance with both a large effect size and a high confidence interval by several orders of magnitude. With such an effect that is clear in that participant, the question is then whether it generalizes to other participants and toward the population.
iEEG research also needs to deal with the complexity related to the precise anatomy. Interpreting results at the group level and generalizing to the population implies defining the homology of brain regions and reporting phenomena at a scale that is coarser than that of the individuals’ observations, similar as it is for instance done in EEG to report on a specific ERP component such as a P300 at a specific electrode and latency. Yet, one should keep in mind that the validity of the simplification depends on the granularity of the anatomical parcellation that is used. This can be compared to smoothing of data along other dimensions: too little smoothing highlights the interindividual variability which then might be interpreted as noise, whereas too much smoothing washes out the effects. The right level of smoothness is an empirical question: establishing the relationship between anatomy and function with iEEG implies a detailed participant-level anatomical investigation, and to go back-and-forth between group-level and individual analysis (even when many participants have been recorded).
Because the electrode positions are variable across patients, recording at sufficiently similar locations in multiple participants requires enough participants and hence time to collect the complete dataset. Whether a sufficiently large sample can be collected depends on whether that particular cortical location is frequently targeted by intracranial surgery. The feasibility to acquire data from identical electrode locations is further reduced when studying coupling between cortical areas, as in that case both electrodes comprising a pair need to be consistently located. Thus, in some cases, it may seem unreasonable to expect iEEG studies to display perfectly identical functional responses in more than a few participants. However, the association of high specificity and high sensitivity legitimates iEEG findings from a (few) participant(s), which can be reported as case(s) study (i.e., with a number of participants that would not be endorsed in M/EEG).
Last, because iEEG relies on the recording of a brain with epilepic activity, a frequent criticism states that no iEEG study can ever claim that its conclusion is a general property of the healthy human brain. To assert that an observation is not incidental and/or due to the medical condition of the participant, it is at least necessary to show that the effect of interest is not particular to one patient, with a particular type of epilepsy or cortical (re)configuration. In this case, two participants with completely different epileptic profiles are a primer to generalization. In fact, the replication in two participants echoes the requirements of NHP electrophysiological studies, which often report results obtained in two specimens (this comparison is further bolstered by the LFP analogy). Beyond two participants, replication across more participants is then “more convincing” that the results are replicable. That is, it increases the likelihood that if other researchers record from the exact same cortical location in the same paradigm, they will observe the reported effect. For that reason, in order to guide the interpretation and inform robustness, we recommend always reporting the number of participants recorded in the exact same region and showing / not showing the effect of interest. Additionally, comparing behavior between patients and healthy controls can serve as a check whether the observed brain activity correlated with this behavior is likely to reflect the normal function of the brain area(s) under study.
5.2.5. Challenges, recommendations and reporting advice
The review of the spectrum of practices shows a gradation from observation made at the single participant level to group studies allowing generalization to the population. With the recent rise of open-data, it has become easier for a researcher to complement data of a single case with data published by another researcher, facilitating replication to scale up findings to multiple cases or even to generalize from a group. Good iEEG practices must favor the prerequisite for combining knowledge. For that reason we recommend always preparing iEEG data so that any single participant can be added without further effort in a larger study (even if it might at first be intended as a single case report). Information needed to aggregate electrodes across participants (i.e., individual MRI and electrode coordinates) must be accessible to allow future use of the data (see section 1.3.8).
Combining data (e.g. across research centers) does not necessarily imply improving statistical power or result robustness. The cornerstone is the harmonization of approaches between centers (e.g. in data acquisition, preprocessing and use of complementary information): it can be more effective to combine a small number of high quality datasets than to use all available data sets (see the context of non-human primate neuroimaging, Autio et al., NeuroImage 2021).
We recommend always including detailed information about individual participants when possible: (i) the implantation schemes, (ii) seizure onset zone, (iii) handedness, (iv) type of epilepsy, (v) possible brain lesion, malformation or previous resection, (vi) neuropsychological impairments, (vii) post-implantation status (e.g. seizure-free after resection).
Given iEEG sensitivity and specificity (i.e., high SNR and spatial specificity), a finding that is not significant at the group level but that is replicated in a subset of the participants can be a serendipitous finding and therefore worth reporting as a non-significant observation of interest.
Some statistical approaches (e.g., combining p-values or correcting for multiple comparisons) imply some prerequisite(s) such as the (in)dependence of the measures (e.g., iEEG channels). Data’s prerequisite(s) inherent to the use of a given statistical method should be explicitly stated.
Although correction for multiple comparisons is required to control the number of false positives (type-I errors), there is currently no good way to deal with how type-II error rates are affected by the spatial sparsity (e.g., the density of surrounding contacts being different for each brain location), the difference between working with a volume-based or a surface-based approach and the related dependence between electrodes. Acknowledging the potential limit of the statistical approach and providing related information should be part of the iEEG good practice to assist future replication.
5.3. Combining recording sites across participants
Combining data across participants implies expressing individual participants’ electrode locations in a space that is common for all participants (see section 2) and consequently aggregating electrodes. This procedure faces two main challenges. The first challenge is the tailored surgical placement of electrodes in each participant. The second challenge is the variability in each individual’s brain anatomy and underlying brain function.
To combine data across participants, three main approaches are followed. The first relies only on spatial normalization, which can either be volume based or surface based. It results in a table with the coordinates ([x y z] positions in mm) in a normalized space (e.g., MNI) for each electrode. In the second approach, the normalization is combined with a parcellation (atlas based or geometrically based), where the labels are mapped to the electrodes. It results in a table containing a list of electrode names and corresponding labels (i.e., name of the parcel). These two approaches can be further complemented using functional information (e.g., a localizer task). In the following sections, we describe the different approaches and discuss their advantages and weaknesses, especially regarding the challenges raised by individuals’ peculiarities.
5.3.1. Based on coordinates
Once the brain and electrode locations of each participant are registered in a standardized space (e.g., MNI, see section 2.2), the electrodes with similar coordinates can be compared between participants. A simple way is to aggregate electrodes based on their distances to each other. However, this approach neglects anatomical information, for instance by aggregating electrodes located on two opposite sides of a sulcus while not aggregating more distant electrodes lying on the same gyrus. To circumvent this issue, aggregating based on electrode coordinates has been sometimes completed by considering broad anatomical landmarks, such as cortical lobes or gyri (see the first paragraph of section 5.3.2.2). That is to aggregate electrodes close to each other and visually belonging to a ROI. While refined as compared to a pure distance-based approach, this alternative often relies on a subjective appreciation of the ROIs which does not favor replicability and remains coarse, especially for those uninitiated in brain anatomy. Nevertheless, working with electrode coordinates facilitates comparison between studies, especially when it comes to combining single cases.
In single case reports we recommend providing electrode coordinates in a normalized space (e.g., MNI, including the exact reference template space and the procedure for normalization and electrode mapping, see section 2.2.2) to make further meta-analysis possible. Electrode coordinates should ideally be complemented with a visualization of their anatomical location in the individual’s brain.
5.3.2. Based on labels
A finer and more objective means to combine electrodes across participants is to make use of a parcellation by aggregating electrodes belonging to the same parcel (see section 2.3). Parcellation can either be derived from an atlas projected on a template brain, or based on a geometrical parcellation of a template brain. The parcels are then defined by the parcellation granularity (i.e., the number and the size of the parcels). When the parcellation is atlas-based, parcels are further informed by the atlas feature(s) (e.g., anatomical, functional).
For multiple case studies and group studies, we recommend aggregating electrodes on the basis of the anatomical information defined by a parcellation and to report all information regarding the procedure applied to label the electrodes (i.e., registration, electrode mapping, parcellation reference and probabilistic approach, as described in section 2.3.2). Because there are more ways to define parcels than there are brain templates, aggregating electrodes using labels adds variability in the literature. However, this approach relies better on brain anatomy than aggregating using coordinates.
5.3.2.1. Atlas based
Registration of the individual’s space to a normalized space allows the researcher to deal with electrode positions in both the normalized space and the individual space. The transformation of the electrodes from individual to normalized space maps the electrodes to the related atlas parcellation(s); inversely, the transformation also permits mapping the atlas parcellation to a delineation of the individual’s brain (see section 2.3). In both cases, the electrode positions are labeled based on the parcellation and therefore can be aggregated across participants. The two approaches lead to similar results as both approaches rely on the same transformation. The normalized space is evidently convenient to visualize electrodes from different participants on the same template brain. However, there are two advantages to going back to individual space. First, it offers the possibility to visually check that the atlas parcellation is correctly mapped and translated into the individual delineation. Individual peculiarity in brain anatomy is not rare (Zilles et al., 1997) and not always well handled by registration algorithms. For instance, Heschl's gyrus is duplicated in a large portion of the healthy population (Marie et al., 2015). Second, visualizing electrodes in their original individual space facilitates evaluating if an electrode is fully or partly belonging to a given parcel. Third, a significant subset of participants may have brain anomalies which might not be well handled by normalization algorithm and need to be visually checked (see section 2.2.2)
To map ECoG electrodes, we recommend working with the tissue described as a mesh (i.e., surface) because grids/strips lie on the surface of the cortex. In practice, when an ECoG strip is inserted between the two hemispheres, working with a surface takes into account the fact that the strip of electrodes are only facing one hemisphere. This type of mapping is less suitable for sEEG because these depth electrodes are located inside the brain, therefore the tissue must be described as a volume (i.e., voxels). An sEEG electrode records activity from the various cortical locations in its 3D vicinity, therefore it is not recommended to project it to one point (or one parcel) of the cortical surface (e.g., located between two sulci, an electrode records activity from both of them). Last, some sEEG shafts penetrate the cortical sheet to target deep subcortical structure(s). Because volume-based mapping relies on volume deformation of the whole brain (i.e., not only on the cortex but also subcortical regions), it is more appropriate for mapping sEEG electrodes on a template brain and for labeling sEEG electrodes (see recommendations in section 2.4).
Electrode localization implies some uncertainty inherent to the procedure itself (see section 2). To take into account this uncertainty and the fact that the surface or the volume of an electrode (respectively in ECoG and in sEEG) can span more than one parcel of an atlas, we encourage the use of a probabilistic approach in electrode labeling to guide the aggregating of electrodes (see section 2.3.2 and 5.4.3).
5.3.2.2. Geometrically defined
Prior to the development and widespread availability of digital atlases, iEEG researchers used to delineate an individual’s brain and to combine recording sites manually (Allison et al., 1994; Halgren et al., 1994; McCarthy et al., 1995; Puce et al., 1999). For instance, electrodes located in/over the gyrus anterior to the central sulcus were labeled ‘motor cortex’, while the ones in/over the gyrus posterior to the central sulcus were labeled ‘somatosensory cortex’. This precise but tedious hand-made labeling was guided by human brain anatomy atlases such as that of Talairach and Tournoux (Talairach and Tournoux, 1988) or Brodmann (Brodmann Korbinian, 1909). With the digital era, automation led to a faster labeling procedure, but possibly also to an overconfidence in the automated results. This becomes more striking when the spatial precision of the electrodes is at a scale that does not match the atlas. First, when an electrode is at the border of two parcels, a binary classification (e.g., this electrode belongs to parcel A but not to parcel B) overlooks the approximation due to the individual’s brain normalization and overlooks the influence of local passive spread of the signals (see probabilistic approach in Section 2.3.2). Second, the granularity of the atlas can vary with brain regions and depends on the original atlas authors' goals and data used to build the atlas. This may lead to some brain regions that are more finely parcellated than others. Consequently, parcels are not equal in size which makes some parcels more likely to be associated with the electrodes.
To objectify geometrical structure homology, Trotta and colleagues came up with a solution example in the context of ECoG by defining isometric surface-based parcels on the brain template to then adjust them to the individual’s cortical anatomy (Trotta et al., 2018). The sampling of the standardized surface with parcels permits the investigator to establish a correspondence between sets of vertices across participants and thus makes it possible to aggregate electrode data over parcels for comparison across participants.
The method introduced by Trotta and colleagues highlights how difficult it is to perform the aggregation of data across participants, and especially the relation between mapping electrodes to parcels and the passive signal spread that affects the spatial selectivity of the electrodes. When the cortical sheet is projected onto a sphere, two neighboring gyri are further away as they are separated by the sulcus in between them, which misrepresents the actual distance between the gyri that is relevant from the point of view of passive signal spread. Furthermore, this method exemplifies the arbitrary spatial scale at which data is aggregated (going from electrodes to parcels), allowing for flexibility in parcel size. Combined with the uncertainty in electrode localization (e.g. an electrode that spans a sulcus can be assigned to two different gyri), the spatial scale at which data is analyzed results in non-trivial consequences for estimates of connectivity and for statistical analysis, as some electrodes may be assigned to two parcels whereas other electrodes are only assigned to a single parcel. Thus, practically it leads to dependency between some parcels, but not between some others (see section 5.3.4).
5.3.3. Based on a functional response or localizer
From a functional perspective, even when electrodes are located at a similar location, slight differences in brain folding and/or brain organization may explain brain response variability across participants. When electrodes are combined across participants, some functional correspondence is assumed or expected between electrodes. Yet, the risk of not examining the results from the single participant analysis is to overlook inter-participant functional differences within parcels and inter-participant functional similarities between parcels. The aggregation of electrodes can be functionally informed to take interindividual variability in anatomy and functional organization into account. While aggregating electrodes only on the basis of functional responses may not be appropriate, functional responses can complement the coordinate-based or label-based aggregation. For instance, if two electrodes from two participants are localized at a distance of more than a few millimeters in a standardized space or in neighboring parcels, they are unlikely to be aggregated on the basis of their anatomical location. If they do show the same response pattern (defined on neural activity or on iES result, see section 1.1.4), it makes sense to aggregate them as their identical response suggests their functional correspondence.
However, this flexibility must come with precaution to avoid double dipping. Aggregating electrodes using functional responses should not be performed using the statistical effect of the response of interest, but to adjust an anatomically-informed aggregation (e.g., probabilistic approach in electrode labeling, see section 2.3). Here, the use of an independent or orthogonal functional localizer avoids such bias as the functional response is defined on a different set of data than the one investigated. For instance, if the research question relates to face processing in the fusiform gyrus, running a functional localizer helps to select in each participant the electrode, located in the vicinity of the fusiform gyrus, that shows the highest SNR in response to faces. In any case, when a functional response is used to aggregate electrodes, it has to be reported.
5.3.4. Challenges, recommendations and reporting advice
- In practice, we recommend making the most of the different approaches to aggregate electrodes across participants.
- First, the aggregation must be based on anatomy as it is the primary objective feature of electrode location. This step corresponds to the labeling of the electrodes (either atlas parcellation or geometric parcellation).
- Second, for each participant the electrodes must be visualized in individual space together with the individual’s delineation and compared to electrode mapping in the normalized space with the template brain delineation. This sanity check permits adjusting electrode labeling to individual anatomical fine peculiarities and to eventually correct the normalization (e.g., an electrode that is located on a specific gyrus in single participant space, may end up at the other side of a sulcus in normalized space).
- Third, the adjustment can be guided thanks to a probabilistic approach in the labeling (see section 2.3) and may be informed by functional data that have to be orthogonal/independent to the effect of interest. For instance, if an electrode is on the edge of a parcel but its centroid is outside, it can be reasonable to incorporate this electrode to that parcel, especially if the response is more similar to the ones observed in that parcel in other participants.
The granularity of a parcellation is critical when iEEG data is aggregated across participants. The number of recorded participants being limited, one can try to use wider regions/parcels which increases the chance of matching electrode locations over participants. However, as part of the iEEG signal is confined to a few millimeters, compiling results in regions/parcels with larger size lessens its specificity. Thus, the size of the ROI indicates how a given effect (e.g., brain response) is highly focal or spans a wide region. More generally, we recommend defining the level of granularity or the choice of an atlas in link with the research question.
- As epilepsy is more prevalent and epilepsy surgery more likely to be successful in some regions (e.g., temporal lobe), electrode density varies over the brain making some regions/parcels more likely to host electrodes and thus more likely used for analysis. Thus, in iEEG, aggregating electrodes across participants faces sampling variability in spatially sparse data. That is, electrode density varies over each participant’s brain and between participants, making participants unlikely to contribute equally to the group analysis.
- We recommend considering the potential confounding effect of having a different number of electrodes per parcel over participants. These confounding effects depend on the quantification of the effect, e.g., for ERPs averaged over electrodes there is no amplitude bias, whereas such a bias can be present if non-linear measures are applied. As with any selection of data points, if some electrode(s) is (are) selected as representative of a given parcel, it should be motivated by the research objective and reported. Overall, such selection process introduces many degrees of freedom (e.g., which possibly valuable data is thrown out) and only applies to statistical scenarios where inter-electrode and inter-participant variance cannot be separately accounted for. However, using a nested model may in principle be enough to account for multiple electrodes per participant (see section 6.9.1).
- In multiple case studies and in group studies, we also recommend reporting the density of electrodes per region/parcel to inform about the spatial sampling bias at the population level. This can be achieved by depicting electrode distributions over the brain with a color code to indicate which electrodes are aggregated together across participants (see also section 2.4). If the electrode density is different over different brain regions, that should be acknowledged and reported, as the statistical results from a densely sampled region will be more robust and are more likely to be representative of the population than those from poorly sampled regions.
- Conversely, in the case where electrodes from all participants are considered together to create a ‘meta participant’ (as in a multiple cases study, see section 5.3.2), some disparity may arise with some participants contributing more than others. While the ‘meta participant’ is not recommended, if this approach would be followed it would require reporting the contribution of each participant, for instance by plotting all electrodes in a normalized space with a color code for each participant.
The variability of brain responses is part of the richness of iEEG data and reflects differences in brain organization. We recommend visualizing the individual response patterns when combining electrodes across participants and to estimate the variance across the combined response. As an example, slight variability in the placement of an electrode within the active cortical area or the choice of the reference could result in an ERP polarity reversal across participants (see section 4.1.2). This illustrates the relevance of visualizing individual data before averaging across individuals.
Parcels from the same participant are not independent for statistical purposes, and even less when the method used to combine electrodes allows for redundancy (i.e., an electrode is mapped to different parcels). This lack of independence must be kept in mind for statistical analysis, especially in the case of connectivity measures as it introduces artificial correlation between parcels. Furthermore, the dependence between parcels should be taken into account in the choice of a method to combine p-values (see section 5.2.2) or when correcting for multiple comparisons (Groppe et al., 2011).
As discussed in section 2.4, electrode projection should not be confused with functional data interpolation. The projection of electrodes permits linking recording sites to the anatomy, and combining sites across participants. The interpolation of the functional outcomes of the iEEG analysis is useful for visualization, yet it is not recommended for grouping data across participants.
5.4. Data visualization
The graphical representation of the statistical results is at the heart of good research practices as it conveys/summarizes both the interpretation of the analysis and the quality of the assessment (Rousselet et al., 2016). For that reason we recommend providing as much information as possible about the distribution (e.g., confidence intervals, individual/single trials data, in experimental design with paired comparisons one should provide the distribution of pairwise differences; for an example see Figure 1 in (Rousselet et al., 2016), for illustrations of statistical summaries si (Wickham, 2016)21, for further discussion see (Weissgerber et al., 2015)). Some of this functionality is available in the “R Analysis and Visualization of iEEG” (RAVE) software (see Figure 4 in (Magnotti, 2020)) and in the MIA toolbox (Figure 10; Dubarry et al. 2022). When it is not possible to depict on the same figure the effect of interest and its representativeness, we recommend providing the necessary information as supplemental material.
The depiction of iEEG results as a volume or on the surface implies in most cases an interpolation in space (otherwise the data is barely visible as it is limited to the single voxel corresponding to the electrode centroid). The extent of that projection should take into account passive signal spread (see section 2.3.2) and should be reported as it can visually bias the representation of density-sparsity of the electrode (see sections 2.4 and 5.3.4).
When all patients from a group study have been combined in a normalized space, we recommend going back-and-forth between the global picture and its individual components. That is, to first explore the data/result in a 4D representation (i.e., space and time) to reveal potential ROI and corresponding single electrodes, and then zoom in on each individual's response profile accompanied by the individual’s localization (see Figure 15 from the free software HiBoP22 available as an online platform23, HiBoP is designed specifically to combine group-level and single patient-level analysis).
6. Perspectives
The previous sections of this article show the current state of iEEG research. In this section we present where we envision research in iEEG to be in ten years from now. In the last decade, iEEG research progressed in many ways, but the directions that were taken might not only be the ones that were anticipated ten years ago. Inherited from the global tendency to set a parallel between scientific advancement and technical/methodological progress, anticipations on iEEG progress commonly tend to overlook what can be done with what is already accessible. While it is certain that the use of new technologies energizes discoveries, achievements are also reached thanks to a better organization of current resources (Frith, 2020; Salo and Heikkinen, 2018; Stengers et al., 2018). Also when it comes to envisaging the most promising steps forward on the basis of what they were ten years ago, it is reasonable to assert that technology indirectly fuels iEEG research as much as directly (e.g., big data servers versus micro-electrodes).
Probably the most remarkable advance of the last decade is the spread of the use of iEEG for fundamental research. Together with the multiplication of iEEG research centers, a multitude of tools have been published and shared with the community. In the same vein, the emergence of open science and the realization of its utility for both the researcher and the common good had fostered the sharing of data and methods. Beside, while the use of new techniques such as micro-electrodes tends to be restricted to some specialized centers, it has brought a cardinal perspective in cognitive neuroscience linking both spatial scales and investigations across species.
6.1. Strengthen patient-centered collaborative work
In the next few years, the trend is likely to see a larger number of centers implanting patients, which might reduce the number of patients implanted in some established centers. Yet, the overall number of patients participating in research is expected to continue to increase as well as the tendency to see the implantations being more individually-tailored. Another tendency among current iEEG centers, pertains to a move from ECoG to sEEG, possibly due to the procedural morbidity of ECoG (Jehi et al., 2021; Tandon et al., 2019). In the long run, a possible scenario that must be envisaged is a decrease in the number of acutely implanted patients, thanks to improved diagnostics and treatment for epilepsy. This positive medical perspective should motivate the storage and the sharing of structured data, to make the most out of the current opportunity to directly record from the human brain (at least for ECoG data if it becomes rare). Concurrently, we may see an increase in the use of chronic implants to complement, or replace, pharmaceutical treatment; an approach which offers opportunities for longitudinal recordings (Henin et al., 2019; M. Aghajan et al., 2017; Meisenhelter et al., 2019; Rao et al., 2017; Topalovic et al., 2020) and naturalistic neuroscience (see section 6.5). These scenarios are more likely to occur if collaborative work is strengthened and if good practices are placed at the heart of iEEG research. It follows that in the near future we expect to see a more efficient use of the current iEEG resources at two levels: between research centers with an expansion to the Open Science framework, and within iEEG centers with more synergistic interactions between the clinic and the research (Figure 2).
6.2. Transparent reporting and data sharing
The rationale for good research practices encompass a wide spectrum of motives. On the one hand research is a common good supported by public funding, which implies duties such as openness and credibility; on the other hand scientific standards and publication guidelines force us to be critical about what we do. At minimum, good research practices include clear reporting as well as the sharing of data and analysis scripts to allow replication (i.e., the same results are found if the same data is analyzed the same way).
Recent initiatives for reproducible M/EEG research promote good practices in reporting by defining recommendations and warning on issues related to data analysis (Keil et al., n.d.; Pernet et al., 2020). It might be argued that the high spatial sensitivity and high SNR of iEEG make iEEG results less dependent on the choice of an analysis method. For instance there is potentially less ambiguity about localizing neural activity as compared to the use of M/EEG: with iEEG the measure does not depend on a specific inverse solution algorithm. Yet, it is crucial to specify how data were processed, especially for referencing and for normalizing the individual brain getting coordinates in a normalized space. Following the M/EEG initiatives, and anticipating the need to grant future iEEG research, the present guidelines aim at paving the way for good practices in iEEG.
Replicable research implies publishing with sufficient detail to make the study/data reusable. It relies also on open data sharing that must follow a community-driven organizational framework to make data understandable. To serve that purpose the Brain Imaging Data Structure specification (BIDS) was created for MEG (Niso et al., 2018), for EEG (Pernet et al., 2019) and subsequently extended to human intracranial electrophysiology (Holdgraf et al., 2019). For now, the iEEG BIDS specification and examples mainly relate to the technical aspects of the recording and not so much the clinical aspects of the participants. In the future, we anticipate the iEEG community will extend the existing BIDS raw format and define BIDS derivatives (e.g., with clinical annotations and details on lesions and impairments), in line with Figure 2 that indicates the interaction between the clinical and research team. We hope that this article will encourage and prime the definition of these improvements to BIDS.
As an aftermath of BIDS systematization, we foresee the multiplication of open access local iEEG libraries (e.g., see 24, 25(Miller, 2019) or (Berezutskaya et al., 2022)). Akin to the ERP-core for EEG (see 26), the BrainMap (see 27) and Brain Activity Atlas (see 28) for fMRI, we further expect the emergence of online meta-databases that will permit examining iEEG results from studies that investigated a certain topic and/or a specific part of the brain. The ERP-core is a free online resource for human ERP research building upon the standardization of ERP paradigms and analysis protocols across studies (Kappenman et al., 2021). The BrainMap is a database of published functional and structural neuroimaging experiments (Vanasse et al., 2018) and the Brain Activity atlas (BAA) is a battery of functional localizers in a large cohort of healthy adults using task-evoked fMRI. Similarly, a recent initiative, the Human Intracranial EEG Platform, is an open-source platform designed for large scale and optimized collection, storage, curation, sharing and analysis of multiscale human iEEG data at the international level (see 29 and 30).
Such meta-database resources will be an efficient way to re-use data, and transfer what has been done from one study to another by different members of the iEEG community. iEEG databases should not be restricted to group studies but also open to single-case studies. It will help test reproducibility (e.g. if there is a channel at a given location what can we expect to record there?), as well as going back and forth between an individual and a population (i.e. taking into account the richness of data variability). Open databases will be pertinent to combine multiple datasets from different protocols to disambiguate/complement results. Last, knowing “what is happening where” helps to refine hypothesis-driven research (e.g., to select an experiment with regard to electrode localization, to build some priors in a Bayesian perspective) and it can reveal the “holes in the map” (i.e., foster exploratory research).
As Open Science practices become more common in general, we expect this to positively impact the sharing of knowledge in iEEG practices. Sharing common iEEG artifacts is one concrete example that would benefit the community (see sections 1.3.4, 1.3.5 and 3.2). Many researchers have gained experience with identifying artifacts in iEEG data, but this knowledge/expertise remains available in somewhat isolated form within smaller communities of researchers (e.g., in a local research lab). A shared database of iEEG artifact templates (both physiological, e.g. spikes, and non-physiological), contributed to by different researchers, would be a valuable resource for the community, and especially for the training of early stage researchers in iEEG analyses. Ideally, in this ‘atlas’, the iEEG signal would be accompanied by other types of recording to validate the artifacts (e.g., EOG or EMG recordings) and to better understand iEEG physiology (i.e. passive field spread). An interest for artifact-contaminated data also applies when developing new data analysis methods. For instance, for machine learning approaches both "positive" and "negative" data samples are needed to prevent bias in classification results (see section 6.9.2). In the making of such database(s), the contribution by various researchers would align with the concept of "team science" and a live database format would allow information to be updated and improved, differently from the static format of regular peer-reviewed publications.
While iEEG can be considered a niche, and as such might be overlooked by researchers working with other approaches, it provides an incomparable opportunity to cross-validate results through a multimodal approach. In the next decade, we anticipate more translational work between approaches (e.g., iEEG and fMRI), as well as within iEEG (e.g. passive recording and electrical stimulation) and with the deep-brain approach used in movement disorders (de Hemptinne et al., 2015; Little et al., 2021). With a great deal of precautions regarding ethical issues (see section 1.3.8), clinical information available about the participants (e.g., electrostimulation, multiscale recordings) may be incorporated, thereby creating multimodal databases similar to those available in the fields of MRI (see for instance the Allen institute initiative, see 31). The results of these may fuel brain modeling such as in the Virtual brain project or biophysical models based on iEEG signal model (see 32 and (Medani et al., 2021)). Last, this multimodal approach is also likely to go beyond imaging data in humans, like interspecies cross-scale measures (Paulk et al., 2021) or the use of additional big data such as genetics (Gao et al., 2020).
6.3. Bringing together the clinic and the research
Bringing together clinicians and researchers creates many opportunities for future research, given their overlapping interests and complementary skills. As participants usually participate in several studies, in addition to the standard clinical procedures, it makes sense to share and discuss findings collectively; in much the same way as clinical meetings often discuss single patients. It is obviously beneficial for the patient, because an exhaustive knowledge of task-induced brain activations can complement or guide functional mapping (Crone et al., 1998b, 1998a; Sinai et al., 2005). It is also desirable for research teams, who might use observations made in that patient in other cognitive paradigms to discriminate between alternative interpretations of their own results. Last but not least, the synergy between clinical and neuroscientific expertise is needed to address the complex relationship between pathology and cognition, for instance the effect of epileptic activity on cognition. Nonetheless, non-clinically validated research (e.g. task, methods) entails a risk if it influences clinical decision making. Clinicians need to be aware/informed of the validation-status of any research result. Research should not modify the clinical electrode placement plan and it should be always clear to the patient that participation in research is not part of the clinical routine (e.g., by itself it should not guide surgery away from major functional networks as it is not certain that it would prevent post-surgical cognitive deficits).
The medical expertise is unique for iEEG as the clinic is by essence multimodal and multidisciplinary. Clinicians start the exploration of the patient before the implantation with different approaches (e.g. semiology, surface recording, imaging, see section 1.1.1). Knowledge about the medical history, pre-op imaging, and iES can potentially shed a light on the results of some research protocols, for instance, when epileptic status influences cognitive performance, to the reorganization of some brain functions or to the effect of direct electrical stimulation.
In parallel, researchers in iEEG can encourage technical and methodological development in the clinic. For instance by pursuing higher standards in data quality, by encouraging the clinical validation and use of high-density probes, or by motivating the application of advanced signal processing. In the past decade, common iEEG hardware has not changed significantly (i.e., amplifiers and electrodes). Considering the recent technical advances with cochlear implants, peripheral nerve recording-stimulation, or deep-brain-stimulation, we see some potential in technical modernization and we anticipate a convergence of clinical and research sides to meet the same standards (e.g., technical and methodological) for a mutual benefit.
6.4. Innovations in electrodes
A brief review of the technical innovations of the last decades shows that many electrode prototypes have been developed but that their use has been limited to few hospitals/centers (Engel et al., 2005). For instance, the multipronged temporal pole electrode array developed to target the temporopolar cortex (Abel et al., 2014). Another example is the depth-strip hybrid operculo-insular electrode that was developed to investigate perisylvian/insular refractory epilepsy (Bouthillier et al., 2012), and whereas this hybrid sEEG-ECoG approach on the same probe was interesting, it did not disseminate in the iEEG community. Cross-scale measurements, however, have had a long history in iEEG. Already in the nineties, a model of hybrid depth electrodes that included high-impedance bipolar contacts between low-impedance sEEG contacts allowed the recording of both single neuron spikes and LFPs (Howard et al., 1996). Since then, different configurations of iEEG probes for single-unit, multi-unit and LFP recording have become more common (Pothof et al., 2016). The two most famous of which are the Behnke-Fried microwires inserted through the macroelectrode shaft and protruding from the tip of a depth sEEG macroelectrode (Fried et al., 1997; Lehongre et al., 2022); and the high density ECoG grids with smaller contacts which increase spatial density of the implant (Bleichner et al., 2016; Branco et al., 2017; Flinker et al., 2011; Muller et al., 2016; Ramsey et al., 2018). The study of epileptiform activity has been further facilitated by the use of dense two-dimensional cortical microelectrode arrays (Schevon et al., 2008) and laminar probes to drive current source density from multi-unit activity recorded across cortical layers (Csercsa et al., 2010; Dougherty et al., 2019; Halgren et al., 2015, 2019). Laminar ‘optodes’ have also been developed to compare the fine hemodynamic response with laminar source current density and multi-units activity (Keller et al., 2009). Last, sEEG recording was used to perform microdialysis by means of a probe inserted through the lumen of the depth, which permitted to directly sample extracellular neurotransmitter concentration in midbrain (Fried et al., 2001). The aftermath of these advances on iEEG probes is twofold. First, they show that technological developments in the field of iEEG may serve the clinical cause. Second, they led to important findings in cognitive neuroscience, especially in domains where animal research has its limits (e.g., language function).
We anticipate that, in the next decade, technical developments will mainly focus on a quantitative orientation with an increase in the electrode density per probe or grid (Chang, 2015; Schippers et al., 2021). As for now, the dissemination of the multiscale probes and probes with higher spatial density beyond the centers where they were developed, is to be expected only if a clear benefit for the patient is established. For instance, the use of multiscale electrodes may spread if it contributes to micro-surgical intervention which in return must lead to an improvement of epilepsy treatment (i.e., diminish epileptic seizure and lessen the risk of brain function deficit as compared to a classic intervention). In direct link with the progress in electrode specification (e.g. micro-electrodes), the acquisition set-up should reach higher standards, especially regarding the acquisition sampling rate to allow seeing single unit activity. The main limits to this renewal of the materials being the cost for the hospital and the regulatory issues (e.g., onerous approval, insurance coverage).
6.5. Naturalistic neuroscience and discovery science
Major neuroscientific conclusions can be reached by investigating neural activity during unconstrained behavior and in naturalistic conditions (di Pellegrino et al., 1992; Glanz et al., 2018; Hamilton and Huth, 2020; Matusz et al., 2019; Rizzolatti and Fabbri-Destro, 2010). Yet, two factors must coexist to make such a discovery possible: 1) high signal-to-noise recordings, with reliable neural responses at a single-trial level (even during free movements); 2) high spatiotemporal resolution, to capture the immediate effect of environmental events or behaviors. iEEG offers such properties and is therefore suited to investigate the neural basis of realistic behavior (Del Vecchio et al., 2020; Podvalny et al., 2017). These characteristics are also critical when considering the development of close-loop experimental settings (Branco et al., 2018a; Zelmann et al., 2020), and related foreseen technological applications for brain computer interface (Moran, 2010). Because iEEG appears as the tool of choice for observation-driven 'discovery neuroscience' (Genon et al., 2018), we forecast that research with ecological approaches will spread in upcoming years.
The prospect of seeing an increase of ecological iEEG research is fostered by the emergence of active wireless devices (i.e., amplifier/transmitter implants) for epilepsy monitoring. We predict that these electrodes will have a drastic impact on iEEG research for three reasons. First, the signal-to-noise ratio of iEEG is good enough to extract meaningful neural signals in unique situations (e.g., single trials, individual movements). Second, iEEG is relatively immune to most artifacts that contaminate scalp signals when participants perform daily life activities (but see recommendations in section 3.2). Last, iEEG does not imply strict restrictions on the participants' motor behavior, in contrast with non-invasive recordings (e.g., fMRI, MEG), for which strict technical aspects prevent unconstrained exploratory neuroscience (e.g., supine position, limited space exploration, altered visual feedback). In that regard, the recent development of wireless iEEG and chronic recordings from implanted devices opens endless perspectives to naturalistic neuroscience (M. Aghajan et al., 2017; Meisenhelter et al., 2019; Stangl et al., 2021; Topalovic et al., 2020) and will likely recast the clinical monitoring. Continuous iEEG monitoring is a unique opportunity to study human brain activity as it happens in daily life (e.g., active reading, conversation) and not only in link with the pathological activity. For researchers, it permits testing, in ecological situations, the validity of functional interpretations drawn from 'classic' lab experiments (see the BrainTV approach (Jerbi et al., 2009b; Lachaux et al., 2007b) or the BCI2000 approach (Schalk et al., 2008)). Surely, wireless devices will require clinical approval, which might be facilitated by the fact that: (i) it permits a continuous recording, even when the patient has to move in the hospital, (ii) wireless devices reduce movement related artifact thanks to the absence of cables/headbox, (iii) the risk of infection is lowered due to the absence of transcutaneous wires.
The ability of participants to report their subjective experience opens a whole new universe of possibilities, as changes of neural activity can be related not only to externally-triggered events but also to internally-generated ones (see for example the investigation of spatially sparse but action specific brain activation during real-life orofacial motor behavior captured with the audiovisual monitoring system (Kern et al., 2019)). This requires that patients have access to their own neural activity in real-time, to detect systematic relationships with their own perceived mental events (such as emotions, mental images, verbalized thoughts, etc.). iEEG offers that opportunity, and a set-up for such neuro-phenomenological investigation has already been proposed more than a decade ago (Jerbi et al., 2009b; Lachaux et al., 2007b). We believe that in the future, this kind of approach will spread in most iEEG centers as more and more groups seek to investigate the neural grounding of naturalistic behavior (including covert mental acts) and the ecological validity of conclusions obtained in constrained laboratory conditions. To go beyond a mere collection of unrelated anecdotes, one could envision that a set of patients are recorded - with the video - during a systematic sequence of natural behaviors under the guidance of an experimenter. There the research would simply engage the patients in a scripted set of activities, as in neuropsychological evaluation. To make the most of this approach, we anticipate that iEEG databases will incorporate video recording of natural behavior by means of techniques to replace the patient with an avatar. Finally, the development of live/online-analysis will permit a closed-loop approach (Zelmann et al., 2020), which will improve research investigations by optimizing stimulation approaches using neurally informed personalized targets, as well as direct electrical stimulation protocols and neuromodulatory therapies.
6.6. Electrical stimulation used for research purposes
Intracranial electrical stimulations (iES) are used clinically to map the organization of functional and epileptic networks in the patient’s brain (see section 1.1.4.1). iES consists of either a high-frequency train of 50 or 60 Hz stimulation lasting a couple of seconds (HFS), or of a series of individual pulses sent typically every second (LFS). Historically, the two types of iES can be traced to the two iEEG approaches: HFS was first implemented in ECoG (Penfield and Boldrey, 1937) and LFS in sEEG (George et al., 2020). Each iES type has its advantages; for example, HFS is more effective to induce seizures, with LFS the false positives are rare, and the brief stimulation artifact in LFS allows visualizing the evolution of induced discharges (Kovac et al., 2016; Mouthaan et al., 2016; Munari et al., 1993; Valentín et al., 2005; Zangaladze et al., 2008). Also, there is a large variability in the use of iES from center to center, which is further increased by contextual factors: the interference of iES with a cognitive process manifest during active engagement of that process, in some instances the effect of iES varies with the timing relative to an external stimuli (Keller et al., 2017), and different brain regions do not have the same responsiveness (Murphey et al., 2009; Trevisi et al., 2018) (see section 1.1.4.1). In line with our forecast of increased interaction between clinic and research, we expect further developments related to iES (see next paragraphs): first the exploitation for research purposes of iES data stemming from clinical procedure (i.e., functional mapping), second in the application of iES during research protocol. In turn, these developments will contribute to a better understanding of the neurophysiology of iES (i.e., the links between ES specs and the underlying electrophysiological outcomes), and to define standards for iES parameters (e.g., current amplitude, duration of the HFS stimulation, inter-stimulation-interval of the LFS).
When an electrical pulse (LFS) is applied at a given iEEG site, this can generate an evoked potential (ERP) at iEEG sites that are anatomically connected to the stimulated site (David et al., 2010; Keller et al., 2014; Matsumoto et al., 2004), such iES related potential (iESRP) is also referred to as Cortico-Cortical Evoked Potentials (CCEP). Analyses of the latency and the amplitude of such iESRPs provide a measure of connectivity between the two sites (see sections 4.3.1 and 4.3.2) and permit drawing neural networks of the patienťs brain. Using a ROI-like approach, this method permits grouping sites over different patients which lie within the same brain region and are connected to the same target regions. Using this approach on iEEG data over different centers, and originally acquired for clinical purposes, a large-scale open initiative has resulted in the construction of a Functional Brain Tractography atlas33 (Trebaul et al., 2018). Until now, due to the lasting HFS artifacts, the investigation of data with high-frequency train stimulation (HFS) remains less investigated (Barborica et al., 2022; Perrone‐Bertolotti et al., 2020). Yet, considering the amount of HFS datasets existing in iEEG centers, and more generally of any iES, we anticipate HFS data to be exploited similarly (e.g. to construct HFS based connectivity atlasses) and expect further investigations comparing stimulation types, and between iES based atlases and structural connectivity (Crocker et al., 2021; Donos et al., 2016; Silverstein et al., 2020).
During the functional mapping in clinic, it is common to observe that iES impacts cognition by either eliciting or impairing a process (see section 1.1.4.1), such as the perception of a stimulus (e.g., disruption of face recognition, hallucination of a place), enhancing (Ezzyat et al., 2018) or disrupting memory (Goyal et al., 2018; Jacobs et al., 2016), or motor behavior (e.g. speech arrest, eye movement). In link with some research questions, iES has been utilized to establish causal inference, not only to relate a structure and a function but also to assess the neurophysiological basis underlying a cognitive process. For instance, iES was used to mimic local neural activity (Jacobs et al., 2012), which can in turn modulate behavioral performance (Alagapan et al., 2019; Kucewicz et al., 2018; Suthana et al., 2012; Titiz et al., 2017) in a task specific manner (Hansen et al., 2018; Jun et al., 2020), and/or shape network dynamics (Beauchamp et al., 2012; Fell et al., 2013; Khambhati et al., 2019; Kim et al., 2018; Solomon et al., 2018). However, these investigations remain sparse. Because iES is the only approach allowing interplay with neural activity in humans with millimeter spatial specificity, we forecast a growing interest for the use of iES during research paradigms.
6.7. From iEEG to the neuroscience of brain lesions
An increasingly diffuse approach in the drug-resistant focal epilepsy surgery is represented by sEEG-guided Radiofrequency-Thermocoagulation (RFTC), i.e. a set of small focal cortical lesions informed by the sEEG recordings that are performed as a therapeutic option immediately before the sEEG electrode removal (Bourdillon et al., 2017; Cossu et al., 2015; Dimova et al., 2017). Beyond their relevant clinical value (Cossu et al., 2015), such a procedure may offer a solution to a longstanding issue in the neuroscience of brain lesions. Indeed, a major goal of system neuroscience is the comprehensive understanding of the structural, functional and connectional modifications following brain lesions. However, the impossibility to compare the post-lesional recordings with pre-lesional ones prevented the achievement of mechanistic insights into post-lesional processes like adaptive neuroplasticity and diaschisis (Fornito et al., 2015).
In this realm, the effects of controlled lesions can be assessed with spatially resolved electrophysiological recordings and compared to prelesional measurements within the same individual (Russo et al., 2021). Following this principle, Russo et al (2021) demonstrated that after RFTC, not only the perilesional area presented an increased delta activity whose amplitude rapidly decayed with distance, but also that sleep-like slow waves were prominent in the perilesional areas but could also percolate through a network of connected areas as predicted by individual patterns of long-range effective connectivity.
Despite the application of RFTC being solely guided by clinical purposes, the collection of pre- and post-lesional iEEG data might enrich and complement the current knowledge about brain functions with causal, focal and well-localized evidence.
6.8. Exploiting white matter signals
In the case of sEEG about 30–40% explore the white matter (WM) (Mercier et al., 2017). To date, these signals have been used mostly to compose the reference channel (see sections 1.3.2 and 3.3.1.2), yet the value of the WM signals is still underexplored. In a recent study, Mercier and coworkers showed that, once accounted for the backpropagation and volume conduction effects from the nearby cortical electrodes, the signals recorded from white matter tracts can also reflect neuronal communication between distant regions of cortex through WM fiber tracts, with small but positive cross-correlation time lag values. Similar conclusions were obtained by Rizzi and colleagues (Rizzi et al., 2021), who showed that the WM recordings increase in the gamma band power along visual stimulations following the neuroanatomical distribution of the optical radiation according to both in-vivo and ex-vivo investigations. Furthermore, even in this study the timing observed in the leads intersecting the optical radiation differed from that observed in the other leads exploring the WM or the gray matter leads, reinforcing the notion that the sEEG signal collected from WM leads has peculiar features related to the functional role of the networks incorporating the specific fibers. Studies have also investigated WM contacts using intracranial electrical stimulation (iES, see sections 1.1.4.1 and 6.6), and showed the relevance of electrically stimulating WM to defined functional connectivity in brain networks (Koubeissi et al., 2013; Solomon et al., 2018).
Such a picture opens up to two main points to be considered in the design of an iEEG experiment and the analysis of the relative data. On one hand, WM leads cannot be defined as being neutral regardless of the neural processes that will be engaged in the patient; consequently, WM contact(s) that compose the reference should not only be anatomically identified but also characterized according to functional criteria (e.g., minimum variance, see section 3.3.1.2). On the other hand, the large amount of data regularly collected from WM leads in sEEG recordings provides valuable information to describe the anatomical and functional correlates of several WM bundles. Given their coherence with tractography and anatomical data, these studies would contribute to characterize the human connectome with an innovative source of information, complementing the neuroimaging techniques with a four-dimensional (i.e., space and time), time-dependent characterization of WM structures.
6.9. Advances analysis methods
6.9.1. Mixed-effects multilevel analysis to take into account spatial sparsity
The field of iEEG increasingly acknowledges the importance of appropriately modeling the correlation structure of the data, e.g., that electrodes should not simply be pooled across participants in statistical group analyses. Improperly accounting for the correlation structure, e.g., that electrodes within an ROI will be more correlated within a participant than between participants, can lead to an inflation of Type I errors (Yu et al., 2021). So-called linear mixed effects (LME) models provide a flexible statistical framework that allows for group analyses and the correlation structure of the data. Such models include both fixed effects, such as experimental conditions, which are constant across individuals, as well as random effects, which refer to factors that take into account variability between participants or observations (see section 5.2.3). Furthermore, they allow the inclusion of nested random effect factors, which allow explicit hierarchical grouping of observations (Galbraith et al., 2010; Krzywinski et al., 2014), e.g., electrodes within a participant. Thereby, such analyses are guarded against individual participants (or electrodes) driving the effects. Furthermore, they can easily account for sparse, missing, or unequal numbers of electrodes per ROI. Additionally, by nature of being a random effects analysis, the results are readily generalizable beyond the participants under study. For these reasons, this type of statistical analysis is becoming increasingly prevalent in iEEG studies (e.g., (Golan et al., 2016; Kadipasaoglu et al., 2014; Schwiedrzik et al., 2018). LME models are straightforwardly implemented in R, e.g., in the well known LME package (Bates et al., 2015), as well as in recent versions of MATLAB (some alternatives exist in Python, for iEEG see (Combrisson et al., 2021)). However, LME modeling is not without caveats. For example, it remains an unresolved question whether to model the data with the maximal random effect structure allowed by the experimental design, which includes random intercepts and random slopes for all within experimental factors, or to target specific statistical hypotheses with more parsimonious models.
6.9.2. Multivariate analysis and machine learning
Like in MRI and M/EEG (Haynes, 2015; King and Dehaene, 2014; Ritchie et al., 2019), multivariate analysis benefits from the high dimensionality of iEEG data because it combines information from all available features (e.g., electrodes and time/frequency points). Moreover, this approach provides a means to deal with the varying sensitivity of the iEEG signal across channels (see section 3.2.4).
Supervised learning methods (using, among others, multivariate pattern analysis, linear discriminant analysis or support vector machine), are generally based on a cross-validation procedure (for an introduction see: (Blankertz et al., 2011; Grootswagers et al., 2017; Guggenmos et al., 2018)). The algorithm initially classifies experimental conditions on a subset of the data by defining decoding weights assigned to each feature (e.g. electrode); the weights reflecting how a feature contributed to maximizing the decoding. Subsequently, it decodes conditions in another subset of the data and provides decoding scores (Anumanchipalli et al., 2019; Moses et al., 2021; Pasley et al., 2012). Decoding analysis can be run on processed data (e.g., frequency analysis), not necessitating a-priori assumptions on specific data features (e.g., frequency range of interest) and allowing for different brain regions (or electrodes) having relevant information in different frequency bands (Baroni et al., 2020; Combrisson et al., 2017; Henin et al., 2021; Iemi et al., 2022; Ter Wal et al., 2020). This permits to caracterise interplay in functional networks (see section 4.3 and 4.4). However, caution is required to reach the full potential of these methods and to avoid pitfalls (Combrisson and Jerbi, 2015). For instance, interpreting the weights is not straightforward. While their transformation back into activation patterns should permit identifying features that distinguish experimental conditions, in some cases it can be misleading (e.g., if a feature provides information about the noise which turns out to help the decoding). How to combine data from multiple participants is also a matter of debate and, as for other types of analyses, no consensus emerged until now (see section 5).
Multivariate analysis of iEEG data is nonetheless at an early stage and we expect a spread in its application to iEEG research, as well as the adoption of other machine learning methods such as time-varying dynamic Bayesian network (Collard et al., 2016) and deep neural networks (Li et al., 2022).
6.10. Network dynamics
Brain functions can overlap in time and/or space. iEEG offers the unique possibility of obtaining electrophysiological recordings with high temporal and spatial resolution (ms over a volume of mm3). This allowed researchers to overcome the “Heisenberg principle” of non-invasive neuroimaging and recording techniques, i.e., the impossibility of having both spatially resolved and dynamic profiles of functioning brain networks (see sections 4.3, 4.4 and (Avanzini et al., 2016)).
Thanks to iEEG, four-dimensional maps (spatial coordinates plus the time dimension) and networks are now increasingly considered as elements to understand brain activity in fundamental neuroscience (Johnson and Knight, 2015; Nakai et al., 2019; Wang et al., 2021) and clinical neuroscience (Bartolomei et al., 2017; Burns et al., 2014; Laufs, 2012; Stam, 2014). The time course of activity (either in terms of amplitude, in terms of spectral magnitude or phase) provides a pivotal factor to make a distinction among different functional implementations of processes in these networks. Such an analytical approach has been successfully applied to studies of conscious perception, in which iEEG revealed that sustained, tonic high-frequency responses are required to make the patient consciously perceive visual (Fisch et al., 2009) and somatosensory (Del Vecchio et al., 2021) stimulations. As the majority of neurons have multidimensional response properties, their ensemble activity leads to representations in a high-dimensional space (Gothard, 2020). Highly-resolved temporal features of network response allow for the identification of and distinguishing between functional roles (Del Vecchio and Avanzini, 2020). We see value in further subdividing the temporal domain into multiple scales (e.g., milliseconds, days/weeks, age), each reflecting and indexing different neural processes (e.g., brain dynamics, plasticity and learning, aging, see (Sonoda et al., 2021)).
Highlights.
We provide good practices for both new and experienced iEEG researchers.
We bridge the patient, clinician and researcher perspective.
Sections include data collection, processing, localization, analysis and statistics.
Each section provides a review, gives recommendations and demonstrates visualization.
We provide an outlook and discuss perspectives on continued iEEG development.
8. Acknowledgements
We dedicate this review paper to the patients that make this research possible, thanks to their participation they move clinical and fundamental neuroscience forward. We thank the organizers and attendees of the online LiveMEEG 2020 conference for providing a welcoming platform to present and discuss some of the ideas that formed the basis for this manuscript. We thank Mariana P. Branco for help with Figure 4. Last, we acknowledge the three anonymous reviewers for thoroughly reading the manuscript and for their insightful comments.
MRM is supported by EU-REA H2020 MSCA - IF 798853.
FT is supported by NIH/NIBIB R01EB026299.
DH is supported by NIMH/NIH R01MH122258.
RTK is supported by NIH/NINDS 2 R01 NS021135, NIH/NINDS 1U19NS107609–01.
NFR and MJV are supported by NIH/NIDCD U01 DC016686 and NWO 17619.
PM is supported by Swiss National Science Foundation grants 167836 and 194507.
LM is supported by the Max Planck Society.
AP is supported by NIH/NIBIB R01EB030896.
CMS is supported by the Emmy Noether Program of the German Research Foundation (SCHW1683/2–1).
7. Glossary
To ensure a consistent nomenclature throughout the manuscript and with other guidelines (e.g., iEEG BIDS and M/EEG COBIDAS), we here provide a short summary of terms related to iEEG research.
- patient, research participant, and subject
After being informed about the research goals and the fact that the research does not aim to contribute to, but will also not impeach, the clinical procedures, the patient may give consent to participate in the research study and from that moment on becomes a participant. This is similar to a healthy person that volunteers or signs up for a study: after that person receives information about the research protocol and she/he consents, that person becomes a participant. Patient stresses the clinical side, participant the experimental side. The term ‘subject’ relates to being (passively) subjected to experimental manipulations; it is a term that we prefer to avoid, as we prefer to highlight the role of an active contributor and stakeholder in the research
- epileptogenic zone
The region(s) that is (are) necessary and sufficient for initiating seizures and whose removal (or disconnection) is indispensable for complete abolition of seizures (Jehi, 2018; Lüders et al., 2019; Rosenow et al., 2016; Talairach and Bancaud, 1966)
- seizure onset zone
The region(s) from which clinical seizures are initiated. Because the overlap of the seizure onset zone (identified by iEEG measurements) and the epileptogenic zone can be imperfect, surgical resection of the seizure onset zone may be insufficient to achieve lasting seizure freedom (Jehi, 2018; Rosenow and Lüders, 2001)
- seizure semiology and symptomatogenic zone
The symptoms experienced by the patient, and the clinical features that they display, during a seizure. The seizure semiology is related to the concept of the symptomatogenic zone (Rosenow and Lüders, 2001), which does not necessarily coincide with the seizure onset zone
- interictal epileptiform discharge and irritative zone
A brief pattern of EEG activity that is used as a marker of epilepsy in epilepsy patients. Interictal epileptiform discharges originate in the irritative zone (Rosenow and Lüders, 2001). As their name implies, interictal epileptiform discharges are distinct from seizures. Here, we avoid the commonly-used term “epileptic spikes” to prevent confusion with neuronal spiking (i.e. action potentials)
- pre/post-implantation imaging
Medical images acquired before and after surgical placement of the iEEG implants to allow an accurate representation of the implants’ placement with respect to internal brain structures. The post-implantation imaging provides the location of individual recording electrode contacts of the implant. Because the implants create imaging artifacts, post-implantation imaging is generally combined with pre-implantation imaging which allows for a high quality rendering of the anatomy. Pre/post-implantation comparison also allows to assess post-implant tissue displacement due to edema or any injury due to the implant; in the latter case, pre/post-implantation imaging can be further completed with a post-explantation scan (i.e., after the electrode implants have been removed). This is also known as pre/post-surgical scan and post-explantation scan, where surgery refers to the implantation. The scans can be MRI and/or X-ray CT
- atlas
An atlas is the result of mapping 3D information (such as population density) on a geometrical template (description + delineation). The represented information can be continuous (such as the amount of rain that falls) but is often restricted to a spatial delineation and geographic description (e.g., roads, cities, municipalities). The results of such a mapping can be considered an atlas once they are shared with the purpose of re-use for another dataset. An atlas is helpful to visualize and interpret spatial information
- parcellation
Delineation of brain regions on the basis of anatomical and/or functional information. A given parcellation is generally defined on an atlas. If an individual brain is geometrically co-registered with the atlas, it is possible to project the parcellation on the individual brain
- Individual space
This refers to a 3-D cartesian coordinate system where the direction of the axes and the origin of the coordinate system (i.e., the point [0, 0, 0]) is defined in relation to anatomical landmarks of the individual (i.e., the single person whose brain activity is being measured). The relationship of these landmarks is preserved across different people, but the relative distances between them is different. The most common example of this in the brain would be a coordinate system defined by the anterior & posterior commissures and the brain’s midline. This is sometimes also referred to as the “native” space or coordinate system, but here we avoid that to prevent confusion with the device coordinate system where the direction of the axes and the origin are expressed relative to the scanner (i.e., the center of the gradient coils in case of MRI)
- iEEG implant / probe / shaft / grid / strip
There are two main types of intracranial electroencephalography (iEEG) implants (a.k.a. intracranial probes): stereo-encephalographic implants (sEEG) are stereotactically inserted intraparenchymal semi-rigid electrode shafts with multiple contacts whose endpoints target particular anatomical structures within the brain itself (e.g. hippocampus), while contacts along the shaft typically target cortical areas. sEEG probes are sited through boreholes drilled through the skull. Electrocorticographic implants (ECoG) consist of either electrode arrays arranged in grids (2D) or strips (1D) that are inserted below the dura mater to line the surface of the brain
- electrodes and electrode contacts
The term was introduced in physics to name an electrical conductor used to make contact with a nonmetallic part of a circuit. Therefore it refers to the sensor that is in contact with the recorded tissue. In ECoG, individual sensors are arranged in arrays in grid or strip configuration. These individual sensors can also be called electrode contacts. A semi-rigid sEEG shaft houses multiple electrode-contacts
- site
In this manuscript we use it as the physical location where an electrode is located on or in the brain
- channel
A single signal from an amplifier or transducer that is digitally sampled. For iEEG this is the output of the analog-to-digital (ADC) conversion from the amplified voltage difference between an electrode contact of interest and a reference electrode. A channel does not have to correspond to a single site or physical electrode. For instance in the case of a bipolar reference: one electrode is re-referenced to its neighbor, the result is generally considered as a channel that is virtually located at mid-distance between the two electrodes
- signal
A time series of the data being recorded by an analog-digital converter, which can include both brain and non-brain activity. After recording to disk, the signal is subjected to offline preprocessing and analysis with the aim to increase the signal feature(s) related to neural activity
- acquisition or recording
Storing a signal for either online or offline analysis
- impedance
Electrical impedance is the resistance to electric current (which is the flow of electric charge), or to changes in electric current for capacitive and inductive impedance. The impedance is of relevance in biopotential measurements, as it impacts the signal quality. A distinction must be made for the impedance of the different elements that play a role in the signal acquisition. There is the impedance of the biological tissue, the impedance of the interface between the tissue and the electrode, the impedance of the electrodes and the wires, and that of the electronic components in the amplifier. Different brain tissues have different impedances, which can vary with tissue integrity (e.g., inflammation) and over time (Sillay et al., 2013)
- volume conduction
Volume conduction refers to the passive signal spread in the conductive tissues and can be modeled with Maxwell’s equations. It is sometimes called passive ‘propagation’; in French it is sometimes referred to as ‘diffusion’ (which is technically not appropriate when it comes to electricity). In this manuscript we restrict the use of ‘propagation’ for the active biochemical processes at the synapses and along the cell membrane of neurons
- active propagation
Biochemical processes corresponding to the propagating electrical impulse traveling along the cell membrane of neurons and nerve fibers
- activity
The biochemical neural activity can be split into postsynaptic potentials (at the dendrites) and action potentials (along the axon). Local Field Potentials recorded with iEEG originate mostly from the postsynaptic potentials; an LFP is the sum of these synchronized inputs that passively spreads and contributes to the signal detected on the electrode. From a neural perspective, post-synaptic potentials are graded potentials (i.e. synaptic potentials, subthreshold oscillations…) that initiate or inhibit action potentials. Action potentials travel along the nerve fiber (i.e., axon). As the neuron membrane depolarization-repolarization lasts a few milliseconds, measuring action potential requires higher frequency sampling than LFPs (generally >10kHz) and therefore dedicated equipment (e.g. amplifier, micro-electrode)
- reference scheme or montage
Reference scheme (or reference montage) refers to the way the online or offline referencing between electrode pairs is performed to amplify, record and/or visualize the potential differences. In a ‘unipolar’ reference scheme one electrode is used as the reference for all other electrodes. In a ‘bipolar’ scheme each electrode is referenced to a neighboring electrode. In a common average reference scheme all electrodes are referenced to the average of all electrodes. It is also possible to have composite reference schemes. The term ‘multipolar’ is to be avoided
- spatial resolution
In the field of neurophysiology, the term "spatial resolution” can pertain to various aspects that are somehow related: spatial specificity for picking up different physiological sources, (spatial) density of the recording electrodes, the spatial extent of a single electrode (i.e., the electrode contact size), and the (spatial) coverage. Rather than using “spatial resolution” in the manuscript, we prefer to explicitly use these specific terms where applicable.Spatial specificity can be defined as “the ability to extract independent time course estimates of electrical brain activity from two separate brain locations in close proximity" (from (Brookes et al., 2010)) The (spatial) density of the recording relates to the distance between recording sites, like in the commonly used expression “high density EEG”. The electrode contact size relates to the spatial extent of the electrode-electrolyte interface over which the biopotential is integrated. Small electrodes pick up focal (spatially specific) activity, whereas large electrodes average over a larger area. The (spatial) coverage refers to the extent of the samping relative to the whole brain (e.g. unilateral/bilateral implant, number of lobes or sublobar region covered). This can be compared to MRI, where the coverage can be configured as a few slices, the whole brain, or even the whole head. An analogy can be drawn to the field of digital photography where spatial resolution depends not only on the number of pixels in the CCD chip, but also on the optical characteristics of the lens (the focal length and aperture) and on the focus of the lens. We employ the term spatial resolution to refer to the spatial specificity, as with iEEG the sampling is always sparse, while the signal can be focused to achieve high spatial specificity (i.e., through re-referencing). In the photography analogy, spatial coverage refers to the field-of-view
- time-frequency decomposition and time-frequency representation
the decomposition refers to the process (analysis), the representation refers to the outcome of that process (figure, statistics…). More generally in this manuscript we mostly consider the methodological aspects and try to use specific terminology and avoid general terms, such as: “activity” (unless it refers to the actual neural activity, not the signal that is being analyzed) or “measures” (not specific enough, unless it relates to the signal itself or to a metric). Other terms could be appropriate when considering the actual activity, for instance Event-Related Synchronization/Desynchronization (ERS/ERD), or Event Related Spectral Perturbation (ERSP). Those terms relate to, or suggest, an active neural process, whereas the results of a time-frequency analysis can be flat, and a change in activity only revealed after baseline correction. Finally, some terminology relates more specifically to the kind of analysis that was performed, for example Event-Related Band Power (ERBP) which is a time-locked analysis focusing on the power in a given frequency band
- oscillations or brain rhythms
The terms “oscillation” or “rhythms” generally are used to reflect the periodic changes in the measured voltages. These were the earliest observed phenomena in neuroelectrophysiology, and can generally be seen in the raw trace. Spectral analysis can reveal distinct peaks in the power that are indicated with greek-letter names (e.g., delta, theta, alpha, beta, gamma). It is useful to consider the distinction between the neurophysiological mechanisms (e.g., brain rhythms), the signals that are recorded, and the results of an analysis on those signals (e.g., oscillation from Fourier transform)
- aperiodic and broadband activity
In contrast to commonly-described oscillations or rhythms centered at a specific frequency, there are also signal features that have no specific timescale and do not have a periodic structure. The raw voltage traces of these changes appear as shifts in the random-walk aspects of the signal that are often treated as “background noise”. Careful parameterization of these broadband signals—often using Fourier-domain approaches that extract spectrally wide features—provides another avenue for analyzing the data in a manner that provides physiological insights not captured by oscillations
- non-sinusoidal oscillation
Common analysis approaches as often implemented (e.g., Fourier- or wavelet-based approaches) decompose the signal into mixtures of simple sine and cosine components of different frequencies. However, most neural oscillations are not sinusoidal; rather they exhibit a rich variety of periodic waveforms that can include the the classic mu-shaped rhythm in somatosensory cortex, V-shaped alpha oscillations in visual cortex, and sawtooth-shaped beta oscillations in motor cortex or theta oscillations in hippocampus. These non-sinusoidal features are easily overlooked using common approaches or confounded for cross-frequency coupling, despite the fact that they might carry critical physiological information
- non-stationarity
This characterizes a time series whose statistical properties are not stable over time. In iEEG analysis this most commonly refers to oscillations, where oscillation power, frequency, and phase are dynamic and change over time. This is especially important given that many oscillations are “bursty”—where non-oscillatory periods are dominated by “aperiodic activity”—and where narrow band filtering can give the appearance of a stable oscillation where one might not exist
- baseline
The baseline is a concept that relates to signal processing, data visualization and statistics. Considering preprocessing and ERP analysis, the baseline window refers to the time interval (often pre-stimulus) in which the baseline DC offset is estimated; this estimate can be used to subtract the constant potential offset. Rather than referring to this as baseline correction, in this manuscript we prefer the term DC offset removal. In time-frequency analysis the baseline window refers to the time interval in which the background spectral activity is estimated, and this period may vary depending on analysis type. The estimated spectral activity can be used in visualization and statistical assessment to express the change with respect to the baseline background oscillatory activity (the foreground being the one of interest).Generally, we think of baseline periods as those where no defined task or process is taking place. However, the brain is never quiescent, so the expectation of the experimenter is that the effect of undefined (ongoing) brain activity is removed due to averaging, revealing the effect of the defined task or process
Footnotes
9. References
- Abel TJ, Rhone AE, Nourski KV, Granner MA, Oya H, Griffiths TD, Tranel DT, Kawasaki H, Howard MA, 2014. Mapping the temporal pole with a specialized electrode array: technique and preliminary results. Physiol. Meas 35, 323–337. 10.1088/0967-3334/35/3/323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramian D, Eklund A, 2019. Refacing: Reconstructing Anonymized Facial Features Using GANS, in: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Presented at the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI), IEEE, Venice, Italy, pp. 1104–1108. 10.1109/ISBI.2019.8759515 [DOI] [Google Scholar]
- Adhikari A, Sigurdsson T, Topiwala MA, Gordon JA, 2010. Cross-correlation of instantaneous amplitudes of field potential oscillations: a straightforward method to estimate the directionality and lag between brain areas. J. Neurosci. Methods 191, 191–200. 10.1016/j.jneumeth.2010.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagapan S, Lustenberger C, Hadar E, Shin HW, Frӧhlich F, 2019. Low-frequency direct cortical stimulation of left superior frontal gyrus enhances working memory performance. NeuroImage 184, 697–706. 10.1016/j.neuroimage.2018.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alday PM, 2019. How much baseline correction do we need in ERP research? Extended GLM model can replace baseline correction while lifting its limits. Psychophysiology 56, e13451. 10.1111/psyp.13451 [DOI] [PubMed] [Google Scholar]
- Alexander DM, Ball T, Schulze-Bonhage A, van Leeuwen C, 2019. Large-scale cortical travelling waves predict localized future cortical signals. PLOS Comput. Biol 15, e1007316. 10.1371/journal.pcbi.1007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison T, McCarthy G, Nobre A, Puce A, Belger A, 1994. Human extrastriate visual cortex and the perception of faces, words, numbers, and colors. Cereb. Cortex N. Y. N 1991 4, 544–554. 10.1093/cercor/4.5.544 [DOI] [PubMed] [Google Scholar]
- Aminoff EM, Li Y, Pyles JA, Ward MJ, Richardson RM, Ghuman AS, 2016. Associative hallucinations result from stimulating left ventromedial temporal cortex. Cortex 83, 139–144. 10.1016/j.cortex.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Mohlberg H, Bludau S, Zilles K, 2020. Julich-Brain: A 3D probabilistic atlas of the human brain’s cytoarchitecture. Science 369, 988–992. 10.1126/science.abb4588 [DOI] [PubMed] [Google Scholar]
- Anumanchipalli GK, Chartier J, Chang EF, 2019. Speech synthesis from neural decoding of spoken sentences. Nature 568, 493–498. 10.1038/s41586-019-1119-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnulfo G, Narizzano M, Cardinale F, Fato MM, Palva JM, 2015. Automatic segmentation of deep intracerebral electrodes in computed tomography scans. BMC Bioinformatics 16, 99. 10.1186/s12859-015-0511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnulfo G, Wang SH, Myrov V, Toselli B, Hirvonen J, Fato MM, Nobili L, Cardinale F, Rubino A, Zhigalov A, Palva S, Palva JM, 2020. Long-range phase synchronization of high-frequency oscillations in human cortex. Nat. Commun 11, 5363. 10.1038/s41467-020-18975-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aru Juhan, Aru Jaan, Priesemann V, Wibral M, Lana L, Pipa G, Singer W, Vicente R, 2015. Untangling cross-frequency coupling in neuroscience. Curr. Opin. Neurobiol 31, 51–61. 10.1016/j.conb.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ, 2005. Unified segmentation. NeuroImage 26, 839–851. 10.1016/j.neuroimage.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Avanzini P, Abdollahi RO, Sartori I, Caruana F, Pelliccia V, Casaceli G, Mai R, Lo Russo G, Rizzolatti G, Orban GA, 2016. Four-dimensional maps of the human somatosensory system. Proc. Natl. Acad. Sci 113, E1936–E1943. 10.1073/pnas.1601889113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccalá LA, Sameshima K, 2001. Partial directed coherence: a new concept in neural structure determination. Biol. Cybern 84, 463–474. 10.1007/PL00007990 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M, 1998. Methods and statistics for combining motif match scores. J. Comput. Biol. J. Comput. Mol. Cell Biol 5, 211–221. 10.1089/cmb.1998.5.211 [DOI] [PubMed] [Google Scholar]
- Bakr SM, Patel A, Zaazoue MA, Wagner K, Lam SK, Curry DJ, Raskin JS, 2021. Standard work tools for dynamic stereoelectroencephalography using ROSA: naming convention and perioperative planning. J. Neurosurg. Pediatr 27, 411–419. 10.3171/2020.8.PEDS20420 [DOI] [PubMed] [Google Scholar]
- Ball T, Kern M, Mutschler I, Aertsen A, Schulze-Bonhage A, 2009. Signal quality of simultaneously recorded invasive and non-invasive EEG. NeuroImage 46, 708–716. 10.1016/j.neuroimage.2009.02.028 [DOI] [PubMed] [Google Scholar]
- Bancaud J,T,J, 1965. La Stereoelectroencephalographie Dans L’ epilepsie: Informations Neurophysiopathologiques Apportees Par L’investigation Fonctionnelle Stereotaxique. Masson, Paris. [Google Scholar]
- Bannier E, Barker G, Borghesani V, Broeckx N, Clement P, Emblem KE, Ghosh S, Glerean E, Gorgolewski KJ, Havu M, Halchenko YO, Herholz P, Hespel A, Heunis S, Hu Y, Hu C, Huijser D, Iglesia Vayá M, Jancalek R, Katsaros VK, Kieseler M, Maumet C, Moreau CA, Mutsaerts H, Oostenveld R, Ozturk‐Isik E, Pascual Leone Espinosa N, Pellman J, Pernet CR, Pizzini FB, Trbalić AŠ, Toussaint P, Visconti di Oleggio Castello M, Wang, Wang, Zhu, 2021. The Open Brain Consent: Informing research participants and obtaining consent to share brain imaging data. Hum. Brain Mapp 42, 1945–1951. 10.1002/hbm.25351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barborica A, Oane I, Donos C, Daneasa A, Mihai F, Pistol C, Dabu A, Roceanu A, Mindruta I, 2022. Imaging the effective networks associated with cortical function through intracranial high‐frequency stimulation. Hum. Brain Mapp 43, 1657–1675. 10.1002/hbm.25749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni F, Morillon B, Trébuchon A, Liégeois-Chauvel C, Olasagasti I, Giraud A-L, 2020. Converging intracortical signatures of two separated processing timescales in human early auditory cortex. NeuroImage 218, 116882. 10.1016/j.neuroimage.2020.116882 [DOI] [PubMed] [Google Scholar]
- Bartolomei F, Lagarde S, Wendling F, McGonigal A, Jirsa V, Guye M, Bénar C, 2017. Defining epileptogenic networks: Contribution of SEEG and signal analysis. Epilepsia 58, 1131–1147. 10.1111/epi.13791 [DOI] [PubMed] [Google Scholar]
- Bassett DS, Sporns O, 2017. Network neuroscience. Nat. Neurosci 20, 353–364. 10.1038/nn.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin J, Lebranchu P, Jerbi K, Kahane P, Orban G, Lachaux J-P, Berthoz A, 2012. Direct recordings in human cortex reveal the dynamics of gamma-band [50–150 Hz] activity during pursuit eye movement control. NeuroImage 63, 339–347. 10.1016/j.neuroimage.2012.07.011 [DOI] [PubMed] [Google Scholar]
- Bastos AM, Schoffelen J-M, 2015. A Tutorial Review of Functional Connectivity Analysis Methods and Their Interpretational Pitfalls. Front. Syst. Neurosci. 9, 175. 10.3389/fnsys.2015.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S, 2015. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Baudena P, Halgren E, Heit G, Clarke JM, 1995. Intracerebral potentials to rare target and distractor auditory and visual stimuli. III. Frontal cortex. Electroencephalogr. Clin. Neurophysiol 94, 251–264. 10.1016/0013-4694(95)98476-o [DOI] [PubMed] [Google Scholar]
- Baxendale S, Wilson SJ, Baker GA, Barr W, Helmstaedter C, Hermann BP, Langfitt J, Reuner G, Rzezak P, Samson S, Smith M-L, 2019. Indications and expectations for neuropsychological assessment in epilepsy surgery in children and adults. Epileptic Disord. Int. Epilepsy J. Videotape 21, 221–234. 10.1684/epd.2019.1065 [DOI] [PubMed] [Google Scholar]
- Beauchamp MS, Sun P, Baum SH, Tolias AS, Yoshor D, 2012. Electrocorticography links human temporoparietal junction to visual perception. Nat. Neurosci 15, 957–959. 10.1038/nn.3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabid AL, Pollak P, Louveau A, Henry S, de Rougemont J, 1987. Combined (Thalamotomy and Stimulation) Stereotactic Surgery of the VIM Thalamic Nucleus for Bilateral Parkinson Disease. Stereotact. Funct. Neurosurg 50, 344–346. 10.1159/000100803 [DOI] [PubMed] [Google Scholar]
- Bénar CG, Papadopoulo T, Torrésani B, Clerc M, 2009. Consensus Matching Pursuit for multi-trial EEG signals. J. Neurosci. Methods 10. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Morrone MC, Tomassini A, 2020. The Common Rhythm of Action and Perception. J. Cogn. Neurosci 32, 187–200. 10.1162/jocn_a_01436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezutskaya J, Vansteensel MJ, Aarnoutse EJ, Freudenburg ZV, Piantoni G, Branco MP, Ramsey NF, 2022. Open multimodal iEEG-fMRI dataset from naturalistic stimulation with a short audiovisual film. Sci. Data 9, 91. 10.1038/s41597-022-01173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, 2019. Changing the Way We Report, Interpret, and Discuss Our Results to Rebuild Trust in Our Research. eNeuro 6, ENEURO.0259–19.2019. 10.1523/ENEURO.0259-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi A, Cendes F, Theodore WH, Gill RS, Koepp MJ, Hogan RE, Jackson GD, Federico P, Labate A, Vaudano AE, Blümcke I, Ryvlin P, Bernasconi N, 2019. Recommendations for the use of structural magnetic resonance imaging in the care of patients with epilepsy: A consensus report from the International League Against Epilepsy Neuroimaging Task Force. Epilepsia epi.15612. 10.1111/epi.15612 [DOI] [PubMed] [Google Scholar]
- Betzel RF, Medaglia JD, Kahn AE, Soffer J, Schonhaut DR, Bassett DS, 2019. Structural, geometric and genetic factors predict interregional brain connectivity patterns probed by electrocorticography. Nat. Biomed. Eng 3, 902–916. 10.1038/s41551-019-0404-5 [DOI] [PubMed] [Google Scholar]
- Bidet-Caulet A, Fischer C, Besle J, Aguera P-E, Giard M-H, Bertrand O, 2007. Effects of Selective Attention on the Electrophysiological Representation of Concurrent Sounds in the Human Auditory Cortex. J. Neurosci 27, 9252–9261. 10.1523/JNEUROSCI.1402-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billig AJ, Herrmann B, Rhone AE, Gander PE, Nourski KV, Snoad BF, Kovach CK, Kawasaki H, Howard MA, Johnsrude IS, 2019. A Sound-Sensitive Source of Alpha Oscillations in Human Non-Primary Auditory Cortex. J. Neurosci 39, 8679–8689. 10.1523/JNEUROSCI.0696-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bím J, De Feo V, Chicharro D, Bieler M, Hanganu-Opatz IL, Brovelli A, Panzeri S, 2019. A Non-negative Measure Of Feature-Related Information Transfer Between Neural Signals (preprint). Neuroscience. 10.1101/758128 [DOI] [Google Scholar]
- Blankertz B, Lemm S, Treder M, Haufe S, Müller K-R, 2011. Single-trial analysis and classification of ERP components — A tutorial. NeuroImage 56, 814–825. 10.1016/j.neuroimage.2010.06.048 [DOI] [PubMed] [Google Scholar]
- Bleichner MG, Freudenburg ZV, Jansma JM, Aarnoutse EJ, Vansteensel MJ, Ramsey NF, 2016. Give me a sign: decoding four complex hand gestures based on high-density ECoG. Brain Struct. Funct 221, 203–216. 10.1007/s00429-014-0902-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichner MG, Vansteensel MJ, Huiskamp GM, Hermes D, Aarnoutse EJ, Ferrier CH, Ramsey NF, 2011. The effects of blood vessels on electrocorticography. J. Neural Eng 8, 044002. 10.1088/1741-2560/8/4/044002 [DOI] [PubMed] [Google Scholar]
- Blenkmann AO, 2017. iElectrodes: A Comprehensive Open-Source Toolbox for Depth and Subdural Grid Electrode Localization. Front. Neuroinformatics 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly M, Jones B, Findlay G, Plumley E, Mensen A, Hermann B, Tononi G, Maganti R, 2017. Altered sleep homeostasis correlates with cognitive impairment in patients with focal epilepsy. Brain 140, 1026–1040. 10.1093/brain/awx017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers S, Himmelbach M, Logothetis N, Karnath H-O, 2012. Direct electrical stimulation of human cortex — the gold standard for mapping brain functions? Nat. Rev. Neurosci 13, 63–70. 10.1038/nrn3140 [DOI] [PubMed] [Google Scholar]
- Bouchard KE, Mesgarani N, Johnson K, Chang EF, 2013. Functional organization of human sensorimotor cortex for speech articulation. Nature 495, 327–332. 10.1038/nature11911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewyn MA, Luck SJ, Farrens JL, Kappenman ES, 2018. How many trials does it take to get a significant ERP effect? It depends. Psychophysiology 55, e13049. 10.1111/psyp.13049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdillon P, Isnard J, Catenoix H, Montavont A, Rheims S, Ryvlin P, Ostrowsky-Coste K, Mauguiere F, Guénot M, 2017. Stereo electroencephalography-guided radiofrequency thermocoagulation (SEEG-guided RF-TC) in drug-resistant focal epilepsy: Results from a 10-year experience. Epilepsia 58, 85–93. 10.1111/epi.13616 [DOI] [PubMed] [Google Scholar]
- Bouthillier A, Surbeck W, Weil AG, Tayah T, Nguyen DK, 2012. The hybrid operculo-insular electrode: a new electrode for intracranial investigation of perisylvian/insular refractory epilepsy. Neurosurgery 70, 1574–1580; discussion 1580. 10.1227/NEU.0b013e318246a3b7 [DOI] [PubMed] [Google Scholar]
- Branco MP, Freudenburg ZV, Aarnoutse EJ, Bleichner MG, Vansteensel MJ, Ramsey NF, 2017. Decoding hand gestures from primary somatosensory cortex using high-density ECoG. NeuroImage 147, 130–142. 10.1016/j.neuroimage.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MP, Freudenburg ZV, Aarnoutse EJ, Vansteensel MJ, Ramsey NF, 2018a. Optimization of sampling rate and smoothing improves classification of high frequency power in electrocorticographic brain signals. Biomed. Phys. Eng. Express 4, 045012. 10.1088/2057-1976/aac3ac [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MP, Gaglianese A, Glen DR, Hermes D, Saad ZS, Petridou N, Ramsey NF, 2018b. ALICE: A tool for automatic localization of intra-cranial electrodes for clinical and high-density grids. J. Neurosci. Methods 301, 43–51. 10.1016/j.jneumeth.2017.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MP, Leibbrand M, Vansteensel MJ, Freudenburg ZV, Ramsey NF, 2018c. GridLoc: An automatic and unsupervised localization method for high-density ECoG grids. NeuroImage 179, 225–234. 10.1016/j.neuroimage.2018.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandmeir NJ, Savaliya S, Rohatgi P, Sather M, 2018. The comparative accuracy of the ROSA stereotactic robot across a wide range of clinical applications and registration techniques. J. Robot. Surg 12, 157–163. 10.1007/s11701-017-0712-2 [DOI] [PubMed] [Google Scholar]
- Bressler SL, Seth AK, 2011. Wiener-Granger causality: a well established methodology. NeuroImage 58, 323–329. 10.1016/j.neuroimage.2010.02.059 [DOI] [PubMed] [Google Scholar]
- Brett M, Johnsrude IS, Owen AM, 2002. The problem of functional localization in the human brain. Nat. Rev. Neurosci 3, 243–249. 10.1038/nrn756 [DOI] [PubMed] [Google Scholar]
- Bridges D, Pitiot A, MacAskill MR, Peirce JW, 2020. The timing mega-study: comparing a range of experiment generators, both lab-based and online. PeerJ 8, e9414. 10.7717/peerj.9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbinian Brodmann, 1909. Vergleichende Lokalisationslehre der Grosshirnrinde. Leipzig: Johann Ambrosius Barth. [Google Scholar]
- Brookes MJ, Zumer JM, Stevenson CM, Hale JR, Barnes GR, Vrba J, Morris PG, 2010. Investigating spatial specificity and data averaging in MEG. NeuroImage 49, 525–538. 10.1016/j.neuroimage.2009.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookshire G, 2021. Re-evaluating rhythmic attentional switching: Spurious oscillations from shuffling-in-time (preprint). Neuroscience. 10.1101/2021.05.07.443101 [DOI] [Google Scholar]
- Brovelli A, Lachaux J-P, Kahane P, Boussaoud D, 2005. High gamma frequency oscillatory activity dissociates attention from intention in the human premotor cortex. NeuroImage 28, 154–164. 10.1016/j.neuroimage.2005.05.045 [DOI] [PubMed] [Google Scholar]
- Bruns A, 2004. Fourier-, Hilbert- and wavelet-based signal analysis: are they really different approaches? J. Neurosci. Methods 137, 321–332. 10.1016/j.jneumeth.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Bruns A, Eckhorn R, 2004. Task-related coupling from high- to low-frequency signals among visual cortical areas in human subdural recordings. Int. J. Psychophysiol. Off. J. Int. Organ. Psychophysiol 51, 97–116. 10.1016/j.ijpsycho.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Bruns A, Eckhorn R, Jokeit H, Ebner A, 2000. Amplitude envelope correlation detects coupling among incoherent brain signals. Neuroreport 11, 1509–1514. [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2012. The economy of brain network organization. Nat. Rev. Neurosci 13, 336–349. 10.1038/nrn3214 [DOI] [PubMed] [Google Scholar]
- Burns SP, Santaniello S, Yaffe RB, Jouny CC, Crone NE, Bergey GK, Anderson WS, Sarma SV, 2014. Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc. Natl. Acad. Sci. U. S. A 111, E5321–5330. 10.1073/pnas.1401752111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A, Chrabaszcz A, Peterson V, Saravanan V, Dastolfo-Hromack C, Lipski WJ, Richardson RM, 2021. Differentiation of speech-induced artifacts from physiological high gamma activity in intracranial recordings (preprint). Neuroscience. 10.1101/2021.04.26.441553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Anastassiou CA, Koch C, 2012. The origin of extracellular fields and currents — EEG, ECoG, LFP and spikes. Nat. Rev. Neurosci 13, 407–420. 10.1038/nrn3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin-Jageman RJ, Cumming G, 2019. Estimation for Better Inference in Neuroscience. eNeuro 6, ENEURO.0205–19.2019. 10.1523/ENEURO.0205-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canolty RT, Cadieu CF, Koepsell K, Ganguly K, Knight RT, Carmena JM, 2012. Detecting event-related changes of multivariate phase coupling in dynamic brain networks. J. Neurophysiol 107, 2020–2031. 10.1152/jn.00610.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canolty RT, Edwards E, Dalal SS, Soltani M, Nagarajan SS, Kirsch HE, Berger MS, Barbaro NM, Knight RT, 2006. High gamma power is phase-locked to theta oscillations in human neocortex. Science 313, 1626–1628. 10.1126/science.1128115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canolty RT, Knight RT, 2010. The functional role of cross-frequency coupling. Trends Cogn. Sci 14, 506–515. 10.1016/j.tics.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F, Casaceli G, Raneri F, Miller J, Lo Russo G, 2016. Implantation of Stereoelectroencephalography Electrodes: A Systematic Review. J. Clin. Neurophysiol 33, 490–502. 10.1097/WNP.0000000000000249 [DOI] [PubMed] [Google Scholar]
- Caruana F, Gerbella M, Avanzini P, Gozzo F, Pelliccia V, Mai R, Abdollahi RO, Cardinale F, Sartori I, Lo Russo G, Rizzolatti G, 2018. Motor and emotional behaviours elicited by electrical stimulation of the human cingulate cortex. Brain J. Neurol 141, 3035–3051. 10.1093/brain/awy219 [DOI] [PubMed] [Google Scholar]
- Caruana F, Gozzo F, Pelliccia V, Cossu M, Avanzini P, 2016. Smile and laughter elicited by electrical stimulation of the frontal operculum. Neuropsychologia 89, 364–370. 10.1016/j.neuropsychologia.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Centracchio J, Sarno A, Esposito D, Andreozzi E, Pavone L, Di Gennaro G, Bartolo M, Esposito V, Morace R, Casciato S, Bifulco P, 2021. Efficient automated localization of ECoG electrodes in CT images via shape analysis. Int. J. Comput. Assist. Radiol. Surg 16, 543–554. 10.1007/s11548-021-02325-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EF, 2015. Towards large-scale, human-based, mesoscopic neurotechnologies. Neuron 86, 68–78. 10.1016/j.neuron.2015.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier J, Anumanchipalli GK, Johnson K, Chang EF, 2018. Encoding of Articulatory Kinematic Trajectories in Human Speech Sensorimotor Cortex. Neuron 98, 1042–1054.e4. 10.1016/j.neuron.2018.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumon M, Puce A, George N, 2021. Statistical power: Implications for planning MEG studies. NeuroImage 233, 117894. 10.1016/j.neuroimage.2021.117894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvel P, Gonzalez-Martinez J, Bulacio J, 2019. Presurgical intracranial investigations in epilepsy surgery, in: Handbook of Clinical Neurology. Elsevier, pp. 45–71. 10.1016/B978-0-444-64142-7.00040-0 [DOI] [PubMed] [Google Scholar]
- Chen S, Tan Z, Xia W, Gomes CA, Zhang X, Zhou W, Liang S, Axmacher N, Wang L, 2021. Theta oscillations synchronize human medial prefrontal cortex and amygdala during fear learning. Sci. Adv 7, eabf4198. 10.1126/sciadv.abf4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Chang EF, 2012. Real-time, time–frequency mapping of event-related cortical activation. J. Neural Eng 9, 046018. 10.1088/1741-2560/9/4/046018 [DOI] [PubMed] [Google Scholar]
- Chiong W, Leonard MK, Chang EF, 2018. Neurosurgical Patients as Human Research Subjects: Ethical Considerations in Intracranial Electrophysiology Research. Neurosurgery 83, 29–37. 10.1093/neuros/nyx361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimbalnik J, Dolezal J, Topçu Ç, Lech M, Marks VS, Joseph B, Dobias M, Van Gompel J, Worrell G, Kucewicz M, 2022. Intracranial electrophysiological recordings from the human brain during memory tasks with pupillometry. Sci. Data 9, 6. 10.1038/s41597-021-01099-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayson PE, Baldwin SA, Rocha HA, Larson MJ, 2021. The data-processing multiverse of event-related potentials (ERPs): A roadmap for the optimization and standardization of ERP processing and reduction pipelines. NeuroImage 245, 118712. 10.1016/j.neuroimage.2021.118712 [DOI] [PubMed] [Google Scholar]
- Cohen Michael X, 2017. Comparison of linear spatial filters for identifying oscillatory activity in multichannel data. J. Neurosci. Methods 278, 1–12. 10.1016/j.jneumeth.2016.12.016 [DOI] [PubMed] [Google Scholar]
- Cohen Michael X., 2017. Multivariate cross-frequency coupling via generalized eigendecomposition. eLife 6, e21792. 10.7554/eLife.21792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MX, 2015. Effects of time lag and frequency matching on phase-based connectivity. J. Neurosci. Methods 250, 137–146. 10.1016/j.jneumeth.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Cohen MX, 2014. Analyzing neural time series data: theory and practice, Issues in clinical and cognitive neuropsychology. The MIT Press, Cambridge, Massachusetts. [Google Scholar]
- Cole S, Voytek B, 2019. Cycle-by-cycle analysis of neural oscillations. J. Neurophysiol 122, 849–861. 10.1152/jn.00273.2019 [DOI] [PubMed] [Google Scholar]
- Cole SR, van der Meij R, Peterson EJ, de Hemptinne C, Starr PA, Voytek B, 2017. Nonsinusoidal Beta Oscillations Reflect Cortical Pathophysiology in Parkinson’s Disease. J. Neurosci. Off. J. Soc. Neurosci 37, 4830–4840. 10.1523/JNEUROSCI.2208-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SR, Voytek B, 2017. Brain Oscillations and the Importance of Waveform Shape. Trends Cogn. Sci 21, 137–149. 10.1016/j.tics.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Colgin LL, Denninger T, Fyhn M, Hafting T, Bonnevie T, Jensen O, Moser M-B, Moser EI, 2009. Frequency of gamma oscillations routes flow of information in the hippocampus. Nature 462, 353–357. 10.1038/nature08573 [DOI] [PubMed] [Google Scholar]
- Collard MJ, Fifer MS, Benz HL, McMullen DP, Wang Y, Milsap GW, Korzeniewska A, Crone NE, 2016. Cortical subnetwork dynamics during human language tasks. NeuroImage 135, 261–272. 10.1016/j.neuroimage.2016.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombet B, Woodman M, Badier JM, Bénar CG, 2015. AnyWave: A cross-platform and modular software for visualizing and processing electrophysiological signals. J. Neurosci. Methods 242, 118–126. 10.1016/j.jneumeth.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Combrisson E, Allegra M, Basanisi R, Ince RAA, Giordano B, Bastin J, Brovelli A, 2021. Group-level inference of information-based measures for the analyses of cognitive brain networks from neurophysiological data (preprint). Neuroscience. 10.1101/2021.08.14.456339 [DOI] [PubMed] [Google Scholar]
- Combrisson E, Jerbi K, 2015. Exceeding chance level by chance: The caveat of theoretical chance levels in brain signal classification and statistical assessment of decoding accuracy. J. Neurosci. Methods 250, 126–136. 10.1016/j.jneumeth.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Combrisson E, Perrone-Bertolotti M, Soto JL, Alamian G, Kahane P, Lachaux J-P, Guillot A, Jerbi K, 2017. From intentions to actions: Neural oscillations encode motor processes through phase, amplitude and phase-amplitude coupling. NeuroImage 147, 473–487. 10.1016/j.neuroimage.2016.11.042 [DOI] [PubMed] [Google Scholar]
- Cooper PS, Baillet S, Maroun REK, Chong TT-J, 2021. Over the rainbow: Guidelines for meaningful use of colour maps in neurophysiology. NeuroImage 245, 118628. 10.1016/j.neuroimage.2021.118628 [DOI] [PubMed] [Google Scholar]
- Corley JA, Nazari P, Rossi VJ, Kim NC, Fogg LF, Hoeppner TJ, Stoub TR, Byrne RW, 2017. Cortical stimulation parameters for functional mapping. Seizure 45, 36–41. 10.1016/j.seizure.2016.11.015 [DOI] [PubMed] [Google Scholar]
- Cosandier-Rimélé D, Merlet I, Badier JM, Chauvel P, Wendling F, 2008. The neuronal sources of EEG: Modeling of simultaneous scalp and intracerebral recordings in epilepsy. NeuroImage 42, 135–146. 10.1016/j.neuroimage.2008.04.185 [DOI] [PubMed] [Google Scholar]
- Cossu M, Fuschillo D, Casaceli G, Pelliccia V, Castana L, Mai R, Francione S, Sartori I, Gozzo F, Nobili L, Tassi L, Cardinale F, Lo Russo G, 2015. Stereoelectroencephalography-guided radiofrequency thermocoagulation in the epileptogenic zone: a retrospective study on 89 cases. J. Neurosurg 123, 1358–1367. 10.3171/2014.12.JNS141968 [DOI] [PubMed] [Google Scholar]
- Crameri F, Shephard GE, Heron PJ, 2020. The misuse of colour in science communication. Nat. Commun 11, 5444. 10.1038/s41467-020-19160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker B, Ostrowski L, Williams ZM, Dougherty DD, Eskandar EN, Widge AS, Chu CJ, Cash SS, Paulk AC, 2021. Local and distant responses to single pulse electrical stimulation reflect different forms of connectivity. NeuroImage 237, 118094. 10.1016/j.neuroimage.2021.118094 [DOI] [PubMed] [Google Scholar]
- Crone NE, Miglioretti DL, Gordon B, Lesser RP, 1998a. Functional mapping of human sensorimotor cortex with electrocorticographic spectral analysis. II. Event-related synchronization in the gamma band. Brain J. Neurol 121 ( Pt 12), 2301–2315. 10.1093/brain/121.12.2301 [DOI] [PubMed] [Google Scholar]
- Crone NE, Miglioretti DL, Gordon B, Sieracki JM, Wilson MT, Uematsu S, Lesser RP, 1998b. Functional mapping of human sensorimotor cortex with electrocorticographic spectral analysis. I. Alpha and beta event-related desynchronization. Brain J. Neurol 121 ( Pt 12), 2271–2299. 10.1093/brain/121.12.2271 [DOI] [PubMed] [Google Scholar]
- Csercsa R, Dombovári B, Fabó D, Wittner L, Eross L, Entz L, Sólyom A, Rásonyi G, Szucs A, Kelemen A, Jakus R, Juhos V, Grand L, Magony A, Halász P, Freund TF, Maglóczky Z, Cash SS, Papp L, Karmos G, Halgren E, Ulbert I, 2010. Laminar analysis of slow wave activity in humans. Brain J. Neurol 133, 2814–2829. 10.1093/brain/awq169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello Oderiz C, von Ellenrieder N, Dubeau F, Eisenberg A, Gotman J, Hall J, Hincapié A-S, Hoffmann D, Job A-S, Khoo HM, Minotti L, Olivier A, Kahane P, Frauscher B, 2019. Association of Cortical Stimulation–Induced Seizure With Surgical Outcome in Patients With Focal Drug-Resistant Epilepsy. JAMA Neurol. 76, 1070. 10.1001/jamaneurol.2019.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Liu Y, Li Yuxing, Guo M, Li Yiming, Li X, Wang T, Zeng X, Ye C, 2019. Semi-Supervised Brain Lesion Segmentation with an Adapted Mean Teacher Model. ArXiv190301248 Cs. [Google Scholar]
- Dalal SS, Baillet S, Adam C, Ducorps A, Schwartz D, Jerbi K, Bertrand O, Garnero L, Martinerie J, Lachaux J-P, 2009. Simultaneous MEG and intracranial EEG recordings during attentive reading. NeuroImage 45, 1289–304. 10.1016/j.neuroimage.2009.01.017 [DOI] [PubMed] [Google Scholar]
- Dalal SS, Edwards E, Kirsch HE, Barbaro NM, Knight RT, Nagarajan SS, 2008. Localization of neurosurgically implanted electrodes via photograph-MRI-radiograph coregistration. J. Neurosci. Methods 174, 106–115. 10.1016/j.jneumeth.2008.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington RB, Hayes AF, 2000. Combining independent p values: extensions of the Stouffer and binomial methods. Psychol. Methods 5, 496–515. 10.1037/1082-989x.5.4.496 [DOI] [PubMed] [Google Scholar]
- David O, Bastin J, Chabardès S, Minotti L, Kahane P, 2010. Studying Network Mechanisms Using Intracranial Stimulation in Epileptic Patients. Front. Syst. Neurosci 4. 10.3389/fnsys.2010.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cheveigné A, 2018. Robust detrending, rereferencing, outlier detection, and inpainting for multichannel data 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cheveigné A, Nelken I, 2019. Filters: When, Why, and How (Not) to Use Them. Neuron 102, 280–293. 10.1016/j.neuron.2019.02.039 [DOI] [PubMed] [Google Scholar]
- de Hemptinne C, Swann NC, Ostrem JL, Ryapolova-Webb ES, San Luciano M, Galifianakis NB, Starr PA, 2015. Therapeutic deep brain stimulation reduces cortical phase-amplitude coupling in Parkinson’s disease. Nat. Neurosci 18, 779–786. 10.1038/nn.3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis LM, 2001. Brain Tumors. N. Engl. J. Med 344, 114–123. 10.1056/NEJM200101113440207 [DOI] [PubMed] [Google Scholar]
- Del Vecchio M, Avanzini P, 2020. La Recherche du Temps Perdu: Timing in Somatosensation. Commentary: Somatosensation in the Brain: A Theoretical Re-evaluation and a New Model. Front. Syst. Neurosci 14, 597755. 10.3389/fnsys.2020.597755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio M, Caruana F, Sartori I, Pelliccia V, Zauli FM, Lo Russo G, Rizzolatti G, Avanzini P, 2020. Action execution and action observation elicit mirror responses with the same temporal profile in human SII. Commun. Biol 3, 80. 10.1038/s42003-020-0793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio M, Fossataro C, Zauli FM, Sartori I, Pigorini A, d’Orio P, Abarrategui B, Russo S, Mikulan EP, Caruana F, Rizzolatti G, Garbarini F, Avanzini P, 2021. Tonic somatosensory responses and deficits of tactile awareness converge in the parietal operculum. Brain awab 384. 10.1093/brain/awab384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Miyakoshi M, Jung T-P, Makeig S, 2015. Grand average ERP-image plotting and statistics: A method for comparing variability in event-related single-trial EEG activities across subjects and conditions. J. Neurosci. Methods 250, 3–6. 10.1016/j.jneumeth.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deman P, Bhattacharjee M, Tadel F, Job A-S, Rivière D, Cointepas Y, Kahane P, David O, 2018. IntrAnat Electrodes: A Free Database and Visualization Software for Intracranial Electroencephalographic Data Processed for Case and Group Studies. Front. Neuroinformatics 12, 40. 10.3389/fninf.2018.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerath NJ, 1949. THE AMERICAN SOLDIER: VOLUME I, ADJUSTMENT DURING ARMY LIFE. By Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM, Jr. VOLUME II, COMBAT AND ITS AFTERMATH. By Stouffer SA, Lumsdaine AA, Lumsdaine MH, Williams RM, Jr., Smith MB, Janis IL, Star SA, Cottrell LS, Jr. Princeton, New Jersey: Princeton University Press, 1949. Vol. I, 599 pp., Vol. II, 675 pp. $7.50 each; $13.50 together. Soc. Forces 28, 87–90. 10.2307/2572105 [DOI] [Google Scholar]
- Desai R, Liebenthal E, Possing ET, Waldron E, Binder JR, 2005. Volumetric vs. surface-based alignment for localization of auditory cortex activation. NeuroImage 26, 1019–1029. 10.1016/j.neuroimage.2005.03.024 [DOI] [PubMed] [Google Scholar]
- Despotović I, Goossens B, Philips W, 2015. MRI segmentation of the human brain: challenges, methods, and applications. Comput. Math. Methods Med. 2015, 450341. 10.1155/2015/450341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hondt F, Lassonde M, Collignon O, Dubarry A-S, Robert M, Rigoulot S, Honoré J, Lepore F, Sequeira H, 2010. Early brain-body impact of emotional arousal. Front. Hum. Neurosci 4, 33. 10.3389/fnhum.2010.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pellegrino G, Fadiga L, Fogassi L, Gallese V, Rizzolatti G, 1992. Understanding motor events: a neurophysiological study. Exp. Brain Res 91, 176–180. 10.1007/BF00230027 [DOI] [PubMed] [Google Scholar]
- Dimova P, de Palma L, Job-Chapron A-S, Minotti L, Hoffmann D, Kahane P, 2017. Radiofrequency thermocoagulation of the seizure-onset zone during stereoelectroencephalography. Epilepsia 58, 381–392. 10.1111/epi.13663 [DOI] [PubMed] [Google Scholar]
- Ding M, Chen Y, Bressler SL, 2006. Granger Causality: Basic Theory and Application to Neuroscience, in: Schelter B, Winterhalder M, Timmer J.(Eds.), Handbook of Time Series Analysis. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, pp. 437–460. 10.1002/9783527609970.ch17 [DOI] [Google Scholar]
- Donoghue T, Haller M, Peterson EJ, Varma P, Sebastian P, Gao R, Noto T, Lara AH, Wallis JD, Knight RT, Shestyuk A, Voytek B, 2020. Parameterizing neural power spectra into periodic and aperiodic components. Nat. Neurosci 23, 1655–1665. 10.1038/s41593-020-00744-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue T, Schaworonkow N, Voytek B, 2021. Methodological considerations for studying neural oscillations. Eur. J. Neurosci ejn.15361. 10.1111/ejn.15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donos C, Mălîia MD, Mîndruţă I, Popa I, Ene M, Bălănescu B, Ciurea A, Barborica A, 2016. A connectomics approach combining structural and effective connectivity assessed by intracranial electrical stimulation. NeuroImage 132, 344–358. 10.1016/j.neuroimage.2016.02.054 [DOI] [PubMed] [Google Scholar]
- Dougherty ME, Nguyen APQ, Baratham VL, Bouchard KE, 2019. Laminar origin of evoked ECoG high-gamma activity. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Int. Conf 2019, 4391–4394. 10.1109/EMBC.2019.8856786 [DOI] [PubMed] [Google Scholar]
- Dubarry A-S, Badier J-M, Trébuchon-Da Fonseca A, Gavaret M, Carron R, Bartolomei F, Liégeois-Chauvel C, Régis J, Chauvel P, Alario F-X, Bénar C-G, 2014. Simultaneous recording of MEG, EEG and intracerebral EEG during visual stimulation: From feasibility to single-trial analysis. NeuroImage 99, 548–558. 10.1016/j.neuroimage.2014.05.055 [DOI] [PubMed] [Google Scholar]
- Dubey A, Ray S, 2019. Cortical Electrocorticogram (ECoG) Is a Local Signal. J. Neurosci 39, 4299–4311. 10.1523/JNEUROSCI.2917-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak D, Fenton AA, 2014. Toward a proper estimation of phase-amplitude coupling in neural oscillations. J. Neurosci. Methods 225, 42–56. 10.1016/j.jneumeth.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra AR, Chan AM, Quinn BT, Zepeda R, Keller CJ, Cormier J, Madsen JR, Eskandar EN, 2012. Individualized localization and cortical surface-based registration of intracranial electrodes 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke D, Aasebø IEJ, Akintoye S, Knight W, Karakasidis A, Mikulan E, Ochang P, Ogoh G, Oostenveld R, Pigorini A, Stahl BC, White T, Zehl L, 2021. Pseudonymisation of neuroimages and data protection: Increasing access to data while retaining scientific utility. Neuroimage Rep. 1, 100053. 10.1016/j.ynirp.2021.100053 [DOI] [Google Scholar]
- Engel AK, Gerloff C, Hilgetag CC, Nolte G, 2013. Intrinsic coupling modes: multiscale interactions in ongoing brain activity. Neuron 80, 867–886. 10.1016/j.neuron.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Engel AK, Moll CKE, Fried I, Ojemann GA, 2005. Invasive recordings from the human brain: clinical insights and beyond. Nat. Rev. Neurosci 6, 35–47. 10.1038/nrn1585 [DOI] [PubMed] [Google Scholar]
- Englot DJ, Ouyang D, Wang DD, Rolston JD, Garcia PA, Chang EF, 2013. Relationship between hospital surgical volume, lobectomy rates, and adverse perioperative events at US epilepsy centers: Clinical article. J. Neurosurg 118, 169–174. 10.3171/2012.9.JNS12776 [DOI] [PubMed] [Google Scholar]
- Etame AB, Fox WC, Sagher O, 2011. Osmotic diuresis paradoxically worsens brain shift after subdural grid placement. Acta Neurochir. (Wien) 153, 633–637. 10.1007/s00701-010-0856-6 [DOI] [PubMed] [Google Scholar]
- Ezzyat Y, Wanda PA, Levy DF, Kadel A, Aka A, Pedisich I, Sperling MR, Sharan AD, Lega BC, Burks A, Gross RE, Inman CS, Jobst BC, Gorenstein MA, Davis KA, Worrell GA, Kucewicz MT, Stein JM, Gorniak R, Das SR, Rizzuto DS, Kahana MJ, 2018. Closed-loop stimulation of temporal cortex rescues functional networks and improves memory. Nat. Commun 9, 365. 10.1038/s41467-017-02753-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahimi Hnazaee M, Wittevrongel B, Khachatryan E, Libert A, Carrette E, Dauwe I, Meurs A, Boon P, Van Roost D, Van Hulle MM, 2020. Localization of deep brain activity with scalp and subdural EEG. NeuroImage 223, 117344. 10.1016/j.neuroimage.2020.117344 [DOI] [PubMed] [Google Scholar]
- Fein G, Raz J, Brown FF, Merrin EL, 1988. Common reference coherence data are confounded by power and phase effects. Electroencephalogr. Clin. Neurophysiol 69, 581–584. 10.1016/0013-4694(88)90171-x [DOI] [PubMed] [Google Scholar]
- Feinsinger A, Pouratian N, Ebadi H, Adolphs R, Andersen R, Beauchamp MS, Chang EF, Crone NE, Collinger JL, Fried I, Mamelak A, Richardson M, Rutishauser U, Sheth SA, Suthana N, Tandon N, Yoshor D, 2022. Ethical commitments, principles, and practices guiding intracranial neuroscientific research in humans. Neuron 110, 188–194. 10.1016/j.neuron.2021.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell J, Klaver P, Lehnertz K, Grunwald T, Schaller C, Elger CE, Fernández G, 2001. Human memory formation is accompanied by rhinal–hippocampal coupling and decoupling. Nat. Neurosci 4, 1259–1264. 10.1038/nn759 [DOI] [PubMed] [Google Scholar]
- Fell J, Staresina BP, Do Lam ATA, Widman G, Helmstaedter C, Elger CE, Axmacher N, 2013. Memory Modulation by Weak Synchronous Deep Brain Stimulation: A Pilot Study. Brain Stimulat 6, 270–273. 10.1016/j.brs.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Fiebelkorn IC, Kastner S, 2019. A Rhythmic Theory of Attention. Trends Cogn. Sci 23, 87–101. 10.1016/j.tics.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch L, Privman E, Ramot M, Harel M, Nir Y, Kipervasser S, Andelman F, Neufeld MY, Kramer U, Fried I, Malach R, 2009. Neural “Ignition”: Enhanced Activation Linked to Perceptual Awareness in Human Ventral Stream Visual Cortex. Neuron 64, 562–574. 10.1016/j.neuron.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. NeuroImage 62, 774–781. 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA, 1970. Statistical methods for research workers, 14th ed., revised and enlarged. ed. Oliver and Boyd, Edinburgh. [Google Scholar]
- Flinker A, Chang EF, Barbaro NM, Berger MS, Knight RT, 2011. Sub-centimeter language organization in the human temporal lobe. Brain Lang. 117, 103–109. 10.1016/j.bandl.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinker A, Korzeniewska A, Shestyuk AY, Franaszczuk PJ, Dronkers NF, Knight RT, Crone NE, 2015. Redefining the role of Broca’s area in speech. Proc. Natl. Acad. Sci. U. S. A 112, 2871–2875. 10.1073/pnas.1414491112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin E, Gross J, Pfeifer J, Fink GR, Timmermann L, 2010. The effect of filtering on Granger causality based multivariate causality measures. NeuroImage 50, 577–588. 10.1016/j.neuroimage.2009.12.050 [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M, 2015. The connectomics of brain disorders. Nat. Rev. Neurosci 16, 159–172. 10.1038/nrn3901 [DOI] [PubMed] [Google Scholar]
- Fried I, MacDonald KA, Wilson CL, 1997. Single neuron activity in human hippocampus and amygdala during recognition of faces and objects. Neuron 18, 753–765. 10.1016/s0896-6273(00)80315-3 [DOI] [PubMed] [Google Scholar]
- Fried I, Wilson CL, MacDonald KA, Behnke EJ, 1998. Electric current stimulates laughter. Nature 391, 650–650. 10.1038/35536 [DOI] [PubMed] [Google Scholar]
- Fried I, Wilson CL, Morrow JW, Cameron KA, Behnke ED, Ackerson LC, Maidment NT, 2001. Increased dopamine release in the human amygdala during performance of cognitive tasks. Nat. Neurosci 4, 201–206. 10.1038/84041 [DOI] [PubMed] [Google Scholar]
- Fries P, 2015. Rhythms for Cognition: Communication through Coherence. Neuron 88, 220–235. 10.1016/j.neuron.2015.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P, 2005. A mechanism for cognitive dynamics: neuronal communication through neuronal coherence. Trends Cogn. Sci 9, 474–480. 10.1016/j.tics.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Friston KJ, 2011. Functional and effective connectivity: a review. Brain Connect. 1, 13–36. 10.1089/brain.2011.0008 [DOI] [PubMed] [Google Scholar]
- Friston KJ (Ed.), 2007. Statistical parametric mapping: the analysis of funtional brain images, 1st ed. ed. Elsevier/Academic Press, Amsterdam ; Boston. [Google Scholar]
- Frith U, 2020. Fast Lane to Slow Science. Trends Cogn. Sci 24, 1–2. 10.1016/j.tics.2019.10.007 [DOI] [PubMed] [Google Scholar]
- Galbraith S, Daniel JA, Vissel B, 2010. A study of clustered data and approaches to its analysis. J. Neurosci. Off. J. Soc. Neurosci 30, 10601–10608. 10.1523/JNEUROSCI.0362-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Peterson EJ, Voytek B, 2017. Inferring synaptic excitation/inhibition balance from field potentials. NeuroImage 158, 70–78. 10.1016/j.neuroimage.2017.06.078 [DOI] [PubMed] [Google Scholar]
- Gao R, van den Brink RL, Pfeffer T, Voytek B, 2020. Neuronal timescales are functionally dynamic and shaped by cortical microarchitecture. eLife 9, e61277. 10.7554/eLife.61277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaser C, Dahnke R, others, n.d. CAT-a computational anatomy toolbox for the analysis of structural MRI data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavaret M, Dubarry A, Carron R, Bartolomei F, 2016. Simultaneous SEEG-MEG-EEG recordings Overcome the SEEG limited spatial sampling. Epilepsy Res. 128, 68–72. 10.1016/j.eplepsyres.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Gelman A, Loken E, 2014. The Statistical Crisis in Science. Am. Sci 102, 460. 10.1511/2014.111.460 [DOI] [Google Scholar]
- Genon S, Reid A, Langner R, Amunts K, Eickhoff SB, 2018. How to Characterize the Function of a Brain Region. Trends Cogn. Sci 22, 350–364. 10.1016/j.tics.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George DD, Ojemann SG, Drees C, Thompson JA, 2020. Stimulation Mapping Using Stereoelectroencephalography: Current and Future Directions. Front. Neurol 11, 320. 10.3389/fneur.2020.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster M, Waterstraat G, Litvak V, Lehnertz K, Schnitzler A, Florin E, Curio G, Nikulin V, 2022. Separating Neural Oscillations from Aperiodic 1/f Activity: Challenges and Recommendations. Neuroinformatics. 10.1007/s12021-022-09581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanz O, Derix J, Kaur R, Schulze-Bonhage A, Auer P, Aertsen A, Ball T, 2018. Real-life speech production and perception have a shared premotor-cortical substrate. Sci. Rep 8, 8898. 10.1038/s41598-018-26801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan T, Davidesco I, Meshulam M, Groppe DM, Mégevand P, Yeagle EM, Goldfinger MS, Harel M, Melloni L, Schroeder CE, Deouell LY, Mehta AD, Malach R, 2017. Increasing suppression of saccade-related transients along the human visual hierarchy. eLife 6, e27819. 10.7554/eLife.27819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan T, Davidesco I, Meshulam M, Groppe DM, Mégevand P, Yeagle EM, Goldfinger MS, Harel M, Melloni L, Schroeder CE, Deouell LY, Mehta AD, Malach R, 2016. Human intracranial recordings link suppressed transients rather than “filling-in” to perceptual continuity across blinks. eLife 5, e17243. 10.7554/eLife.17243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Martinez J, Bulacio J, Alexopoulos A, Jehi L, Bingaman W, Najm I, 2013. Stereoelectroencephalography in the “difficult to localize” refractory focal epilepsy: Early experience from a North American epilepsy center: SEEG in the United States. Epilepsia 54, 323–330. 10.1111/j.1528-1167.2012.03672.x [DOI] [PubMed] [Google Scholar]
- Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YO, Handwerker DA, Hanke M, Keator D, Li X, Michael Z, Maumet C, Nichols BN, Nichols TE, Pellman J, Poline J-B, Rokem A, Schaefer G, Sochat V, Triplett W, Turner JA, Varoquaux G, Poldrack RA, 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044. 10.1038/sdata.2016.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, 2020. Multidimensional processing in the amygdala. Nat. Rev. Neurosci 21, 565–575. 10.1038/s41583-020-0350-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Miller J, Watrous AJ, Lee SA, Coffey T, Sperling MR, Sharan A, Worrell G, Berry B, Lega B, Jobst BC, Davis KA, Inman C, Sheth SA, Wanda PA, Ezzyat Y, Das SR, Stein J, Gorniak R, Jacobs J, 2018. Electrical Stimulation in Hippocampus and Entorhinal Cortex Impairs Spatial and Temporal Memory. J. Neurosci 38, 4471–4481. 10.1523/JNEUROSCI.3049-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados A, Vakharia V, Rodionov R, Schweiger M, Vos SB, O’Keeffe AG, Li K, Wu C, Miserocchi A, McEvoy AW, Clarkson MJ, Duncan JS, Sparks R, Ourselin S, 2018. Automatic segmentation of stereoelectroencephalography (SEEG) electrodes post-implantation considering bending. Int. J. Comput. Assist. Radiol. Surg 13, 935–946. 10.1007/s11548-018-1740-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandchamp R, Delorme A, 2011. Single-trial normalization for event-related spectral decomposition reduces sensitivity to noisy trials. Front. Psychol 2, 236. 10.3389/fpsyg.2011.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt RE, Pflieger ME, Ossadtchi AE, 2012. Connectivity measures applied to human brain electrophysiological data. J. Neurosci. Methods 207, 1–16. 10.1016/j.jneumeth.2012.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Zamboni G, Khan A, Li L, Bonifacio G, Sundaresan V, Schulz UG, Kuker W, Battaglini M, Rothwell PM, Jenkinson M, 2016. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. NeuroImage 141, 191–205. 10.1016/j.neuroimage.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis JC, Metcalf NV, Corbetta M, Shulman GL, 2021. Lesion Quantification Toolkit: A MATLAB software tool for estimating grey matter damage and white matter disconnections in patients with focal brain lesions. NeuroImage Clin. 30, 102639. 10.1016/j.nicl.2021.102639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootswagers T, Wardle SG, Carlson TA, 2017. Decoding Dynamic Brain Patterns from Evoked Responses: A Tutorial on Multivariate Pattern Analysis Applied to Time Series Neuroimaging Data. J. Cogn. Neurosci 29, 677–697. 10.1162/jocn_a_01068 [DOI] [PubMed] [Google Scholar]
- Groppe DM, 2017. Combating the scientific decline effect with confidence (intervals). Psychophysiology 54, 139–145. 10.1111/psyp.12616 [DOI] [PubMed] [Google Scholar]
- Groppe DM, Bickel S, Dykstra AR, Wang X, Mégevand P, Mercier MR, Lado FA, Mehta AD, Honey CJ, 2017. iELVis: An open source MATLAB toolbox for localizing and visualizing human intracranial electrode data. J. Neurosci. Methods 281, 40–48. 10.1016/j.jneumeth.2017.01.022 [DOI] [PubMed] [Google Scholar]
- Groppe DM, Bickel S, Keller CJ, Jain SK, Hwang ST, Harden C, Mehta AD, 2013. Dominant frequencies of resting human brain activity as measured by the electrocorticogram. NeuroImage 79, 223–233. 10.1016/j.neuroimage.2013.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe DM, Urbach TP, Kutas M, 2011. Mass univariate analysis of event-related brain potentials/fields I: a critical tutorial review. Psychophysiology 48, 1711–1725. 10.1111/j.1469-8986.2011.01273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, 2014. Analytical methods and experimental approaches for electrophysiological studies of brain oscillations. J. Neurosci. Methods 228, 57–66. 10.1016/j.jneumeth.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Baillet S, Barnes GR, Henson RN, Hillebrand A, Jensen O, Jerbi K, Litvak V, Maess B, Oostenveld R, Parkkonen L, Taylor JR, van Wassenhove V, Wibral M, Schoffelen J-M, 2013. Good practice for conducting and reporting MEG research. NeuroImage 65, 349–363. 10.1016/j.neuroimage.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara R, Velazquez JLP, Nenadovic V, Wennberg R, Senjanovic G, Dominguez LG, 2005. Phase synchronization measurements using electroencephalographic recordings: what can we really say about neuronal synchrony? Neuroinformatics 3, 301–314. 10.1385/NI:3:4:301 [DOI] [PubMed] [Google Scholar]
- Guggenmos M, Sterzer P, Cichy RM, 2018. Multivariate pattern analysis for MEG: A comparison of dissimilarity measures. NeuroImage 173, 434–447. 10.1016/j.neuroimage.2018.02.044 [DOI] [PubMed] [Google Scholar]
- Gulbinaite R, İlhan B, VanRullen R, 2017. The Triple-Flash Illusion Reveals a Driving Role of Alpha-Band Reverberations in Visual Perception. J. Neurosci. Off. J. Soc. Neurosci 37, 7219–7230. 10.1523/JNEUROSCI.3929-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Ferreira D, Fink K, Westman E, Granberg T, 2019. Repeatability and reproducibility of FreeSurfer, FSL-SIENAX and SPM brain volumetric measurements and the effect of lesion filling in multiple sclerosis. Eur. Radiol 29, 1355–1364. 10.1007/s00330-018-5710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Seth AK, Kendrick KM, Zhou C, Feng J, 2008. Partial Granger causality--eliminating exogenous inputs and latent variables. J. Neurosci. Methods 172, 79–93. 10.1016/j.jneumeth.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Gupta D, Hill NJ, Adamo MA, Ritaccio A, Schalk G, 2014. Localizing ECoG electrodes on the cortical anatomy without post-implantation imaging. NeuroImage Clin. 6, 64–76. 10.1016/j.nicl.2014.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren E, Baudena P, Clarke JM, Heit G, Liégeois C, Chauvel P, Musolino A, 1995a. Intracerebral potentials to rare target and distractor auditory and visual stimuli. I. Superior temporal plane and parietal lobe. Electroencephalogr. Clin. Neurophysiol 94, 191–220. 10.1016/0013-4694(94)00259-n [DOI] [PubMed] [Google Scholar]
- Halgren E, Baudena P, Clarke JM, Heit G, Marinkovic K, Devaux B, Vignal JP, Biraben A, 1995b. Intracerebral potentials to rare target and distractor auditory and visual stimuli. II. Medial, lateral and posterior temporal lobe. Electroencephalogr. Clin. Neurophysiol 94, 229–250. 10.1016/0013-4694(95)98475-n [DOI] [PubMed] [Google Scholar]
- Halgren E, Baudena P, Heit G, Clarke JM, Marinkovic K, Clarke M, 1994. Spatio-temporal stages in face and word processing. I. Depth-recorded potentials in the human occipital, temporal and parietal lobes [corrected]. J. Physiol. Paris 88, 1–50. 10.1016/0928-4257(94)90092-2 [DOI] [PubMed] [Google Scholar]
- Halgren E, Kaestner E, Marinkovic K, Cash SS, Wang C, Schomer DL, Madsen JR, Ulbert I, 2015. Laminar profile of spontaneous and evoked theta: Rhythmic modulation of cortical processing during word integration. Neuropsychologia 76, 108–124. 10.1016/j.neuropsychologia.2015.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren M, Ulbert I, Bastuji H, Fabó D, Erőss L, Rey M, Devinsky O, Doyle WK, Mak-McCully R, Halgren E, Wittner L, Chauvel P, Heit G, Eskandar E, Mandell A, Cash SS, 2019. The generation and propagation of the human alpha rhythm. Proc. Natl. Acad. Sci. U. S. A 116, 23772–23782. 10.1073/pnas.1913092116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton LS, Chang DL, Lee MB, Chang EF, 2017. Semi-automated Anatomical Labeling and Inter-subject Warping of High-Density Intracranial Recording Electrodes in Electrocorticography. Front. Neuroinformatics 11, 62. 10.3389/fninf.2017.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton LS, Edwards E, Chang EF, 2018. A Spatial Map of Onset and Sustained Responses to Speech in the Human Superior Temporal Gyrus. Curr. Biol 28, 1860–1871.e4. 10.1016/j.cub.2018.04.033 [DOI] [PubMed] [Google Scholar]
- Hamilton LS, Huth AG, 2020. The revolution will not be controlled: natural stimuli in speech neuroscience. Lang. Cogn. Neurosci 35, 573–582. 10.1080/23273798.2018.1499946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton LS, Oganian Y, Hall J, Chang EF, 2021. Parallel and distributed encoding of speech across human auditory cortex. Cell 184, 4626–4639.e13. 10.1016/j.cell.2021.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoun S, Baalbaki M, Haddad R, Saaybi S, El Ayoubi NK, Yamout BI, Khoury SJ, Hourani R, 2018. Gadolinium effect on thalamus and whole brain tissue segmentation. Neuroradiology 60, 1167–1173. 10.1007/s00234-018-2082-5 [DOI] [PubMed] [Google Scholar]
- Hansen N, Chaieb L, Derner M, Hampel KG, Elger CE, Surges R, Staresina B, Axmacher N, Fell J, 2018. Memory encoding-related anterior hippocampal potentials are modulated by deep brain stimulation of the entorhinal area. Hippocampus 28, 12–17. 10.1002/hipo.22808 [DOI] [PubMed] [Google Scholar]
- Hari R, Puce A, 2017. MEG-EEG primer. Oxford University Press, New York, NY. [Google Scholar]
- Hartkens T, Hill DLG, Castellano-Smith AD, Hawkes DJ, Maurer CR, Martin AJ, Hall WA, Liu H, Truwit CL, 2003. Measurement and analysis of brain deformation during neurosurgery. IEEE Trans. Med. Imaging 22, 82–92. 10.1109/TMI.2002.806596 [DOI] [PubMed] [Google Scholar]
- Hastreiter P, Rezk-Salama C, Soza G, Bauer M, Greiner G, Fahlbusch R, Ganslandt O, Nimsky C, 2004. Strategies for brain shift evaluation. Med. Image Anal 8, 447–464. 10.1016/j.media.2004.02.001 [DOI] [PubMed] [Google Scholar]
- Haufe S, DeGuzman P, Henin S, Arcaro M, Honey CJ, Hasson U, Parra LC, 2018. Elucidating relations between fMRI, ECoG, and EEG through a common natural stimulus. NeuroImage 179, 79–91. 10.1016/j.neuroimage.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes J-D, 2015. A Primer on Pattern-Based Approaches to fMRI: Principles, Pitfalls, and Perspectives. Neuron 87, 257–270. 10.1016/j.neuron.2015.05.025 [DOI] [PubMed] [Google Scholar]
- He BJ, Zempel JM, Snyder AZ, Raichle ME, 2010. The temporal structures and functional significance of scale-free brain activity. Neuron 66, 353–369. 10.1016/j.neuron.2010.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard NA, Rubin-Delanchy P, 2018. Choosing between methods of combining $p$-values. Biometrika 105, 239–246. 10.1093/biomet/asx076 [DOI] [Google Scholar]
- Henin S, Shankar A, Borges H, Flinker A, Doyle W, Friedman D, Devinsky O, Buzsáki G, Liu A, 2020. Spatiotemporal dynamics between interictal epileptiform discharges and ripples during associative memory processing (preprint). Neuroscience. 10.1101/2020.07.22.216416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henin S, Shankar A, Hasulak N, Friedman D, Dugan P, Melloni L, Flinker A, Sarac C, Fang M, Doyle W, Tcheng T, Devinsky O, Davachi L, Liu A, 2019. Hippocampal gamma predicts associative memory performance as measured by acute and chronic intracranial EEG. Sci. Rep 9, 593. 10.1038/s41598-018-37561-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henin S, Turk-Browne NB, Friedman D, Liu A, Dugan P, Flinker A, Doyle W, Devinsky O, Melloni L, 2021. Learning hierarchical sequence representations across human cortex and hippocampus. Sci. Adv 7, eabc4530. 10.1126/sciadv.abc4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes D, Miller KJ, Noordmans HJ, Vansteensel MJ, Ramsey NF, 2010. Automated electrocorticographic electrode localization on individually rendered brain surfaces. J. Neurosci. Methods 185, 293–298. 10.1016/j.jneumeth.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Hermes D, Miller KJ, Vansteensel MJ, Aarnoutse EJ, Leijten FSS, Ramsey NF, 2012. Neurophysiologic correlates of fMRI in human motor cortex. Hum. Brain Mapp 33, 1689–1699. 10.1002/hbm.21314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes D, Miller KJ, Wandell BA, Winawer J, 2015. Stimulus Dependence of Gamma Oscillations in Human Visual Cortex. Cereb. Cortex 25, 2951–2959. 10.1093/cercor/bhu091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes D, Nguyen M, Winawer J, 2017. Neuronal synchrony and the relation between the blood-oxygen-level dependent response and the local field potential. PLOS Biol. 15, e2001461. 10.1371/journal.pbio.2001461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero JL, Khuvis S, Yeagle E, Cerf M, Mehta AD, 2018. Breathing above the brain stem: volitional control and attentional modulation in humans. J. Neurophysiol 119, 145–159. 10.1152/jn.00551.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CS, Rach S, Vosskuhl J, Strüber D, 2014. Time-frequency analysis of event-related potentials: a brief tutorial. Brain Topogr. 27, 438–450. 10.1007/s10548-013-0327-5 [DOI] [PubMed] [Google Scholar]
- Hill DL, Maurer CR, Maciunas RJ, Barwise JA, Fitzpatrick JM, Wang MY, 1998. Measurement of intraoperative brain surface deformation under a craniotomy. Neurosurgery 43, 514–526; discussion 527–528. 10.1097/00006123-199809000-00066 [DOI] [PubMed] [Google Scholar]
- Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK, 2012. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat. Neurosci 15, 884–890. 10.1038/nn.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdgraf C, Appelhoff S, Bickel S, Bouchard K, D’Ambrosio S, David O, Devinsky O, Dichter B, Flinker A, Foster BL, Gorgolewski KJ, Groen I, Groppe D, Gunduz A, Hamilton L, Honey CJ, Jas M, Knight R, Lachaux J-P, Lau JC, Lee-Messer C, Lundstrom BN, Miller KJ, Ojemann JG, Oostenveld R, Petridou N, Piantoni G, Pigorini A, Pouratian N, Ramsey NF, Stolk A, Swann NC, Tadel F, Voytek B, Wandell BA, Winawer J, Whitaker K, Zehl L, Hermes D, 2019. iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Sci. Data 6, 102. 10.1038/s41597-019-0105-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Kühn AA, 2015. Lead-DBS: A toolbox for deep brain stimulation electrode localizations and visualizations. NeuroImage 107, 127–135. 10.1016/j.neuroimage.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Howard MA, Volkov IO, Granner MA, Damasio HM, Ollendieck MC, Bakken HE, 1996. A hybrid clinical-research depth electrode for acute and chronic in vivo microelectrode recording of human brain neurons. Technical note. J. Neurosurg 84, 129–132. 10.3171/jns.1996.84.1.0129 [DOI] [PubMed] [Google Scholar]
- Hu L, Xiao P, Zhang ZG, Mouraux A, Iannetti GD, 2014. Single-trial time-frequency analysis of electrocortical signals: baseline correction and beyond. NeuroImage 84, 876–887. 10.1016/j.neuroimage.2013.09.055 [DOI] [PubMed] [Google Scholar]
- Hughes AM, Whitten TA, Caplan JB, Dickson CT, 2012. BOSC: a better oscillation detection method, extracts both sustained and transient rhythms from rat hippocampal recordings. Hippocampus 22, 1417–1428. 10.1002/hipo.20979 [DOI] [PubMed] [Google Scholar]
- Huigen E, Peper A, Grimbergen CA, 2002. Investigation into the origin of the noise of surface electrodes. Med. Biol. Eng. Comput 40, 332–338. 10.1007/BF02344216 [DOI] [PubMed] [Google Scholar]
- Hunter JD, Hanan DM, Singer BF, Shaikh S, Brubaker KA, Hecox KE, Towle VL, 2005. Locating chronically implanted subdural electrodes using surface reconstruction. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol 116, 1984–1987. 10.1016/j.clinph.2005.03.027 [DOI] [PubMed] [Google Scholar]
- Hurtado JM, Rubchinsky LL, Sigvardt KA, 2004. Statistical method for detection of phase-locking episodes in neural oscillations. J. Neurophysiol 91, 1883–1898. 10.1152/jn.00853.2003 [DOI] [PubMed] [Google Scholar]
- Hyafil A, Giraud A-L, Fontolan L, Gutkin B, 2015. Neural Cross-Frequency Coupling: Connecting Architectures, Mechanisms, and Functions. Trends Neurosci. 38, 725–740. 10.1016/j.tins.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Iemi L, Gwilliams L, Samaha J, Auksztulewicz R, Cycowicz YM, King J-R, Nikulin VV, Thesen T, Doyle W, Devinsky O, Schroeder CE, Melloni L, Haegens S, 2022. Ongoing neural oscillations influence behavior and sensory representations by suppressing neuronal excitability. NeuroImage 247, 118746. 10.1016/j.neuroimage.2021.118746 [DOI] [PubMed] [Google Scholar]
- Ince RA, Paton AT, Kay JW, Schyns PG, 2021. Bayesian inference of population prevalence. eLife 10, e62461. 10.7554/eLife.62461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince RAA, Giordano BL, Kayser C, Rousselet GA, Gross J, Schyns PG, 2017. A statistical framework for neuroimaging data analysis based on mutual information estimated via a gaussian copula. Hum. Brain Mapp 38, 1541–1573. 10.1002/hbm.23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Kahana MJ, 2010. Direct brain recordings fuel advances in cognitive electrophysiology. Trends Cogn. Sci 14, 162–171. 10.1016/j.tics.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Kahana MJ, Ekstrom AD, Fried I, 2007. Brain Oscillations Control Timing of Single-Neuron Activity in Humans. J. Neurosci 27, 3839–3844. 10.1523/JNEUROSCI.4636-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Lega B, Anderson C, 2012. Explaining How Brain Stimulation Can Evoke Memories. J. Cogn. Neurosci 24, 553–563. 10.1162/jocn_a_00170 [DOI] [PubMed] [Google Scholar]
- Jacobs J, Miller J, Lee SA, Coffey T, Watrous AJ, Sperling MR, Sharan A, Worrell G, Berry B, Lega B, Jobst BC, Davis K, Gross RE, Sheth SA, Ezzyat Y, Das SR, Stein J, Gorniak R, Kahana MJ, Rizzuto DS, 2016. Direct Electrical Stimulation of the Human Entorhinal Region and Hippocampus Impairs Memory. Neuron 92, 983–990. 10.1016/j.neuron.2016.10.062 [DOI] [PubMed] [Google Scholar]
- Jeffery ND, Bate ST, Safayi S, Howard MA, Moon L, Jeffery U, 2018. When neuroscience met clinical pathology: partitioning experimental variation to aid data interpretation in neuroscience. Eur. J. Neurosci 47, 371–379. 10.1111/ejn.13847 [DOI] [PubMed] [Google Scholar]
- Jehi L, 2018. The Epileptogenic Zone: Concept and Definition. Epilepsy Curr. 18, 12–16. 10.5698/1535-7597.18.1.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehi L, Morita-Sherman M, Love TE, Bartolomei F, Bingaman W, Braun K, Busch RM, Duncan J, Hader WJ, Luan G, Rolston JD, Schuele S, Tassi L, Vadera S, Sheikh S, Najm I, Arain A, Bingaman J, Diehl B, de Tisi J, Rados M, Van Eijsden P, Wahby S, Wang X, Wiebe S, 2021. Comparative Effectiveness of Stereotactic Electroencephalography Versus Subdural Grids in Epilepsy Surgery. Ann. Neurol 90, 927–939. 10.1002/ana.26238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. NeuroImage 62, 782–790. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Jensen O, Colgin LL, 2007. Cross-frequency coupling between neuronal oscillations. Trends Cogn. Sci 11, 267–269. 10.1016/j.tics.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Jerbi K, Freyermuth S, Dalal S, Kahane P, Bertrand O, Berthoz A, Lachaux J-P, 2009a. Saccade Related Gamma-Band Activity in Intracerebral EEG: Dissociating Neural from Ocular Muscle Activity. Brain Topogr. 22, 18–23. 10.1007/s10548-009-0078-5 [DOI] [PubMed] [Google Scholar]
- Jerbi K, Freyermuth S, Minotti L, Kahane P, Berthoz A, Lachaux J, 2009b. Chapter 12 Watching Brain TV and Playing Brain Ball, in: International Review of Neurobiology. Elsevier, pp. 159–168. 10.1016/S0074-7742(09)86012-1 [DOI] [PubMed] [Google Scholar]
- Jobst BC, Bartolomei F, Diehl B, Frauscher B, Kahane P, Minotti L, Sharan A, Tardy N, Worrell G, Gotman J, 2020. Intracranial EEG in the 21st Century. Epilepsy Curr. 20, 180–188. 10.1177/1535759720934852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EL, Knight RT, 2015. Intracranial recordings and human memory. Curr. Opin. Neurobiol 31, 18–25. 10.1016/j.conb.2014.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SR, 2016. When brain rhythms aren’t “rhythmic”: implication for their mechanisms and meaning. Curr. Opin. Neurobiol 40, 72–80. 10.1016/j.conb.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Lee SA, Kim JS, Jeong W, Chung CK, 2020. Task-dependent effects of intracranial hippocampal stimulation on human memory and hippocampal theta power. Brain Stimulat. 13, 603–613. 10.1016/j.brs.2020.01.013 [DOI] [PubMed] [Google Scholar]
- Jwa A, Poldrack R, 2021. The Spectrum of Data sharing Policies in Neuroimaging Data Repositories (preprint). PsyArXiv. 10.31234/osf.io/cnuy7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadipasaoglu CM, Baboyan VG, Conner CR, Chen G, Saad ZS, Tandon N, 2014. Surface-based mixed effects multilevel analysis of grouped human electrocorticography. NeuroImage 101, 215–224. 10.1016/j.neuroimage.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Kajikawa Y, Schroeder CE, 2011. How Local Is the Local Field Potential? Neuron 72, 847–858. 10.1016/j.neuron.2011.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakisaka Y, Kubota Y, Wang ZI, Piao Z, Mosher JC, Gonzalez-martinez J, Jin K, Alexopoulos AV, Burgess RC, 2012. Utility of simultaneous depth and MEG recording may provide complementary information regarding the epileptogenic region 14, 1–6. 10.1684/epd.2012.0517 [DOI] [PubMed] [Google Scholar]
- Kamiński MJ, Blinowska KJ, 1991. A new method of the description of the information flow in the brain structures. Biol. Cybern 65, 203–210. 10.1007/BF00198091 [DOI] [PubMed] [Google Scholar]
- Kanner AM, 2016. Management of psychiatric and neurological comorbidities in epilepsy. Nat. Rev. Neurol 12, 106–116. 10.1038/nrneurol.2015.243 [DOI] [PubMed] [Google Scholar]
- Kappenman ES, Farrens JL, Zhang W, Stewart AX, Luck SJ, 2021. ERP CORE: An open resource for human event-related potential research. NeuroImage 225, 117465. 10.1016/j.neuroimage.2020.117465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz CN, Patel K, Talakoub O, Groppe D, Hoffman K, Valiante TA, 2020. Differential Generation of Saccade, Fixation, and Image-Onset Event-Related Potentials in the Human Mesial Temporal Lobe. Cereb. Cortex 30, 5502–5516. 10.1093/cercor/bhaa132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil A, Debener S, Gratton G, Junghöfer M, Kappenman ES, Luck SJ, Luu P, Miller GA, Yee CM, n.d. Committee report: Publication guidelines and recommendations for studies using electroencephalography and magnetoencephalography 21. [DOI] [PubMed] [Google Scholar]
- Keller CJ, Cash SS, Narayanan S, Wang C, Kuzniecky R, Carlson C, Devinsky O, Thesen T, Doyle W, Sassaroli A, Boas DA, Ulbert I, Halgren E, 2009. Intracranial microprobe for evaluating neuro-hemodynamic coupling in unanesthetized human neocortex. J. Neurosci. Methods 179, 208–218. 10.1016/j.jneumeth.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CJ, Davidesco I, Megevand P, Lado FA, Malach R, Mehta AD, 2017. Tuning face perception with electrical stimulation of the fusiform gyrus: Tuning Face Perception with ES of the FG. Hum. Brain Mapp 38, 2830–2842. 10.1002/hbm.23543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CJ, Honey CJ, Mégevand P, Entz L, Ulbert I, Mehta AD, 2014. Mapping human brain networks with cortico-cortical evoked potentials. Philos. Trans. R. Soc. B Biol. Sci 369, 20130528. 10.1098/rstb.2013.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis S, Sorensen L, Darvas F, Sayres C, O’Neill K, Brown RB, House P, Ojemann J, Greger B, 2016. Multi-scale analysis of neural activity in humans: Implications for micro-scale electrocorticography. Clin. Neurophysiol 127, 591–601. 10.1016/j.clinph.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Ken S, Di Gennaro G, Giulietti G, Sebastiano F, De Carli D, Garreffa G, Colonnese C, Passariello R, Lotterie J-A, Maraviglia B, 2007. Quantitative evaluation for brain CT/MRI coregistration based on maximization of mutual information in patients with focal epilepsy investigated with subdural electrodes. Magn. Reson. Imaging 25, 883–888. 10.1016/j.mri.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Kern M, Aertsen A, Schulze-Bonhage A, Ball T, 2013. Heart cycle-related effects on event-related potentials, spectral power changes, and connectivity patterns in the human ECoG. NeuroImage 81, 178–190. 10.1016/j.neuroimage.2013.05.042 [DOI] [PubMed] [Google Scholar]
- Kern M, Bert S, Glanz O, Schulze-Bonhage A, Ball T, 2019. Human motor cortex relies on sparse and action-specific activation during laughing, smiling and speech production. Commun. Biol 2, 118. 10.1038/s42003-019-0360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern M, Schulze-Bonhage A, Ball T, 2021. Blink- and saccade-related suppression effects in early visual areas of the human brain: Intracranial EEG investigations during natural viewing conditions. NeuroImage 230, 117788. 10.1016/j.neuroimage.2021.117788 [DOI] [PubMed] [Google Scholar]
- Kerr NL, 1998. HARKing: hypothesizing after the results are known. Personal. Soc. Psychol. Rev. Off. J. Soc. Personal. Soc. Psychol. Inc 2, 196–217. 10.1207/s15327957pspr0203_4 [DOI] [PubMed] [Google Scholar]
- Khambhati AN, Kahn AE, Costantini J, Ezzyat Y, Solomon EA, Gross RE, Jobst BC, Sheth SA, Zaghloul KA, Worrell G, Seger S, Lega BC, Weiss S, Sperling MR, Gorniak R, Das SR, Stein JM, Rizzuto DS, Kahana MJ, Lucas TH, Davis KA, Tracy JI, Bassett DS, 2019. Functional control of electrophysiological network architecture using direct neurostimulation in humans. Netw. Neurosci 3, 848–877. 10.1162/netn_a_00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee C, Knowlton R, Rozzelle C, Blount JP, 2011. Safety and utility of supplemental depth electrodes for localizing the ictal onset zone in pediatric neocortical epilepsy: Clinical article. J. Neurosurg. Pediatr 8, 49–56. 10.3171/2011.4.PEDS10519 [DOI] [PubMed] [Google Scholar]
- Kim K, Ladenbauer J, Babo-Rebelo M, Buot A, Lehongre K, Adam C, Hasboun D, Lambrecq V, Navarro V, Ostojic S, Tallon-Baudry C, 2019. Resting-State Neural Firing Rate Is Linked to Cardiac-Cycle Duration in the Human Cingulate and Parahippocampal Cortices. J. Neurosci 39, 3676–3686. 10.1523/JNEUROSCI.2291-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Schedlbauer A, Rollo M, Karunakaran S, Ekstrom AD, Tandon N, 2018. Network-based brain stimulation selectively impairs spatial retrieval. Brain Stimulat. 11, 213–221. 10.1016/j.brs.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J-R, Dehaene S, 2014. Characterizing the dynamics of mental representations: the temporal generalization method. Trends Cogn. Sci 18, 203–210. 10.1016/j.tics.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger K, Hummel C, Stefan H, 1998. Postoperative multichannel magnetoencephalography in patients with recurrent seizures after epilepsy surgery. Acta Neurol. Scand 98, 1–7. 10.1111/j.1600-0404.1998.tb07370.x [DOI] [PubMed] [Google Scholar]
- Kleen JK, Scott RC, Holmes GL, Roberts DW, Rundle MM, Testorf M, Lenck-Santini P-P, Jobst BC, 2013. Hippocampal interictal epileptiform activity disrupts cognition in humans. Neurology 81, 18–24. 10.1212/WNL.0b013e318297ee50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Ghosh SS, Avants B, Yeo BTT, Fischl B, Ardekani B, Gee JC, Mann JJ, Parsey RV, 2010. Evaluation of volume-based and surface-based brain image registration methods. NeuroImage 51, 214–220. 10.1016/j.neuroimage.2010.01.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W, Sauseng P, Hanslmayr S, Gruber W, Freunberger R, 2007. Event-related phase reorganization may explain evoked neural dynamics. Neurosci. Biobehav. Rev 31, 1003–1016. 10.1016/j.neubiorev.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Klinge C, Eippert F, Röder B, Büchel C, 2010. Corticocortical connections mediate primary visual cortex responses to auditory stimulation in the blind. J. Neurosci. Off. J. Soc. Neurosci 30, 12798–12805. 10.1523/JNEUROSCI.2384-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koessler L, Cecchin T, Colnat-Coulbois S, Vignal J-P, Jonas J, Vespignani H, Ramantani G, Maillard LG, 2015. Catching the Invisible: Mesial Temporal Source Contribution to Simultaneous EEG and SEEG Recordings. Brain Topogr. 28, 5–20. 10.1007/s10548-014-0417-z [DOI] [PubMed] [Google Scholar]
- Kopell NJ, Gritton HJ, Whittington MA, Kramer MA, 2014. Beyond the connectome: the dynome. Neuron 83, 1319–1328. 10.1016/j.neuron.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewska A, Crainiceanu CM, Kuś R, Franaszczuk PJ, Crone NE, 2008. Dynamics of event-related causality in brain electrical activity. Hum. Brain Mapp 29, 1170–1192. 10.1002/hbm.20458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosciessa JQ, Grandy TH, Garrett DD, Werkle-Bergner M, 2020. Single-trial characterization of neural rhythms: Potential and challenges. NeuroImage 206, 116331. 10.1016/j.neuroimage.2019.116331 [DOI] [PubMed] [Google Scholar]
- Koubeissi MZ, Kahriman E, Syed TU, Miller J, Durand DM, 2013. Low-frequency electrical stimulation of a fiber tract in temporal lobe epilepsy: Koubeissi et al: Electrical Stimulation in TLE. Ann. Neurol. n/a-n/a 10.1002/ana.23915 [DOI] [PubMed] [Google Scholar]
- Kovac S, Kahane P, Diehl B, 2016. Seizures induced by direct electrical cortical stimulation – Mechanisms and clinical considerations. Clin. Neurophysiol 127, 31–39. 10.1016/j.clinph.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Kovac S, Vakharia VN, Scott C, Diehl B, 2017. Invasive epilepsy surgery evaluation. Seizure 44, 125–136. 10.1016/j.seizure.2016.10.016 [DOI] [PubMed] [Google Scholar]
- Kovach CK, 2011. Manifestation of ocular-muscle EMG contamination in human intracranial recordings 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach CK, Gander PE, 2016. The demodulated band transform. J. Neurosci. Methods 261, 135–154. 10.1016/j.jneumeth.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev D, Spreer J, Honegger J, Zentner J, Schulze-Bonhage A, Huppertz H-J, 2005. Rapid and fully automated visualization of subdural electrodes in the presurgical evaluation of epilepsy patients. AJNR Am. J. Neuroradiol 26, 1078–1083. [PMC free article] [PubMed] [Google Scholar]
- Kramer MA, Eden UT, 2013. Assessment of cross-frequency coupling with confidence using generalized linear models. J. Neurosci. Methods 220, 64–74. 10.1016/j.jneumeth.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraskov A, Stögbauer H, Grassberger P, 2004. Estimating mutual information. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 69, 066138. 10.1103/PhysRevE.69.066138 [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PSF, Baker CI, 2009. Circular analysis in systems neuroscience: the dangers of double dipping. Nat. Neurosci 12, 535–540. 10.1038/nn.2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelbach ML, Jenkinson N, Owen SLF, Aziz TZ, 2007. Translational principles of deep brain stimulation. Nat. Rev. Neurosci 8, 623–635. 10.1038/nrn2196 [DOI] [PubMed] [Google Scholar]
- Krucoff MO, Chan AY, Harward SC, Rahimpour S, Rolston JD, Muh C, Englot DJ, 2017. Rates and predictors of success and failure in repeat epilepsy surgery: A meta-analysis and systematic review. Epilepsia 58, 2133–2142. 10.1111/epi.13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Altman N, Blainey P, 2014. Points of significance: nested designs. For studies with hierarchical noise sources, use a nested analysis of variance approach. Nat. Methods 11, 977–978. 10.1038/nmeth.3137 [DOI] [PubMed] [Google Scholar]
- Kucewicz MT, Berry BM, Kremen V, Miller LR, Khadjevand F, Ezzyat Y, Stein JM, Wanda P, Sperling MR, Gorniak R, Davis KA, Jobst BC, Gross RE, Lega B, Stead SM, Rizzuto DS, Kahana MJ, Worrell GA, 2018. Electrical Stimulation Modulates High γ Activity and Human Memory Performance. eneuro 5, ENEURO.0369–17.2018. 10.1523/ENEURO.0369-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuś R, Kamiński M, Blinowska KJ, 2004. Determination of EEG activity propagation: pair-wise versus multichannel estimate. IEEE Trans. Biomed. Eng 51, 1501–1510. 10.1109/TBME.2004.827929 [DOI] [PubMed] [Google Scholar]
- Lachaux J-P, Axmacher N, Mormann F, Halgren E, Crone NE, 2012. High-frequency neural activity and human cognition: Past, present and possible future of intracranial EEG research. Prog. Neurobiol 98, 279–301. 10.1016/j.pneurobio.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux J-P, Fonlupt P, Kahane P, Minotti L, Hoffmann D, Bertrand O, Baciu M, 2007a. Relationship between task-related gamma oscillations and BOLD signal: new insights from combined fMRI and intracranial EEG. Hum. Brain Mapp 28, 1368–1375. 10.1002/hbm.20352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux J-P, Hoffmann D, Minotti L, Berthoz A, Kahane P, 2006. Intracerebral dynamics of saccade generation in the human frontal eye field and supplementary eye field. NeuroImage 30, 1302–1312. 10.1016/j.neuroimage.2005.11.023 [DOI] [PubMed] [Google Scholar]
- Lachaux J-P, Jerbi K, Bertrand O, Minotti L, Hoffmann D, Schoendorff B, Kahane P, 2007b. A Blueprint for Real-Time Functional Mapping via Human Intracranial Recordings. PLoS ONE 2, e1094. 10.1371/journal.pone.0001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux JP, Rodriguez E, Martinerie J, Varela FJ, 1999. Measuring phase synchrony in brain signals. Hum. Brain Mapp 8, 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos P, Shah AS, Knuth KH, Ulbert I, Karmos G, Schroeder CE, 2005. An oscillatory hierarchy controlling neuronal excitability and stimulus processing in the auditory cortex. J. Neurophysiol 94, 1904–1911. 10.1152/jn.00263.2005 [DOI] [PubMed] [Google Scholar]
- Langers DRM, 2014. Assessment of tonotopically organised subdivisions in human auditory cortex using volumetric and surface-based cortical alignments. Hum. Brain Mapp 35, 1544–1561. 10.1002/hbm.22272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante RA, Tang W, Peled N, Vallejo DI, Borzello M, Dougherty DD, Eskandar EN, Widge AS, Cash SS, Stufflebeam SM, 2017. The interactive electrode localization utility: software for automatic sorting and labeling of intracranial subdural electrodes. Int. J. Comput. Assist. Radiol. Surg 12, 1829–1837. 10.1007/s11548-016-1504-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascano AM, Perneger T, Vulliemoz S, Spinelli L, Garibotto V, Korff CM, Vargas MI, Michel CM, Seeck M, 2016. Yield of MRI, high-density electric source imaging (HD-ESI), SPECT and PET in epilepsy surgery candidates. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol 127, 150–155. 10.1016/j.clinph.2015.03.025 [DOI] [PubMed] [Google Scholar]
- Laufs H, 2012. Functional imaging of seizures and epilepsy: evolution from zones to networks. Curr. Opin. Neurol 25, 194–200. 10.1097/WCO.0b013e3283515db9 [DOI] [PubMed] [Google Scholar]
- LaViolette PS, Rand SD, Ellingson BM, Raghavan M, Lew SM, Schmainda KM, Mueller W, 2011. 3D visualization of subdural electrode shift as measured at craniotomy reopening. Epilepsy Res. 94, 102–109. 10.1016/j.eplepsyres.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Van Quyen M, Foucher J, Lachaux J, Rodriguez E, Lutz A, Martinerie J, Varela FJ, 2001. Comparison of Hilbert transform and wavelet methods for the analysis of neuronal synchrony. J. Neurosci. Methods 111, 83–98. 10.1016/s0165-0270(01)00372-7 [DOI] [PubMed] [Google Scholar]
- Le Van Quyen M, Staba R, Bragin A, Dickson C, Valderrama M, Fried I, Engel J, 2010. Large-scale microelectrode recordings of high-frequency gamma oscillations in human cortex during sleep. J. Neurosci. Off. J. Soc. Neurosci 30, 7770–7782. 10.1523/JNEUROSCI.5049-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Stephen KJ, n.d. Biopotential Electrode Sensors in ECG/EEG/EMG Systems. [Google Scholar]
- Leeman-Markowski B, Hardstone R, Lohnas L, Cowen B, Davachi L, Doyle W, Dugan P, Friedman D, Liu A, Melloni L, Selesnick I, Wang B, Meador K, Devinsky O, 2021. Effects of hippocampal interictal discharge timing, duration, and spatial extent on list learning. Epilepsy Behav. 123, 108209. 10.1016/j.yebeh.2021.108209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehongre K, Lambrecq V, Whitmarsh S, Frazzini V, Cousyn L, Soleil D, Fernandez-Vidal S, Mathon B, Houot M, Lemaréchal J-D, Clemenceau S, Hasboun D, Adam C, Navarro V, 2022. Long-term deep intracerebral microelectrode recordings in patients with drug-resistant epilepsy: Proposed guidelines based on 10-year experience. NeuroImage 254, 119116. 10.1016/j.neuroimage.2022.119116 [DOI] [PubMed] [Google Scholar]
- Leszczyński M, Barczak A, Kajikawa Y, Ulbert I, Falchier AY, Tal I, Haegens S, Melloni L, Knight RT, Schroeder CE, 2020. Dissociation of broadband high-frequency activity and neuronal firing in the neocortex. Sci. Adv 6, eabb0977. 10.1126/sciadv.abb0977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Anumanchipalli GK, Mohamed A, Lu J, Wu J, Chang EF, 2022. Dissecting neural computations of the human auditory pathway using deep neural networks for speech (preprint). Neuroscience. 10.1101/2022.03.14.484195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindig T, Kotikalapudi R, Schweikardt D, Martin P, Bender F, Klose U, Ernemann U, Focke NK, Bender B, 2018. Evaluation of multimodal segmentation based on 3D T1-, T2- and FLAIR-weighted images - the difficulty of choosing. NeuroImage 170, 210–221. 10.1016/j.neuroimage.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Lisman J, 2005. The theta/gamma discrete phase code occuring during the hippocampal phase precession may be a more general brain coding scheme. Hippocampus 15, 913–922. 10.1002/hipo.20121 [DOI] [PubMed] [Google Scholar]
- Little S, Gilron R, Perrone R, Wilt R, Starr P, 2021. Using cortico-basal ganglia network physiology for chronic embedded adaptive DBS in movement disorders. Brain Stimulat. 14, 1726. 10.1016/j.brs.2021.10.456 [DOI] [Google Scholar]
- Lobier M, Siebenhühner F, Palva S, Palva JM, 2014. Phase transfer entropy: a novel phase-based measure for directed connectivity in networks coupled by oscillatory interactions. NeuroImage 85 Pt 2, 853–872. 10.1016/j.neuroimage.2013.08.056 [DOI] [PubMed] [Google Scholar]
- Lopes da Silva F, 2013. EEG and MEG: relevance to neuroscience. Neuron 80, 1112–1128. 10.1016/j.neuron.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Lopes da Silva FH, 2006. Event-related neural activities: what about phase? Prog. Brain Res 159, 3–17. 10.1016/S0079-6123(06)59001-6 [DOI] [PubMed] [Google Scholar]
- Lorio S, Fresard S, Adaszewski S, Kherif F, Chowdhury R, Frackowiak RS, Ashburner J, Helms G, Weiskopf N, Lutti A, Draganski B, 2016. New tissue priors for improved automated classification of subcortical brain structures on MRI. NeuroImage 130, 157–166. 10.1016/j.neuroimage.2016.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughin TM, 2004. A systematic comparison of methods for combining p-values from independent tests. Comput. Stat. Data Anal 47, 467–485. 10.1016/j.csda.2003.11.020 [DOI] [Google Scholar]
- Lozano-Soldevilla D, Ter Huurne N, Oostenveld R, 2016. Neuronal Oscillations with Non-sinusoidal Morphology Produce Spurious Phase-to-Amplitude Coupling and Directionality. Front. Comput. Neurosci 10, 87. 10.3389/fncom.2016.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders H, Akamatsu N, Amina S, Baumgartner C, Benbadis S, Bermeo‐Ovalle A, Bleasel A, Bozorgi A, Carreño M, Devereaux M, Fernandez‐Baca Vaca G, Francione S, García Losarcos N, Hamer H, Holthausen H, Jamal Omidi S, Kalamangalam G, Kanner A, Knake S, Lacuey N, Lhatoo S, Lim S, Mani J, Matsumoto R, Miller J, Noachtar S, Palmini A, Park J, Rosenow F, Shahid A, Schuele S, Steinhoff B, Szabo CÁ, Tandon N, Terada K, Van Emde Boas W, Widdess‐Walsh P, Kahane P, 2019. Critique of the 2017 epileptic seizure and epilepsy classifications. Epilepsia epi.14699. 10.1111/epi.14699 [DOI] [PubMed] [Google Scholar]
- Aghajan MZ, Schuette P, Fields TA, Tran ME, Siddiqui SM, Hasulak NR, Tcheng TK, Eliashiv D, Mankin EA, Stern J, Fried I, Suthana N, 2017. Theta Oscillations in the Human Medial Temporal Lobe during Real-World Ambulatory Movement. Curr. Biol 27, 3743–3751.e3. 10.1016/j.cub.2017.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Hagiwara A, Hori M, Andica C, Haruyama T, Kuramochi M, Nakazawa M, Koshino S, Irie R, Kamagata K, Wada A, Abe O, Aoki S, 2019. Effect of Gadolinium on the Estimation of Myelin and Brain Tissue Volumes Based on Quantitative Synthetic MRI. AJNR Am. J. Neuroradiol 40, 231–237. 10.3174/ajnr.A5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotti JF, 2020. RAVE: Comprehensive open-source software for reproducible analysis and visualization of intracranial EEG data 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahvash M, König R, Wellmer J, Urbach H, Meyer B, Schaller K, 2007. Coregistration of digital photography of the human cortex and cranial magnetic resonance imaging for visualization of subdural electrodes in epilepsy surgery. Neurosurgery 61, 340–344; discussion 344–345. 10.1227/01.neu.0000303992.87987.17 [DOI] [PubMed] [Google Scholar]
- Makeig S, Debener S, Onton J, Delorme A, 2004. Mining event-related brain dynamics. Trends Cogn. Sci 8, 204–210. 10.1016/j.tics.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Manning JR, Jacobs J, Fried I, Kahana MJ, 2009. Broadband shifts in local field potential power spectra are correlated with single-neuron spiking in humans. J. Neurosci. Off. J. Soc. Neurosci 29, 13613–13620. 10.1523/JNEUROSCI.2041-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolov R, Solanas A, 2012. Assigning and combining probabilities in single-case studies. Psychol. Methods 17, 495–509. 10.1037/a0029248 [DOI] [PubMed] [Google Scholar]
- Marie D, Jobard G, Crivello F, Perchey G, Petit L, Mellet E, Joliot M, Zago L, Mazoyer B, Tzourio-Mazoyer N, 2015. Descriptive anatomy of Heschl’s gyri in 430 healthy volunteers, including 198 left-handers. Brain Struct. Funct 220, 729–743. 10.1007/s00429-013-0680-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Mikutta C, Leonard MK, Hungate D, Koelsch S, Shamma S, Chang EF, Millán J.del R, Knight RT, Pasley BN, 2018. Neural Encoding of Auditory Features during Music Perception and Imagery. Cereb. Cortex 28, 4222–4233. 10.1093/cercor/bhx277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto R, Nair DR, LaPresto E, Najm I, Bingaman W, Shibasaki H, Lüders HO, 2004. Functional connectivity in the human language system: a cortico-cortical evoked potential study. Brain 127, 2316–2330. 10.1093/brain/awh246 [DOI] [PubMed] [Google Scholar]
- Matusz PJ, Dikker S, Huth AG, Perrodin C, 2019. Are We Ready for Real-world Neuroscience? J. Cogn. Neurosci 31, 327–338. 10.1162/jocn_e_01276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, Schwalb JM, Kennedy SH, 2005. Deep Brain Stimulation for Treatment-Resistant Depression. Neuron 45, 651–660. 10.1016/j.neuron.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Mayer A, Schwiedrzik CM, Wibral M, Singer W, Melloni L, 2016. Expecting to See a Letter: Alpha Oscillations as Carriers of Top-Down Sensory Predictions. Cereb. Cortex 26, 3146–3160. 10.1093/cercor/bhv146 [DOI] [PubMed] [Google Scholar]
- Mazaheri A, Jensen O, 2006. Posterior activity is not phase-reset by visual stimuli. Proc. Natl. Acad. Sci 103, 2948–2952. 10.1073/pnas.0505785103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B, 2001. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B. Biol. Sci 356, 1293–1322. 10.1098/rstb.2001.0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J, 1995. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). NeuroImage 2, 89–101. 10.1006/nimg.1995.1012 [DOI] [PubMed] [Google Scholar]
- Mazzola L, Mauguière F, Isnard J, 2019. Functional mapping of the human insula: Data from electrical stimulations. Rev. Neurol. (Paris) 175, 150–156. 10.1016/j.neurol.2018.12.003 [DOI] [PubMed] [Google Scholar]
- McCarthy G, Nobre AC, Bentin S, Spencer DD, 1995. Language-related field potentials in the anterior-medial temporal lobe: I. Intracranial distribution and neural generators. J. Neurosci. Off. J. Soc. Neurosci 15, 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty MJ, Woolnough O, Mosher JC, Seymour J, Tandon N, 2022. The Listening Zone of Human Electrocorticographic Field Potential Recordings. eNeuro 9, ENEURO.0492–21.2022. 10.1523/ENEURO.0492-21.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medani T, Garcia-Prieto J, Tadel F, Schrader S, Antonakakis M, Joshi AA, Engwer C, Wolters CH, Mosher JC, Leahy RM, 2021. Realistic head modeling of electromagnetic brain activity: an integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions, in: Bosmans H, Zhao W, Yu L.(Eds.), Medical Imaging 2021: Physics of Medical Imaging. Presented at the Physics of Medical Imaging, SPIE, Online Only, United States, p. 135. 10.1117/12.2580935 [DOI] [Google Scholar]
- Medani T, Garcia-Prieto J, Tadel F, Schrader S, Joshi A, Engwer C, Wolters CH, Mosher JC, Leahy RM, 2020. Realistic head modeling of electromagnetic brain activity: An integrated Brainstorm pipeline from MRI data to the FEM solution. ArXiv201101292 Phys. [Google Scholar]
- Medina Villalon S, Paz R, Roehri N, Lagarde S, Pizzo F, Colombet B, Bartolomei F, Carron R, Bénar C-G, 2018. EpiTools, A software suite for presurgical brain mapping in epilepsy: Intracerebral EEG. J. Neurosci. Methods 303, 7–15. 10.1016/j.jneumeth.2018.03.018 [DOI] [PubMed] [Google Scholar]
- Mégevand P, Groppe DM, Goldfinger MS, Hwang ST, Kingsley PB, Davidesco I, Mehta AD, 2014. Seeing scenes: topographic visual hallucinations evoked by direct electrical stimulation of the parahippocampal place area. J. Neurosci. Off. J. Soc. Neurosci 34, 5399–5405. 10.1523/JNEUROSCI.5202-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégevand P, Mercier MR, Groppe DM, Zion Golumbic E, Mesgarani N, Beauchamp MS, Schroeder CE, Mehta AD, 2020. Crossmodal Phase Reset and Evoked Responses Provide Complementary Mechanisms for the Influence of Visual Speech in Auditory Cortex. J. Neurosci. Off. J. Soc. Neurosci 40, 8530–8542. 10.1523/JNEUROSCI.0555-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenhelter S, Testorf ME, Gorenstein MA, Hasulak NR, Tcheng TK, Aronson JP, Jobst BC, 2019. Cognitive tasks and human ambulatory electrocorticography using the RNS System. J. Neurosci. Methods 311, 408–417. 10.1016/j.jneumeth.2018.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler SL, 2019. Does data cleaning improve brain state classification? J. Neurosci. Methods 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloni L, Schwiedrzik CM, Rodriguez E, Singer W, 2009. (Micro)Saccades, corollary activity and cortical oscillations. Trends Cogn. Sci 13, 239–245. 10.1016/j.tics.2009.03.007 [DOI] [PubMed] [Google Scholar]
- Mercier MR, Bickel S, Megevand P, Groppe DM, Schroeder CE, Mehta AD, Lado FA, 2017. Evaluation of cortical local field potential diffusion in stereotactic electro-encephalography recordings: A glimpse on white matter signal. NeuroImage 147, 219–232. 10.1016/j.neuroimage.2016.08.037 [DOI] [PubMed] [Google Scholar]
- Mercier MR, Foxe JJ, Fiebelkorn IC, Butler JS, Schwartz TH, Molholm S, 2013. Auditory-driven phase reset in visual cortex: Human electrocorticography reveals mechanisms of early multisensory integration. NeuroImage 79, 19–29. 10.1016/j.neuroimage.2013.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier MR, Molholm S, Fiebelkorn IC, Butler JS, Schwartz TH, Foxe JJ, 2015. Neuro-oscillatory phase alignment drives speeded multisensory response times: an electro-corticographic investigation. J. Neurosci. Off. J. Soc. Neurosci 35, 8546–8557. 10.1523/JNEUROSCI.4527-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesgarani N, Cheung C, Johnson K, Chang EF, 2014. Phonetic Feature Encoding in Human Superior Temporal Gyrus. Science 343, 1006–1010. 10.1126/science.1245994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurers T, Bild R, Do K-M, Prasser F, 2021. A scalable software solution for anonymizing high-dimensional biomedical data. GigaScience 10, giab068. 10.1093/gigascience/giab068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmann S, Treder MS, Griffiths B, Kerrén C, Roux F, Wimber M, Rollings D, Sawlani V, Chelvarajah R, Gollwitzer S, Kreiselmeyer G, Hamer H, Bowman H, Staresina B, Hanslmayr S, 2018. Data-driven re-referencing of intracranial EEG based on independent component analysis (ICA). J. Neurosci. Methods 307, 125–137. 10.1016/j.jneumeth.2018.06.021 [DOI] [PubMed] [Google Scholar]
- Mikulan E, Russo S, Parmigiani S, Sarasso S, Zauli FM, Rubino A, Avanzini P, Cattani A, Sorrentino A, Gibbs S, Cardinale F, Sartori I, Nobili L, Massimini M, Pigorini A, 2020. Simultaneous human intracerebral stimulation and HD-EEG, ground-truth for source localization methods. Sci. Data 7, 127. 10.1038/s41597-020-0467-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulan E, Theoharis A, Russo S, Zauli FM, Sartori I, Parmigiani S, Sarasso S, Del Vecchio M, Avanzini P, Pigorini A, 2021. Scalp EEG prediction of intracranial high-frequency responses to median nerve stimulation: insights from simultaneous recordings. Brain Stimulat. 14, 1640–1641. 10.1016/j.brs.2021.10.169 [DOI] [Google Scholar]
- Miller KJ, 2019. A library of human electrocorticographic data and analyses. Nat. Hum. Behav 3, 1225–1235. 10.1038/s41562-019-0678-3 [DOI] [PubMed] [Google Scholar]
- Miller KJ, 2010. Broadband spectral change: evidence for a macroscale correlate of population firing rate? J. Neurosci. Off. J. Soc. Neurosci 30, 6477–6479. 10.1523/JNEUROSCI.6401-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KJ, denNijs M, Shenoy P, Miller JW, Rao RPN, Ojemann JG, 2007a. Real-time functional brain mapping using electrocorticography. NeuroImage 37, 504–507. 10.1016/j.neuroimage.2007.05.029 [DOI] [PubMed] [Google Scholar]
- Miller KJ, Hermes D, Honey CJ, Hebb AO, Ramsey NF, Knight RT, Ojemann JG, Fetz EE, 2012. Human motor cortical activity is selectively phase-entrained on underlying rhythms. PLoS Comput. Biol 8, e1002655. 10.1371/journal.pcbi.1002655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KJ, Honey CJ, Hermes D, Rao RP, denNijs M, Ojemann JG, 2014. Broadband changes in the cortical surface potential track activation of functionally diverse neuronal populations. NeuroImage 85, 711–720. 10.1016/j.neuroimage.2013.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KJ, Huiskamp G, van Blooijs D, Hermes D, Gebbink TA, Ferrier CH, van Rijen PC, Gosselaar P, Ramsey NF, Leijten FSS, 2019. An observation of anatomical clustering in inputs to primary motor cortex in cortico-cortical brain surface evoked potentials, in: 2019 7th International Winter Conference on Brain-Computer Interface (BCI). Presented at the 2019 7th International Winter Conference on Brain-Computer Interface (BCI), IEEE, Gangwon, Korea (South), pp. 1–2. 10.1109/IWW-BCI.2019.8737326 [DOI] [Google Scholar]
- Miller KJ, Makeig S, Hebb AO, Rao RPN, denNijs M, Ojemann JG, 2007b. Cortical electrode localization from X-rays and simple mapping for electrocorticographic research: The “Location on Cortex” (LOC) package for MATLAB. J. Neurosci. Methods 162, 303–308. 10.1016/j.jneumeth.2007.01.019 [DOI] [PubMed] [Google Scholar]
- Miller KJ, Sorensen LB, Ojemann JG, den Nijs M, 2009. Power-Law Scaling in the Brain Surface Electric Potential. PLoS Comput. Biol 5, e1000609. 10.1371/journal.pcbi.1000609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein J, Mormann F, Fried I, Koch C, 2009. Neuronal shot noise and Brownian 1/f2 behavior in the local field potential. PloS One 4, e4338. 10.1371/journal.pone.0004338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minotti L, Montavont A, Scholly J, Tyvaert L, Taussig D, 2018. Indications and limits of stereoelectroencephalography (SEEG). Neurophysiol. Clin 48, 15–24. 10.1016/j.neucli.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Mitra PP, Pesaran B, 1999. Analysis of dynamic brain imaging data. Biophys. J 76, 691–708. 10.1016/S0006-3495(99)77236-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran D, 2010. Evolution of brain-computer interface: action potentials, local field potentials and electrocorticograms. Curr. Opin. Neurobiol 20, 741–745. 10.1016/j.conb.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, O’Brien TJ, Cook MJ, Murphy M, Bowden SC, 2004. A computer-generated stereotactic “Virtual Subdural Grid” to guide resective epilepsy surgery. AJNR Am. J. Neuroradiol 25, 77–83. [PMC free article] [PubMed] [Google Scholar]
- Moses DA, Metzger SL, Liu JR, Anumanchipalli GK, Makin JG, Sun PF, Chartier J, Dougherty ME, Liu PM, Abrams GM, Tu-Chan A, Ganguly K, Chang EF, 2021. Neuroprosthesis for Decoding Speech in a Paralyzed Person with Anarthria. N. Engl. J. Med 385, 217–227. 10.1056/NEJMoa2027540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher CP, Wei Y, Kamiński J, Nandi A, Mamelak AN, Anastassiou CA, Rutishauser U, 2020. Cellular Classes in the Human Brain Revealed In Vivo by Heartbeat-Related Modulation of the Extracellular Action Potential Waveform. Cell Rep. 30, 3536–3551.e6. 10.1016/j.celrep.2020.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher JC, Funke ME, 2020. Towards Best Practices in Clinical Magnetoencephalography: Patient Preparation and Data Acquisition. J. Clin. Neurophysiol 37, 498–507. 10.1097/WNP.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouthaan BE, van ‘t Klooster MA, Keizer D, Hebbink GJ, Leijten FSS, Ferrier CH, van Putten MJAM, Zijlmans M, Huiskamp GJM, 2016. Single Pulse Electrical Stimulation to identify epileptogenic cortex: Clinical information obtained from early evoked responses. Clin. Neurophysiol 127, 1088–1098. 10.1016/j.clinph.2015.07.031 [DOI] [PubMed] [Google Scholar]
- Mukamel R, Fried I, 2012. Human Intracranial Recordings and Cognitive Neuroscience. Annu. Rev. Psychol 63, 511–537. 10.1146/annurev-psych-120709-145401 [DOI] [PubMed] [Google Scholar]
- Mukamel R, Gelbard H, Arieli A, Hasson U, Fried I, Malach R, 2005. Coupling between neuronal firing, field potentials, and FMRI in human auditory cortex. Science 309, 951–954. 10.1126/science.1110913 [DOI] [PubMed] [Google Scholar]
- Muller L, Hamilton LS, Edwards E, Bouchard KE, Chang EF, 2016. Spatial resolution dependence on spectral frequency in human speech cortex electrocorticography. J. Neural Eng 13, 056013. 10.1088/1741-2560/13/5/056013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munari C, Kahane P, Tassi L, Francione S, Hoffmann D, Russo GL, Benabid AL, 1993. Intracerebral Low Frequency Electrical Stimulation: a New Tool for the Definition of the “Epileptogenic Area”?, in: Meyerson BA, Broggi G, Martin-Rodriguez J, Ostertag C, Sindou M.(Eds.), Advances in Stereotactic and Functional Neurosurgery 10. Springer Vienna, Vienna, pp. 181–185. 10.1007/978-3-7091-9297-9_42 [DOI] [PubMed] [Google Scholar]
- Murphey DK, Maunsell JHR, Beauchamp MS, Yoshor D, 2009. Perceiving electrical stimulation of identified human visual areas. Proc. Natl. Acad. Sci 106, 5389–5393. 10.1073/pnas.0804998106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murta T, Chaudhary UJ, Tierney TM, Dias A, Leite M, Carmichael DW, Figueiredo P, Lemieux L, 2017. Phase–amplitude coupling and the BOLD signal: A simultaneous intracranial EEG (icEEG) - fMRI study in humans performing a finger-tapping task. NeuroImage 146, 438–451. 10.1016/j.neuroimage.2016.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Sugiura A, Brown EC, Sonoda M, Jeong J, Rothermel R, Luat AF, Sood S, Asano E, 2019. Four‐dimensional functional cortical maps of visual and auditory language: Intracranial recording. Epilepsia 60, 255–267. 10.1111/epi.14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narizzano M, Arnulfo G, Ricci S, Toselli B, Tisdall M, Canessa A, Fato MM, Cardinale F, 2017. SEEG assistant: a 3DSlicer extension to support epilepsy surgery. BMC Bioinformatics 18, 124. 10.1186/s12859-017-1545-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulin VV, Nolte G, Curio G, 2011. A novel method for reliable and fast extraction of neuronal EEG/MEG oscillations on the basis of spatio-spectral decomposition. NeuroImage 55, 1528–1535. 10.1016/j.neuroimage.2011.01.057 [DOI] [PubMed] [Google Scholar]
- Nir Y, Fisch L, Mukamel R, Gelbard-Sagiv H, Arieli A, Fried I, Malach R, 2007. Coupling between neuronal firing rate, gamma LFP, and BOLD fMRI is related to interneuronal correlations. Curr. Biol CB 17, 1275–1285. 10.1016/j.cub.2007.06.066 [DOI] [PubMed] [Google Scholar]
- Niso G, Gorgolewski KJ, Bock E, Brooks TL, Flandin G, Gramfort A, Henson RN, Jas M, Litvak V, Moreau T, J., Oostenveld R, Schoffelen J-M, Tadel F, Wexler J, Baillet S, 2018. MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Sci. Data 5, 180110. 10.1038/sdata.2018.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte G, Bai O, Wheaton L, Mari Z, Vorbach S, Hallett M, 2004. Identifying true brain interaction from EEG data using the imaginary part of coherency. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol 115, 2292–2307. 10.1016/j.clinph.2004.04.029 [DOI] [PubMed] [Google Scholar]
- Nolte G, Ziehe A, Nikulin VV, Schlögl A, Krämer N, Brismar T, Müller K-R, 2008. Robustly estimating the flow direction of information in complex physical systems. Phys. Rev. Lett 100, 234101. 10.1103/PhysRevLett.100.234101 [DOI] [PubMed] [Google Scholar]
- Norman Y, Yeagle EM, Harel M, Mehta AD, Malach R, 2017. Neuronal baseline shifts underlying boundary setting during free recall. Nat. Commun 8, 1301. 10.1038/s41467-017-01184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek BA, Ebersole CR, DeHaven AC, Mellor DT, 2018. The preregistration revolution. Proc. Natl. Acad. Sci. U. S. A 115, 2600–2606. 10.1073/pnas.1708274114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttin B, Cosyns P, Demeulemeester H, Gybels J, Meyerson B, 1999. Electrical stimulation in anterior limbs of internal capsules in patients with obsessive-compulsive disorder. The Lancet 354, 1526. 10.1016/S0140-6736(99)02376-4 [DOI] [PubMed] [Google Scholar]
- Oehrn CR, Baumann C, Fell J, Lee H, Kessler H, Habel U, Hanslmayr S, Axmacher N, 2015. Human Hippocampal Dynamics during Response Conflict. Curr. Biol CB 25, 2307–2313. 10.1016/j.cub.2015.07.032 [DOI] [PubMed] [Google Scholar]
- Ojemann GA, Ojemann J, Ramsey NF, 2013. Relation between functional magnetic resonance imaging (fMRI) and single neuron, local field potential (LFP) and electrocorticography (ECoG) activity in human cortex. Front. Hum. Neurosci 7, 34. 10.3389/fnhum.2013.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojemann GA, Whitaker HA, 1978. Language localization and variability. Brain Lang. 6, 239–260. 10.1016/0093-934x(78)90061-5 [DOI] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE, 1997. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. NeuroImage 6, 156–167. 10.1006/nimg.1997.0289 [DOI] [PubMed] [Google Scholar]
- on behalf of the MAGNIMS Study Group and Alzheimer’s Disease Neuroimaging Initiative, de Sitter A, Visser M, Brouwer I, Cover KS, van Schijndel RA, Eijgelaar RS, Müller DMJ, Ropele S, Kappos L, Rovira Á, Filippi M, Enzinger C, Frederiksen J, Ciccarelli O, Guttmann CRG, Wattjes MP, Witte MG, de Witt Hamer PC, Barkhof F, Vrenken H, 2020. Facing privacy in neuroimaging: removing facial features degrades performance of image analysis methods. Eur. Radiol 30, 1062–1074. 10.1007/s00330-019-06459-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill GC, Tewarie P, Vidaurre D, Liuzzi L, Woolrich MW, Brookes MJ, 2018. Dynamics of large-scale electrophysiological networks: A technical review. NeuroImage 180, 559–576. 10.1016/j.neuroimage.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, Schoffelen J-M, 2011. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci 2011, 156869. 10.1155/2011/156869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenveld R, Oostendorp TF, 2002. Validating the boundary element method for forward and inverse EEG computations in the presence of a hole in the skull. Hum. Brain Mapp 17, 179–192. 10.1002/hbm.10061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JP, Wells WM, Golby AJ, 2006. Using Surface Normals to Localize Subdural Intracranial Electrodes Placed During Neurosurgery, in: 3rd IEEE International Symposium on Biomedical Imaging: Macro to Nano, 2006. Presented at the 3rd IEEE International Symposium on Biomedical Imaging: Macro to Nano, 2006., IEEE, Arlington, Virginia, USA, pp. 331–334. 10.1109/ISBI.2006.1624920 [DOI] [Google Scholar]
- Ossandón T, Vidal JR, Ciumas C, Jerbi K, Hamamé CM, Dalal SS, Bertrand O, Minotti L, Kahane P, Lachaux J-P, 2012. Efficient “pop-out” visual search elicits sustained broadband γ activity in the dorsal attention network. J. Neurosci. Off. J. Soc. Neurosci 32, 3414–3421. 10.1523/JNEUROSCI.6048-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo H, Ochi A, Imai K, Akiyama T, Fujimoto A, Go C, Dirks P, Donner EJ, 2008. High-frequency oscillations of ictal muscle activity and epileptogenic discharges on intracranial EEG in a temporal lobe epilepsy patient. Clin. Neurophysiol 119, 862–868. 10.1016/j.clinph.2007.12.014 [DOI] [PubMed] [Google Scholar]
- Oya H, Howard MA, Magnotta VA, Kruger A, Griffiths TD, Lemieux L, Carmichael DW, Petkov CI, Kawasaki H, Kovach CK, Sutterer MJ, Adolphs R, 2017. Mapping effective connectivity in the human brain with concurrent intracranial electrical stimulation and BOLD-fMRI. J. Neurosci. Methods 277, 101–112. 10.1016/j.jneumeth.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya H, Poon PWF, Brugge JF, Reale RA, Kawasaki H, Volkov IO, Howard MA, 2007. Functional connections between auditory cortical fields in humans revealed by Granger causality analysis of intra-cranial evoked potentials to sounds: Comparison of two methods. Biosystems 89, 198–207. 10.1016/j.biosystems.2006.05.018 [DOI] [PubMed] [Google Scholar]
- Ozker M, Doyle W, Devinsky O, Flinker A, 2022. A cortical network processes auditory error signals during human speech production to maintain fluency. PLOS Biol. 20, e3001493. 10.1371/journal.pbio.3001493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-D, Bernasconi F, Salomon R, Tallon-Baudry C, Spinelli L, Seeck M, Schaller K, Blanke O, 2018. Neural Sources and Underlying Mechanisms of Neural Responses to Heartbeats, and their Role in Bodily Self-consciousness: An Intracranial EEG Study. Cereb. Cortex 28, 2351–2364. 10.1093/cercor/bhx136 [DOI] [PubMed] [Google Scholar]
- Pasley BN, David SV, Mesgarani N, Flinker A, Shamma SA, Crone NE, Knight RT, Chang EF, 2012. Reconstructing Speech from Human Auditory Cortex. PLoS Biol. 10, e1001251. 10.1371/journal.pbio.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M, Govaart GH, Schettino A, 2021. Making ERP research more transparent: Guidelines for preregistration. Int. J. Psychophysiol. Off. J. Int. Organ. Psychophysiol 164, 52–63. 10.1016/j.ijpsycho.2021.02.016 [DOI] [PubMed] [Google Scholar]
- Paulk AC, Yang JC, Cleary DR, Soper DJ, Halgren M, O’Donnell AR, Lee SH, Ganji M, Ro YG, Oh H, Hossain L, Lee J, Tchoe Y, Rogers N, Kiliç K, Ryu SB, Lee SW, Hermiz J, Gilja V, Ulbert I, Fabó D, Thesen T, Doyle WK, Devinsky O, Madsen JR, Schomer DL, Eskandar EN, Lee JW, Maus D, Devor A, Fried SI, Jones PS, Nahed BV, Ben-Haim S, Bick SK, Richardson RM, Raslan AM, Siler DA, Cahill DP, Williams ZM, Cosgrove GR, Dayeh SA, Cash SS, 2021. Microscale Physiological Events on the Human Cortical Surface. Cereb. Cortex N. Y. N 1991 31, 3678–3700. 10.1093/cercor/bhab040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield W, Boldrey E, 1937. SOMATIC MOTOR AND SENSORY REPRESENTATION IN THE CEREBRAL CORTEX OF MAN AS STUDIED BY ELECTRICAL STIMULATION. Brain 60, 389–443. 10.1093/brain/60.4.389 [DOI] [Google Scholar]
- Penfield W, Jasper HH, Penfield W, 1954. Epilepsy and the functional anatomy of the human brain. Little, Brown, Boston. [Google Scholar]
- Penfield W, Perot P, 1963. THE BRAIN’S RECORD OF AUDITORY AND VISUAL EXPERIENCE: A FINAL SUMMARY AND DISCUSSION. Brain 86, 595–696. 10.1093/brain/86.4.595 [DOI] [PubMed] [Google Scholar]
- Penfield W, Rasmussen T, 1950. The Cerebral Cortex of Man: A Clinical Study of Localization of Function. Macmillan. [Google Scholar]
- Penny WD, Duzel E, Miller KJ, Ojemann JG, 2008. Testing for nested oscillation. J. Neurosci. Methods 174, 50–61. 10.1016/j.jneumeth.2008.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet C, 2016. Null hypothesis significance testing: a short tutorial. F1000Research 4, 621. 10.12688/f1000research.6963.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet C, Garrido MI, Gramfort A, Maurits N, Michel CM, Pang E, Salmelin R, Schoffelen JM, Valdes-Sosa PA, Puce A, 2020. Issues and recommendations from the OHBM COBIDAS MEEG committee for reproducible EEG and MEG research. Nat. Neurosci 23, 1473–1483. 10.1038/s41593-020-00709-0 [DOI] [PubMed] [Google Scholar]
- Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R, 2019. EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci. Data 6, 103. 10.1038/s41597-019-0104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone‐Bertolotti M, Alexandre S, Jobb A, De Palma L, Baciu M, Mairesse M, Hoffmann D, Minotti L, Kahane P, David O, 2020. Probabilistic mapping of language networks from high frequency activity induced by direct electrical stimulation. Hum. Brain Mapp 41, 4113–4126. 10.1002/hbm.25112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaran B, Vinck M, Einevoll GT, Sirota A, Fries P, Siegel M, Truccolo W, Schroeder CE, Srinivasan R, 2018. Investigating large-scale brain dynamics using field potential recordings: analysis and interpretation. Nat. Neurosci 21, 903–919. 10.1038/s41593-018-0171-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantoni G, Hermes D, Ramsey N, Petridou N, 2021. Size of the spatial correlation between ECoG and fMRI activity. NeuroImage 242, 118459. 10.1016/j.neuroimage.2021.118459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton TW, Bentin S, Berg P, Donchin E, Hillyard SA, Johnson R, Miller GA, Ritter W, Ruchkin DS, Rugg MD, Taylor MJ, 2000. Guidelines for using human event-related potentials to study cognition: Recording standards and publication criteria. Psychophysiology 37, 127–152. 10.1111/1469-8986.3720127 [DOI] [PubMed] [Google Scholar]
- Pieters TA, Conner CR, Tandon N, 2013. Recursive grid partitioning on a cortical surface model: an optimized technique for the localization of implanted subdural electrodes. J. Neurosurg 118, 1086–1097. 10.3171/2013.2.JNS121450 [DOI] [PubMed] [Google Scholar]
- Pizzo F, Roehri N, Medina Villalon S, Trébuchon A, Chen S, Lagarde S, Carron R, Gavaret M, Giusiano B, McGonigal A, Bartolomei F, Badier JM, Bénar CG, 2019. Deep brain activities can be detected with magnetoencephalography. Nat. Commun 10, 971. 10.1038/s41467-019-08665-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podvalny E, Yeagle E, Mégevand P, Sarid N, Harel M, Chechik G, Mehta AD, Malach R, 2017. Invariant Temporal Dynamics Underlie Perceptual Stability in Human Visual Cortex. Curr. Biol 27, 155–165. 10.1016/j.cub.2016.11.024 [DOI] [PubMed] [Google Scholar]
- Pothof F, Bonini L, Lanzilotto M, Livi A, Fogassi L, Orban GA, Paul O, Ruther P, 2016. Chronic neural probe for simultaneous recording of single-unit, multi-unit, and local field potential activity from multiple brain sites. J. Neural Eng 13, 046006. 10.1088/1741-2560/13/4/046006 [DOI] [PubMed] [Google Scholar]
- Prasser F, Kohlmayer F, Lautenschläger R, Kuhn KA, 2014. ARX--A Comprehensive Tool for Anonymizing Biomedical Data. AMIA Annu. Symp. Proc. AMIA Symp 2014, 984–993. [PMC free article] [PubMed] [Google Scholar]
- Prime D, Woolfe M, O’Keefe S, Rowlands D, Dionisio S, 2020. Quantifying volume conducted potential using stimulation artefact in cortico-cortical evoked potentials. J. Neurosci. Methods 337, 108639. 10.1016/j.jneumeth.2020.108639 [DOI] [PubMed] [Google Scholar]
- Puce A, Allison T, McCarthy G, 1999. Electrophysiological studies of human face perception. III: Effects of top-down processing on face-specific potentials. Cereb. Cortex N. Y. N 1991 9, 445–458. 10.1093/cercor/9.5.445 [DOI] [PubMed] [Google Scholar]
- Ramantani G, Maillard L, Koessler L, 2016. Correlation of invasive EEG and scalp EEG. Seizure 41, 196–200. 10.1016/j.seizure.2016.05.018 [DOI] [PubMed] [Google Scholar]
- Ramsey NF, Salari E, Aarnoutse EJ, Vansteensel MJ, Bleichner MG, Freudenburg ZV, 2018. Decoding spoken phonemes from sensorimotor cortex with high-density ECoG grids. NeuroImage 180, 301–311. 10.1016/j.neuroimage.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Leonard MK, Kleen JK, Lucas BA, Mirro EA, Chang EF, 2017. Chronic ambulatory electrocorticography from human speech cortex. NeuroImage 153, 273–282. 10.1016/j.neuroimage.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Crone NE, Niebur E, Franaszczuk PJ, Hsiao SS, 2008. Neural correlates of high-gamma oscillations (60–200 Hz) in macaque local field potentials and their potential implications in electrocorticography. J. Neurosci. Off. J. Soc. Neurosci 28, 11526–11536. 10.1523/JNEUROSCI.2848-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JB, Kaplan DM, Klein C, 2019. Decoding the Brain: Neural Representation and the Limits of Multivariate Pattern Analysis in Cognitive Neuroscience. Br. J. Philos. Sci 70, 581–607. 10.1093/bjps/axx023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière D, Geffroy D, Denghien I, Souedet N, Cointepas Y, 2009. BrainVISA: an extensible software environment for sharing multimodal neuroimaging data and processing tools. NeuroImage 47, S163. 10.1016/S1053-8119(09)71720-3 [DOI] [Google Scholar]
- Rizzi M, Sartori I, Vecchio MD, Zauli FM, Berta L, Lizio D, De Benedictis A, Sarubbo S, Mariani V, Al-Orabi K, Avanzini P, 2021. Tracing in Vivo the Dorsal Loop of the Optic Radiation: Convergent Perspectives From Tractography and Electrophysiology Compared to a Neuroanatomical Ground Truth. (preprint). In Review 10.21203/rs.3.rs-589114/v1 [DOI] [PubMed] [Google Scholar]
- Rizzolatti G, Fabbri-Destro M, 2010. Mirror neurons: from discovery to autism. Exp. Brain Res 200, 223–237. 10.1007/s00221-009-2002-3 [DOI] [PubMed] [Google Scholar]
- Roach BJ, Mathalon DH, 2008. Event-related EEG time-frequency analysis: an overview of measures and an analysis of early gamma band phase locking in schizophrenia. Schizophr. Bull 34, 907–926. 10.1093/schbul/sbn093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins K, Truong D, Appelhoff S, Delorme A, Makeig S, 2021. Capturing the nature of events and event context using hierarchical event descriptors (HED). NeuroImage 245, 118766. 10.1016/j.neuroimage.2021.118766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW, Hartov A, Kennedy FE, Miga MI, Paulsen KD, 1998. Intraoperative brain shift and deformation: a quantitative analysis of cortical displacement in 28 cases. Neurosurgery 43, 749–758; discussion 758–760. 10.1097/00006123-199810000-00010 [DOI] [PubMed] [Google Scholar]
- Rocher L, Hendrickx JM, de Montjoye Y-A, 2019. Estimating the success of re-identifications in incomplete datasets using generative models. Nat. Commun 10, 3069. 10.1038/s41467-019-10933-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehri N, Medina Villalon S, Jegou A, Colombet B, Giusiano B, Ponz A, Bartolomei F, Bénar C-G, 2021. Transfer, Collection and Organisation of Electrophysiological and Imaging Data for Multicentre Studies. Neuroinformatics 19, 639–647. 10.1007/s12021-020-09503-6 [DOI] [PubMed] [Google Scholar]
- Rosenberg JR, Halliday DM, Breeze P, Conway BA, 1998. Identification of patterns of neuronal connectivity--partial spectra, partial coherence, and neuronal interactions. J. Neurosci. Methods 83, 57–72. 10.1016/s0165-0270(98)00061-2 [DOI] [PubMed] [Google Scholar]
- Rosenow F, Bast T, Czech T, Feucht M, Hans VH, Helmstaedter C, Huppertz H-J, Noachtar S, Oltmanns F, Polster T, Seeck M, Trinka E, Wagner K, Strzelczyk A, 2016. Revised version of quality guidelines for presurgical epilepsy evaluation and surgical epilepsy therapy issued by the Austrian, German, and Swiss working group on presurgical epilepsy diagnosis and operative epilepsy treatment. Epilepsia 57, 1215–1220. 10.1111/epi.13449 [DOI] [PubMed] [Google Scholar]
- Rosenow F, Lüders H, 2001. Presurgical evaluation of epilepsy. Brain J. Neurol 124, 1683–1700. 10.1093/brain/124.9.1683 [DOI] [PubMed] [Google Scholar]
- Roussel P, Godais GL, Bocquelet F, Palma M, Hongjie J, Zhang S, Giraud A-L, Mégevand P, Miller K, Gehrig J, Kell C, Kahane P, Chabardés S, Yvert B, 2020. Observation and assessment of acoustic contamination of electrophysiological brain signals during speech production and sound perception. J. Neural Eng 17, 056028. 10.1088/1741-2552/abb25e [DOI] [PubMed] [Google Scholar]
- Rousselet GA, 2012. Does Filtering Preclude Us from Studying ERP Time-Courses? Front. Psychol 3, 131. 10.3389/fpsyg.2012.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselet GA, Foxe JJ, Bolam JP, 2016. A few simple steps to improve the description of group results in neuroscience. Eur. J. Neurosci 44, 2647–2651. 10.1111/ejn.13400 [DOI] [PubMed] [Google Scholar]
- Russo S, Pigorini A, Mikulan E, Sarasso S, Rubino A, Zauli FM, Parmigiani S, d’Orio P, Cattani A, Francione S, Tassi L, Bassetti CLA, Lo Russo G, Nobili L, Sartori I, Massimini M, 2021. Focal lesions induce large-scale percolation of sleep-like intracerebral activity in awake humans. NeuroImage 234, 117964. 10.1016/j.neuroimage.2021.117964 [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Ross IB, Mamelak AN, Schuman EM, 2010. Human memory strength is predicted by theta-frequency phase-locking of single neurons. Nature 464, 903–907. 10.1038/nature08860 [DOI] [PubMed] [Google Scholar]
- Sadaghiani S, Brookes MJ, Baillet S, 2022. Connectomics of human electrophysiology. NeuroImage 247, 118788. 10.1016/j.neuroimage.2021.118788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Reimer J, Penn R, Ojakangas CL, Hatsopoulos NG, 2010. Fast and slow oscillations in human primary motor cortex predict oncoming behaviorally relevant cues. Neuron 65, 461–471. 10.1016/j.neuron.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo P, Heikkinen HLT, 2018. Slow Science: Research and Teaching for Sustainable Praxis. Confero Essays Educ. Philos. Polit 6, 87–111. 10.3384/confero.2001-4562.181130 [DOI] [Google Scholar]
- Samaha J, Cohen MX, 2022. Power spectrum slope confounds estimation of instantaneous oscillatory frequency. NeuroImage 250, 118929. 10.1016/j.neuroimage.2022.118929 [DOI] [PubMed] [Google Scholar]
- Sammaritano M, de Lotbinière A, Andermann F, Olivier A, Gloor P, Quesney LF, 1987. False lateralization by surface EEG of seizure onset in patients with temporal lobe epilepsy and gross focal cerebral lesions: False Lateralization by Surface EEG. Ann. Neurol 21, 361–369. 10.1002/ana.410210408 [DOI] [PubMed] [Google Scholar]
- Sanai N, Mirzadeh Z, Berger MS, 2008. Functional Outcome after Language Mapping for Glioma Resection. N. Engl. J. Med 358, 18–27. 10.1056/NEJMoa067819 [DOI] [PubMed] [Google Scholar]
- Santiuste M, Nowak R, Russi A, Tarancon T, Oliver B, Ayats E, Scheler G, Graetz G, 2008. Simultaneous magnetoencephalography and intracranial EEG registration: technical and clinical aspects. J. Clin. Neurophysiol. Off. Publ. Am. Electroencephalogr. Soc 25, 331–9. 10.1097/WNP.0b013e31818e7913 [DOI] [PubMed] [Google Scholar]
- Sauseng P, Klimesch W, Gruber WR, Hanslmayr S, Freunberger R, Doppelmayr M, 2007. Are event-related potential components generated by phase resetting of brain oscillations? A critical discussion. Neuroscience 146, 1435–1444. 10.1016/j.neuroscience.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Schalk G, Leuthardt EC, Brunner P, Ojemann JG, Gerhardt LA, Wolpaw JR, 2008. Real-time detection of event-related brain activity. NeuroImage 43, 245–249. 10.1016/j.neuroimage.2008.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaworonkow N, Voytek B, 2021. Enhancing oscillations in intracranial electrophysiological recordings with data-driven spatial filters. PLoS Comput. Biol 17, e1009298. 10.1371/journal.pcbi.1009298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer HJ, Sander T, Trahms L, 2006. The influence of amplifier, interface and biological noise on signal quality in high-resolution EEG recordings. Physiol. Meas 27, 109–117. 10.1088/0967-3334/27/2/002 [DOI] [PubMed] [Google Scholar]
- Scheffer-Teixeira R, Tort AB, 2016. On cross-frequency phase-phase coupling between theta and gamma oscillations in the hippocampus. eLife 5, e20515. 10.7554/eLife.20515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevon CA, Ng SK, Cappell J, Goodman RR, McKhann G, Waziri A, Branner A, Sosunov A, Schroeder CE, Emerson RG, 2008. Microphysiology of epileptiform activity in human neocortex. J. Clin. Neurophysiol. Off. Publ. Am. Electroencephalogr. Soc 25, 321–330. 10.1097/WNP.0b013e31818e8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers A, Vansteensel MJ, Freudenburg ZV, Leijten FSS, Ramsey NF, 2021. Detailed somatotopy of tongue movement in the human sensorimotor cortex: A case study. Brain Stimulat. 14, 287–289. 10.1016/j.brs.2021.01.010 [DOI] [PubMed] [Google Scholar]
- Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, Hoshi M, Ilg R, Schmid VJ, Zimmer C, Hemmer B, Mühlau M, 2012. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. NeuroImage 59, 3774–3783. 10.1016/j.neuroimage.2011.11.032 [DOI] [PubMed] [Google Scholar]
- Schmidt Paul, 2017. Bayesian inference for structured additive regression models for large-scale problems with applications to medical imaging. Ludwig-Maximilians-Universität München. 10.5282/EDOC.20373 [DOI] [Google Scholar]
- Schreiber, null, 2000. Measuring information transfer. Phys. Rev. Lett 85, 461–464. 10.1103/PhysRevLett.85.461 [DOI] [PubMed] [Google Scholar]
- Schroeder CE, Wilson DA, Radman T, Scharfman H, Lakatos P, 2010. Dynamics of Active Sensing and perceptual selection. Curr. Opin. Neurobiol 20, 172–176. 10.1016/j.conb.2010.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Kremers WK, Wiste HJ, Gunter JL, Vemuri P, Spychalla AJ, Kantarci K, Schultz AP, Sperling RA, Knopman DS, Petersen RC, Jack CR, 2021. Changing the face of neuroimaging research: Comparing a new MRI de-facing technique with popular alternatives. NeuroImage 231, 117845. 10.1016/j.neuroimage.2021.117845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiedrzik CM, Sudmann SS, Thesen T, Wang X, Groppe DM, Mégevand P, Doyle W, Mehta AD, Devinsky O, Melloni L, 2018. Medial prefrontal cortex supports perceptual memory. Curr. Biol CB 28, R1094–R1095. 10.1016/j.cub.2018.07.066 [DOI] [PubMed] [Google Scholar]
- Sebastiano F, Di Gennaro G, Esposito V, Picardi A, Morace R, Sparano A, Mascia A, Colonnese C, Cantore G, Quarato PP, 2006. A rapid and reliable procedure to localize subdural electrodes in presurgical evaluation of patients with drug-resistant focal epilepsy. Clin. Neurophysiol 117, 341–347. 10.1016/j.clinph.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Seeber M, Cantonas L-M, Hoevels M, Sesia T, Visser-Vandewalle V, Michel CM, 2019. Subcortical electrophysiological activity is detectable with high-density EEG source imaging. Nat. Commun 10, 753. 10.1038/s41467-019-08725-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehatpour P, Molholm S, Schwartz TH, Mahoney JR, Mehta AD, Javitt DC, Stanton PK, Foxe JJ, 2008. A human intracranial study of long-range oscillatory coherence across a frontal–occipital–hippocampal brain network during visual object processing. Proc. Natl. Acad. Sci 105, 4399–4404. 10.1073/pnas.0708418105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaganesan K, Whitehead E, DeAlwis PM, Schindler MK, Inati, Souheil, Saad, Z.S., Ohayon, J.E., Cortese, I.C.M., Smith, B., Steven Jacobson, null, Nath, A., Reich, D.S., Inati, Sara, Nair, G., 2019. Robust, atlas-free, automatic segmentation of brain MRI in health and disease. Heliyon 5, e01226. 10.1016/j.heliyon.2019.e01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth AK, 2010. A MATLAB toolbox for Granger causal connectivity analysis. J. Neurosci. Methods 186, 262–273. 10.1016/j.jneumeth.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Shah AS, Bressler SL, Knuth KH, Ding M, Mehta AD, Ulbert I, Schroeder CE, 2004. Neural dynamics and the fundamental mechanisms of event-related brain potentials. Cereb. Cortex N. Y. N 1991 14, 476–483. 10.1093/cercor/bhh009 [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM, 2002. BrainSuite: an automated cortical surface identification tool. Med. Image Anal 6, 129–142. 10.1016/s1361-8415(02)00054-3 [DOI] [PubMed] [Google Scholar]
- Sherman MA, Lee S, Law R, Haegens S, Thorn CA, Hämäläinen MS, Moore CI, Jones SR, 2016. Neural mechanisms of transient neocortical beta rhythms: Converging evidence from humans, computational modeling, monkeys, and mice. Proc. Natl. Acad. Sci. U. S. A 113, E4885–4894. 10.1073/pnas.1604135113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirhatti V, Borthakur A, Ray S, 2016. Effect of Reference Scheme on Power and Phase of the Local Field Potential. Neural Comput. 28, 882–913. 10.1162/NECO_a_00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum J, Hermes D, Foster BL, Dastjerdi M, Rangarajan V, Winawer J, Miller KJ, Parvizi J, 2013. A Brain Area for Visual Numerals. J. Neurosci 33, 6709–6715. 10.1523/JNEUROSCI.4558-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M, Donner TH, Engel AK, 2012. Spectral fingerprints of large-scale neuronal interactions. Nat. Rev. Neurosci 13, 121–134. 10.1038/nrn3137 [DOI] [PubMed] [Google Scholar]
- Sillay KA, Rutecki P, Cicora K, Worrell G, Drazkowski J, Shih JJ, Sharan AD, Morrell MJ, Williams J, Wingeier B, 2013. Long-Term Measurement of Impedance in Chronically Implanted Depth and Subdural Electrodes During Responsive Neurostimulation in Humans. Brain Stimulat. 6, 718–726. 10.1016/j.brs.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Silverstein BH, Asano E, Sugiura A, Sonoda M, Lee M-H, Jeong J-W, 2020. Dynamic tractography: Integrating cortico-cortical evoked potentials and diffusion imaging. NeuroImage 215, 116763. 10.1016/j.neuroimage.2020.116763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai A, Bowers CW, Crainiceanu CM, Boatman D, Gordon B, Lesser RP, Lenz FA, Crone NE, 2005. Electrocorticographic high gamma activity versus electrical cortical stimulation mapping of naming. Brain J. Neurol 128, 1556–1570. 10.1093/brain/awh491 [DOI] [PubMed] [Google Scholar]
- Smith ME, Halgren E, Sokolik M, Baudena P, Musolino A, Liegeois-Chauvel C, Chauvel P, 1990. The intracranial topography of the P3 event-related potential elicited during auditory oddball. Electroencephalogr. Clin. Neurophysiol 76, 235–248. 10.1016/0013-4694(90)90018-F [DOI] [PubMed] [Google Scholar]
- Solomon EA, Kragel JE, Gross R, Lega B, Sperling MR, Worrell G, Sheth SA, Zaghloul KA, Jobst BC, Stein JM, Das S, Gorniak R, Inman CS, Seger S, Rizzuto DS, Kahana MJ, 2018. Medial temporal lobe functional connectivity predicts stimulation-induced theta power. Nat. Commun 9, 4437. 10.1038/s41467-018-06876-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda M, Silverstein BH, Jeong J-W, Sugiura A, Nakai Y, Mitsuhashi T, Rothermel R, Luat AF, Sood S, Asano E, 2021. Six-dimensional dynamic tractography atlas of language connectivity in the developing brain. Brain awab 225. 10.1093/brain/awab225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Betzel RF, 2016. Modular Brain Networks. Annu. Rev. Psychol 67, 613–640. 10.1146/annurev-psych-122414-033634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R, 2005. The human connectome: A structural description of the human brain. PLoS Comput. Biol 1, e42. 10.1371/journal.pcbi.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam CJ, 2014. Modern network science of neurological disorders. Nat. Rev. Neurosci 15, 683–695. 10.1038/nrn3801 [DOI] [PubMed] [Google Scholar]
- Stam CJ, Nolte G, Daffertshofer A, 2007. Phase lag index: assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources. Hum. Brain Mapp 28, 1178–1193. 10.1002/hbm.20346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangl M, Topalovic U, Inman CS, Hiller S, Villaroman D, Aghajan ZM, Christov-Moore L, Hasulak NR, Rao VR, Halpern CH, Eliashiv D, Fried I, Suthana N, 2021. Boundary-anchored neural mechanisms of location-encoding for self and others. Nature 589, 420–425. 10.1038/s41586-020-03073-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengers I, Muecke S, ProQuest (Firme), 2018. Another science is possible: a manifesto for slow science. [Google Scholar]
- Stolk A, 2018. Integrated analysis of anatomical and electrophysiological human intracranial data. Nat. Protoc 13, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolk A, Griffin S, van der Meij R, Dewar C, Saez I, Lin JJ, Piantoni G, Schoffelen J-M, Knight RT, Oostenveld R, 2018. Integrated analysis of anatomical and electrophysiological human intracranial data. Nat. Protoc 13, 1699–1723. 10.1038/s41596-018-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahnen D, Kapanaiah SKT, Bygrave AM, Kätzel D, 2021. Lack of redundancy between electrophysiological measures of long-range neuronal communication. BMC Biol. 19, 24. 10.1186/s12915-021-00950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme C, Novotny E, Zubal IG, Duncan JS, 2001. Estimating tissue deformation between functional images induced by intracranial electrode implantation using anatomical MRI. NeuroImage 13, 561–576. 10.1006/nimg.2000.0692 [DOI] [PubMed] [Google Scholar]
- Sudhyadhom A, Haq IU, Foote KD, Okun MS, Bova FJ, 2009. A high resolution and high contrast MRI for differentiation of subcortical structures for DBS targeting: The Fast Gray Matter Acquisition T1 Inversion Recovery (FGATIR). NeuroImage 47, T44–T52. 10.1016/j.neuroimage.2009.04.018 [DOI] [PubMed] [Google Scholar]
- Surbeck W, Bouthillier A, Weil AG, Crevier L, Carmant L, Lortie A, Major P, Nguyen DK, 2011. The combination of subdural and depth electrodes for intracranial EEG investigation of suspected insular (perisylvian) epilepsy: Invasive EEG Investigations of Insular Epilepsies. Epilepsia 52, 458–466. 10.1111/j.1528-1167.2010.02910.x [DOI] [PubMed] [Google Scholar]
- Suthana N, Haneef Z, Stern J, Mukamel R, Behnke E, Knowlton B, Fried I, 2012. Memory Enhancement and Deep-Brain Stimulation of the Entorhinal Area. N. Engl. J. Med 366, 502–510. 10.1056/NEJMoa1107212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadel F, Baillet S, Mosher JC, Pantazis D, Leahy RM, 2011. Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci 2011, 879716. 10.1155/2011/879716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Bancaud J, 1966. Lesion, “Irritative” Zone and Epileptogenic Focus. Stereotact. Funct. Neurosurg 27, 91–94. 10.1159/000103937 [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P, 1988. Co-planar stereotaxic atlas of the human brain: 3-dimensional proportional system: an approach to cerebral imaging. Georg Thieme, Stuttgart; ; New York. [Google Scholar]
- Talakoub O, Marquez-Chin C, Popovic MR, Navarro J, Fonoff ET, Hamani C, Wong W, 2017. Reconstruction of reaching movement trajectories using electrocorticographic signals in humans. PloS One 12, e0182542. 10.1371/journal.pone.0182542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallon-Baudry, null, Bertrand, null, 1999. Oscillatory gamma activity in humans and its role in object representation. Trends Cogn. Sci 3, 151–162. 10.1016/s1364-6613(99)01299-1 [DOI] [PubMed] [Google Scholar]
- Tallon-Baudry C, Bertrand O, Delpuech C, Permier J, 1997. Oscillatory gamma-band (30–70 Hz) activity induced by a visual search task in humans. J. Neurosci. Off. J. Soc. Neurosci 17, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon N, Tong BA, Friedman ER, Johnson JA, Von Allmen G, Thomas MS, Hope OA, Kalamangalam GP, Slater JD, Thompson SA, 2019. Analysis of Morbidity and Outcomes Associated With Use of Subdural Grids vs Stereoelectroencephalography in Patients With Intractable Epilepsy. JAMA Neurol. 76, 672–681. 10.1001/jamaneurol.2019.0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao JX, Hawes-Ebersole S, Baldwin M, Shah S, Erickson RK, Ebersole JS, 2009. The accuracy and reliability of 3D CT/MRI co-registration in planning epilepsy surgery. Clin. Neurophysiol 120, 748–753. 10.1016/j.clinph.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Tass P, Rosenblum MG, Weule J, Kurths J, Pikovsky A, Volkmann J, Schnitzler A, Freund H-J, 1998. Detection of n : m Phase Locking from Noisy Data: Application to Magnetoencephalography. Phys. Rev. Lett 81, 3291–3294. 10.1103/PhysRevLett.81.3291 [DOI] [Google Scholar]
- Taylor KN, Joshi AA, Hirfanoglu T, Grinenko O, Liu P, Wang X, Gonzalez-Martinez JA, Leahy RM, Mosher JC, Nair DR, 2021. Validation of semi-automated anatomically labeled SEEG contacts in a brain atlas for mapping connectivity in focal epilepsy. Epilepsia Open 6, 493–503. 10.1002/epi4.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Wal M, Platonov A, Cardellicchio P, Pelliccia V, LoRusso G, Sartori I, Avanzini P, Orban GA, Tiesinga PHE, 2020. Human stereoEEG recordings reveal network dynamics of decision-making in a rule-switching task. Nat. Commun 11, 3075. 10.1038/s41467-020-16854-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielscher A, Antunes A, Saturnino GB, 2015. Field modeling for transcranial magnetic stimulation: A useful tool to understand the physiological effects of TMS? Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Int. Conf 2015, 222–225. 10.1109/EMBC.2015.7318340 [DOI] [PubMed] [Google Scholar]
- Tibon R, Geerligs L, Campbell K, 2022. Bridging the big (data) gap: levels of control in small- and large-scale cognitive neuroscience research. Trends Neurosci. S0166223622000625. 10.1016/j.tins.2022.03.011 [DOI] [PubMed] [Google Scholar]
- Titiz AS, Hill MRH, Mankin EA, M Aghajan Z, Eliashiv D, Tchemodanov N, Maoz U, Stern J, Tran ME, Schuette P, Behnke E, Suthana NA, Fried I, 2017. Theta-burst microstimulation in the human entorhinal area improves memory specificity. eLife 6, e29515. 10.7554/eLife.29515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalovic U, Aghajan ZM, Villaroman D, Hiller S, Christov-Moore L, Wishard TJ, Stangl M, Hasulak NR, Inman CS, Fields TA, Rao VR, Eliashiv D, Fried I, Suthana N, 2020. Wireless Programmable Recording and Stimulation of Deep Brain Activity in Freely Moving Humans. Neuron 108, 322–334.e9. 10.1016/j.neuron.2020.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Valderrama A, Oostenveld R, Vansteensel MJ, Huiskamp GM, Ramsey NF, 2010. Gain of the human dura in vivo and its effects on invasive brain signal feature detection. J. Neurosci. Methods 187, 270–279. 10.1016/j.jneumeth.2010.01.019 [DOI] [PubMed] [Google Scholar]
- Tort ABL, Komorowski R, Eichenbaum H, Kopell N, 2010. Measuring phase-amplitude coupling between neuronal oscillations of different frequencies. J. Neurophysiol 104, 1195–1210. 10.1152/jn.00106.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth E, Fabó D, Entz L, Ulbert I, Erőss L, 2016. Intracranial neuronal ensemble recordings and analysis in epilepsy. J. Neurosci. Methods 260, 261–269. 10.1016/j.jneumeth.2015.09.028 [DOI] [PubMed] [Google Scholar]
- Trebaul L, Deman P, Tuyisenge V, Jedynak M, Hugues E, Rudrauf D, Bhattacharjee M, Tadel F, Chanteloup-Foret B, Saubat C, Reyes Mejia GC, Adam C, Nica A, Pail M, Dubeau F, Rheims S, Trébuchon A, Wang H, Liu S, Blauwblomme T, Garcés M, De Palma L, Valentin A, Metsähonkala E-L, Petrescu AM, Landré E, Szurhaj W, Hirsch E, Valton L, Rocamora R, Schulze-Bonhage A, Mindruta I, Francione S, Maillard L, Taussig D, Kahane P, David O, 2018. Probabilistic functional tractography of the human cortex revisited. NeuroImage 181, 414–429. 10.1016/j.neuroimage.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebaul L, Rudrauf D, Job A-S, Mălîia MD, Popa I, Barborica A, Minotti L, Mîndruţă I, Kahane P, David O, 2016. Stimulation artifact correction method for estimation of early cortico-cortical evoked potentials. J. Neurosci. Methods 264, 94–102. 10.1016/j.jneumeth.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trébuchon A, Chauvel P, 2016. Electrical Stimulation for Seizure Induction and Functional Mapping in Stereoelectroencephalography: J. Clin. Neurophysiol 33, 511–521. 10.1097/WNP.0000000000000313 [DOI] [PubMed] [Google Scholar]
- Trevisi G, Eickhoff SB, Chowdhury F, Jha A, Rodionov R, Nowell M, Miserocchi A, McEvoy AW, Nachev P, Diehl B, 2018. Probabilistic electrical stimulation mapping of human medial frontal cortex. Cortex 109, 336–346. 10.1016/j.cortex.2018.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta MS, Cocjin J, Whitehead E, Damera S, Wittig JH, Saad ZS, Inati SK, Zaghloul KA, 2018. Surface based electrode localization and standardized regions of interest for intracranial EEG. Hum. Brain Mapp 39, 709–721. 10.1002/hbm.23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuyisenge V, Trebaul L, Bhattacharjee M, Chanteloup-Forêt B, Saubat-Guigui C, Mîndruţă I, Rheims S, Maillard L, Kahane P, Taussig D, David O, 2018. Automatic bad channel detection in intracranial electroencephalographic recordings using ensemble machine learning. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol 129, 548–554. 10.1016/j.clinph.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher D, Klimes P, Cimbalnik J, Roman R, Pail M, Brazdil M, Jurak P, n.d. Stereo-electroencephalography (SEEG) reference based on low-variance signals 4. [DOI] [PubMed] [Google Scholar]
- Vakharia VN, Sparks R, Rodionov R, Vos SB, Dorfer C, Miller J, Nilsson D, Tisdall M, Wolfsberger S, McEvoy AW, Miserocchi A, Winston GP, O’Keeffe AG, Ourselin S, Duncan JS, 2018. Computer-assisted planning for the insertion of stereoelectroencephalography electrodes for the investigation of drug-resistant focal epilepsy: an external validation study. J. Neurosurg 1–10. 10.3171/2017.10.JNS171826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentín A, Alarcón G, Honavar M, García Seoane JJ, Selway RP, Polkey CE, Binnie CD, 2005. Single pulse electrical stimulation for identification of structural abnormalities and prediction of seizure outcome after epilepsy surgery: a prospective study. Lancet Neurol. 4, 718–726. 10.1016/S1474-4422(05)70200-3 [DOI] [PubMed] [Google Scholar]
- van ’t Klooster MA, Zijlmans M, Leijten FSS, Ferrier CH, van Putten MJAM, Huiskamp GJM, 2011. Time-frequency analysis of single pulse electrical stimulation to assist delineation of epileptogenic cortex. Brain J. Neurol 134, 2855–2866. 10.1093/brain/awr211 [DOI] [PubMed] [Google Scholar]
- Vanasse TJ, Fox PM, Barron DS, Robertson M, Eickhoff SB, Lancaster JL, Fox PT, 2018. BrainMap VBM: An environment for structural meta-analysis. Hum. Brain Mapp 39, 3308–3325. 10.1002/hbm.24078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRullen R, 2016. Perceptual Cycles. Trends Cogn. Sci 20, 723–735. 10.1016/j.tics.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Vanrullen R, 2011. Four common conceptual fallacies in mapping the time course of recognition. Front. Psychol 2, 365. 10.3389/fpsyg.2011.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela F, Lachaux JP, Rodriguez E, Martinerie J, 2001. The brainweb: phase synchronization and large-scale integration. Nat. Rev. Neurosci 2, 229–239. 10.1038/35067550 [DOI] [PubMed] [Google Scholar]
- Vaz AP, Yaffe RB, Wittig JH, Inati SK, Zaghloul KA, 2017. Dual origins of measured phase-amplitude coupling reveal distinct neural mechanisms underlying episodic memory in the human cortex. NeuroImage 148, 148–159. 10.1016/j.neuroimage.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidailhet M, Pollak P, 2005. Deep brain stimulation for dystonia: Make the lame walk. Ann. Neurol 57, 613–614. 10.1002/ana.20491 [DOI] [PubMed] [Google Scholar]
- Vidailhet M, Vercueil L, Houeto J-L, Krystkowiak P, Benabid A-L, Cornu P, Lagrange C, Tézenas du Montcel S, Dormont D, Grand S, Blond S, Detante O, Pillon B, Ardouin C, Agid Y, Destée A, Pollak P, 2005. Bilateral Deep-Brain Stimulation of the Globus Pallidus in Primary Generalized Dystonia. N. Engl. J. Med 352, 459–467. 10.1056/NEJMoa042187 [DOI] [PubMed] [Google Scholar]
- Vidal JR, Freyermuth S, Jerbi K, Hamamé CM, Ossandon T, Bertrand O, Minotti L, Kahane P, Berthoz A, Lachaux J-P, 2012. Long-distance amplitude correlations in the high γ band reveal segregation and integration within the reading network. J. Neurosci. Off. J. Soc. Neurosci 32, 6421–6434. 10.1523/JNEUROSCI.4363-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal JR, Ossandón T, Jerbi K, Dalal SS, Minotti L, Ryvlin P, Kahane P, Lachaux J-P, 2010. Category-Specific Visual Responses: An Intracranial Study Comparing Gamma, Beta, Alpha, and ERP Response Selectivity. Front. Hum. Neurosci 4, 195. 10.3389/fnhum.2010.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieluf S, Amengual‐Gual M, Zhang B, El Atrache R, Ufongene C, Jackson MC, Branch S, Reinsberger C, Loddenkemper T, 2021. Twenty‐four‐hour patterns in electrodermal activity recordings of patients with and without epileptic seizures. Epilepsia 62, 960–972. 10.1111/epi.16843 [DOI] [PubMed] [Google Scholar]
- Vinck M, Oostenveld R, van Wingerden M, Battaglia F, Pennartz CMA, 2011. An improved index of phase-synchronization for electrophysiological data in the presence of volume-conduction, noise and sample-size bias. NeuroImage 55, 1548–1565. 10.1016/j.neuroimage.2011.01.055 [DOI] [PubMed] [Google Scholar]
- Vinck M, van Wingerden M, Womelsdorf T, Fries P, Pennartz CMA, 2010. The pairwise phase consistency: a bias-free measure of rhythmic neuronal synchronization. NeuroImage 51, 112–122. 10.1016/j.neuroimage.2010.01.073 [DOI] [PubMed] [Google Scholar]
- Vinding MC, Oostenveld R, 2022. Sharing individualised template MRI data for MEG source reconstruction: A solution for open data while keeping subject confidentiality. NeuroImage 254, 119165. 10.1016/j.neuroimage.2022.119165 [DOI] [PubMed] [Google Scholar]
- von Ellenrieder N, Beltrachini L, Muravchik CH, 2012. Electrode and brain modeling in stereo-EEG. Clin. Neurophysiol 123, 1745–1754. 10.1016/j.clinph.2012.01.019 [DOI] [PubMed] [Google Scholar]
- Voytek B, Secundo L, Bidet-Caulet A, Scabini D, Stiver SI, Gean AD, Manley GT, Knight RT, 2010. Hemicraniectomy: a new model for human electrophysiology with high spatio-temporal resolution. J. Cogn. Neurosci 22, 2491–2502. 10.1162/jocn.2009.21384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliemoz S, Spinelli L, Pellise D, Ives JR, 2010. A New Ground and Reference Technique for Invasive EEG Recordings. Am. J. Electroneurodiagnostic Technol 50, 50–58. 10.1080/1086508X.2010.11079753 [DOI] [PubMed] [Google Scholar]
- Wacker M, Witte H, 2013. Time-frequency techniques in biomedical signal analysis. a tutorial review of similarities and differences. Methods Inf. Med 52, 279–296. 10.3414/ME12-01-0083 [DOI] [PubMed] [Google Scholar]
- Wang HE, Bénar CG, Quilichini PP, Friston KJ, Jirsa VK, Bernard C, 2014. A systematic framework for functional connectivity measures. Front. Neurosci 8, 405. 10.3389/fnins.2014.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Korzeniewska A, Usami K, Valenzuela A, Crone NE, 2021. The Dynamics of Language Network Interactions in Lexical Selection: An Intracranial EEG Study. Cereb. Cortex 31, 2058–2070. 10.1093/cercor/bhaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yan J, Wen J, Yu T, Li X, 2016. An Intracranial Electroencephalography (iEEG) Brain Function Mapping Tool with an Application to Epilepsy Surgery Evaluation. Front. Neuroinformatics 10. 10.3389/fninf.2016.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber TL, Milic NM, Winham SJ, Garovic VD, 2015. Beyond bar and line graphs: time for a new data presentation paradigm. PLoS Biol. 13, e1002128. 10.1371/journal.pbio.1002128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer J, von Oertzen J, Schaller C, Urbach H, König R, Widman G, Van Roost D, Elger CE, 2002. Digital photography and 3D MRI-based multimodal imaging for individualized planning of resective neocortical epilepsy surgery. Epilepsia 43, 1543–1550. 10.1046/j.1528-1157.2002.30002.x [DOI] [PubMed] [Google Scholar]
- Wen H, Liu Z, 2016. Separating Fractal and Oscillatory Components in the Power Spectrum of Neurophysiological Signal. Brain Topogr. 29, 13–26. 10.1007/s10548-015-0448-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Rangarajan G, Ding M, 2013. Multivariate Granger causality: an estimation framework based on factorization of the spectral density matrix. Philos. Transact. A Math. Phys. Eng. Sci 371, 20110610. 10.1098/rsta.2011.0610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, 2005. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J. Evol. Biol 18, 1368–1373. 10.1111/j.1420-9101.2005.00917.x [DOI] [PubMed] [Google Scholar]
- Whitmer D, Worrell G, Stead M, Lee IK, Makeig S, n.d. Utility of independent component analysis for interpretation of intracranial EEG. Front. Hum. Neurosci 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibral M, Priesemann V, Kay JW, Lizier JT, Phillips WA, 2017. Partial information decomposition as a unified approach to the specification of neural goal functions. Brain Cogn. 112, 25–38. 10.1016/j.bandc.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Wickham H, 2016. ggplot2: Elegant Graphics for Data Analysis, 2nd ed. 2016. ed, Use R! Springer International Publishing : Imprint: Springer, Cham. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Widmann A, Schröger E, 2012. Filter effects and filter artifacts in the analysis of electrophysiological data. Front. Psychol 3, 233. 10.3389/fpsyg.2012.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann A, Schröger E, Maess B, 2015. Digital filter design for electrophysiological data--a practical approach. J. Neurosci. Methods 250, 34–46. 10.1016/j.jneumeth.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Wilcox RR, Rousselet GA, 2018. A Guide to Robust Statistical Methods in Neuroscience. Curr. Protoc. Neurosci. 82. 10.1002/cpns.41 [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg Ij, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B, 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Webster MA, Brooks JC, Tracey I, Smith SM, Nichols TE, 2016. Non-parametric combination and related permutation tests for neuroimaging. Hum. Brain Mapp 37, 1486–1511. 10.1002/hbm.23115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler PA, Vollmar C, Krishnan KG, Pfluger T, Brückmann H, Noachtar S, 2000. Usefulness of 3-D reconstructed images of the human cerebral cortex for localization of subdural electrodes in epilepsy surgery. Epilepsy Res. 41, 169–178. 10.1016/S0920-1211(00)00137-6 [DOI] [PubMed] [Google Scholar]
- Wolpert N, Tallon-Baudry C, 2021. Coupling between the phase of a neural oscillation or bodily rhythm with behavior: Evaluation of different statistical procedures. NeuroImage 236, 118050. 10.1016/j.neuroimage.2021.118050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Morris N, Lu Q, Elston RC, 2009. Choosing an optimal method to combine P-values. Stat. Med 28, 1537–1553. 10.1002/sim.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ngo GH, Greve D, Li J, He T, Fischl B, Eickhoff SB, Yeo BTT, 2018. Accurate nonlinear mapping between MNI volumetric and FreeSurfer surface coordinate systems. Hum. Brain Mapp 39, 3793–3808. 10.1002/hbm.24213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AI, Wang X, Doyle WK, Halgren E, Carlson C, Belcher TL, Cash SS, Devinsky O, Thesen T, 2012. Localization of dense intracranial electrode arrays using magnetic resonance imaging. NeuroImage 63, 157–165. 10.1016/j.neuroimage.2012.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Guindani M, Grieco SF, Chen L, Holmes TC, Xu X, 2021. Beyond t test and ANOVA: applications of mixed-effects models for more rigorous statistical analysis in neuroscience research. Neuron S0896–6273(21)00845-X. 10.1016/j.neuron.2021.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangaladze A, Sharan A, Evans J, Wyeth DH, Wyeth EG, Tracy JI, Chervoneva I, Sperling MR, 2008. The effectiveness of low-frequency stimulation for mapping cortical function. Epilepsia 49, 481–487. 10.1111/j.1528-1167.2007.01307.x [DOI] [PubMed] [Google Scholar]
- Zaveri HP, Duckrow RB, Spencer SS, 2006. On the use of bipolar montages for time-series analysis of intracranial electroencephalograms. Clin. Neurophysiol 117, 2102–2108. 10.1016/j.clinph.2006.05.032 [DOI] [PubMed] [Google Scholar]
- Zaveri HP, Duckrow RB, Spencer SS, 2000. The effect of a scalp reference signal on coherence measurements of intracranial electroencephalograms. Clin. Neurophysiol 7. [DOI] [PubMed] [Google Scholar]
- Zelano C, Jiang H, Zhou G, Arora N, Schuele S, Rosenow J, Gottfried JA, 2016. Nasal Respiration Entrains Human Limbic Oscillations and Modulates Cognitive Function. J. Neurosci 36, 12448–12467. 10.1523/JNEUROSCI.2586-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelmann R, Paulk AC, Basu I, Sarma A, Yousefi A, Crocker B, Eskandar E, Williams Z, Cosgrove GR, Weisholtz DS, Dougherty DD, Truccolo W, Widge AS, Cash SS, 2020. CLoSES: A platform for closed-loop intracranial stimulation in humans. NeuroImage 223, 117314. 10.1016/j.neuroimage.2020.117314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhigalov A, Arnulfo G, Nobili L, Palva S, Palva JM, 2017. Modular co-organization of functional connectivity and scale-free dynamics in the human brain. Netw. Neurosci 1, 143–165. 10.1162/NETN_a_00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhigalov A, Arnulfo G, Nobili L, Palva S, Palva JM, 2015. Relationship of Fast- and Slow-Timescale Neuronal Dynamics in Human MEG and SEEG. J. Neurosci 35, 5385–5396. 10.1523/JNEUROSCI.4880-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Schleicher A, Langemann C, Amunts K, Morosan P, Palomero-Gallagher N, Schormann T, Mohlberg H, Bürgel U, Steinmetz H, Schlaug G, Roland PE, 1997. Quantitative analysis of sulci in the human cerebral cortex: development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Hum. Brain Mapp. 5, 218–221. [DOI] [PubMed] [Google Scholar]
- Zion Golumbic EM, Ding N, Bickel S, Lakatos P, Schevon CA, McKhann GM, Goodman RR, Emerson R, Mehta AD, Simon JZ, Poeppel D, Schroeder CE, 2013. Mechanisms underlying selective neuronal tracking of attended speech at a “cocktail party.” Neuron 77, 980–991. 10.1016/j.neuron.2012.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]