Abstract
To translate artificial intelligence (AI) algorithms into clinical practice requires generalizability of models to real-world data. One of the main obstacles to generalizability is data shift, a data distribution mismatch between model training and real environments. Explainable AI techniques offer tools to detect and mitigate the data shift problem and develop reliable AI for clinical practice. Most medical AI is trained with datasets gathered from limited environments, such as restricted disease populations and center-dependent acquisition conditions. The data shift that commonly exists in the limited training set often causes a significant performance decrease in the deployment environment. To develop a medical application, it is important to detect potential data shift and its impact on clinical translation. During AI training stages, from premodel analysis to in-model and post hoc explanations, explainability can play a key role in detecting model susceptibility to data shift, which is otherwise hidden because the test data have the same biased distribution as the training data. Performance-based model assessments cannot effectively distinguish the model overfitting to training data bias without enriched test sets from external environments. In the absence of such external data, explainability techniques can aid in translating AI to clinical practice as a tool to detect and mitigate potential failures due to data shift.
©RSNA, 2023
Quiz questions for this article are available in the supplemental material.
Introduction
Over the past decade, deep learning has dominated the artificial intelligence (AI) research area, including in health care. There have been many AI algorithms, especially deep learning methods for medical image analysis, that achieved state-of-the-art results on public datasets. For example, there have been numerous Grand Challenges (https://grand-challenge.org/challenges/) in medical image analysis spanning various anatomy and imaging modalities. Algorithms in these challenge leaderboards have reported high accuracy on the held-out test set, and performance has sometimes surpassed that of human experts. However, few of these AI algorithms have been applied in clinical practice.
One of the main challenges is the lack of AI applications that work well across multiple institutions or heterogeneous populations. It is often seen that a deep learning model trained with data from one hospital fails at other hospitals (1,2). The underlying cause arises from differences between the limited data on which the AI was trained and the data encountered in real-world clinical practice. This problem is known as data shift.
For AI to learn a general model requires a training set that covers the full spectrum of the disease and its visualization at imaging, as influenced by the image acquisition machine and conditions. Training sets are often acquired at one hospital or a small number of hospitals or are limited because of the patient selection criteria, which introduce biases that do not reflect the broader spectrum.
An AI model performs analysis on the basis of imaging features. In a training set, each feature will be derived from a probability distribution based on the disease spectrum in the dataset. Machine learning algorithms in effect estimate this probability distribution for different disease states from the training data and classify it accordingly. There is an implicit assumption that the probability distributions reflect those that will be seen in clinical practice; however, if the spectrum of disease is different, then the performance of the AI will vary from that seen in training.
The training sets of the models are usually collected from a few data sources with specific clinical conditions, then the trained models are applied to a different more variable environment in clinical practice. Both the training and test sets have probability distributions that may differ from the unknown real distribution of the target disease spectrum because they are collected from the restricted environments. This change in distribution is known as data shift and can degrade AI performance when transitioning from training to real-world application.
For example, we have obtained a training set of three-dimensional lung high-resolution CT images from patients at a single institution with idiopathic pulmonary fibrosis (IPF), the most common type of interstitial lung disease (ILD). We trained a binary classification model to predict progressive versus stable (nonprogressive) IPF from a single image. The training set was gathered from the IPF disease spectrum. If applied to a more general ILD spectrum, this training set may introduce a spurious association with IPF-specific patterns. The model may fail if applied to the spectrum of non-IPF ILD, such as inflammatory myopathy–ILD.
There are other examples in the literature of models developing spurious associations from the training data that result in a significant performance decrease in the test environments. For example, a melanoma classification model for dermoscopic skin lesions was trained on images with visual aids such as skin markings or rulers, but then was tested on images without the visual aids and performance dropped (3). Overcoming data shift is an important challenge in translating AI to clinical practice.
However, susceptibility to data shift is difficult to detect during the model training. Explainability refers to an ability to analyze data and models to assess whether they are generally applicable to and consistent with domain knowledge, the expert knowledge of the disease process and clinical imaging modality. In health care, with its limited datasets, explainability can offer tools that can be helpful to detect potential failures due to data shift in translating AI to clinical application.
The goals of this article are (a) to clarify terms that are relevant to the data shift problem, (b) to describe data shift and its negative impact on translation of AI into clinical practice, and (c) to show the role of explainability in detecting and mitigating the data shift problem.
Terminology Related to Data Shift
In this section, we review several relevant terms that have different meanings and underlying assumptions between the machine learning literature and clinical literature.
Training Data, Test Data, and External Test Data
Training data is a set of observations used to train a model, and test data is a set of previously unseen observations used for model assessment. Any sample from test data should not be used for training or model selection. The training error refers to the average error over the training data, and the test error is the prediction error over the test set.
Many classic machine learning algorithms and techniques assume independent and identically distributed (IID) data (4). Suppose we would like to develop a model with a given set of observations, drawn from an unknown distribution. We typically assume that this dataset is IID, and the model will be applied to data from the same probability distribution. We split the dataset into the training set and held-out test set, fit the model on the training samples, and evaluate its performance on the test samples.
With the IID assumption, samples in the training data and test data are drawn independently and identically from the same distribution. Test error, also referred to as generalization error, can be used to estimate an average generalization error of a new test set from the same distribution. These underlying assumptions are commonly violated, such as when going from controlled training data to real-world clinical practice.
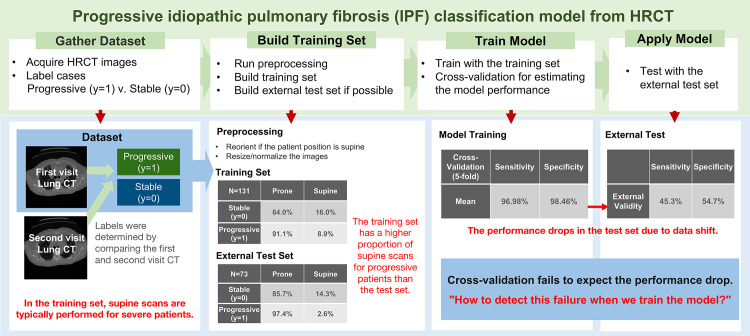
Let’s revisit the progressive IPF classification problem. Patients were imaged at total lung capacity (TLC) in the prone position using standard diffuse lung disease CT protocols. However, the particular hospital where the training data were collected frequently imaged the patient in the supine position when the patient had severe symptoms. Although the images were reoriented to standardize the orientation of the anatomy as viewed on the image, the training set can introduce a spurious association based on the patient positioning.
If the proportion of supine imaging within the progressive cases is not the same in practice, this data shift can affect the model performance. The distribution of the training data has bias on the patient positioning that may not be reflected in the distribution seen in clinical practice, and the model may fail when applied at institutions that image patients with IPF in the prone position only. This failure is difficult to detect when we train the model, as the test error is also derived from data from the same hospital and does not estimate the prediction errors of samples from an external source.
We will work through the definitions introduced in the article with this example to aid in understanding the concepts introduced. Figure 1 shows schematics of this example to crystalize each concept. To demonstrate model validity on external sources, we need new test sets from external independent sources with different environments. We will refer to this test set as the external test data to distinguish it from the test set generated from the original source.
Figure 1.
Diagram of the binary classification model for predicting progressive versus stable (nonprogressive) IPF from three-dimensional lung high-resolution CT (HRCT) images. Patients were imaged at total lung capacity (TLC) in the prone position using standard diffuse lung disease CT protocols, and a few patients were imaged in the supine position owing to their severe symptoms. A notable data shift in this example is the change in the proportion of supine images for patients with progressive IPF. The model shows performance drop due to data shift.
Overfitting and Generalization
Overfitting and generalization have been significant topics in the machine learning literature (4,5). Overfitting is a condition where the test error is significantly larger than the training error. This happens because the model memorizes some patterns in the training set that are not replicated in the test set. When the algorithm successfully avoids overfitting, and the test error is similar to the training error, we say that the model has generalized well. If our progressive IPF model shows similar performance on the test set, we consider that this model has generalized well to this test set.
Generalization is an important topic in practical applications, and there are many machine learning techniques (eg, cross-validation, dropout, early stopping, regularization) that aim to prevent overfitting (6). However, a well-generalized model trained from one data source is not guaranteed to work well on data from another external source. Although the model is assessed as generalizing well, this assessment is under the independent and identically distributed (IID) assumptions described earlier. Many techniques for preventing overfitting also assume that the training set and test set are generated from the same environment and do not ensure that the models work well in other environments. The term generalizability is often confused with external validity—the prediction capability on an external source—in the clinical literature (7).
Data Shift and Out-of-Distribution Generalization
External validity can be linked to the term out-of-distribution (OOD) generalizability (8). OOD generalization, also known as domain generalization, assumes that the model is trained with data drawn from a set of available data sources, but there exists a larger set of data sources including unseen data sources. External test data may include some of these unseen data sources.
The entire set of data sources can be thought of as all the environments where we want to apply the model, commonly larger than the environments in which the model is trained. The goal of OOD generalization is to maintain model performance across the entire set of data sources. Intuitively, OOD generalization attempts to learn features that are invariant throughout the different data sources in the entire set of data sources.
Data shift means any situation of mismatch between the distribution of the training dataset and the distribution of the data where the model will be applied (9). When the distribution of the training set of data sources is shifted from the distribution of the entire set of data sources, learning the bias only in the training data will cause a sharp drop in the performance of a machine learning algorithm when applied to the entire set of data sources (10). This poor OOD generalizability occurs especially when the label (eg, disease type, disease severity, or the location of the lesions) and the input image of the training data distribution have spurious associations. In the IPF classification example, the goal of OOD generalization could be to guide the model to learn the patterns of lung fibrosis instead of the CT table, which can inform the patient position.
Data shift is one of the major obstacles because it is not easily detected or addressed with the classic techniques for preventing overfitting, which assume independent and identically distributed (IID) data. In this article, we promote explainability as a tool to detect and mitigate the data shift problem.
Explainability
Explainability is one of the core elements of AI applications in clinical practice (11,12). However, the definition of explainability is unclear in the machine learning community because of the lack of mathematical rigor (11). In this article, we follow the later definition suggested by Roscher et al (11), originally from Montavon et al (13): “An explanation is the collection of features of the interpretable domain, that have contributed for a given example to produce a decision (eg, classification or regression).”
The term interpretation means realizing abstract concepts from the model in a human-readable format. The predicted class of the IPF model given by the probability is an example of interpretation. When interpretation helps in understanding the model’s prediction, it is called explanation. A heat map that highlights the important location for the model’s decision is an interpretation and also an explanation. If we can interpret the model’s decision using human knowledge, we can say that this model has explainability.
Explanations are widely used to justify the AI results, provide better control, improve models, and discover new patterns (14). This article focuses on explainability with the objective of improving a model, especially its OOD generalizability.
Data Shift and Its Impact on Clinical Translation of AI
The Challenge of Translation and Notable Failures
Two review articles about the notable failure of machine learning algorithms for coronavirus disease (COVID-19) reveal how challenging it is to develop reliable AI for health care. The systematic review by Wynants et al (1) demonstrates that none of the 169 studies of COVID-19–related algorithms published in 2020 were reliable enough to translate to clinical practice. To evaluate the reliability of models as clinical applications, they applied the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modeling Studies (CHARMS) checklist (15) and assessed the risk of bias using the Prediction Model Risk of Bias Assessment Tool (PROBAST) (16). They found that all models were at high risk of bias, assessed as unreliable to apply in clinical practice.
The review by Roberts et al (2) on 415 articles for COVID-19 machine learning models published in 2020 reached a similar conclusion. After quality screening using the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) (17), 62 articles remained to analyze their reliability by assessing the risks of bias with PROBAST. Again, none of the articles reached their threshold of robustness and reproducibility for clinical use.
Failures Due to Data Shift Are Not Easy to Detect
From both reviews, data shift was the predominant cause of the high risks of bias. Although many articles considered overfitting and showed their generalizability to the test set, none of the models were assessed as having potential data shift problems, for example, population difference, nonrepresentative selection of control patients, or exclusion of some patients. Without a rich number of external datasets from the various environments, data shift failure is difficult to detect with classic methods such as cross-validation. We propose use of explainable AI to detect potential model failures due to data shift.
Data Shift in Medical Imaging and Its Negative Impact on Clinical Use
Castro et al (18) categorizes the possible data shifts in medical imaging into five types: (a) population shift, (b) prevalence shift, (c) acquisition shift, (d) annotation shift, and (e) manifestation shift. Those concepts are commonly observed and have negative impacts on OOD generalization.
Population Shift.—Demographic or other differences related to the characteristics of model training and external populations can be categorized as a population shift. If characteristics such as age, ethnicity, or habits are correlated with the target class outputs, this shift will have a negative impact on OOD generalization. Martin et al (19) raised ethical concerns about implementing the polygenic risk scores for breast cancer and diabetes based on genome-wide association experiments where the data are Eurocentric. Such models may perform worse in non-European populations owing to population shift.
As another example, the spatial distribution of prostate cancer lesions within the prostate gland may be related to the characteristics of the population, such as age and ethnicity. Black and White population groups showed different disease patterns (20). Thus, the ethnicity ratios of the dataset can affect the prostate cancer model and make it susceptible to changes in the underlying distribution patterns if applied in a setting where the patient ethnicity ratios are different.
Prevalence Shift.—Prevalence shift relates to class balance between datasets. The prevalence of disease influences classifier performance measures. The systematic review by Roberts et al (2) reported prevalence shift across the datasets used for 62 studies about the diagnosis or prognosis of COVID-19. They compared the case-control balance of the training set and test set from each study.
The training set and test set within each study had similar class balances. However, datasets across the different studies showed inhomogeneous class balances and thus the potential for prevalence shift. Many studies showed high risks of bias from results of the Prediction Model Risk of Bias Assessment Tool (PROBAST) because of this bias.
Acquisition Shift.—Acquisition shift is the most common distribution bias that occurs in medical image analysis. An acquisition condition means any parameter for acquisition or reconstruction of the images, such as CT protocol, radiation dose level, reconstructed section thickness, and kernel. Acquisition shift refers to differences in the distributions of the data arising from different acquisition conditions. Datasets gathered from different imaging protocols will have acquisition shift.
When this acquisition bias has unexpected associations with the labels, the model can be fit to the acquisition bias instead of disease-related patterns. The progressive IPF example shows a degradation of model performance arising from acquisition shift, where the training data had a spurious association with patient positioning (Fig 1). The proportion of patients with progressive IPF imaged in the supine position shows differences between the training and test sets. This data shift worsened the OOD generalizability of the classification model.
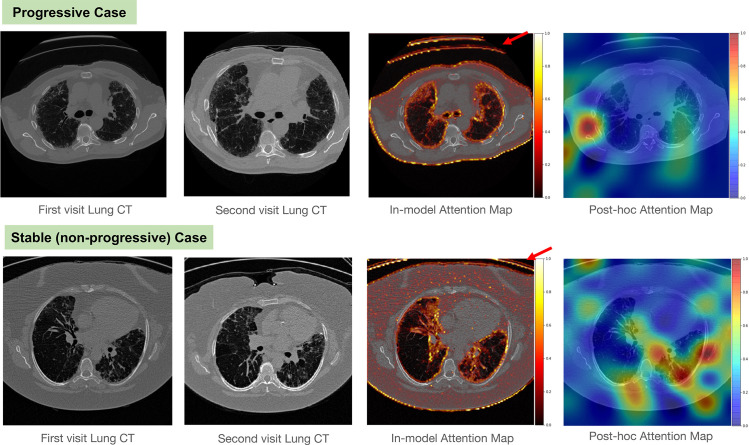
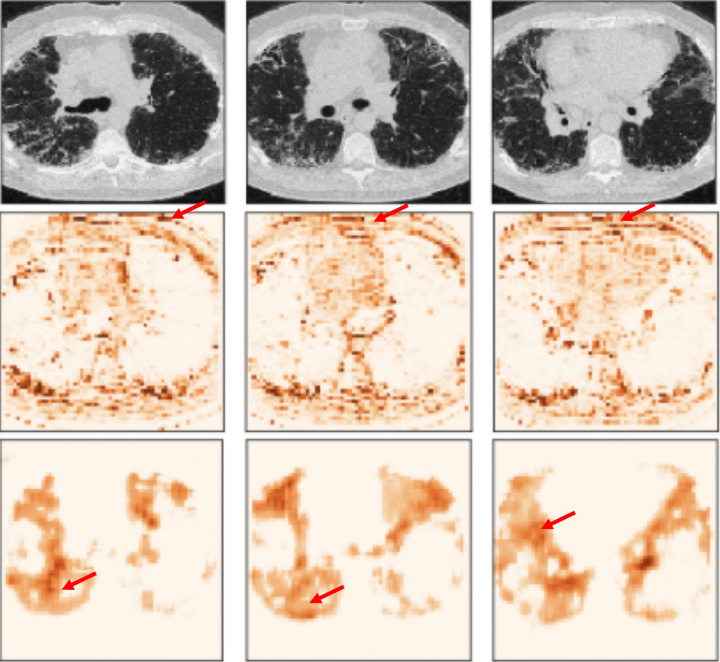
Cross-validation is a popular method to estimate the generalized performance of the model. It iteratively resamples the data to train and test from different portions and averages the performances from each iteration. However, cross-validation is weak at detecting data shift. In Figure 1, the cross-validation performance of the model was about 100% for both sensitivity and specificity, but the performance dropped to 45.3% for sensitivity and 54.7% for specificity in the independent test set. The model prediction was incorrect for all test images in the supine position. Figure 2 shows example cases from the single-institution IPF dataset.
Figure 2.
Example cases from the IPF dataset. Top row: case of progressive IPF; bottom row: stable (nonprogressive) case. From left to right, the columns show the first-visit image, second-visit image, in-model attention map overlay on the first-visit image, and post hoc attention map overlay on the first-visit image. Note that the in-model attention map highlights the CT table (arrow), and the post hoc attention map hints that the model focused on the wrong area. These two visualizations imply that the model may be overfitted to acquisition shift and may fail in clinical practice.
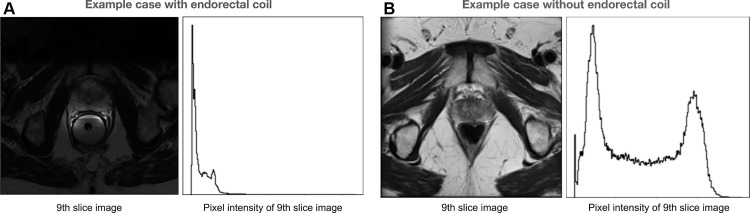
The prostate MRI dataset PROMISE12 from Litjens et al (21) shows acquisition shift. This dataset is constructed with T2-weighted images and corresponding prostate segments from four centers, each of which has different acquisition techniques. Notably, the images acquired with and without an endorectal coil are mixed in this dataset. We can observe that the existence of the endorectal coil affects the shape of the prostate and the intensity histogram of the T2-weighted images.
Figure 3 shows example cases of subjects with and without an endorectal coil. It shows differences in image intensity ranges and prostate shape distortion caused by the endorectal coil. If a prostate segmentation model is trained with images mostly acquired without an endorectal coil, the performance may decrease when the endorectal coil is present.
Figure 3.
Acquisition shift in example cases of subjects from the public PROMISE12 dataset (21). (A) Axial T2-weighted image and intensity histogram in an example case with an endorectal coil. This case shows shape distortion because of the endorectal coil. (B) Axial T2-weighted image and intensity histogram in an example case without an endorectal coil.
Annotation Shift.—Annotation shift includes any differences in annotations generated from different annotators or data sources. This includes differences arising from image segmentation methods: automatic, semiautomatic, and manual (22). One of the well-known examples of annotation shift is the interobserver variability from manual segmentation. Because of the annotator’s experience level or annotation instructions, observers can annotate differently for the same datum.
Walsh et al (23) reported significant interobserver variability among thoracic radiologists for a diagnosis of IPF or usual interstitial pneumonia (UIP) at CT. They documented that there is disagreement in the IPF categorization of cases (UIP, possibly UIP, and inconsistent with UIP) based on differences in interpretation of lung CT patterns (presence of honeycombing, traction bronchiectasis, and emphysema). Thus, different teams of annotators can cause variations in how training sets and test sets might be labeled, which in turn will impact AI performance.
Manifestation Shift.—Manifestation shift occurs when there are differences in anatomic manifestation of the disease label in the datasets. We see again an example of manifestation shift in the IPF problem. Diagnosis of IPF is based on the presence of the UIP pattern on CT images (24). The patterns of UIP are refined to patterns of definite UIP, probable UIP, indeterminate for UIP, or an alternative diagnosis based on the CT features.
For example, the diagnostic criteria for definite UIP include honeycombing with or without peripheral traction bronchiectasis or bronchiolectasis, and the diagnostic criteria for probable UIP include a reticular pattern with peripheral traction bronchiectasis or bronchiolectasis. Thus, a training set of patients with IPF may or may not have honeycombing and bronchiectasis patterns on their CT images. If the presence of these features changes between the training set and test set populations, then a manifestation shift has occurred and classifier performance may be affected.
Explainability and Mitigating Data Shift
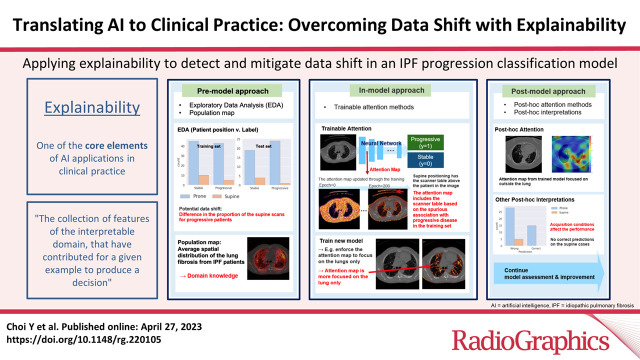
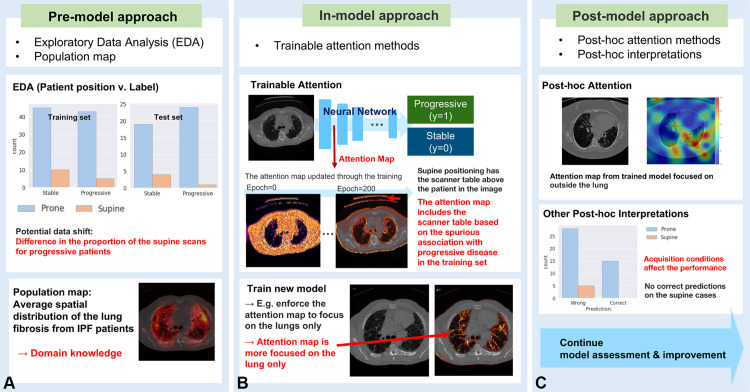
AI systems with proper explanations can detect data bias, which may cause model failures in external datasets. This section introduces some approaches to explainability based on the stage of model training (25): the (a) premodel approach, (b) in-model approach, and (c) postmodel approach. Figure 4 illustrates how to apply the three approaches in each stage of the model training, with examples from the progressive IPF classification model.
Figure 4.
Example of applying explainability to detect and mitigate data shift in the IPF progression classification model. (A) Premodel approach. (B) In-model approach. (C) Postmodel approach.
The premodel approach includes exploratory data analysis to address the contribution of data features to model decisions. The in-model approach is for the models equipped with interpretable methods, such as attention methods. Attention methods are a group of techniques that provide a visual representation of image regions that are most influential in the model prediction, known as the attention map. When the model contains the attention mechanism inside, we consider the model to use the in-model approach.
The postmodel approach includes methods implemented after model training to generate post hoc interpretations combined with domain knowledge; post hoc attention is an example in this category. Figure 2 shows examples of the in-model and postmodel attention maps of the IPF classification model. In this section, we describe use—with examples—of explainability approaches for detecting data shift at each training stage.
Premodel Approach
Exploratory Data Analysis
Exploratory data analysis (EDA) explores the statistics and visualizations of the characteristics of the dataset, such as the demographics, the prevalence of the disease, the acquisition conditions, and the association between the characteristics and the disease labels. EDA should be a primary approach to finding the potential data shift.
Population Maps
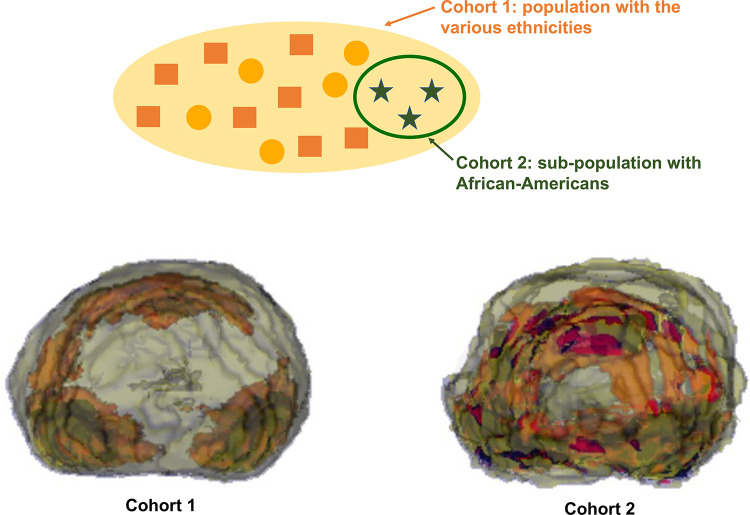
A map of averaged spatial locations of target lesion contours from a given dataset, called a population map (26), is a premodel analysis that can visualize population bias. A population map of prostate cancers shows use of a population map to detect potential population shift. From the prostate gland and prostate tumor contours on the T2-weighted MR images from a University of California–Los Angeles resection cohort, we generated the population map to visualize the averaged spatial locations of prostate cancers of the dataset. We built two datasets with different demographics to see whether the population map can reveal the population shift in this study.
Figure 5 shows that the population maps from the two datasets are significantly different. In this study, the first dataset includes various ethnicities: 65% White, 5% Black, and 30% other (Asian, Hispanic, others, and missing). The second dataset is a subset of the first dataset and includes African Americans only (Fig 5). When we have multiple datasets, we can generate a population map for each to check for any significant difference in the mean trend of the tumor locations. The inconsistent spatial distribution between the population maps means that there is a potential population shift between the datasets.
Figure 5.
Population maps generated from the prostate MRI dataset of the resection cohort from the University of California, Los Angeles. Top: diagram of the study cohorts. Bottom: population maps of each cohort. Each color indicates the area with a given range of probabilities: blue, 0%–1%; yellow, 1%–6%; red, 6%–8%; and black, 8%–13.6%.
In-Model Approach
Attention Methods in Deep Learning
Attention methods are one of the most popular approaches in explainable AI. Any kind of approach that provides an attention map for a deep learning model can be categorized as an attention method. Trainable attention methods are prevalent approaches in explainable AI. These approaches incorporate attention maps into networks to emphasize important regions in the image during training.
Attention maps function in convolutional neural networks (CNNs) as layers to focus network emphasis on regions of significance and serve to improve performance. The CNN training will include attention gating, in which the image is multiplied by an attention map to emphasize more relevant image regions for model prediction. The in-model attention map and the attention-gated image are used to improve model performance during training. Trainable attention maps have been demonstrated in image classification (27), image segmentation (28), and medical image analysis (29,30). Attention maps provide a natural fit for medical images, permitting the network to emphasize specific localized regions of interest learned from the data.
Trainable attention maps are illustrated in the IPF progression classification example in Figure 2. The attention map values closer to 1 (white to yellow) represent the more important areas for the model than the values closer to 0 (red to black). Note that the in-model attention maps highlight the edge of the lung, edge of the body, and CT table. This is a hint that the model overfit to the acquisition bias.
Furthermore, we can restrict the attention map to more focus on a given specific area to mitigate the data shift problem. We can guide the in-model attention map by encouraging the attention map to be similar to the image subarea known as generally important on the basis of domain knowledge (25,31,32). Figure 4B shows an example of using this attention-guiding approach. When we detected that the model has been overfitted to the data shift, we can consider bringing the domain knowledge to mitigate the problem.
As shown in Figure 4B, we implemented the population map of the lung fibrosis region distribution from the IPF patients as domain knowledge to help the progressive IPF model. We guided the IPF model by restricting the trainable attention to be focused on the higher-value area from the population map. The new attention map shows that the model now focused on the area within the lung.
As another example, Yu et al (31) suggested that the IPF diagnosis model from high-resolution CT images demonstrates use of trainable attention for detecting acquisition bias. This is similar to the progressive IPF classification example, but a different problem. This model distinguishes IPF from non-IPF among subjects from the University of California, Los Angeles.
Notably, the approach from Yu et al (31) suggests guiding the in-model attention maps with the location important for diagnosis of IPF. They employed the averaged spatial locations of lung fibrosis from IPF patients from the previous research (33) for identifying important locations. Two models were trained with different amounts of guidance.
Figure 6 shows the processed CT images and the attention maps of each image. The arrows indicate the significant areas for the models and shows that the attention region of the first model emphasized the area outside the lung. This suggests that the model may be overfitted to the acquisition biases in the training data. Meanwhile, the second model focused on plausible lung regions.
Figure 6.
Example cases and corresponding attention maps from two guided attention models for diagnosing IPF from high-resolution CT images. Each column is for each case. Top: CT images. Center: attention maps from the model with area under the curve (AUC) = 0.972. Arrow = higher-attention area of this model. This model focuses outside the lung, raising concern about overfitting to the acquisition shift. Bottom: attention maps from the model with AUC = 0.943. Arrow = focusing of this model on higher attention inside the lung. This model focuses on appropriate areas for distinguishing IPF.
The authors built the test set with cases not used for training but coming from the same data sources as used for training. The test set performance of the first model was higher than that of the second model, demonstrating that test set performance may not reveal a lack of OOD generalizability of the model. Instead, the visual interpretations with domain knowledge showed the risk of failure due to data shift. We need a test set from a new data source to measure the OOD performance. To overcome this data requirement, OOD generalization methods are being actively studied (8,34).
Postmodel Approach
Post Hoc Attention Methods
Post hoc attention methods are techniques for generating an attention map from an already trained network. The gradient-weighted class activation mapping method (grad-CAM) from Selvaraju et al (35) is one of the well-known post hoc attention approaches. This method uses the gradients of any target label to calculate the heat map highlighting the local area in the image, which is important for the model to predict the label.
Figure 2 provides examples of post hoc attention maps, which highlight the influential area for the model decision making with grad-CAM. It allowed identification of the acquisition-bias problem in the IPF progression classification model. The attention map of the biased model focused on the CT table, while the attention map of the unbiased model emphasized the area inside the lung.
Post Hoc Interpretations
Post hoc interpretations with domain knowledge can provide explanations about factors that cause model failures. Post hoc interpretation refers to any information extracted from learned models (36). The post hoc attention maps and the case studies from model predictions are also included in this category. The association between the CT-based AI prediction and the reconstruction conditions of the CT image is an example of post hoc interpretation. The patterns on CT images can vary with the CT reconstruction conditions.
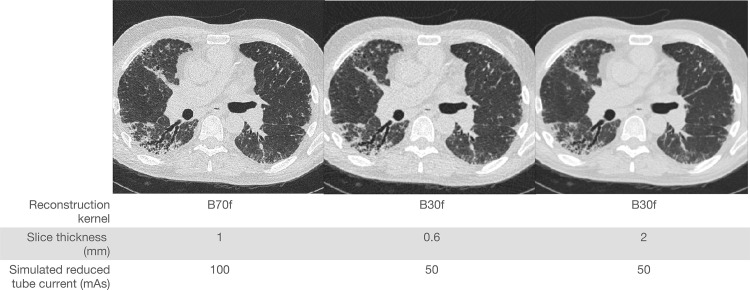
For example, Chong et al (37) illustrated the impact of technical parameters on texture patterns found in fibrotic interstitial lung disease (ILD). Example CT images from the multireconstruction dataset of Chong et al (37) are shown in Figure 7. Figure 7 illustrates the CT images reconstructed differently from CT raw sinogram data collected from one patient. When the model can use the patterns or features impacted by the CT reconstruction conditions, we can identify spurious associations between the CT reconstruction conditions and the model predictions to detect potential acquisition bias.
Figure 7.
CT images reconstructed differently from CT raw sinogram data collected from one patient. CT protocol information is given for each reconstruction image.
The post hoc interpretations from the IPF-ILD diagnosis model from Yu (38) hint that the models can be influenced by the acquisition bias. The author used a dataset of paired CT images with varying acquisition parameters from five multicenter trials. Each pair of CT images was reconstructed differently with the different acquisition parameters from a single raw data acquisition. Data with one condition of the three conditions were used as the training set, and the other two datasets with the remaining conditions were used as the external test for evaluating the OOD generalization performance.
The performance of the external test decreased, and this performance drop was not shown in cross-validation of the training set. Yu (38) shows the statistical analysis of the performance decrease on the external test and the acquisition conditions and shows that the section thickness and the effective tube current–time product (known as the effective milliamperage-seconds) influence the OOD generalization of the model.
Conclusion
Many state-of-the-art medical AI algorithms are susceptible to performance drops due to differences between the model training and real environments. This is known as the data shift problem and raises concerns about the reliability of AI in clinical practice. This data shift issue is challenging because it is often not apparent during model training. To make a model reliable in various environments, it is important to recognize possible data shift and to prevent the model from overfitting to the distribution bias. We described examples to crystallize the negative impact of the five common types of data shifts in medical imaging: population shift, prevalence shift, acquisition shift, annotation shift, and manifestation shift.
We highlighted the importance of explainability as a prerequisite for translating AI to clinical application. Explainability plays a central role in detecting this hidden model failure by providing human-understandable explanations about what contributes to the model’s decision making. We introduced techniques for explainability and reviewed example uses of explanations for detecting the data shift problem. The examples demonstrate that explainability can reveal potential data shift issues at the model training stage. With domain knowledge, the explanations provide information for sanity checks and robust model selection.
Funding.—Supported by funds from the Integrated Diagnostics Shared Resource, Department of Radiological Sciences and Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California, Los Angeles, and by grant R21-HL140465 from the National Heart, Lung, and Blood Institute.
Presented as an education exhibit at the 2021 RSNA Annual Meeting.
Disclosures of conflicts of interest.—: J.G.G. Founder of MedQIA. All other authors, the editor, and the reviewers have disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- IPF
- idiopathic pulmonary fibrosis
- OOD
- out-of-distribution
- UIP
- usual interstitial pneumonia
References
- 1. Wynants L , Van Calster B , Collins GS , et al . Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal . BMJ 2020. ; 369 : m1328 . [Published correction appears in BMJ 2020;369:m2204.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roberts M , Driggs D , Thorpe M , et al . Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans . Nat Mach Intell 2021. ; 3 ( 3 ): 199 – 217 . [Google Scholar]
- 3. Bevan PJ , Atapour-Abarghouei A . Skin Deep Unlearning: Artefact and Instrument Debiasing in the Context of Melanoma Classification . Proceedings of the 39th International Conference on Machine Learning . Proc Mach Learn Res 2022. ; 162 : 1874 – 1892 . [Google Scholar]
- 4. Hastie T , Tibshirani R , Friedman JH . The elements of statistical learning: data mining, inference, and prediction . New York, NY: : Springer; , 2009. ; 219–260 , 389 – 416 . [Google Scholar]
- 5. Goodfellow I , Bengio Y , Courville A . Deep learning . Cambridge, Mass: : MIT Press; , 2016. ; 98 – 168 . [Google Scholar]
- 6. James G , Witten D , Hastie T , Tibshirani R . An introduction to statistical learning . New York, NY: : Springer; , 2013. ; 15–58 , 175 – 202 . [Google Scholar]
- 7. Yu AC , Eng J . One algorithm may not fit all: how selection bias affects machine learning performance . RadioGraphics 2020. ; 40 ( 7 ): 1932 – 1937 . [DOI] [PubMed] [Google Scholar]
- 8. Ye H , Xie C , Cai T , Li R , Li Z , Wang L . Towards a theoretical framework of out-of-distribution generalization . Adv Neural Inf Process Syst 2021. ; 34 : 23519 – 23531 . [Google Scholar]
- 9. Quiñonero-Candela J , Sugiyama M , Schwaighofer A , Lawrence ND , eds. Dataset shift in machine learning . Cambridge, Mass: : MIT Press; , 2008. ; 1 – 28 . [Google Scholar]
- 10. Dou Q , Coelho de Castro D , Kamnitsas K , Glocker B . Domain generalization via model-agnostic learning of semantic features . Adv Neural Inf Process Syst 2019. ; 32 . https://papers.nips.cc/paper/2019/hash/2974788b53f73e7950e8aa49f3a306db-Abstract.html . [Google Scholar]
- 11. Roscher R , Bohn B , Duarte MF , Garcke J . Explainable machine learning for scientific insights and discoveries . IEEE Access 2020. ; 8 : 42200 – 42216 . [Google Scholar]
- 12. Amann J , Blasimme A , Vayena E , Frey D , Madai VI; . Precise4Q consortium. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective . BMC Med Inform Decis Mak 2020. ; 20 ( 1 ): 310 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montavon G , Samek W , Müller KR . Methods for interpreting and understanding deep neural networks . Digit Signal Process 2018. ; 73 : 1 – 5 . [Google Scholar]
- 14. Adadi A , Berrada M . Peeking inside the black-box: a survey on explainable artificial intelligence (XAI) . IEEE Access 2018. ; 6 : 52138 – 52160 . [Google Scholar]
- 15. Moons KG , de Groot JA , Bouwmeester W , et al . Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist . PLoS Med 2014. ; 11 ( 10 ): e1001744 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wolff R , Whiting P , Mallett S , Riley R , Westwood M , Kleijnen J . PROBAST: Prediction Model Risk of Bias Assessment Tool . Presented at the Evidence Synthesis Network: Systematic Reviews of Prognostic Studies—New Approaches to Prognostic Reviews and Qualitative Evidence Synthesis, University of Manchester , Manchester, England: , May 27, 2014 . [Google Scholar]
- 17. Mongan J , Moy L , Kahn CE Jr . Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers . Radiol Artif Intell 2020. ; 2 ( 2 ): e200029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castro DC , Walker I , Glocker B . Causality matters in medical imaging . Nat Commun 2020. ; 11 ( 1 ): 3673 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin AR , Kanai M , Kamatani Y , Okada Y , Neale BM , Daly MJ . Clinical use of current polygenic risk scores may exacerbate health disparities . Nat Genet 2019. ; 51 ( 4 ): 584 – 591 . [Published correction appears in Nat Genet 2021;53(5):763.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundi D , Kryvenko ON , Carter HB , Ross AE , Epstein JI , Schaeffer EM . Pathological examination of radical prostatectomy specimens in men with very low risk disease at biopsy reveals distinct zonal distribution of cancer in black American men . J Urol 2014. ; 191 ( 1 ): 60 – 67 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Litjens G , Toth R , van de Ven W , et al . Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge . Med Image Anal 2014. ; 18 ( 2 ): 359 – 373 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tingelhoff K , Moral AI , Kunkel ME , et al . Comparison between Manual and Semi-automatic Segmentation of Nasal Cavity and Paranasal Sinuses from CT Images . In: 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society , Lyon, France , August 22–26, 2007 . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2007. ; 5505 – 5508 . [DOI] [PubMed] [Google Scholar]
- 23. Walsh SL , Calandriello L , Sverzellati N , Wells AU , Hansell DM ; UIP Observer Consort . Interobserver agreement for the ATS/ERS/JRS/ALAT criteria for a UIP pattern on CT . Thorax 2016. ; 71 ( 1 ): 45 – 51 . [DOI] [PubMed] [Google Scholar]
- 24. Raghu G , Remy-Jardin M , Myers JL , et al . Diagnosis of idiopathic pulmonary fibrosis: an official ATS/ERS/JRS/ALAT clinical practice guideline . Am J Respir Crit Care Med 2018. ; 198 ( 5 ): e44 – e68 . [DOI] [PubMed] [Google Scholar]
- 25. Sinha A , Dolz J . Multi-scale self-guided attention for medical image segmentation . IEEE J Biomed Health Inform 2021. ; 25 ( 1 ): 121 – 130 . [DOI] [PubMed] [Google Scholar]
- 26. Nagarajan MB , Raman SS , Lo P , et al . Building a high-resolution T2-weighted MR-based probabilistic model of tumor occurrence in the prostate . Abdom Radiol (NY) 2018. ; 43 ( 9 ): 2487 – 2496 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W , Shen J . Deep visual attention prediction . IEEE Trans Image Process 2018. ; 27 ( 5 ): 2368 – 2378 . [DOI] [PubMed] [Google Scholar]
- 28. Ren M , Zemel RS . End-to-End Instance Segmentation with Recurrent Attention . In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) , Honolulu, HI , July 21–26, 2017 . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2017. ; 293 – 301 . [Google Scholar]
- 29. Guan Q , Huang Y , Zhong Z , Zheng Z , Zheng L , Yang Y . Thorax disease classification with attention guided convolutional neural network . Pattern Recognit Lett 2020. ; 131 : 38 – 45 . [Google Scholar]
- 30. Schlemper J , Oktay O , Schaap M , et al . Attention gated networks: learning to leverage salient regions in medical images . Med Image Anal 2019. ; 53 : 197 – 207 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu W , Zhou H , Choi Y , Goldin JG , Teng P , Kim GH . An automatic diagnosis of idiopathic pulmonary fibrosis (IPF) using domain knowledge-guided attention models in HRCT images . In: Mazurowski MA , Drukker K , eds. Proceedings of SPIE: Medical Imaging 2021—Computer-aided Diagnosis . Vol 11597 . Bellingham, Wash: : International Society for Optics and Photonics; , 2021. ; 115971Y . [Google Scholar]
- 32. Yu W , Zhou H , Choi Y , Goldin JG , Kim GH . Mga-Net: Multi-Scale Guided Attention Models for an Automated Diagnosis of Idiopathic Pulmonary Fibrosis (IPF) . In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) , Nice, France: , April 13–16, 2021 . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2021. ; 1777 – 1780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim HG , Tashkin DP , Clements PJ , et al . A computer-aided diagnosis system for quantitative scoring of extent of lung fibrosis in scleroderma patients . Clin Exp Rheumatol 2010. ; 28 ( 5 Suppl 62 ): S26 – S35 . [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Z , Liu J , He Y , et al . Towards out-of-distribution generalization: a survey . arXiv preprint arXiv:2108.13624. https://arxiv.org/abs/2108.13624. Posted August 31, 2021. Accessed February 2022 . [Google Scholar]
- 35. Selvaraju RR , Cogswell M , Das A , Vedantam R , Parikh D , Batra D . Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization . In: 2017 IEEE International Conference on Computer Vision (ICCV) , Venice, Italy: , October 22–29, 2017 . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2017. ; 618 – 626 . [Google Scholar]
- 36. Lipton ZC . The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery . Queue 2018. ; 16 ( 3 ): 31 – 57 . [Google Scholar]
- 37. Chong DY , Kim HJ , Lo P , et al . Robustness-driven feature selection in classification of fibrotic interstitial lung disease patterns in computed tomography using 3D texture features . IEEE Trans Med Imaging 2016. ; 35 ( 1 ): 144 – 157 . [DOI] [PubMed] [Google Scholar]
- 38. Yu W , Zhou H , Choi Y , et al . Multi-scale, domain knowledge-guided attention + random forest: a two-stage deep learning-based multi-scale guided attention model to diagnose idiopathic pulmonary fibrosis from computed tomography images . Med Phys 2023. ; 50 ( 2 ): 894 – 905 . [DOI] [PMC free article] [PubMed] [Google Scholar]