Abstract
BCG vaccines are substrains of Mycobacterium bovis derived by attenuation in vitro. After the original attenuation (1908 to 1921), BCG strains were maintained by serial propagation in different BCG laboratories (1921 to 1961). As a result, various BCG substrains developed which are now known to differ in a number of genetic and phenotypic properties. However, to date, none of these differences has permitted a direct phenotype-genotype link. Since BCG strains differ in their abilities to synthesize methoxymycolic acids and since recent work has shown that the mma3 gene is responsible for O-methylation of hydroxymycolate precursors to form methoxymycolic acids, we analyzed methoxymycolate production and mma3 gene sequences for a genetically defined collection of BCG strains. We found that BCG strains obtained from the Pasteur Institute in 1927 and earlier produced methoxymycolates in vitro but that those obtained from the Pasteur Institute in 1931 and later all failed to synthesize methoxymycolates, and furthermore, the mma3 sequence of the latter strains differs from that of Mycobacterium tuberculosis H37Rv by a point mutation at bp 293. Site-specific introduction of this guanine-to-adenine mutation into wild-type mma3 (resulting in the replacement of glycine 98 with aspartic acid) eliminated the ability of this enzyme to produce O-methylated mycolic acids when the mutant was cloned in tandem with mma4 into Mycobacterium smegmatis. These findings indicate that a point mutation in mma3 occurred between 1927 and 1931, and that this mutant population became the dominant clone of BCG at the Pasteur Institute.
BCG vaccines are attenuated substrains of Mycobacterium bovis that were grown in vitro for much of the first half of the 20th century (9). In clinical trials, the protective efficacy has varied considerably, leading to speculation that prolonged growth in vitro has resulted in overattenuated vaccines (3). Based on a recent analysis of gene content of various BCG strains, it is now apparent that during prolonged in vitro passage, BCG strains have lost polygenic regions both at the Pasteur Institute and at other BCG laboratories (5). However, the genetic deletions described have not been directly linked to phenotypic changes; therefore, their implication in the attenuation process remains unknown.
An important and defining characteristic of mycobacteria is their capacity to synthesize long-chain β-hydroxy, α-alkyl fatty acids, known as mycolic acids (2). One type of mycolic acid, containing an α-methyl branched methyl ether, is known as methoxymycolic acid. It has long been known that certain BCG strains, such as BCG-Pasteur, are incapable of synthesizing methoxymycolates (18). Recently, Yuan and colleagues were able to implicate the mma3 gene in methoxymycolate synthesis by complementing BCG-Pasteur with the wild-type mma3 gene from Mycobacterium tuberculosis (27). Importantly, hyperexpression of mma3 not only resulted in methoxymycolate production but also altered cell wall function and growth in macrophages, the cells where BCG resides after vaccination.
Unfortunately, the results obtained by Yuan were difficult to interpret in a phylogenetic context because of the choice of BCG strains analyzed and the comparison of genetic sequences generated by different laboratories (7). As well, because of the large number of genetic differences observed between BCG strains and M. tuberculosis H37Rv, it was not possible to determine precisely the genetic lesion associated with impaired methoxymycolate production. We have therefore undertaken a blinded assessment of methoxymycolate production across a genetically characterized collection of BCG strains and compared this phenotype to the mma3 gene sequence for each of these strains. Because of previous work which enabled us to document the historical propagation of BCG strains from the Pasteur Institute (4), we are able to show here that a single nucleotide polymorphism occurred in mma3 between 1927 and 1931, resulting in loss of methoxymycolic acid production in BCG substrains obtained after that period.
MATERIALS AND METHODS
Genetic analysis of the mma3 sequence from BCG strains.
Mycobacteria listed in Table 1 were grown for 14 to 21 days in 7H9 medium supplemented with OADC enrichment (Difco), and whole genomic DNA was extracted as previously described (24). To produce high-quality sequence data for the entire 879-bp mma3 gene (nucleotides 737271 to 738149 of the M. tuberculosis H37Rv genome, searchable at the TubercuList website [http://bioweb.pasteur.fr/GenoList/TubercuList/]), we designed two pairs of primers to obtain sequence spanning beyond the gene. Primers 5′-CGCGTTGTAGGCGAACTTGA-3′ (forward) and 5′-GATGTGCCATGCACCGTGT-3′ (reverse) amplified the 5′ portion of mma3, while primers 5′-CGGCCATTCTCGTCATGTTCT-3′ (forward) and 5′-ACTGGGCCAACTTCAGCGAG-3′ (reverse) were used to amplify the 3′ portion. PCR mixtures contained 50 ng of genomic DNA, 20 mM Tris-acetate (pH 9.0), 10 mM ammonium sulfate, 75 mM potassium acetate, 0.05% Tween 20, 2.5 mM MgSO4, 4 nmol of deoxynucleoside triphosphates, 25 pmol of each primer, and 1 U of Tfl polymerase (Promega) in a final volume of 50 μl. The PCR consisted of a 5-min denaturation at 94°C followed by 35 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 2 min, with a final extension cycle of 72°C for 10 min. Amplification was confirmed by agarose gel electrophoresis, and PCR products were purified with a kit (Qiagen). Purified samples were sequenced using ABI Prism Big Dye Terminator Cycle Sequencing (Perkin-Elmer Applied Biosystems).
TABLE 1.
Bacterial strains used in this analysis
| Strain | Source | Yr strain was obtained from Pasteur Institute |
|---|---|---|
| M. tuberculosis H37Rv | ATCCa | NAb |
| M. bovis 2122 | Glyn Hewinson, Surrey, England | NA |
| M. smegmatis mc2155 | W. R. Jacobs, Jr., New York, N.Y. (22) | NA |
| BCG Russia | ATCC | 1924 |
| BCG Moreau | Primary seed lot lyophilized in Rio de Janeiro, 1968 | 1925 |
| BCG Tokyo | Japanese Anti-Tuberculosis Association | 1925 |
| BCG Sweden | Statens Serum Institut, Copenhagen, Denmark | 1926 |
| BCG Birkhaug | ATCC | 1927 |
| BCG Danish 1331 | ATCC | 1931 |
| BCG Prague | Statens Serum Institut | 1931 to Denmark, then 1947 to Prague |
| BCG Glaxo | ATCC | 1931 to Denmark, then 1953 to England |
| BCG Tice | ATCC | 1934 |
| BCG Frappier | ATCC | 1937 |
| BCG Connaught | Connaught Laboratories | 1937 to Frappier, then 1948 to Connaught |
| BCG Phipps | ATCC | 1938 |
| BCG Pasteur 1173P2B | Institut Pasteur | Lyophilized in 1961 |
ATCC, American Type Culture Collection.
NA, not applicable.
For rapid identification of the single mutation detected, genomic DNA was amplified by PCR for the 5′ 562 bp of mma3. Purified PCR products were then digested with 5 U of SalI (Promega) for 1 h at 37°C. Samples were run in duplicate on a 2% agarose gel and scored as wild type or mutant based on restriction pattern.
Analysis of mycolic acid profiles in BCG strains.
The 13 BCG strains were grown in 7H9–ADC–0.05% Tween, and mycolic acid methyl esters were prepared and analyzed by one-dimensional thin-layer chromatography (TLC) as previously described (27). TLC plates were visualized by immersion in ceric ammonium molybdate dip and heating to 120°C for 20 min. Ceric ammonium molybdate dip is prepared by dissolving 1 g of ceric sulfate monohydrate and 25 g of ammonium molybdate in 450 ml of distilled water. With stirring, 50 ml of concentrated sulfuric acid is carefully added, and the solution is stirred for 1 h at room temperature.
Mutagenesis of mma3 at bp 293.
The mma3 and mma4 genes of M. tuberculosis H37Rv (TubercuList Rv0643c and Rv0642c) were PCR amplified from genomic DNA using primers gctctagaGATGGCCACCTGCTGAAG (forward) and gcgcaagcttGGGCTTATGC-GTCTGCTC (reverse), which contain nonhomologous sequences at each 5′ end (lowercase letters) that are used to add XbaI and HindIII sites, respectively. This PCR product was digested with XbaI and HindIII and cloned into the XbaI and HindIII sites of pUC19 to make plasmid pBGS93. The entire insert was sequenced to ensure that no mutations were introduced by PCR amplification. In addition to the standard M13 forward and reverse sequencing primers, the following custom primers were used for sequencing the insert: 3R1, GCTTCGATGGTTACGATGCGG (R indicates reverse; F indicates forward); mut3.1 (see below), 4F1 GAGACGATCGAGGAGCATGTG; 4R1, GGTAGCTGACGCTGCTCTGG; and 4F2, GACCCGACCCGAACTTAC. A single-base G-to-A mutation was introduced at base 293 of the mma3 coding sequence using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and mutagenic primers mut3.1 (CGACAGGGTCAAGtCGACGACGTTGACGTCAACGTCGTCGaCTTGACCCTGTCG) (forward) and mut3.2 (CGACAGGGTCAAGtCGACGACGTTGAC) (reverse) (mutated bases are shown in lowercased). The resulting pUC19 clone containing the mutated mma3 and wild-type mma4 genes, designated pBGS95, was sequenced using the primers listed above to ensure that the desired mutation was present and that no unwanted mutations were introduced. The XbaI-to-HindIII fragments were excised from pBGS93 and pBGS95 and introduced into the same sites of the Escherichia coli-mycobacterial shuttle vector pMV206_Hyg(11) to produce pBGS94 and pBGS96, respectively. To boost expression of mma3 and mma4 in Mycobacterium smegmatis, the hsp60 promoter from pMV261 was introduced in front of mma3 (11). Plasmids pBGS94 and pBGS96 were digested with XbaI, the resulting overhangs were filled in with Klenow fragment, and the products were digested with KpnI and then ligated to the 418-bp KpnI-PvuII fragment containing the hsp60 promoter from pMV261 to produce plasmids pBGS99 and pBGS100, respectively.
The mma4 gene alone was subcloned from pMV206_mma4 into pMV206_Hyg as a 1.7-kb BamHI fragment to make pBGS16 (26). The orientation of the mma4 insert in pBGS16 is such that PvuII digestion yields 4.7- and 1.2-kb fragments.
Plasmids pMV206_Hyg, pBGS16, pBGS99, and pBGS100 were introduced into M. smegmatis mc2155 by electroporation, cultures were labeled with [1-14C]acetate, and mycolic acid methyl esters were prepared and analyzed by two-dimensional argentation TLC as described previously (11).
RESULTS
Sequence analysis of mma3 in 13 different strains of M. bovis BCG.
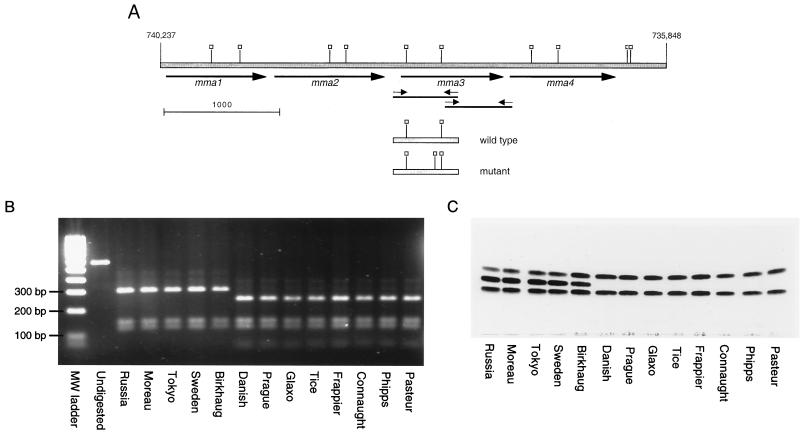
To elucidate the molecular basis for the loss of production of methoxymycolic acids by certain strains of BCG, we obtained an extensively characterized collection of BCG isolates with a defined history (5) (Table 1). The mma3 gene from each of these 13 strains was obtained by PCR amplification of genomic DNA, and these products were sequenced in their entirety (Fig. 1A). A comparison of the nucleotide sequence of the mma3 gene between BCG strains, M. bovis 2122, and M. tuberculosis H37Rv revealed that all nucleotides in the gene were identical across all strains tested with the exception of position 293 of the 879-nucleotide open reading frame. At this position, M. tuberculosis H37Rv, M. bovis 2122, and BCG strains obtained from the Institut Pasteur from 1924 to 1927 (strains Russia, Moreau, Tokyo, Sweden, and Birkhaug) had the same sequence as that reported for M. tuberculosis H37Rv (6). In the eight BCG strains obtained in 1931 or later, a G-to-A base substitution was seen at position 293. This transition results in an amino acid change from glycine (Gly, encoded by GGC) to aspartic acid (Asp, encoded by GAC) at codon 98. Unlike the findings of previous studies (7, 27), no other differences were observed in BCG strains. To confirm these results, SalI digestion of the 562-bp PCR product representing the 5′ portion of the gene resulted in two clearly distinct DNA fragment patterns, based on the G-to-A base substitution causing an extra SalI cleavage site (see Fig. 1A and B).
FIG. 1.
Genetic and phenotypic analysis of BCG methoxymycolic acid production. (A) Map of the gene locus responsible for the production of methoxymycolates. The shaded bar represents genomic DNA, with numerical locations on the chromosome of strain H37Rv indicated (6). The heavy arrows indicate methyltransferase open reading frames. Small arrows indicate primers used to amplify the 5′ and 3′ regions of mma3. Thin solid lines represent the PCR products. Open squares, SalI sites. Shown below the genomic regions are the amplicons from wild-type and mutant mma3 5′ regions, with SalI sites indicated (note the additional site in the mutant mma3). Scale marker, 1,000 bp. (B) Purified 562-bp PCR products containing the 5′ end of the mma3 gene from the 13 BCG strains were digested with SalI. Without digestion, a 562-bp product is seen. After incubation, the wild-type strains have three fragments of 303, 144, and 115 bp, while the mutant strains have four fragments of 246, 144, 115, and 57 bp. (C) TLC analysis of purified mycolic acids from various BCG substrains. The same BCG strains shown in the PCR-RFLP gel were analyzed by TLC for production of mycolic acids as described in Materials and Methods. The three bands obtained represent (from bottom to top) ketomycolates, methoxymycolates, and α-mycolates. It is seen in this figure that eight BCG strains lack methoxymycolates. These are the same strains that gave the extra SalI band; they represent strains of BCG obtained from the Pasteur Institute in 1931 or later.
Analysis of mycolic acid production among BCG strains.
We examined the mycolic acid subclass production among the collection of 13 BCG strains by simple TLC analysis of the methyl esters of saponified total cell lysates as shown in Fig. 1C. Samples are resolved based upon the polarity of the functional groups involved so that the fastest-migrating band represents the α-mycolate subclass (with a single cis cyclopropane in the distal position); the middle, more-variable band represents methoxymycolic acids containing a more polar α-methyl-methyl ether; and the slowest-migrating band represents ketomycolates, which contain the most-polar functional group, an α-methyl branched ketone. The strains analyzed fell into two broad classes: those that produced all three mycolic acids and those that produced approximately 50% α-mycolate and 50% ketomycolate. There was a perfect correlation between BCG strains that produced MMAS-3 (G98D) and those that failed to produce the middle band representing methoxymycolates. With the stain used in Fig. 1C, we did not observe the production of hydroxymycolic acids, which would run more slowly than ketomycolates in this TLC system. A very small amount of hydroxymycolate could be detected in [14C]acetate-radiolabeled mycolates; however, there was no correlation between the amount present and the mma3 mutation (data not shown).
Mutagenesis of mma3 and functional analysis of MMAS-3(G98D).
Glycine 98 lies in a region of MMAS-3 which is highly conserved among all fatty acyl methyltransferases identified to date (Fig. 2). Immediately N-terminal of Gly98 is an S-adenosylmethionine (SAM)-binding motif shared by a wide variety of SAM-dependent methyltransferases (13). Sequence alignment of the cyclopropane fatty acid synthase from E. coli and all six mycolic acid methyltransferase sequences for which a function has been described revealed that Gly 98 was absolutely conserved. We hypothesized that the mutation of this small, neutral residue to negatively charged aspartic acid would be sufficient to inactivate MMAS-3 activity.
FIG. 2.
Alignment of the pertinent region of amino acid sequence from various methyltransferases. The starting amino acid number is shown at the left. An arrow indicates the G98D mutation observed in the BCG MMAS-3 sequence that appears to correlate with production of methoxymycolic acids by M. tuberculosis. Sequences are MMAS-3 (TubercuList (http://bioweb.pasteur.fr/GenoList/TubercuList/) accession number Rv0643c), CMAS-1 (Rv3392c), CMAS-2 (Rv0503c), MMAS-1 (Rv0645c), MMAS-2 (Rv0644c), and MMAS-4 (Rv0642c), as well as the cyclopropane fatty acid synthase from E. coli (CFAS; SwissProt [http://www.ebi.ac.uk/cgi-bin/swissfetch] accession number P30010).
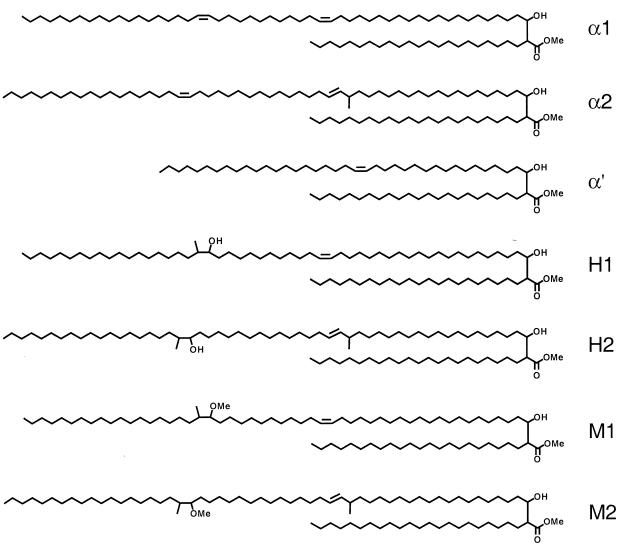
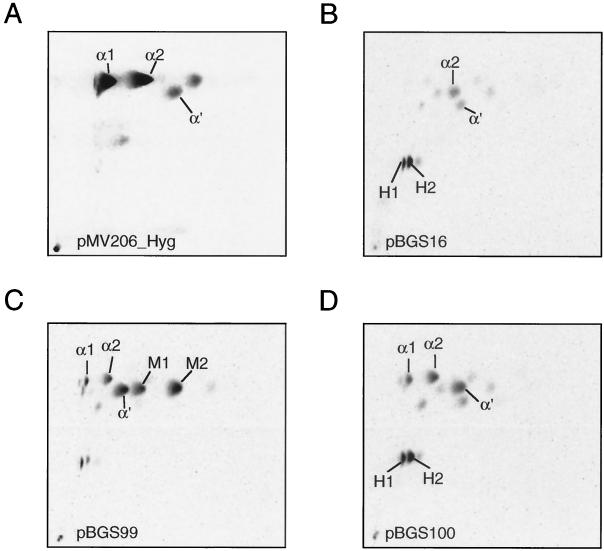
In order to assess the functional relevance of this mutation, we introduced this mutation specifically into the mma3 gene derived from M. tuberculosis by site-directed mutagenesis (see Materials and Methods). Assessing MMAS-3 function is possible only in a heterologous background that produces the appropriate hydroxymycolate precursor. In M. tuberculosis this hydroxymycolate precursor is produced by the MMAS-4 protein (26). This background can be created by expressing the MMAS-4 protein in the heterologous host M. smegmatis (Fig. 3). Introduction of mma4 alone gives a strictly hydroxymycolate profile showing H1 and H2 (Fig. 4B), while introduction of both mma4 and mma3 simultaneously gives a mixture of two isomers of methoxymycolate (one derived from the α1 series with a proximal cis-olefin, M1, and one derived from the α2 series with a proximal trans-olefin and allylic methyl branch, M2) (Fig. 4C). Introduction of an identical construct bearing the mutation resulting in the G98D amino acid substitution results in failure of the O-methyltransferase portion of the reaction, and the mycolate profile looks identical to that resulting from expression of MMAS-4 alone (Fig. 4D and B, respectively). This result supports the notion that the G98D mutation directly results in loss of the O-methyltransferase function of MMAS-3.
FIG. 3.
Structures and biosynthetic relationships of mycolic acids produced by heterologous expression of MMAS-4 and MMAS-3 in M. smegmatis. α1, α2, and α′ are the normal complement of mycolic acids. H1 and H2 result from the action of MMAS-4 on precursors to the normal mycolic acids, and M1 and M2 result from the action of MMAS-3 upon H1 and H2, respectively.
FIG. 4.
Two-dimensional TLC of M. smegmatis expressing various methyltransferases. (A) Wild-type mc2155 containing the empty vector pMV206_Hyg; (B) mc2155 expressing MMAS-4; (C) mc2155 expressing MMAS-4 and wild-type MMAS-3; (D) mc2155 expressing MMAS-4 and MMAS-3(G98D). Designations are as explained in the legend to Fig. 3.
DISCUSSION
In previous work it has been shown that BCG vaccines vary in terms of genetic composition due to evolution both at the Pasteur Institute and in other BCG laboratories. Differences documented to date include deletions (5), variable restriction fragment length polymorphism (RFLP) patterns when strains are probed for IS6110 (4) or direct repeats (12), and variable numbers of intergenic repeats as determined by the length of PCR amplicons (10, 16). To date, none of these genetic differences among BCG strains has been specifically linked to a phenotypic change. The work described in this report documents one more genetic event that has occurred during the serial propagation of BCG vaccine. In this case, an important phenotypic alteration has resulted, as BCG vaccines obtained from the Pasteur Institute after 1927 are unable to synthesize methoxymycolic acids. This indicates that between 1927 and 1931, a mutation in mma3 occurred which permitted the mutant population to overgrow the parent methoxymycolate-producing population. In previous studies, sequences were obtained from strains of ill-defined history (27) or previously constructed cosmid libraries (7). Unlike these investigators, we used a historically defined collection of strains which were minimally passaged after receipt, and we performed direct sequencing to demonstrate that only a single nucleotide polymorphism occurred during serial propagation of BCG strains.
In light of the biosynthetic relationship between hydroxy- and methoxymycolates, mma3 mutants might be expected to produce hydroxymycolic acids. However, in M. smegmatis, overproduction of hydroxymycolates alters colony surface morphology (26) and severely impairs growth (B. G. Schroeder, unpublished data). This suggests that cells that acquire a mutation in mma3 are under selective pressure to reduce levels of hydroxymycolates. This could be accomplished either by reducing MMAS-4 activity or by increasing the rate of oxidation of hydroxymycolate to ketomycolate. The fact that we do not observe an increase in hydroxymycolate levels in mma3 mutants of BCG supports the notion that one or both of these mechanisms may be in use.
From 1908 to 1921, BCG was grown on glycerinated potato medium with bile, but from 1921 to 1932, the Pasteur Institute maintained three lineages of BCG, one on the conventional medium, one on glycerinated potato without bile, and one on potato Sauton medium (20). After 1932, the potato Sauton medium became the standard means of propagating BCG, with 2 to 3 passages per year on potato glycerine. Thus, somewhere between 1921 and 1932, the principal lineage of BCG was changed from a bile-containing medium to one without this natural detergent. Although we have previously shown that uptake of chenodeoxycholate (a bile component) was not appreciably different between an organism that produces a wild-type complement of mycolic acids and one that produces only α- and methoxymycolates (27), there is good reason to think that production of only α- and ketomycolates does significantly alter the permeability properties. The MIC of rifampin for an organism that produces only α- and ketomycolates is slightly lower than that for an organism that produces the normal set of three mycolic acids (27). Permeability to glucose and sensitivity to ampicillin are affected in apparently opposite ways, with ketomycolate production contributing to higher rates of glucose uptake but increased resistance to ampicillin (27). These results make it difficult to reconstruct the original basis for selection of the mutant. Whatever the reason for the selection of the mma3 mutant, it is now clear that all BCG strains obtained after the era in which it occurred do not produce methoxymycolic acids.
After vaccination, it is thought that BCG vaccines become incorporated in macrophages, where they survive and replicate for an unknown duration. Reports of disseminated BCG-itis in AIDS patients 30 years after vaccination suggest that, at least for some persons, the vaccine remains viable in the host long after immunization (1). The importance of the survival of BCG vaccine in the host was suggested in the 1950s by Dubos, who observed that animal inoculation with M. tuberculosis H37Ra, which does not replicate in mice, is associated with limited protection against subsequent challenge with virulent M. tuberculosis (8). Thus, the survival and growth of BCG vaccines in macrophages may in part contribute to their ability to provide protective immunity. In previous work, it was shown that changes in methoxymycolate production by BCG vaccines result in impaired survival in macrophage cell culture. Although these analyses employed overproducers of MMAS-3, with no isogenic mma3 knockouts studied, the comparison of a strain which produces only α- and ketomycolates (designated “Pasteur” in that report [27]) to one with all three classes of these molecules (designated “Connaught”) suggests the possibility that the loss of the ability to produce methoxymycolic acid may be directly related to the ability of these strains to grow in macrophages.
It is tempting to scrutinize the historical record from 1927 to 1931 in order to determine whether a change in vaccine properties in vivo occurred. However, it should be noted that during the era in question, there was also a loss of a deletion region (RD2) (17), and expression of secreted proteins MPB70 and MPB83 appears to be greater in strains obtained prior to that era (19). Before 1931, there was considerable concern that the newly derived vaccine might revert to virulence, with many investigators alleging that they could dissociate virulent from avirulent forms of BCG (21). After 1931, these efforts could no longer be replicated, and in fact, observational studies from that period suggested that BCG had instead become less virulent (14). An autopsy study performed on children who died of other causes found that, prior to 1929, live BCG vaccine could be cultured out of mesenteric nodes up to 6 months after vaccination, while autopsies from 1930 onwards were no longer able to detect viable vaccine (28). Thus, the fact that BCG strains changed between 1927 and 1931 was observed and recorded by investigators from that era, although the reason for these observed changes remains to be confirmed in light of the number of changes that can be dated to this era. Whether these mutations have impacted on vaccine efficacy and virulence is difficult to infer from the present data. For instance, in countries which have experienced different vaccines, BCG-Russia was reported to be more virulent than BCG-Prague (25) and BCG-Sweden had higher rates of dissemination than BCG-Glaxo (15). In both examples, the methoxymycolate-producing strain was the more virulent of the two. However, BCG-Pasteur and BCG-Glaxo are both mma3 mutants, and in a randomized trial between them, BCG-Pasteur was the more virulent and more protective vaccine (23). Thus, the ability of BCG strains to synthesize methoxymycolic acids is likely but one of a number of determinants of how BCG vaccines behave in vivo. The implications of the mma3 mutation for selection of BCG strains used in immunization programs remain to be determined.
REFERENCES
- 1.Armbruster C, Junker W, Vetter N, Jaksch G. Disseminated bacille Calmette-Guérin infection in an AIDS patient 30 years after BCG vaccination. J Infect Dis. 1990;162:1216. doi: 10.1093/infdis/162.5.1216. [DOI] [PubMed] [Google Scholar]
- 2.Barry C E, III, Lee R E, Mdluli K, Sampson A E, Schroeder B G, Slayden R A, Yuan Y. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 3.Behr M A, Small P M. Has BCG attenuated to impotence? Nature. 1997;389:133–134. doi: 10.1038/38151. [DOI] [PubMed] [Google Scholar]
- 4.Behr M A, Small P M. A historical and molecular phylogeny of BCG strains. Vaccine. 1999;17:915–922. doi: 10.1016/s0264-410x(98)00277-1. [DOI] [PubMed] [Google Scholar]
- 5.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. . (Erratum, 396:190.) [DOI] [PubMed] [Google Scholar]
- 7.Dubnau E, Marrakchi H, Smith I, Daffe M, Quemard A. Mutations in the cmaB gene are responsible for the absence of methoxymycolic acid in Mycobacterium bovis BCG Pasteur. Mol Microbiol. 1998;29:1526–1528. [PubMed] [Google Scholar]
- 8.Dubos R J, Pierce C H, Schaefer W B. Anti-tuberculosis immunity induced in mice by vaccination with living cultures of attenuated tubercle bacilli. J Exp Med. 1953;97:207–220. doi: 10.1084/jem.97.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fine P E. Bacille Calmette-Guérin vaccines: a rough guide. Clin Infect Dis. 1995;20:11–14. doi: 10.1093/clinids/20.1.11. [DOI] [PubMed] [Google Scholar]
- 10.Frothingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 11.George K M, Yuan Y, Sherman D R, Barry C E., III The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis: identification and functional analysis of CMAS-2. J Biol Chem. 1995;270:27292–27298. doi: 10.1074/jbc.270.45.27292. [DOI] [PubMed] [Google Scholar]
- 12.Howard S T, Laszlo A, Johnson W M. Genetic identification of Mycobacterium bovis BCG by restriction fragment length polymorphism analysis of the direct-repeat region. J Clin Microbiol. 1997;35:965–968. doi: 10.1128/jcm.35.4.965-968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingrosso D, Fowler A V, Bleibaum J, Clarke S. Sequence of the d-aspartyl/l-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989;264:20131–20139. [PubMed] [Google Scholar]
- 14.Jensen K A. Practice of the Calmette vaccination. Acta Tuberc Scand. 1946;20:1–45. [Google Scholar]
- 15.Kroger L, Brander E, Korppi M, Wasz-Hockert O, Backman A, Kroger H, Launiala K, Katila M L. Osteitis after newborn vaccination with three different Bacillus Calmette-Guérin vaccines: twenty-nine years of experience. Pediatr Infect Dis J. 1994;13:113–116. doi: 10.1097/00006454-199402000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Magdalena J, Supply P, Locht C. Specific differentiation between Mycobacterium bovis BCG and virulent strains of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:2471–2476. doi: 10.1128/jcm.36.9.2471-2476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minnikin D E, Minnikin S M, Dobson G, Goodfellow M, Portaels F, van den Breen L, Sesardic D. Mycolic acid patterns of four vaccine strains of Mycobacterium bovis BCG. J Gen Microbiol. 1983;129:889–891. doi: 10.1099/00221287-129-3-889. [DOI] [PubMed] [Google Scholar]
- 19.Oettinger T, Jorgensen M, Ladefoged A, Haslov K, Andersen P. Development of the Mycobacterium bovis BCG vaccine: review of the historical and biochemical evidence for a genealogical tree. Tuber Lung Dis. 1999;79:243–250. doi: 10.1054/tuld.1999.0206. [DOI] [PubMed] [Google Scholar]
- 20.Osborn T W. Changes in BCG strains. Tubercle. 1983;64:1–13. doi: 10.1016/0041-3879(83)90044-2. [DOI] [PubMed] [Google Scholar]
- 21.Petroff S A. A new analysis of the value and safety of protective immunization with BCG. Am Rev Tuberc. 1929;20:275–296. [Google Scholar]
- 22.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 23.ten Dam H G. BCG vaccination. In: Reichman L B, Hershfield E S, editors. Tuberculosis: a comprehensive international approach. New York, N.Y: Marcel Dekker; 1993. pp. 251–274. [Google Scholar]
- 24.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitkova E, Galliova J, Krepela K, Kubin M. Adverse reactions to BCG. Cent Eur J Public Health. 1995;3:138–141. [PubMed] [Google Scholar]
- 26.Yuan Y, Barry C E., III A common mechanism for the biosynthesis of methoxy and cyclopropyl mycolic acids in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:12828–12833. doi: 10.1073/pnas.93.23.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Zhu Y, Crane D D, Barry C E., III The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol Microbiol. 1998;29:1449–1458. doi: 10.1046/j.1365-2958.1998.01026.x. [DOI] [PubMed] [Google Scholar]
- 28.Zeyland J, Piasecka-Zeyland E. Sur la vitalité du BCG dans l'organisme vacciné. Ann Inst Pasteur. 1936;56:46–51. [Google Scholar]