Abstract
The HARO7 gene of the methylotrophic, thermotolerant yeast Hansenula polymorpha was cloned by functional complementation. HARO7 encodes a monofunctional 280-amino-acid protein with chorismate mutase (EC 5.4.99.5) activity that catalyzes the conversion of chorismate to prephenate, a key step in the biosynthesis of aromatic amino acids. The HARO7 gene product shows strong similarities to primary sequences of known eukaryotic chorismate mutase enzymes. After homologous overexpression and purification of the 32-kDa protein, its kinetic parameters (kcat = 319.1 s−1, nH = 1.56, [S]0.5 = 16.7 mM) as well as its allosteric regulatory properties were determined. Tryptophan acts as heterotropic positive effector; tyrosine is a negative-acting, heterotropic feedback inhibitor of enzyme activity. The influence of temperature on catalytic turnover and the thermal stability of the enzyme were determined and compared to features of the chorismate mutase enzyme of Saccharomyces cerevisiae. Using the Cre-loxP recombination system, we constructed mutant strains carrying a disrupted HARO7 gene that showed tyrosine auxotrophy and severe growth defects. The amount of the 0.9-kb HARO7 mRNA is independent of amino acid starvation conditions but increases twofold in the presence of methanol as the sole carbon source, implying a catabolite repression system acting on HARO7 expression.
Methylotrophic yeasts have gained increasing recognition in basic research as well as in applied biotechnology in the last few years. Most of them are ascomycetes of the genera Hansenula, Pichia, and Candida (38), with Hansenula polymorpha (synonym, Pichia angusta) representing the most prominent member (for a review, see reference 26). Utilization of methanol as sole source of carbon and energy by H. polymorpha is generally accompanied by strong proliferation of microbodies, so-called peroxisomes, and high-level induction of peroxisomal matrix enzymes required for C1 metabolism (49, 67). The first step in the methanol-utilizing pathway is the oxidation of methanol to formaldehyde and H2O2, catalyzed by the MOX-encoded methanol oxidase (EC 1.1.3.13) (37). Additional important gene products involved in methanol assimilation are a dihydroxyacetone synthase (EC 2.2.1.3) encoded by the DAS gene, a catalase (EC 1.11.1.6) encoded by the CAT1 gene, and a formate dehydrogenase activity (EC 1.2.1.2) which is the FMD gene product (9, 31, 33). In the presence of glucose, expression of these genes is subject to a repression system, whereas upon methanol utilization the promoters of these genes are strongly induced (12). The tightly regulated strength of genes involved in methanol metabolism forms the basis for the biotechnological and commercial use of H. polymorpha in recombinant gene expression systems. In recent years, a tractable vector-host system has been developed using either homologous or heterologous metabolic genes as selectable markers in combination with defined mutant strains and taking advantage of the strong promoters of genes that are part of the methanol-utilizing machinery (14, 20). Integration of autonomously replicating plasmids into the chromosomal DNA can be achieved, yielding up to 100 tandemly repeated copies of the transforming DNA that are mitotically stable in the H. polymorpha genome (19, 34). Additionally, H. polymorpha is able to grow at temperatures of up to 48°C, with an optimal growth temperature of 37°C, which is unusual for methylotrophic yeasts (40).
In contrast to the specialized methanol-utilizing pathway of methylotrophic yeasts, biosynthesis of aromatic amino acids is a common feature of most living organisms. Chorismic acid, the end product of the shikimate pathway, is formed in seven invariable enzyme-catalyzed reactions starting with compounds of primary metabolism, erythrose 4-phosphate and phosphoenolpyruvate (27). Conversion of chorismate to anthranilate initiates the biosynthetic branch resulting in l-tryptophan, whereas intramolecular rearrangement of the enolpyruvyl side chain of chorismate to yield prephenate is the initial step in the synthesis of l-tyrosine and l-phenylalanine (68). The latter reaction is unique, as it is the only Claisen rearrangement identified so far in primary metabolism (18). Generally, the conversion of chorismate to prephenate is catalyzed by chorismate mutases (EC 5.4.99.5) which have been identified and characterized in archea, bacteria, fungi, and plants (for a review, see reference 50). Crystallographic data for three natural enzymes have led to a classification based on structural elements as well as primary sequence information. AroH class chorismate mutases are α/β-barrel proteins, as is the trimeric Bacillus subtilis enzyme (6), whereas the AroQ class comprises all-helix bundle polypeptides that are often part of a bifunctional enzyme like the chorismate mutase domain of the Escherichia coli chorismate mutase-prephenate dehydratase activity (8, 39). Eukaryotic chorismate mutases are also classified in the latter class on the basis of conservation of crucial catalytic residues and related tertiary structure (43, 71). Whereas a number of prokaryotic genes encoding chorismate mutase activities have been cloned to date, only few sequences that originate from eukaryotic organisms and code for chorismate mutase enzymes are available. The best-studied eukaryotic enzyme with respect to structure, allosteric regulation, and mechanism of catalytic turnover is that of the baker's yeast Saccharomyces cerevisiae (42, 56, 57, 65). Recently, additional data for the chorismate mutase enzyme of the filamentous fungus Aspergillus nidulans which is encoded by the aroC gene have been made available (35). The A. nidulans chorismate mutase was found to be similar in catalytic and structural properties to the well-characterized enzyme of S. cerevisiae. Nevertheless, different mechanisms for allosteric regulation upon effector binding have been proposed for these two chorismate mutases.
To extend the eukaryotic subclass of AroQ enzymes, we here present the cloning and characterization of the HARO7 gene coding for a chorismate mutase activity of the methylotrophic yeast H. polymorpha, an organism closely related to S. cerevisiae. haro7Δ disruption strains were constructed by establishing the Cre-loxP recombination system of bacteriophage P1 (63) in this yeast to constitute HARO7 as a new marker gene to the vector-host expression system of H. polymorpha for biotechnological applications (G. Gellissen, G. Braus, R. Pries, S. Krappmann, and A. W. Strasser, German patent application 19919124.7, April 1999). Transcriptional expression patterns of HARO7 were monitored with respect to different environmental stimuli like amino acid starvation conditions or alternative carbon sources, indicating that HARO7 expression is the target of a catabolite repression system but not of the general control of amino acid biosynthesis. Additionally, the enzyme was overexpressed and purified to homogeneity, taking advantage of an H. polymorpha expression system. Kinetic assays as well as regulatory analyses of the chorismate mutase indicated that catalytic activity is tightly regulated in an allosterical manner and that this enzyme of H. polymorpha has a higher optimal temperature for catalytic turnover than its counterpart from S. cerevisiae despite lower thermal stability.
MATERIALS AND METHODS
Materials.
Chorismic acid as barium salt was purchased from Sigma (St. Louis, Mo.). 5-Fluoroorotic acid was obtained from Toronto Research Chemicals Inc. (Toronto, Ontario, Canada). l-Tyrosine for supplementation was obtained as free base (SigmaUltra, >99%) from Sigma-Aldrich Chemie GmbH (Steinheim, Germany) and alternatively from Fluka (Neu-Ulm, Germany) in BioChemika grade (>99%; foreign amino acids, <0.3%). Protein solutions were concentrated by using stirred cells (volumes of 180 and 10 ml) with PM-10 ultrafiltration membranes from Millipore (Eschborn, Germany). The Mini 2D sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) system and Bradford protein assay solution for determination of protein concentrations were from Bio-Rad Laboratories (Hercules, Calif.). Platinum Pfx DNA polymerase from Life Technologies GmbH (Karlsruhe, Germany) was used for PCRs. All other chemicals were supplied by Fluka or Sigma-Aldrich.
Strains, media, growth conditions, and transformation procedures.
Plasmid DNA was propagated in Escherichia coli DH5α (70). S. cerevisiae RH2185 (MATa suc2-Δ9 ura3-52 leu2-3 leu2-112 his4-519 aro7::LEU2 GAL2) (59) with the genetic background of the laboratory strain X2180-1A (MATa gal2 SUC2 mal CUP1) was used as recipient for cloning of a DNA fragment containing the HARO7 gene of H. polymorpha from a genomic library. The uracil-auxotrophic H. polymorpha strain RB11 (odc1) was obtained from Rhein Biotech GmbH (Düsseldorf, Germany) and has been described by Weydemann et al. (69). H. polymorpha RH2408 (odc1 FMD promoter::HARO7 URA3) was used for homologous overexpression and purification of the HARO7 gene product. Mutant strains RH2409 (odc1 haro7::loxP-ODC1MX-loxP) and RH2410 (odc1 haro7::loxP) are derivatives of H. polymorpha RB11 carrying a disrupted HARO7 gene. E. coli cells were grown in LB medium (44) supplemented with ampicillin (100 μg · ml−1) at 37°C. Complex medium for growth of yeasts was YEPD (1% yeast extract, 2% peptone, 2% glucose). Selective medium contained 0.14% yeast nitrogen base (without amino acids and ammonium sulfate) and 0.5% ammonium sulfate. Carbon sources were either 2% glucose, 1% glycerol, or 0.7% methanol. Supplements were added according to Guthrie and Fink (25). In contrast to S. cerevisiae, which was cultivated at 30°C, H. polymorpha strains were propagated at 37°C, which is the optimal temperature for growth of this yeast. Transformation of E. coli was performed as described by Inoue et al. (32). S. cerevisiae was transformed by a modification of the protocol of Elble (13). For transformation of H. polymorpha, an electroporation procedure was used (15).
Isolation and analyses of nucleic acids.
For isolation of plasmid DNA from bacterial strains, a plasmid purification system from Qiagen (Hilden, Germany) was used. Genomic DNA from yeasts was isolated according to Hoffmann and Winston (30) and analyzed by Southern blotting (62) or diagnostic PCR (52) using oligonucleotides OLSK57 (5′-CAATGCCAGCAATATGGAGACG-3′) and RP1 (5′-GAACTAGAATTCGAGAATAATTAAAG-3′). Total RNA from H. polymorpha cultures was prepared according to Cross and Tinkelenberg (7), and transcript levels were determined by Northern hybridization (48) using a Bio-Imaging analyzer from Fuji Photo Film Co. Ltd. (Tokyo, Japan). Transcript length was determined using a 0.16- to 1.77-kb RNA ladder from Life Technologies, Inc. Sequencing reactions were carried out using a BigDye sequencing kit (28) and analyzed on an ABI PRISM 310 genetic analyzer (PE Biosystems, Foster City, Calif.).
Cloning techniques; construction of genomic library and plasmids.
Standard techniques for cloning and manipulation of recombinant DNA were applied (44). For identification of the HARO7 gene, a genomic library was constructed by ligating partially Sau3A-digested DNA of H. polymorpha RB11 in the BamHI restriction site of shuttle vector pRS426 (61). The ligation products were transformed in E. coli to yield a library pool of approximately 100,000 independent clones from which plasmid DNA was isolated. Plasmid pME1524 contains a 5-kb genomic Sau3A fragment containing the HARO7 gene of H. polymorpha RB11 in pRS426. Subcloning generated plasmid pME1525, which carries a 1.8-kb ApaI/HindIII fragment, originating from pME1524, in pRS426. For working purposes, plasmid pME1526, which carries the 1.8-kb ApaI/HindIII fragment of pME1524 in the bacterial plasmid pBluescript II KS (Stratagene (La Jolla, Calif.), was constructed. Overexpression of the HARO7 gene product in RB11 was achieved using plasmid pME1686. This plasmid carries the entire open reading frame of HARO7 amplified from pME1525 using oligonucleotides OLSK50 (5′-TATAGAATTCATGGACTTTATGAAGCC-3′, EcoRI site underlined) and RP1 in a PCR; the resulting EcoRI fragment was cloned in the expression vector pFPMT121 (Rhein Biotech) to yield an FMD promoter::HARO7::MOX terminator expression cassette in a plasmid autonomously replicating in H. polymorpha. For characterization of a haro7Δ mutant strain, a disruption cassette was constructed. To this end, the 5′ flanking sequence of HARO7 was amplified from pME1524 using OLSK34 (5′-ATATAGATCTACAAAAACTAAACAGG-3′, BglII site underlined) as reverse primer and cloned as a 2-kb SalI/BglII fragment. The 3′ region flanking HARO7 was cloned as a 2.5-kb BglII/NotI fragment amplified in a PCR with OLSK35 (5′-ATATAGATCTGATGCGACGCAGAAAAGC-3′, BglII site underlined) as forward primer using a partial BglII genomic sublibrary of RB11 cloned in pBluescript II KS (BamHI) as template. Both flanking regions were cloned in pBluescript II KS (SalI/NotI) to yield vector pME1687. A loxP-ODC1MX-loxP cassette as a 1.6-kb BamHI fragment was cloned in the BglII site of this disruption vector. This cassette was constructed from plasmid pUG-ODC1 and is a derivative of the loxP-kanMX-loxP module (24) where the kanamycin resistance gene was replaced by the ODC1 gene of H. polymorpha. The resulting vector from which a 6-kb disruption cassette was released by KpnI digestion is pME1688. For forcing recombination between the loxP sites in strain RH2409, we used plasmid pME1690, which carries the coding sequence of the Cre recombinase (64) as a PCR fragment fused to the FMD promoter as well as the HARO7 gene as an Eco72I/SspI fragment cloned in the SmaI site of pFPMT121. Plasmid pME1689 carries part of the coding region (exon III) of the ACT gene from H. polymorpha DL1-L (W. K. Hong, H. A. Kang, J. H. Shon, E. S. Choi, and S. K. Rhee, unpublished data [GenBank accession no. AF085278]) as a BamHI/ScaI fragment.
Overexpression and purification of H. polymorpha chorismate mutase.
H. polymorpha RH2408 (odc1 FMD promoter::HARO7 URA3) generally was grown at 37°C as shake flask (750 ml) culture in YNB supplemented with 3% glycerol as sole carbon source for FMD promoter derepression. Cells were harvested at an optical density at 546 nm (OD546) of 7 to 8, washed twice with 50 mM potassium phosphate buffer (pH 7.6), and stored in 1 ml of buffer per g of wet cells at −20°C in the presence of protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 0.2 mM EDTA, 1 mM dl-dithiothreitol). For purification, 20 to 30 g of cells was thawed and passed three times through a French pressure cell (18,000 lb/in2). Cell debris was sedimented by centrifugation at 30,000 × g for 20 min. The chorismate mutase enzyme was purified as described by Schmidheini et al. (54), with the modification that for buffer changing or desalting pooled fractions after the first anion-exchange run, gel filtration on a Superdex 75pg column was applied. Chorismate mutase was detected by SDS-PAGE (36) and by enzymatic activity assays. Native PAGE was performed as described by Andersson et al. (1), using a gradient of 4 to 15% polyacrylamide. Protein concentrations were measured by the Bradford assay (5).
Enzyme assays and data evaluation.
Chorismate mutase activity was measured as described previously (54). Enzymatic reactions were carried out at 37°C; as effector concentrations in substrate saturation assays, 100 μM tyrosine and 10 μM tryptophan were chosen. Enzymatic activity was measured spectrophotometrically, determining the concentration of phenylpyruvate. Since absorbance of phenylpyruvate is temperature dependent due to a keto-enol equilibrium, the assay was standardized by keeping the spectrophotometer cell at 30°C. Evaluation of kinetic data was performed as described previously (reference 35 and references therein). Thermal stabilities were determined according to Segel (60), and 3-deoxy-d-arabinoheptulosonic acid (DAHP) synthase activities were measured as described by Teshiba et al. (66).
Sequence alignments.
Sequence analyses were performed using the LASERGENE Biocomputing software from DNASTAR (Madison, Wis.). Alignments were created based on the Lipman-Pearson method (41).
Nucleotide sequence accession number.
The sequence obtained from plasmid pME1525 for the HARO7 gene has been assigned GenBank accession no. AF204738.
RESULTS
The HARO7 gene of H. polymorpha codes for a chorismate mutase enzyme.
The HARO7 gene from the methylotrophic yeast H. polymorpha was cloned by functional complementation of an S. cerevisiae aro7Δ mutant strain. Strains of S. cerevisiae with a deleted ARO7 gene are devoid of endogenous chorismate mutase activity and generally are unable to grow on medium lacking tyrosine or phenylalanine. S. cerevisiae RH2185 (aro7::LEU2 ura3-52) (59) was transformed with genomic DNA of H. polymorpha RB11 (odc1) (69) cloned into the high-copy-number plasmid pRS426 (61). Transformants were selected for viability on minimal medium YNB lacking tyrosine and phenylalanine. One colony appeared after 5 days of growth. A plasmid (pME1524) isolated from this clone was able to complement the auxotrophy of the recipient aro7Δ S. cerevisiae strain. The recipient strain harboring this plasmid grew more slowly than the positive control. Restriction analysis of this plasmid indicated that it contained a genomic DNA insert 5 kb in length. Subcloning of this fragment revealed a 1.7-kb ApaI/Sau3A fragment that was able to complement the Tyr/Phe auxotrophy of S. cerevisiae RH2185 when recloned into pRS426. The recipient strain transformed with this subclone plasmid (pME1525) grew at a rate similar to that of an S. cerevisiae wild-type strain. The DNA insert of pME1525 was subjected to sequence analyses. The genomic sequence is 1,648 bp in length and includes an open reading frame of 843 bp with 281 codons with the capacity to encode a polypeptide with a calculated Mr of 32,067. The 5′ flanking region of the genomic fragment spans 342 nucleotides, whereas the 3′ region is 467 bp in length. Conserved splicing motifs described for yeast (51) are not present within the coding region of the identified gene, indicating the absence of any intron sequences. Upon alignment, the deduced amino acid sequence of the gene exhibited high degrees of identity and similarity to genes of known chorismate mutases of other eukaryotic organisms (Fig. 1). The best alignment was to the enzyme of S. cerevisiae, with 54% identity and 70% similarity when conservative changes are taken into account. With the described primary sequence of chorismate mutase from Schizosaccharomyces pombe, 43% identical and 63% similar residues, including conservative replacements, were found. Comparison to chorismate mutase of the filamentous fungus A. nidulans revealed 43% identity and 65% similarity. The plastidic chorismate mutase of Arabidopsis thaliana is less related; the deduced amino acid sequence is 35% identical to the mature plant enzyme and 58% similar when conservative exchanges are included. Because of this strong similarity to described chorismate mutases and the functional complementation of a chorismate mutase-deficient S. cerevisiae strain, the isolated gene from H. polymorpha was named HARO7, in analogy to the homologous ARO7 gene of S. cerevisiae.
FIG. 1.
Multiple sequence alignment of deduced eukaryotic chorismate mutases including the amino acid sequence of H. polymorpha chorismate mutase. Sources of sequences: HpCM, H. polymorpha (accession no. AF204738); ScCM, S. cerevisiae (accession no. M24517) (54); SpCM, Schizosaccharomyces pombe (accession no. Z98529) (K. Oliver, D. Harris, B. G. Barrell, M. A. Rajandream, and V. Wood, unpublished data); AnCM, A. nidulans (accession no. AF133241) (35); AtCM, Arabidopsis thaliana, mature plastidic enzyme (accession no. Z26519) (10). Conserved residues are boxed in black.
Disruption of HARO7 in H. polymorpha results in tyrosine auxotrophy.
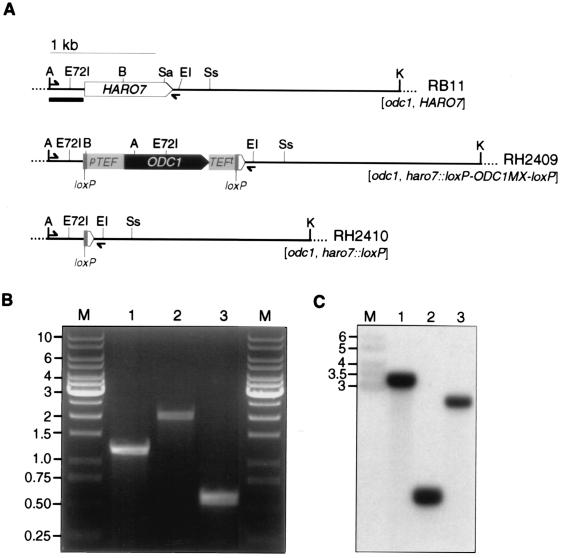
Gene replacement mutant strains were constructed via homologous integration to characterize the HARO7 gene in more detail (Fig. 2A). Therefore, H. polymorpha RB11 was transformed with a disruption cassette consisting of a loxP-ODC1MX-loxP module flanked by 5′ and 3′ homologous sequences of the HARO7 locus. In this construct, 93% of the HARO7 coding sequence is replaced by the marker cassette expressing orotidine-5′-phosphate decarboxylase, which is encoded by the ODC1 gene (46). Transformants were selected on minimal medium supplemented with tyrosine and phenylalanine but lacking uracil. Auxotrophic mutants were identified by replica plating on minimal medium without supplements. Out of approximately 1,000 Ura+ transformants, five independent, auxotrophic clones were isolated, in line with the low frequency of homologous recombination reported previously for H. polymorpha (16). Retransformation with a DNA fragment comprising the HARO7 coding sequence restored prototrophy of these strains, whereas in negative control experiments no transformants were able to grow. A descendant without the ODC1 expression cassette was obtained from one of these clones (RH2409), taking advantage of the loxP recombination sites in the disruption construct (53). For this purpose, H. polymorpha RH2409 was transformed with the autonomously replicating plasmid pME1690, carrying the cre coding sequence inserted between the inducible FMD promoter and the MOX termination region and the HARO7 coding sequence as a marker gene in addition to the S. cerevisiae URA3 gene. Transformants were selected on minimal medium and propagated for 24 h on glycerol-containing medium to derepress expression of the Cre recombinase driven by the FMD promoter. Cured clones in which the ODC1 cassette had been removed by forced homologous recombination between the flanking loxP sites were counterselected on supplemented medium in the presence of 5-fluoroorotic acid (4). One strain (RH2410) isolated by this procedure showed uracil auxotrophy.
FIG. 2.
Construction of haro7Δ mutant strains of H. polymorpha using the Cre-loxP recombination system. (A) Physical maps of the HARO7, haro7::loxP-ODC1MX-loxP, and haro7::loxP loci. Coding sequences of HARO7 and ODC1 are schematically drawn as tipped boxes, promoter and termination sequences of the TEF2 gene from Ashbya gossypii are shown as light gray boxes, and loxP sites are marked in dark gray. Primer positions chosen for diagnostic PCR are indicated by half arrows. The horizontal bar represents the HARO7 promoter-specific probe used in Southern analysis. A, ApaI; B, BglII; E, EcoRI; E72I, Eco72I; K, KpnI; Sa, SalI; Ss, SspI. (B) Diagnostic PCR on genomic DNA of wild-type H. polymorpha HARO7 strain RB11 (lane 1) and haro7Δ mutant strains RH2409 (lane 2) and RH2410 (lane 3) with HARO7-specific 5′ and 3′ oligonucleotides. M, marker DNA fragments of the indicated size in kilobases. (C) Southern hybridization of a specific HARO7 promoter probe on ApaI/KpnI-digested genomic DNA of strains RB11 (lane 1), RH2409 (lane 2), and RH2410 (lane 3). M, nonspecifically hybridizing DNA marker fragments with the indicated length in kilobases.
The correct genotype of both haro7Δ mutant strains was confirmed by diagnostic PCR with oligonucleotides specific for HARO7 5′ and 3′ flanking regions (Fig. 2B). Whereas in the wild-type H. polymorpha HARO7 strain RB11 a 1.2-kb fragment was amplified from genomic DNA, insertion of the loxP-ODC1MX-loxP cassette resulted in a 2-kb amplicon. Removal of the ODC1 expression cassette in strain RH2410 was indicated in the PCR approach by amplification of a 0.5-kb DNA fragment. In Southern analysis, a HARO7 promoter-specific probe was hybridized to ApaI/KpnI-digested genomic DNA of H. polymorpha RB11, RH2409, or RH2410 (Fig. 2C). For RB11, a 3.2-kb signal corresponding to the wild-type HARO7 locus was observed. Insertion of the marker module introduced an additional ApaI site in RH2409, thus shortening the hybridizing signal to 0.8 kb. Due to the removal of this ApaI site within the ODC1 gene, a 1.6-kb signal was detected in Southern analysis of RH2410. As expected, the haro7Δ strains showed no growth on solid, nonsupplemented medium but grew on minimal medium supplemented with standard concentrations of tyrosine, phenylalanine, and uracil. No growth was observed on complex medium YEPD even in the presence of tyrosine and phenylalanine or on synthetic complete medium. Surprisingly, both strains grew slowly but reproducibly on minimal medium containing tyrosine as sole amino acid. Therefore, the haro7Δ mutant strains of H. polymorpha are auxotrophic for the aromatic amino acid tyrosine but bradytrophic for phenylalanine. In liquid cultures of minimal medium, retarded growth was observed only when tyrosine was added at five times the concentration used for supplementation in solid medium. In summary, both mutant strains showed growth defects that depended on the composition of the growth medium, indicating the importance of chorismate mutase activity for growth of H. polymorpha. This conclusion is supported by the fact that strains retransformed with the HARO7 coding sequence, either as a linear DNA fragment or on a plasmid, did not show any of the growth defects described above and were able to grow on complex medium.
HARO7 expression is regulated transcriptionally upon methanol utilization but not upon amino acid starvation.
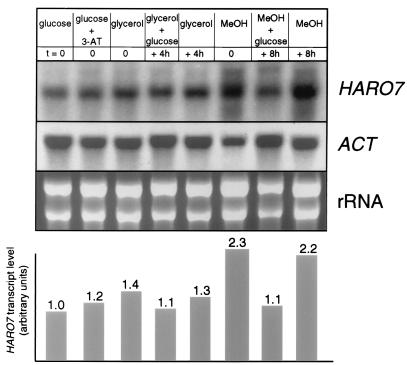
In silico analysis of the flanking 5′ region of HARO7 identified two sequence elements that resemble conserved binding sites for yeast transcription factors. One motif (5′-CACGTG-3′, positions −140 to −135 relative to the translational start codon AUG) matches a binding site for Pho4p (5′-CANNTG-3′) (17), the ultimate effector for phosphate utilization in S. cerevisiae. Additionally, an upstream regulatory sequence specific for H. polymorpha was identified in the HARO7 promoter region. This sequence element (5′-TTGCCACCGGAA-3′, positions −275 to −264) is similar to the core region of a binding site for Mbp1p (5′-TTGCACCGCAA-3′) within the promoter of the MOX gene encoding the peroxisomal methanol oxidase of this methylotrophic yeast (22, 37). A similar motif is found in the promoter of the H. polymorpha CAT1 gene, which codes for a peroxisomal catalase (5′-TCGCACCGCAA-3′) (9). In contrast, no conserved sequence elements directing 3′-end formation were identified in the HARO7 gene fragment of pME1525. The putative Mbp1p-binding element in the HARO7 promoter region implies a transcriptional regulation of HARO7 expression upon methanol utilization. To monitor transcription of the HARO7 gene, steady-state transcript levels were quantified in Northern hybridization analyses. The length of the HARO7-encoded mRNA was determined using an RNA size standard as approximately 0.9 kb (not shown). HARO7 transcription was monitored with respect to availability of different carbon sources, using expression of the ACT gene of H. polymorpha as an internal standard (Fig. 3). To this end, RB11 was cultured in minimal medium containing glycerol as a nonfermentable carbon source or methanol as an inducer of the methanol-utilizing metabolic pathway in H. polymorpha (11). Cells were harvested at mid-exponential phase of growth and 4 or 8 h later for total RNA preparation. In addition, glucose was added to identical cultures grown in the presence of glycerol or methanol, respectively, and cultivation was continued for 4 and 8 h prior to RNA preparation. Northern analysis using ACT transcript levels as internal standard revealed different expression patterns of HARO7 transcription. Transcript levels increased slightly but reproducibly when cells were grown in glycerol-containing medium. Furthermore, methanol as carbon source had a more pronounced effect on HARO7 transcription, with transcript levels increasing by a factor of 2 compared to glucose-grown cells. These effects induced by the nonoptimal carbon sources were diminished when glucose was added to the medium, implying repression of HARO7 transcription.
FIG. 3.
Expression pattern of HARO7 under amino acid starvation conditions or in the presence of different carbon sources. H. polymorpha RB11 was grown in minimal medium containing glucose (lane 1), glucose and 1 mM 3-AT (lane 2), glycerol (lane 3), or methanol (MeOH; lane 6) to mid-log phase (OD546 ≈ 1.2, t = 0), and total RNA was prepared. Additionally, glucose was added to cultures of RB11 grown in glycerol and methanol medium, respectively, and incubation was continued for 4 or 8 h (lanes 4 and 7) before RNA preparation. As a control, RNA was prepared from cultures after prolonged cultivation in the absence of additional glucose (lanes 5 and 8). For Northern analysis, 20 μg of total RNA was loaded in each lane and probed successively with probes specific for HARO7 and ACT of H. polymorpha. Ethidium bromide-stained total RNA is included as a control. Quantified steady-state levels of HARO7 transcripts are shown in the histogram after standardization with respect to ACT transcript levels. The values are averages of two independent experiments with a standard deviation not exceeding 20%.
Chorismate mutase is a key enzyme in aromatic amino acid biosynthesis. Therefore, transcript levels of HARO7 mRNA were quantified under conditions of amino acid starvation. H. polymorpha RB11 was grown in minimal medium supplemented with the false feedback inhibitor 3-amino-1,2,4-triazole (3-AT) at 1 mM to induce histidine starvation and to derepress the general control system of amino acid biosynthesis (3, 28). Specific DAHP synthase activities (EC 4.1.2.15) determined in crude extracts of RB11 grown in the absence or presence of 3-AT, respectively, were used as a control and showed an increase by a factor of 2 (data not shown), indicating that the general control of amino acid biosynthesis had been induced by the false feedback inhibitor (66). The specific chorismate mutase activity was unaffected by the absence or presence of 3-AT (data not shown). Total RNA was prepared from cultures in mid-log phase (OD546 ≈ 1.2) and subjected to Northern analysis. Quantification of signal strength revealed no significant increase of HARO7 transcript levels after the shift to starvation conditions (Fig. 3). We conclude, therefore, that HARO7 transcription is not triggered by the general control system of amino acid biosynthesis.
Chorismate mutase of H. polymorpha is allosterically regulated by tyrosine and tryptophan.
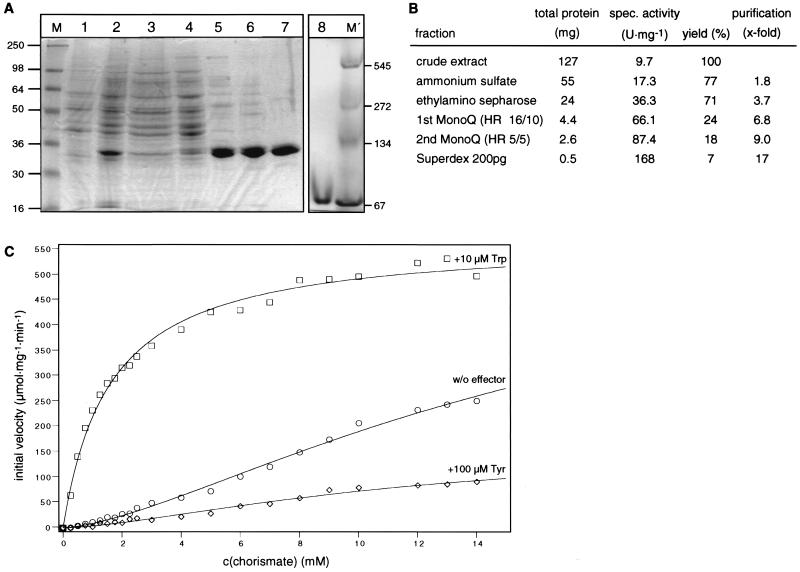
The HARO7 gene product is a key enzyme in the biosynthesis of aromatic amino acids. Chorismate mutase activity has to be regulated stringently to control the flux through the branch point. As no regulation of expression is evident with respect to amino acid availability, we were interested in whether certain amino acids might influence catalytic activity. To determine its enzymatic properties, the HARO7 gene product was overproduced and purified. The HARO7 coding sequence was cloned into the expression vector pFPMT121, where it is fused to the promoter of the H. polymorpha FMD gene coding for formate dehydrogenase (31) and flanked by the termination region of the MOX gene (37). H. polymorpha RB11 was transformed with this expression plasmid (pME1686) and sequentially grown in selective and rich media to obtain mitotically stable transformants (19). One clone (RH2408) analyzed by Southern hybridization was identified to harbor approximately 50 copies of the expression construct ectopically integrated into the genome of the host strain (data not shown). Cultivation in minimal medium containing glycerol as sole carbon source derepressed the FMD-driven expression of HARO7. The resulting chorismate mutase activity was purified to homogeneity from this overexpressing strain by the purification procedure described in Materials and Methods (Fig. 4A and B). In a gradient PAGE under nondenaturating conditions, the purified protein displayed an apparent molecular mass of approximately 70 kDa (Fig. 4A). This indicates that the native enzyme consists of two protomers combined to form a dimeric quaternary structure. Kinetic stop assays for determination of catalytic parameters were performed at 37°C, which is the optimal temperature for growth of H. polymorpha (Fig. 4C; see below). In the absence of effectors, the enzyme showed positive cooperativity toward its substrate chorismate, resulting in a sigmoid substrate saturation curve. An [S]0.5 value of 16.7 mM and a maximal turnover rate of 319.1 s−1 per active site were determined. The calculated Hill coefficient, nH, of 1.56 clearly supports positive cooperativity. Additionally, the regulatory properties of the enzyme were determined by kinetic assays in the presence of allosteric effectors. Tryptophan at 10 μM acts as strong heterotropic positive effector of enzymatic activity due to increased affinity of the enzyme for its substrate. A loss of cooperativity was observed (nH = 0.97), resulting in Michaelis-Menten-type kinetics with a Km of 1.6 mM and a maximum turnover value of 303.8 s−1. In contrast, tyrosine inhibits chorismate mutase activity: the turnover rate decreased to 89.3 s−1, and an [S]0.5 value of 12.0 mM was calculated at a 100 μM concentration of this heterotropic effector. An nH of 1.32 indicates retention of cooperativity.
FIG. 4.
Purification of chorismate mutase of H. polymorpha and enzymatic properties. (A) SDS-polyacrylamide gel of the purification of chorismate mutase of H. polymorpha (left) and native PAGE (right). Lanes: M, marker proteins with the indicated molecular masses in kilodaltons; 1, crude extract of RB11; 2, crude extract of RH2409; 3, supernatant of ammonium sulfate precipitation; 4, ethylamino-Sepharose pool; 5, first MonoQ pool; 6, second MonoQ pool; 7, Superdex 200pg pool; 8, purified Haro7p in a nondenaturing polyacrylamide gel (gradient 4 to 15%); M', marker proteins with the indicated native molecular masses in kilodaltons. A total of 5 μg protein was loaded on each lane. (B) Purification protocol for H. polymorpha chorismate mutase. Enzyme assays were performed in the presence of 500 μM tryptophan and 1 mM chorismate for 10 min. (C) Substrate saturation plot of enzyme assays. Purified H. polymorpha chorismate mutase was assayed with 10 μM tryptophan, without (w/o) effector, or in the presence of 100 μM tyrosine, as indicated. Catalytic turnover was carried out for 1 min, and data were fitted to functions describing cooperative or Michaelis-Menten-type saturation (solid lines). Specific activities are mean values of at least five independent measurements with a standard deviation not exceeding 20%.
In summary, the HARO7-encoded chorismate mutase enzyme of H. polymorpha is strictly regulated in its activity. Whereas HARO7 transcription is constitutive with respect to amino acid starvation and derepressed in the presence of methanol, catalytic turnover is triggered in an allosteric manner by homotropic and heterotropic effectors specific for the biosynthetic pathway of aromatic amino acids.
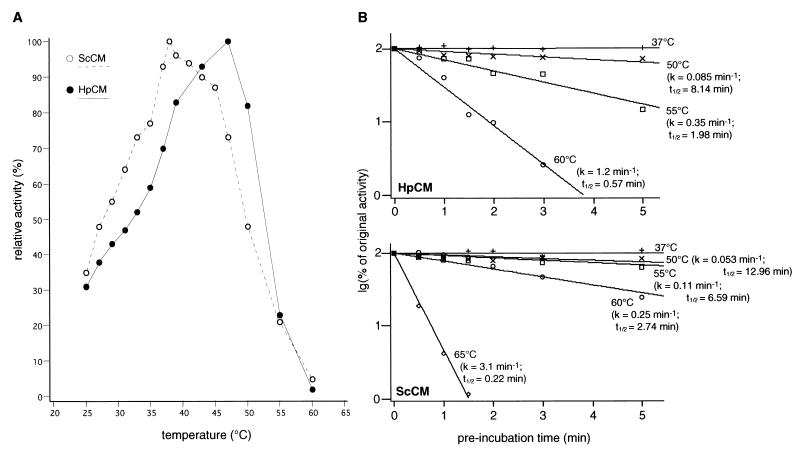
Unliganded H. polymorpha chorismate mutase shows a higher optimal temperature for catalytic turnover than its S. cerevisiae counterpart despite a lower thermal stability.
Kinetic stop assays with the unliganded enzyme were carried out to characterize the temperature profile of catalytic activity of the HARO7-encoded chorismate mutase, and purified yeast chorismate mutase from S. cerevisiae was subjected to identical assays for comparison (Fig. 5A). For the enzyme derived from the thermotolerant yeast H. polymorpha, maximum enzymatic activity was achieved at a temperature of 48°C. In comparison, the S. cerevisiae enzyme shows a decrease in catalytic turnover at temperatures higher than 38°C. With respect to the different maxima of catalytic turnover at elevated temperatures, we were interested in the stability of both enzymes upon incubation at different temperatures. To determine the rate constants in the decrease of catalytic activity due to irreversible denaturation, aliquots of both enzymes were preincubated for different time periods at the specified temperatures before residual chorismate activity was determined in stop assays at low temperature with 2 mM substrate and no effectors present (Fig. 5B). Both enzymes displayed thermal stability at 37°C. After preincubation at 50°C, a decrease in catalytic activity was determined for the H. polymorpha chorismate mutase, with a calculated half-life (t1/2) at this temperature of approximately 8 min. At 55°C, the t1/2 decreased to 2 min; in preincubation experiments at 60°C, catalytic activity displayed a significant decrease over the recorded time period, with a t1/2 of nearly 1 min. For the S. cerevisiae chorismate mutase, t1/2s of 7 and 2.74 min were determined at 55 and 60°C, respectively. After preincubation at 65°C, also for this enzyme there was a sharp drop in catalytic activity, with a deduced t1/2 of 13 s. In conclusion, the chorismate mutase of H. polymorpha displays a lower thermal stability in comparison to its S. cerevisiae homologue, as deduced from the higher rate constants of inactivation at the temperatures used.
FIG. 5.
Temperature profiles and thermal stabilities of chorismate mutases from H. polymorpha and S. cerevisiae. (A) Determination of optimum temperatures for catalytic turnover. Catalytic activities were quantified with purified chorismate mutase enzymes of S. cerevisiae (ScCM) and H. polymorpha (HpCM) in the absence of effectors. Enzyme samples in buffer were preincubated for 5 min at the indicated temperature before chorismate was added to a final concentration of 1 mM to start catalytic turnover. Reactions were stopped after 10 min. Each measurement was performed twice with a standard deviation not exceeding 20%; 100% activity equals 13.1 U (μmol · mg−1 · min−1) for ScCM and 5.6 U for HpCM, respectively. (B) Analysis of rate constants for thermal inactivation. Enzyme samples of purified chorismate mutase of H. polymorpha (HpCM) and S. cerevisiae (ScCM) were preincubated in the absence of effectors for different time periods at the indicated temperature and chilled on ice, and residual activities were determined in stop assays at 37°C (HpCM) and 30°C (ScCM), with 2 mM substrate concentration and 2 min of catalytic turnover after 1 min of incubation at the assay temperature; 100% of original activity equals 49.8 U for ScCM and 15.4 U for HpCM, respectively, and each measurement was performed twice with a standard deviation not exceeding 20%.
DISCUSSION
Chorismic acid, the formerly called elusive branch-point compound (21), is an intermediate of several metabolic pathways, like biosynthesis of ubiquinone and other quinones, 4-aminobenzoate, or aromatic amino acids, in which it is the last common compound of a branched biosynthetic cascade. Conversion of chorismate to prephenate, finally resulting in tyrosine and phenylalanine, is an unusual chemical reaction in primary metabolism that is accelerated by chorismate mutase enzymes up to a factor of 106. In addition, eukaryotic chorismate mutases, especially, have been established as model enzymes for allosteric regulation of catalytic turnover.
We have cloned the H. polymorpha HARO7 gene coding for the chorismate mutase activity of this methylotrophic yeast. This newly identified enzyme extends the number of described sequences that constitute chorismate mutases. The H. polymorpha chorismate mutase is highly similar to the enzyme of the related yeast S. cerevisiae, placing the HARO7 gene product into the AroQ class of chorismate mutases. Additionally, from alignment of the published primary sequences of chorismate mutases a consolidated consensus sequence can be deduced, indicating invariant residues for catalytic activity as well as for regulatory properties. For the yeast enzyme derived from S. cerevisiae, several residues have been identified and characterized in detail with respect to enzymatic function (23, 58, 59). Almost all of these specific amino acids are conserved in the H. polymorpha enzyme, except for the effector-binding residue at position 143. In S. cerevisiae, this position corresponds to Thr145, whereas a methionine is found at this position in the H. polymorpha enzyme. Nevertheless, overall alignment with other enzymes reveals that this particular position is variable in primary sequence. In contrast, a highly conserved region is present within the primary structures, spanning from Cys148 to Phe167 in the H. polymorpha enzyme. Based on crystal structures of the S. cerevisiae enzyme, this protein segment constitutes a helix (helix 8) that is part of the active site as well as of the regulatory site at the dimer interface and that contributes to the strong hydrophobic interaction between the monomers. The general importance of this secondary structure element accounts for its strictly conserved primary sequence. In the global alignment of cloned eukaryotic chorismate mutases, the H. polymorpha enzyme has a unique C-terminal extension. Molecular modeling studies based on the crystal structures determined for the S. cerevisiae enzyme imply an additional turn in the C-terminal helix (not shown), but functionality of this extension with respect to catalytic or regulatory properties remains to be elucidated.
Using a loxP-ODC1MX-loxP cassette, we were able to construct an H. polymorpha strain disrupted in its HARO7 locus (RH2409). Retransformation of HARO7, either as linear DNA fragment or plasmid bound, restored growth of the disruptant on the complex medium YEPD. This clearly supports the idea that the observed growth defect of strain RH2409 is linked to its haro7Δ genotype and not to a background mutation. Surprisingly, the mutant strain showed auxotrophy for tyrosine but not for phenylalanine, with no residual chorismate mutase activity detectable in crude extracts of the disruption strain. One explanation, that the HARO7 gene might encode a bifunctional enzyme like a chorismate mutase-prephenate dehydrogenase activity (T protein), was ruled out because the HARO7 gene was not able to complement a tyr1 mutant strain of S. cerevisiae which lacks prephenate dehydrogenase activity (45). We conclude, therefore, that the HARO7-encoded activity is the only chorismate mutase enzyme in H. polymorpha and that no other redundant catalytic activity is encoded by a homologous gene. This is in agreement with results determined by Bode and Birnbaum, who found no evidence for the occurrence of isoenzymic chorismate mutases in yeasts, among them H. polymorpha (2). The reasons for the unexpected Phe+ phenotype remain obscure, but we speculate that spontaneous, noncatalytic rearrangement of chorismate to prephenate is sufficient in H. polymorpha to feed the tyrosine-specific branch, implying a higher affinity of prephenate to the dehydrogenase activity than to the dehydratase enzyme. Alternatively, H. polymorpha might be able to synthesize phenylalanine via additional routes or from exogenous tyrosine, but both possibilities are unlikely since no catalytic activities sufficient for such pathways have been described so far. By transient expression of the Cre recombinase, we were able to rescue the genetic marker in the haro7 disruption strain RH2409 due to excision of the ODC1 expression construct. The resulting strain, RH2410, was identical in growth phenotypes to its progenitor but in addition required uracil. With respect to biotechnological applications, this strain is a new suitable recipient in the vector-host system of H. polymorpha, as it is able to harbor two expression plasmids carrying different metabolic marker genes. Furthermore, we have demonstrated for the first time that the Cre-loxP recombination system can be applied in H. polymorpha, providing an efficient tool for repeated marker rescue following gene disruptions.
Taking advantage of the vector-host system of H. polymorpha, we were able to overexpress the HARO7 gene in a homologous way and to purify the encoded chorismate mutase to homogeneity in order to characterize its enzymatic properties in detail. Catalytic activity of a given enzyme is generally linked to temperature. We have shown that the H. polymorpha chorismate mutase reaches maximal turnover at a temperature 10°C higher than that of the related yeast S. cerevisiae. In experiments like these, elevated turnover based on increased enzyme-substrate collisions is superimposed by irreversible denaturation of the enzyme. We therefore addressed the question of thermal stability of both enzymes. Surprisingly, the H. polymorpha enzymes displayed higher rate constants of inactivation upon incubation at elevated temperatures in comparison to its S. cerevisiae counterpart. We therefore conclude that the higher optimum temperature for the former is based mainly on increased turnover at the catalytic site. In its allosteric regulation of catalytic activity the H. polymorpha chorismate mutase fits well in the theory of concerted transition as proposed by Monod et al. in 1965 (47). The substrate chorismate acts as homotropic, positive effector that shifts the equilibrium between tense and relaxed state to the more active relaxed state, indicated by a sigmoid curvature of initial velocities as determined in saturation assays as well as by an nH of 1.56. Additionally, this value for nH indicates that the enzyme contains at least two binding sites for the substrate. Furthermore, regulation of catalytic turnover is achieved by heterotropic effects of two aromatic amino acids. Tyrosine, one end product of the chorismate mutase-specific branch, reduces catalytic efficiency by a factor of 2.6 (kcat/Km = 7.44 versus 19.1 mM−1 · s−1), whereas tryptophan, the final product of the opposite branch, increases the kcat/Km by a factor of 9.8 to 188.1 mM−1 · s−1 and abolishes cooperativity. This activating effect of tryptophan is based on increased affinity for the substrate, indicated by the Km of 1.6 mM, and accounts for an allosteric K system. The overall modulation range of catalytic efficiency via both allosteric effectors is given by a factor of 25. Altering catalytic efficiency is one mode of regulation for a given enzyme. An additional and more general way to tune the flux through a metabolic pathway is based on altered expression levels of a gene product. We have shown for the HARO7 gene that transcription is not increased upon the environmental signal of amino acid starvation, as induced by the false feedback inhibitor 3-AT. This is not unusual for chorismate mutase-encoding genes, as neither ARO7 from S. cerevisiae nor aroC from A. nidulans is the target of a cross-pathway control system acting on amino acid starvation in fungi (35, 55). In contrast to this constitutive expression pattern, HARO7 transcription is induced twofold upon methanol utilization, a specific metabolic feature of H. polymorpha, whereas glycerol as nonoptimal carbon source slightly derepresses HARO7 transcription. Both effects are abolished in the presence of glucose, which accounts for a repression system acting on HARO7 transcription. This is the first example of transcriptional regulation of a eukaryotic chorismate mutase-encoding gene. As methanol utilization is accompanied by drastically increased expression of enzymes specific for this pathway, this mode of chorismate mutase expression might reflect the general need for larger amino acid pools in the yeast cell. We have identified a putative binding site for the MOX-binding factor (Mbf1p) in the HARO7 promoter region. This sequence elements differs from the characterized upstream activating sequence found in the MOX promoter by one transversion and one insertion. Nevertheless, this conserved motif is a promising candidate for a positive, cis-acting element triggering HARO7 transcription.
ACKNOWLEDGMENTS
This work was supported by Rhein Biotech GmbH and by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the Volkswagen-Stiftung, and the Niedersächsischen Vorab der Volkswagen-Stiftung, Forschungsstelle für nachwachsende Rohstoffe.
We thank Markus Hartmann for determination of DAHP synthase activities, Silke Busch for critical reading of the manuscript, and all other members of the laboratory for helpful discussions.
REFERENCES
- 1.Andersson L O, Borg H, Mikaelsson M. Molecular weight estimation of proteins by electrophoresis in polyacrylamide gel of graded porosity. FEBS Lett. 1972;20:199–202. doi: 10.1016/0014-5793(72)80793-2. [DOI] [PubMed] [Google Scholar]
- 2.Bode R, Birnbaum D. Regulation of chorismate mutase activity of various yeast species by aromatic acids. Antonie Leeuwenhoek. 1991;59:9–13. doi: 10.1007/BF00582113. [DOI] [PubMed] [Google Scholar]
- 3.Bode R, Schüssler K, Schmidt H, Hammer T, Birnbaum D. Occurrence of the general control of amino acid biosynthesis in yeasts. J Basic Microbiol. 1990;30:31–35. doi: 10.1002/jobm.3620300109. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chook Y M, Ke H, Lipscomb W N. Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc Natl Acad Sci USA. 1993;90:8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross F R, Tinkelenberg A. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 8.Davidson B E, Hudson G S. Chorismate mutase-prephenate dehydrogenase from Escherichia coli. Methods Enzymol. 1987;142:440–450. doi: 10.1016/s0076-6879(87)42055-7. [DOI] [PubMed] [Google Scholar]
- 9.Didion T, Roggenkamp R. Targeting signal of the peroxisomal catalase in the methylotrophic yeast Hansenula polymorpha. FEBS Lett. 1992;303:113–116. doi: 10.1016/0014-5793(92)80500-g. [DOI] [PubMed] [Google Scholar]
- 10.Eberhard J, Raesecke H-R, Schmid J, Amrhein N. Cloning and expression in yeast of a higher plant chorismate mutase. FEBS Lett. 1993;334:233–236. doi: 10.1016/0014-5793(93)81718-f. [DOI] [PubMed] [Google Scholar]
- 11.Eggeling L, Sahm H. Regulation of alcohol oxidase synthesis in Hansenula polymorpha: oversynthesis during growth on mixed substrates and induction by methanol. Arch Microbiol. 1981;127:119–124. doi: 10.1007/BF00428015. [DOI] [PubMed] [Google Scholar]
- 12.Egli H, van Dijken J P, Veenhuis M, Harder W, Fiechter A. Methanol metabolism in yeasts: regulation of the synthesis of catabolytic enzymes. Arch Microbiol. 1980;124:115–121. [Google Scholar]
- 13.Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 14.Faber K N, Haima P, Gietl C, Harder W, Ab G, Veenhuis M. Methylotrophic yeasts as factories for the production of foreign proteins. Yeast. 1995;11:1331–1344. doi: 10.1002/yea.320111402. [DOI] [PubMed] [Google Scholar]
- 15.Faber K N, Haima P, Harder W, Veenhuis M, Ab G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr Genet. 1994;25:305–310. doi: 10.1007/BF00351482. [DOI] [PubMed] [Google Scholar]
- 16.Faber K N, Swaving G J, Faber F, Ab G, Harder W, Veenhuis M, Haima P. Chromosomal targeting of replicating plasmids in the yeast Hansenula polymorpha. J Gen Microbiol. 1992;138:2405–2416. doi: 10.1099/00221287-138-11-2405. [DOI] [PubMed] [Google Scholar]
- 17.Fisher F, Goding C R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem B. The mechanism of the Claisen rearrangement: déjà vu all over again. Angew Chem Int Ed Engl. 1996;35:936–945. [Google Scholar]
- 19.Gatzke R, Weydemann U, Janowicz Z A, Hollenberg C P. Stable multicopy integration of vector sequences in Hansenula polymorpha. Appl Microbiol Biotechnol. 1995;43:844–849. doi: 10.1007/BF02431917. [DOI] [PubMed] [Google Scholar]
- 20.Gellissen G, Hollenberg C P, Janowicz Z A. Gene expression in methylotrophic yeasts. In: Smith A, editor. Gene expression in recombinant microorganisms. New York, N.Y: Marcel Dekker; 1994. pp. 395–439. [Google Scholar]
- 21.Gibson F. The elusive branch-point compound of aromatic amino acid biosynthesis. Trends Biochem Sci. 1999;24:36–38. doi: 10.1016/s0968-0004(98)01330-9. [DOI] [PubMed] [Google Scholar]
- 22.Gödecke S, Eckart M, Janowicz Z A, Hollenberg C P. Identification of sequences responsible for transcriptional regulation of the strongly expressed methanol oxidase-encoding gene in Hansenula polymorpha. Gene. 1994;139:35–42. doi: 10.1016/0378-1119(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 23.Graf R, Dubaquié Y, Braus G H. Modulation of the allosteric equilibrium of yeast chorismate mutase by variation of a single amino acid. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guthrie C, Fink G R, editors. Methods in enzymology. 15. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. p. 15. [Google Scholar]
- 26.Hansen H, Hollenberg C P. Hansenula polymorpha (Pichia angusta) In: Wolf K, editor. Nonconventional yeasts in Biotechnology. Berlin, Germany: Springer-Verlag; 1996. pp. 293–311. [Google Scholar]
- 27.Haslam E. The shikimate pathway. London, England: Butterworth & Co.; 1974. [Google Scholar]
- 28.Heiner C R, Hunkapiller K L, Chen S, Glass J I, Chen E Y. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 1998;8:557–561. doi: 10.1101/gr.8.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilton J L, Kearney P C, Ames B N. Mode of action of the herbicide 3-amino-1,2,4-triazole (amitrole): inhibition of an enzyme of histidine biosynthesis. Arch Biochem Biophys. 1965;112:544–547. doi: 10.1016/0003-9861(65)90093-7. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 31.Hollenberg C P, Janowicz Z A. DNA molecules coding for FMDH control region and structured gene for a protein having FMDH-activity and their uses. European patent EP 0299108-A1. 1989. [Google Scholar]
- 32.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 33.Janowicz Z, Eckart M, Drewke C, Roggenkamp R, Hollenberg C P, Maat J, Ledeboer A M, Visser C, Verrips C T. Cloning and characterization of the DAS gene encoding the major methanol assimilatory enzyme from the methylotrophic yeast Hansenula polymorpha. Nucleic Acids Res. 1985;13:3043–3062. doi: 10.1093/nar/13.9.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janowicz Z A, Melber K, Merckelbach A, Jacobs E, Harford N, Comberbach M, Hollenberg C P. Simultaneous expression of the S and L surface antigens of hepatitis B, and formation of mixed particles in the methylotrophic yeast, Hansenula polymorpha. Yeast. 1991;7:431–443. doi: 10.1002/yea.320070502. [DOI] [PubMed] [Google Scholar]
- 35.Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus G H. The aroC gene of Aspergillus nidulans codes for a monofunctional, allosterically regulated chorismate mutase. J Biol Chem. 1999;274:22275–22282. doi: 10.1074/jbc.274.32.22275. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Ledeboer A M, Edens L, Maat J, Visser C, Bos J W, Verrips C T, Janowicz Z, Eckart M, Roggenkamp R, Hollenberg C P. Molecular cloning and characterization of a gene coding for methanol oxidase in Hansenula polymorpha. Nucleic Acids Res. 1985;13:3063–3082. doi: 10.1093/nar/13.9.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J-D, Komagata K. Taxonomic study of methanol-assimilating yeasts. J Gen Appl Microbiol. 1980;26:133–158. [Google Scholar]
- 39.Lee A, Stewart J D, Clardy J, Ganem B. New insight into the catalytic mechanism of chorismate mutases from structural studies. Chem Biol. 1995;2:195–203. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 40.Levine D W, Cooney C L. Isolation and characterization of thermotolerant methanol-utilizing yeasts. Appl Microbiol. 1973;26:982–990. doi: 10.1128/am.26.6.982-990.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipman D J, Pearson W R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Zheng X, Schnappauf G, Braus G, Karplus M, Lipscomb W N. Yeast chorismate mutase in the R state: simulations of the active site. Proc Natl Acad Sci USA. 1998;95:14640–14645. doi: 10.1073/pnas.95.25.14640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacBeath G, Kast P, Hilvert D. A small, thermostable, and monofunctional chorismate mutase from the archeon Methanococcus jannaschii. Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- 44.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 45.Mannhaupt G, Stucka R, Pilz U, Schwarzlose C, Feldmann H. Characterization of the prephenate dehydrogenase-encoding gene, TYR1, from Saccharomyces cerevisiae. Gene. 1989;85:303–311. doi: 10.1016/0378-1119(89)90422-8. [DOI] [PubMed] [Google Scholar]
- 46.Merckelbach A, Gödecke S, Janowicz Z A, Hollenberg C P. Cloning and sequencing of the ura3 locus of the methylotrophic yeast Hansenula polymorpha and its use for the generation of a deletion by gene replacement. Appl Microbiol Biotechnol. 1993;40:361–364. doi: 10.1007/BF00170393. [DOI] [PubMed] [Google Scholar]
- 47.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 48.Rave N, Crkvenjakov R, Bödtker H. Identification of procollagen mRNAs transferred to diazobenzoyloxymethyl paper from formaldehyde agarose gels. Nucleic Acids Res. 1979;6:3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roggenkamp R, Janowicz Z, Stanikowski B, Hollenberg C P. Biosynthesis and regulation of the peroxisomal methanol oxidase from the methylotrophic yeast Hansenula polymorpha. Mol Gen Genet. 1984;194:489–493. doi: 10.1007/BF00425563. [DOI] [PubMed] [Google Scholar]
- 50.Romero R M, Roberts M F, Phillipson J D. Chorismate mutase in microorganisms and plants. Phytochemistry. 1995;40:1015–1025. doi: 10.1016/0031-9422(95)00010-5. [DOI] [PubMed] [Google Scholar]
- 51.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene Expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 52.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H E, Arnheim N. Enzymatic amplification of β-globin genomic structures and restriction sites analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 53.Sauer B. Functional expression of the Cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidheini T, Mösch H-U, Evans J N S, Braus G. Yeast allosteric chorismate mutase is locked in the activated state by a single amino acid substitution. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 55.Schmidheini T, Mösch H-U, Graf R, Braus G H. A GCN4 protein recognition element is not sufficient for GCN4-dependent regulation of transcription in the ARO7 promoter of Saccharomyces cerevisiae. Mol Gen Genet. 1990;224:57–64. doi: 10.1007/BF00259451. [DOI] [PubMed] [Google Scholar]
- 56.Schmidheini T, Sperisen P, Paravicini G, Hütter R, Braus G. A single point mutation results in a constitutively activated and feedback-resistant chorismate mutase of Saccharomyces cerevisiae. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnappauf G, Krappmann S, Braus G H. Tyrosine and tryptophan act through the same binding site at the dimer interface of yeast chorismate mutase. J Biol Chem. 1998;273:17012–17017. doi: 10.1074/jbc.273.27.17012. [DOI] [PubMed] [Google Scholar]
- 58.Schnappauf G, Lipscomb W N, Braus G H. Separation of inhibition and activation of the allosteric yeast chorismate mutase. Proc Natl Acad Sci USA. 1998;95:2868–2873. doi: 10.1073/pnas.95.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnappauf G, Sträter N, Lipscomb W N, Braus G. A glutamate residue in the catalytic center of the yeast chorismate mutase restricts enzyme activity to acidic conditions. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segel I H. Enzyme kinetics. Classics Library Edition, New York: Wiley; 1975. pp. 926–929. , N.Y. [Google Scholar]
- 61.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 63.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 64.Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- 65.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Mechanism of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 66.Teshiba S, Furter R, Niederberger P, Braus G, Paravicini G, Hütter R. Cloning of the ARO3 gene of Saccharomyces cerevisiae and its regulation. Mol Gen Genet. 1986;205:353–357. doi: 10.1007/BF00430450. [DOI] [PubMed] [Google Scholar]
- 67.Veenhuis M, van Dijken J P, Pilon S A F, Harder W. Development of crystalline peroxisomes in methanol-grown cells of the yeast Hansenula polymorpha and its relation to environmental conditions. Arch Microbiol. 1978;117:153–163. doi: 10.1007/BF00402303. [DOI] [PubMed] [Google Scholar]
- 68.Weiss U, Edwards J M. The biosynthesis of aromatic amino acids. New York, N.Y: John Wiley & Sons; 1980. [Google Scholar]
- 69.Weydemann U, Keup P, Piontek M, Strasser A W, Schweden J, Gellissen G, Janowicz Z A. High-level secretion of hirudin by Hansenula polymorpha—authentic processing of three different preprohirudins. Appl Microbiol Biotechnol. 1995;44:377–385. doi: 10.1007/BF00169932. [DOI] [PubMed] [Google Scholar]
- 70.Woodcock D M. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue Y, Lipscomb W N, Graf R, Schnappauf G, Braus G. The crystal structure of allosteric chorismate mutase at 2.2 Å resolution. Proc Natl Acad Sci USA. 1994;91:10814–10818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]