Abstract
In an attempt to dissect the virulence regulatory mechanism in Vibrio vulnificus, we tried to identify the V. cholerae transmembrane virulence regulator toxRS (toxRSVc) homologs in V. vulnificus. By comparing the sequences of toxRS of V. cholerae and V. parahaemolyticus (toxRSVp), we designed a degenerate primer set targeting well-conserved sequences. Using the PCR product as an authentic probe for Southern blot hybridization, a 1.6-kb BglII-HindIII fragment and a 1.2-kb HindIII fragment containing two complete open reading frames and one partial open reading frame attributable to toxRVv, toxSVv, and htpGVv were cloned. ToxRVv shared 55.0 and 63.0% sequence homology with ToxRVc and ToxRVp, respectively. ToxSVv was 71.5 and 65.7% homologous to ToxSVc and ToxSVp, respectively. The amino acid sequences of ToxRSVv showed transmembrane and activity domains similar to those observed in ToxRSVc and ToxRSVp. Western blot analysis proved the expression of ToxRVv in V. vulnificus. ToxRSVv enhanced, in an Escherichia coli background, the expression of the V. vulnificus hemolysin gene (vvhA) fivefold. ToxRSVv also activated the ToxRVc-regulated ctx promoter incorporated into an E. coli chromosome. A toxRVv null mutation decreased hemolysin production. The defect in hemolysin production could be complemented by a plasmid harboring the wild-type gene. The toxRVv mutation also showed a reversed outer membrane protein expression profile in comparison to the isogenic wild-type strain. These results demonstrate that ToxRVv may regulate the virulence expression of V. vulnificus.
Vibrio vulnificus is a halophilic estuarine bacterium that causes fatal septicemia and necrotizing wound infections. Primary septicemia occurs following ingestion of raw seafood contaminated with V. vulnificus. V. vulnificus preferentially affects persons with underlying hepatic diseases, a heavy alcohol drinking habit, and other immunocompromised conditions. Primary septicemia shows a rapidly progressing and fulminant course, which results in a high mortality rate of over 50% despite aggressive antimicrobial and supportive shock therapies (38, 39, 48).
The optimal natural habitat of V. vulnificus is an estuary. The bacterium normally flourishes in estuarine seawater, shellfish, and plankton during warm months (8, 35, 50, 54). V. vulnificus is concentrated in oysters and probably in other shellfish as well (21, 49). V. vulnificus opportunistically causes primary septicemia when contaminated shellfish is eaten raw by susceptible patients. This opportunist should experience a very dramatic change in environmental parameters during the infection process. Successful infection by pathogenic bacteria, in general, is established by coordinate expression of various virulence factors in vivo. Expression of virulence factors is controlled by environmental cues. Pathogenic bacteria possess elegant regulatory systems that sense and react to fluctuations in environmental parameters such as temperature, osmolarity, pH, iron concentration, CO2 concentration, etc. (12, 27).
Many pathogens employ novel signal transduction systems in regulating the virulence gene expression (12). Toxigenic V. cholerae has the toxRS system for that purpose (30). The genes toxR and toxS are clustered in an operon and encode transmembrane proteins ToxR and ToxS, respectively (10, 29, 32). ToxR regulates expression of multiple V. cholerae virulence factors such as the cholera toxin (ctx), toxin-coregulated pilus (tcp), and accessory colonization factor (acf) genes (9, 47). The activity of ToxR is further enhanced by ToxS, which interacts with the former protein in the periplasmic space and is thought to stabilize it (10, 36). The toxRS system seems to play universally important roles in the survival and host-microorganism interaction of Vibrio species. V. parahaemolyticus and V. fischeri also have homologs of the V. cholerae toxRS (toxRSVc) system (25, 43). In the present study, we identified the toxRS homolog in V. vulnificus and showed the functional homology of ToxRSVv with ToxRSVc. We also observed that ToxRVv regulates the production of hemolysin, the most potent exotoxin produced by the organism (14).
MATERIALS AND METHODS
Bacterial strains, media, plasmids, and reagents.
The Escherichia coli and V. vulnificus strains and plasmids used in this study are listed in Table 1. V. vulnificus type strain ATCC 29307 was used for the toxRSVv cloning experiment. For mutant construction and functional studies of ToxRSVv, we used the highly virulent clinical isolate V. vulnificus MO6-24/O. E. coli and V. vulnificus strains were grown in LB medium (28) and 2.5% NaCl brain heart infusion medium (6), respectively. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; tetracycline, 20 μg/ml; kanamycin, 30 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| V. vulnificus | ||
| ATCC 29307 | Type strain | ATCCa |
| MO6-24/O | Clinical isolate | J. G. Morris, Jr. |
| CMM981 | MO6-24/O with toxR null mutation | This study |
| E. coli strains | ||
| DH5α | F−recA1; restriction negative | Laboratory collection |
| VM2 | SY327 [1NFVM1 F(ctx′-lacZ+)] | 29 |
| SY327 λ pir | D(lac pro) argE(Am) rif nalA recA56 λ pir lysogen | 30 |
| SM10 λ pir | thi thr leu tonA lacY supE recA::RP4-2-Tcr:Mu Kmr λ pir lysogen | 30 |
| INVαF′ | Competent cell line used for PCR TA cloning | Invitrogen |
| XL2 blue-MRF′ | Competent cell line used for genomic library construction | Stratagene |
| Plasmids | ||
| pCRII | PCR TA cloning vector | Invitrogen |
| pBAD24 | PBAD expression vector | 14 |
| pACYC184 | Tcr Cmr | Laboratory collection |
| pUC18, pUC19 | Apr α-lac | Laboratory collection |
| pGEX-2T | GST fusion expression vector | Pharmacia |
| pGEX-3X | GST fusion expression vector | Pharmacia |
| pNQ705 | Suicide vector with R6K origin; Cmr | 32 |
| pLAFR3 | IncP broad-host-range plasmid; Tcr | Laboratory collection |
| pCVD702 | 3.4-kb fragment containing vvh cloned into HindIII-EcoRI sites of pBR325 | 55 |
| pJL961 | 3.4-kb vvh fragment in pCVD702 cloned into SalI site of pACYC184 | This study |
| pCMM9 | pCRII containing PCR-amplified 864-bp fragment carrying parts of toxRVv and toxSVv | This study |
| pCMM12 | pCRII containing PCR-amplified 1,450-bp fragment carrying entire toxRSVv genes | This study |
| pCMM51 | pUC19 containing 1,582-bp HindIII-BglII fragment carrying part of toxRVv and entire toxS gene | This study |
| pCMM74 | pUC19 containing 1,115-bp HindIII fragment carrying parts of toxRVv and htpGVv genes | This study |
| pCMM601 | toxRSVv cloned into EcoRI site of pBAD24 | This study |
| pCMM610 | toxRVv cloned into EcoRI site of pBAD24 | This study |
| pCMM641 | toxRSVc cloned into KpnI-EcoRI sites of pBAD24 | This study |
| pCMM651 | toxRVc cloned into KpnI-EcoRI sites of pBAD24 | This study |
| pCMM700 | KpnI-HindIII fragment of pCMM9 cloned into pNQ705 | This study |
| pCMM709 | toxRVv cloned into BamHI-EcoRI sites of pGEX-3X | This study |
| pCMM820 | toxSVv cloned into BamHI-EcoRI sites of pGEX-2T | This study |
| pCMM2001 | 2.7-kb HindIII-HindIII-BglII fragment containing toxRSVv operon and part of htpGVv cloned into pLAFR3 | This study |
ATCC, American Type Culture Collection.
DNA manipulations were performed as previously described (44) and in accordance with the recommendations of the restriction enzyme manufacturer (Boehringer Mannheim GmbH, Mannheim, Germany). To reduce the replication errors accompanying PCR, the Expand High Fidelity PCR System (Boehringer Mannheim) was used for cloning.
Amplification of a putative toxRSVv DNA fragment by degenerate PCR.
A DNA fragment used as an authentic probe for the screening of plasmid libraries was amplified by degenerate PCR. By comparing the toxRS genes of V. cholerae and V. parahaemolyticus, we designed a set of degenerate primers (sense primer SCH961 [5′-CGATTAGGNAGCAACGAAAGCCG-3′] and antisense primer SCH962 [5′-GTCACTNCCCCANTANAACC-3′]) encompassing parts of toxR and toxS. Chromosomal DNA of V. vulnificus ATCC 29307, prepared as described elsewhere (24), was used as the template for PCR amplification. PCR was performed with a water-cooled thermal cycler (Single Block Easycycler; Ericomp Inc., San Diego, Calif.).
The amplified DNA band on a 1.0% agarose gel was cut out, and DNA was eluted by the QIAEX II Gel Extraction Kit (QIAGEN, Hilden, Germany). The extracted DNA fragments were cloned into the pCRII vector using the Original TA Cloning Kit (Invitrogen, NV Leek, The Netherlands). The resulting plasmid was designated pCMM9 and used for DNA sequencing and restriction mapping. Sequencing was done by the double-strand dideoxy-chain termination method using the Sequenase Version 2.0 DNA Sequencing Kit (USB, Cleveland, Ohio).
Cloning of toxRSVv from plasmid libraries.
The chromosomal DNA of V. vulnificus ATCC 29307 was fully digested with various restriction enzymes, electrophoresed, and transferred to nitrocellulose membranes (Bio-Rad, Hercules, Calif.). Southern blot analysis was performed as described by Sambrook et al. (44). The insert DNA of pCMM9 and the two fragments resulting from its HindIII digestion were used as probes. The 222-bp PCR product of the vvh hemolysin gene was used as a control probe (24). The probes were labeled by [32P]dCTP (DuPont NEN, Boston, Mass.) using the nick translation system or the random priming system of Promega (Madison, Wis.), depending upon the probe length.
On the basis of the Southern blot analysis results, 4.0-kb BglII and 1.2-kb HindIII fragments hybridizing with the probes were used for plasmid library construction. DNA fragments of appropriate sizes were cut from the agarose gel after electrophoresis and extracted with the QIAEX II Gel Extraction Kit (QIAGEN). The insert DNA fragments were ligated to pUC19 cut by appropriate enzymes and treated with shrimp alkaline phosphatase (Boehringer Mannheim). The resulting ligates were transformed into ultracompetent E. coli XL2 blue-MRF′ cells (Stratagene, La Jolla, Calif.). The libraries were screened by the colony blot hybridization method (44). The longer (ca. 600-bp) EcoRI-HindIII fragment from pCMM9 was used as the probe in a screening of the HindIII library, while the shorter (ca. 300-bp) HindIII-EcoRI fragment was used to clone the 1.6-kb HindIII-BglII fragment from the BglII library. The plasmid clones with appropriate insert sizes were tested by restriction mapping and Southern blot analysis. Most probable candidate clones were used for DNA sequencing as described above. DNA and deduced amino acid sequences were analyzed by MacDNASIS software version 3.6 (Hitachi Software Engineering Co., San Bruno, Calif.).
Expression of ToxR and ToxS as fusion proteins.
Expression of the toxRVv and toxSVv genes as proteins was tested by using the Glutathione S-Transferase (GST) Gene Fusion System (Amersham Pharmacia Biotech, Piscataway, N.J.). For this study, fragments encoding toxRVv and toxSVv were amplified by PCR and cloned into the pCRII TA cloning vector. For directional cloning into GST fusion vectors at the BamHI-EcoRI site, the primers were designed to contain BamHI or EcoRI restriction sites at their 5′ regions. toxRVv was amplified by primers GSTR1 (5′-CGGGATCCTCATGAAGTAATACCGGCA-3′) and GSTR2 (5′-CGGAATTCTTATTTACAGATAGAACC-3′). toxSVv was amplified by primers GSTS1 (5′-CGGGATCCATGAAACTTCGAATTGCT-3′) and GSTS2 (5′-CGGAATTCTCAGTTAGAAAACAGTAC-3′). Boldface and underlined letters denote restriction enzyme recognition sites and the putative start or stop sites of the genes, respectively. toxRVv was cloned into pGEX-3X, and toxSVv was cloned into pGEX-2T. The resulting plasmids were transformed into E. coli DH5α, and the transformants were induced by isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma, St. Louis, Mo.) in accordance with the manufacturer's protocol. The expressed GST-ToxR and GST-ToxS proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the bacterial lysate and subsequent Coomassie staining.
Production of anti-ToxR serum.
For every batch culture, expression of the fusion protein was confirmed by SDS-PAGE and Coomassie staining. After induction, the culture was harvested by centrifugation. To prepare a bacterial lysate for affinity column chromatography, the pellet was resuspended in lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.5 mM EDTA [pH 8.0], 1 mM benzamidine HCl, 0.2 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 0.1% β-mercaptoethanol) and sonicated (Vibra Cell Sonicator Model CV26; Sonics & Materials Inc., Danbury, Conn.) on an ice bed. After sonication, the GST-ToxRVv fusion protein in the lysate was purified by the affinity chromatographic method recommended by the manufacturer (Amersham Pharmacia Biotech).
A New Zealand White rabbit was immunized intradermally with the eluted fusion protein completely mixed with same volume of complete Freund's adjuvant (Gibco BRL, Gaithersburg, Md.) at a 2-week interval. One month after the second immunization, a final booster injection was carried out using the pure fusion protein in SDS-PAGE gel slices. The gel slices containing the pure fusion protein band were frozen with liquid nitrogen and ground in a mortar. The resulting powder was suspended with phosphate-buffered saline (PBS) and injected intradermally. One week after the last booster immunization, the serum antibody titer was tested.
Western blot analysis of ToxRVv expression.
ToxR expression in the V. vulnificus and E. coli backgrounds was analyzed by Western blot analysis using the rabbit antiserum as the primary polyclonal antibody. E. coli VM2 harboring pCMM610 was used as the positive control. Protein bands separated by SDS-PAGE were transferred to nitrocellulose paper (Bio-Rad). The paper was first treated with blocking buffer (5% skim milk, 0.1% Tween 20 in PBS) for 1 h and then washed three times with washing buffer (1% skim milk, 0.5% bovine serum albumin, 0.1% Tween 20 in PBS) for 10 min each time. The antiserum diluted 1:500 was incubated with the washed paper strip for 1 h with gentle agitation. After washing, alkaline phosphatase-conjugated anti-rabbit immunoglobulin G antibody (Sigma) diluted 1:15,000 was reacted for 1 h. After thorough washing, the strip was dipped in a 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Sigma) substrate solution. All reaction and washing steps were performed at room temperature.
Stimulation of ctx promoter by ToxRVv.
The functional similarity of ToxRVv to its V. cholerae counterpart was tested in an E. coli background. PBAD expression plasmids pCMM601 and pCMM610 were transformed into E. coli VM2 harboring a ctx::lacZ fusion in the chromosome (30). E. coli VM2 transformed with either pCMM610 (toxRVv) or pCMM601 (toxRSVv) was induced by 0.2% arabinose as described by Guzman et al. (15). β-Galactosidase assays (28) of the culture aliquots collected at appropriate time intervals were carried out in triplicate. The same experiments were done with pCMM641 (toxRSVc) and pCMM651 (toxRVc) as positive controls.
Interaction of ToxRSVv with vvhA in an E. coli background.
Regulation of vvhA gene expression by ToxRVv or ToxRSVv was investigated in an E. coli background. E. coli DH5α harboring pJL961, a derivative of pACYC184 containing a 3.4-kb vvhA fragment from pCVD702 (56), was transformed with pCMM601, pCMM610, pCMM641, or pCMM651. The cotransformants were grown in NZY broth (15) containing both chloramphenicol and ampicillin. Induction of the PBAD promoter by arabinose was done as described above. At appropriate time intervals after induction, culture supernatants were collected for hemolysin assay. Hemolysin activity in the supernatant was assayed in triplicate by the tube method as previously described (45). Western blot analysis of the ToxRVv levels in the E. coli background was performed to address whether coexpression of ToxSVv increased ToxRVv stability and contributed to significant hemolysin production while the expression of ToxRVv alone could not induce hemolysin production.
Construction of a toxR mutant.
To investigate the physiological role of toxR in hemolysin production by V. vulnificus, a toxR mutant of strain MO6-24/O was constructed by inserting a suicide vector into the chromosome using the strategy originally described by Miller and Mekalanos (31). A KpnI-HindIII toxRVv fragment from pCMM9 was ligated to suicide vector pNQ705 (33). The toxRVv fragment lacks about 100 and 300 bp at the 5′ and 3′ ends, respectively, of the toxRVv open reading frame. The ligated DNA was used to transform competent cells of E. coli SY327 λ pir. Chloramphenicol-resistant (Cmr) transformants were selected, and plasmid DNA was analyzed by restriction mapping to identify a plasmid, designated pCMM700, carrying the correct insert. Plasmid pCMM700 was transformed into E. coli SM10 λ pir and subsequently transferred to V. vulnificus MO6-24/O by conjugation. Cmr transconjugants, which contain the mobilized plasmid integrated into the genome by homologous recombination, were selected on thiosulfate-citrate-bile-sucrose (TCBS) agar plates containing chloramphenicol at 2 μg/ml. Insertional mutation was confirmed by Southern hybridization and PCR analysis of the chromosomal DNAs of the mutant and the wild type (26, 31). The truncated toxRVv insert was labeled with the nonradioactive digoxigenin labeling kit (Boehringer Mannheim) and used as the probe for Southern blot hybridization. The difference in hemolysin production between the mutant and parent wild-type strains was determined by culturing both strains in 2.5% NaCl heart infusion broth at 37°C and 220 rpm. Hemolytic activity in the supernatant was measured as described above.
OMP profile.
Changes in the outer membrane protein (OMP) profile of the toxRVv mutant were tested. OMPs were purified by a method reported elsewhere (7). Briefly, bacterial cells were harvested by centrifugation at 3,000 × g for 15 min. The pellets were washed three times with 0.8 M sucrose in 50 mM Tris-HCl buffer (pH 8.0). Spheroplasts were made by suspending the pellets in 50 mM Tris-HCl buffer (pH 8.0) containing lysozyme at 10 μg/ml and 2.5 mM EDTA and incubated at 37°C for 30 min. Spheroplasts were pelleted by centrifugation at 8,000 × g for 15 min. The supernatants were collected and pelleted at 30,000 × g for 30 min. The resulting pellets were treated with 1.5% Sarkosyl at room temperature for 20 min. After this treatment, OMPs were pelleted at 30,000 × g for 30 min. The OMP preparations were analyzed by SDS–12% PAGE.
For determination of the amino-terminal amino acid sequences of the two major OMP bands, the OMPs from V. vulnificus MO6-24/O were subjected to SDS–12% PAGE and then blotted onto a Westran polyvinylidene difluoride transfer membrane (Schleicher & Schuell Co.). After staining with Coomassie blue for 60 s or less, the two major bands were excised, destained with 50% methanol, and subjected to amino-terminal sequencing with an Applied Biosystems automated sequencer. This analysis was performed at the Korea Basic Science Institute in Daejun City, Korea.
Nucleotide sequence accession number.
The complete toxRSVv and partial htpGVv DNA sequences have been deposited in the GenBank database under accession number AF166120.
RESULTS
Identification and analysis of toxRSVv.
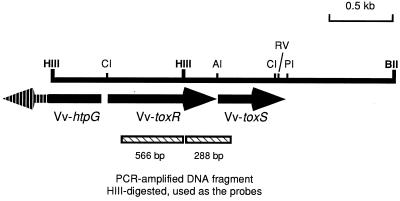
By using degenerate primers, ca. 900-bp DNA fragments were amplified from the chromosomal DNAs of V. vulnificus ATCC 29307, V. cholerae ATCC 14033, and V. parahaemolyticus ATCC 17802. All three of the Vibrio species gave the same size of amplification product. The ca. 900-bp DNA fragment amplified from V. vulnificus ATCC 29307 was cloned into the pCRII vector and sequenced. Restriction and DNA sequence analysis showed that the insert was 854 nucleotides in length and had a HindIII site yielding 566- and 288-bp fragments after digestion with the restriction enzyme (Fig. 1). The DNA sequence of the insert showed 61.2, 46.9, and 47.8% homologies to the corresponding regions in the toxRS genes of V. parahaemolyticus, V. cholerae, and V. fischeri, respectively. This insert was used, with or without HindIII digestion, as the authentic probe in further studies to identify the chromosomal copy of toxRSVv.
FIG. 1.
Restriction map of the 2.8-kb HindIII-HindIII-BglII DNA fragment cloned from V. vulnificus ATCC 29307. The fragment was cloned as separate 1.2-kb HindIII and 1.6-kb HindIII-BglII fragments. The arrows indicate the locations and directions of transcription of two complete open reading frames (toxRVv and toxSVv) and one partial open reading frame (htpGVv). Restriction sites are abbreviated as follows: AI, Alw44I; BII, BglII; CI, ClaI; HIII, HindIII; PI, PvuI; RV, EcoRV. The hatched boxes indicate fragments of the PCR-amplified DNA product used as the authentic probes. The whole product or the two fragments resulting from HindIII digestion were labeled by [32P]dCTP and used in the genomic Southern blot hybridization analysis.
Southern blot hybridization analysis of V. vulnificus chromosomal DNA showed that a 4.0-kb BglII-digested fragment hybridized with the 854-bp probe. DNA fragments of ca. 4.0 kb were eluted from the agarose gel and ligated into the BamHI site of pUC19. The resulting chromosomal DNA library was transformed into ultracompetent E. coli XL-2 blue MRF′ cells, and the transformants were screened by the colony blot hybridization method. Unfortunately, the 4.0-kb target fragments appeared to be lethal to the host E. coli cells. Thus, we took advantage of the information that the PCR-amplified DNA fragment has a HindIII restriction site inside. The 4.0-kb BglII fragments were digested with HindIII and cloned into the BamHI-HindIII site of pUC18 and the HindIII-BamHI site of pUC19. A 1.6-kb HindIII-BglII fragment was successfully cloned, while the 2.4-kb BglII-HindIII fragment of the opposite side could not be cloned by any means. The 1.6-kb DNA fragment hybridized only with the smaller 288-bp fragment of the PCR-amplified insert produced by digestion with HindIII (Fig. 1). Southern blot analysis of HindIII-digested chromosomal DNA revealed a 1.2-kb HindIII fragment that seemed to contain the 5′ part of the toxRSVv operon. The 1.2-kb HindIII fragment was cloned into pUC19 and hybridized only with the larger 566-bp fragment after digestion with HindIII (Fig. 1). The sequences of the two clones showed homologies to those of the toxRS genes of V. cholerae, V. parahaemolyticus, and V. fischeri.
The 2.8-kb DNA fragment could be reconstituted by ligation of the 1.2-kb HindIII and 1.6-kb HindIII-BglII fragments at the HindIII site. The intact toxRSVv fragment in the chromosome could also be identified and cloned by PCR. Sequence analysis of the DNA fragment revealed two complete open reading frames and one partial open reading frame (Fig. 2).
FIG. 2.
Nucleotide sequence of the locus containing the toxRSVv genes. The arrows indicate the directions of transcription of open reading frames. The partial open reading frame of htpGVv runs opposite to the toxRSVv operon. Putative Shine-Dalgarno sequences are underlined. The underlined, italicized sequences were designated the putative initiation or stop codons of the open reading frames.
The ATG start codon of toxRVv could be assigned to positions 449 through 451. A putative ribosome-binding Shine-Dalgarno sequence was found 4 nucleotides upstream of the ATG codon (Fig. 2). The termination codon was assigned to position 1,319. toxRVv shared 50.5 and 60.8% nucleotide sequence homology with toxRVc and toxRVp, respectively. The deduced amino acid sequence of ToxRVv showed 55.0 and 63.0% homology with ToxRVc and ToxRVp. A putative DNA-binding domain was found at the amino-terminal region of ToxRVv (indicated by solid underlining in Fig. 3). The region showed very high identity to the DNA-binding regions of ToxRVc and ToxRVp, as well as to the other well-known DNA-binding domains of OmpR, VirG, PhoP, and PhoB (25, 32, 37). A very hydrophobic region, which could be designated the membrane-spanning domain (32), composed of 13 amino acids with an α-helical configuration inferred by computer analysis, was noted in the central part of ToxRVv (indicated by broken underlining in Fig. 3). The deduced amino acid sequence of the intervening region between the DNA-binding and transmembrane domains of ToxRVv was not very well conserved with respect to that of ToxRVc or ToxRVp. The carboxy-terminal region, which is supposed to act as the signal sensor and interact with ToxS (10, 36), showed high similarity to that of ToxRVc or ToxRVp.
FIG. 3.
Comparison of ToxRVv with ToxRVc and ToxRVp at the deduced amino acid sequence level. The amino acid sequences were aligned for maximum homology using the MacDNASIS software (Hitachi). Dashes indicate missing sequences. Identical sequences are indicated by a black background. Solid underlining indicates the region supposed to be the transcription activation domain that binds to target promoter sequences (24, 31). Broken underlining indicates the putative transmembrane domain of ToxRVv (24, 31).
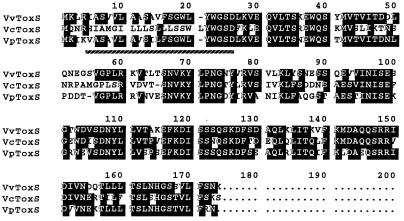
toxSVv started downstream of toxRVv with an intergenic space of 12 bp, whereas those of toxRSVc and toxRSVp are 9 and 11 bp (10, 25), and toxSVf initiates inside the termination codon of toxRVf (43). The initiation codon of toxSVv could be assigned to position 1,334. A putative ribosomal binding site was observed at the intergenic sequence (Fig. 2). The deduced amino acid sequence of ToxSVv showed 71.5 and 65.7% homology with ToxSVc and ToxSVp, respectively (Fig. 4). The very hydrophobic region at the amino terminus (underlined with a broken line) was presumed to be a transmembrane domain, as was reported in ToxSVc and ToxSVp (10, 25).
FIG. 4.
Comparison of ToxSVv with ToxSVc and ToxSVp at the deduced amino acid sequence level. The amino acid sequences were aligned for maximum homology using the MacDNASIS software (Hitachi). Dashes indicate missing sequences. Identical sequences are indicated by a black background. Broken underlining indicates the putative transmembrane domain of ToxSVv (24, 28).
A partial open reading frame showing high identity to htpGVc was found upstream and oriented in the opposite direction of the toxRSVv operon (Fig. 2). The 72-amino-acid derived sequence was well (ca. 90%) conserved with respect to those of htpGVc and htpGVf (Fig. 5). The intergenic space between htpGVv and toxRVv was 222 bp, in comparison with 189 bp for V. cholerae and 230 bp for V. fischeri.
FIG. 5.
Comparison of HtpGVv with HtpGVc (31) and HtpGVf (42) at the deduced amino acid sequence level. The amino acid sequences were aligned for maximum homology using the MacDNASIS software (Hitachi). Dashes indicate missing sequences. Identical sequences are indicated by a black background.
Expression of ToxR and ToxS.
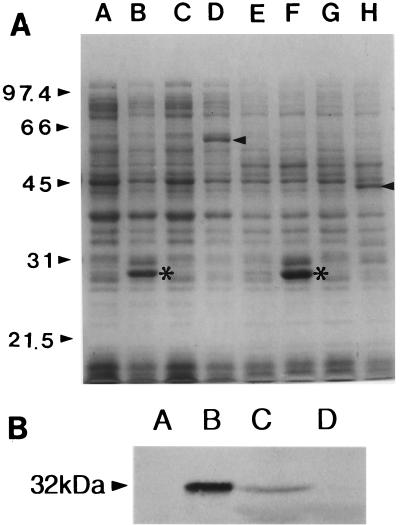
We tested whether the putative toxRVv and toxSVv open reading frames could express proteins of the expected sizes. We expressed the genes as GST fusion proteins. When fusion plasmids pCMM709 (toxRVv) and pCMM820 (toxSVv) were induced with IPTG, ToxRVv and ToxSVv were successfully expressed. The molecular masses of ToxRVv and ToxSVv were estimated to be 32 and 19 kDa, respectively, on the gel (Fig. 6A). The molecular masses of ToxR and ToxS were estimated to be the same by DNA sequence analysis.
FIG. 6.
Observation of toxRSVv expression by SDS-PAGE (A) and Western blot analysis (B). (A) Expression of ToxRVv and ToxSVv as GST fusion proteins. The molecular masses of ToxR and ToxS were estimated to be 32 and 19 kDa, respectively. Molecular size markers are indicated on the left. In lanes A through H, each pair represents lysates of E. coli DH5α containing corresponding plasmids before and after IPTG induction as follows: A and B, pGEX-3X; C and D, pCMM709; E and F, pGEX-2T; G and H, pCMM820. Arrowheads indicate expressed fusion proteins. Asterisks indicate GST bands expressed after induction. (B) Western blot analysis of ToxRVv expression in V. vulnificus MO6-25/O. Lanes: A, E. coli DH5α harboring pCMM610 without arabinose induction; B, same as lane A except that the cells were induced by 0.2% arabinose; C, V. vulnificus MO6-24/O, the isogenic wild-type strain; D, V. vulnificus CMM981, the toxRVv null mutant. The molecular mass on the left is that of ToxRVv.
The expression of ToxR in V. vulnificus ATCC 29307 was further confirmed by Western blot analysis (Fig. 6B). V. vulnificus expressed a ToxR antibody-reacting protein of the same size as that produced by E. coli carrying pCMM610 (pBAD24::toxR) after arabinose induction. But the concentration of ToxRVv in the V. vulnificus strain appeared much less than in E. coli harboring the high-copy-number expression plasmid. This difference could be attributed to the single-copy existence of the gene in the chromosome.
Stimulation of the ctx promoter by ToxRVv.
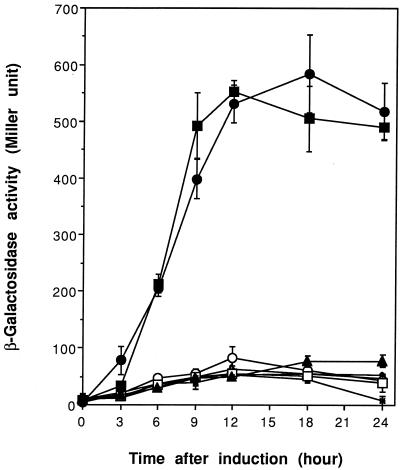
The V. cholerae ctx promoter is known to be activated by ToxR, even in an E. coli background (30). In order to verify that the putative ToxRVv protein is functionally related to ToxRVc, we tested whether ToxRVv could cross-activate the ctx promoter incorporated in the chromosome of E. coli VM2 (30). This E. coli strain harbors a recombinant lambda phage containing the upstream regulatory region of the ctx operon and the nucleotides encoding the first 23 amino acids of the A subunit of cholera toxin fused to the β-galactosidase coding region (lacZ). In the present study, we placed toxRVv (pCMM610), toxRSVv (pCMM601), toxRVc (pCMM651), and toxRSVc (pCMM641) under the control of the promoter PBAD. Using these constructs, we could compare the ctx activation potency of ToxRVv to that of its native counterpart, ToxRVc. The transformants harboring ToxR expression vectors were evaluated both before and after arabinose induction. E. coli VM2, either with or without the vector pBAD24, provided negative controls. ToxRVv could activate the ctx promoter with an efficiency comparable to that of native ToxRVc (Fig. 7). Stimulation of the ctx promoter by either ToxRSVv or ToxRVv showed no significant difference in almost all of the experiments. However, the induction of ToxRSVc resulted in the greatest expression of the ctx promoter. The β-galactosidase activity induced by ToxRSVc was usually 100 to 150 U higher than in other induction experiments at the same time point, while ToxRSVv showed no such enhancement.
FIG. 7.
Cross-activity of toxRVc and toxRVv in the stimulation of the ctx promoter in an E. coli background. Both genes were cloned into expression vector pBAD24 and transformed into E. coli VM2 with a ctx promoter-lacZ fusion in the chromosome. Cultures were induced by 0.2% arabinose at time zero. Each error bar indicates the standard error of the mean of triplicate experiments. Symbols: × and ⧫, E. coli VM2 without plasmids, before and after induction, respectively; ▵ and ▴, VM2 carrying pBAD24, before and after induction, respectively; ○ and ●, VM2 carrying pCMM610, before and after induction, respectively; □ and ■, VM2 carrying pCMM651, before and after induction, respectively.
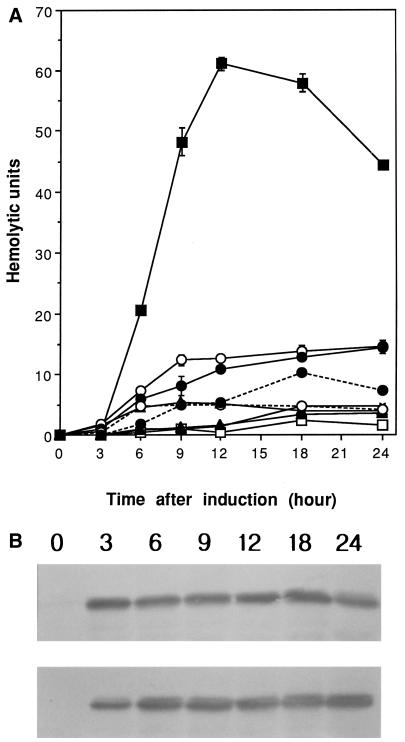
Stimulation of vvhA gene expression by ToxRSVv in E. coli.
The effect of ToxRSVv on the expression of vvhA gene was examined in an E. coli background. The vvhA gene was subcloned into pACYC184, which is compatible with the pBAD24 vector, to construct pJL961. Plasmid pJL961 was cotransformed with an expression vector bearing toxRSVv, toxRVv, toxRSVc, or toxRVc under the control of the PBAD promoter into E. coli DH5α. Only ToxRSVv could significantly stimulate hemolysin production from the vvhA gene in the E. coli background (Fig. 8A). Repeated experiments showed that toxRSVv induction increased hemolysin production 5- to 10-fold. In contrast to the results of ctx promoter induction experiments, both ToxRVv and ToxSVv were required to stimulate hemolysin production.
FIG. 8.
Stimulation of the vvhA gene by ToxRSVv in an E. coli background. Compatible plasmid pJL961 carrying the 3.4-kb vvhA gene fragment was cotransformed with a plasmid carrying toxRSVv (pCMM601), toxRVv (pCMM610), toxRSVc (pCMM641), or toxRVc (pCMM651) under the control of the PBAD promoter. (A) Hemolytic activity in the culture supernatant was assayed at the indicated time intervals. Cultures were induced with 0.2% arabinose. Each error bar indicates the standard error of the mean of triplicate experiments. Symbols: ×, pJL961 only; □ and ■, pCMM601 before and after induction, respectively; ▵ and ▴, pCMM610 before and after induction, respectively; ○ and ● with solid line, pCMM641 before and after induction, respectively; ○ and ● with broken line, pCMM651 before and after induction, respectively. (B) ToxRVv levels in the bacterial cells were measured by Western blot analysis. Top, pCMM610 after induction; bottom, pCMM601 after induction. The numbers above the photographs are the times after induction.
These results indicated that stable conformation of ToxRVv resulting from the interaction with ToxSVv (10, 36) might be a prerequisite for activator activity on the vvhA gene. Neither ToxRSVc nor ToxRVc could activate vvhA gene expression, indicating that the interaction between ToxRSVv and the vvhA promoter might be more restrictive than that between ToxRSVc and the ctx promoter. Western blot analysis of the ToxRVv levels in E. coli cotransformed with pJL961 and pCMM601 or pCMM610 showed that this was not the case. ToxRVv levels after arabinose induction in the presence of ToxSVv were not significantly different from those without the coexpression of ToxSVv (Fig. 8B). Expression levels of ToxRVv did not increase with culture time. The expression level at 3 h after induction did not differ from that at 24 h. Full expression of ToxRVv preceded hemolysin production. At 3 h, when hemolysin was not detected, the ToxRVv level already approached a plateau while significant hemolytic activity in the supernatant was detected after 9 h.
Effect of a toxRVv mutation on hemolysin production.
A V. vulnificus toxRVv mutant strain was constructed by inserting a suicide plasmid into the chromosomal copy of toxRVv by homologous recombination. Null mutation was confirmed by Western blot analysis of the whole-cell lysate using rabbit anti-ToxRVv serum (Fig. 6B).
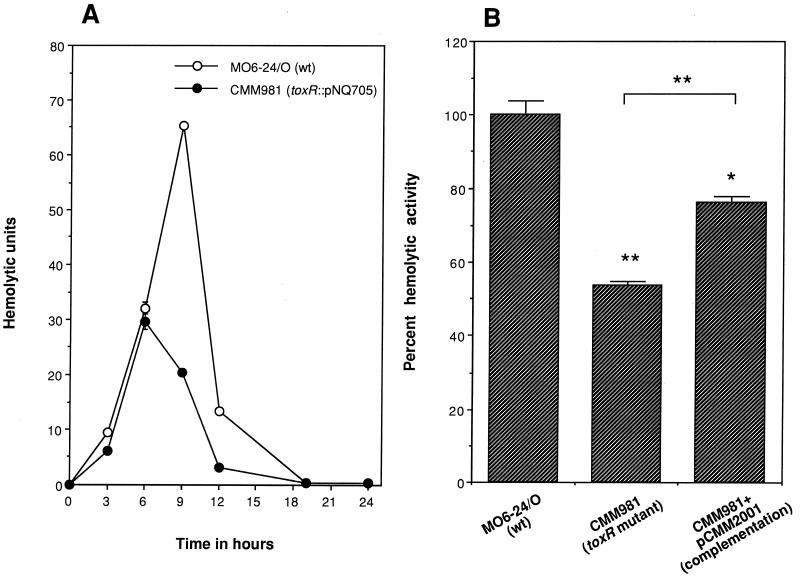
The resulting isogenic toxRVv mutant (CMM981) and the wild-type parent strain (MO6-24/O) were compared for hemolysin production. The toxRVv mutation decreased hemolysin production. The hemolysin production profile of the mutant was similar to that of the parent strain between 3 and 6 h of culture growth. The difference in hemolysin production became evident after then. By 9 h of incubation, the mutant produced threefold less hemolysin than the wild type (Fig. 9A). The hemolysin production defect of the mutant was complemented by the wild-type toxRSVv encoded by pCMM2001. The peak hemolysin production of the mutant, decreased to 50% of the wild-type level, was restored to 75% by complementation (Fig. 9B). A similar incomplete-complementation result obtained with plasmid-encoded wild-type genes was reported for V. parahaemolyticus. Lin et al. reported that the production of thermostable direct hemolysin (TDH) in a toxRVp mutant was restored to only 72% of the wild-type level by a plasmid carrying wild-type toxRSVp (25). These results indicate that ToxRVv should play a significant role in the regulation of hemolysin production by V. vulnificus during the late exponential and early stationary growth phases.
FIG. 9.
Effect of toxRVv mutation on V. vulnificus hemolysin production and its complementation by the wild-type (wt) gene. (A) Hemolysin production profiles of the toxRVv mutant (CMM981) and isogenic wild-type strain MO6-24/O. Peak hemolysin production by the mutant decreased to about half of the level of the isogenic wild-type strain. (B) Partial complementation of the hemolysin production defect of the mutant in trans by plasmid-encoded wild-type toxRSVv. Hemolysin production was significantly restored by complementation. Percent peak hemolysin production of the mutant before or after complementation was compared with that of the wild type. The symbols * and ** indicate statistically significant differences from the wild type by the Student t test at P < 0.005 and P < 0.001, respectively. The mean and the standard error of the mean were calculated from triplicate experiments.
Effect of a toxRVv mutation on the OMP profile.
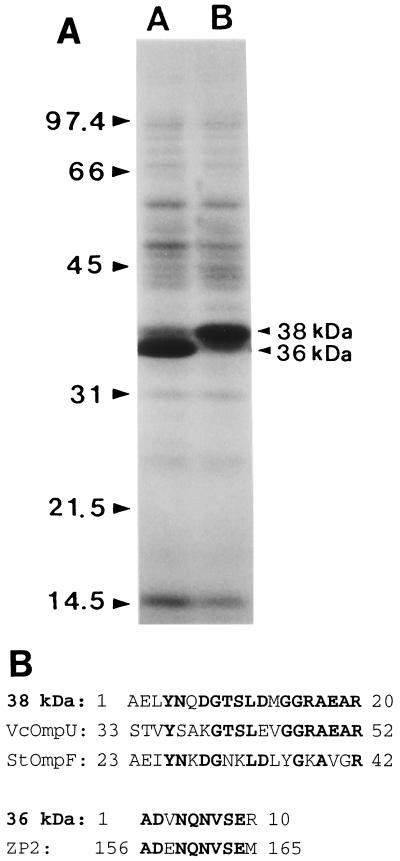
In V. cholerae, ToxR was reported to activate ompU expression and reciprocally inhibit ompT expression (9, 31, 47). One of the most prominent phenotypic changes after the toxRVc mutation was the alteration in OMP expression (31). Mutation of toxRVv also resulted in a change in the OMP profile, but the pattern of change was different. V. vulnificus MO6-24/O showed two major OMP bands of 36 and 38 kDa on SDS-PAGE. The toxRVv mutation resulted in reciprocal changes in the expression of the two major OMP bands. V. cholerae OmpT and OmpU show molecular masses of 40 and 38 kDa. Contrary to the case with V. cholerae, the high-molecular-mass OMP was positively regulated by toxRVv and the toxR mutation increased the expression of the low-molecular-mass OMP (Fig. 10A). It is noteworthy that stationary cells grown at 25°C, rather than 37°C, showed a more prominent difference in the OMP profile.
FIG. 10.
Effect of toxRVv mutation on OMP expression. (A) OMP profiles of toxRVv mutant CMM981 and isogenic wild-type strain MO6-24/O. V. vulnificus strains were grown in 2.5% NaCl heart infusion at 25°C to stationary phase. OMPs were prepared and analyzed by SDS–12% PAGE. The two major bands appeared to have molecular masses of 38 and 36 kDa. Lanes: A, CMM981; B, MO6-24/O. The values on the left are molecular masses in kilodaltons. (B) Amino-terminal amino acid sequences of the two major OMP bands and their similarities to proteins in the sequence databases. Boldface letters indicate identical sequences. Amino acid residue numbers are shown at the beginning and end of each sequence. ZP2, mouse zona pollucida sperm-binding protein 2 precursor.
The amino-terminal amino acid sequences of the two major OMP bands were determined, and the GenBank and EMBL sequence databases were searched for similar sequences. The sequence of the first 20 amino acids of the 38-kDa OMP showed high similarity to those of OmpU of V. cholerae and OmpF of Salmonella typhimurium. The sequence of the first 10 amino acids of the 36-kDa OMP showed no similarity to any sequence in the prokaryotic protein databases but did show high similarity to the mouse zona pellucida sperm-binding protein 2 precursor (Fig. 10B).
DISCUSSION
In the present study, we cloned and sequenced the toxRSVv operon from V. vulnificus type strain ATCC 29307. We also proved that ToxRVv is expressed in V. vulnificus at the expected size and showed that ToxRSVv could upregulate hemolysin production in both the E. coli and V. vulnificus backgrounds.
Pathogenic Vibrio species such as V. cholerae, V. parahaemolyticus, and V. vulnificus cycle between two distinct environments: the human body and the estuarine environment. Maintaining life in such different environments requires many different gene products and a finely tuned regulatory system for the genes. The virulence regulation of V. cholerae is under the master control of a cascade regulatory system in which cytoplasmic membrane proteins ToxR and TcpP control expression of the AraC-type transcriptional activator ToxT (2, 9, 16, 17, 18, 19). Many virulence factors, such as cholera toxin and the toxin-coregulated pilus, are under the control of this ToxR-TcpP regulon. More than 20 V. cholerae genes proved to constitute the ToxR-TcpP regulon (47). Several environmental factors, such as temperature, medium pH, osmolarity, and aeration are known to serve as the regulatory signals of the ToxR regulon (31, 40, 51).
ToxR and TcpP of V. cholerae are cytoplasmic membrane proteins with a cytoplasmic amino-terminal DNA-binding domain and a periplasmic carboxy-terminal domain that interacts with the other transmembrane proteins ToxS and TcpH, respectively (10, 16, 31, 32). The regulatory network in the ToxR and TcpP regulon seems to be multilayered and hierarchical. Most of the genes in the regulon are controlled by a second AraC type of transcription activator, ToxT, which is located at a novel pathogenicity island and is itself positively regulated by ToxR and TcpP (10, 16, 19, 20, 34). ToxT then directly activates a number of virulence genes, such as those producing the toxin-coregulated pilus and accessory colonization factors (9, 47). Recently, the global regulator cyclic AMP (cAMP)-cAMP receptor protein (CRP) was also reported to influence the expression of the ToxR regulon under various environmental conditions (46, 47). Our previous report that V. vulnificus hemolysin production was influenced by temperature and/or salinity change and was inhibited by exogenous glucose (S. E. Lee, C. M. Kim, S. Y. Kim, S. J. Kim, P. Y. Ryu, H. C. Lee, J. S. Oh, S. S. Chung, and J. H. Rhee, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. B-172, p. 84, 1998) suggests the possible existence of a multilayered regulatory network in V. vulnificus.
In 1993, Lin et al. discovered that V. parahaemolyticus has a homolog of the V. cholerae toxRS operon which mediates environmentally induced regulation of the TDH (25). Cherwonogrodzky et al. (3, 4, 5) reported that TDH production by V. parahaemolyticus could also be modulated by cultural parameters, as observed in V. cholerae. These observations encouraged the Nishibuchi group to speculate on the existence of the ToxRVc-like signal-transducing regulator in V. parahaemolyticus (25). In that report, they showed that ToxRVp promoted the expression of the tdh gene. Subsequently, Reich and Schoolnik cloned other toxRS genes from V. fischeri (43). The existence of a ToxT homolog system in V. parahaemolyticus and V. fischeri has yet to be reported. In the present study, we found that the ToxRSVv proteins were homologous to previously reported ToxRS proteins. The operon was found in all of the clinical and environmental V. vulnificus isolates studied by colony blot and Southern hybridization assays. Low-stringency colony blot hybridization analyses showed the presence of toxRSVv homologs in various Vibrio spp. (J. H. Rhee, S. H. Shin, and S. E. Lee, unpublished data). These findings suggest that the toxRS operon played important roles in the survival of numerous Vibrio species throughout evolution. Many virulence factors of Vibrio spp. might have come under the control of ToxR serendipitously. This hypothesis was also proposed by Lin et al. (25) on the basis of the finding that the tdh genes of V. parahaemolyticus appeared to be transferred among strains as transposon-like units (52) and were discovered only in subpopulations of the bacterium, while the toxRSVp genes were harbored by every V. parahaemolyticus strain tested.
As was the case for V. parahaemolyticus TDH production, production of V. vulnificus hemolysin was affected by changes in environmental parameters (Lee et al., 98th Gen. Meet. Am. Soc. Microbiol.). The experimental results gave us a strong clue to the existence of toxRS homologs in V. vulnificus. The most remarkable parameters that change in the course of the infection process should be temperature and salinity (osmolarity). The optimal temperature and salinity of estuaries that give the highest isolation rate for V. vulnificus have been reported to be about 25°C (35, 53) and over 23‰ (50), respectively. When V. vulnificus was cultured at a salinity of 2.5%, it produced the hemolysin poorly. More hemolysin is produced at 37°C than at 25°C. At a salinity of 0.9%, the hemolysin was produced more efficiently than at 2.5%. Shifting from 2.5% salinity and 25°C (reflecting the estuarine environment) to 0.9% salinity and 37°C (representing the human internal milieu) resulted in a 2.5-fold increase in hemolysin production (Lee et al., 98th Gen. Meet. Am. Soc. Microbiol.). These findings contradict reports that cholera toxin and TDH are produced more efficiently at 30°C than at 37°C and under high osmolarity (4, 13, 31). V. vulnificus responded to the temperature and/or salinity shift by increasing hemolysin production. This suggests that V. vulnificus senses environmental parameter changes during the infection process and can become more virulent in the human body. The results also suggested that V. vulnificus should have a common signal recognition-transduction system that senses changes in temperature and/or salinity and modulates hemolysin production. The ToxRSVv system was identified as the strongest candidate for the signal transduction system and proved to play an important role in the modulation of hemolysin production in the present study. However, the operational mode of the ToxRSVv system may be different from that of V. cholerae or V. parahaemolyticus because temperature and osmolarity affect downstream response contradictorily. This contradiction was also noted in the regulation of OMP expression by ToxRS. In V. vulnificus, the high-molecular-mass (38-kDa) OMP was positively regulated and the low-molecular-mass (36-kDa) OMP was negatively regulated by ToxRSVv. On the other hand, V. cholerae OmpU (low molecular mass [38 kDa]) was positively regulated and OmpT (40 kDa) was negatively regulated by ToxRVc. The 38-kDa OMP of V. vulnificus was found to be a homolog of OmpUVc by amino-terminal amino acid sequencing. The amino-terminal amino acid sequence of the 36-kDa OMP showed no homology to known OMPs in the molecular biological databases. The question of whether the OMP serves as a functional homolog of OmpTVc should be answered by further molecular biological studies.
The finding that hemolysin production is under the control of a transmembrane signal transducer may explain the raison d'être of the exotoxin. Wright and Morris proposed that hemolysin might not play a major role in the pathogenesis of V. vulnificus septicemia (55). They reported that the 50% lethal dose of the vvh-inactivated mutant for mice was no different from that of the wild-type strain. Whether the mouse septicemia model exactly reproduces the human disease needs further investigation. Previously, our group showed that hemolysin induced vasodilatation independently of nitric oxide synthase at a concentration more than 100-fold lower than that needed to produce cytotoxicity or hemolysis (22, 23). Onset of septic shock usually coincides with or precedes the earlier skin manifestations such as petechia, ecchymosis, or vesicles (38, 39). Hemolysin is highly cytotoxic and was proposed to play a significant role in producing typical skin manifestations (14). Pharmacological vasodilatation and hypotension seem to occur prior to hemolysin-induced overt damage of endothelial cells and surrounding tissues. We suppose that in vivo hemolysin production should be tightly regulated at the time and place needed. We propose that the ToxRSVv system is part of the regulatory system.
It is widely accepted that ToxS stabilizes ToxR in a conformation that is optimal for transcriptional activation, possibly as a heterodimer or as a ToxR homodimer (10, 11, 36). In the present study, we found that ToxRSVv functionally mimics ToxRSVc in stimulating the ctx promoter in an E. coli background. This finding is supported by the significant homologies of the proteins. ToxRSVc showed the highest promoting activity, as expected. ToxRVv alone was as potent as ToxRVc in promoting the ctx gene. The effect of ToxRSVv was not superior to that of ToxRVv. In contrast, in the E. coli background, only overexpression of toxRSVv, but not toxRVv alone, toxRSVc, or toxRVc alone, resulted in significant activation of the vvhA gene. These results suggest an important role of ToxSVv in the optimal activation of the vvhA promoter but not in that of the ctx promoter. For optimal activity on the vvhA promoter, ToxRVv may need the greater stabilization provided by ToxSVv binding. But this possibility was ruled out by Western blot analyses of ToxRVv levels in the E. coli background. The amount of ToxRVv was not affected by the presence or absence of ToxSVv when the former protein was overexpressed. Under physiological conditions, ToxR forms a heterodimer with ToxS and become stabilized. ToxR-ToxR homodimers were also formed when ToxR was overexpressed (36). The homodimers were supposed to be as resistant as heterodimers to destruction, since overexpressed ToxRVv alone showed a band intensity similar to that of overexpressed ToxRSVv in the present study. These results indicate that ToxSVv might play an active role in regulating vvhA expression other than mere stabilization of ToxRVv. Pfau and Taylor (42) showed that the DNA-binding and transcription activities could be separated and that chaperone-like activity of ToxS may be required for the formation of the transcription activation complex but not the ToxR-DNA complex at the ctx promoter. At the vvhA promoter region, proper formation of a ToxR-ToxSVv complex might be a prerequisite for both DNA binding and formation of the transcription activation complex. Another explanation for the fact that only ToxRSVv could activate the vvhA promoter in the E. coli background is that ToxRVv might not be a major transcription activator of the vvhA gene. Under physiological conditions, full stabilization of ToxRVv by ToxSVv would take effect in a small-scale activation or fine tuning of the vvhA promoter while some other major transcription factor(s) is responsible for regulating hemolysin production on a large scale(s). The hypothesis comes from the result that the toxRVv null mutation decreased hemolysin production to only half of that of the isogenic wild-type strain. Another factor, such as CRP, would play the major role as a transcription activator. We recently showed that a vvhA-luxCDABE fusion reporter requires cAMP and CRP in an E. coli background (1). In that report, we also showed that a putative chemical crp mutant of V. vulnificus did not produce hemolysin. ToxRVv would play an auxiliary role along with CRP as a fine tuner of vvh expression, while cAMP and CRP determine whether the switch of the hemolysin gene should be turned on or off. The presence of 0.5% glucose in the culture medium totally shuts off hemolysin production (1; Lee et al., 98th Gen. Meet. Am. Soc. Microbiol.). In the 5′ untranslated region of vvhBA, a region showing homology to the consensus CRP binding sequence was noted (56). We are currently investigating other factors acting at the promoter region of vvhA.
ToxRVc binds to tandem TTTTGAT repeats present in the upstream region of the ctx promoter (32, 41). The ToxRVc binding site of the toxT upstream DNA has no TTTTGAT repeats but is rich in inverted-repeat elements (17, 18). In the vvhA upstream DNA sequence, neither TTTTGAT repeats nor the inverted repeats present around the toxT upstream ToxRVc-binding region were observed in the vvhA promoter region (56). To elucidate the mechanism by which the vvhA promoter is activated by ToxRVv, the ToxRVv-binding sites at the promoter region should be mapped and their interaction with the transcription activator needs to be investigated by using an in vitro transcription assay system.
ACKNOWLEDGMENTS
We are grateful to John J. Mekalanos; J. Glenn Morris, Jr.; and Debra Milton for kindly providing us with V. vulnificus MO6-24/O, E. coli VM2, toxRSVc, and pNQ705, respectively. J.H.R. thanks Bonnie Bassler, Colin Manoil, and Nancy Trun for their encouragement during and after the 1996 Advanced Bacterial Genetics Course of the Cold Spring Harbor Laboratory.
This work was supported in part by a grant from the 1997 Basic Medical Research Fund from the Ministry of Education awarded to J.H.R.
REFERENCES
- 1.Bang Y B, Lee S E, Rhee J H, Choi S H. Evidence that expression of the Vibrio vulnificus hemolysin gene is dependent on cyclic AMP and cyclic AMP receptor protein. J Bacteriol. 1999;181:7639–7642. doi: 10.1128/jb.181.24.7639-7642.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 3.Cherwonogrodzky J W, Clark A G. Effect of pH on the production of the Kanagawa hemolysin by Vibrio parahaemolyticus. Infect Immun. 1981;34:115–119. doi: 10.1128/iai.34.1.115-119.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherwonogrodzky J W, Clark A G. Production of the Kanagawa hemolysin by Vibrio parahaemolyticus in a synthetic medium. Infect Immun. 1982;37:60–63. doi: 10.1128/iai.37.1.60-63.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherwonogrodzky J W, Clark A G. Effect of d-tryptophan on hemolysin production in Vibrio parahaemolyticus. J Clin Microbiol. 1984;20:909–911. doi: 10.1128/jcm.20.5.909-911.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung S S, Park J H, Rhee J H. Bacteriological characteristics of Vibrio vulnificus. Kor J Infect Dis. 1986;18:55–62. [Google Scholar]
- 7.Dai J, Lee Y, Wong H. Effects of iron limitation on production of a siderophore, outer membrane proteins, and hemolysin and on hydrophobicity, cell adherence, and lethality for mice of Vibrio parahaemolyticus. Infect Immun. 1992;60:2952–2956. doi: 10.1128/iai.60.7.2952-2956.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePaola A, Capers G M, Alexander D. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast Appl Environ Microbiol. 1994;60:984–988. doi: 10.1128/aem.60.3.984-988.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiRita V J. Three-component regulatory system controlling virulence in Vibrio cholerae. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 351–365. [Google Scholar]
- 10.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 11.Dziejman M, Mekalanos J J. Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol Microbiol. 1994;13:485–494. doi: 10.1111/j.1365-2958.1994.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 12.Dziejman M, Mekalanos J J. Two-component signal transduction and its role in the expression of bacterial virulence factors. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 305–317. [Google Scholar]
- 13.Evans D J, Jr, Richardson S H. In vitro production of choleragen and vascular permeability factor by Vibrio cholerae. J Bacteriol. 1968;96:126–130. doi: 10.1128/jb.96.1.126-130.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray L D, Kreger A S. Mouse skin damage caused by cytolysin from Vibrio vulnificus and by V. vulnificus infection. J Infect Dis. 1987;155:236–241. doi: 10.1093/infdis/155.2.236. [DOI] [PubMed] [Google Scholar]
- 15.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in toxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 18.Higgins D E, DiRita V J. Genetic analysis of the interaction between Vibrio cholerae transcription activator ToxR and toxT promoter DNA. J Bacteriol. 1996;178:1080–1087. doi: 10.1128/jb.178.4.1080-1087.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaolis D K, Johnson J A, Bailey C C, Boedecker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly M T, Dinuzzo A. Uptake and clearance of Vibrio vulnificus from Gulf Coast oysters (Crassostrea virginica) Appl Environ Microbiol. 1985;50:1548–1549. doi: 10.1128/aem.50.6.1548-1549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kook H, Lee S E, Baik Y H, Chung S S, Rhee J H. Vibrio vulnificus hemolysin dilates rat thoracic aorta by activating guanylate cyclase. Life Sci. 1996;59:41–47. doi: 10.1016/0024-3205(96)00292-5. [DOI] [PubMed] [Google Scholar]
- 23.Kook H, Rhee J H, Lee S E, Kang S Y, Chung S S, Cho K W, Baik Y H. Activation of particulate guanylyl cyclase by Vibrio vulnificus hemolysin. Eur J Pharmacol. 1999;365:267–272. doi: 10.1016/s0014-2999(98)00870-x. [DOI] [PubMed] [Google Scholar]
- 24.Lee S E, Kim S Y, Kim S J, Kim H S, Shin J H, Choi S H, Chung S S, Rhee J H. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J Clin Microbiol. 1998;36:2887–2892. doi: 10.1128/jcm.36.10.2887-2892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 27.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 33.Milton D L, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anlguillarum. J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogierman M A, Manning P A. Homology of TcpN, a putative regulatory protein of Vibrio cholerae, to the AraC family of transcriptional activators. Gene. 1992;116:93–97. doi: 10.1016/0378-1119(92)90634-2. [DOI] [PubMed] [Google Scholar]
- 35.Oliver J D, Earner R A, Cerans D R. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl Environ Microbiol. 1983;45:985–998. doi: 10.1128/aem.45.3.985-998.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottemann K M, Mekalanos J J. The ToxR protein of Vibrio cholerae forms homodimers and heterodimers. J Bacteriol. 1996;178:156–162. doi: 10.1128/jb.178.1.156-162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottemann K M, DiRita V J, Mekalanos J J. ToxR proteins with substitutions in residues conserved with OmpR fail to activate transcription from the cholera toxin promoter. J Bacteriol. 1992;174:6807–6814. doi: 10.1128/jb.174.21.6807-6814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paik K W, Moon B, Park C W, Kim K T, Ji M S, Choi S K, Rew J S, Yoon C M. Clinical characteristics of ninety-two cases of Vibrio vulnificus infections. Kor J Infect Dis. 1995;27:355–365. [Google Scholar]
- 39.Park S D, Shon H S, Joh N J. Vibrio vulnificus septicemia in Korea: clinical and epidemiologic findings in seventy patients. J Am Acad Dermatol. 1991;24:397–403. doi: 10.1016/0190-9622(91)70059-b. [DOI] [PubMed] [Google Scholar]
- 40.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfau J D, Taylor R K. Genetic footprint of the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol. 1996;20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 42.Pfau J D, Taylor R K. Mutation in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J Bacteriol. 1998;180:4724–4783. doi: 10.1128/jb.180.17.4724-4733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Shinoda S, Miyoshi S, Yamanaka H, Miyoshi N N. Some properties of Vibrio vulnificus hemolysin. Microbiol Immunol. 1985;29:583–590. doi: 10.1111/j.1348-0421.1985.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 46.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 48.Tacket C D, Brenner R, Blake P A. Clinical features and epidemiological study of Vibrio vulnificus infections. J Infect Dis. 1984;149:558–561. doi: 10.1093/infdis/149.4.558. [DOI] [PubMed] [Google Scholar]
- 49.Tamplin M L, Capers G M. Persistence of Vibrio vulnificus in tissues of Gulf Coast oysters, Crassostrea virginica, exposed to seawater disinfected with UV light. Appl Environ Microbiol. 1992;58:1506–1510. doi: 10.1128/aem.58.5.1506-1510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamplin M L, Rodrick G E, Blake N J, Cuba T. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl Environ Microbiol. 1982;44:1466–1470. doi: 10.1128/aem.44.6.1466-1470.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;90:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terai A, Baba K, Shirai H, Yoshida O, Takeda Y, Nishibuchi M. Evidence for insertion sequence-mediated spread of the thermostable direct hemolysin gene among Vibrio species. J Bacteriol. 1991;173:5036–5046. doi: 10.1128/jb.173.16.5036-5046.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tilton, R. C., and R. W. Ryan. Clinical and ecological characteristics of Vibrio vulnificus in the northeastern United States. Diagn. Microbiol. Infect. Dis. 6:109–117. [DOI] [PubMed]
- 54.Wright A C, Hill R T, Johnson J A, Roghman M-C, Colwell R R, Morris J G., Jr Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl Environ Microbiol. 1996;62:717–724. doi: 10.1128/aem.62.2.717-724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright A C, Morris J G., Jr The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect Immun. 1991;59:192–197. doi: 10.1128/iai.59.1.192-197.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto K, Wright A C, Kaper J B, Morris J G., Jr The cytolysin gene of Vibrio vulnificus: sequence and relationship to V. cholerae El Tor hemolysin gene. Infect Immun. 1990;58:2706–2709. doi: 10.1128/iai.58.8.2706-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]