Abstract
We isolated and characterized spontaneous mutants with defects in the 147-amino-acid Salmonella protein FliJ, which is a cytoplasmic component of the type III flagellar export apparatus. These mutants, including ones with null mutations, have the ability to form swarms on motility agar plates after prolonged incubation at 30°C; i.e., they display a leaky motile phenotype. One mutant, SJW277, which formed significantly bigger swarms than the others, encoded only the N-terminal 73 amino acids of FliJ, one-half of the protein. At 30°C, overproduction of this mutant protein improved, to wild-type levels, both motility and the ability to export both rod/hook-type (FlgD; hook capping protein) and filament-type (FliC; flagellin) substrates. At 42°C, however, export was inhibited, indicating that the mutant FliJ protein was temperature sensitive. Taking advantage of this, we performed temperature upshift experiments, which demonstrated that FliJ is directly required for the export of FliC. Co-overproduction of FliJ and either of two export substrates, FliE or FlgG, hindered their aggregation in the cytoplasm. We conclude that FliJ is a general component of the flagellar export apparatus and has a chaperone-like activity for both rod/hook-type and filament-type substrates.
The Salmonella flagellum is a complex structure; most of its components lie beyond the cytoplasmic membrane (8) and, therefore, have to be exported. With the exception of the basal-body P- and L-ring proteins, these components utilize a specialized export system called the type III flagellar export apparatus. Many other type III systems exist for the export of virulence factors (4).
The substrate specificity of the flagellar export apparatus changes on completion of the hook structure (9–11), and on the basis of this difference, its export substrates are divided into two classes. One is the rod/hook-type export class, comprising FliE (basal-body protein); FlgB, FlgC, and FlgF (proximal rod proteins); FlgG (distal rod protein); FlgE (hook protein); FlgD (hook capping protein); and FliK (hook length control protein). The other is the filament-type export class, comprising FlgK (first hook-filament junction protein), FlgL (second hook-filament junction protein), FlgM (anti-sigma factor), FliC (flagellin), and FliD (filament capping protein).
Six membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, and FliR) were found to be required for all flagellar protein substrates tested to be translocated across the plane of the cytoplasmic membrane (11). These membrane components are believed to be located in a patch of membrane within the annular pore of the basal-body MS ring (2). Two soluble proteins (FliH and FliI) were also found to be required. Recently, we provided evidence for a number of interactions among export components and between export components and their substrates (12).
FliJ is another cytoplasmic protein implicated in export: mutants with defects in FliJ fail to export rod/hook-type substrates into the periplasm. However, for technical reasons, we could not test whether FliJ also might be required for the export of filament-type substrates. This possibility was raised by our subsequent finding that FliJ binds to both filament-type substrates and rod/hook-type substrates (12). We also found that FliJ interacts with the soluble components FliH and FliI and the soluble cytoplasmic domains FlhAC and FlhBC of the export apparatus, strengthening the argument that FliJ itself is involved in the export process.
FliJ is a small protein consisting of 147 amino acids (19). It shares several structural features with the Salmonella flagellar proteins FliS, FlgN, and FliT, which are putative chaperones for the filament-type substrates FliC, FlgK and FlgL, and FliD, respectively (3, 26), and with other members of the type III cytoplasmic chaperone family, including the Yersinia Syc (for specific Yop chaperones) proteins (4, 23). These chaperones are small proteins (ca. 15 to 20 kDa) with a high proportion of charged residues; they are predicted to have a large amount of α-helical structure, including potential amphipathic helices toward their C termini.
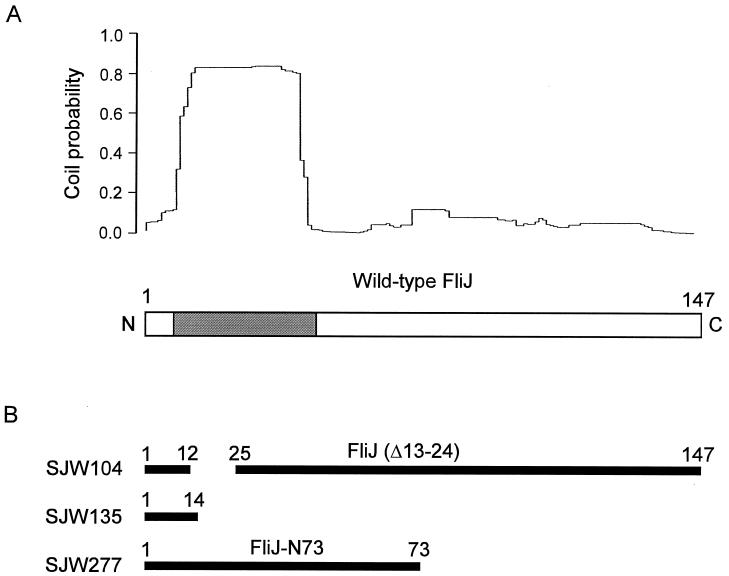
In addition to possessing these characteristics, FliJ has a sequence with a high probability of α-helical coiled-coil structure near its N terminus (24) (Fig. 1A), which suggests that FliJ is likely to engage in such an interaction, either with itself, to form a homodimer, or with its substrates.
FIG. 1.
(A) Estimation of the total (dimer plus trimer) probability that a given residue of the 147-amino-acid wild-type sequence of FliJ will participate in an α-helical coiled-coil structure. The probability for residues 14 through 42 is ≥80%. Data were calculated by the MultiCoil program of Wolf et al. (24). In the schematic representation of the wild-type FliJ sequence (bottom), the region with a high probability of having coiled-coil structure is shaded. (B) Mutant FliJ proteins analyzed in this study. Sequence present is shown in solid bars, with the beginning and ending residues indicated. SJW104 has an in-frame deletion. SJW135 and SJW277 are nonsense mutants. The SJW104 and SJW277 FliJ products are referred to throughout this paper as FliJ(Δ13–24) and FliJ-N73, respectively.
Stephens et al. (17) hypothesized that FliJ of Caulobacter crescentus might be a chaperone. We tentatively proposed (11) that FliJ of Salmonella might be a chaperone specific for rod/hook-type export substrates, based on its role in their export. In the present study, we showed that FliJ is also required for the export of filament-type substrates and that it prevents aggregation of export substrates in the cytoplasm.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. The fliJ mutants were spontaneous, essentially nonflagellated derivatives of wild-type Salmonella strain SJW1103 (25) and were mapped by transduction using bacteriophage P22. L broth (LB), M9, and soft tryptone agar plates were used as described previously (10, 11). Ampicillin and Casamino Acids (CA) were added to the media at final concentrations of 100 μg/ml and 0.3%, respectively.
TABLE 1.
Strains and plasmids used in this study.
| Strain or plasmid | Genotype or encoded protein | Source or reference |
|---|---|---|
| Strainsa | ||
| SJW1103 | Wild type for flagellation and motility | 25 |
| SJW1368 | Δ(cheW-flhD) | 15 |
| SJW1364 | flhA | 11 |
| SJW104 | fliJ (spontaneous); deletion of amino acids 13–24 | 11 |
| SJW135 | fliJ (spontaneous); truncation at amino acid 14 | This study |
| SJW277 | fliJ (spontaneous); truncation at amino acid 73 | This study |
| SJW1103gK | flgK::Tn10 | 11 |
| SJW135gK | fliJ flgK::Tn10 | This study |
| Plasmids | ||
| pTrc99A | Vector | Pharmacia |
| pET22b | Vector | Novagen |
| pMM404 | pTrc99A/FliJ wild type | This study |
| pRCJ104 | pTrc99A/FliJ(Δ13–24) | This study |
| pRCJ277 | pTrc99A/FliJ-N73 | This study |
| pMM406 | pTrc99A/N-His-FliJ wild type | 12 |
| pMM410 | pTrc99A/N-His-FliJ-N73 | This study |
| pMM411 | pTrc99A/N-His-FliJ(Δ13–24) | This study |
| pMM450 | pTrc99A/FliJ His4opal | This study |
| pMM451 | pET22b-FliJ His4opal | This study |
| pMM1004 | pTrc99A/N-His-FLAG-FliE | This study |
| pMM1004iJ | pTrc99A/N-His-FLAG-FliE + N-His-FliJ | This study |
| pMM203 | pTrc99A/N-His-FLAG-FlgG | This study |
| pMM203iJ | pTrc99A/N-His-FLAG-FlgG + N-His-FliJ | This study |
All strains are Salmonella and are derived from wild-type strain SJW1103.
DNA techniques.
PCR and cloning were performed as described previously (11). DNA sequencing was carried out with the modified T7 DNA polymerase Sequenase (U.S. Biochemical Corp., Cleveland, Ohio) and various synthetic primers.
Preparation of soluble cellular fractions and culture supernatants.
Overnight 60-μl cultures of SJW135gK carrying pTrc99A, pMM404, pRCJ104, or pRCJ277 were inoculated into 3-ml volumes of M9 plus CA medium containing ampicillin and incubated at either 30 or 42°C with shaking. With pRCJ104 and pRCJ277, isopropyl-β-d-thiogalactopyranoside (IPTG) was also added at the concentrations indicated (see Fig. 4). When the cells reached an optical density at 600 nm (OD600) of 1.0 to 1.2, 1.5 ml of each culture was centrifuged and the culture supernatants were collected. The cells of each culture were resuspended in 300 μl of B-PER reagent (Pierce, Rockford, Ill.) by up-and-down pipetting. After 1 min of vortexing, centrifugation was carried out at 20,800 × g for 5 min and the pellet was discarded, leaving the soluble intracellular fraction (cytoplasm plus periplasm) in the supernatant. The proteins in the intracellular fraction and the culture supernatant were each precipitated with 10% trichloroacetic acid, suspended in Tris-saturated sodium dodecyl sulfate (SDS) loading buffer, and boiled for 3 min as described previously (11).
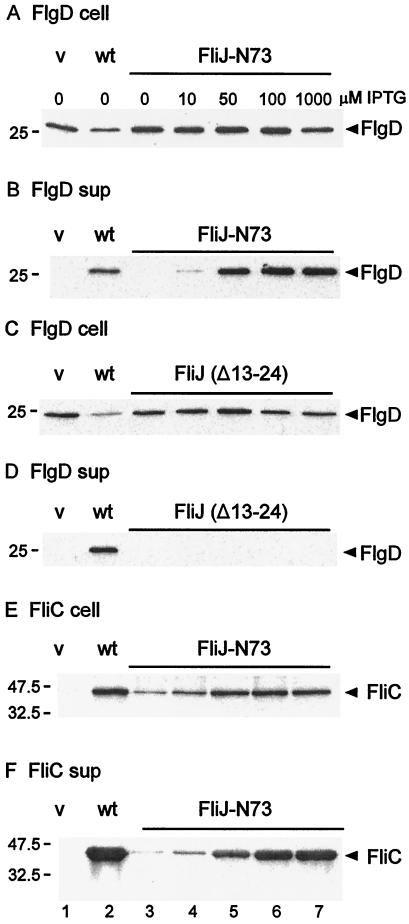
FIG. 4.
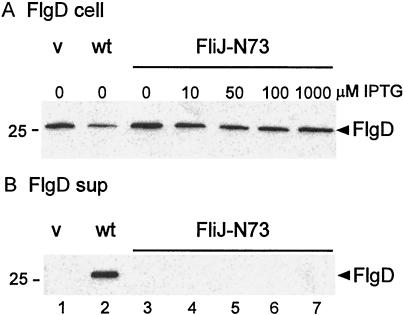
Effects of FliJ expression on intracellular levels of the rod/hook-type protein FlgD and the filament-type protein FliC (A, C, and E) and their export to the culture supernatant (sup) (B, D, and F). Strain SJW135gK (fliJ flgK::Tn10) was transformed with pTrc99A (vector [v]), pMM404 (wild-type [wt] FliJ), and either pRCJ277 (FliJ-N73) (A, B, E, and F) or pRCJ104 [FliJ(Δ13–24)] (C and D). Transformants were grown until late log phase in medium containing IPTG at the concentrations indicated. The samples were subjected to SDS-PAGE and immunoblotted with anti-FlgD (A to D) or anti-FliC (E and F). The positions of molecular mass markers (in kilodaltons) are shown on the left.
Immunoblotting.
Immunoblotting with polyclonal anti-FlgD and anti-FliC antibodies and monoclonal anti-FLAG (Sigma, St. Louis, Mo.) and anti-His (Pierce) antibodies was performed as described previously (11).
Temperature upshift experiment.
A 60-μl volume of an overnight culture of SJW135gK carrying pTrc99A, pMM404, or pRCJ277 was inoculated into 3 ml of LB medium containing ampicillin (and, in the case of pRCJ277, 1 mM IPTG). The culture was grown at 30°C with shaking until the cells reached an OD600 of 1.2 to 1.4 and then centrifuged, and the pelleted cells were washed twice with M9 plus CA medium containing ampicillin, prewarmed to 42°C. The cells were then suspended in 3 ml of the same medium and incubated at 42°C for 60 min in the absence of IPTG. Intracellular and supernatant fractions were prepared as described above and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting with anti-FliC antibody.
Pulse-labeling with [35S]methionine.
Overnight cultures (60 μl) of SJW1368 [Δ(cheW-flhD)] carrying pMM1004, pMM1004iJ, pMM203, or pMM203iJ were inoculated into 3-ml volumes of LB containing ampicillin and incubated at 37°C with shaking. When the cells reached an OD600 of 0.8, 1.5 ml of each culture was centrifuged. The cells were washed twice with M9 medium containing ampicillin. Constant numbers of cells were resuspended in M9 medium containing ampicillin, and 150 μl of culture was incubated at 37°C for 20 min. Then 1 μl of 1.5-μCi/μl [35S]methionine (Amersham Pharmacia, Piscataway, N.J.) was added, and incubation was continued for another 5 min. After centrifugation, the pelleted cells were suspended in 30 μl of 50 mM Tris-HCl (pH 8.0) containing 1 mM EDTA and 1% SDS and heated at 100°C for 3 min. A 470-μl volume of TNE (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1 mM EDTA) containing 0.1% Thesit (Roche Molecular Biochemicals, Indianapolis, Ind.), 2.5 μl of a 1-mg/ml solution of a trypsin inhibitor (Sigma), and 1 μl of a solution of anti-FLAG antibody were added. After overnight incubation of the cells at 4°C on a rotator, 5 mg of protein A-Sepharose CL-4B (Pharmacia, Piscataway, N.J.) was added and incubation was continued for another 1 h at 4°C on a rotator. After centrifugation, the pellets were washed twice with 500 μl of TNE containing 0.1% Thesit and once with 500 μl of 50 mM Tris-HCl (pH 8.0). The pellets were resuspended in 50-μl volumes of SDS loading buffer and boiled for 3 min. The immunoprecipitated samples were separated by SDS-PAGE and then transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). After the membranes were dried, they were exposed to X-ray film.
Cell fractionation into aggregated and soluble materials.
Volumes (60 μl) of overnight cultures of SJW1368 [Δ(cheW-flhD)] carrying pMM1004, pMM1004iJ, pMM203, or pMM203iJ were inoculated into 3-ml volumes of LB containing ampicillin, and the cells were grown at 37°C with shaking. Then IPTG was added to a final concentration of 1 mM, and incubation was continued for another 3 h. A 1.5-ml volume of each culture was centrifuged. The pelleted cells were resuspended in 300 μl of B-PER reagent by up-and-down pipetting. After 1 min of vortexing, centrifugation was carried out at 20,800 × g for 5 min. The supernatants (soluble fraction) and pellets (insoluble fraction) were collected separately. The pellets were each resuspended in 300 μl of 50 mM Tris-HCl (pH 8.0)–0.5% NaCl.
RESULTS
Isolation of spontaneous fliJ mutants.
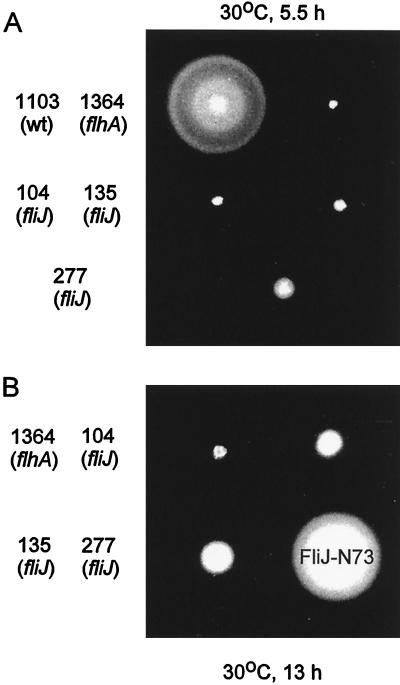
In an attempt to understand the role of FliJ in the export process, we isolated a set of 13 spontaneous fliJ mutants. Those used explicitly in this study are listed in Table 1. All of the fliJ mutations resulted in severe inhibition of swarming on motility agar plates (Fig. 2A). However, after prolonged incubation, the mutants all formed distinct swarms (Fig. 2B), indicating that they had a leaky motile phenotype; this was most pronounced in the case of SJW277. In contrast, a mutant with a defect in FlhA, which is a general component of the flagellar export apparatus (11), did not swarm at all.
FIG. 2.
Swarming ability of fliJ mutants incubated at 30°C on semisolid tryptone agar plates for 5.5 h (A) or 13 h (B). All strain numbers carry the prefix SJW, which has been omitted for clarity. SJW1103 is wild type (wt) for motility and chemotaxis. SJW1364 is defective in FlhA, a component essential for export. The other strains are spontaneous fliJ mutants, all of which (most notably SJW277, encoding FliJ-N73) show some swarming ability by 13 h.
When examined in liquid medium, most cells of strain SJW277 swam, but they did so poorly. Dark-field microscopy revealed that only about one or two flagella were present per cell. For the other fliJ strains, only a small fraction (ca. 1%) of the cells were motile; again, those that were motile had only one or two flagella. Cells of wild-type strain SJW1103 were well flagellated and vigorously motile.
Sequence analysis of the fliJ mutants.
The fliJ mutant alleles were amplified by PCR, and the amplified fragments were sequenced by using various internal primers.
All of the fliJ mutations were fairly severe. They consisted of in-frame deletion, nonsense, and frameshift mutations; there were no missense mutations. The mutations for those strains used in the present study are represented graphically in Fig. 1B. The most severe mutation was that in SJW135, which generated an ochre (TAA) stop codon at position 15, and so presumably SJW135 is a null mutant. Since this strain, like the others, displayed a leaky motile phenotype (Fig. 2B), we conclude that FliJ, though important, is not absolutely required for the flagellum-specific export process. We also conclude that the mutation in SJW135 does not have a polar effect on the only downstream gene in the operon, fliK, since this would result in a polyhook phenotype (long hooks without filaments) and total failure to swarm (16, 18).
The fliJ mutation in SJW277, the strain which swarmed significantly better than the others (Fig. 2B), created an amber (TAG) stop codon at position 74, resulting in an N-terminal fragment of 73 amino acids (FliJ-N73) that lacks the C-terminal 73 amino acids; the fragment thus consists of exactly half of the wild-type sequence.
Complementation effects of mutant FliJ proteins on motility.
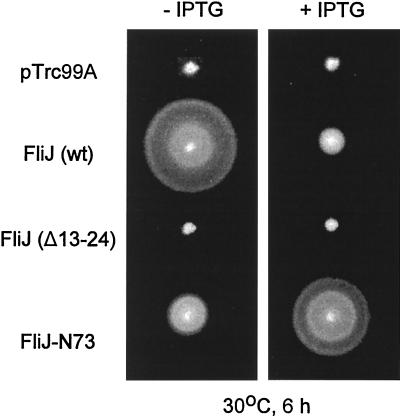
The motility phenotype of SJW277 suggested that the N-terminal region of FliJ is important for its function but that its C-terminal region might be dispensable to some degree. To test this hypothesis, we cloned into pTrc99A the fliJ alleles from SJW1103 (wild type), SJW104 [which contains a 12-codon in-frame deletion near the 5′ end of the gene and encodes FliJ(Δ13–24) (Fig. 1B)], and SJW277 (encoding FliJ-N73) to give pMM404, pRCJ104, and pRCJ277, respectively. These plasmids were introduced into SJW135 (fliJ), and the resulting transformants were inoculated on plates of soft tryptone agar with or without 0.1 mM IPTG. In the absence of IPTG, pRCJ277 (FliJ-N73) complemented the motility of SJW135 to a considerable degree (Fig. 3, left panel), causing significantly better motility than that of SJW277 (Fig. 2A); even when uninduced, pTrc99A-based plasmids generally result in higher-than-chromosomal expression levels (13). Addition of IPTG improved the complementation to almost the wild-type level. pRCJ104, in contrast, did not complement at all, even in the presence of IPTG, indicating that damage to the N-terminal sequence of FliJ has severe consequences. pMM404 (wild-type FliJ) complemented fully in the absence of IPTG but, consistent with previous observations (12), complemented poorly in the presence of IPTG; i.e., it exerted a negative or inhibitory multicopy effect.
FIG. 3.
Swarming ability of the fliJ-null mutant SJW135 transformed with pTrc99A, pMM404 (wild-type [wt] FliJ), pRCJ104 [FliJ(Δ13–24)], or pRCJ277 (FliJ-N73). Transformants were incubated at 30°C for 6 h on semisolid tryptone agar plates with (+) or without (−) 0.1 mM IPTG.
Complementation effects of mutant FliJ proteins on the export of flagellar components.
We next examined whether the stimulation of motility caused by overproduction of FliJ-N73 was a consequence of stimulation of the export of flagellar components. We used SJW135gK (fliJ flgK::Tn10) as the host to prevent filament assembly and thus facilitate detection of the export of substrates. We chose the hook capping protein FlgD as an example of a rod/hook-type substrate and FliC (flagellin) as an example of a filament-type substrate (11). SJW135gK was transformed with pTrc99A, pMM404 (wild-type FliJ), pRCJ277 (FliJ-N73), and pRCJ104 [FliJ(Δ13–24)], and intracellular fractions and culture supernatants were prepared. After SDS-PAGE, immunoblotting was carried out with anti-FlgD and anti-FliC antibodies.
With pRCJ277 (FliJ-N73), the intracellular level of FlgD was independent of the IPTG concentration and was comparable to that of cells transformed with vector alone or with a plasmid encoding wild-type FliJ (Fig. 4A). However, the amount of FlgD exported into the culture medium increased with the IPTG concentration so that at 50 μM IPTG the amount of exported FlgD exceeded the wild-type level (Fig. 4B, cf. lanes 5 and 2). These results suggest that FliJ-N73 is directly involved in the export of rod/hook-type substrates.
No FlgD export was seen with SJW135gK carrying pRCJ104 [FliJ(Δ13–24)] (Fig. 4D), even though intracellular levels of the protein were higher than those of the wild type (Fig. 4C), in which FlgD is presumably depleted by the export process. Note that failure to export any of the substrates that assemble prior to penetration of the outer membrane (i.e., the rod proteins) will by itself result in failure of FlgD to be exported into the external medium; however, in a previous study (11), we have shown that the presence of an FliJ(Δ13–24) allele results in the failure of FlgD to be exported even into the periplasm.
With SJW135gK carrying pRCJ277, the intracellular levels of flagellin (FliC) increased with the IPTG concentration (Fig. 4E), reaching a constant level by 50 μM. This increase is presumably a result of export of the anti-sigma factor, FlgM, when substrate specificity switches upon completion of the hook (5, 6). The extracellular levels of flagellin also increased with the IPTG concentration (Fig. 4F) but never fully reached the wild-type level, even at 1 mM IPTG. Comparing the IPTG dependencies of intracellular and exported FliC levels in Fig. 4E and F, we conclude that a large part, but probably not all, of the stimulation of FliC export is a consequence of enhanced FliC synthesis. Thus, although overproduction of FliJ-N73 results in stimulation of the export of a filament-type protein (FliC) as well as that of a rod/hook-type protein (FlgD), it is difficult to judge from these experiments the extent to which the effect of FliJ on FliC export is direct.
With pRCJ104 [FliJ(Δ13–24)], no FliC was detected in either the intracellular fraction or the culture supernatant, regardless of the IPTG concentration (data not shown). The absence of this protein in the intracellular fraction presumably reflects a failure to export the flagellum-specific anti-sigma factor FlgM and to activate fliC gene expression (5, 6).
High temperature inhibits the export function of FliJ-N73.
Preliminary work had indicated that the swarm behavior of fliJ mutants, including SJW277, displayed some temperature sensitivity (data not shown). To examine the effect of high temperature on the function of FliJ-N73 with respect to protein export, we took SJW135gK/pRCJ277 cell cultures that had been grown at 42°C in the presence of various IPTG concentrations, prepared the intracellular fractions and culture supernatants, and performed immunoblotting with anti-FlgD. With FliJ-N73, intracellular levels of FlgD (Fig. 5A, lanes 3 to 7) were independent of the IPTG concentration and were higher than those with uninduced wild-type FliJ (lane 2). With FliJ-N73, no FlgD was detected in the culture supernatants, even at 1 mM IPTG (Fig. 5B, lanes 3 to 7), in contrast to the situation with wild-type FliJ (lane 2). We conclude that although overproduced FliJ-N73 can support a wild-type level of export function at 30°C, it cannot function at 42°C.
FIG. 5.
Export properties of FliJ-N73 at 42°C. Strain SJW135gK (fliJ flgK::Tn10) was transformed with pTrc99A (vector [v]), pMM404 (wild-type [wt] FliJ), or pRCJ277 (FliJ-N73). Transformants were grown at 42°C to late log phase in medium containing IPTG at the concentrations indicated. The intracellular fractions (A) and culture supernatants (sup) (B) were subjected to SDS-PAGE and immunoblotted with anti-FlgD. The position of a molecular mass marker (in kilodaltons) is shown on the left.
Temperature shift experiment with cells carrying pRCJ277.
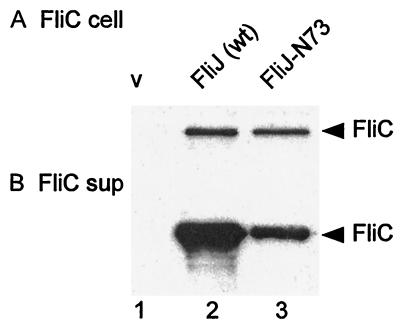
Because flagellin synthesis (5, 6) and export (10) are dependent on the completion of hook assembly, we were concerned that the stimulation of flagellin export by FliJ-N73 (Fig. 4F) might be an indirect effect caused by stimulation of hook protein export and assembly and FlgM export. We were able to take advantage of the temperature sensitivity of FliJ-N73 to examine whether FliJ directly stimulates flagellin export.
SJW135gK cells transformed with either pTrc99A, pMM404, or pRCJ277 were grown at 30°C in LB (plus 1 mM IPTG for pRCJ277 only). The cells were then washed twice in M9 plus CA containing ampicillin (prewarmed to 42°C), resuspended in the same medium, and incubated at 42°C for 60 min in the absence of IPTG. The intracellular fraction and culture supernatant were then subjected to SDS-PAGE and immunoblotting with anti-FliC antibody. Even though the intracellular level of FliC for cells transformed with pRCJ277 (Fig. 6A, lane 3) was the same as that of cells transformed with pMM404 (lane 2), the amount in the culture supernatant was much lower in the former (Fig. 6B, lane 3) than the latter (lane 2), establishing that FliJ is directly involved in the export of filament-type substrates.
FIG. 6.
Temperature shift experiment to measure FliJ-mediated export of FliC at 42°C. SJW135gK (fliJ flgK::Tn10) cells carrying the pTrc99A vector (v), pMM404 (wild-type [wt] FliJ), or pRCJ277 (FliJ-N73) were grown at 30°C (in the presence of 1 mM IPTG for pRCJ277 only), washed, and grown at 42°C for 60 min in the absence of IPTG. The intracellular fractions (A) and culture supernatants (B) were subjected to SDS-PAGE and immunoblotted with anti-FliC antibody.
Steady-state levels of FliJ-N73 are lower than those of wild-type FliJ.
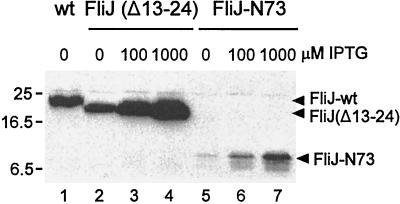
The need for IPTG induction with FliJ-N73 in order to reach wild-type levels of substrate export and swarming may mean that the truncated protein is less functional or less stable. To examine this issue, we first attempted to use anti-FliJ antibody, but we found that the intracellular levels of FliJ-N73 were too low to be detected. We therefore constructed pTrc99A-based plasmids pMM406, pMM411, and pMM410, which encode N-terminally His-tagged versions of wild-type FliJ, FliJ(Δ13–24), and FliJ-N73, respectively. We transformed SJW135 with these plasmids and examined the intracellular levels of the His-tagged forms of FliJ by immunoblotting with anti-His antibody (Fig. 7). Even after induction with 1 mM IPTG, the amount of FliJ-N73 (lane 7) was considerably lower than that of wild-type FliJ produced under noninducing conditions (lane 1). We conclude that the primary reason for the reduced efficacy of FliJ-N73 is its low intracellular level. Pulse-chase data (not shown) also indicate that it is less stable than wild-type FliJ.
FIG. 7.
Steady-state intracellular levels of forms of FliJ in SJW135 (fliJ) cells carrying the plasmids pMM406 (N-His-tagged wild-type [wt] FliJ), pMM411 [N-His-tagged FliJ(Δ13–24)], or pMM410 (N-His-tagged FliJ-N73), induced at the IPTG concentrations shown and immunoblotted with anti-His antibody. The positions of molecular mass markers (in kilodaltons) are shown to the left.
The intracellular level of FliJ(Δ13–24) under noninducing conditions (lane 2) was only slightly less than that of wild-type FliJ and increased with induction (lanes 3 and 4); this establishes that FliJ(Δ13–24) is intrinsically incapable of sustaining substrate export (Fig. 4) or swarming (Fig. 2 and 3).
Effect of the level of full-length FliJ on motility and protein export.
Studies of FliJ-N73 yielded valuable information about the roles of FliJ, especially its role in the export of filament-type proteins. However, we also wanted to know the effect of subnormal levels of wild-type FliJ on export and motility. Manipulation of FliJ levels by using IPTG was pointless since uninduced cells transformed with pMM404 (carrying the wild-type gene) swarmed as well as wild-type SJW1103 cells (cf. Fig. 3, left panel, and Fig. 2A).
We therefore decided to lower the uninduced level of FliJ by introducing an opal codon into the gene and relying on the ca. 2% natural misreading of this as a Trp codon (13, 14). In previous studies, there had been suitable Trp codons in the gene of interest that could be replaced by an opal codon; this was not the case with fliJ. At position 4, which in the wild-type gene is a His codon, we therefore created an opal codon (i.e., we made an H4opal substitution). This should then result in a null product (H4opal) 98% of the time and a conservative His→Trp substitution (H4W) 2% of the time.
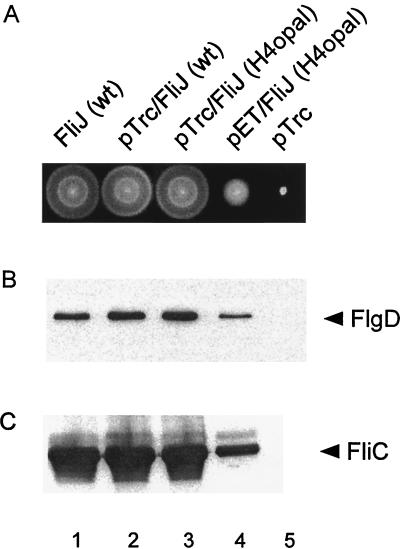
The resulting pTrc99A-based plasmid, pMM450, restored wild-type swarming (Fig. 8A) and wild-type FlgD and flagellin export levels (Fig. 8B and C, lanes 3) to SJW135 (fliJ) cells, even under noninducing conditions. We attribute this to the fact that the level of uninduced expression from a pTrc99A-based plasmid is about 20-fold higher than the level of chromosomal expression (13), almost offsetting the 50-fold decrease resulting from the opal mutation.
FIG. 8.
(A) Effect of FliJ expression on swarming of cells on semisolid tryptone agar at 30°C for 6 h. Colony 1, wild-type (wt) SJW1103/pTrc99A; colony 2, SJW1103/pMM404 (pTrc99A encoding wild-type FliJ); colony 3, SJW135 (fliJ)/pMM450 (pTrc99A encoding FliJ H4opal); colony 4, SJW135/pMM451 (pET22b encoding FliJ H4opal; colony 5, SJW135/pTrc99A. (B and C) Effect of FliJ expression on export of FlgD (hook capping protein) and FliC (flagellin) into the culture supernatant, detected in both cases with the appropriate antibody. Lanes 1, SJW1103gK (flgK)/pTrc99A; lanes 2, SJW1103gK/pMM404 (pTrc99A encoding wild-type FliJ); lanes 3, SJW135gK (fliJ flgK)/pMM450 (pTrc99A encoding FliJ H4opal); lanes 4, SJW135gK/pMM451 (pET22b encoding FliJ H4opal); lanes 5, SJW135gK/pTrc99A. No induction by IPTG was used.
We therefore introduced the H4opal allele into pET22b to give plasmid pMM451. The chromosomal level of FliJ production was too low to estimate directly, but using the hook protein FlgE as an example, we estimated that the level of pET22b-based expression is approximately threefold higher than that from the chromosome (data not shown). In combination with the 50-fold decrease resulting from the opal mutation, we therefore estimated that there would be about a 16-fold decrease in the FliJ level with pMM451 compared to that evident with chromosomal expression. In agreement with this expectation, SJW135 cells transformed with pMM451 swarmed poorly compared to wild-type cells (Fig. 8A), and export levels of FlgD and flagellin were reduced about 10-fold (Figs. 8B and C, lanes 4).
We conclude that in wild-type cells, FliJ is produced in amounts that are in excess of need, but not by a large factor. Measurement of the wild-type copy number of FliJ is in progress.
FliJ affects inclusion body formation by export substrates.
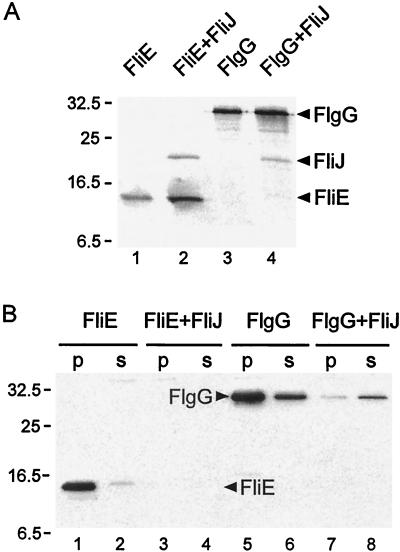
FliJ shares several structural features with type III cytoplasmic chaperones (see the introduction). It might, therefore, have an activity which prevents export substrates from premature aggregation in the cytoplasm. To investigate this possibility, we used FliE and FlgG as export substrates, because overproduction of these proteins results in formation of inclusion bodies (reference 12 and data not shown). We constructed pMM1004, pMM1004iJ, pMM203, and pMM203iJ, which are pTrc99A-based plasmids encoding N-His-FLAG-FliE, N-His-FLAG-FliE plus N-His-FliJ, N-His-FLAG-FlgG, and N-His-FLAG-FlgG plus N-His-FliJ, respectively (Table 1). Where fliJ was present, it was the downstream gene.
We first tested whether FliJ affects the synthesis of these export substrates by carrying out radiolabeling experiments with [35S]methionine, using SJW1368 [Δ(cheW-flhD)] as the host to avoid contributions from chromosomal gene expression. After being radiolabeled, the cells were spun down, dissolved in SDS buffer, and diluted, and the FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody. N-His-FLAG-FliE and FlgG synthesis was not inhibited by N-His-FliJ (Fig. 9A, lanes 2 and 4); in fact, levels were slightly higher in the presence of this protein than in its absence (lanes 1 and 3). N-His-FliJ was also detected (lanes 2 and 4), indicating that it coimmunoprecipitated with the FLAG-tagged proteins; it was not detected in their absence (data not shown). This result supports our previous observation, by affinity blotting, that FliJ interacts with export substrates (12).
FIG. 9.
Inhibition by FliJ of aggregation of flagellar substrates FlgG (a rod protein) and FliE (a basal-body protein). SJW1368 [Δ(cheW-flhD)], a mutant defective in the master operon, was transformed with pTrc99A-based plasmid pMM1004 (encoding N-His-FLAG-FliE), pMM1004iJ (N-His-FLAG-FliE + N-His-FliJ), pMM203 (N-His-FLAG-FlgG), or pMM203iJ (N-His-FLAG-FlgG + N-His-FliJ). (A) Cells were grown in the absence of IPTG, washed, preincubated in fresh buffer, and radiolabeled with [35S]methionine for 5 min in the presence of IPTG. The sample was then boiled in SDS buffer, immunoprecipitated with anti-FLAG antibody, subjected to SDS-PAGE, and autoradiographed. (B) Cells were solubilized with B-PER reagent and fractionated by centrifugation at 20,800 × g for 5 min. The soluble (s) and pellet (p) fractions were then subjected to SDS-PAGE and immunoblotted with anti-FLAG antibody. The level of FliE is below the detection threshold in lanes 3 and 4. The positions of molecular mass markers (in kilodaltons) are shown on the left.
We next examined the effect of FliJ on the levels and state of aggregation of FliE and FlgG in the cytoplasm. SJW1368 [Δ(cheW-flhD)] cells carrying pMM1004, pMM1004iJ, pMM203, or pMM203iJ were solubilized with B-PER reagent and fractionated by centrifugation. The soluble and pellet fractions were then subjected to SDS-PAGE and immunoblotted with anti-FLAG antibody. For cells with pMM1004 or pMM203, most of the FliE or FlgG was found in the pellet fraction (Fig. 9B, lanes 1 and 5), confirming previous observations that these proteins form inclusion bodies when overproduced. However, in the presence of FliJ (plasmids pMM1004iJ and pMM203iJ), the amount of aggregated material was severely decreased, to below the detection threshold in the case of FliE (lane 3) and to close to the detection threshold in the case of FlgG (lane 7). The level of FlgG in the soluble fraction was also reduced by FliJ (cf. lanes 6 and 8), but not by so much; in fact, the amount in the soluble fraction (lane 8) now exceeded that in the pellet fraction (lane 7). The level of FliE in the soluble fraction (lane 4), as with that in the pellet fraction (lane 3), was too low to be detected.
Thus, our data strongly suggest that FliJ prevents export substrates from premature aggregation. The decrease in the soluble amounts presumably indicates that they are susceptible to protease digestion, perhaps because, of the export components, only FliJ is being overproduced.
In parallel experiments with FlgD and FlgE, which are stable in the cytoplasm and do not form aggregates even when overproduced, co-overproduction of FliJ had no effect on their intracellular levels (data not shown).
DISCUSSION
Most flagellar components are translocated across the cytoplasmic membrane via the type III flagellar export apparatus. We have previously established that at least six membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, and FliR) and two cytoplasmic proteins (FliH and FliI) constitute this apparatus (11).
In this study, we examined the role of a third cytoplasmic protein, FliJ, in the export process. We provided evidence that FliJ is a general component of the flagellar export apparatus and has a chaperone-like activity in terms of preventing substrate aggregation.
All fliJ mutants display a leaky motile phenotype.
All of the spontaneous fliJ mutants that we isolated displayed a leaky motile phenotype (Fig. 2). This was true even of SJW135, which encodes only the N-terminal 14 amino acids and so presumably can be regarded as a null mutant. In other words, although FliJ is required for efficient export of substrates, export can occur, with a low probability, in its absence.
The N-terminal 73 amino acids of FliJ are necessary and sufficient for full function.
One mutant, SJW277, formed significantly larger swarms than the rest (Fig. 2). Its fliJ allele encodes just the N-terminal 73 amino acids (FliJ-N73) (Fig. 1B). Overproduction of this polypeptide resulted in stimulation of both motility and export of flagellar components to wild-type levels (Fig. 3 and 4). Thus, the N-terminal 73 amino acids of FliJ can supply full function. This idea is strongly supported by the fact that an N-terminally His-tagged variant of FliJ-N73, encoded by plasmid pMM410, complemented swarming to wild-type levels even without induction (T. Minamino and R. M. Macnab, unpublished results). We do not understand the role of the C-terminal region of FliJ in its function but suggest that it may contribute to maintaining the proper conformation and stability of the N-terminal region.
The N-terminal region is essential for FliJ function. Overproduction of a version of FliJ lacking residues 13 to 24 did not improve either its motility (Fig. 3) or its export (Fig. 4).
FliJ acts as a general component in the export pathway.
We originally suggested that FliJ might be a chaperone specific for rod/hook-type proteins (11) but then found that FliJ bound to both rod/hook-type and filament-type proteins (12). This raised the question of whether FliJ might also be required for export of filament-type proteins.
We found that FliJ-N73 overproduction stimulated the export of flagellin (FliC) as well as that of the hook-capping protein (FlgD) (Fig. 4). However, this result should be treated with caution, since the stimulation of FliC export could be attributed at least in part to stimulation of hook completion (which is needed for export of filament-type substrates to commence) and to stimulation of FliC synthesis (Fig. 4E). (The latter effect is presumably a consequence of export of the anti-sigma factor FlgM, which can only occur after completion of hook assembly [5, 6].)
We therefore took advantage of the fact that FliJ-N73 behaves as a wild type when overproduced at 30°C (Fig. 3 and 4) but as a mutant when produced at 42°C (Fig. 5). Recall that export of filament-type proteins is dependent on a switch in the substrate specificity that occurs only on completion of the hook structure. In a temperature shift experiment, we found that even though hook assembly had been completed at the permissive temperature and that intracellular levels of FliC were normal, a shift to the restrictive temperature severely inhibited FliC export (Fig. 6). Therefore, we feel confident in concluding that FliJ is a general component of the flagellar export apparatus, which is required for efficient export of both rod/hook-type and filament-type export substrates.
FliJ inhibits substrate aggregation in the cytoplasm.
Several export substrates, including FliE and FlgG, form aggregates when overproduced. Co-overproduction of FliJ greatly reduced aggregation (Fig. 9B). Even though the co-overproduction of FliJ did not reduce FliE or FlgG synthesis, it did cause a decrease in their steady-state intracellular levels. Because monomeric flagellar proteins are not completely folded and their termini are disordered (20–22), they may be more sensitive to proteolysis than if they are aggregated. Thus, at least under overproduction conditions, FliJ is unable to protect the substrates, perhaps because other essential components are also not being overproduced.
The molecular characteristics of FliJ versus those of other type III chaperones.
In the flagellar export pathway, FlgN and FliT specifically bind to FlgK and FlgL and to FliD, respectively (3), and are important for their export; FliS is believed to play a similar role for FliC (26). They are therefore believed to be specific chaperones for these proteins. Similarly, in the type III Yersinia Yop export pathway, some of the Yop proteins associate with specific chaperones called Syc proteins in order to be maintained in an export-competent conformation (1, 23). These chaperones have several common features: small size, high charge, acidic pI (ca. 4.5 to 5.2), and the potential to form an amphipathic α-helix that has been proposed to mediate interactions with their targets.
FliJ has similar features: a molecular mass of 17 kDa; 31 mol% charged (D-E-K-R) residues, although with a slightly basic pI of 7.8; 86 mol% predicted α-helical residues; and a predicted amphipathic α-helix in the region of residues 85 to 105. However, it has an additional, distinctive characteristic. We noted above that the N-terminal region of FliJ is essential for its function. Within this region is a strongly predicted, sharply bounded, coiled-coil structure (>80% probability for residues Ala-14 through Leu-42) (Fig. 1A). The flagellar and Syc chaperones differ considerably in terms of their propensity to form coiled-coil structures, especially if analyzed by the more stringent pairwise correlation algorithm of Wolf et al. (24) rather than by the simpler, individual probability algorithm of Lupas et al. (7). The Syc chaperones show essentially no predicted coiled-coil structure. Both FlgN and FliT do have predicted coiled-coil structure near the central portion of their sequences, FlgN at around the 40% probability level and FliT at around the 20% level; FliS shows zero probability throughout its structure. Thus, the data for FliJ are striking, in terms of both the strength of the prediction (>80%) and the N-terminal location of the predicted region. We do not understand the significance of these differences. It should, however, be remembered that a lack of coiled-coil structure is not necessarily equivalent to a lack of amphipathic helical interactions. One obvious question regarding FliJ is whether the predicted coiled coil is within a homodimer. Experiments addressing these and related questions are under way.
What are the relative roles of FliJ and of FlgN and FliT?
Unlike FlgN and FliT, FliJ does not display substrate specificity, because it binds to both rod/hook-type and filament-type export substrates (12) and is necessary for their transport (this study). Therefore, we conclude that FliJ is a general flagellar chaperone. FliJ binds to the other cytoplasmic export components, FliH and FliI, including the cytoplasmic domains of FlhA (FlhAC) and FlhB (FlhBC) (12). Perhaps its role is more intimately related to the export process itself and proteins like FlgN, FliS, and FliT play an earlier role in protecting their substrates in the time between their synthesis and their export.
ACKNOWLEDGMENTS
We thank May Kihara and Gabrielle Miller for technical assistance and Gillian Fraser for helpful discussions.
This work was supported by USPHS grant AI12202.
REFERENCES
- 1.Cornelis G R. The Yersinia deadly kiss. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 3.Fraser G M, Bennett J C Q, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 4.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 6.Kutsukake K. Excretion of the anti-sigma factor through a flagellar structure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 7.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 8.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 9.Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci Biotechnol Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- 10.Minamino T, González-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab R M. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 11.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minamino T, Macnab R M. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 13.Muramoto K, Makishima S, Aizawa S-I, Macnab R M. Effect of hook subunit concentration on assembly and control of length of the flagellar hook of Salmonella. J Bacteriol. 1999;181:5808–5813. doi: 10.1128/jb.181.18.5808-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramoto K, Makishima S, Aizawa S-I, Macnab R M. Effect of the cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J Mol Biol. 1998;277:871–882. doi: 10.1006/jmbi.1998.1659. [DOI] [PubMed] [Google Scholar]
- 15.Ohnishi K, Ohto Y, Aizawa S-I, Macnab R M, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson-Delafield J, Martinez R J, Stocker B A D, Yamaguchi S. A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype—superhooks. Arch Mikrobiol. 1973;90:107–120. doi: 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- 17.Stephens C, Mohr C, Boyd C, Maddock J, Gober J, Shapiro L. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J Bacteriol. 1997;179:5355–5365. doi: 10.1128/jb.179.17.5355-5365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T, Iino T. Role of the flaR gene in flagellar hook formation in Salmonella spp. J Bacteriol. 1981;148:973–979. doi: 10.1128/jb.148.3.973-979.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vonderviszt F, Ishima R, Akasaka K, Aizawa S-I. Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum? J Mol Biol. 1992;226:575–579. doi: 10.1016/0022-2836(92)90616-r. [DOI] [PubMed] [Google Scholar]
- 21.Vonderviszt F, Kanto S, Aizawa S-I, Namba K. Terminal regions of flagellin are disordered in solution. J Mol Biol. 1989;209:127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- 22.Vonderviszt F, Uedaira H, Kidokoro S-I, Namba K. Structural organization of flagellin. J Mol Biol. 1990;214:97–104. doi: 10.1016/0022-2836(90)90149-g. [DOI] [PubMed] [Google Scholar]
- 23.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 24.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 26.Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]