Abstract
The morphologic assessment of uterine leiomyosarcoma (LMS) may be challenging, and diagnostic immunohistochemistry (IHC) is currently lacking. We evaluated the genomic landscape of 167 uterine LMS by targeted next-generation sequencing (NGS) to identify common genomic alterations. IHC corresponding to these genomic landmarks was applied to a test cohort of 16 uterine LMS, 6 smooth muscle tumors of uncertain malignant potential (STUMP), and 6 leiomyomas with NGS data, as well as a validation cohort of 8 uterine LMS, 12 STUMPs, 21 leiomyomas and leiomyoma variants, 7 low-grade endometrial stromal sarcomas, and 2 diagnostically challenging uterine smooth muscle tumors. IHC was individually interpreted by three pathologists blinded to NGS data. Overall, 94% of LMS had ≥1 genomic alteration involving TP53, RB1, ATRX, PTEN, CDKN2A, or MDM2, with 80% having alterations in ≥2 of these genes. In the test cohort, an initial panel of p53, Rb, PTEN, and ATRX was applied, followed by a panel of DAXX, MTAP and MDM2 in cases without abnormalities. Abnormal p53, Rb, PTEN, and ATRX IHC was seen in 75%, 88%, 44%, and 38% of LMS, respectively. Two or more abnormal IHC results among these markers were seen in 81% of LMS. STUMPs demonstrated only one IHC abnormality involving these markers. No IHC abnormalities were seen in leiomyomas. In the validation cohort, abnormal p53, Rb, and PTEN IHC were seen in LMS, while rare STUMP or leiomyoma with bizarre nuclei showed IHC abnormalities involving only one of the markers. Abnormalities in ≥2 markers were present in both diagnostically challenging smooth muscle tumors, confirming LMS. Concordance was excellent among pathologists in the interpretation of IHC (Kappa: 0.97) and between IHC and NGS results (Kappa: 0.941). Uterine LMS have genomic landmark alterations for which IHC surrogates exist, and a diagnostic algorithm involving molecular-based IHC may aid in the evaluation of unusual uterine smooth muscle tumors.
Keywords: Uterine leiomyosarcoma, genomic profiling, genomic landmarks, immunohistochemistry
Introduction:
Leiomyosarcoma (LMS) is the most common malignant uterine mesenchymal neoplasm, constituting approximately 1% of gynecologic cancers1–3. It is characterized by an aggressive and often lethal disease course1,2. While there are several chemotherapy regimens that can achieve objective responses in patients with metastatic disease, complete responses are uncommon, and time to disease progression is relatively short2,4.
Its diagnosis is solely based on morphologic evaluation of nuclear atypia, tumor necrosis, and mitotic index. Any two of the following features are diagnostic of conventional or spindle cell LMS: diffuse moderate to severe cytologic atypia, tumor necrosis, and ≥10 mitoses per 10 high power fields5. A lower diagnostic threshold is used for epithelioid and myxoid uterine LMS6,7.
Despite established morphologic criteria, LMS remains a challenging diagnosis in some uterine smooth muscle tumors. Evaluation of tumor necrosis is subjective, with only a moderate degree of reproducibility among gynecologic pathologists8. Tumor heterogeneity with foci resembling leiomyoma or admixtures of different smooth muscle cell types may complicate interpretation of histologic criteria9,10. Morphology is also unreliable in predicting outcomes in smooth muscle tumors of uncertain malignant potential (STUMP) as a subset demonstrates aggressive behavior11–13.
Limited data suggest that molecular biomarkers may aid the diagnostic evaluation of uterine LMS. The genetic landscape of uterine LMS is characterized by common alterations in cell cycle regulators, chromosomal instability, and whole genome duplication14–16. Uterine LMS contain alterations affecting the functions of p53, Rb, and PTEN proteins, along with common alterations of the alternative telomere lengthening pathway16–19. These findings have led to studies investigating the role of immunohistochemistry (IHC) for several proteins, including p16, p53, p21, Ki-67, p21, stathmin 1, BCL2, PHH3, ATRX, and DAXX in uterine smooth muscle tumor classification20–28. However, the role of IHC in improving the diagnosis and prognostication of these lesions remains limited in clinical practice.
In this study, we evaluated the genetic landscape of uterine LMS by targeted DNA-based next-generation sequencing (NGS) to determine common genetic abnormalities that may be detectable by IHC. We then assessed a panel of IHC in LMS, STUMP, and leiomyomas while blinded to the sequencing data to determine its correlation with mutation and copy number alteration (CNA) status. We also applied the IHC panel to diagnostically challenging uterine smooth muscle tumors and mimickers of smooth muscle neoplasia, including low-grade endometrial stromal sarcomas (LGESS), to assess its utility in clinical practice.
Methods and Materials:
Case Selection
After obtaining Institutional Review Board approval, the pathology archives of Memorial Sloan Kettering Cancer Center (MSKCC) were retrospectively searched for all primary or metastatic uterine LMS that were previously assessed by NGS as part of the clinical work up. All consecutively sequenced uterine smooth muscle tumors (n=179), including LMS (n=167), STUMP (n=6), and leiomyoma (n=6), successfully profiled between 2014 and 2019 were included to characterize the genetic landscape of uterine LMS and to develop an IHC panel of possible surrogate markers that may correlate with mutation and CNA status determined by NGS. Sequenced LMS with available complete sets of hematoxylin-and-eosin-stained slides were histologically re-reviewed blinded to the NGS profile, and percentages of spindled, epithelioid, and myxoid morphologies were recorded. A test cohort comprising a subset of profiled tumors (n=28) were randomly chosen with stratification used to ensure representation of LMS (n=16), STUMP (n=6), and leiomyoma (n=6) for IHC correlation. IHC validation was performed retrospectively on a separate cohort of uterine smooth muscle tumors (n=41) and LGESS (n=7) that were reviewed at MSKCC in 2020 and did not have NGS data. Available clinical data, including follow-up, of the validation cohort were recorded. All cases undergoing NGS and/or IHC were histologically confirmed by expert gynecologic pathologists at MSKCC.
Targeted Massively Parallel Sequencing and Genomic Data Extraction
Tumor and matched peripheral blood samples were subjected to targeted DNA NGS using the MSK Integrated Mutational Profiling of Actionable Cancer Targets (MSK-IMPACT), a clinically validated NGS assay that targets all coding exons and selected regulatory regions and introns of 410 [n=58 (LMS, n=56; STUMP, n=2)] or 468 [n=121 (LMS, n=111; STUMP, n=4; leiomyoma, n=6)] key cancer-associated genes29,30. DNA was sequenced to an average of 622 (range, 179–1211)-fold sequence coverage. All patient-level clinical and genomic data are available at the cBioPortal.
The genomic data, including somatic mutations, CNA, and structural variants, were extracted as previously described30. GISTIC 2.0 (version 2.0.23) was used to analyze the broad copy number data31. The mutational data included chromosomal location, base-pair change, protein change, predicted functional impact of the mutation, and the associated variant frequency. Data on allele-specific CNA and ploidy were extracted using the ‘facets’ R package (version 0.5.14)32. This package is an allele-specific copy-number analysis pipeline for NGS data that is optimized for the MSK-IMPACT assay, which utilizes heterozygous sites of both a tumor and a normal sample to measure allelic imbalance between the two samples and allows for evaluation of loss of heterozygosity (LOH), copy neutral LOH, and ploidy as well as providing an integer copy number assessment of a specific gene.
Immunohistochemistry (IHC)
After identification of common uterine LMS-associated genetic alterations detected by MSK-IMPACT, a panel of commercially available antibodies (n=7) were selected for IHC confirmation of mutation and CNA status in the test and validation cohorts. The cases were randomly chosen for the test cohort among uterine smooth muscle tumors with available formalin-fixed, paraffin-embedded tissue that were subjected to clinical NGS. For the validation cohort, a random selection of uterine smooth muscle tumors and LGESS that were evaluated as part of routine clinical practice were chosen. Primary antibodies included p53, ATRX, DAXX, Rb, PTEN, mTAP, and MDM2 (Table 1). IHC testing was performed as an initial panel of p53, ATRX, Rb, and PTEN with subsequent testing for mTAP and MDM2 in cases with wild-type p53 expression and staining for DAXX in cases with retained ATRX expression (see rationale in the results section). IHC interrogation of CDKN2C alterations, which can be seen in a small subset of uterine LMS was not included in the panel due to the lack of a clinically validated immunohistochemical biomarker.
Null (complete absence of expression) or over-expression (diffuse nuclear or cytoplasmic expression) of p53 was considered a mutant pattern. A wild-type p53 pattern was defined as heterogeneous nuclear expression of variable intensity. Null expression (complete absence of nuclear ATRX, DAXX, Rb, and cytoplasmic PTEN staining) was considered mutant pattern for these proteins. Wild-type patterns were defined as positive nuclear ATRX, DAXX, and Rb or cytoplasmic PTEN expression. Since Rb expression in non-mutant tissue is often weak and heterogenous, positivity of any intensity or extent was considered wild-type pattern. Evaluation of CDKN2A abnormalities was performed using anti-mTAP, a surrogate marker in which absence of cytoplasmic expression was considered mutant pattern, while cytoplasmic positivity of any extent or intensity was considered wild-type pattern. Diffuse nuclear MDM2 expression was considered mutant pattern, while absence of expression was considered wild-type pattern (Figure 1). All cases were reviewed by three gynecologic pathologists (AMB, LHE, SC) who were blinded to the genomic profile of the samples and independently scored and evaluated the IHC. Slides for which disagreement existed between at least two pathologists were documented.
Figure 1. Molecular-based immunohistochemical (IHC) profiles of uterine leiomyosarcoma (LMS).
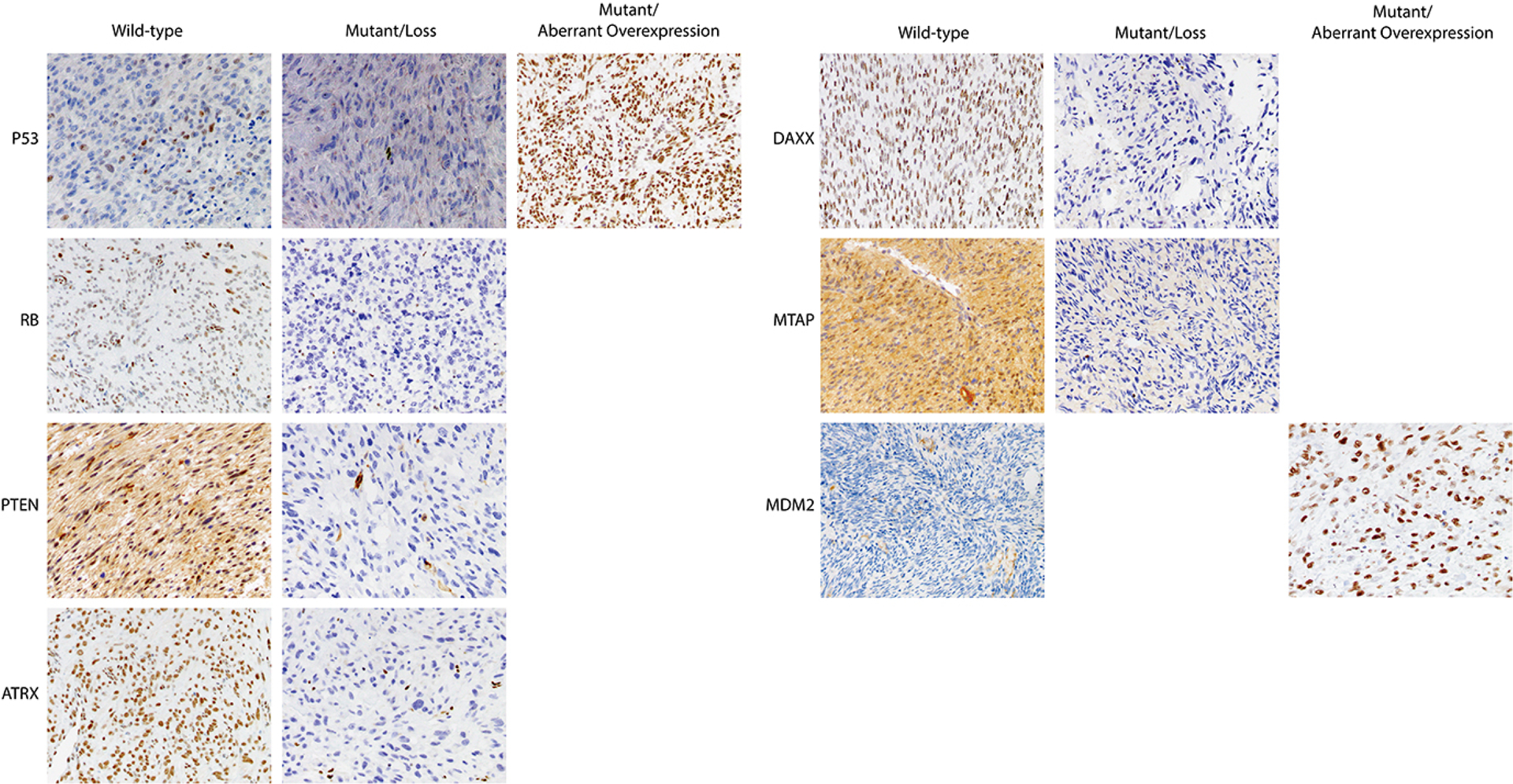
Wild-type and mutant expression patterns of p53, Rb, PTEN, ATRX, DAXX, MTAP, and MDM2 are shown.
Statistical Analysis
Evaluation for mutual exclusivity/co-occurrence was performed using the ‘DISCOVER’ R package (v0.9)33. Concordance analysis between NGS and IHC was performed using the Nam’s score and McNemar test. Inter-assay agreement was measured using the Cohen’s Kappa index, and interobserver agreement was measured using the Fleiss Kappa index.
Results:
Genomic Landscape of Uterine Leiomyosarcoma (LMS)
Among uterine LMS, 98% (n=163/167) showed at least one genomic alteration among the genes included in MSK-IMPACT. At least two genomic alterations were identified in 96% (n=156/163) of samples, while only 4% (n=7/163) showed only one genomic alteration. The median fraction of genome altered in these tumors was 0.331 (Interquartile range [IQR], 0.216 – 0.519). The median number of somatic mutations in these tumors was three (IQR, 1 – 4).
The most altered genes in uterine LMS were TP53 (n=119/167, 71%), followed by RB1 (n=85/167, 51%), ATRX (n=64/167, 38%), PTEN (n=40/167, 24%), MED12 (n=27/167, 16%), CDKN2A (n=14/167, 8%), DAXX (n=7/167, 4%), CDKN2C (n=7/167, 4%), CIC (n=7/167, 4%) and MDM2 (n=5/167, 3%). Alterations involving one or more of these genes were seen in 96% (n=160/167) of uterine LMS (Figure 2A). Other alterations involved AMER1 (n=6/167, 4%), BCOR (n=10/167, 6%), and JAK2 (n=5/167, 3%). Alterations in TP53, ATRX, and MED12 mainly consisted of single nucleotide variants (SNV); mutations in these genes were observed in 61%, 34%, and 17% of uterine LMS, respectively. In contrast, alterations in RB1, PTEN, and CDKN2A mainly consisted of homozygous deletions of these genes (Figure 2A). Homozygous deletions and SNVs in these genes were mutually exclusive (DISCOVER P: <0.001). Alterations in TP53, RB1, ATRX, PTEN, and MED12 showed tendency towards co-occurrence, while alterations in TP53 showed mutual exclusivity with CDKN2A and MDM2 alterations (DISCOVER P: <0.001 and 0.002, respectively) (Figure 2B). Alterations in ATRX and DAXX were also mutually exclusive (DISCOVER P: 0.048) (Figure 2B). Rare LMS (n=3/167, 2%) had homozygous deletion of CDKN2C in absence of alterations in the above referenced genes. A previous study suggested that CDKN2C and CIC are often co-deleted in LMS34; however, we observed this phenomenon in only one case in our cohort.
Figure 2. Somatic mutations and copy number alterations (CNA) in uterine leiomyosarcoma (LMS).
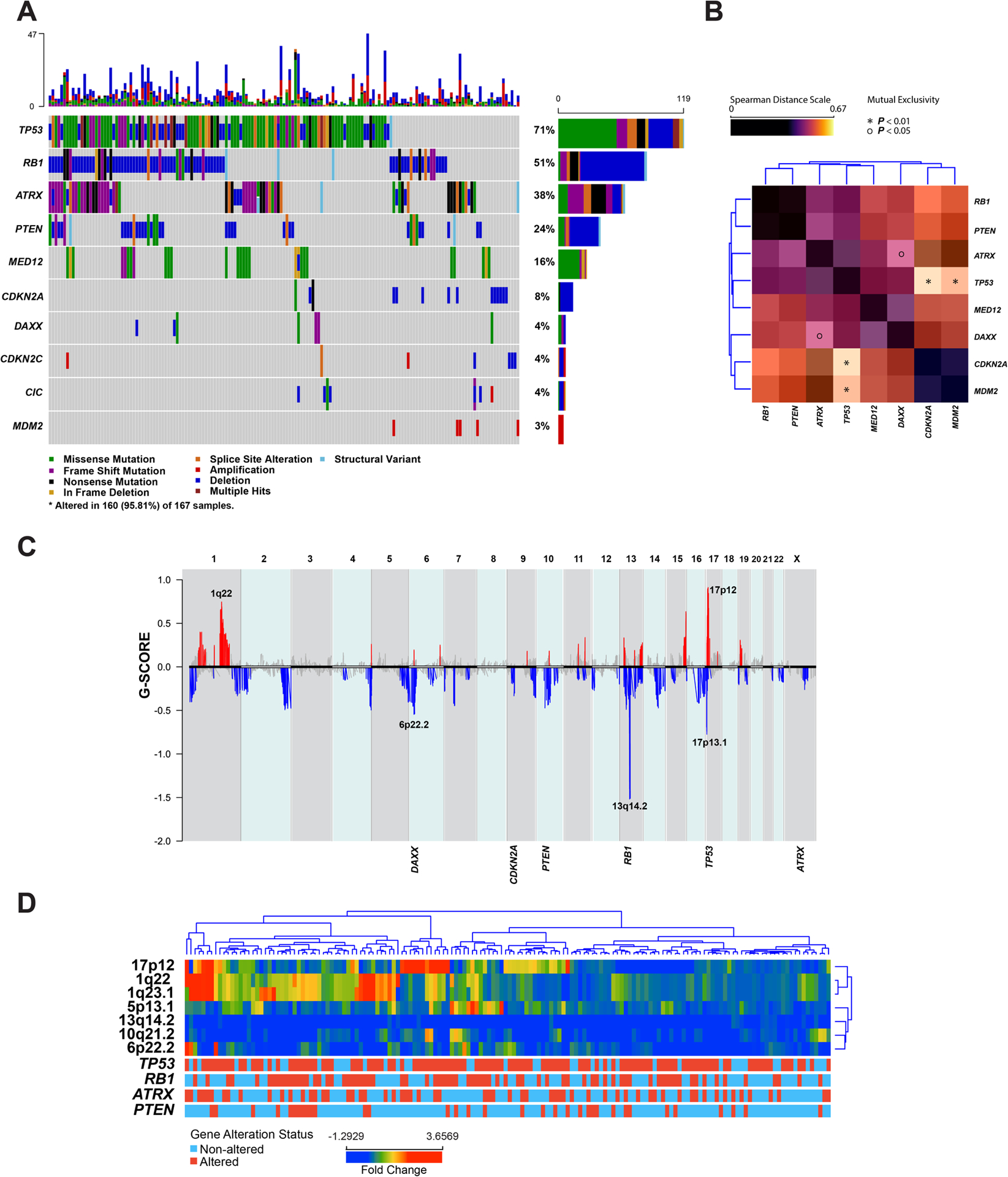
(A) Oncoprint depicting the most recurrent genomic alterations in uterine LMS. Each column represents a tumor with the top bar graph depicting the number and distribution of alterations per sample. Rows show alterations for each gene. The right bar graph shows the number and distribution of alterations for each gene. Mutation types and clinicopathologic features are color-coded according to the legend. (B) Spearman distance matrix showing the somatic interaction of commonly altered genes in uterine LMS. The * and O correspond to p-value as determined by DISCOVER algorithm. The colors correspond to the spearman distance in accordance with the legend. (C) Summary of CNA in uterine LMS based on GISTIC results. Loci and genes depicted in blue represent deletions, and the ones depicted in red represent gains/amplifications. The Y-axis represents G-score which considers the amplitude of the aberration as well as the frequency of its occurrence across samples. (D) Unsupervised hierarchical clustering of the most commonly altered genomic regions in uterine LMS.
Uterine LMS showed frequent CNA; the most common chromosome arm level alterations were gains of chromosome arms 1q, 5p, 8q, and 19p as well as losses of 2p, 2q, 10p, 10q, 16q, and 19q. The most common focal CNA included gains/amplifications of 1q22 and 17p12. The most common focal losses included 6p22, 13q14, and 17p13 (Figure 2C). Unsupervised hierarchical clustering based on focal copy number loci showed uterine LMS divided into four distinct groups defined by 1q22 gains, 17p12 gains, 5p13.1 gains, and the absence of the characteristics mentioned above (Figure 2D).
Rare tumors (n=8/167, 5%) showed intrachromosomal rearrangements resulting in predicted non-functional fusion transcripts involving commonly altered tumor suppressor genes of uterine LMS including ATRX (n=3), RB1 (n=3), and PTEN (n=1). Rearrangements as well as intragenic duplication of exons 1 and 2 of the TP53 gene with a breakpoint within exon 2 (n=1) (Supplemental Figure 1) were also detected. The intrachromosomal rearrangements mainly consisted of inversion events (n=5), intragenic duplications (n=2), and one intragenic deletion. In addition, one case showed a translocation between exon 8 of the PTEN gene on chromosome 10 with the LINC01205 gene on chromosome 3.
Overall, 94% (n=157/167) of samples had at least one genomic alteration involving either TP53, RB1, ATRX, PTEN, CDKN2A, or MDM2, with 80% (n=133/167) having alterations in at least two of these genes. Together, these alterations were considered genomic landmarks of uterine LMS.
Correlation of Genomic Landmarks and Morphologic Features in Uterine Leiomyosarcoma (LMS)
Among the sequenced LMS, 65 tumors had complete sets of hematoxylin-and-eosin stained slides available for morphologic assessment blinded to the NGS data. Among them, 51% were exclusively spindled (n=33/65), 30% were predominantly spindled with any amount of myxoid matrix (n=19/65), and 10% were spindled with any epithelioid features (n=7/65). Among tumors with any degree of epithelioid histology, 9% showed ≥50% epithelioid appearance and were thus considered epithelioid LMS (n=6/65). None of the tumors with any myxoid matrix were diagnosed as myxoid LMS.
In this subset of cases, 53 (82%) had alterations involving at least two of the landmark genes. As expected, TP53 was the most commonly altered gene (n=44/65, 68%) followed by RB1 (n=40/65, 62%), PTEN (n=23/65, 36%), ATRX (n=22/65, 34%), MED12 (n=7/65, 11%), DAXX (n=6/65, 9%), CDKN2C (n=5/65, 8%), CDKN2A (n=4/65, 6%) and MDM2 (n=2/65, 3%). The distribution of the genomic alterations did not show any correlation with presence of any morphologic features (P >0.05 for all comparisons) (Supplemental Table 1).
Immunohistochemical Interrogation of Genomic Landmarks in Uterine Smooth Muscle Tumors
An IHC panel was designed to evaluate expression of proteins corresponding to TP53, RB1, ATRX, PTEN, CDKN2A, or MDM2 genes. A randomly selected test cohort of 28 uterine smooth muscle tumors consisting of LMS (n=16), STUMP (n=6), and leiomyoma (n=6; including 3 cases of benign metastasizing leiomyoma) that were previously sequenced by MSK-IMPACT as described above were evaluated by IHC targeting the genomic landmarks of uterine LMS. An initial panel of p53, Rb, PTEN, and ATRX IHC was performed on the entire test cohort (n=28) and interpreted independently by three gynecologic pathologists blinded to the NGS data. For all tumors, including those that did not harbor any alterations in the initial IHC panel of p53, Rb, PTEN, and ATRX, additional IHC testing of DAXX, MTAP, and MDM2 was performed. Subsequently, these additional IHC markers were also evaluated in cases harboring the corresponding genomic alterations.
Among uterine LMS, IHC abnormalities in p53, Rb, PTEN, and ATRX were seen in 75% (n=12/16), 88% (n=14/16), 44% (n=7/16), and 38% (n=6/16) of tumors, respectively (Figure 3). Two or more abnormal IHC results among these four markers were seen in 81% of uterine LMS (n=13/16). Abnormal ATRX IHC expression was seen in one STUMP (n=1/6, 17%), and another STUMP showed null pattern staining with p53 (n=1/6, 17%). No abnormalities in these four markers were seen in leiomyomas.
Figure 3. Evaluation of concordance between next generation sequencing (NGS) results and immunohistochemistry (IHC) results.
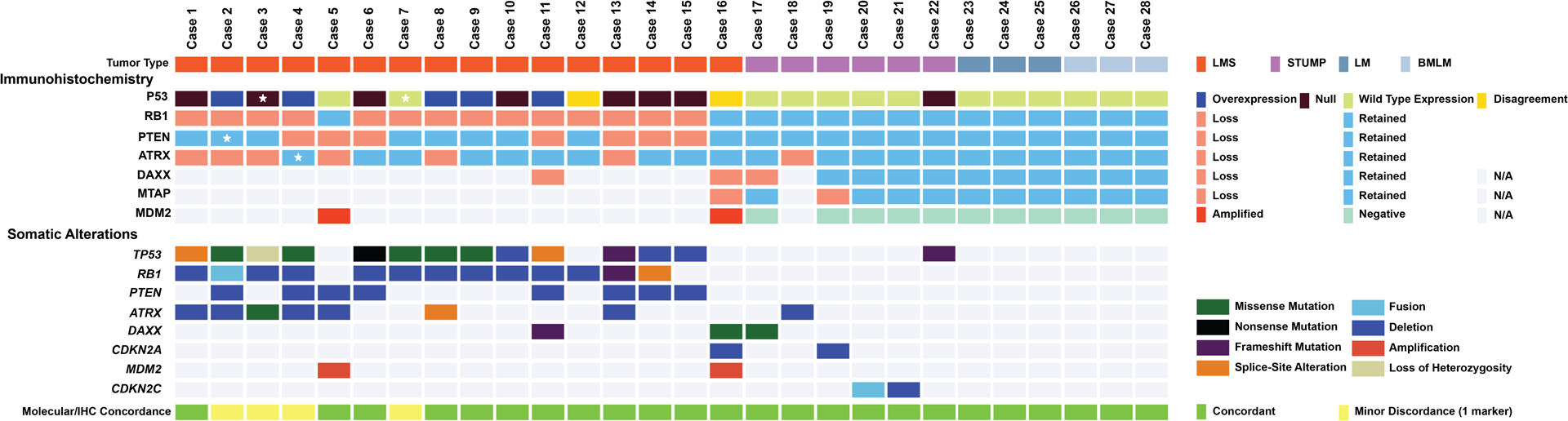
Oncoprint depicting the genomic alterations in a selected cohort of uterine smooth muscle tumors consisting of leiomyosarcoma (LMS), smooth muscle tumor of uncertain malignant potential (STUMP), leiomyoma (LM), and benign metastasizing leiomyoma (BMLM). Each column represents a tumor sample with bottom rows depicting the somatic alterations based on NGS and the top rows depicting the corresponding IHC results. The last row shows concordance status between NGS and IHC with yellow-colored boxes showing minor discordance (one marker discordance); the discordant marker is marked with a star in the corresponding IHC row. For two samples, the reviewing pathologists disagreed on the interpretation of the p53 IHC. N/A: not applicable.
For all tumors, including LMS (n=1), STUMP (n=5), and leiomyomas (n=6), that did not harbor any alterations in the initial IHC panel of p53, Rb, PTEN, and ATRX, additional IHC testing of DAXX, MTAP, and MDM2 was performed. Abnormal expression of all three markers were observed in the single LMS (case 16). Among the remaining STUMP cases, two demonstrated only one IHC abnormality involving DAXX (case 17) or MTAP (case 19). No DAXX, MTAP, or MDM2 IHC abnormalities were seen in leiomyomas (Figure 3).
Concordance was excellent among three gynecologic pathologists in the interpretation of IHC results (Fleiss Kappa: 0.97, X2 P: <0.0001). There was disagreement only in the interpretation of p53 IHC in two uterine LMS (cases 12 and 16), in which the p53 IHC was interpreted as wild-type by one pathologist and aberrant by the other two pathologists (Supplemental Figure 2). Interpretation of all remaining markers was concordant among all pathologists.
NGS testing showed that all uterine LMS (n=16, 100%) in the test cohort demonstrated alterations in TP53, RB1, PTEN, ATRX, DAXX, CDKN2A, and/or MDM2. Except for one tumor (case 12), all uterine LMS showed alterations in at least two of these landmark genes. Most (n=15/16, 94%) had alterations involving at least one of the TP53, RB1, PTEN, and ATRX genes; the remaining case (case 16) did not harbor alterations in any of these genes and instead had abnormalities in DAXX, CDKN2A, and MDM2. TP53 (n=13/16, 81%) and RB1 (n=13/16, 81%) were the most commonly altered genes in the test cohort of uterine LMS, followed by PTEN (n=8/16, 50%) and ATRX (n=7/16, 44%). Alterations in RB1 were homozygous deletions (n=10/13, 77%) or mutations (n=2/13, 15%). One tumor (case 2) had an RB1 intrachromosomal rearrangement consisting of an inversion of exons 1–20 of the gene forming a non-functional fusion of RB1 with the promoter of LRCC63 (Supplemental Figure 1).
Four STUMP (n=4/6, 67%) each harbored one alteration in TP53, ATRX, DAXX, and CDKN2A, respectively. Two remaining STUMP each showed an alteration involving CDKN2C (n=2/6, 33%), while leiomyomas (n=6), including benign metastasizing leiomyomas, did not harbor alterations in any of the landmark genes (Figure 3).
Excellent concordance was observed between IHC and genomic profiles (Cohen’s kappa: 0.941, X2 P: <0.0001) (Figure 3). Only four IHC results in four uterine LMS (cases 2–4 and 7) were discordant with their corresponding genomic profiles. One LMS (case 2) demonstrated retained cytoplasmic PTEN staining (wild-type pattern) despite harboring a PTEN deletion. Another LMS (case 4) showed retained nuclear ATRX staining (wild-type pattern) but had an ATRX deletion. P53 staining was wild-type in one LMS (case 7) with a TP53 mutation, while aberrant (overexpression) in another LMS (case 3) harboring LOH of the TP53 gene (Figure 3).
Upon correlation of the IHC results with corresponding genomic profiles of the test cohort, MDM2 and DAXX IHC were each performed in two LMS (cases 5 and 11) (Figure 3). Case 5 harbored MDM2 amplification confirmed by MDM2 IHC, while case 11 harbored a frameshift mutation involving DAXX which was also confirmed by DAXX IHC.
The variant allele frequency of the mutations in the test cohort was compared with the estimated tumor purity and showed excellent correlation with a B coefficient of 0.85 (R2: 0.46, Pearson P: 0.008), with ATRX mutations being the only outliers. Evaluation of the total and allele specific total copy number of TP53 by ‘facets’ in these tumors showed LOH and copy neutral LOH in 9 of 10 samples with TP53 mutations and homozygous deletions in another 3 tumors, supporting bi-allelic inactivation of landmark tumor suppressor genes in uterine LMS (Supplemental Figure 3).
Validation of Immunohistochemistry
p53, Rb, PTEN, and ATRX IHC was evaluated in routine clinical cases, including uterine LMS (n=8), STUMP (n=12), leiomyoma and leiomyoma variants (n=21), and LGESS (n=7). Leiomyoma variants included cellular leiomyoma (n=1) and leiomyoma with bizarre nuclei (n=13; fumarate hydratase deficient, n=10, fumarate hydratase retained, n=3) (Figure 4). Six LMS (Cases 29–34) demonstrated abnormal p53 and Rb; two cases had abnormal PTEN expression, one of which also showed abnormal ATRX staining. IHC abnormalities were observed in a subset of STUMP (n=4/12, 25%), including abnormal p53 (n=1), Rb (n=2), and PTEN (n=1) expression. Among leiomyoma and leiomyoma variants (n=21), two fumarate hydratase deficient tumors each showed one alteration in either p53 or Rb expression (Figure 4). No IHC abnormalities were seen among LGESS.
Figure 4. Validation of immunohistochemistry (IHC) panel in uterine smooth muscle tumors and endometrial stromal sarcomas.
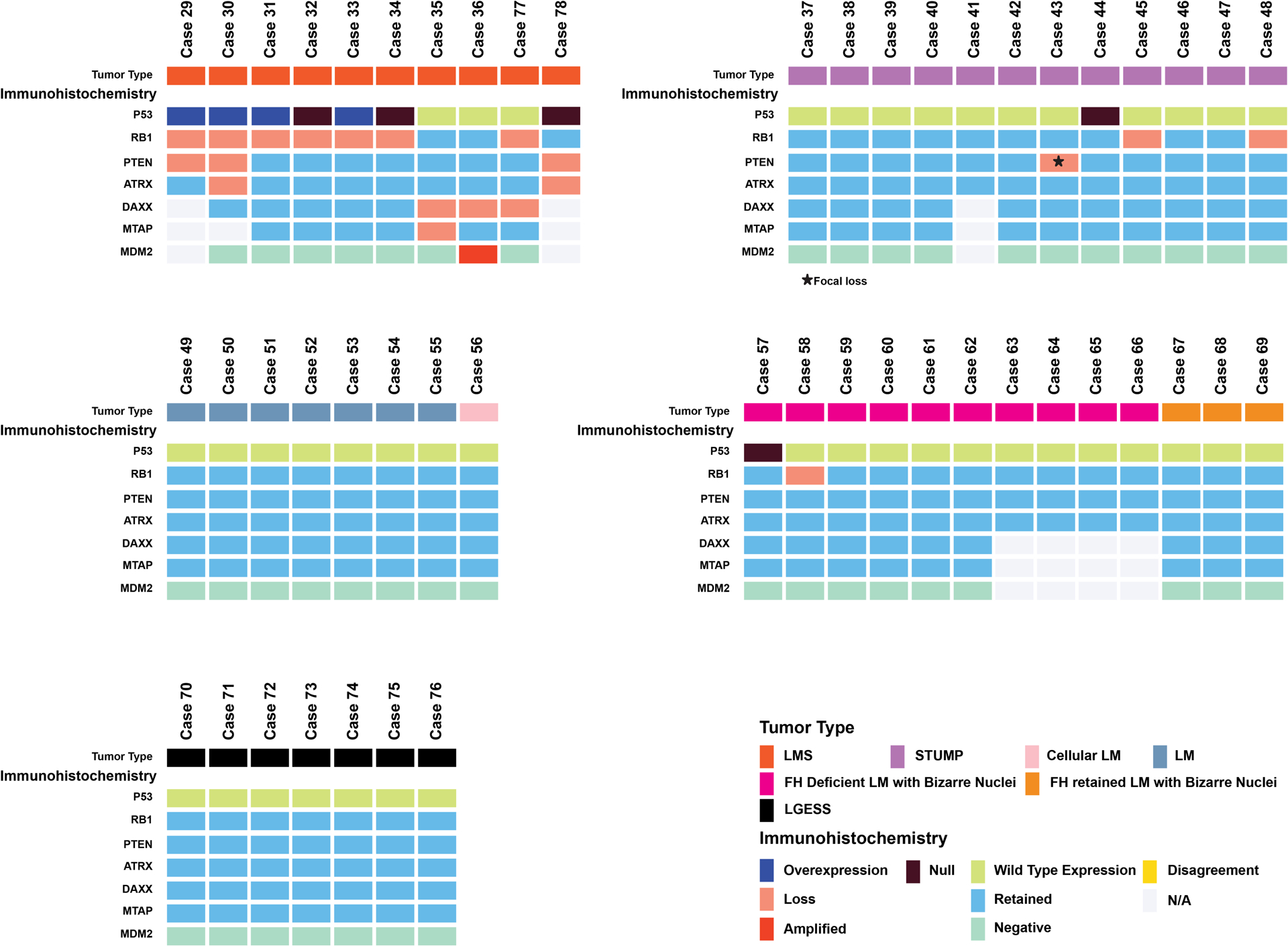
Oncoprint depicting the results of p53, Rb, PTEN, and ATRX in a routine clinical cases including leiomyosarcoma (LMS), smooth muscle tumor of uncertain malignant potential (STUMP), leiomyoma (LM), LM variants including cellular LM, fumarate hydratase deficient LM with bizarre nuclei and fumarate hydratase retained LM with bizarre nuclei, and low-grade endometrial stromal sarcoma (LGESS). Uterine LMS (with the exception of cases 77 and 78) fulfilled morphologic criteria for malignancy.
The IHC panel was then applied to two diagnostically challenging uterine smooth muscle tumors (Cases 77 and 78) for which NGS data was unavailable. One patient (Case 77) had a 9 cm uterine submucosal smooth muscle tumor with epithelioid (<50%) and spindled morphology, mild nuclear atypia, no tumor necrosis, and a mitotic index of 7 mitotic figures per 10 high power fields and thus did not fulfill morphologic criteria for epithelioid LMS (Figure 5A). No IHC abnormalities were detected in ATRX, p53, or PTEN. However, loss of Rb and DAXX IHC expression was observed, leading to a diagnosis of LMS (Figure 5B, 5C). Another patient (Case 78) had FIGO stage 4 uterine LMS with spindled, myxoid, and epithelioid features that demonstrated marked nuclear atypia, tumor necrosis, a mitotic index of 51 mitotic figures per 10 high power fields (Figure 5D). IHC demonstrated loss of PTEN and ATRX expression (Figure 5E, 5F) with null p53 and retained RB expression. However, a polypectomy performed eight years prior demonstrated a spindle cell proliferation with mild nuclear atypia, focal myxoid features, no necrosis, and a mitotic index 6 per 10 high power fields (Figure 5G). IHC performed on the polypectomy specimen demonstrated loss of PTEN and ATRX (Figure 5H, 5I) with aberrant null p53 expression in the atypical spindle cell proliferation, suggesting LMS by molecular-based IHC, despite insufficient morphologic criteria for malignancy. Overall sensitivity and specificity of using at least two abnormal IHC findings were 92% and 100%, respectively (Supplemental Table 2).
Figure 5. Utility of molecular-based immunohistochemistry (IHC) in diagnostically challenging uterine smooth muscle tumors.
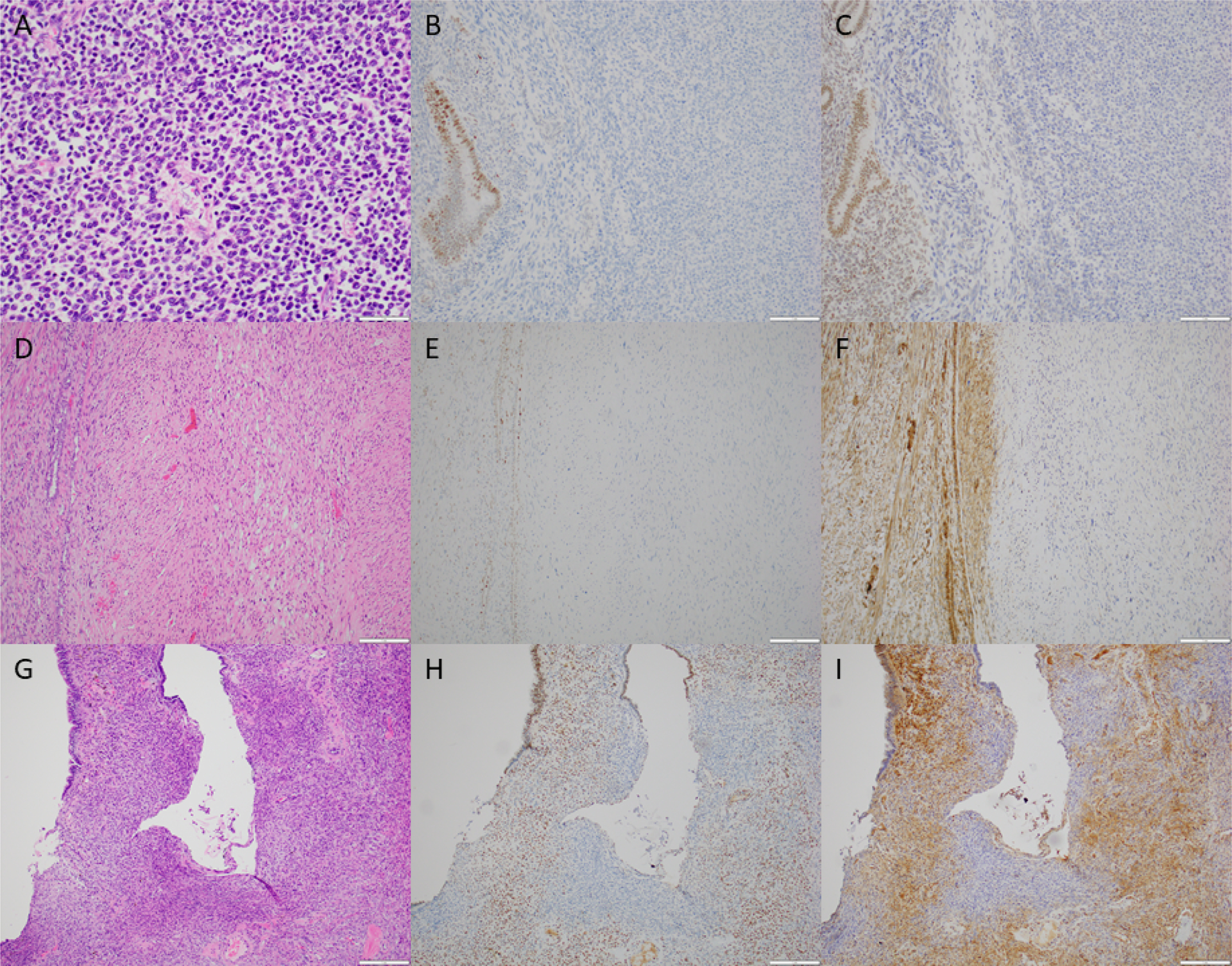
(A) Case 77 demonstrating epithelioid cells with mild nuclear atypia, no tumor necrosis, and low mitotic activity. (B) Loss of Rb and (C) DAXX IHC expression in the tumor (center and right) with positive internal control (endometrial glands and stroma, left). (D) Hysterectomy specimen from Case 78 demonstrating marked nuclear atypia. (E) Loss of cytoplasmic PTEN and (F) loss of nuclear ATRX IHC expression in the tumor (center and right) with positive internal control (myometrium, left). (G) A prior polypectomy demonstrating atypical spindle cells surrounding an endometrial gland. (H) Loss of cytoplasmic PTEN and (I) loss of nuclear ATRX in the atypical spindle cell proliferation with positive internal control (surrounding endometrial stroma and glands).
Discussion:
In this study, we characterized the genomic profile of uterine LMS and identified surrogate IHC markers that may assist in the diagnostic evaluation of challenging uterine smooth muscle tumors. Through NGS analysis, we showed that uterine LMS is frequently characterized by genomic alterations affecting cell cycle regulators namely RB1 deletion, TP53 and cell cycle regulatory pathway alterations (TP53, CDKN2A, CDKN2C and MDM2), and alternate lengthening of telomeres through ATRX or DAXX alterations and deletions of PTEN35. These genetic abnormalities followed a non-redundant manner leading to mutual exclusivity of alterations affecting the same pathway (i.e. between alterations of TP53, MDM2, and CDKN2A as well as ATRX and DAXX). We also confirmed that alterations of these genes result in abnormal protein expression determined by IHC (with the exception of CDKN2C for which no clinically validated immunohistochemical biomarker was available)15,18,35. Excellent concordance of p53, RB, ATRX, PTEN, DAXX, MTAP, and MDM2 IHC was observed when correlated to genomic profiles of uterine LMS. Any two abnormalities in an initial IHC panel of p53, RB, ATRX, and PTEN was informative in approximately 81% of LMS, which increases to 88% with the addition of DAXX, MTAP, and MDM2. IHC and/or NGS profiles of uterine LMS were distinct from STUMP, leiomyoma including morphologic variants, and LGESS. Notably, STUMP and leiomyoma variants demonstrated no or at most one abnormality in uterine LMS genomic landmarks. Molecular-based IHC may be useful in challenging uterine smooth muscle tumors that may not fulfill morphologic criteria for malignancy.
The vast majority of uterine LMS have at least two landmark genomic alterations with TP53 and RB1 alterations being the most commonly observed pair15. The synergistic effect of the co-inactivation of Rb and p53 proteins has been well-established in other malignancies,36 such as small cell carcinoma37, prostate carcinoma38, ovarian cancer,39 and osteosarcoma40. Comparison of uterine LMS with STUMP and leiomyoma shows that any uterine smooth muscle tumor that harbors two or more of the landmark alterations by NGS or IHC is invariably LMS, while rare LMS may only harbor one alteration or show abnormality in expression of just one IHC marker. The absence of abnormalities in any of the markers, particularly in a smooth muscle tumor lacking sufficient morphologic features for malignancy, renders a diagnosis of LMS highly unlikely.
The landmark alterations involving a combination of TP53, RB1, ATRX, PTEN, DAXX, CDKN2A, and MDM2 are ubiquitous in solid tumors; however, in the context of uterine mesenchymal tumors, these alterations are highly enriched in uterine LMS. Mohammad et al. recently showed wild-type p53 IHC patterns among high-grade endometrial stromal sarcoma (HGESS), LGESS, inflammatory myofibroblastic tumors, and sarcomas with PDGFB fusion. Only 1 of 11 NTRK fusion-positive sarcomas showed aberrant p53 expression, and that tumor demonstrated marked nuclear pleomorphism41. High-grade Mullerian adenosarcoma with or without stromal overgrowth may harbor TP53, ATRX and/or MDM2 alterations42,43, and malignant perivascular epithelioid cell tumors (PEComa) may harbor alterations of TP53, ATRX, or RB144. Given this genomic overlap, interpretation of mutational profiles and surrogate IHC results are informative only in the appropriate histologic context and when myogenic differentiation is confirmed. HGESS, LGESS, NTRK, and PDGFB fusion-positive sarcomas can largely be excluded based on absence of desmin expression without confirmation of landmark alterations by mutational profiling or surrogate IHC. ALK IHC and confirmation of ALK fusion are helpful in identification of ALK-rearranged inflammatory myofibroblastic tumor and myofibroblastic sarcomas. High-grade adenosarcoma with sarcomatous overgrowth usually demonstrates foci of biphasic growth, periglandular stromal condensation, and phyllodes-like architecture. Diagnostic criteria for uterine PEComa is controversial, but absence of melanocytic marker expression, TSC1/TSC2 alterations, and TFE3 rearrangements favors smooth muscle neoplasia45.
Among differential diagnoses, the distinction between LMS and STUMP or leiomyoma with bizarre nuclei is likely the most challenging in practice. Leiomyomas with bizarre nuclei definitionally demonstrate unifocal, multifocal, or diffuse marked cytologic atypia; low mitotic activity; and no tumor necrosis. A subset of sporadic leiomyomas with bizarre nuclei harbor somatic FH mutations or homozygous deletions46. Rare FH-deficient tumors may also harbor TP53 mutations46; in the absence of other alterations involving LMS-associated landmark genes, these TP53-mutant leiomyomas with bizarre nuclei that are also FH-deficient would not be categorized as LMS by our proposed IHC panel. Biallelic inactivation of FH has been reported in uterine LMS47–49, and some FH-deficient uterine smooth muscle tumors demonstrate elevated mitotic activity (≥5 mitotic figures/10 high power fields) that would be considered STUMP. Application of our diagnostic algorithm in these unusual tumors and acquisition of long-term clinical follow-up require additional study. A recent study also reported TP53 and RB1 alterations in rare FH-proficient leiomyomas with bizarre nuclei, but no outcome data was provided46. By our IHC panel, these tumors would have likely been classified as LMS, and clinical follow-up of those patients would be of great interest.
While most uterine LMS easily fulfill morphologic criteria for malignancy, the utility of the proposed IHC panel is maximized when applied to diagnostically challenging smooth muscle tumors, particularly those that would otherwise be diagnosed as STUMP. In our study, surrogate IHC was especially informative in two smooth muscle tumors that showed some worrisome histologic features but did not fulfill diagnostic criteria for malignancy. One tumor showed <50% epithelioid morphology, no atypia or necrosis, and a mitotic index of 7 mitotic figures per 10 high power; loss of Rb and DAXX IHC expression was observed which led to a diagnosis of LMS. In a patient with known advanced stage uterine LMS, a prior endometrial polypectomy specimen showed a mildly atypical spindle cell proliferation with focal myxoid features, no necrosis, and 6 mitotic figures per 10 high power fields; loss of PTEN and ATRX along with aberrant null p53 expression suggested the presence of LMS by molecular-based IHC at the time of polypectomy, despite insufficient morphologic criteria for malignancy. Given the study findings, we now routinely performed this IHC panel on STUMP and lesions such as these that would otherwise be misclassified as benign.
IHC and NGS results appear equivalent in identifying alterations of the landmark alterations in uterine LMS, highlighting the utility of IHC as a surrogate for genomic alterations. While prior studies have suggested using p16 as a surrogate marker for CDKN2A alterations, we suggest utilizing MTAP IHC for this purpose. The absence of p16 labeling can be seen in CDKN2A non-altered tumors; furthermore, in cases with absent p16 labeling, the gene inactivation mechanism is unknown. In contrast, since the MTAP gene is often co-deleted with CDKN2A, absence of IHC labelling for MTAP is a more reliable surrogate marker of CDKN2A gene deletion50.
Prior studies have attempted to use adjunct tools for the diagnosis of uterine smooth muscle tumors. Abnormal p53 and p16 protein expression in uterine LMS has been shown in multiple studies and have been applied with varying degrees of success for distinguishing uterine LMS from its mimics either alone or in combination with proliferation markers such as Ki67 and MCM223,51–53. Croce et al. used comparative genomic hybridization to risk stratify STUMP and showed that high-risk STUMP and uterine LMS have similar array-CGH landscapes54. More recently, Schaefer et al. used a combination of p53, Rb, p16, and PTEN IHC for evaluation of uterine and soft tissue LMS; they showed abnormal expression of p53 and Rb in >80% of all LMS and abnormal PTEN expression in >40% of the samples and further established that these abnormalities often co-occur15.
Based on our results, we propose a diagnostic algorithm for challenging uterine smooth muscle tumors with atypical features (Figure 6). A smooth muscle tumor can be classified as LMS if it fulfills the morphologic criteria for malignancy. In the setting of a smooth muscle tumor with any unusual features, such as STUMP, for which morphology is not conclusive, IHC may be performed. The data presented here suggest a first round of IHC for p53, ATRX, Rb, and PTEN is often informative. Abnormal results in any two or more of these markers should prompt a diagnosis of LMS. If less than two markers are abnormal, then IHC for MDM2, DAXX, and MTAP can be pursued. Given mutual exclusivity of TP53 alterations with CDKN2A and MDM2 alterations, MTAP and MDM2 IHC should be performed in p53 wild-type cases. ATRX and DAXX alterations are also mutually exclusive, so DAXX IHC should be performed in the setting of retained ATRX expression. After these evaluation steps, any uterine smooth muscle tumor with two or more abnormal IHC results likely represents LMS. Tumors with one abnormal IHC result may represent STUMP or LMS, and those with no abnormal IHC results are likely either leiomyoma or STUMP. An important research question is whether STUMP with one abnormal IHC result compared to no abnormal IHC results have a higher risk for recurrence or more aggressive clinical behavior.
Figure 6.
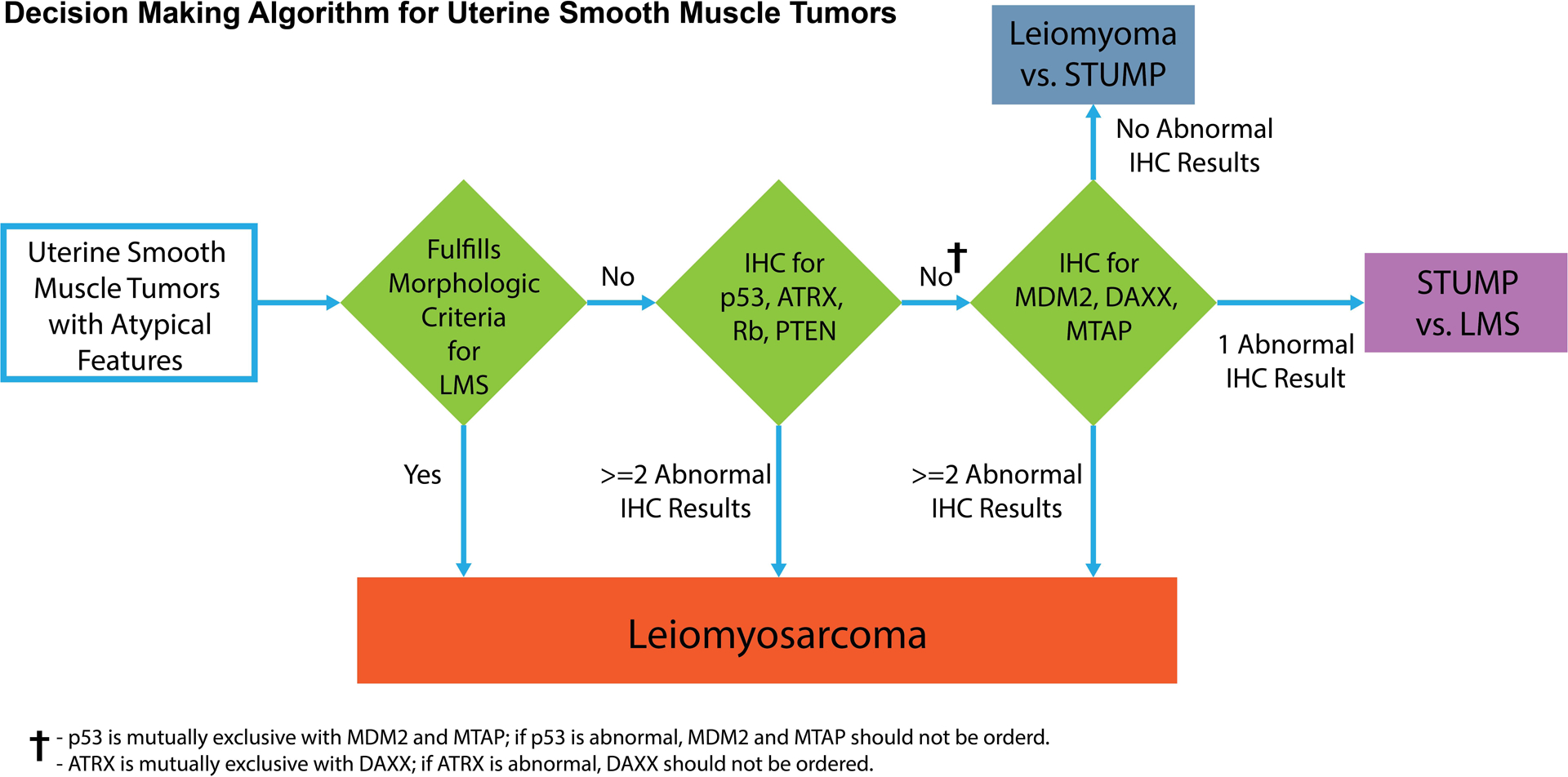
Proposed decision support algorithm for diagnosis of uterine smooth muscle neoplasms with atypical features
Careful interpretation of IHC is paramount with the use of this diagnostic algorithm (Supplemental Table 3). In brief, aberrant p53 expression is defined by strong and diffuse nuclear or cytoplasmic staining (overexpression) or complete absence of staining (null). The complete absence of nuclear ATRX, DAXX, Rb, and cytoplasmic PTEN and mTAP staining is required for designation as a mutant pattern for these proteins. Diffuse nuclear MDM2 expression is considered mutant pattern. Given the frequent presence of non-neoplastic inflammatory or endothelial cells within the tumor, interpretation of these IHC markers must only be made in tumor cells. Evaluation of ATRX, DAXX, PTEN, and mTAP staining patterns is usually straightforward given that complete loss of expression in tumor cells is readily apparent in the presence of an internal positive control.
However, interpretational difficulties may occur in assessing p53, RB1, and MDM2 staining. Wild-type p53 expression may show variable extent ranging from staining of few tumor nuclei to staining of many tumor nuclei, the latter which can be confused with aberrant overexpression. However, the expression pattern of p53 in the tumor should always be compared to the internal control, and any variability in the intensity of nuclear staining should be interpreted as wild-type. The complete absence of p53 staining of the tumor cells should be considered a mutant pattern if the internal control staining is adequate, and its recognition is usually straightforward except in limited tissue samples. Interpretation of Rb is often difficult as wild-type Rb often shows faint and heterogenous staining. In our experience, loss of Rb expression is determined when there is complete absence of nuclear staining of any intensity in the tumor cells. This often requires review of multiple fields at high magnification, and careful interpretation of staining in tumor cells only given that nuclear staining is expected in endothelial and inflammatory cells (Supplemental Figure 4). MDM2 immunohistochemistry is more frequently used in the evaluation of lipomatous soft tissue tumors and may be variable in extent and intensity of staining. In this study, however, nuclear MDM2 staining was diffuse and seen only in LMS harboring MDM2 amplification; MDM2 expression was otherwise completely negative in the remaining tumors in the test and validation cohorts. Patchy, focal, or rare nuclear MDM2 staining may be interpreted as equivocal, and MDM2 amplification should be confirmed by fluorescence in situ hybridization.
In conclusion, we have shown that uterine LMS have several genomic landmark alterations involving TP53, RB1, PTEN, ATRX, DAXX, CDKN2A, and MDM2 for which IHC surrogates exist. Correlation between p53, RB, PTEN, ATRX, DAXX, MTAP, and MDM2 abnormalities detected by IHC and NGS profiles is excellent. Excellent reproducibility is observed in the interpretation of these IHC markers among pathologists. A diagnostic algorithm involving molecular-based IHC can aid in the evaluation of unusual uterine smooth muscle tumors. Future work on larger and independent cohorts is needed to validate the utility of this diagnostic algorithm.
Supplementary Material
Funding:
The study was supported in part by the MSK Cancer Center Support Grant P30 CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest:
S.C. is a paid consultant for AstraZeneca. M.L.H. is an advisory board member for Glaxo Smith Kline; a paid consultant for Lilly, Exact Sciences, Research to Practice, and UpToDate; and reports spouse employment by Sanofi. No conflicts of interests were disclosed by the other authors.
Ethics Approval and Consent to Participate:
Institutional Review Board approval was obtained at Memorial Sloan Kettering Cancer Center (MSKCC).
Data Availability Statement:
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References:
- 1.Cui R, Wright J, Hou J. Uterine leiomyosarcoma: a review of recent advances in molecular biology, clinical management and outcome. BJOG. 2017;124:1028–1037. [DOI] [PubMed] [Google Scholar]
- 2.George S, Serrano C, Hensley ML, Ray-Coquard I. Soft tissue and uterine leiomyosarcoma. J. Clin. Oncol 2018;36:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mastrangelo G, Coindre JM, Ducimetière F, et al. Incidence of soft tissue sarcoma and beyond: a population-based prospective study in 3 European regions. Cancer. 2012;118:5339–5348. [DOI] [PubMed] [Google Scholar]
- 4.Friedman CF, Hensley ML. Options for adjuvant therapy for uterine leiomyosarcoma. Curr. Treat. Options Oncol 2018;19:1–11. [DOI] [PubMed] [Google Scholar]
- 5.Moch H Female genital tumours: WHO Classification of Tumours, Volume 4. WHO Classification of Tumours. 2020;4 [Google Scholar]
- 6.Chapel DB, Nucci MR, Quade BJ, Parra-Herran C. Epithelioid leiomyosarcoma of the uterus: modern outcome-based appraisal of diagnostic criteria in a large institutional series. Am. J. Surg. Pathol 2021;46:464–475. [DOI] [PubMed] [Google Scholar]
- 7.Toledo G, Oliva E. Smooth muscle tumors of the uterus: a practical approach. Arch. Pathol. Lab. Med 2008;132:595–605. [DOI] [PubMed] [Google Scholar]
- 8.Lim D, Alvarez T, Nucci MR, et al. Interobserver variability in the interpretation of tumor cell necrosis in uterine leiomyosarcoma. Am. J. Surg. Pathol 2013;37:650–658. [DOI] [PubMed] [Google Scholar]
- 9.Yanai H, Wani Y, Notohara K, Takada Si, Yoshino T. Uterine leiomyosarcoma arising in leiomyoma: clinicopathological study of four cases and literature review. Pathol. Int 2010;60:506–509. [DOI] [PubMed] [Google Scholar]
- 10.Mittal KR, Chen F, Wei JJ, et al. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod. Pathol 2009;22:1303–1311. [DOI] [PubMed] [Google Scholar]
- 11.Gadducci A, Zannoni GF. Uterine smooth muscle tumors of unknown malignant potential: a challenging question. Gynecol. Oncol 2019;154:631–637. [DOI] [PubMed] [Google Scholar]
- 12.Oliva E. Practical issues in uterine pathology from banal to bewildering: the remarkable spectrum of smooth muscle neoplasia. Mod. Pathol 2016;29:S104–S120. [DOI] [PubMed] [Google Scholar]
- 13.Ip PP, Cheung AN. Pathology of uterine leiomyosarcomas and smooth muscle tumours of uncertain malignant potential. Best Pract. Res. Clin. Obstet. Gynaecol 2011;25:691–704. [DOI] [PubMed] [Google Scholar]
- 14.Astolfi A, Nannini M, Indio V, et al. Genomic database analysis of uterine leiomyosarcoma mutational profile. Cancers (Basel). 2020;12:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer IM, Lundberg MZ, Demicco EG, et al. Relationships between highly recurrent tumor suppressor alterations in 489 leiomyosarcomas. Cancer. 2021;127:2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensley ML, Chavan SS, Solit DB, et al. Genomic landscape of uterine sarcomas defined through prospective clinical sequencing. Clin. Cancer Res 2020;26:3881–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liau J-Y, Tsai J-H, Jeng Y-M, Lee J-C, Hsu H-H, Yang C-Y. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am. J. Surg. Pathol 2015;39:236–244. [DOI] [PubMed] [Google Scholar]
- 18.Chudasama P, Mughal SS, Sanders MA, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat. Comm 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazar AJ, McLellan MD, Bailey MH, et al. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int. J. Gynecol. Pathol 2008;27:326–332. [DOI] [PubMed] [Google Scholar]
- 21.Mills AM, Ly A, Balzer BL, et al. Cell cycle regulatory markers in uterine atypical leiomyoma and leiomyosarcoma: immunohistochemical study of 68 cases with clinical follow-up. Am. J. Surg. Pathol 2013;37:634–642. [DOI] [PubMed] [Google Scholar]
- 22.Ip PP, Lim D, Cheung AN, Oliva E. Immunoexpression of p16 in uterine leiomyomas with infarct-type necrosis: an analysis of 35 cases. Histopathol. 2017;71:743–750. [DOI] [PubMed] [Google Scholar]
- 23.O’neill C, McBride H, Connolly L, McCluggage W. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathol. 2007;50:851–858. [DOI] [PubMed] [Google Scholar]
- 24.Ünver NU, Acikalin MF, Öner Ü, Ciftci E, Ozalp SS, Colak E. Differential expression of P16 and P21 in benign and malignant uterine smooth muscle tumors. Arch. Gynecol. Obstet 2011;284:483–490. [DOI] [PubMed] [Google Scholar]
- 25.Allen M-ML, Douds JJ, Liang SX, Desouki MM, Parkash V, Fadare O. An immunohistochemical analysis of stathmin 1 expression in uterine smooth muscle tumors: differential expression in leiomyosarcomas and leiomyomas. Int. J. Clin. Exp. Pathol 2015;8:2795. [PMC free article] [PubMed] [Google Scholar]
- 26.Chow KL, Tse KY, Cheung CL, et al. The mitosis-specific marker phosphohistone-H3 (PHH 3) is an independent prognosticator in uterine smooth muscle tumours: an outcome-based study. Histopathol. 2017;70:746–755. [DOI] [PubMed] [Google Scholar]
- 27.Cao C, Omar J, De Cotiis D, Richard S, Rosenblum N, Chan J. Digital quantification of Ki-67 and PHH3 in the classification of uterine smooth muscle tumors. Gynecol. Oncol 2021;162:S132. [DOI] [PubMed] [Google Scholar]
- 28.Slatter TL, Hsia H, Samaranayaka A, et al. Loss of ATRX and DAXX expression identifies poor prognosis for smooth muscle tumours of uncertain malignant potential and early stage uterine leiomyosarcoma. J. Pathol. Clin. Res 2015;1:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 2017;23:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2. 0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131–e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canisius S, Martens JW, Wessels LF. A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol. 2016;17:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams EA, Sharaf R, Decker B, et al. CDKN2C-null leiomyosarcoma: a novel, genomically distinct class of TP53/RB1–wild-type tumor with frequent CIC genomic alterations and 1p/19q-codeletion. JCO Precis. Oncol 2020;4:955–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mäkinen N, Aavikko M, Heikkinen T, et al. Exome sequencing of uterine leiomyosarcomas identifies frequent mutations in TP53, ATRX, and MED12. PLoS Genet. 2016;12:e1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2(2):103–112. [DOI] [PubMed] [Google Scholar]
- 37.Kim K-B, Dunn CT, Park K-S. Recent progress in mapping the emerging landscape of the small-cell lung cancer genome. Exp. Mol. Med 2019;51:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamid AA, Gray KP, Shaw G, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur. Urol 2019;76:89–97. [DOI] [PubMed] [Google Scholar]
- 39.López-Reig R, López-Guerrero JA. The hallmarks of ovarian cancer: proliferation and cell growth. Eur. J. Cancer Suppl 2020;15:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Azevedo JWV, Fernandes TAAdM, Fernandes JV, et al. Biology and pathogenesis of human osteosarcoma. Oncol. Lett 2020;19:1099–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohammad N, Stewart CJ, Chiang S, et al. p53 immunohistochemical analysis of fusion-positive uterine sarcomas. Histopathol. 2021;78:805–813. [DOI] [PubMed] [Google Scholar]
- 42.Howitt BE, Sholl LM, Dal Cin P, et al. Targeted genomic analysis of Müllerian adenosarcoma. J. Pathol 2015;235:37–49. [DOI] [PubMed] [Google Scholar]
- 43.Hodgson A, Amemiya Y, Seth A, Djordjevic B, Parra-Herran C. High-grade Müllerian adenosarcoma. Am. J. Surg. Pathol 2017;41:1513–1522. [DOI] [PubMed] [Google Scholar]
- 44.Bennett JA, Ordulu Z, Pinto A, et al. Uterine PEComas: correlation between melanocytic marker expression and TSC alterations/TFE3 fusions. Mod. Pathol 2022;35:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soslow RA. Melanocytic marker expression and TSC alterations/TFE3 fusions in uterine PEComas. Mod. Pathol 2022;35:449–450. [DOI] [PubMed] [Google Scholar]
- 46.Bennett JA, Weigelt B, Chiang S, et al. Leiomyoma with bizarre nuclei: a morphological, immunohistohemical and molecular analysis of 31 cases. Mod. Pathol 2017;30:1476–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiuru M, Lehtonen R, Arola J, et al. Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. 2002;62:4554–4557. [PubMed] [Google Scholar]
- 48.Makinen N, Kampjarvi K, Frizzell N, et al. Characterization of MED12, HMGA2, and FH alterations reveals molecular variability in uterine smooth muscle tumors. Mol. Cancer 2017;16:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liegl-Atzwanger B, Heitzer El, Flicker K, et al. Exploring chromosomal abnormalities and genetic changes in uterine smooth muscle tumors. Mod. Pathol 2016;29:1262–1277. [DOI] [PubMed] [Google Scholar]
- 50.Powell EL, Leoni LM, Canto MI, et al. Concordant loss of MTAP and p16/CDKN2A expression in gastroesophageal carcinogenesis: evidence of homozygous deletion in esophageal noninvasive precursor lesions and therapeutic implications. Am. J. Surg. Pathol 2005;29:1497–1504. [DOI] [PubMed] [Google Scholar]
- 51.Schaefer IM, Hornick JL, Sholl LM, Quade BJ, Nucci MR, Parra-Herran C. Abnormal p53 and p16 staining patterns distinguish uterine leiomyosarcoma from inflammatory myofibroblastic tumour. Histopathol. 2017;70:1138–1146. [DOI] [PubMed] [Google Scholar]
- 52.Delgado B, Dreiher J, Braiman D, Meirovitz M, Shaco-Levy R. P16, Ki67, P53, and WT1 Expression in uterine smooth muscle tumors: an adjunct in confirming the diagnosis of malignancy in ambiguous cases. Int. J. Gynecol. Pathol 2021;40:257–262. [DOI] [PubMed] [Google Scholar]
- 53.Keyhanian K, Lage JM, Chernetsova E, Sekhon H, Eslami Z, Islam S. Combination of MCM2 with Ki67 and p16 immunohistochemistry can distinguish uterine leiomyosarcomas. Int. J. Gynecol. Pathol 2020;39:354–361. [DOI] [PubMed] [Google Scholar]
- 54.Croce S, Ribeiro A, Brulard C, et al. Uterine smooth muscle tumor analysis by comparative genomic hybridization: a useful diagnostic tool in challenging lesions. Mod. Pathol 2015;28:1001–1010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


