Abstract
The thymus is a highly specialized organ that plays an indispensable role in the establishment of self-tolerance, a process characterized by the “education” of developing T-cells. To provide competent T-cells tolerant to self-antigens, medullary thymic epithelial cells (mTECs) orchestrate negative selection by ectopically expressing a wide range of genes, including various tissue-restricted antigens (TRAs). Notably, recent advancements in the high-throughput single-cell analysis have revealed remarkable heterogeneity in mTECs, giving us important clues for dissecting the mechanisms underlying TRA expression. We overview how recent single-cell studies have furthered our understanding of mTECs, with a focus on the role of Aire in inducing mTEC heterogeneity to encompass TRAs.
Keywords: mTEC, Aire, single-cell analysis, heterogeneity, tissue-restricted antigens
1. Introduction
In the thymus, the cortex and the medulla coordinate to elicit proficient T-cells that can effectively eliminate foreign pathogens while simultaneously exhibiting tolerance towards self-components. Basically, cortical thymic epithelial cells (cTECs) orchestrate positive selection, while medullary thymic epithelial cells (mTECs) orchestrate negative selection as well as the induction of regulatory T-cells (Tregs) (1, 2). To effectively screen for an extensive array of self-reactive T-cell clones, mature mTECs are capable of “ectopically” or “promiscuously” expressing nearly 90% of the coding genome, including thousands of tissue-restricted antigens (TRAs) (3–5). Due to their exceptional characteristics, the thymic medulla has been proposed to function as “a mosaic of epithelial self” by mirroring extra-thymic tissues (6, 7).
Since long before, scientists have been aware of the heterogeneity among mTECs as evidenced by histological examinations through electron microscopy. This is exemplified by the identification of multiple subsets such as myoid cells (8), ciliated cells, and secretory cells (9). Besides these morphological variations, further analysis disclosed the nature of promiscuous gene expression (pGE) among mTECs (7), and a seminal study identified autoimmune regulator (Aire) as the key molecule for pGE (10). Notably, individual mTECs do not uniformly express TRA genes in the existence of Aire, but it is estimated that only 1-3% of mTECs would express a particular TRA at any given time (4, 5, 11–13). Consequently, there exist two kinds of heterogeneity among mTECs; (i) inter-populational heterogeneity among mTECs based on the morphology and biological properties and (ii) intra-populational heterogeneity among Aire-expressing mTECs associated with pGE. Although recent transcriptomic analyses have provided various insights into the biology of mTECs, the complexity of mTEC heterogeneity impedes understanding of how mTECs orchestrate the presentation of TRAs as a whole. In this review, we will attempt to unravel this enigma by summarizing the findings in mTECs and Aire brought to us by recent single-cell studies.
2. The “two-faced” role of Aire in mTECs
Depending on their surface markers, mTECs have been generally categorized into two distinct subsets; MHC-IIlowCD80low (mTEClow) and MHC-IIhighCD80high (mTEChigh), the latter being considered a mature subset. Along with the high expression of molecules related to antigen presentation, mTEChigh is distinguished by the unique expression of Aire in the form of dot morphology in the nucleus (14–16). Initially, the human AIRE gene was positionally cloned as the causative gene for autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), which is characterized by organ-specific autoimmune disease with an autosomal recessive inheritance (17, 18). As for the pathogenesis of the disease, Anderson et al. first demonstrated that TRAs are enriched among the down-regulated genes in mTECs from Aire-knockout (Aire-KO) mice utilizing microarray analysis (10). Based on this result, they suggested that Aire is responsible for the establishment of central tolerance by promoting TRA expression from mTECs. Consistent with this finding, subsequent transcriptomic studies by high-throughput RNA-seq analysis reported that nearly 4,000 genes, including a large number of TRAs, were down-regulated in Aire-KO mTEChigh compared with those from wild-type (WT) mTEChigh (4, 19).
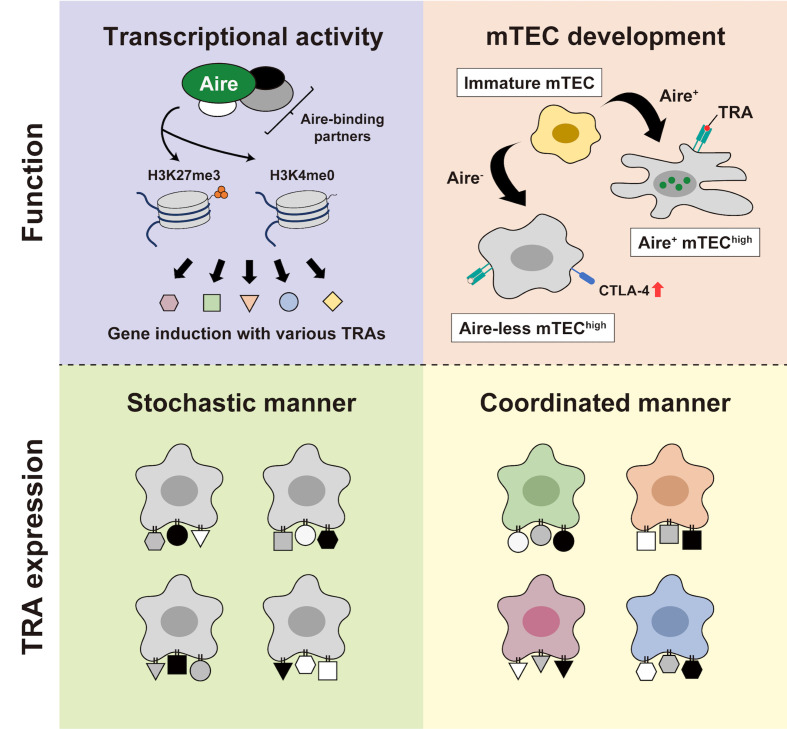
Then, how does a single Aire gene elicit such a dynamic alteration in the transcriptome? The current prevailing view is that Aire operates as a transcription factor to promote a broad array of transcriptional targets, not by recognizing particular DNA sequences but by involving epigenetic mechanisms. It has been suggested that Aire preferentially engages in the interaction with repressive chromatin states, such as H3K4me0 and H3K27me3 (4, 20–23), in cooperation with various partner molecules (24–29) ( Figure 1 , upper left). Upon being recruited to such a chromatin state, Aire is considered to promote the transcription of TRA genes by releasing stalled polymerase II from their promoters (30, 31). Furthermore, a recent study using ChIP-seq demonstrated that Aire-containing complexes are preferentially located on super-enhancers, chromatin stretches hosting high densities of general and cell-type-specific transcription factors, to induce TRAs (32). However, the function of Aire turned out to extend beyond simply serving as a transcriptional regulator of TRA genes, suggesting that it plays a pivotal role in the development/differentiation of mTECs ( Figure 1 , upper right). This notion was first derived from observing morphological alteration in the medullary components in Aire-KO mice (33, 34). Aire-KO mice were reported to exhibit increased numbers of mTECs with a globular cell shape (35, 36) and a near absence of Hassall’s corpuscle-like structures (36, 37), along with increased percentages of mTEChigh (38–41). Recently, we and another group have also reported that mTEChigh from Aire-KO mice ectopically expresses CTLA-4 at a high level (42, 43). Thus, the Aire-KO mTEChigh is not only defective in TRA expressions but also impaired in their normal development, which would be required for tolerogenic function.
Figure 1.
The “two-faced” role of Aire. (Upper left) Aire orchestrates its transcriptional function by employing epigenetic mechanisms. Through coordinating with various binding partners, Aire preferentially interacts with repressive chromatin marks, such as H3K4me0 and H3K27me3, thereby derepressing gene expression. (Upper right) Aire’s participation in the mTEC development. In the presence of Aire, mTEChigh attains a fully developed capability with abundant TRA expression. Conversely, Aire-deficiency leads to the altered development of mTEChigh with ectopic CTLA-4 expression and a defect in TRA expression. (Lower left and lower right) Depicting two potential paradigms for TRA expression; a stochastic manner without any coherent biological relevance (left) and a coordinated manner based on the biological properties of particular types of mTEC (right).
Besides this “two-faced” mode regarding the role of Aire (i.e., transcriptional activity and mTEC development), there also exist two potential paradigms in debate for the way in which mTECs present TRAs to T-cells in the thymus. The first is a pattern in which individual mTECs express various TRAs in a completely “stochastic” manner, lacking any coherent functional/developmental relevance ( Figure 1 , lower left). The second is a pattern in which TRAs are expressed by particular mTECs in a “coordinated” manner based on their biological properties ( Figure 1 , lower right). Comparing these two paradigms with the two-faced function of Aire, the stochastic model seems to be more closely linked with pGE due to Aire’s genuine transcriptional activity, whereas the coordinated model appears to pertain to the engagement of Aire in mTEC development. So far, the role of Aire has been basically discussed in a dichotomous manner; “(genuine) transcriptional activity” versus “mTEC development” and “stochastic gene induction” versus “coordinated gene induction.” However, upon considering the findings of prior studies, it seems reasonable to assume that Aire fulfills both of these capacities. We discuss below the multifaceted nature of Aire as revealed by the single-cell analyses.
3. Heterogeneity of mTECs revealed by single-cell analysis
Like mTEChigh, the mTEClow fraction also appears to consist of non-homogenous subsets. Using ontogenetic analysis and reaggregate thymic organ culture (RTOC) system, Gray et al. demonstrated that the mTEClow compartment comprises progenitors developing into mature mTEChigh (38). For this developmental process of mTECs, they require multiple TNF receptor superfamily signals, including RANK, CD40 and lymphotoxin-β receptor (44–51). Furthermore, fate-mapping and lineage-tracing studies revealed that the mTEClow fraction also holds mTECs in a terminally differentiated stage, termed “post-Aire” mTECs, that have progressively lost their expression of MHC-II, CD80, and Aire (52, 53). These studies suggested that the classification into mTEChigh and mTEClow is insufficient to capture the biology of mTECs.
In addition to these notions, recent advancements in transcriptomic analysis via single-cell RNA-seq (scRNA-seq) have extended our understanding of mTEC heterogeneity. We have summarized previous reports utilizing single-cell analysis, specifically those pertaining to TECs, with their significance in Table 1 . scRNA-seq has now become a widely employed technique for identifying cell types or cell states in the field of immunology (70). Indeed, scRNA-seq studies have uncovered multiple mTEC subpopulations based on their transcriptome, exemplified by the identification of a novel mTEC subset reminiscent of tuft cells in the small intestine (56, 57). Based on the transcriptome similarity computed by scRNA-seq, Bornstein et al. divided mTECs into four subgroups referring to the previously established mTEC subsets; mTEC I (CCL21+ mTEClow), mTEC II (mature mTEChigh), mTEC III (post-Aire mTECs), and mTEC IV (thymic tuft cells) (56). Other single-cell studies also reproduced the identification of thymic tuft cells, not only in mice (59, 61, 62, 64–69) but also in the human thymic stroma (60, 63). Notably, the development of thymic tuft cells is dependent on the transcription factor Pou2f3, which serves as a master regulator of intestinal tuft cells (56, 57). The identification of thymic tuft cells reminded us of the model that the thymus comprises “a mosaic of epithelial self.” Besides scRNA-seq, single-cell ATAC-seq (scATAC-seq) has been recently developed as a means to infer the cis- and trans- gene regulatory mechanisms in single cells in a massively parallel way (71).
Table 1.
Reports of single-cell approach to TECs including both scRNA-seq and scATAC-seq analyses.
| Authors | Organism | Platform | Subject | Significance |
|---|---|---|---|---|
| Sansom et al., 2014 (4) | Mouse | Fluidigm C1 Smart-seq |
MHC-IIhigh mTECs | Identifying Aire-induced genes and uncovering their sparse yet strong expressions within individual mTECs |
| Brennecke et al., 2015 (5) | Mouse | Smart-Seq2 | MHC-IIhigh mTECs, Ceacam1+ mTECs, Klk5+ mTECs, Tspan8+ mTECs | Demonstrating coordinated patterns in TRA expression with their probable association with chromatin accessibility |
| Meredith et al., 2015 (13) | Mouse | CEL-Seq | MHC-IIhigh mTECs | Detecting diminutive co-expression clusters in Aire’s target genes, characterizing their fashion as “ordered stochasticity” |
| Miragaia et al., 2018 (54) | Mouse | Fluidigm C1 Smart-seq2 |
Total mTECs | Analyzing the acquisition process of TRA expression in correlation with the developmental stages of mTECs |
| Kernfeld et al., 2018 (55) |
Mouse | Drop-seq | Total TECs | Uncovering the heterogeneity and transcriptomic dynamics of TECs during thymus organogenesis in embryos |
| Bornstein et al., 2018 (56) | Mouse | MARS-seq | Total mTECs | The identification of thymic tuft cells as a novel subset of mTECs, with evidence of their dependency on Pou2f3 |
| Miller et al., 2018 (57) | Mouse | Smart-seq2 | Thymic tuft cells | |
| Zeng et al., 2019 (58) | Human | Chromium STRT-seq |
Total TECs | Characterizing TEC populations during embryonic and fetal stages with the anticipation of intercellular interactions |
| Dhalla et al., 2020 (59) | Mouse | Chromium | Total mTECs, TSPAN8+ mTECs, GP2+ mTECs |
Identifying ordered co-expression patterns among TRA genes with uncertain biological significance in mTECs |
| Park et al., 2020 (60) | Human | Chromium | Total TECs | Construction of the comprehensive cell atlas of the thymus, comprising TECs, across the prenatal and postnatal stages |
| Baran-Gale et al., 2020 (61) | Mouse | Smart-seq2 | Total TECs | Unveiling the effect of aging on the TEC composition associated with the disruption of progenitor differentiation |
| Wells et al., 2020 (62) | Mouse | Chromium | Total TECs | Proposing a branching development into Aire- and Ccl21a-expressing mTECs from transit-amplifying TECs |
| Bautista et al., 2021 (63) | Human | Chromium | Total TECs | Characterizing thymic stromal cells across different stages of life and detecting unique subsets among mTECs |
| Nishijima et al., 2022 (64) | Mouse | Chromium | Total TECs | Revealing composition of mTECs dramatically alters in Aire-deficient mice, leading to the impairment of TRA expression |
| Gao et al., 2022 (65) | Mouse | Chromium | Total TECs | Profiling the temporal dynamics of TEC development and transcriptomics throughout embryonic to adult stages |
| Miyao et al., 2022 (66) | Mouse | Chromium RamDA-seq |
Total TECs (RNA & ATAC) RTOC (RNA) |
Demonstrating that transit-amplifying Aire+ mTECs serve as precursors of functionally mature Aire+ mTECs |
| Nusser et al., 2022 (67) | Mouse | CEL-Seq2 | Total TECs | Identifying two distinct types of principal TEC progenitors by utilizing a CRISPR-Cas9-based barcoding system |
| Michelson et al., 2022 (68) | Mouse | Chromium | MHC-IIhigh mTECs (ATAC), MHC-IIlowPdpn-CD104- mTECs (RNA) |
Suggesting mTECs co-opt lineage-defining transcription factors to mirror extra-thymic cell types for TRA expression |
| Liang et al., 2022 (69) | Mouse | Chromium | Total TECs | Demonstrating TEC-specific deletion of Furin leads to a decrease in the number of mTECs, including thymic tuft cells |
4. Altered composition of mTECs due to Aire deficiency
Michelson et al. have recently extended the idea described above by demonstrating that mTECs utilize lineage-defining transcription factors to mirror the extra-thymic cell types (68). They conducted a large set of single-cell experiments, including scATAC-seq and scRNA-seq, to investigate the mechanisms of TRA expressions in mTECs. They categorized mTECs into various subpopulations based on their resemblance to particular extra-thymic tissues, such as keratinocytes, tuft cells, microfold cells, neuroendocrine/secretory cells, ciliated cells, and myocytes. They called these populations “mimetic cells” and demonstrated that the majority of them are in the “post-Aire” stage. Similar to thymic tuft cells, they discovered that microfold mTECs are nearly eliminated in mice lacking Sox8 or Spib, transcription factors that characterize the corresponding subset (i.e., microfold cells) (68). Thus, their findings support the concept of coordinated machinery underlying the expression of TRAs ( Figure 1 , lower right). Several studies have also reported matched mTEC clusters for mimetic cells, albeit with slight variations in terminology ( Table 2 ). Importantly, Aire-KO mice showed the reduction of certain mimetic cell clusters to varying degrees, suggesting that Aire is partially and variably required for the development of these subpopulations (68).
Table 2.
mTEC subpopulations mirroring extra-thymic cell types identified by single-cell analysis.
| Subset | Organism | Gene signature | Other description | Reference |
|---|---|---|---|---|
| Post-Aire mTEC | Mouse | Spink5, Ivl, Krt10, Krt80, Pigr, Ly6d, Cnfn, Flg | mTEC-III, Keratinocyte mTEC, Corneocyte-like mTEC, Spink5+ cell | (56, 59–69) |
| Human | KRT1, KRT10, IVL | |||
| Thymic tuft cell | Mouse | Pou2f3, Dclk1, Avil, Lrmp, Trpm5, Il25, Gng13, L1cam, Sox9, Chat | mTEC-IV, Tuft-like mTEC |
(56, 59–69) |
| Human | POU2F3, DCLK1, GNAT3, GNB3, PLCB2, OVOL3 | |||
| Microfold mTEC | Mouse | Sox8, Spib, Gp2, Ccl6, Ccl9, Ccl20, Ccr5, Tnfaip2, Tnfrsf11b | Gp2-preffered mTEC, Gp2+ TEC, Ccl6+ cell |
(59, 65, 66, 68) |
| Neuroendocrine/ Secretory cell |
Mouse | Snap25, Stxbp5l, Car8, Cd177 | Structural TEC, TEC (neuro) |
(60, 61, 63, 66, 68) |
| Human | NEUROD1, NEUROG1, CHGA, BEX1 | |||
| Ciliated cell | Mouse | Dynlrb2, Dnah12, Spag16, Wdr34, Bbs7, Tppp3, Fam183b | Cilia-TEC | (59, 63, 66, 68) |
| Human | ATOH1, GFI1, LHX3, FOXJ1 | |||
| Myoid cell | Mouse | Myog, Myl1, Actc1 | Muscle mTEC, TEC (myo) |
(60, 63, 68) |
| Human | MYOD1, MYOG, DES | |||
| Lung mTEC | Mouse | Aqp4, Aqp5, Muc5b, Slc12a2, Bpifa1 | Bpifa1+ cell | (65, 68) |
| Ionocyte | Mouse | Slc12a2, Atp6v1b1 | (63, 68) | |
| Human | CFTR, FOXI1, ASCL3, CLCNKB | |||
| Enterocyte/ Hepatocyte mTEC |
Mouse | Reg3g, Saa1, Saa3, Aldob | (68) | |
| Myelin+ cell | Human | SOX10, MPZ, MBP, S100A1 | (63) |
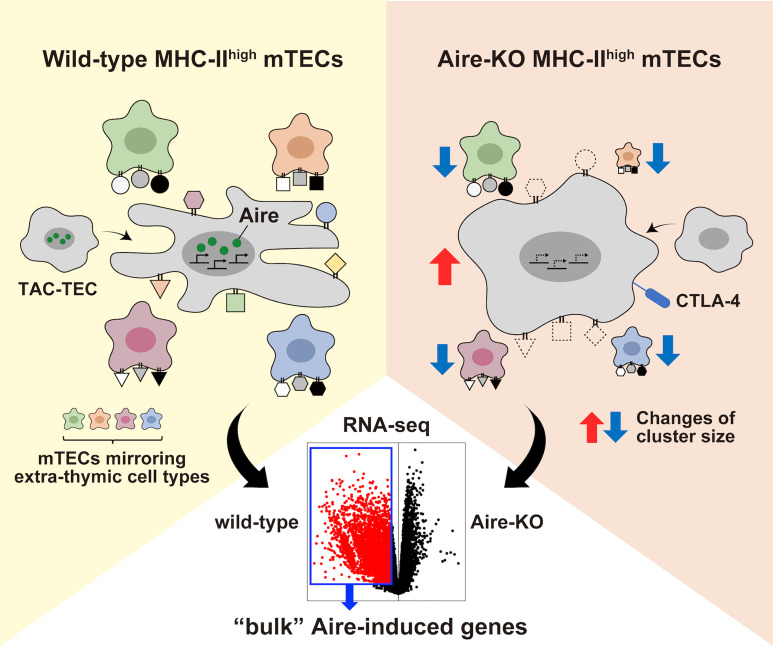
Our previous scRNA-seq study further focused on transcriptome changes in the primary Aire-expressing mTECs caused by the lack of Aire (64). Interestingly, a comprehensive analysis of WT and Aire-KO mice revealed a significant alteration in the composition of mTECs, as also documented in other reports (56, 68). The most striking change was the presence of clusters unique to Aire-KO mTECs that emerged instead of Aire-expressing mTEChigh in WT mice; “Aire-less” mTEChigh, hereafter. These clusters were reminiscent of morphologically abnormal mTECs observed in the medulla in Aire-KO mice (35, 36), and they exhibited high expression of Ctla4 (42). Besides the defect of TRA genes in Aire-less mTEChigh compared with Aire-expressing mTEChigh, a comparison of these two groups identified a signature of “keratinocyte differentiation,” suggesting the impairment of normal developmental process in Aire-less mTEChigh (64). Thus, recent studies utilizing single-cell techniques and Aire-KO mice have indicated that Aire deficiency leads to the impaired development of mTECs at the Aire-expressing stage itself, as well as subpopulations mimicking the extra-thymic tissues to a variable extent. However, the exact mechanisms through which Aire promotes the development of various mTEC subpopulations remain elusive.
5. Aire-induced genes revisited by single-cell resolution
As aforementioned, Sansom et al. reported the down-regulation of nearly 4,000 genes in Aire-KO mTEChigh in comparison to the wild-type counterpart (4). They classified these genes as “Aire-induced genes” and further defined a group of genes whose expression is entirely dependent on Aire as “Aire-dependent genes.” Although these gene lists have been extensively employed in subsequent studies with the assumption that Aire directly regulates their transcription within Aire-expressing mTECs, the following single-cell studies have provided an alternative interpretation.
Regarding the mTECs in WT mice, a previous single-cell analysis demonstrated that MHC-IIhigh fraction comprises Aire-expressing mTECs and various mTEC subsets mirroring extra-thymic cell types (i.e., mimetic cells) (68). Furthermore, it has been reported that Aire-expressing mTECs in active cell-cycle, known as transit-amplifying TECs (TAC-TECs), also exist within the mTEChigh population (62, 66). While these cells were suggested to be the precursors of quiescent and functionally mature Aire-expressing mTEChigh, they were incapable of expressing sufficient levels of TRA genes together with Aire-induced genes (66).
Notably, compositional changes are taking place in the mTEChigh fraction from Aire-KO mice. First, Aire-KO mTECs harbor “Aire-less” mTEChigh, which shows the defect of TRA expressions (64), altered globular morphology (35, 36) along with the enlarged cell numbers (38–40, 64), and abnormal Ctla4 expression (42, 43). Second, several mimetic cell clusters were reported to decrease in various degrees (e.g., ciliated mTECs, neuroendocrine/secretory mTECs, and microfold mTECs) (68). In contrast to functionally mature Aire-expressing mTEChigh, the effect of Aire deficiency in the TAC-TEC cluster seems to be minimal, as we observed no significant changes in their distribution and cell numbers in Aire-KO mice (64).
When the transcriptomes of mTEChigh from WT and Aire-KO mice are compared by bulk RNA-seq, differentially upregulated genes in WT mTEChigh would be detected as Aire-induced genes ( Figure 2 ). Therefore, “bulk” Aire-induced genes do not constitute a single group of genes brought by the genuine transcriptional activity of Aire. We suggest that the “bulk” Aire-induced genes consist of two contexts in which Aire promotes the genes via distinct regulatory mechanisms; (i) genes induced by Aire’s genuine transcriptional activity and (ii) genes preferentially expressed in mimetic cells. In other words, the higher a mimetic cell cluster’s dependence on Aire, the more genes characteristic of the corresponding cluster will be detected as Aire-induced genes.
Figure 2.
The landscape of the bulk Aire-induced genes. Illustrating the compositional changes between wild-type MHC-IIhigh mTECs and Aire-KO MHC-IIhigh mTECs. Besides the defect in the transcriptional activity in primary Aire-expressing mTEChigh, the altered composition of various mTEC subpopulations in mTEChigh contributes to the difference in the transcriptome detected by the bulk RNA-seq.
6. Stochastic “and” coordinated manner in TRAs?
Initially, single-cell analyses with single-cell PCR were applied to MHC-IIhigh mTECs, focusing on certain TRAs to dissect the mechanism underlying TRA expressions (11, 12, 72). Although these studies emphasized the stochasticity of pGE in individual mTECs, later scRNA-seq experiments utilizing a few hundred MHC-IIhigh mTECs concluded that Aire-induced genes fall into some sort of coordinated patterns besides its stochasticity (4, 5, 13). Furthermore, a recent large-scale scRNA-seq analysis (total of 6,894 mTECs) also noted an ordered co-expression pattern in TRAs, confirmed by focusing on particular Aire-induced TRAs (i.e., Tspan8 and Gp2), while the biological significance of co-expression remained inexplicable (59). It should be noted that these arguments are predicated upon the assumption that TRA induction is primarily brought by a single mechanism. However, as discussed above, it seems reasonable to posit that Aire induces TRA expression through a combination of (i) a stochastic pattern resulting from the genuine transcriptional activity of Aire and (ii) a coordinated pattern established by mTECs mirroring extra-thymic cell types (i.e., mimetic cells). Given this two-faced role of Aire, analyses utilizing total mTEChigh or focusing on specific TRAs highly expressed within a particular mimetic cell cluster may not be sufficient. Instead, it might be essential to classify mTECs accurately into subclusters in advance and then investigate the significance of Aire-induced genes or TRA genes within individual clusters. In this regard, single-cell analysis, especially the droplet-based approach, is an ideal tool to dissect inter-populational heterogeneity underlying mTECs.
However, since the sequencing depth in each cell is relatively low in the droplet-based method, it may not be sufficient to detect pGE in mTECs due to its sparse nature. To capture the dynamics of the transcriptome in single mTECs, another plate-based approach, such as Smart-seq (73) and RamDA-seq (74), would be better than the droplet-based method as it offers much higher sensitivity and near-complete full-length transcript coverage. Furthermore, the method employed for cell isolation would significantly affect the outcome of downstream analysis. Indeed, a previous study utilizing scRNA-seq for FACS-sorted MHC-IIlowPdpn−CD104–-mTEClow fraction (68) detected various mimetic cells at a higher resolution compared with other studies utilizing the total TEC population (62, 64–66). On the other hand, the protocol of TEC isolation itself would also have a significant impact on the result of single-cell analysis (75, 76). Given that a previous imaging study has reported ~1.1x106 Aire+ mTECs in 5 weeks old mice (77), it is possible that there still exists unknown TEC populations. The integration of these techniques and methodologies will lead to a better understanding of inter-populational heterogeneity in TECs and intra-populational heterogeneity in Aire-expressing mTECs.
7. Shared and distinct features between mouse and human mTECs
Although previous scRNA-seq studies have demonstrated the similarities between mouse and human mTECs, certain differences also exist between them. Representative mTEC clusters corresponding to CCL21+ mTEClow, AIRE-expressing mTEChigh and post-AIRE mTECs were identified among human TECs (60, 63). Several mimetic cell clusters, such as thymic tuft cells, myoid cells and ciliated cells have also been detected both in mouse and human thymic stroma ( Table 1 ). Nevertheless, the composition of mimetic cell clusters varies between mice and humans. For example, the cluster of thymic tuft cells was relatively small and indistinct in human mTECs compared with mouse mTECs (60, 63). In contrast, Bautista et al. reported the presence of myelin-expressing mTECs in humans that have not been reported in the mouse thymi (63). Furthermore, the TAC-TEC cluster has not been captured in the previous studies related to human TECs (60, 63). However, we also need to consider the dynamics of TEC composition associated with aging and other stress (61) when comparing mouse and human TECs. Since the single-cell studies on human TECs are currently limited, future studies employing larger sample sizes, higher sequencing depth, and more sophisticated analytical tools will be crucial for elucidating the full spectrum of cellular heterogeneity in human TECs.
8. Single-cell analysis beyond mTECs
8.1. Aire expression from antigen-presenting cells in the periphery
Other than mTECs, Gardner et al. characterized Aire-expressing hematopoietic population in peripheral lymphoid tissues as extrathymic Aire-expressing cells (eTACs), showing the characteristics of MHC-IIhigh antigen-presenting cells (78, 79). A recent study utilizing single-cell multi-omics (scRNA-seq + scATAC-seq) further divided eTACs into two distinct types; migratory dendritic cell (DC)-type and RORγt+ group 3 innate lymphoid cell (ILC3)-type, as they termed the latter Janus cells (JCs) (80). The result was also corroborated by three back-to-back studies that identified Aire-expressing lineage among RORγt+ cells by scRNA-seq (81–84). Notably, Dobeš et al. demonstrated that Aire deficiency in ILC3-type eTACs (i.e., JCs) leads to the impaired generation of Candida-specific T-cell response, providing new insight into the mechanism underlying mucocutaneous candidiasis of APECED patients and extra-thymic function of Aire (85).
Given that the transcriptional impact of Aire varies with cell type (86), it is questionable whether the presence of Aire elicits the expression of TRAs in hematopoietic cells similar to its effect in mTECs. Wang et al. suggested the similarity in the transcriptome between JCs and mTECs by calculating the cosine similarity score and demonstrated the enrichment of TRAs in JCs, while the effect of Aire deficiency was not mentioned (80). Another study revealed that Aire deficiency impacted the transcriptome under the heat-killed C. albicans (HKCA)-stimulated condition in JCs, resulting in the impaired induction of genes encoding cytokines (Il6, Il18 and Bmp2), C. albicans-sensing receptors (Clec7a), cell adhesion molecules (Vcam1 and Cadm1), costimulatory molecules (Cd86) and enzymes involved in proinflammatory response (Ptgs2) (85). However, the transcriptome changes were not as pronounced as those observed in Aire-KO mTECs. In contrast, we noted no considerable effect on the transcriptome of DC-type eTACs as a result of the lack of Aire (87). Despite these equivocal findings regarding the transcriptional activity of Aire in antigen-presenting cells, a comprehensive study of Aire in distinct cell types and its effect may improve our understanding in the biology of Aire.
8.2. AIRE expression in thymic epithelial tumors
Recently, two groups independently reported the scRNA-seq analysis of thymic epithelial tumors (TETs) (88, 89). TETs are classified into six types of thymomas (Type A, AB, B1-B3 thymomas, and micronodular thymoma with lymphoid stroma) and thymic carcinoma according to the current WHO classification (90). While there is no doubt that TETs originate from TECs, it is still elusive whether each histological type originates from cTECs or mTECs. A previous immunohistochemical study reported that most thymomas showed characteristics of bi-lineage differentiation toward mTECs and cTECs, suggesting that they derive from a common progenitor cell (91). Indeed, the scRNA-seq data showed that each TET contained varying proportions of neoplastic TECs resembling normal cTECs and mTECs within the tumor, containing a small number of AIRE-expressing cells (88, 89). In great contrast, our study combining immunohistochemical analysis for AIRE with published scRNA-seq data demonstrated that most thymic carcinomas express AIRE protein and harbor the molecular characteristics of several subpopulations in mTECs, but not cTECs, suggesting their cell of origin as mTECs (92). A recent report also demonstrated that a tuft cell-like signature was prevalent in thymic carcinomas (93).
Patients with thymomas often develop autoimmunity, most typically myasthenia gravis (MG), whereas thymoma is recorded in only 10-15% of all patients with MG (94–96). Besides thymomas, thymic hyperplasia (thymic follicular hyperplasia), which forms abundant germinal centers in the medulla, can also cause MG. Of note, autoimmunity is significantly rare in patients with thymic carcinomas (94, 95). Although the role of AIRE and mTECs in the pathogenesis of MG is still elusive, the absence or the insufficiency of AIRE expression from thymoma tissues has been discussed with the development of autoimmunity in thymoma patients in several studies (97–100). However, our immunohistochemical approach revealed that most type B thymomas, in which paraneoplastic autoimmunity is most frequent, harbored focal but intense AIRE staining in the area showing differentiation to mTECs (92). Furthermore, we did not see a correlation between the incidence of MG and the expression of AIRE protein in our TET cohort, raising a possibility that some other factors than AIRE might be responsible for the development of MG in thymomas. In this regard, it is quite important to disclose whether AIRE is also functional in the neoplastic milieu. While the information on AIRE-dependent genes in humans is currently unavailable, single-cell data accumulated so far from human TECs may help to identify the putative AIRE’s targets. Furthermore, spatial transcriptomics may provide novel insights into the biology of TETs by capturing their abnormal architecture and altered gene expressions simultaneously.
9. Conclusions and perspectives
Although recent advances in the single-cell analysis have expanded our understanding of mTECs, it is an issue of great importance for further study to reveal how a single Aire gene participates in such a multilayered task outlined above and how it functions even in the extra-thymic cellular context. It should also be noted that scRNA-seq analysis of embryofetal and neonatal mice draws dramatically different distributions of TECs from adult samples (56, 65). Considering the distinct profiles of Tregs generated in early life (101) and the importance of Aire expression during the perinatal period (102), it would be worthwhile to focus on Aire’s actions during early life to see any possible difference from those seen in adults at a single-cell resolution. It is also crucial to clarify how the TRAs from Aire-expressing mTECs and mimetic clusters share roles for inducing immune tolerance. Particularly, the benefit of stochastic TRA expression from a small population in Aire-expressing mTECs is of great interest. We hope that our review will contribute to a better understanding of mTEC biology and illuminate a new perspective on the Aire.
Author contributions
MinM wrote the manuscript draft in consultation with KT and TO for improvement. HY and MitM supervised the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported in part by JSPS KAKENHI Grant Numbers 19H03699, 22H02892, 22K19437 (to MitM) and 23K14478 (to MinM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol (2014) 14(6):377–91. doi: 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol (2009) 9(12):833–44. doi: 10.1038/nri2669 [DOI] [PubMed] [Google Scholar]
- 3. Kadouri N, Nevo S, Goldfarb Y, Abramson J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat Rev Immunol (2020) 20(4):239–53. doi: 10.1038/s41577-019-0238-0 [DOI] [PubMed] [Google Scholar]
- 4. Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single-cell genomics reveal the aire dependency, relief from polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res (2014) 24(12):1918–31. doi: 10.1101/gr.171645.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Kuchler R, et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol (2015) 16(9):933–41. doi: 10.1038/ni.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farr AG, Rudensky A. Medullary thymic epithelium: a mosaic of epithelial “self”? J Exp Med (1998) 188(1):1–4. doi: 10.1084/jem.188.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol (2001) 2(11):1032–9. doi: 10.1038/ni723 [DOI] [PubMed] [Google Scholar]
- 8. Bockman DE. Myoid cells in adult human thymus. Nature (1968) 218(5138):286–7. doi: 10.1038/218286a0 [DOI] [PubMed] [Google Scholar]
- 9. Khosla S, Ovalle WK. Morphology and distribution of cystic cavities in the normal murine thymus. Cell Tissue Res (1986) 246(3):531–42. doi: 10.1007/BF00215193 [DOI] [PubMed] [Google Scholar]
- 10. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science (2002) 298(5597):1395–401. doi: 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- 11. Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A. (2008) 105(2):657–62. doi: 10.1073/pnas.0707486105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A. (2008) 105(41):15854–9. doi: 10.1073/pnas.0808069105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol (2015) 16(9):942–9. doi: 10.1038/ni.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsumoto M. The role of autoimmune regulator (Aire) in the development of the immune system. Microbes infection/Institut Pasteur. (2009) 11(12):928–34. doi: 10.1016/j.micinf.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 15. Akiyoshi H, Hatakeyama S, Pitkanen J, Mouri Y, Doucas V, Kudoh J, et al. Subcellular expression of autoimmune regulator (AIRE) is organized in a spatiotemporal manner. J Biol Chem (2004) 279(32):33984–91. doi: 10.1074/jbc.M400702200 [DOI] [PubMed] [Google Scholar]
- 16. Pitkänen J, Vähämurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. characterization of nuclear targeting and transcriptional activation domain. J Biol Chem (2001) 276(22):19597–602. doi: 10.1074/jbc.M008322200 [DOI] [PubMed] [Google Scholar]
- 17. Aaltonen J, Björses P, Perheentupa J, Horelli–Kuitunen N, Palotie A, Peltonen L, et al. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet (1997) 17(4):399–403. doi: 10.1038/ng1297-399 [DOI] [PubMed] [Google Scholar]
- 18. Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet (1997) 17(4):393–8. doi: 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- 19. St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol (2015) 195(2):498–506. doi: 10.4049/jimmunol.1500558 [DOI] [PubMed] [Google Scholar]
- 20. Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U.S.A. (2008) 105(41):15878–83. doi: 10.1073/pnas.0808470105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koh AS, Kingston RE, Benoist C, Mathis D. Global relevance of aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A. (2010) 107(29):13016–21. doi: 10.1073/pnas.1004436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Org T, Chignola F, Hetényi C, Gaetani M, Rebane A, Liiv I, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep (2008) 9(4):370–6. doi: 10.1038/embor.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Org T, Rebane A, Kisand K, Laan M, Haljasorg U, Andreson R, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet (2009) 18(24):4699–710. doi: 10.1093/hmg/ddp433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshida H, Bansal K, Schaefer U, Chapman T, Rioja I, Proekt I, et al. Brd4 bridges the transcriptional regulators, aire and p-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci U S A. (2015) 112(32):E4448–57. doi: 10.1073/pnas.1512081112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell (2010) 140(1):123–35. doi: 10.1016/j.cell.2009.12.030 [DOI] [PubMed] [Google Scholar]
- 26. Pitkanen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem (2000) 275(22):16802–9. doi: 10.1074/jbc.M908944199 [DOI] [PubMed] [Google Scholar]
- 27. Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits p-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol (2007) 27(24):8815–23. doi: 10.1128/MCB.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Waterfield M, Khan IS, Cortez JT, Fan U, Metzger T, Greer A, et al. The transcriptional regulator aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat Immunol (2014) 15(3):258–65. doi: 10.1038/ni.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chuprin A, Avin A, Goldfarb Y, Herzig Y, Levi B, Jacob A, et al. The deacetylase Sirt1 is an essential regulator of aire-mediated induction of central immunological tolerance. Nat Immunol (2015) 16(7):737–45. doi: 10.1038/ni.3194 [DOI] [PubMed] [Google Scholar]
- 30. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. (2012) 109(2):535–40. doi: 10.1073/pnas.1119351109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zumer K, Saksela K, Peterlin BM. The mechanism of tissue-restricted antigen gene expression by AIRE. J Immunol (2013) 190(6):2479–82. doi: 10.4049/jimmunol.1203210 [DOI] [PubMed] [Google Scholar]
- 32. Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator aire binds to and activates super-enhancers. Nat Immunol (2017) 18(3):263–73. doi: 10.1038/ni.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumoto M, Nishikawa Y, Nishijima H, Morimoto J, Matsumoto M, Mouri Y. Which model better fits the role of aire in the establishment of self-tolerance: the transcription model or the maturation model? Front Immunol (2013) 4:210. doi: 10.3389/fimmu.2013.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsumoto M. Contrasting models for the roles of aire in the differentiation program of epithelial cells in the thymic medulla. Eur J Immunol (2011) 41(1):12–7. doi: 10.1002/eji.201041024 [DOI] [PubMed] [Google Scholar]
- 35. Gillard GO, Dooley J, Erickson M, Peltonen L, Farr AG. Aire-dependent alterations in medullary thymic epithelium indicate a role for aire in thymic epithelial differentiation. J Immunol (2007) 178(5):3007–15. doi: 10.4049/jimmunol.178.5.3007 [DOI] [PubMed] [Google Scholar]
- 36. Yano M, Kuroda N, Han H, Meguro-Horike M, Nishikawa Y, Kiyonari H, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J Exp Med (2008) 205(12):2827–38. doi: 10.1084/jem.20080046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X, Laan M, Bichele R, Kisand K, Scott HS, Peterson P. Post-aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front Immunol (2012) 3(March):19. doi: 10.3389/fimmu.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing aire. J Exp Med (2007) 204(11):2521–8. doi: 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hubert FX, Kinkel SA, Crewther PE, Cannon PZ, Webster KE, Link M, et al. Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutation present with only a mild autoimmune phenotype. J Immunol (2009) 182(6):3902–18. doi: 10.4049/jimmunol.0802124 [DOI] [PubMed] [Google Scholar]
- 40. Kuroda N, Mitani T, Takeda N, Ishimaru N, Arakaki R, Hayashi Y, et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of aire-deficient mice. J Immunol (2005) 174(4):1862–70. doi: 10.4049/jimmunol.174.4.1862 [DOI] [PubMed] [Google Scholar]
- 41. Nishikawa Y, Nishijima H, Matsumoto M, Morimoto J, Hirota F, Takahashi S, et al. Temporal lineage tracing of aire-expressing cells reveals a requirement for aire in their maturation program. J Immunol (2014) 192(6):2585–92. doi: 10.4049/jimmunol.1302786 [DOI] [PubMed] [Google Scholar]
- 42. Morimoto J, Matsumoto M, Miyazawa R, Yoshida H, Tsuneyama K, Matsumoto M. Aire suppresses CTLA-4 expression from the thymic stroma to control autoimmunity. Cell Rep (2022) 38(7):110384. doi: 10.1016/j.celrep.2022.110384 [DOI] [PubMed] [Google Scholar]
- 43. Michelson DA, Benoist C, Mathis D. CTLA-4 on thymic epithelial cells complements aire for T cell central tolerance. Proc Natl Acad Sci U S A. (2022) 119(48):e2215474119. doi: 10.1073/pnas.2215474119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, Maruyama Y, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity (2008) 29(3):423–37. doi: 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 45. Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, et al. RANK signals from CD4 + 3- inducer cells regulate development of aire-expressing epithelial cells in the thymic medulla. J Exp Med (2007) 204(6):1267–72. doi: 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Irla M, Hugues S, Gill J, Nitta T, Hikosaka Y, Williams IR, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity (2008) 29(3):451–63. doi: 10.1016/j.immuni.2008.08.007 [DOI] [PubMed] [Google Scholar]
- 47. Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity (2008) 29(3):438–50. doi: 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 48. Akiyama T, Maeda S, Yamane S, Ogino K, Kasai M, Kajiura F, et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science (2005) 308(5719):248–51. doi: 10.1126/science.1105677 [DOI] [PubMed] [Google Scholar]
- 49. Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration and the control of autoimmunity require lympho-epithelial cross talk via LTβR. J Exp Med (2003) 198(5):757–69. doi: 10.1084/jem.20030794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Venanzi ES, Gray DH, Benoist C, Mathis D. Lymphotoxin pathway and aire influences on thymic medullary epithelial cells are unconnected. J Immunol (2007) 179(9):5693–700. doi: 10.4049/jimmunol.179.9.5693 [DOI] [PubMed] [Google Scholar]
- 51. White AJ, Nakamura K, Jenkinson WE, Saini M, Sinclair C, Seddon B, et al. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J Immunol (2010) 185(8):4769–76. doi: 10.4049/jimmunol.1002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishikawa Y, Hirota F, Yano M, Kitajima H, Miyazaki J, Kawamoto H, et al. Biphasic aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J Exp Med (2010) 207(5):963–71. doi: 10.1084/jem.20092144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Metzger TC, Khan IS, Gardner JM, Mouchess ML, Johannes KP, Krawisz AK, et al. Lineage tracing and cell ablation identify a post-aire-expressing thymic epithelial cell population. Cell Rep (2013) 5(1):166–79. doi: 10.1016/j.celrep.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miragaia RJ, Zhang X, Gomes T, Svensson V, Ilicic T, Henriksson J, et al. Single-cell RNA-sequencing resolves self-antigen expression during mTEC development. Sci Rep (2018) 8(1):685. doi: 10.1038/s41598-017-19100-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity (2018) 48(6):1258–70 e6. doi: 10.1016/j.immuni.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature (2018) 559(7715):622–6. doi: 10.1038/s41586-018-0346-1 [DOI] [PubMed] [Google Scholar]
- 57. Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature (2018) 559(7715):627–31. doi: 10.1038/s41586-018-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zeng Y, Liu C, Gong Y, Bai Z, Hou S, He J, et al. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity (2019) 51(5):930–48 e6. doi: 10.1016/j.immuni.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 59. Dhalla F, Baran-Gale J, Maio S, Chappell L, Hollander GA, Ponting CP. Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J (2020) 39(1):e101828. doi: 10.15252/embj.2019101828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (2020) 367(6480):eaay3224. doi: 10.1126/science.aay3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baran-Gale J, Morgan MD, Maio S, Dhalla F, Calvo-Asensio I, Deadman ME, et al. Ageing compromises mouse thymus function and remodels epithelial cell differentiation. eLife (2020) 9:e56221. doi: 10.7554/eLife.56221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wells KL, Miller CN, Gschwind AR, Wei W, Phipps JD, Anderson MS, et al. Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. eLife (2020) 9 :e60188. doi: 10.7554/eLife.60188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bautista JL, Cramer NT, Miller CN, Chavez J, Berrios DI, Byrnes LE, et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun (2021) 12(1):1096. doi: 10.1038/s41467-021-21346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nishijima H, Matsumoto M, Morimoto J, Hosomichi K, Akiyama N, Akiyama T, et al. Aire controls heterogeneity of medullary thymic epithelial cells for the expression of self-antigens. J Immunol (2022) 208(2):303–20. doi: 10.4049/jimmunol.2100692 [DOI] [PubMed] [Google Scholar]
- 65. Gao H, Cao M, Deng K, Yang Y, Song J, Ni M, et al. The lineage differentiation and dynamic heterogeneity of thymic epithelial cells during thymus organogenesis. Front Immunol (2022) 13:805451. doi: 10.3389/fimmu.2022.805451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miyao T, Miyauchi M, Kelly ST, Terooatea TW, Ishikawa T, Oh E, et al. Integrative analysis of scRNA-seq and scATAC-seq revealed transit-amplifying thymic epithelial cells expressing autoimmune regulator. eLife (2022) 11:e73998. doi: 10.7554/eLife.73998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nusser A, Sagar, Swann JB, Krauth B, Diekhoff D, Calderon L, et al. Developmental dynamics of two bipotent thymic epithelial progenitor types. Nature (2022) 606(7912):165–71. doi: 10.1038/s41586-022-04752-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell (2022) 185(14):2542–58 e18. doi: 10.1016/j.cell.2022.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liang Z, Zhang Z, Zhang Q, Dong X, Yang X, Zhang J, et al. The proprotein convertase furin regulates the development of thymic epithelial cells to ensure central immune tolerance. iScience (2022) 25(10):105233. doi: 10.1016/j.isci.2022.105233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ginhoux F, Yalin A, Dutertre CA, Amit I. Single-cell immunology: past, present, and future. Immunity (2022) 55(3):393–404. doi: 10.1016/j.immuni.2022.02.006 [DOI] [PubMed] [Google Scholar]
- 71. Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol (2019) 37(8):925–36. doi: 10.1038/s41587-019-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pinto S, Michel C, Schmidt-Glenewinkel H, Harder N, Rohr K, Wild S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A. (2013) 110(37):E3497–505. doi: 10.1073/pnas.1308311110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks G-J, Larsson AJM, et al. Single-cell RNA counting at allele and isoform resolution using smart-seq3. Nat Biotechnol (2020) 38(6):708–14. doi: 10.1038/s41587-020-0497-0 [DOI] [PubMed] [Google Scholar]
- 74. Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun (2018) 9(1):619. doi: 10.1038/s41467-018-02866-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xing Y, Hogquist KA. Isolation, identification, and purification of murine thymic epithelial cells. J Vis Exp (2014) 90:e51780. doi: 10.3791/51780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Seach N, Wong K, Hammett M, Boyd RL, Chidgey AP. Purified enzymes improve isolation and characterization of the adult thymic epithelium. J Immunol Methods (2012) 385(1-2):23–34. doi: 10.1016/j.jim.2012.07.023 [DOI] [PubMed] [Google Scholar]
- 77. Sakata M, Ohigashi I, Takahama Y. Cellularity of thymic epithelial cells in the postnatal mouse. J Immunol (2018) 200(4):1382–8. doi: 10.4049/jimmunol.1701235 [DOI] [PubMed] [Google Scholar]
- 78. Gardner JM, Devoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, et al. Deletional tolerance mediated by extrathymic aire-expressing cells. Science (2008) 321(5890):843–7. doi: 10.1126/science.1159407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gardner JM, Metzger TC, McMahon EJ, Au-Yeung BB, Krawisz AK, Lu W, et al. Extrathymic aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4(+) T cells. Immunity (2013) 39(3):560–72. doi: 10.1016/j.immuni.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang J, Lareau CA, Bautista JL, Gupta AR, Sandor K, Germino J, et al. Single-cell multiomics defines tolerogenic extrathymic aire-expressing populations with unique homology to thymic epithelium. Sci Immunol (2021) 6(65):eabl5053. doi: 10.1126/sciimmunol.abl5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gardner JM, Liston A. RORγt-lineage APCs: the aire apparent. Sci Immunol (2022) 7(78):eade9240. doi: 10.1126/sciimmunol.ade9240 [DOI] [PubMed] [Google Scholar]
- 82. Kedmi R, Najar TA, Mesa KR, Grayson A, Kroehling L, Hao Y, et al. A RORγt(+) cell instructs gut microbiota-specific t(reg) cell differentiation. Nature (2022) 610(7933):737–43. doi: 10.1038/s41586-022-05089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lyu M, Suzuki H, Kang L, Gaspal F, Zhou W, Goc J, et al. ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature (2022) 610(7933):744–51. doi: 10.1038/s41586-022-05141-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Akagbosu B, Tayyebi Z, Shibu G, Paucar Iza YA, Deep D, Parisotto YF, et al. Novel antigen-presenting cell imparts t(reg)-dependent tolerance to gut microbiota. Nature (2022) 610(7933):752–60. doi: 10.1038/s41586-022-05309-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dobeš J, Ben-Nun O, Binyamin A, Stoler-Barak L, Oftedal BE, Goldfarb Y, et al. Extrathymic expression of aire controls the induction of effective TH17 cell-mediated immune response to candida albicans. Nat Immunol (2022) 23(7):1098–108. doi: 10.1038/s41590-022-01247-6 [DOI] [PubMed] [Google Scholar]
- 86. Guerau-de-Arellano M, Mathis D, Benoist C. Transcriptional impact of aire varies with cell type. Proc Natl Acad Sci U S A. (2008) 105(37):14011–6. doi: 10.1073/pnas.0806616105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miyazawa R, Nagao JI, Arita-Morioka KI, Matsumoto M, Morimoto J, Yoshida M, et al. Dispensable role of aire in CD11c+ conventional dendritic cells for antigen presentation and shaping the transcriptome. Immunohorizons (2023) 7(1):140–58. doi: 10.4049/immunohorizons.2200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yasumizu Y, Ohkura N, Murata H, Kinoshita M, Funaki S, Nojima S, et al. Myasthenia gravis-specific aberrant neuromuscular gene expression by medullary thymic epithelial cells in thymoma. Nat Commun (2022) 13(1):4230. doi: 10.1038/s41467-022-31951-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xin Z, Lin M, Hao Z, Chen D, Chen Y, Chen X, et al. The immune landscape of human thymic epithelial tumors. Nat Commun (2022) 13(1):5463. doi: 10.1038/s41467-022-33170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. WHO Classification of Tumours Editorial Board . Thoracic tumours. 5th ed Vol. 5. . Lyon (France: International Agency for Research on Cancer; (2021). [Google Scholar]
- 91. Ströbel P, Hartmann E, Rosenwald A, Kalla J, Ott G, Friedel G, et al. Corticomedullary differentiation and maturational arrest in thymomas. Histopathology (2014) 64(4):557–66. doi: 10.1111/his.12279 [DOI] [PubMed] [Google Scholar]
- 92. Matsumoto M, Ohmura T, Hanibuchi Y, Ichimura-Shimizu M, Saijo Y, Ogawa H, et al. AIRE illuminates the feature of medullary thymic epithelial cells in thymic carcinoma. Cancer Med (2023). doi: 10.1002/cam4.5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yamada Y, Simon-Keller K, Belharazem-Vitacolonnna D, Bohnenberger H, Kriegsmann M, Kriegsmann K, et al. A tuft cell-like signature is highly prevalent in thymic squamous cell carcinoma and delineates new molecular subsets among the major lung cancer histotypes. J Thorac Oncol (2021) 16(6):1003–16. doi: 10.1016/j.jtho.2021.02.008 [DOI] [PubMed] [Google Scholar]
- 94. Radovich M, Pickering CR, Felau I, Ha G, Zhang H, Jo H, et al. The integrated genomic landscape of thymic epithelial tumors. Cancer Cell (2018) 33(2):244–58.e10. doi: 10.1016/j.ccell.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marx A, Willcox N, Leite MI, Chuang WY, Schalke B, Nix W, et al. Thymoma and paraneoplastic myasthenia gravis. Autoimmunity (2010) 43(5-6):413–27. doi: 10.3109/08916930903555935 [DOI] [PubMed] [Google Scholar]
- 96. Gilhus NE, Verschuuren JJ. Myasthenia gravis: subgroup classification and therapeutic strategies. Lancet Neurol (2015) 14(10):1023–36. doi: 10.1016/S1474-4422(15)00145-3 [DOI] [PubMed] [Google Scholar]
- 97. Marx A, Hohenberger P, Hoffmann H, Pfannschmidt J, Schnabel P, Hofmann HS, et al. The autoimmune regulator AIRE in thymoma biology: autoimmunity and beyond. J Thorac Oncol (2010) 5(10 Suppl 4):S266–72. doi: 10.1097/JTO.0b013e3181f1f63f [DOI] [PubMed] [Google Scholar]
- 98. Ströbel P, Murumägi A, Klein R, Luster M, Lahti M, Krohn K, et al. Deficiency of the autoimmune regulator AIRE in thymomas is insufficient to elicit autoimmune polyendocrinopathy syndrome type 1 (APS-1). J Pathol (2007) 211(5):563–71. doi: 10.1002/path.2141 [DOI] [PubMed] [Google Scholar]
- 99. Suzuki E, Kobayashi Y, Yano M, Fujii Y. Infrequent and low AIRE expression in thymoma: difference in AIRE expression among WHO subtypes does not correlate with association of MG. Autoimmunity (2008) 41(5):377–82. doi: 10.1080/08916930801987573 [DOI] [PubMed] [Google Scholar]
- 100. Scarpino S, Di Napoli A, Stoppacciaro A, Antonelli M, Pilozzi E, Chiarle R, et al. Expression of autoimmune regulator gene (AIRE) and T regulatory cells in human thymomas. Clin Exp Immunol (2007) 149(3):504–12. doi: 10.1111/j.1365-2249.2007.03442.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science (2015) 348(6234):589–94. doi: 10.1126/science.aaa7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Guerau-de-Arellano M, Martinic M, Benoist C, Mathis D. Neonatal tolerance revisited: a perinatal window for aire control of autoimmunity. J Exp Med (2009) 206(6):1245–52. doi: 10.1084/jem.20090300 [DOI] [PMC free article] [PubMed] [Google Scholar]