Abstract
Highly washed membrane preparations from cells of the hyperthermophilic archaeon Pyrococcus furiosus contain high hydrogenase activity (9.4 μmol of H2 evolved/mg at 80°C) using reduced methyl viologen as the electron donor. The enzyme was solubilized with n-dodecyl-β-d-maltoside and purified by multistep chromatography in the presence of Triton X-100. The purified preparation contained two major proteins (α and β) in an approximate 1:1 ratio with a minimum molecular mass near 65 kDa and contained ∼1 Ni and 4 Fe atoms/mol. The reduced enzyme gave rise to an electron paramagnetic resonance signal typical of the so-called Ni-C center of mesophilic NiFe-hydrogenases. Neither highly washed membranes nor the purified enzyme used NAD(P)(H) or P. furiosus ferredoxin as an electron carrier, nor did either catalyze the reduction of elemental sulfur with H2 as the electron donor. Using N-terminal amino acid sequence information, the genes proposed to encode the α and β subunits were located in the genome database within a putative 14-gene operon (termed mbh). The deduced sequences of the two subunits (Mbh 11 and 12) were distinctly different from those of the four subunits that comprise each of the two cytoplasmic NiFe-hydrogenases of P. furiosus and show that the α subunit contains the NiFe-catalytic site. Six of the open reading frames (ORFs) in the operon, including those encoding the α and β subunits, show high sequence similarity (>30% identity) with proteins associated with the membrane-bound NiFe-hydrogenase complexes from Methanosarcina barkeri, Escherichia coli, and Rhodospirillum rubrum. The remaining eight ORFs encode small (<19-kDa) hypothetical proteins. These data suggest that P. furiosus, which was thought to be solely a fermentative organism, may contain a previously unrecognized respiratory system in which H2 metabolism is coupled to energy conservation.
Hydrogenases catalyze the reversible reduction of protons to hydrogen gas. They are found in a wide variety of microorganisms and enable them to use H2 as a source of reductant under either aerobic or anaerobic conditions. Alternatively, fermentative-type organisms utilize hydrogenase to dispose of reductant without the need of terminal electron acceptors other than protons (1, 3). Hydrogenases can be divided into two major types, depending on the metals they contain (5). The so-called iron-only hydrogenases have high specific activities and usually function to evolve H2. Their catalytic site is comprised of a novel 6Fe cluster (26, 29). The active site of nickel- and iron-containing hydrogenases (NiFe-hydrogenases), on the other hand, consists of a binuclear NiFe center (12, 36). The NiFe-hydrogenases are less active than their Fe-only counterparts, and their physiological role is usually to oxidize H2. In aerobic H2-oxidizing bacteria, NiFe-hydrogenases can function both as cytoplasmic, NAD-reducing enzymes and as part of conventional membrane-bound (MB) respiratory chains where O2 is the terminal electron acceptor (4, 9). In contrast, in anaerobic respiratory systems, the role of hydrogenase is poorly understood. For example, the methanogen Methanosarcina barkeri contains an MB NiFe-hydrogenase as part of a multiprotein complex (18, 25), the components of which show high sequence similarity to a NiFe-hydrogenase-containing complex present in the photosynthetic bacterium Rhodospirillum rubrum (13, 14). Both of these MB systems are thought to be involved in energy conservation, but the pathways of electron transfer and the precise role of the hydrogenases and of the associated proteins are unclear. In addition, three of the four MB NiFe-hydrogenases present in Escherichia coli are also thought to be involved in energy conservation (7, 8, 34).
In this study, we focused on the metabolism of H2 by the anaerobic archaeon Pyrococcus furiosus, an obligate organotroph that grows optimally near 100°C (11). This fermentative organism utilizes sugars via a modified ADP-dependent Embden-Meyerhof pathway (16), while amino acids derived from peptides are metabolized via transaminases and a suite of 2-keto acid oxidoreductases (2). In both pathways energy is conserved via substrate-level phosphorylation. The coenzyme A (CoA) derivatives that are generated are converted to organic acids directly by a novel pair of enzymes, acetyl-CoA synthetases I and II, which simultaneously convert ADP and phosphate to ATP (23). The major end products of fermentation are acetate, H2, and CO2; some other organic acids are also produced when peptides are the growth substrate. The oxidation of amino acid-derived 2-keto acids and of glyceraldehyde-3-phosphate and pyruvate in the glucose fermentation pathway are all carried out by ferredoxin-dependent oxidoreductases. It has been proposed (22) that the oxidation of reduced ferredoxin is coupled to the reduction of NADP via ferredoxin: NADP oxidoreductase (FNOR [19]) and that NADPH then serves as the electron donor to two cytoplasmic H2-evolving hydrogenases (I and II) (10, 21, 28). The reason why two such enzymes are present is not understood (21).
P. furiosus also reduces elemental sulfur (S0) to H2S. This process decreases the amount of H2 produced and has a stimulatory effect on growth, as indicated by an increase in cell density and growth rate (11). Moreover, during growth on maltose, the cell yield per gram of substrate used is 50% higher if S0 is present in the medium (35). This suggests that the reduction of S0 by P. furiosus is not merely a means of disposing of excess reductant but rather is an energy-conserving process. To date three enzymes that are capable of reducing S0 to H2S have been purified from P. furiosus. These are the aforementioned FNOR, also referred to as sulfide dehydrogenase (19), and the H2-evolving hydrogenases, otherwise known as sulfhydrogenases (20, 21). However, all three of these enzymes are located in the cytoplasm, and it seems unlikely that they would be involved in energy conservation.
In an effort to determine whether P. furiosus contains an MB sulfur reductase system analogous to that found in the S0-respiring mesophile Wolinella succinogenes (15, 30), we sought to obtain a membrane fraction from cell extracts that lacked the H2-dependent, S0 reduction activity of the cytoplasmic sulfhydrogenases. Surprisingly, even after repeated washings with buffers containing high salt concentrations, the membrane of P. furiosus still contained high hydrogenase (H2 evolution) activity. The purification and characterization of this integral MB hydrogenase is described herein. The enzyme is of the NiFe type, functions to evolve H2 but does not reduce S0, and is distinct from the well-characterized cytoplasmic enzyme. It appears to be part of a large multienzyme complex, the components of which show high sequence similarity to the respiratory-linked, MB NiFe hydrogenases found in some methanogens and photosynthetic bacteria and to the nonenergy-conserving formate hydrogen lyase system (hydrogenase 3) of E. coli (34).
MATERIALS AND METHODS
Growth of the organism.
P. furiosus (DSM 3638) was grown in a 600-liter fermentor at 90°C under pH-controlled conditions in the absence of S0, using maltose (Sigma Chemical Co., St. Louis, Mo.), tryptone (United States Biochemical, Cleveland, Ohio), and yeast extract (United States Biochemical) as carbon sources, each at a concentration of 5 g/liter, as described previously (10).
Membrane isolation and hydrogenase purification.
All procedures for membrane isolation and enzyme purification were carried out at 23°C under anaerobic conditions. All solutions were repeatedly degassed with and maintained under a positive pressure of Ar. The buffer used throughout was 50 mM Tris (pH 8.0) containing 2 mM sodium dithionite unless otherwise stated. Cell extracts of P. furiosus were prepared by suspending 150 g (wet weight) of frozen cells in 450 ml of buffer containing 4 mM sodium dithionite and 50 μg of DNase I (Sigma). The cell suspension was sonicated for 90 min (Branson, Danbury, Conn.) under a constant flow of Ar. Unbroken cells were removed by centrifugation (5,000 × g; 15 min). The membranes were isolated by ultracentrifugation at 120,000 × g for 2.0 h, suspended in buffer, and then subjected to successive washes with buffer containing 1.0, 2.0, and 4.0 M NaCl. After each wash, the suspension was centrifuged for 2 h at 120,000 × g and the membrane fraction was resuspended in an anaerobic chamber (Vacuum Atmospheres, Hawthorne, Calif.). The final membrane preparation was suspended to a protein concentration of 12 mg/ml in buffer without NaCl. n-Dodecyl-β-d-maltoside (1.5%, wt/vol) was then added, and the membranes were extracted using a tissue homogenizer under anaerobic conditions. The resulting suspension was centrifuged at 120,000 × g for 2.0 h, and the supernatant was loaded on to a column (5.0 by 6.1 cm) of DEAE High-Capacity (Amersham Pharmacia Biotech) equilibrated with 50 mM Tris (pH 8.45), 4 mM sodium dithionite, and 0.05% Triton X-100 (buffer A) containing 2.0 M urea. The proteins were eluted at a flow rate of 10 ml/min with a 2.4-liter linear gradient from 0 to 1.0 M NaCl in buffer A containing 2.0 M urea. The fractions eluting between 250 and 350 mM NaCl contained the hydrogenase activity, and these were pooled and loaded onto a column (1.6 by 30 cm) of hydroxyapatite (HAP; American International Chemical, Natick, Mass.) equilibrated with buffer A (pH 7.5) at a flow rate of 6 ml/min. The protein was eluted with a linear gradient (600 ml) from 0 to 500 mM potassium phosphate in buffer A (pH 7.5). The protein eluted when 60 mM phosphate was applied to the column. The hydrogenase-containing fractions were subsequently loaded onto a column (1.6 by 7.5 cm) of Q-Sepharose High Performance (Amersham Pharmacia Biotech) equilibrated with buffer A (pH 7.5) at a flow rate of 3 ml/min. Proteins were eluted with a linear gradient (600 ml) from 0 to 1.0 M NaCl in buffer A (pH 7.5). Fractions containing hydrogenase activity eluted between 150 and 200 mM NaCl, and these were pooled, concentrated, and stored as pellets in liquid N2.
Analytical methods.
Hydrogenase activity was determined at 80°C by H2 evolution using dithionite-reduced methyl viologen (2 mM) (10) or NADPH (2 mM) (22) as the electron donor. The H2 produced was measured by gas chromatography (model GC-8A; Shimadzu). One unit of hydrogenase activity is defined as the production of 1 μmol of H2 produced/min. Hydrogenase-catalyzed H2 evolution was also measured using reduced ferredoxin as the electron donor, which was generated by the pyruvate ferredoxin oxidoreductase (POR) reaction (22). The 2-ml assay mixture contained pyruvate (5 mM), CoA (0.1 mM), thiamine pyrophosphate (0.4 mM), MgCl2 (2.5 mM), ferredoxin (100 μg), and POR (75 μg) in 100 mM EPPS [N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid] buffer (pH 8.0). The H2 oxidation of the hydrogenase was determined spectrophotometrically by measuring the reduction of benzyl viologen (2 mM) at 578 nm. The assay buffer (50 mM EPPS buffer, pH 8.4) was saturated with H2, and the assays were performed at 80°C in serum-stoppered cuvettes. One unit of H2 oxidation activity corresponded to 1 μmol of H2 consumed/min. To measure the effects of pH on hydrogenase activity, a mixture of MES [2-(N-morpholino)ethanesulfonic acid; 50 mM], MOPS [3-(N-morpholino)propanesulfonic acid; 50 mM], and Bicine [N,N-bis-(2-hydroxyethyl)glycine; 50 mM] was used for the pH range of 5.5 to 9.0; for the pH 10.0 to 12.0, a mixture of CHES [2-N-(cyclohexylamino)ethanesulfonic acid; 50 mM] and CAPS [3-(cyclohexylamino)propanesulfonic acid; 50 mM] was used. S0 reduction activity was measured in 8-ml vials which contained 2 ml of EPPS buffer (pH 8.4) and 0.5 g of elemental sulfur under an H2 headspace at 80°C. The amount of H2S produced was measured periodically using a gas chromatograph (17). One unit of activity is defined as the production of 1 μmol of H2S/min. Glutamate dehydrogenase (GDH) activity was measured according to Robb et al. (31).
Protein concentrations were determined with a detergent-compatible Lowry assay kit, using bovine serum albumin as the standard (Bio-Rad, Hercules, Calif.). Metal analysis was performed using inductively coupled plasma emission spectroscopy at the Chemical Analysis Laboratory (University of Georgia, Athens). Protein purity was determined by denaturing gel electrophoresis using 10% Tricine gels (Novex, Carlsbad, Calif.) with Tricine (pH 8.3) as the running buffer according to the manufacturer's instructions. For NH2-terminal sequence analysis, the protein was applied to a denaturing gradient 4 to 12% NuPAGE gel (Novex) and subsequently blotted onto a polyvinylidene difluoride membrane using NuPAGE transfer buffer at pH 7.2 according to the manufacturer's instructions. The membrane was stained with Coomassie blue R-250, and bands were excised from the membranes. The protein bands were sequenced with an Applied Biosystems model 477 sequencer at the Molecular Genetics Instrumentation Facility (University of Georgia, Athens).
Electron paramagnetic resonance (EPR) spectra were recorded on a Bruker 300E spectrometer equipped with an Oxford Instruments ITC flow cryostat and interfaced to an ESP 3220 computer. Samples were prepared by concentrating the purified protein using a 5-ml Hi-Trap Q column equilibrated with 50 mM Tris buffer (pH 8.0). The protein was eluted with buffer A containing 500 mM NaCl. The sample was subsequently desalted in an anaerobic chamber (Vacuum Atmospheres, Hawthorne, Calif.) using a Microcon-30 concentrator (Amicon, Beverly, Mass.) with 100 mM EPPS buffer (pH 8.0) and 2 mM sodium dithionite. The protein was then transferred to an EPR tube under anaerobic conditions and rapidly frozen in a heptane-liquid N2 mixture.
RESULTS AND DISCUSSION
Identification of an MB hydrogenase in P. furiosus.
Our initial objective was to prepare by ultracentrifugation membranes of P. furiosus that lacked the cytoplasmic hydrogenases so that they could be assayed for S0 reduction activity. However, as shown in Table 1, such membranes retained about 30% of the H2 evolution activity that was initially present in the cell extract, even after extensive washing with buffer containing high salt concentrations (up to 4.0 M NaCl). As discussed below, this membrane-associated activity was sensitive to inactivation by O2 (air), and all washing and purification steps were carried out under anaerobic conditions. To investigate whether the H2 evolution activity in the membranes was due to contaminating amounts of the cytoplasmic hydrogenases, the activity of GDH, a known cytoplasmic protein (31), was measured (Table 1). Only negligible amounts of GDH activity were detected in the 2.0 and 4.0 M NaCl washes and in the final membrane preparation, showing that this enzyme is efficiently removed by the washing procedure. Separate assays confirmed that the decrease in GDH activity was not due to inhibition of the enzyme by the high salt concentrations (data not shown). That the hydrogenase in the purified membranes was distinct from the cytoplasmic hydrogenases was suggested by its catalytic properties. As shown in Table 1, the membrane-associated enzyme did not evolve H2 using NADPH as the electron donor, nor did it catalyze the reduction of S0 with H2 as the source of reductant. Both of these activities, which are characteristics of the cytoplasmic hydrogenases (20, 21), decreased in parallel with that of GDH activity as the membranes were successively washed. The MB hydrogenase did exhibit H2 oxidation activity using benzyl viologen as the artificial electron acceptor (Table 1), but the activity was very low. For example, the ratio of the H2 evolution to H2 oxidation activity was approximately 2,350 (Table 1), which compares with values near 3 for the cytoplasmic hydrogenases when they are assayed under the same conditions (10, 21).
TABLE 1.
Hydrogenase and GDH activities during purification of membranes from cell extracts of P. furiosus
| Fraction | H2 evolution
|
H2 oxidation, benzyl viologen
|
Sulfur reduction
|
GDH
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Methyl viologen
|
NADPH
|
|||||||||
| Activity (U) | Sp act (U/mg) | Activity (U) | Sp act (U/mg) | Activity (U) | Sp act (U/mg) | Activity (U) | Sp act (U/mg) | Activity (U) | Sp act (U/mg) | |
| Cell extract | 1,330 | 10 | 117 | 0.9 | 104 | 0.8 | 2.6 | 0.04 | 721 | 1.2 |
| Cytoplasm | 864 | 90 | 83 | 8.9 | 64 | 6.7 | 1.0 | 0.20 | 425 | 2.6 |
| Tris wash | 269 | 30 | 31 | 2.0 | 13 | 1.6 | <0.01 | <0.001 | 94 | 1.2 |
| 1.0 M NaCl | 37 | 3 | <0.01 | <0.001 | 10 | 0.8 | <0.01 | <0.001 | 42 | 0.8 |
| 2.0 M NaCl | <0.01 | <0.001 | <0.01 | <0.001 | <0.01 | <0.001 | <0.01 | <0.001 | 21 | 0.1 |
| 4.0 M NaCl | 1.1 | 3.5 | <0.01 | <0.001 | <0.01 | <0.001 | <0.01 | <0.001 | <0.01 | <0.001 |
| Washed membranes | 435 | 9.4 | <0.01 | <0.001 | 0.2 | 0.004 | <0.01 | <0.001 | 21 | 0.3 |
Purification of the MB hydrogenase.
To investigate procedures to solubilize the MB hydrogenase, the salt-washed membranes were extracted with a variety of zwitterionic, ionic, and nonionic detergents. Treatments with CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate}, Triton X-100, n-octyl-β-d-glucoside, and sodium deoxycholate were not as effective as using n-dodecyl-β-d-maltoside. About 80% of the total hydrogenase activity was released into the supernatant fraction after ultracentrifugation (120,000 × g for 2.0 h) when the washed membranes were treated with n-dodecyl-β-d-maltoside at a concentration of 1.5% (wt/vol). The enzyme was further purified by ion-exchange chromatography and HAP, using buffers containing 0.05% Triton X-100. We observed aggregation leading to a substantial loss of activity on the first ion-exchange step unless the buffer also contained 2.0 M urea. No activity was lost when the enzyme preparation was incubated with 2.0 M urea (at 23°C) even after 48 h. After three chromatography steps, the purified MB hydrogenase had a specific activity in the H2 evolution assay of 31 U/mg (Table 2). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) revealed the presence of two major protein bands, α and β, with apparent masses of 40 and 20 kDa, respectively, and one minor protein band with a molecular mass of 55 kDa (Fig. 1). The electrophoretic behavior of the MB enzyme is different from that of the cytoplasmic hydrogenase I and hydrogenase II, as both of these are heterotetramers containing subunits with masses of 50, 43, 33, and 31 kDa and 52, 39, 30, and 24 kDa, respectively (21, 28). Densitometric analysis of gels stained with Coomassie blue showed that the α and β proteins were present in a ratio of 1:0.92, suggesting that they are subunits of the same complex. This was also suggested by the fact that the two bands could not be separated by additional ion-exchange, HAP, and gel filtration chromatography using the purified MB hydrogenase preparation.
TABLE 2.
Purification of the MB hydrogenase of P. furiosusa
| Step | Protein (mg) | Activity (U) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Washed membranes | 1,920 | 7,690 | 4.0 | 1 | 100 |
| Detergent extract | 1,060 | 5,890 | 5.6 | 1.4 | 77 |
| DEAE-High Capacity | 272 | 3,910 | 14.1 | 3.5 | 51 |
| HAP | 54 | 1,300 | 24.1 | 6.0 | 17 |
| Q-Sepharose | 15 | 535 | 30.6 | 7.7 | 7.0 |
Using 150 g (wet weight) of cells.
FIG. 1.

SDS-polyacrylamide gel of the purified MB hydrogenase of P. furiosus. Marker proteins with the indicated molecular masses are in the left lane, and the purified hydrogenase is in the right lane. The two subunits of the hydrogenase (α and β) are indicated.
The N-terminal sequence of the α subunit of the MB hydrogenase was MKKVEYWVKI-. This sequence matched exactly the translated N terminus of an open reading frame (ORF) in the P. furiosus genome database (http://comb5-156.umbi.umd.edu/) which would encode a protein of 47,930 Da (427 residues). The N-terminal sequence of the β subunit was SKAEMVANKI-. With the exception of an N-terminal methionine, which is presumably cleaved in vivo, this sequence was identical to the translated N terminus of an ORF near that predicted to encode the α subunit (see below), and this would correspond to a protein of mass 17,491 Da (149 residues). These analyses therefore indicate that the solubilized MB hydrogenase is composed of two subunits in a 1:1 ratio with a combined mass of about 65 kDa. There was no significant sequence similarity between the ORFs predicted to encode the α and β subunits of the MB hydrogenase and any of the subunits of the cytoplasmic hydrogenases from P. furiosus (21, 28), confirming that they are indeed distinct enzymes. However, as discussed below, the sequences of the α and β subunits are highly similar to two subunits of the MB hydrogenase complexes of the methanogen M. barkeri (EchE and -C, respectively), of the purple photosynthetic bacterium R. rubrum (CooH and -L, respectively), and of E. coli (hydrogenase 3; HycE and -G, respectively) (6, 8, 18, 33, 34). The N-terminal sequence of the third protein band evident on the SDS-gel near 55 kDa (Fig. 1) was MDKLKLYVAG-, which matches exactly the translated N-terminal region of an ORF in the genome that would encode a very hydrophobic protein with a molecular mass of 38,619 Da (339 residues). The ORF is not part of the putative hydrogenase operon (see below), and whether the corresponding protein, which has no obvious motifs, that copurifies with the α and β subunits of the MB hydrogenase is functionally related is not clear.
Molecular and catalytic properties of the MB hydrogenase.
The properties of the hydrogenase of P. furiosus were investigated in its MB and solubilized states. Its sensitivity to inactivation by O2 increased upon solubilization. With the washed membranes, the time required for the enzyme (0.3 mg/ml in buffer A) to lose 50% of its H2 evolution activity (t1/2 value) was about 4 days in air, but this decreased to about 3 h with the purified enzyme (data not shown). This sensitivity may explain the significant loss of activity observed during the final purification steps (Table 2). The thermal stability of the enzyme also decreased with purification. For example, the t1/2 of the enzyme within the membranes (0.3 mg/ml in buffer A) at 100°C was 2 h, compared with a t1/2 of only 30 min for the purified enzyme. Accordingly, the optimum temperature for H2 evolution (measured over an 8-min period) was 90°C for the washed membranes but 80°C for the purified enzyme. The two forms of the enzyme showed similar responses to pH. H2 evolution activity (at 80°C) was confined to a rather narrow range between 6.0 and 8.5, with a pH optimum around pH 7.0 (data not shown).
While the purified MB hydrogenase both evolved and oxidized (albeit poorly) H2 with viologen dyes as electron carriers, it would not utilize NAD, NADH, NADP, or NADPH (up to 4 mM). Moreover, the enzyme would not utilize P. furiosus ferredoxin as an electron carrier. This is the primary electron acceptor for the fermentative pathways in this organism (2), but the MB hydrogenase preparation did not evolve H2 from reduced ferredoxin. This was the case when ferredoxin was reduced with sodium dithionite or with POR from P. furiosus using pyruvate as the electron donor. Unwashed membranes and cytosolic preparations of freshly lysed cells did catalyze ferredoxin-dependent H2 evolution activity using pyruvate (via POR) as the electron donor. The activities were 0.5 and 1.5 U/mg, respectively. However, after washing with buffer containing 2.0 and 4.0 M NaCl, the membrane preparation did not catalyze any detectable H2 production using the pyruvate/POR system, even when incubated for more than 30 min at 80°C. The activity of the same sample using methyl viologen as the electron carrier under standard assay conditions was 7 U/mg, indicating that the hydrogenase was active. Like the MB hydrogenase, the cytoplasmic hydrogenases do not evolve H2 from reduced ferredoxin, and in vivo they are thought to oxidize NADPH which is reduced by FNOR (21, 22). Thus, the loss of the ferredoxin-dependent hydrogenase activity upon washing the membranes is presumably due to removal of various cytoplasmic proteins, rather than to loss of a subunit or cofactor that mediates electron transfer between reduced ferredoxin and the MB hydrogenase. Similarly, the lack of activity with ferredoxin as the electron carrier for the purified MB enzyme does not appear to be a result of the solubilization procedure.
The purified MB hydrogenase preparation contained 0.85 mol of Ni and 4.4 mol of Fe per mol of protein by direct chemical analyses, assuming a molecular mass of 65 kDa. EPR spectroscopy of the reduced enzyme confirmed the presence of a redox-active Ni site. The enzyme reduced by sodium dithionite gave rise to a rhombic-type signal with g values of 2.39, 2.16, and 2.05 that could be observed up to 50 K (data not shown). This spectrum is very reminiscent of that seen from several other NiFe-hydrogenases and corresponds to the so-called Ni-C form, which represents an intermediate redox state of Ni in this type of hydrogenase (5, 27). The reduced MB hydrogenase gave rise to a more complex spectrum near g = 2 at low temperature (8 K) which likely arises from one or more iron-sulfur centers (data not shown), in accordance with the metal analyses.
MB hydrogenase operon sequence analysis.
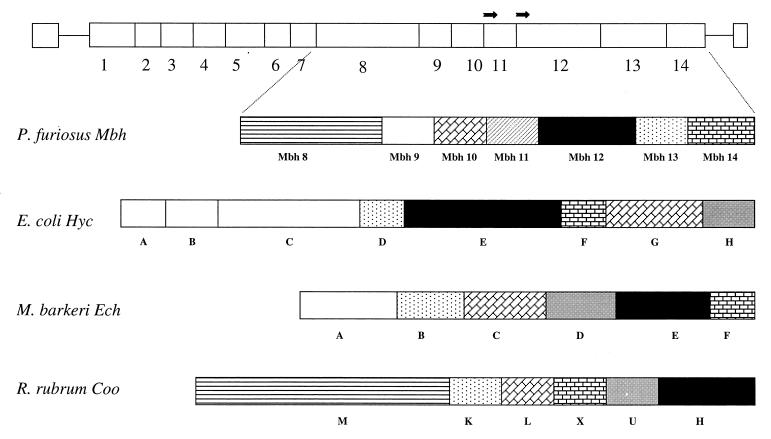
Analysis of the sequences surrounding the genes proposed to code for the two subunits of the MB hydrogenase revealed the presence of a large putative mbh operon which contained a total of 14 ORFs spanning 8.02 kb (nucleotides 1337413 to 1345430 [http://comb5-156.umbi.umd.edu/]). As shown in Fig. 2, the two subunits of the purified enzyme correspond to Mbh 11 (β) and Mbh 12 (α). An operon arrangement is suggested by the absence of intervening sequences between the 14 genes, and all appear to have a strong ribosome binding site ∼8 bp upstream of each start codon. Moreover, all would be transcribed in the same direction, and there is an AT-rich region (data not shown) which could contain the transcriptional start site upstream of the first ORF (Mbh 1).
FIG. 2.
Proposed operon encoding the MB hydrogenase complex of P. furiosus. The horizontal arrows near Mbh 11 and Mbh 12 indicate positions of the N-terminal amino acid sequences obtained from the purified complex. Also shown are the gene arrangements of the hyc, ech, and coo operons from E. coli, M. barkeri, and R. rubrum, respectively (8, 13, 18). Genes showing significant sequence similarity have the same shading.
The 14 ORFs in the putative operon can be divided into two categories. Mbh 1 to 7 and Mbh 9 code for small proteins (9 to 19 kDa) that are mostly hydrophobic in character, contain one or no Cys residue, and show sequence similarity only to conserved hypothetical proteins. On the other hand, Mbh 8, 10, 13, and 14, together with Mbh 11 and 12, resemble proteins encoded by the ech operon from M. barkeri (18, 25), the coo operon from R. rubrum (13), and/or the hyc operon from E. coli (7, 33), all of which contain structural genes for MB hydrogenase complexes. As shown in Fig. 2, the six genes are arranged similarly, although not identically, in the four operons. Mbh 12 encodes the catalytic subunit of the P. furiosus complex. This is one (α) of the two subunits of the purified H2-evolving enzyme, and it shows high similarity (35 to 49%) to the complete sequences of the catalytic subunits of the other three MB hydrogenases. This includes two Cys-X-X-Cys motifs near the N and C termini. These motifs coordinate the NiFe site of the structurally characterized (cytoplasmic) NiFe-hydrogenase of Desulfovibrio gigas (36). However, Mbh 12 shows a much lower sequence similarity (20%) to the latter enzyme than it does to the MB hydrogenases. Interestingly, the C-terminal motif in these MB enzymes, DPCXSCTXR, contains a terminal Arg residue instead of the His residue found in cytoplasmic NiFe-hydrogenases (32). Maturation of the D. gigas and other NiFe-hydrogenases involves proteolytic cleavage at this His residue (upon Ni insertion) (24). Although there is no direct evidence to support a similar maturation process with P. furiosus hydrogenase, the molecular mass of its α subunit would decrease from 47,930 to 42,944 Da if the C terminus is processed. The lower value corresponds to the size (∼42 kDa) of the α subunit as estimated by SDS-PAGE, suggesting that processing after the Arg residue does take place.
The second subunit (β) of the purified P. furiosus complex, Mbh 11 (17,491 Da), lacks Cys residues and, of the three MB hydrogenase complexes, shows sequence similarity (39%) only with the 160-amino-acid N-terminal region of HycE (65 kDa) of E. coli hydrogenase 3 (Fig. 2). Similarly, Mbh 8 (54,976 Da, 511 residues), which appears to contain eight membrane-spanning helices, has sequence similarity (21%) only to the CooM protein (133 kDa) from R. rubrum. Of the remaining proteins of the MB hydrogenase operon, Mbh 10 (18,512 Da, 170 residues) contains five Cys residues in an atypical motif and shows sequence similarity (27 to 59%) to CooL, EchC, and HycG, while Mbh 13 (35,385 Da, 322 residues) lacks Cys and shows similarity (39 to 54%) to HycD, CooK, and EchB. Finally, Mbh 14 (15,684 Da, 140 residues) shows similarity (32 to 41%) to HycF, EchF, and CooX. All four of these proteins contain eight Cys residues arranged in a 8Fe-ferredoxin-like motif, suggesting that they all contain two [4Fe-4S] clusters and are involved in electron transfer to and from the catalytic subunit of their respective hydrogenases. Thus, like the α subunit (Mbh 12), Mbh 10, 13, and 14 are analogs of proteins found in all three of the mesophilic hydrogenase complexes, but Mbh 8 and Mbh 11 are found together only in P. furiosus.
Physiological role of the MB hydrogenase.
P. furiosus has always been considered to be a fermentative organism, and so the presence of an MB hydrogenase raises the fundamental question of whether this enzyme is part of a respiratory system. This is suggested by the high sequence similarity between the purified enzyme, as well as several of the proteins postulated to be part of the membrane complex (Fig. 2), with analogous proteins of the MB hydrogenase complexes of E. coli (hyc encoding hydrogenase 3 [8, 34]), M. barkeri (18), and R. rubrum (13, 14). The complexes in the latter two organisms, although not that in E. coli, are thought to play a role in energy conservation through proton translocation by as yet unknown mechanisms. Another hydrogenase system in E. coli, hydrogenase 4 encoded by the hyf operon, is thought to have an energy-conserving function as a formate hydrogenlyase system comprised of hydrogenase and formate dehydrogenase (7). However, there is no significant similarity between the components of the hyf operon and those within the operon proposed to encode the MB hydrogenase system of P. furiosus. In addition, we have been unable to detect formate dehydrogenase activity (NAD[P] or viologen linked) in the cell extracts of P. furiosus used to prepare the MB hydrogenase (data not shown).
On the other hand, some of the other proteins that appear, from the operon analysis, to be associated with P. furiosus MB hydrogenase may play a role in energy conservation. For example, the sequences of Mbh 10, 13, and 14 show 45, 42, and 38% similarity, respectively, to those of the NuoB, NuoH, and NuoI subunits of the proton-translocating NADH dehydrogenase complex that is part of the aerobic respiratory complex of E. coli (37). Similarly, Mbh 2, 5, 6, 7, and 9 show weak sequence similarity to other NADH dehydrogenase structural subunits and/or an Na+/H+ antiporter, although no indications of the functions of Mbh 1 to 7 and 9 (Fig. 2) are evident from sequence analyses. There is also no evidence that the catalytic subunit, Mbh 12, is involved in proton translocation per se, nor is there any indication of a leader peptide, suggesting that it is facing the cytoplasmic side of the membrane.
The nature of the physiological electron carrier for the MB hydrogenase of P. furiosus is not known since neither the purified enzyme nor washed membranes interacted with either NAD(P)H or P. furiosus ferredoxin. Moreover, washed membranes did not reduce S0 with H2, suggesting that a S0-reducing system is not present in the membranes or, if it is, that it is not associated with the MB hydrogenase. Thus, at present we can only conclude that P. furiosus appears to contain an as yet unexplored pathway of H2 metabolism that may serve a role in energy conservation. The pathways of electron transfer between the oxidative fermentative pathways and the cytoplasmic and MB hydrogenases, how (or if) they are regulated, and their relationships to the pathway(s) of S0 reduction all remain to be elucidated.
ACKNOWLEDGMENTS
This research was supported by grants from the Department of Energy (FG05-95ER20175 and under contract DE-AC05-96OR22464 with Lockheed Martin Energy Research Corp.) and the National Science Foundation (MCB 9809060).
We thank Angeli L. Menon and Amy M. Grunden for assistance with sequence analyses.
REFERENCES
- 1.Adams M W W. The structure and mechanism of iron-hydrogenases. Biochim Biophys Acta. 1990;1020:115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- 2.Adams M W W, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archaea. Adv Protein Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 3.Adams M W W, Mortenson L E. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. J Biol Chem. 1984;259:7045–7055. [PubMed] [Google Scholar]
- 4.Albracht S P J. Intimate relationships of the large and the small subunits of all nickel hydrogenases with two nuclear-encoded subunits of mitochondrial NADH: ubiquinone oxidoreductase. Biochim Biophys Acta. 1993;1144:221–224. doi: 10.1016/0005-2728(93)90176-g. [DOI] [PubMed] [Google Scholar]
- 5.Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Andrews S C, Berks B C, McClay J, Ambler A, Quail M A, Golby P, Guest R. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 8.Böhm R, Sauter M, Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 9.Bowien B, Schlegel H G. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu Rev Microbiol. 1981;35:405–452. doi: 10.1146/annurev.mi.35.100181.002201. [DOI] [PubMed] [Google Scholar]
- 10.Bryant F O, Adams M W W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J Biol Chem. 1989;264:5070–5079. [PubMed] [Google Scholar]
- 11.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 12.Fontecilla-Camps J C. The active site of Ni-Fe hydrogenases: model chemistry and crystallographic results. J Biol Inorg Chem. 1996;1:91–98. [Google Scholar]
- 13.Fox J D, He Y, Shelver D, Roberts G P, Ludden P W. Characterization of the region encoding the CO-induced hydrogenase from Rhodospirillum rubrum. J Bacteriol. 1996;178:6200–6208. doi: 10.1128/jb.178.21.6200-6208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox J D, Kerby R L, Roberts G P, Ludden P W. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter K O. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1999;22:353–381. [Google Scholar]
- 16.Kengen S W M, Stams A J M, de Vos W M. Sugar metabolism of hyperthermophiles. FEMS Microbiol Rev. 1996;18:119–137. [Google Scholar]
- 17.Kim C, Woodward C A, Kaufman E N, Adams M W W. Stability and sulfur-reduction activity in non-aqueous phase liquids of the hydrogenase from the hyperthermophile Pyrococcus furiosus. Biotechnol Bioeng. 1999;65:108–113. [PubMed] [Google Scholar]
- 18.Kunkel A, Vorholt J A, Thauer R K, Hedderich R. An Escherichia coli hydrogenase-3 type hydrogenase in methanogenic archaea. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- 19.Ma K, Adams M W W. Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J Bacteriol. 1994;176:6509–6517. doi: 10.1128/jb.176.21.6509-6517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma K, Schicho R N, Kelly R M, Adams M W W. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci USA. 1993;90:5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K, Weiss R, Adams M W W. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol. 2000;182:1864–1871. doi: 10.1128/jb.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma K, Zhou Z H, Adams M W W. Hydrogen production from pyruvate by enzymes purified from the hyperthermophilic archaeon, Pyrococcus furiosus. FEMS Microbiol Lett. 1994;122:245–250. [Google Scholar]
- 23.Mai X, Adams M W W. Purification and identification of two reversible and ADP-dependent acetyl-coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menon N K, Robbins J, DerVartanian M, Patil D, Peck H D, Menon A L, Robson R L, Przybyla A E. Carboxy-terminal processing of the large subunit of [NiFe] hydrogenases. FEBS Lett. 1993;331:91–95. doi: 10.1016/0014-5793(93)80303-c. [DOI] [PubMed] [Google Scholar]
- 25.Meuer J, Bartoschek S, Koch J, Kunkel A, Hedderich R. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur J Biochem. 1999;265:325–335. doi: 10.1046/j.1432-1327.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 26.Nicolet Y, Piras C, Legrand P, Hatchikian C E, Fontecilla-Camps J C. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 27.Niu S, Thomson L M, Hall M B. Theoretical characterization of the reaction intermediates in a model of the nickel-iron hydrogenase of Desulfovibrio gigas. J Am Chem Soc. 1999;121:4000–4007. [Google Scholar]
- 28.Pedroni P, Della Volpe A, Galli G, Mura G M, Pratesi C, Grandi G. Characterization of the locus encoding the [Ni-Fe] sulfhydrogenase from the archaeon Pyrococcus furiosus: evidence for a relationship to bacterial sulfite reductases. Microbiology. 1995;141:449–458. doi: 10.1099/13500872-141-2-449. [DOI] [PubMed] [Google Scholar]
- 29.Peters J W, Lanzilotta W N, Lemon B J, Seefeldt L C. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 30.Ringel M, Gröb R, Kröger A, Schauder R. Growth of Wolinella succinogenes with elemental sulfur in the absence of polysulfide. Arch Microbiol. 1996;165:62–64. [Google Scholar]
- 31.Robb F T, Park J B, Adams M W W. Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 32.Rossmann R, Sauter M, Lottspeich F, Böck A. Maturation of the large subunit (HycE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg 537. Eur J Biochem. 1994;220:377–384. doi: 10.1111/j.1432-1033.1994.tb18634.x. [DOI] [PubMed] [Google Scholar]
- 33.Sauter M, Böhm R, Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 34.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 35.Schicho R N, Ma K, Adams M W W, Kelly R M. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1993;175:1823–1830. doi: 10.1128/jb.175.6.1823-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volbeda A, Charon M-H, Piras C, Hatchikian E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 37.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli: organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]