Abstract
The synchronization of circadian clock depends on a central pacemaker located in the suprachiasmatic nuclei. However, the potential feedback of peripheral signals on the central clock remains poorly characterized. To explore whether peripheral organ circadian clocks may affect the central pacemaker, we used a chimeric model in which mouse hepatocytes were replaced by human hepatocytes. Liver humanization led to reprogrammed diurnal gene expression and advanced the phase of the liver circadian clock that extended to muscle and the entire rhythmic physiology. Similar to clock-deficient mice, liver-humanized mice shifted their rhythmic physiology more rapidly to the light phase under day feeding. Our results indicate that hepatocyte clocks can affect the central pacemaker and offer potential perspectives to apprehend pathologies associated with altered circadian physiology.
A model of liver humanization in mice demonstrate that hepatocyte clocks can feed back on the central circadian clock.
INTRODUCTION
To adapt homeostasis to the changing environment present on Earth, organisms from bacteria to mammals have evolved a timing system that anticipates these changes. This endogenous timing system, called the circadian clock, orchestrates most aspects of physiology and behavior. The mammalian circadian system is hierarchically organized. A central clock localized in the suprachiasmatic nuclei (SCN) of the hypothalamus is daily synchronized by light via the retinohypothalamic tract and coordinates the peripheral clocks localized in peripheral tissues. The SCN synchronizes most aspects of circadian physiology and is required to keep phase coherence between the different peripheral organs (1, 2). Nonetheless, how the SCN orchestrates this phase coherence is still only partially understood.
In the absence of SCN, the individual peripheral organs continue to show robust rhythms, indicating the existence of self-sustained cellular clocks (2, 3). In all mammalian cells, this rhythmicity is generated by molecular machinery consisting of interconnected transcriptional and translational feedback loops in which the transcription factor brain and muscle Aryl hydrocarbon receptor nuclear translocator–like 1 (BMAL1, encoded by the Arntl gene) plays a critical role. Abolishing BMAL1 activity in mouse hepatocytes demonstrated that around 35% of liver rhythmic genes depend on a functional circadian clock, whereas the other rhythmic genes are regulated by systemic and/or feeding cues (4, 5). Nevertheless, disrupting the hepatocyte clock has no impact on the central clock in the SCN (6). On the other hand, restoring Bmal1 expression and a functional circadian clock specifically in hepatocytes in a full-body Bmal1 knockout (KO) animal does not affect the global rhythmic activity and behavior of these animals (7). Therefore, while the central clock synchronizes peripheral clocks, there is so far no evidence that peripheral clocks can affect the central clock in the SCN.
In addition, recent evidence further suggests that the circadian clocks of different cell types communicate with each other inside the same organ. For example, astrocytes’ circadian clock can entrain neuronal clock in the SCN (8) and is important to determine the circadian period (9). Moreover, the disruption of the circadian clock in hepatocytes modulates the rhythmic gene expression of other liver cell types (10) and even the synchronization of the circadian clock in other tissues in response to feeding cues (11). These studies highlight the importance of the communication between the clocks in the different cell types inside the same tissue or other more distant tissues (12).
To decipher further the impact of these cellular communications in the organization of liver circadian physiology, we implanted in mouse liver human hepatocytes that are hypothesized to show different physiological properties and responses to systemic and feeding cues compared to mouse hepatocytes (13, 14), as well as different rhythmic functions as shown in nonhuman primates (15). Here, we show that implantation of human hepatocytes in mice [liver-humanized mice (LHM)] results in a global loss of rhythmicity in liver gene expression in comparison to control animals implanted with mouse hepatocytes [liver-“murinized” mice (LMM)]. While most circadian clock genes were still rhythmic in the liver of LHM, they displayed an advanced phase and lower amplitude in both mouse and human hepatocytes, suggesting that human hepatocytes can entrain the remaining mouse hepatocytes in the liver of LHM. Notably, we found that these human hepatocytes also regulate rhythmic genes in the muscle and affect global rhythmic behavior and metabolism, suggesting potential feedback of the implanted human hepatocytes on the hypothalamus and the central clock in the SCN. Last, during imposed day feeding, LHM adapted more quickly to the light phase, confirming that the engrafted human hepatocytes affect the entrainment property of the central circadian pacemaker. Together, the chimeric LHM provide a yet uncharacterized mechanism by which perturbation of the liver clock might directly alter global circadian physiology.
RESULTS
Human hepatocytes in a rodent environment impose specific rhythmic gene expression to surrounding liver cells
It is now established that most cells in the body contain self-sustained molecular clocks that can be synchronized by both signals from the central clock in the SCN and systemic cues including feeding rhythm (12). Recently, it has been shown that the disruption of the circadian clock specifically in the hepatocyte (10, 11) or the presence of a functional clock only in hepatocytes in a clock-depleted animal (7) modulates the rhythmic expression in other cell types inside the same tissue or other tissues, as well as their response to feeding. However, while transplanting SCN cells with different circadian properties in an intact animal can transfer the donor’s SCN circadian properties and affect animal behavior (1), there is no description of such effects for peripheral tissues.
To answer this question, we used chimeric LHM in which mice livers are repopulated with human hepatocytes that are expected to have their own circadian properties and capacities to respond to systemic signals, as described for humans and another diurnal primate (14, 15). In this study, we used the FRG-triple KO mouse model that exhibits a mutated Fumarylacetoacetate Hydrolase (Fah) and severe immunodeficiency (Rag2−/− and Il2rg−/−) (13). After transplantation with primary human hepatocytes, the engrafted cells can repopulate the liver up to 80% due to positive selection over rodent hepatocytes functionally impaired for FAH (13). Further improvement of this model was obtained through backcrossing FRG-KO mice with the nonobese diabetic (NOD) mice, which are not diabetic because of the absence of B and T cells (16) and can more easily accept xenografts. The resulting model FRGN mice were engrafted with human hepatocytes from a donor or murine hepatocyte from NOD mice to produce, respectively, liver-humanized (Hu-FRGN, LHM) or control-murinized mice (Mu-FRGN, LMM) (Fig. 1A) (17). Circulating levels of human albumin (hAlb) in LHM were used to corroborate the extent of human hepatocyte repopulation and were on average 6414 ± 412 μg/ml in LHM, while no hAlb could be detected in LMM (fig. S1A). Over the course of the entire study, only mice with circulating levels of hAlb > 3500 μg/ml [expecting to correspond to human hepatocyte repopulation >70% (18)] were used. An additional functional readout of successful liver humanization was gathered through the measure of triglycerides (TG) content in low-density lipoprotein (LDL). While rodents transport TG at low levels mainly in very low-density lipoprotein (VLDL), TG in human serum are typically enriched in liver-born VLDL and LDL. Unlike LMM, the LHM lipoprotein profile displayed enrichment of TG in LDL, confirming the extent of liver humanization and in agreement with previous observations (fig. S1B) (17).
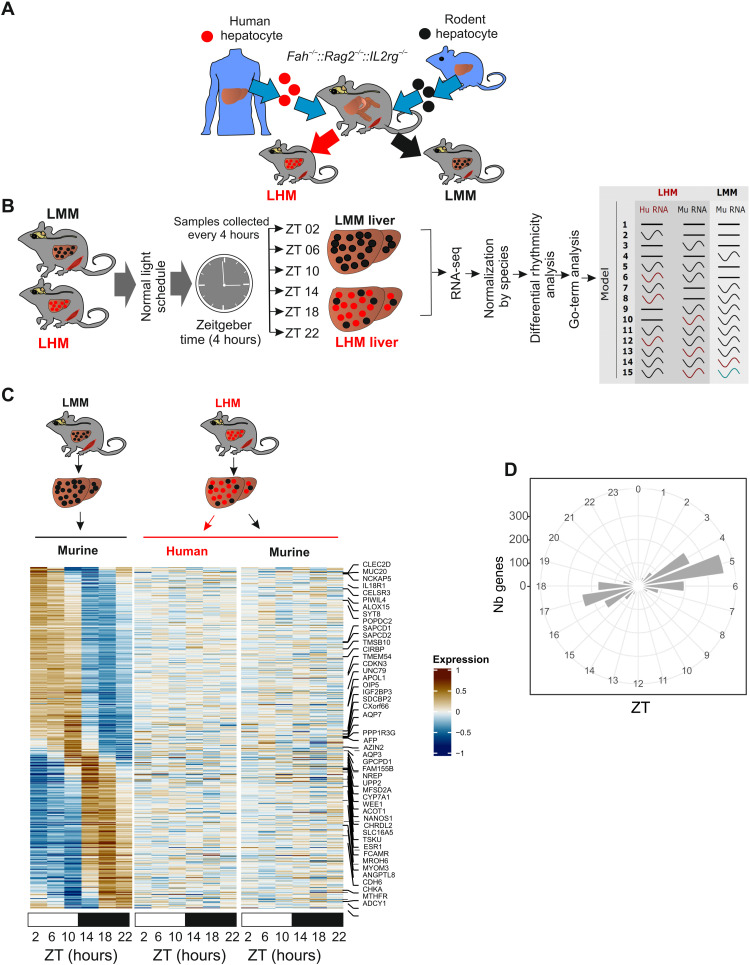
Fig. 1. Engraftment of human hepatocyte affects liver rhythmic gene expression.
(A) Model for humanized Fah−/−, Rag2−/−, Il2rg−/− (FRG-KO) that can be repopulated with primary human (red) or murine (black) hepatocytes to produce LHM or LMM mice. (B) Experimental design for liver tissue collection before RNA extraction, sequencing, and analysis according to gene expression rhythmic properties. Alteration of rhythmic gene expression of murine transcript in the liver of LMM (black, Mu RNA) and both murine (black, Mu-RNA) and human transcript (red, Hu-RNA) in the liver of LHM mice is assessed by model selection (models 1 to 15): black line, stable transcription; black wave, rhythmic transcription; red or green waves, rhythmic profiles with different rhythmic parameters (i.e., phase and/or amplitude). (C) Heatmaps of normalized rhythmic mRNA levels (BICW > 0.3, log2 amplitude > 0.5) in the liver of LMM (black) and LHM (black) murine and human transcript (red) in the liver of LMM and LHM. RNA presented here belonged to model 4 where genes were rhythmic only in LMM. (D) Radial plot of the distribution peak phase of expression for genes rhythmic only in the liver of LMM (model 4, Nb genes, number of genes).
To decipher the impact of the transplantation of human hepatocytes on the rhythmic physiology of the mouse liver, we analyzed liver rhythmic gene expression in LMM and LHM. Both LMM and LHM mice, engrafted on the same week and exposed to the same nutritional and pharmacologic treatment (see Materials and Methods) and housing conditions, were sacrificed every 4 hours, and liver tissue was collected before RNA extraction and sequencing. Because of the high-depth RNA sequencing (RNA-seq), we can discriminate between human and nonhuman RNA orthologs within LHM liver samples, allowing independent normalization for the two species before the analysis of differential rhythmicity of gene expression using our recently developed dryR method (5) (Fig. 1B and fig. S1C). dryR assigns groups of genes to various models of rhythmic behavior and species. For instance, genes assigned to model 4 are rhythmic only in LMM, while those in models 2, 3, 5, and 6 are arrhythmic in LMM but display rhythm in LHM with similar or different species-specific rhythmic parameters (Fig. 1B).
The analysis reveals that the largest group (model 4) is composed of genes that lose rhythmicity specifically in LHM animals, for both human and mouse transcripts (Fig. 1, C and D; fig. S1D; and table S1). These genes show the classical biphasic pattern observed for rhythmic genes regulated by systemic or feeding cues (Fig. 1D) (5). These rhythmic genes are mainly involved in protein synthesis and ribosome biogenesis, suggesting an attenuated activation of the mechanistic target of the rapamycin (mTOR) pathway (table S2). In a comparison with the genes differentially expressed in the liver of Raptor or Tsc1 KO mice, the main positive and negative regulators of the target of rapamycin complex 1 (TORC1), respectively (19), confirm that the genes differentially expressed in LHM are indeed enriched in genes transcriptionally regulated by the mTOR pathway (fig. S1E). Additional analysis of the phosphorylation of ribosomal protein S6 (a bona fide TORC1 target) confirms the absence of the expected rhythmic activation of this pathway in the liver of LHM (fig. S1F) (20). The mTOR pathway being recognized as a key sensor of systemic and nutritional signals, this suggests that the human hepatocytes are unable to respond to such signals when placed in a mouse environment. Alternatively, the human hepatocytes can also respond to these signals in a different manner that does not result in the rhythmic activation of the mTOR pathway. The inappropriate response of human hepatocytes to mouse signals is also illustrated by the blunted response to growth hormone (GH), an important regulator of the sex-biased liver function and metabolism (21). Endogenous mouse GH is high, while insulin-like growth factor 1 (IGF-1) is low in LHM (fig. S1G). This shows that human hepatocytes cannot secrete IGF-1 in response to GH, and, in turn, the low level of IGF-1 is not sufficient to exert its inhibitory action on GH secretion, resulting in constantly high GH levels. As additional evidence of the disruption of GH signaling, the sex-biased gene expression and the activation of sex-biased activation of the signal transducer and activator of transcription 5 (STAT5) pathway, two recognized readouts of the GH signaling (22, 23), are also perturbed, with an increase of female-biased signaling and a decrease of the male-biased signaling, likely as a result of the constantly high GH level (fig. S1H). The IGF-1/mTOR pathway being important for the synchronization of the circadian clock by systemic signals (24), this blunted response could contribute to the lack of entrainment of human hepatocytes.
On the other hand, hundreds of genes mainly involved in angiogenesis and telomere maintenance are only rhythmic in human hepatocytes (model 2, fig. S1D), suggesting that these cells can express their own specific rhythmic physiological program different from the one of mouse hepatocytes (fig. S2, A and B, and tables S1 and S2). Of particular interest is model 14, in which 110 genes are found to share similar phase advances in both human and rodent cells in LHM compared to the liver of LMM (Fig. 2, A and B, and table S1). Unexpectedly, this group shows a clear enrichment of circadian clock–related pathways (table S2) and includes most circadian clock and clock target genes (Fig. 2C). This phase advance of clock genes expression associated with a lower amplitude mirrors the presence of a functional clock only in hepatocytes in a clock-depleted animal (7), suggesting that the human hepatocytes clock in a mouse liver operates like an independent oscillator not responding to synchronizing rhythmic systemic signals. To support this hypothesis, we compared our results with the rhythmic genes present in the liver of wild type (WT), Bmal1 KO, and Bmal1 KO with a specific rescue of Bmal1 only in the liver [liver-rescue (RE)], an example of an independent hepatocyte oscillator in a circadian clock–depleted animal (7). The genes that lost rhythmicity in the liver of LHM (model 4) are the only group of genes that present an enrichment in genes rhythmic only in WT animals (fig. S2C). This supports the idea that these genes are indeed mainly regulated by systemic signals originating from other organs in a circadian clock–dependent fashion but independent of the hepatocyte circadian clock. It confirms that the human hepatocytes cannot respond to mouse synchronizing rhythmic systemic signals and operate largely independently from the rest of the organism. The fact that the mouse hepatocytes in LHM mice show the same circadian behavior suggests that the human hepatocytes can entrain the remaining mouse cells in the liver of LHM.
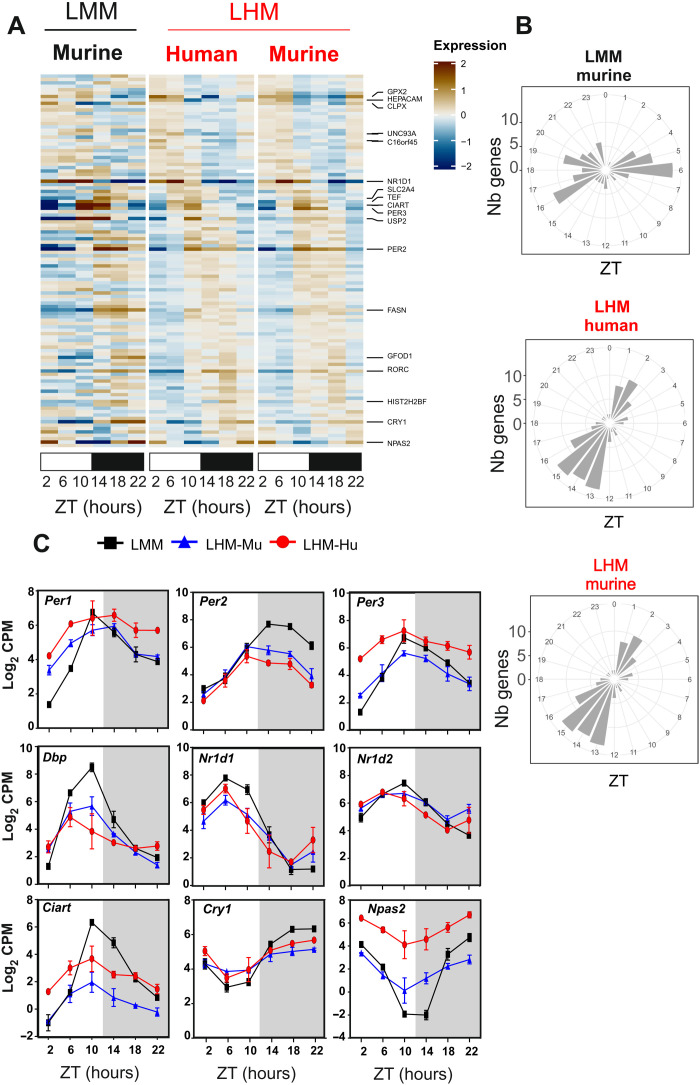
Fig. 2. Engraftment of human hepatocyte affects the phase of the liver circadian clock.
(A) Heatmaps of normalized rhythmic mRNA levels (BICW > 0.3, log2 amplitude > 0.5) in the liver of LMM (black) and LHM (red) murine and human transcript (red) in the liver of LMM and LHM. Genes presented here belonged to model 14 where both LHM-Mu and LHM-Hu ortholog transcripts share the same phase, which is different from LMM. (B) Radial plot distribution of the peak phase of expression for rhythmic genes in the liver of LMM and murine and human orthologs in the liver of LHM from model 14. (C) Rhythmic expression of circadian clock genes for murine (black, LMM) and murine (blue, LHM-Mu) and human (red, LHM-Hu) RNA orthologs in the humanized liver. Data are expressed as means ± SEM of Log2 counts per million reads mapped (CPM) (n = 12 mice per group, 2 points per replicate). For statistical details, see table S1.
The muscle circadian clock of LHM is phase advanced
To determine whether this entrainment by human hepatocytes is specific to the mouse cells in the liver, we performed a similar RNA-seq analysis on the skeletal muscle (quadriceps) of LMM and LHM (Fig. 3A). While the biggest group of genes (model 4: 32%) shows similar rhythms in LHM and LMM, a similar and important proportion of genes (model 2: 26% and model 3: 28%, respectively) are only rhythmic in the muscle of LHM and LMM mice, respectively, showing that human hepatocytes in LHM have a strong influence on the rhythmicity of other tissues (fig. S3A and table S1). The genes that lose rhythmicity in LHM (model 3) are mainly involved in protein-lipid and cholesterol metabolisms, showing the impact of the human hepatocytes on the global physiology and metabolism of the animals (fig. S3, B and C, and table S3). Notably, the group of genes displaying different circadian parameters (model 5: 13%) includes almost all the circadian clock genes and bona fide clock target genes (Fig. 3, B and C; fig. S3A; and tables S1 and S3). While we did not find a clear effect on the amplitude, all these clock genes show an advanced phase of expression more important than the one we found in the liver (Fig. 3D). This result demonstrates that the engraftment of human hepatocytes in mouse liver can unexpectedly phase advance the circadian clock of skeletal muscle.
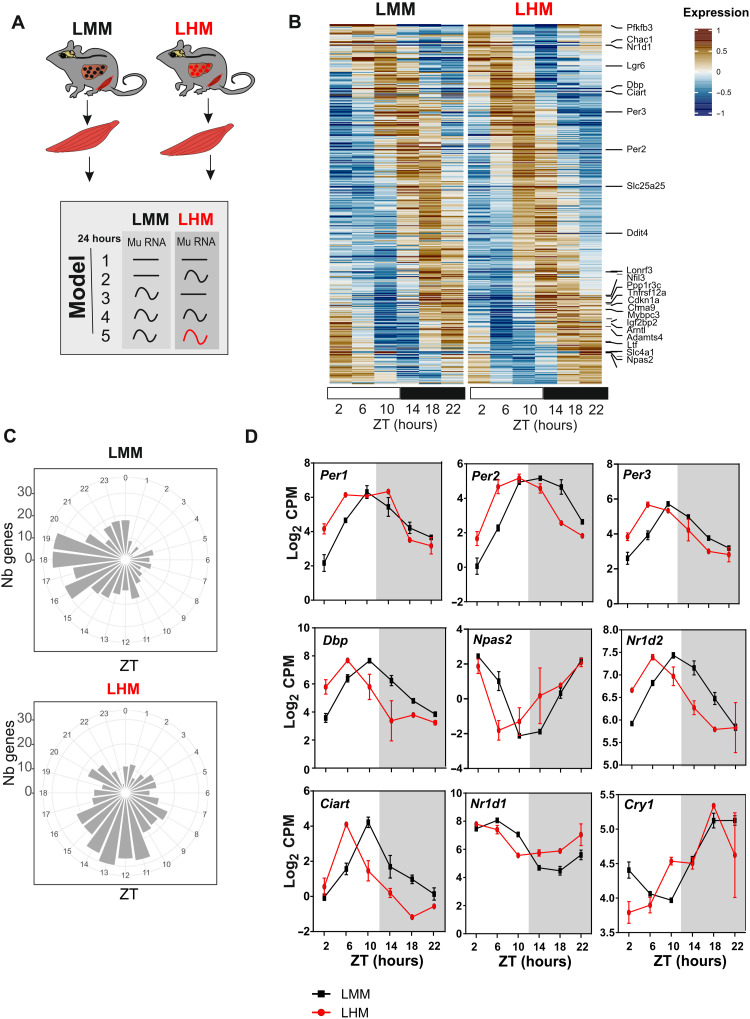
Fig. 3. The muscle circadian clock of LHM is phase advanced.
(A) Experimental design for muscle tissue collection before RNA extraction, sequencing, and analysis according to rhythmic properties and alteration of rhythmic gene expression of murine transcript in the muscle of LMM (black) and LHM (red) assessed by model selection (models 1 to 5): black line, stable transcription; black wave, rhythmic transcription; red wave, rhythmic profiles with different rhythmic parameters (i.e., phase and/or amplitude). (B) Heatmaps of normalized rhythmic muscle mRNA levels (BICW > 0.5, log2 amplitude > 0.5) in LMM (black) and LHM (red) from model 5 where LMM and LHM muscle transcript are rhythmic but with different phases. (C) Radial plot distribution of the peak phase of expression for rhythmic genes in the liver of LMM and murine and human orthologs in the liver of LHM from model 5. (D) Rhythmic expression of circadian clock genes in the muscle of LMM and LHM. Data are expressed as means ± SEM (n = 11 to 12 animals per group). For statistical details, see table S1.
Human hepatocytes advance the phase of circadian metabolism and behavior of LHM
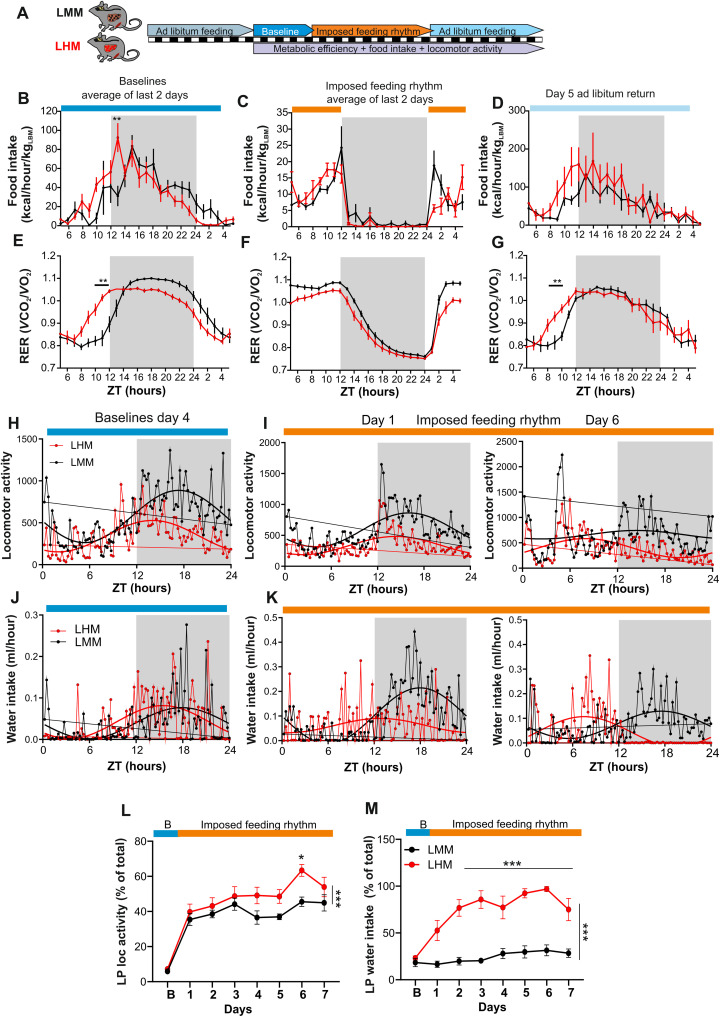
Because feeding cues act as the main driver of the phase of peripheral clocks, disconnecting them from the entrainment by the SCN (25), we hypothesized that the advanced phase of the muscle circadian clock could be a consequence of a change of rhythmic metabolic and behavioral output driven by the engrafted human hepatocytes. Therefore, we performed a comprehensive characterization of the rhythmic physiology of LMM and LHM. Under a normal light/dark cycle, LHM notably display an approximately 2-hour phase advance of locomotor activity, feeding behavior, respiratory exchange ratio, and fat oxidation compared to LMM (Fig. 4, A to H). The amplitude of the rhythmic locomotor activity and food intake is also decreased (fig. S4, A to D). Of note, this lot of LMM and LHM did not significantly differ in average cumulative food intake (fig. S4B) but show a slightly reduced body weight and body fat (fig. S4E). This suggests that the change in entrainment properties did not result from major metabolic impairment. Together, this confirms that the phase advance observed in the muscle of LHM is a consequence of liver humanization, which, by modulating the liver-brain axis, might indirectly affect on muscle physiology either by changing locomotor/muscular activity, feeding rhythms, autonomic nervous output, or endocrine signals.
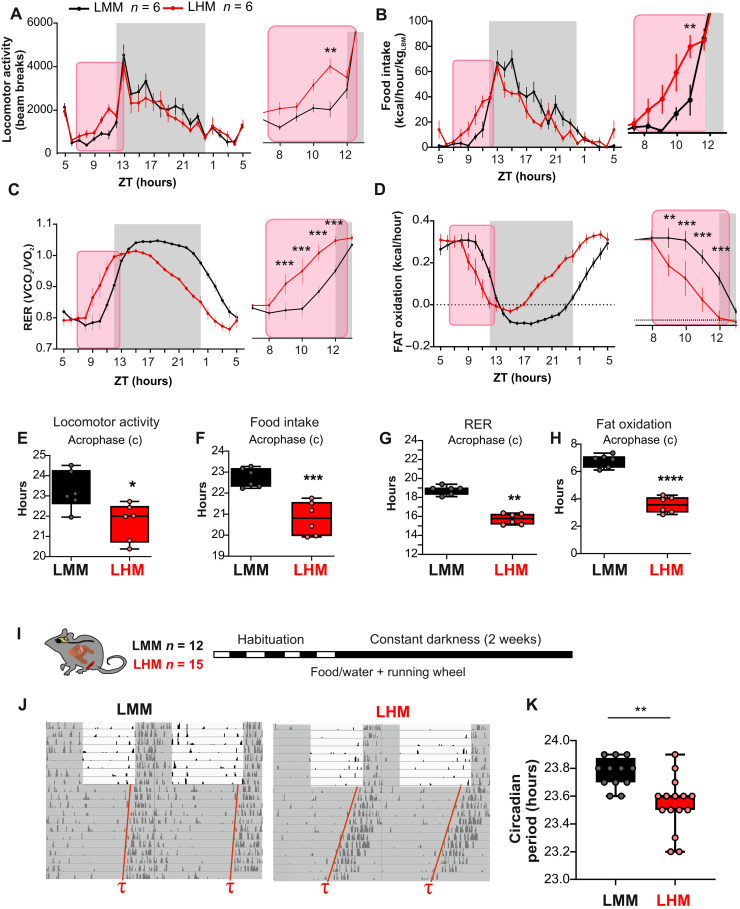
Fig. 4. Human hepatocytes advance the phase of circadian metabolism and behavior of LHM.
(A to D) Metabolic evaluation of LMM and LHM represented as 3-day average spontaneous locomotor activity (A), food intake (B, kcal/hour per kg of lean body weight), respiratory exchange ratio (RER, C), and fat oxidation (D). Inserts show magnification of the ZT8 to ZT12 time window. (E to H) Cosinor analysis of rhythmic parameters acrophase (c) for locomotor activity (E), food intake (F), RER (G), and fat oxidation (H). (I) Experimental design for determining the circadian period of LMM and LHM via recording the circadian running wheel activity in constant darkness. (J) Representative actogram and circadian period in LMM and LHM. (K) Circadian period of locomotor activity in LHM and LMM (n = 12 and 15 for LMM and LHM, respectively). Codes for statistical values: *P < 0.05, **P < 0.01, ***P < 0.0001.
As described in the case of the familial advanced sleep–phase syndrome and the related animal models, this phase advance observed in light/dark conditions could be a consequence of a short circadian period (26, 27). Accordingly, humans with shorter circadian periods show an early chronotype (28), and animal KO for the Clock gene that show a slightly shorter period (29) present also an advanced phase of behavior and metabolism under light/dark conditions (30). Thus, we determined the circadian period of the animals via the recording of their circadian wheel running activity in constant darkness (Fig. 4I). Notably, LHM show a significantly shorter period (Fig. 4, J and K), demonstrating feedback signals from human hepatocytes to the central clock in the SCN.
Engrafted human hepatocytes affect rhythmic gene expression in the hypothalamus
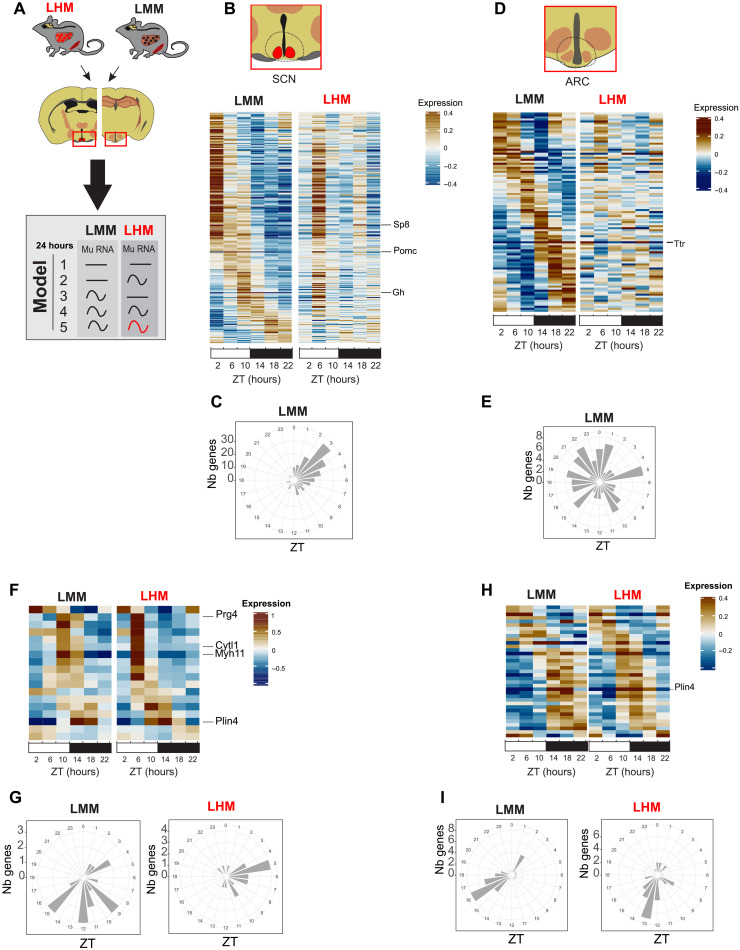
To decipher how the engrafted human hepatocytes can extend their influence beyond the liver and muscle to the brain centers involved in the control of metabolism and circadian rhythms, we performed a time-resolved RNA-seq experiment on the hypothalamic brain punches of the Arcuate nucleus (ARC) and the SCN (Fig. 5A). Relative enrichment of the SCN-enriched gene Aralkylamine N-Acetyltransferase (Aanat) or ARC-enriched Agouti-related peptide (Agrp) confirms dissection accuracy (fig. S5A). The majority (58 and 53%, model 4) of the rhythmic genes keep their rhythmicity in the SCN and the ARC, respectively, including most circadian clock genes (fig. S5, B and C, and table S1). However, the second largest group of genes representing 30 and 26% of the rhythmic genes (model 3) loses rhythmicity in the SCN and the ARC of LHM, respectively, showing that engraftment of human hepatocytes can unexpectedly affect the rhythmic gene expression in the hypothalamus (Fig. 5, B to E; fig. S5, B and C; and table S1). These genes that lose rhythmicity in the SCN and ARC of LHM are enriched for genes encoding proteins involved in ion transport, critical for the function of neurons (tables S4 and S5). In addition, a small subset of 2 and 10% of rhythmic genes in the SCN and the ARC, respectively, exhibit different circadian parameters between LMM and LHM (model 5; Fig. 5, F to I). In contrast to liver and muscle, only a few circadian clock genes, including Nr1d1 and Dbp, are included in these groups, both showing a phase advance in LHM animals (fig. S5, D and E). Nevertheless, additional analysis shows that the genes in models 4 and 5 exhibit both an enrichment for genes controlled by the circadian regulator CLOCK (31). This suggests that only a subset of circadian clock–controlled genes is phase advanced in the hypothalamus, potentially because of conflicting light signals synchronizing the SCN clock (fig. S5F) (32). Together, these results show that the human hepatocytes in LHM can affect the function of the hypothalamus, resulting in a substantial loss of rhythmicity or a change of rhythmicity of gene expression in the SCN and the ARC.
Fig. 5. Engrafted human hepatocytes affect rhythmic gene expression in the hypothalamus.
(A) Experimental design for SCN and ARC samples collection before RNA extraction, sequencing, and analysis according to rhythmic properties and alteration of rhythmic gene expression of murine transcript in the liver of LMM (black) and LHM (red) assessed by model selection (models 1 to 5): black line, stable transcription; black wave, rhythmic transcription; red wave, rhythmic profiles with different rhythmic parameters (i.e., phase and/or amplitude). (B to E) Heatmaps of normalized rhythmic mRNA levels (BICW > 0.5, log2 amplitude > 0.5) and radial plot of the distribution of the peak phase of expression of the cycling genes and in the SCN (B and C) and ARC (D and E) in LMM (black) and LHM (red) from genes that lost rhythmicity in LHM (model 3). (F to I) Heatmaps of normalized rhythmic mRNA levels (BICW > 0.5, log2 amplitude > 0.5) and radial plot of the distribution of the peak phase of expression of the cycling genes and in the SCN (F and G) and ARC (H and I) in LMM (black) and LHM (red) from genes that show altered rhythmic parameters in LHM (model 5).
Engraftment of human hepatocytes reveals the ability of hepatic signals to feedback on the central pacemaker
While the SCN is required for the coordination of feeding and drinking rhythms (33), a functional SCN counteracts and delays the synchronization of mice to day feeding. Consequently, SCN-lesioned animals synchronized their circadian peripheral clocks more rapidly to day feeding (3, 34). On the other hand, feeding rhythm is a very potent synchronizing signal for peripheral tissues and can uncouple peripheral oscillators from the central clock (25). Because LHM show an advanced phase of physiology and a global loss of rhythmic gene expression in the SCN, we speculate that the engraftment of human hepatocyte can affect the capacity of the SCN to oppose the synchronization of animal physiology to feeding during the light phase.
To test this hypothesis, LMM and LHM were subjected to a 5-day measurement of baseline rhythmic physiology under ad libitum feeding, followed by a 7-day imposed feeding during the light phase before returning to ad libitum feeding (Fig. 6A). As described above, we observed an advance in the phase of feeding and metabolic outputs at baseline conditions (Fig. 6, B and E, and fig. S6A), which were abolished during an imposed feeding regimen (Fig. 6, C and F, and fig. S6A). Notably, upon day 5 of return to ad libitum feeding, the advanced phase in LHM reappeared (Fig. 6, D and G, and fig. S6A), showing that this phase advance is an intrinsic property of the SCN of LHM. While around 40% of the locomotor activity of LMM shift to the day, as expected as an adaptation to feeding during the light phase, they kept the circadian component of their locomotor activity at night, even after 6 days of this regimen (Fig. 6, H, I, and L, and fig. S6B). However, LHM lose this circadian component and shift rapidly most of their daily locomotor activity to the light phase (Fig. 6, H, I, and L, and fig. S6B). Notably, this particular group of LMM and LHM did not significantly differ in body weight and body fat (fig. S6C), suggesting that the change in entrainment properties can be observed even in the absence of difference in body composition. More notably, while the drinking activity of LMM, which is imposed by the central clock, is essentially restricted to the dark phase, it is shifted very rapidly to the light phase in LHM. Thus, approximately 90% of drinking activity during the light phase was observed already after 3 days (Fig. 6, J, K, and M, and fig. S6B). This observation shows that the implantation of human hepatocyte modifies the entrainment properties of the SCN of LHM, which becomes more sensitive to feeding cues and let them adapt more rapidly to the daytime feeding paradigm, reminiscent of the adaptation observed in SCN lesioned animals (34).
Fig. 6. Engraftment of human hepatocytes reveals the ability of hepatic signals to feedback on the central pacemaker.
(A) Experimental design for the imposed feeding regimen during the light phase. (B to G) Two-day average analysis of rhythmic food intake (B to D) and RER (E and F) during the baseline period (B and E), imposed daylight feeding (C and F), or day 5 after return to ad libitum feeding (D and G) in LMM (black) and LHM (red) (n = 6 per groups). (H to K) Representative of average distribution and nonlinear cosinor fitting of baseline day 4 (H and J) and after 1 and 6 days of imposed light phase feeding rhythm (I and K) for locomotor activity (H and I) and water consumption (J and K). (L and M) Evolution of locomotor activity (L) and water intake (M) during the light phase (LP) after transitioning from baseline (blue) to imposed feeding during the light phase (orange) as a percentage of the baseline value. Data are expressed as means ± SEM (n = 6 animals per condition). *P < 0.05, **P < 0.01, ***P < 0.0001.
DISCUSSION
The data presented here show that the engraftment of human hepatocytes can affect the diurnal physiology and behavior of grafted mice. Notably, the human hepatocytes advance not only the phase of surrounding mouse liver cells but also the phase of distant muscle cells, likely through the change in global rhythmic physiology and behavior. Even more unexpected is the discovery that the human hepatocytes also influence the rhythmic property of hypothalamic centers, probably by affecting the rhythmic gene expression in the SCN and the ARC. These effects resulted in a shorter circadian period, a phase-advanced activity under a light-dark cycle, including the feeding rhythm, and a faster synchronization to feeding cues. This suggests that a circadian oscillator in peripheral organs with different circadian properties can feedback on brain centers to control circadian physiology and behavior. While keeping the central and peripheral clock in phase is clearly of paramount importance, there are still some uncertainties as to the relative independence or total subordination of peripheral oscillators versus the central clock. Uncertainties also exist on how a peripheral organ rhythm can possibly modulate or even entrain the central clock. Although the SCN clock is required to maintain synchrony between the peripheral organs (2, 3, 35), the hepatocyte oscillator can work autonomously (7), and the synchrony between the hepatocytes persists in SCN-lesioned mice (3). However, while the hepatocyte clock can modulate the circadian physiology of other liver cells or distant organs (10, 11), no impact on the central circadian clock has been reported. Hence, our data suggest that the hierarchy between SCN and hepatocytes in the control of circadian physiology can be challenged since peripheral oscillators with different circadian rhythms and responses to systemic cues affect the central oscillator.
Several mechanisms could be involved. The liver-brain axis could encompass rhythmic secretion of liver-borne molecules acting centrally and/or peripherally together with the nervous pathway connecting these two organs. However, while several food-related hormones or metabolites modulate the synchronization of peripheral clocks or feeding behavior in response to nutrient signals, none of them have been shown to affect the central circadian clock and modulate the circadian period of the animals (36, 37). Of importance, the hepatocytes in LMM and LHM result from the migration of injected hepatocytes in the spleen and transfer to the liver, followed by a positive selection of human hepatocytes and healthy murine hepatocytes (13, 17, 18). In this procedure, LMM and LHM retain the architecture of the liver-brain vagal connection. Hence, it is possible that the nervous connection between the liver and the brain may have routed hepatic information able to modify the central control of circadian physiology. In turn, modified central control will impede peripheral organs’ function through the hypothalamic-pituitary-adrenal axis and/or the autonomic nervous system, resulting in peripheral tissue synchronization.
The misalignment of central and peripheral clocks due to shift work, jet lag, or desynchrony between food and light signals (abnormal feeding pattern) has been implicated in various diseases including sleep disorders, psychiatric disorders, and cardiometabolic disease, including atherosclerosis, obesity, diabetes, and nonalcoholic fatty liver disease (38). Conversely, the metabolic syndrome is associated with disruption of circadian rhythms and sleep (39). Mice exposed to a high-fat diet exhibit an alteration of their circadian clocks and feeding behavior (40) and circadian desynchrony affecting phase coherence between peripheral organs and communication between peripheral and central clocks (41). Moreover, a high-fat diet induces an advance of phase of the liver’s circadian clock (42) and alters the response of the central clock to the light signals (43). Likely, these changes are rather a consequence of the metabolic syndrome caused by this regimen than the high-fat diet per se. Female or male mice from inbred lines that did not develop metabolic syndrome did not display alteration of their circadian and feeding behavior (44, 45). Liver diseases in humans were shown to alter the function of the peripheral and central circadian clocks. Liver cirrhosis was associated with sleep disturbances, delayed phase of diurnal skin temperature cycle (46, 47), and shifted plasma melatonin and cortisol profiles, two hormonal phase markers of the human SCN clock (48). Nevertheless, assuming that this phenomenon is somehow related to what we observed in LHM, the phase of this animal is advanced, and they present a shorter period, in opposite to the phase delay observed in humans with cirrhosis. One hypothesis could be that, like for light signals (49), nocturnal (mouse) and diurnal (human) animals integrate also differently liver signals. Further research is required to test this hypothesis. In conclusion, our study provides evidence that the implantation of hepatocytes with an independent self-sustained circadian clock with different responses to systemic cues can advance behavioral and metabolic rhythms through action on hypothalamic centers.
While many studies in rodents have underscored the implication of the hepatocyte circadian clock on animal physiology, we describe here in mice with hepatocyte humanization the ability of peripheral cells to feedback to the central clock to the extent of redefining the free-running period and the phase of behavior and metabolism. Our study illustrates the benefit of chimeric animals to decrypt the impact of peripheral tissues in the control of circadian physiology. By taking advantage of the species-specific difference in cellular physiology, LHM allow probing hierarchical organization of the circadian system, which would otherwise be inaccessible in a homogeneous rodent or human context. While our study does not provide a simple molecular mechanism by which human hepatocytes are capable of entraining tissues, our results force us to reconsider the importance of peripheral cues as Zeitgebers for behavior and physiology. The reciprocal relationship between central and peripheral clocks should be considered in future studies aimed at the dissection of circadian physiology and metabolism. We believe that our results beg to reconsider the importance of the hepatic signals as Zeitgebers. In that view, it is formally possible that the cardiometabolic diseases associated with disrupted circadian rhythms could primarily originate in peripheral tissues, which, in turn, will promote desynchrony in the other tissues.
MATERIALS AND METHODS
Animal ethics
All animal experiments were performed with the approval of the Animal Care Committee of the Université Paris Cité (CEB-25-2016; number: 004667.02) and the veterinary office of the Canton of Vaud, Switzerland (authorization VD 3170). Chimeric animals were generated at Yecuris Corporation (Tualatin, OR, USA) or Karolinska Institutet (Stockholm, Sweden). This study was approved by the animal care committee and complies with the Declaration of Helsinki, and ethical approvals (2010/678-31/3 and S82-13) were obtained from local authorities. All animal work was conducted according to approved Institutional Animal Care and Use Committee (Yecuris Corporation) protocol DN000024 and National Health Institutes (NIH) Office of Laboratory Animal Welfare (OLAW) assurance no. A4664-01. The protocols follow the NIH Guide for the Care and Use of Laboratory Animals.
Animals
Fah−/−, Rag2−/−, Il2rg−/− mice (FRG) were crossed with NOD mouse strain to create FRGN mice (Yecuris, Tualatin, OR, USA), whose livers can be fully repopulated with human or murine hepatocytes, as described (13, 16–18). Human liver tissue and hepatocytes were obtained through the Liver Tissue Cell Distribution System, and the studies were exempted by the Institutional Review Board 0411142 since no human subjects were involved (University of Pittsburgh). Human and murine hepatocytes were transplanted via splenic injections that allowed to repopulate the mouse liver by maintaining on 2-(2-nitro-4-fluoromethylbenzoyl) cyclohexane-1,3-dione [CuRx Nitisinone (NTBC), catalog no. 20-0026] in the drinking water, a medication that prevents hepatocyte death when FAH deficiency is present. Engraftment is sustained over the life of the animal with an appropriate regimen of NTBC and prophylactic treatment of sulfamethoxazole (SMX)/trimethoprin (TMP) antibiotics (20-0037). Because of the impaired immune system of these stains, they are susceptible to respiratory infections. Mice received prophylactic antibiotic treatment cycles to minimize respiratory infections. Treatments with a combination of SMX/TMP for 5 days twice a month greatly reduce the occurrence of common respiratory infections and subsequent secondary infections. The final concentration of SMX/TMP drinking water is 640 μg/ml, 128 μg/ml (TMP) (CuRx SMX/TMP antibiotic, catalog no. 20-0037, Yecuris corporation), and 3% dextrose (Sigma-Aldrich, ref. D9434). The homozygous deletion of Fah in FRGN mice produces a fumaryloacetoacetate dehydrogenase deficiency that disrupts tyrosine metabolism, leading to the buildup of hepatotoxic metabolite, fumarylacetoacetate. With the administration of NTBC the intracellular accumulation of furmarylacetoacetate is blocked, and the mice live normal immune-deficient mouse lifespans. Mice are cycling on NTBC (8 mg/liter) treatment with SMX/TMP antibiotic and 3% of dextrose for 3 days every 5 weeks. To maintain hydration of the mice during the extended period off NTBC, it is best to use sterile drinking water that contains 3% dextrose. Because dextrose provides an opportunistic environment for bacterial growth, new containers of sterile dextrose water need to be prepared every week, as described previously (13). Chimeric animals were generated at Yecuris Corporation (Tualatin, OR, USA) from ten-week-old male mice FRGN repopulated with human hepatocytes (LHH, Hu-FRGN) or murine hepatocytes (LMM, Mu-FRGN) (catalog nos. 10-0013 and 10-0009, YECURIS corporation, Oregon, USA). Mice were maintained on PicoLab High Energy Mouse Diet 5LJ5 (LabDiet). Only mice with human hepatocyte repopulation of >70% (corresponding to circulating levels of hAlb > 3.5 mg/ml) were used in this study.
Detailed engraftment for the experimental group is as follows: Experimental group presented in Figs. 1 and 2 and figs. S1 (A, C, D, E, and H) and S2 is composed of 10 mice transplanted with human female donor HHF13022 plus 2 mice transplanted with human male donor HHM30017 (n = 12, LHM-F1) and 12 mice transplanted with NOD male murine donor (n = 12, LMM-F1). The experimental group presented in Fig. 3 and figs. S1F and S3 is composed of 5 mice transplanted with human male donor HHM30017 plus 10 mice transplanted with human female donor HHF13022 (n = 15, LHM-F1) and 12 mice transplanted with male NOD murine donor (n = 12, LMM-F1). The experimental group presented in Fig. 4 (A to H) and figs. S1B and S4 is composed of 6 mice transplanted with human female donor HHF17006 plus 2 mice transplanted with human male donor HHM30017 (n = 12, LHM-F1) and 12 mice transplanted with NOD male murine donor (n = 12, LMM-F1). The experimental group in Fig. 4 (I to K) is composed of 5 mice transplanted with human male donor HHM30017 plus 10 mice transplanted with human female donor HHF13022 (n = 15, LHM-F1) and 12 mice transplanted with male NOD murine donor (n = 12, LMM-F1). The experimental group presented in Figs. 5 and 6 and figs. S1G, S5, and S6 is composed of 12 mice transplanted with human female donor HHF13022 (n = 12, LHM-F1) and 12 mice transplanted with male NOD murine donor (n = 12, LMM-F1). All the mouse hepatocytes from NOD mice used to produce LMM were from male animals.
Time-resolved tissue collection
Except otherwise noticed, mice were kept under diurnal lighting conditions (12-hour light/12-hour dark) with unrestricted access to food and water. Mice were sacrificed at 4-hour intervals over 24 hours corresponding to Zeitgeber time [ZT, represents the time when the light was switched on (ZT0) or off (ZT12)] 2, ZT6, ZT10, ZT14, ZT18, and ZT22 by decapitation, and tissues were dissected, snap-frozen in liquid nitrogen, pulverized to reduce potential local effect caused by the engraftment procedure, and stored at −80°C until further processing.
Serum samples
Blood samples were collected in serum collection tubes and allowed to clot for 1 hour at 4°C on ice. Tubes were centrifuged at 10,000g for 5 min at 4°C, and serum was stored immediately at −80°C. During the data collection, serum samples were aliquoted and thawed no more than twice.
hAlb measurement
After 1000x or 10,000x dilution with tris-buffered saline, hAlb concentration was measured with the hAlb ELISA Quantitation Kit (Bethyl, E80-129), according to the manufacturer’s protocol, to monitor the progress of humanization of transplanted mice. Circulating hAlb (1000 μg/ml) correlates with ~20% engraftment of human hepatocytes, 2000 μg/ml with ~40%, and mice with hAlb (4000 μg/ml) showed approximately ~80% of human hepatocytes in the repopulated liver (18).
Plasma rodent GH and IGF-1 measure
The plasma rodent GH (ref. 22-GHOMS-E01) and rodent IGF-1 (ref. 22-IG1MS-E01) content of each sample were measured in duplicate with a colorimetric assay provided by ALPCO Corporation. The absorbance from each sample was measured in duplicate using a spectrophotometric microplate reader. The intra- and inter-assay coefficients of variation for these kits were around 0.3 to 8%. Plasma samples were tested in duplicate within one assay, and the results were expressed in terms of the standards supplied (nanograms per milliliter).
Quantification of plasma lipoprotein lipids
Lipoproteins were separated from 2.5 μl of individual plasma samples by size exclusion chromatography using a Superose 6 PC 3.2/300 column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Lipoproteins were eluted as a fraction appearing in the exclusion volume of the sepharose column that contained VLDL, then LDL, and lastly HDL. TG concentrations were calculated after integration of the individual chromatograms (50, 51), generated by the enzymatic-colorimetric reaction with the respective following kits, TG GPO-PAP (Roche Diagnostics, Mannheim, Germany).
Indirect calorimetry metabolic analysis
The indirect calorimetry system carries out preclinical noninvasive and fully automated measurement of food and water intake, O2 consumption, CO2 production, respiratory quotient (as an indicator of glycolic or oxidative metabolic status), and whole energy expenditure, together with tridimensional (X, Y, and Z) spontaneous activity and fine movement (Phenomaster, TSE Systems GmbH), as described. Mice were monitored for whole energy expenditure or heat, oxygen consumption and carbon dioxide production, respiratory exchange rate (RER = VCO2/VO2, where V is volume), and locomotor activity using calorimetric cages with bedding, food, and water (Labmaster, TSE Systems GmbH, Bad Homburg, Germany). The ratio of gases was determined through an indirect open-circuit calorimeter. This system monitors O2 and CO2 concentration by volume at the inlet ports of a tide cage through which a known flow of air is being ventilated (0.4 liter/min) and compared regularly to a reference empty cage. For optimum analysis, the flow rate was adjusted according to the animal body weights to set the differential in the composition of the expired gases between 0.4 and 0.9% (Labmaster, TSE Systems GmbH, Bad Homburg, Germany). The flow was previously calibrated with an O2 and CO2 mixture of known concentrations (Air Liquide, S.A. France). Oxygen consumption, carbon dioxide production, and energy expenditure were recorded every 15 min for each animal during the entire experiment. Whole energy expenditure was calculated using the Weir equation respiratory gas exchange measurements. Food consumption was measured as the instrument combines a set of highly sensitive feeding and drinking sensors for automated online measurements. Unless noted otherwise, mice had free access to food and water ad libitum. To allow measurement of every ambulatory movement, each cage was embedded in a frame with an infrared light beam–based activity monitoring system with the online measurement at 100 Hz. The sensors for gases and detection of movement operated efficiently in both light and dark phases, allowing continuous recording. Mice were monitored for body weight and composition at the entry and exit of the experiment. Body mass composition (lean tissue mass, fat mass, free water, and total water content) was analyzed using an Echo Medical systems’ EchoMRI (Whole Body Composition Analyzers, EchoMRI, Houston, USA), according to the manufacturer’s instructions. Briefly, mice were weighed before they were put in a mouse holder and inserted into an MRI analyzer. Readings of body composition were given within 1 min. Data analysis was performed on Excel XP using the extracted raw value of VO2 consumed, VCO2 production (express in milliliters per hour), and energy expenditure (kilocalories per hour). Subsequently, each value was expressed by total body weight extracted from the EchoMRI analysis.
Determination of circadian period
Mice were housed individually in cages equipped with a running-wheel (Phenome Technologies). After a 2-week period of habituation in light-dark conditions, circadian activity was recorded in constant darkness for 2 weeks. Data were acquired and analyzed with the Clocklab software (Actimetrics). The circadian period was determined using the chi-square periodogram method.
Imposed light period feeding regimen
Mice were individually housed for at least 2 weeks, and body weight was monitored daily. Mice were then housed in cages allowing for a measure of metabolic efficiency, food intake, water intake, and locomotor activity (Phenomaster system described previously). After 10 days of ad libitum feeding and baseline recording, mice were imposed strict daylight feeding for 8 days (food available from ZT0 to ZT12) and returned to the normal regimen for 8 days.
RNA extraction and sequencing
Liver RNA was isolated using the RNeasy kit from QIAGEN according to the manufacturer’s recommendations. The integrity and the concentration of each RNA sample were assessed using an Agilent 2100 bioanalyzer. cDNA was prepared following the NEBNext Ultra II Directional RNA Library Prep Kit protocol (New England Biolabs). The libraries were sequenced using paired-end 150–base pair (bp) sequencing on a HiSeq 4000 System (Illumina). Muscle, ARC, and SCN RNA were isolated using the RNAdvance Tissue kit from Beckman Coulter according to the manufacturer’s recommendations. The integrity and the concentration of each RNA sample were assessed using Agilent Fragment Analyzer-96 and with Quant-iT Ribogreen from Life Technologies. Libraries were prepared following the TruSeq Stranded mRNA Kit protocol from Illumina and sequenced using paired-end 126-bp sequencing on a HiSeq 2500 System (Illumina).
Analysis of rhythmicity
STAR 2.4.0i was used to map RNA-seq reads onto a chimeric genome consisting of the Mus musculus (GRCm38/mm10) and Homo sapiens (GRCh38/hg19) reference sequences and to quantify the number of uniquely mapped reads per gene and organism. For the liver data, human and mouse genes with, on average, more than 10 read counts in LMM or LHM were retained. Human and mouse genes were matched using the mouse-human orthologs database from Ensembl. To assess differential rhythmicity and mean differences of gene expression in RNA-seq raw count data, we used the dryR method based on a model selection framework with generalized linear models (5). Genes with a BICW (likelihood to belong to the chosen rhythmic model) larger than 0.3 (liver) or 0.5 (muscle, SCN, and ARC) and a log2 amplitude > 0.5 in at least one condition were used for the functional and gene set enrichment analysis.
Functional and gene set enrichment analysis
GO analysis
Gene set enrichment analysis was performed using enrichR (52) with Gene Ontology (GO) Biological Process (2021) or GO Molecular Function (2021) databases. Terms with adjusted P value < 0.1, combined score > 50, and odds ratio > 6 were selected.
mTOR targets
A supplementary table with differential expression analysis was downloaded from (19). Genes were considered as differentially expressed between WT and, respectively, Raptor KO and Tsc1 KO mice at ZT12 if absolute log2 fold change (FC) was larger than 1 and P value smaller than 0.05. Statistical overrepresentation of those gene sets in genes up-regulated (log2FC > 1, dryR chosen model mean 2) or down-regulated (log2FC < 1 and dryR chosen model mean 2) in LHM versus LMM liver was computed using a hypergeometric test.
System-driven genes
A supplementary table for rhythmicity analysis was downloaded from (7). For each category (WT, Bmal1 KO, and Bmal1 Liver-RE), genes were defined as rhythmic if the P value was smaller than 0.05. Statistical overrepresentation of those gene sets in the different rhythmic models of LHM and LMM (BICW > 0.5 and log2 amplitude > 0.5) was assessed using a hypergeometric test.
CLOCK-target genes
The CLOCK-target gene set was retrieved from Alhopuro et al. (31). We used hypergeometric testing to calculate the overrepresentation of the gene set within the statistical models identified by dryR. A threshold of BICW > 0.5 was applied to the data and for the liver dataset an additional threshold on amplitude log2FC > 0.5.
Sex-biased gene expression and activation of the STAT5 pathway
To predict the activities of transcription factors and sex-biased genes, we used ISMARA (53) on the count data after the variance stabilizing transformation. Target genes for sex-biased STAT5 targets and sex-biased genes in mouse liver were taken from Zhang et al. (22) and Weger et al. (23), respectively.
Detection of phospho-S6 and S6 ribosomal protein in mouse extracts
Liver from LMM and LHM were resuspended into lysis buffer [phosphate-buffered saline, 150 mM NaCl (pH 7.5), 1% Triton X-100, 2 mM NaF, 2 mM Na₃VO₄, and protease inhibitors], sonicated (10 s, 10% power), and lysed for 30 min at 4°C under agitation. The lysate was centrifuged for 20 min at 15,000g (4°C), and the sample protein concentration was determined using Bradford’s method. Sixty micrograms of each sample was separated by SDS–polyacrylamide gel electrophoresis (4 to 12%, Invitrogen) followed by a transfer onto a nitrocellulose membrane (0.22 μm; GE Healthcare) for 1 hour at 4°C. A ponceau staining of the membrane was done to ensure equal protein loading. Membranes were then blocked in 5% bovine serum albumin (BSA) in Tris-buffered saline containing 0.1% Tween-20 (TBST) [tris 20 mM, 150 mM NaCl (pH 7.6), and 0.1% Tween-20] for 1 hour and incubated overnight with phospho-S6 ribosomal protein antibody (#2211, Cell Signaling Technology) in 1% BSA TBST at 4°C. The next day, membranes were washed three times with TBST before incubation with secondary antibody for 1 hour at room temperature. Membranes were then washed three more times, and the signal was detected by chemiluminescence using ECL Prime reagent (GE Healthcare) on an Amersham Imager 600 detection system (GE Healthcare, France). Membranes were later stripped using stripping buffer [tris 60 mM (pH 6.8), 2% SDS, and 100 mM β-mercaptoethanol] for 30 min at room temperature, blocked in 5% BSA TBST for 1 hour, and reprobed with S6 ribosomal protein antibody (#2317, Cell Signaling Technology) using the same protocol as above. Signal analyses were performed using ImageJ software taking as standard reference a fixed threshold of fluorescence.
Statistical analysis
All statistical comparisons were performed with Prism 9 (GraphPad Software, La Jolla, CA, USA). All the data were analyzed using either a Student’s t test (paired or unpaired) with equal variances, one-way analysis of variance (ANOVA) or two-way ANOVA. Cosinor parameter (acrophase, mean, period, and amplitude) results were analyzed by simple comparisons (unpaired t test). In all cases, the significance threshold was automatically set at P < 0.05. ANOVA analyses were followed by the Bonferroni post hoc test for specific comparisons only when overall ANOVA revealed a significant difference (at least P < 0.05).
Acknowledgments
We would like to thank all the consortium members of the EU-funded research project HUMAN (Health and the Understanding of Metabolism, Aging and Nutrition) who have directly and indirectly contributed to the discussion of the results. We thank O. Kacanski for administrative support; I. Le Parco, L. Maingault, A. Dauvin, A. Djemat, F. Michel, M. Boa, and D. Quintas for animals’ care; and S. Allithi for genotyping. We are grateful to U. Schibler for critical input in the manuscript. We acknowledge the technical platform Functional and Physiological Exploration platform (FPE) of the Université de Paris (BFA-UMR 8251), the animal core facility Buffon of the Université de Paris/Institut Jacques Monod, and A. Kaabi (Nestlé Research) for animal experiments.
Funding: This work has been financed by the HUMAN project (grant agreement no. 602757) within the EU’s Seventh Framework Program (FP7) for research, technological development, and demonstration. F.G. receives support from the Institute for Molecular Bioscience, The University of Queensland. M.Q. was supported by a postdoctoral contract from the Galician Government type A (Xunta de Galicia, ED481B2014/039-0) and type B (Xunta de Galicia, ED481B2018/004). M.Q. is currently funded by a research contract “Miguel Servet” (CP21/00108) from the ISCIII and cofunded by the European Union. We acknowledge funding supports from the Centre National la Recherche Scientifique (CNRS) and the Université Paris Cité.
Author contributions: Conceptualization: A.-S.D., M.Q., P.P., F.G., and S.L.; investigation: A.-S.D., M.Q., C.G., J.C., R.G.P.D., J.B., B.D.W., A.C., S.M., J.P., M.K., L.-L.V., M.E.M., M.P., and F.G.; formal data analysis: C.G., B.D.W., and E.C.; resources: J.B. and E.M.W.; funding acquisition: P.P., F.G., and S.L.; supervision: A.-S.D., P.P., F.G., and S.L.; writing—original draft: F.G. and S.L.; writing—review and editing: all authors. Competing interests: J.B. was formerly President and Chief Executive Officer of Yecuris Corporation. E.M.W. was formerly Senior Scientist at Yecuris Corporation. C.G., B.D.W., A.P., S.M., J.P., M.K., and F.G. are or were employees of Société des Produits Nestlé SA. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq raw files have been deposited in NCBI’s Gene Expression Omnibus (GSE212079; https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212079).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S5
Other Supplementary Material for this manuscript includes the following:
Data S1 to S5
REFERENCES AND NOTES
- 1.M. R. Ralph, R. G. Foster, F. C. Davis, M. Menaker, Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 (1990). [DOI] [PubMed] [Google Scholar]
- 2.S. H. Yoo, S. Yamazaki, P. L. Lowrey, K. Shimomura, C. H. Ko, E. D. Buhr, S. M. Siepka, H. K. Hong, W. J. Oh, O. J. Yoo, M. Menaker, J. S. Takahashi, PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. Sinturel, P. Gos, V. Petrenko, C. Hagedorn, F. Kreppel, K. F. Storch, D. Knutti, A. Liani, C. Weitz, Y. Emmenegger, P. Franken, L. Bonacina, C. Dibner, U. Schibler, Circadian hepatocyte clocks keep synchrony in the absence of a master pacemaker in the suprachiasmatic nucleus or other extrahepatic clocks. Genes Dev. 35, 329–334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.B. Kornmann, O. Schaad, H. Reinke, C. Saini, U. Schibler, Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb. Symp. Quant. Biol. 72, 319–330 (2007). [DOI] [PubMed] [Google Scholar]
- 5.B. D. Weger, C. Gobet, F. P. A. David, F. Atger, E. Martin, N. E. Phillips, A. Charpagne, M. Weger, F. Naef, F. Gachon, Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. U.S.A. 118, e2015803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. A. Lamia, K. F. Storch, C. J. Weitz, Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.K. B. Koronowski, K. Kinouchi, P. S. Welz, J. G. Smith, V. M. Zinna, J. Shi, M. Samad, S. Chen, C. N. Magnan, J. M. Kinchen, W. Li, P. Baldi, S. A. Benitah, P. Sassone-Corsi, Defining the independence of the liver circadian clock. Cell 177, 1448–1462.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Brancaccio, M. D. Edwards, A. P. Patton, N. J. Smyllie, J. E. Chesham, E. S. Maywood, M. H. Hastings, Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O. Barca-Mayo, M. Pons-Espinal, P. Follert, A. Armirotti, L. Berdondini, D. de Pietri Tonelli, Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 8, 14336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D. Guan, Y. Xiong, T. M. Trinh, Y. Xiao, W. Hu, C. Jiang, P. Dierickx, C. Jang, J. D. Rabinowitz, M. A. Lazar, The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science 369, 1388–1394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G. Manella, E. Sabath, R. Aviram, V. Dandavate, S. Ezagouri, M. Golik, Y. Adamovich, G. Asher, The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat. Metab. 3, 829–842 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. B. Koronowski, P. Sassone-Corsi, Communicating clocks shape circadian homeostasis. Science 371, eabd0951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.H. Azuma, N. Paulk, A. Ranade, C. Dorrell, M. al-Dhalimy, E. Ellis, S. Strom, M. A. Kay, M. Finegold, M. Grompe, Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 25, 903–910 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. Jiang, P. Li, X. Ruan, Y. Ma, K. Kawai, H. Suemizu, H. Cao, Comparative transcriptomics analyses in livers of mice, humans, and humanized mice define human-specific gene networks. Cell 9, 2566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.L. S. Mure, H. D. le, G. Benegiamo, M. W. Chang, L. Rios, N. Jillani, M. Ngotho, T. Kariuki, O. Dkhissi-Benyahya, H. M. Cooper, S. Panda, Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, eaao0318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. M. Wilson, J. Bial, B. Tarlow, G. Bial, B. Jensen, D. L. Greiner, M. A. Brehm, M. Grompe, Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res. 13, 404–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. E. Minniti, M. Pedrelli, L. L. Vedin, A. S. Delbès, R. G. P. Denis, K. Öörni, C. Sala, C. Pirazzini, D. Thiagarajan, H. J. Nurmi, M. Grompe, K. Mills, P. Garagnani, E. C. S. Ellis, S. C. Strom, S. H. Luquet, E. M. Wilson, J. Bial, K. R. Steffensen, P. Parini, Insights from liver-humanized mice on cholesterol lipoprotein metabolism and LXR-agonist pharmacodynamics in humans. Hepatology 72, 656–670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.E. C. Ellis, S. Nauglers, P. Parini, L.-M. Mörk, C. Jorns, H. Zemack, A. L. Sandblom, I. Björkhem, B.-G. Ericzon, E. M. Wilson, S. C. Strom, M. Grompe, Mice with chimeric livers are an improved model for human lipoprotein metabolism. PLOS ONE 8, e78550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.X. Bai, Y. Liao, F. Sun, X. Xiao, S. Fu, Diurnal regulation of oxidative phosphorylation restricts hepatocyte proliferation and inflammation. Cell Rep. 36, 109659 (2021). [DOI] [PubMed] [Google Scholar]
- 20.C. Jouffe, G. Cretenet, L. Symul, E. Martin, F. Atger, F. Naef, F. Gachon, The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. M. Lichanska, M. J. Waters, How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet. 24, 41–47 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Y. Zhang, E. V. Laz, D. J. Waxman, Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 32, 880–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. D. Weger, C. Gobet, J. Yeung, E. Martin, S. Jimenez, B. Betrisey, F. Foata, B. Berger, A. Balvay, A. Foussier, A. Charpagne, B. Boizet-Bonhoure, C. J. Chou, F. Naef, F. Gachon, The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 29, 362–382.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.P. Crosby, R. Hamnett, M. Putker, N. P. Hoyle, M. Reed, C. J. Karam, E. S. Maywood, A. Stangherlin, J. E. Chesham, E. A. Hayter, L. Rosenbrier-Ribeiro, P. Newham, H. Clevers, D. A. Bechtold, J. S. O’Neill, Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177, 896–909.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.F. Damiola, N. Le Minh, N. Preitner, B. Kornmann, F. Fleury-Olela, U. Schibler, Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Xu, Q. S. Padiath, R. E. Shapiro, C. R. Jones, S. C. Wu, N. Saigoh, K. Saigoh, L. J. Ptá̌ek, Y. H. Fu, Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Y. Xu, K. L. Toh, C. R. Jones, J. Y. Shin, Y. H. Fu, L. J. Ptá̌ek, Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 128, 59–70 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. A. Brown, D. Kunz, A. Dumas, P. O. Westermark, K. Vanselow, A. Tilmann-Wahnschaffe, H. Herzel, A. Kramer, Molecular insights into human daily behavior. Proc. Natl. Acad. Sci. U.S.A. 105, 1602–1607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. P. Debruyne, E. Noton, C. M. Lambert, E. S. Maywood, D. R. Weaver, S. M. Reppert, A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477 (2006). [DOI] [PubMed] [Google Scholar]
- 30.K. L. Eckel-Mahan, V. R. Patel, R. P. Mohney, K. S. Vignola, P. Baldi, P. Sassone-Corsi, Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 5541–5546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. Alhopuro, M. Björklund, H. Sammalkorpi, M. Turunen, S. Tuupanen, M. Biström, I. Niittymäki, H. J. Lehtonen, T. Kivioja, V. Launonen, J. Saharinen, K. Nousiainen, S. Hautaniemi, K. Nuorva, J. P. Mecklin, H. Järvinen, T. Orntoft, D. Arango, R. Lehtonen, A. Karhu, J. Taipale, L. A. Aaltonen, Mutations in the circadian gene CLOCK in colorectal cancer. Mol. Cancer Res. 8, 952–960 (2010). [DOI] [PubMed] [Google Scholar]
- 32.P. Xu, S. Berto, A. Kulkarni, B. Jeong, C. Joseph, K. H. Cox, M. E. Greenberg, T. K. Kim, G. Konopka, J. S. Takahashi, NPAS4 regulates the transcriptional response of the suprachiasmatic nucleus to light and circadian behavior. Neuron 109, 3268–3282.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.F. K. Stephan, I. Zucker, Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. Saini, A. Liani, T. Curie, P. Gos, F. Kreppel, Y. Emmenegger, L. Bonacina, J. P. Wolf, Y. A. Poget, P. Franken, U. Schibler, Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev. 27, 1526–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Izumo, M. Pejchal, A. C. Schook, R. P. Lange, J. A. Walisser, T. R. Sato, X. Wang, C. A. Bradfield, J. S. Takahashi, Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife 3, e04617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. Landgraf, A. H. Tsang, A. Leliavski, C. E. Koch, J. L. Barclay, D. J. Drucker, H. Oster, Oxyntomodulin regulates resetting of the liver circadian clock by food. eLife 4, e06253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. L. Bookout, M. H. M. de Groot, B. M. Owen, S. Lee, L. Gautron, H. L. Lawrence, X. Ding, J. K. Elmquist, J. S. Takahashi, D. J. Mangelsdorf, S. A. Kliewer, FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat. Med. 19, 1147–1152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C. Vetter, H. S. Dashti, J. M. Lane, S. G. Anderson, E. S. Schernhammer, M. K. Rutter, R. Saxena, F. A. J. L. Scheer, Night shift work, genetic risk, and type 2 diabetes in the UK Biobank. Diabetes Care 41, 762–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.P. Zimmet, K. G. M. M. Alberti, N. Stern, C. Bilu, A. el-Osta, H. Einat, N. Kronfeld-Schor, The circadian syndrome: Is the metabolic syndrome and much more! J. Intern. Med. 286, 181–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Kohsaka, A. D. Laposky, K. M. Ramsey, C. Estrada, C. Joshu, Y. Kobayashi, F. W. Turek, J. Bass, High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 41.K. A. Dyar, D. Lutter, A. Artati, N. J. Ceglia, Y. Liu, D. Armenta, M. Jastroch, S. Schneider, S. de Mateo, M. Cervantes, S. Abbondante, P. Tognini, R. Orozco-Solis, K. Kinouchi, C. Wang, R. Swerdloff, S. Nadeef, S. Masri, P. Magistretti, V. Orlando, E. Borrelli, N. H. Uhlenhaut, P. Baldi, J. Adamski, M. H. Tschöp, K. Eckel-Mahan, P. Sassone-Corsi, Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. S. Pendergast, K. L. Branecky, W. Yang, K. L. J. Ellacott, K. D. Niswender, S. Yamazaki, High-fat diet acutely affects circadian organisation and eating behavior. Eur. J. Neurosci. 37, 1350–1356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Mendoza, P. Pevet, E. Challet, High-fat feeding alters the clock synchronization to light. J. Physiol. 586, 5901–5910 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. T. Palmisano, J. M. Stafford, J. S. Pendergast, High-fat feeding does not disrupt daily rhythms in female mice because of protection by ovarian hormones. Front. Endocrinol. 8, 44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.T. N. Buckley, O. Omotola, L. A. Archer, C. R. Rostron, E. P. Kamineni, J. D. Llanora, J. M. Chalfant, F. Lei, E. Slade, J. S. Pendergast, High-fat feeding disrupts daily eating behavior rhythms in obesity-prone but not in obesity-resistant male inbred mouse strains. Am. J. Physiol. Regul. Integr. Comp. Physiol. 320, R619–R629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Garrido, D. Saccardo, M. de Rui, E. Vettore, A. Verardo, P. Carraro, N. di Vitofrancesco, A. R. Mani, P. Angeli, M. Bolognesi, S. Montagnese, Abnormalities in the 24-hour rhythm of skin temperature in cirrhosis: Sleep-wake and general clinical implications. Liver Int. 37, 1833–1842 (2017). [DOI] [PubMed] [Google Scholar]
- 47.J. Cordoba, J. Cabrera, L. Lataif, P. Penev, P. Zee, A. T. Blei, High prevalence of sleep disturbance in cirrhosis. Hepatology 27, 339–345 (1998). [DOI] [PubMed] [Google Scholar]
- 48.S. Montagnese, B. Middleton, A. R. Mani, D. J. Skene, M. Y. Morgan, On the origin and the consequences of circadian abnormalities in patients with cirrhosis. Am. J. Gastroenterol. 105, 1773–1781 (2010). [DOI] [PubMed] [Google Scholar]
- 49.E. Challet, Minireview: Entrainment of the suprachiasmatic clockwork in diurnal and nocturnal mammals. Endocrinology 148, 5648–5655 (2007). [DOI] [PubMed] [Google Scholar]
- 50.P. Parini, L. Johansson, A. Broijersen, B. Angelin, M. Rudling, Lipoprotein profiles in plasma and interstitial fluid analyzed with an automated gel-filtration system. Eur. J. Clin. Invest. 36, 98–104 (2006). [DOI] [PubMed] [Google Scholar]
- 51.M. Pedrelli, P. Davoodpour, C. Degirolamo, M. Gomaraschi, M. Graham, A. Ossoli, L. Larsson, L. Calabresi, J. Å. Gustafsson, K. R. Steffensen, M. Eriksson, P. Parini, Hepatic ACAT2 knock down increases ABCA1 and modifies HDL metabolism in mice. PLOS ONE 9, e93552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. V. Kuleshov, M. R. Jones, A. D. Rouillard, N. F. Fernandez, Q. Duan, Z. Wang, S. Koplev, S. L. Jenkins, K. M. Jagodnik, A. Lachmann, M. G. McDermott, C. D. Monteiro, G. W. Gundersen, A. Ma'ayan, Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.P. J. Balwierz, M. Pachkov, P. Arnold, A. J. Gruber, M. Zavolan, E. van Nimwegen, ISMARA: Automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 24, 869–884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S5
Data S1 to S5