Abstract
The dicarboxylate transport (Dct) system of Sinorhizobium meliloti, which is essential for a functional nitrogen-fixing symbiosis, has been thought to transport only dicarboxylic acids. We show here that the permease component of the Dct system, DctA, can transport orotate, a monocarboxylic acid, with an apparent Km of 1.7 mM and a Vmax of 163 nmol min−1 per mg of protein in induced cells. DctA was not induced by the presence of orotate. The transport of orotate was inhibited by several compounds, including succinamic acid and succinamide, which are not dicarboxylic acids. The dicarboxylic acid maleate (cis-butenedioic acid) was not an inhibitor of orotate transport, which suggests that it was not recognized by DctA. However, maleate was an excellent inducer of DctA expression. Our evaluation of 17 compounds as inducers and inhibitors of transport suggests that substrates recognized by S. meliloti DctA must have appropriately spaced carbonyl groups and an extended conformation, while good inducers are more likely to have a curved conformation.
Soil bacteria belonging to the genera Sinorhizobium, Rhizobium, Bradyrhizobium, and Azorhizobium can form symbiotic associations with leguminous plants. The bacteria elicit the formation of specialized root organs called nodules, in which they reduce atmospheric dinitrogen, and provide the resulting ammonia to the plant. Symbiotic nitrogen fixation requires a large energy input. To provide this energy, the host plant supplies organic compounds such as sucrose, which are transported to the nodules and converted to substrates supplied to the bacteroids (17). The tricarboxylic acid (TCA) cycle intermediates succinate, malate, and fumarate are likely to be the major carbon sources for rhizobial bacteroids in the nodule (20). It is thought that these compounds are imported into the bacteroids using the rhizobial dicarboxylate transport (Dct) system, which in Sinorhizobium meliloti is encoded by three genes located on megaplasmid II, dctA, dctB, and dctD (20). The dctA gene codes for a high-affinity permease. dctA mutants produce nodules that are symbiotically ineffective, and bacteroids from these nodules are unable to transport dicarboxylates (4, 20). The dctB and dctD genes encode a two-component regulatory system, which activates the transcription of dctA in response to the presence of dicarboxylates in the periplasm, where the sensor domain of DctB is located (12, 18). S. meliloti DctA participates in the regulation of its induction—dctA::phoA fusions are induced to a very high level unless there is an active DctA protein in the cell (22).
Succinate, malate, fumarate, and aspartate are considered to be substrates for the Dct system (19). Other compounds, including d-lactate, 2-methylsuccinate, 2,2- or 2,3-dimethylsuccinate, acetoacetate, β-hydroxybutyrate, mercaptosuccinate, α-ketoglutarate, and itaconate, are either substrates or potential substrates for DctA (4, 10, 11). Recently, a study examining fluoroorotic acid (FOA) resistance in Salmonella enterica serovar Typhimurium and Escherichia coli showed that one class of resistant mutants can be complemented by an E. coli gene that encodes a protein with 94% sequence identity with S. meliloti DctA (2). An implication of this work is that enteric DctA is able to transport orotate, a cyclic monocarboxylate, although this was not shown directly.
Given the importance of the Dct system to establishing a nitrogen-fixing association between rhizobia and their host plants, detailed knowledge of the regulation and function of DctA is essential. The main aims of this work were to test orotate as a potential substrate of the S. meliloti Dct system and to define its relationship to the transport of dicarboxylates and other potential DctA substrates, including compounds similar to orotate. Orotate was transported by the S. meliloti Dct system with an affinity greater than that toward aspartate and with a very high capacity. Orotate uptake was inhibited by various compounds not previously shown to be DctA substrates. Several substrates recognized by DctA were not inducers of DctA, and not all inducers of DctA-dependent transport were recognized as competitive inhibitors of DctA-mediated transport.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains KUR1349 and KUR1351 were supplied by R. Kelln, and S. meliloti strains Rm8002, Rm8384, and RmF726 were supplied by T. Finan. S. meliloti strains were grown at 30°C either on minimal mannitol medium containing NH4 (MM NH4) (16) or on M9 medium (21) modified by replacing Na2HPO4 with 8.7 g of K2HPO4 per liter. The M9 medium was supplemented with either 20 mM mannitol or 0.2% aspartate and 5 ml of 1% yeast extract (Difco) per liter. E. coli strains were grown at 37°C in either Luria-Bertani (LB) or M9 minimal salts (13) medium with 0.2% glycerol, malate, fumarate, succinate, or aspartate as the carbon source, supplemented with carbamoyl aspartate (100 μg/ml) and thiamine (2 μg/ml). Antibiotics for S. meliloti were added at 200 μg/ml (streptomycin), 40 μg/ml (kanamycin), or 10 μg/ml (tetracycline). For E. coli, tetracycline was used at 25 μg/ml. FOA was added at 1, 2, 4, or 50 μg/ml for S. meliloti and 50, 100, or 200 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| S. meliloti | ||
| Rm8002 | Rm1021 phoA | 9 |
| Rm8384 | Rm8002 fix-384::TnphoA (dctA::phoA) | 9 |
| RmF726 | Rm1021 ΔΩ5079-5149::Tn5-233 | 22 |
| E. coli | ||
| KUR1349 | pyrB usp-4 | 2 |
| KUR1351 | KUR1349 out-2 | 2 |
| S17-1 | pro hsdR recA [RP4-2(Tc::Mu) (Km::Tn7)] | 14 |
| JM109 | lac-proAB | 21 |
| Plasmids | ||
| pTH32 | 2.2-kb EcoRI dctA subclone from pTH24 into pRK7813-1 | 22 |
| pTH2C6 | pTH24 dctA::TnphoA dctB+ dctD+ | 22 |
| pCPP33 | IncP LacZ; Tcr | 7 |
| pSM100 | 1.5-kb BamHI dctA subclone from pTH32 into pCPP33 | This work |
Subcloning of S. meliloti dctA.
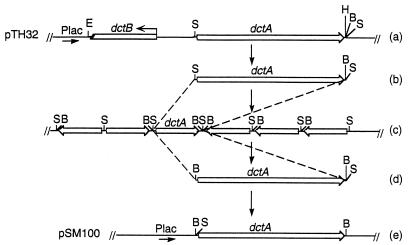
Plasmid pTH32, containing S. meliloti dctA (22), is able to complement Rm8384, a strain carrying TnphoA inserted into chromosomal dctA. pTH32 is unable to complement RmF726, a strain with a deletion of the entire dct operon. The complementation only in Rm8384 can be explained by the presence of the DctB and DctD regulatory proteins in Rm8384 but not in RmF726. However, since dctA was oriented correctly downstream of the E. coli lacZ promoter present in pTH32, the lack of dctA expression in RmF726(pTH32) also suggests that the 780 bp of S. meliloti DNA in pTH32 separating the lacZ promoter and dctA contains a transcriptional terminator. To allow us to assess the substrate specificity of DctA without requiring an inducer to be present, we constructed pSM100, a plasmid in which a 1.5-kb DNA fragment that contained only dctA was placed adjacent to a lacZ promoter, as shown in Fig. 1. pSM100 was transformed into E. coli S17-1 and then mated by conjugation into S. meliloti strains Rm8002, Rm8384, and RmF726. RmF726(pSM100) grew at a normal rate using succinate as a sole carbon source, suggesting that the lacZ promoter in pSM100 was able to activate expression of DctA to a physiologically significant level.
FIG. 1.
Construction of pSM100. Plasmid pTH32 was digested with SmaI, and the DNA fragments were separated on a 0.8% agarose gel. The 1.5-kb fragment was isolated (b) and ligated to itself to generate concatemers (c), which were then digested with BamHI to yield fragments that included a 1.5-kb fragment with BamHI ends (d). This BamHI fragment was ligated into the BamHI site of the broad-host-range vector pCPP33 (7), which contains lacZα (e). The ligation mixture was transformed into JM109, and possible recombinant clones were selected by screening on LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), isopropyl-β-d-thiogalactopyranoside (IPTG), and tetracycline. One clone containing the 1.5-kb fragment in the correct orientation was named pSM100.
Transport assays with whole cells of S. meliloti.
To induce transport, cells were cultured either in M9 medium containing 0.2% aspartate or in M9 medium with mannitol and then incubated for 4 h in M9 medium containing mannitol and an inducer. To assess transport, the cells were chilled, washed twice in M9 salts, and resuspended in M9 medium containing mannitol. An aliquot of cells was added to a vial with shaking at 30°C and warmed for 2 min. Labeled substrates were added and 100-μl aliquots were taken at intervals. The uptake was measured using 50 μM [14C]succinate (Moravek Biochemicals, Brea, Calif.) containing 1 μCi ml−1 or 450 μM [3H]orotate (Moravek Biochemicals) containing 1 μCi ml−1. Samples were filtered through 0.45-μm-pore-size nitrocellulose membrane filters (NitroBind), washed twice with 3 ml of M9 salts, and dried. Their radioactivity was measured in Scintisafe Econol scintillation fluid (Fisher Scientific, Pittsburgh, Pa.) using a Packard Tri-Carb 2100 TR scintillation counter. A 10-μl aliquot was spotted on a dry filter and counted to measure total radioactivity in the assay culture. Experiments were repeated at least twice.
Transport assays with whole cells of E. coli.
The cells were cultured in LB. To assay transport, the cells were chilled, washed three times in 50 mM potassium phosphate (pH 6.0) plus 5 mM MgSO4, and resuspended in the same buffer. An aliquot of cells was added to a vial with shaking at 37°C and warmed for 2 min. Labeled substrates were added and 100-μl aliquots were taken at intervals. The uptake was measured using 25 mM orotate containing 1 μCi ml−1. Samples were filtered through 0.45-μm-pore-size nitrocellulose membrane filters (NitroBind), washed twice with 3 ml of 50 mM potassium phosphate–5 mM MgSO4 (pH 6.0), and dried. Their radioactivity was measured as described above. A 10-μl aliquot spotted on a dry filter and counted to measure total radioactivity in the assay culture was used to normalize measurements. Measurements were generally made several times; all were repeated at least twice.
Alkaline phosphatase assay.
The alkaline phosphatase assay was done as described by Yarosh et al. (22). Five-milliliter cultures grown in M9 medium containing 1.5 mM glucose for 24 to 48 h were centrifuged and resuspended in 1 ml of M9 salts. Then 0.1 ml of culture was used to inoculate 5 ml of M9-glucose medium or M9 medium containing 1.5 mM concentrations of various potential inducers and incubated for 4 h. For all strains carrying plasmids, the medium was supplemented with 5 μg of tetracycline ml−1. The cells were then centrifuged, washed, and resuspended in 1 M Tris HCl to an absorbance of 0.1 to 0.3 (600 nm). After equilibration at 30°C, 0.1 ml of p-nitrophenyl-phosphate (4 mg/ml) was added to 0.9 ml of the cell suspension and the tubes were incubated for 30 min, after which 0.1 ml of 1 M KH2PO4 was added and the absorbances at 420 and 600 nm were measured. Units of alkaline phosphatase activity were calculated using the formula 1,000 × [A420 − (1.5 × A600)]/[time in minutes × A600). Assuming a molar extinction coefficient of 16,000 for p-nitrophenol, 1 U is equal to 0.062 nmol of p-nitrophenol-phosphate hydrolyzed per min at a cell optical density at 600 nm of 1. Experiments were repeated at least three times.
Protein assay.
The total protein concentration was measured with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), using bovine serum albumin as the standard.
RESULTS
Transport of FOA by the Dct system of S. meliloti.
FOA is an analog of orotic acid, an intermediate in pyrimidine metabolism. Once it has entered the cell, FOA can be converted to fluorouridine monophosphate, a powerful inhibitor of thymidylate synthetase, which is needed for thymidine synthesis and ultimately for DNA replication. To test the ability of DctA to transport orotate, we compared the sensitivities of S. meliloti wild-type strain Rm8002 and dctA mutant strains to FOA. Growth of Rm8002 on MM NH4 was inhibited very strongly by FOA at 2 to 4 mg/liter. A mutant that lacked the entire dct region (RmF726) and a mutant bearing a Tn5 insertion in dctA (Rm8384) were both resistant to FOA at concentrations under 50 to 100 mg/liter, which is consistent with the idea that DctA has a role in importing FOA. When pSM100, a plasmid containing a dctA gene expressed under the control of the E. coli lac promoter, was introduced into RmF726, the resulting strain was sensitive to FOA at 0.1 mg/liter. When succinate, malate, or fumarate replaced mannitol as the carbon source in MM NH4, sensitivity to FOA in strains Rm8002 and RmF726(pSM100) decreased significantly, and 50 mg of FOA per liter was required to block the growth of these strains completely. This indicated that these dicarboxylates can compete effectively with FOA for the available DctA and block FOA transport.
S. meliloti DctA can mediate transport of dicarboxylates and FOA into E. coli.
Baker et al. (2) showed that E. coli was sensitive to FOA and that a mutation in dctA led to FOA resistance, but they did not actually determine the mutation's effect on orotate uptake. We measured the uptake of radioactive orotate and found that, consistent with the inhibition results, strain KUR1349 has a fourfold-higher level of [3H]orotate transport than its dctA mutant, KUR1351 [133 versus 28 pmol (min · mg of protein)−1]. KUR1351(pSM100) imported 83 pmol (min · mg of protein)−1, showing that S. meliloti dctA was able to function in E. coli KUR1349(pCPP33), KUR1351(pCPP33), and KUR1351(pSM100) were plated on minimal glycerol medium with carbamoyl aspartate as the pyrimidine source in the presence of various concentrations of FOA and tetracycline. Growth of KUR1349(pCPP33) was stopped by FOA at 50 mg/liter, while its dctA derivative KUR1351(pCPP33) could grow on glycerol plates containing 200 mg of FOA/liter. KUR1351(pSM100) was unable to grow on this medium with FOA at 100 mg/liter. Sensitivity to FOA thus correlates with the level of FOA uptake.
Although KUR1351(pSM100) could import orotate, it could not use fumarate or malate as the sole carbon source. We interpret this to mean that DctA activity was too low to support the high level of transport needed to support growth on these DctA substrates, even though enough FOA was able to enter the cell to interfere with DNA replication. Slower growth on malate and fumarate is seen when a plasmid copy of E. coli dctA is used to rescue a chromosomal dctA mutant (3). When KUR1351(pSM100) was placed on malate, fumarate, or aspartate medium, isolated colonies were found. Plasmid DNA isolated from these colonies was transformed into KUR1351, producing strains that grew on either malate, fumarate, or aspartate. We are characterizing these apparent plasmid mutations.
Transport of succinate and orotate by S. meliloti.
A comparison of the uptake of radioactive succinate or radioactive orotate by different S. meliloti strains is shown in Table 2. Uninduced cells of Rm8002 showed a low rate of succinate and orotate uptake, but this level was higher than the very low rate of uptake of both substrates by the deletion strain RmF726. When Rm8002 was grown in the presence of aspartate to induce DctA and then shifted to media without inducer in order to measure the uptake of the transport substrate, the uptake of both succinate and orotate increased considerably. In contrast, growth in the presence of orotate did not induce either succinate or orotate transport. This implies that while DctA was able to recognize and transport orotate, the regulatory proteins DctB and DctD were not able to respond to the presence of orotate by inducing dctA gene expression.
TABLE 2.
Transport of succinate and orotate by S. meliloti strains
| Strain | Uptake
[nmol (min · mg of protein)−1]
|
|
|---|---|---|
| Succinate (0.05 mM) | Orotate (0.45 mM) | |
| Rm8002a | 4.7 | 4.1 |
| Rm8002b | 104.2 | 26.7 |
| Rm8002(pSM100) | 76.9 | 15.0 |
| RmF726 | 0 | 0 |
| RmF726(pSM100) | 29.3 | 18.3 |
| Rm8002c | 5.9 | 3.5 |
Cultures were grown in M9-mannitol unless otherwise noted. The cultures were washed and resuspended in M9 medium without a carbon source. Assays were performed using 0.05 mM [14C]succinate and 0.45 mM [3H]orotate.
The culture was grown in M9 medium containing 0.2% aspartate.
The culture was grown in M9-mannitol with 0.2% orotate.
The presence of plasmid pSM100, which contains dctA, allowed RmF726 to transport both succinate and orotate. This result was in agreement with the observation reported above that RmF726(pSM100) was two- to fourfold more sensitive to FOA than was the parental strain Rm8002. Uninduced cells of Rm8002(pSM100) have a higher rate of uptake for both succinate and orotate than uninduced cells of Rm8002.
Determination of apparent Km values for orotate uptake.
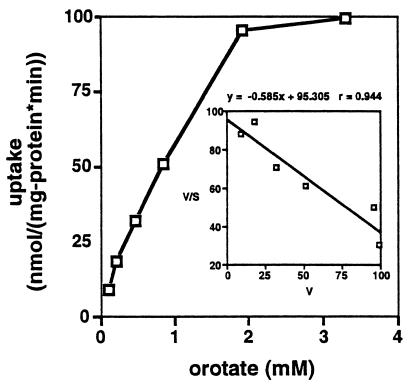
Orotate transport by Rm8002 was measured at orotate concentrations ranging from 25 μM to 3.3 mM to assess the affinity of DctA for orotate and the capacity of DctA to transport orotate (Fig. 2). Transport was not saturated at 3.3 mM orotate, but because of orotate's low solubility (1.7 g/liter, 10 mM), this was the highest concentration of orotate we could test. The apparent Km and Vmax for orotate transport were 1.7 mM and 163 nmol (min · mg of protein)−1, respectively. It has been reported that the apparent Km for succinate transport is 15 μM, while that for aspartate transport is 10 mM (19). Thus, the affinity of the Dct system for orotate was only about 1% of that for succinate and about six times that for aspartate.
FIG. 2.
Determination of apparent Km values for orotate uptake. S. meliloti Rm8002 cultures were grown in M9 medium with 0.2% aspartate as the carbon source. The cultures were washed and resuspended in M9 medium without a carbon source. Assays were performed using [3H]orotate. The uptake rate (V) at different concentrations of orotate (S) is plotted. The insert shows the Eadie-Hofstee plot of V/S versus V that was used to calculate an apparent Km value.
Testing the interaction of various compounds with S. meliloti DctABD.
To compare the relative affinities of DctA for succinate, malate, and orotate, the transport of succinate and orotate by induced cells of strain Rm8002 was measured in the presence of malate (Table 3). Succinate transport was halved in the presence of an equal concentration of malate but was depressed very little by a fourfold excess of orotate. In contrast, malate effectively inhibited orotate uptake, even when added at 1/10 the concentration of orotate. These results suggest that DctA had a significantly lower affinity for orotate than for malate or succinate.
TABLE 3.
Inhibition of succinate and orotate uptake by orotate, succinate, and malatea
| Inhibitor concn (mM) | Uptake [nmol (min · mg of
protein)−1]
|
||
|---|---|---|---|
| Orotate (0.45 mM) inhibited by malate | Succinate (0.05 mM) inhibited by:
|
||
| Orotate | Malate | ||
| 0.000 | 26.7 | 104.2 | 104.2 |
| 0.025 | 20.1 | ||
| 0.050 | 9.4 | 54.6 | |
| 0.100 | 4.8 | 95.5 | |
| 0.200 | 97.4 | ||
S. meliloti Rm8002 cultures were grown in M9 medium with 0.2% aspartate as a carbon source. The cultures were washed and resuspended in M9 medium without a carbon source. Assays were performed using 50 μM [14C]succinate and 450 μM [3H]orotate with the carbon substrate tested as inhibitor present at the concentrations shown.
The observation that orotate was a substrate for DctA led us to test various compounds that had not previously been considered as substrates because of their lack of resemblance to a dicarboxylic acid. Some of these compounds do not support good growth of wild-type S. meliloti, so we used their ability to inhibit orotate transport to assess whether they could be recognized by DctA. The observation that a compound inhibits transport does not guarantee that it can also be transported, but it has been found in studies of the glutamate transporter family, of which DctA is a member, that most competitive inhibitors are substrates (15). We also tested these compounds for their ability to induce succinate transport, since the sensory domain of DctB is periplasmic and some compounds unable to be transported might still be recognized as inducers by DctB. We tested the inhibition of orotate uptake and induction of succinate uptake by 17 compounds that might potentially interact with the Dct proteins. These compounds were selected by their structural similarity to C4-dicarboxylates or to orotate or by their potential importance as substrates for bacteroid nitrogen fixation. The results of the inhibition studies are summarized in Table 4 and those of the induction studies are summarized in Table 5. Benzoate, α-ketoglutarate, threonine, propionate, carnitine, cis-aconitate, γ-hydroxybutyric acid, and α-ketobutyrate neither inhibited orotate transport nor induced succinate transport.
TABLE 4.
Inhibition of orotate uptake by different compoundsa
| Inhibitor | Inhibition (%) at concn (mM) of
inhibitor
|
|||
|---|---|---|---|---|
| 0.1 | 0.5 | 1 | 10 | |
| Aspartate | 2 | 60 | ||
| Fumarate | 78 | |||
| l-Malate | 83 | 97 | ||
| d-Malate | 14 | 42 | 60 | 95 |
| Succinate | 80 | |||
| Succinamic acid | 40 | 27 | 54 | 90 |
| Succinamideb | 3 | 53 | ||
| Chlorosuccinate | 48 | 78 | 86 | 96 |
| Itaconate | 44 | 44 | 51 | 74 |
| Mesaconic acid | 20 | 56 | 70 | 74 |
| Carbamoyl aspartate | 46 | 76 | 79 | 94 |
S. meliloti Rm8002 cultures were grown in M9 medium with 0.2% aspartate as the carbon source. The cultures were washed and resuspended in M9 medium without a carbon source. Assays were performed using 25 μM [3H]orotate with the carbon substrate tested as the inhibitor.
0.4 and 2 mM succinamide.
TABLE 5.
Induction of succinate uptake and a dctA::TnphoA fusion
| Inducer | Induction
(%) of:
|
|
|---|---|---|
| Succinate uptakea | Alkaline phosphatase-specific activityb | |
| Mannitola Glucoseb | 100 | 100 |
| Aspartate | 1,102 | 850 |
| Fumarate | 568 | |
| Maleate | 1,368 | 354 |
| l-Malate | 1,038 | |
| d-Malate | 1,084 | 617 |
| Succinate | 799 | 531 |
| Succinamic acid | 685 | |
| Succinamide | 182 | |
| Chlorosuccinate | 742 | |
| Asparagine | 188 | |
| Itaconate | 203 | |
| Mesaconic acid | 50 | 148 |
| Carbamoyl aspartate | 173 | |
S. meliloti Rm8002 cultures were incubated in M9-mannitol overnight, distributed into separate tubes that contained a 20 mM concentration of the potential inducer, and incubated for 4 h. The cultures were then washed and resuspended in M9 medium without a carbon source. Assays were initiated by adding 50 μM [14C]succinate. Levels are reported relative to the activity of mannitol-grown cells [147 nmol (min · mg of protein)−1].
S. meliloti Rm8002(pTH2C6) cultures were grown and induced as described in Materials and Methods. Alkaline phosphatase activity was measured and compared to the levels in glucose-grown cultures (42.5 U).
The TCA cycle intermediates succinate, l-malate, and fumarate significantly inhibited orotate transport at relatively low concentrations. Several other compounds, including d-malate, chlorosuccinate, carbamoyl aspartate, succinamic acid, succinamide, itaconate, and mesaconic acid, also reduced the rate of orotate uptake by S. meliloti but only when higher levels of the competitor were added (Table 5). Succinamic acid and succinamide are not dicarboxylic acids; succinamic acid has one carboxyl group and succinamide has no carboxyl groups at all. The least efficient inhibitor of orotate transport within this group was aspartate, which inhibited orotate transport significantly only when tested at 10 mM. Watson et al. (19) found that aspartate was not able to inhibit succinate uptake even at very high concentrations, probably because the affinity of DctA for succinate was so much greater than its affinity for aspartate. Maleate (cis-butenedioic acid) did not compete with orotate at all.
Most of the compounds tested induced succinate uptake by DctA, at least to some extent (Table 5). The best inducer, maleate, did not compete with orotate for transport using DctA (Table 4). As reported earlier by Watson et al. (19), aspartate was also an excellent inducer of transport, as were both l- and d-malate. Succinate and fumarate were less effective. Succinamic acid was as effective as the latter TCA cycle intermediates, but succinamide led to only a twofold induction. Because cells were incubated for several hours in the presence of an inducer before assay, we cannot eliminate the possibility that the induction observed with the amidated C4 compounds was actually due to succinate generated by deamidation. However, the rate of this hypothetical deamidation was not high enough to allow Rm8002 to grow on either succinamic acid or succinamide as the carbon source. Asparagine was also a weak inducer, as might be expected if its structure is considered to be a variant of aspartate, an excellent inducer, or of succinamic acid. Itaconate and carbamoyl aspartate were also weak inducers.
The DctA transporter is the only major transporter of succinate in S. meliloti. However, to show that inducers like maleate and d-malic acid were actually inducing dctA expression, we measured the induction of alkaline phosphatase activity in Rm8002 carrying a dctA::TnphoA fusion on a plasmid. Aspartate, succinate, d-malate, maleate, and mesaconic acid were able to induce alkaline phosphatase activity (Table 5). The presence of maleate or d-malate did not restore the ability of dctA mutants to grow on succinate medium, suggesting that the induction of succinate uptake by maleate and d-malate was not the result of activating an alternate succinate carrier. Succinate transport in Rm726 remained at background levels even when cells had been previously exposed to maleate, d-malate, or aspartate.
The addition of mesaconic acid to cultures of Rm8002 growing on mannitol as the major carbon source halved the cells' ability to subsequently import succinate (Table 5). One possible explanation for this inhibition, that mesaconic acid inhibited cell growth during the induction period, was tested directly by comparing the number of CFU in cultures incubated for 4 to 6 h in M9-mannitol medium with and without 20 mM mesaconic acid. No difference was seen in either the viable titer or the protein content of the two cultures. Mesaconic acid was able to directly inhibit orotate uptake to some extent (Table 4). While it is possible that mesaconic acid interacts with DctA so that it cannot import orotate, we washed the cells to remove inducers (and potential competitors) prior to the transport assay and we did not expect the concentration of mesaconic acid to be high during the transport assay. If mesaconate inhibited transport by direct interaction with DctA, its action must persist. An alternative explanation is that mesaconate binds to DctB and interferes with the signaling that leads to the measurable background seen in uninduced Rm8002 cells (Table 2).
DISCUSSION
The rhizobial DctABD Dct system is essential for symbiotic nitrogen fixation in several different bacterium-plant interactions. This has been attributed to DctA's ability to transport dicarboxylic acids, which are thought to be a major carbon and energy source for nitrogen-fixing bacteria. Previous work showed that the dicarboxylic acids succinate, malate, fumarate, and aspartate are DctA substrates (5, 6, 19). DctA is the major, if not the only, dicarboxylate transporter in S. meliloti, in contrast to E. coli, which has several Dct systems. In E. coli, dctA has been linked to the transport of orotic acid (2). We show that S. meliloti DctA can transport orotic acid and its analog FOA, which suggests that orotate transport is not an idiosyncrasy of the E. coli protein but is conserved over a significant evolutionary distance. Orotate is a cyclic monocarboxylic acid that is also much larger than previously recognized substrates, but it is a good substrate for transport, with an apparent Km of 1.7 mM and a Vmax of 163 nmol (min · mg of protein)−1 in induced cells of S. meliloti.
Cells of Rm8002 grown in M9-orotate had the same rate of succinate transport as cells grown in M9-mannitol, which means that orotate was not recognized as an inducer of DctA. Transport and induction are mediated by separate components of the Dct system, DctA and DctBD, respectively, although recent work suggests that there is some change in transport substrate specificity when DctA is induced by DctBD (12). This may be reflected in our data (Table 2), in which the ratio of succinate transport to orotate transport in wild-type cells was near 4 when the induction was mediated by DctBD but was less than 2 for DctA expressed from the lacZ promoter in the absence of DctBD.
Our data show that DctA and DctBD differ in the substrates that they recognize. In contrast to orotate, which is a good substrate but not an inducer of DctA, maleate and asparagine were recognized as inducers of DctA but not as inhibitors of DctA transport. In this they resemble aspartate, which had been recognized as the strongest inducer among compounds previously tested but is not a very good substrate for transport, with a Km about 600-fold less than those of the TCA cycle dicarboxylic acids succinate, malate, and fumarate (19). Orotate can be considered a representative compound that appears to interact with DctA but not with DctB. In contrast, maleate (and, to some extent, aspartate and asparagine) appears to interact much better with DctB than with DctA. The difference between the recognition specificities of DctA and DctB is not consistent with the idea that DctB recognizes a dicarboxylate-bound form of DctA (12, 22).
These data also suggest that the assignment of DctA as a Dct system is not entirely accurate. Maleate is a dicarboxylic acid that was not recognized by DctA. On the other hand, the monocarboxylic acids orotate and succinamate were recognized by DctA, and succinamide, a C4 compound that has no carboxyl groups, was also able to inhibit orotate transport. We sought some pattern in the compounds studied that might be more predictive of substrate or inducer function. The strong inducing ability of maleate, a cis-butene dicarboxylic acid, and the comparatively weak activity of its trans isomer, fumarate, indicate that DctB prefers compounds with the C4 backbone bent into a C shape. Although they are expected to be less rigid than maleate, aspartate and malate, two excellent inducers of DctA, also have a C-type bend in their C4 backbones with both carboxyl groups on the same side of the molecule (1). In contrast, DctA had a low affinity for aspartate and did not recognize either maleate or asparagine, indicating that it preferred an extended C4 backbone. A similar distinction between “stretched” and “folded” versions of glutamate analogs has been noted for substrates of the well-studied glutamate transporters (15).
α-Ketobutyric acid and γ-hydroxybutyric acid were not good inhibitors of orotate transport, suggesting that one carboxyl group is not sufficient for a C4 compound to be recognized by DctA. Since succinamide was able to inhibit orotate transport by DctA, it appears that a minimum criterion for recognition by DctA is two C O groups separated by two carbon atoms. The compounds in Table 4, which all inhibit orotate transport, meet this standard. Substituting a carboxyl group for each amide to yield succinamate and succinamate leads to a successive increase in both the ability of the substrate to induce DctA and its affinity for DctA. Adding length to the structure also can affect the affinity. α-Ketoglutarate, which can be thought of as a C4 compound with properly positioned C O groups that contains an additional carboxyl group on the carbonyl carbon, was not recognized by DctA. Additions to a core succinate structure generally decrease the derivative's ability to be recognized by either DctA or DctB, but as noted above, the effect can differ for DctA and DctB. Substituting chlorine for hydrogen in succinate to yield chlorosuccinate does not change the ability of the compound to be recognized by DctA and DctB. On the other hand, substituting a methyl group at this position to yield methylsuccinate blocks recognition by Rhizobium leguminosarum DctB (12), while substituting an amino group to yield aspartate increases the affinity. Itaconate, which has a methylene group at this position, could still induce DctA and be recognized by it. It is reasonable to ask whether the difference in recognition specificity of DctA and DctB might have any functional significance and in particular why two substrates, maleate and aspartate, that are so poorly recognized by the transporter, are excellent inducers. One possibility that we are considering is that DctB might also regulate the transport of other molecules, like phthalic acid, which is imported into Burkholderia by other permeases. Phthalate, 1,2-benzene dicarboxylic acid, is relatively common in the soil.
Several new compounds should be added to the list of those that can be recognized by S. meliloti DctA, including orotate, succinamide, succinamate, and carbamoyl aspartate. These might play a role in symbiotic metabolism, although in general, their binding to DctA is not very strong. Kim and Chae (8) suggested that nitrogen exchange between the host plant and bacteria might involve the exchange of malonamate; a similar role might be proposed for succinamate.
ACKNOWLEDGMENTS
We thank Turlough Finan and Rod Kelln for strains and helpful discussions.
This work was supported by grant 98353056553 from the U.S. Department of Agriculture Competitive Research Grants Office. K.N.R. was supported by a Summer Undergraduate Fellowship from the Plant Biochemistry Research and Training Center through grant DEFG0694ER20160 from the U.S. Department of Energy.
REFERENCES
- 1.Ash J E, Boyett R E, Towm W G. Communities on the Web: Chem.Web.com—the World Wide Club for the chemical community. Trends Anal Chem. 1998;17:54–58. http://cwgen.chemweb.com/nci3d// . [Online.] http://cwgen.chemweb.com/nci3d//. . [Google Scholar]
- 2.Baker K E, Ditullio K P, Neuhard J, Kelln R A. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli requires the dicarboxylate transport protein encoded by dctA. J Bacteriol. 1996;178:7099–7105. doi: 10.1128/jb.178.24.7099-7105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies S J, Golby P, Omrani D, Broad S A, Harrington V L, Guest J R, Kelly D J, Andrews S C. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J Bacteriol. 1999;181:5624–5635. doi: 10.1128/jb.181.18.5624-5635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelke T, Jagadish M N, Puhler A. Biochemical and genetical analysis of Rhizobium meliloti mutants defective in C4-dicarboxylate transport. J Gen Microbiol. 1987;133:3019–3029. [Google Scholar]
- 5.Engelke T, Jording D, Kapp D, Pühler A. Identification and sequence analysis of the Rhizobium meliloti dctA gene encoding the C4-dicarboxylate carrier. J Bacteriol. 1989;171:5551–5560. doi: 10.1128/jb.171.10.5551-5560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finan T M, Wood J M, Jordan D C. Succinate transport in Rhizobium leguminosarum. J Bacteriol. 1981;148:193–202. doi: 10.1128/jb.148.1.193-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H-C, He S Y, Bauer D W, Collmer A. The Pseudomonas syringae pv. syringae 61 hrpHproduct, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y S, Chae H Z. A model of nitrogen flow by malonamate in Rhizobium japonicum-soybean symbiosis. Biochem Biophys Res Commun. 1990;169:692–699. doi: 10.1016/0006-291x(90)90386-2. [DOI] [PubMed] [Google Scholar]
- 9.Long S, McCune S, Walker G C. Symbiotic loci of Rhizobium meliloti identified by random TnphoAmutagenesis. J Bacteriol. 1988;170:4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAllister C F, Lepo J E. Succinate transport by free-living forms of Rhizobium japonicum. J Bacteriol. 1983;153:1155–1162. doi: 10.1128/jb.153.3.1155-1162.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKay I A, Dilworth M J, Glenn A R. C4-dicarboxylate metabolism in free-living and bacteroid forms of Rhizobium leguminosarumMNF3841. J Gen Microbiol. 1988;134:1433–1440. [Google Scholar]
- 12.Reid C J, Poole P S. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J Bacteriol. 1998;180:2660–2669. doi: 10.1128/jb.180.10.2660-2669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Simon R, Priefer U, Puhler A. A broad host-range mobilization system for in vivogenetic engineering; transposon mutagenesis of Gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 15.Slotboom D J, Konings W N, Lolkema J S. Structural features of the glutamate transporter family. Microbiol Mol Biol Rev. 1999;63:293–307. doi: 10.1128/mmbr.63.2.293-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somerville J E, Kahn M L. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J Bacteriol. 1983;156:168–176. doi: 10.1128/jb.156.1.168-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streeter J G. Transport and metabolism of carbon and nitrogen in legume nodules. Adv Bot Res. 1991;18:129–187. [Google Scholar]
- 18.Watson R J. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB, and dctD. Mol Plant-Microbe Interact. 1990;3:174–181. doi: 10.1094/mpmi-3-174. [DOI] [PubMed] [Google Scholar]
- 19.Watson R J, Rastogi V K, Chan Y-K. Aspartate transport in Rhizobium meliloti. J Gen Microbiol. 1993;139:1315–1323. [Google Scholar]
- 20.Watson R J, Chan Y-K, Wheatcroft R, Yang A-F, Han S. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988;170:927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 22.Yarosh O K, Charles T C, Finan T M. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]