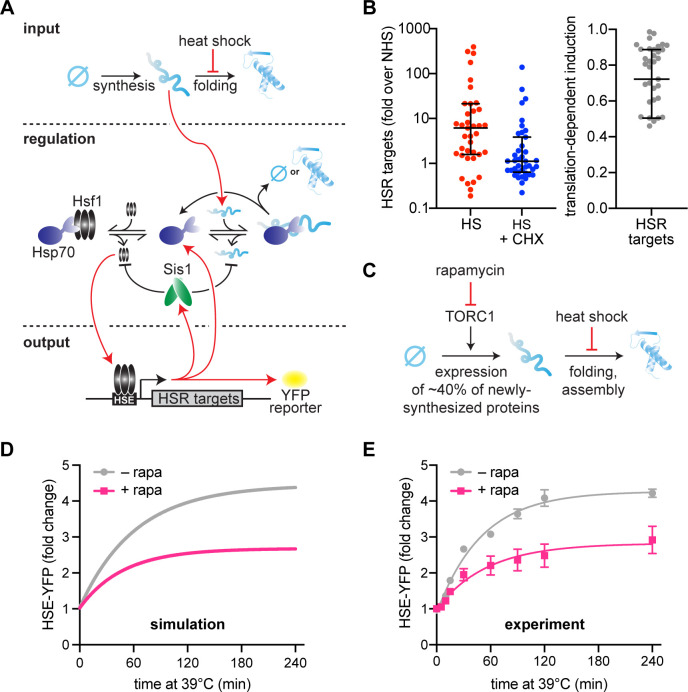
Figure 1. Newly synthesized proteins drive full activation of the heat shock response (HSR).
(A) Schematic of the mathematical model. Heat shock stalls folding of newly synthesized proteins so newly synthesized unfolded proteins accumulate. Hsp70 is recruited to unfolded proteins (red arrow) in turn releasing Hsf1 to induce its transcriptional targets. Among these targets are Hsp70 and Sis1 (and HSE-YFP, our ectopic reporter of Hsf1 activity). Of note, Sis1 has two known roles. It substantiates Hsf1–Hsp70 binding and substantiates Hsp70-misfolded protein interactions. (B) Hsf1 target gene transcript levels, relative to non-heat shock levels. Cells were treated with 200 µg/ml cycloheximide (CHX; blue) and subjected to 42°C heat shock for 20 min. Untreated control (red) plotted for comparison (red). Raw data from Triandafillou et al., 2020. One minus the transcript levels of CHX-treated over untreated cells is the portion of transcriptional induction which depends on ongoing translation (right graph). Each data point represents an Hsf1 target gene. (C) Model of rapamycin effect. Heat shock stalls newly synthesized protein folding, and newly synthesized unfolded proteins trigger the HSR. Upstream, rapamycin inhibits target of rapamycin complex 1 (TORC1) which inhibits ribosomal protein production, causing a 40% decrease in newly synthesized proteins. (D) HSE-YFP levels during heat shock. Cells are pretreated with 10 µg/ml rapamycin 5 min before heat shock (pink) and compared to untreated control (gray). Each time point represents the average of the three biological replicates. Error bar represents the standard deviation of three replicates. (E) Simulation of HSE-YFP levels during heat shock with or without rapamycin.
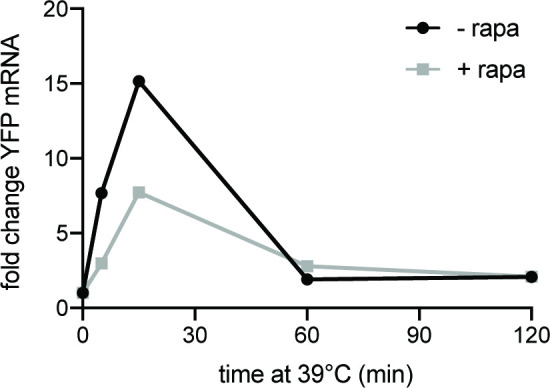
Figure 1—figure supplement 1. Rapamycin reduces HSE-YFP at the mRNA level RNA levels during heat shock relative to non-heat shock, measured by qRT-PCR.