Abstract
The successive emergence of SARS-CoV-2 Omicron variants has completely changed the modalities of use of therapeutic monoclonal antibodies. Recent in vitro studies indicated that only Sotrovimab has maintained partial activity against BQ.1.1 and XBB.1. In the present study, we used the hamster model to determine whether Sotrovimab retains antiviral activity against these Omicron variants in vivo. Our results show that at exposures consistent with those observed in humans, Sotrovimab remains active against BQ.1.1 and XBB.1, although for BQ.1.1 the efficacy is lower than that observed against the first globally dominant Omicron sublineages BA.1 and BA.2.
Keywords: SARS-CoV-2, Omicron variants, Monoclonal antibody, Sotrovimab, Hamster model
Therapeutic monoclonal antibodies (mAbs) targeting the SARS-CoV-2 spike protein have been widely used during the current COVID-19 pandemic, particularly in immunocompromised patients in whom vaccination induces an inadequate immune response. However, almost all clinically approved mAbs have lost part or all of their neutralizing activity against the different sub-lineages of the Omicron variant that have successively spread globally and contain several mutations in the spike protein associated with potential escape from humoral immunity and higher transmissibility (Campbell et al., 2021; Cao et al., 2022; Cox et al., 2022). While Bebtelovimav, Cilgavimab, Imdevimab and Sotrovimab retained some activity against BA.5, only Sotrovimab maintains partial activity against BQ.1.1 and XBB.1, that are BA.5 and BA.2 respective subvariants with increasing incidence in the USA and Europe (Arora et al., 2023; Imai et al., 2022; Planas et al., 2022; Touret et al., 2022; Wang et al., 2023). This loss of activity is partially related to R346T and N460K spike mutations harbored by both BQ1.1 and XBB.1 variants and to K444T spike mutation carried by the BQ1.1 variant (Wang et al., 2023). It has been shown that despite its partial loss of in vitro neutralizing activity against BA.2 and BA.5, Sotrovimab exhibits antiviral activity in Syrian hamsters against these Omicron variants (Park et al., 2022; Uraki et al., 2022). It is therefore urgent to determine whether this mAb retains activity in vivo against BQ.1.1 and XBB.1 at exposures similar to those observed in humans.
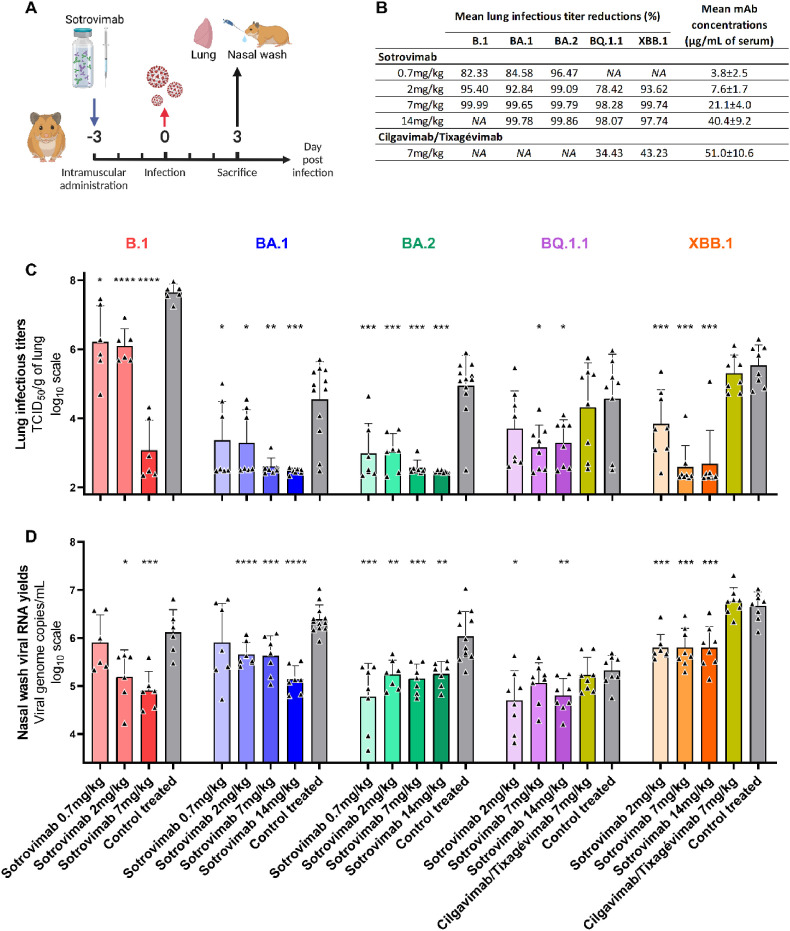
In this study, the efficacy of Sotrovimab against four clinical strains of Omicron variant (BA.1, BA.2, BQ.1.1 and XBB.1) was assessed in a hamster model using an ancestral B.1 strain as reference. Three days before the intranasal infection, groups of animals received pre-exposure prophylaxis by intramuscular injection of increasing doses of Sotrovimab and were compared with control treated animals receiving an isotype control mAb (Palivizumab) (Fig. 1 A). Overall, results showed that Sotrovimab exhibited an antiviral activity against all viruses, although less marked for BQ.1.1, in agreement with recently published in vitro data based on a VeroE6/TMPRSS2 cell assay with replicating virus (Touret et al., 2022). Indeed, globally, the higher the doses of Sotrovimab administered, the more we observed a reduction of infectious titers in the lungs, with a strong reduction observed with all Omicron variants from 7 mg/kg. When compared with corresponding control treated animals, mean reductions ranged between 82.33 and 99.99% for the ancestral strain, between 84.58 and 99.78% for BA.1, between 96.47 and 99.86% for BA.2, between 78.42% and 98.28% for BQ.1.1 and between 93.62% and 99.74% for XBB.1 (Fig. 1B–C). This decrease was significant for all doses of Sotrovimab used against all viruses (p values ranging between <0.0001 and 0.0369), except with the dose of 2 mg/kg for the BQ.1.1. Furthermore, administration of Sotrovimab led to a reduction of viral RNA yields in nasal washes for all doses with all viruses (p ranged between <0.0001 and 0.0227), except with the dose of 0.7 mg/kg for the ancestral strain and the BA.1 variant, and the dose of 7 mg/kg for the BQ.1.1 (Fig. 1D). In addition, clinical monitoring of animals showed at 3 days post-infection (dpi) significantly higher normalized weight with the dose of 7 mg/kg for the B.1 strain, the dose of 2 and 14 mg/kg for the BA.2 variant and the dose of 2, 7 and 14 mg/kg for the BQ.1.1 variant when compared with corresponding control treated groups (Fig S1).
Fig. 1.
In vivo efficacy of Sotrovimab and Cilgavimab/Tixagévimab against BA.1, BA.2, BQ.1.1 and XBB.1 Omicron variants.
(A) Experimental timeline. (B) Mean lung reduction infectious titer reduction compared to control treated animals and mean serum concentrations of mAbs at 3dpi. (C) Lung infectious titers and (D) viral RNA yields in nasal washes at 3dpi (Data represent mean ± SD of individual data). ****, ***, ** and * symbols indicate that the average value for the group is significantly lower than that of the control treated group with a p-value <0.0001, ranging between 0.0001 and 0.001, 0.001–0.01, and 0.01–0.05, respectively.
Mean serum concentrations of Sotrovimab at 3dpi were measured and ranged between 3.8 and 40.4 μg/mL (Fig. 1B and S2). Pharmacokinetic data published by the US Food Drug Administration in humans indicate that mean serum concentrations peak at 143 μg/mL and remain above 40 μg/mL twenty-nine days after intravenous administration of 500 mg Sotrovimab (COMET-ICE trial (NCT04545060)). These exposures are higher than those measured in groups of animals in which antiviral activity against the BQ1.1 and the XBB.1 variants is obtained, suggesting that Sotrovimab may retain some activity against these variants in treated patients.
Finally, a group of animals was also treated with 7 mg/kg of Cilgavimab/Tixagévimab and infected with BQ.1.1 or XBB.1. No antiviral activity was obtained at this dose (Fig. 1), and despite mean serum mAb concentrations above the geometric mean of 37.2 μg/mL observed twenty-nine days after the administration of 600 mg of this mAb cocktail during the TACKLE trial (NCT04723394) (Fig. 1B and S2B). These results are consistent with recently published in vitro data that showed that several spike mutations carried by the BQ.1.1 and XBB.1 variants confer resistance to Cilgavimab (R346T for BQ1.1 and XBB.1, K444T for BQ.1.1, and V445P for XBB.1) as well as to Tixagévimab (F486S for XBB.1) (Wang et al., 2023).
Our data demonstrate that Sotrovimab retains some antiviral activity in vivo against the BQ.1.1 variant, although at a lower level than that observed against the first globally dominant BA.1 and BA.2 Omicron sublineages, in agreement with recently published in vitro data (Touret et al., 2022). In addition, treatment with S309 (parent antibody of Sotrovimab; 10 or 30 mg/kg) in mice or with Sotrovimab (10 mg/kg) in non-human primates also conferred protection against challenge with BQ.1.1 and corroborated our results (Addetia et al., 2023; Hérate et al., 2023). Regarding the XBB.1 variant, in our model, sotrovimab maintains a significant in vivo activity, comparable to that observed against the BA.1 and BA.2 Omicron sublineages. These results are in agreement with the activities observed in vitro using infected VeroE6/TMPRSS2 cells (Touret et al., 2022). While comparing exposure data in animals with those observed in humans, these findings support the potential efficacy of Sotrovimab against BQ.1.1 and underscore the urgent need to evaluate this mAb in humans. The clinical impact of Sotrovimab treatment against XBB.1 will need to be documented in more relevant models such as non-human primates and in humans. The constant antigenic evolution of SARS-CoV-2 reinforces the need for therapeutic antibodies and antiviral molecules with broad-spectrum activity and that may be used alone or in combination.
1. Methods
1.1. Cell line
VeroE6/TMPRSS2 cells were cultured at 37 °C with 5% CO2 in minimal essential medium (MEM) supplemented with 1% Penicillin/Streptomycin, 1% non-essential amino acids, 7% of heat-inactivated fetal bovine serum (FBS) and 2% of G-418 disulfate 50 mg/mL (all from ThermoFisher Scientific).
1.2. Antibodies
Sotrovimab (Xevudy), Palivizumab (Synagis) and Cilgavimab/Tixagévimab (Evusheld) were obtained from pharmacy of the University hospital of La Timone, Marseille, France.
1.3. Virus isolates
SARS-CoV-2 B.1 strain BavPat1 was obtained from Pr. C Drosten through EVA GLOBAL (https://www.european-virus-archive.com/) and contains the D614G mutation. SARS-CoV-2 Omicron BA.1 (B.1.1.529) was isolated the 1st of December 2021 in Marseille, France. The full genome sequence has been deposited on GISAID: EPI_ISL_7899754. The strain, called 2021/FR/1514, is available through EVA GLOBAL (www.european-virus-archive.com, ref: 001V-04436). SARS-CoV-2 Omicron BA.2 strain hCoV-19/France/NAQ-HCL022005338701/2022 was obtained from Pr. B Lina and the sequence is available on GISAID: EPI_ISL_9426119. SARS-CoV-2 Omicron BQ.1.1 (hCoV-19/France/IDF-IPP50823/2022) was isolated by the National Reference Center for Respiratory Viruses hosted by Institut Pasteur (Paris, France) and headed by Pr. Sylvie van der Werf, from a specimen provided by Dr Beate Heym, Laboratoire des Centres de Santé et d'Hôpitaux d'IDF, 75020 Paris, France. The sequence is available on GISAID: EPI_ISL_15195982. SARS-CoV-2 Omicron XBB.1.1 (hCoV-19/France/PAC-HCL022171892001/2022) was obtained from Pr. B Lina and the sequence is available on GISAID: EPI_ISL_15619797. All viral stocks were made by propagation in Vero E6/TMPRSS2 cells. All experiments with infectious viruses were performed in a biosafety level 3 laboratory.
1.4. Study design
In vivo experiments were approved by the local ethical committee (C2EA—14) and the French ‘Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation’ (APAFIS#35014). Three-week-old female Syrian hamsters (Mesocricetus auratus) were provided by Janvier Labs. Animals were maintained in ISOcage P -Bioexclusion System (Techniplast) with unlimited access to water/food and 14h/10h light/dark cycle. Animals were weighed and monitored daily for the duration of the study to detect the appearance of any clinical signs of illness, suffering or distress. Intramuscular administrations (in the hindlimb), infections, nasal washes and euthanasia (cervical dislocation) were performed under general anesthesia (isoflurane, Isoflurin®, Axience).
Group size was calculated with an effect size of 2 and a power of 80%, resulting in 6–12 animals/group. For untreated groups of hamsters (receiving an isotype control mAb): n = 6 for the B.1 strain, n = 12 for BA.1 and BA.2 variants, and n = 8 for the BQ.1.1 and XBB.1.1 variants. For groups treated by Sotrovimab: n = 6 for the B.1 strain, n = 7 for BA.1 and BA.2 variants, and n = 8 for the BQ.1.1 and XBB.1.1 variants. For groups treated by Sotrovimab and sacrificed before infection: n = 6. For groups treated by Cilgavimab/Tixagévimab: n = 8 for the BQ.1.1 and XBB.1.1 variants. Sample sizes were maximized within the capacity of the BSL3 housing and virus stock availability. Animals were randomly assigned to groups, but confounders were not controlled. Since, the same experimenters carried out infection/treatment/clinical follow-up, it was impossible to perform a blind trial. Predefined humane endpoints (>20% weight loss, moribund and a scoring >10 calculated according to a clinical evaluation scale) were set as exclusion criteria. No animals were excluded from the study.
1.5. Experiment conduction
Four-to six-week-old anesthetized animals were intramuscularly inoculated with either a solution of Sotrovimab (0.7, 2, 7 or 14 mg/kg), Cilgavimab/Tixagévimab (7 mg/kg) or Palivizumab (7 mg/kg; an isotype control mAb for ‘control treated’ animals) diluted in 0.9% sodium chloride. Three days later, they were intranasally infected with 50 μL containing 1x104 (B.1), 1x105 (BA.1 and BA.2), 2x105 (BQ.1.1) and 3x104 (XBB.1.1) TCID50 of virus in 0.9% sodium chloride solution. To monitor exposure during the entire period in which the animals are infected, three additional groups of 6 uninfected animals were sacrificed three days after the intramuscular administration of Sotrovimab (2, 7 or 14 mg/kg).
Nasal washes were performed at 3dpi: 150 μl of 0.9% sodium chloride solution were instilled in the nasal cavities and transferred into 1.5 mL tubes containing 0.5 mL of 0.9% sodium chloride solution. Tubes were then centrifuged at 16,200g for 10 min and stored at −80 °C. The animals were euthanised immediately following the completion of the nasal washes at 3 dpi. Lung samples were collected immediately after euthanasia. Left pulmonary lobes were washed with 10 mL of 0.9% sodium chloride solution, blotted with filter paper, weighed, transferred into 2 mL tubes containing 1 mL of 0.9% sodium chloride solution and 3 mm glass beads, crushed using a Tissue Lyser machine (Retsch MM400) for 20min at 30 cycles/s and centrifuged 10 min at 16,200g. Supernatant media were transferred into 1.5 mL tubes, centrifuged 10 min at 16,200g and stored at −80 °C. Two milliliters of blood were harvested in a 5 mL serum tube. Blood was centrifuged for 10 min at 3,500g. Sera were transferred into 1.5 mL tubes and stored at −20 °C.
1.6. Tissue-culture infectious dose 50 (TCID50) assay
96-well culture plates containing confluent VeroE6/TMPRSS2 cells were inoculated with 150 μL per well of four-fold serial dilutions of each lung clarified homogenate samples. Each dilution was performed in sextuplicate. After 5 days of incubation, plates were read for the absence or presence of cytopathic effect in each well. Infectious titers were estimated using the method characterized by Reed & Muench (Reed, 1938).
1.7. Quantitative real-time RT-PCR (RT-qPCR) assays
Nucleic acid from 100 μL of nasal wash (spiked with bacteriophage MS2 (internal control)) were extracted using QIAamp 96 DNA kit and Qiacube HT robot (both from Qiagen). MS2 viral genome detection was performed as previously described(Driouich et al., 2021). Viral RNA yields were measured using a real time RT-qPCR assay targeting the RdRp coding region as previously described (Driouch et al., 2022; Driouich et al., 2021; Touret et al., 2021): Briefly, viral RNA was quantified by real-time RT-qPCR (GoTaq 1 step RT-qPCR kit, Promega). Quantification was provided by serial dilutions of an appropriate RNA standard. RT-qPCR reactions were performed on QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems) and analyzed using QuantStudio 12K Flex Applied Biosystems software v1.2.3.
1.8. Monoclonal antibodies quantification
To estimate the exposure of animals to monoclonal antibodies, we measured the level of human serum IgG antibodies directed against the S1 domain of the spike protein of the SARS-CoV-2 using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Euroimmun) following manufacturer instructions. Results were expressed in binding antibody units per mL (BAU/mL) following manufacturer instructions and converted to μg/mL using blank plasma from untreated/infected animals spiked with known quantities of monoclonal antibodies.
1.9. Interpretation of the results
Graphical representations and statistical analyses were realized using Graphpad Prism 7 software. Two-sided statistical analysis were performed using Shapiro–Wilk normality test, Fisher's exact test, Student t-test, Mann–Whitney test and Welch's test. P-values lower than 0.05 were considered statistically significant. Lung infectious titers reductions were estimated as follows: lung infectious titer of an animal divided by the mean titer of the untreated group infected with the corresponding virus. Experimental timelines (Fig. 1A) were generated on biorender.com.
Author contribution
Conceptualisation: JSD, XDL, AN, Data curation, formal analysis, investigation: JSD, OB, FT, AN, Funding acquisition, supervision: XDL, AN, Validation, writing – original draft: JSD, AN, Writing – review & editing: OB, FT, XDL.
Inclusion and diversity
We support inclusive, diverse and equitable conduct of research.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments and funding
We thank Michela Scaffidi and Gregory Moureau for their technical contribution. We thank Pr. S. Van der Werf, Pr. B. Lina and Pr. C. Drosten for providing the SARS-CoV-2 strains. This work was performed in the framework of the Preclinical Study Group of the Emerging Infectious Diseases, the French research agency (ANRS-MIE). It was supported by the ANRS-MIE (BIOVAR and PRI projects of the EMERGEN research program) and by the European Commission (European Virus Archive Global project (EVA GLOBAL, grant agreement No 871029) of the Horizon 2020 research and innovation program). The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105638.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data used for the research can be found in the Supplementary Data file.
References
- Addetia A., Piccoli L., Case J.B., Park Y.J., Beltramello M., Guarino B., Dang H., Pinto D., Scheaffer S., Sprouse K., Bassi J., Silacci-Fregni C., Muoio F., Dini M., Vincenzetti L., Acosta R., Johnson D., Subramanian S., Saliba C., Giurdanella M., Lombardo G., Leoni G., Culap K., McAlister C., Rajesh A., Dellota E., Zhou J., Farhat N., Bohan D., Noack J., Lempp F.A., Cameroni E., Whitener B., Giannini O., Ceschi A., Ferrari P., Franzetti-Pellanda A., Biggiogero M., Garzoni C., Zappi S., Bernasconi L., Kim M.J., Schnell G., Czudnochowski N., Franko N., Logue J.K., Yoshiyama C., Stewart C., Chu H., Schmid M.A., Purcell L.A., Snell G., Lanzavecchia A., Diamond M., Corti D., Veesler D. Therapeutic and vaccine-induced cross-reactive antibodies with effector function against emerging Omicron variants. bioRxiv. 2023 doi: 10.1101/2023.01.17.523798. [DOI] [Google Scholar]
- Arora P., Kempf A., Nehlmeier I., Schulz S.R., Jack H.M., Pohlmann S., Hoffmann M. Omicron sublineage BQ.1.1 resistance to monoclonal antibodies. Lancet Infect. Dis. 2023;23:22–23. doi: 10.1016/S1473-3099(22)00733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T., Morgan O., le Polain de Waroux O. vol. 26. 2021. (Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., Wang Y., Wang P., Zhao L., Sun H., Yu L., Yang S., Niu X., Xiao T., Gu Q., Shao F., Hao X., Xu Y., Jin R., Shen Z., Wang Y., Xie X.S. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2022 doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M., Peacock T.P., Harvey W.T., Hughes J., Wright D.W., Consortium C.-G.U., Willett B.J., Thomson E., Gupta R.K., Peacock S.J., Robertson D.L., Carabelli A.M. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2022;1–13 doi: 10.1038/s41579-022-00809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouch J.S., Cochin M., Touret F., Petit P.R., Gilles M., Moureau G., Bathelemy K., Laprie C., Wattanakul T., Chotsiri P., Hoglund R.M., Tarning J., Fraisse L., Sjo P., Mowbray C.E., Escudie F., Scnadale I., Chatelain E., de Lamballerie X., Solas C., Nougairede A. Pre-clinical evaluation of antiviral activity of nitazoxanide against SARS-CoV-2. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J.S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.R., Piorkowski G., Barthelemy K., Laprie C., Coutard B., Guedj J., de Lamballerie X., Solas C., Nougairede A. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérate C., Marlin R., Touret F., Dereuddre-Bosquet N., Donati F., Relouzat F., Junges L., Galhaut M., Dehan O., Sconosciutti Q., Nougairède A., de Lamballerie X., van der Werf S., Grand R.L. Sotrovimab retains activity against SARS-CoV-2 Omicron variant BQ.1.1 in a non-human primate model. bioRxiv. 2023 doi: 10.1101/2023.02.15.528538. 2023.2002.2015.528538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ito M., Kiso M., Yamayoshi S., Uraki R., Fukushi S., Watanabe S., Suzuki T., Maeda K., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Halfmann P.J., Kawaoka Y. Efficacy of antiviral agents against Omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Pinto D., Walls A.C., Liu Z., De Marco A., Benigni F., Zatta F., Silacci-Fregni C., Bassi J., Sprouse K.R., Addetia A., Bowen J.E., Stewart C., Giurdanella M., Saliba C., Guarino B., Schmid M.A., Franko N.M., Logue J.K., Dang H.V., Hauser K., di Iulio J., Rivera W., Schnell G., Rajesh A., Zhou J., Farhat N., Kaiser H., Montiel-Ruiz M., Noack J., Lempp F.A., Janer J., Abdelnabi R., Maes P., Ferrari P., Ceschi A., Giannini O., de Melo G.D., Kergoat L., Bourhy H., Neyts J., Soriaga L., Purcell L.A., Snell G., Whelan S.P.J., Lanzavecchia A., Virgin H.W., Piccoli L., Chu H.Y., Pizzuto M.S., Corti D., Veesler D. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science. 2022;378:619–627. doi: 10.1126/science.adc9127. [DOI] [PubMed] [Google Scholar]
- Planas D., Bruel T., Staropoli I., Guivel-Benhassine F., Porrot F., Maes P., Grzelak L., Prot M., Mougari S., Planchais C., Puech J., Saliba M., Sahraoui R., Femy F., Morel N., Dufloo J., Sanjuan R., Mouquet H., Andre E., Hocqueloux L., Simon-Loriere E., Veyer D., Prazuck T., Pere H., Schwartz O. Resistance of Omicron subvariants BA.2.75.2, BA.4.6 and BQ.1.1 to neutralizing antibodies. bioRxiv. 2022 doi: 10.1101/2022.11.17.516888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J.a.M.H. A simple method of estimating fifty per cent endpoint. Am. J. Epidemiol. 1938;27(3):493–497. [Google Scholar]
- Touret F., Driouich J.S., Cochin M., Petit P.R., Gilles M., Barthelemy K., Moureau G., Mahon F.X., Malvy D., Solas C., De Lamballerie X., Nougairede A. Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2. Antivir. Res. 2021;193 doi: 10.1016/j.antiviral.2021.105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F., Giraud E., Bourret J., Donati F., Tran-Rajau J., Chiaravalli J., Lemoine F., Agou F., Simon-Lorière E., van der Werf S., de Lamballerie X. Enhanced neutralization escape to therapeutic monoclonal antibodies by SARS-CoV-2 Omicron sub-lineages. bioRxiv. 2022 doi: 10.1101/2022.12.22.521201. 2022.2012.2022.521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraki R., Kiso M., Iida S., Imai M., Takashita E., Kuroda M., Halfmann P.J., Loeber S., Maemura T., Yamayoshi S., Fujisaki S., Wang Z., Ito M., Ujie M., Iwatsuki-Horimoto K., Furusawa Y., Wright R., Chong Z., Ozono S., Yasuhara A., Ueki H., Sakai-Tagawa Y., Li R., Liu Y., Larson D., Koga M., Tsutsumi T., Adachi E., Saito M., Yamamoto S., Hagihara M., Mitamura K., Sato T., Hojo M., Hattori S.I., Maeda K., Valdez R., team I.s., Okuda M., Murakami J., Duong C., Godbole S., Douek D.C., Maeda K., Watanabe S., Gordon A., Ohmagari N., Yotsuyanagi H., Diamond M.S., Hasegawa H., Mitsuya H., Suzuki T., Kawaoka Y. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022;607:119–127. doi: 10.1038/s41586-022-04856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Lekthan S., Li Z.T., Liu L.Y., Guo Y.C., Huang Y.M., Bowen A.D., Liu M.C., Wang M.P., Yu J., Valdez R., Lauring A.S., Sheng Z.Z., Wang H.H., Gordon A., Liu L.H., Ho D.D. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for the research can be found in the Supplementary Data file.