Abstract
Background/Aims
Real-time polymerase chain reaction (RT-PCR) is a fast and simple method for the simultaneous detection of clarithromycin (CLR) resistance and Helicobacter pylori. We evaluated the effectiveness of RT-PCR compared to that of the rapid urease test (RUT) and assessed its value in verifying CLR resistance.
Methods
A total of 70 specimens with confirmed H. pylori infection in culture were enrolled and analyzed in this prospective study. All specimens were subjected to RT-PCR assay using fluorescence melting peak signals to detect H. pylori infection and CLR resistances caused by either A2142G or A2143G mutations in the 23S ribosomal RNA gene (23S rRNA). The results were compared to those of RUT and antimicrobial susceptibility culturing tests to investigate the efficacy of RT-PCR.
Results
Among the 70 specimens analyzed, the positivity rate was 97.1% (68/70) with RT-PCR and 82.9% (58/70) with RUT. CLR resistance (minimum inhibitory concentration >1.0 μg/mL) was confirmed in 18.6% (13/70), and fluorescence melting curve analysis showed that 84.6% (11/13) had point mutations in 23S rRNA. Ten specimens had only A2143G mutation, and one specimen contained both A2142G and A2143G mutations.
Conclusions
RT-PCR assay was found to be more efficient than RUT in detecting H. pylori infection and could effectively verify CLR resistance compared to the antimicrobial susceptibility culturing test. Considering the high sensitivity and accessibility of RT-PCR method, it could be used to easily detect CLR-resistant H. pylori, thus helping clinicians select suitable treatment regimen and improve the eradication rate.
Keywords: Clarithromycin resistance, Helicobacter pylori, Culture, Rapid urease test, Real-time polymerase chain reaction
INTRODUCTION
Helicobacter pylori is a Gram-negative, spiral-shaped bacterium that colonizes the gastric mucosa and may cause atrophic gastritis, peptic ulcers, gastric cancer, and mucosal-associated lymphoid tissue lymphoma. Therefore, eradication of H. pylori is important not only for patients infected with the disease but also for asymptomatic, healthy individuals. With a global increase in antibiotic resistance, the success rate of H. pylori eradication is decreasing.1-3 The main cause of treatment failure has been reported as clarithromycin (CLR) resistance, and among Korean patients, CLR resistance has gradually increased from 13.7% in 2003 to 16.7% in 2006 and 23.7% in 2012.4 Thus, it is important to detect CLR resistance and select an appropriate treatment regimen to improve the H. pylori eradication rate.
The rapid urease test (RUT) is the most common method for detecting H. pylori infection. It yields results quickly, has high sensitivity and specificity, and is economic and simple to use. However, it requires a high density of bacteria and cannot detect antibiotic resistance.5 Culture and antibiotic susceptibility tests can confirm antibiotic resistance regardless of genetic mutations, but culturing bacterial takes at least 5 to 7 days. Additionally, because the culture conditions and composition environments may vary depending on the institution and are difficult to construct, the culture success rate ranges from 50% to 80%.6 However, the real-time polymerase chain reaction (RT-PCR) assay is a fast and simple method for the simultaneous detection of CLR resistance and H. pylori.7 Although it is a sensitive and specific method for detecting H. pylori infection in gastric tissue samples, there is a lack of studies comparing the RT-PCR assay, RUT, and antibiotic susceptibility test using conventional culture.
Therefore, we evaluated whether the RT-PCR assay is more efficient than RUT for detecting H. pylori and whether it is appropriate for identifying CLR resistance with the associated mutations.
MATERIALS AND METHODS
1. Study design and population
This prospective study was conducted between December 2019 and December 2020 at Asan Medical Center, Seoul, Korea. This study was approved by the Institutional Review Board of Asan Medical Center (IRB number: 2020-0082) and conducted in accordance with the standards of the Declaration of Helsinki. The patients, who were between 19 and 80 years old and had no history of H. pylori eradication, were enrolled after providing written informed consent. Patients with a history of gastric surgery, hematologic disease with bleeding tendency, or antibiotics administration within the past month were excluded.
All patients underwent serum H. pylori immunoglobulin G (IgG) testing and esophagogastroduodenoscopy with pairs of mucosal biopsies from gastric antrum and corpus for each RUT and culture with antimicrobial susceptibility tests. Of these, the specimens from patients who were positive for either antrum or body on RUT were used for the RT-PCR assay with U-TOPTM HPy & ClaR Detection Kit (SeaSun Biomaterials, Daejeon, Korea). Finally, the 70 specimens that were identified as having H. pylori infection in culture, which was considered a confident method, were analyzed to investigate the H. pylori detection of RT-PCR assay and RUT, as well as the presence of CLR resistances by detecting A2142G or A2143G mutations in the 23S ribosomal RNA gene (23S rRNA) (Supplementary Fig. 1).
2. H. pylori culture
The tissues obtained from gastric antrum and corpus during esophagogastroduodenoscopy were first placed in a sterile Eppendorf tube and then placed in a vacuum bottle containing dry ice. The Eppendorf tubes were stored in a –80°C deep freezer and the tissues were allowed to thaw at room temperature before analysis. The specimens were inoculated onto an H. pylori isolation medium (Brucella broth agar supplemented with 5% sheep blood and containing 10 μg/L vancomycin, 5 μg/L trimethoprim, 5 μg/L amphotericin B, and 2.5 IU polymyxin B) and incubated at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 5 to 7 days. Suspected H. pylori colonies were identified based on morphology, Gram staining, and urease positive reaction.
The minimum inhibitory concentrations (MICs) of antibiotics were determined using the serial 2-fold agar dilution method as described previously.8 Bacteria were sub-cultured for 48 hours on Mueller–Hinton agar supplemented with 5% defibrinated sheep blood. The bacterial suspension was adjusted to 1×107 colony-forming units and inoculated to each antibiotic-containing agar dilution plate followed by incubation for 3 days at 37°C under microaerophilic conditions. The MIC of antibiotics was evaluated after 72 hours. A standard H. pylori strain (ATCC 43504) was used as the control. Resistance breakpoint for CLR was set as >1 μg/mL.9
3. RT-PCR for detection of H. pylori infection and CLR resistance
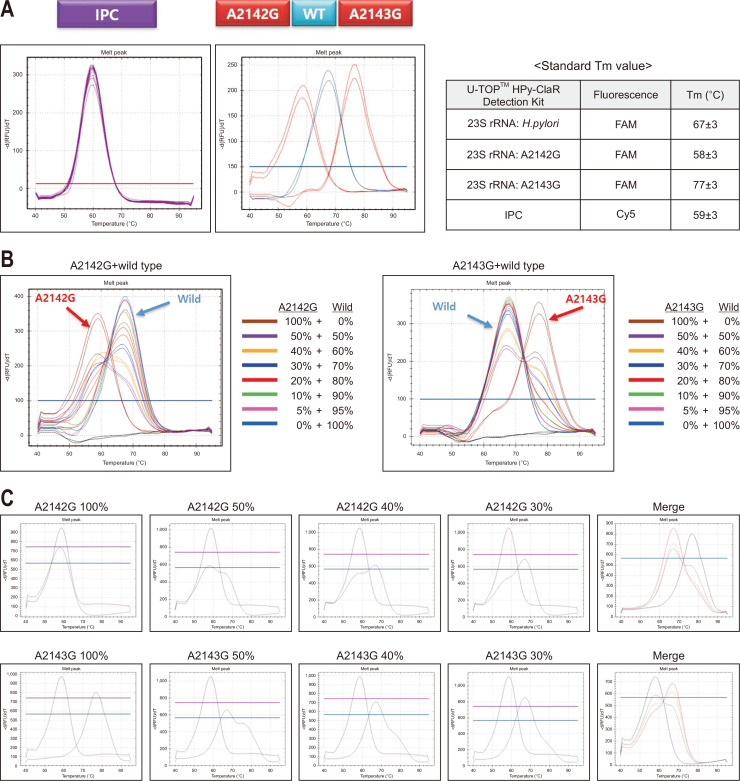
According to the manufacturer's instruction (QIAamp DNA Tissue Kit; Qiagen, Hilden, Germany), the genomic DNA was extracted from the leftover biopsied samples after performing the RUT. The quality and quantity of the genomic DNA samples were evaluated using a Nanodrop spectrophotometers (Thermo Fisher, Waltham, MA, USA), with a >15 ng/μL cutoff value of appropriate bacterial concentration for the test. RT-PCR for detection of H. pylori and CLR resistances was performed with the U-TOPTM HPy & ClaR Detection Kit (SeaSun Biomaterials) using the CFX96 Real-Time PCR detection system V1.6 (Bio-Rad, Hercules, CA, USA). Specific primers were used to amplify the H. pylori genes. The polymerase nucleic acid probe-based qPCR method was used for analyzing the mutations status of H. pylori. Data were analyzed using the SS AnalyzerII V1.0 (SeaSun Biomaterials, Seoul, Korea) software. Infection and CLR resistance were differentiated by the fluorescence melting peak signal (Tm) (Fig. 1A). For cases with mixed infection of wild type and mutant H. pylori, melt curve analysis was further performed to distinguish two different peaks according to each percentage, and the presence of point mutation was differentiated using the gradient difference (Fig. 1B and C).
Fig. 1.
Representative results of internal positive control (IPC), Helicobacter pylori-positive, A2142G mutation, and A2143G mutation using U-TOPTM HPy & ClaR Detection Kit. (A) Examples of melting peak signal (Tm) with standard value. (B) Tm results according to the percentage of wild type and mutant (A2142G and A2143G point mutations) in mixed-infection samples, and (C) examples with different percentage.
4. Statistical analysis
Descriptive statistics for the categorical variables were summarized as proportions, and the continuous variables were summarized using the medians (interquartile range). The McNemar test was applied to calculate the diagnostic sensitivity of each test. The sensitivity (%) was calculated as follows: 100×true positives/(true positives+false negatives). The true positives and false negatives were determined depending on whether the H. pylori infection was confirmed by culture. The specificity (%) of the method was calculated as follows: 100×true negatives/(false positives+true negatives), where true negatives and false positives were defined according to H. pylori infection determined by combining the results of culture and H. pylori IgG antibody test. All statistical analyses were conducted using SPSS version 24 (IBM Corp., Armonk, NY, USA).
RESULTS
1. Clinical characteristics and diagnostic tests
A total of 70 specimens from 39 patients were included in this study. The median age was 62 years (interquartile range, 58 to 70 years), and 87% of the patients (34/39) were male. Indications of test were 56% (22/39) of dysplasia and 44% (17/39) of early gastric cancer. The RUT results showed 72% (23/32) antrum positivity and 92% (35/38) body positivity, whereas the RT-PCR results showed 94% (30/32) antrum positivity and 100% (38/38) body positivity. The results of H. pylori IgG antibody test, RUT, and RT-PCR of the 70 samples are summarized in Supplementary Table 1.
2. Comparison of H. pylori detection rate using RUT and RT-PCR
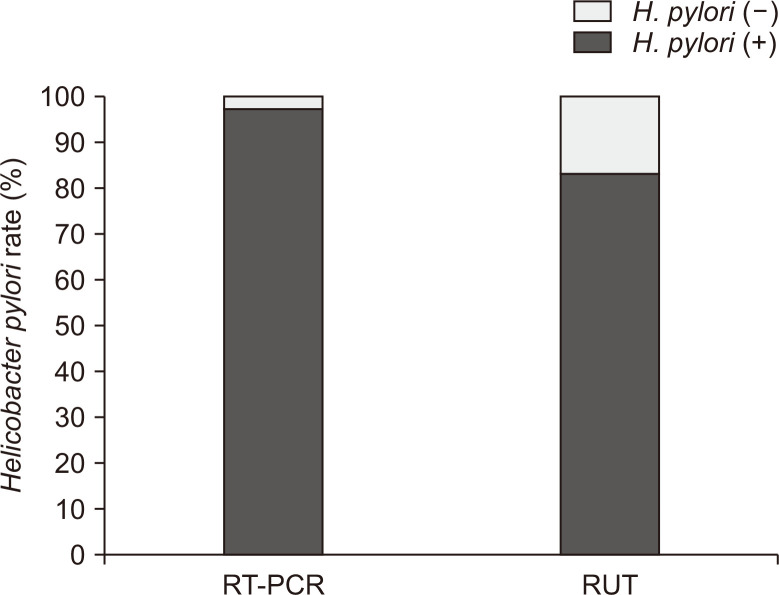
Among the 70 specimens, RT-PCR and RUT assays showed sensitivities of 97.1% (68/70) and 82.9% (58/70) (Fig. 2), and specificities of 100% (8/8) and 62.5% (5/8), respectively. Table 1 shows the accordance of each H. pylori diagnostic test according to the stomach location, namely, the antrum and body. The RUT and RT-PCR assay showed accordance of 70% (21/30) for the antrum and 92% (35/38) for the body. The RUT and culture represented 72% (23/32) and 92% (35/38) of accordance for the antrum and body, respectively. In the case of RT-PCR and culture, the accordance was 94% (30/32) for the antrum and 100% (38/38) for the body. Consequently, the body specimens showed higher accordance between the two tests than the antrum specimens.
Fig. 2.
Comparison of Helicobacter pylori detection rates between the rapid urease test (RUT) and real-time polymerase chain reaction (RT-PCR) assay in culture-positive specimens.
Table 1.
Accordance among RUT, RT-PCR, and Culture for Helicobacter pylori Diagnosis According to Its Location in the Stomach of Culture-Positive Specimens (n=70)
| Variable | RUT | RT-PCR | ||||
|---|---|---|---|---|---|---|
| Antrum | Body | Antrum | Body | |||
| Culture (n=70) | Antrum (n=32) | 23 (72) | 30 (94) | |||
| Body (n=38) | 35 (92) | 38 (100) | ||||
| RT-PCR (n=68) | Antrum (n=30) | 21 (70) | * | * | ||
| Body (n=38) | 35 (92) | * | * | |||
Data are presented as number (%).
RUT, rapid urease test; RT-PCR, real-time polymerase chain reaction.
*Comparisons of the same test (RT-PCR).
3. CLR resistance by culture and RT-PCR
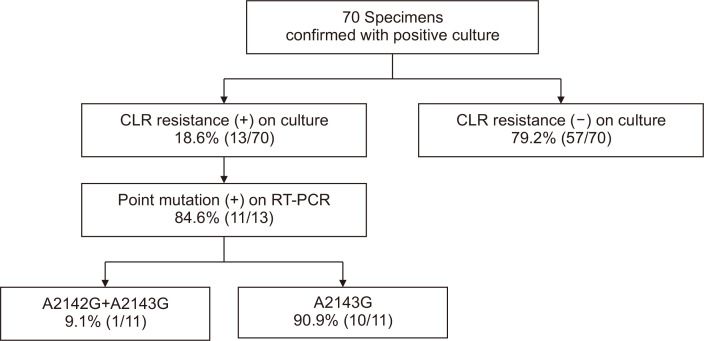
Fig. 3 shows the CLR resistance in culture and the proportions of point mutations. Among the 70 specimens, 13 (18.6%) were CLR resistant (MIC >1.0 μg/mL) and 57 (82.3%) were CLR susceptible. Of the CLR-resistant specimens, 11 (84.6%) had 23S ribosomal RNA point mutations, detected using fluorescence melting curve analysis. The A2143G mutation was observed in 10 specimens, and notably, one specimen showed A2142G and A2143G double mutations. The MIC of CLR and the mutation types of each specimen are summarized in Table 2.
Fig. 3.
Clarithromycin (CLR) resistance and proportion of point mutations (A2142G and A2143G) confirmed using agar dilution test and real-time polymerase chain reaction (RT-PCR).
Table 2.
MIC of CLR and Mutation Types Detected Using RT-PCR Assay on CLR-Resistant Specimens
| Specimen number | CLR MIC, mg/L | Mutation type |
|---|---|---|
| 1 | >128 | A2142G, A2143G |
| 2 | >128 | A2143G |
| 3 | 64 | A2143G |
| 4 | 64 | A2143G |
| 5 | 32 | A2143G |
| 6 | 16 | A2143G |
| 7 | 16 | A2143G |
| 8 | 16 | A2143G |
| 9 | 16 | A2143G |
| 10 | 16 | A2143G |
| 11 | 8 | A2143G |
| 12 | 8 | - |
| 13 | 8 | - |
MIC, minimum inhibitory concentration; CLR, clarithromycin; RT-PCR, real-time polymerase chain reaction.
RT-PCR was performed using the U-TOPTM HPy & ClaR Detection Kit (SeaSun Biomaterials).
DISCUSSION
Although several PCR-based H. pylori detection methods with high sensitivity and specificity have recently been reported, few studies have clinically focused on the comparison between PCR and other methods such as RUT and standard susceptibility culturing tests. In this study, we investigated whether the RT-PCR assay is more efficient than the RUT for detecting H. pylori and whether it is more appropriate for identifying CLR resistance compared to the susceptibility culturing test. Among the 70 specimens with confirmed H. pylori infection in culture, 97.1% and 82.9% were positively confirmed using RT-PCR and RUT, respectively. The CLR resistance in culture was confirmed in 18.6% of the specimens, and 84.6% of them had A2142G and A2143G mutations in the 23S rRNA gene, as detected using the fluorescence melting curve analysis. Additionally, H. pylori IgG antibody test was performed using all specimens, of which, four were found to be negative. The sensitivity of H. pylori IgG antibody test was 94.3% (66/70), similar to those reported in previous studies.10,11
According to previous studies, the results of H. pylori detection and CLR resistance of RT-PCR-based assays were similar to those of the commercialized DPO-PCR (SeeplexⓇ ClaR-H. pylori ACE Detection; Seegene Institute of Life Sciences, Seoul, Korea) and conventional PCR.12,13 RT-PCR-based assay is a simple and accurate method for detecting H. pylori and mutation types of CLR resistance that can be analyzed using a small formalin-fixed, paraffin-embedded specimen after amplification. As it is a one-step analysis, it can avoid cross-contamination that can occur during electrophoresis. Moreover, RT-PCR allows easy and accurate interpretation of mutation types through the fluorescence melting peak signal. In this study, two H. pylori-positive specimens using culture did not show bacterial detection using RT-PCR. We believe that the detection failure was caused by using the leftover tissues on RUT without controlling the incubation times. In fact, neither specimen (each had a bacterial concentration of 8 ng/μL and 5.3 ng/μL) satisfied the cutoff value of appropriate bacterial concentration for the RT-PCR test. In previous studies, when the leftover tissue following RUT was used within 4 hours after inoculation, the H. pylori could be cultured more successfully.14 It is expected that if the RT-PCR assay were performed using fresh gastric tissue, the detection rate might increase. We need further randomized controlled trials with large numbers of patients for further investigations.
CLR resistance affects the eradication rate and is considered the main cause of treatment failure. In previous studies, most point mutations causing CLR resistance by preventing the macrolide from binding were located in A2143G (69.8%), A2142G (11.7%), and A2142C (2.6%).15 Additionally, other mutations such as A2115G, A2142T, G2141A, and T2182C can also influence CLR resistance.15,16 The mutation distributions vary in different regions. In the United States of America, the positivity rates of A2142G and A2143G mutations are 48% to 53% and 39% to 45%, respectively, whereas the A2142C mutation is 0% to 7%. Similarly, in Europe, the A2142G and A2143G mutations are 23% to 33% and 44% to 67%, respectively, and A2142C mutation has been reported as 2% to 10%.17,18 On the other hand, in Japan and China, over 90% and 100%, respectively, of CLR resistance cases were found to have A2143G mutations, although the number of patients was relatively small.19,20 Additionally, most CLR resistance cases in South Korea were confirmed to have the A2143G mutation.21,22
Currently, CLR resistance can detect A2142G and A2143G using this RT-PCR kit. In our study, two specimens that were identified as having CLR resistance in antimicrobial susceptibility tests showed neither resistance nor A2142G or A2143G mutations in RT-PCR. One of these was a quality control failed specimen (concentration of 6.7 ng/μL), whereas the other had suitable quality and quantity for performing RT-PCR. We speculated that the quality and quantity of the samples leftover from RUT might be not suitable for performing RT-PCR. It is also possible that the specimen may have had mutations other than A2142G and A2143G, causing CLR resistance in the antimicrobial susceptibility test. Further analysis is needed to verify the presence of other, abovementioned mutations and to investigate the mutation types that affect clinically significant CLR resistance.
There are several limitations to this study. First, the sample size was relatively small for analyzing the RT-PCR predictive values and accuracy. Second, there was no comparative group to demonstrate that using leftover tissue of RUT as fresh tissue was insufficient for conducting RT-PCR. However, our study did establish that RT-PCR is more appropriate than RUT and agrees well with the antimicrobial susceptibility culturing test.
In conclusion, the RT-PCR assay may be an alternate method for RUT to detect H. pylori infection and can simultaneously and effectively verify the presence of CLR resistance. It is expected that the H. pylori eradication rate will increase if clinicians select treatment regimens based on these results.
Funding Statement
ACKNOWLEDGEMENTS This work was supported by a grant from SK Chemical Research Fund of the Korean Society of Gastroenterology in 2020 and the Korean College of Helicobacter and Upper Gastrointestinal Research Foundation (grant number: KCHUGR-202002501).
Footnotes
SUPPLEMENTARY MATERIALS
Supplementary materials can be accessed at https://doi.org/10.5009/gnl220076.
CONFLICTS OF INTEREST
J.Y.A. is an editorial board member of the journal but was not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflicts of interest relevant to this article were reported.
AUTHOR CONTRIBUTIONS
Study concept and design: J.Y.A., J.C. Data acquisition: J.H.N., J.Y.A., J.C., Y.S.P., H.K.N., J.H.L., K.W.J., D.H.K., K.D.C., H.J.S., G.H.L., H.Y.J., J.M.K. Data analysis and interpretation of the data: J.H.N., J.Y.A., J.C. Drafting of the manuscripts: J.H.N., J.Y.A., J.C. Critical revision of the article for intellectual content: J.H.N., J.Y.A., J.C. Obtained funding: J.Y.A. Approval of final manuscript: all authors.
REFERENCES
- 1.Fock KM, Katelaris P, Sugano K, et al. Second Asia-Pacific Consensus Guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24:1587–1600. doi: 10.1111/j.1440-1746.2009.05982.x. [DOI] [PubMed] [Google Scholar]
- 2.Kim SG, Jung HK, Lee HL, et al. Guidelines for the diagnosis and treatment of Helicobacter pylori infection in Korea, 2013 revised edition. J Gastroenterol Hepatol. 2014;29:1371–1386. doi: 10.1111/jgh.12607. [DOI] [PubMed] [Google Scholar]
- 3.Malfertheiner P, Megraud F, O'Morain CA, et al. Management of Helicobacter pylori infection: the Maastricht IV/Florence Consensus Report. Gut. 2012;61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 4.Chung JW, Lee GH, Han JH, et al. The trends of one-week first-line and second-line eradication therapy for Helicobacter pylori infection in Korea. Hepatogastroenterology. 2011;58:246–250. [PubMed] [Google Scholar]
- 5.Yakoob J, Jafri W, Abid S, et al. Role of rapid urease test and histopathology in the diagnosis of Helicobacter pylori infection in a developing country. BMC Gastroenterol. 2005;5:38. doi: 10.1186/1471-230X-5-38.27ef4687f7d347b0b93c836e4ea4f40a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerqueira L, Fernandes RM, Ferreira RM, et al. Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J Clin Microbiol. 2013;51:1887–1893. doi: 10.1128/JCM.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung JW, Lee GH, Jeong JY, et al. Resistance of Helicobacter pylori strains to antibiotics in Korea with a focus on fluoroquinolone resistance. J Gastroenterol Hepatol. 2012;27:493–497. doi: 10.1111/j.1440-1746.2011.06874.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Ahn JY, Choi KD, et al. Nationwide antibiotic resistance mapping of Helicobacter pylori in Korea: a prospective multicenter study. Helicobacter. 2019;24:e12592. doi: 10.1111/hel.12592. [DOI] [PubMed] [Google Scholar]
- 10.Vaira D, Gatta L, Ricci C, Miglioli M. Review article: diagnosis of Helicobacter pylori infection. Aliment Pharmacol Ther. 2002;16 Suppl 1:16–23. doi: 10.1046/j.1365-2036.2002.0160s1016.x. [DOI] [PubMed] [Google Scholar]
- 11.Loy CT, Irwig LM, Katelaris PH, Talley NJ. Do commercial serological kits for Helicobacter pylori infection differ in accuracy? A meta-analysis. Am J Gastroenterol. 1996;91:1138–1144. [PubMed] [Google Scholar]
- 12.Jung DH, Kim JH, Jeong SJ, et al. Peptide nucleic acid probe-based analysis as a new detection method for clarithromycin resistance in Helicobacter pylori. Gut Liver. 2018;12:641–647. doi: 10.5009/gnl18111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nahm JH, Kim WK, Kwon Y, Kim H. Detection of Helicobacter pylori with clarithromycin resistance-associated mutations using peptide nucleic acid probe-based melting point analysis. Helicobacter. 2019;24:e12634. doi: 10.1111/hel.12634. [DOI] [PubMed] [Google Scholar]
- 14.Gong EJ, Ahn JY, Jung DK, et al. Isolation of Helicobacter pylori using leftover tissue in the rapid urease test kit. Helicobacter. 2020;25:e12733. doi: 10.1111/hel.12733. [DOI] [PubMed] [Google Scholar]
- 15.Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hultén K, Gibreel A, Sköld O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/AAC.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alarcón T, Domingo D, Prieto N, López-Brea M. Clarithromycin resistance stability in Helicobacter pylori: influence of the MIC and type of mutation in the 23S rRNA. J Antimicrob Chemother. 2000;46:613–616. doi: 10.1093/jac/46.4.613. [DOI] [PubMed] [Google Scholar]
- 18.van Doorn LJ, Glupczynski Y, Kusters JG, et al. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45:1500–1504. doi: 10.1128/AAC.45.5.1500-1504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato S, Fujimura S, Udagawa H, et al. Antibiotic resistance of Helicobacter pylori strains in Japanese children. J Clin Microbiol. 2002;40:649–653. doi: 10.1128/JCM.40.2.649-653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan ZJ, Su WW, Tytgat GN, Dankert J, van der Ende A. Assessment of clarithromycin-resistant Helicobacter pylori among patients in Shanghai and Guangzhou, China, by primer-mismatch PCR. J Clin Microbiol. 2002;40:259–261. doi: 10.1128/JCM.40.1.259-261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An B, Moon BS, Kim H, et al. Antibiotic resistance in Helicobacter pylori strains and its effect on H. pylori eradication rates in a single center in Korea. Ann Lab Med. 2013;33:415–419. doi: 10.3343/alm.2013.33.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung WC, Jung SH, Oh JH, et al. Dual-priming oligonucleotide-based multiplex PCR using tissue samples in rapid urease test in the detection of Helicobacter pylori infection. World J Gastroenterol. 2014;20:6547–6553. doi: 10.3748/wjg.v20.i21.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.