Abstract
Computer analysis of the archaeal genome databases failed to identify orthologues of all of the bacterial cobamide biosynthetic enzymes. Of particular interest was the lack of an orthologue of the bifunctional nucleoside triphosphate (NTP):5′-deoxyadenosylcobinamide kinase/GTP:adenosylcobinamide-phosphate guanylyltransferase enzyme (CobU in Salmonella enterica). This paper reports the identification of an archaeal gene encoding a new nucleotidyltransferase, which is proposed to be the nonorthologous replacement of the S. enterica cobU gene. The gene encoding this nucleotidyltransferase was identified using comparative genome analysis of the sequenced archaeal genomes. Orthologues of the gene encoding this activity are limited at present to members of the domain Archaea. The corresponding ORF open reading frame from Methanobacterium thermoautotrophicum ΔH (MTH1152; referred to as cobY) was amplified and cloned, and the CobY protein was expressed and purified from Escherichia coli as a hexahistidine-tagged fusion protein. This enzyme had GTP:adenosylcobinamide-phosphate guanylyltransferase activity but did not have the NTP:AdoCbi kinase activity associated with the CobU enzyme of S. enterica. NTP:adenosylcobinamide kinase activity was not detected in M. thermoautotrophicum ΔH cell extract, suggesting that this organism may not have this activity. The cobY gene complemented a cobU mutant of S. enterica grown under anaerobic conditions where growth of the cell depended on de novo adenosylcobalamin biosynthesis. cobY, however, failed to restore adenosylcobalamin biosynthesis in cobU mutants grown under aerobic conditions where de novo synthesis of this coenzyme was blocked, and growth of the cell depended on the assimilation of exogenous cobinamide. These data strongly support the proposal that the relevant cobinamide intermediates during de novo adenosylcobalamin biosynthesis are adenosylcobinamide-phosphate and adenosylcobinamide-GDP, not adenosylcobinamide. Therefore, NTP:adenosylcobinamide kinase activity is not required for de novo cobamide biosynthesis.
De novo cobamide (Cba) biosynthesis is believed to be restricted to procaryotes (13, 29). Synthesis of the corrin ring occurs via the anaerobic pathway found in Salmonella enterica, Propionibacterium freundenreichii subsp. shermanii (31), and Bacillus megaterium (25), or via the aerobic pathway found in Pseudomonas denitrificans (4). The main differences between these pathways appear to be the timing of cobalt insertion (2, 11, 23, 46) and ring contraction (31, 34, 36, 45). Work on these organisms has given considerable insight into the details of the Cba biosynthetic pathway and has set the framework for comparison with other organisms (3, 26, 33).
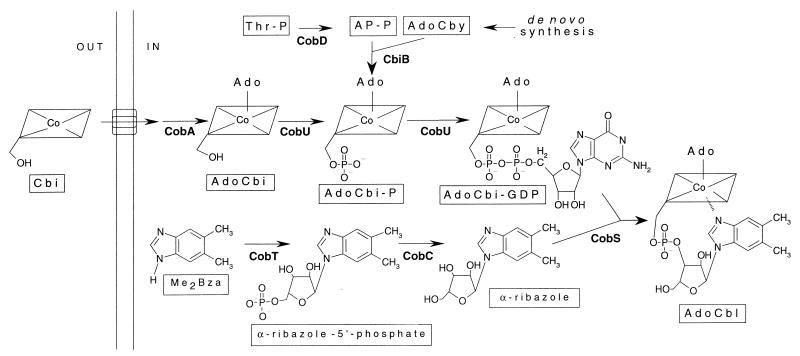
At present, knowledge of the Cba biosynthetic pathway in archaea is limited (6, 10, 32, 37). Cba biosynthesis is essential to several archaea. For example, methanogenic archaea require Cba for methanogenesis (14, 40), and Archaeoglobus fulgidus may require Cba for DNA synthesis based on the presence of a gene encoding a putative Cba-dependent ribonucleotide reductase (18). The availability of the genome sequences of six archaea (9, 17, 18, 35) (accession numbers AJ248283.1 and AP000063.1) has provided a unique opportunity to learn how these procaryotes synthesize Cba. The existence of orthologues to most of the bacterial Cba biosynthetic enzymes leads to the conclusion that Cba biosynthesis in archaea occurs via pathways similar to those found in bacteria (9, 17–20, 35). Of interest to us was the absence of an orthologue for the bifunctional nucleoside triphosphate (NTP):5′-deoxyadenosylcobinamide (AdoCbi) kinase/GTP:5′-deoxyadenosylcobinamide-phosphate (AdoCbi-P) guanylyltransferase enzyme found in bacteria (CobU in S. enterica). This enzyme plays an essential role in Cba biosynthesis in bacteria, and its absence in the archaeal genomes suggested that a different protein performs this activity. The archaeal enzyme responsible for the synthesis of AdoCbi-GDP could also be bifunctional but distinct from CobU. Alternatively, two different proteins could have evolved to perform these functions, or selective pressures may have resulted in the elimination of one of the activities. The possibility of only requiring guanylyltransferase function can be supported by earlier results which suggest that the kinase activity of CobU is not required for de novo adenosylcobalamin (AdoCbl) biosynthesis in S. enterica (8). Brushaber et al. (8) proposed that AdoCbi was not a de novo Cba intermediate based on the finding that the CobD enzyme of S. enterica decarboxylates threonine phosphate (Thr-P) to generate aminopropanol-phosphate (AP-P). These authors proposed that AP-P is attached to 5′-deoxyadenosylcobyric acid (AdoCby) by the cobinamide (CbiB) synthase enzyme to generate AdoCbi-P, not AdoCbi as previously thought (Fig. 1). If this idea were correct, there would be no need for an AdoCbi kinase activity during de novo AdoCbl biosynthesis. These authors also proposed that the well-documented kinase activity of CobU (24) would be required only for the salvaging of unphosphorylated Cbi from the environment (Fig. 1) (8).
FIG. 1.
Late steps of AdoCbl biosynthesis in S. enterica serovar Typhimurium. The model shows the late steps of AdoCbl biosynthesis via the de novo biosynthetic pathway of the corrin ring (CobD, CbiB sequence), or via the Cbi assimilatory pathway (CobA, CobU kinase sequence). Enzymes catalyzing each step are in boldface and shown below each corresponding reaction. The enzyme not found in archaea is CobU. At the left, OUT is the periplasm and IN is the cytoplasm. The transport system that translocates Cbi across the cytoplasmic membrane is illustrated by the overlapping open rectangles. Intermediates and final product are identified by boxed abbreviations below each compound. Abbreviations: AP-P, aminopropanol phosphate; AdoCby, adenosylcobyric acid; AdoCbl, adenosylcobalamin; AdoCbi, adenosylcobinamide; AdoCbi-P, adenosylcobinamide-P; AdoCbi-GDP, adenosylcobinamide-GDP; Cbi, cobinamide; Me2Bza, 5,6-dimethylbenzimidazole; Thr-P, threonine phosphate.
This paper reports the identification of the NTP:AdoCbi-P nucleotidyltransferase from the methanogenic archaeon Methanobacterium thermoautotrophicum ΔH, which is referred to as CobY. This enzyme was shown to have guanylyltransferase activity that converted AdoCbi-P to AdoCbi-GDP but lacked NTP:AdoCbi kinase activity. A plasmid carrying the wild-type allele of cobY complemented an S. enterica cobU mutant during de novo AdoCbl biosynthesis but failed to complement under growth conditions that demanded assimilation of exogenous Cbi. This was strong in vivo evidence that an AdoCbi kinase was not needed for Cba biosynthesis and that AdoCbi was not an intermediate of de novo AdoCbl biosynthesis.
MATERIALS AND METHODS
Bacterial strains.
All S. enterica serovar Typhimurium LT2 strains used were derivatives of TR6583 (metE205 ara-9). Overexpression of six-His-tagged CobY (H6CobY) protein was performed in Escherichia coli strain BL21(λDE3) (Novagen, Madison, Wis.).
Synthesis of AdoCbi and AdoCbi-P.
AdoCbi was synthesized using a modification of a previously reported protocol (24). The reaction mixture contained 200 mM Tris-HCl buffer (pH 8.0) at 37°C, 5 mM MgCl2, 420 μM CoCl2, 800 μM ATP, and 150 μM (CN)2Cbi in a final volume of 50 ml. The reaction mixture was made anoxic by bubbling with oxygen-free N2 gas for 1 h at 25°C. A 1-h degassing period was used to ensure complete removal of molecular oxygen, since the Co[I] nucleophile is very reactive. Five-milliliter samples of the reaction mixture were added to anoxic serum vials containing 25 mg of potassium borohydride. The vial was incubated at 25°C for approximately 10 min or until the solution turned a grey-green color indicative of Co[III]→Co[I] reduction of Cbi (38). Ten micrograms of purified ATP:corrinoid adenosyltransferase (CobA) enzyme isolated from S. enterica as described elsewhere (38) was added to each 5-ml sample, and the complete reaction was incubated at 37°C for 3 to 4 h while constantly flushing with oxygen-free N2 gas. The reaction was terminated by exposing the reaction mixture to air to oxidize Co[I]. Particulates were removed by filtration using 0.2-μm-pore-size, 25-mm-diameter disposable syringe filters (Nalgene, Rochester, N.Y.). The filtered reaction mixture was loaded onto a 5-ml LiChroprep RP-18 (EM Separations, Gibbstown, N.J.) column (6 by 1 cm) equilibrated with doubled-distilled water. The column was washed with 50 ml of double-distilled water, and AdoCbi was eluted within a 10-column volume 0 to 100% methanol linear gradient. Elution of AdoCbi was monitored using UV-visible spectroscopy to identify AdoCbi by its characteristic spectrum (data not shown).
AdoCbi-P was synthesized in a 500-μl reaction mixture containing 100 mM Tris-HCl (pH 8.5) at 25°C, 25 mM MgCl2, 4 mM ATP, 400 μM AdoCbi, and 40 μg of purified CobU protein. The reaction mixture was incubated at 25°C for 2 h, and the reaction was stopped by heating the reaction mixture to 70°C for 10 min. AdoCbi-P was purified using the chromatographic procedure described above. All manipulations during AdoCbi or AdoCbi-P synthesis and purification were performed in dim light to minimize photolysis of the C-Co bond. Quantitation of AdoCbi and AdoCbi-P was performed by conversion of both substrates to their dicyano derivatives and subsequent use of the molar extinction coefficient of (CN)2Cbi (ɛ367 30,800 M−1 cm−1) in 0.1 M KCN, pH 10.0 (15). Authentic (CN)2Cbi was purchased from Sigma (St. Louis, Mo.).
Conditions for the in vitro assembly of AdoCbl by archaeal enzymes starting from AdoCbi or AdoCbi-P.
Five grams of M. thermoautotrophicum ΔH cells was resuspended in 10 ml of buffer A (50 mM Tris-HCl [pH 8.5] at 4°C, 1 mM dithiothreitol, 680 mM glycerol, 0.1 M NaCl, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride). The cell suspension was passed three times through a French pressure cell at ∼104 kPa. Soluble and insoluble materials were separated by centrifugation at 36,000 × g for 30 min at 4°C in a Beckman J21 centrifuge. The supernatant was dialyzed extensively against buffer A at 4°C. NTP:AdoCbi kinase/NTP:AdoCbi-P nucleotidyltransferase activity assays were performed in 20-μl reaction mixtures containing 90 μg of protein, 50 mM Tris-HCl (pH 8.5) at 25°C, 10 mM MgCl2, 50 μM AdoCbi, 2 mM GTP, 1 mM nicotinate mononucleotide, 0.1 mM 5,6-dimethylbenzimidazole, and 2 mM CTP, UTP, or ATP. Reaction mixtures were incubated for 4 h at 25°C. The reaction temperature of 25°C was chosen to slow the reaction. A kinetic analysis of the reaction would require an incubation temperature that would allow sufficient time for manipulations. This temperature is 25°C for CobU but has not been determined for CobY. Reactions were stopped by heating reaction mixtures to 98°C for 10 min followed by centrifugation at 14,000 × g for 10 min to pellet denatured protein.
Detection of in vitro-synthesized Cbl.
The presence of cobalamin (Cbl) in reaction mixtures was assessed using strain JE212 [metE205 ara-9 Δ299(hisG-cob)], a Cbl auxotroph. Five microliters of each in vitro AdoCbl assembly reaction mixture was spotted onto an agar overlay containing cells of strain JE212. Minimal medium (44) was supplemented with 11 mM glucose and 0.1 mM histidine. Growth of strain JE212 around the application site was indicative of the presence of Cbl. The same procedures for in vitro AdoCbl assembly and AdoCbl detection were used to assess AdoCbi-P nucleotidyltransferase activity, except that AdoCbi-P replaced AdoCbi and ATP, GTP, CTP, and UTP were tested individually. All reactions described above were performed under dim lighting.
Biochemical assays for GTP:AdoCbi-P guanylyltransferase activity.
The assay for GTP:AdoCbi-P guanylyltransferase activity was a modification of previously reported protocols (5, 24). The reaction mixtures (20 μl) contained 50 mM Tris-HCl (pH 8.5) at 25°C, 680 mM glycerol, 0.1 M NaCl, 1.25 mM Tris(2-carboxyethyl)phosphine hydrochloride (Pierce Chemical Co., Rockford, Ill.), 10 mM MgCl2, 100 μM GTP, and 1 μCi of [α-32P]GTP (800 Ci/mmol; NEN Life Science Products, Boston, Mass.). Either 7 ng of CobU enzyme or 41 ng of CobY was included in the reaction mixture in the presence or absence of 25 μM AdoCbi-P. Complete reaction mixtures were incubated at 25°C for 20 min. Reactions were terminated by the addition of 5 μl of 100 mM KCN followed by a 10-min incubation at 80°C. Samples were centrifuged for 30 s at 14,000 × g in a Marathon 16KM microcentrifuge (Fisher, Itasca, Ill.), and 5 μl of the reaction mix was spotted onto cellulose thin-layer chromatography (TLC) plates (PolygramCEL 400; Macherey-Nagel, Düren, Germany). Products and reactants were separated using ascending TLC with a solvent system of isobutyric acid-water-ammonium hydroxide (66:33:1) (24, 27). The solvent front was allowed to migrate 10 cm; the TCL plate was dried and analyzed using a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). All reactions described above were performed under dim lighting.
Amplification and cloning of cobY (ORF MTH1152).
Primers MTH1152-NdeI (5′-AGGAGGATAAATCATATGAATAGGATGATG-3′) and MTH1152-HindIII (5′-TCTTCAAGCTTCCCCAGGATG-3′) were used to amplify open reading frame (ORF) MTH1152 from chromosomal DNA of M. thermoautotrophicum ΔH (a gift from R. Wolfe, University of Illinois, Urbana-Champaign). The primer bases in boldface type changed from the reported sequence to introduce an NdeI or HindIII restriction site. The base underlined and in boldface type shows the change of the start codon from TTG to ATG. The amplified product was cloned directly into pGEM-T Vector System I as instructed by the manufacturer (Promega, Madison, Wis.) to generate plasmid pCOBY1. cobY was subcloned into plasmid pET-15b (Novagen) by way of a NdeI-SalI fragment of pCOBY1 ligated to the compatible overhangs of an NdeI-XhoI digest of plasmid pET-15b to generate plasmid pCOBY4. This resulted in the introduction of a six-histidine tag to the amino terminus of CobY. The same primers and amplified product were directly cloned into the T7 overexpression plasmid pT7-7 (39), using the NdeI and HindIII restriction sites to generate plasmid pCOBY3. Plasmids pCOBY3 and pCOBY1 were sequenced to ensure that no PCR-induced base changes had occurred. Two additional primers were used in this sequencing: MTH1152-Seq1 (5′-AACTCTGACCTCCCACTT3′) and MTH1152-seq2 (5′TCTGGTACTGCGACACA3′). All primers were from Integrated DNA Technology, Inc. (Coralville, Iowa).
Overexpression and purification of H6CobY.
The H6CobY protein was overexpressed in E. coli strain BL21(λDE3). Purification of H6CobY was achieved by nickel affinity chromatography as instructed by the manufacturer. Modifications were made to the elution conditions where elution of H6CobY was achieved by a linear gradient of 100 to 400 mM imidazole in elution buffer. Fractions containing H6CobY were pooled and dialyzed against 50 mM Tris-Cl (pH 8.0) at 4°C with 5 mM EDTA, 0.1 M NaCl, and 0.68 M glycerol for 4 h. Additional dialysis was performed against 50 mM Tris-Cl (pH 8.0) at 4°C with 1 mM dithiothreitol, 0.1 M NaCl, and 0.68 M glycerol for 12 h.
Complementation of cobU function by cobY.
S. enterica strain JE873 is unable to synthesize AdoCbl de novo due to the deletion of the cobUST genes, the final three genes in the AdoCbl biosynthetic operon (30). To test for complementation of Cbl biosynthesis, plasmids containing cobU+ (pJO52), cobY+ (pCOBY3), or the control plasmid (pT7-7) were transformed into derivatives of JE873 containing a plasmid encoding cobST+ (pJO30) or the control plasmid pSU18 (22). The resulting strains were JE4750(pSU18/pT7-7), JE4751(pSU18/pCOBY3), JE4752(pSU18/pJO52), JE4753(pJO30/pT7-7), JE4754(pJO30/pCOBY3), and JE4755(pJO30/pJO52). Four independent colonies of each strain were patched onto Luria-Bertani–ampicillin (50 μg/ml)–chloramphenicol (10 μg/ml) agar, grown for 4 h at 37°C, and replica printed onto minimal medium agar plates containing ampicillin (25 μg/ml) and chloramphenicol (5 μg/ml), with or without 15 nM (CN)2Cbl. Plates were incubated for 16 h at 37°C anaerobically in an ANA-PAK system (Scott Laboratories, Inc., Fiskeville, R.I.), using a BBL GasPak anaerobic system (Becton Dickinson, Cockeysville, Md.). Growth of the strains after 16 h indicated de novo AdoCbl biosynthesis.
RESULTS
Archaea lack an orthologue of the S. enterica CobU enzyme.
The protein coding sequences of the six sequenced archaeal genomes were analyzed for orthologues of known Cba biosynthetic enzymes from S. enterica and P. denitrificans, using the National Center for Biotechnology Information BLAST 2.0 and PSI-BLAST programs (1). The results obtained were similar to those previously reported (9, 17–20, 35) (accession numbers AJ248283.1 and AP000063.1), with the additional identification of ORF PH0377 (Pyrococcus horikoshii), ORF AF2024 (A. fulgidus), ORF MJ0955 (Methanococcus jannaschii), ORF MTH1587 (M. thermoautotrophicum ΔH), ORF PAB0026 (Pyrococcus abyssi), and ORF 2035 (Aeropyrum pernix) as the genes encoding orthologues of CobD, the Thr-P decarboxylase in S. enterica (8).
Because of the essential role of cobamides in the metabolism of several archaea, it was not surprising to find that M. jannaschii, M. thermoautotrophicum ΔH, and A. fulgidus contained orthologues to nearly all of the bacterial Cba biosynthetic enzymes (data not shown) (9, 18, 35). In P. horikoshii, P. abyssi, and A. pernix, however, our search identified orthologues to only four or five AdoCba biosynthetic enzymes, suggesting that either these organisms are unable to perform complete de novo AdoCba biosynthesis or different Cba biosynthetic enzymes have evolved in these procaryotes. Our search indicated that none of the archaea encoded an orthologue of CobU, the bifunctional NTP:AdoCbi kinase/GTP:AdoCbi-P guanylyltransferase of S. enterica (30). This result was intriguing because we found orthologues to biosynthetic enzymes that act immediately upstream and downstream of CobU in the Cba biosynthetic pathway.
M. thermoautotrophicum ΔH contains NTP:AdoCbi-P nucleotidyltransferase activity but lacks NTP:AdoCbi kinase activity.
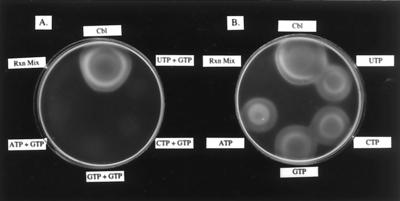
NTP:AdoCbi kinase and NTP:AdoCbi-P nucleotidyltransferase activities were measured using a bioassay that detected the presence of AdoCbl in reaction mixtures containing either AdoCbi or AdoCbi-P as substrates for enzymes present in cell extracts (CE) of M. thermoautotrophicum ΔH. If AdoCbl was synthesized in vitro, the Cbl auxotrophy of tester strain JE212 was corrected, resulting in growth around the point of application of the sample. AdoCbl synthesis was not detected in any of the reaction mixtures containing AdoCbi as substrate (Fig. 2A), suggesting that M. thermoautotrophicum ΔH extracts were unable to convert AdoCbi to AdoCbl in vitro. In contrast, AdoCbi-P was converted to AdoCbl by the same CE regardless of the source of nucleoside monophosphate donor (Fig. 2B), suggesting that M. thermoautotrophicum ΔH contained AdoCbi-P nucleotidyltransferase activity. Possible explanations for the inability to synthesize AdoCbl from AdoCbi were the lack of AdoCbi kinase activity in the CE, lability of the kinase activity to oxygen inactivation, or enzyme inactivation due to the conditions used in the assay. Previous findings in S. enterica (8) made the first explanation attractive. Briefly, these findings suggested that the NTP:AdoCbi kinase activity of CobU is not required for de novo AdoCbl biosynthesis. If this idea were correct, the guanylyltransferase activity of CobU would be sufficient under conditions where growth of the cell depends on de novo synthesis of AdoCbl.
FIG. 2.
In vitro synthesis of AdoCbl by M. thermoautotrophicum ΔH CE. (A) AdoCbl biosynthesis from AdoCbi requires AdoCbi kinase and AdoCbi-P nucleotidyltransferase activities; (B) AdoCbl biosynthesis from AdoCbi-P requires only AdoCbi-P nucleotidyltransferase activities. Both panels show reaction mixtures spotted onto top agar seeded with cells of a Cbl auxotroph (strain JE212). Rxn mix, reaction mixture without CE. Faint growth seen in reaction mixtures in plate A was due to background cobamide in the CE.
Identification of the gene encoding NTP:AdoCbi-P nucleotidyltransferase in archaea.
To identify genes encoding NTP:AdoCbi-P nucleotidyltransferase activity, we took a comparative genomics approach. We analyzed ORFs encoded by the three or four genes on either side on an identified Cba biosynthetic enzyme orthologue for motifs that would be suggestive of nucleotidyltransferase function. This was rational given the common occurrence of clustering and/or operon organization of genes encoding enzymes of a particular pathway in procaryotes.
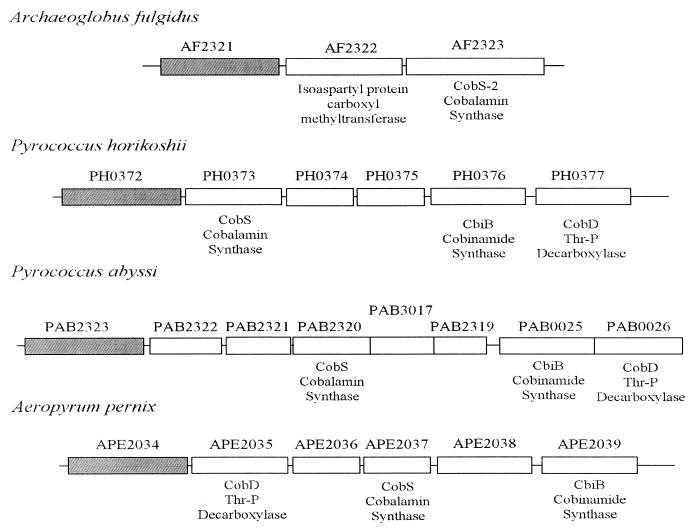
Analysis of the M. jannaschii and M. thermoautotrophicum ΔH genome sequences showed that genes encoding proposed Cba biosynthetic enzymes were distributed randomly throughout their genomes without notable clustering or operon organization (9, 35). A. fulgidus, however, displayed significant clustering and possible operon organization of many of the Cba biosynthetic genes, but no ORF of interest was identified in this cluster (18). Comparison of the location of genes flanking cobS-2 (AF2323) in A. fulgidus and cobS (PH0373) in P. horikoshii (Fig. 3) identified ORFs AF2321 (A. fulgidus) and PH0372 (P. horikoshii) as ORFs potentially encoding a previously unidentified Cba biosynthetic enzyme. This was based on their similarity to each other and their proximity to ORFs encoding the cobalamin(5′-phosphate) synthase (CobS in S. enterica) enzyme. Orthologues of ORFs AF2321 and PH0372 were also identified in M. thermoautotrophicum ΔH (MTH1152) and M. jannaschii (MJ1117). The amino-terminal regions of all predicted proteins showed homology to the amino-terminal regions of sugar 1-phosphate nucleotidyltransferases (Fig. 4) (43), or XDP-sugar pyrophosphorylases, as they are more commonly referred to (7). Recently, all four of these ORFs were placed together in a cluster of orthologous groups, and their homology to sugar 1-phosphate nucleotidyltransferases was noted (20).
FIG. 3.
ORFs surrounding putative cobS orthologues in A. fulgidus, P. horikoshii, P. abysii, and A. pernix. Reported ORF designation is indicated above each rectangle, with the reported annotation below. We have changed the annotation for the CobD orthologue to Thr-P decarboxylase based on the demonstrated biochemical function of this protein in S. enterica serovar Typhimurium. ORFs not annotated are reported as conserved hypotheticals. Grey boxes indicate putative AdoCbi-P nucleotidyltransferases that are orthologous. The overlapping boxes shown for P. abysii indicate proposed overlapping ORFs in the sequenced genome (accession number AJ248283.1).
FIG. 4.
Homology of the amino-terminal domain of archaeal orthologues to the amino-terminal domain of members of the sugar 1-phosphate nucleotidyltransferase superfamily. The amino-terminal domains of the six putative archaeal AdoCbi-P nucleotidyltransferases are aligned with three members of the sugar 1-phosphate nucleotidyltransferase superfamily encoded by E. coli. MobA, molybdopterin-guanine dinucleotide biosynthesis protein A (16); GlmA, N-acetylglucosamine-1-phosphate uridylyltransferase (7); RffH, glucose-1-phosphate thymidylyltransferase (21). Boldface residues are amino acids that show similarity in seven or more sequences. Boldface and underlined residues are amino acids conserved in all nine sequences. Superscript numbers indicated amino acid number in each protein.
Recently, the genome sequences of two more archaea, Pyrococcus abyssi (R. Heilig; accession number AJ248283.1) and Aeropyrum pernix (T. Tanaka et al.; accession number AP000063.1), were released. Again, we did not find an orthologue of CobU in these archaea, but CobY orthologues were present in both organisms encoded by ORFs PAB2323 and APE2034, respectively, and show the same amino-terminal sequence similarity as sugar 1-phosphate nucleotidyltransferases (data not shown). In both cases, these ORFs were found near the gene encoding the CobS, CobD, and CbiB orthologues in each organism (Fig. 3).
The H6MTH1152 protein has GTP:AdoCbi-P guanylyltransferase activity.
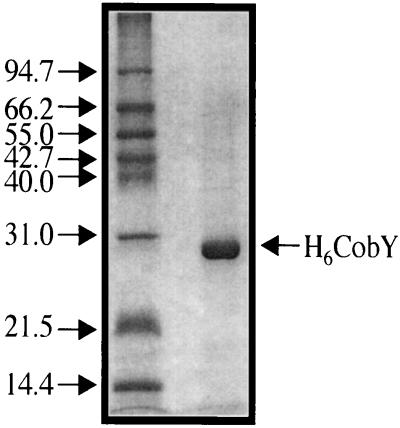
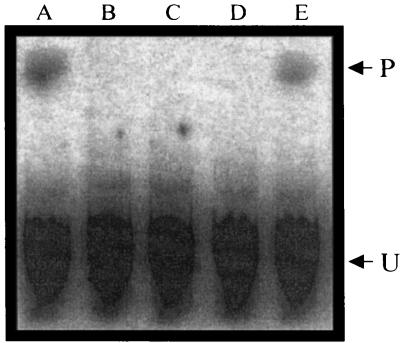
To permit biochemical analysis of the protein encoded by MTH1152, the gene was cloned into a vector that introduced a hexahistidine tag at the amino terminus of the protein. This tagged protein (H6MTH1152) was subsequently purified to near homogeneity by nickel affinity chromatography (Fig. 5). Purified H6MTH1152 was tested for GTP:AdoCbi-P guanylyltransferase activity by monitoring formation of the product AdoCbi-GDP. This product was detected only when AdoCbi-P was included in the reaction mixture (Fig. 6, lane E). Heating the purified protein to 65°C for 10 min prior to starting the reaction did not abolish activity (data not shown). Since under these conditions CobU enzyme from S. enterica is inactivated within 30 s (M. G. Thomas and J. C. Escalante-Semerena, unpublished results), and since E. coli CobU enzyme is highly similar to S. enterica CobU (91% similarity, 82% identity), it was unlikely that the activity shown in Fig. 6 was due to background levels of E. coli CobU. This result strongly suggested that ORF MTH1152 of M. thermoautotrophicum ΔH encoded the AdoCbi-P nucleotidyltransferase, hereafter referred to as CobY (encoded by the gene cobY). The specific activity of H6CobY for GTP:AdoCbi-P guanylyltransferase activity was 17 ± 3 nmol/min/mg. Under identical assay conditions, CobU had a specific activity of 194 ± 6 nmol/min/mg. It is difficult to compare these activities directly because the optimal assay conditions for CobY activity are yet to be determined. It is anticipated that the optimal activity for CobY would be obtained at 65°C, the physiologically relevant temperature for M. thermoautotrophicum ΔH, not 25°C as tested here. Furthermore, as will be discussed in more detail below, the enzymatic mechanisms and structures of CobU and CobY are anticipated to be very different, thereby complicating direct comparison of the their activities. Even though we showed that CobY had guanylyltransferase activity, we refer to the enzyme as an NTP:AdoCbi-P nucleotidyltransferase until a more thorough substrate specificity analysis is performed.
FIG. 5.
Denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified H6CobY protein. Numbers on the left indicate molecular weight size standards in kilodaltons. Five micrograms of H6CobY was electrophoresed.
FIG. 6.
Detection of the AdoCbi-P guanylyltransferase activity of H6CobY. Representative TLC separation and PhosphorImager visualization of products and reactants of guanylyltransferase reaction mixtures. P (product), (CN)2Cbi-GDP; U, unincorporated [α-32P]GTP. Reaction mixtures contained only CobU, AdoCbi-P, and [α-32P]GTP (lane A), [α-32P]GTP (lane B), AdoCbi-P and [α-32P]GTP (lane C), H6CobY and [α-32P]GTP (lane D), and H6CobY, AdoCbi-P, and [α-32P]GTP (lane E).
H6CobY was also tested for direct transfer of GDP from GTP to AdoCbi. If CobY catalyzed this transfer, the need for an NTP:AdoCbi kinase would be eliminated. H6CobY did not produce any detectable AdoCbi-GDP under the following substrate and enzyme concentrations: 135 μM AdoCbi, 100 μM GTP, and 0.6 μg of H6CobY (data not shown). Additionally, H6CobY did not have any detectable NTP:AdoCbi kinase activity when either 100 μM ATP or GTP was used as a γ-phosphate donor, suggesting that this enzyme was not bifunctional (data not shown). Similar results were found when assaying CE of E. coli that expressed CobY without the hexahistidine tag; therefore, the presence of the affinity tag does not appear to affect CobY activity.
cobY complements an S. enterica cobU mutant.
Identification of CobY as an NTP:AdoCbi-P nucleotidyltransferase that does not have NTP:AdoCbi kinase activity allowed us to assess the hypothesis that in S. enterica, the NTP:AdoCbi kinase activity is not required for de novo AdoCbl biosynthesis. To test this hypothesis, we used an S. enterica strain that carried two relevant mutations. First, the cobU, cobS, and cobT genes were deleted, abolishing the strain's ability to perform the late steps of AdoCbl biosynthesis (Fig. 1). Second, the strain had a mutation in metE, the Cbl-independent methionine synthase, thus rendering growth of the strain dependent on the Cbl-dependent methionine synthase, MetH. Growth on medium lacking methionine would indicate that de novo Cbl biosynthesis was restored in the strains tested.
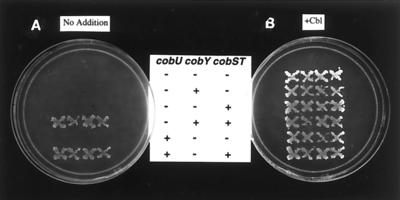
A plasmid containing a wild-type allele of cobY or cobU was introduced into this strain in the presence or absence of plasmid pJO30 (cobST+). Expression of cobU (from plasmid pJO52) and cobY (from plasmid pCOBY3) was under the control of the T7 promoter and ribosome-binding site. It was assumed that residual expression of these genes in the absence of T7 RNA polymerase would allow us to assess complementation. Under anaerobic growth conditions, where de novo AdoCbl biosynthesis can occur in S. enterica, complementation of AdoCbl biosynthesis was observed when either cobY or cobU was provided in trans (Fig. 7). Under aerobic growth conditions (where AdoCbi synthesis does not occur, but Cbl can be synthesized from exogenously supplied Cbi), only the strain containing the plasmid carrying cobU+ was able to synthesize AdoCbl from exogenously supplied Cbi (data not shown). This result was consistent with the finding that cobY did not have AdoCbi kinase activity. This was the first in vivo evidence suggesting that de novo AdoCbl biosynthesis in S. enterica does not require the AdoCbi kinase activity, in good agreement with the model that the NTP:AdoCbi kinase activity of CobU functions in Cbi salvaging (8). Lack of complementation by cobY+ was not due to sensitivity of CobY to oxygen inactivation, because the same strain grew under aerobic conditions on medium where cobyric acid substituted for Cbi (K. R. Brushaber and J. C. Escalante-Semerena, unpublished results). Our working hypothesis predicts this should be the case, since cobyric acid should be converted to AdoCbi-P, not AdoCbi. Generation of AdoCbi-P renders the lack of a kinase activity in CobY irrelevant.
FIG. 7.
Complementation of a S. enterica cobU mutant strain by cobY. Complementation was assessed by introducing plasmids into strain JE873, a Cbl auxotroph, containing cobY or cobU in the presence or absence of a plasmid containing cobST. Plate A was incubated under conditions that demanded de novo AdoCbl biosynthesis; plate B shows the growth response to exogenously added Cbl. The genotypes of the plasmids carried by each strain are indicated in the center. Plates show four independent colonies of each strain tested.
DISCUSSION
The Cba biosynthetic enzyme in the methanogenic archaeon M. thermoautotrophicum ΔH that catalyzes the conversion of AdoCbi-P to AdoCbi-GDP has been identified. Biochemical and genetic data strongly support the conclusion that this enzyme, referred to as CobY, is the nonorthologous replacement of CobU GTP:AdoCbi-P guanylyltransferase activity. CobY, however, does not possess the NTP:AdoCbi kinase activity also seen with CobU (24, 41) and its orthologue in P. dentrificans (5). Interestingly, under the assay conditions tested, NTP:AdoCbi kinase activity could not be detected in CE of M. thermoautotrophicum ΔH, suggesting that this species may not possess this activity. The finding that cobY complemented an S. enterica cobU mutant supports the previously proposed model that NTP:AdoCbi kinase activity is not required for de novo AdoCbl biosynthesis. These findings strongly support the idea that relevant Cbi intermediates during Cba biosynthesis in S. enterica, and most likely M. thermoautotrophicum ΔH and other procaryotes, are AdoCbi-P and AdoCbi-GDP, not AdoCbi as previously thought.
CobU and CobY are nonorthologous enzymes.
Comparison of the amino acid sequences of CobU and CobY gave the first evidence that CobU and CobY do not share a common ancestor. In support of this conclusion, biochemical analysis of CobU determined the enzyme catalyzes GTP:AdoCbi-P guanylyltransferase activity via a double displacement mechanism wherein CobU first forms a covalently linked CobU-GMP intermediate and subsequently transfers the GMP moiety onto AdoCbi-P (24, 41). CobY, however, shows amino acid sequence similarity with a superfamily of enzymes that have been studied extensively and proceed via a single displacement mechanism (12). It is anticipated that CobY will form a tertiary complex with GTP and AdoCbi-P followed by direct transfer of the GMP moiety to AdoCbi-P without an enzyme-linked intermediate. In support of this, initial attempts to detect a CobY-GMP intermediate were unsuccessful (data not shown). Therefore, the enzymatic mechanism of AdoCbi-P nucleotidyltransferase activity of CobU and CobY are likely different.
Further support for the conclusion that CobU and CobY have distinct evolutionary origins comes from the structural analysis of CobU (41, 42) and the structural analysis of GlmU (7), a member of the sugar 1-phosphate nucleotidyltransferase superfamily, to which CobY belongs. CobU was found to be structurally and topologically similar to the RecA protein from E. coli (42). The topology of these proteins is, to date, restricted to these two enzymes (42), although they have a Rossmann fold commonly found in nucleotide-binding proteins (28). This suggests that CobU and RecA share a common nucleotide-binding ancestor. CobY, on the other hand, will most likely be structurally and topologically similar to the nucleotidyltransferase domain of GlmU. This domain, while having a fold similar to a Rossmann fold, is topologically and structurally distinct from CobU. In fact, structural homologues of the nucleotidyltransferase domain of GlmU could not be found (7). Therefore, CobY does not share a common ancestor with CobU. Further biochemical and structural analysis of CobY is in progress.
Why have two enzymes evolved to catalyze the same reaction?
Since cobY complements an S. enterica cobU mutant, this bacterium, and likely other Cba-synthesizing bacteria, does not require NTP:AdoCbi kinase activity during de novo AdoCbl biosynthesis. Then, what is the function of the kinase activity of CobU? And why would M. thermoautotrophicum ΔH and other archaea not have a comparable NTP:AdoCbi kinase? One hypothesis that has been put forward for S. enterica is that the kinase activity of CobU is essential during the assimilation of exogenous Cbi, not during de novo synthesis (8). The in vivo data reported herein support this conclusion since cobY complemented a cobU mutant during de novo AdoCbl biosynthesis but not during Cbi assimilation. It could be hypothesized that M. thermoautotrophicum ΔH, and possibly other archaea, cannot assimilate exogenous Cbi because it lacks NTP:AdoCbi kinase activity. A report in the literature appears to contradict this conclusion (37). Stupperich et al. showed M. thermoautotrophicum Marburg can assimilate exogenous Cbi into Coα-(5-hydroxybenzimidazolyl)cobamide (37), the endogenous Cba of this archaeon. These results suggest that the Marburg strain may contain the AdoCbi kinase activity that was not detected in the ΔH strain. This apparent assimilation of cobinamide, however, could be achieved via an alternative pathway that circumvents the need for an AdoCbi kinase enzyme. For example, the 1-amino-2-propanol moiety of AdoCbi may be removed by an amide hydrolase, generating AdoCby, which would be converted to AdoCbi-P by the addition of AP-P by the cobinamide-phosphate synthase (Fig. 1). The resulting AdoCbi-P product would enter the pathway to Coα-(5-hydroxybenzimidazolyl)cobamide (Fig. 1). Current work is exploring these possibilities.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM40313 to J.C.E.-S.
We thank M. Fonseca for purified CobA enzyme.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Miller W, Lipmann D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balachandran S, Vishwakarma R A, Monaghan S M, Prelle A, Stamford N P, Leeper F J, Battersby A R. Biosynthesis of porphyrins and related macrocycles. Part 42. Pulse labelling experiments concerning the timing of cobalt insertion during vitamin B12 biosynthesis. J Chem Soc Perkin Trans. 1994;1994:487–491. [Google Scholar]
- 3.Battersby A R. How nature builds the pigments of life: the conquest of vitamin B12. Science. 1994;264:1551–1557. doi: 10.1126/science.8202709. [DOI] [PubMed] [Google Scholar]
- 4.Blanche F, Cameron B, Crouzet J, Debussche L, Thibaut D, Vuilhorgne M, Leeper F J, Battersby A R. Vitamin B12: how the problem of its biosynthesis was solved. Angew Chem Int Ed Engl. 1995;34:383–411. [Google Scholar]
- 5.Blanche F, Debussche L, Famechon A, Thibaut D, Cameron B, Crouzet J. A bifunctional protein from Pseudomonas denitrificans carries cobinamide kinase and cobinamide phosphate guanylyltransferase activities. J Bacteriol. 1991;173:6052–6057. doi: 10.1128/jb.173.19.6052-6057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanche F, Robin C, Couder M, Faucher D, Cauchois L, Cameron B, Crouzet J. Purification, characterization, and molecular cloning of S-adenosyl-l-methionine:uroporphyrinogen III methyltransferase from Methanobacterium ivanovii. J Bacteriol. 1991;173:4637–4645. doi: 10.1128/jb.173.15.4637-4645.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K, Pompeo F, Dixon S, Mengin-Lecreulx D, Cambillau C, Bourne Y. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 1999;18:4096–4107. doi: 10.1093/emboj/18.15.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brushaber K R, O'Toole G A, Escalante-Semerena J C. CobD, a novel enzyme with l-threonine-O-3-phosphate decarboxylase activity is responsible for the synthesis of (R)-1-amino-propanol O-2-phosphate, a proposed new intermediate in cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:2684–2691. doi: 10.1074/jbc.273.5.2684. [DOI] [PubMed] [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1072. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Eisenreich W, Bacher A. Biosynthesis of 5-hydroxybenzimidazolylcobamid (factor III) in Methanobacterium thermoautotrophicum. J Biol Chem. 1991;266:23840–23849. [PubMed] [Google Scholar]
- 11.Fazzio T G, Roth J R. Evidence that the CysG protein catalyzes the first reaction specific to B12 synthesis in Salmonella typhimurium, insertion of cobalt. J Bacteriol. 1996;178:6952–6959. doi: 10.1128/jb.178.23.6952-6959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey P A, Richard J P, Ho H T, Brody R S, Sammons R D, Sheu K F. Stereochemistry of selected phosphotransferases and nucleotidyltransferases. Methods Enzymol. 1982;87:213–235. doi: 10.1016/s0076-6879(82)87016-x. [DOI] [PubMed] [Google Scholar]
- 13.Friedmann H C, Thauer R K, editors. Macrocyclic tetrapyrrole biosynthesis in bacteria. Vol. 3. New York, N.Y: Academic Press, Inc.; 1992. [Google Scholar]
- 14.Harms U, Thauer R K. Identification of the active histidine in the corrinoid protein MtrA of the energy-conserving methyltransferase complex from Methanobacterium thermoautotrophicum. Eur J Biochem. 1997;250:783–788. doi: 10.1111/j.1432-1033.1997.00783.x. [DOI] [PubMed] [Google Scholar]
- 15.Hogenkamp H P C. The chemistry of cobalamins and related compounds. In: Babior B M, editor. Cobalamin: biochemistry and pathophysiology. New York, N.Y: John Wiley & Sons; 1975. pp. 21–74. [Google Scholar]
- 16.Iobbi-Nivol C, Palmer T, Whitty P W, McNairn E, Boxer D H. The mob locus of Escherichia coli K12 required for molybdenum cofactor biosynthesis is expressed at very low levels. Microbiology. 1995;141:1663–1671. doi: 10.1099/13500872-141-7-1663. [DOI] [PubMed] [Google Scholar]
- 17.Kawarabayasi Y, Sawada M, H. H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 18.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavag A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V, Mushegian A R, Galperin M Y, Walker D R. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 20.Makarova K S, Aravind L, Galperin M Y, Grishin N V, Tatusov R L, Wolf Y I, Koonin E V. Comparative genomics of the archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 21.Marolda C L, Valvano M A. Genetic analysis of the dTDO-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez E, Bartolomé B, de la Cruz F. pACY184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 23.Müller G, Zipfel F, Hliney K, Savvidis E, Hertle R, Traub-Eberhard U, Scott A I, Williams H J, Stolowich N J, Santander P J, Warren M, Blanche F, Thibaut D. Timing of cobalt insertion in vitamin B12 biosynthesis. J Am Chem Soc. 1991;113:9893–9895. [Google Scholar]
- 24.O'Toole G A, Escalante-Semerena J C. Purification and characterization of the bifunctional CobU enzyme of Salmonella typhimurium LT2. Evidence for a CobU-GMP intermediate. J Biol Chem. 1995;270:23560–23569. doi: 10.1074/jbc.270.40.23560. [DOI] [PubMed] [Google Scholar]
- 25.Raux E, Lanois A, Warren M J, Rambach A, Thermes C. Cobalamin (vitamin B12) biosynthesis: identification and characterization of a Bacillus megaterium cobI operon. Biochem J. 1998;335:159–166. doi: 10.1042/bj3350159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rondon M R, Trzebiatowski J R, Escalante-Semerena J C. Biochemistry and molecular genetics of cobalamin biosynthesis. Prog Nucleic Acid Res Mol Biol. 1997;56:347–384. doi: 10.1016/s0079-6603(08)61010-7. [DOI] [PubMed] [Google Scholar]
- 27.Ronzio R A, Barker H A. Enzymic synthesis of guanosine diphosphate cobinamide by extracts of propionic acid bacteria. Biochemistry. 1967;6:2344–2354. doi: 10.1021/bi00860a009. [DOI] [PubMed] [Google Scholar]
- 28.Rossmann M G, Argos P. A comparison of the heme binding pocket in globins and cytochrome b5. J Biol Chem. 1975;250:7525–7532. [PubMed] [Google Scholar]
- 29.Roth J R, Lawrence J G, Bobik T A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 30.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santander P J, Roessner C A, Stolowich N, Holderman M T, Scott A I. How corrinoids are synthesized without oxygen: nature's first pathway to vitamin B12. Chem Biol. 1997;4:659–666. doi: 10.1016/s1074-5521(97)90221-0. [DOI] [PubMed] [Google Scholar]
- 32.Scherer P, Höllriegl V, Krug C, Bokel M, Renz P. On the biosynthesis of 5-hydroxybenzimidazolylcobamide (vitamin B12-factor III) in Methanosarcina barkeri. Arch Microbiol. 1984;138:354–359. [Google Scholar]
- 33.Scott A I. How nature synthesizes vitamin B12. A survey of the last four billion years. Angew Chemie Int Ed Engl. 1993;32:1223–1243. [Google Scholar]
- 34.Scott A I. On the duality of the mechanism of ring contraction in vitamin B12 biosynthesis. Heterocycles. 1994;39:471–476. [Google Scholar]
- 35.Smith D R, Doucette-Stamm L A, Deloughery C, Hongmei L, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, G. S, Goyal A, Pietrokovski S, Church G M, Daniels C H, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer J B, Stolowich N J, Roessner C A, Min C, Scott A I. Biosynthesis of vitamin B12: ring contraction is preceded by incorporation of molecular oxygen into precorrin-3. J Am Chem Soc. 1993;115:11610–11611. [Google Scholar]
- 37.Stupperich E, Steiner I, Eisinger H J. Substitution of Coα-(5-hydroxybenzimidazolyl)cobamide (factor III) by vitamin B12 in Methanobacterium thermoautotrophicum. J Bacteriol. 1987;169:3076–3081. doi: 10.1128/jb.169.7.3076-3081.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh S-J, Escalante-Semerena J C. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Wiley Interscience; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 40.Thauer R K. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 41.Thompson T, Thomas M G, Escalante-Semerena J C, Rayment I. Three-dimensional structure of the guanylylated form of adenosylcobinamide kinase/adenosylcobinamide-phosphate guanylyltransferase enzyme of Salmonella typhimurium at 2.3Å resolution. Biochemistry. 1999;38:12995–13004. doi: 10.1021/bi990910x. [DOI] [PubMed] [Google Scholar]
- 42.Thompson T B, Thomas M G, Escalante-Semerena J C, Rayment I. Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined at 2.3Å resolution. Biochemistry. 1998;37:7686–7695. doi: 10.1021/bi973178f. [DOI] [PubMed] [Google Scholar]
- 43.Thorson J S, Kelly T M, Liu H-W. Cloning, sequencing, and overexpression in Escherichia coli of the α-d-glucose-1-phosphate cytidyltransferase gene isolated from Yersinia pseudotuberculosis. J Bacteriol. 1994;176:1840–1849. doi: 10.1128/jb.176.7.1840-1849.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification, and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 45.Wang J, Stolowich N J, Santander P J, Ho P J, Scott A I. Biosynthesis of vitamin B12: concerning the identity of the two-carbon fragment eliminated during anaerobic formation of cobyrinic acid. Proc Natl Acad Sci USA. 1996;93:14320–14322. doi: 10.1073/pnas.93.25.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren M J, Bolt E L, Ropessner C A, Scott A I, Spencer J B, Woodcock S C. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem J. 1994;302:837–844. doi: 10.1042/bj3020837. [DOI] [PMC free article] [PubMed] [Google Scholar]