Abstract
Background and Aims:
Interferon (IFN) signaling is critical to the pathogenesis of alcohol-associated hepatitis (AH), yet the mechanisms for activation of this system are elusive. We hypothesize that host-derived 5S rRNA pseudogene (RNA5SP) transcripts regulate IFN production and modify immunity in AH.
Approach and Results:
Mining of transcriptomic datasets revealed that in patients with severe alcohol-associated hepatitis (sAH), hepatic expression of genes regulated by IFNs was perturbed and gene sets involved in IFN production were enriched. RNA5SP transcripts were also increased and correlated with expression of type I IFNs. Interestingly, inflammatory mediators upregulated in sAH, but not in other liver diseases, were positively correlated with certain RNA5SP transcripts. Real-time quantitative PCR demonstrated that RNA5SP transcripts were upregulated in peripheral blood mononuclear cells (PBMCs) from patients with sAH. In sAH livers, increased 5S rRNA and reduced nuclear MAF1 (MAF1 homolog, negative regulator of RNA polymerase III) protein suggested a higher activity of RNA polymerase III (Pol III); inhibition of Pol III reduced RNA5SP expression in monocytic THP-1 cells. Expression of several RNA5SP transcript-interacting proteins was downregulated in sAH, potentially unmasking transcripts to immunosensors. Indeed, siRNA knockdown of interacting proteins potentiated the immunostimulatory activity of RNA5SP transcripts. Molecular interaction and cell viability assays demonstrated that RNA5SP transcripts adopted Z-conformation and contributed to ZBP1-mediated caspase-independent cell death.
Conclusions:
Increased expression and binding availability of RNA5SP transcripts was associated with hepatic IFN production and inflammation in sAH. These data identify RNA5SP transcripts as a potential target to mitigate inflammation and hepatocellular injury in AH.
INTRODUCTION
Alcohol-associated hepatitis (AH), a distinct clinical syndrome with variable degrees of severity, can lead to severe complications and is associated with high mortality.[1] AH is characterized by steatosis, cell death, inflammatory cell infiltration, and fibrosis.[2,3] Damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) activate immune cells, resulting in inflammation. The immunologic mechanisms involved in the non-resolving hepatic inflammation characterizing AH are not completely understood.
Three types of interferons (IFNs) (I, II, and III) impart context and cell type-specific, overlapping, but often distinct, transcriptional output through regulating expression of interferon-stimulated genes (ISGs).[4] IFN signaling and its components are critical to modulating liver immunity.[5,6] Indeed, hepatic IFN and ISG expression is increased in mice after ethanol exposure.[7,8] IRF3, a critical transcription factor regulating expression of IFNs, is activated in the livers of ethanol-fed mice and patients with AH. IRF3 exhibits cell type-specific (eg, hepatocytes vs. nonparenchymal cells) roles in the pathogenesis of ethanol-induced liver injury in murine models of alcohol-associated liver disease (ALD).[7–10] Deficiency of IRF3 or its activators, cyclic guanosine monophosphate-AMP synthase (cGAS) and stimulator of interferon genes (STING), protects against ethanol-induced liver injury in mice.[7–9] While these data suggest a role for IFN signaling in ALD, the potential impact of IFN signaling on the pathogenesis of AH in patients is still uncertain.
Defining how the IFN system is activated in AH is essential to developing therapeutic approaches. Host-derived nucleic acids can activate IFN production and inflammation after being recognized by various immune sensing receptors.[11–13] For instance, double-stranded RNA derived from host retroelements, forming left-handed Z-form nucleic acids (Z-NAs), acts as a Zα-domain ligand and activates Zα domain-containing Z-DNA binding protein 1 (ZBP1, also known as DAI or DLM-1).[14] ZBP1, an ISG, induces type I IFN production, activates innate immune responses, and engages in inflammation and multiple modes of cell death.[15] More than 500 5S ribosomal RNA pseudogenes (RNA5SPs, RNA5SP20–RNA5SP536) are transcribed in humans. RNA5SP transcripts are strongly induced and their interacting proteins are downregulated by viral infection, allowing these unshielded transcripts to activate the innate immune sensor RIG-I to promote IFN production and immune responses.[16] The Z-conformation exists in a large variety of noncoding RNAs and can be induced/recognized by Zα domain-containing proteins.[17] How RNA5SP transcripts contribute to the pathogenesis of AH and whether they are able to adopt Z-conformation to activate ZBP1 has not been studied.
We hypothesized that RNA5SP transcripts promote IFN production, modify immunity, and serve as ligands of ZBP1 in AH. We discovered that a group of RNA5SP transcripts, produced via RNA polymerase III (Pol III), is specifically upregulated in severe AH (sAH) compared with liver diseases of other etiologies, and associated with IFN production and hepatic inflammation. Our findings suggest that RNA5SP transcripts can serve as a novel type of host-derived DAMP that contributes to the pathogenesis of AH.
MATERIALS AND METHODS
Transcriptomic datasets
GSE28619: DNA microarray profiling in livers from healthy controls (n = 7) and patients with sAH (n = 15),[18] generating List 1 representing the genes changed in sAH (cutoff: ≥ 1.2). GSE143318: RNA sequencing using livers from healthy controls (n = 5) and explanted livers from patients with acute sAH (n = 5),[19] generating List 2 representing the genes changed in sAH (cutoff: ≥ 2.0). DbGaP_phs001807: RNA sequencing profiling hepatic gene expression in nondiseased human livers (healthy controls, n = 10), early stage of alcohol-associated steatohepatitis (ASH, n = 12), sAH (n = 18, generating List 3 representing the genes changed in sAH, cutoff: ≥ 2.0), explanted livers from sAH (AH_Expl, n = 10, generating List 4 representing the genes changed in sAH, cutoff: ≥ 2.0), NAFLD (n = 8), noncirrhotic HCVinfection (n = 9), and compensated HCV-related cirrhosis (HCV_Cirr, n = 9).[20]
Human tissues
Liver specimens from patients with sAH and healthy control donors were provided by the Clinical Resources for Alcoholic Hepatitis Investigations at Johns Hopkins University. Peripheral blood mononuclear cells (PBMCs) from patients with sAH or non-NASH were obtained from the Northern Ohio Alcohol Center biorepository (NCT03224949). The studies were approved by the IRBs of all participating institutions for the collection of human samples. Those patients with Modified Maddrey’s Discriminant Function (mDF) > 32 or Model for End-stage Liver Disease (MELD) score > 20 were considered to have sAH.
For additional and detailed materials and methods, please refer to Supporting Information (http://links.lww.com/HEP/A53).
RESULTS
Gene expression is regulated by IFNs and IFN production increased in sAH livers
To understand whether IFNs play a role in the development of sAH, 4 lists of genes representing changes in hepatic gene expression in sAH were generated from 3 transcriptomic datasets. Concordantly changed genes (466 upregulated and 369 downregulated) were interrogated using Interferome (http://www.interferome.org/), a program enabling reliable identification of interferon-regulated genes (IRGs). Overall, 93.1% (434/466) of concordantly upregulated and 68.8% (254/369) of concordantly downregulated genes were identified as IRGs; nearly all identified IRGs are known to be regulated by type I or type II, but not type III IFNs (Figure 1A). There were a large number of nonconcordantly changed genes among the 4 lists of genes, likely a result of different methods used for transcriptomic profiling between the datasets. In DbGaP_phs001807, as expected, large proportions of perturbed genes were also IRGs in liver diseases of other etiologies; however, more perturbed IRGs were found in sAH livers (including sAH and AH_Expl) (Supporting Figure 1A, http://links.lww.com/HEP/A54). Upset plot visualization of the IRGs identified the largest intersection of sAH and AH_Expl livers, compared with other intersections of the diseased livers (Supporting Figure 1B, 1C, http://links.lww.com/HEP/A54 and Supporting Table 5, http://links.lww.com/HEP/A59). Gene set enrichment analysis (GSEA) revealed that genes involved in IFN production were significantly enriched in sAH livers compared with healthy controls (GSE143318) (Figure 1B, Supporting Figure 2, http://links.lww.com/HEP/A54).
FIGURE 1.
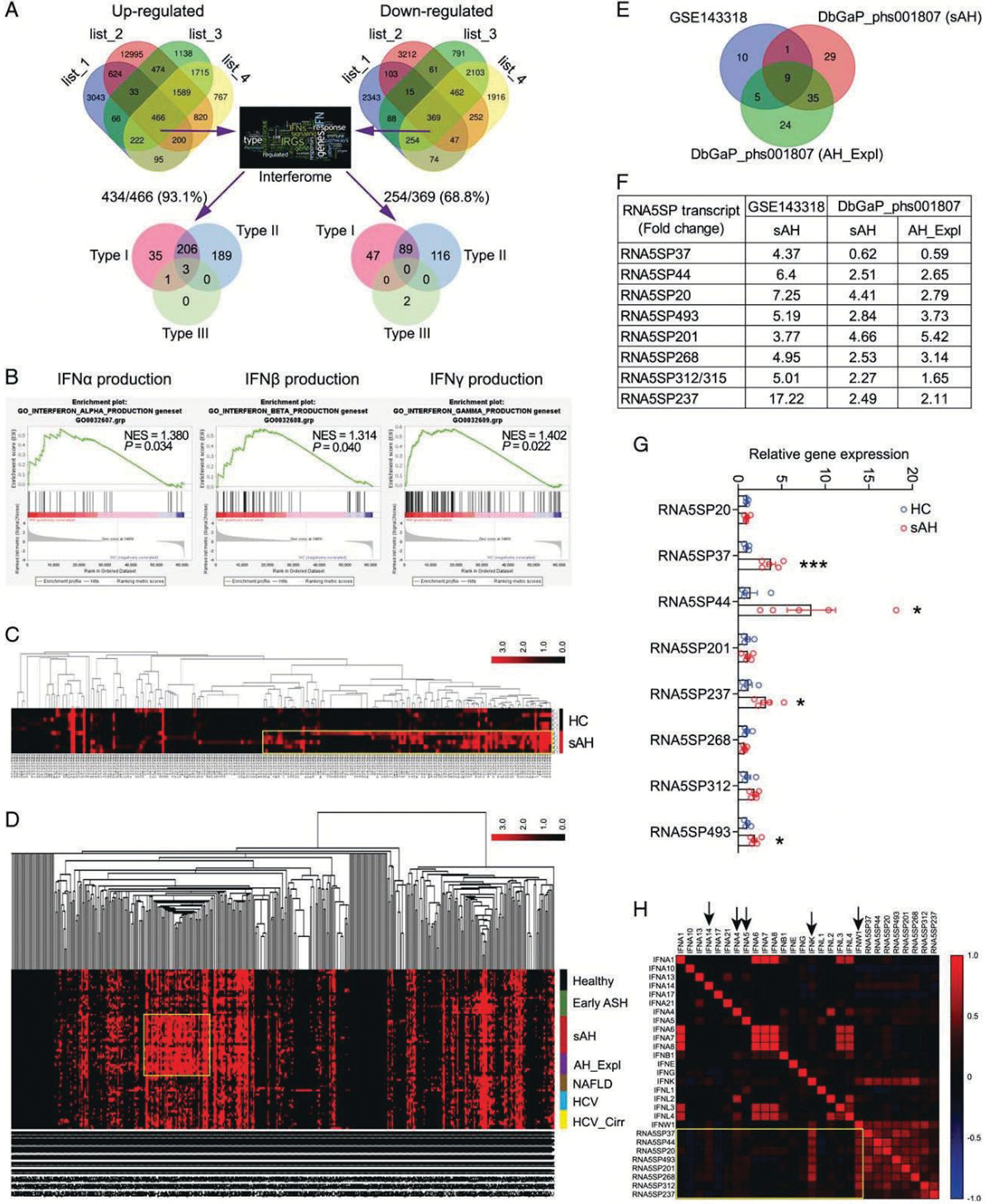
Gene expression in sAH livers is regulated by IFNs which correlate with RNA5SP expression. (A) The concordantly changed genes were interrogated against Interferome. List 1 to 4 are changed genes in sAH livers compared with healthy controls. (B) GSEA analysis of GSE143318 identified the increased production of IFNs in sAH livers. (C) Cluster analysis of 219 RNA5SP transcripts in GSE143318. Transcripts with TPM values of zero were removed. (D) Cluster analysis of 485 RNA5SP transcripts in DbGaP_phs001807. (E) Venn diagram showing the significantly and commonly changed RNA5SP transcripts in sAH livers. (F) Fold changes of the RNA5SP transcripts from (E). (G) Quantitative PCR validation of the upregulation of RNA5SP transcripts in sAH livers. *p < 0.05, ***p < 0.001. (H) Correlation matrix of the TPMs of RNA5SP transcripts and IFNs in sAH livers from GSE143318 and DbGaP_phs001807, plotted based on Spearman correlation test (n = 33). Abbreviations: GSEA, Gene Set Enrichment Analysis; HC, healthy control; IFN, interferon; RNA5SP, 5S rRNA pseudogene; sAH, severe alcohol-associated hepatitis.
Host-derived nucleic acids can activate IFN production. A cluster of RNA5SP transcripts with higher expression was identified in sAH livers from GSE143318 (Figure 1C). Moreover, cluster analysis of DbGaP_phs001807 identified that some RNA5SP transcripts were uniquely upregulated in sAH livers (including sAH and AH_Expl), but not in livers from healthy controls, early stage of ASH, NAFLD, noncirrhotic HCV, and cirrhotic HCV (Figure 1D). Nine commonly and significantly altered RNA5SP transcripts were identified from these 2 datasets, with the sequence of RNA5SP312 identical to RNA5SP315 (Figure 1E, F). Increased expression of the majority of these RNA5SPs in sAH livers was confirmed by quantitative PCR (Figure 1G). Correlation analysis of gene expression levels in GSE143318 and DbGaP_phs001807 revealed that expression of RNA5SP transcripts was strongly correlated with type I IFNs, especially IFNK and IFNW1 (Figure 1H). We were unable to identify RNA5SP expression in GSE28619 as it is a DNA array dataset. Taken together, these results suggest that IFN production is increased in sAH livers and perturbs expression of IRGs; these changes were associated with increased expression of host-derived RNA5SP transcripts.
RNA5SP transcripts correlate with inflammatory mediators uniquely upregulated in sAH livers
Various cytokines and chemokines constitute the local and systemic inflammatory environments in AH. To better understand the link between RNA5SP activation and hepatic inflammation, we performed a correlation analysis using the dataset DbGaP_phs001807.[20] Hepatic RNA5SP transcripts correlated with expression of many cytokines and chemokines. For instance, transcripts of RNA5SP237 and RNA5SP268 were positively correlated with expression of IL17B and IL17C, respectively, both of which can stimulate release of TNF and IL1B in monocytic cells (Supporting Figure 3, http://links.lww.com/HEP/A55).[21] Interestingly, among the many inflammatory mediators, hepatic expression of IL17D (stimulating the production of other cytokines such as IL6, IL8, and granulocyte-macrophage colony-stimulating factor[22]), IL18 (a proinflammatory cytokine), and CCL28 (a chemokine with immunomodulatory activity[23]) mRNAs was increased only in sAH (including sAH and AH_Expl) (Figure 2A). In contrast, IL1B mRNA was increased in liver diseases of multiple etiologies (DbGaP_phs001807) (Figure 2A). In this set of diseased livers (DbGaP_phs001807), expression of RNA5SP493 and RNA5SP44 was positively correlated with expression of IL17D, IL18, and CCL28, but not IL1B, mRNAs (Figure 2B). These findings suggest that increased expression of RNA5SPs is associated with hepatic inflammatory responses.
FIGURE 2.
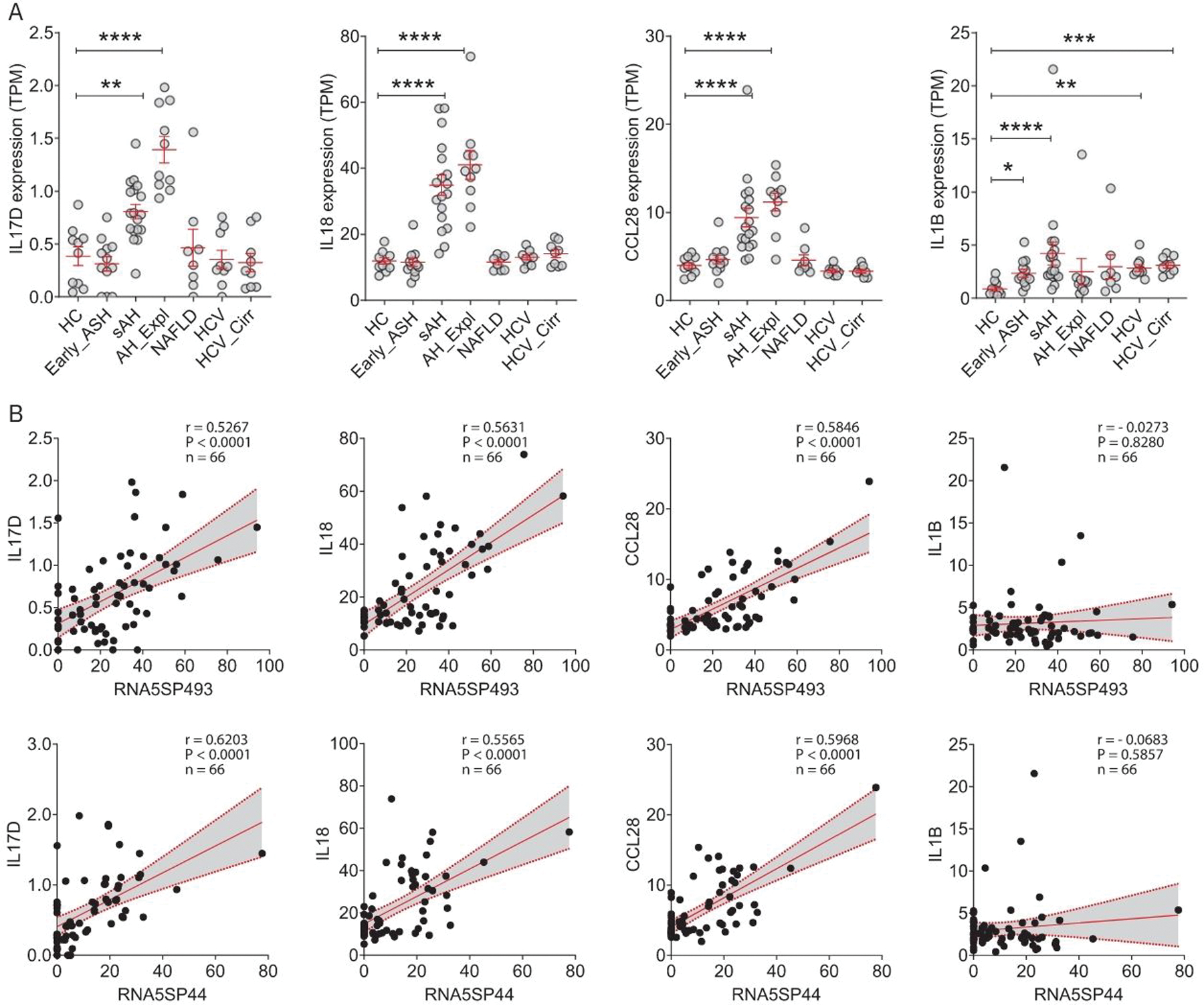
RNA5SP transcripts correlate with expression of inflammatory mediators that are upregulated uniquely in severe alcohol-associated hepatitis livers. (A) Expression of the indicated genes, represented by TPMs from DbGaP_phs001807. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 versus HC. (B) Spearman correlation analysis of the TPMs of RNA5SP transcripts and inflammatory mediators in diseased livers from DbGaP_phs001807 (n = 66). Abbreviations: HC, healthy control; RNA5SP, 5S rRNA pseudogene; TPM, transcripts per million.
Hepatic expression of Pol III transcripts is increased in sAH and expression of RNA5SP transcripts is regulated by Pol III
Pol III transcribes a variety of short noncoding RNAs, including 5S rRNA, and is implicated in regulation of immune functions.[24] Expression of mRNAs for components of Pol III machinery did not change in terms of transcripts per million (TPM) in sAH livers (GSE143318), except for POLR3C, which was moderately increased (Figure 3A). However, expression of MAF1 protein, the Pol III corepressor, was diminished in sAH livers (Figure 3B, C) and MAF1 nuclear localization, required for its corepressor activity, was decreased in both parenchymal and nonparenchymal cells in sAH livers (Figure 3D).[25] It is noteworthy that MAF1 expression showed uneven staining and variation across liver sections from patients with sAH and healthy controls (Supporting Figure 4, http://links.lww.com/HEP/A56). Further, abundance of 5S rRNA was increased in sAH livers compared with healthy controls (Figure 3E), consistent with elevated transcriptional output from Pol III. RNA5SP expression in human monocytic THP-1 cells was increased in response to poly (I:C), an agent known to activation of Pol III-dependent transcription (Figure 3F).[26] This increase was prevented by pharmacological inhibition of Pol III activity (Figure 3G). Collectively, these results suggest that increased expression of RNA5SP transcripts in livers from patients with sAH is related to activity of Pol III.
FIGURE 3.
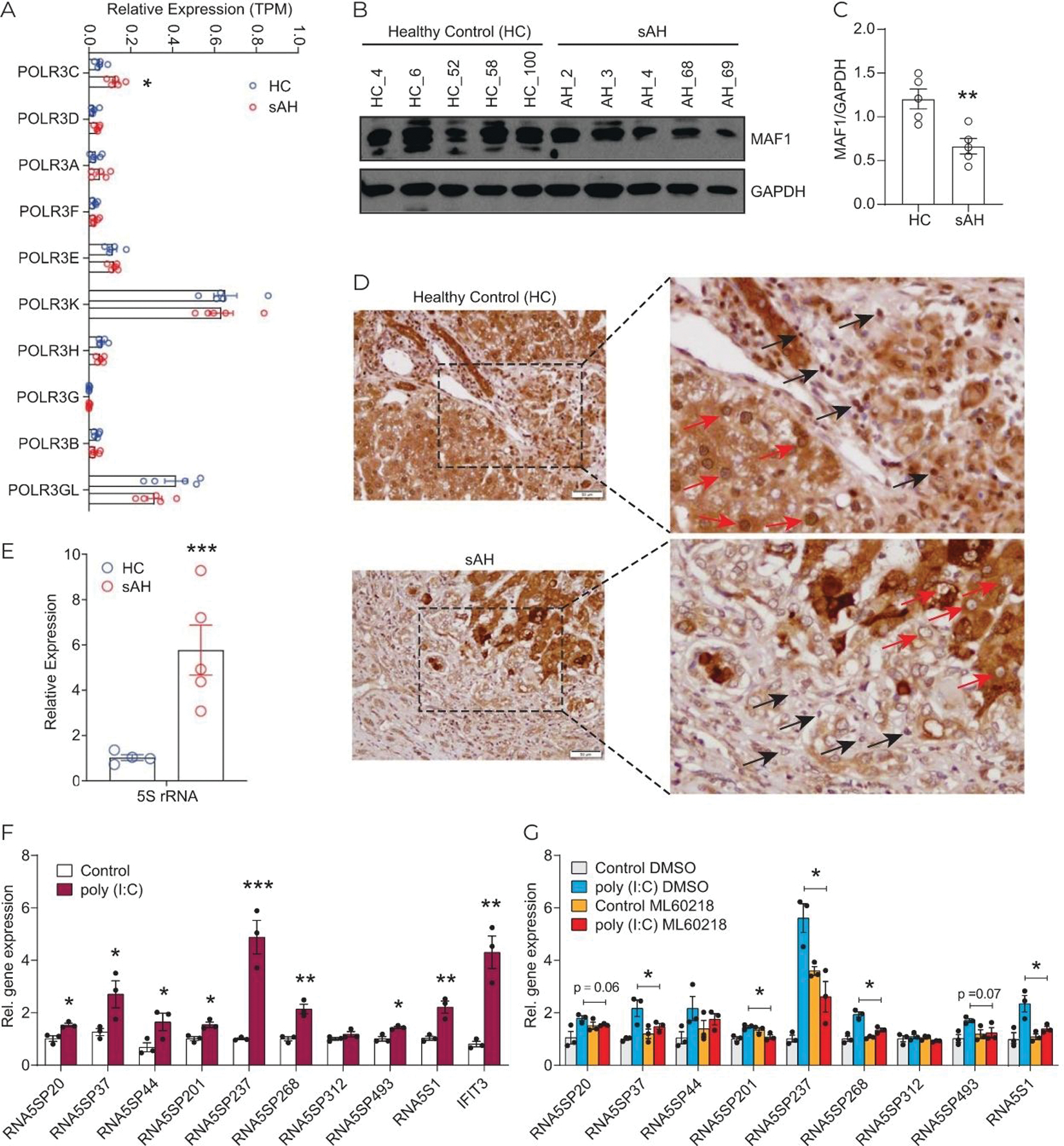
Activation of RNA Pol III promotes RNA5SP expression. (A) Expression of the components of RNA Pol III machinery, represented by TPMs from GSE143318. (B) Western blot of MAF1 expression in healthy control and sAH livers. (C) Quantification of the western blot in using ImageJ. (D) Immunohistochemistry of hepatic MAF1 expression. Red arrows: Nuclear staining of MAF1 in parenchymal cells; black arrows: in nonparenchymal cells. (E) Quantitative PCR of 5S rRNA expression. (F) Expression of RNA5SP transcripts was increased by poly (I:C). THP-1 cells were transfected with poly (I:C) for 24 hours, and gene expression was determined by quantitative PCR. (G) The Pol III inhibitor, ML60218, inhibited poly (I:C)-induced RNA5SP expression. (F and G): Data are mean ± SEM of 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. (G): Only comparisons made were between poly (I:C) DMSO versus poly (I:C) ML60218. Abbreviations: Pol III, RNA polymerase III; sAH, severe alcohol-associated hepatitis; TPM, transcripts per million.
Peripheral monocytes from patients with sAH express higher levels of RNA5SP transcripts
To assess in which cell types expression of RNA5SPs was increased, we performed gene deconvolution of the bulk RNA sequencing transcriptomes of sAH livers (DbGaP_phs001807).[27,28] The cluster of RNA5SP transcripts uniquely upregulated in sAH livers was attributed to multiple cell types; both myeloid and hepatocyte clusters were identified (Figure 4A). In AH, immune cells from the peripheral blood infiltrate the liver. PBMCs from patients with AH exhibit increased sensitivity to stimulation by a variety of DAMPs and PAMPs.[29] We asked whether RNA5SP transcripts in PBMCs were increased in patients with sAH. Two transcripts, RNA5SP44 and RNA5SP268, were specifically increased in PBMCs of patients with sAH compared with healthy controls and patients with NASH (Figure 4B). In purified CD14+/CD16+ monocytes of patients with sAH, these 2 transcripts were also increased (Figure 4C) and positively correlated with IFNA1 mRNA (Figure 4D). In contrast to the increase of multiple RNA5SP transcripts in sAH livers, only 2 highly expressed transcripts were identified in PBMCs, suggesting that diverse expression of RNA5SPs in sAH is likely due to expression by multiple types of resident liver cells, as well as infiltrating monocytes, which is in accordance with the gene deconvolution analysis.
FIGURE 4.
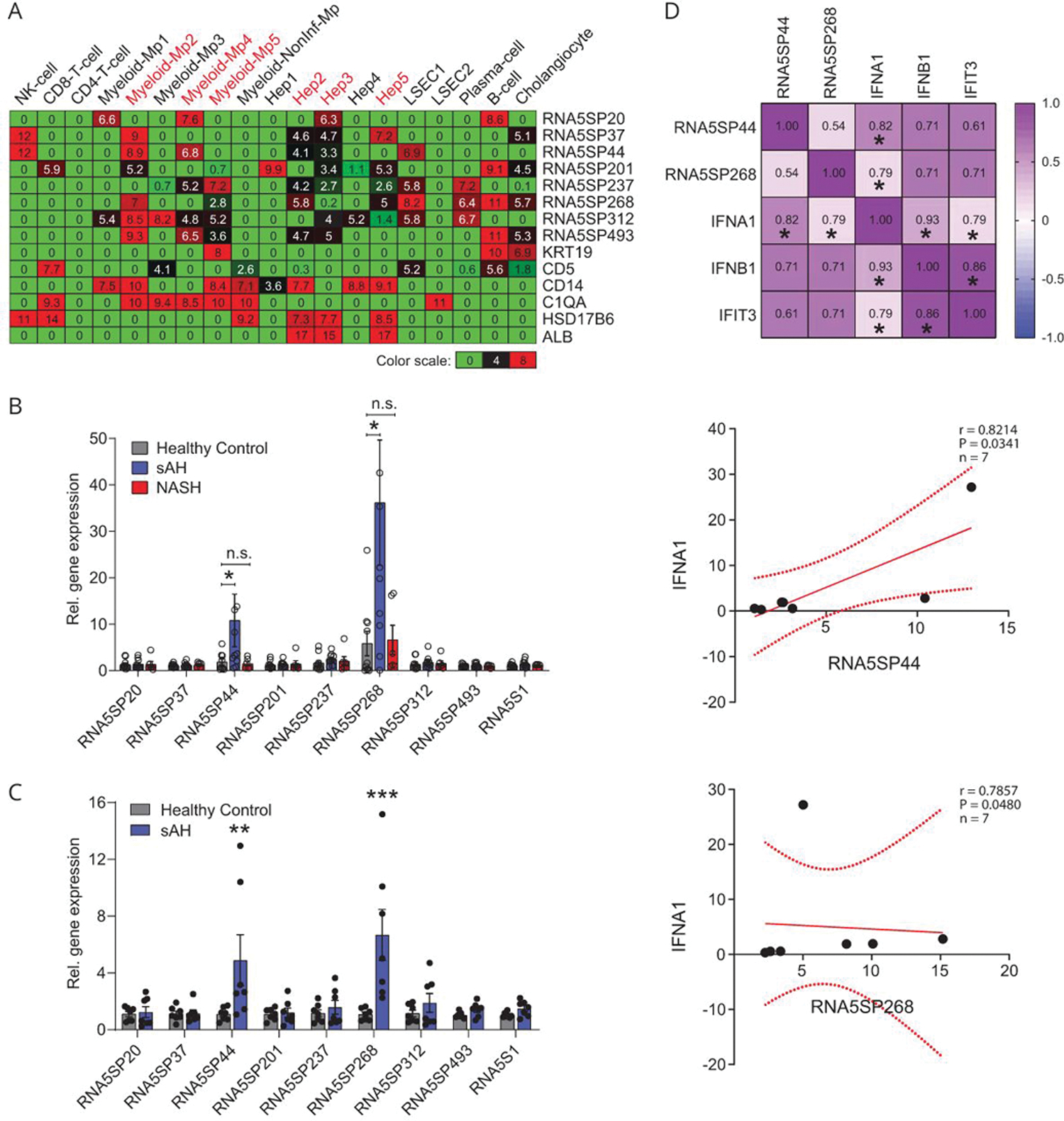
RNA5SP transcripts are increased in PBMCs of patients with sAH. (A) Gene deconvolution analysis to impute cell type-specific expression of RNA5SP transcripts in sAH livers, which was performed on the transcriptomes of sAH livers (from DbGaP_phs001807) using CIBERSORTx. The values in the heatmap are the imputed expression values for each gene (row) and each cell type (columns). (B) Quantitative PCR quantification of RNA5SP expression in human PBMCs. Healthy controls and sAH: n = 10; NASH: n = 6. (C) Quantitative PCR quantification of RNA5SP expression in CD14+/CD16+ monocytes isolated from PBMCs of healthy controls or patients with sAH (n = 7). Data are mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001 versus healthy control. (D) Top: Spearman correlation matrix. The numbers represent Spearman r; *p < 0.05, n = 7. Middle and bottom: scatter plots of Spearman correlation of the indicated genes. All correlation graphs were based on relative expression determined by quantitative PCR in CD14+/CD16+ monocytes from patients with sAH. Abbreviations: Hep 1–5, hepatocyte cluster 1 to 5; LSEC, liver sinusoidal endothelial cell; myeloid-Mp1–5, myeloid-derived cell clusters 1 to 5; NonInf-Mp, noninflammatory macrophage cluster; PBMC, peripheral blood mononuclear cell; sAH, severe alcohol-associated hepatitis.
RNA5SP transcript-interacting proteins are downregulated in sAH livers
Loss of interacting proteins unmasks RNA5SP transcripts, enabling them to act as DAMPs to activate immune responses in virial infection.[16] We hypothesized that sAH would also decrease expression of the interacting proteins. Due to the similarity of nucleotide sequences (Supporting Figure 5A, http://links.lww.com/HEP/A57), it is possible that multiple RNA5SP transcripts adopt the same binding partners. We predicted the interacting proteins for the 9 RNA5SP transcripts upregulated in sAH livers (Figure 1F, Supporting Figure 5A, http://links.lww.com/HEP/A57) using catRAPID, an algorithm to estimate the binding propensity of protein-RNA pairs. In total, 869 potential candidates were next matched to 4521 downregulated genes from List_2 (GSE143318) to identify potential interacting proteins downregulated in sAH livers. Among 10 identified candidates, zinc finger CCCH-type and G-patch domain containing (ZGPAT), epithelial splicing regulatory protein 2 (ESRP2), and APOBEC1 complementation factor (A1CF) were chosen because of their highly significant downregulation (Figure 5A). Thiosulfate sulfurtransferase (TST), a primary interacting protein for RNA5SP141 transcript,[16] was also included for further analysis. Expression of ZGPAT, ESRP2, and A1CF, but not TST, mRNAs was downregulated in sAH (including sAH and AH_Expl), but not in early ASH, NAFLD, HCV, and HCV cirrhosis (DbGaP_phs001807) (Figure 5B). RNA pull-down using 3′-end labeled transcripts of RNA5SP44 and RNA5SP268, followed by LC-MS/MS, was performed to profile RNA5SP transcript-interacting proteins. ZGPAT, ESRP2, and TST, but not A1CF, among a total of 247 interacting proteins, were identified in the pull-down (Figure 5C, Supporting Table 4 http://links.lww.com/HEP/A58).
FIGURE 5.
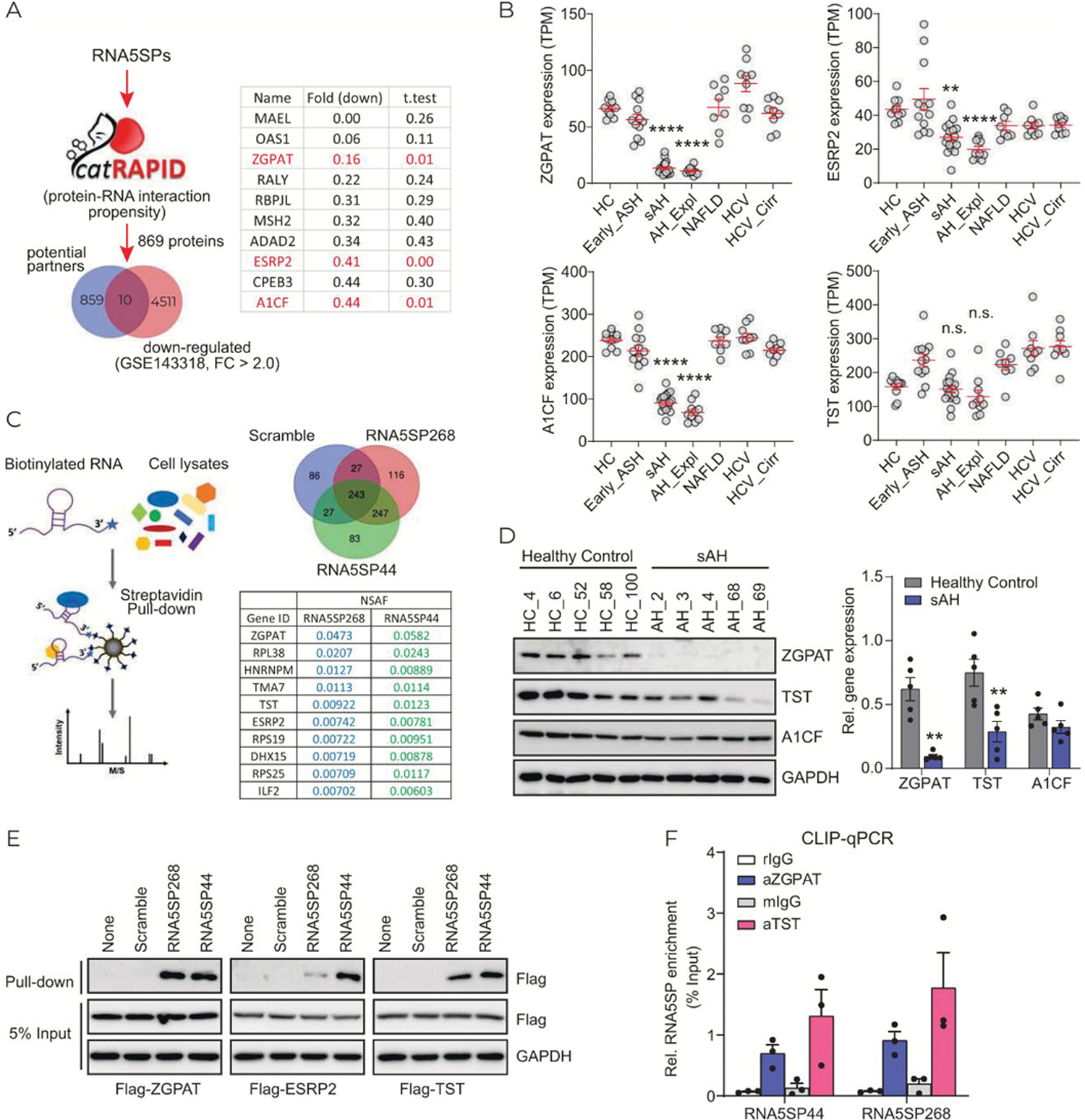
Expression of RNA5SP transcript-interacting proteins is decreased in sAH livers. (A) RNA-protein binding propensity analysis. The increased RNA5SP transcripts in sAH livers, shown in Figure 1F, were interrogated in CatRAPID. In total of 869 potential binding proteins were matched to 4521 downregulated genes in sAH livers from List_2 (GSE143318) and demonstrated in Venn diagram. The candidates were listed in the table, with fold changes and t tests. (B) TPMs of the indicated genes from DbGaP_phs001807. (C) Left: the schematic showing 3′-end biotinylated RNA pull-down assay followed by LC-MS/MS to identify RNA interacting proteins. Right: Venn diagram showing the numbers of identified interacting proteins of the 3′-end biotinylated in vitro-transcribed RNAs; RNA5SP44 and RNA5SP268 transcripts’ representative interacting proteins and the normalized spectral abundance factors (NSAFs) are shown in the table. (D) Western blot (left) and its quantification using ImageJ (right). (E) Binding of the biotinylated in vitro-transcribed RNAs to the indicated flag-tagged proteins which were overexpressed in HEK293T cells, was accessed by streptavidin pull-down followed by western blot. (F) CLIP-quantitative PCR showing the association of RNA5SP transcripts with endogenous proteins in THP-1 cells. rIgG and mIgG, control Igs of rabbit or mouse origin. Data are mean ± SEM of 3 independent experiments. (B and D): **p < 0.01, ****p < 0.0001 versus healthy control.
Expression of ZGPAT and TST proteins was downregulated in patients with sAH; however, A1CF protein did not change (Figure 5D). Downregulation of ESRP2 protein in sAH livers has been reported.[19] RNA pull-down assays demonstrated that exogenous ZGPAT, ESRP2, and TST interacted with RNA5SP44 and RNA5SP268 transcripts, although ESRP2 showed a weaker interaction with RNA5SP268 compared with RNA5SP44 (Figure 5E). The interactions with TST and ZGAPT were further validated by CLIP-quantitative PCR (Figure 5F). ESRP2 was not detectable in THP-1 cells (data not shown).
There were more upregulated genes than downregulated genes in sAH livers (Figure 1A, Supporting Figure 1A, http://links.lww.com/HEP/A54). For potential RNA5SP transcript-interacting proteins, compared with the 10 identified candidate genes downregulated in GSE143318 (Figure 5A), we identified 117 upregulated genes in our in silico analysis (Supporting Figure 6A, http://links.lww.com/HEP/A57). Pathway and process enrichment analysis showed that they have diverse molecular functions (Supporting Figure 6B, http://links.lww.com/HEP/A57). Most of these genes were not identified in our proteomics analysis; we only identified 2 proteins, SF3B1 and SARNP, likely to impact availability of RNA5SP transcripts (Supporting Figure 6A, http://links.lww.com/HEP/A57). However, the genes coding for these 2 proteins were not consistently upregulated in sAH livers in the DbGaP_phs001807 dataset (supporting Figure 6C, http://links.lww.com/HEP/A57). Based on the limited evidence for upregulation of RN5SP-binding proteins, we postulated that the unmasking of RNA5SP transcripts overwhelmed the masking in the pathogenesis of sAH.
Taken together, these data indicate that multiple RNA5SP transcript-interacting proteins are downregulated in sAH, potentially exposing RNA5SP transcripts as “non-self” to immune surveillance, leading to activation of immune responses.
RNA5SP transcripts are immunostimulatory and this property is potentiated upon loss of interacting proteins in THP-1 cells
To determine the potential immunostimulatory impact of RNA5SP transcripts, THP-1 cells were transfected with in vitro-transcribed RNA5SP268 RNA. The RNA5SP268 transcript has a secondary conformation very close to RNA5S1 (Supporting Figure 5B, http://links.lww.com/HEP/A57). The RNA5SP268 transcript potently increased expression of mRNAs for IFNB1, IL1B, and TNF, as well as IFIT3, an ISG, compared with scrambled RNA (Figure 6A). Transfection of RNA5SP268 transcript also induced IRF3 dimerization, a critical step for induction of IFN expression (Figure 6B). siRNA-mediated knockdown of ZGPAT and TST strikingly enhanced IFNB1 mRNA expression triggered by transfection of RNA5SP268 transcript (Figure 6C, D), suggesting that these transcript-interacting proteins shielded RNA5SP transcripts and prevented their immunostimulatory activity. These findings support the concept that host-derived RNAs can activate innate immunity upon loss of endogenous chaperones.[30]
FIGURE 6.
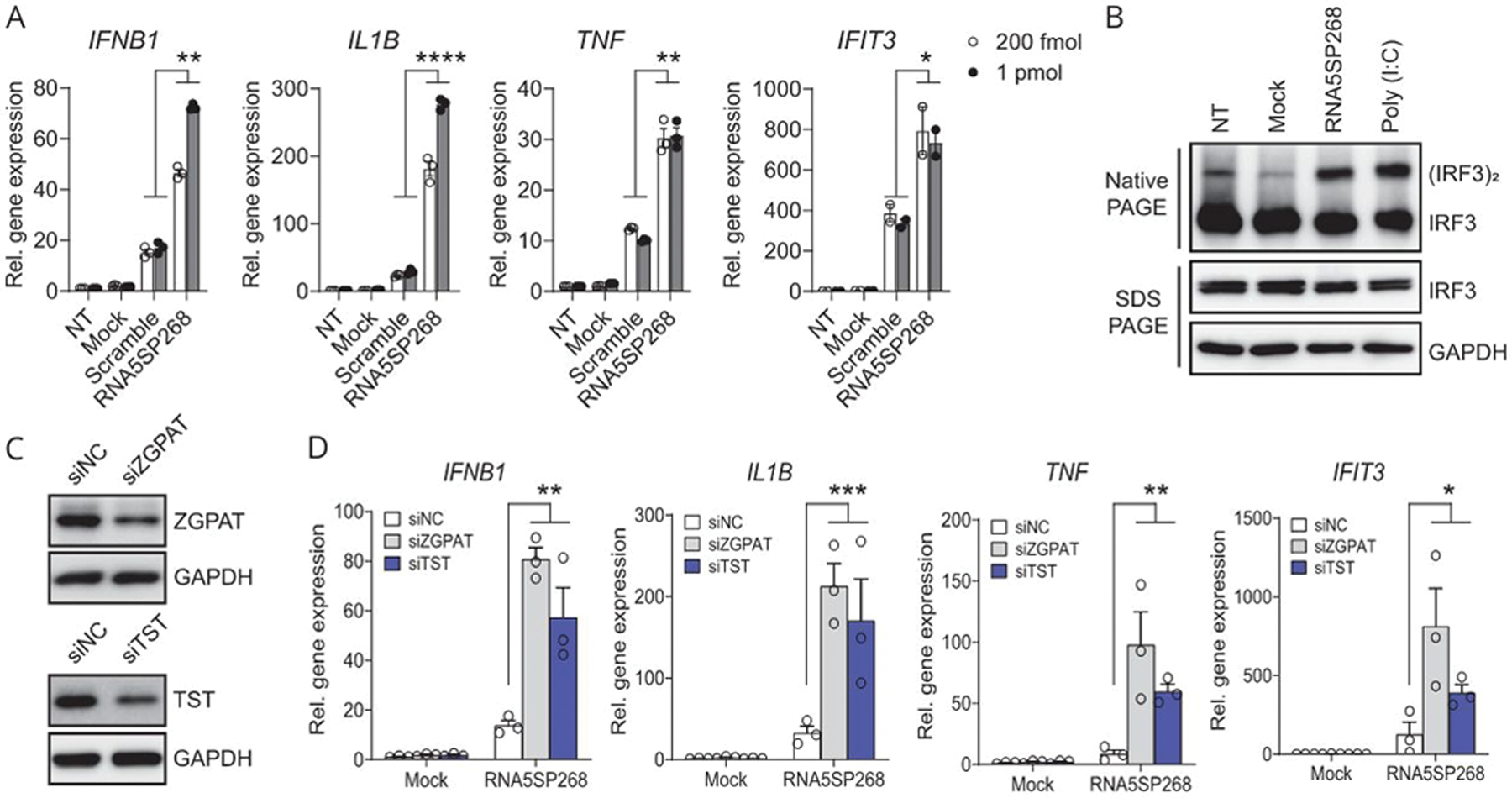
RNA5SP268 transcript is immunostimulatory and loss of RNA5SP transcript-interacting proteins potentiates inflammatory responses. (A) Quantitative real-time PCR of the indicated gene expression in THP-1 cells NT, mock-transfected, or transfected with in vitro-transcribed scrambled or RNA5SP268 RNA for 24 hours. (B) Native PAGE and SDS-PAGE of whole cell lysates from THP-1 cells transfected with 125 pmol of in vitro-transcribed RNA5SP268 RNA. A 100 ng/mL of poly (I:C) was transfected in parallel as a positive control. Western blot was performed with the indicated antibodies. (C) Western blot of THP-1 cells transfected with the indicated siRNAs (50 nM, 48 hours). (D) Quantitative real-time PCR quantification of the indicated gene expression in THP-1 cells in which 50 nM of siRNAs were transfected for 24 hours, followed by mock transfection or transfection of 200 fmol of in vitro-transcribed RNA5SP268 RNA for another 24 hours. (A and D): Mean ± SEM of 3 independent experiments. IFIT3 in (A): Two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Abbreviations: NC, negative control; NT, nontransfected.
RNA5SP transcripts contribute to ZBP1-mediated cell death
Zα domain-containing proteins sense the Z-conformation that is found in a variety of large and small RNA structures.[17,31] In an RNA EMSA assay, a Z-NAs antibody recognizing both Z-DNA and Z-RNA,[32] robustly retarded the mobility of in vitro-transcribed RNA5SP44 and RNA5SP268 transcripts, indicating the presence of Z-forming domains within these RNA5SP transcripts (Figure 7A). ZBP1 contains Zα domains and regulates programmed cell death in response to ligation to Z-NAs.[14,15,33] ZBP1 protein expression in sAH livers was strongly increased (Figure 7B). Wild-type ZBP1, but not its mutant (mZα1–2) lacking Z-NA binding capacity, interacted with endogenous RNA5SP44 and RNA5SP268 transcripts from THP-1 cells (Figure 7C). Pull-down followed by dot blot assays showed that immunopurified wild-type, but not mZα1–2, ZBP1 directly interacted with in vitro-transcribed RNA5SP44 and RNA5SP268 transcripts (Figure 7D). Consistently, internally biotinylated RNA5SP44 and RNA5SP268 transcripts also interacted with wild-type, but not mZα1–2, ZBP1 in RNA pull-down assays (Figure 7E). Wild-type, but not mZα1–2, ZBP1, combined with co-expression of RNA5SP44 or RNA5SP268 transcripts, induced caspase-independent cell death in THP-1 cells (Figure 7F). Phosphorylation of MLKL at serine358, an event associated with caspase-independent necroptotic cell death, was also increased under these conditions (Figure 7G).[34] Taken together, these results indicate that RNA5SP transcripts are capable of forming Z-conformation and activating ZBP1 to promote caspase-independent cell death, consistent with a possible role of RNA5SP transcripts in promoting hepatocellular death in sAH.
FIGURE 7.
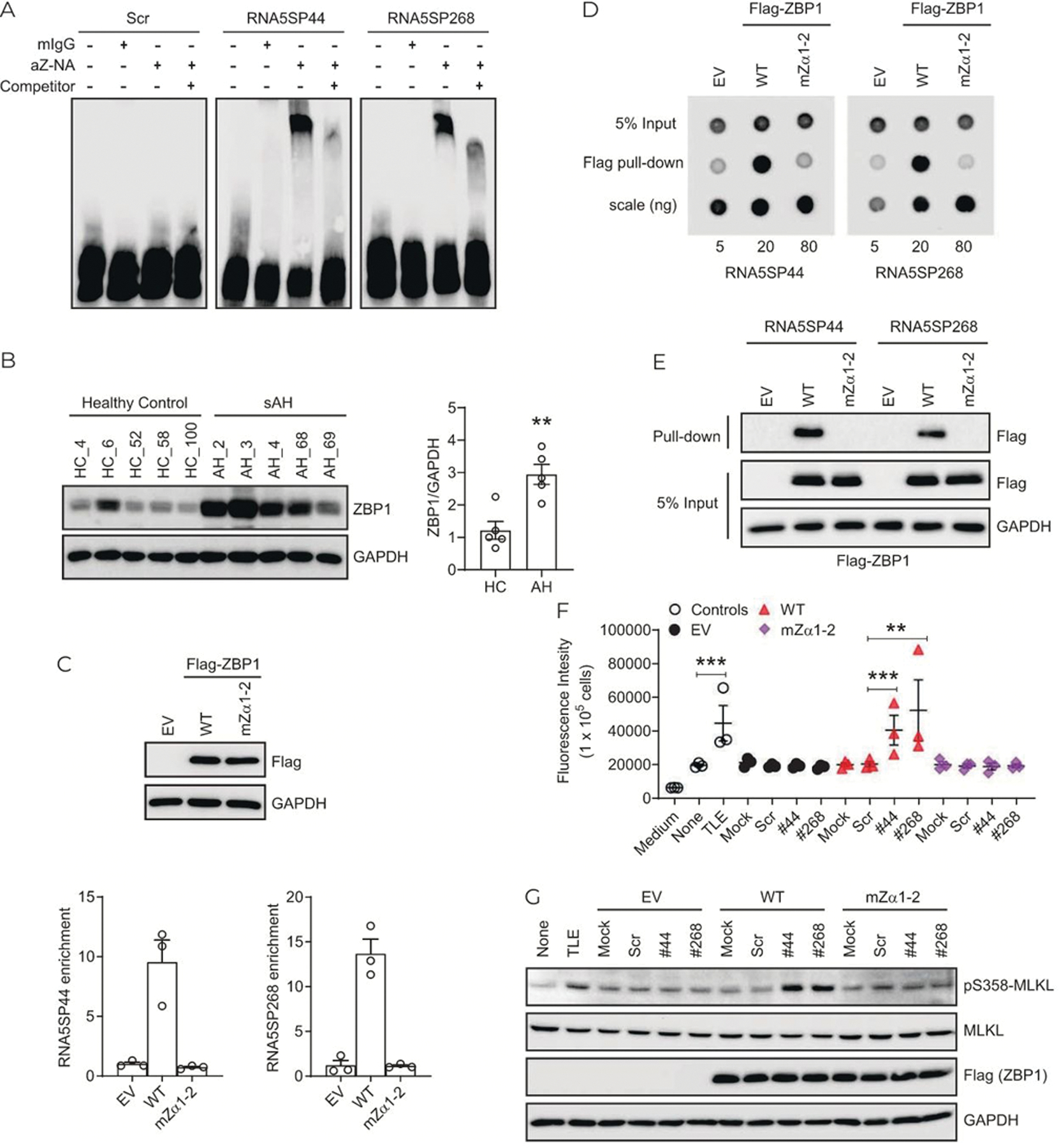
RNA5SP transcripts contribute to ZBP1-mediated cell death. (A) REMSA assay. A 3′-end biotinylated in vitro-transcribed RNA5SP transcripts were incubated with the Z-NAs antibody or a mouse IgG (mIgG) isotype control. A 160-fold nonbiotinylated RNA5SP transcripts (competitor) were used to compete for the binding. (B) Western blot (left) and the quantification using ImageJ (right). **p < 0.01. (C) RNA immunoprecipitation to detect the interaction between endogenous RNA5SP transcripts and ZBP1 in THP-1 cells overexpressing. Empty vector (EV), flag-tagged wild-type ZBP1 (WT), or Zα-domain-mutated ZBP1 (mZα1–2). Top: Western blot of ZBP1 expression. Bottom: The enriched RNAs in the immunoprecipitation were quantified in quantitative PCR and normalized to EV. Data are mean ± SEM of 3 independent experiments. (D) Dot blot to examine the binding of immunopurified flag-ZBP1 or its Zα-domain mutant with 3′-end biotinylated in vitro-transcribed RNA5SP RNAs. The detection was performed with streptavidin antibody against RNAs isolated from anti-flag pull-down. (E) RNA pull-down assays as described in Figure 5E. (F) Cell death was measured by the incorporation of cell impermeant SYTOX Green (488/523 nm). THP-1 cells (1 × 105/well) were transfected with 20 ng of EV or ZBP1, along with 50 fmol of scrambled or RNA5SP RNAs in 96-well plates for 24 hours, and then treated with emricasan (5 μM) for 16 hours. TLE (TNF, 50 ng/mL; LCL-161, 5 μM; emricasan, 5 μM; 16 hours) was used as a positive control. Data are mean ± SEM of 3 independent experiments, **p < 0.01, ***p < 0.001. (G) Western blot using whole cell lysates of THP-1 cells as treated in (F). Abbreviations: RNA5SP, 5S rRNA pseudogene; sAH, severe alcohol-associated hepatitis.
DISCUSSION
AH is a disease with high mortality and leads to substantial public health burden; effective therapies are still unavailable due to the incomplete understanding of the molecular mechanisms for disease progression. DAMPs and PAMPs play a fundamental role in the pathogenesis of AH. In this study, we demonstrate that RNA5SP transcripts, a form of host-derived nucleic acids, can serve as novel DAMPs, associated with the proinflammatory profile of livers of patients with sAH. Specifically, RNA5SP transcripts promoted IFN production and inflammatory responses and contributed to ZBP1-mediated cell death (summarized in Figure 8).
FIGURE 8.
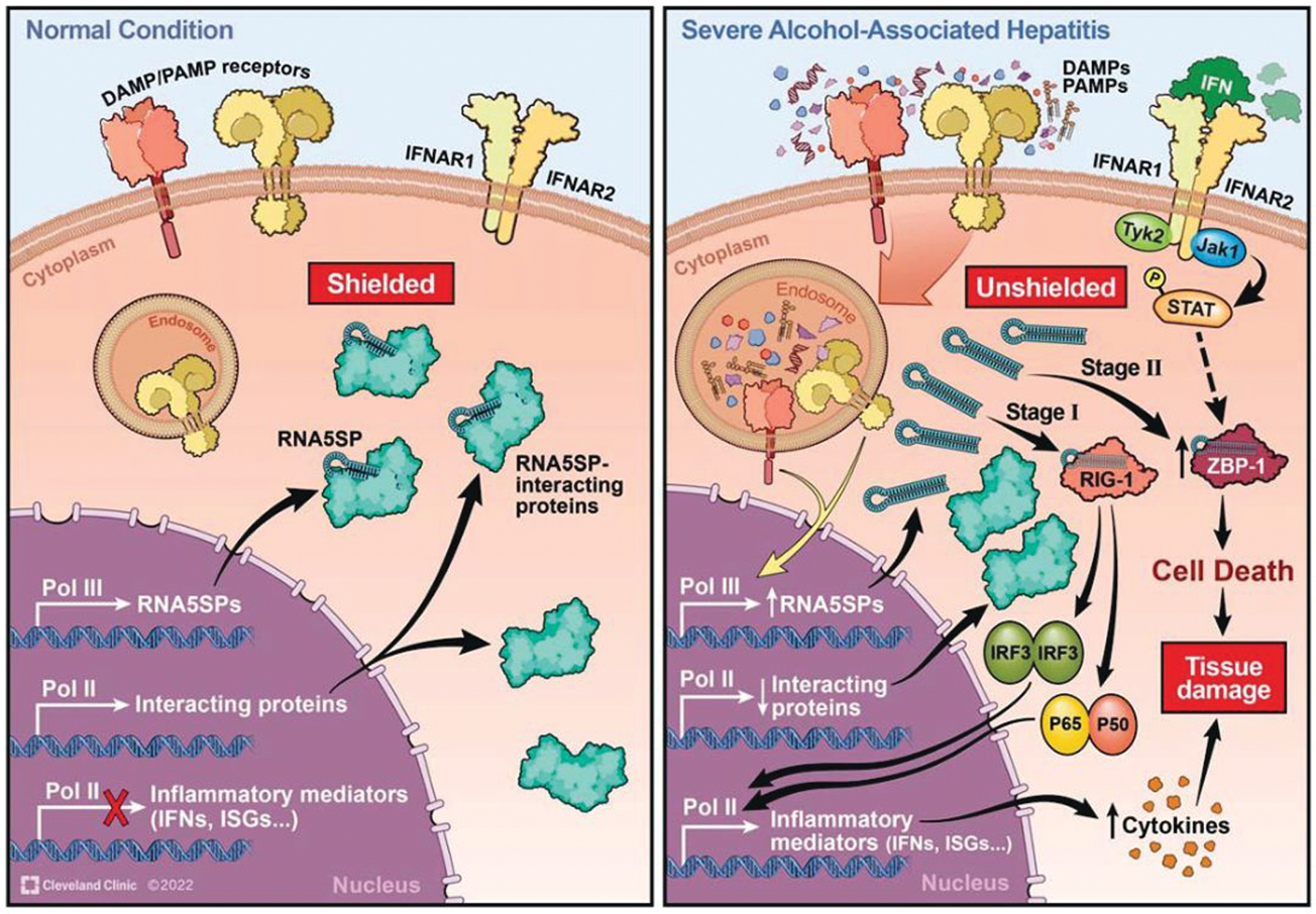
Schematic depicting RNA5SP transcripts serving as an endogenous stimulant of interferon production and inflammatory responses in monocytic cells. Under normal conditions, RNA5SP transcripts are shielded by interacting proteins and immuno-silent. In severe alcohol-associated hepatitis, RNA5SP transcript-interacting proteins are downregulated; the unshielded RNA5SP transcripts can be recognized by RIG-I and promote expression of inflammatory factors in host cells (stage 1). In the presence of ZBP1, the sensing of RNA5SP transcripts by its Zα domains activates ZBP1-mediated cell death (stage 2). Abbreviations: IFN, interferon; DAMP, damage-associated molecular pattern; ISG, interferon-stimulated gene; PAMP, pathogen-associated molecular pattern; RIG-I, retinoic acid-inducible gene I; RNA5SP, 5S rRNA pseudogene; ZBP1, Zα domain-containing Z-DNA binding protein 1.
Hepatic gene expression patterns are uniquely impacted in patients with sAH. Indeed, multiple RNA5SP transcripts were increased only in sAH livers compared with liver diseases of different etiologies (Figure 1D). We confirmed that these RNA5SP transcripts are immunostimulatory and can promote IFN expression. Further, the distinct inflammatory profile of sAH, characterized by increased expression of IL17D, IL18, and CCL28 mRNAs, was positively correlated with expression of RNA5SP493 and RNA5SP44 transcripts (Figure 2). This study demonstrates that RNA5SPs are not only involved in the response to viral infection, but also critically associated with alcohol-associated inflammation.[16] These data, combined with previous reports on the specific contributions of the nontranscriptional activity of IRF3 in promoting ethanol-induced liver injury in mice,[7] indicate that additional components of IFN signaling, such as the upstream molecular pattern activators and receptors, should be studied to strategically and therapeutically target IFN signaling in ALD.[8,9]
Chronic alcohol administration disrupts Pol III machinery.[35] PAMPs and DAMPs may trigger signaling pathways to regulate Pol III-driven transcription. A CK2/MAF1/Pol III axis, downstream of toll-like receptors, is critical to the immunostimulatory function of dendritic cells.[26] We found that the toll-like receptor 3 ligand, poly (I:C), induced expression of RNA5SP transcripts in monocytic cells and that nuclear localization of MAF1 was almost completely absent in sAH livers. Taken together, these data support the concept that dysregulation of Pol III-mediated transcription is a disease contributing factor and that Pol III is involved in immune responses in sAH.[24,36]
We found that multiple RNA5SP transcripts were perturbed in sAH and it is likely that they function together to participate in the pathologic process. Inhibition of single or a few RNA5SP transcripts might be not sufficient enough to impair their overall pathologic function. The mechanisms for induction of specific RNA5SP transcripts in sAH are not understood. We could not identify typical promoter regions within the RNA5SPs upregulated in sAH through interrogating the Pol III CHIP-sequencing datasets from Gene Expression Omnibus (GEO) (data not shown), suggesting that RNA5SP expression is likely regulated through promoter-independent transcription, the nonconventional function of Pol III.[37] Our finding that Pol III activates RNA5SP expression adds to the mechanisms by which Pol III functions in innate immunity and the pathogenesis of ALD. Owing to the ubiquitous activity of Pol III, RNA5SPs appear to be expressed in abundance in varieties of cell types. Indeed, gene deconvolution analysis and our data in PBMCs from patients suggest that multiple cell types, especially myeloid cells, contribute to the accumulation of RNA5SP transcripts in sAH livers.
RNA5SP transcripts adopt 5′-triphosphates which can be recognized by RIG-I to elicit immune responses upon loss of interacting proteins.[16] We demonstrate that RNA5SP transcripts can form Z-conformation to contribute to ZBP1-mediated caspase-independent cell death. A large variety of noncoding RNAs adopt Z-conformations by sequences other than CpGs.[17] It is noteworthy that nucleic acids capable of Z-formation may not necessarily exist as a preformed structure in host cells but can be induced/converted when ligating to Zα-containing proteins.[38] Increased accumulation of ZBP1 in sAH may be due to infiltration of cells constitutively expressing ZBP1 and/or induction of ZBP1 expression by liver resident cells. Temporal and spatial regulation of hepatic expression of ZBP1, an ISG, still needs to be investigated.[14,39] We propose that the regulatory role of RNA5SP transcripts in cell death depends on availability and/or induction of ZBP1 under pathologic conditions (Figure 8). It is likely that other proteins also become available to engage in ZBP1-mediated cell death in ALD. For instance, RIPK3, involved in the formation of necrosomes with ZBP1 to mediate necroptosis,[14] is only expressed in hepatic nonparenchymal cells under normal conditions,[40] but is induced in hepatocytes in murine models of ALD and livers of patients with ALD.[41,42]
We propose the following working model in monocytic cells (Figure 8) for the contribution of RNA5SP transcripts in the progression of AH. Stage 1: unshielded RNA5SP transcripts are recognized by RIG-I to promote expression of inflammatory factors such as IFNs in host cells, before the induction of ZBP1, an ISG. Stage 2: these unshielded pseudogene transcripts ligate ZBP1 to activate cell death when ZBP1 (and other related regulators) becomes physically and functionally available (Figure 8). While we have focused on immune cells, it is highly likely that RNA5SP transcripts regulate function of other cell types in the liver, because (1) both liver parenchymal and nonparenchymal cells produce IFNs, although there is specificity in the type of IFN expressed; (2) Pol III is ubiquitous but how it activates RNA5SP expression is still elusive; (3) the function of RNA5SP transcripts depends on their unmasking/availability based on expression of interacting proteins; however, it is still unclear how or if the interacting proteins are perturbed in different cell types in sAH; and (4) how the immunosensors for RNA5SP transcripts such as ZBP1 and RIG-I are temporally and spatially induced in specific cell types in the liver is not defined. Thus, the complex regulation, activity, and biological consequences of RNA5SP transcripts in different cell types can vary, which needs further investigation.
In summary, we demonstrated that RNA5SP transcripts are important contributors to the inflammatory milieu in sAH. The pathologic effects of RNA5SP transcripts were associated with their unmasking, due to decreased expression of transcript-interacting proteins and increased availability of effector molecules, such as ZBP1, recognizing the RNA5SP transcripts. Host-derived nucleic acids and their biological effects on cell fate and innate immunity are potentially interesting targets for development of therapeutic interventions and/or diagnostic biomarkers in patients with ALD. Further understanding of the availability, localization, structure, recognition, and immunostimulatory properties of host-derived nucleic acids in discriminating “self” from “non-self” would shed light on the pathogenesis of inflammatory diseases.[43]
Supplementary Material
ACKNOWLEDGMENTS
The authors sincerely thank Dr Zhaoli Sun at Johns Hopkins University for the human liver specimens, as well as Dr Belinda Willard and Dr Dongmei Zhang at the Lerner Research Institute’s Proteomics & Metabolomics Core for the mass spectrometry service.
Funding information
This work was in part supported by NIH P30DK097948 through the Cleveland Digestive Diseases Research Core Center (DDRCC) Pilot/Feasibility program (Pilot, J. W.), K01AA029474 (J.W.), K99AA028048 (A. K.), K99AA029146 (X.W.), P50AA024333 (L. E.N, S.D., D.S.A.), K08AA028794 (N.W), R01GM119174 (S.D.), R01DK113196 (S.D.), U01AA021890 (L.E.N.), U01AA026976 (S.D.), R56HL141744, U01DK061732 (S.D.), R21AR071046 (S.D.), R01AA027456 (L.E.N.), and U01AA026938 (L.E.N). The acquisition of human liver specimens was supported by R24 AA025017 (Z.S.). The Fusion Lumos LC-MS instrument was purchased via the NIH shared instrument grant 1S10OD023436-01.
Abbreviations:
- A1CF
APOBEC1 complementation factor
- ALD
alcohol-associated liver disease
- cGAS
cyclic guanosine monophosphate-AMP synthase
- CK2
casein kinase 2
- CLIP-qPCR
cross-linking immunoprecipitation and qPCR
- DAMPs
damage-associated molecular patterns
- ESRP2
epithelial splicing regulatory protein 2
- GSEA
Gene Set Enrichment Analysis
- IFIT3
interferon-induced protein with tetratricopeptide repeats 3
- IFN
interferon
- IRF3
interferon regulatory factor 3
- IRGs
interferon-regulated genes
- ISG
interferon-stimulated gene
- MAF1
MAF1 homolog, negative regulator of RNA polymerase III
- MLKL
mixed lineage kinase domain like pseudokinase
- PAMPs
pathogen-associated molecular patterns
- PBMCs
peripheral blood mononuclear cells
- Pol III
RNA polymerase III
- RIG-I
retinoic acid-inducible gene I
- RIPK3
receptor interacting serine/threonine kinase 3
- RNA5SP
5S rRNA pseudogene
- sAH
severe alcohol-associated hepatitis
- STING
stimulator of interferon genes
- TPM
transcripts per million
- TST
thiosulfate sulfurtransferase
- ZBP1
Zα domain-containing Z-DNA binding protein 1
- ZGPAT
zinc finger CCCH-type and G-patch domain containing
- Z-NAs
Z-form nucleic acids
Footnotes
CONFLICT OF INTEREST
D.S.A. advises Incyte. The remaining authors report no conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study is not research involving human subjects and does not require patient-informed consent, which was determined by the Institutional Review Board (IRB) at the Cleveland Clinic. The use of human blood cells was covered by the NIH P50AA024333 Clinical Core IRB Exempt protocol (08–361) to L.E.N.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website, www.hepjournal.com.
DATA AVAILABILITY
The datasets used and/or analyzed in the current study are available from NCBI. The index numbers have been described in the manuscript.
REFERENCES
- 1.European Association for the Study of the Liver. EASL clinical practice guidelines: management of alcohol-related liver disease. J Hepatol. 2018;69:154–81. [DOI] [PubMed] [Google Scholar]
- 2.Crabb DW, Im GY, Szabo G, Mellinger JL, Lucey MR. Diagnosis and treatment of alcohol-associated liver diseases: 2019 practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2020;71:306–33. [DOI] [PubMed] [Google Scholar]
- 3.Gao B, Ahmad MF, Nagy LE, Tsukamoto H. Inflammatory pathways in alcoholic steatohepatitis. J Hepatol. 2019;70: 249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow KT, Gale M Jr. SnapShot: interferon signaling. Cell. 2015; 163:e1801. [DOI] [PubMed] [Google Scholar]
- 5.Zhai Y, Qiao B, Gao F, Shen X, Vardanian A, Busuttil RW, et al. Type I, but not type II, interferon is critical in liver injury induced after ischemia and reperfusion. Hepatology. 2008;47:199–206. [DOI] [PubMed] [Google Scholar]
- 6.Lercher A, Bhattacharya A, Popa AM, Caldera M, Schlapansky MF, Baazim H, et al. Type I interferon signaling disrupts the hepatic urea cycle and alters systemic metabolism to suppress T cell function. Immunity. 2019;51:1074–87.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanz-Garcia C, Poulsen KL, Bellos D, Wang H, McMullen MR, Li X, et al. The non-transcriptional activity of IRF3 modulates hepatic immune cell populations in acute-on-chronic ethanol administration in mice. J Hepatol. 2019;70:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luther J, Khan S, Gala MK, Kedrin D, Sridharan G, Goodman RP, et al. Hepatic gap junctions amplify alcohol liver injury by propagating cGAS-mediated IRF3 activation. Proc Natl Acad Sci USA. 2020;117:11667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci USA. 2013; 110:16544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrasek J, Dolganiuc A, Csak T, Nath B, Hritz I, Kodys K, et al. Interferon regulatory factor 3 and type I interferons are protective in alcoholic liver injury in mice by way of crosstalk of parenchymal and myeloid cells. Hepatology. 2011; 53:649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartok E, Hartmann G. Immune sensing mechanisms that discriminate self from altered self and foreign nucleic acids. Immunity. 2020;53:54–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Shim YR, Seo W, Kim MH, Choi WM, Kim HH, et al. Mitochondrial double-stranded RNA in exosome promotes interleukin-17 production through toll-like receptor 3 in alcohol-associated liver injury. Hepatology. 2020;72:609–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, Xu MJ, Koritzinsky EH, Zhou Z, Wang W, Cao H, et al. Mitochondrial DNA-enriched microparticles promote acute-on-chronic alcoholic neutrophilia and hepatotoxicity. JCI Insight. 2017;2:e92634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao H, Wachsmuth L, Kumari S, Schwarzer R, Lin J, Eren RO, et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature. 2020;580:391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kesavardhana S, Kanneganti TD. ZBP1: a STARGTE to decode the biology of Z-nucleic acids in disease. J Exp Med. 2020;217: e20200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang JJ, Sparrer KMJ, van Gent M, Lassig C, Huang T, Osterrieder N, et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol. 2018;19:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols PJ, Bevers S, Henen M, Kieft JS, Vicens Q, Vogeli B. Recognition of non-CpG repeats in Alu and ribosomal RNAs by the Z-RNA binding domain of ADAR1 induces A-Z junctions. Nat Commun. 2021;12:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Affo S, Dominguez M, Lozano JJ, Sancho-Bru P, Rodrigo-Torres D, Morales-Ibanez O, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyun J, Sun Z, Ahmadi AR, Bangru S, Chembazhi UV, Du K, et al. Epithelial splicing regulatory protein 2-mediated alternative splicing reprograms hepatocytes in severe alcoholic hepatitis. J Clin Invest. 2020;130:2129–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argemi J, Latasa MU, Atkinson SR, Blokhin IO, Massey V, Gue JP, et al. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat Commun. 2019;10:3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, et al. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc Natl Acad Sci USA. 2000;97:773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol. 2002; 169:642–6. [DOI] [PubMed] [Google Scholar]
- 23.Mohan T, Deng L, Wang BZ. CCL28 chemokine: an anchoring point bridging innate and adaptive immunity. Int Immunopharmacol. 2017;51:165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeganeh M, Hernandez N. RNA polymerase III transcription as a disease factor. Genes Dev. 2020;34:865–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, et al. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reverendo M, Arguello RJ, Polte C, Valecka J, Camosseto V, Auphan-Anezin N, et al. Polymerase III transcription is necessary for T cell priming by dendritic cells. Proc Natl Acad Sci USA. 2019;116:22721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim A, Wu X, Allende DS, Nagy LE. Gene deconvolution reveals aberrant liver regeneration and immune cell infiltration in alcohol-associated hepatitis. Hepatology. 2021;74:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim A, Bellar A, McMullen MR, Li X, Nagy LE. Functionally diverse inflammatory responses in peripheral and liver monocytes in alcohol-associated hepatitis. Hepatol Commun. 2020;4:1459–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–24.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Ascenzo L, Leonarski F, Vicens Q, Auffinger P. ‘Z-DNA like’ fragments in RNA: a recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016;44:5944–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarling DA, Calhoun CJ, Feuerstein BG, Sena EP. Cytoplasmic microinjection of immunoglobulin Gs recognizing RNA helices inhibits human cell growth. J Mol Biol. 1990;211:147–60. [DOI] [PubMed] [Google Scholar]
- 33.Karki R, Lee S, Mall R, Pandian N, Wang Y, Sharma BR, et al. ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci Immunol. 2022;7:eabo6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–27. [DOI] [PubMed] [Google Scholar]
- 35.Zhong S, Machida K, Tsukamoto H, Johnson DL. Alcohol induces RNA polymerase III-dependent transcription through c-Jun by co-regulating TATA-binding protein (TBP) and Brf1 expression. J Biol Chem. 2011;286:2393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graczyk D, White RJ, Ryan KM. Involvement of RNA polymerase III in immune responses. Mol Cell Biol. 2015;35:1848–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessler AC, Maraia RJ. The nuclear and cytoplasmic activities of RNA polymerase III, and an evolving transcriptome for surveillance. Nucleic Acids Res. 2021;49:12017–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maelfait J, Liverpool L, Bridgeman A, Ragan KB, Upton JW, Rehwinkel J. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017;36:2529–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J, Kumari S, Kim C, Van TM, Wachsmuth L, Polykratis A, et al. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dara L, Liu ZX, Kaplowitz N. Questions and controversies: the role of necroptosis in liver disease. Cell Death Discov. 2016;2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Ni HM, Dorko K, Kumer SC, Schmitt TM, Nawabi A, et al. Increased hepatic receptor interacting protein kinase 3 expression due to impaired proteasomal functions contributes to alcohol-induced steatosis and liver injury. Oncotarget. 2016;7: 17681–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roychowdhury S, McMullen MR, Pisano SG, Liu X, Nagy LE. Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology. 2013;57:1773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed in the current study are available from NCBI. The index numbers have been described in the manuscript.


