Abstract
Background:
Hypertension is common in older individuals and is a major risk factor for cardiovascular disease. Blood DNA methylation (DNAm) profiles have been used to derive metrics of biological age that capture age-related physiological change, disease risk, and mortality. The relationships between hypertension and DNAm-based biological age metrics have yet to be carefully described.
Methods:
Among 4,419 women enrolled in the prospective Sister Study cohort, DNAm data generated from whole blood samples collected at baseline were used to calculate three biological age metrics (PhenoAgeAccel, GrimAgeAccel, DunedinPACE). Women were classified as hypertensive at baseline if they had high blood pressure (systolic blood pressure ≥ 140 mmHg or diastolic blood pressure ≥ 90 mmHg) or reported current use of antihypertensive medication. New incident cases of hypertension during follow-up were identified via self-report on annual health questionnaires.
Results:
All three DNAm metrics of biological age were positively associated with prevalent hypertension at baseline (per 1-SD increase; PhenoAgeAccel, adjusted OR: 1.16, 95% CI: 1.05, 1.28; GrimAgeAccel, adjusted OR: 1.28, 95% CI: 1.14, 1.45; DunedinPACE, adjusted OR: 1.16 95% CI: 1.03, 1.30). Among 2,610 women who were normotensive at baseline, women with higher biological age were more likely to be diagnosed with incident hypertension (per 1-SD increase; PhenoAgeAccel, adjusted HR: 1.09, 95% CI: 0.97, 1.23; GrimAgeAccel, adjusted HR: 1.16, 95% CI: 0.99, 1.36; DunedinPACE, adjusted HR: 1.16 95% CI: 1.01, 1.33).
Conclusions:
Methylation-based biological age metrics increase before a hypertension diagnosis and appear to remain elevated in the years after clinical diagnosis and treatment.
Keywords: Hypertension, Blood pressure, DNA methylation, Epigenetic clocks, Biological age
Gracphical Abstract
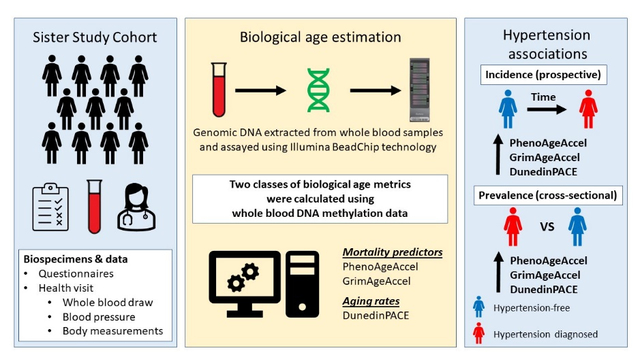
Introduction
Hypertension is a major risk factor for cardiovascular diseases, including coronary heart disease, myocardial infarction, and stroke.1–3 The incidence of hypertension increases with age and affects nearly 75% of the population aged over 60 years.4 Genome-wide changes in DNA methylation (DNAm) occur with aging and may reflect molecular changes that underlie hypertension development.5–7 However, few of the previously reported associations between DNAm at individual genomic positions and blood pressure replicate across populations.8 Recently, genome-wide DNAm data have been used to derive biological age metrics, or epigenetic clocks, that correlate with age and predict lifespan and functionality.9–11 Although epigenetic clocks are related to factors associated with cardiovascular health,12–19 epigenetic clock associations with hypertension have not been carefully examined in published studies.
The methodologies used to develop epigenetic clocks have become increasingly sophisticated. For example, the PhenoAge and GrimAge epigenetic clocks were designed by selecting sets of cytosine-phosphate-guanine (CpG) sites where DNAm correlates with mortality-associated clinical and molecular markers.9,10 The PhenoAge epigenetic clock is based on a set of CpGs where blood DNAm is predictive of a previously developed clinical ‘phenotypic age’ risk score based on 9 blood measures (albumin, creatinine, glucose, C-reactive protein, alkaline phosphatase, white blood cell count, lymphocyte percent, red blood cell width and volume) and chronological age.9 The GrimAge epigenetic clock was developed by first identifying independent sets of CpGs that were predictive of 7 circulating proteins (adrenomedullin, beta-2-microglobulin, cystatin C, growth differentiation factor 15, leptin, plasma activator inhibitor-1, and tissue metalloproteinase-1) and smoking history (pack years), and then combining these DNAm predicted proteins and smoking metrics with chronological age and sex.10 The PhenoAge and GrimAge epigenetic clocks provide biological age predictions that are strongly correlated with chronological age. These estimated biological ages are used, in turn, to estimate “age acceleration:” the difference (calculated using residuals from linear models regressing biological age on chronological age) between a person’s predicted biological age and their known chronological age. More recently, the DunedinPACE epigenetic clock was designed by selecting CpGs where DNAm is predictive of the previously developed “Pace of aging” metric.11,20 The Pace of Aging metric reflects 19 biomarkers of the cardiovascular, metabolic, renal, hepatic, immune, dental, and pulmonary systems that were serially measured in a birth cohort over the course of twenty years.11 By design, DunedinPACE has a mean value of 1 which reflects the average amount of aging in a year observed in the original cohort study. For convenience and in order to distinguish it from the age acceleration metrics produced by the PhenoAge and GrimAge clocks, we refer to DunedinPACE as a measure of “aging rates.” These three epigenetic clocks have varying associations with lifestyle factors, environmental exposure, and disease incidence, suggesting that they capture different aspects of aging.11,21–29
Here, using data from a racially diverse, prospective cohort of nearly 4,500 women, we examine biological age associations with hypertension prevalence and incidence. By examining both cross-sectional and prospective associations between epigenetic clocks and hypertension, we are able to determine whether changes in biological age precede or follow the clinical onset of hypertension.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request. More information about obtaining data from the Sister Study can be found at: https://sisterstudy.niehs.nih.gov/English/coll-data.htm.
Study population
The Sister Study is a prospective cohort of 50,884 women enrolled between 2003–2009. The study was designed to identify environmental and biological factors associated with breast cancer and other chronic diseases.30 Eligible women were aged between 35 and 74 years, living in the United States or Puerto Rico, and did not have a personal history of breast cancer but had a biological sister (full or half) previously diagnosed. As part of study enrollment, women completed an extensive computer-assisted telephone interview to ascertain demographic and lifestyle characteristics, medication use, and health information. During an enrollment home visit, trained medical examiners followed standardized protocols to collect whole blood samples and measure height, weight, and resting blood pressure.30,31 Participants are recontacted annually and are asked to self-report any major changes in health; the annual response rate is nearly 90%. Every two to three years, a more comprehensive questionnaire is mailed to ask about changes in lifestyle, exposure, and health. Written informed consent was collected at the home visit and the Institutional Review Board of the National Institutes of Health oversees the study.
Whole blood DNA methylation assessment and quality control
Two samples of women were selected for DNAm profiling.32 Women were eligible for DNAm profiling if they self-identified as non-Hispanic White or Black, did not have prevalent cancer at the time of the blood draw, and had baseline blood available. In 2014, blood DNA samples from 2,878 self-identified non-Hispanic White women were assayed on the Infinium HumanMethylation450 BeadChip. This sample was selected using a case-cohort design and included 1,542 women who had developed breast cancer in the years following their enrollment.33,34 In 2019, blood DNA samples from 2,166 self-identified Black (Hispanic or non-Hispanic) and non-Hispanic White women were assayed on the Infinium MethylationEPIC BeadChip, which included 541 samples that were selected in the earlier sample. This sample was also selected using a case-cohort design for breast cancer and included 999 women who had developed breast cancer in the years following enrollment. For women with available DNAm data from both BeadChips, data from the HumanMethylation450 BeadChip were used.
For both samples, genomic DNA was extracted from whole blood aliquots using an automated system (AutoPure, Gentra Systems) in the NIEHS Molecular Genetics Core Facility or using DNAQuik at BioServe Biotechnologies LTD (Beltsville, MD). Extracted DNA was bisulfate-converted using the EZ DNA Methylation Kit (Zymo Research, Orange County, CA). Samples were tested for complete bisulfate conversion, and converted DNA was analyzed using Illumina’s Infinium BeadChip protocols using high-throughput robotics to minimize batch effects. Methylation analysis was carried out at the NIH Center for Inherited Disease Research (Baltimore, MD) in 2014 and at the National Cancer Institute (Bethesda, MD) in 2019.
DNAm data was preprocessed using the ENmix software pipeline, which included background correction, dye-bias correction, inter-array normalization, and probe-type bias correlation.35–37 Samples were excluded if they did not meet quality control measure including bisulfate intensity < 4,000 or < 4,750 for the 450k and EPIC samples, had greater than 5% of probes with low quality methylation values (detection P > 0.000001, < 3 beads, or values outside 3 times the interquartile range), or were outliers for their methylation beta value distributions. In total, 4,911 samples passed quality control, including 428 duplicates across the two arrays, resulting in available DNAm data for 4,483 women (Figure S1A). For all samples that passed quality control, peripheral immune cell composition was derived using the Salas et al. reference panels and used to estimate circulating proportions of granulocytes, monocytes, B cells, CD8+ and CD4+ T cells, and natural killers.38
Biological age metric calculation
Epigenetic age acceleration represents the interval between a person’s chronological age and their predicted biological age. It is calculated as the residuals from linear regression models where the DNAm-predicted age is treated as the dependent variable and chronological age is treated as the independent variable. Thus, age acceleration values are, by design, uncorrelated with chronological age. Age acceleration values can be either positive or negative; positive values represent accelerated aging and are associated with greater risk of mortality whereas negative values represent decelerated aging and are associated with a lower risk of mortality.9,10 The epigenetic age acceleration values based on the PhenoAge and GrimAge epigenetic clocks are called PhenoAgeAccel and GrimAgeAccel and were obtained separately for the HumanMethylation450 and MethylationEPIC subsamples using an online calculator (https://dnamage.genetics.ucla.edu/home).
The DunedinPACE metric was developed by identifying genomic positions where DNAm predicts age-related changes in cardiovascular, metabolic, renal, hepatic, immune, dental, and pulmonary function and is proposed to reflect rate of aging.11 DunedinPACE values are always positive; values greater than one represent faster rates of aging whereas values less than one represent slower rates of aging. The DunedinPACE metric was calculated separately for the HumanMethylation450 and MethylationEPIC subsamples using the methylAge function within the ENmix R package on Bioconductor (https://www.bioconductor.org/packages/release/bioc/html/ENmix.html)
Blood pressure assessment and hypertension classification
At the enrollment home visit, resting systolic and diastolic blood pressure (SBP, DBP) were measured in a sitting position three times by trained medical examiners using an aneroid sphygmomanometer (model 760 & 775X; American Diagnostic Corporation). Measurements were taken from alternating arms, starting with the left arm using a left-right-left protocol, approximately two minutes apart. Examinations were scheduled, whenever possible, in the morning. The average of the second and third blood pressure measurements were used in this study. Less than 2% of the study population was missing one or two of the blood pressure measurements; for those women, the second measurement or the single value was used. Women were classified as hypertensive at baseline if they met any of the following criteria: SBP ≥ 140 mmHg, DBP ≥ 90 mmHg, or self-reported current use of antihypertensive medication.
Over the course of study follow-up, women had the opportunity to report an incident diagnosis of hypertension via either annual health update or detailed triennial follow-up questionnaire. On the annual health update, women responded to the question: “Since (last date of contact), has a doctor or other health professional told you that you had any of the following conditions? … Hypertension?” Women who responded yes were also asked to self-report the month and year of the diagnosis. Women who reported a hypertension diagnosis via the triennial questionnaire responded to the question: “Has a doctor or other health professional ever told you that you had hypertension or high blood pressure?” Among those responding yes, women reported when the diagnosis occurred. Participants also reported any new use of antihypertensive medications. Of the 422 women reporting an incident hypertension diagnosis, 305 (72%) reported new use of antihypertensive medication. In all analyses of incident hypertension, women classified as hypertensive at enrollment were excluded.
Statistical analysis
Among the 4,483 women with available DNAm data, 4,419 women remained after excluding those with extreme outlier biological age metrics values, defined as outside 4 standard deviations from the mean, and those missing baseline blood pressure or medication use data (Figure S1B). Among the 2,847 women classified as hypertensive-free at baseline, 2,610 had available information on hypertension status through September 30, 2019 (Data Release 9.1) (Figure S1C).
To account for the case-cohort designs used to sample women for DNAm assessment, all analyses incorporated inverse probability of selection weights so the results are generalizable to the full sample of Black and non-Hispanic White women enrolled in the Sister Study cohort.39 For example, women sampled because they later developed breast cancer were given smaller weight values (~1) whereas women who remained breast cancer-free were assigned larger values (~20). Sample characteristics are described using weighted means and standard deviations or weighted proportions overall and stratified by hypertension status at baseline. Associations between the biological age metrics and participant characteristics were estimated using weighted linear regression models treating the biological age metrics as the dependent variable.
Associations between prevalent hypertension and the biological age metrics (per 1-standard deviation increase) were estimated using weighted logistic regression models. In the models, hypertension status at baseline was treated as the dependent variable and the biological age metrics as the independent variables. Associations were examined in minimally-adjusted models that included only baseline chronological age and methylation platform (HumanMethylation450, MethylationEPIC) and fully-adjusted models that additionally accounted for established hypertension risk factors that are associated with DNAm, including: self-reported race (non-Hispanic White, Black), body mass index (kg/m2), alcohol consumption (drinks/wk.), smoking history (total pack years), physical activity (metabolic equivalent tasks/wk.), diet quality (dietary approaches to stop hypertension index score), and menopause status (premenopausal, postmenopausal). Associations were reported for the full sample population overall, and sensitivity analyses were performed among only the women who remained cancer-free and adjusting for peripheral immune cell composition. In the full sample, we also report associations stratified by time since blood draw, menopause status, body mass index, and self-reported race. To determine whether biological age was more strongly associated with the physical state of high blood pressure or antihypertensive medication use, associations were estimated using separate logistic regression models treating as dependent variables either high blood pressure (women with SBP ≥ 140 or DBP ≥ 90 vs women with lower blood pressure; adjusted for antihypertensive medication use) or antihypertensive medication use (current users vs non-users; adjusted for blood pressure) and the biological age metrics as the independent variables.
In a separate analysis, among women who reported no current use of antihypertensive medication, cross-sectional associations with blood pressure measurements collected at baseline were estimated using separate weighted linear regression models that were minimally and fully-adjusted where the blood pressure measurement (SBP, DBP) was treated as the dependent variable and the biological age metric as the independent variable.
Prospective hypertension associations with the three biological age metrics were estimated using weighted Cox regression models. In all models, age was treated as the primary timescale;40 left truncation was determined by age at blood draw and right censoring was determined by age of the hypertension event or end of study follow-up (September 30, 2019). Because women with a history of breast cancer experience higher rates of hypertension in the years following diagnosis,41 women were censored for development of hypertension at the time of their breast cancer diagnosis. Associations with incident hypertension were examined in minimally and fully-adjusted models using the covariate sets listed earlier; baseline SBP and DBP were also included in the fully adjusted covariate set. Associations were examined for the population overall and stratified by baseline blood pressure (SBP < 120 & DBP < 80 vs 120 < SBP < 140 or 80 < DBP < 90), time since blood draw, menopause status, body mass index, and self-reported race. The size of the association estimates and confidence intervals were used to determine meaningful associations. All analyses were conducted using Stata version 17 (College Station, TX).
Results
Participant characteristics and biological age metrics
At enrollment, women in the study sample had a weighted mean age of 56 years (standard deviation: 9 years), 90% self-identified as non-Hispanic White, and 68% were postmenopausal (Table 1). Of the 4,419 women included, 1,572 (36%) were classified as having prevalent hypertension based on baseline blood pressure measurements and antihypertensive medication use. Of these, 1,491 (95%) reported current antihypertensive medication use and 264 (17%) had high blood pressure, defined as either SBP ≥ 140 mmHg or DBP ≥ 90 mmHg. Higher chronological age was positively associated with hypertension prevalence before and after adjustment for established hypertension risk factors (Table S1). Compared to the 2,847 women who were classified as normotensive at baseline, the women with prevalent hypertension were older (weighted mean: 60 years vs 54 years), had a higher mean body mass index (weighted mean: 31 kg/m2 vs 26 kg/m2), had a higher mean personal smoking history (weighted mean: 9 pack years vs 6 pack years), and were more likely to self-identify as Black (weighted proportion: 15% vs 7%) (Table 1).
Table 1.
Survey weighted participant characteristics overall and stratified by baseline hypertension status (N=4,419)
| Characteristic | Overall (N=4,419) |
No Hypertension (N=2,847) |
Hypertension (N=1,572) |
|---|---|---|---|
|
| |||
| Age, mean years (SD) | 56 (9) | 54 (8) | 60 (8) |
| Household income, mean thousand dollars (SD) | 43 (28) | 44 (27) | 42 (30) |
| Body mass index, mean kg/m2 (SD) | 28 (6) | 26 (5) | 31 (7) |
| Physical activity, mean METs (SD) | 51 (32) | 53 (31) | 47 (31) |
| DASH, mean score (SD) | 24 (5) | 24 (5) | 24 (5) |
| Smoking history, mean pack-years (SD) | 7 (13) | 6 (11) | 9 (16) |
| Alcohol use, mean drinks/wk. (SD) | 2.8 (5) | 2.8 (4) | 2.8 (6) |
| Parity, mean number (SD) | 1.9 (1) | 1.8 (1) | 2.1 (1) |
| SBP, mean mmHg (SD) | 115 (14) | 111 (11) | 123 (15) |
| DBP, mean mmHg (SD) | 72 (9) | 71 (8) | 76 (10) |
| Race (%) | |||
| Non-Hispanic White | 90 | 93 | 85 |
| Black | 10 | 7 | 15 |
| Education (%) | |||
| High school/GED or less | 15 | 13 | 20 |
| Some college | 33 | 31 | 38 |
| College graduate or more | 52 | 56 | 42 |
| Smoking status (%) | |||
| Never | 54 | 56 | 52 |
| Former | 37 | 35 | 40 |
| Current | 9 | 9 | 8 |
| Menopause status (%) | |||
| Premenopausal | 32 | 40 | 18 |
| Postmenopausal | 68 | 60 | 82 |
| Hormone therapy1 (%) | |||
| Non-user | 86 | 86 | 87 |
| Currently using | 14 | 14 | 13 |
| Hormone birth control (%) | |||
| Non-user | 96 | 94 | 99 |
| Currently using | 4 | 6 | 1 |
Among postmenopausal women
The PhenoAge and GrimAge epigenetic clocks were positively correlated with chronological age (Figure S2). In contrast, the age acceleration metrics based of these clocks were, as expected, independent of chronological age (Figure 1A). The DunedinPACE metric was also independent of chronological age (Figure 1A). All three biological age metrics were positively correlated with each other (Figure 1B). The strongest correlation was for the GrimAgeAccel and DunedinPACE metrics (weighted ρ: 0.62), whereas the weakest was for the PhenoAgeAccel and DunedinPACE metrics (weighted ρ: 0.37). There was no association between PhenoAgeAccel and self-reported race; however, GrimAgeAccel and DunedinPACE were higher in Black women (Figure 1C) and all three metrics were higher among women with body mass indexes greater than 25 kg/m2 (Figure 1D). The biological age metrics showed little association with reproductive factors, including exogenous hormone use, parity, or menopause status; however, the metrics were inversely associated with educational attainment and positively associated with smoking status (Table S2).
Figure 1. Biological age metric characteristics.
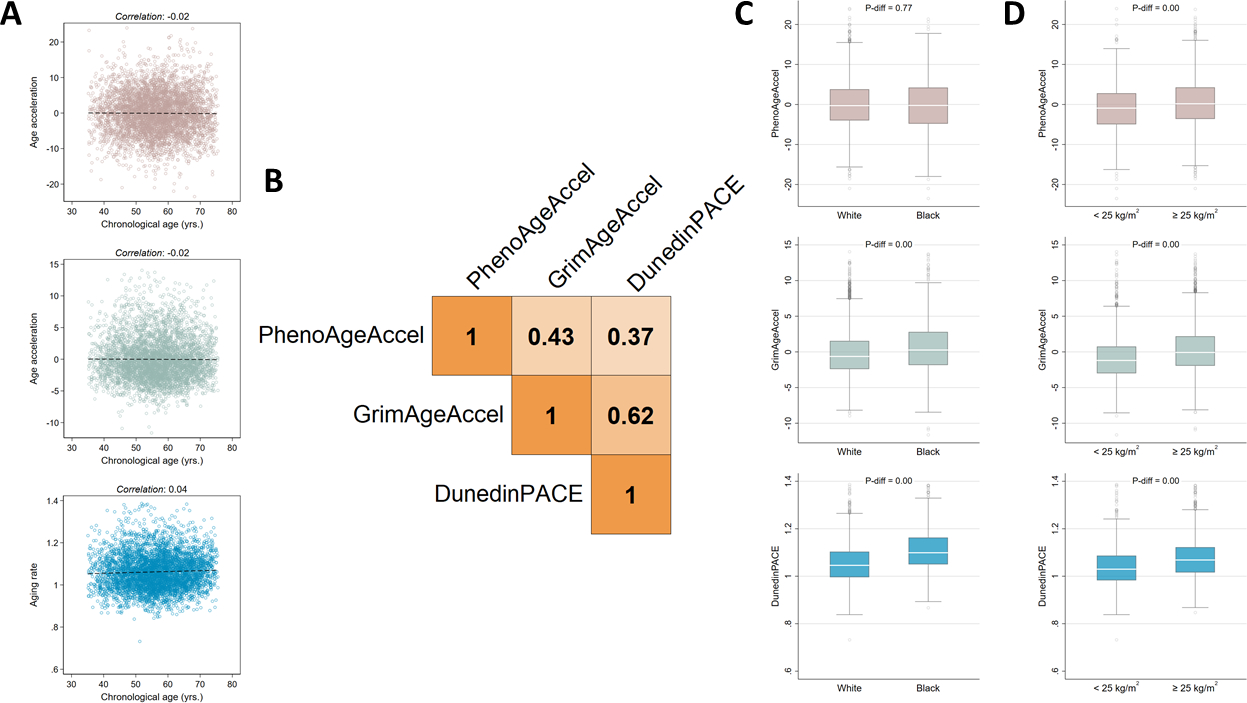
Scatter plots, linear fit lines and Pearson correlation coefficients for the relationship between the three biological age metrics and chronological age (A). A Pearson correlation coefficient matrix for the relationships between the three biological age metrics (B). Box plots for the three biological age metrics, stratified by self-reported race (C) and body mass index group (D).
Biological age and prevalent hypertension at baseline
In minimally adjusted models, all three biological age metrics were positively associated with prevalent hypertension and remained so after accounting for lifestyle and behavioral risk factors (per 1-SD increase; PhenoAgeAccel, adjusted OR: 1.16, 95% CI: 1.05, 1.28, P= 0.004; GrimAgeAccel, adjusted OR: 1.28, 95% CI: 1.14, 1.45, P< 0.001; DunedinPACE, adjusted OR: 1.16, 95% CI: 1.03, 1.30, P= 0.01) (Figure 2; Table S3). In sensitivity analyses, associations were similar when restricting the analysis to women who remained breast cancer-free over follow-up (Table S4). Adjustment for blood cell composition slightly attenuated the associations (Table S5). Biological age associations with prevalent hypertension were consistent across different times since diagnosis for PhenoAgeAccel and GrimAgeAccel; however, DunedinPACE associations appeared stronger for women diagnosed within 7.5 years of the baseline blood draw (Table S6). Hypertension associations were similar when stratified by menopause status, body mass index, or race (Figure S3). Because women could be classified as hypertensive based on either current medication use or their measured blood pressure at enrollment, we also examined these two groups separately. The biological age associations appeared to be stronger among those defined by medication use than among those defined by high blood pressure at enrollment (Figure S4).
Figure 2. Associations for the biological age metrics and prevalent hypertension.
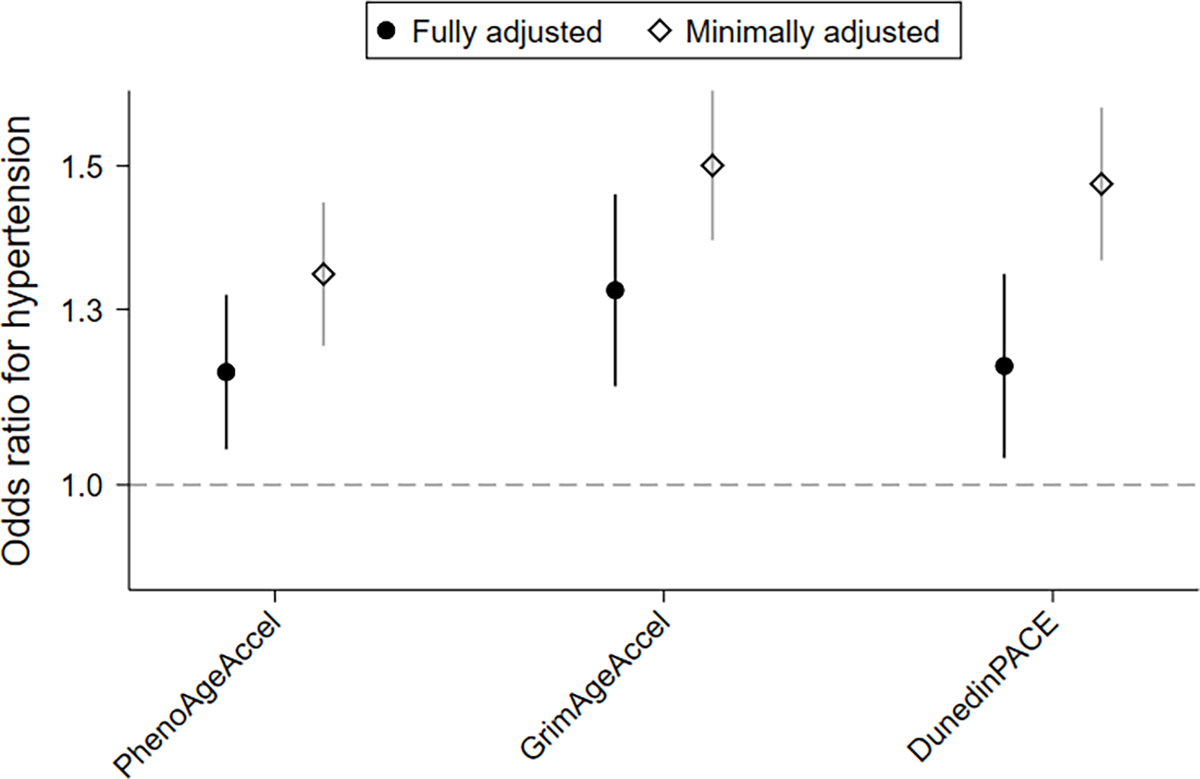
Prevalent hypertension associations estimated using separate weighted logistic regression models treating hypertension status at baseline as the outcome and biological age metrics as predictors. Fully adjusted models include age, self-reported race, body mass index, alcohol consumption, smoking history, menopause status, physical activity, dietary index, and methylation platform; minimally adjusted only include age and methylation platform.
Biological age and blood pressure at baseline
Among the 2,928 women at baseline who were not currently taking antihypertensive medication, the weighted mean SBP was 112 mmHg and the weighted mean DBP was 71 mmHg. In minimally adjusted models that did not account for established hypertension risk factors, all three biological age metrics were positively associated with systolic and diastolic blood pressure (Table S7). However, in fully adjusted models the associations were substantially shifted towards the null.
Biological age and hypertension incidence
The 2,847 women who were classified as normotensive at baseline were followed up for an average of 10 years for the development of new-onset hypertension; 422 of whom reported an incident hypertension diagnosis. In minimally-adjusted analysis all three metrics were positively associated with hypertension incidence and remained so after accounting for lifestyle and behavioral risk factors (PhenoAgeAccel, adjusted HR: 1.09, 95% CI: 0.97, 1.23, P= 0.16; GrimAgeAccel, adjusted HR: 1.16, 95% CI: 0.99, 1.36, P= 0.07; DunedinPACE, adjusted HR: 1.16, 95% CI: 1.01, 1.33, P= 0.04) (Figure 3; Table S8). Further adjustment for blood cell composition did not appreciably alter associations (Table S9).
Figure 3. Prospective associations for the biological age metrics and hypertension incidence.
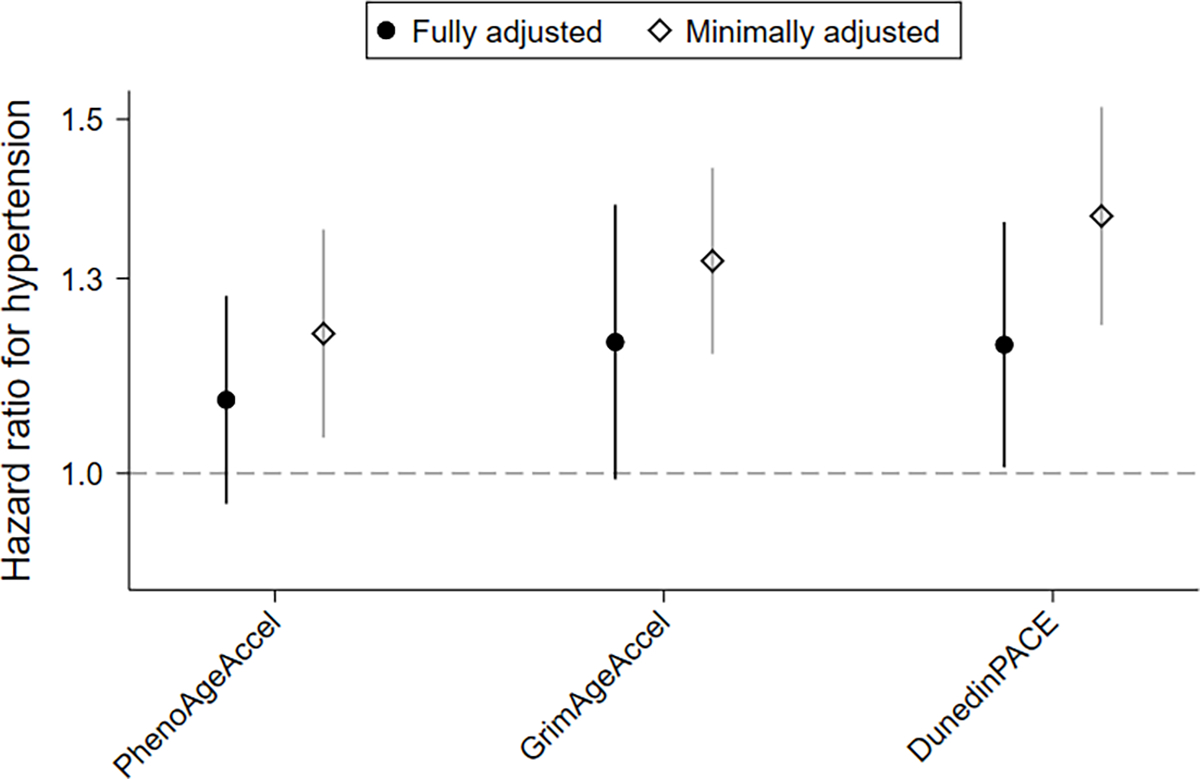
Hypertension incidence associations estimated using weighted Cox regression models treating new-onset hypertension as the outcome. Using age as the time scale, fully adjusted models include self-reported race, body mass index, alcohol consumption, smoking history, menopause status, physical activity, dietary index, blood pressure, and methylation platform; minimally adjusted only include methylation platform.
We also examined associations with hypertension incidence after stratifying by various characteristics. Although there was little difference in the strengths of the hypertension incidence associations for PhenoAgeAccel and GrimAgeAccel by baseline blood pressure, DunedinPACE associations with hypertension incidence appeared stronger for women who had blood pressure measurements in the higher end of the normal range at baseline (among women with 120 < SBP < 140 or 80 < DBP < 90, HR: 1.27, 95% CI: 1.05, 1.54, P= 0.01; among women with SBP < 120 & DBP < 80, HR: 1.08, 95% CI: 0.87, 1.34, P= 0.47) (Figure S5). When analyses were stratified by follow-up time, the association with GrimAgeAccel appeared stronger for incident hypertension diagnosed within the years immediately following blood draw (within 3 years, HR: 1.43, 95% CI: 1.12, 1.83, P= 0.004; after 3 years, HR: 1.10, 95% CI: 0.91, 1.33, P= 0.34) (Figure S6). When analyses were stratified by participant characteristics, hypertension incidence associations with GrimAgeAccel were stronger for women with lower body mass indexes (< 25 kg/m2, HR: 1.33, 95% CI: 1.03, 1.72, P= 0.03; ≥ 25 kg/m2, HR: 1.12, 95% CI: 0.92, 1.37, P= 0.26) (Figure S7).
Discussion
Based on a racially diverse sample of nearly 4,500 women enrolled in a prospective cohort, we examined associations between methylation-based biological age and both prevalent and incident hypertension. Compared to normotensive women, women classified as hypertensive at baseline had higher PhenoAgeAccel, GrimAgeAccel and DunedinPACE. Associations were slightly attenuated after adjustment for blood cell composition. Our group has previously reported that circulating leukocyte proportions are altered in women with hypertension;32 thus, associations between biological age and prevalent hypertension could be partly explained by differences in blood cell composition. In cross-sectional analyses of blood pressure at enrollment, none of the biological age metrics were associated after adjustment for lifestyle and behavioral risk factors. Finally, among women who were normotensive at study enrollment, the incidence of new-onset hypertension in the following years was positively associated with all three biological age metrics. Together, these observations suggest that increases in biological age precede the development of hypertension and appear to remain elevated after clinical diagnosis and treatment.
All three biological age metrics were elevated in women with hypertension. In our classification, we included both women on hypertensive medication and women who had high blood pressure at baseline. Stratification of these women into two groups revealed that the biological age associations appeared to be stronger among women who had their blood pressure under control through use of medication. Our findings are consistent with an earlier study among men enrolled in the Normative Aging Study that observed biological age was higher for those using antihypertensive medications.42 The fact that biological age metrics remain elevated despite successful treatment lends support to the hypothesis that these metrics are not directly responding to increased blood pressure, but rather are reflective of the underlying age-related molecular conditions giving rise to hypertension.
To begin testing this hypothesis, we estimated associations between the biological age metrics and direct blood pressure measurements. Prior cross-sectional studies have reported that systolic blood pressure, analyzed as a continuous variable, is positively correlated with both PhenoAgeAccel and GrimAgeAccel.9,10,16–18 In minimally-adjusted models we found similar positive associations for both PhenoAgeAccel and GrimAgeAccel but, after adjusting for established cardiovascular disease risk factors, found little evidence of systolic blood pressure associations with either metric. A similar pattern of association was observed for DunedinPACE. These observations suggest that the biological age metrics may be more closely associated with cardiovascular disease risk factors than to blood pressure itself.
Biological age metrics correlate with lifestyle factors and health scores that in turn are associated with hypertension incidence.12–16,21–23 To our knowledge, no prospective study has directly examined the relationship between biological age and incident hypertension. We found that all three biological age metrics were higher among women who went on to be diagnosed with hypertension suggesting that increases in biological age occur in the years preceding the clinical onset of hypertension. This interpretation is supported by the stratified analyses in which the metrics become elevated only in the years directly preceding diagnosis (GrimAgeAccel) and among women with baseline blood pressure measurements in the higher end of the normal range (DunedinPACE). Notably, the biological age associations remained after adjustments for established hypertension risk factors, supporting the hypothesis that blood DNAm profiles capture additional information related to hypertension risk.
This study is not without limitations. Important differences in blood pressure regulation and hypertension have been reported by sex,43,44 so the findings from this study may not extend to men. In our analysis of hypertension incidence, we relied on self-reported information which may result in some misclassification of outcomes. However, this misclassification is probably small, as women were asked about a new hypertension diagnosis on an annual basis and women reporting a new diagnosis of hypertension were also likely to report the initiation of antihypertensive medication on the detailed follow-up questionnaire. Finally, our study used the higher diagnostic cut-point of SBP ≥ 140 mmHg or DBP ≥ 90 mmHg in determining hypertension status at enrollment and so may have missed some cases that would have been captured using lower cut-points. These were the recommended thresholds at the time of study enrollment and would have been used in community practice during follow-up. Despite these limitations, this study is strengthened by the large and diverse sample, the combination of both prospective and case-control designs, and the examination of different biological age metrics.
Perspectives
Here, we find that women with accelerated biological age are at increased risk of developing incident hypertension. This risk persists even after adjustment for known hypertension risk factors, suggesting that DNAm biological age is capturing a unique aspect of hypertension risk. We also note that the biological age metrics were elevated even after clinical diagnosis and successful hypertension treatment. While the clinical utility of biological age metrics for hypertension prediction is unknown, our findings help clarify the molecular links between aging and hypertension.
Supplementary Material
Pathophysiological Novelty and Relevance.
What is new?
Women classified as hypertensive at baseline had higher biological age than normotensive women
Among women who were normotensive at baseline, higher biological age was associated with higher risk of developing hypertension
What is relevant?
Biological age metrics appear to detect unique aspects of hypertension risk that are not related to common behavioral and lifestyle risk factors
Successful treatment of hypertension through the use of antihypertensive medications may not address the age-related molecular changes that give rise to hypertension
Clinical/Pathophysiological implications?
Biological age metrics reflect the age-related biological consequences of hypertension
Biological age metrics could be useful for hypertension risk stratification
Sources of funding:
This research was supported by the Intramural Research Program of the National Institutes of Health (Z01-ES049033, Z01-ES049032, Z01-ES044005).
Non-standard abbreviations:
- CpG
cytosine-phosphate-guanine
- DBP
diastolic blood pressure
- DNAm
DNA methylation
- SBP
systolic blood pressure
Footnotes
Disclosures: The authors report no competing interests
References
- 1.Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah AD, Denaxas S, White IR, Caulfield MJ, Deanfield JE, Smeeth L, Williams B, Hingorani A & Hemingway H Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1.25 million people. Lancet 383, 1899–1911 (2014). 10.1016/S0140-6736(14)60685-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Cai X, Zhang J, Mai W, Wang S, Hu Y, Ren H & Xu D Prehypertension and Incidence of ESRD: a systematic review and meta-analysis. Am J Kidney Dis 63, 76–83 (2014). 10.1053/j.ajkd.2013.07.024 [DOI] [PubMed] [Google Scholar]
- 3.Lawes CM, Bennett DA, Feigin VL & Rodgers A Blood pressure and stroke: an overview of published reviews. Stroke 35, 1024 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ostchega Y, Fryar CD, Nwankwo T & Nguyen DT Hypertension Prevalence Among Adults Aged 18 and Over: United States, 2017–2018. NCHS Data Brief, 1–8 (2020). [PubMed] [Google Scholar]
- 5.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102, 10604–10609 (2005). 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madrigano J, Baccarelli A, Mittleman MA, Sparrow D, Vokonas PS, Tarantini L & Schwartz J Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics 7, 63–70 (2012). 10.4161/epi.7.1.18749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P & Baccarelli A Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130, 234–239 (2009). 10.1016/j.mad.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irvin MR, Jones AC, Claas SA & Arnett DK DNA Methylation and Blood Pressure Phenotypes: A Review of the Literature. Am J Hypertens 34, 267–273 (2021). 10.1093/ajh/hpab026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10, 573–591 (2018). 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L & Horvath S DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11, 303–327 (2019). 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife 11 (2022). 10.7554/eLife.73420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemke E, Vetter VM, Berger N, Banszerus VL, Konig M & Demuth I Cardiovascular health is associated with the epigenetic clock in the Berlin Aging Study II (BASE-II). Mech Ageing Dev 201, 111616 (2022). 10.1016/j.mad.2021.111616 [DOI] [PubMed] [Google Scholar]
- 13.Pottinger TD, Khan SS, Zheng Y, Zhang W, Tindle HA, Allison M, Wells G, Shadyab AH, Nassir R, Martin LW et al. Association of cardiovascular health and epigenetic age acceleration. Clin Epigenetics 13, 42 (2021). 10.1186/s13148-021-01028-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce BT, Gao T, Zheng Y, Ma J, Hwang SJ, Liu L, Nannini D, Horvath S, Lu AT, Bai Allen N et al. Epigenetic Age Acceleration Reflects Long-Term Cardiovascular Health. Circ Res 129, 770–781 (2021). 10.1161/CIRCRESAHA.121.318965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nannini DR, Joyce BT, Zheng Y, Gao T, Liu L, Yoon G, Huan T, Ma J, Jacobs DR Jr., Wilkins JT et al. Epigenetic age acceleration and metabolic syndrome in the coronary artery risk development in young adults study. Clin Epigenetics 11, 160 (2019). 10.1186/s13148-019-0767-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ammous F, Zhao W, Ratliff SM, Mosley TH, Bielak LF, Zhou X, Peyser PA, Kardia SLR & Smith JA Epigenetic age acceleration is associated with cardiometabolic risk factors and clinical cardiovascular disease risk scores in African Americans. Clin Epigenetics 13, 55 (2021). 10.1186/s13148-021-01035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lind L, Ingelsson E, Sundstrom J, Siegbahn A & Lampa E Methylation-based estimated biological age and cardiovascular disease. Eur J Clin Invest 48 (2018). 10.1111/eci.12872 [DOI] [PubMed] [Google Scholar]
- 18.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 9, 419–446 (2017). 10.18632/aging.101168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao L, Zan G, Liu C, Xu X, Li L, Chen X, Zhang Z & Yang X Associations Between Blood Pressure and Accelerated DNA Methylation Aging. J Am Heart Assoc 11, e022257 (2022). 10.1161/JAHA.121.022257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belsky DW, Caspi A, Houts R, Cohen HJ, Corcoran DL, Danese A, Harrington H, Israel S, Levine ME, Schaefer JD et al. Quantification of biological aging in young adults. Proc Natl Acad Sci U S A 112, E4104–4110 (2015). 10.1073/pnas.1506264112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kresovich JK, Martinez Lopez AM, Garval EL, Xu Z, White AJ, Sandler DP & Taylor JA Alcohol consumption and methylation-based measures of biological age. The Journals of Gerontology: Series A 76, 2107–2111 (2021). 10.1093/gerona/glab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kresovich JK, Park YM, Keller JA, Sandler DP & Taylor JA Healthy eating patterns and epigenetic measures of biological age. Am J Clin Nutr 115, 171–179 (2022). 10.1093/ajcn/nqab307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kresovich JK, Garval EL, Martinez Lopez AM, Xu Z, Niehoff NM, White AJ, Sandler DP & Taylor JA Associations of Body Composition and Physical Activity Level With Multiple Measures of Epigenetic Age Acceleration. Am J Epidemiol 190, 984–993 (2021). 10.1093/aje/kwaa251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kresovich JK, Xu Z, O’Brien KM, Weinberg CR, Sandler DP & Taylor JA Methylation-based biological age and breast cancer risk. J Natl Cancer Inst 111, 1051–1058 (2019). 10.1093/jnci/djz020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kresovich JK, Harmon QE, Xu Z, Nichols HB, Sandler DP & Taylor JA Reproduction, DNA methylation and biological age. Human Reproduction 34, 1965–1973 (2019). 10.1093/humrep/dez149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kresovich JK, Xu Z, O’Brien KM, Weinberg CR, Sandler DP & Taylor JA Epigenetic mortality predictors and incidence of breast cancer. Aging (Albany NY) 11, 11975–11987 (2019). 10.18632/aging.102523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, Taylor JA & Sandler DP Air pollution, particulate matter composition and methylation-based biologic age. Environ Int 132, 105071 (2019). 10.1016/j.envint.2019.105071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White AJ, Kresovich JK, Xu Z, Sandler DP & Taylor JA Shift work, DNA methylation and epigenetic age. Int J Epidemiol 48, 1536–1544 (2019). 10.1093/ije/dyz027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence KG, Kresovich JK, O’Brien KM, Hoang TT, Xu Z, Taylor JA & Sandler DP Association of Neighborhood Deprivation With Epigenetic Aging Using 4 Clock Metrics. JAMA Netw Open 3, e2024329 (2020). 10.1001/jamanetworkopen.2020.24329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D’Aloisio AA, Suarez LM, Kleeberger CA, Shore DL, DeRoo LA, Taylor JA, Weinberg CR & Team SSR The Sister Study Cohort: Baseline Methods and Participant Characteristics. Environ Health Perspect 125, 127003 (2017). 10.1289/EHP1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SH, Van Hee VC, Bergen S, Szpiro AA, DeRoo LA, London SJ, Marshall JD, Kaufman JD & Sandler DP Long-Term Air Pollution Exposure and Blood Pressure in the Sister Study. Environ Health Perspect 123, 951–958 (2015). 10.1289/ehp.1408125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kresovich JK, Xu Z, O’Brien KM, Parks CG, Weinberg CR, Sandler DP & Taylor JA Peripheral Immune Cell Composition is Altered in Women Before and After a Hypertension Diagnosis. Hypertension 80, 43–53 (2023). 10.1161/HYPERTENSIONAHA.122.20001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kresovich JK, Xu Z, O’Brien KM, Shi M, Weinberg CR, Sandler DP & Taylor JA Blood DNA methylation profiles improve breast cancer prediction. Mol Oncol (2021). 10.1002/1878-0261.13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kresovich JK, O’Brien KM, Xu Z, Weinberg CR, Sandler DP & Taylor JA Prediagnostic Immune Cell Profiles and Breast Cancer. JAMA Netw Open 3, e1919536 (2020). 10.1001/jamanetworkopen.2019.19536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z, Niu L, Li L & Taylor JA ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res 44, e20 (2016). 10.1093/nar/gkv907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Langie SA, De Boever P, Taylor JA & Niu L RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics 18, 4 (2017). 10.1186/s12864-016-3426-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu L, Xu Z & Taylor JA RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics 32, 2659–2663 (2016). 10.1093/bioinformatics/btw285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salas LA, Zhang Z, Koestler DC, Butler RA, Hansen HM, Molinaro AM, Wiencke JK, Kelsey KT & Christensen BC Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. Nat Commun 13, 761 (2022). 10.1038/s41467-021-27864-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien KM, Lawrence KG & Keil AP The Case for Case-Cohort: An Applied Epidemiologist’s Guide to Reframing Case-Cohort Studies to Improve Usability and Flexibility. Epidemiology 33, 354–361 (2022). 10.1097/EDE.0000000000001469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiebaut AC & Benichou J Choice of time-scale in Cox’s model analysis of epidemiologic cohort data: a simulation study. Stat Med 23, 3803–3820 (2004). 10.1002/sim.2098 [DOI] [PubMed] [Google Scholar]
- 41.Kwan ML, Cheng RK, Iribarren C, Neugebauer R, Rana JS, Nguyen-Huynh M, Shi Z, Laurent CA, Lee VS, Roh JM et al. Risk of Cardiometabolic Risk Factors in Women With and Without a History of Breast Cancer: The Pathways Heart Study. J Clin Oncol, JCO2101738 (2022). 10.1200/JCO.21.01738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X, Colicino E, Shen J, Just AC, Nwanaji-Enwerem JC, Wang C, Coull B, Lin X, Vokonas P, Zheng Y, Hou L, Schwartz J & Baccarelli AA Accelerated DNA methylation age and the use of antihypertensive medication among older adults. Aging (Albany NY) 10, 3210–3228 (2018). 10.18632/aging.101626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joyner MJ, Wallin BG & Charkoudian N Sex differences and blood pressure regulation in humans. Exp Physiol 101, 349–355 (2016). 10.1113/EP085146 [DOI] [PubMed] [Google Scholar]
- 44.Gillis EE & Sullivan JC Sex Differences in Hypertension: Recent Advances. Hypertension 68, 1322–1327 (2016). 10.1161/HYPERTENSIONAHA.116.06602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


