Abstract
Children born preterm are at heightened risk of neurodevelopmental impairments, including Autism Spectrum Disorder (ASD). The placenta is a key regulator of neurodevelopmental processes, though the underlying molecular mechanisms remain unclear. Here, we employed a multi-omic approach to identify placental transcriptomic and epigenetic modifications related to ASD diagnosis at age ten, among children born preterm. Working with the Extremely Low Gestational Age (ELGAN) cohort, we hypothesized that a pro-inflammatory placental environment would be predictive of ASD diagnosis at age ten. Placental messenger RNA (mRNA) expression, CpG methylation, and microRNA (miRNA) expression were compared among 368 ELGANs (28 children diagnosed with ASD and 340 children without ASD). A total of 111 genes displayed expression levels in the placenta that were associated with ASD. Within these ASD-associated genes is an ASD regulatory complex comprising key genes that predicted ASD case status. Genes with expression that predicted ASD case status included Ewing Sarcoma Breakpoint Region 1 (EWSR1) (OR: 6.57 (95%CI: 2.34, 23.58)) and Bromodomain Adjacent To Zinc Finger Domain 2A (BAZ2A) (OR: 0.12 (95%CI: 0.03, 0.35)). Moreover, of the 111 ASD-associated genes, nine or 8.1% displayed associations with CpG methylation levels, while 14 or 12.6% displayed associations with miRNA expression levels. Among these, LRR Binding FLII Interacting Protein 1 (LRRFIP1) was identified as being under the control of both CpG methylation and miRNAs, displaying an OR of 0.42 (95% CI: 0.17, 0.95). This gene, as well as others identified as having functional epimutations, plays critical roles in immune system regulation and inflammatory response. In summary, a multi-omic approach was used to identify functional epimutations in the placenta that are associated with the development of ASD in children born preterm, highlighting future avenues for intervention.
Keywords: Multi-Omic, Autism Spectrum Disorder, Preterm Birth, Epigenetics, MicroRNA, CpG Methylation, messenger RNA, Placenta
Lay Summary:
Children who are born preterm have an increased risk for neurodevelopmental impairments including autism spectrum disorder (ASD). The placenta is known to play a pivotal role in child development, and is posited to regulate neurodevelopment as well. Our research reveals a significant number of placental genes expressed in relation to ASD, and that many of these genes are under the control of epigenetic processes. This research is important for understanding how the placenta may contribute to the development of ASD later in life in children born preterm.
Introduction:
Children born preterm are at an increased risk for neurodevelopment impairments, including complex developmental conditions such as Autism Spectrum Disorder (ASD) (Anderson, 2014; Chen et al., 2019). Increasing evidence suggests an association exists between the in utero environment, altered placental function, and child health later in life (Goyal, Limesand, & Goyal, 2019; Green & Arck, 2020; O’Donnell & Meaney, 2017). The current literature establishes a connection between intrauterine inflammation and preterm birth (Green & Arck, 2020; Humberg et al., 2020; Outcomes, 2007). Recent studies have demonstrated links between inflammation and early- and later-life neurodevelopmental outcomes, such as ASD (Jiang, Cowan, Moonah, & Petri, 2018; Leviton, Gressens, Wolkenhauer, & Dammann, 2015; Meltzer & Van de Water, 2017; Vohr, Poggi Davis, Wanke, & Krebs, 2017). The relationship between in utero inflammation and ASD risk later in life is also supported in preclinical models, where immune system dysregulation during pregnancy is tied to behavioral features characteristic of ASD (Favrais et al., 2011; Hagberg, Gressens, & Mallard, 2012; Hagberg et al., 2015; Patterson, 2011). While the in utero environment likely contributes to ASD, the specific components of the placental cellular machinery (e.g., transcriptomic, epigenomic, or proteomic) and key target genes underlying this association in children born preterm remains understudied.
Numerous studies have examined individual components of the placental cellular machinery to identify key genes and pathways as they relate to child health later in life (Bangma, Hartwell, Santos, O’Shea, & Fry, 2021; Oldenburg et al., 2021; Peng et al., 2017; Tilley et al., 2017). For example, our previous work uncovered novel associations between placental CpG methylation of key genes and cognitive outcomes at 10 years of age (Tilley et al., 2018). Still, there is limited research into how the various components of the placental cellular machinery interact with one another to regulate developmental processes. A multi-omic approach can provide a more robust understanding of the cellular contributions to later-life disease, specifically employing assessments of differential expression of mRNA, miRNAs, and CpG methylation (Hodyl, Roberts, & Bianco-Miotto, 2016; Schepici, Cavalli, Bramanti, & Mazzon, 2019). To address this, our research team recently employed a multi-omic kernel aggregation analysis of placental transcriptomic and epigenomic datasets to predict intellectual and social impairment, two neurological outcomes that are associated with ASD (Santos et al., 2020). Still, a gap exists in identifying the specific components of the placental cellular machinery, and key target genes, associated with ASD. This research aims to fill this gap by evaluating associations between the components of the placental cellular machinery and ASD, using a multi-omic approach.
The placenta is critical for healthy fetal development, as it serves as the regulator of the uterine environment by transporting nutrients and gas to the fetus, secreting hormones, and removing waste (Gude, Roberts, Kalionis, & King, 2004; Guttmacher, Maddox, & Spong, 2014). Abnormal placental function during pregnancy is tied to adverse later-life outcomes in the offspring including heart disease, obesity, and impaired neurodevelopment (Barker, Bull, Osmond, & Simmonds, 1990; Bronson & Bale, 2016). In particular, alterations in the placental transcriptome have been linked to brain damage and to the development of adverse neurodevelopmental outcomes (Bangma et al., 2021; Freedman et al., 2022; Marable, Roell, Kuban, O’Shea, & Fry, 2022; Oldenburg et al., 2021) supporting the placenta-brain-axis. Still, the role of the placental epigenome in regulating these transcriptome-level changes is not well established.
In this study, we conducted a multi-omic analysis integrating mRNA expression data, CpG methylation data, and miRNA expression data from the placenta. These were studied in relation to the development of ASD at 10 years of age in children from the Extremely Low Gestational Age Newborn (ELGAN) study, with replication in the Rhode Island Child Health Study (RICHS). Given the complexity of this developmental disorder, we used a multi-tiered approach to identify gene expression profiles in the placenta associated with ASD status, and to establish whether these genes were under the control of CpG methylation or miRNAs. This study is among the first to integrate numerous components of the placental cellular machinery to predict the onset of ASD in children.
Methods:
ELGAN Recruitment and Study Participants
The process for recruitment for the ELGAN study has been described in detail elsewhere (O’Shea et al., 2009). From 2002 to 2004, in 14 hospitals in five states in the United States, women who gave birth before 28 weeks gestational age were asked to participate in the study (O’Shea et al., 2009). The study was approved by the Institutional Review Board at each participating institution. After delivery, a trained nurse measured demographic and pregnancy variables using a structured questionnaire. The ELGAN cohort consists of 1,506 infants, from a total of 1,249 mothers recruited. Of this group, 1,405 placentas were collected. Subsequent analysis was restricted to those placentas that were available for ELGANs who were assessed at age 10 and placentas that had sufficient quality and quantity for the CpG methylation, mRNA sequencing, and miRNA sequencing resulted in a total of 368 samples for analysis. For this prospective case-control study, the exposures of interest are the placental transcriptomic and epigenomic measures, and the outcome of interest is ASD diagnosis at age 10, among infant born extremely preterm. A total of 28 placental samples were from ASD cases, and 340 placentas were from non-cases for the gene expression and CpG methylation analysis. For the miRNA analysis, a total of 27 placental samples were from ASD cases, and 339 placentas were from non-cases.
ASD diagnosis in ELGANs
All participants were assessed for ASD at 10 years of age (Joseph, O’Shea, et al., 2017). Diagnostic assessment of ASD was conducted with three well-validated measures, administered sequentially. First, the Social Communication Questionnaire (SCQ) was administered to screen for potential ASD, using a score ≥ 11 to increase sensitivity relative to the standard criterion score of ≥ 15 (Joseph, Korzeniewski, et al., 2017; Joseph, O’Shea, et al., 2017). For children who screened positive on the SCQ criterion, we conducted the Autism Diagnostic Interview–Revised (ADI-R) with the primary caregiver (Lord, Rutter, & Le Couteur, 1994). All children who met ADI-R criteria for ASD, or who had a prior clinical diagnosis of ASD and/or exhibited symptoms of ASD during cognitive testing according to the site psychologist were then assessed with the Autism Diagnostic Observation Schedule, Second Version (ADOS-2) Module 1, 2, or 3, depending on language level (no speech/single words, phrase speech, or fluent speech, respectively). The ADOS-2 served as the criterion measure of ASD in this study. All ADOS-2 administrations were independently scored by a second rater with autism diagnostic and ADOS-2 expertise, as detailed in Joseph 2017 (Joseph, O’Shea, et al., 2017).
General cognitive and language abilities
General cognitive ability (or IQ) was assessed with the School-Age Differential Ability Scales-II (DAS-II) Verbal (VIQ) and Nonverbal Reasoning (NVIQ) scales (Elliot, 2007). Language ability was evaluated with the Oral and Written Language Scales (OWLS) Oral Expression and Listening Comprehension subtests, which assess production and comprehension of elaborated sentences (Carrow-Woolfolk, 1995).
Sample Collection
Placental collection for the ELGAN study has been described previously (Onderdonk et al., 2008). Briefly, placentas were collected upon delivery and placed in a sterilized basin to transport to the sampling room for biopsy. The chorion was exposed via retraction of the amnion. A sample (<1 g) was removed by applying traction to the chorion and trophoblast tissue from the base of the chorion. Samples were placed in a sterile two-mL cryovial and subsequently submerged into liquid nitrogen and transferred to a −80°C freezer for long-term storage. Placental tissue samples were batch-shipped frozen to the University of North Carolina at Chapel Hill for processing. These samples were first processed by placing the placental biopsy-containing cryotubes on dry ice. Frozen tissue samples were sliced into roughly 0.02g segments using a sterile dermal curette, and washed in 1x PBS, to reduce blood contamination (Fisher Scientific, Waltham, MA). Washed samples were snap frozen immediately in homogenization tubes and placed back to dry ice to preserve sample integrity. Tissue segments were homogenized using a sterile stainless-steel bead (Qiagen, Germantown, MD) in RLT+ lysis buffer (Qiagen) with TissueLyserII (Qiagen). Following, samples were clarified by spinning to collect cellular debris and the bead. Homogenated samples were stored at −80°C until nucleic acid extraction.
Placental DNA and RNA Extraction and Quantification
The homogenized placental samples previously collected and frozen were thawed and used for nucleic acid extraction. Nucleic acids were extracted using an AllPrep DNA/RNA/miRNA Universal kit (Qiangen, Germany). The quantity and quality of DNA and RNA were analyzed using the NanoDrop 1000 spectrophotometer. The nucleic acid integrity was verified by the Agilent 2100 BioAnalyzer.
RNA sequencing
RNA molecules 18 nucleotides or greater were extracted using the AllPrep DNA/RNA/miRNA Universal kit (Qiagen), as previously noted. mRNA expression was determined using the Illumina QuantSeq 3’ mRNA-Seq Library Prep Kit, which has high strand specificity and can quantify transcripts with lower range RNA integrity numbers (RINs). mRNA-sequencing libraries were pooled and sequenced (single-end 50 bp) on one lane of the Illumina Hiseq 2500. mRNAs were quantified through pseudo-alignment with Salmon v.14.0 (Patro, Duggal, Love, Irizarry, & Kingsford, 2017), mapped to the GENCODE Release 31 (GRCh37) reference transcriptome. Raw and processed signature data are available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository under GEO series GSE154829 (NCBI, 2020).
Placental CpG methylation
DNA sequences previously extracted were bisulfate-converted using the EZ DNA methylation kit (Zymo Research, Irvine, CA), then quantified using the Infinium MethylationEPIC BeadChip (Illumina, San Diego), which measures CpG loci at a single nucleotide resolution, which has been previously described (Addo et al., 2019; Clark et al., 2019; Santos et al., 2019). Quality control and normalization were performed resulting in 3,747 CpG probes from downstream data processing and statistical analysis. Methylation is represented as the average methylation level at a single CpG site.
Differential expression testing of the ASD-associated mRNA
Similar to our prior genome-wide mRNA and miRNA analyses (Eaves et al., 2020; Payton et al., 2020) universally lowly expressed transcripts were excluded, requiring that >25% of the samples be expressed at signals above the overall median signal intensity. This resulted in n = 11,229 mRNA transcripts being included in the models. QA/QC was conducted on the count data in order to identify any subjects with outlier values. This was done utilizing hierarchical clustering and principal components, with the hclust and prcomp functions, respectively. This resulted in the removal of n= four outlier subjects that were clearly differentiated from the others, resulting in the final n=368.
The DESeq2 package (v 1.24.0) was used to normalize the count data, resulting in variance stabilized counts (Ashrap et al., 2020). The SVA package (v 3.32.1) was used to account for potential batch effects and sources of sample heterogeneity with control probes empirically estimated using default parameters. Three surrogate variables were calculated and included as covariates in the statistical model. A model was constructed based on covariates that were significantly associated (p < 0.05) or had known relationships with placental miRNA and/or mRNA expression levels and ASD. This was intended to capture potential sources of bias while mitigating loss of precision. This resulted in the inclusion of the following covariates: birthweight-for-gestational age (binned according to z-score normalized range: < −2; −2 ≥ and < −1; −1 ≥ and < 1; ≥1), gestational days (continuous), maternal age (continuous), maternal education (binned according to years of education: < 12; 12; 13–15; 16; > 16 years), race (categorical: white/black/other), and sex (categorical: male/female). Statistical methods employing negative binomial generalized linear models within DESeq2 were used to identify miRNAs and mRNAs with differential expression according to ASD, controlling for covariates listed previously as well as three surrogate variables. The p-values were adjusted using the Benjamini and Hochberg (BH) procedure to account for multiple testing. Differentially expressed mRNAs were defined as those with a false discovery rate (BH- adjusted p) < 0.1.
Identifying placental mRNAs that are predictors of ASD case status
For mRNAs identified as significantly differentially expressed in the DESeq2-based analysis, their capacity to predict ASD case status was estimated using logistic regression models to derive adjusted odds ratios (ORs). For calculating the adjusted ORs across all participants (n=366), each participant’s mRNA expression was first categorized as high (> median) or low (≤ median) for each of the 111 significantly differentially expressed genes. Then, an adjusted logistic regression model was fit with ASD as the dependent variable, and the binary high or low gene variable as the independent variable. The same covariates were incorporated as for the differential expression analysis (i.e., birthweight-for-gestational age, gestational age, race, maternal age, infant sex, and maternal education). Significance was set at p-value <0.05.
Identification of miRNA targets of the ASD-associated genes and miRNA differential expression analysis
Following the identification of ASD-associated genes, miRNAs predicted to target these mRNAs were identified using an in silico approach utilizing TargetScan(Vikram, 2018). TargetScan searches for the presence of sites that match the seed region of each miRNA. miRNA-mRNA interactions were queried, based on algorithms that identify potential matches between 3’-untranslated mRNA regions and miRNA seed sequences(V. Agarwal, G. W. Bell, J. W. Nam, & D. P. Bartel, 2015; Rager et al., 2014). The interactions were filtered for high predicted confidence, as defined by those with a context plus score < −0.4. The context plus score controls for factors influence miRNA targeting, including local adenine and uracil content, target site abundance, supplementary pairing, miRNA binding site type and location, and seed-pairing stability (Garcia et al., 2011).
The predicted miRNAs generated using TargetScan were then queried for differential expression in the placenta by ASD status. Differential expression of miRNAs was queried using similar methods to those described above with mRNA, including filtering out universally lowly expressed counts, QA/QC process, the same model specification and utilizing the DeSeq2 package. QA/QC identified n= two additional outliers in the miRNA data, resulting in n=366 subjects included in the miRNA models. Differentially expressed miRNAs were defined as those with a false discovery rate (BH- adjusted p) < 0.1.
Identification of CpG methylation sites of the ASD-associated genes and differential methylation analysis
To evaluate potential epigenetic control by CpG methylation, CpG sites mapped to ASD-associated genes were identified using the Infinium MethylationEPIC v1.0 B4 Manifest file (MethylationEPIC). In testing the hypothesis that placental methylation of CpG sites annotated to ASD-associated genes is associated with ASD diagnosis at age ten, we implemented a logistic regression model at each of the probes. The general model includes whether a child was diagnosed with ASD at age 10 and the average methylation level (M-value) for a probe located within a CpG site mapped to an ASD-associated gene, as measured in placental tissue. A beta coefficient was also incorporated into the model, representing the log(OR) of ASD diagnosis at age 10 corresponding to a one-unit increase in methylation at the respective CpG probe. An error term and a matrix of potential confounders representing adjustments made for birth weight, gestational age, maternal age, maternal education, race, and sex was also included in the model. Potential epigenetic control was then evaluated for CpG sites significantly associated with ASD diagnosis at age ten using a Pearson correlation test of CpG methylation levels (M-values) and mRNA expression levels (variance-stabilized counts). ASD-associated mRNAs were considered under putative epigenetic control by CpG methylation if CpG-mRNA pairings were significant (p < 0.05) and inversely (r<0) correlated. All statistical analysis was conducted in R (v.4.0.3).
Network analysis
To understand the relationship among the proteins encoded by the genes that were differentially expressed in relation to ASD in the placenta, a network analysis was created, generating the visual representation of the ASD regulatory complex. The list of 111 differentially expressed genes was analyzed using the Search Tool for Retrieval of Interacting Genes/Proteins (STRING), a web-based software for understanding protein-protein interaction networks and functional enrichment (STRING, 2022). The resulting network was graphed using Cytoscape, an open-source software platform for visualizing complex interactions within networks (Shannon et al., 2003). Five clusters were noted, as detailed in Supplemental Tables 2, 4 and 6. These clusters represent proteins encoded by the ASD-associated genes. Specifically, those that were: 1) CpG methylation-controlled; 2) miRNA-controlled; 3) CpG methylation- and miRNA-controlled; 4) epigenetic independent; and 5) predictors of ASD case status.
Biological pathway enrichment
To identify the functional effects of the differentially expressed genes in relation to ASD, QIAGEN’s Ingenuity Pathway Analysis (IPA), was employed coupled with a search of the current literature. Two gene sets were analyzed from the 111 genes expressed with relation to ASD; one comprising genes for which expression in the placenta was higher in children later diagnosed with ASD (n= four), and one comprising the genes for which expression in the placenta was lower in children later diagnosed with ASD (n=107). The resulting analyses were used to classify the members of the ASD-associated genes in terms of their biological functions and processes.
Replication of data from the RICHS Cohort
Data from the replication cohort were derived from the Rhode Island Child Health Study (RICHS) Recruitment for the RICHS has been described elsewhere (Bhattacharya et al., 2022;E. Kennedy et al., 2020). Briefly, enrolled mother-infant pairs from the Women & Infants Hospital in Providence, Rhode Island from 2010–2013. Mothers without life-threatening conditions at least 18 years of age were eligible, with singleton pregnancies without congenital/chromosomal abnormalities at or after 37 weeks of gestation. Additionally, RICHS oversampled for large-for-gestational age (>90th birth weight percentile) and small-for-gestational age (<10th birth weight percentile) infants. All participants provided written informed consent and all protocols were approved by the IRBs at the Women & Infant Hospital of Rhode Island and Emory University. The replication cohort uses data from n= 119 participants. RNA sequencing data provided for this study was evaluated using the HiSeq 2500 platform (Illumina) and expression analysis was performed using DESeq2. Data provided by RICHS for miRNA expression were sequenced by Omega Bioservices (Norcross, Georgia) and analyzed using DESeq2 (E. M. Kennedy et al., 2021). Data provided by this study for site-specific measures of placental DNA methylation were evaluated using the Illumina Infinium Human MethylationEPIC BreadChip (Illumina), and analyzed using the minfi Bioconductor package in R (Tian et al., 2020). A regression analysis was run to test for replication of CpG-mRNA and miRNA-mRNA expression pairs that were statistically significant in the ELGAN cohort.
Results:
Demographic characteristics of the ELGANs.
Transcriptomic and epigenomic analyses were conducted in n=368 ELGAN subjects for the present study. General characteristics of the ELGAN participants, including ASD diagnosis, are summarized in Table 1. Among the 368 ELGANs, 28 (8%) of the infants were diagnosed with ASD at age 10. The average gestational age for infants was 26.0 weeks. Mean birthweight was significantly different between ASD cases and non-cases, 747 versus 841 grams, respectively. The overall average age of participating mothers at birth was 29.7 years. The prevalence of fetal growth restriction/SGA, defined as birthweight z-score, was the most significantly different variable between ASD cases and non-cases in the ELGAN sample. Because birthweight was significantly different between ASD cases and non-cases, it was included in subsequent models as a confounder, as was birthweight-for-gestational age which normalizes birthweight within gestational age categories.
Table 1. Demographic characteristics of the ELGAN study participants (n=368).
Subjects with missing data on were not included in the percentage calculation.
| Overall (n=368) | Non-Cases (n=340) | ASD Cases (n=28) | p-value* | |
|---|---|---|---|---|
| N (%), or | N (%), or | N (%), or | ||
| Mean [SD] | Mean [SD] | Mean [SD] | ||
| Gestational Age (wks) | 26.0 [1.29] | 26.0 [1.29] | 25.5 [1.26] | 0.06 |
| Gestational Age (wks) | 0.07 | |||
| 23–24 weeks | 83 (22.6%) | 72 (21.2%) | 11 (39.3%) | |
| 25–26 weeks | 159 (43.2%) | 148 (43.5%) | 11 (39.3%) | |
| 27 weeks | 126 (34.2%) | 120 (35.3%) | 6 (21.4%) | |
| Birthweight (grams) | 834 [189] | 841 [188] | 747 [172] | 0.01 |
| Birthweight (grams) | 0.08 | |||
| <= 750 | 137 (37.2%) | 120 (35.3%) | 17 (60.7%) | |
| 751–1000 | 163 (44.3%) | 154 (45.3%) | 9 (32.1%) | |
| 1001–1250 | 61 (16.6%) | 59 (17.4%) | 2 (7.14%) | |
| >1250 | 7 (1.90%) | 7 (2.06%) | 0 (0.00%) | |
| Child Sex | 0.02 | |||
| Female | 175 (47.6%) | 168 (49.4%) | 7 (25.0%) | |
| Male | 193 (52.4%) | 172 (50.6%) | 21 (75.0%) | |
| Multiple Births | 0.77 | |||
| Yes | 134 (37.9%) | 125 (38.2%) | 9 (33.3%) | |
| No | 220 (62.1%) | 202 (61.8%) | 18 (66.7%) | |
| Maternal Age (years) | 29.7 [6.6] | 29.7 [6.7] | 29.8 [5.1] | 0.89 |
| Maternal Education | 0.27 | |||
| High School | 139 (38.9%) | 125 (37.8%) | 14 (53.8%) | |
| Some College | 77 (21.6%) | 73 (22.1%) | 4 (15.4%) | |
| College or Greater | 141 (39.5%) | 133 (40.2%) | 8 (30.8%) | |
| Public Insurance | 0.32 | |||
| Yes | 123 (33.8%) | 111 (39.2%) | 12 (44.4%) | |
| No | 241 (66.2%) | 226 (67.1%) | 15 (55.6%) | |
| Race | 0.43 | |||
| White | 228 (62.3%) | 213 (63.0%) | 15 (53.6%) | |
| Non-white | 138 (37.3%) | 125 (37.0%) | 13 (46.4%) | |
| Ethnicity | 0.72 | |||
| Non-Hispanic | 336 (91.3%) | 311 (91.5%) | 25 (89.3%) | |
| Hispanic | 32 (8.7%) | 29 (8.5%) | 3 (10.7%) | |
| Maternal Smoking Status | 1.0 | |||
| Yes | 39 (10.8%) | 37 (11.0%) | 2 (7.7%) | |
| No | 322 (89.2%) | 298 (89.0%) | 24 (92.3%) | |
| Maternal pre-pregnancy BMI | 0.47 | |||
| Underweight | 26 (7.3%) | 24 (7.3%) | 2 (7.7%) | |
| Normal | 192 (54.1%) | 181 (55.0%) | 11 (42.3%) | |
| Overweight | 65 (18.3%) | 60 (18.2%) | 5 (19.2%) | |
| Obese | 72 (20.3%) | 64 (19.5%) | 8 (30.8%) | |
| Birthweight Z-Score: | 0.04 | |||
| 1 (< −2) | 20 (5.43%) | 16 (4.71%) | 4 (14.3%) | |
| 2 (< −1) | 41 (11.1%) | 39 (11.5%) | 2 (7.14%) | |
| 3 (<= 1) | 267 (72.6%) | 245 (72.1%) | 22 (78.6%) | |
| 4 (> 1) | 40 (10.9%) | 40 (11.8%) | 0 (0.00%) | |
P-values were generated using T-tests for continuous variables and Chi-squared tests or Fisher’s exact tests for categorical variables, as appropriate.
The cohort comprises 175 (47.6%) females and 193 (52.4%) males. Demographic characteristics of the ELGANs stratified by sex are provided in Supplemental Table 1. No statistically significant differences in demographic characteristics were observed between males and females in this study. Among the ASD cases, 21 (75%) were male and seven (25%) were female (Table 1).
In relation to the cognitive and language characteristics of the 28 ELGANs with ASD, mean scores for ADOS-2 Social Affect, Restricted and Repetitive Behaviors, DAS-II IQ and OWLS language ability scores are presented in Table 2. Across ADOS-2 modules, there are 10 Social Affect items, yielding total scores from 0–20, and four Restricted and Repetitive Behavior scores, yielding total scores from 0–8. Statistically significant differences were observed between ASD cases and non-cases for each of the measures presented (Table 2).
Table 2.
Summary of cognitive and language characteristics of the ASD cases
| ASD Cases (n=28) Mean (SD) |
Non-Cases (n=340) Mean (SD) |
p-value | |
|---|---|---|---|
| ADOS-21 Social Affect Score | 13.1 (4.77) | 0 (0.0) | <0.001 |
| ADOS-2 Restricted and Repetitive Behaviors Score | 4.96 (1.74) | 0 (0.0) | <0.001 |
| ADOS-2 Total Score | 18.7 (4.75) | 0 (0.0) | <0.001 |
| DAS-II Verbal IQ | 48.9 (20.6) | 94.9 (16.7) | <0.001 |
| DAS-II Nonverbal IQ | 62.5 (25.2) | 91.6 (14.8) | <0.001 |
| OWLS Oral expression | 49.2 (15.2) | 91.3 (16.5) | <0.001 |
| OWLS Listening comprehension | 50.0 (13.5) | 90.5 (15.4) | <0.001 |
| OWLS Oral language composite | 48.6 (13.5) | 89.9 (15.7) | <0.001 |
Of ASD cases, eight were administered Module 1 (minimal speech), three were administered Module 2 (phrase speech), and 16 were administered Module 3 (fluent speech)
Genome-wide RNA sequencing-based analysis identified 111 mRNAs in the placenta associated with ASD.
We used a multi-tiered analytical approach to identify mRNAs in the placenta in ELGANs associated with ASD, followed by those that were also under epigenetic control (Figure 1). Differential expression analysis of the RNA sequencing data resulted in the identification of 111 genes that were significantly (BH p-value<0.1) differentially expressed in the placental of children who developed ASD versus non-cases (Supplemental Table 2). Of these 111 genes, DESeq2 analysis highlighted four (3.6%) that displayed increased expression and 107 (96.4%) that displayed decreased expression in relation to ASD (Figure 2).
Figure 1.
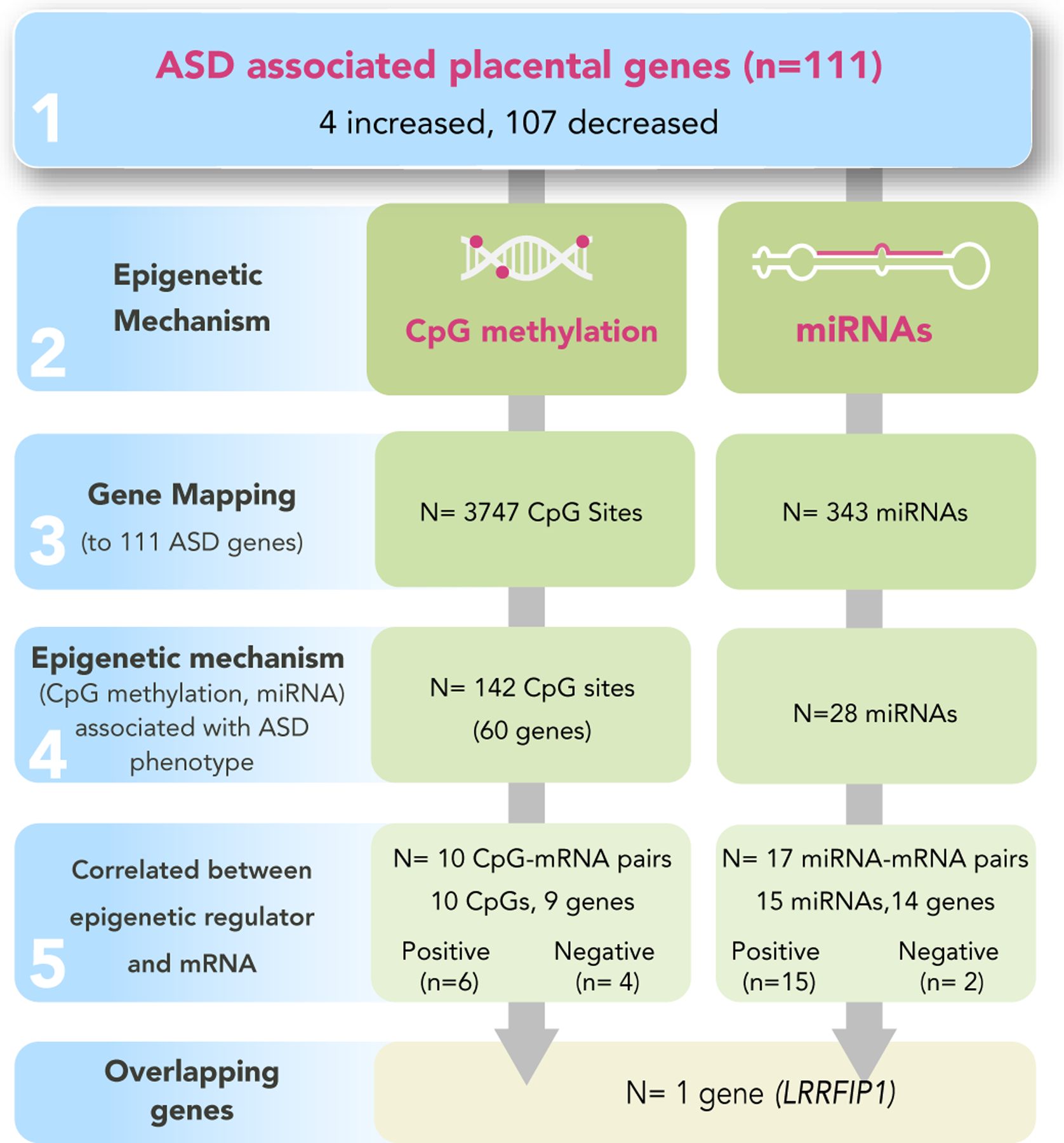
Flowchart of workflow
Figure 2. A volcano plot of the 111 ASD-associated mRNAs in the placenta.
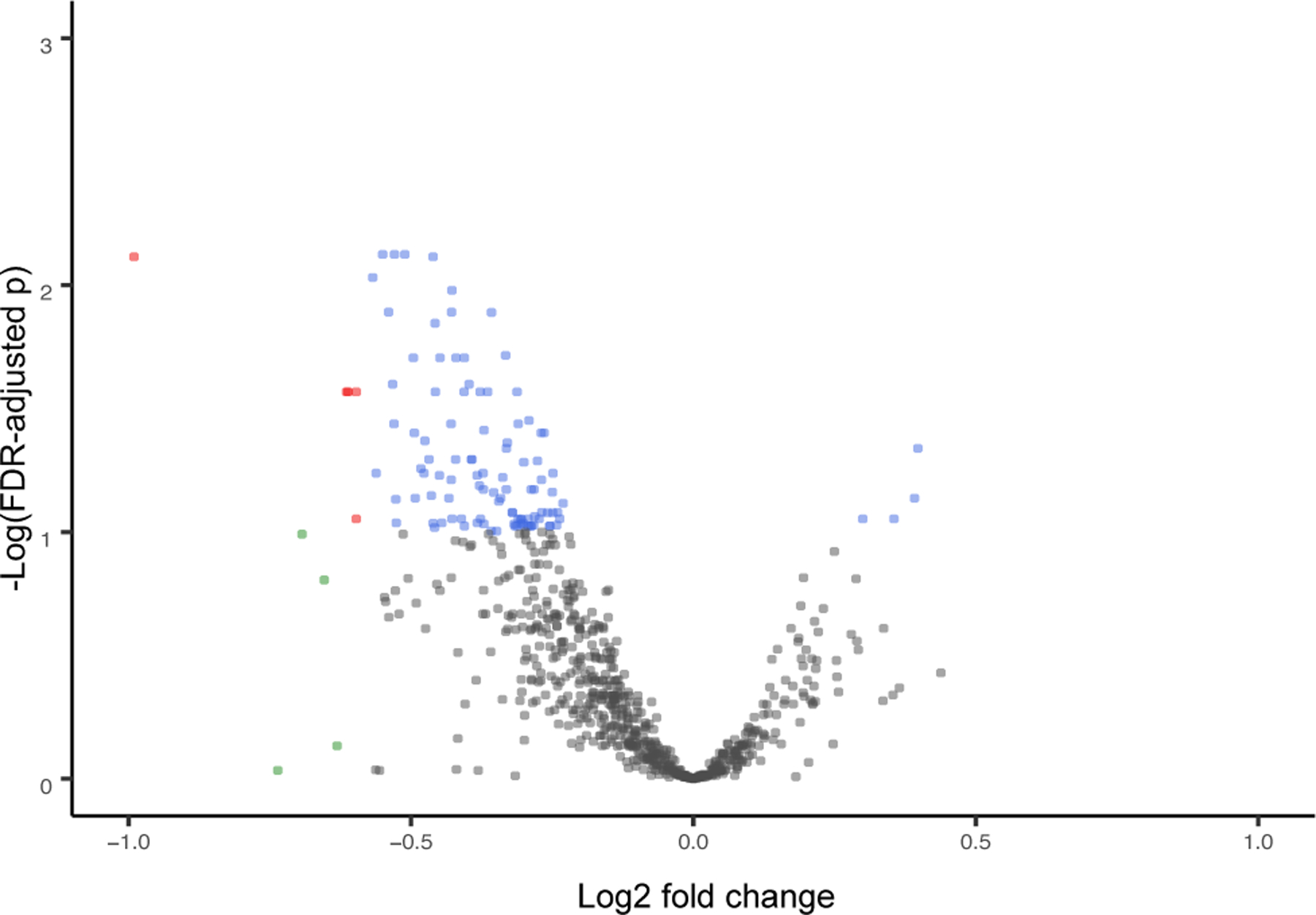
Red indicates genes where the fold change (FC) >1.5 or <−1.5 and FDR p<0.1. Green indicates genes where the FC >1.5 or <−1.5. Blue indicates FDR p<0.1. Gray indicates genes that were not significant.
The relationship between the expression levels of each of these 111 ASD-associated genes and their capacity to predict ASD case status was quantified using logistic regression models resulting in adjusted odds ratios (Supplemental Table 2). A total of 15 genes were identified as having statistically significant (p-value <0.05) ORs with expression concordant to the DESeq2 fold changes. The ORs ranged from 0.16–6.57. The highest ORs were identified for the following: Ewing Sarcoma Breakpoint Region 1 (EWSR1) (OR: 6.57 (95%CI: 2.34, 23.58)), Activating Transcription Factor 7 Interacting Protein (ATF7IP) (OR: 3.45 (95%CI: 1.39, 9.83)), and DEAD-Box Helicase 59 (DDX59) (OR: 2.84 (95%CI: 1.18, 7.60)). High ORs indicate that high placental mRNA levels are predictive of ASD status at age 10. Conversely, the lowest ORs were identified for the following: Adaptor Related Protein Complex 3 Subunit Delta 1 (AP3D1) (OR: 0.19 (95%CI: 0.06, 0.48)), Mitochondrially Encoded Cytochrome C Oxidase III (MT-CO3) (OR: 0.19 (95%CI: 0.06, 0.48)), and Bromodomain Adjacent To Zinc Finger Domain 2A (BAZ2A) (OR: 0.12 (95%CI: 0.03, 0.35)). Low ORs indicate that low placental mRNA levels are predictive of ASD status at age 10.
A biological functional analysis of the 111 ASD-associated genes highlighted roles in immunological disease, inflammatory response, chromatin regulation, and cancer (Supplemental Table 2). Further, network analysis was conducted to understand the relationships among the proteins encoded by these genes (Figure 3). Among the 111 genes, 90 displayed known protein-protein interactions and are termed the ASD regulatory complex (Figure 3). Several of the ASD-associated genes are noteworthy for their roles in inflammation and immune response including Collagen Type IV Alpha 1 Chain (COL4A1), Annexin A1 (ANXA1), DEK Proto-Oncogene (DEK), ATPase Plasma Membrane Ca2+ Transporting 1 (ATP2B1), Ribosomal Protein L32 (RPL32), Heat Shock Protein 90 Beta Family Member 1 (HSP90B1), Eukaryotic Translation Elongation Factor 1 Alpha 1 (EEF1A1), BBX High Mobility Group Box Domain Containing (BBX), and LRR Binding FLII Interacting Protein 1 (LRRFIP1) (Supplemental Table 2).
Figure 3. Network analysis identified the ASD Regulatory Complex.
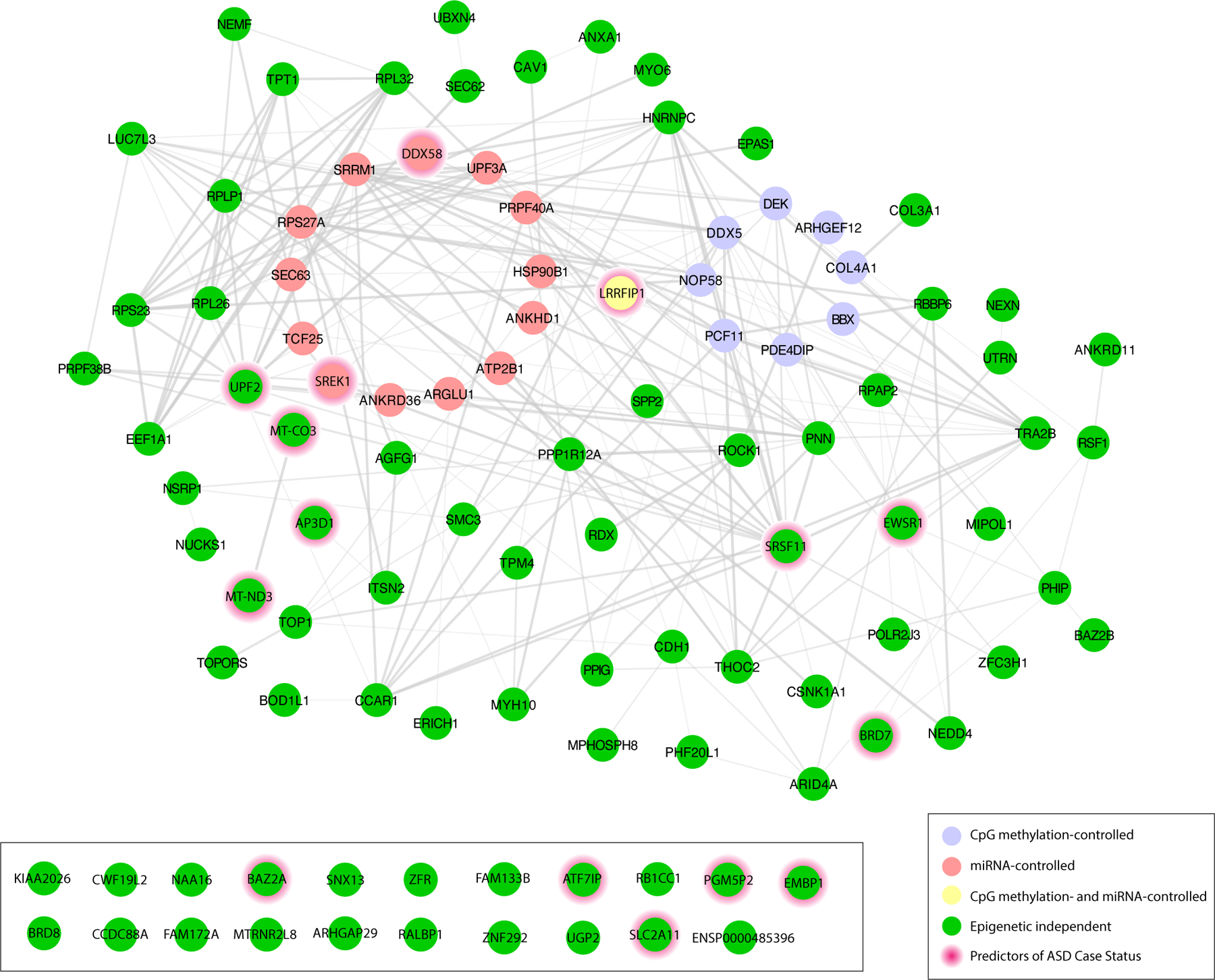
Network analysis of the 111 ASD-associated genes identified the complex (n= 90). Genes are represented in five clusters represented as differentially expressed genes under predicted epigenetic control as follows: CpG methylation-controlled (Purple); miRNA-controlled (Red); CpG methylation- and miRNA-controlled (Yellow); Epigenetic independent (Green); Predictors of ASD Case Status (Red halo)
Targeted analysis of CpG sites mapped to ASD-associated mRNAs identified 10 CpG sites differentially methylated in relation to ASD.
To investigate epigenetic control of placental transcription by CpG methylation, we performed targeted CpG methylation analysis of the 3,747 CpG sites that mapped to the 111 ASD-associated genes. Of these, 142 CpG sites displayed differential DNA methylation in relation to ASD mapping to 60 of the 111 ASD-associated genes (Figure 4a, Supplemental Table 3). As CpG methylation can control expression of mRNAs, we performed a correlation analysis between the M-values of the ASD-associated CpG sites and variance-stabilized expression levels of ASD-associated genes. This analysis identified 10 CpG-mRNA pairs that were significantly associated (Supplemental Table 4), representing nine unique genes. Specifically, the expression levels of nine of the 111 (8.1%) of the ASD-associated genes were correlated with CpG methylation levels either within or near sites of the genes, suggesting potential epigenetic control of gene expression associated with ASD. An example of one of the significant pairs, cg10493270-ARHGEF12, is illustrated in Figure 4b.
Figure 4. CpG sites (n= 142) associated with ASD at age 10 in ELGANs.
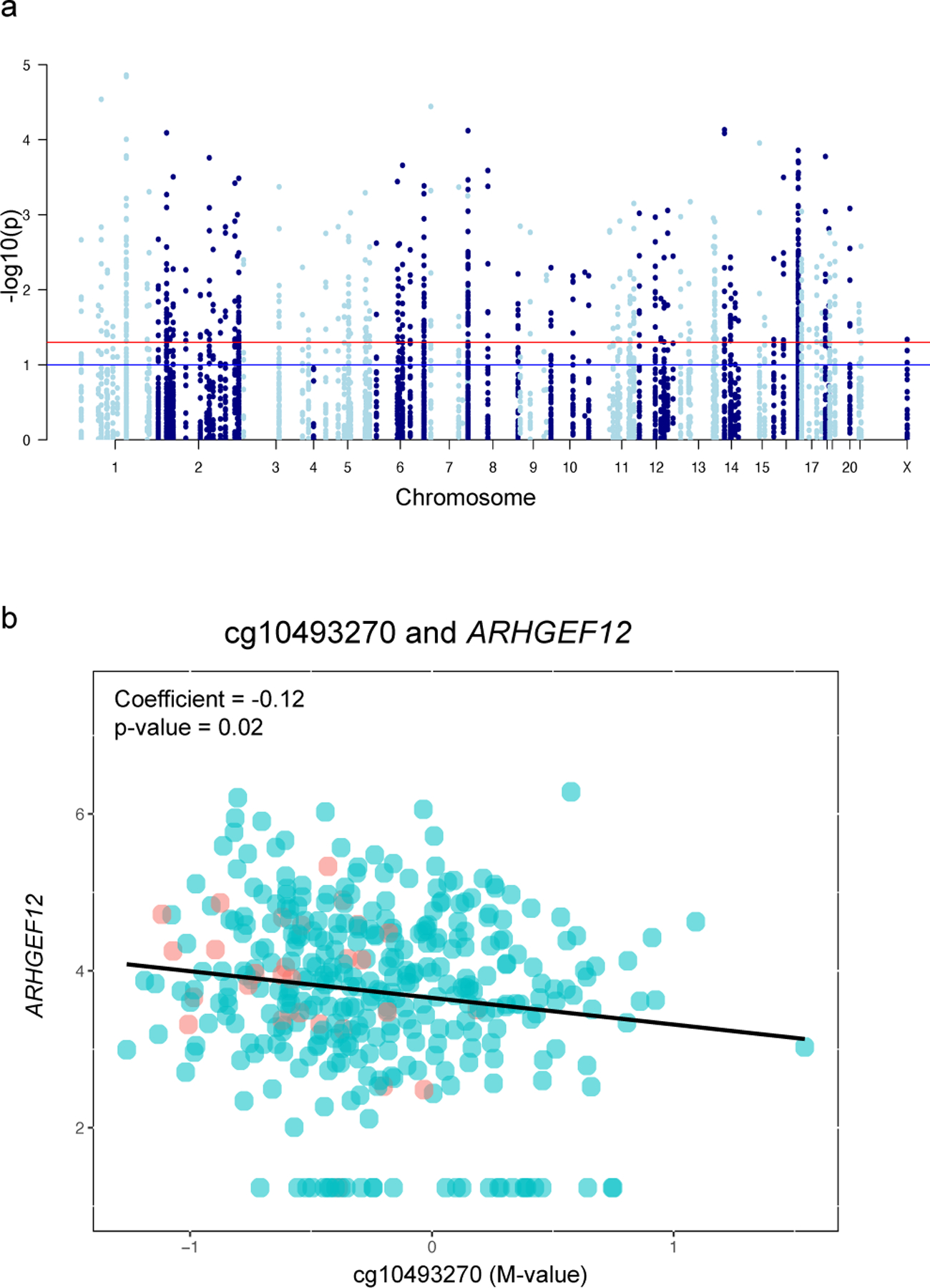
(a) Manhattan plot of identified CpGs mapped to chromosomal location in the placenta. (b) Correlation analysis of a representative pair for cg10493270 and ARHGEF12. Red indicates ASD case status and blue indicates non-case status.
miRNA-based analysis identified 17 miRNAs with differential expression in relation to ASD and ASD-associated transcript level changes.
To examine the relationship between miRNA expression and mRNA abundance, TargetScan (V. Agarwal, G. W. Bell, J.-W. Nam, & D. P. Bartel, 2015) was used to identify miRNAs known to target ASD- associated mRNAs. 769 miRNAs were identified as potential regulators of the 111 ASD-associated genes. Of these, a total of 343 miRNAs had count levels above background filtering levels within our dataset, and were thus able to be evaluated for differential expression. Of these, 28 miRNAs displayed differential expression levels between placentas from ELGANs who were later diagnosed with ASD versus non-cases (Figure 5a, Supplemental Table 5). Further, through a correlation analysis, 17 mRNA-miRNA pairs were identified where 15 unique miRNAs displayed a strong correlation with their target mRNAs, representing 14 unique genes (Supplemental Table 6). Together, 14 of the 111 (12.6%) ASD-associated genes were predicted to be regulated by miRNAs, suggesting potential post-transcriptional control of placental gene expression associated with ASD. An example of the correlated miRNA-mRNA pair, miR-27b-3p-ANKRD36 is displayed in Figure 5b.
Figure 5. miRNAs (n= 28) differentially expressed in relation to ASD-associated genes.
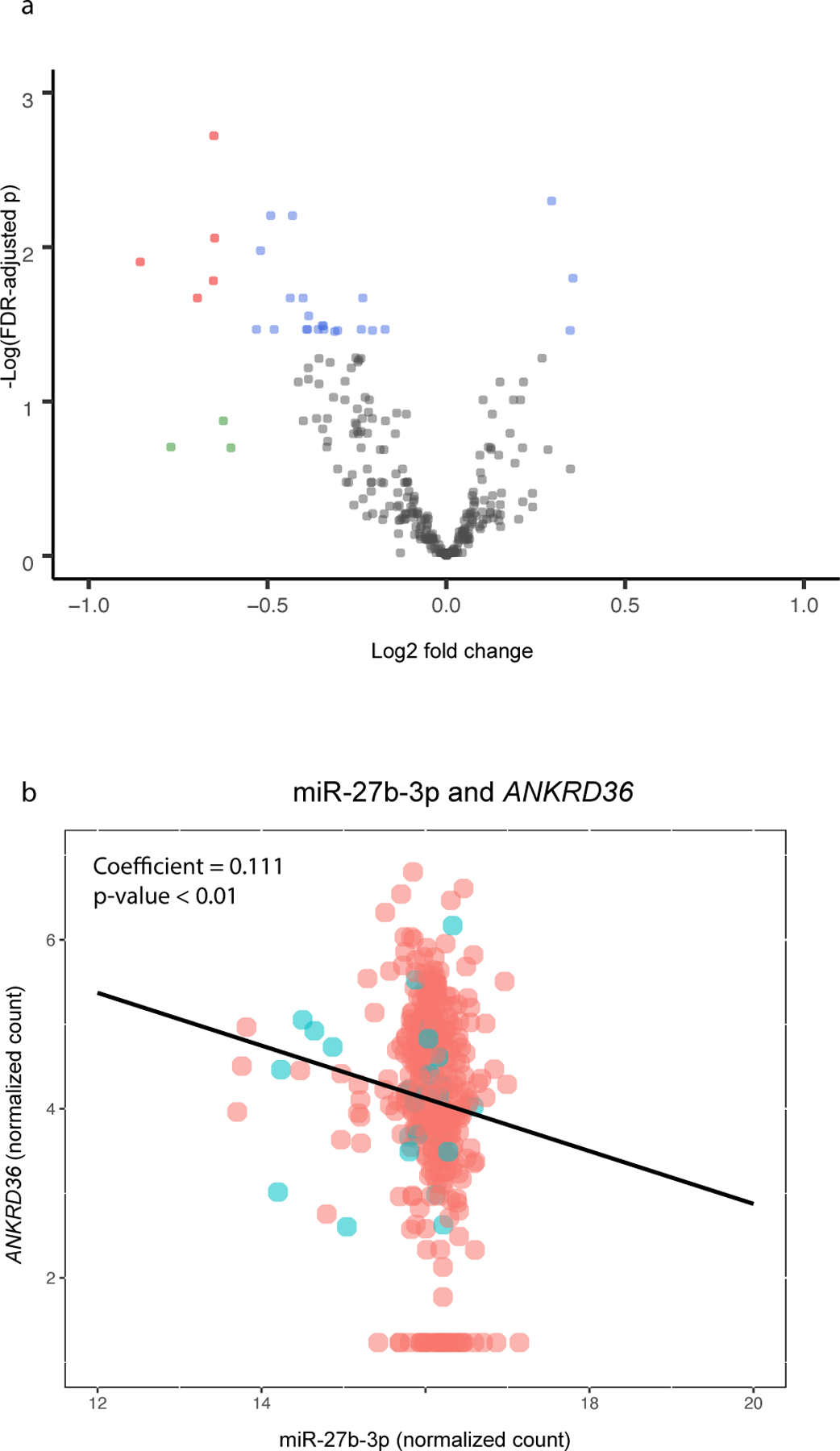
(a) Volcano plot demonstrating miRNAs associated with ASD across human placenta samples. Red indicates genes where the fold change (FC) >1.5 or <−1.5 and FDR p<0.1. Green indicates genes where the FC >1.5 or <−1.5. Blue indicates FDR p<0.1. Gray indicates genes that were not significant. (b) Scatterplot demonstrating a correlated mRNA-miRNA pair: mmiR-27b-3p-ANKRD36. Red indicates ASD case status and blue indicates non-case status.
ASD-associated gene LRRFIP1 is controlled via both miRNA and CpG methylation change.
Comparing the genes that were under either CpG methylation or miRNA control revealed that the expression of one ASD-associated gene was correlated with both miRNA expression levels and CpG methylation levels, namely LRR Bindings FLII Interacting Protein 1 (LRRFIP1). LRRFIP1 demonstrated decreased expression in relation to ASD diagnosis at age 10 (Supplemental Table 2). LRRFIP1 expression was significantly correlated with cg23517743 methylation, and miR-744–3p expression, both demonstrating a positive correlation between the epigenetic modifier and gene expression (Supplemental Tables 4 and 6). Interestingly, the expression of LRRFIP1 also demonstrated a predictive capacity for ASD case status with an OR of 0.42 (95% CI: 0.17, 0.95) (Supplemental Table 2).
Replication cohort.
A cohort that would allow for complete replication of all data (particularly the ASD diagnosis) in this study is not available. For those components of the study that could be compared, the RICHS study (n= 199) was used to assess CpG methylation-mRNA pairs and miRNA-mRNA pairs. Of the 10 CpG-mRNA expression pairs that displayed significant associations in the ELGAN cohort, five of the pairs also shared similar directionality in their association from the RICHS cohort yet did not reach statistical significance (Supplemental Table 4). An example of one of the displayed pairs, cg10493270-ARHGEF12 is illustrated in Supplemental Figure 1. Of the 17 miRNA-mRNA expression pairs that displayed significant associations within the ELGAN cohort, seven of the pairs showed similar directionalities in their associations in the replication cohort with miR-27b-3p-ANKRD36 and miR-199b-5p-SRRM1 showing statistical significance (Supplemental Table 6, Supplemental Figure 2).
Discussion:
The placenta is a critical, yet transient, organ linking the early life environment of the fetus to later in life child outcomes. Our data support that the analysis of -omic data measuring levels of components of the placental cellular machinery at the time of birth reflects a placental “recording” of the early life environment (Clark et al., 2019). This suggests that transcriptomic and epigenomic signatures measured in placental tissue may provide insight into the origins of later in life disease. The current study employed a multi-omic approach to identify associations between various components of the placental cellular machinery and ASD diagnosis later in life in ELGANs. Using a genome-wide differential expression analysis, a total of 111 ASD-associated genes were identified. Within these genes was the ASD regulatory complex including genes with expression levels that serve as predictors of ASD case status. By integrating data on the expression of the 111 ASD-associated mRNAs, with the expression of miRNAs, and the methylation levels of CpG sites, it was observed that roughly 12% and 8% of the ASD-associated genes displayed functional epimutations with expression under the epigenetic control of miRNAs and CpGs, respectively. In contrast to traditional mutations, epimutations result in chemical changes to DNA that do not change the DNA coding sequence. These data support our prior work that highlights epigenetic control of the placental transcriptome (Clark et al., 2021; Eaves et al., 2020), and extends this work to identify transcriptomic predictors of ASD status.
Interestingly, among the genes that were predictors of ASD status was EWSR1 and ATF7IP. For both genes, high expression of EWSR1 or ATF7IP in the placenta is associated with increased odds (OR~7 and OR~3, respectively) of developing ASD at age 10. Importantly, EWSR1 is involved in cell signaling, RNA processing and transport (Aynaud et al., 2020). Further, EWSR1 and ATF7IP are involved in chromatin regulation and organization (Aynaud et al., 2020; L. Liu et al., 2009). Given our a priori hypothesis that ASD-associated genes would be involved in inflammation and immune response, it is noteworthy that identified genes that strongly predict ASD case status are involved in chromatin regulation. DDX59 (OR ~3) is implicated in nervous system development and function and is one that was highlighted in our recent article (Santos et al., 2020). Further, among the ASD predictors were UPF2 Regulator Of Nonsense Mediated MRNA Decay (UPF2) and Serine And Arginine Rich Splicing Factor 11 (SRSF11). Interestingly, in support of these findings, these genes have been highlighted as candidate ASD risk genes within the Simons Foundation Autism Research Initiative (SFARI) database (Banerjee-Basu & Packer, 2010). Further, impaired UPF2 function has been associated with neurodevelopmental dysfunction by activating the immune response (Johnson et al., 2019). Highlighting the potential relationship of the altered expression of these genes in the placenta and brain damage in the ELGAN neonate, four of the ASD predictors were also associated with cerebral white matter damage in neonates, namely, BAZ2A, MT-CO3, Bromodomain Containing 7 (BRD7) and Phosphoglucomutase 5 Pseudogene 2 (PGM5P2) (Marable et al., 2022).
An analysis of the ASD-associated genes that are under epigenetic control revealed that several that were correlated with CpG methylation levels also have known roles in inflammation and immune response. These genes include BBX (Moffitt et al., 2017), COL4A1 (Hetmanczyk-Sawicka et al., 2020; Kauerhof et al., 2019), and DEK (Kappes et al., 2008; Mor-Vaknin et al., 2006). Similarly, several of the ASD-associated genes under miRNA control have known roles in inflammation and/or immune-related processes, including ATP2B1 (Allantaz et al., 2007) and HSP90B1 (Graustein et al., 2018; He et al., 2019; Stengel et al., 2020). Interestingly, a single gene was identified to be associated with ASD and under the control of both miRNA expression and CpG methylation, namely LRRFIP1. In terms of the predictive capacity of this gene, high expression of LRRFIP1 in the placenta was associated with lower odds of a child developing ASD at age 10 suggesting a protective role for this gene. Studies have demonstrated that LRRFIP1 plays multiple roles in the regulation of biological systems including immune response to microorganisms, as well as auto-immunity, signal transduction pathways and transcriptional regulations of genes (Takimoto, 2019). LRRFIP1 plays a role in regulating inflammation, which may cause physiological vulnerabilities during development. Specifically, LRRFIP1 plays a role in repressing Tumor Necrosis Factor Alpha (TNF-α), a pro-inflammatory cytokine with diverse roles in the regulation of immune and inflammatory responses (Suriano et al., 2005; Takimoto, 2019). Furthermore, it has been characterized as a transcriptional repressor to an upstream element of Epidermal Growth Factor Receptor (EGFR) gene (Takimoto, 2019). Interestingly, LRRFIP1 largely plays a role in enhances the interactions of certain proteins to increase the transcription of type 1 interferon (INF) gene, leading to NF-κB activation (Y. Liu et al., 2015; Plourde et al., 2013). In the present study, LRRFIP1 displayed lower expression in placentas collected from children who went on to develop ASD, suggesting the potential for enhanced TNF-α and EGFR expression. Notably, these data suggest that low placental expression of LRRFIP1 in relation to later-life ASD diagnosis may be enhanced by epimutations, given that CpG methylation and miRNA expression were positively correlated with LRRFIP1 expression. Studies investigating genetic influences specific to autism-associated behaviors have also found that LRRFIP1 is associated with quantitative trait nucleotides (QTN) or quantitative trait loci (QTL) associated with spoken language (Devlin, 2015; Hu, Devlin, & Debski, 2019).
This study examined the relationship between placental omics data and ASD risk later in life, in a cohort of infants all born extremely preterm. The relationship between preterm birth and ASD is complex, where perinatal environmental factors may lead to altered inflammatory and immune signaling which may lead to preterm birth (Green & Arck, 2020; Humberg et al., 2020). Still, not all children within the ELGAN cohort developed ASD, suggesting other factors beyond preterm birth, such as perinatal inflammation, are likely contributing factors to disease risk. Our analyses still leave open the possibility of inflammation persistence as a driver of ASD. It is plausible that inflammatory signals, beginning as early as the first trimester, could initiate a cascade of inflammatory events resulting in dysregulation of placental development, postnatal inflammation and subsequent disrupted fetal brain development. Preclinical models provide support for this concept, showing that intrauterine inflammation can lead to neuroinflammation that persists postnatally and is associated with disrupted brain development (Favrais et al., 2011). There is a large body of epidemiologic literature describing the relationship between prenatal inflammation, postnatal inflammation, and impaired neurodevelopment, reviewed in Jian 2018, Leviton 2015, Meltzer 2017, and Vohr 2017 (Jiang et al., 2018; Leviton et al., 2015; Meltzer & Van de Water, 2017; Vohr et al., 2017). Across epidemiologic studies, there are three primary stages during which inflammation has been associated with ASD risk. First, related to the prenatal/in utero period, fetal head growth measurements during the second and third trimester of pregnancy provide evidence of disrupted brain development as a possible marker of ASD (Bonnet-Brilhault et al., 2018; Caly et al., 2021). Further supporting this in utero linkage, fetal growth restriction in the ELGAN cohort was associated with an increased risk of ASD (Korzeniewski et al., 2017). Second, related to measurements at birth, the data from this study support a link between functional epimutations in the placenta and ASD. Specifically, genes involved in various biological processes including inflammation and immune response, as well as chromatin regulation were identified as strong predictors of ASD risk. Lastly, relating to the postnatal period, disrupted signaling of inflammatory markers in circulating blood collected within the first weeks of life is associated with increased risk of ASD in ELGANs (Korzeniewski et al., 2018). Further support for inflammation persistence postnatally, our research group has recently demonstrated strong associations between placental CpG methylation and intermittent or sustained systemic inflammation over the first two postnatal weeks of life within ELGANs (Eaves et al., 2022). Thus, when considering the early-life cascade of events, ASD risk is tied to inflammatory signals present during gestation, at delivery assessed in the placenta, as well as within the first few weeks of life, at least among infants born before 28 weeks. This work contributes to understanding these complex pre- and perinatal signals, by uncovering a novel link between placental immune and inflammation-related mRNAs, their epigenetic mediators, and ASD diagnosis later in life. Future research should investigate factors (e.g., fetal genotype, chemical exposures, maternal stress) that influence interindividual placental responses associated with later life ASD status.
This study employs multi-omic methods to further the understanding of the genes and pathways in the placenta that may play a role in the onset of ASD later in life. This study is not without limitations. This study focuses on a cohort of children born extremely preterm, which limits the generalizability of the results for children born at term. Still, the conservation of directionality in expression control of the miRNA-mRNA and CpG-mRNA pairs in the RICHS cohort, which consists of all term (<30 weeks gestational age) infants, lends support for the underling molecular mechanisms controlling the expression of these placental genes. Another limitation is the potential for selection bias due to the exclusion of children who died at birth or before age 10, or who were lost to follow-up exists. In spite of this, the cohort remains of sufficient size to identify key associations. While the results highlight genes with potential predictive capacity for ASD case status, these results should be validated in a larger and more diverse cohort, and their potential clinical role should be validated in subsequent studies. It is also important to note that an observational study cannot establish a causal relationship between placental omics data (miRNA and CpG methylation) and the development of ASD later in life. This research also has many strengths. Integrating data provided through RNA sequencing and DNA methylation analyses in the placenta allows for a novel multi-omic approach toward understanding epigenetic drivers of ASD. Future research can investigate which perinatal factors are associated with the altered expression of genes involved in the ASD regulatory complex to identify potential modifiable factors associated with later-life ASD diagnosis. Our findings contribute toward the mechanistic insights on ASD in children born preterm, and provide future avenues for research with an eye ultimately on intervention.
Supplementary Material
Supplemental Table 1. Demographic characteristics of the ELGAN study subjects stratified by sex (n= 368).
Supplemental Table 2. ASD-associated mRNAs. Significance was defined as BH p values <0.1. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg); chr (chromosome); OR (odds ratio); CI (confidence interval). Associated pathways identified from Ingenuity Pathway Analysis and Genecards. Bold OR p-values indicate the 15 predictor genes of ASD case status.
Supplemental Table 3. CpG sites significantly associated with ASD. These CpG sites (n = 142), mapped to genes associated with ASD (n=111). Significance defined as BH p values <0.1. Estimate represents the log(odds ratio) of developing ASD given degree of methylation at defined CpG site. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg)
Supplemental Table 4. CpG-mRNA pairs. A total of 10 predicted CpG-mRNA expression pairs from targeted analysis, constituting 10 unique CpGs and nine unique mRNAs. Bold text represents replication in additional cohort.
Supplemental Table 5. miRNA significantly associated with ASD. miRNAs (n= 28) identified with (A) upregulated and (B) downregulated expression mapped to genes associated with ASD (n=111). Significance defined as BH p-values <0.1. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg).
Supplemental Table 6. miRNA-mRNA pairs. A total of 17 predicted miRNA-mRNA expression pairs, constituting 15 unique miRNA and 14 unique mRNAs. Bold text represents replication in additional cohort, * = statistically significant in replication cohort p<0.1.
Supplemental Figure 1. Scatterplot demonstrating representative CpG-mRNA pair between cg10493270 and ARHGEF12 in RICHS cohort.
Supplemental Figure 2. Scatterplot demonstrating representative mRNA-miRNA pair between miR-27b-3b and ANKRD26 in RICHS cohort.
Acknowledgements
We would like to thank the ELGAN Study Investigators and study participants for their contributions to this research.
Funding
This research was funded by grants from the National Institutes of Health (UH3OD023348, T32ES007018, and R01HD092374)
Footnotes
Competing interests
The authors declare that they have no competing interests
Ethics approval and consent to participate
This study included written consent for participation by study subjects and was approved by the Institutional Review Board at UNC-Chapel Hill (IRB #16–2535).
Availability of data and materials
All data generated or analyzed in this study are included in this published article. DNA Methylation data have been deposited in Gene Expression Omnibus [GEO: GSE167885] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167885) mRNA and miRNA sequencing data have been deposited in Gene Expression Omnibus [GEO: GSE154829] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154829)
References
- Addo KA, Bulka C, Dhingra R, Santos HP Jr., Smeester L, O’Shea TM, & Fry RC (2019). Acetaminophen use during pregnancy and DNA methylation in the placenta of the extremely low gestational age newborn (ELGAN) cohort. Environ Epigenet, 5(2), dvz010. doi: 10.1093/eep/dvz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam J-W, & Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, e05005. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, … Pascual V (2007). Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med, 204(9), 2131–2144. doi: 10.1084/jem.20070070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PJ (2014). Neuropsychological outcomes of children born very preterm. Semin Fetal Neonatal Med, 19(2), 90–96. doi: 10.1016/j.siny.2013.11.012 [DOI] [PubMed] [Google Scholar]
- Ashrap P, Watkins DJ, Mukherjee B, Boss J, Richards MJ, Rosario Z, … Meeker JD (2020). Maternal blood metal and metalloid concentrations in association with birth outcomes in Northern Puerto Rico. Environ Int, 138, 105606. doi: 10.1016/j.envint.2020.105606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aynaud MM, Mirabeau O, Gruel N, Grossetete S, Boeva V, Durand S, … Zinovyev A (2020). Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep, 30(6), 1767–1779 e1766. doi: 10.1016/j.celrep.2020.01.049 [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, & Packer A (2010). SFARI Gene: an evolving database for the autism research community. Dis Model Mech, 3(3–4), 133–135. doi: 10.1242/dmm.005439 [DOI] [PubMed] [Google Scholar]
- Bangma JT, Hartwell H, Santos HP Jr., O’Shea TM, & Fry RC (2021). Placental programming, perinatal inflammation, and neurodevelopment impairment among those born extremely preterm. Pediatr Res, 89(2), 326–335. doi: 10.1038/s41390-020-01236-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Bull AR, Osmond C, & Simmonds SJ (1990). Fetal and placental size and risk of hypertension in adult life. BMJ, 301(6746), 259–262. doi: 10.1136/bmj.301.6746.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Freedman AN, Avula V, Harris R, Liu W, Pan C, … Santos HP Jr. (2022). Placental genomics mediates genetic associations with complex health traits and disease. Nat Commun, 13(1), 706. doi: 10.1038/s41467-022-28365-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet-Brilhault F, Rajerison TA, Paillet C, Guimard-Brunault M, Saby A, Ponson L, … Roux S (2018). Autism is a prenatal disorder: Evidence from late gestation brain overgrowth. Autism Res, 11(12), 1635–1642. doi: 10.1002/aur.2036 [DOI] [PubMed] [Google Scholar]
- Bronson SL, & Bale TL (2016). The Placenta as a Mediator of Stress Effects on Neurodevelopmental Reprogramming. Neuropsychopharmacology, 41(1), 207–218. doi: 10.1038/npp.2015.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly H, Rabiei H, Coste-Mazeau P, Hantz S, Alain S, Eyraud JL, … Ben-Ari Y (2021). Machine learning analysis of pregnancy data enables early identification of a subpopulation of newborns with ASD. Sci Rep, 11(1), 6877. doi: 10.1038/s41598-021-86320-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrow-Woolfolk E (1995). OWLS, Oral and Written Language Scales. : NCS Pearson Incorporated. [Google Scholar]
- Chen LW, Wang ST, Wang LW, Kao YC, Chu CL, Wu CC, … Huang CC (2019). Behavioral characteristics of autism spectrum disorder in very preterm birth children. Mol Autism, 10, 32. doi: 10.1186/s13229-019-0282-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Eaves LA, Gaona AR, Santos HP Jr., Smeester L, Bangma JT, … Fry RC (2021). Pre-pregnancy BMI-associated miRNA and mRNA expression signatures in the placenta highlight a sexually-dimorphic response to maternal underweight status. Sci Rep, 11(1), 15743. doi: 10.1038/s41598-021-95051-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Martin E, Bulka CM, Smeester L, Santos HP, O’Shea TM, & Fry RC (2019). Associations between placental CpG methylation of metastable epialleles and childhood body mass index across ages one, two and ten in the Extremely Low Gestational Age Newborns (ELGAN) cohort. Epigenetics, 14(11), 1102–1111. doi: 10.1080/15592294.2019.1633865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin CA (2015). Phenotype-Genotype Associations in Concordant and Discordant Monozygotic and Dizygotic Twins Based on Quantitative Trait and Case-Control Association Analyses. (M.S.). The George Washington University, Ann Arbor. Retrieved from http://libproxy.lib.unc.edu/login?url=https://www.proquest.com/dissertations-theses/phenotype-genotype-associations-concordant/docview/1722526576/se-2 [Google Scholar]
- http://VB3LK7EB4T.search.serialssolutions.com/?genre=dissertations&atitle=&author=Devlin%2C+Christine+A.&volume=&issue=&spage=&date=2015&rft.btitle=&rft.jtitle=&issn=&isbn=978-1-339-09685-8&sid=ProQuest+Dissertations+%26+Theses+Global_ ProQuest Central; ProQuest Dissertations & Theses Global database. (1600605)
- Eaves LA, Enggasser AE, Camerota M, Gogcu S, Gower WA, Hartwell H, … Fry RC (2022). CpG methylation patterns in placenta and neonatal blood are differentially associated with neonatal inflammation. Pediatr Res. doi: 10.1038/s41390-022-02150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LA, Phookphan P, Rager JE, Bangma J, Santos HP Jr., Smeester L, … Fry RC (2020). A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics, 12(17), 1543–1558. doi: 10.2217/epi-2020-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot C (2007). Differential Ability Scales®-l|(DAS-ll).
- Favrais G, van de Looij Y, Fleiss B, Ramanantsoa N, Bonnin P, Stoltenburg-Didinger G, … Gressens P (2011). Systemic inflammation disrupts the developmental program of white matter. Ann Neurol, 70(4), 550–565. doi: 10.1002/ana.22489 [DOI] [PubMed] [Google Scholar]
- Freedman AN, Eaves LA, Rager JE, Gavino-Lopez N, Smeester L, Bangma J, … Fry RC (2022). The placenta epigenome-brain axis: placental epigenomic and transcriptomic responses that preprogram cognitive impairment. Epigenomics, 14(15), 897–911. doi: 10.2217/epi-2022-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, & Bartel DP (2011). Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol, 18(10), 1139–1146. doi: 10.1038/nsmb.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal D, Limesand SW, & Goyal R (2019). Epigenetic responses and the developmental origins of health and disease. J Endocrinol, 242(1), T105–T119. doi: 10.1530/JOE-19-0009 [DOI] [PubMed] [Google Scholar]
- Graustein AD, Misch EA, Musvosvi M, Shey M, Shah JA, Seshadri C, … Hawn TR (2018). Toll-like receptor chaperone HSP90B1 and the immune response to Mycobacteria. PLoS One, 13(12), e0208940. doi: 10.1371/journal.pone.0208940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ES, & Arck PC (2020). Pathogenesis of preterm birth: bidirectional inflammation in mother and fetus. Semin Immunopathol, 42(4), 413–429. doi: 10.1007/s00281-020-00807-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gude NM, Roberts CT, Kalionis B, & King RG (2004). Growth and function of the normal human placenta. Thromb Res, 114(5–6), 397–407. doi: 10.1016/j.thromres.2004.06.038 [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Maddox YT, & Spong CY (2014). The Human Placenta Project: placental structure, development, and function in real time. Placenta, 35(5), 303–304. doi: 10.1016/j.placenta.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg H, Gressens P, & Mallard C (2012). Inflammation during fetal and neonatal life: implications for neurologic and neuropsychiatric disease in children and adults. Ann Neurol, 71(4), 444–457. doi: 10.1002/ana.22620 [DOI] [PubMed] [Google Scholar]
- Hagberg H, Mallard C, Ferriero DM, Vannucci SJ, Levison SW, Vexler ZS, & Gressens P (2015). The role of inflammation in perinatal brain injury. Nat Rev Neurol, 11(4), 192–208. doi: 10.1038/nrneurol.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He GL, Luo Z, Shen TT, Yang J, Li P, Luo X, & Yang XS (2019). Inhibition of HSP90beta by ganetespib blocks the microglial signalling of evoked pro-inflammatory responses to heat shock. Int J Biochem Cell Biol, 106, 35–45. doi: 10.1016/j.biocel.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Hetmanczyk-Sawicka K, Iwanicka-Nowicka R, Fogtman A, Ciesla J, Wlodarski P, Zyzynska-Granica B, … Lugowska A (2020). Changes in global gene expression indicate disordered autophagy, apoptosis and inflammatory processes and downregulation of cytoskeletal signalling and neuronal development in patients with Niemann-Pick C disease. Neurogenetics, 21(2), 105–119. doi: 10.1007/s10048-019-00600-6 [DOI] [PubMed] [Google Scholar]
- Hodyl NA, Roberts CT, & Bianco-Miotto T (2016). Cord Blood DNA Methylation Biomarkers for Predicting Neurodevelopmental Outcomes. Genes (Basel), 7(12). doi: 10.3390/genes7120117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Devlin CA, & Debski JJ (2019). ASD Phenotype-Genotype Associations in Concordant and Discordant Monozygotic and Dizygotic Twins Stratified by Severity of Autistic Traits. Int J Mol Sci, 20(15). doi: 10.3390/ijms20153804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humberg A, Fortmann I, Siller B, Kopp MV, Herting E, Gopel W, … Priming Immunity at the beginning of life, C. (2020). Preterm birth and sustained inflammation: consequences for the neonate. Semin Immunopathol, 42(4), 451–468. doi: 10.1007/s00281-020-00803-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang NM, Cowan M, Moonah SN, & Petri WA Jr. (2018). The Impact of Systemic Inflammation on Neurodevelopment. Trends Mol Med, 24(9), 794–804. doi: 10.1016/j.molmed.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Stoica L, Liu Y, Zhu PJ, Bhattacharya A, Buffington SA, … Costa-Mattioli M (2019). Inhibition of Upf2-Dependent Nonsense-Mediated Decay Leads to Behavioral and Neurophysiological Abnormalities by Activating the Immune Response. Neuron, 104(4), 665–679 e668. doi: 10.1016/j.neuron.2019.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RM, Korzeniewski SJ, Allred EN, O’Shea TM, Heeren T, Frazier JA, … Investigators, E. S. (2017). Extremely low gestational age and very low birthweight for gestational age are risk factors for autism spectrum disorder in a large cohort study of 10-year-old children born at 23–27 weeks’ gestation. Am J Obstet Gynecol, 216(3), 304 e301–304 e316. doi: 10.1016/j.ajog.2016.11.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RM, O’Shea TM, Allred EN, Heeren T, Hirtz D, Paneth N, … Kuban KC (2017). Prevalence and associated features of autism spectrum disorder in extremely low gestational age newborns at age 10 years. Autism Res, 10(2), 224–232. doi: 10.1002/aur.1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, … Ferrando-May E (2008). DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol, 28(10), 3245–3257. doi: 10.1128/MCB.01921-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauerhof AC, Nicolas N, Bhushan S, Wahle E, Loveland KA, Fietz D, … Fijak M (2019). Investigation of activin A in inflammatory responses of the testis and its role in the development of testicular fibrosis. Hum Reprod, 34(8), 1536–1550. doi: 10.1093/humrep/dez109 [DOI] [PubMed] [Google Scholar]
- Kennedy E, Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, … Marsit CJ (2020). Copper associates with differential methylation in placentae from two US birth cohorts. Epigenetics, 15(3), 215–230. doi: 10.1080/15592294.2019.1661211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Hermetz K, Burt A, Everson TM, Deyssenroth M, Hao K, … Marsit CJ (2021). Placental microRNA expression associates with birthweight through control of adipokines: results from two independent cohorts. Epigenetics, 16(7), 770–782. doi: 10.1080/15592294.2020.1827704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski SJ, Allred EN, Joseph RM, Heeren T, Kuban KCK, O’Shea TM, … Investigators, E. S. (2017). Neurodevelopment at Age 10 Years of Children Born <28 Weeks With Fetal Growth Restriction. Pediatrics, 140(5). doi: 10.1542/peds.2017-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski SJ, Allred EN, O’Shea TM, Leviton A, Kuban KCK, & investigators E. s. (2018). Elevated protein concentrations in newborn blood and the risks of autism spectrum disorder, and of social impairment, at age 10 years among infants born before the 28th week of gestation. Transl Psychiatry, 8(1), 115. doi: 10.1038/s41398-018-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton A, Gressens P, Wolkenhauer O, & Dammann O (2015). Systems approach to the study of brain damage in the very preterm newborn. Front Syst Neurosci, 9, 58. doi: 10.3389/fnsys.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ishihara K, Ichimura T, Fujita N, Hino S, Tomita S, … Nakao M (2009). MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem, 284(8), 5165–5174. doi: 10.1074/jbc.M807098200 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zou Z, Zhu B, Hu Z, Zeng P, & Wu L (2015). LRRFIP1 Inhibits Hepatitis C Virus Replication by Inducing Type I Interferon in Hepatocytes. Hepat Mon, 15(5), e28473. doi: 10.5812/hepatmon.15(5)2015.28473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, & Le Couteur A (1994). Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord, 24(5), 659–685. doi: 10.1007/BF02172145 [DOI] [PubMed] [Google Scholar]
- Marable CA, Roell K, Kuban K, O’Shea TM, & Fry RC (2022). Placental transcriptional signatures associated with cerebral white matter damage in the neonate. Front Neurosci, 16, 1017953. doi: 10.3389/fnins.2022.1017953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer A, & Van de Water J (2017). The Role of the Immune System in Autism Spectrum Disorder. Neuropsychopharmacology, 42(1), 284–298. doi: 10.1038/npp.2016.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MethylationEPIC, v. B. M. F. I. Retrieved from https://support.illumina.com/array/array_kits/infinium-methylationepic-beadchip-kit/downloads.html
- Moffitt AB, Ondrejka SL, McKinney M, Rempel RE, Goodlad JR, Teh CH, … Dave SS (2017). Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J Exp Med, 214(5), 1371–1386. doi: 10.1084/jem.20160894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N, Punturieri A, Sitwala K, Faulkner N, Legendre M, Khodadoust MS, … Markovitz DM (2006). The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol, 26(24), 9484–9496. doi: 10.1128/MCB.01030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. (2020). GEO Gene Expression Omnibus. Placental Genomic and Epigenomic Signatures in Infants Borns at Extremely Low Gestational Age. Retrieved from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154829.
- O’Donnell KJ, & Meaney MJ (2017). Fetal Origins of Mental Health: The Developmental Origins of Health and Disease Hypothesis. Am J Psychiatry, 174(4), 319–328. doi: 10.1176/appi.ajp.2016.16020138 [DOI] [PubMed] [Google Scholar]
- O’Shea TM, Allred EN, Dammann O, Hirtz D, Kuban KC, Paneth N, … Investigators, E. s. (2009). The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum Dev, 85(11), 719–725. doi: 10.1016/j.earlhumdev.2009.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KS, Eaves LA, Smeester L, Santos HP, O’Shea TM, & Fry RC (2021). Development of the genomic inflammatory index (GII) to assess key maternal antecedents associated with placental inflammation. Placenta, 111, 82–90. doi: 10.1016/j.placenta.2021.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, & Extremely Low Gestational Age Newborns Study, I. (2008). Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am J Obstet Gynecol, 198(1), 110 e111–117. doi: 10.1016/j.ajog.2007.05.044 [DOI] [PubMed] [Google Scholar]
- Outcomes, I. o. M. U. C. o. U. P. B. a. A. H. (2007). Preterm Birth: Causes, Consequences, and Prevention. In Behrman RE & Butler AS (Eds.), Preterm Birth: Causes, Consequences, and Prevention. Washington (DC). [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, & Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods, 14(4), 417–419. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson PH (2011). Maternal infection and immune involvement in autism. Trends Mol Med, 17(7), 389–394. doi: 10.1016/j.molmed.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton A, Clark J, Eaves L, Santos HP Jr., Smeester L, Bangma JT, … Rager JE (2020). Placental genomic and epigenomic signatures associated with infant birth weight highlight mechanisms involved in collagen and growth factor signaling. Reprod Toxicol, 96, 221–230. doi: 10.1016/j.reprotox.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Deyssenroth MA, Di Narzo AF, Lambertini L, Marsit CJ, Chen J, & Hao K (2017). Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum Mol Genet, 26(17), 3432–3441. doi: 10.1093/hmg/ddx265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plourde M, Vohl MC, Bellis C, Carless M, Dyer T, Dolley G, … Perusse L (2013). A variant in the LRRFIP1 gene is associated with adiposity and inflammation. Obesity (Silver Spring), 21(1), 185–192. doi: 10.1002/oby.20242 [DOI] [PubMed] [Google Scholar]
- Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, … Fry RC (2014). Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ Mol Mutagen, 55(3), 196–208. doi: 10.1002/em.21842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HP Jr., Bhattacharya A, Joseph RM, Smeester L, Kuban KCK, Marsit CJ, … Fry RC (2020). Evidence for the placenta-brain axis: multi-omic kernel aggregation predicts intellectual and social impairment in children born extremely preterm. Mol Autism, 11(1), 97. doi: 10.1186/s13229-020-00402-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HP Jr., Bhattacharya A, Martin EM, Addo K, Psioda M, Smeester L, … Fry RC (2019). Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics, 14(8), 751–765. doi: 10.1080/15592294.2019.1614743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepici G, Cavalli E, Bramanti P, & Mazzon E (2019). Autism Spectrum Disorder and miRNA: An Overview of Experimental Models. Brain Sci, 9(10). doi: 10.3390/brainsci9100265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11), 2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel ST, Fazio A, Lipinski S, Jahn MT, Aden K, Ito G, … Rosenstiel P (2020). Activating Transcription Factor 6 Mediates Inflammatory Signals in Intestinal Epithelial Cells Upon Endoplasmic Reticulum Stress. Gastroenterology, 159(4), 1357–1374 e1310. doi: 10.1053/j.gastro.2020.06.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRING. (2022). Retrieved from https://string-db.org/
- Suriano AR, Sanford AN, Kim N, Oh M, Kennedy S, Henderson MJ, … Sullivan KE (2005). GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol Cell Biol, 25(20), 9073–9081. doi: 10.1128/MCB.25.20.9073-9081.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto M (2019). Multidisciplinary Roles of LRRFIP1/GCF2 in Human Biological Systems and Diseases. Cells, 8(2). doi: 10.3390/cells8020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian FY, Everson TM, Lester B, Punshon T, Jackson BP, Hao K, … Marsit CJ (2020). Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: An epigenome-wide study of two U.S. birth cohorts. Environ Int, 137, 105508. doi: 10.1016/j.envint.2020.105508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SK, Joseph RM, Kuban KCK, Dammann OU, O’Shea TM, & Fry RC (2017). Genomic biomarkers of prenatal intrauterine inflammation in umbilical cord tissue predict later life neurological outcomes. PLoS One, 12(5), e0176953. doi: 10.1371/journal.pone.0176953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SK, Martin EM, Smeester L, Joseph RM, Kuban KCK, Heeren TC, … Fry RC (2018). Placental CpG methylation of infants born extremely preterm predicts cognitive impairment later in life. PLoS One, 13(3), e0193271. doi: 10.1371/journal.pone.0193271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A (2018). TargetScanHuman. Retrieved from http://www.targetscan.org/vert_72/
- Vohr BR, Poggi Davis E, Wanke CA, & Krebs NF (2017). Neurodevelopment: The Impact of Nutrition and Inflammation During Preconception and Pregnancy in Low-Resource Settings. Pediatrics, 139(Suppl 1), S38–S49. doi: 10.1542/peds.2016-2828F [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Demographic characteristics of the ELGAN study subjects stratified by sex (n= 368).
Supplemental Table 2. ASD-associated mRNAs. Significance was defined as BH p values <0.1. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg); chr (chromosome); OR (odds ratio); CI (confidence interval). Associated pathways identified from Ingenuity Pathway Analysis and Genecards. Bold OR p-values indicate the 15 predictor genes of ASD case status.
Supplemental Table 3. CpG sites significantly associated with ASD. These CpG sites (n = 142), mapped to genes associated with ASD (n=111). Significance defined as BH p values <0.1. Estimate represents the log(odds ratio) of developing ASD given degree of methylation at defined CpG site. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg)
Supplemental Table 4. CpG-mRNA pairs. A total of 10 predicted CpG-mRNA expression pairs from targeted analysis, constituting 10 unique CpGs and nine unique mRNAs. Bold text represents replication in additional cohort.
Supplemental Table 5. miRNA significantly associated with ASD. miRNAs (n= 28) identified with (A) upregulated and (B) downregulated expression mapped to genes associated with ASD (n=111). Significance defined as BH p-values <0.1. Abbreviations: miRNA (microRNA); BH (Benjamini-Hochberg).
Supplemental Table 6. miRNA-mRNA pairs. A total of 17 predicted miRNA-mRNA expression pairs, constituting 15 unique miRNA and 14 unique mRNAs. Bold text represents replication in additional cohort, * = statistically significant in replication cohort p<0.1.
Supplemental Figure 1. Scatterplot demonstrating representative CpG-mRNA pair between cg10493270 and ARHGEF12 in RICHS cohort.
Supplemental Figure 2. Scatterplot demonstrating representative mRNA-miRNA pair between miR-27b-3b and ANKRD26 in RICHS cohort.
Data Availability Statement
All data generated or analyzed in this study are included in this published article. DNA Methylation data have been deposited in Gene Expression Omnibus [GEO: GSE167885] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167885) mRNA and miRNA sequencing data have been deposited in Gene Expression Omnibus [GEO: GSE154829] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154829)


