Abstract
Purpose
In men with metastatic castration-resistant prostate cancer (mCRPC), prostate-specific membrane antigen (PSMA)-targeted radioligand therapy has drastically improved clinical outcomes. A liquid biopsy characterizing PSMA expression could be useful in guiding optimal therapy.
Experimental Design
We conducted a retrospective analysis of the prospective multicenter PROPHECY (Prospective CiRculating PrOstate Cancer Predictors in HighEr Risk mCRPC StudY) trial of men with mCRPC (n = 118) treated with abiraterone (abi) or enzalutamide (enza). Circulating tumor cells (CTC) were enriched (CTC/mL) and characterized for PSMA protein expression/heterogeneity at baseline and progression. We utilized proportional hazards modeling of the association between PSMA-positive (PSMA+) CTC enumeration with overall survival (OS) and progression-free survival (PFS).
Results
Overall, 97 men with mCRPC had evaluable blood samples for baseline CTC PSMA detection; 78 men (80%) had detectable CTCs. Of these, 55% (43/78) of men had any PSMA CTC detection, 21% (16/78) had ≥2 PSMA+ CTCs/mL, and 19% (8/43) were 100% PSMA+. At progression on abi/enza, 88% (50/57) of men had detectable CTCs, 68% (34/50) had any PSMA CTCs, and 12% (4/34) had 100% PSMA+ CTCs. Among paired cases (n = 57), PSMA+ CTC detection increased slightly after abi/enza progression. Using an optimal cutoff of ≥2 PSMA+ CTCs/mL, median OS was 26, 21, and 11 months for men without CTCs, PSMA− CTCs, and PSMA+ CTCs. Adjusting for prior abi/enza therapy, Halabi clinical risk score, and CTC enumeration, the HRs for OS and PFS for PSMA+ CTC+ were 3.0 [95% confidence interval (CI) = 1.1–7.8] and 2.3 (95% CI = 0.9–5.8).
Conclusions
We observed PSMA CTC heterogeneity between and within patients with mCRPC over time during abi/enza progression. CTC PSMA enumeration was adversely prognostic independent of clinical factors and disease burden. Further validation is warranted in the context of PSMA-targeted therapies.
Introduction
Prostate-specific membrane antigen (PSMA) is a type II transmembrane glycoprotein receptor that is overexpressed in most prostate cancers. The level of PSMA expression may increase with higher grade disease and castration resistance (1, 2), but may decrease due to lineage plasticity, such as during the neuroendocrine (NE) transformation (3). Because of the limited sensitivity and specificity of conventional imaging with CT, MRI, and bone scan in detecting metastatic disease, particularly in the setting of low PSA values (4), PSMA-based PET imaging can improve lesion detection utilizing radioligands such as 68Ga-PSMA-11 and 18F-DCFPyL which are currently FDA-approved radioligands for PSMA-targeted PET imaging. In addition, PSMA targeting with 177Lu-PSMA-617 radioligand therapy demonstrated longer imaging-based progression-free survival (PFS) and overall survival (OS) compared with the standard of care in men with PSMA-positive (PSMA+) advanced metastatic castration-resistant prostate cancer (mCRPC), according to the prospective VISION phase III trial (5). However, while PET imaging is used for patient selection, not all patients respond to 177Lu-PSMA-617 therapy, with nearly 50% of men failing to have durable responses over time despite PSMA+ PET imaging, and a significant heterogeneity in PFS and OS has been observed (5, 6, 7).
PSMA avidity and outcome heterogeneity could be impacted by many biological factors such as disease status, castration-sensitive versus-resistant disease, genotype, type of prior therapy, and timing and location of metastatic disease such as liver versus lymph node or bone metastases, as well as lineage plasticity and loss of androgen receptor (AR) dependence (2, 8, 9). Prior work has established the clear heterogeneity of PSMA expression in circulating tumor cells (CTC) from men with mCRPC in the context of chemotherapy (10). However, the dynamics of PSMA alterations over time in AR-based therapy settings, on the other hand, remain poorly understood.
The utility of CTC PSMA expression may lie both in prognostication and also in identifying patients with greater PSMA CTC homogeneity who may benefit from PSMA-targeted therapy. Such noninvasive testing may permit a greater degree of precision medicine and patient selection than PET imaging by capturing the actively disseminating cells and their phenotype at a given time, which can then be monitored longitudinally in response to therapy. Prior work using the Epic Sciences CTC platform has established that CTC enumeration is highly prognostic in men with mCRPC, that CTC AR-V7 detection can identify men unlikely to benefit from AR-targeted therapies, and that CTC NE prostate cancer phenotypes can identify men with a very poor prognosis in this setting (11-14, 15). Here, we sought to investigate the prognostic association of PSMA+ CTC detection and heterogeneity with clinical outcomes in men with mCRPC during treatment with potent AR inhibitors such as abiraterone (abi) or enzalutamide (enza), and to quantify the changing cell heterogeneity of PSMA CTC detection over time. To accomplish this, we conducted an analysis of the prospective multicenter PROPHECY (Prospective CiRculating PrOstate Cancer Predictors in HighEr Risk mCRPC StudY) clinical trial (NCT02269982) by utilizing the Epic Sciences CTC platform.
Materials and Methods
Patients and eligibility
In this prospective study, the PROPHECY clinical trial (NCT02269982) is a multicenter trial investigating clinical outcomes among men with progressive mCRPC on standard-of-care abi and enza treatment (N = 118). Study details such as eligibility criteria and definitions of high-risk disease requiring ≥2 poor prognosis clinical factors were described previously (15), and disease progression at entry followed Prostate Cancer Clinical Trials Working Group (PCWG2) criteria (16). All patients with mCRPC provided their written informed permission. The Institutional Review Board approval was obtained from Duke University (Durham, NC) or Weill Cornell Medical College (New York, NY) in compliance with the Declaration of Helsinki ethical principles.
Blood samples for CTC analysis and immunophenotyping were collected at baseline prior to abi/enza therapy, and again upon disease progression as determined by the treating physician, typically based on imaging or clinical parameters rather than based solely on PSA levels over time. Imaging was performed every 3 months during therapy to determine response and progression according to RECIST 1.1 and PCWG3 criteria.
The PSMA CTC test (Epic Sciences) was developed using LNCaP (high PSMA), 22Rv1 (low PSMA), and PC3 (neg PSMA) cells spiked into normal blood. Nucleated blood cells were plated onto glass slides (~3M cells) and subjected to immunofluorescence staining using antibodies targeting cytokeratins (CK), CD45, PSMA, and 4′,6-diamidino-2-phenylindole (DAPI), followed by high-resolution scanning and CTC identification by a multiparametric digital pathology algorithm (Fig. 1A). CTC candidates were confirmed independently by two trained technicians at Epic Sciences. The 4-color assay evaluated PSMA protein expression [Abcam anti-PSMA (ERP6253) rabbit mAb] on individual CTCs, which were CK+, CD45−, and with an intact DAPI nucleus, demonstrating tumor-related morphologies. For each patient, the prevalence of CTC enumeration was represented as “CTC per mL” of blood.
Figure 1.
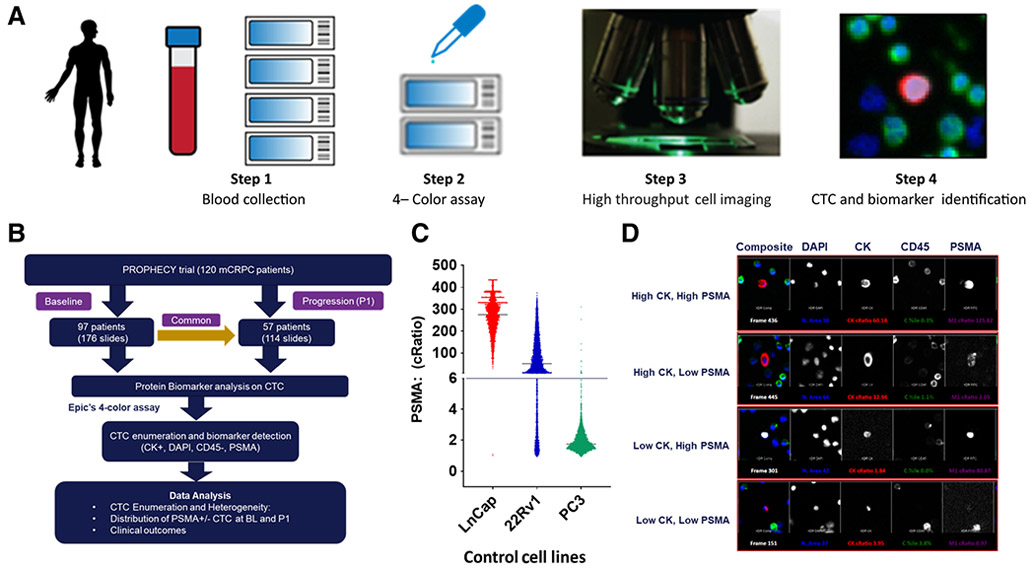
Epic Sciences PSMA detection platform identifies CTC in PROPHECY trial cohorts. A, Step 1: a whole blood sample was collected from the patient and shipped overnight to the Epic facility. RBCs were lysed; nucleated cells were resuspended; approximately 3M nucleated cells from each sample were plated on a microscope slide. Step 2: The slides were stained with a cocktail of antibodies: pan CK, CD45, and DAPI to assess CTC enumeration, and the fourth channel to evaluate PSMA protein expression. Step 3: The stained slides were scanned by Epic’s rapid automated fluorescent scanning method. Step 4: Epic’s proprietary algorithm analyzes cellular parameters, including PSMA expression and cell morphology, to differentiate candidate CTCs from surrounding white blood cells (WBC). Candidate CTCs were identified and displayed in a report and are confirmed by two trained technicians. B, Workflow of the prospective PROPHECY study, a total of 154 mCRPC blood samples were used for CTC enumeration and PSMA expression detection, including baseline (N = 97) and paired progression (N = 57). C, In vitro, PSMA protein expression was evaluated using LNCaP-positive cells, PC3-negative cells, and 22Rv1-medium control cells. cRatio represents the ratio of a cell’s intensity on a specified channel divided by the average of the median cell intensities for all frames in the same channel. D, For example: CTCs expressing high CK high PSMA, high CK low PSMA, low CK high PSMA, and low CK low PSMA in patients with mCRPC were displayed as a CTC gallery (10×).
Data analysis
Patient samples were analyzed for frequency of cell types at calculated cell counts per mL of blood, and univariate distributions of CTC biomarkers were compared at the patient level for each diagnostic category.
The primary objective was to assess the prognostic association of PSMA CTC detection and heterogeneity with clinical outcomes in men with mCRPC who were enrolled in the PROPHECY trial. The primary endpoint was PFS, defined as the time until either radiographic progression using PCWG2 and RECIST 1.1 criteria, clinical progression, or death. OS and response rates (PSA and radiographic) were secondary clinical outcomes. Details regarding the study design have been published elsewhere (15).
We utilized the maximum rank statistical method to find cut-off points for PSMA positivity that corresponded to the largest discrepancy between the positive and negative PSMA CTC groups (10). The primary analysis was based on the optimal cut-off point of two or more CTCs per mL whole blood. In another exploratory analysis, PSMA CTC positivity was based on the observed median of ≥1. Men who had at least one PSMA+ CTC were considered PSMA+ (positive status). The proportional hazards model was used to investigate the prognostic value of PSMA CTC enumeration, PSMA CTCs with OS and PFS adjusting for CellSearch CTC number (≥5), prior therapy (abi/enza), and Halabi and colleagues prognostic factors (risk score; ref. 17), including PSA level, alkaline phosphatase, lactate dehydrogenase (LDH), opioid analgesic use, Eastern Cooperative Oncology Group performance status, albumin, hemoglobin, and metastatic site (visceral, bone, node only). The Kaplan–Meier product-limit approach was used to estimate the PFS and OS distributions by the PSMA+ CTCs. No P values will be reported as these analyses were exploratory.
Data availability
The data generated in this study are available upon request from the corresponding author.
Results
In this prospective study, 118 men with mCRPC were enrolled in the PROPHECY trial from five academic medical centers between May 2015 and January 2017. A total of 154 mCRPC blood samples were used for the CTC enumeration and PSMA protein expression detection, including baseline (N = 97) and paired progression (N = 57) on abi or enza. The CONSORT diagram of mCRPC men treated with abi or enza from the PROPHECY trial (NCT02269982) is shown in Fig. 1B. Table 1 summarizes the patient clinical characteristics for the overall cohort for CTC biomarker detection and was based on the presence of two or more PSMA+ CTCs at baseline which is based on the optimal threshold for PSMA positivity in CTC for PFS and OS discrimination as described below.
Table 1.
The table summarizes the patient characteristics for the overall cohort for CTC biomarker detection. The prognostic association between PSMA+ CTCs and clinical outcomes were performed, and the optimal threshold for PSMA positivity CTC was ≥2 CTC/mL.
| Baseline characteristics | All men (n = 97) | CTC = 0 (n = 19) | PSMA negative (<2 CTCs/mL optimal cutoff) (n = 62) | PSMA positive (≥2 CTCs/mL, optimal cutoff) (n = 16) |
|---|---|---|---|---|
| Age, median years (range) | 73 (44–92) | 71 (57–92) | 74 (44–86) | 71 (48–87) |
| Race: White/Black/other (%) | 83/12/5 | 84/11/5 | 84/11/5 | 75/19/6 |
| Gleason sum 8–10 (%) | 63 | 74 | 61 | 56 |
| Karnofsky score ≥90 (%) | 74 | 74 | 81 | 50 |
| High-risk features | ||||
| Hemoglobin <12 g/dL (%) | 38 | 37 | 34 | 56 |
| Elevated alkaline phosphatase (%) | 43 | 11 | 47 | 69 |
| Elevated serum LDH (%) | 33 | 21 | 23 | 88 |
| Prior abiraterone or enzalutamide (%) | 36 | 47 | 36 | 25 |
| Presence of liver or lung metastasis (%) | 26 | 26 | 27 | 19 |
| Presence of clinically significant pain requiring opiates (%) | 28 | 26 | 24 | 44 |
| CellSearch CTC ≥5 cells per 7.5 mL (%) | 43 | 16 | 39 | 94 |
| Radiographic progression at entry (%) | 78 | 68 | 76 | 100 |
| PSA doubling time <3 months (%) | 62 | 47 | 66 | 63 |
| Prior docetaxel for mHSPC (%) | 19 | 16 | 18 | 25 |
| M1 stage at diagnosis (%) | 29 | 26 | 24 | 50 |
| >20 bone metastases (%) | 33 | 5 | 32 | 69 |
| Median baseline PSA ng/mL (range) | 18 | 16 | 14 | 58 |
| 0.08–4,190 | 0.23–482 | 0.08–1,100 | 0.32–4,190 | |
| Epic nuclear AR-V7 positive (%) | 8 | 0 | 7 | 25 |
Abbreviation: mHSPC, metastatic hormone-sensitive prostate cancer.
CTC enumeration and PSMA expression at baseline and progression
In this study, we performed CTC enumeration and PSMA expression using the Epic PSMA assay, which detects protein expression at a single CTC level. Positive and negative cell line controls spiked into normal blood were used as a reference during each batch of clinical samples immunostaining for assay optimization. The PSMA protein expression distributions in process control cell line cells are shown in Fig. 1C (LNCaP used as a positive control, PC3 as a negative cell, and 22Rv1 as a medium cell control). cRatio represents the ratio of a cell’s intensity on a specified channel divided by the average of the median cell intensities for all frames in the same channel (cRatio cutoff for CK was 3, and for PSMA was 6). CTCs expressing high CK, high PSMA; high CK, low PSMA; low CK, high PSMA; and low CK, low PSMA in patients with mCRPC were displayed as a CTC gallery in Fig. 1D.
The detection of Epic defined CTCs in patients at pretreatment baseline was 80% (78/97), and of these, 55% (43/78) of men harbored PSMA+ CTCs (minimum of at least one CTC). In addition, 21% (16/78) of men had ≥2 PSMA+ CTCs/mL which was established as the optimal cutoff for OS prognostication. Nineteen percent (8/43) of these cases had homogeneous PSMA expression on CTCs, with 100% CTC PSMA positivity (Fig. 2A and B). All the other analyzed patients with mCRPC at baseline detected prominent levels of tumor heterogeneity in CTC enumeration and PSMA CTC protein expression detection (Supplementary Fig. S1). Similarly, at disease progression on abi/enza, 88% (50/57) of men had ≥1 detectable CTC, of whom 68% (34/50) had detectable PSMA+ CTCs (≥1), and 46% (23/50) had ≥2 PSMA+ CTCs/mL and 12% had 100% PSMA+ CTCs (Fig. 2C and D).
Figure 2.
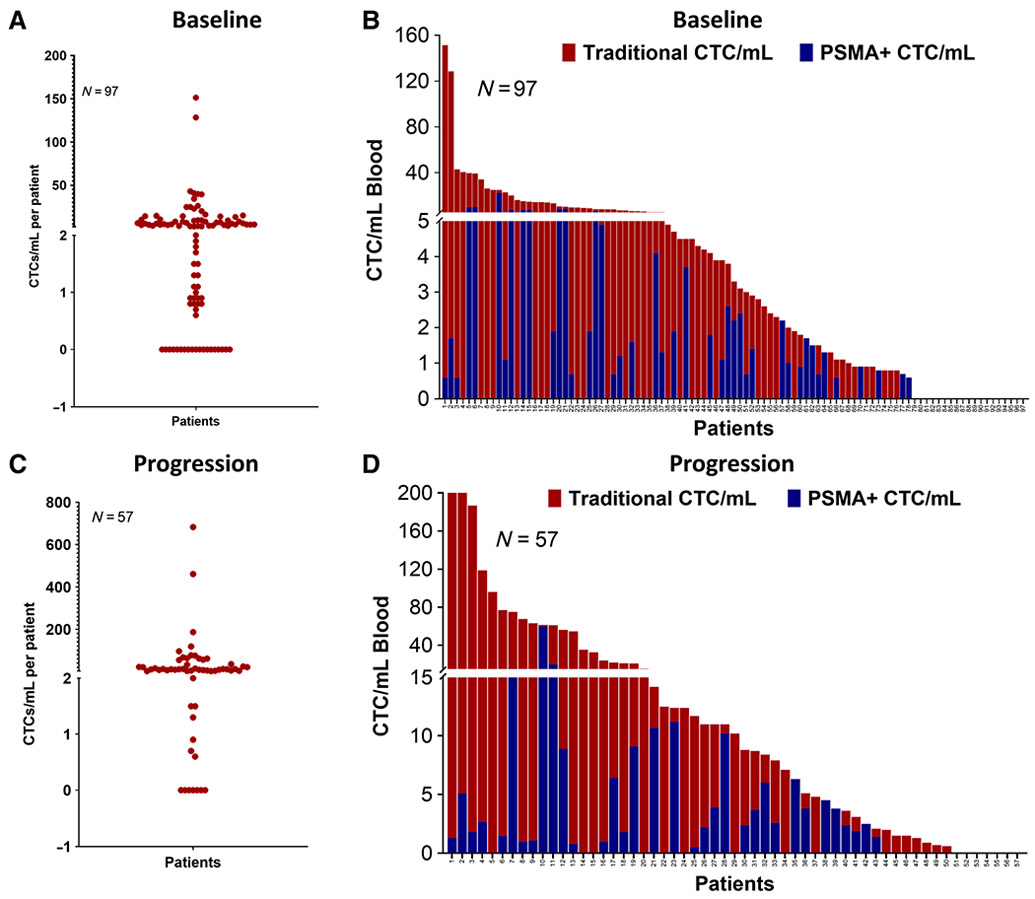
CTC enumeration and PSMA protein expression heterogeneity in pre- and post-abi/enza therapy. A, Dot plot of CTC enumeration at baseline expressed as CTC/mL (N = 97). B, Bar plot of traditional CTC/mL (red) and PSMA+ CTC/mL (blue) at baseline (N = 97). C, Graph depicting CTC enumeration during abi/enza progression as CTC/mL (N = 57). D, A bar graph depicting the traditional CTC (red) and PSMA+ CTC/mL (blue) at progression (N = 57).
PSMA+ CTC expression in paired intraindividual baseline and progression samples was compared (n = 57; Table 2). At baseline, 47/57 (82%) patients were CTC+, and 24/47 (51%) were PSMA+ (at least one CTC), with 6/24 (25%) patients having more than 50% PSMA+ CTCs. Similarly, at progression, 50/57 (88%) patients had CTCs, 34/50 (68%) had PSMA+ CTCs, and 13/34 (38%) had >50% PSMA+ CTCs (Fig. 3A and B). As a result, PSMA+ CTC expression increased slightly in most patients who progressed on either abi or enza. The case-by-case comparison of all 57 cases at baseline and progression, including traditional CTC/mL and PSMA+ CTC/mL at baseline and progression, is shown in Fig. 3C and D. Furthermore, a head-to-head comparison of the common baseline and progression on abi/enza is summarized in Supplementary Table S1.
Table 2.
CTC enumeration and PSMA+ CTC expression at baseline (N = 97) and progression (N = 57) samples from PROPHECY study. All the CTC parameters (mean, median, and range) were represented in CTC/mL.
| Baseline Traditional CTC/mL | Baseline PSMA+ CTC/mL | Progression Traditional CTC/mL | Progression PSMA+ CTC/mL | |
|---|---|---|---|---|
| Overall | N = 97 | N = 57 | ||
| Mean | 9.57 | 1.44 | 42.53 | 3.82 |
| Median | 3.30 | 0.00 | 10.20 | 1.30 |
| Range | 0–151.4 | 0–22 | 0–682.7 | 0–59.7 |
| CTC negative | 19 (20%) | 7 (12%) | ||
| CTC positive | 78 (80%) | 50 (88%) | ||
| PSMA+ CTC cases (N) | 43/78 (55%) | 34/50 (68%) | ||
| Mean | 15.1 | 2.7 | 64.4 | 6.4 |
| Median | 5.4 | 1.7 | 13.3 | 3.2 |
| Range | 0.9–151.4 | 0.5–22 | 0.9–682.7 | 0.5–59.7 |
| PSMA− CTC cases (N) | 35/78 (45%) | 16/50 (32%) | ||
| Mean | 8.03 | 0.00 | 14.67 | 0.00 |
| Median | 4.50 | 0.00 | 5.95 | 0.00 |
| Range | 0.8–40.6 | 0.8–96.10 |
Figure 3.
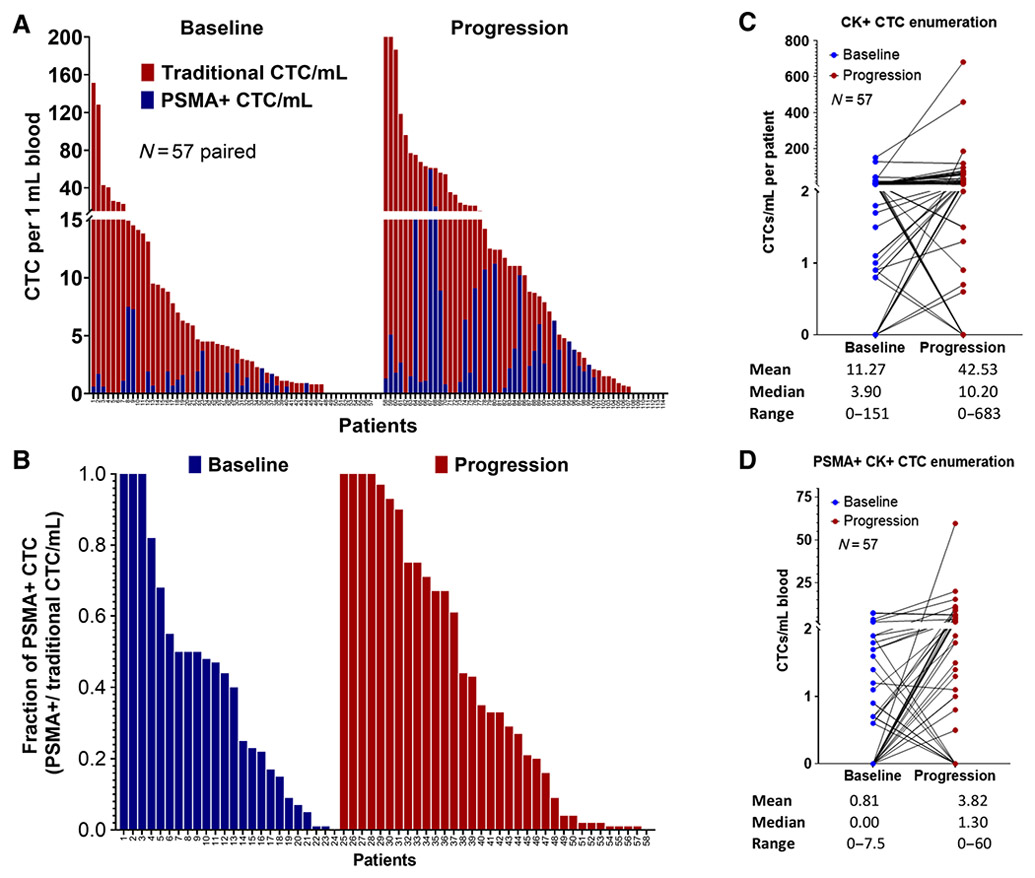
PSMA positivity comparison between common baseline and progression cases. A, Head-to-head comparison of CTC enumeration and PSMA+ CTC/mL at baseline and progression in 57 common mCRPC cases. B, Fraction of PSMA+ CTC expression comparison between baseline and progression samples (PSMA+ CTC/traditional CTC). C, Case-by-case comparison of CTC/mL at baseline and progression (N = 57). D, Case-by-case comparison of PSMA+ CTC/mL at baseline and progression (N = 57). Blue = baseline, Red = progression.
Clinical outcomes: relationship between PSMA+ CTC enumeration with OS and PFS
The prevalence of PSMA and traditional CTC positivity at baseline and progression, based on the observed median and optimal cutoffs is represented in Supplementary Table S2. We identified ≥2 CTCs per mL whole blood as the optimal discriminating threshold for PSMA+ CTC detection for both PFS and OS in univariate analysis.
The median OS in months for men with no detectable CTCs (CTC0; reference), detectable CTCs without PSMA CTC detection (PSMA− CTC), and detectable CTCs with PSMA CTC detection (PSMA+ CTC, using the optimal 2/mL cutoff), respectively, were 25.7 [95% confidence interval (CI) = 19.8–not reachable (NR)], 20.7 (95% CI = 17.7–30.2), and 11.2 (95% CI = 8.0–22.4); univariate HR = 1.2 (95% CI = 0.7–2.3) and HR = 3.4; 95% CI = 1.6–7.0 for men with CTCs that lacked PSMA detection (PSMA− CTC) and CTCs that had at least two positive PSMA CTCs/mL (PSMA+ CTC) as compared with the reference group of men with mCRPC who lacked CTC detection (CTC0), respectively (Fig. 4A). For CTC0 patients (reference), PSMA− CTC, and PSMA+ CTC, the median PFS in months were 7.6 (95% CI = 6.7–16.5), 5.6 (95% CI = 3.6–9.0), and 4.4 (95% CI = 1.8–8.5); univariate HR = 1.5 (95% CI = 0.9–2.6) and HR = 2.8; 95% CI = 1.4–5.8 for PSMA− CTC and PSMA+ CTC versus CTC = 0, respectively (Fig. 4B). Both PSMA+ and PSMA− groups were CTC positive.
Figure 4.
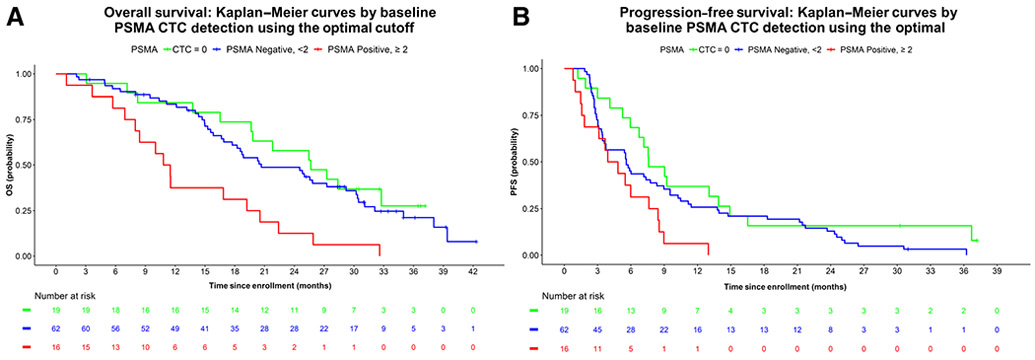
Depicts the association between PSMA+ CTC enumeration and OS and PFS. The associations of PSMA+ CTC enumeration with OS and PFS were explored using the proportional hazard model. A, The median OS in months for CTC = 0 (reference), PSMA− CTC, and PSMA+ CTC, respectively, were 25.7 (95% CI = 19.8–NR), 20.7 (95% CI = 17.7–30.2), and 11.2 (95% CI = 8.0–22.4); univariate HR = 1.2 (95% CI = 0.7–2.3) and HR 3.4; 95% CI = 1.6–7.0 for PSMA− CTC and PSMA+ CTC versus CTC = 0, respectively. B, For CTC0 (reference), PSMA− CTC, and PSMA+ CTC, the median PFS in months were 7.6 (95% CI = 6.7–16.5), 5.6 (95% CI = 3.6–9.0), and 4.4 (95% CI = 1.8–8.5); univariate HR = 1.5 (95% CI = 0.9–2.6) and HR = 2.8; 95% CI = 1.4–5.8 for PSMA− CTC and PSMA+ CTC versus CTC = 0, respectively.
The prognostic importance of PSMA positivity was retained in multivariable analysis of OS adjusting for prior therapy, Halabi risk score, and CellSearch CTC enumeration. The adjusted HRs for OS for the PSMA− CTC and PSMA+ CTC groups using the optimal cutoff were 1.4 (95% CI = 0.6–2.8) and 3.0 (95% CI = 1.1–7.8), respectively, as compared with the CTC0 reference group. Similarly, the adjusted HRs for PFS for the PSMA− CTC and PSMA+ CTC groups using the optimal cutoff of two or more PSMA+ CTCs/mL whole blood was 1.5 (95% CI = 0.8–2.9) and 2.3 (95% CI = 0.9–5.8), respectively, and presented in Tables 3 and 4.
Table 3.
Multivariable analysis of PSMA in predicting OS using the optimal cutoff for PSMA CTC positivity adjusting for prior therapy, CellSearch CTC and Halabi risk score.
| Cutoff (≥) | HR (95% confidence interval) | |
|---|---|---|
| PSMA continuous | NA | 1.10 (1.00–1.20) |
| PSMA continuous (log2(x+1)) | NA | 1.25 (0.94–1.66) |
| PSMA negative (observed median cutoff) | 1 | 1.35 (0.63–2.88) |
| PSMA positive (observed median cutoff) | 1 | 1.73 (0.77–3.92) |
| PSMA negative (optimal cutoff) | 2 | 1.35 (0.64–2.83) |
| PSMA positive (optimal cutoff) | 2 | 2.96 (1.13–7.79) |
Table 4.
Multivariable analysis of PSMA predicting PFS using the optimal cutoff for PSMA CTC positivity adjusting for prior therapy, CellSearch CTC and Halabi risk score.
| Cutoff (≥) | HR (95% confidence interval) | |
|---|---|---|
| PSMA continuous | NA | 1.05 (0.97–1.15) |
| PSMA continuous (log2(x+1)) | NA | 1.30 (0.97–1.73) |
| PSMA negative (observed median cutoff) | 1 | 1.44 (0.75–2.79) |
| PSMA positive (observed median cutoff) | 1 | 1.90 (0.92–3.93) |
| PSMA negative (optimal cutoff) | 2 | 1.51 (0.80–2.87) |
| PSMA positive (optimal cutoff) | 2 | 2.28 (0.90–5.76) |
We further analyzed PFS and OS outcomes in men based on PSMA CTC homogeneity (100% PSMA CTC expression) as compared with PSMA CTC heterogeneity (<100% PSMA CTC expression). The median OS in months for CTC0 patients (reference), CTC+, PSMA− CTC, CTC+ PSMA CTC+ but heterogeneous (<100%), and CTC+ PSMA 100%, respectively, were 25.7 (95% CI = 19.8–NR), 24.5 (95% CI = 16.7–30.4), 15.6 (95% CI = 14.4–20.7), and 35.0 [95% CI = 16.9–Not Available (NA)]; univariate HR = 1.3 (95% CI = 0.7–2.6), HR = 2.0 (95% CI = 1.0–3.8) and HR 0.7; 95% CI = 0.2–2.1 for CTC+, PSMA− CTC, CTC+ PSMA CTC+ but heterogeneous (<100%), and CTC+ PSMA 100% versus CTC = 0, respectively. For PFS outcomes, median PFS CTC0 (reference), CTC+, PSMA− CTC, CTC+ PSMA CTC+ but heterogeneous (<100%), and CTC+ PSMA 100%, respectively, were 7.6 (95% CI = 6.7–16.5), 6.0 (95% CI = 3.5–11.4), 5.5 (95% CI = 3.6–7.6), and 5.7 (95% CI = 3.0–NA) months, respectively. The univariate HR for PFS for these groups relative to CTC0 patients was 1.4 (95% CI = 0.8–2.6), HR = 2.0 (95% CI = 1.1–3.6) and HR = 1.5; 95% CI = 0.7–3.6 for CTC+, PSMA− CTC, CTC+ PSMA CTC+ but heterogeneous (<100%), and CTC+ PSMA 100% versus CTC = 0, respectively. See Supplementary Figs. S2A and S2B and Supplementary Tables S3A and S3B.
Secondary clinical outcomes: relationship of PSMA+ CTC enumeration with confirmed PSA declines and objective radiographic responses
We analyzed the proportion of men who achieved a 50% or greater PSA decline from baseline, confirmed on a subsequent value 4 or more weeks later, with abi or enza, based on pretreatment PSMA CTC detection using the optimal threshold of two or more PSMA+ CTCs. PSA50 confirmed declines were observed in 50% of men (9/18) who were CTC0, 27.5% of men who were CTC+ PSMA− (14/51), and 15.4% of men who were CTC+ PSMA+ (2/13). Likewise, using RECIST 1.1, the proportion of RECIST evaluable men who achieved an objective partial response with abi or enza was 28.6% of men who were CTC0 (2/7), 22.2% of men who were CTC+ PSMA− (6/27), and 33.3% of men who were CTC+ PSMA+ (2/6). See Supplementary Fig. S3A and S3B. Thus, PSMA CTCs were prognostic for worse OS and PFS outcomes with abi or enza, lower rates of confirmed PSA declines, but no differences in objective radiographic responses in this study.
Association of AR-V7, chromosomal instability, and neuroendocrine prostate cancer (NEPC) CTC phenotypes with PSMA expression
Finally, we explored whether PSMA CTC detected differed in groups of men who harbored a NEPC CTC phenotype, given prior work suggesting that NEPC transformation may lead to loss of PSMA expression. We analyzed coexpression of CTC PSMA with biomarkers associated with poor outcome and aggressive clinical behavior in castration-resistant prostate cancer, including detection of AR-V7 nuclear protein in CTCs, CTC chromosomal instability (CIN) immunophenotype, and NEPC phenotypes in CTCs at baseline and over time during progression on abi or enza treatment as detailed in prior prospective validation studies of men with mCRPC (12, 13, 15). CTC biomarker group incidence was compared by the PSMA optimal cutoff for CTC AR-V7, CIN, and NE detection at baseline (97 cases).
In pretreatment baseline samples, 15/16 (94%) CIN-positive cases, 6/16 (37.5%) NE-positive cases, and 4/16 (25%) AR-V7–positive cases were PSMA+ according to the optimal threshold CTC cutoff (n = 16 cases PSMA+). Interestingly, a trend of lower PSMA positivity and thus greater PSMA negativity within the NEPC CTC+ patients was observed in comparison with PSMA+/NE− group, with the majority of NEPC+ patients having <50% PSMA CTC detection (Supplementary Fig. S4). Similarly, at progression as per optimal threshold cutoff (n = 23 cases PSMA+), 11/23 (48%) AR-V7–positive, 4/23 (17%) NE-positive cases, and 13/23 (57%) CIN-positive cases were PSMA+ (Supplementary Table S3). Thus, we observed PSMA expression heterogeneity regardless of CTC-CIN, NEPC, or AR-V7 phenotype and a general reduction of CTC PSMA detection in many patients with evidence of poor prognosis NEPC or AR-V7 CTC biomarker detection.
Discussion
In this retrospective analysis of CTC PSMA detection in the multicenter prospective PROPHECY study, we identified several important and clinically relevant findings that may have implications for clinical research and patient care. The first is that PSMA detection was heterogeneous among men with mCRPC, with some men exhibiting uniform PSMA+ disease and some men lacking PSMA detection entirely despite having a large number of CTCs. This heterogeneity was also present after progression on abi or enza therapy, with a modest increase in PSMA+ CTCs and the percentage of patients with any PSMA CTC detection in this post-AR inhibitor mCRPC setting. Second, we found that using an optimal cut-off point of two or more PSMA+ CTCs/mL of whole blood, PSMA+ CTCs were independently prognostic of both PFS and OS in the context of AR inhibitor therapy in men with mCRPC. Thus, PSMA+ CTCs may confer added risk above and beyond standard CTC enumeration and clinical risk prognostic factors including prior abi or enza therapy. These data suggest that PSMA CTC detection and characterization is adversely and independently prognostic in an AR inhibitor context in men with mCRPC. Such patients currently may benefit from PSMA Lu177 radioligand therapy, and thus a liquid biopsy assay that more completely characterizes this PSMA detection heterogeneity could be useful in identifying men who may have differential outcomes with PSMA-targeted therapy, such as Lu177-PSMA-617 radioligand therapy. Further validation of this CTC biomarker in such a novel context of use setting is now planned to demonstrate such critical predictive clinical utility.
PSMA-targeted radionuclide therapy has significantly improved outcomes in men with mCRPC and is now being tested in earlier disease settings. However, there is clear outcome heterogeneity despite the utility of PSMA PET imaging and patient selection, with some men exhibiting long-term durable responses and survival, and some men exhibiting a lack of any clinical benefit. PSMA PET characteristics such as standardized uptake value (SUV) mean (whole body) may identify differential outcomes with such therapy, but these assessments are not readily available in the community (18). In addition, posttreatment PSA levels may also capture the heterogeneity of PFS and OS outcomes following PSMA-Lu177 therapy (6). A liquid biopsy assay to detect and characterize PSMA expression and heterogeneity at baseline and over time could be helpful to guide optimal therapy and identify patients most likely to benefit.
Our retrospective analysis of PROPHECY suggests that PSMA expression was effectively detected and quantitated at a single CTC level, with a high prevalence of detection at disease progression or abi or enza. Lower detection of PSMA+ CTCs was also clearly identified in men with poor prognostic NEPC phenotypes as measured by the prospectively validated CTC NEPC assay (12). We quantified PSMA CTC heterogeneity in men with mCRPC before and following progression on abi or enza therapy, finding an increase in PSMA+ CTC identification following AR inhibitor therapy in the majority of patients, and a decrease in expression in patients with NEPC or AR-V7–positive CTC phenotypes. Preservation of CTC PSMA expression in patients with CIN is of interest and suggests that men with mCRPC and DNA repair deficiencies such as BRCA2 loss may have PSMA avid disease as has been reported previously. The CTC and PSMA CTC enumerations were adversely prognostic and given the known prognostic impact of PSMA PET detection and intensity on outcomes, this assay could be useful in selecting patients with mCRPC for PSMA-targeted therapies in the early stages of disease progression (19, 20). Such personalization of prognostic outcomes based on CTC biomarkers may facilitate optimal treatment selection in the future. For example, we hypothesize that men with more abundant PSMA CTC expression homogeneity will actually have improved outcomes with PSMA-targeting radioligand therapy as compared with men with CTCs lacking PSMA or having greater PSMA heterogeneity. While it is known that patterns of spread such as to visceral organs such as liver may negatively impact response and benefits of PSMA radioligand therapy, a liquid biopsy to more fully capture this loss of PSMA expression may lead to combination radioligand or treatment approaches, or alternatives for patients predicted to not respond to this therapy, pending prospective validation (18). The relationship between PSMA CTC enumeration and heterogeneity characterization with clinical outcomes in the context of PSMA-directed therapy will be further explored in future prospective research.
The limitations of this work include the lack of external validation in the context of PSMA-targeted therapy and PSMA PET/CT characteristics, and this work is ongoing in the phase II PRINCE trial (NCT03658447) of pembrolizumab plus Lu177-PSMA-617 radioligand therapy and other trials. The optimal cut-off point of 2 CTC/mL is not yet externally validated. The prospective validation studies are now planned in the context of PSMA-targeted therapy to evaluate the optimal cut-off point for PSMA CTC enumeration and heterogeneity characterization at baseline and over time for prediction of survival and PFS benefits. The potential for future predictive studies of PSMA CTC detection may be to complement PSMA PET/CT imaging to further optimize care delivery and monitoring of PSMA-directed therapies, pending future validation efforts in this context. Prior work suggests that AR inhibition may modulate PSMA detection on imaging, and our findings support this in some men, but further studies are needed using PSMA CTC detection earlier in the course of potent AR inhibition rather than at progression (21). Such work should provide a greater basis for optimal patient selection for PSMA-directed therapeutics and decisions on further therapy based on clinical benefits and early pharmacodynamic measures of efficacy such as PSMA CTC enumeration.
Supplementary Material
Translational Relevance.
In men with metastatic castration-resistant prostate cancer, we found that prostate-specific membrane antigen (PSMA) detection on circulating tumor cells (CTC) was heterogeneous both between patients and within the same patients over time, increasing during treatment with the potent androgen receptor (AR) inhibitors abiraterone or enzalutamide. PSMA CTC detection using the optimal prognostic threshold of ≥2 PSMA-positive CTCs detected per mL of whole blood was independently associated with shorter overall and progression-free survival after adjusting for clinical risk factors, CTC enumeration, and prior AR therapy exposure. PSMA CTC detection was also independent of AR-V7–positive CTC detection or neuroendocrine CTC phenotype. Our findings suggest a liquid biopsy technique that detects and identifies PSMA protein expression and heterogeneity over time at a single cell level could be useful in identifying individuals who have worse outcomes with existing hormonal therapies and, pending future predictive validation, those men who may benefit the most from PSMA-targeted therapy.
Acknowledgments
We wish to thank the Prostate Cancer Foundation and Movember for their financial support of this Global Treatment Sciences Challenge Award (A.J. Armstrong, Duke), and the U.S. Department of Defense Prostate Cancer Clinical Trial Consortium for infrastructural support for this multicenter study. A.J. Armstrong was supported by a Prostate Cancer Foundation grant, an NIH R01 (1R01CA233585-01) and the DCI P30 CA014236 as well as Duke Cancer Institute shared resources for biostatistics, Flow Cytometry, and Sequencing and Genomic Technologies. This work was partially funded by Department of Defense grants W81XWH-13-PCRP-CCA, W81XWH-17-2-0021 and W81XWH-14-2-0179 (D.J. George, A.J. Armstrong, Duke), W81XWH-15-1-0467 (S. Halabi, Duke), W81XWH-18-1-0278 (S. Halabi, Duke), W81XWH-14-2-0159 (D.M. Nanus, Weill Cornell), W81XWH-15-2-0018 (R.Z. Szmulewitz, Chicago), W81XWH-15-2-0018 (H.I. Scher, MSKCC), and W81XWH-16-PCRP-CCRSA (E.S. Antonarakis, Johns Hopkins). D.C. Danilla and H.I. Scher were also supported in part by NCI grants P30CA008748 and the MSKCC Sidney Kimmel Center for Prostate and Urologic Cancers. We wish to thank the study coordinators at Weill Cornell, Duke University, Johns Hopkins, University of Chicago, and Memorial Sloan Kettering Cancer Center. We wish to acknowledge the dedication of our patients to provide blood samples at no clear benefit to them but for the benefit of all patients with prostate cancer.
Authors’ Disclosures
S. Halabi reports grants from Epic Sciences during the conduct of the study, as well as other support from Sanofi, BMS, Aveo Oncology, and Janssen outside the submitted work. A. Tubbs reports personal fees and non-financial support from Epic Sciences, Inc. during the conduct of the study, as well as personal fees and non-financial support from Epic Sciences, Inc. outside the submitted work. D.J. George reports personal fees from Advanced Accelerator Applications, American Association for Cancer Research, AVEO Pharmaceuticals, Brown and James Atty, Eisai, IdeoOncology, Medscape Education, Merck Sharp & Dohme, Michael J Hennessey & Associates, Millennium Medical Publishing, Myovant Sciences, Nektar Therapeutics, Propella Therapeutics, Seattle Genetics, UroGPO, UroTo-day, WebMD, WilmerHale Atty, and Xcures; grants and personal fees from Astellas, AstraZeneca, Bayer H/C Pharma, Exelixis, Janssen Pharma, Pfizer, and Sanofi; and grants from BMS, Novartis, and Calithera outside the submitted work. D.M. Nanus reports grants from Prostate Cancer Foundation during the conduct of the study, as well as personal fees from Janssen Oncology outside the submitted work. E.S. Antonarakis reports grants and personal fees from Janssen, Sanofi, Bayer, Bristol Myers Squibb, Curium, Merck, Pfizer, AstraZeneca, and Clovis; grants from Johnson & Johnson, Novartis, and Celgene; and personal fees from Amgen, Blue Earth, Exact Sciences, Invitae, and Eli Lilly outside the submitted work. D.C. Danila reports grants from Prostate Cancer Foundation during the conduct of the study, as well as personal fees from Angle LLT and BioView outside the submitted work. In addition, D.C. Danila reports research support from U.S. Department of Defense, American Society of Clinical Oncology, Prostate Cancer Foundation, Stand Up 2 Cancer, Amgen, Janssen Research & Development, Astellas, Medivation, Agensys, Genentech, and CreaTV, as well as consultant for Angle LLT, Axiom LLT, Janssen Research & Development, AstraZeneca, BioView LTD, Clovis, Astellas, Medivation, Pfizer, Agensys, and Merck. R.Z. Szmulewitz reports personal fees from Novartis during the conduct of the study, as well as personal fees from Astellas, Janssen, and Bayer outside the submitted work. R. Wenstrup reports other support from Epic Sciences during the conduct of the study. A.J. Armstrong reports personal fees from Epic Sciences during the conduct of the study. A.J. Armstrong also reports grants and personal fees from Pfizer, Astellas, Bayer, Janssen, Merck, BMS, AstraZeneca, and Forma; personal fees from Ideaya; and grants from Constellation outside the submitted work. No disclosures were reported by the other authors.
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Prior presentation: Presented in part at the 2022 European Society of Medical Oncology.
References
- 1.Bakht MK, Derecichei I, Li Y, Ferraiuolo RM, Dunning M, Oh SW, et al. Neuroendocrine differentiation of prostate cancer leads to PSMA suppression. Endocr Relat Cancer 2018;26:131–46. [DOI] [PubMed] [Google Scholar]
- 2.Paschalis A, Sheehan B, Riisnaes R, Rodrigues DN, Gurel B, Bertan C, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol 2019;76:469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020;26:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaittanis C, Andreou C, Hieronymus H, Mao N, Foss CA, Eiber M, et al. Prostate-specific membrane antigen cleavage of vitamin B9 stimulates oncogenic signaling through metabotropic glutamate receptors. J Exp Med 2018; 215:159–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor O, de Bono J, Chi KN, Fizazi K, Herrmann K, Rahbar K, et al. Lutetium-177-PSMA-617 for metastatic castration-resistant prostate cancer. N Engl J Med 2021;385:1091–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong AJ, Sartor O, Saad F, Czernin J, Shore ND, Kendi AT, et al. Association between prostate-specific antigen decline and clinical outcomes in patients with metastatic castration-resistant prostate cancer in the VISION trial. Ann Oncol 2022;33:S1169–70. [Google Scholar]
- 7.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal 2003;43:121–37. [Google Scholar]
- 8.Sokoloff RL, Norton KC, Gasior CL, Marker KM, Grauer LS. A dual monoclonal sandwich assay for prostate-specific membrane antigen: levels in tissues, seminal fluid, and urine. Prostate 2000;43:150–7. [DOI] [PubMed] [Google Scholar]
- 9.Gafita A, Calais J, Grogan TR, Hadaschik B, Wang H, Weber M, et al. Nomograms to predict outcomes after 177Lu-PSMA therapy in men with metastatic castration-resistant prostate cancer: an international, multicentre, retrospective study. Lancet Oncol 2021;22: 1115–25. [DOI] [PubMed] [Google Scholar]
- 10.Autio KA, Dreicer R, Anderson J, Garcia JA, Alva A, Hart LL, et al. Safety and efficacy of BIND-014, a docetaxel nanoparticle targeting prostate-specific membrane antigen for patients with metastatic castration-resistant prostate cancer: a Phase 2 clinical trial. JAMA Oncol 2018;4:1344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S, Halabi S, Kemeny G, Anand M, Giannakakou P, Nanus DM, et al. Circulating tumor cell genomic evolution and hormone therapy outcomes in men with metastatic castration-resistant prostate cancer. Mol Cancer Res 2021; 19:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown LC, Halabi S, Schonhoft JD, Yang Q, Luo J, Nanus DM, et al. Circulating tumor cell chromosomal instability and neuroendocrine phenotype by immunomorphology and poor outcomes in men with mCRPC treated with abiraterone or enzalutamide. Clin Cancer Res 2021;27:4077–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher HI, Armstrong AJ, Schonhoft JD, Gill A, Zhao JL, Barnett E, et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer 2021;150:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong AJ, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, Danila DC, et al. Prospective multicenter study of circulating tumor cell AR-V7 and taxane versus hormonal treatment outcomes in metastatic castration-resistant prostate cancer. JCO Precis Oncol 2020;4:PO.20.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. J Clin Oncol 2019;37:1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the prostate cancer clinical trials working group. J Clin Oncol 2008;26:1148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halabi S, Lin CY, Small EJ, Armstrong AJ, Kaplan EB, Petrylak D, et al. Prognostic model predicting metastatic castration-resistant prostate cancer survival in men treated with second-line chemotherapy. J Natl Cancer Inst 2013;105:1729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillip K, Jacob H, Kambiz R, Ayse TK, Xiao X, Phillip K, et al. [68Ga]Ga-PSMA-11 PET baseline imaging as a prognostic tool for clinical outcomes to [177Lu]Lu-PSMA-617 in patients with mCRPC: a VISION substudy. J Clin Oncol 40: 16s, 2022. (suppl; abstr 5002). [Google Scholar]
- 19.Paschalis A, Sheehan B, Riisnaes R, Rodrigues DN, Gurel B, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol 2019;76:469–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gafita A, Calais J, Grogan TR, Hadaschik B, Wang H, et al. Nomograms to predict outcomes after 177Lu-PSMA therapy in men with metastatic castration-resistant prostate cancer: an international, multicentre, retrospective study. Lancet Oncol 2021;22:1115–25. [DOI] [PubMed] [Google Scholar]
- 21.Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Australas Med J 1869;14:146–9. [Google Scholar]
- 22.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.


