Abstract
Two chromosome partitioning proteins, Soj (ParA) and Spo0J (ParB), regulate the initiation of sporulation in Bacillus subtilis. In a spo0J null mutant, sporulation is inhibited by the action of Soj. Soj negatively regulates expression of several sporulation genes by binding to the promoter regions and inhibiting transcription. All of the genes known to be inhibited by Soj are also activated by the phosphorylated form of the transcription factor Spo0A (Spo0A∼P). We found that, in a spo0J null mutant, Soj affected sporulation, in part, by decreasing the level of Spo0A protein. Soj negatively regulated transcription of spo0A and associated with the spo0A promoter region in vivo. Expression of spo0A from a heterologous promoter in a spo0J null mutant restored Spo0A levels and partly bypassed the sporulation and gene expression defects. Soj did not appear to significantly affect phosphorylation of Spo0A. Thus, in the absence of Spo0J, Soj inhibits sporulation and sporulation gene expression by inhibiting accumulation of the activator protein Spo0A and by acting downstream of Spo0A to inhibit gene expression directly.
Bacillus subtilis can sporulate in response to starvation and high cell density. Sporulation involves an asymmetric cell division and requires DNA replication and chromosome segregation. Spo0A is a critical transcription factor in early sporulation. Cells initiate sporulation when they accumulate a threshold amount of phosphorylated Spo0A (Spo0A∼P) (3, 6, 12). Many signals that regulate sporulation do so by affecting the accumulation of Spo0A∼P. Spo0A obtains phosphate from a set of proteins called the phosphorelay (2). A balance of kinases and phosphatases regulates phosphate input into the phosphorelay and the phosphorylation state of Spo0A (3, 30–32).
Two proteins involved in chromosome partitioning, Soj and Spo0J, also regulate the initiation of sporulation, perhaps in response to chromosome structure or partitioning (4, 14, 28). Soj and Spo0J are members of the ParA-SopA and ParB-SopB protein families, respectively. Many ParA- and ParB-type proteins are encoded on low-copy-number plasmids and are required for accurate plasmid partitioning (29, 42). Chromosome-encoded family members have also been identified, several of which function in chromosome partitioning (14, 22, 26). Members of the ParA-SopA families can act as transcriptional repressors and are themselves often regulated by members of the ParB-SopB family (7, 9, 24, 27, 36).
Spo0J is required both for accurate chromosome partitioning and for sporulation (14). Spo0J binds to a series of sites (parS sites) in the origin-proximal 20% of the chromosome (22), clustering them into a large focus (10, 21, 23). Defects in spo0J perturb chromosome partitioning. Cultures of spo0J null mutant cells produce approximately 1% anucleate cells, a frequency approximately 20- to 100-fold greater than that of wild-type cells (14).
A null mutation in spo0J also causes an approximately 300-fold defect in sporulation relative to wild-type cells (14, 28, 34). spo0J null mutants are blocked early in sporulation (stage 0) and have decreased expression of at least three Spo0A∼P-activated, sporulation genes (14, 28). The defects in sporulation and gene expression caused by Δspo0J are completely suppressed by the deletion of soj. Deletion of soj alone has little effect on sporulation. These results suggest that Soj is a negative regulator of sporulation and is itself antagonized by Spo0J (14). This may represent a signaling pathway that regulates sporulation in response to aspects of chromosome organization or partitioning.
Soj negatively regulates transcription of several early sporulation genes. Histidine-tagged Soj inhibits transcription from the spoIIG promoter in vitro (4). In addition, formaldehyde crosslinking has shown that Soj associates with several Spo0A∼P-dependent promoters in vivo, including spoIIA, spoIIE, and spoIIG, and that this association correlates with decreased gene expression (36).
We have found that Soj also affects sporulation and Spo0A∼P-dependent gene expression by decreasing levels of Spo0A protein. Soj negatively regulated transcription of spo0A and associated with the spo0A promoter region in vivo. Expression of spo0A from a heterologous promoter restored Spo0A levels and partly bypassed the sporulation and gene expression defects of a spo0J null mutant.
MATERIALS AND METHODS
Strains.
The B. subtilis strains used are listed in Table 1. All strains are derivatives of JH642 (33), which is referred to as wild type. Standard procedures were used for transformations and strain construction (11). The Δspo0J::spc and Δ(soj-spo0J)::spc deletion-insertions were previously described (14). The Pspac-spo0A+ and Pspac-spo0Asad67 constructs were previously described (15). The spoIIG-lacZ fusion (17) is located in the specialized transducing phage SPβ. A cat marker ∼90% linked (by transformation) to spo0A (13) was used to transfer the rvtA11 mutation (38).
TABLE 1.
B. subtilis strains used
| Strain | Genotype | Reference |
|---|---|---|
| JH642 | Wild type (trpC2 pheA1) | |
| AG1332 | amyE::(spo0A-129-lacZ neo) | |
| AG1334 | amyE::(spo0A-78-lacZ neo) | |
| AG1431 | spo0FΔS spo0BΔPst-phe+ | 19 |
| AG1468 | Δspo0J::spc | 14 |
| AG1505 | Δ(soj-spo0J)::spc | 14 |
| DZR158 | amyE::(Pspac-spo0A+cat) | 15 |
| DZR160 | amyE::(Pspac-spo0A+cat) spo0A::erm | 15 |
| JQ266 | amyE::(spo0A-129-lacZ neo) Δspo0J::spc | |
| JQ270 | amyE::(spo0A-129-lacZ neo) Δ(soj-spo0J)::spc | |
| JQ279 | amyE::(Pspac-spo0A+cat) Δ(soj-spo0J)::spc | |
| JQ280 | amyE::(Pspac-spo0A+cat) Δspo0J::spc | |
| JQ284 | SPβ::(spoIIG-lacZ erm cat::neo) | 15 |
| JQ302 | SPβ::(spoIIG-lacZ erm cat::neo) Δspo0J::spc | |
| JQ303 | SPβ::(spoIIG-lacZ erm cat::neo) Δ(soj-spo0J)::spc | |
| JQ304 | SPβ::(spoIIG-lacZ erm cat::neo) amyE::(Pspac-spo0A+cat) | |
| JQ305 | SPβ::(spoIIG-lacZ erm cat::neo) amyE::(Pspac-spo0A+cat) Δspo0J::spc | |
| JQ306 | SPβ::(spoIIG-lacZ erm cat::neo) amyE::(Pspac-spo0A+cat) Δ(soj-spo0J)::spc | |
| JQ390 | rvtA11-cat | 19 |
| JRL783 | spo0FΔS spo0BΔPst-phe+rvtA11-cat | 19 |
| JRL791 | rvtA11-cat ΔkinC::spc | 19 |
| JRL792 | spo0FΔS spo0BΔPst-phe+rvtA11-cat ΔkinC::erm | 19 |
| KI1775 | rvtA11-cat Δspo0J::spc | 13 |
| KI1947 | rvtA11-cat Δspo0J::spc ΔkinC::erm | 19 |
| KI1968 | amyE::(Pspac-spo0A+cat) Δspo0J::spc | |
| KI1969 | amyE::(Pspac-spo0Asad67 cat) Δspo0J::spc | |
| SIK25 | amyE::(Pspac-spo0Asad67 cat) |
The transcriptional fusion of the spo0A sporulation promoter Ps to lacZ [spo0As(−129)−lacZ] was constructed in the amyE vector pKS2 and was integrated into the B. subtilis chromosome by double crossover at the amyE locus. The endpoints of the fusion were 129 bp upstream of the Ps transcription start site and the BglII restriction site within the spo0A coding region.
Routine growth and maintenance of E. coli and B. subtilis was done in Luria-Bertani (LB) medium. For sporulation experiments, the rich sporulation broth, 2×SG (20) was used. IPTG (isopropyl-β-d-thiogalactopyranoside) was used at 0.5 mM to induce expression of spo0A from Pspac. Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol (Cm) at 5 μg/ml, spectinomycin (Spc) at 100 μg/ml, neomycin (Neo) at 5 μg/ml, and erythromycin and lincomycin together (MLS) at 0.5 and 12.5 μg/ml, respectively.
Protein extracts and Western blot analysis.
Cells were grown in 2×SG, and samples of 1 to 5 ml were taken 2 h after the end of exponential growth and centrifuged to collect cells. Pspac-spo0A expression was induced with 0.5 mM IPTG three to four doublings before the end of exponential growth. Cell pellets were frozen and stored at −70°C. Pellets were resuspended in ice cold buffer (10 mM Tris, pH 8; 100 mM NaCl; 10 mM KCl) with 1 mM phenazine methosulfate (PMSF) and lysed by sonication on ice for four to six pulses of 10 s each (Branson Sonifier, Power 4, duty cycle 100%). Cell debris was pelleted by centrifugation in a microcentrifuge (10 min, 13,000 rpm, 4°C), and the supernatant was collected for further use. Total soluble protein was determined by the Bio-Rad Protein Assay (Bio-Rad) using bovine serum albumin as the standard. Sodium dodecyl sulfate (SDS) sample buffer was added, and the extracts were vortexed for 20 s and briefly centrifuged prior to electrophoresis on SDS–12% polyacrylamide gels.
Proteins were transferred to polyvinylidene difluoride membrane (Schleicher and Schuell) for 45 min at 450 mA using a semidry electroblotter (Owl Scientific). Membranes were blocked by incubation in Tris-buffered saline with Tween 20 (TBST [37]) plus 5% dried milk (Carnation). The primary antibody was a polyclonal rabbit antisera raised against purified Spo0A tagged with six histidines. The antibody was diluted 1:1,000 in blocking solution. The secondary antibody was 125I-labeled goat anti-rabbit immunoglobulin G (DuPont-New England Nuclear), used at 0.3 μCi/ml. The 125I signal was detected using a PhosphorImager (Molecular Dynamics) and quantified using ImageQuant software (Molecular Dynamics).
β-Galactosidase assays.
Cells were grown and treated as described in the figure legends. β-Galactosidase activity was determined as described previously (16, 25). The specific activity is calculated as the change in A420 per minute per milliliter of culture per unit of optical density at 600 nm times 1,000.
Spore assays.
Cells were grown in 2×SG at 37°C, and spores were assayed 20 to 24 h after the end of exponential growth. The number of viable cells per milliliter of culture was determined as the total number of CFU on LB plates. The number of spores per milliliter of culture was determined as the number of CFU on plates after heat treatment (80°C for 20 min). The percent sporulation is 100 times the ratio of spores per milliliter to viable cells per milliliter.
Formaldehyde cross-linking and immunoprecipitations.
Two hours after the end of exponential growth in 2×SG, 10-ml samples of culture were taken for analysis. Generally, cross-linking and sample preparations were performed as described earlier (22, 36) and are based on previously published chromatin immunoprecipitation assays (8, 39, 40). Samples were treated with sodium phosphate (10 mM final concentration) and formaldehyde (1% final concentration) for 3 min at room temperature. Cells were pelleted and washed twice with 10 ml of phosphate-buffered saline (pH 7.3) (1), resuspended in 500 μl of solution A (10 mM Tris [pH 8], 20% sucrose, 50 mM NaCl, 10 mM EDTA) containing 20 mg of lysozyme per ml, and incubated at 37°C for 30 min. Then, 0.5 ml of 2× IP buffer (100 mM Tris, pH 7; 300 mM NaCl; 2% Triton X-100) and PMSF (to final concentration of 1 mM) were added, and the cells were incubated for 10 min at 37°C. DNA was sheared to an average size of 500 to 1,000 bp by sonication. Insoluble debris was removed by centrifugation, and the supernatant was transferred to a fresh microfuge tube. To determine the amount of DNA immunoprecipitated relative to the amount of total DNA prior to immunoprecipitation, 100 μl of supernatant was removed and saved for later analysis (“total” DNA control).
Protein and protein-DNA complexes were immunoprecipitated (overnight, 4°C) by incubation with affinity-purified polyclonal anti-Soj antibodies followed by incubation with 30 μl of a 50% protein A-Sepharose slurry (1 h, room temperature). Complexes were collected by centrifugation and washed seven times with 1× IP buffer (twice-diluted 2× IP buffer described above) and twice with 1 ml of TE (10 mM Tris, pH 8; 0.1 mM EDTA). The slurry was resuspended in 100 μl of TE. The 100 μl of “total” DNA control was mixed with 100 μl of TE and brought to a final concentration of 0.1% SDS. Formaldehyde crosslinks of both immunoprecipitated and total DNA samples were reversed by incubation at 65°C for 6 h. Samples were used for PCR without further treatment.
PCR was performed with Vent DNA polymerase (New England Biolabs) using serial dilutions of the immunoprecipitate and the total DNA as templates. Oligonucleotide primers were typically 20 to 25 bases in length, and the amplified fragments ranged from ∼250 to 550 bp in size. Sequences of all primers are available upon request. Relative amounts of Soj-DNA complexes were determined by comparing the intensity of bands in the linear range of the PCR from both the immunoprecipitate and the “total” DNA control. Gels were photographed onto Polaroid 665 film, and the negatives were scanned and processed using Adobe Photoshop software.
RESULTS AND DISCUSSION
Soj decreases the level of Spo0A protein in a spo0J null mutant.
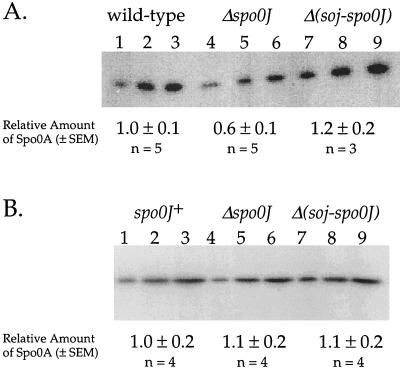
Deletion of spo0J causes a decrease in expression of at least three early sporulation genes that require the Spo0A transcription factor for activation (14). We found that Spo0A protein levels were lower in the spo0J null mutant. We measured the amount of Spo0A protein by quantitative Western blots (see Materials and Methods). A spo0J null mutant had approximately 45% less Spo0A protein than the wild-type cells (Fig. 1A). The amount of Spo0A was restored to wild-type levels in a Δ(soj-spo0J) double mutant. This indicates that the decrease in Spo0A protein levels in the spo0J mutant was dependent on soj, that Soj negatively regulates accumulation of Spo0A protein, and that this effect of Soj is antagonized by Spo0J.
FIG. 1.
Quantitative 125I Western blot of Spo0A in strains containing mutations in spo0J and soj. Strains were grown in 2×SG medium. Samples were harvested 2 h after the end of exponential growth. Three different amounts of protein were loaded for each sample to show the linearity of the detection system. Quantitation of Spo0A protein levels compiled from independent experiments is shown below the gel. Numbers are the mean ± the standard error of the mean, normalized such that wild-type cells have 1 unit of Spo0A protein. (A) Spo0A expressed from the native promoter. Proteins in each lane were isolated from the following strains. Lanes 1 to 3, AG1113 (wild type); lanes 4 to 6, KI1778 (Δspo0J); lanes 7 to 9, KI1798 [Δ(soj-spo0J)]. The total amounts of protein loaded were 2 (lanes 1, 4, and 7), 4 (lanes 2, 5, and 8) and 6 (lanes 3, 6, and 9) μg. (B) Spo0A expressed from the Pspac promoter. Proteins in each lane are isolated from the following strains. Lanes 1 to 3, DZR160 (Pspac-spo0A); lanes 4 to 6, JQ280 (Pspac-spo0A Δspo0J); lanes 7 to 9, JQ279 [Pspac-spo0A Δ(soj-spo0J)]. The total amounts of protein loaded were 3 (lanes 1, 4, and 7), 6 (lanes 2, 5, and 8), and 9 (lanes 3, 6, and 9) μg.
The effect of spo0J and soj on Spo0A protein levels was dependent on the spo0A promoter. We placed expression of spo0A under control of the LacI-repressible–IPTG-inducible promoter, Pspac (43), in a strain otherwise null for spo0A. Expression of spo0A from Pspac bypassed the effect of Spo0J and Soj on Spo0A protein levels. In the presence of IPTG, Pspac-spo0A permitted production of similar levels of Spo0A in spo0J+, Δspo0J, and Δsoj-spo0J strains (Fig. 1B). Furthermore, the amount of Spo0A present in early sporulation was similar to that found in wild-type cells expressing spo0A from its native promoters. These data indicate that Soj regulates the accumulation of Spo0A protein when spo0A is expressed from its own promoter and that Soj might affect transcription of spo0A. Results discussed below indicate that the effect of Soj on Spo0A protein levels contributes to the sporulation defect in the Δspo0J mutant.
Soj cross-links to the spo0A promoter region in a Spo0J-regulated manner.
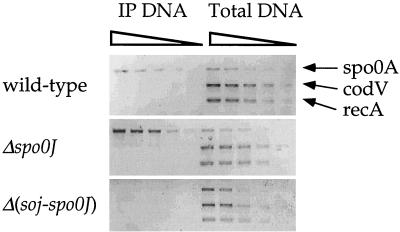
Soj associates with several sporulation promoters in vivo (36) and at least one of these promoters in vitro (4). We tested for the association of Soj with the spo0A promoter region in vivo by using a cross-linking–immunoprecipitation assay (see Materials and Methods). Briefly, formaldehyde was added to cultures to cross-link proteins and DNA. Cells were lysed, and the DNA was sheared into fragments with an average size of 500 to 1,000 bp. We used affinity-purified polyclonal antibodies to immunoprecipitate Soj and DNA associated with Soj. After thorough washing, the cross-links were reversed and the presence of a particular region of the chromosome in the immunoprecipitate was detected by PCR.
We found that Soj was associated with the spo0A promoter region in wild-type cells and in a spo0J null mutant (Fig. 2). Cross-linking was done 2 h after the end of stationary phase, a time when the effect of Soj on transcription of sporulation genes is readily apparent. In the experiments shown (Fig. 2), dilutions of total DNA (before immunoprecipitation) and dilutions of DNA from the immunoprecipitates were used for PCR. This allowed for the comparison of relative amounts of specific regions in the immunoprecipitate from wild-type and spo0J mutant cells (36). There was approximately fourfold-more specific DNA in the immunoprecipitates from the spo0J null mutant than from wild-type cells (Fig. 2). This difference was not caused by differences in the amount of DNA present in the immunoprecipitation nor in the amount of Soj present in the cells. All the immunoprecipitations were done from approximately equal amounts of sheared DNA (Fig. 2, righthand panels), and there is slightly less Soj protein present in spo0J null mutants than in wild-type cells (S. Venkatasubramanyam and A. D. Grossman, unpublished results). The presence of DNA in the immunoprecipitate was dependent on Soj because little or no DNA was detected in immunoprecipitates from a Δ(soj-spo0J) double mutant (Fig. 2). These results indicate that Soj specifically associates with DNA near the spo0A promoter and that this association increases in the absence of Spo0J.
FIG. 2.
Association of Soj with the spo0A promoter region in vivo. Wild-type (JH642), Δspo0J (AG1468), and Δ(soj-spo0J) (AG1505) strains were grown in 2×SG medium at 37°C. Two hours after the end of exponential growth, formaldehyde was added to cross-link the protein and DNA. Protein-DNA complexes were immunoprecipitated by using anti-Soj antibodies. The presence of a given promoter region was assayed by PCR amplification with primers designed to amplify the promoter regions of spo0A, and codV and recA were used as controls. Lanes labeled IP DNA are DNA products from PCR assays performed on a dilution series (4, 2, 1, 0.5, and 0.25 μl) of the immunoprecipitated material. Lanes labeled Total DNA are DNA products from PCR assays performed on a dilution series (the equivalent of 1/250, 1/500, 1/1,000, 1/2,000, and 1/4,000 μl) of sample DNA taken prior to immunoprecipitation.
These results are consistent with previous analysis of the in vivo association of Soj with the promoter regions of the spoIIA, spoIIE, and spoIIG loci (36). In all cases, the association of Soj with DNA increases in the absence of Spo0J and correlates with decreased gene expression. All of these promoters are also activated by Spo0A∼P, but association of Soj with the promoter regions in vivo was independent of spo0A (data not shown). It is possible that Soj interacts with a binding site similar to that for Spo0A, but in vitro experiments have indicated that Soj binding to DNA is complicated and that Soj is not simply recognizing the same sequences as Spo0A (4).
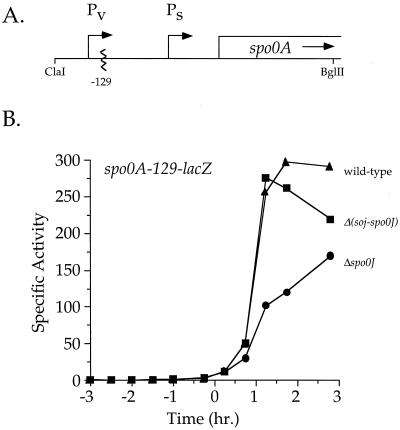
Soj inhibits transcription of the sigma-H-dependent promoter of spo0A.
To determine whether Soj affects transcription of spo0A, we examined expression of spo0A-lacZ fusions. Transcription of spo0A is directed from two promoters (Fig. 3A): a sigma-A-dependent promoter (Pv) expressed during growth and a sigma-H and Spo0A∼P-dependent promoter (Ps) expressed early during sporulation (5, 35, 41). Transcription from spo0A Ps was reduced approximately 60% in the spo0J mutant and was fully restored in the Δ(soj-spo0J) double mutant (Fig. 3B). Expression from the Pv promoter was not affected by Spo0J or Soj (data not shown). These results show that, in a Δspo0J background, Soj inhibits transcription from the spo0APs promoter.
FIG. 3.
spo0J and soj regulate the expression of the spo0A sporulation promoter. (A) spo0A is expressed from two promoters: sigma-A-activated Pv and sigma-H-activated Ps. (B) β-Galactosidase activity expressed from the spo0A-Ps-129-lacZ fusion. Strains were grown in 2×SG medium, and samples were taken for the determination of β-galactosidase specific activity. Time zero indicates the time at which the culture left exponential growth. Symbols: ▴, wild-type (AG1332); ●, Δspo0J (JQ266); ■, Δ(soj-spo0J) (JQ270).
Previous results indicated that mutations in spo0J and soj had little if any effect on transcription of spo0A from the complete Pv+Ps promoter region (4). We found that expression of spo0A(Pv+Ps)-lacZ was reduced approximately 25% in a spo0J null mutant (data not shown). This effect is quite small and is consistent with approximately equal contributions from each promoter to the total amount of spo0A transcript (5). Since the effect of spo0A protein levels appears larger than the effect on transcription, we suspect that transcripts initiating from Ps might be translated more efficiently than those initiating from Pv.
Restoring Spo0A protein to wild-type levels in a Δspo0J background partly restores sporulation and early sporulation gene expression.
We have shown that in the absence of Spo0J, Soj causes an ∼45% decrease in Spo0A protein levels. It is possible that this decrease causes part, none, or all of the sporulation defect in Δspo0J cells. To distinguish between these possibilities, we examined whether restoring Spo0A protein to near wild-type levels, by expressing it from Pspac, could also restore sporulation in a spo0J null mutant.
Expressing spo0A from Pspac partly rescued sporulation in Δspo0J cells, increasing sporulation approximately 10-fold relative to Δspo0J cells expressing spo0A from the endogenous promoters (Table 2). In wild-type or Δ(soj-spo0J) backgrounds, Pspac-spo0A+ did not affect sporulation. The amount of Spo0A protein that accumulated in Pspac-spo0A+ strains varied from ∼15% less than to ∼40% greater than wild type, without affecting either the degree of sporulation rescue or the kinetics of Spo0A-activated gene expression. Therefore, it is unlikely that the 10-fold rescue of sporulation was caused by an overexpression of spo0A. Thus, the decrease in Spo0A levels observed in Δspo0J cells causes part, but not all, of the sporulation defect in these cells.
TABLE 2.
Expression of spo0A from the Pspac promoter partially rescues the sporulation defect of a spo0J null mutant
| Strain | Relevant genotype | % Sporulationa |
|---|---|---|
| JQ284 | Wild type | 78 |
| JQ302 | Δspo0J | 0.28 |
| JQ305 | Δspo0J Δspo0A amyE::(Pspac-spo0A+) | 5.4 |
| JQ304 | Δspo0A amyE::(Pspac-spo0A+) | 56 |
| JQ303 | Δ(soj-spo0J) | 63 |
| JQ306 | Δ(soj-spo0J) Δspo0A amyE::(Pspac-spo0A+) | 82 |
All strains were grown in 2×SG sporulation medium. The Pspac promoter was induced by adding 0.5 mM IPTG to the cultures ∼3 to 4 doublings before the end of the exponential growth phase. Similar results were obtained in four experiments.
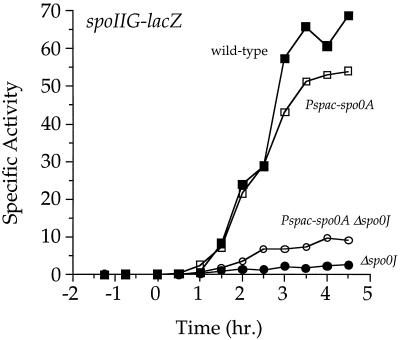
Transcription of spo0A from Pspac also partially restored expression of spoIIE and spoIIG in the Δspo0J mutant. spoIIA, spoIIE, and spoIIG are all transcribed early in sporulation from Spo0A∼P-activated promoters and code for proteins that are essential for spore formation. In a Δspo0J mutant, Pspac-spo0A+ caused a three- to fourfold increase in expression (as judged by fusions to lacZ) of spoIIG (Fig. 4) and spoIIE (data not shown) relative to a strain expressing spo0A from its native promoters. Pspac-spo0A+ had no detectable effect on spoIIA promoter activity (data not shown). In wild-type or Δ(soj-spo0J) backgrounds, Pspac-spo0A+ had no effect on the timing or level of activity from any of the spoII promoters tested. These results show that, in Δspo0J cells, Soj represses spoIIE and spoIIG expression partly through its effect on Spo0A levels. However, it is clear from these results that Soj also inhibits transcription of spoIIA, spoIIE, and spoIIG in a manner independent of its effect on Spo0A levels. At least part of this inhibition appears to be direct as Soj associates with these promoter regions in vivo (36) and inhibits transcription from the spoIIG promoter in vitro (4).
FIG. 4.
Expression of spoIIG-lacZ in strains expressing spo0A from the native promoters or the Pspac promoter. Strains were grown in 2×SG medium, and samples were taken for the determination of β-galactosidase specific activity. Strains expressing spo0A from Pspac were treated with 0.5 mM IPTG 3 to 4 h before the end of exponential growth. Time zero indicates the time at which the culture left exponential growth. Symbols: ■, wild-type (JQ284); ●, Δspo0J (JQ302); □, Pspac-spo0A (JQ304); ○, Pspac-spo0A Δspo0J (JQ305).
A constitutively active form of Spo0A does not fully bypass the sporulation defect of the spo0J mutant.
Soj inhibits expression from at least four sporulation promoters (spo0APs, spoIIA, spoIIE, and spoIIG), and part of this effect appears to be independent of its effect on Spo0A protein levels. Many signals that regulate the initiation of sporulation act by affecting phosphorylation of Spo0A (3, 12). We used a constitutively active mutant form of Spo0A to show that Soj does not significantly impair phosphorylation of Spo0A.
If Soj regulates sporulation by decreasing Spo0A phosphorylation, then a mutant form of Spo0A (called Spo0ASad67) (15) that can activate gene expression and sporulation in an unphosphorylated state ought to bypass the sporulation defect of a Δspo0J mutant. Expression of spo0Asad67 from the Pspac promoter permits wild-type sporulation levels in cells that cannot phosphorylate Spo0A and allows Spo0A∼P-activated gene expression in the absence of the regulatory signals that are normally required (15).
We compared sporulation in Δspo0J cells carrying either Pspac-spo0A+ or Pspac-spo0Asad67. In the presence of IPTG, both Pspac-spo0A and Pspac-spo0Asad67 rescued sporulation to the same degree, i.e., about sevenfold (Table 3). Pspac-spo0Asad67 was no more efficient at bypassing the effects of Δspo0J on sporulation than was Pspac-spo0A+. This result strongly indicates that Soj does not inhibit sporulation by decreasing the phosphorylation state of Spo0A.
TABLE 3.
Effects of the sad67 allele of spo0A on sporulation
| Strain | Relevant genotype | Presence (+) or absence (−) of IPTG | % Sporulationa |
|---|---|---|---|
| DZR158 | amyE::(Pspac-spo0A) | − | 64 |
| DZR158 | amyE::(Pspac-spo0A) | + | 72 |
| SIK25 | amyE::(Pspac-spo0Asad67) | − | 57 |
| SIK25 | amyE::(Pspac-spo0Asad67) | + | 95 |
| KI1968 | amyE::(Pspac-spo0A) Δspo0J | − | 0.17 |
| KI1968 | amyE::(Pspac-spo0A) Δspo0J | + | 1.4 |
| KI1969 | amyE::(Pspac-spo0Asad67) Δspo0J | − | 0.34 |
| KI1969 | amyE::(Pspac-spo0Asad67) Δspo0J | + | 2.9 |
All strains were grown in 2×SG sporulation medium. IPTG (0.5 mM) was added one to two doublings before the end of the exponential growth phase. Similar results were obtained in three experiments.
rvtA11 allele of spo0A and sporulation defect of Δspo0J cells.
The notion that Soj and Spo0J do not affect the phosphorylation state of Spo0A appears to contradict previous findings that indicated Soj and Spo0J might affect the pathway leading to phosphorylation of Spo0A (14). In wild-type cells, the phosphorylation pathway consists of at least three histidine kinases (KinA, KinB, and KinC), Spo0F, Spo0B, and Spo0A (2). The kinases autophosphorylate and donate phosphate to Spo0F. Phosphate is then transferred from Spo0F to Spo0B and finally from Spo0B to Spo0A. Mutations in spo0F and/or spo0B abolish sporulation (34), indicating that the kinases normally cannot directly phosphorylate Spo0A sufficiently to activate sporulation. A mutation in spo0A, rvtA11, suppresses the sporulation defect caused by spo0F and spo0B mutations (38), and this suppression depends on kinC (Table 4) (18, 19), indicating that Spo0ArvtA11 can obtain phosphate directly from KinC.
TABLE 4.
Effects of the rvtA11 allele of spo0A on sporulation
| Strain | Relevant genotype | % Sporulationa |
|---|---|---|
| JH642 | Wild type | 62 |
| JQ390 | rvtA11 | 60 |
| JRL791 | rvtA11 ΔkinC | 51 |
| AG1431 | Δspo0B Δspo0F | <10−5 |
| JRL783 | Δspo0B Δspo0F rvtA11 | 43 |
| JRL792 | Δspo0B Δspo0F rvtA11 ΔkinC | 1.6 × 10−5 |
| AG1468 | Δspo0J | 0.25 |
| KI1775 | Δspo0J rvtA11 | 20 |
| KI1947 | Δspo0J rvtA11 ΔkinC | 8.3 |
Strains were grown in 2×SG sporulation medium. Similar results were obtained in three experiments.
Previously we showed that rvtA11 significantly bypasses the sporulation defect caused by a null mutation in spo0J (14), indicating that Δspo0J might block sporulation by decreasing activity of the phosphorelay. The implication that Δspo0J affects sporulation by decreasing phosphorylation of Spo0A is contradictory to results with the spo0Asad67 mutants described above.
To explore this apparent contradiction, we determined if kinC is required for the rvtA11 bypass of Δspo0J. If the phosphorelay is inhibited in the spo0J mutant, then the bypass in sporulation caused by rvtA11 should depend on kinC. We found that even in a kinC null mutant, rvtA11 was able to partly suppress the sporulation defect of the spo0J mutant (Table 4). In the absence of kinC, the sporulation efficiency of the rvtA11 Δspo0J strain decreased threefold, indicating that the spo0J null mutation causes a small effect on the phosphorelay but that most of the suppression by rvtA11 was independent of KinC. These results are consistent with those obtained with the spo0Asad67 mutation and reinforce the conclusion that the Soj-dependent decrease in sporulation observed in Δspo0J cells is not caused by a decrease in phosphorylation of Spo0A. We do not know how rvtA11 suppresses the sporulation defect of a spo0J mutant better than the constitutively active Pspac-spo0Asad67.
Summary.
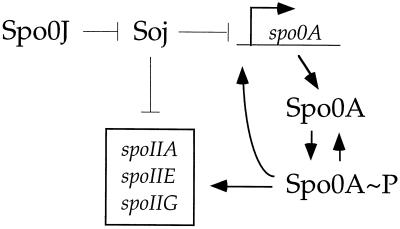
We have used genetic methods to clarify the role of the partitioning protein, Soj, in regulating the initiation of sporulation. Our current view of the effects of Soj on sporulation gene expression is summarized in Fig. 5. In addition to its apparently direct effect on transcription of several spoII genes (4, 36), Soj also negatively regulates expression of spo0A, which is itself required to activate transcription of the spoII genes. Restoring Spo0A to wild-type levels partially bypasses the sporulation defect in Δspo0J cells. Soj does not appear to regulate sporulation significantly by affecting phosphorylation of Spo0A. These results are consistent with Soj acting as a transcriptional repressor of several Spo0A∼P-activated genes, including spo0A, spoIIA, spoIIE, and spoIIG (Fig. 5).
FIG. 5.
A model for the regulation of gene expression by Spo0J, Soj, and Spo0A. Soj negatively regulates expression of spo0A Ps, spoIIA, spoIIE, and spoIIG. Spo0A∼P stimulates expression from these same promoters. Soj acts directly and indirectly through its effect on spo0APs. Spo0J antagonizes Soj.
Spo0J probably antagonizes the activity of Soj by maintaining Soj localization at the cell poles and away from the promoter regions of Spo0A∼P-activated genes (24, 36). This sequestration appears to be related to the putative ATPase activity of Soj. To date, it has not been possible to detect Soj ATPase activity in vitro, either in the presence or in the absence of Spo0J (4, 36). We suspect that the activity of Spo0J is somehow associated with its role in organizing the origin region of the chromosome (22, 23) and/or its function in chromosome partitioning (14). It will be challenging to determine how this regulation occurs.
ACKNOWLEDGMENTS
We thank K. Siranosian for constructing the spo0A-lacZ fusions, K. Ireton for several strains and for doing preliminary experiments, and K. Bacon for comments on the manuscript. J.D.Q. was supported, in part, by an NSF predoctoral fellowship. This work was also supported, in part, by NIH Public Health Services grant GM41934 to A.D.G.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 2.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 3.Burkholder W F, Grossman A D. Regulation of the initiation of endospore formation in Bacillus subtilis. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 151–166. [Google Scholar]
- 4.Cervin M A, Spiegelman G B, Raether B, Ohlsen K, Perego M, Hoch J A. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 5.Chibazakura T, Kawamura F, Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991;173:2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung J D, Stephanopoulos G, Ireton K, Grossman A D. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M A, Martin K A, Austin S J. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 8.Dedon P C, Soults J A, Allis C D, Gorovsky M A. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- 9.Friedman S A, Austin S J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988;19:103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- 10.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 11.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. [Google Scholar]
- 12.Hoch J A. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction system. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 129–144. [Google Scholar]
- 13.Ireton K, Grossman A D. Coupling between gene expression and DNA synthesis early during development in Bacillus subtilis. Proc Natl Acad Sci USA. 1992;89:8808–8812. doi: 10.1073/pnas.89.18.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ireton K, Gunther IV N W, Grossman A D. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 16.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney T J, York K, Youngman P, Moran C P., Jr Genetic evidence that RNA polymerase associated with sigma-A factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayashi Y. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J Bacteriol. 1995;177:176–182. doi: 10.1128/jb.177.1.176-182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leighton T J, Doi R H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971;252:268–272. [PubMed] [Google Scholar]
- 21.Lewis P J, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 22.Lin DC-H, Grossman A D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 23.Lin DC-H, Levin P A, Grossman A D. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marston A L, Errington J. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 25.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Mohl D A, Gober J W. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 27.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 28.Mysliwiec T H, Errington J, Vaidya A B, Bramucci M G. The Bacillus subtilis spo0J gene: evidence for involvement in catabolite repression of sporulation. J Bacteriol. 1991;173:1911–1919. doi: 10.1128/jb.173.6.1911-1919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordström K, Austin S J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 30.Perego M. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 1998;6:366–370. doi: 10.1016/s0966-842x(98)01350-x. [DOI] [PubMed] [Google Scholar]
- 31.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glasser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 32.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 34.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Predich M, Nair G, Smith I. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing ςH. J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quisel J D, Lin DC-H, Grossman A D. Control of development by altered localization of a transcription factor in B. subtilis. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 37.Ronson C W, Nixon B T, Ausubel F M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987;49:579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- 38.Sharrock R A, Rubenstein S, Chan M, Leighton T. Intergenic suppression of spo0 phenotypes by the Bacillus subtilis mutation rvtA. Mol Gen Genet. 1984;194:260–264. doi: 10.1007/BF00383525. [DOI] [PubMed] [Google Scholar]
- 39.Solomon M J, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 41.Strauch M A, Trach K A, Day J, Hoch J A. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie. 1992;74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]
- 42.Williams D R, Thomas C M. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- 43.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]