Abstract
BmpA, BmpB, BmpC, and BmpD are homologous Borrelia burgdorferi lipoproteins of unknown functions, encoded by the bmp genes of paralogous chromosomal gene family 36. At least some of the Bmp proteins are immunogens in infected vertebrate hosts. The genetic organization of the bmp region has been characterized for a variety of B. burgdorferi sensu lato strains by Southern hybridization, PCR amplification, and DNA sequencing. All four bmp genes were present in the same relative order in all B. burgdorferi sensu lato low- and high-passage-number isolates. While there were no differences in the relative orders of the bmp genes in these species, variations in DNA sequence in the bmpD-bmpC and bmpC-bmpA intergenic regions were significantly more common than in the corresponding 3′ bmpD and bmpC coding regions. The genetic structure of the chromosomal region containing the bmp genes thus appears to be well conserved across different species of B. burgdorferi, but variations in DNA fine structure that prevent PCR primer annealing may occur in this region and make Southern hybridization much more reliable than PCR for detection of the presence of these genes. Our results also suggest that bmp gene products may be used as reagents in the preparation of vaccines and diagnostic assays to protect against and diagnose Lyme disease produced by B. burgdorferi sensu lato.
DNA sequencing of Borrelia burgdorferi B31 has identified open reading frames that encode at least 105 lipoproteins belonging to several redundant gene families (6). Paralogous plasmid and chromosomal genes encoding lipoproteins are, in general, characteristic of the B. burgdorferi genome (6) and include bmp (1, 17, 19), erp (21), the 2.9 gene family (16), and vls, the B. hermsii vmp homologue (24). While DNA of any of these gene families could be a substrate for stochastic genetic rearrangement to yield variation in gene expression and/or antigenic variation (14), this phenomenon has not yet been demonstrated to occur in B. burgdorferi infections.
In B. burgdorferi B31, bmp genes (paralogous gene family 36) are located in tandem in the chromosome in the order bmpD bmpC bmpA bmpB in a region extending from nucleotides 391932 to 396563 (6). They are also present in the chromosomes of other B. burgdorferi strains (1, 17, 19). In B. burgdorferi B31, DNA sequence homologies among bmp genes range from 56 to 64%. DNA sequence analysis has suggested that bmpC is preceded by two promoters (1), that bmpD and bmpA are preceded by individual promoters (17, 19), and that bmpB is preceded by no promoter (19). The putative bmpA promoter is located within the bmpC coding sequence (1, 19). Although the functions of the proteins encoded by the bmp genes are unknown, Borrelia organisms in culture synthesize mRNAs of all four bmp genes (17; E. Dobrikova, V. Gorbacheva, and F. C. Cabello, unpublished data) and antibodies to BmpA, BmpC, and BmpD proteins are present in infected hosts (1, 2, 17, 19). These data suggest that the functions of these proteins may be necessary for in vitro and in vivo growth and that at least three members of this family may have a role in virulence (4).
Very few genes of B. burgdorferi that are involved in virulence have been identified as a result of obtaining the complete sequence of this organism (6). Analysis of a B. burgdorferi chromosomal region whose genes code for exposed, putatively in vivo-induced and clearly immunogenic lipoproteins may therefore be relevant to Borrelia virulence. The presence of Bmp proteins on the surface of B. burgdorferi, the tandem arrangement of their genes in the chromosome of Borrelia, and their overlapping transcriptional signals suggest that these proteins may be virulence related and that the expression of their genes may be coregulated (1, 17, 19).
It is not known whether bmp genes are present in the genomes of all isolates of B. burgdorferi sensu lato, but at least bmpC and bmpA have been identified in B. garinii and B. afzelii (1, 17, 19). To provide a basis for understanding the role that chromosomally encoded Bmp proteins might play in the biology of B. burgdorferi and in the pathogenesis of Lyme disease, and to evaluate the usefulness of Bmp proteins as reagents for diagnosis of Lyme disease produced by different Borrelia strains (1, 18), the structures of the bmp regions in several Borrelia species were analyzed using DNA hybridization, PCR amplification, and DNA sequencing. There were no differences in the relative order of the bmp genes in these species, but variations in DNA sequence were significantly more common in intergenic regions than in coding regions.
Bacterial strains and culture.
B. burgdorferi B31 (ATCC 35210) and 297 (20); 10 B. burgdorferi sensu stricto strains recently isolated from skin biopsies and blood samples from patients with Lyme disease and passaged only once (10); B. garinii G25 and N34 (from R. Marconi); B. afzelii Ip3 (9), ACA1 (3), VS461 (9), and VS486 (from J. Benach); B. bissettii 25015 (formerly B. burgdorferi sensu lato group DN127) (22); B. andersonii 21038 (from R. Marconi); B. japonica H014 (13); and B. hermsii (from R. Johnson) were grown at 32 to 34°C in BSK-H medium supplemented with 7% rabbit serum (Sigma Chemical Co., St. Louis, Mo.) (7). Strains were cloned by two rounds of limiting dilution in BSK-H medium or by subsurface agarose colony isolation (5). Cell concentration was determined by counting cells stained with acridine orange under fluorescence microscopy (23) or by counting viable cells on agarose plates (5). Comparable results were obtained by both techniques.
Southern hybridization.
Total DNA from each Borrelia strain was purified from a mid-log-phase culture (8) and digested overnight with SwaI and/or HindIII (New England Biolabs, Inc., Beverly, Mass.) according to the manufacturer's instructions. The resulting DNA fragments were separated by agarose gel electrophoresis (1% agarose in Tris-acetate-EDTA [TAE] buffer), stained with ethidium bromide, and transferred by capillary action to a nylon membrane (Magna Graph; Micron Separation, Inc., Westboro, Mass.) (1). DNA probes were generated by PCR using partial (P) primers (Fig. 1) to amplify the central part of each bmp gene. Templates for these reactions were pUC19-based plasmids containing different DNA segments of the bmp region of B. burgdorferi 297 (2). A DNA probe targeting the flaB gene for use as a control was obtained by PCR amplification by using total DNA purified from B. burgdorferi 297 as a template and appropriate primers (5′-CTAGTGGGTACAGAATTAATCGAGC-3′ and 5′-GCCTGCGCAATCATTGCCATTGC-3′) (11). DNA probes were purified, labeled with digoxigenin-11-dUTP by the random primer method according to the instructions of the manufacturer (Boehringer Mannheim, Indianapolis, Ind.), hybridized to DNA blots at 65°C, and washed under high-stringency conditions in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer at 68°C (1). Bound probes were detected colorimetrically using nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) technology according to the manufacturer's instructions.
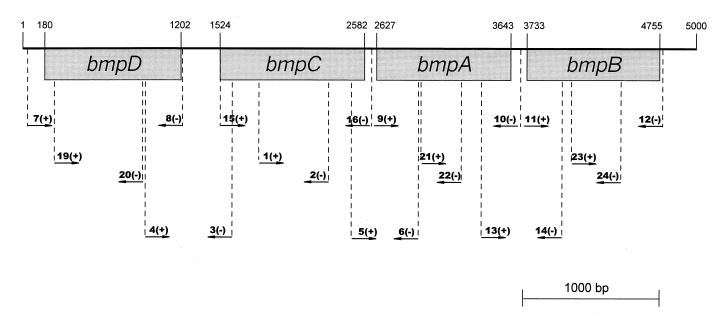
FIG. 1.
Schematic representation of the PCR primer binding sites in the bmp chromosomal region. The sequence of this region was created on the basis of sequences of bmpD from B. burgdorferi JD1 (accession no. U35450) (17) and bmpC, -A, and -B genes from B. burgdorferi 297 (accession no. U49938) (2) using the program Primer 3 Output (Center for Genome Research). Position 1 corresponds to nucleotide 396706, and position 5000 corresponds to nucleotide 391707 on the B. burgdorferi B31 chromosomal map (6). The relative locations of primer binding sites are indicated by arrows. W primer pairs are for amplification of the entire indicated coding sequence, and P primer pairs are for amplification of partial regions of the indicated coding sequence. All primers with the suffix (+) are plus-strand primers, and those with the suffix (−) are minus-strand primers (i.e., reverse complement of the gene sequences). Primer sequences (5′→3′) for the indicated regions were as follows. bmpD: 7(+), GAATGGCTGAAGCAAATAAAGC(W); 8(−), CAAATCAGCTCAATAAAAATC (W); 19(+), CTGATGATGGCAAGTCGGAG(P); 20(−), ACGCCTATACCAGAAAGCCC (P). bmpC: 15(+), GGCAAGGGCATATGTTTAAAAGATTTATTTTTATTA (W); 16(−), CGCAGATCTCCCCTTTACAAACAAAGC (W); 1(+), GATGAGGCAATGACTGAGGA (P); 2(−), GCAGCGTCATAAACTCCAAGACC (P). bmpA: 9(+), TGTAAAGGGGAAATAGTTTATG (W); 10(−), TTCAAACAAAACCAATGTG (W); 21(+), CCAAGGTTGCGGCTCTTC (P); 22(−), CTTCTACCAGCTTCAAGGTCAG (P). bmpB: 11(+), AAACACATTGGTTTTGTTTG (W); 12(−), TCTTTCTATTTCAAAAGTTTATAAC (W); 23(+), TGGTGATGATGTTCAGATTCC (P); 24(−), TTTGCTGCCTCAATAACACC (P). bmpD to bmpC: 3(+), AGGCCGCAAAAGAGTTGGG; 4(−), GCTACCATGAGCCAAAACACC. bmpC to bmpA: 5(+), TGATCGGGGGTTAAAGGAAGG; 6(−), TGAAGAGCCGCAACCTTGGC. bmpA to bmpB: 13(+), GGCCTTAAAGAAGGAGTTGTGGG; 14(−), CCAAATCAAGTCTGAGCC.
PCR.
Primers to amplify full-length coding regions and flanking regions of bmp genes (whole [W] primers) or partial internal regions of each bmp gene (P primers) were designed on the basis of nucleotide sequences of the bmpD region of B. burgdorferi JD1 (GenBank accession no. U35450) (17) and the bmpC, bmpA, and bmpB regions of B. burgdorferi 297 (accession no. U49938) (2) by using the program Primer 3 Output (Center for Genome Research, Whitehead Institute for Biochemical Research, Cambridge, Mass.) (Fig. 1) and synthesized (GenoSys Biotechnology, The Woodlands, Tex.). Primers specific for 16S rRNA genes (5′-GAATTTTACAATCTTTCGACC-3′ and 5′-GGGGAATAATTATCTCTAAC-3′) (10) and the flaB gene (see above) (17) were a gift from I. Schwartz. PCR amplifications were performed in a Rapid Cycler (Idaho Technology, Idaho Falls) according to the manufacturer's recommendations, with the final mixture in 10-μl glass capillary tubes containing a 200 μM concentration of each deoxynucleoside triphosphate, 2 mM MgCl2, 50 mM Tris-HCl (pH 8.3), 0.5 mg of bovine serum albumin/ml, 0.5 to 1% Ficoll (Idaho Technology); 0.25 U of Taq polymerase (Gibco BRL, Gaithersburg, Md.), and 0.5 μM concentrations of primers. Chromosomal DNA was initially denatured for 5 s at 96°C, followed by a total of 30 cycles of 94°C for 0 s, 60°C for 1 s, and 72°C for 30 s and a final cycle of 2 min at 72°C. PCR-generated products were purified and sequenced by the dideoxy chain termination method using a dye terminator-Taq cycle sequencing kit and a model 377 DNA sequencer (Perkin-Elmer, Foster City, Calif.). DNA sequences were aligned and analyzed with Assembly-LIGN and MacVector software (IBI, New Haven, Conn.), Clustal W 1.7, and Laling CCM Search Launcher (Human Genome Center, Baylor, Tex.). Some manual refinement of the alignments was performed.
Detection of bmp genes in B. burgdorferi sensu lato.
Southern hybridization results are summarized in Table 1. DNA hybridization patterns in Southern blots with PCR-generated probes specific for each bmp gene indicated that a single copy of each bmp gene was present in the genomic DNA of B. burgdorferi sensu lato. Differences in hybridization patterns of B. burgdorferi strains consisted of variations in the lengths of the DNA restriction fragments hybridizing with the DNA probes and in band intensity. For example, total DNA from B. burgdorferi sensu lato strains digested with SwaI and hybridized with a P probe specific for bmpC yielded the expected single 1,271-bp band with B. burgdorferi B31; a single 3,750-bp band with B. garinii, B. afzelii, B. afzelii, and B. japonica; a single 1,750-bp band with B. andersoni; and a major 750-bp band and a minor 550-bp band with B. bissettii. The additional fragment in B. bissettii is likely generated by an alternative restriction site for SwaI present inside bmpC in this Borrelia species. This possibility is supported by the facts that amplification with primers specific for the central partial coding region of bmpC in this strain generated only one product (Table 1) and that the sum of the molecular masses of the fragments observed during the hybridization was equal to the molecular mass of the expected fragment containing only one copy of bmpC. B. hermsii total DNA digested in the same manner used as a control failed to hybridize with any of the bmp DNA probes used in these experiments. A DNA probe for the highly conserved B. burgdorferi flaB gene (11) hybridized to all borrelial total DNAs examined (both B. burgdorferi sensu lato and B. hermsii) and gave the expected pattern and sizes of amplicons (data not shown). This result indicated that the lack of hybridization of B. hermsii total DNA with bmp DNA probes was unlikely to be due to technical problems related to hybridization itself.
TABLE 1.
Detection of bmp genes in different Borrelia burgdorferi species and isolates by PCR and Southern hybridization
| Species and isolate | Result by indicated test for:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
bmpD
|
bmpC
|
bmpA
|
bmpB
|
bmpD–bmpC
|
bmpC–bmpA
|
bmpA–bmpB
|
|||||||||
| Southern | PCRa
|
Southern | PCR
|
Southern | PCR
|
Southern | PCR
|
PCR | PCR | PCR | |||||
| W | P | W | P | W | P | W | P | ||||||||
| B. burgdorferi | |||||||||||||||
| 297 | NDb | + | + | ND | + | + | ND | + | + | ND | + | + | + | + | + |
| B31 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| B. garinii | |||||||||||||||
| G25 | + | − | ± | + | − | + | + | − | − | + | − | + | + | − | + |
| N34 | ND | − | + | ND | − | + | ND | − | − | ND | − | + | + | − | + |
| B. afzelii | |||||||||||||||
| Ip3 | + | − | + | + | − | + | + | − | − | + | − | + | + | − | + |
| ACA1 | ND | − | + | ND | − | + | ND | − | − | ND | − | + | + | − | + |
| VS461 | ND | − | + | + | − | + | ND | − | − | ND | − | + | + | − | + |
| VS486 | ND | − | + | ND | − | + | ND | − | − | ND | − | + | + | − | + |
| B. bissettii 25015 | + | − | − | + | − | + | + | ± | − | + | − | + | ± | − | + |
| B. andersonii 21038 | + | − | + | + | − | + | + | − | + | + | − | + | ± | + | + |
| B. japonica H014 | + | − | − | + | − | + | + | − | − | + | − | + | − | − | − |
| B. hermsii | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Detection of genes or intergenic sequences with primers to amplify entire (W) or partial (P) coding regions of the indicated bmp genes. See the text for details.
ND, not determined.
PCR analysis of the bmp genes and their intergenic noncoding regions.
PCR amplification of borrelial DNA was done using three groups of primers (Fig. 1). W primers were designed to analyze full-length coding regions of bmp genes and generated amplicons for bmp genes only from B. burgdorferi sensu stricto isolates as well as B. bissettii, from which bmpA was weakly amplified (Table 1). All products obtained with W primer sets were of the expected sizes (data not shown). No products were obtained from B. hermsii DNA with any W primers.
P primers were designed to amplify a partial internal region of each bmp gene. Amplicons corresponding to bmpC and bmpB were obtained from all B. burgdorferi sensu lato strains (Table 1), amplicons corresponding to bmpD were obtained from all B. burgdorferi sensu lato strains but not B. bissettii and B. japonica (Table 1), and amplicons corresponding to bmpA were obtained only from B. burgdorferi sensu stricto and B. andersonii (Table 1). Amplicons had the same size in all tested B. burgdorferi strains when they were obtained with each pair of P primers. For example, all bmpC amplicons of B. burgdorferi sensu lato obtained using P primers 1 and 2 were the predicted size of 484 bp; comparable results were obtained with amplicons from bmpB and bmpD (data not shown). Similar PCR results were obtained with DNAs from cloned and uncloned Borrelia strains (data not shown). No products were obtained from B. hermsii DNA with any P primers. In these experiments, primers specific for 16S rRNA genes yielded amplicons from all borrelial DNA, including B. hermsii DNA (data not shown), suggesting that the failure to amplify bmp genes from B. hermsii DNA and some B. burgdorferi sensu lato DNAs (e.g., bmpA) was not due to template degradation or to the presence of PCR inhibitors and suggested the existence of DNA sequence heterogeneity in this region.
As an additional step in revealing possible DNA sequence heterogeneity in the bmp cluster, a third group of primers was designed to amplify bmp intergenic regions. These intergenic primer sets amplified regions extending from the 3′ end of the upstream gene to the 5′ end of the downstream gene of each bmp. B. burgdorferi sensu stricto and B. andersonii DNAs generated amplicons with single primer pairs designed to amplify bmpD-bmpC, bmpC-bmpA, and bmpA-bmpB intergenic regions (Table 1). In other B. burgdorferi sensu lato strains, only bmpD-bmpC and bmpA-bmpB regions were amplified (Table 1). B. japonica DNA did not generate a product when it was amplified with primers targeting any of the intergenic regions. Other primer sets composed of combinations of different plus- and minus-strand primers (Fig. 1) were used to provide further characterization of the bmp region. These additional primer sets generated amplicons from all B. burgdorferi sensu lato DNAs except that of B. japonica. In each case, the products obtained were of the expected sizes and overlapped throughout the entire bmp region. This result ruled out the possibility that the observed negative PCR amplifications were caused by the presence of extensive deletions in these regions and suggested that the failure to amplify was due to sequence polymorphism. None of the primers directed at amplifying bmp intergenic regions generated any product from B. hermsii DNA.
PCR analysis of the bmp chromosomal region in B. burgdorferi low-passage-number strains.
Ten B. burgdorferi sensu stricto strains recently isolated from skin biopsies and blood samples from patients with Lyme disease and passaged only once (10) were used to characterize the genetic structure of the bmp chromosomal region in low-passage-number B. burgdorferi strains. All primer sets directed at amplifying full-length and partial coding regions of each bmp gene and of the bmp intergenic regions yielded single DNA products identical in size to those amplified from B. burgdorferi 297 and B31. Although continuous in vitro cultivation can cause significant changes in the B. burgdorferi genome (15), low-passage-number B. burgdorferi sensu stricto strains recently isolated from patients with Lyme disease were identical to B. burgdorferi 297 and B31 in terms of both PCR pattern and amplicon size.
DNA sequence analysis of bmp intergenic regions in B. burgdorferi sensu lato.
To identify the molecular basis for the polymorphism observed in the bmp region during PCR amplification, and to assess the genetic divergence in this region among different strains of the B. burgdorferi sensu lato complex, PCR products containing bmpD-bmpC and bmpC-bmpA intergenic regions were sequenced and compared (Fig. 2). In those cases when the primer sets designed to amplify the intergenic region did not yield an amplification product, combinations of different primers were used to amplify DNA sequences of an increased size that included the regions of interest. Because it was not possible to obtain amplification of B. japonica DNA with any available primer set, this isolate was not subjected to further analysis.
FIG. 2.
Alignment of intergenic BmpD-bmpC (A) and bmpC-bmpA (B) sequences from selected B. burgdorferi sensu lato strains. Gaps introduced by alignments are indicated by hyphens, nucleotides nonidentical to those in B. burgdorferi 297 sequence are shaded, and translation start and stop codons are indicated by uppercase letters. Bb, B. burgdorferi 297; Bg, B. garinii G25; Bafz, B. afzelii Ip3; Bbis, B. bissettii 25015; Band, B. andersoni 21038.
The bmpD-bmpC intergenic region analyzed comprised approximately 80 bases upstream from the bmpD stop codon to approximately 90 bases downstream from the bmpC start codon (Fig. 2A). The DNA sequence of this region of B. burgdorferi 297 was 97% identical to that of B. burgdorferi B31 (6). Nucleotide identities to B. burgdorferi 297 for this region were 82.2, 85.1, 87.0, and 88.0% for B. afzelii, B. garinii, B. bissettii, and B. andersonii, respectively. Coding sequences were relatively conserved among different strains, with few single-base replacements. Nucleotide insertions, deletions, and substitutions were concentrated in a few clusters (Fig. 2A) and were significantly greater in number per 100 nucleotides in the bmpD-bmpC intergenic region that in the 3′ bmpD coding region preceding this intergenic region (P < 0.02, Kruskal-Wallis analysis of variance with Dunn multiple-comparison post-test). The most important characteristic of the intergenic region was the presence of two deletions: one of 12 nucleotides detected in a B. garinii-derived amplicon and another of 18 nucleotides found in a B. bissetti amplicon (Fig. 2A). Only minor differences at a few positions were found when DNA sequences were compared between two Borrelia strains of the same genotype; for example, bmpD-bmpC sequences of B. afzelii ACA1 and B. afzelii Ip3 were 98.3% identical. However, the region immediately upstream of the bmpC start codon was highly polymorphic between strains of different genotypes and contained various numbers of thymidines in runs of thymidines (Fig. 2A).
The bmpC-bmpA intergenic region analyzed comprised approximately 150 bases upstream from the bmpC stop codon and approximately 310 bases downstream from the bmpA start codon (Fig. 2B). Here, too, nucleotide insertions, deletions, and substitutions were significantly greater in number per 100 nucleotides in the bmpC-bmpA intergenic region than in the preceding 3′ bmpC coding region (P < 0.02, Kruskal-Wallis analysis of variance with Dunn multiple-comparison post-test). Numbers of insertions, deletions, and substitutions per 100 nucleotides were not significantly different between bmpD-bmpC and bmpC-bmpA intergenic regions. The presence of these variations in the DNA sequence was confirmed by sequencing both strands of several independently generated PCR amplicons.
This study shows that bmp genes are widely distributed among all strains of B. burgdorferi sensu lato and are not present in B. hermsii. These results are consistent with previous reports about the species-specific nature of the members of this family (1, 17, 19). The similarity of amplicon size obtained with each primer set used also suggests conservation of these genes among B. burgdorferi sensu lato strains. In all cases of positive amplification, a single product was obtained, confirming previous results demonstrating that only one copy of these genes is present in the borrelial genome (1, 17, 19). Our data also demonstrate conservation of bmp gene order in the borrelial chromosome (bmpD bmpC bmpA bmpB).
The genetic structure of the bmp chromosomal region appears to be similar in low- and high-passage-number B. burgdorferi sensu stricto strains. Whether this region undergoes any changes while the spirochetes are maintained in zoonotic cycles involving ticks and small rodents or during tick feeding and human infection is not known. The observed conservation of the size and order of the bmp genes is surprising, as their DNA sequence similarity could potentially generate the homology needed for recombination events to alter this order as happens with B. burgdorferi plasmid-borne members of paralogous gene families (15, 16). This difference in genetic behavior between chromosomally and plasmid-located paralogous genes may reflect alternative biological roles of their gene products.
Despite the constancy of the overall genetic organization of the bmp cluster in B. burgdorferi sensu lato, DNA sequence variations existed over the entire region. These variations were significantly higher in noncoding regions, where there was a tendency for them to be clustered at particular points. Our observation that primers designed to target entire bmp coding regions generated amplicons only in B. burgdorferi sensu stricto strains suggested that primer annealing (and therefore amplification) was prevented either by deletions in the central regions of these genes or by DNA sequence variations in regions flanking these genes. Our subsequent successful amplification of bmp genes in some B. burgdorferi sensu lato isolates using alternative primer combinations with different plus- and minus-strand primers generated amplicons that overlapped the entire bmp region with sizes corresponding to those of the B. burgdorferi 297 sequence. This result indicated that the previous failure to generate amplicons was not due to deletions or insertions in bmp coding sequences and implied that lack of amplification was due to the DNA sequence variations in flanking regions (Fig. 2). Although no amplicons were generated from B. japonica DNA, Southern hybridization analysis indicated that all bmp genes were present in the genome of this species. DNA sequence variations appear to be more pronounced in B. japonica than in B. burgdorferi sensu lato strains and may reflect evolutionary divergency (12).
In summary, the genetic structure of the chromosomal region containing the bmp genes appears to be well conserved across different species of B. burgdorferi, but variations in DNA fine structure that prevent PCR primer annealing may occur in this region and make Southern hybridization much more reliable than PCR for detection of the presence of these genes. Our results also suggest that bmp gene products may be used as reagents in the preparation of vaccines and diagnostic assays to protect against and diagnose Lyme disease produced by B. burgdorferi sensu lato.
Nucleotide sequence accession numbers.
Nucleotide sequences of bmpD-bmpC and bmpC-bmpA intergenic regions have been deposited in GenBank under accession numbers U49934, AF222434, AF222435, AF222436, AF222437, AF222438, AF222439, AF222440, and AF222441.
Acknowledgments
We gratefully acknowledge C. Pavia, R. Johnson, R. Marconi, J. Benach, and D. Liveris for providing us with different Borrelia isolates. We also thank Ira Schwartz for Borrelia 16S ribosomal DNA and flagellin primers and for stimulating discussions regarding DNA sequence analysis. L. Aron provided us with recombinant plasmid DNA containing different DNA segments of the bmp region of the B. burgdorferi 297 chromosome. S. Newman advised us on the use of programs needed for the DNA sequence analysis and in the preparation of the manuscript. H. V. Harrison and M. Steinberg made important contributions in the preparation of the manuscript.
Funds for this work were provided by Public Health Service grants R01 AI43063 and R44 AI36004 to F. C. Cabello.
REFERENCES
- 1.Aron L, Alekhun M, Schwartz I, Perlee L, Godfrey H P, Cabello F C. Cloning and DNA sequence analysis of bmpC, a gene encoding a potential membrane lipoprotein of Borrelia burgdorferi. FEMS Microbiol Lett. 1994;123:75–82. doi: 10.1111/j.1574-6968.1994.tb07204.x. [DOI] [PubMed] [Google Scholar]
- 2.Aron L, Toth C, Godfrey H P, Cabello F C. Identification and mapping of a chromosomal gene cluster of Borrelia burgdorferi containing genes expressed in vivo. FEMS Microbiol Lett. 1996;145:309–314. doi: 10.1111/j.1574-6968.1996.tb08594.x. [DOI] [PubMed] [Google Scholar]
- 3.Åsbrink E, Hederstedt B, Hovmark A. A spirochetal etiology of acrodermatitis chronica atrophicans Herxheimer. Acta Dermato-Venereol. 1984;64:506–512. [PubMed] [Google Scholar]
- 4.de Silva A M, Fikrig E, Hodzic E, Kantor F S, Telford III S R, Barthold S W. Immune evasion by tickborne and host-adapted Borrelia burgdorferi. J Infect Dis. 1998;177:395–400. doi: 10.1086/514200. [DOI] [PubMed] [Google Scholar]
- 5.Dever L L, Jorgensen J H, Barbour A G. In vitro antimicrobial susceptibility testing of Borrelia burgdorferi: a microdilution MIC method and time-kill studies. J Clin Microbiol. 1992;30:2692–2697. doi: 10.1128/jcm.30.10.2692-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White W, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richarson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 7.Indest K J, Ramamoorthy R, Sole M, Gilmore R D, Johnson B J B, Philipp M T. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeFebre R B, Foley J W, Thierman A B. Rapid and simplified protocol for isolation and characterization of leptospiral chromosomal DNA for taxonomy and diagnosis. J Clin Microbiol. 1985;30:606–608. doi: 10.1128/jcm.22.4.606-608.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liveris D, Gazumyan A, Schwartz I. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1995;33:589–595. doi: 10.1128/jcm.33.3.589-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, McKenna D, Nadelman R B, Novakowski J, Wormser G P, Schwartz I. Genetic diversity of Borrelia burgdorferi in Lyme disease in patients as determined by culture versus direct PCR with clinical specimens. J Clin Microbiol. 1999;37:565–569. doi: 10.1128/jcm.37.3.565-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marconi R T, Garon C F. Development of polymerase reaction primer sets for diagnosis of Lyme disease for species-specific identification of Lyme disease isolates by 16S rRNA signature nucleotide analysis. J Clin Microbiol. 1992;30:2830–2834. doi: 10.1128/jcm.30.11.2830-2834.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marconi R T, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analysis of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuzawa T, Okada Y, Yanigahara Y, Sato N. Antigenic properties of Borrelia burgdorferi isolated from Ixodes ovatus and Ixodes persulcatus in Hokkaido, Japan. J Clin Microbiol. 1991;29:1568–1573. doi: 10.1128/jcm.29.8.1568-1573.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Microbiology. 1994;141:1321–1329. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 15.Norris S J, Howell J K, Garza S A, Ferdows M S, Barbour A G. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun. 1995;63:2206–2212. doi: 10.1128/iai.63.6.2206-2212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramamoorthy R, Povinelli L, Philipp M T. Molecular characterization, genomic arrangement, and expression of bmpD, a new member of the bmp class of genes encoding membrane proteins of Borrelia burgdorferi. Infect Immun. 1996;64:1259–1264. doi: 10.1128/iai.64.4.1259-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson W J, Burgdorfer W, Schrumpf M E, Karstens R H, Schwan T G. Antibody to a 39-kilodalton Borrelia burgdorferi antigen (P39) as a marker for infection in experimentally and naturally inoculated animals. J Clin Microbiol. 1991;29:236–243. doi: 10.1128/jcm.29.2.236-243.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson W J, Cieplak W, Schrumpf M E, Barbour A G, Schwan T G. Nucleotide sequence and analysis of the gene in Borrelia burgdorferi encoding the immunogenic P39 antigen. FEMS Microbiol Lett. 1994;119:381–388. doi: 10.1111/j.1574-6968.1994.tb06917.x. [DOI] [PubMed] [Google Scholar]
- 20.Steere A C, Grodzicki R L, Kornblatt A N, Craft J E, Barbour A G, Bergdorfer W, Schmid G P, Johnson E, Malawista S E. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 21.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strle F, Picken R N, Cheng Y, Cimperman J, Maraspin V, Lotric-Furlan S, Ruzic-Sabljic E, Picken M M. Clinical findings for patients with Lyme borreliosis caused by Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities to strain 25015. Clin Infect Dis. 1997;25:273–280. doi: 10.1086/514551. [DOI] [PubMed] [Google Scholar]
- 23.West S S. Quantitative microscopy in bacteriology. Ann N Y Acad Sci. 1969;158:111–122. [Google Scholar]
- 24.Zhang J-R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]