Abstract
The technique of mRNA differential display was used to identify simultaneously two metabolic genes involved in the degradation of cyclohexanone in a new halotolerant Brevibacterium environmental isolate. In a strategy based only on the knowledge that cyclohexanone oxidation was inducible in this strain, the mRNA population of cells exposed to cyclohexanone was compared to that of control cells using reverse transcription-PCR reactions primed with a collection of 81 arbitrary oligonucleotides. Three DNA fragments encoding segments of flavin monooxygenases were isolated with this technique, leading to the identification of the genes of two distinct cyclohexanone monooxygenases, the enzymes responsible for the oxidation of cyclohexanone. Each monooxygenase was expressed in Escherichia coli and characterized. This work validates the application of mRNA differential display for the discovery of new microbial metabolic genes.
It is now widely accepted that the diversity of microorganisms extends far beyond the few thousand species in culture collections (15). This diversity of microbes and their metabolism constitutes a vast source of enzymes and genes for biotechnology applications. The identification of useful metabolic genes has traditionally proceeded either through a direct genetic approach or by the reverse genetics approach, starting with the purification of the enzyme of interest followed by identification of its gene through the use of antibodies or amino acid sequence information obtained from the pure protein.
Although both strategies are routinely used, they are often limited by technical problems. The direct genetic approach can be used only for organisms that have a developed genetic system or whose genes can be expressed in heterologous hosts. The reverse genetics approach requires purification of the protein of interest, which often takes a long time, and the successful amplification of a DNA probe from degenerate primers, a technique that sometimes fails. Recently mRNA techniques have made it possible to access regulated genes directly without the purification of their gene products and in the absence of a genetic system. These approaches are based on comparison of the mRNA population between two cultures or tissues and identification of the subset of genes whose mRNA is more abundant under conditions of induction. These techniques rely on the hybridization of labeled mRNAs onto arrays of DNA on membranes (4) or DNA microarrays (9), large-scale sample sequencing of expressed sequence tag libraries (28), or the sampling of mRNA by the production of randomly amplified DNA fragments by reverse transcription (RT) followed by PCR (RT-PCR) (19, 20, 35). Because it can easily be done by individual scientists at low cost, the latter approach has been used extensively since it was first described.
Two variations of this RT-PCR method have been published. The first, called differential display (DD) (19, 20), begins with the synthesis of cDNAs by RT of mRNA using a poly(dT) primer that hybridizes to the poly(A) tail of eukaryotic messages. Synthesis of the second DNA strand is initiated at random sites under low-stringency conditions using an oligonucleotide of arbitrary sequence. Subsequent exponential amplification by PCR yields a series of DNA fragments in a process essentially identical to that of random amplification of polymorphic DNA (RAPD) (37). This technique is the most widely used, accounting for more than 90% of published applications of DD. However, it is limited to eukaryotes since archaeal and bacterial mRNAs lack stable poly(A) tails. The second variation of DD uses an arbitrary oligonucleotide primer to initiate RT of the message at random sites. It is independent of poly(A) tails and can be used for both eukaryotic and prokaryotic cells (35). Very few applications of DD to prokaryotes have been published (1, 10, 18, 29, 30, 38–40). In this work, we used mRNA DD to directly identify the key genes involved in the degradation of cyclohexanone in an environmental Brevibacterium strain, a high-G+C gram-positive bacterium. We show how this technique has a great potential for gene discovery targeted to metabolic pathways, particularly in newly isolated microorganisms.
MATERIALS AND METHODS
Isolation of cyclohexanone-degrading Brevibacterium sp. strain HCU.
Selection for a halotolerant bacterium able to degrade cyclohexanol and cyclohexanone was performed on agar plates of a halophilic minimal medium (per liter: agar, 15 g; NaCl, 100 g, MgSO4, 10 g; KCl, 2 g; NH4Cl, 1 g; KH2PO4, 50 mg; FeSO4, 2 mg; Tris HCl, 8 g [pH 7]) containing traces of yeast extract and Casamino Acids (0.005% each) under vapors of cyclohexanone at 30°C. The inoculum was the resuspension of sludge from an industrial aerobic wastewater bioreactor. After a 2-week incubation period, beige colonies were observed and streaked to purity on the same plates under the same conditions. Taxonomic identification was performed by PCR amplification of 16S rRNA gene (rDNA) using primers corresponding to conserved regions of the 16S rDNA molecule (5′-GAGTTTGATCCTGGCTCAG and 5′-TACCTTGTTACGACTT) (17). The sequence of the amplified 1,481-bp fragment was determined and compared to entries in the GenBank database using the BlastN software.
Induction of the cyclohexanone degradation pathway.
Inducibility of the cyclohexanone pathway was tested by respirometry in low-salt medium. One colony of strain HCU was inoculated in 300 ml of S12 mineral medium [50 mM KHPO4 buffer (pH 7.0), 10 mM (NH4)2SO4, 2 mM MgCl2, 0.7 mM CaCl2, 50 μM MnCl2, 1 μM FeCl3, 1 μM ZnCl3, 1.72 μM CuSO4, 2.53 μM CoCl2, 2.42 μM Na2MoO2, 0.36 nM FeSO4] containing 0.005% yeast extract. When the optical density of the culture at 600 nm reached 0.5, the culture was split in two flasks. One flask received 10 mM acetate, and the other received 10 mM cyclohexanone. Each flask was incubated for 6 h at 30°C to allow for the induction of the cyclohexanone degradation genes. The cultures were then chilled on ice, harvested by centrifugation, and washed three times with ice-cold S12 mineral medium. Cells were finally resuspended to an optical density at 600 nm of 2.0 and kept on ice until assayed.
Half a milliliter of each culture was placed in a water-jacketed respirometry cell equipped with an oxygen electrode (Yellow Springs Instruments Co., Yellow Springs, Ohio) and containing 5 ml of air-saturated S12 medium at 30°C. After establishing the baseline for the respiration (oxygen consumption) of each cell suspension, acetate or cyclohexanone (10 mM) was added, and the rate of O2 consumption was further monitored.
Isolation of total cellular RNA.
A 20-ml culture of Brevibacterium sp. strain HCU was grown and split as described earlier. The cyclohexanone oxidation pathway was induced in one of the two subcultures by addition of 10 mM cyclohexanone, and the culture was incubated further at 30°C for 4 h. Each 10-ml culture was chilled rapidly in an ice-water bath and transferred to a 15-ml tube. Cells were collected by centrifugation for 2 min at 12,000 × g in a rotor chilled to −4°C. The supernatants were discarded, the pellets resuspended in 0.7 ml of an ice-cold solution of 1% sodium dodecyl sulfate (SDS) and 100 mM sodium acetate at pH 5 and transferred to a 2-ml tube containing 0.7 ml of aqueous phenol (pH 5) and 0.3 ml of 0.5 mm zirconia beads (Biospec Products, Bartlesville, Okla.). The tubes were placed in a Bead Beater (Biospec) and disrupted at 2,400 beats/min for 2 min.
Following disruption of the cells, the liquid phases were transferred to new microcentrifuge tubes and separated by centrifugation for 3 min at 15,000 × g. The aqueous phase containing total RNA was extracted twice with phenol (pH 5) and twice with phenol-chloroform-isoamyl alcohol (pH 7.5) until a precipitate was no longer visible at the phenol/water interface. Nucleic acids were recovered from the aqueous phase by precipitation with 3 volumes of ethanol, and the pellet was resuspended in 0.5 ml of diethyl pyrocarbonate-treated water. DNA was digested with 6 U of RNase-free DNase (Boehringer Mannheim, Indianapolis, Ind.) for 1 h at 37°C. The total RNA solution was extracted twice with phenol-chloroform-isoamyl alcohol (pH 7.5), recovered by ethanol precipitation, and resuspended in 1 ml of diethyl pyrocarbonate-treated water to an approximate concentration of 0.2 mg/ml. The absence of DNA in the RNA preparation was verified by the fact that RAPD DNA fragments could not be generated by the Taq polymerase (Perkin-Elmer, Foster City, Calif.) unless the reverse transcriptase was also present (data not shown).
RT-PCR oligonucleotide set.
A set of 81 primers was used for RT-PCRs. The primer sequence was CGGAGCAGATCGAWXYZ, where WXYZ represent all combinations of the three bases A, G, and C at the last four positions of the 3′ end. The choice of the invariant 13-bp sequence was arbitrary. A subset of these primers was used in the out-PCR experiments (see below).
Generation of RAPD patterns from arbitrarily reverse transcribed total RNA.
Arbitrarily amplified DNA fragments were generated from the total RNA of control and induced cells by following a protocol adapted from that of Wong et al. (38). A series of 81 parallel RT and PCR amplification reactions each using a single oligonucleotide was performed on the total RNA from the control and induced cells. The RT reactions contained 2 to 10 ng of total RNA, 100 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, Wis.), 0.5 mM each deoxynucleoside triphosphate, (dNTP) and 1 mM each oligonucleotide primer in a total volume of 50 μl of 1× MMLV buffer provided by the manufacturer. Reactions were prepared on ice and incubated at 37°C for 1 h.
A 5-μl aliquot of each RT reaction was used as the template in a 50-μl PCR containing the same primer as used in the RT reaction (0.25 μM), dNTPs (0.2 mM each), magnesium acetate (4 mM), and 2.5 U of the Taq DNA polymerase Stoffel fragment in the PCR buffer as indicated by the manufacturer (Perkin-Elmer, Foster City, Calif.). The following temperature program was used: 94°C (5 min), 40°C (5 min), and 72°C (5 min) for 1 cycle followed by 40 cycles of 94°C (1 min), 60°C (1 min), and 72°C (5 min).
RAPD fragments were separated by electrophoresis on vertical acrylamide gels (15 cm by 15 cm by 1.5 mm, 6% acrylamide, 29/1 acryl-bisacrylamide, 100 mM Tris, 90 mM borate, 1 mM EDTA [pH 8.3]). The products from the control and induced mRNA reactions directed by the same primer were analyzed side by side so that their banding patterns could be compared. Electrophoresis was performed at 1 V/cm. DNA fragments were visualized by silver staining using a Plus One DNA silver staining kit in the Hoefer automated gel stainer (Amersham Pharmacia Biotech, Piscataway, N.J.).
Reamplification of the differentially expressed DNA.
Stained gels were rinsed extensively for 1 h with distilled water. Bands generated from the RNA of cyclohexanone-induced cells but not from the RNA of control cells were excised from the gel and placed in a tube containing 50 μl of 10 mM KCl and 10 mM Tris-HCl (pH 8.3) and heated to 95°C for 1 h to allow some of the DNA to diffuse out of the gel. To reamplify the eluted DNA, 5-μl aliquots of serial dilutions of the eluate over a 200-fold range were used as the template for a new PCR using the Taq polymerase. The primer used for each reamplification (0.25 μM) was the one that had generated the pattern in the RT-PCR experiment. The reamplification conditions were 94°C (1 min), 60°C (1 min), and 72°C (5 min) for 40 cycles.
Each reamplified fragment was cloned into the blue/white cloning vector pCR2.1 (Invitrogen, San Diego, Calif.) and sequenced using the universal forward and reverse primers. The nucleotide sequence of the cloned fragments was compared against entries in the nonredundant GenBank database using the BLASTX program (National Center for Biotechnology Information).
Extension of monooxygenase sequences by out-PCR.
To obtain the complete nucleotide sequence of both monooxygenase genes, kilobase-long DNA fragments extending beyond the sequences identified by DD were generated by a technique we named out-PCR. Genomic DNA was copied at arbitrary sites in 10 separate 50-μl PCRs using the long-range rTth XL DNA polymerase (Perkin-Elmer) and one of any 10 arbitrary primers described above. The reaction included in a 1× solution of the rTth XL buffer provided by the manufacturer, 1.2 mM magnesium acetate, 0.2 mM each dNTP, genomic DNA (10 to 100 ng), and 1 U of rTth XL polymerase. For the first PCR, cycle annealing was performed at 45°C to allow arbitrary priming of the genomic DNA and DNA replication was performed for 15 min at 72°C. At this point, each reaction was split into two separate tubes. One of the two tubes was kept unchanged and used as a control, while the other tube received a specific primer (0.4 μM) corresponding to the end of the sequence to be extended and directed toward the outside of the fragment. For example, to extend the sequence of the first monooxygenase, two primers were designed, one diverging from the 5′ end of the differentially displayed fragment 1 (5′-GATCCACCAAGTTCCTCC-3′) and one diverging from 3′ end of the differentially displayed fragment 3 (5′-CCCGGTAAATCACGTGAGTACCACG-3′). Thirty additional PCR cycles were performed under stringent conditions (annealing at 60°C), and the two reactions were analyzed side by side by agarose electrophoresis. The low annealing stringency conditions of the first PCR cycle led to the generation of RAPD patterns similar for both tubes (37). Bands present in the reaction having received the specific primer but not in the reaction containing the arbitrary primer alone potentially correspond to the sequence to be extended. They were excised from the gel, melted in 0.5 ml of H2O, and used as template in a set of new PCRs containing the specific and arbitrary primer. The reamplified bands were cloned into the pCR2.1 vector (Invitrogen) and sequenced. Sequence assembly was performed with the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.).
Expression of monooxygenase genes.
The monooxygenase genes were cloned in the multiple cloning site of the N-terminal His6 expression vector pQE-30 (Qiagen). Each gene was amplified by PCR from chromosomal DNA using primers corresponding to the ends of the gene and engineered to introduce a restriction site (underlined) not present in the gene. The oligonucleotides 5′-GAAAGATCGAGGATCCATGCCAATTACACAAC-3′ and 5′-TCGAGCAAGCTTGGCTGCAA-3′ were used for the monooxygenase 1 gene; 5′-TCGAAGGAGGAGGCATGCATGACGTCAACC-3′ and 5′-CAGCAGGGACAAGCTTAGACTCGACA-3′ were used for the monooxygenase 2 gene.
The resulting plasmids (pPCB1 and pPCB2) were introduced into Escherichia coli DH10B (Gibco BRL, Gaithersburg, Md.) containing a pACYC184 derivative (Tetr) with the lacIq gene cloned in the EcoRI site of the chloramphenicol acetyltransferase gene to provide a tighter repression of the gene to be expressed. Expression of the His6-tagged proteins was achieved by growing the cells carrying the expression plasmids in 1 liter of Luria-Bertani (LB) broth (23) containing ampicillin (100 μg/ml) and tetracycline (10 μg/ml) at 28°C. Because both monooxygenases are flavoproteins, riboflavin (1 μg/ml) was also added to the medium. When the absorption at 600 nm reached 0.5, 1 mM isopropyl-thio-β-galactoside (IPTG) was added to the culture. Cells were harvested 4 h later, resuspended in 2 ml of 300 mM NaCl–5% glycerol–20 mM Tris-HCl (pH 8.0) (buffer A) containing 10 mM EDTA and 100 μg of lysozyme and disrupted by three freeze-thaw cycles. Nucleic acids were digested by addition of MgCl2 (20 mM), RNase A and DNase I (10 μg of each). The particulate fraction was removed by centrifugation at 15,000 × g, and the supernatant was mixed for 1 h at 4°C with 150 μl of a metal chelation agarose (Ni-nitrilotriacetic acid Superflow; Qiagen) saturated with Ni(II) and equilibrated buffer A containing 5 mM imidazole. The resin was washed batchwise with a series of 10-ml volumes of buffer A containing 5, 10, 15, 20, 40, 80, 150, and 300 mM imidazole, respectively. The bound proteins were eluted with an imidazole concentration between 80 and 150 mM. Eluted proteins were concentrated by ultrafiltration with a Centricon device (cutoff, 10,000 Da; Amicon), and the buffer was replaced by buffer A.
Enzymatic assays.
The cyclohexanone monooxygenase activity of each overexpressed enzyme was assayed spectrophotometrically at 340 nm by monitoring the oxidation of NADPH. The spectrophotometer cuvette contained 50 mM Tris-HCl, 50 mM potassium acetate (pH 7) at 30°C, 0.3 mM NADPH, and 20 to 50 μg of homogenous monooxygenase. The reaction was initiated by the addition of 1 mM cyclohexanone. The concentration of purified enzymes was measured using the extinction coefficient of flavin adenine dinucleotide (FAD) at 445 nm (12.3/mM/cm).
Confirmation of the oxidation of cyclohexanone into caprolactone was determined in separate experiments by gas chromatography (GC)-mass spectrometry (MS) on an HP 5890 gas chromatograph with HP 5971 mass selective detector equipped with an HP-1 capillary column (Hewlett-Packard). Prior to analysis, samples were acidified to pH 3 by HCl, extracted by dichloromethane three times, dried with MgSO4, and filtered.
The substrate specificity of each enzyme (25 μg/ml) was tested spectrophotometrically as described above except that it was done in 100 mM glycine-NaOH buffer (pH 9) at room temperature.
Nucleotide sequence accession numbers.
The nucleotide sequences of the two cyclohexanone monooxygenase genes have been deposited in the GenBank database under accession no. AF257214 and AF257215.
RESULTS
Isolation of Brevibacterium epidermidis HCU.
A halotolerant bacterial strain capable of degrading cyclohexanone and cyclohexanol in the presence of 10 to 15% NaCl was isolated from an industrial wastewater treatment plant using cyclohexanone as the sole carbon source. 16S rDNA typing showed that this halotolerant cyclohexanone-utilizing strain (HCU) is very closely related (99% identity) to B. epidermidis strain NCDO 2286 (accession no. X76565; J. Cai, unpublished results), which is a member of the high-G+C gram-positive bacteria. Halotolerance is a characteristic of members of the Brevibacterium genus (5), and Brevibacterium sp. strain HCU grew in the presence of up to 15% NaCl (data not shown), although NaCl was not required for growth. Cyclohexanone (up to 0.4%) supported growth as a sole carbon and energy source, although traces of yeast extract (0.005%) were required.
Other substrates that supported growth include cyclopentanol, cyclohexanol, ethanol, 1-propanol, 1-butanol, glycerol, acetate, propionate, butyrate, lactate, succinate, glucose, fructose, yeast extract, and Casamino Acids. No growth was observed on cyclohexane, cycloheptanone, cycloheptanol, cyclooctanone, benzene, benzoate, phenol, or toluene. The doubling times of Brevibacterium sp. strain HCU were 1.6 h on LB and 3.0 h on S12 with 10 mM cyclohexanone.
Bacteria reported to grow on cycloalkanones belong to several deeply separated phyla including high-G+C gram-positive bacteria (Nocardia and Arthrobacter) (14, 26), alpha proteobacteria (Xanthobacter) (21, 34), beta proteobacteria (P. C. Brzostowicz and P. E. Rouvière, unpublished results), and gamma proteobacteria (Pseudomonas and Acinetobacter [8, 11, 16]). The gene sequences of only three enzymes involved in cyclohexanone degradation have been reported. These were found in the gamma proteobacterium Acinetobacter sp. strain NCIB 9871 (3, 16). They include the gene of the cyclohexanone monooxygenase (chnB) (3). Possibly because of differences in the G+C content of the two strains (63% for Brevibacterium versus 45% for Acinetobacter) (5, 31), codon preference, and sequence divergence, no hybridization was detected on a dot blot of Brevibacterium HCU DNA using the PCR-amplified Acinetobacter chnB gene as a probe (data not shown). Similarly, oligonucleotide primers designed against the Acinetobacter monooxygenase gene failed to amplify its Brevibacterium homologues (data not shown). We then used Brevibacterium sp. strain HCU to develop the application of mRNA differential display to identify metabolic genes from prokaryotic species.
Induction of cyclohexanone degradation genes.
The degradation of cyclohexanone in Brevibacterium sp. strain HCU is inducible. SDS-polyacrylamide gel electrophoresis showed the synthesis of new proteins in response to cyclohexanone addition (data not shown). Monitoring cell respiration using an oxygen electrode showed an increase in oxygen consumption following the addition of cyclohexanone in cells previously exposed to cyclohexanone but not in control cells grown on acetate (Fig. 1B and C). This indicated that the enzymes responsible for cyclohexanone degradation are induced only in cells preexposed to cyclohexanone. The viability of the control cells was shown by the increase in respiration following the addition of acetate (Fig. 1A). We then tried to identify the genes of some of the induced oxidative enzymes by looking for mRNA synthesized only in response to the addition of cyclohexanone.
FIG. 1.
Inducibility of cyclohexanone degradation in Brevibacterium sp. strain HCU. Oxygen consumption of cultures grown on acetate or cyclohexanone was measured before and after addition of acetate or cyclohexanone (indicated by arrow). Measurements (individual dots) were taken at 3-s intervals. Cyclohexanone-grown but not acetate-grown cells can oxidize cyclohexanone, indicating the inducibility of this pathway.
Identification of cyclohexanone-induced gene sequences.
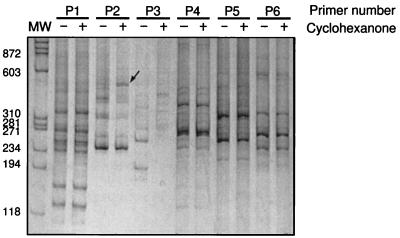
Arbitrarily amplified DNA fragments were generated from total RNA of cyclohexanone-induced and control cells in 81 parallel RT-PCR experiments each using a different primer. We focused on a subset of 23 reactions that showed a strong change in the expression of one or more bands. Two- to threefold changes in the intensity of silver-stained bands were ignored. RT-PCR amplifications from the same RNA preparations were repeated for these 23 primers. Seven of these reactions reproduced the differential amplification of nine of the bands previously observed. The DNA from these bands was reamplified by PCR for subsequent cloning. A typical experiment for a few of the 81 pairs of RT-PCR reactions is presented in Fig. 2. The RAPD DNA fragments generated by each primer from control and induced RNA were analyzed side by side by polyacrylamide gel electrophoresis and visualized with silver stain. In most reactions, the patterns of fragments generated from control and induced samples were identical (Fig. 2, primer 1 [P1], P4, P5, and P6). About 5% of the reactions yielded completely unrelated patterns (Fig. 2, P3). In some of the reactions additional bands were present, reflecting the differential expression of a gene, as was the case for the reactions using P2. This is shown by the strong increase in intensity of a band in the RAPD pattern generated from cyclohexanone-induced RNA compared to that generated from the control RNA.
FIG. 2.
Comparison of randomly amplified fragments for 6 of the 81 arbitrary primers tested (labeled P1 to P6). Each primer was used to reverse transcribe and amplify DNA fragments from total RNA of control (− lanes) and cyclohexanone-induced cells (+ lanes). Most often the RAPD pattern is the same for both RNA samples (P1 and P4 to P6). For about 20% of the primers tested, additional bands generated from RNA of cyclohexanone-induced cells are observed (P2, band marked by arrow). Unrelated DNA patterns (P3) are observed in 5% of the cases. The first lane contains molecular DNA weight markers (MW) with band size (in base pairs) indicated.
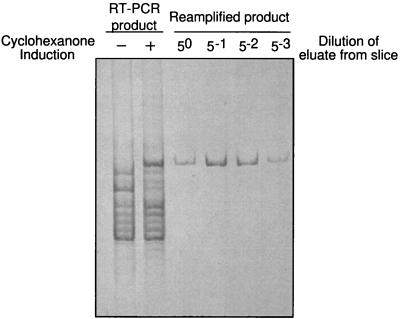
The nine DNA fragments, which could be differentially expressed repeatedly, were eluted from the polyacrylamide and reamplified using the primer initially used to generate the fragment. Confirmation of amplification of the correct PCR product was validated by comparison with the original differentially expressed band. The reamplification of the largest differentially expressed band present in Fig. 2 (P2) is shown in Fig. 3. Since the DNA fragments were amplified using the same oligonucleotide primer at both ends, they cannot be sequenced directly but must first be cloned. For each cloning, 10 transformants were picked and the insert in their plasmid was sequenced using universal primers.
FIG. 3.
Reamplification of a differentially expressed DNA band. Gel slices containing differentially expressed bands were excised from the silver-stained polyacrylamide gels and resuspended in buffer (see Materials and Methods). The band was reamplified by PCR using serial dilutions of the elution buffer as the template and the original primer used in the RT-PCR. The gel shows the reamplification of the differentially expressed band from P2 in Fig. 2. Left two lanes, RT-PCR patterns generated from RNA of control and cyclohexanone-induced cells by P2 (same as in Fig. 2). Right four lanes, PCR reamplification of the differentially expressed band using serial dilutions of eluted DNA from silver stained slice as the template. The products of these reamplifications were cloned and sequenced.
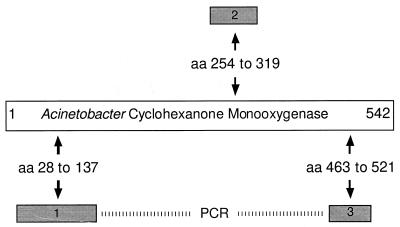
Out of the nine cloned DNA fragments, three DNA fragments coded for protein sequences homologous to the ends and the middle of the Acinetobacter cyclohexanone monooxygenase (Fig. 4) (GenBank accession no. A28550), the first enzyme in the degradation pathway of cyclohexanone (3).
FIG. 4.
Homologies of amino acid sequences encoded by three differentially expressed DNA fragments to the amino acid sequence of the Acinetobacter cyclohexanone monooxygenase. Three of nine reproducibly differentially expressed bands, generated by three distinct arbitrary primers, encode protein fragments homologous to the cyclohexanone monooxygenase from Acinetobacter. Numbers refer to the position of homology on the Acinetobacter protein sequence. The physical linkage of fragments 1 and 3 was demonstrated by PCR (dashed bar). Fragment 2 could not be linked to fragment 1 or 3. Further sequence of the DNA between fragments 1 and 3 showed that they do not belong to the same gene as fragment 2. These three differentially expressed bands identified two distinct cyclohexanone monooxygenase genes in Brevibacterium sp. strain HCU.
Three fragments were homologous to DNA gyrase, ribonucleotide reductase, and 23S rDNA genes, respectively. The translated sequence of the other three fragments showed no homology to proteins in sequence databases. Since we had identified three sequences corresponding exactly to the gene targeted by this experiment, we did not further test if the other remaining six DNA fragments represented truly differentially expressed genes or were false positives.
Cloning the cyclohexanone monooxygenase genes.
To show the physical linkage of the short DNA fragments identified by DD, primers directed outward were designed from each sequence (Fig. 4). Pairwise combinations of these primers were used in PCRs using chromosomal DNA as the template.
No product was obtained by PCR amplification of genomic DNA either using primers matching the 3′ end of fragment 1 and the 5′ end of fragment 2 or using primers matching the 3′ end of fragment 2 and the 5′ end of fragment 3. However, a fragment of the expected size was amplified using primers corresponding to the 3′ end of fragment 1 and the 5′ end of fragment 3, showing linkage of these two sequences (Fig. 4). DNA sequence of the region linking fragments 1 and 3 did not include that of fragment 2, indicating that this fragment corresponded to a second, distinct cyclohexanone monooxygenase gene. The occurrence of these two genes explains the failures of PCR amplification between fragments 1 and 2 and fragments 2 and 3, respectively.
To rule out that the two cyclohexanone monooxygenase genes were amplified from two different species in one culture, Brevibacterium sp. strain HCU was streaked to purity repeatedly on LB plates. Fragments from both genes could be amplified from the total DNA of a single colony, indicating that the strain contains two distinct cyclohexanone monooxygenase isozymes.
We generated DNA fragments extending the known partial sequence of each monooxygenase gene by out-PCR and determined the complete sequences of the genes encoding the two putative monooxygenases. Both were homologous to the Acinetobacter cyclohexanone monooxygenase sequence. Sequence analysis of the DNA region surrounding both monooxygenase genes showed that they are part of two unlinked gene clusters that include other genes involved in the degradation of cyclohexanone (Brzostowicz and Rouvière, unpublished).
Overexpression and activity of the monooxygenases.
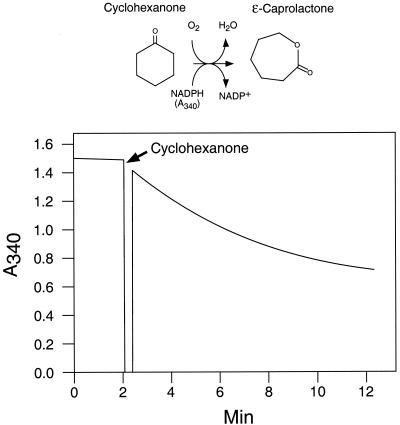
Each monooxygenase was overexpressed in E. coli with an N-terminal His6 tag and purified to homogeneity on Ni-agarose. Following concentration by ultrafiltration, each purified protein had the characteristic yellow color of flavoproteins. Thin-layer chromatography analysis of the protein solutions which were denatured by boiling (33) showed that both enzymes contained FAD and not flavin mononucleotide (data not shown). The enzymatic activity (oxidation of cyclohexanone by O2 with the simultaneous oxidation of a reduced NAD) was first shown spectrophotometrically for both enzymes by monitoring the cyclohexanone-dependent oxidation of NADPH (Fig. 5). No activity was observed using NADH instead of NADPH. GC-MS analysis confirmed the production of caprolactone from cyclohexanone by both purified enzymes. Analysis of the oxidation of cyclohexanone showed the disappearance of the cyclohexanone peak and the appearance of a new peak comigrating with that of a caprolactone standard. MS analysis of that peak showed the expected molecular ion with m/z 114 (data not shown).
FIG. 5.
Enzymatic activity of the cloned cyclohexanone monooxygenase 1. The His6-tagged cyclohexanone monooxygenase 1 was overexpressed, purified to homogeneity, and concentrated by ultrafiltration to 1 mg/ml (see Materials and Methods). Activity was measured spectrophotometrically by monitoring decrease of absorption at 340 nm, which corresponds to the oxidation of the cosubstrate NADPH, in the presence of 20 μg of monooxygenase. The production of caprolactone was confirmed by GC-MS analysis (data not shown).
The substrate specificity of each isozyme was tested using a variety of cyclic ketones (Table 1). Both enzymes had weak activity with cycloheptanone and could not oxidize cyclic ketones with more than seven carbons. Both enzymes were also able to oxidize derivatives of C4, C5, and C6 cyclic ketones, albeit with distinct specific activities. The most dramatic differences were for 2-methylcyclohexanone, preferentially used by monooxygenase 1, and 1,3-cyclohexanedione, cyclohex-2-ene-1-one, and cyclopentanone, preferentially used by monooxygenase 2. Since cyclopentanone could be used as a sole carbon and energy source, it must be solely metabolized by monooxygenase 2. Under the assay conditions, the turnover values of the His6-tagged enzymes shown in Table 1 were comparable to the turnover values of around 400 min−1 for the Acinetobacter and Xanthobacter cyclohexanone monooxygenases (2, 7, 33).
TABLE 1.
Substrate specificity of the two Brevibacterium cyclohexanone monooxygenasesa
| Substrate | Concn (mM) | Turnover value (min−1)
|
|
|---|---|---|---|
| Monooxygenase 1 | Monooxygenase 2 | ||
| Cyclobutanone | 0.1 | 235 | 92 |
| 0.5 | 171 | 96 | |
| Cyclopentanone | 0.1 | 1.2 | 90 |
| 0.5 | 7.0 | 120 | |
| 2-Methylcyclopentanone | 0.1 | 40 | 120 |
| 0.5 | 110 | 110 | |
| Cyclohexanone | 0.1 | 160 | 100 |
| 0.5 | 290 | 100 | |
| 2-Methylcyclohexanone | 0.1 | 250 | 37 |
| 0.5 | 260 | 97 | |
| Cyclohex-2-ene-1-one | 0.1 | 2.3 | 64 |
| 0.5 | 1.9 | 80 | |
| 1,2-Cyclohexanedione | 0.1 | 9 | 7.6 |
| 0.5 | 52 | 34 | |
| 1,3-Cyclohexanedione | 0.1 | 0.3 | 18 |
| 0.5 | 1.2 | 60 | |
| 1,4-Cyclohexanedione | 0.1 | 130 | 53 |
| 0.5 | 210 | 88 | |
| Cycloheptanone | 0.1 | 4.5 | 3.9 |
| 0.5 | 18 | 8.6 | |
| Cyclooctanone | 0.1 | 0.9 | 1.3 |
| 0.5 | 0.4 | 1.3 | |
| Cyclodecanone | 0.1 | ND | 1.8 |
| 0.5 | 1 | 1.2 | |
| Cycloundecanone | 0.1 | 1.2 | 0.6 |
| 0.5 | 1.4 | 0.9 | |
| Cyclododecanone | 0.1 | 1 | 1.2 |
| 0.5 | 0.9 | 1.8 | |
Substrates were tested as provided by the manufacturers. Enzymes were added at 0.5 μM, and the rates of substrate-dependent NADPH oxidation are expressed by the turnover value. For each monooxygenase and under the assay conditions, a turnover of 100 represent a specific activity of 2 μmol/min/mg of enzyme. Activities below 2.0 min−1 could reflect contaminants in the substrate preparations which themselves could be used as substrate for the enzyme.
Relationship of the two Brevibacterium enzymes to other flavin monooxygenases.
Sequence comparison using the BLASTX program against the nonredundant GenBank database showed that the two Brevibacterium cyclohexanone monooxygenases are part of the large family of flavin-dependent monooxygenases. Sequences with the highest similarity are the Acinetobacter cyclohexanone monooxygenase (A28550) and the Rhodococcus steroid monooxygenase (AB010439) (3, 24). The activities of both proteins have been characterized biochemically. A multiple alignment of these four sequences is shown in Fig. 6. Within this group of sequences, the level of amino acid identity lies between 35 and 40%, with overall similarity around 60%. Specific relationships within the group are difficult to establish and depend on the alignment program and parameters chosen. It is worth noting that the two Brevibacterium isozymes are never the closest relatives. A phylogenetic tree derived from the alignment shown in Fig. 6 indicates that monooxygenase 1 is more closely related to the Rhodococcus enzyme whereas monooxygenase 2 is the most distantly related member of this cluster (data not shown). The sequence divergence of the two Brevibacterium isozymes explains the fact that multiclonal antibodies raised against each enzyme do not react with the other (data not shown).
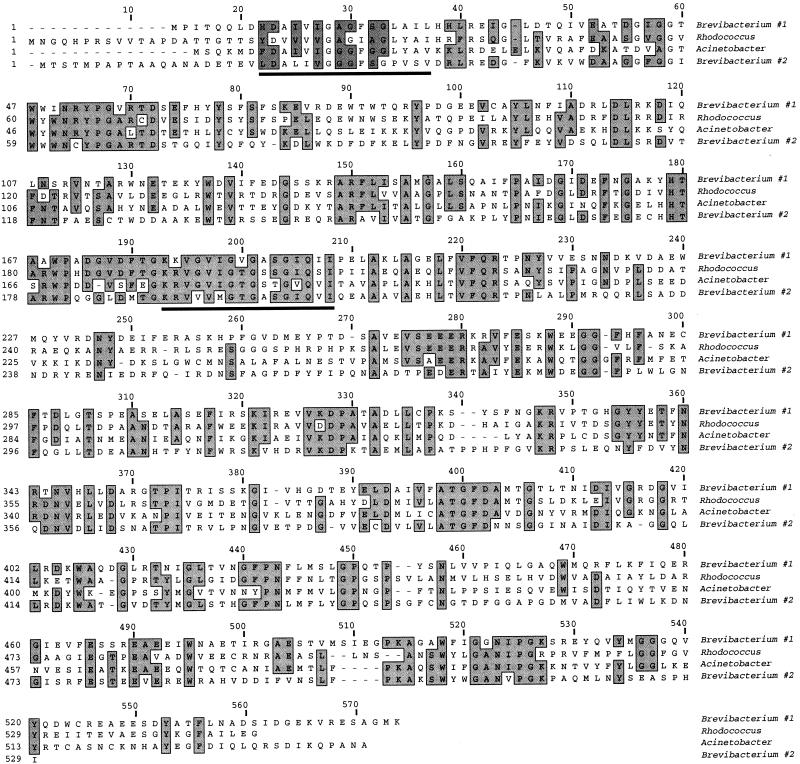
FIG. 6.
Sequences of the two Brevibacterium cyclohexanone monooxygenases aligned with sequences of their closest homologues identified in GenBank, from Acinetobacter (A28550) and Rhodococcus (AB010439). Alignment was performed with the ClustalW program. The FAD binding motif (around position 30) and the NADP binding motif (around position 200) centered around nucleotide signature GXGXXG are underlined.
Homology is also found between the two Brevibacterium monooxygenases and several putative proteins identified by genomic sequencing including six sequences from Mycobacterium tuberculosis (AL021287, Z80108, AL123456, AL021942, AL021309, and Z83864), Pseudomonas fluorescens (AF090329), Rhizobium sp. (AE000078), Sphingomonas sp. (AJ223219), and Emericella nidulans (U34740). These sequences share between 20 and 30% amino acid identity with Brevibacterium monooxygenases. Finally, homology extending up to alignment position 400 (Fig. 6) is found with the conserved family of mammalian liver N-hydroxylating dimethylaniline monooxygenases (AL021026).
Conserved and characterized motifs within this family include the nucleotide binding motif GXGXXG at the flavin binding pocket located at approximately position 30 (25, 32) and the NADP binding pocket around position 200 (Fig. 6). Conservation of these domains extends to the surrounding charged and hydrophobic amino acids. A third motif (DX5FATGYX4P) identified in the group of N-hydroxylating flavin-containing monooxygenases (32) is also present, albeit in a degenerate form (EX6ATGF) at approximately position 400. In addition, many of the conserved amino acids between alignment positions 415 and 455 are also conserved in the microbial homologues.
DISCUSSION
In the course of studies on the biodegradation of environmental pollutants in high-salt environments, we isolated a Brevibacterium strain capable of degrading cyclohexanol and cyclohexanone in the presence of 10 to 15% NaCl. This biodegradation pathway has been characterized biochemically in several organisms (8, 13, 34). Cyclohexanol is oxidized into cyclohexanone by a NAD-dependent dehydrogenase. A flavin NADPH-dependent monooxygenase introduces an oxygen atom in the ring by a Bayer-Villiger reaction to yield the C6 lactone (caprolactone) which is subsequently hydrolyzed into hydroxycaproate. The hydroxy group is then oxidized into a carboxylic group by two NAD-dependent dehydrogenases to yield adipic acid (7, 8, 26, 33, 36).
As with the oxidation pathways of many xenobiotic compounds, the degradation of cyclohexanone is inducible. We used this fact to identify the genes involved in cyclohexanone degradation using mRNA DD, a technique that compares the mRNA sampled by arbitrary RT-PCR amplification between control and induced cells. This technique has been applied to bacterial gene discovery only a few times so far (1, 10, 18, 29, 30, 38–40) and only in one case has it been used to identify metabolic genes (10).
Previous applications of DD have used a small set of primers to generate many bands that were analyzed by long high-resolution sequencing gels. We have tried the converse approach, i.e., using a larger set of 81 primers and analyzing the RT-PCR patterns on relatively short, thick polyacrylamide urea gels. These gels do not have the resolution of sequencing gels and do not allow the detection of faint bands. However, as shown in this work, these gels proved to be sufficient for this initial investigation of bacterial DD. In our experiments, each primer generated a RAPD pattern of approximately 10 DNA fragments on average (Fig. 2 and 3). In theory, a set of 81 primers should generate around 800 independent bands. Assuming (i) a genome size of about 3 Mbp for Brevibacterium sp. HCU, as is the case for its close relative Brevibacterium lactofermentens (Corynebacterium glutamicum) (6), (ii) an average of one gene per kilobase, (iii) an average of three genes per operon, and (iv) the expression of 50% of the operons, the mRNA population may contain around 500 distinct multicistronic mRNA species at any given time. (v) Assuming finally an equal probability of amplifying rare messages as of amplifying more abundant messages after 40 cycles of PCR (22), the probability of not sampling a specific mRNA in an RT-PCR generating 800 RAPD bands is (1-1/500)800 i.e., 20%. Conversely, the probability of sampling a specific operon is around 80% for a genome of 3 Mbp.
Despite the simplifications made above and others such as the different hybridization efficiencies for primers with different melting temperatures or the abundance of the various parts of a multicistronic message following mRNA processing, these calculations indicate that identification of induced genes using a large set of arbitrary primers can compensate for low gel resolution. The advantage of this technique is that the entire experiment (81 × 2 PCRs) can be analyzed on seven 24-well gels. Such gels are easy to cast and handle, and they are fast to run. Furthermore, the use of nonradioactive visualization greatly simplifies the procedure.
In this experiment, we were able to identify simultaneously two distinct cyclohexanone monooxygenase genes in Brevibacterium sp. strain HCU. These two genes could not be linked by PCR using a combination of outward primers. Further analysis of the sequences surrounding each gene showed that they are not part of the same operon (Brzostowicz and Rouvière, unpublished). None of the six other DNA bands identified by differential display showed homology to other metabolic genes expected to be involved in the oxidation of cyclohexanone and cyclohexanol, i.e., a hydrolase and three dehydrogenases. These enzymes belong to well-characterized and recognizable gene families (8, 12, 16).
Why were these genes sampled when other genes of the pathway were not? First, the experimental conditions used may not allow an exhaustive sampling of the mRNA population that is calculated. Second, the hypothesis that the message abundance can be overcome by a large number of cycles (22) may not be correct. The success of this differential display experiment may be due to the fact the flavin monooxygenases are very abundant enzymes because of their low specific activity. Their specific activity is ∼100 min−1 or 2 U/mg, compared to ∼10,000 min−1 or ∼300 U/mg for typical dehydrogenases. Because we wanted to avoid false positives, we analyzed only bands showing a strong differential expression (more than 10-fold induction). Thus, from 81 separate RT-PCRs generating ∼800 bands, only nine bands were further analyzed with only three of them sampling a cyclohexanone monooxygenase gene as identified by sequence similarity. This bias in our selection of the bands possibly increased our chances of identifying strongly induced mRNAs. More generally, degradative metabolic pathways are usually strongly induced and are likely to yield strong differentially expressed bands. They may thus be more readily identified by differential display.
The two cyclohexanone monooxygenase genes identified in Brevibacterium HCU belong to the family of flavin NADPH-dependent monooxygenases. The closest relatives of the two enzymes, the Acinetobacter cyclohexanone monooxygenase and a Rhodococcus steroid monooxygenase, show important divergence, with amino acid identity between 35 and 40%. The significance of the two isozymes of the cyclohexanone monooxygenase is not known, although it may be related to their different substrate specificities. In particular, monooxygenase 1 has very low activity with cyclopentanone whereas monooxygenase 2 oxidizes that substrate readily (Table 1). Whether cyclohexanone is a natural substrate for Brevibacterium is not known. Rather, the activity of these enzymes in nature might be toward natural products that include cyclohexanone as a substructure, such as some steroids or polyketides. This may explain the presence of two related isozymes of distinct specificity. In Nocardia globerula CL1, two distinct cyclohexanone monooxygenases have also been found (27).
The identification of these two monooxygenase genes using mRNA DD strongly supports the use of this technique for bacterial applications. The strength of the technique lies in the fact that only a physiological characterization of the desired biochemistry is needed. The identification of metabolic pathways from environmental isolates by DD should be particularly useful for several reasons. (i) This technique can be performed with bacterial isolates for which genetic systems have not been developed, which is generally the case for environmental microbes, or for which the enzymes are usually not expressed in heterologous hosts. (ii) DD can succeed where techniques based on sequence homologies (Southern blotting and PCR amplification from degenerate primers) fail, because of significant divergence within a gene family. (iii) Metabolic genes are very often expressed only when they are required for the growth of the organism. Therefore, by setting a very low baseline expression level when the genes are not induced, these genes lend themselves to this comparative analysis. (iv) Metabolic genes are often strongly expressed, and performing the RT-PCR analysis under conditions where abundant messages can be predominantly amplified (less than 20 PCR cycles) may bias the sampling of their message and increase the probability of their identification. (v) Genes for bacterial metabolic pathways are almost always clustered in operons coding for all or part of the metabolic pathway. Sampling one of the genes from the operon allows the identification of the entire operon. (vi) Metabolic pathways always include at least some genes that belong to well-characterized gene families encoding enzymes involved in redox, hydrolytic, activation, and condensation reactions. Even though the complete sequencing of microbial genomes still uncovers around 40% of open reading frames with no sequence similarities to other genes, within metabolic gene clusters the fraction of genes with no homologues in the databases may be much lower since the metabolic genes have received the largest part of biochemical, physiological, and genetic research. Consequently, as was observed in the present analysis, it is easy to distinguish differentially expressed genes likely to be involved in the metabolism studied from the false positives that often appear in DD experiments. Finally, this analysis can, in principal, be applied to microbial enrichments enabling one to identify several related genes from several organisms in one environment.
In conclusion, we have shown that mRNA DD can be used successfully to identify prokaryotic genes encoding targeted enzymes and biochemical pathways. Because this technique is applicable to all organisms, we anticipate that as the protocols for DD become streamlined, it will become a widespread tool for the discovery of new metabolic genes.
ACKNOWLEDGMENTS
We thank Vasantha Nagarajan for initially suggesting the use of differential display for the discovery of metabolic genes and for constant scientific discussions. We also thank Ivan Turner, Sr., Sylvia Stack, Ray Jackson, and Tom Miller for assistance with protein purification, GC-MS analysis, DNA sequencing and oligonucleotide synthesis.
REFERENCES
- 1.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 2.Branchaud B P, Walsh C T. Functional group diversity in enzymatic oxygenation reactions catalyzed by bacterial flavin-containing cyclohexanone monooxygenase. J Am Chem Soc. 1985;107:2153–2161. [Google Scholar]
- 3.Chen Y C, Peoples O P, Walsh C T. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J Bacteriol. 1988;170:781–789. doi: 10.1128/jb.170.2.781-789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang S E, Blattner F R. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins M D. The genus Brevibacterium. In: Balows A, editor. The prokaryotes. 2nd ed. II. New York: Springer-Verlag; 1992. pp. 1351–1354. [Google Scholar]
- 6.Correia A, Martin J F, Castro J M. Pulsed-field gel electrophoresis analysis of the genome of amino acid producing corynebacteria: chromosome sizes and diversity of restriction patterns. Microbiology. 1994;140:2841–2847. doi: 10.1099/00221287-140-10-2841. [DOI] [PubMed] [Google Scholar]
- 7.Donoghue N A, Norris D B, Trudgill P W. The purification and properties of cyclohexanone oxygenase from Nocardia globerula CL1 and Acinetobacter NCIB 9871. Eur J Biochem. 1976;63:175–192. doi: 10.1111/j.1432-1033.1976.tb10220.x. [DOI] [PubMed] [Google Scholar]
- 8.Donoghue N A, Trudgill P W. The metabolism of cyclohexanol by Acinetobacter NCIB 9871. Eur J Biochem. 1975;60:1–7. doi: 10.1111/j.1432-1033.1975.tb20968.x. [DOI] [PubMed] [Google Scholar]
- 9.Duggan D J, Bittner M, Chen Y, Meltzer P, Trent J M. Expression profiling using cDNA microarrays. Nat Genet. 1999;21(1 Suppl.):10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 10.Fleming J T, Yao W H, Sayler G S. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl Environ Microbiol. 1998;64:3698–3706. doi: 10.1128/aem.64.10.3698-3706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin M, Trudgill P W. Metabolism of cyclopentanol by Pseudomonas NCIB 9872. Biochem J. 1972;129:595–603. doi: 10.1042/bj1290595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin M, Trudgill P W. Purification and properties of cyclopentanone oxygenase of Pseudomonas NCIB 9872. Eur J Biochem. 1976;63:199–209. doi: 10.1111/j.1432-1033.1976.tb10222.x. [DOI] [PubMed] [Google Scholar]
- 13.Griffin M, Trudgill P W. Purification of cyclopentanone oxygenase from Pseudomonas NCIB 9872. Biochem Soc Trans. 1974;1:1255–1258. doi: 10.1111/j.1432-1033.1976.tb10222.x. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa Y, Hamano K, Obata H, Tokuyama T. Microbial degradation of cycloheptanone. Agric Biol Chem. 1982;46:1139–1143. [Google Scholar]
- 15.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwaki H, Hasegawa Y, Teraoka M, Tokuyama T, Bergeron H, Lau P C. Identification of a transcriptional activator (ChnR) and a 6-oxohexanoate dehydrogenase (ChnE) in the cyclohexanol catabolic pathway in Acinetobacter sp. strain NCIMB 9871 and localization of the genes that encode them. Appl Environ Microbiol. 1999;65:5158–5162. doi: 10.1128/aem.65.11.5158-5162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kullen M J, Klaenhammer T R. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol Microbiol. 1999;33:1152–1161. doi: 10.1046/j.1365-2958.1999.01557.x. [DOI] [PubMed] [Google Scholar]
- 19.Liang P, Averboukh L, Pardee A B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 21.Magor A M, Warburton J, Trower M K, Griffin M. Comparative study of the ability of three Xanthobacter species to metabolize cycloalkanes. Appl Environ Microbiol. 1986;52:665–671. doi: 10.1128/aem.52.4.665-671.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathieu-Daude F, Welsh J, Vogt T, McClelland M. DNA rehybridization during PCR: the ‘Cot effect’ and its consequences. Nucleic Acids Res. 1996;24:2080–2086. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Morii S, Sawamoto S, Yamauchi Y, Miyamoto M, Iwami M, Itagaki E. Steroid monooxygenase of Rhodococcus rhodochrous: sequencing of the genomic DNA, and hyperexpression, purification, and characterization of the recombinant enzyme. J Biochem (Tokyo) 1999;126:624–631. doi: 10.1093/oxfordjournals.jbchem.a022494. [DOI] [PubMed] [Google Scholar]
- 25.Nishiya Y, Imanaka T. Analysis of interaction between the Arthrobacter sarcosine oxidase and the coenzyme flavin adenine dinucleotide by site-directed mutagenesis. Appl Environ Microbiol. 1996;62:2405–2410. doi: 10.1128/aem.62.7.2405-2410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris D B, Trudgill P W. Metabolism of cyclohexanol by Nocardia globerula CL1. Biochem J. 1971;121:363–370. doi: 10.1042/bj1210363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norris D B, Trudgill P W. Multiple forms of cyclohexanone oxygenase from Nocardia globerula CL1. Eur J Biochem. 1976;63:193–198. doi: 10.1111/j.1432-1033.1976.tb10221.x. [DOI] [PubMed] [Google Scholar]
- 28.Rafalski J A, Hanafey M, Miao G H, Ching A, Lee J-M, Dolan M, Tingey S. New experimental and computational approaches to the analysis of gene expression. Acta Biochim Pol. 1998;45:929–934. [PubMed] [Google Scholar]
- 29.Rindi L, Lari N, Garzelli C. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem Biophys Res Commun. 1999;258:94–101. doi: 10.1006/bbrc.1999.0591. [DOI] [PubMed] [Google Scholar]
- 30.Rivera-Marrero C A, Burroughs M A, Masse R A, Vannberg F O, Leimbach D L, Roman J, Murtagh J J., Jr Identification of genes differentially expressed in Mycobacterium tuberculosis by differential display PCR. Microb Pathog. 1998;25:307–316. doi: 10.1006/mpat.1998.0235. [DOI] [PubMed] [Google Scholar]
- 31.Shanley M S, Harrison A, Parales R E, Kowalchuk G, Mitchell D J, Ornston L N. Unusual G + C content and codon usage in catIJF, a segment of the ben-cat supra-operonic cluster in the Acinetobacter calcoaceticus chromosome. Gene. 1994;138:59–65. doi: 10.1016/0378-1119(94)90783-8. [DOI] [PubMed] [Google Scholar]
- 32.Stehr M, Diekmann H, Smau L, Seth O, Ghisla S, Singh M, Macheroux P. A hydrophobic sequence motif common to N-hydroxylating enzymes. Trends Biochem Sci. 1998;23:56–57. doi: 10.1016/s0968-0004(97)01166-3. [DOI] [PubMed] [Google Scholar]
- 33.Trower M K, Buckland R M, Griffin M. Characterization of an FMN-containing cyclohexanone monooxygenase from a cyclohexane-grown Xanthobacter sp. Eur J Biochem. 1989;181:199–206. doi: 10.1111/j.1432-1033.1989.tb14711.x. [DOI] [PubMed] [Google Scholar]
- 34.Trower M K, Buckland R M, Higgins R, Griffin M. Isolation and characterization of a cyclohexane-metabolizing Xanthobacter sp. Appl Environ Microbiol. 1985;49:1282–1289. doi: 10.1128/aem.49.5.1282-1289.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welsh J, Chada K, Dalal S S, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willetts A. Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 1997;15:55–62. doi: 10.1016/S0167-7799(97)84204-7. [DOI] [PubMed] [Google Scholar]
- 37.Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuk M H, Harvill E T, Miller J F. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol Microbiol. 1998;28:945–959. doi: 10.1046/j.1365-2958.1998.00850.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J P, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]