Summary
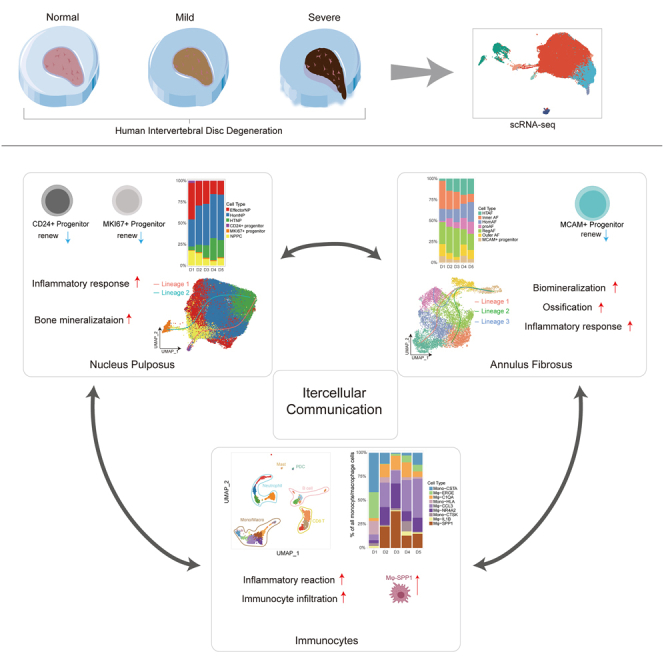
The complexity of the human intervertebral disc (IVD) has hindered the elucidation of the microenvironment and mechanisms underlying IVD degeneration (IVDD). Here we determined the landscapes of nucleus pulposus (NP), annulus fibrosus (AF), and immunocytes in human IVD by scRNA-seq. Six NP subclusters and seven AF subclusters were identified, whose functional differences and distribution during different stages of degeneration (Pfirrmann I-V) were investigated. We found MCAM+ progenitor in AF, as well as CD24+ progenitor and MKI67+ progenitor in NP, forming a lineage trajectory from CD24+/MKI67+ progenitors to EffectorNP_⅓ during IVDD. There is a significant increase in monocyte/macrophage (Mφ) in degenerated IVDs (p = 0.044), with Mφ-SPP1 exclusively found in IVDD but not healthy IVDs. Further analyses of the intercellular crosstalk network revealed interactions between major subpopulations and changes in the microenvironment during IVDD. Our results elucidated the unique characteristics of IVDD, thereby shedding light on therapeutic strategies.
Subject areas: Biopsy sample, Specialized functions of cells, Transcriptomics
Graphical abstract
Highlights
-
•
sRNA-seq reveals heterogeneity in human intervertebral disc degeneration (IVDD)
-
•
Progenitors and cell lineage trajectories in NP and AF were identified
-
•
Monocyte/macrophage (Mφ) especially Mφ-SPP1 significantly increases during IVDD
-
•
Intercellular crosstalk network reveals interactions and changes during IVDD
Biopsy sample; Specialized functions of cells; Transcriptomics
Introduction
Intervertebral disc degeneration (IVDD) is considered the leading cause of neck and low back pain, which is a major cause of disabling health conditions that cause a burden on global healthcare and a significant impact on socio-economic development.1 The degeneration of the intervertebral disc (IVD) has been observed as early as age 11, and IVD degenerative diseases are much more common in the aging adult population.2 IVDD is believed to be caused by multiple factors, such as aging, obesity, environment/occupation, and genetic inheritance.3 Even though IVDD is prevalent worldwide, the present clinical prevention and therapeutic strategies, including both conservative therapies and surgical treatments, mainly focus on symptomatic relief rather than re-establishing homeostasis and ameliorating the degeneration of the IVD.4
IVD is composed of the following three distinct components: the central gelatinous nucleus pulposus (NP), circumjacent annulus fibrosus (AF), and cartilaginous endplate (CEP). IVD constitutes the fundamental building block of the spine, mainly transmits and absorbs mechanical loadings onto the spine, and helps to maintain a motional segment that allows flexion, extension, bending, and rotation.5 A healthy NP is a highly hydrated tissue that is rich in proteoglycans. It distributes tensions and pressure evenly on the AF.6 A healthy AF, including the inner AF and outer AF, is a highly organized fibrous structure consisting of nearly 20 concentric lamellae of collagen fibers.6,7 IVDD is a complicated process that is driven by the loss of viable disc NP and AF cells, activation of matrix-degrading enzymes, disorders of catabolism and anabolism, and increase of inflammation.5,8 Early development of disc degeneration typically occurs in NP.9 Degenerative NP is characterized by the apoptosis of NP cells, decrease in proteoglycan and collagen II in the extracellular matrix (ECM), and gradual loss of its normal physiological ability to transmit pressure and maintain the disc height of the spine.10 Degenerative AF shows AF cells degeneration and the content change of collagen-I, collagen-II, and aggrecan, which results in highly disorganized lamellae fibers with rim lesions, de-lamination, and radial fissures.11 With the progression of IVDD, vascular in-growth of the disc increases the inflammatory cells infiltration and cytokines secretion.12
Recently, studies have reported the heterogeneity of IVD cells. Comparative microarray and bulk RNA-sequencing (RNA-seq) have shown novel marker genes and phenotypes of NP and AF cells.13 Minogue BM et al. characterized the NP and IVD transcriptional profiles to differentiate between NP, AF, and articular cartilage cells.14 The NP cells are believed to be derived from the notochord cells, which are assumed to be the precursors of all NP cells.15,16 Nucleus pulposus progenitor cells (NPPCs) were found to exist in IVD because of the expression of stemness-related markers such as Sox2, Oct3/4, Nanog, and CD133.17 Besides, during the degenerated IVDs, infiltrated immune cells such as CD68+ macrophages (Mφ), neutrophils, and T lymphocytes (CD4+, CD8+) are present.18 Infiltrating immunocytes and inflammatory cytokines play important roles in different grades of IVDD.5 However, the exact cell lineages in IVD and intercellular interaction mechanisms underlying the progression of IVDD have not been completely elucidated.
Given the complexity and poor understanding of human IVDD, in the present study, we performed single-cell RNA-sequencing (scRNA-seq) to identify various cell types and presented the landscapes of IVD during the degeneration process. Through the high-precision and unbiased resolution for distinguishing cell populations of scRNA-seq, we determined the heterogeneity of IVD and identified different cell subsets of NP cells, AF cells, immunocytes, and their corresponding marker genes. We analyzed the various cell populations and their distribution in different degeneration grades. Importantly, we identified CD24+ progenitor and MKI67+ progenitor in NP and MCAM+ progenitor in AF. Our results revealed cell lineage trajectories of NP subpopulations during IVDD progression grades: stemming from CD24+ progenitor or MKI67+ progenitor to EffectorNP_1 or EffectorNP_3. Furthermore, a total of nine macrophage subclusters were identified. Mφ-SPP1 was not found in the healthy IVDs; however, it was increased in degenerated IVDs. Results suggested important functions of Mφ-SPP1 in the tissue self-repair and inflammatory reactions during IVDD. Finally, the Intercellular-crosstalk mechanism between major subpopulations and microenvironment changes during the degeneration process was elucidated. Our results revealed a comprehensive landscape of NP cells, AF cells, and immunocytes in human IVD and laid a foundation for elucidating the biological mechanisms underlying IVDD. The present study provides potential diagnostic and therapeutic markers and options for human IVD degenerative diseases.
Results
Single-cell RNA-sequencing revealed cellular heterogeneous landscape in human intervertebral disc
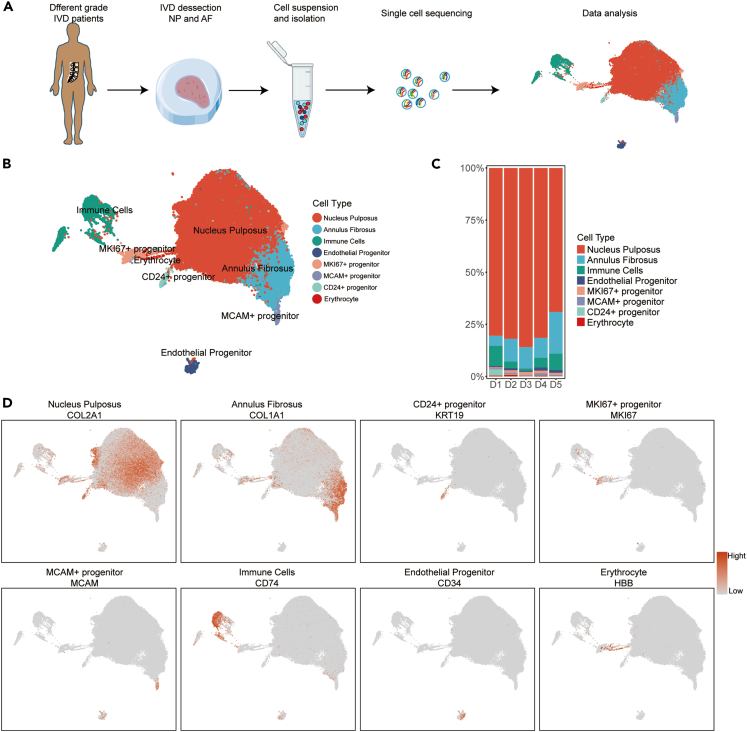
To define cell subtypes of human IVDs and determine genome-wide gene expression patterns, we initially obtained 14 IVD tissues (Table S1), which included two healthy human IVDs (Pfirrmann I named D1) and 12 degenerated human IVDs (Pfirrmann II-V named D2-D5, respectively) for scRNA-seq on 10x Genomics Chromium System (Figure 1A). We sequenced a total of 91145 individual IVD cells and retained 73562 IVD cells for subsequent analysis after extensive filtration.
Figure 1.
Single-cell RNA sequencing analysis of human IVD cells
(A) Schematic workflow of the experimental strategy to isolate cells from human NP and AF for subsequent single-cell RNA sequencing.
(B) Uni-form Mani-fold Approximation and Projection (UMAP) showing the single-cell transcriptomic landscape of human IVD cells. Eight clusters were visualized.
(C) Distribution of cell clusters (defined in B) in human D1-D5 (Pfirrmann grade I-V) IVDs.
(D) The Signature gene expression of representative marker genes for cell clusters defined in B.
A total of 4944 differentially expressed genes (DEGs) were found that identified human IVD cell populations (Table S3). Eight putative cell clusters were identified in IVD samples (Figure 1B), including NP cells, AF cells, immune cells, endothelial progenitor, erythrocytes, and three progenitor clusters of MKI67+ progenitor, MCAM+ progenitor, and CD24+ progenitor. The proportion and distribution of these cell clusters in different degenerated grades of IVDs were shown in Figure 1C. Results showed that the proportion of NP cells decreased and AF cells increased in D5 IVDs compared with those of D1-D4 IVDs (Figure 1C). The immune cells were significantly increased in D5 IVDs (Figure 1C).
NP cells highly expressed chondrocyte-specific genes of COL2A1, ACAN, and ECM genes of COMP and FMOD (Figure 1D and Table S3). AF cells highly expressed cartilage extracellular genes, namely COL1A1, CRTAC1, ASPN, and matrix metalloproteinase (MMP) 2 (Figure 1D and Table S3). Immune cells were identified in all degenerated grades by the markers of CD74, SRGN, LAPTM5, and HLA-DRA (Figure 1D and Table S3). The endothelial progenitor was also found in IVDs (Figure 1B), which was consistent with previous studies.19 Endothelial progenitor highly expressed genes of PECAM1, CDH5, CD34, and KDR (Figure 1D and Table S3). Erythrocytes were identified by genes of HBA1 and HBB (Figure 1D and Table S3).
Additionally, we found three progenitor clusters. CD24+ progenitor and MKI67+ progenitor highly expressed the NP cell-associated genes of COL2A1, ACAN, COMP, and FMOD (Table S3), and were speculated as NP cell progenitors. CD24+ progenitor highly expressed the signature markers of CD24 and KRT19, and MKI67+ progenitor highly expressed MKI67 and CENPF (Figure 1D). The MCAM+ progenitor highly expressed MCAM (Figure 1D) and AF cell-associated genes of COL1A1 and THY1 (Table S3). Thus, it was speculated to be the progenitor of AF cells. The expression of some widely reported marker genes of IVD cells was also detected (Table S3). Taken together, these results showed the overall pattern of cell cluster landscape and transcriptome status of human IVDs at a single-cell level.
Identification of nucleus pulposus subpopulations and cell lineage trajectories during the intervertebral disc degeneration process
To determine the heterogeneity of NP and functions of NP subpopulations during IVDD progression, we performed PCA to cluster NP cells (Figure S1). The NP cells were divided into four parts (part 1-part 4) along the axes of PC1 and PC2 according to Gene Ontology (GO) functions of ECM organization, notochord development, inflammation, regulation of cell-cell adhesion, ossification, regulation of myeloid cell differentiation and hemopoiesis (Figure S1A). Therefore, the part 1 cells were enriched in inflammation, part 2 cells were enriched in inflammation and ossification, part 3 cells were enriched in notochord development and myeloid cell differentiation, and part 4 cells were enriched in ECM organization and notochord development. The distribution of four parts in different degenerated grades of IVDs is shown in Figure S1B. The proportion of cells in part 3 and part 4 decreased with IVDD progression. However, the proportion of cells in part 1 and part 2 increased in the higher degeneration grade of IVD.
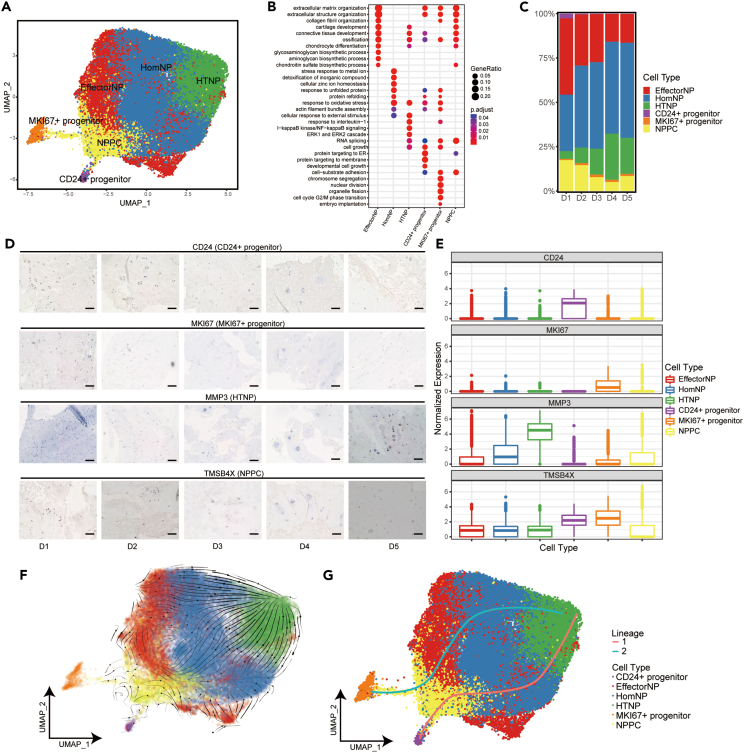
We further performed runUMAP no linear dimension reduction analysis to distinguish these four parts of NP cells, which resulted in a total of six putative cell clusters (Figure 2A) of effector NP cells (EffectorNP), homeostasis NP cells (HomNP), hypertrophic NP cells (HTNP), NPPC, and two progenitors (CD24+/MKI67+ progenitor) mentioned previously. A total of 983 DEGs were found among the six NP subclusters (Table S4 and Figure 2). The expression of representative markers for these cell clusters and heatmap of the DEGs were shown in Figure S2. EffectorNP was enriched in ECM organization and cartilage development (Figure 2B). HomNP focused on stress response to metal ion, unfolded protein, and detoxification of inorganic compound (Figure 2B). HTNP had the capacity of cellular response to external stimulus and inflammatory reaction (Figure 2B). NPPC was enriched in chondrocyte differentiation, extracellular structure organization, ossification, and some intracellular regulatory responses (Figure 2B). CD24+ progenitors were enriched in protein synthesis, cellular pluripotency, and ECM metabolism (Figure 2B). Compared with CD24+ progenitors, MKI67+ progenitors were much more enriched in cell division and multiplication, epithelial-mesenchymal transition (EMT), and inflammatory response (Figures 2B and S3). The functional comparison and DEGs of CD24+ progenitor and MKI67+ progenitor were shown in Figure S3.
Figure 2.
Identification of NP subpopulations and their distribution in different degeneration grades of human IVD
(A) UMAP plot showing the single-cell transcriptomic landscape of human NP cells. Six subpopulations were identified.
(B) GO (gene ontology) functions of six NP subpopulations defined in A.
(C) Distribution of six NP subpopulations (defined in A) in human D1-D5 IVDs.
(D) Representative immunohistochemistry staining of signature markers of the indicated cell clusters in the NP from human D1-D5 IVD tissues. Scale bar, 10 μm.
(E) Signature gene expression level of marker genes is shown for the six NP subpopulations defined in A.
(F) RNA velocity-based pseudotime analysis inferred from UMAP embedding of NP subclusters.
(G) Two cell lineage trajectories from CD24+/MKI67+ progenitor to HTNP based on slingshot pseudotime analysis.
The proportion and distribution of NP subclusters in different degenerated grades of IVDs were shown in Figure 2C. No significant variation in the percentage of HomNP was found between different degeneration grades (Figure 2C). However, the percentage of HTNP increased, but NPPC and EffectorNP decreased progressively as the IVDD grade increased, indicating that the inflammatory reaction increased, and ECM metabolism and NP cell differentiation decreased in the degenerated IVDs. These results suggested that HTNP plays an important role in the progression of NP degeneration. However, NPPC and EffectorNP can regulate the NP-associated functions to inhibit the degeneration. Furthermore, we performed immunohistochemistry assay of tissue sections from five patients (Pfirrmann grade D1-D5), and confirmed the distribution tendency of NPPC and HTNP during IVDD (Figures 2D and 2E). Interestingly, the two NP progenitors showed different trends during IVDD. The proportion of CD24+ progenitors was considerably higher in D1 than in D2-5 (Figure 2C). However, the MKI67+ progenitor was significantly lower in D1 than in D2-5 (Figure 2C). The immunohistochemical assay also confirmed these results (Figures 2D and 2E). We further performed RNA velocity and Slingshot analysis to determine the relationship and cell lineage trajectories of NP subclusters. As expected, results showed two trajectories from two progenitor clusters of CD24+/MKI67+ progenitor to HTNP (Figures 2F and 2G).
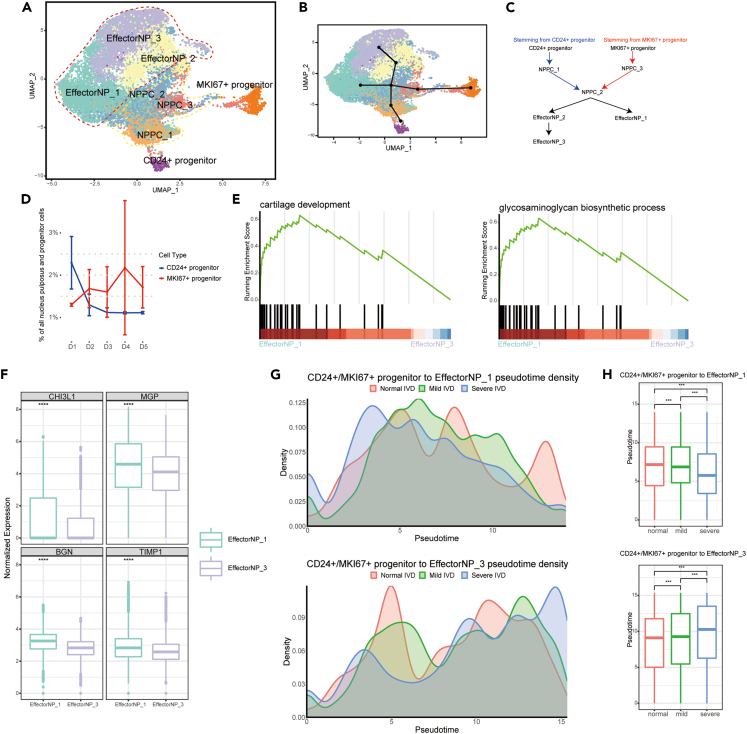
To further distinguish the differentiation pathways and how the lineage trajectories going from CD24+/MKI67+ progenitor to HTNP, we divided NPPC into NPPC_1-3 subclusters, and EffectorNP into EffectorNP_1-3 subclusters (Figure 3A). The GO functions and heatmap of DEGs of NPPC_1-3 and EffectorNP_1-3 were shown in Figure S4. NPPC_1 cluster was enriched in ECM organization, chondrocyte differentiation, and cartilage development. NPPC_2 cluster was enriched in protein/RNA synthesis and ATP biosynthetic process. NPPC_3 cluster mainly focused on cell division and protein/RNA synthesis. EffectorNP_1 cluster was enriched in ECM organization, glycosaminoglycan biosynthetic process, and chondrocyte differentiation. EffectorNP_2 cluster regulated hemostasis and cell adhesion. EffectorNP_3 cluster focused on aging, osteoblast differentiation, and bone mineralization. Furthermore, we performed Slingshot to find lineage by fitting minimum spanning trees based on NP clusters. We found four cell lineage trajectories of NP subclusters: stemming from CD24+/MKI67+ progenitor to NPPC_2 and then to EffectorNP_1 or EffectorNP_3 (Figures 3B and 3C). As previously described, the proportion of CD24+ progenitor was high in D1 and decreased progressively in D2-D5 (Figure 3D). The proportion of MKI67+ progenitors was low in D1 and high in D2-D5 (Figure 3D). Through the gene set enrichment analysis, we found that comparing with EffectorNP_3 cluster, the EffectorNP_1 cluster had more function of cartilage development and glycosaminoglycan biosynthetic process (Figures 3E and 3F). These results predicted distinct differentiation pathways from NP progenitors to EffectorNP clusters. In normal IVDs, CD24+ progenitors play important roles. But in degenerated IVDs, MKI67+ progenitors can play important roles. The differentiation pathways from progenitors can develop to EffectorNP_1 which had glycosaminoglycan biosynthesis and cartilage development, or EffectorNP_3 which had more function of bone mineralization and aging. We further compared the enrichment of different pathways in normal (D1), mild (D2-D3), and severe (D4-D5) degenerated IVDs through pseudotime analysis. Consistent with the above results, the trajectories from CD24+/MKI67+ progenitor to EffectorNP_1 was much more enriched in normal and mild degenerated IVDs (Figures 3G and 3H). The trajectories to EffectorNP_3 were much more enriched in severe degenerated IVDs (Figures 3G and 3H).
Figure 3.
The trajectories of NP subclusters during the degeneration process of human IVD
(A) UMAP plot showing the subclusters of NP progenitors, NPPC and EffectorNP.
(B) Slingshot-based pseudotime trajectories inferred from UMAP embedding of NP sub-celltypes.
(C) Four possible trajectories from CD24+ progenitor or MKI67+ progenitor to EffectorNP_1 or EffectorNP_3 cluster based on slingshot pseudotime analysis.
(D) Relative frequencies of CD24+ progenitor and MKI67+ progenitor in D1-D5 IVDs.
(E) GSEA showing enrichment of pathways between EffectorNP_1 and EffectorNP_3 cluster.
(F) Boxplot showing the expression levels of representative marker genes of EffectorNP_1 and EffectorNP_3 cluster (p < 0.001, Wilcoxon test).
(G) The cell density distribution of pseudotime along trajectories from CD24+/MKI67+ progenitor to EffectorNP_1 or EffectorNP_3 in normal (D1), mild (D2-3) and severe (D4-5) degenerated IVDs. (H) Boxplot showing the distribution of differentiation trajectories in normal, mild, and severe degenerated IVDs (p < 0.001, Kolmogorov-Smirnov test).
Identification of annulus fibrosus subpopulations and cell lineage trajectories during the intervertebral disc degeneration process
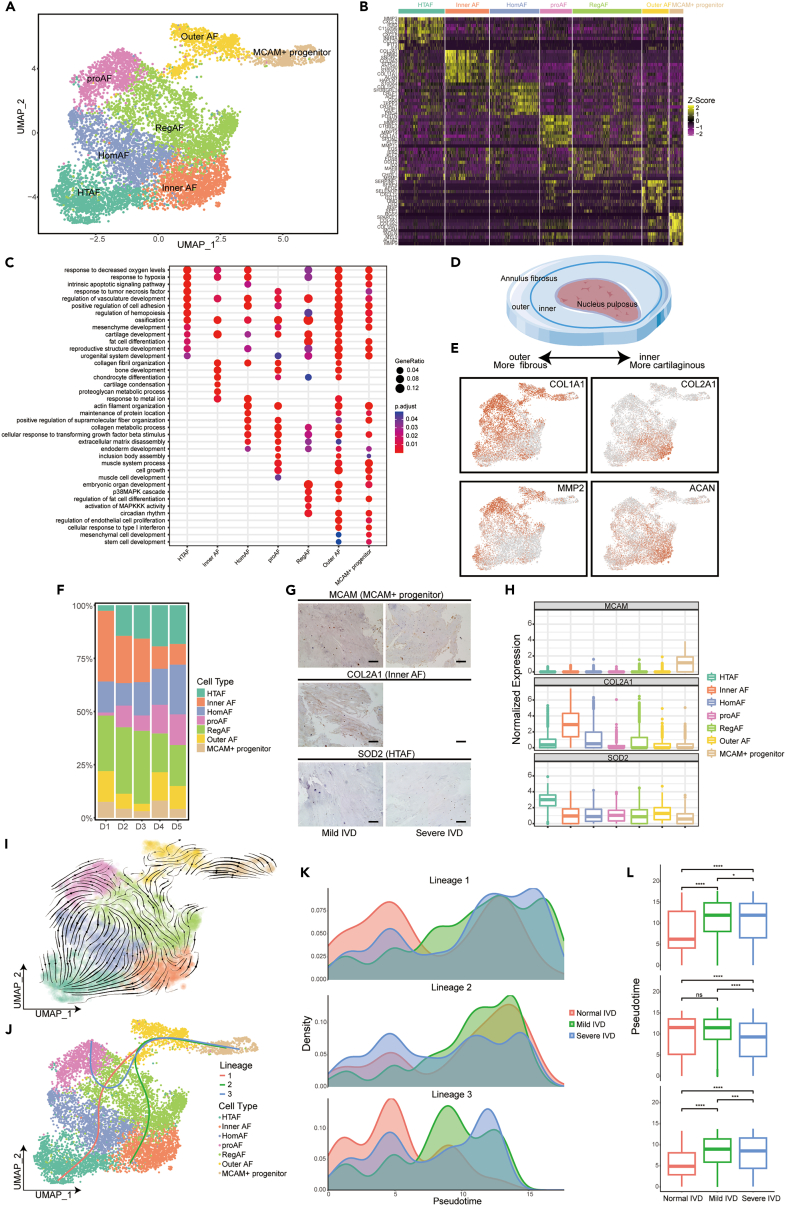
AF subpopulations were identified to determine the heterogeneity of AF and functions of AF subpopulations in IVDD. A total of seven putative AF cell clusters (Figure 4A) were identified, including three empirically defined populations of homeostasis AF (HomAF), InnerAF and OuterAF, and three other populations, namely hypertrophic AF (HTAF), regulatory AF (RegAF), proinflammatory AF (proAF), and MCAM+ progenitor. A total of 2965 DEGs were found among the seven AF subclusters (Table S5). The heatmap of DEGs was shown in Figure 4B. HomAF was enriched in collagen fibril metabolism and organization, response to the metal ion, and maintenance of protein location (Figure 4C). HTAF preferentially responded to decreased oxygen levels, hypoxia and tumor necrosis factor (TNF), and intrinsic apoptotic signaling pathway (Figure 4C). RegAF was enriched in extracellular response and chondrocyte differentiation, and proAF preferred signaling pathways of collagen metabolic process, ossification, and regulation of cell adhesion (Figure 4C). The MCAM+ progenitor had the capacity for stem cell development and cell growth and was defined as the progenitor of AF cells (Figure 4C). Consistent with previous reports on the spatiality and morphology of AF,20 the InnerAF, highly expressing COL2A1 and ACAN, mainly focused on cartilage development and collagen fibril organization (Figures 4C–4E). The OuterAF, highly expressing COL1A1 and MMP2, was enriched in ossification and bone development (Figures 4C–4E).
Figure 4.
Identification of AF subpopulations and their distribution in different degeneration grades of human IVD
(A) UMAP plot showing the single-cell transcriptomic landscape of human AF cells. Seven subpopulations were identified.
(B) Heatmap showing the scaled expression of differentially expressed genes of AF subpopulations defined in A.
(C) GO (gene ontology) functions of seven AF subpopulations defined in A.
(D) Diagrammatic sketch of InnerAF and OuterAF.
(E) The expression level of curated feature genes for InnerAF and OuterAF on the UMAP map.
(F) Distribution of seven AF subpopulations (defined in A) in mild and severe degenerated human IVDs.
(G) Representative immunohistochemistry staining of signature markers of the indicated AF clusters in the mild (D2-3) and severe (D3-4) degenerated human IVD tissues. Scale bar 10 μm.
(H) Boxplot of relative gene expression level of the indicated marker genes for the AF subpopulations defined in A.
(I) RNA velocity-based pseudotime analysis inferred from UMAP embedding of AF subclusters.
(J) Three cell lineage trajectories from MCAM+ progenitor to HTAF, proAF, or inner AF based on slingshot pseudotime analysis.
(K) The cell density distribution of pseudotime along trajectories from MCAM + progenitor to HTAF, proAF, or inner AF in normal, mild, and severe degenerated IVDs.
(L) Boxplot showing the distribution of differentiation trajectories in normal, mild, and severe degenerated IVDs (p < 0.001, Kolmogorov-Smirnov test).
The distribution of AF subclusters in D1-D5 degenerated IVDs was shown in Figure 4F. No significant variation was found about HomAF, RegAF, and OuterAF. However, the percentage of HTAF and proAF was increased, and the percentage of InnerAF was decreased gradually with IVDD progression (Figure 4F). We then performed an immunohistochemistry assay to validate the distribution of major cell types in mild and severe degenerated AF (Figure 4G). Results confirmed the existence of MCAM+ progenitor in AF (Figures 4G and 4H). Compared with mild degenerated AF, InnerAF cells significantly decreased, and HTAF increased in severe degenerated AF (Figures 4G and 4H). Our results suggested that ECM organization and cartilage development decreased, whereas ossification, immune response, and response to extracellular environment increased in the degenerated AF.
We further investigated the relationship and cell lineage trajectories of AF subclusters through RNA velocity and Slingshot analysis. Three cell lineage trajectories of AF subclusters were found: stemming from MCAM+ progenitor to HTAF, innerAF, or proAF (Figures 4I and 4J), indicating distinct differential pathways of AF cells. As shown in Figures 4K and 4L, the trajectories from MCAM+ progenitor to HTAF or proAF were much more enriched in mild and severe degenerated IVDs, which inclined to hypoxia, apoptosis, ossification, and inflammation. Consistent with the above results, the trajectory from MCAM+ progenitor to innerAF was much more enriched in normal and mild degenerated IVDs (Figures 4K and 4L).
Taken together, the results of NP and AF subpopulations landscape and their distribution analysis revealed a set of markers that can be used in combination to classify NP and AF cell phenotypes, and reconstruct the cell-type lineage that recapitulates factors contributing to IVDD progression.
Identification of immunocyte subpopulations and functional definition during intervertebral disc degeneration progression
Although potential resident stem/progenitor cells, macrophages, T lymphocytes, and mast cells are associated with the injury and healing environment of degenerated IVDs,20 the functions of immunocytes in human IVDs have not been thoroughly studied. Here, immunocyte composition, functional states, developmental trajectory, and intercellular interactions of immune cells in human IVDD were explored and compared systematically.
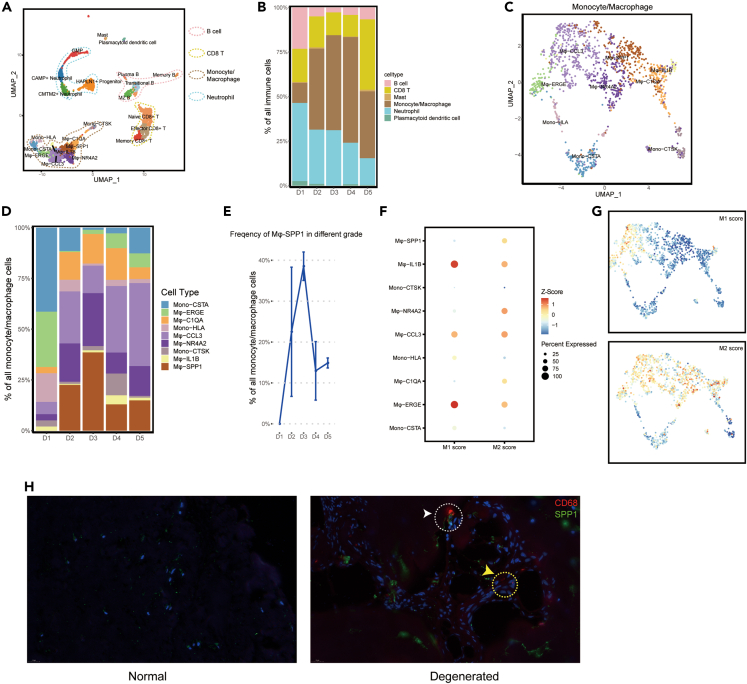
We identified B cells (including plasma B cells, marginal zone B cells, translational B cells, and memory B cells), CD8 T cells (including memory CD8+ T cells, effector CD8+ T cells, and naive CD8+ T cells), neutrophils (including HAPLN1+ progenitor, CMTM2+ neutrophils, CAMP+ neutrophils, and graulocyte-monocyte progenitor), mast cells, plasmacytoid dendritic cells, and monocytes/macrophages were present in the IVDs (Figure 5A). A total of 3719 DEGs were found (Table S6). The proportion and distribution of the aforementioned cells were significantly different in the degenerated D2-D5 IVDs compared with the normal D1 IVDs (Figure 5B). Mature normal IVD tissues are avascular and aneural.2 Vascularization in children consists of vessels across the CEP into the AF; in adults, these vessels disappear, but a rich anastomotic network of vessels remains in the periphery of the disc.21 The vascular network adjacent to CEP and outer AF provide exchanges of nutrients and metabolites for IVD.22 As the disc begins to degenerate, the cells synthesize and release neurogenic factors of NGF and BDNF that promote neural and vascular ingrowths into the disc tissues.23 These would explain the obvious difference in immunocyte distribution between D1 and D2-D5 IVDs.
Figure 5.
Landscape of immune cells in human IVD and their distribution in different degeneration grades of IVD
(A) UMAP plot showing the single-cell transcriptomic landscape of immune cells. Six mian clusters were identified.
(B) Distribution of cell clusters (defined in A) in human D1-D5 (Pfirrmann grade I-V) IVDs.
(C) UMAP plot showing the nine subclusters of Monocyte/Macrophage.
(D) Distribution of subclusters (defined in C) in human D1-D5 IVDs.
(E) The relative frequencies of Mφ-SPP1 in human D1-D5 IVDs.
(F) Gene enrichment for subclusters (defined in C) in comparisons with the classical cell type M1 and M2 macrophage. Bubble size represents the proportion of upregulated and downregulated genes of clusters based on the gene signature on the x axis. The color of the circle represents the directional FDR. Red, enrichment of upregulated genes in the specific cell subsets; blue, depletion of downregulated genes.
(G) Expression level of related genes for the classical M1 and M2 macrophage on the UMAP map. The color of the plots represents the M1/M2 score. Redder color represents higher score.
(H) Multicolor immunofluorescence staining to validate the macrophage and Mφ-SPP1 in normal and degenerated IVD tissues. Red represents CD68 staining of macrophage. Green represents SPP1 staining. Scale bar 20 μm.
Remarkably, the percentage of monocytes/macrophages was significantly increased in degenerated IVDs (p = 0.044, Figure 5B). Therefore, monocyte/macrophage subpopulations were further investigated and distinguished into nine subclusters (Figure 5C), namely Mφ-SPP1, Mφ-CCL3, Mφ-IL1B, Mφ-ERGE, Mφ-NR4A2, Mφ-C1QA, Mono-HLA, Mono-CTSK, and Mono-CSTA. A total of 8064 DEGs was found (Table S7). Further, we analyzed the percentage and distribution of these macrophage subclusters in different degenerated IVDs. The D2-D5 IVDs showed increased percentages of Mφ-SPP1, Mφ-CCL3, Mφ-NR4A2, and Mφ-C1QA (Figure 5D). Interestingly, Mφ-SPP1 was not present in the normal D1 IVDs, whereas increased in D2-D3 IVDs and decreased in D4-D5 IVDs (Figures 5D and 5E). This indicated that Mφ-SPP1 might play crucial roles in the mild degeneration grade. The enrichment of classical M1/M2 macrophage subsets showed that these macrophage subclusters simultaneously resembled the signatures for M1 and M2 macrophages (Figures 5F and 5G). Mφ-SPP1 and Mφ-NR4A2 showed more anti-inflammation and tissue repair functions of M2 macrophages (Figures 5F and 5G), whereas Mφ-CCL3 showed more pro-inflammation function of M1 macrophages (Figures 5F and 5G). We further performed an immunofluorescence assay to validate the distribution of macrophages and Mφ-SPP1 subcluster (Figure 5H). Results revealed that no macrophages were found in D1 IVD (Figure 5H). However, macrophages and the Mφ-SPP1 subcluster infiltrated into degenerated IVD (Figure 5H).
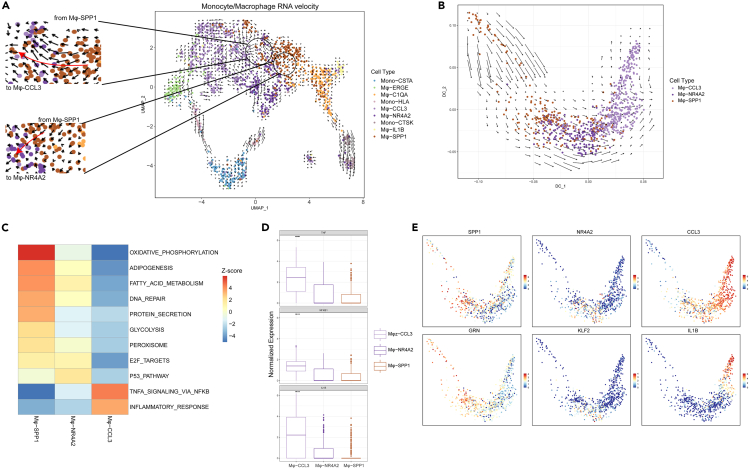
RNA velocity analysis showed that Mφ-SPP1 exhibited a strong directional flow toward Mφ-CCL3 and Mφ-NR4A2 (Figure 6A). A diffusion map of their global transcriptomes showed that Mφ-SPP1, Mφ-NR4A2, and Mφ-CCL3 formed a continuum with distinct expression features (Figures 6B-6E). A clear directional flow from Mφ-SPP1 toward Mφ-NR4A2 and then toward Mφ-CCL3 was observed (Figure 6B). Comparing their expression gene set enrichment, high levels of the inflammatory factors of TNF-α and interleukin 1-β were presented in Mφ-CCL3, and exhibited high inflammatory responses (Figures 6C-6E). These results suggested that macrophages were inclined to play anti-inflammatory roles and tissue repair functions in the mild degenerated IVDs, but then played more proinflammatory roles with IVDD progression.
Figure 6.
RNA velocity analysis of Monocyte/Macrophage subclusters
(A) RNA velocities of Monocyte/Macrophage subclusters are visualized on the UMAP. The black circles were amplified showing the directional flow from Mφ-SPP1 toward Mφ-CCL3 and Mφ-NR4A2.
(B) RNA velocities are visualized on the diffusion map projection of the continuous connection of Mφ-SPP1, Mφ-NR4A2, and Mφ-CCL3.
(C) GSEA showing enrichment of pathwaysdifference between Mφ-SPP1, Mφ-NR4A2, and Mφ-CCL3.
(D) Relative expression level of representative marker genes of Mφ-CCL3.
(E) The expression level of representative marker genes for Mφ-SPP1, Mφ-NR4A2, and Mφ-CCL3 visualized on the diffusion map.
The cell-crosstalk reveals mechanisms of cellular interactions during intervertebral disc degeneration progression
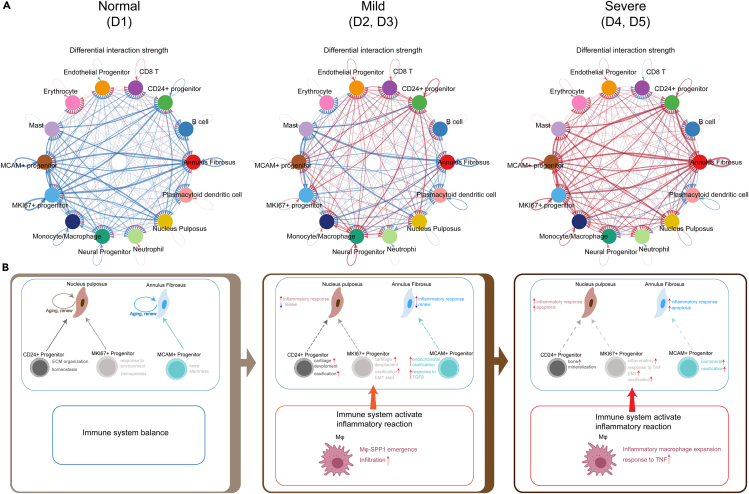
To further investigate the intercellular interactions between NP, AF, progenitors, and immune cells in human IVDD, we analyzed a cell-cell communication network using the CellChat R package on the basis of the ligand-receptor interactions. For the comparative analysis, the degeneration grades of IVD were divided into three groups: normal (D1), mild (D2, D3), and severe (D4, D5). The DEGs regulated by ligands in the different subclusters of IVDD grades are listed in Table S8. Compared with D1, the results of bidirectional signaling interactions and the ligand-receptor interaction strength in mild and severe degenerated IVDs revealed the presence of much more regulated cellular communications (Figure 7A and Figure S5). These results suggested that the intercellular interactions became more frequent, and the microenvironment in the IVD became more complex with the increasing IVDD grades. To explore changes in the microenvironment during IVD degradation, we focused on the intercellular interactions between major cell clusters that are mentioned above. The GO functions regulated by other subcluster ligands in different IVDD grades are shown in Figure S6. Taking together our previous results, the crosstalk interactions of major cell clusters and microenvironment changes in different degenerated grades of IVDs are shown in Figure 7B.
Figure 7.
Overview of the intercellular crosstalk networks among NP cells, AF cells, progenitors, and immune cells in the degeneration process of human IVD
(A) Overview of the intercellular crosstalk networks among NP, AF, progenitors, and immune cells in normal (D1), mild (D2-3), and severe (D4-5) degenerated IVDs. Dots indicate cell clusters. The thickness of the directed line indicates the relative quantity of significant ligand-receptor pairs between any two pairs of cell clusters. Red represents the interaction amounts increased. Blue represents the interaction amounts decreased.
(B) Diagrammatic sketch showing the crosstalk interactions of major cell clusters and microenvironment changes in different degeneration grades of IVDs.
In the normal IVD, NP cells were enriched in ECM organization, chondrocyte differentiation, and cartilage development, whereas AF cells were enriched in ECM organization, cartilage development (Figure S6). The NP and AF cells showed the homeostasis of aging and renewal (Figure 7B). Simultaneously, CD24+ progenitor was enriched in ECM organization and cell homeostasis, MKI67+ progenitor responded to the environment and hemopoiesis, and MCAM+ progenitor was associated with cell stemness (Figures 7B and S6). Considering that the healthy adult IVD is almost entirely avascular, the microenvironment of NP and AF in the IVD was in homeostasis.
In the mild-degenerated IVDs, the NP cells were preferably involved in intercellular interactions of vasculogenesis and macrophage activation, but less involved in collagen fibril organization and chondrocyte differentiation (Figure S6). The AF cells were preferably involved in vasculogenesis and ECM disassembly, but less involved in bone morphogenesis and cartilage development (Figure S6). These revealed that inflammatory responses were increased, and cell self-renewal was decreased in the mild degenerated NP and AF (Figure 7B). As mentioned earlier, macrophages infiltrated into IVD tissues, and Mφ-SPP1 emergence exhibited tissue repair functions in the microenvironment of the IVD; CD24+/MKI67+ progenitors were enriched in cartilage development and ossification; MCAM+ progenitor was enriched in endochondral ossification, and response to transforming growth factor β (Figures 7B and S6).
In the severe-degenerated IVDs, the cellular communications between NP cells, AF cells, immune cells, and progenitors were significantly increased (Figure 7A). The regulated GO functions revealed that ECM organization in the NP cells was decreased, and chondrocyte differentiation disappeared (Figure S6). NP cells were involved in the extrinsic apoptotic signaling pathway and response to biotic stimulus, and AF cells were involved in the regulation of the apoptotic signaling pathway and the regulation of angiogenesis (Figure S6). CD24+ progenitor was associated with the positive regulation of leukocyte migration and bone mineralization; MKI67+ progenitor was associated with the positive regulation of lymphocyte activation, response to TNF, and EMT; MCAM+ progenitor was enriched in ossification, the regulation of vasculature development, and response to TNF (Figure S6). The immune system was further activated by increased inflammatory reactions and Mφ response to TNF (Figure 7B).
Discussion
As degenerative IVD disease (DDD) prevalence is common, finding effective and innovative treatment for preventing or reversing IVDD and improving the clinical outcomes for patients is important.24 An in-depth understanding of the biological basis and pathophysiological process of IVDD is needed to explore new treatment strategies.25 However, the homeostasis and microenvironment of IVD have not been completely elucidated because of its complex multifactorial process and cellular heterogeneity. In the present study, we investigated the NP and AF of IVD at different Pfirrmann grades at a single-cell resolution on the basis of comprehensive IVD samples. We identified cell markers and signatures for verifying each hypothesized subcluster of NP and AF. We identified subclusters of NP and AF and presented their distribution in different IVD grades. Notably, we found CD24+/MKI67+ progenitor in NP and MCAM+ progenitor in AF, forming a lineage trajectory from CD24+/MKI67+ progenitor to EffectorNP_⅓ during IVDD. We revealed distinct subpopulations of immune responders in different grades of IVD. Interestingly, we found a significant increase in monocyte/macrophage (Mφ) in degenerated IVDs, with Mφ-SPP1 exclusively found in IVDD but not healthy IVDs. RNA velocity revealed a clear directional flow from Mφ-SPP1 toward Mφ-NR4A2 and Mφ-CCL3. These results suggested an important role of Mφ-SPP1 in tissue self-repair and inflammatory reactions during IVDD. Finally, the intercellular crosstalk network revealed interactions between major subpopulations and changes in the microenvironment during IVDD. Our study extends the findings of distinguishing molecular and cellular features of IVD and further extends the in-depth understanding of cellular heterogeneity and microenvironment interactions during IVDD, shedding light on therapeutic strategies for human DDD.
Recently, several studies have reported the cellular composition and heterogeneity of NP and AF at the single-cell level.26,27,28 However, due to the complex IVD structure, the precise description of cell populations and microenvironment interactions based on unbiased samples during human IVDD progression is yet to be studied. In the present study, apart from NP and AF cells, immune cells and endothelial progenitor cells were also found in the IVDs. Besides, the proportion of immune cells was increased in the severe degenerated IVDs. Mature disc tissues are normally avascular and aneural.29,30 As the disc begins to degenerate, changes in tissue integrity allow for increased vascularity and neural in-growth into the disc during IVDD progression, which becomes a source of inflammatory reactions and peripheral neuropathy producing pain, weakness, and numbness.29 Our findings suggested that inflammation and neural in-growths play important roles in IVDD and pathophysiological progression. Immune cells and endothelial progenitor cells were also found in D1 IVDs, because that the D1 samples were obtained from two 6-and 7-year-old children (Table S1), and the vascular channels were present in the AF of children and gradually disappeared after adulthood.30
The healthy NP is a gelatinous and highly hydrated structure consisting of NP cells and ECM.2 NP cells are chondrocyte-like cells that are derived from embryonic notochordal cells.3 ECM mainly consists of proteoglycans, collagens, and aggrecan. During degeneration, NP cells transit to nuclear chondrocytes with a decrease in proteoglycan synthesis and an increase in inflammatory cytokines expression, and ECM transit from proteoglycans and collagen type II to a more fibrous tissue primarily consisting of collagen type I.31 Although studies have reported the single-cell transcriptome profiling of NP, the distribution of NP subclusters and cell lineage development during IVDD progression remains undefined.32,33 In the present study, six NP subclusters were identified. We found two major clusters of HomoNP and EffectorNP, playing NP-subjective functions of ECM organization and cartilage development. NPPC had capacities similar to notochordal cells and showed a high proportion in the D1 IVDs and gradually decreased with IVDD. HTNP, enriching for response to inflammation, presented a gradual increase from D1 to D5 IVDs, which coincides with the inflammation in IVDD. Interestingly, we found two NP progenitors, CD24+ progenitor that has been reported in previous studies,3 and a cluster of MKI67+ progenitor that was remarkably capable of cell division. Consistent with previous studies, the percentage of CD24+ progenitor decreased significantly with IVDD progression.34 MKI67+ progenitor showed more characterization of EMT, G2/M checkpoint, and inflammatory responses. EMT is involved in tissue damage repair and the development of tissue fibrosis.35 Thus, MKI67+ progenitor was hypothesized as a subcluster contributing to the self-repair of inflammation-related NP tissues. The functional characteristics and mechanism of MKI67+ progenitor and EMT in IVDD need further investigation. Importantly, we found four cell lineage trajectories originating from CD24+ or MKI67+ progenitor to EffectorNP_⅓. The trajectories from CD24+/MKI67+ progenitor to EffectorNP_1 were much more enriched in normal IVDs, and the trajectories to EffectorNP_3 were much more enriched in severe degenerated IVDs. Our data suggested that NP subclusters were inclined toward ossification differentiation and inflammatory responses with the severity of IVDD. These results suggested potential targeted pathways of prevention and intervention therapy for DDD.
AF comprises outer AF and inner AF and functions to contain NP and maintain NP pressurization under compressive loads.36 The outer AF contains fibroblast-like, thin, and elongated cells, whereas the inner AF cells are spherical and appear similar to articular chondrocytes.36 During IVDD, cell type, matrix composition, and the microstructure of outer AF and inner AF exhibit serial changes, accompanied by cracks and fissures in the AF, innervation, and vascularization.37 However, the heterogeneity and changes in AF cells during degeneration are barely well-documented. In the present study, we identified six subclusters and described changes in them during IVDD. The proportion of inner AF subclusters was high in the D1 IVDs and progressively declined with IVDD. HTAF and proAF, mainly enriched in inflammation response and vasculature development regulation, were remarkably low in D1 IVDs and gradually increased with IVDD. These results were consistent with previous studies about the inflammatory response and fibrocartilaginous metaplasia of AF during IVDD.23 MCAM is regarded as a classical surface marker of mesenchymal stem cells. Notably, MCAM+ progenitor, highly expressing MCAM, ACTA2, MYL9, and TAGLN, was identified in AF. The trajectory from MCAM+ progenitor to innerAF was much more enriched in normal and mild degenerated IVDs. Whereas, trajectories from MCAM+ progenitor to HTAF or proAF were much more enriched in mild and severe degenerated IVDs. MCAM+ progenitor was capable of cell differentiation, proliferation, and stem cell characteristics, providing a potential treatment strategy against AF injury and IVDD.
Several studies have highlighted the important roles of proinflammatory mediators and inflammation responses during IVDD.23,38 Upon degeneration, NP and AF become inflamed, and this inflammatory milieu is associated with a cascade of degenerative events that may eventually result in discogenic pain in patients.23,38 However, the landscape of immune cells and their relationship in the development of IVDD have not been completely understood. Here we presented a comprehensive landscape of immunocytes in IVD, where B cells, CD8 T cells, mast cells, monocytes/macrophages, neutrophils, and plasmacytoid dendritic cells were identified. The percentage of macrophages in D1 was low but remarkably increased in the degenerated IVDs, which caught our attention. Macrophages exist around the degenerated endplate, the outside area of the AF, and in the inner AF and NP.39,40 We distributed macrophages into nine subclusters. The proportions of Mφ-CCL3, Mφ-NR4A2, and Mφ-SPP1 were significantly increased in the degenerated IVD. Interestingly, Mφ-SPP1 was exclusively found in IVDD but not healthy IVDs. RNA velocity confirmed the directional flow from Mφ-SPP1 to Mφ-CCL3 and Mφ-NR4A2, indicating Mφ-SPP1 as an inflammatory regulator during IVDD and tissue self-repair. Yang et al. reported that infiltrating macrophages interacted with native disc cells leading to increasingly inflammatory conditions and promoting catabolism.41 The spontaneous resorption of herniated IVD has been discovered in clinical cases.42,43 Based on the capacity of proinflammatory response and phagocytosis, macrophages are involved in the resorption of herniated disc tissues.44 The polarization capacity of Mφ-SPP1 to Mφ-CCL3 and Mφ-NR4A2 indicates its potential functions in spontaneous hernia resorption.
The healthy and degenerated IVD microenvironment are distinct, which affect both the viability and biological activity of resident disc cells and exogenous cells.45 In particular, studies have highlighted the major role of numerous proinflammatory mediators and immune cells not only in inflammation but also in IVDD development in general.37,46 However, biological interactions between native disc cells and IVD microenvironment, and how they affect IVDD progression are still less well-documented. Therefore, we further explored cell communication between NP cells, AF cells, progenitors, and inflammatory cells both under the healthy and degenerated disc microenvironment. Interestingly, the intercellular communications between these cells were progressively increased and inclined toward inflammatory responses and ossification with IVDD progression. Based on our results, together with previous studies, we proposed a degenerated model of IVD. In a healthy IVD, NP cells, AF cells, and progenitors are in a circulation state of spontaneous aging and renewing with the immune microenvironment in balance. The IVD is in a homeostasis state. In the onset or mild degeneration of IVD, the immune system gets activated, and the cellular interaction between native disc cells and the microenvironment increases. The NP and AF cells are in a renewing and inflammatory response state. In the severe degenerated IVD, the immune microenvironment significantly worsens, leading to an imbalanced microenvironment, and the cellular interaction between native disc cells and the immune system further increases. The cellular interactions are enriched in inflammatory responses, bone mineralization, ossification, and apoptosis. The IVD reaches an imbalanced state. This model can be used to improve the knowledge of cell communications and mechanisms that correlate IVD microenvironment and degeneration, providing possible ideas for DDD treatment. However, the cellular interactions and mechanisms need to be further confirmed and elucidated in future studies.
To conclude, our study not only presented the landscape and heterogenicity of IVD at a single-cell resolution but also presented the changes and development in the major subclusters in both healthy and degenerated IVDs. Moreover, we revealed distinct subpopulations of immune responders in different degeneration grades of IVD and investigated cell communications between the NP, AF, and microenvironment during IVDD. Our findings expand our knowledge of the biological basis and pathophysiological process of IVDD, opening possibilities for re-establishing IVD homeostasis to prevent degeneration. These findings will help develop improved diagnostic and treatment strategies against DDD.
Limitations of the study
Due to its special structure and physiological function, the degeneration of IVD has been found to be closely related to spatial structure and biomechanics. Spatial chondrocyte organization was found to be a strong indicator for local tissue degeneration in the IVD.47 The different biomechanical distribution in IVD forming a mechanical environment of cells, which is important for cell biological functions.6 Although our study found the intercellular communications between cell subpopulations and biological microenvironment variations during the IVDD progression, the distribution of subclusters and cellular biological functions at different spatial location within IVD have not been elucidated. Integrating spatial whole-transcriptomic and single-cell transcriptomics analysis of human IVD during the degeneration need to be further explored in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw scRNA-seq data of IVDD samples | This paper. | GSA: PRJCA014236 |
| Antibodies | ||
| Antibodies of immunohistochemistry assay see Table S2 | This paper. | N/A |
| Software and algorithms | ||
| cellranger | 10x Genomic | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| STAR | Dobin et al.48 | https://github.com/alexdobin/STAR |
| Seurat | Hao et al.49 | https://satijalab.org/seurat |
| DoubletFinder | McGinnis et al.50 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Harmony | Korsunsky et al.51 | https://github.com/immunogenomics/harmony |
| clusterprofiler | Wu et al.52 | https://github.com/YuLab-SMU/clusterProfiler |
| slingshot | Street et al.53 | https://github.com/kstreet13/slingshot |
| velocyto | La Manno et al.54 | http://velocyto.org |
| Velocyto.R | La Manno et al.54 | https://github.com/velocyto-team/velocyto.R |
| CellChat | Jin et al.55 | https://github.com/sqjin/CellChat |
| nichenetR | Browaeys et al.56 | https://github.com/saeyslab/nichenetr |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dandan Wang, E-mail: ddwang@sdfmu.edu.cn.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human IVD tissue specimens
A total of 14 surgical specimens of human NP and AF tissues were obtained from patients who required discectomy for scoliosis (n = 2, Pfirrmann grade I), lumbar disc herniation and spondylolysis (n = 12, Pfirrmann grade II–V). The cases of all included participants were confirmed with magnetic resonance imaging (MRI) according to the Pfirrmann grading system before surgery.57 The clinical characteristics of the patients are listed in Table S1. All specimens were immersed in tissue storage solution (Miltenyi Biotec) immediately and were then sent to Berry Genomics Co. Ltd. for subsequent scRNA-seq. Tissue samples used for immunohistochemistry and immunofluorescence were obtained from five other patients who required percutaneous endoscopic discectomy for lumbar disc herniation (Pfirrmann grade I–V). This study was approved by the Ethics Committee of Shandong First Medical University and Shandong Academy of Medical Sciences (ethical approval document number R202204290100). Informed consent was obtained from all participants included in the study.
Method details
Single-cell RNA sequencing
Human IVD scRNA-seq libraries were prepared using the Chromium Single Cell 3ʹ Reagent Kits v3 (10x Genomics) according to the manufacturer’s protocols. Briefly, the cells were washed with 0.04% bovine serum albumin (BSA) Dulbecco’s phosphate-buffered saline (PBS) three times and then resuscitated to a concentration of 700–1200 cells/μL (viability ≥85%). A single-cell gel bead in the emulsion was generated by combining barcoded Single Cell 3ʹ v3 Gel Beads, a Master Mix containing cells, and partitioning oil onto chromium chip B. The gel beads were dissolved immediately, and cell lysate was mixed with the primer that contains an Illumina TruSeq Read 1 (Read 1 sequencing primer), 16 nucleotide (nt) 10× Barcode, 12 nt unique molecular identifiers (UMI), 30 nt poly (dT) and reverse transcription reagents. Barcoded, full-length complementary DNA (cDNA) was amplified by a polymerase chain reaction to generate sufficient mass for library construction. For gene expression library construction, 50 ng of amplified cDNA was fragmented and end-repaired, and double-size was selected with SPRIselect beads. After quality control, the libraries were sequenced on an Illumina NovaSeq 6000 platform to generate 150-bp paired-end reads, according to the manufacturer’s protocols (Berry Genomics).
Immunohistochemistry and immunofluorescence assay
The tissue sections were deparaffinized with dimethyl benzene and rehydrated with gradient ethanol accordingly. The slides were boiled in antigen retrieval buffer containing 0.01 M sodium citrate-hydrochloric acid (pH = 6.0) for 15 min. After rinsing with PBS, the slides were rinsed in 3% peroxidase quenching solution (Invitrogen), and then 3% BSA (Servicebio, China) to block endogenous peroxidase. After washing with PBS, the slides were incubated with primary antibody (Table S2) at 4°C overnight. After washing, the slides were incubated with a biotinylated secondary antibody (Zhongshan Golden Bridge Biotech, Beijing, China) at room temperature for 30 min. The visualization signal was developed using 3, 3′-diaminobenzidine (DAB) solution. All slides were then counterstained with hematoxylin for 15 min. The slides were finally observed, and images were captured using an inverted fluorescence microscope (ZEISS, Oberkohen, Germany).
For immunofluorescence assay, after incubating with primary antibody (Table S2) overnight, the slides were treated with horseradish peroxidase (HRP)-conjugated secondary antibody (Servicebio, China) at room temperature for 50 min. After washing, the slides were treated with tyramide signal amplification (Servicebio, China) for 10 min. The slides were then washed extensively with PBS, stained with 4′,6-diamidino-2-phenylindole, and images were captured using a laser scanning confocal microscope (NIKON, Japan).
Quantification and statistical analysis
Processing scRNA-seq raw data
The Cell Ranger software (10x Genomic) was used for alignment and quantification of each sample’s scRNA-seq raw data. In brief, 10x cell barcodes and UMI were extracted and adjusted for each record. GRCh38 human genome was used as a reference genome to align reads by STAR.48 For quality control, DoubletFinder50 was used to remove doublet. Seurat49 was used to filter out cells with less than 200 genes and more than 10% mitochondrial gene expression. The high-quality count matrix was generated. Top 2000 high-variable genes generated by FindVariableGenes were used to perform principal component analysis (PCA) with default parameters. We integrated all samples using Seurat and removed batch effects in all data sets using the Harmony R package51 by default parameters. After determining the k-nearest neighbors of every cell, the cells were classified into different clusters. The FindClusters function was performed to cluster cells based on the Louvain algorithm. No linear dimension reduction was performed (runUMAP function) based on cell embedding extracted from harmony results. The cell types were identified according to the marker genes based on FindAllMarkers function (min.pct = 0.25, test.use = MAST58).
Pseudotime and RNA velocity
To determine the cell lineage of subclusters, Slingshot53 was used to find lineage by fitting minimum spanning trees based on clusters through the getLineages function. After constructing the smooth lineages, we inferred the pseudo time of each cell by fitting the principal curves through the getCurves function. The spliced reads and unspliced reads for each gene in every cell were re-counted by velocyto.54 Output matrix was delivered to velocity.R R package. We calculated the velocity of each cell on every gene. The velocity was projected on Uniform Manifold Approximation and Projection (UMAP) embedding that was extracted from Seurat.
Single-cell communication
The cell-cell communication network was identified by the CellChat R package as previously described.55 The cellular communications between different degeneration grades in IVD were analyzed. Briefly, we construct cellchat object base on the expression matrix and meta information from Seurat by using createCellChat function. Then, merge cellchat object of different grade sample by mergeCellChat function. After these, we use compareInteractions to compare different interactions between different grades and netVisual_diffInteraction function to visualize results.
The nichenetR R package56 was used to identify the changes in the downstream genes caused by ligand-receptor interactions. We used nichenet_seuratobj_aggregate function to get the downstream genes based on Seurat object. The function analysis the differential expression genes between grades of target cluster (set by reciver parameter) regulated by source clusters (set by sender parameter, here we set to “all”).
Statistics
Wilcoxon test was performed for comparison of the percentage of subclusters between healthy and degenerated IVDs, and for comparison of relative expression of marker genes between subclusters. Kolmogorov-Smirnov test was used to compare differentiation trajectories in normal, mild and severe degenerated IVDs. All statistical analyses were performed using the R software, and P < 0.05 was considered statistically significant.
Acknowledgments
This study was supported by the National Science Fund for Distinguished Young Scholars of the National Natural Science Foundation of China (grant number 32122022); and Academic Promotion Project of Shandong First Medical University (grant number 2019QL003).
Author contributions
XSC, JRY, and DDW designed the research, acquired funding, and revised the article; DDW, SNC, and LYX performed the experiment; ZZL, YZC, and HZL analyzed the data; WMH and LW collected IVD samples and analyzed clinical data; DDW and ZZL wrote the article.
Declaration of interests
The authors declare that there are no competing interests.
Published: April 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106692.
Contributor Information
Dandan Wang, Email: ddwang@sdfmu.edu.cn.
Xiaoshu Chen, Email: chenxshu3@mail.sysu.edu.cn.
Jian-Rong Yang, Email: yangjianrong@mail.sysu.edu.cn.
Supplemental information
The C01-C02 represent the two patients who required discectomy for scoliosis (Pfirrmann grade I). The R01-R12 represent the twelve patients who required percutaneous endoscopic discectomy, open surgery for lumbar disc herniation, or spondylolysis (Pfirrmann grade II-V).
The table contains seven sheets. Different sheets represent the DEGs of nucleus pulposus, annulus fibrosus, CD24+ progenitor, MKI67+ progenitor, MCAM+ progenitor, monocyte_macrophage and Mφ-SPP1 that are regulated by ligands in other different subclusters respectively.
Data and code availability
-
•
Raw scRNA-seq data of all samples were uploaded to the Genome Sequence Archive (GSA) of the National Genomics Data Center (PRJCA014236), which are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
-
•
Scripts for data analyses in this study have been uploaded to Github (https://github.com/Mpaperlee/IVDD_ana).
-
•
Any additional information required to analysis data reported in this paper is available from the lead contact upon request.
References
- 1.Wang F., Cai F., Shi R., Wang X.H., Wu X.T. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthritis Cartilage. 2016;24:398–408. doi: 10.1016/j.joca.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Dowdell J., Erwin M., Choma T., Vaccaro A., Iatridis J., Cho S.K. Intervertebral disk degeneration and repair. Neurosurgery. 2017;80:S46–S54. doi: 10.1093/neuros/nyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z., Zheng Z., Qi J., Wang J., Zhou Q., Hu F., Liang J., Li C., Zhang W., Zhang X. CD24 identifies nucleus pulposus progenitors/notochordal cells for disc regeneration. J. Biol. Eng. 2018;12:35. doi: 10.1186/s13036-018-0129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Q., Xiao Q., Ge J., Wu C., Wang Y., Yu H., Yang H., Zou J. Bioinformatics-based research on key genes and pathways of intervertebral disc degeneration. Cartilage. 2021;13:582S–591S. doi: 10.1177/1947603520973247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risbud M.V., Shapiro I.M. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vergroesen P.P.A., Kingma I., Emanuel K.S., Hoogendoorn R.J.W., Welting T.J., van Royen B.J., van Dieën J.H., Smit T.H. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage. 2015;23:1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Nakamichi R., Ito Y., Inui M., Onizuka N., Kayama T., Kataoka K., Suzuki H., Mori M., Inagawa M., Ichinose S., et al. Mohawk promotes the maintenance and regeneration of the outer annulus fibrosus of intervertebral discs. Nat. Commun. 2016;7:12503. doi: 10.1038/ncomms12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C.Q., Jiang L.S., Dai L.Y. Programmed cell death in intervertebral disc degeneration. Apoptosis. 2006;11:2079–2088. doi: 10.1007/s10495-006-0290-7. [DOI] [PubMed] [Google Scholar]
- 9.Vadalà G., Russo F., Battisti S., Stellato L., Martina F., Del Vescovo R., Giacalone A., Borthakur A., Zobel B.B., Denaro V. Early intervertebral disc degeneration changes in asymptomatic weightlifters assessed by t1rho-magnetic resonance imaging. Spine. 2014;39:1881–1886. doi: 10.1097/BRS.0000000000000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi Y., Ma J., Chen Y. PTEN promotes intervertebral disc degeneration by regulating nucleus pulposus cell behaviors. Cell Biol. Int. 2020;44:583–592. doi: 10.1002/cbin.11258. [DOI] [PubMed] [Google Scholar]
- 11.Chu G., Shi C., Lin J., Wang S., Wang H., Liu T., Yang H., Li B. Biomechanics in annulus fibrosus degeneration and regeneration. Adv. Exp. Med. Biol. 2018;1078:409–420. doi: 10.1007/978-981-13-0950-2_21. [DOI] [PubMed] [Google Scholar]
- 12.Cunha C., Silva A.J., Pereira P., Vaz R., Gonçalves R.M., Barbosa M.A. The inflammatory response in the regression of lumbar disc herniation. Arthritis Res. Ther. 2018;20:251. doi: 10.1186/s13075-018-1743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai D., Nakai T., Mochida J., Alini M., Grad S. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine. 2009;34:1448–1456. doi: 10.1097/BRS.0b013e3181a55705. [DOI] [PubMed] [Google Scholar]
- 14.Minogue B.M., Richardson S.M., Zeef L.A., Freemont A.J., Hoyland J.A. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 2010;12:R22. doi: 10.1186/ar2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter C.J., Matyas J.R., Duncan N.A. The three-dimensional architecture of the notochordal nucleus pulposus: novel observations on cell structures in the canine intervertebral disc. J. Anat. 2003;202(Pt 3):279–291. doi: 10.1046/j.1469-7580.2003.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risbud M.V., Schaer T.P., Shapiro I.M. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev. Dynam. 2010;239:2141–2148. doi: 10.1002/dvdy.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erwin W.M., Islam D., Eftekarpour E., Inman R.D., Karim M.Z., Fehlings M.G. Intervertebral disc-derived stem cells: implications for regenerative medicine and neural repair. Spine. 2013;38:211–216. doi: 10.1097/BRS.0b013e318266a80d. [DOI] [PubMed] [Google Scholar]
- 18.Shamji M.F., Setton L.A., Jarvis W., So S., Chen J., Jing L., Bullock R., Isaacs R.E., Brown C., Richardson W.J. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010;62:1974–1982. doi: 10.1002/art.27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Cosamalón J., del Valle M.E., Calavia M.G., García-Suárez O., López-Muñiz A., Otero J., Vega J.A. Intervertebral disc, sensory nerves and neurotrophins: who is who in discogenic pain? J. Anat. 2010;217:1–15. doi: 10.1111/j.1469-7580.2010.01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torre O.M., Mroz V., Bartelstein M.K., Huang A.H., Iatridis J.C. Annulus fibrosus cell phenotypes in homeostasis and injury: implications for regenerative strategies. Ann. N. Y. Acad. Sci. 2019;1442:61–78. doi: 10.1111/nyas.13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fucs P., Silva M., Pinhal M., Meves R., Yamada H.H. Spinal infections in children: a review. Int. Orthop. 2012;36:387–395. doi: 10.1007/s00264-011-1388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields A.J., Ballatori A., Liebenberg E.C., Lotz J.C. Contribution of the endplates to disc degeneration. Curr. Mol. Biol. Rep. 2018;4:151–160. doi: 10.1007/s40610-018-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A.N., Jacobsen H.E., Khan J., Filippi C.G., Levine M., Lehman R.A., Jr., Riew K.D., Lenke L.G., Chahine N.O. Inflammatory biomarkers of low back pain and disc degeneration: a review. Ann. N. Y. Acad. Sci. 2017;1410:68–84. doi: 10.1111/nyas.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knezevic N.N., Candido K.D., Vlaeyen J.W.S., Van Zundert J., Cohen S.P. Low back pain. Lancet. 2021;398:78–92. doi: 10.1016/S0140-6736(21)00733-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Kang J., Guo X., Zhu D., Liu M., Yang L., Zhang G., Kang X. Intervertebral disc degeneration models for pathophysiology and regenerative therapy -benefits and limitations. J. Invest. Surg. 2022;35:935–952. doi: 10.1080/08941939.2021.1953640. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Han S., Kong M., Tu Q., Zhang L., Ma X. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthritis Cartilage. 2021;29:1324–1334. doi: 10.1016/j.joca.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Calió M., Gantenbein B., Egli M., Poveda L., Ille F. The cellular composition of bovine coccygeal intervertebral discs: a comprehensive single-cell RNAseq analysis. Int. J. Mol. Sci. 2021;22:4917. doi: 10.3390/ijms22094917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan Y., He J., Zhu J., Xu Z., Wang Z., Yan J., Hu O., Bai Z., Chen L., Xie Y., et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res. 2021;9:37. doi: 10.1038/s41413-021-00163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binch A.L.A., Cole A.A., Breakwell L.M., Michael A.L.R., Chiverton N., Cross A.K., Le Maitre C.L. Expression and regulation of neurotrophic and angiogenic factors during human intervertebral disc degeneration. Arthritis Res. Ther. 2014;16:416. doi: 10.1186/s13075-014-0416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fournier D.E., Kiser P.K., Shoemaker J.K., Battié M.C., Séguin C.A. Vascularization of the human intervertebral disc: a scoping review. JOR Spine. 2020;3:e1123. doi: 10.1002/jsp2.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu C., Luo S., Wei L., Wu H., Gu W., Zhou W., Sun B., Hu B., Zhou H., Liu Y., et al. Integrated transcriptome and proteome analyses identify novel regulatory network of nucleus pulposus cells in intervertebral disc degeneration. BMC Med. Genom. 2021;14:40. doi: 10.1186/s12920-021-00889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S., Zhang Y., Zhang X., Zhang H., Meng S., Kong M., Liu X., Ma X. Single-cell RNA sequencing of the nucleus pulposus reveals chondrocyte differentiation and regulation in intervertebral disc degeneration. Front. Cell Dev. Biol. 2022;10:824771. doi: 10.3389/fcell.2022.824771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu J., Li W., Yang S., Yang P., Yan Q., Wang S., Lai K., Bai X., Wu C., Ding W., et al. Single-cell transcriptome profiling reveals multicellular ecosystem of nucleus pulposus during degeneration progression. Adv. Sci. 2022;9:e2103631. doi: 10.1002/advs.202103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson S.M., Ludwinski F.E., Gnanalingham K.K., Atkinson R.A., Freemont A.J., Hoyland J.A. Notochordal and nucleus pulposus marker expression is maintained by sub-populations of adult human nucleus pulposus cells through aging and degeneration. Sci. Rep. 2017;7:1501. doi: 10.1038/s41598-017-01567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M., Luan F., Zhao Y., Hao H., Zhou Y., Han W., Fu X. Epithelial-mesenchymal transition: an emerging target in tissue fibrosis. Exp. Biol. Med. 2016;241:1–13. doi: 10.1177/1535370215597194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waxenbaum J.A., Reddy V., Futterman B. StatPearls. 2022. Anatomy, back, intervertebral discs. [Google Scholar]
- 37.Navone S.E., Marfia G., Giannoni A., Beretta M., Guarnaccia L., Gualtierotti R., Nicoli D., Rampini P., Campanella R. Inflammatory mediators and signalling pathways controlling intervertebral disc degeneration. Histol. Histopathol. 2017;32:523–542. doi: 10.14670/HH-11-846. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues L.M.R., Oliveira L.Z.d., Silva M., Accardo C.d.M., Giglio A.B.D., Pinhal M. Inflammatory biomarkers in sera of patients with intervertebral disc degeneration. Einstein. 2019;17:eAO4637. doi: 10.31744/einstein_journal/2019AO4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto Y., Kokubo Y., Nakajima H., Honjoh K., Watanabe S., Matsumine A. Distribution and polarization of hematogenous macrophages associated with the progression of intervertebral disc degeneration. Spine. 2022;47:E149–E158. doi: 10.1097/BRS.0000000000004222. [DOI] [PubMed] [Google Scholar]
- 40.Ling Z., Liu Y., Wang Z., Zhang Z., Chen B., Yang J., Zeng B., Gao Y., Jiang C., Huang Y., et al. Single-cell RNA-seq analysis reveals macrophage involved in the progression of human intervertebral disc degeneration. Front. Cell Dev. Biol. 2021;9:833420. doi: 10.3389/fcell.2021.833420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Liu B., Liu Y., He D., Xing Y., An Y., Tian W. Secreted factors from intervertebral disc cells and infiltrating macrophages promote degenerated intervertebral disc catabolism. Spine. 2019;44:E520–E529. doi: 10.1097/BRS.0000000000002953. [DOI] [PubMed] [Google Scholar]
- 42.Yang X., Zhang Q., Hao X., Guo X., Wang L. Spontaneous regression of herniated lumbar discs: report of one illustrative case and review of the literature. Clin. Neurol. Neurosurg. 2016;143:86–89. doi: 10.1016/j.clineuro.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Cvetanovich G.L., Hsu A.R., Frank R.M., An H.S., Andersson G.B. Spontaneous resorption of a large cervical herniated nucleus pulposus. Am. J. Orthoped. 2014;43:E140–E145. [PubMed] [Google Scholar]
- 44.Ohba T., Haro H. TWEAK and TSLP in disc degeneration and spontaneous hernia resorption. JOR Spine. 2020;3:e1068. doi: 10.1002/jsp2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vadalà G., Ambrosio L., Russo F., Papalia R., Denaro V. Interaction between mesenchymal stem cells and intervertebral disc microenvironment: from cell therapy to tissue engineering. Stem Cell. Int. 2019;2019:2376172. doi: 10.1155/2019/2376172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altun I. Cytokine profile in degenerated painful intervertebral disc: variability with respect to duration of symptoms and type of disease. Spine J. 2016;16:857–861. doi: 10.1016/j.spinee.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Bonnaire F.C., Danalache M., Sigwart V.A., Breuer W., Rolauffs B., Hofmann U.K. The intervertebral disc from embryonic development to disc degeneration: insights into spatial cellular organization. Spine J. 2021;21:1387–1398. doi: 10.1016/j.spinee.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 48.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 57.Pfirrmann C.W., Metzdorf A., Zanetti M., Hodler J., Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 58.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The C01-C02 represent the two patients who required discectomy for scoliosis (Pfirrmann grade I). The R01-R12 represent the twelve patients who required percutaneous endoscopic discectomy, open surgery for lumbar disc herniation, or spondylolysis (Pfirrmann grade II-V).
The table contains seven sheets. Different sheets represent the DEGs of nucleus pulposus, annulus fibrosus, CD24+ progenitor, MKI67+ progenitor, MCAM+ progenitor, monocyte_macrophage and Mφ-SPP1 that are regulated by ligands in other different subclusters respectively.
Data Availability Statement
-
•
Raw scRNA-seq data of all samples were uploaded to the Genome Sequence Archive (GSA) of the National Genomics Data Center (PRJCA014236), which are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
-
•
Scripts for data analyses in this study have been uploaded to Github (https://github.com/Mpaperlee/IVDD_ana).
-
•
Any additional information required to analysis data reported in this paper is available from the lead contact upon request.