Abstract
Nicotinamide phosphoribosyltransferase (NAMPT) is a key metabolic enzyme in NAD+ synthesis pathways and is found upregulated in several tumors, depicting NAD(H) lowering agents, like the NAMPT inhibitor FK866, as an appealing approach for anticancer therapy. Like other small molecules, FK866 triggers chemoresistance, observed in several cancer cellular models, which can prevent its clinical application. The molecular mechanisms sustaining the acquired of resistance to FK866 were studied in a model of triple negative breast cancer (MDA-MB-231 parental – PAR), exposed to increasing concentrations of the small molecule (MDA-MB-231 resistant – RES). RES cells are not sensitive to verapamil or cyclosporin A, excluding a potential role of increased efflux pumps activity as a mechanism of resistance. Similarly, the silencing of the enzyme Nicotinamide Riboside Kinase 1 (NMRK1) in RES cells does not increase FK866 toxicity, excluding this pathway as a compensatory mechanism of NAD+ production. Instead, Seahorse metabolic analysis revealed an increased mitochondrial spare respiratory capacity in RES cells. These cells presented a higher mitochondrial mass compared to the FK866-sensitive counterparts, as well as an increased consumption of pyruvate and succinate for energy production. Interestingly, co-treatment of PAR cells with FK866 and the mitochondrial pyruvate carrier (MPC) inhibitors UK5099 or rosiglitazone, as well as with the transient silencing of MPC2 but not of MPC1, induces a FK866-resistant phenotype. Taken together, these results unravel novel mechanisms of cell plasticity to counteract FK866 toxicity, that, besides the previously described LDHA dependency, rely on mitochondrial rewiring at functional and energetic levels.
Keywords: FK866, NAMPT, MPC2, Drug resistance, Mitochondrial fitness, Metabolic adaptation
Introduction
Reprogramming of energy metabolism has been considered a hallmark of cancer, as neoplastic diseases are characterized by a deregulated control of cell growth and division, that require adjustments of energy metabolism [1]. The metabolic rewiring of cancer cells is often sustained by an increased glycolysis, in the presence of oxygen, known as the Warburg effect, that allows both ATP and biomass production due to an increased glucose uptake [2]. Still, aerobic glycolysis does not always emerge due to dysfunctional or impaired mitochondria functioning, as initially proposed by Warburg. Instead, hybrid glycolytic/OXPHOS phenotypes provide tumor tissues a higher level of metabolic flexibility, as mitochondria rewiring allow the utilization of carbon sources other than glucose for energy production [3,4]. Moreover, the metabolic state of cancer cells dictate the rate of mitochondrial biogenesis and turnover, and consequently the mitochondrial mass, which often is a positive regulator of tumorigenesis [5]. Of note, the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) is a positive regulator of mitochondrial biogenesis as it mediates the expression of nuclear encoded mitochondrial genes to enhance mitochondrial ATP production. PGC-1α expression positively correlates with breast cancer invasiveness and metastasis, emphasizing its role in tumorigenesis behind bioenergetics [6].
Nicotinamide adenine dinucleotide (NAD+) is fundamental for energy metabolism, as it is a cofactor for enzymes involved in glycolysis, fatty acid oxidation and the tricarboxylic acids (TCA) cycle [7], [8], [9]. Additionally, NAD+ plays a critical role as substrate for NAD+-consuming enzymes like sirtuins, poly-ADP-ribose polymerases (PARPs) and CD38/157, that among others, sustain DNA repair, genomic integrity and regulation of transcription and signalling [10], [11], [12]. NAD+ can be synthetized from several precursors as nicotinamide (NAM), tryptophan, nicotinic acid (NA) and nicotinamide riboside (NR) [13,14]. Among the enzymes involved in the synthesis pathways, the nicotinamide phosphoribosyltransferase (NAMPT), that catalyzes the conversion of nicotinamide (NAM) to nicotinamide mononucleotide (NMN), is considered the rate-limiting enzyme of the NAD+ “salvage” pathway [15], [16], [17].
Due to its central role in NAD+ synthesis and bioenergetics, NAMPT has been found to be upregulated in many solid tumors, revealing the significance of the development of NAMPT inhibitors for cancer treatment [18], [19], [20], [21]. Moreover, NAMPT inhibitors like FK866 (APO866) have been extensively used in preclinical cancer research to mimic cellular NAD(H) shortage, characteristic of the high energetic demand of rapidly proliferating cancer cells [22]. Acquired resistance to FK866 was observed in several FK866-resistant cell line models, mainly due to mutation on NAMPT or NAD+-related genes, through the exploitation of NAMPT-independent NAD+-synthesis pathways or by modulating the expression of multidrug resistance proteins [23], [24], [25], [26].
We previously showed that cancer cells’ acquirement of resistance to NAMPT inhibition, including MDA-MB-231, occurred in absence of target gene mutation or acquirement of multi drug resistance mechanisms, and invariably induces dependency of LDHA [25]. Nonetheless, the exploitation of additional NAD+-producing pathways as well as the efficiency of mitochondrial metabolism were not investigated. Thus, the aim of the present study was to unravel the potential role of NAD+-producing enzymes and mitochondria in sustaining metabolic adaptation of triple negative breast cancer cells to NAD(H) shortage, that can prevent the success of NAMPT inhibitors for clinical application.
Materials and methods
Cell lines culture conditions
MDA-MB-231 (PAR) human breast cancer cell line was obtained from American Type Culture Collection (ATCC). MDA-MB-231 FK866-resistant cell line (RES) was developed by increased exposure to FK866 during a 3 month period of time, as previously described [25]. Cell lines were cultured in DMEM (Gibco) and supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 U/mL penicillin-streptomycin (all from Lonza). The culture media of RES cells was additionally supplemented with 100 nM of FK866. Cells were maintained at 37 °C under humidified conditions with 5% CO2.
Drug treatments and in vitro cell viability
PAR and RES cells were treated with FK866 (sc-205325, Santa Cruz Biotechnology), CHS-828 (200484-11-3, Cayman Chemical), verapamil (V4629, Sigma), antimycin A (A8674, Sigma), 5-FU (F6627, Sigma), cisplatin (PHR1624, Sigma), olaparib (HY-10162, MedChemExpress), UK5099 (5048170002, Sigma) and rosiglitazone (R2408, Sigma) for 48 h. In vitro drug sensitivity was assessed by OzBlue Cell Viability kit (OzBiosciences) after 2 h of incubation, according to the manufacturer instructions.
Cell proliferation assay
For proliferation assays, 3.0 × 104 cells were seeded and counted every 24 h using 0.2% trypan blue exclusion to determine the number of viable cells. Doubling time was calculated at the exponential growth phase, between 48 and 72 h of seeding.
Determination of NAD(H) and ATP/AMP levels
For intracellular NAD(H) content determination, cells were lysed in 0.6 M perchloric acid (PCA) (for NAD+) or in 0.1 M NaOH (for NADH) at 4 °C. Cell extracts were centrifuged for 3 min at 16000 x g and the supernatants were collected for the determination of NAD(H) content, as described [27]. NAD(H) values were normalized to protein content, determined through the Bradford assay.
To evaluate the ATP/AMP ratio as a marker of cellular energy status, intracellular ATP and AMP content was assayed spectrophotometrically at 340 nm. ATP was measured in a medium containing 0.02 mL of neutralized perchloric extract, 50 mM Tris-HCl pH 8.0, 1 mM NADP, 0.5 mM MgCl2, 5 mM glucose, and 4 µg of purified hexokinase/glucose-6-phosphate dehydrogenase in 1 mL final volume, before and after the addition of the hexokinase/glucose-6-phosphate dehydrogenase. For AMP assay, 0.02 mL of neutralized perchloric extract was added to an assay medium containing 50 mM Tris-HCl, pH 8.0, 0.2 mM NADH, 0.5 mM MgCl2, 1 mM phosphoenolpyruvate, 0.2 mM ATP, 5 µg of purified pyruvate kinase/lactate dehydrogenase mixture, and 2 µg of purified adenylate kinase [28]. ATP and AMP concentrations were normalized to protein content, determined through the Bradford assay.
For measurements of total ATP levels, the CellTiter-Glo Luminescent Cell Viability Assay (G7571, Promega) was used, following the manufacturer's protocol. Protein content of the cell lysate was measured by the Bradford method and used for normalization of the luminescence values.
Irradiation (X-ray)
PAR and RES cells were irradiated with X-rays using Xstrahl RS225 (195 MeV – 10 mA) with a dose rate of 1.6 Gy per minute (dose ranging from 0 to 8 Gy). Immediately after irradiation, cells were collected and seeded for the colony formation assay.
Colony formation assay
Colony formation assay was performed by seeding 200 cells per well per condition in 6 well plates. Two days after, cells were treated with the described concentration of FK866, CH868, 5FU, cisplatin or olaparib. A week after the treatment (with either small molecules/chemotherapeutics or irradiation), colonies were fixed with 4% PFA and stained with crystal violet (C6158, Sigma) for counting.
RNA extraction and real-time qPCR
TriZOL reagent (Invitrogen) was used for total RNA extraction, according to the manufacturer's instructions. cDNA synthesis was carried out with RevertAid RT Reverse Transcription Kit (K1691, ThermoScientific) following manufacturer's instructions. Real-time quantitative PCR (RT-qPCR) analyses were performed in triplicate using the ExcelTaq™ 2X qPCR Master Mix (SMOBIO) on a CFX96 Real-Time PCR Detection System (Bio-Rad). The sequences of the primers employed in the study are present in Table 1. ΔCq method was used for samples’ mRNA quantification, with RPLP0 or β-actin as housekeeping genes.
Table 1.
List of primers used to evaluate the expression of genes of interest by RT-qPCR analysis.
| Gene | Primers (5’ to 3’) |
|---|---|
| NAMPT | FW: TTATGGAACGAAAGATCCTG |
| RV: CAAAAGCATCTTTTTCATGGTC | |
| NAPRT | FW:CTATGCCTTGGCTTTTCCCC |
| RV:GAAGACCTTGCGGATCTCCT | |
| QPRT | FW:CCCTCTGGGTCACACATCTT |
| RV:GTGCTCATTATCACCGCAGA | |
| NMRK1 | FW:TGTGTTTGGGAGGACTCGAA |
| RV:CACTGCAATTTGGGAGGTGT | |
| MPC1 | FW:CAGTGGGCGGATGACATTTG |
| RV:GCGTGGCATGCAAACAGAAG | |
| MPC2 | FW:ACCTACCACCGGCTCCTCGAT |
| RV:CCAGCACACACCAACCCCCATT | |
| PPARGC1A | FW: CCAAAGGATGCGCTCTCGTTCA |
| RV: CGGTGTCTGTAGTGGCTTGACT | |
| TFAM | FW: CCGAGGTGGTTTTCATCTGT |
| RV: AGTCTTCAGCTTTTCCTGCG | |
| RPLP0 | FW:CATTCTCGCTTCCTGGAG |
| RV:CTTGACCTTTTCAGCAAGTGG | |
| ACTB | FW: CTGGAACGGTGAAGGTGACA |
| RV: AGGGACTTCCTGTAACAATGCA |
Protein extraction and western blot analysis
Samples were lysed for 10 min on ice with RIPA lysis buffer supplemented with Protease Inhibitor Cocktail (ThermoScientific), followed by sonication and clarification. SDS-polyacrylamide gel loaded with equal amounts of proteins was used for protein separation, followed by the transference to a PVDF membrane. The primary antibodies used were: NAMPT/Visfatin (A300-779A, Bethyl Laboratories), NAPRT (ab211529, Abcam), QAPRT (TA501520, OriGene), NMRK1 (sc-398852, SantaCruz Biotecnhologies), MPC1 (PA5116938, Cell Signalling), MPC2 (ab236584, Abcam), PDH1Ea (sc-377092, SantaCruz Biotechnologies), TOMM20 (HPA011562, Sigma), β-actin (BK4970S CST) and Calnexin (ab133615, Abcam). β-actin and Calnexin were used as protein loading controls. The anti-mouse and anti-rabbit secondary antibodies (111-035-003) used were obtained from Jackson Immunoresearch Laboratories. Signal was revealed with chemiluminescence detection kit reagents (Amersham ECL Select, GE Healthcare) at ChemiDoc (BioRad). The relative density of protein bands was analyzed using the ImageJ software and represented as the fold change of the corresponding control. A representative western blot of three independent biological experiments is shown.
shRNA and siRNA transfection
HEK293T cells were used to produce lentiviral vector particles (LVP). Cells were co-transfected with a Δ891 and a VSV-G encoding vector along with the shNMRK1 plasmid. PLKO vector was used as the transfer plasmid. Viral supernatants were collected, filtrated, and quantified 72 h post-transfection. Selection with puromycin (1 µg/mL) was performed on RES cells stably transduced with shNMRK1.
Downregulation of MPC1 and/or MPC2 in PAR cells was performed with transient transfection of siMPC1 (sc-95332, Santa Cruz Biotechnology) and/or siMPC2 (sc-78813, Santa Cruz Biotechnology) using INTERFERin in vitro siRNA miRNA transfection reagent (Polyplus), according to the manufacturer's instructions.
Glycolysis rate and mitochondrial respiration
The glycolytic rate, spare respiratory capacity and the mitochondrial respiration were assessed in both PAR and RES cells, respectively with the Seahorse Glycolysis Rate assay kit (103344-100, Agilent) and with the Seahorse MitoStress kit (103015-100, Agilent), following the manufacturer's instructions. Shortly, cells were seeded in the seahorse assay plates for 48 h in Seahorse DMEM supplemented with 10 mM glucose, 2 mM glutamine, 1 mM pyruvate (all from Agilent). The day before the assay, a cartridge was hydrated using Seahorse calibrant XF (Agilent) and incubated overnight at 37 °C in a CO2 deprived atmosphere. The oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) were determined on a XFe96 Seahorse Bioanalyzer (Agilent). The final concentration of injected compounds was: 1 µM oligomycin; 2 µM FCCP (carbonyl cyanide-4(trifluoromethoxy) phenylhydrazone); 2 µM UK5099; 5 µM of each rotenone and antimycin A; 10 mM glucose; and 50 mM 2-DG (2-deoxy-glucose). OCR and ECAR were normalized to the samples’ protein content using the Bradford reagent.
In other experiments, to assess the contribution of the pathways leaded by respiratory complex I or II, the OCR was measured with an amperometric electrode (Unisense Microrespiration, Unisense A/S, Denmark) in a closed chamber, at 25 °C. For each experiment, 2 × 105 cells were resuspended in phosphate buffer saline (PBS) and permeabilized with 0.03 mg/mL digitonin for 1 min. To activate the pathway composed by respiratory complexes I, III, and IV, 10 mM pyruvate plus 5 mM malate were added. To induce the pathway formed by complexes II, III, and IV 20 mM succinate plus 100 μM rotenone (the specific inhibitor of complex I) were employed [29].
DNA extraction
DNA was extracted from PAR and RES cells to determine mitochondrial mass by estimating the mitochondrial DNA (mtDNA) to nuclear DNA (nuclearDNA) ratio. Cells were lysed in 100 µL of lysis buffer (100 mM Tris-HCl pH 8, 200 mM NaCl, 5 mM EDTA, 0.2% SDS, 0.2 mg/mL proteinase K freshly added) for 2 h at 56 °C. Debris removal was obtained by centrifugation at 14000 rpm for 10 min at room temperature, followed by DNA precipitation with 500 µL of isopropanol. After centrifugation at 14000 rpm for 5 min at room temperature, the pellet was washed with 70% ethanol. Subsequently, samples were centrifuged at 14000 rpm for 5 min at room temperature, the supernatant was discarded, and the DNA pellet was dried at room temperature. Finally, the DNA pellet was resuspended in 50 μl DNase-free water and one hundred nanograms was used for further analysis.
Mitochondrial mass determination
To determine the mitochondrial mass of PAR and RES cells, the ratio between mitochondrial and nuclear DNA as well as confocal analysis using a mitochondrial probe were used.
The nuclear-encoded gene, human B2M, and the mitochondrial-encoded gene, human tRNALeu were used to calculate the relative mtDNA:nuclearDNA ratio. RT-qPCR was performed in triplicate using the ExcelTaq™ 2X qPCR Master Mix (SMOBIO) on a CFX96 Real-Time PCR Detection System (Bio-Rad) and the 2−ΔΔCt method was used to calculate the gene expression. The sequences of the used primers were: B2M FW: 5’-TGCTGTCTCCATGTTTGATGTATCT-3’, B2M RV: 5’-TCTCTGCTCCCCACCTCTAAGT-3’, tRNALeu FW: 5’-CACCCAAGAACAGGGTTTGT-3’, and tRNALeu RV: 5’-TGGCCATGGGTATGTTGTTA-3’.
Further assessment of PAR and RES cells mitochondrial mass was determined by incubating cells with 100 nM of MitoTracker Green FM (Life Technologies, M7514) and Hoechst (1:5000) for mitochondria and nuclei staining, respectively. 200 cells per well were seeded in 384-well optic glass plates (CellCarrier384, Perkin Elmer) and Z-stacks were acquired with a Leica TCS SP8 laser scanning confocal system, installed on a Leica DMi8 inverted fluorescence microscope, using a HC PL APO 20x/0.75 CS2 objective. The acquisition parameters were kept constant throughout all the conditions.
After single cell segmentation, the mean intensity of the green fluorescent MitoTracker signal was quantified on the sum projection of all the stacks comprising the total area of each cell.
Cell segmentation and signal intensity measurement has been performed using a semi-automatic macro in Fiji/ImageJ [30]. Representative images of PAR and RES cells were linearly adjusted only for brightness and contrast. At least 300 nuclei were analyzed per condition.
Mitochondrial substrate dependencies
Mitochondrial substrate utilization was assessed with Biolog MitoPlates S-1 (Biolog, Hayward, CA, USA, #14105), as suggested by the manufacturer. MitoPlates were incubated in Omnilog® at 37 °C for 1 h with Biolog Mitochondria Assay Solution (MAS) containing Biolog redox dye MC and saponin (Sigma-Aldrich, 84510, 20 µg/µL) to allow substrate rehydration and cell permeabilization. Viable cell number was previously determined by trypan blue exclusion and resuspended in Biolog MAS, after washing with PBS 1x. Cells were added to the MitoPlates and the kinetic reading of the rate of purple color formation was measured in Omnilog® system (OD590nm) for 4 h. The background was corrected for the blank sample (no substrate).
Respiratory complexes activity
To determine the activity of respiratory complexes I, II, III and IV, 50 μg of total protein homogenate was used [31]. The reactions were followed for 5 min, with 1 min-interval collecting data, and absorbance values were measured in a spectrophotometer. Complex I (NADH-ubiquinone reductase) activity was assayed in a medium containing 50 mM Tris–HCl pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.6 mM NADH, 0.8 mM ferricyanide, and 50 µM antimycin A. The reduction of ferrocyanide in the presence of NADH was followed at 420 nm (ε = 1 mM−1⋅cm−1). Similarly, Complex II (succinate dehydrogenase) activity was assayed in similar conditions but using 20 mM succinate instead of NADH. Complex III (cytochrome c reductase) activity was assayed in a 50 mM Tris buffer pH7.4, 5 mM KCl, 2 mM MgCl2, 0.5 M NaCN, 0.03% oxidized Cyt C, 0.6 mM NADH, and 10 mM succinate. The reduction of cytochrome c in the presence of NADH was followed at 550 nm (ε = 19.1 mM−1⋅cm−1). For Complex IV (cytochrome c oxidase) activity assessment, the oxidation of reduced cytochrome c was followed at 550n m (ε = 19.1 mM−1⋅cm−1), in a 50 mM Tris buffer pH7.4, 5 mM KCl, 2 mM MgCl2, containing 0.03% reduced cytochrome c.
ATP synthesis through Fo‐F1 ATP Synthase
To evaluate the ATP synthesis through the Fo-F1 ATP synthase activity, cells were incubated for 10 min at 37 °C in a medium containing: 100mM Tris-HCl pH 7.4, 100 mM KCl, 1 mM EGTA, 2.5 mM EDTA, 5 mM MgCl2, 0.2 mM di(adenosine-5) penta-phosphate, 0.6 mM ouabain, ampicillin (25 μg/mL), 5 mM KH2PO4, and 10 mM pyruvate plus 5 mM malate, used as respiratory substrates. ATP synthesis started after the addition of 0.2 mM ADP and was monitored for 2 min in a luminometer (Glomax 20/20, Promega) by the luciferin/luciferase chemiluminescent method (luciferin/luciferase ATP bioluminescence assay kit CLSII, Roche, Basel, Switzerland). An ATP standard solution between 10−10 and 10−7 M was used for calibration. To assess the specificity of the method for ATP-synthase assay, ATP synthesis was also evaluated in the presence of 10 μM Oligomycin, a specific inhibitor of ATP synthases Fo moiety [28].
Determination of the mitochondrial membrane potential (MMP, Δψm)
The lipophilic cationic probe TMRE (ab113852, Abcam) was used to determine the mitochondrial membrane potential (Δψm). TMRE rapidly equilibrated between cellular compartments due to differential membrane potential, and a decrease in fluorescence indicated reduced Δψm. Upon 48 h treatment with FK866 and/or UK5099, cells were rising with PBS and TMRE (800 nM) was added directly to the assay wells. After 45 min’ incubation at 37 °C, the fluorescent signal was assessed in a microplate at the Spark (Tecan) plate reader, with excitation at 544 nm and emission at 590 nm. The uncoupling agent FCCP (20 µM) was used as negative control.
Statistical analysis
Data was expressed as mean ± standard error of mean (SEM) of three independent biological experiments conducted in technical triplicates, unless indicated otherwise. Statistical significance between control and test groups was determined using Student's t-test or one-way ANOVA, considering values of *p < 0.05, **p < 0.01, and ***p < 0.001 as significant. Data were plotted by Graph prism 6 software.
Results
Resistance to FK866 is not dependent on canonical mechanism of pharmacoresistance
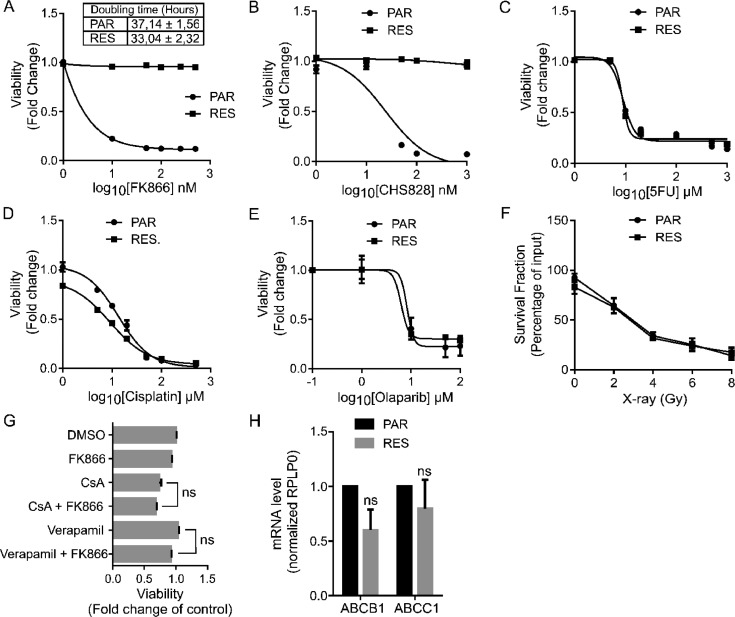
FK866-resistant MDA-MB-231 cells (RES) were obtained by exposing the MDA-MB-231 parental cells (PAR) to increasing concentrations of FK866, for over a 3-month period of time. As previously reported by our group, the same NAD(H) level was observed for PAR and RES cells. FK866 treatment in PAR cells led to an almost complete depletion of NAD(H) while RES cells were able to maintain about half of the NAD(H) starting amount [25]. RES cells did not show changes in the doubling time during the growth exponential phase and were splitted with the same ratio (Fig. 1A). Cell viability was assessed after 48 h of FK866 treatment using the resazurin-based fluorescent OzBlue reagent (Fig. 1A-E) and by colony formation assay (Supplementary Fig. 1A-E). The complete insensitiveness of RES cells to FK866 did not allow the calculation of the EC50, even with increasing FK866 time exposure (Supplementary Fig. 1F), while for PAR cells it was calculated as 4.7 nM (Fig. 1A), which is aligned with our previous data [25]. Similarly, RES cells presented cross-resistance to the NAMPT inhibitor CHS-828 (Fig. 1B), but not to common chemotherapeutic drugs like 5-fluoruracil (5FU), cisplatin, to the PARPs inhibitor olaparib and to X-ray exposure (Fig. 1C–F), confirming a NAMPT-dependent resistance.
Fig. 1.
Resistance to FK866 is not dependent on canonical mechanisms of pharmacoresistance Comparison of PAR and RES cells’ doubling time, as well as their sensitivity to NAMPT inhibitors, chemotherapeutics, X-rays radiation and to efflux pumps inhibition. (A) Doubling time of PAR and RES cells was calculated between 48 and 72 h after cell seeding, at the exponential growth phase, as an indication of the proliferative ratio of these cells. Sensitivity of PAR and RES cells to the small molecules (A) FK866 and (B) CH828 assessed by OzBlue assay after 48 hours of exposure to 0.1–500 nM of the compounds. Cross-resistance of PAR and RES cells to the chemotherapeutics (C) 5-Fluoruracil (5FU) and (D) cisplatin after 48 hours of treatment, and to (E) olaparib after 6 days of treatment. Viability assessed through OzBlue assay. (F) Colony formation ability of irradiated PAR and RES cells was assessed 7 days after the exposure to 2-8 Gy of X-rays. (G) RES cells sensitization to FK866 upon 48 hours of concomitant treatment with the efflux pumps inhibitors verapamil and cyclosporine A (CsA) and (H) expression of the efflux pumps ABCB1 and ABCC1. Final concentrations: FK866 – 20 nM; CsA – 5 µM; Verapamil – 5 µM. All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (ns- non significant) between DMSO and experimental conditions.
The development of drug resistance to NAMPT inhibitors can be conferred by gene mutation [32]. However, NAMPT coding sequence was not mutated (data not shown) in the resistant cell line, as previously reported for other FK866-resistant models [23]. The treatment with inhibitors of drug efflux pumps, like verapamil or cyclosporin A (CsA), failed to sensitize RES cells to FK866, and the expression levels of the ABCB1 and ABCC1 were not significantly modulated between PAR and RES cells (Fig. 1G-H). Thus, the role of the multi-drug resistance mechanisms in the acquirement of resistance to FK866 in this cell model was not further investigated. Therefore, RES cells developed alternative mechanisms, other than target mutation or activation of multi drug efflux pumps to acquire FK866 pharmacoresistance.
Resistance to FK866 is not dependent on compensatory NAD+ biosynthesis pathways
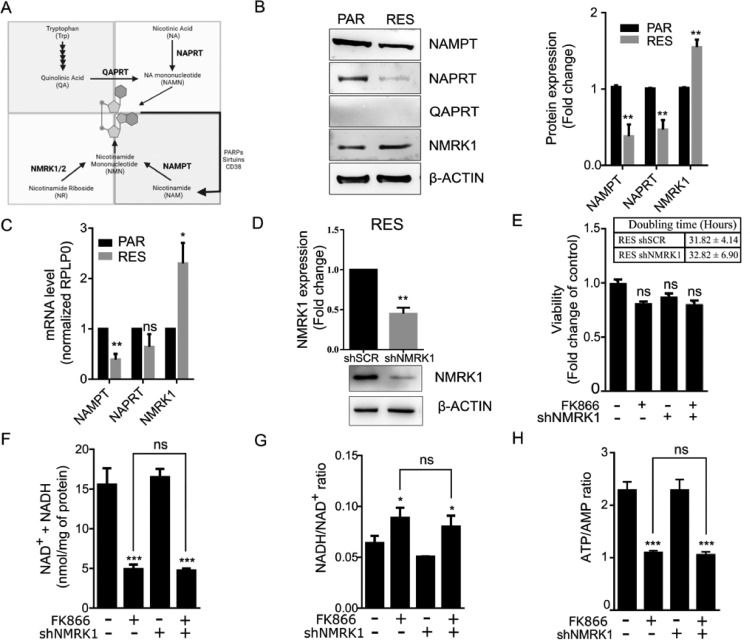
Intracellular NAD(H) biosynthesis is mainly sustained by the salvage pathway from nicotinamide, with NAMPT being the rate-limiting enzyme of the reaction. Additional NAD+ biosynthesis pathways use nicotinic acid, tryptophan, and nicotinamide riboside, as substrates for NAPRT, QAPRT and NMRK, respectively (Fig. 2A).
Fig. 2.
Resistance to FK866 is not dependent on compensatory NAD+ biosynthesis pathways Overview of NAD(H) biosynthesis pathways and genetic differences in this pathway between PAR and RES. Characterization of RES shNMRK1 cells. (A) In the de novo NAD+ synthesis, the amino acid tryptophan is converted to nicotinic acid mononucleotide (NAMN) through a series of reactions culminating with the formation of quinolinic acid (QA), the substrate of QA phosphoribosyl transferase (QPRT). NAMN can be also produced through the Press-Heindler pathway, with nicotinic acid (NA) being used as substate for nicotinic acid phosphoribosyl transferase (NAPRT). NAMN is further converted to NAD+ by NMN adenyl transferases (NMNAT) and NAD synthetase (NADSYN). Instead, nicotinamide phosphoribosyl transferase (NAMPT) that catalyze the conversion of nicotinamide (NAM) to nicotinamide mononucleotide (MNM), is the rate-limiting enzyme of NAD+ synthesis pathways. NAM levels are sustained by dietary input, as well as by the regeneration through NAD+-consuming enzymes like PARPs, sirtuins and CD38/157. NMN is also the product of the conversion of nicotinamide riboside (NR) by nicotinamide riboside kinase (NMRK), and is the substrate of NMNAT for NAD+ synthesis. (B) Western blot showing endogenous levels of NAMPT, NAPRT, QAPRT and NMRK1 in PAR and RES cells (β-ACTIN was used as a loading control). Representative image and quantification of 3 replicates are shown. (C) RT-qPCR to determine NAMPT, NAPRT and NMRK1 expression in PAR and RES cells. RPLP0 was used as a house-keeping gene. (D) Development of NMRK1 stable silenced RES cells (RES shNMRK1). NMRK1 knockdown was assessed by western blot. The effect of NMRK1 silencing to FK866 (20 nM) sensitivity was assessed by (E) cell viability and proliferation assays, (F) total NAD(H) levels quantification, (G) NADH/NAD+ and (H) ATP/AMP ratios determination, after 48 hours of exposure. All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (ns- not significant, **P < 0.01, ***P < 0.001) between DMSO and experimental conditions.
Given the role of these enzymes in the maintenance of intracellular NAD(H), we hypothesized that upon NAMPT inhibition through FK866, cells could sustain intracellular NAD(H) levels by compensatory mechanisms of NAD+ biosynthesis.
The acquirement of resistance to FK866 leads to a down-regulation of both NAMPT and NAPRT expression levels and to an up-regulation of NMRK1. QAPRT expression was not detected (Fig. 2B). Similar results were observed for NAMPT, NAPRT and NMRK1 transcripts levels (Fig. 2C).
Since both NMRK1 and NAMPT catalyze the production of the precursor of NAD+ Nicotinamide Mononucleotide (NMN), we hypothesized that NMRK1 up-regulation could compensate NAMPT down-regulation. Thus, we generated a RES cell line stably silenced for NMRK1 (Fig. 2D). NMRK1 silenced cells did not change their doubling time and, after treatment for 48 h with 100 nM of FK866, cell viability was not affected (Fig. 2E). Upon treatment, the total NAD(H) levels decreased in resistant cells independently of the NMRK1 silencing while the NADH/NAD+ ratio increased. Instead, the ATP/AMP ratio followed the same fashion as the total NAD(H) levels (Fig. 2F, G, H).
Therefore, the silencing of NMRK1 did not sensitize RES cells to FK866 in any of the assays tested (Fig. 2D-H), leading to the conclusion that compensatory NMRK1-mediated NAD+ biosynthesis is not relevant in the resistant phenotype.
FK866 persistent exposure induces an increase of spare respiratory capacity in RES cells
NAMPT inhibitors’ mechanism of action leads to an overall decrease of intracellular NAD(H), which is an essential coenzyme for the glycolytic enzymes. To understand if a modulation of the glycolytic flux occurs in RES, we performed a seahorse analysis using the Glycolytic Rate Assay, to determine the glycolytic proton efflux rate (glycoPER), an indirect measure of glycolysis. In this assay, the ECAR sustained by the extrusion of protons into the extracellular media from the conversion of glucose to pyruvate, was quantified. Additionally, the OCR was used to subtract the mitochondrial-derived CO2 contribution to the extracellular acidification.
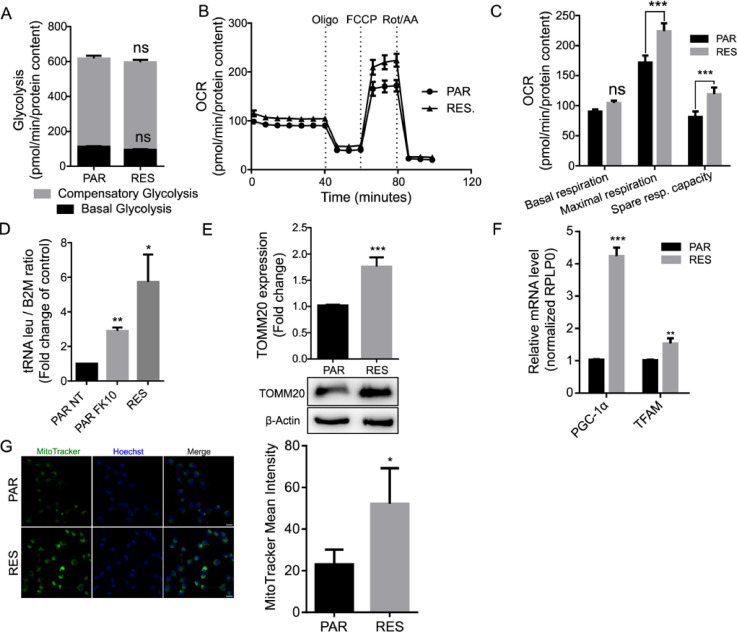
Basal glycolytic level was measured as well as compensatory glycolysis, by the acute injection of rotenone and antimycin A, complex I and III inhibitors, respectively. The injection of 2-Deoxy-D-glucose (2DG), a glucose analog and inhibitor of glycolysis first enzyme hexokinase, shuts down glucose oxidation and consequently the proton efflux, allowing to determine the contribution of other sources of extracellular acidification that are not attributed to glycolysis or mitochondrial TCA. In this assay, we did not observe a significant difference of either basal or compensatory glycolysis between PAR and RES cells (Fig. 3A).
Fig. 3.
FK866 continuous exposure induces increase of spare respiratory capacity and mitochondrial mass in RES cells Metabolic evaluation of glycolytic rate and mitochondrial function. (A) Glycolysis rate assay of PAR and RES cells to determine basal and compensatory glycolysis. (B) Representative MitoStress test to determine (C) basal respiration, maximal respiration and spare respiratory capacity of PAR and RES cells. Abbreviations: Oligo - Oligomycin; FCCP -Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Rot/AA - Rotenone and Antimycin A. RES cells present a higher mitochondrial mass. (D) RT-qPCR to determine tRNAleu and B2M expression in PAR cells (non treated - NT - and treated for 48 hours with 10 nM of FK866) and RES cells. The ratio between these genes reflect the mitDNA/nDNA ratio (mitochondrial versus nuclear DNA). RPLP0 was used as a house-keeping gene. (E) Western blot showing upregulation of TOMM20 in RES cells (β-ACTIN was used as a loading control). Representative image and quantification of 3 replicates are shown. (F) RT-qPCR to determine PGC1-α and TFAM expression in PAR and RES cells. RPLP0 was used as a house-keeping gene. (G) Mitotracker Green FM staining of PAR and RES cells’ mitochondria (Green - left) and Hoechst (Blue- middle) staining of cells nuclei. Composite image as a merge of the green and blue channels with scaling bar of 20 µm (right). Quantification of the MitoTracker mean intensity of at least 300 nuclei of PAR and RES cells is shown. All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (ns- non significant, *P<0.05, **P < 0.01, ***P < 0.001) between PAR and RES cells.
We then assessed the mitochondrial function using the Seahorse MitoStress Assay, in which the OCR is measured after injection of modulators of cellular respiration. The first injection of oligomycin (ATP synthase inhibitor) decreases the electron flow through the ETC, resulting in a reduction of mitochondrial respiration and consequently OCR. The following injection of FCCP have the opposite effect: being an uncoupling agent, it leads to proton leakage into the mitochondria, that is counteracted by an increase activity of complex I-IV, resulting on an increased OCR. This injection is used to calculate the spare respiratory capacity, an indirect measure of the ability of cells to respond to increase metabolic demands. Lastly, rotenone and antimycin A are concomitantly injected to inhibit the mitochondrial dependent oxygen consumption.
Seahorse analysis revealed that RES cells present a significant higher maximum respiratory capacity than PAR. As both cell lines show the same basal OCR values, the spare respiratory capacity (difference between maximum and basal respiration) is higher in RES, suggesting the induction of an adaptation of the mitochondrial function to the continuous exposure to FK866 (Figs. 3B-C, Supplementary 2A, B).
Mitochondrial function is modulated in RES cells
The increased mitochondrial spare respiratory capacity in FK866-resistant cells pinpoints a potential role of mitochondrial rewiring during the acquirement of resistance to the small molecule. In fact, several mitochondrial stress adaptations, like increased mitochondrial biogenesis, have been described to mediate drug resistance [33].
Interestingly, in RES cells we also observed an increase of the tRNAleu/B2M ratio, that are respectively mitochondrial and nuclear genes, in comparison with PAR. The acute exposure of PAR cells to 10 nM of FK866 for 48 h also led to an increment of the mitDNA/nuclearDNA ratio, suggesting that a positive modulation of the mitochondrial DNA is an immediate mechanism of cell adaptation to NAMPT inhibition (Fig. 3D). Aligned with this data, RES cells showed an increased expression of the nuclear gene encoding the translocase of outer mitochondrial membrane 20 (TOMM20), an indirect measure of mitochondrial mass [34] (Fig. 3E).
Additionally, we also observed an augmented expression of nuclear genes involved in the activation of mitochondrial biogenesis as the peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) and mitochondrial transcription factor A (TFAM) in RES cells (Fig. 3F). To confirm the molecular data, we performed a mitochondrial staining using the green fluorescent probe MitoTracker Green, which localizes to mitochondria independently of its membrane potential. Image-based quantitative analysis of the probe mean intensity confirmed a 2-fold increase of the mitochondrial mass in RES cells compared with PAR cells (Fig. 3G).
Collectively, these data indicate that acute FK866 treatment induces an immediate cell response leading to increased amount of mitochondrial to nuclear DNA ratio, that during long term FK866 administration results in increased mitochondrial mass.
RES cells show a dependency on pyruvate as carbon source for the TCA
Similarly to mitochondrial function modulation, rearrangement of mitochondrial bioenergetics is also observed in chemoresistance [33]. To dig deeper into the role of mitochondrial energetic hubs involved in the adaptation to FK866 exposure, we performed a mitochondrial substrate dependency assay (Mitoplate S-1). This assay measures the real time kinetics of electron flow rates upon oxidation of substrates that produce NADH or FADH2. The rate of substrate oxidation is a measure of the reduction of a tetrazolium redox dye (MC) that acts as a terminal electron acceptor.
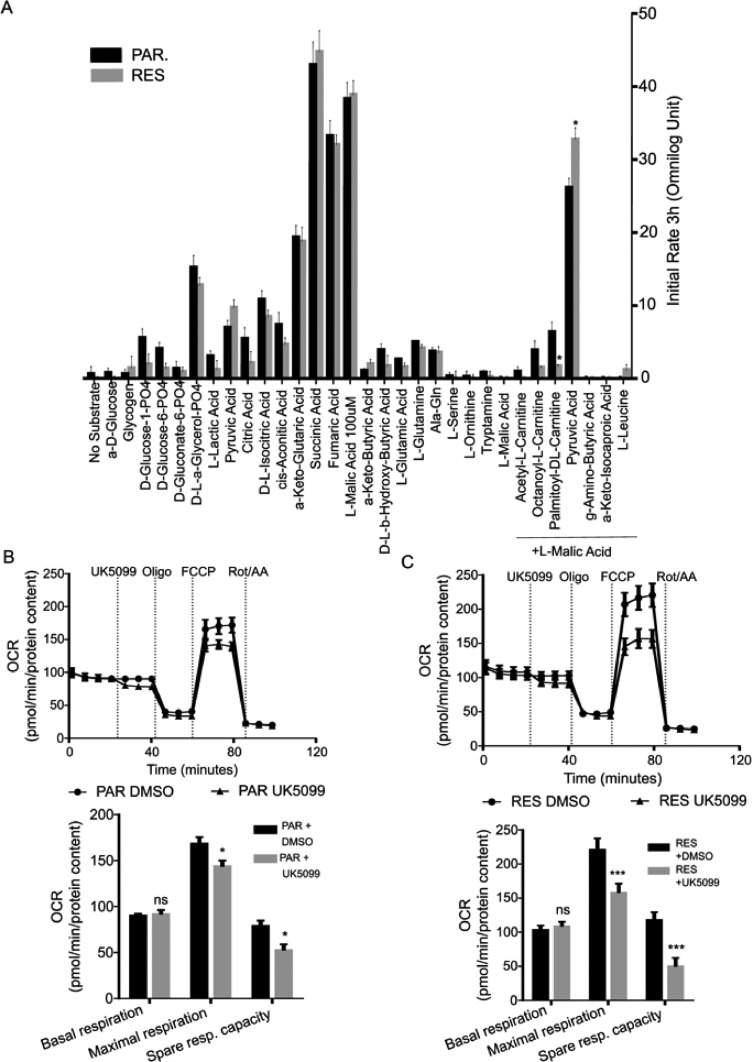
PAR and RES cells were permeabilized with saponin and incubated in a Mitoplate S-1 already containing several mitochondrial substrates and the redox dye. The initial rate of substrate oxidation during the first three hours of the assay was used to identify mitochondrial metabolic dependencies.
Both cell lines presented an almost null background (no substrate) that was subtracted to every tested condition. The TCA substrates α-keto-glutaric acid, succinic acid, fumaric acid and malic acid presented the highest levels of MC reduction, as they directly enter TCA to produce NADH and FADH2. These conditions are the positive control of the assay, assuring the successful cell permeabilization without mitochondrial disruption. However, no differences were observed between the cell models tested. Of notice, RES cells presented a decrease in fatty acids oxidation in comparison with PAR, that is significant for the combination of palmitoyl-DL-carnitine and malic acid (Fig. 4A).
Fig. 4.
RES cells show a dependency on pyruvate as carbon source for the TCA Assessment of mitochondrial dependencies. (A)The Mitoplate S-I assay from Omnilog was used to determine the initial mitochondrial substrate oxidation rate (3 hours of assay) between PAR and RES cells. Representative MitoStress test to determine basal respiration, maximal respiration and spare respiratory capacity of (B) PAR and (C) RES cells upon acute injection of UK5099 (2 µM). All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (ns- non significant, *P<0.05, **P < 0.01, ***P < 0.001) between PAR and RES cells.
Instead, RES cells show a significant increase in pyruvic acid oxidation when in combination with lower doses of L-malic acid [35]. We also observed a tendency of pyruvic acid alone to increase the initial electron flow rate in RES cells when compared with PAR cells (Fig. 4A). This result indicates a shift of mitochondrial substrate's dependencies, namely towards the utilization of pyruvate, upon FK866 long term exposure.
To confirm these observations, we performed a mitochondrial function assay using the Seahorse Mitostress kit, in the presence of UK5099, a MPC inhibitor, that will prevent the MPC mediated import of pyruvate into the mitochondria. This compound was acutely injected after the measurement of the cellular basal OCR levels, followed by the sequential addition of oligomycin, FCCP and rotenone/antimycin A. We observed that PAR cells are sensitive to MPC inhibition (Fig. 4B) but RES cells showed a stronger decrease of the maximal respiration in the cells exposed to UK5099 in comparison to the vehicle DMSO control, confirming the importance of the MPC-dependent pyruvate import for mitochondrial functioning (Fig. 4C).
We hypothesized that an increase in pyruvate oxidation rate could be related with an increased expression of the pyruvate dehydrogenase (PDH) complex or with an increase of the pyruvate import to the mitochondria. However, RES cells did not show a modulation of the PDH1Ea subunit nor of the mitochondrial pyruvate carrier 1 and 2 (MPC1 and MPC2) when compared to PAR cells (Supplementary Fig. 3A-B).
Taken together, these data unravel the metabolic dependency of pyruvate as carbon source for the mitochondria in long term exposure to FK866.
The uncoupling of glycolysis and mitochondrial respiration favored by MPC2 blockage induces a FK866 resistant-like phenotype in PAR cells
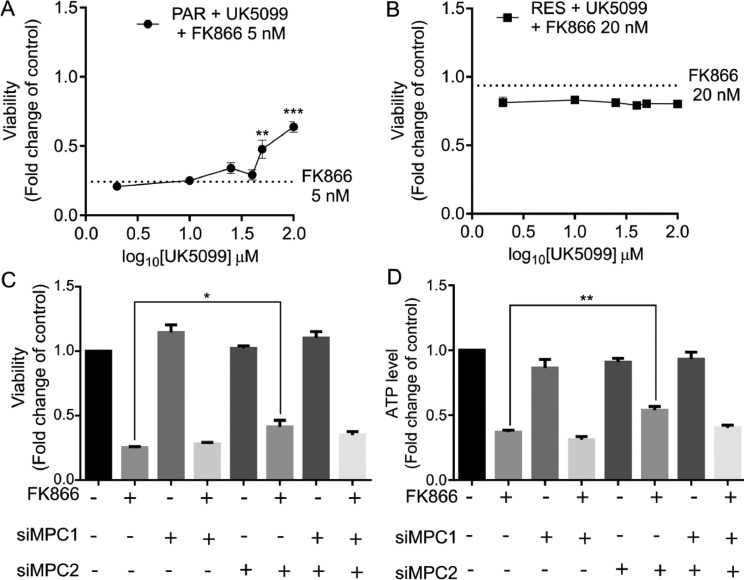
To gain some insights on whether MPC modulates the acquirement of resistance to FK866 in triple negative breast cancer, PAR and RES cells were exposed to increasing concentrations of UK5099 for 48 h. Both cells showed the same sensitivity to the drug (IC50 > 100 µM, Supplementary Fig. 4A). However, the treatment of the parental cells with UK5099 in combination with FK866 lead to a significant increase in cell viability when compared to FK866 single administration (Fig. 5A).
Fig. 5.
The uncoupling of glycolysis and mitochondrial respiration favored by MPC2 blockage induce a FK866 resistant-like phenotype in PAR cells. Co-treatment of (A) PAR and (B) RES cells with UK5099 (0.1–100) µM and FK866 as well as with FK866 single administration (Final concentrations of FK866: 5 nM for PAR cells and 20 nM for RES). Viability assessed by OzBlue assay after 48 h of exposure and compared with DMSO condition. Effect of MPC1 (siMPC) and/or MPC2 (siMPC2) transient silencing in PAR cells in terms of (C) cell viability and (D) relative ATP level, in combination with FK866 5 nM. All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (*P<0.05, **P < 0.01, ***P < 0.001) between control and experimental conditions.
Instead, no significant differences were observed for the co-treatment of RES cells with both small molecules (Fig. 5B). Similar results were obtained for the treatment with rosiglitazone, a previously described MPC inhibitor (Supplementary Fig. 4B-C).
To understand if the acquirement of a FK866-resistant like phenotype in the PAR cells was specifically dependent on pyruvate transport, cells were transiently silenced for MPC1 and/or MPC2 and viability and ATP levels were assessed after 48 h of FK866 treatment. FK866 treatment reduced both viability and ATP levels in the control condition. Interestingly, the treatment of FK866 upon the silencing of MPC2 partially but significantly rescued the FK866 toxic effect observed in the control condition. The same trend was obtained for the ATP level measurements. Instead, the silencing of MPC1 or MPC1 and MPC2 did not lead to a significant rescue of FK866 toxicity for the analyzed parameters, even if the dual silencing approach showed an increasing trend (Fig. 5C-D). Taken together, this data unravels the pivotal importance of regulating the pyruvate import to mitochondria via MPC2 as a mediator of the adaptation of triple negative breast cancer cells to FK866 exposure.
Acquired resistance to FK866 induces dependency on succinate, even in pyruvate-deficiency conditions
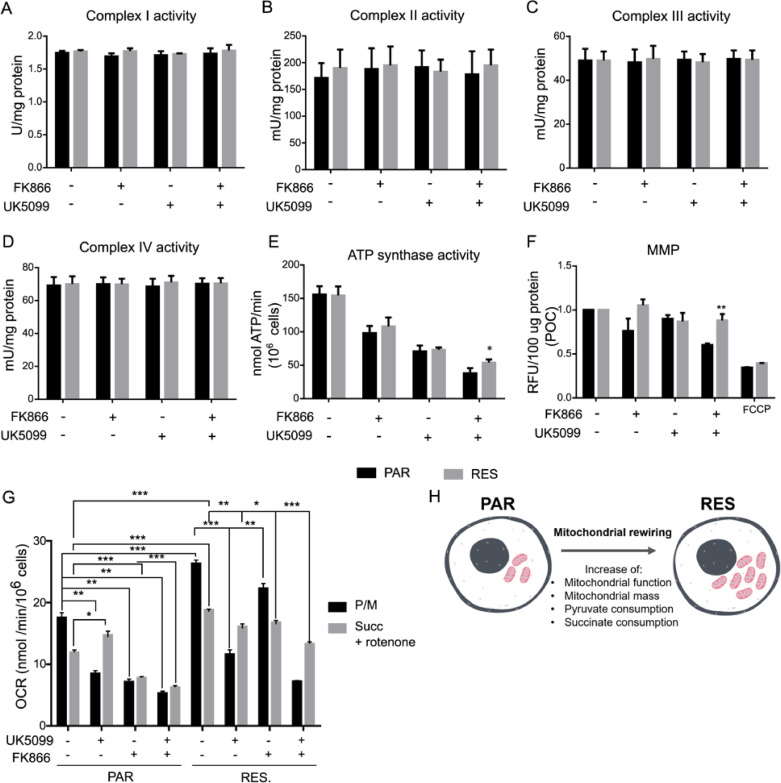
To investigate the mitochondrial bioenergetic dependencies, we questioned if the observed mitochondrial function in RES cells was associated with a differential activity of the ETC complexes. The ETC complexes uses NADH and FADH2, produced trough the oxidation of carbon sources on the TCA, and generate the MMP to sustain mitochondrial ATP production.
Cells were treated with FK866, UK5099 or the combination of both for 48 h, and enzymatic activity of the uncoupled complexes was assessed and normalized to the sample protein content. Complexes I-IV presented the same enzymatic activity for all the treatments performed and among the two cell lines (Fig. 6A–D). Conversely, the activity of ATP synthase (Complex V), induced by the pyruvate/malate addition, decreased similarly in PAR and RES cells, during either the single-treatment with FK866 or UK5099.
Fig. 6.
FK866 long-term exposure model increases succinate consumption. Assessment of Complex I-IV and ATP synthase activities, and Mitochondrial Membrane Potential (MMP). (A – D) The activities of Complex I-IV in PAR and RES cells were measured upon treatment with FK866 and/or UK5099. (E) ATP synthase activity and mitochondrial membrane potential (MMP) were determined in the same conditions, with FCCP being used as positive control for MMP measurement. Determination of mitochondrial substrate consumption. (G) OCR measurements of cells permeabilized with digitonin and treated with UK5099 and/or FK866, in the presence of pyruvate/malate (P/M) or succinate/rotenone (Succ + Rotenone) were also determined. (H) Schematic representation of the PAR cells adaptation by mitochondrial rewiring upon NAD(H) shortage: in RES cells an increased mitochondrial function and mass, and consequently of pyruvate and succinate consumption, is observed. Final concentrations: FK866 – 10 nM for PAR cells and 100 nM for RES; UK5099 – 100 µM for both cell lines. All the experiments were performed in triplicate. Repeated measures one-way ANOVA was used to calculate statistical significance (*P<0.05, **P < 0.01, ***P < 0.001) between PAR and RES cells.
Instead, combining the exposure of cells to both small molecules, we observed that Complex V activity was less affected in RES cells compared to PAR cells (Fig. 6E, Supplementary Fig. 4). We further confirmed that the increased ATP synthase activity is sustained by the maintenance of the MMP, determined spectrophotometrically through the measurement of TMRE fluorescence. This dye accumulates in active mitochondria according with their membrane potential. The co-treatment of RES cells with FK866 and UK5099 did not decrease MMP, indicating a continued mitochondrial ATP production.
Instead, in PAR cells we observed a significant decrease of the fluorescent counts in the same condition, associated with a diminished ability of these cells to produce energy in the mitochondria when uncoupled from glycolysis. As the ionophore uncoupler of the oxidative phosphorylation, FCCP was used as positive control. FCCP determined a decrease of MMP for both cell lines (Fig. 6F). These data show that the long-term exposure to FK866 increases the fitness of mitochondria in ATP synthesis through an increase in ATP synthase activity, that is not only sustained by the MPC-dependent pyruvate import, but potentially by alternative metabolic mechanisms that maintain the MMP in pyruvate-deprivation conditions.
Thus, we decided to investigate mitochondrial substrate dependence in the presence of pyruvate/malate and succinate/rotenone during the silencing and inhibition of MPC subunits to identify the pathways led by Complex I and Complex II, respectively (Fig. 6G). The OCR measurements revealed that mitochondria of RES cells presented a higher OCR than PAR cells, showing a preferential utilization of pyruvate/malate. Reduction of pyruvate entrance into the PAR cells’ mitochondria, by pharmacological means, decreased the pyruvate plus malate-induced OCR but increased the succinate-dependent respiration, suggesting an attempt to compensate the oxygen consumption decrement through Complex I. In the presence of FK866, however, mitochondria are deprived of the ability to activate both Complex I and II, either in the presence or absence of UK5099. In RES cell's mitochondria, MPC blockage reduced pyruvate dependent OCR, and did not increase succinate utilization, that is already higher than in PAR. Interestingly, FK866 treatment was not affecting neither pyruvate nor succinate dependent OCR, further confirming the insensitivity of these cells to NAMPT inhibition. Upon cotreatment with UK5099 and FK866, we still observed succinate exploitation that sustained OCR. Therefore, resistant cells show a higher mitochondrial function than parental ones, sustained by both pyruvate and succinate oxidation.
Discussion
Metabolic rearrangement offers cancer cells the plasticity to respond and adapt to toxic insults [36]. In this study, we used a triple negative breast cancer cell line that is resistant to the NAMPT inhibitor FK866 to dissect molecular mechanisms sustaining the cellular adaptation to prolonged NAD(H) shortage.
Previously reported FK866-resistant cell models often present NAMPT mutations that prevent inhibitors’ binding to its catalytic site [23,32]. Still, in RES cells, NAMPT is not mutated. Accordingly, this model shows cross-resistance to CHS-828, another NAMPT inhibitor [37], but not for some classes of chemotherapeutics commonly used in the clinics for cancer treatment, like 5-fluoruracil (antimetabolite), cisplatin (alkylating agent) and olaparib (PARPs inhibitor). FK866-resistant colon cancer cells, present an increased sensitivity to the DNA damage inducer γ-irradiation of the resistant cells in comparison with their sensitivity counterparts, likely due to mutations of PARP and PARG genes [26]. However, PAR and RES cells present the same colony formation ability upon X-rays exposure which indicates a similar DNA repair system. These data are supported by the observed similar sensitivity of PAR and RES cells to the DNA damage repair protein PARP inhibitor olaparib. Still, the mutational status of NAD+-related enzymes was not further investigated, so we cannot exclude their role in the mechanism of acquirement of resistance to FK866.
Canonical mechanisms of pharmacoresistance differ depending on the drug but can often include the increase of transporter pumps, as described for other FK866 resistant models [23,24,38]. The treatment with the drug efflux pumps’ inhibitors verapamil and cyclosporine A, did not sensitize RES cells to FK866, excluding the role of multi-drug resistance (MDR) mechanisms in the acquired resistance to FK866 in these cells. In line with these data, a modulation of the ABCB1 and ABCC1 levels in RES cells was not observed. Thus, RES cells present a NAMPT-dependent acquired resistance to FK866, that is not dependent on the canonical mechanisms of acquired resistance.
Importantly, cancer cells’ dependency on other enzymes for NAD(H) synthesis like QAPRT or NAPRT can bypass the NAMPT inhibitors’ dependent NAD(H) depletion [25], [39], [40], [41], [42]. In our model, QAPRT is not expressed but NMRK1 expression is positively modulated in RES, in opposition to NAPRT, that presents the inverse trend. This data is aligned with a recent study that showed that dependence of NAMPT is subject to resistance by NMRK1-dependent synthesis of NAD(H), which can explain the failure of NAMPT inhibitors [43]. However, the stable silencing of NMRK1 in RES cells did not sensitize these cells to FK866 treatment, and NAD(H) and ATP were kept unaltered.
Interestingly, we found that the acquisition of resistance to FK866 determines specific mitochondrial adaptation. Real-time cell metabolic analysis revealed that RES cells have an increased spare respiratory capacity (SRC) sustained by a higher maximal respiration (Fig. 6H). Indeed, SRC is a critical component of cellular bioenergetics as it correlates with mitochondrial plasticity and its ability to adapt in response to energetic stress conditions [44,45], with several studies reporting a positive correlation between high levels of SRC and resistance to anti-cancer drugs as cisplatin and 5FU likely due to a shift toward a mitochondrial metabolism [46,47]. Still, to our knowledge, this is the first time that an increased SRC has been associated with FK866 resistance. Similarly, mitochondrial-mediated chemoresistance has also been associated with an increase in the mitochondrial mass [48], [49], [50]. Aligned with these studies, we observed an increase in the mitochondrial DNA versus nuclear upon parental cells’ treatment with FK866, revealing a FK866-depedent increase of mitochondrial mass. The mtDNA/nDNA ratio was further augmented once cells became fully resistant. An increase of the expression of PGC-1α, TFAM and Sirt1 transcripts and of TOMM20 protein levels were also observed. These results corroborate the hypothesis that prolonged NAD(H) shortage led to an increased mitochondrial biogenesis and mass (Fig. 6H).
Alterations on mitochondria physiology imply the maintenance of the import of carbon sources to sustain the TCA cycle, ETC and ultimately ATP synthesis. Using a Mitoplate-SI, we questioned the mitochondrial substrate dependences of RES cells and revealed that these cells metabolize pyruvate in combination with low doses of malic acid faster than the parental ones, but this phenotype was not recapitulated by either pyruvate or malic acid alone. In fact, it is described that malic acid acts as a spark for the TCA cycle to have optimal oxidation rates [35], which confirms that RES cells present a higher dependence on pyruvate metabolism.
Mitochondrial dependence on pyruvate was further confirmed through the inhibition of its uptake by the mitochondrial pyruvate carrier (MPC). RES cells treated with the MPC inhibitor UK5099 failed to achieve the same levels of maximal respiration as the control, which implied a lower spare respiratory capacity. Although not strictly correlated with differential activity, as the expression of both subunits of the MPC complex was not altered between the parental and resistant cells, the increased pyruvate oxidation seems not to be associated with an augmented pyruvate uptake through MPC.
Given the role of MPC for the mitochondrial adaptation to NAD lowering agents observed in RES cells, we questioned the effect UK5099 treatment on the sensitivity to FK866 in these cell lines. The cotreatment with the MPC inhibitor and FK866 led to an increase of cellular viability, counteracting FK866 toxicity in PAR cells. Moreover, by transient silencing of MPC1 and/or MPC2, we were able to associate the resistant phenotype observed with the subunit 2 of MPC (MPC2) and not with MPC1. Instead, sensitivity to FK866 was not modulated upon MPC inhibition in RES cells. These data revealed that MPC inhibition is likely important for the immediate/chronic response to FK866 exposure in triple negative breast cancer but does not entirely sustain the resistant phenotype. The blocking of pyruvate entrance into the pyruvate would rather stimulate LDHA activity, allowing the recycling of NAD+ cytoplasmatic levels, as previously observed in RES cells [25].
Additionally, we observed that complexes I-IV activities, measured in an uncoupled way, were not altered either with the single or combination treatment of FK866 and UK5099, confirming that the respiratory complexes are not the target of these molecules, and that the FK866 and UK5099 effect of OxPhos function depend on the altered pyruvate and NAD(H) availability. We showed that the combination of FK866 and UK5099 treatments in RES cells led to a higher activity of ATP synthase (Complex V), and consequently ATP production, compared with the parental cells. The increased ATP synthase activity is sustained by the maintenance of the mitochondrial membrane potential (MMP), in the same conditions. Collectively, this data show that RES cells’ mitochondria are adapted to maintain MMP and ATP synthesis in MPC-blockage dependent pyruvate-deficiency conditions. Of note, the pyruvate import to the mitochondria is coupled with the symport of one proton [22]. Blocking MPC would prevent not only the import of pyruvate but also of the proton, which favor the maintenance of a higher MMP and ultimately ATP production by Complex V.
Further analysis of mitochondrial function revealed that RES cells present higher basal OCR than PAR cells in the presence of succinate, suggesting a mitochondrial adaptation during NAD(H) shortage (Fig. 6H). Indeed, upon MPC blockage, PAR cells can exploit succinate utilization as carbon source for the TCA. Taken together, our data suggest that acquired resistance to NAMPT inhibition is likely developed through several steps. Initially, the blockage of MPC2, and thus the uncoupling of glycolysis and mitochondrial respiration, results in higher pyruvate cytoplasmatic accumulation. In alignment with previous work from our group, cytoplasmatic pyruvate can be used by LDHA, to keep NAD+ levels high in the cytoplasm. The lack of pyruvate leads mitochondria to adapt to be able to keep OxPhos, namely through the exploitation of succinate as a carbon source for the TCA and increase of mitochondrial mass. In the last stages of the acquisition of a stable FK866 resistant cell line, both pyruvate and succinate are used to sustain ATP synthesis.
Taken together, our data show that mitochondrial plasticity provides a metabolic advantage to cells’ adaptation to NAD(H) shortage.
Conclusions
In this work, we developed a model of triple negative breast cancer cells that are resistant to FK866, to dissect the molecular adaptations of cells to NAD(H) shortage. We observed that the acquired resistance to NAMPT inhibition is sustained by an overall metabolic rewiring sustained by mitochondrial adaptations. These cells presented a higher pyruvate and succinate consumption than the parental ones, that maintain an increased mitochondrial spare respiratory capacity, mass and biogenesis.
Taken together, these data give some insights on the mechanisms developed by cancer cells in response to NAD(H) shortage, which can challenge the NAMPT inhibitors usage in the clinics.
Ethics statement
No human studies, animal studies and potentially identifiable human images or data are presented in this manuscript.
Funding
This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No 813284 to SB, AN and AP.
CRediT authorship contribution statement
Agata Sofia Assuncao Carreira: Conceptualization, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Silvia Ravera: Writing – review & editing. Chiara Zucal: Data curation, Validation, Writing – review & editing. Natthakan Thongon: Data curation, Validation, Writing – review & editing. Caffa Irene: Data curation, Validation, Writing – review & editing. Cecilia Astigiano: Validation, Writing – review & editing. Nadia Bertola: Validation, Writing – review & editing. Arianna Buongiorno: Validation, Writing – review & editing. Michela Roccuzzo: Validation, Writing – review & editing. Alessandra Bisio: Validation, Writing – review & editing. Barbara Pardini: Data curation, Validation, Writing – review & editing. Alessio Nencioni: Funding acquisition, Resources, Supervision, Writing – review & editing. Santina Bruzzone: Funding acquisition, Resources, Supervision, Writing – review & editing. Alessandro Provenzani: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the CIBIO Core Facilities (High throughput Screening and Advanced Imaging) for their technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100903.
Contributor Information
Agata Sofia Assuncao Carreira, Email: agata.carreira@unitn.it.
Silvia Ravera, Email: silvia.ravera@unige.it.
Chiara Zucal, Email: chiara.zucal@unitn.it.
Natthakan Thongon, Email: th.natthakan@gmail.com.
Caffa Irene, Email: Irene.Caffa@unige.it.
Cecilia Astigiano, Email: cecilia.astigiano@gmail.com.
Nadia Bertola, Email: nadia.bertola@gmail.com.
Arianna Buongiorno, Email: aribuongi00@gmail.com.
Michela Roccuzzo, Email: michela.rocuzzo@unitn.it.
Alessandra Bisio, Email: alessandra.bisio@unitn.it.
Barbara Pardini, Email: barbara.pardini@iigm.it.
Alessio Nencioni, Email: alessio.nencioni@unige.it.
Santina Bruzzone, Email: santina.bruzzone@unige.it.
Alessandro Provenzani, Email: alessandro.provenzani@unitn.it.
Appendix. Supplementary materials
Data availability
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Heiden M.G.V., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu L., et al. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017;77:1564–1574. doi: 10.1158/0008-5472.CAN-16-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasan K., Werner M., Chandel N.S. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32:341–352. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diering M., Mitchell, Freeman Mitochondria and cancer. Physiol. Behav. 2018;176:139–148. [Google Scholar]
- 6.LeBleu V.S., et al. Phosphorylation to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopp G., Hottiger E., Hopp A.K., et al. Regulation of glucose metabolism by NAD+ and ADP-ribosylation. Cells. 2019;8:1371. doi: 10.3390/cells8080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu M., et al. NAD kinase sustains lipogenesis and mitochondrial metabolism through fatty acid synthesis. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.110157. [DOI] [PubMed] [Google Scholar]
- 9.Gameiro P.A., Laviolette L.A., Kelleher J.K., Iliopoulos O., Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2013;288:12967–12977. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T., Kraus W.L. SIRT1-dependent regulation of chromatin and transcription: linking NAD+ metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta - Prot. Proteomics. 2010;1804:1666–1675. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X.N., et al. Discovery of an NAD+analogue with enhanced specificity for PARP1. Chem. Sci. 2022;13:1982–1991. doi: 10.1039/d1sc06256e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan K.A., Chini C.C.S., Chini E.N. The Multi-faceted Ecto-enzyme CD38: roles in immunomodulation, cancer, aging, and metabolic diseases. Front. Immunol. 2019;10:1–12. doi: 10.3389/fimmu.2019.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahm F., et al. The endogenous tryptophan metabolite and NAD+ precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res. 2013;73:3225–3234. doi: 10.1158/0008-5472.CAN-12-3831. [DOI] [PubMed] [Google Scholar]
- 14.Preiss J., Handler P. Biosynthesis of diphosphopyridine nucleotide. J. Biol. Chem. 1958;233:488–492. [PubMed] [Google Scholar]
- 15.Bieganowski P., Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a preiss-handler independent route to NAD in fungi and humans it became clear that Sir2 and Sir2-related enzymes termed Sirtuins deacetylate lysine residues with consumpt. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 16.Ghanem M.S., Monacelli F., Nencioni A. Advances in nad-lowering agents for cancer treatment. Nutrients. 2021;13(5):1665. doi: 10.3390/nu13051665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 18.Audrito V., et al. Tumors carrying BRAF-mutations over-express NAMPT that is genetically amplified and possesses oncogenic properties. J. Transl. Med. 2022;20:1–13. doi: 10.1186/s12967-022-03315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicka-Gutaj N., et al. Nicotinamide phosphorybosiltransferase overexpression in thyroid malignancies and its correlation with tumor stage and with survivin/survivin DEx3 expression. Tumor Biol. 2015;36:7859–7863. doi: 10.1007/s13277-015-3506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi T.Q., et al. Overexpression of Nampt in gastric cancer and chemopotentiating effects of the Nampt inhibitor FK866 in combination with fluorouracil. Oncol. Rep. 2011;26:1251–1257. doi: 10.3892/or.2011.1378. [DOI] [PubMed] [Google Scholar]
- 21.Zhou S.J., Bi T.Q., Qin C.X., Yang X.Q., Pang K. Expression of NAMPT is associated with breast invasive ductal carcinoma development and prognosis. Oncol. Lett. 2018;15:6648–6654. doi: 10.3892/ol.2018.8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson A.E., et al. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) with OT-82 induces DNA damage, cell death, and suppression of tumor growth in preclinical models of Ewing sarcoma. Oncogenesis. 2020;9 doi: 10.1038/s41389-020-00264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino Y., Sato A., Uchiumi F., Tanuma S.-I. Cross resistance to diverse anticancer nicotinamide phosphoribosyltransferase inhibitors induced by FK866 treatment. Oncotarget. 2018;9:16451–16461. doi: 10.18632/oncotarget.24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino Y., et al. Association of ABC transporter with resistance to FK866, a NAMPT inhibitor, in human colorectal cancer cells. Anticancer Res. 2019;39:6457–6462. doi: 10.21873/anticanres.13859. [DOI] [PubMed] [Google Scholar]
- 25.Thongon N., et al. Cancer cell metabolic plasticity allows resistance to NAMPT inhibition but invariably induces dependence on LDHA. Cancer Metabol. 2018;6 doi: 10.1186/s40170-018-0174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino Y., Sato A., Uchiumi F., Tanuma S.ichi. Genomic and tumor biological aspects of the anticancer nicotinamide phosphoribosyltransferase inhibitor FK866 in resistant human colorectal cancer cells. Genomics. 2019;111:1889–1895. doi: 10.1016/j.ygeno.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Bruzzone S., et al. Catastrophic NAD+ depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS One. 2009;4:e7897. doi: 10.1371/journal.pone.0007897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappelli E., et al. The passage from bone marrow niche to bloodstream triggers the metabolic impairment in Fanconi Anemia mononuclear cells. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa F., et al. The human fetal and adult stem cell secretome can exert cardioprotective paracrine effects against cardiotoxicity and oxidative stress from cancer treatment. Cancers. 2021;13 doi: 10.3390/cancers13153729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindelin J., et al. Fiji : an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaroli A., et al. Photobiomodulation and oxidative stress : 980nm diode laser light regulates mitochondrial activity and reactive oxygen species production. Oxid Med Cell Longev. 2021 doi: 10.1155/2021/6626286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Elkins K., Oh A., Ho Y.-C., Wu J. Structural basis for resistance to diverse classes of NAMPT inhibitors. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin P. Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management. J. Hematol. Oncol. 2022;15:97. doi: 10.1186/s13045-022-01313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Contino S., et al. Presenilin 2-dependent maintenance of mitochondrial oxidative capacity and morphology. Front. Physiol. 2017;8:1–11. doi: 10.3389/fphys.2017.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Björntorp P. The Oxidation of Fatty Acids Combined with Albumin by Isolated Rat Liver. Mitochondria. 1966;241:1537–1543. [PubMed] [Google Scholar]
- 36.Morandi A., Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. BBA - Rev. Cancer. 2017;1868:1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Olesen U.H., et al. Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem. Biophys. Res. Commun. 2008;367 doi: 10.1016/j.bbrc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Zheng H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8:59950–59964. doi: 10.18632/oncotarget.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J., et al. Identification of novel resistance mechanisms to NAMPT inhibition via the de novo NAD+ biosynthesis pathway and NAMPT mutation. Biochem. Biophys. Res. Commun. 2017;491:681–686. doi: 10.1016/j.bbrc.2017.07.143. [DOI] [PubMed] [Google Scholar]
- 40.Piacente F., et al. Nicotinic acid phosphoribosyltransferase regulates cancer cell metabolism, susceptibility to NAMPT inhibitors and DNA repair. Cancer Res. 2017;3079 doi: 10.1158/0008-5472.CAN-16-3079. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien T., et al. Supplementation of nicotinic acid with NAMPT inhibitors results in loss of in vivo efficacy in NAPRT1-deficient tumor models. Neoplasia (United States) 2013;15:1314–1329. doi: 10.1593/neo.131718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shames D.S., et al. Loss of NAPRT1 expression by tumor-specific promoter methylation provides a novel predictive biomarker for NAMPT inhibitors. Clin. Cancer Res. 2013;19:6912–6923. doi: 10.1158/1078-0432.CCR-13-1186. [DOI] [PubMed] [Google Scholar]
- 43.Chowdhry S., et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature. 2019;569:570–575. doi: 10.1038/s41586-019-1150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchetti P., Fovez Q., Germain N., Khamari R., Kluza J. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020;34:13106–13124. doi: 10.1096/fj.202000767R. [DOI] [PubMed] [Google Scholar]
- 45.Pfleger J., He M., Abdellatif M. Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell Death Dis. 2015;6:1–14. doi: 10.1038/cddis.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teh J.T., Zhu W.L., Newgard C.B., Casey P.J., Wang M. Respiratory capacity and reserve predict cell sensitivity to mitochondria inhibitors: Mechanism-based markers to identify metformin-responsive cancers. Mol. Cancer Ther. 2019;18:693–705. doi: 10.1158/1535-7163.MCT-18-0766. [DOI] [PubMed] [Google Scholar]
- 47.Salunkhe S., et al. Metabolic rewiring in drug resistant cells exhibit higher OXPHOS and fatty acids as preferred major source to cellular energetics. Biochim. Biophys. Acta - Bioenergy. 2020;1861 doi: 10.1016/j.bbabio.2020.148300. [DOI] [PubMed] [Google Scholar]
- 48.Farge T., et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7:716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y., et al. ABT737 reverses cisplatin resistance by targeting glucose metabolism of human ovarian cancer cells. Int. J. Oncol. 2018;53:1055–1068. doi: 10.3892/ijo.2018.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farnie G., Sotgia F., Lisanti M.P. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget. 2015;6:30472–30486. doi: 10.18632/oncotarget.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.