Abstract
MLL rearranged (MLLr) leukemias are associated with a poor prognosis and a limited response to conventional therapies. Moreover, chemotherapies result in severe side effects with significant impairment of the immune system. Therefore, the identification of novel treatment strategies is mandatory.
Recently, we developed a human MLLr leukemia model by inducing chromosomal rearrangements in CD34+ cells using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. This MLLr model authentically mimics patient leukemic cells and can be used as a platform for novel treatment strategies. RNA sequencing of our model revealed MYC as one of the most important key drivers to promote oncogenesis. However, in clinical trials the BRD4 inhibitor JQ-1 leading to indirect blocking of the MYC pathway shows only modest activity. We and others previously reported that epigenetic drugs targeting MAT2A or PRMT5 promote cell death in MLLr cells. Therefore, we use these drugs in combination with JQ-1 leading to augmented anti-leukemic effects. Moreover, we found activation of T, NK and iNKT cells, release of immunomodulatory cytokines and downregulation of the PD-1/PD-L1 axis upon inhibitor treatment leading to improved cytotoxicity.
In summary, the inhibition of MYC and MAT2A or PRMT5 drives robust synergistic anti-leukemic activity in MLLr leukemia. Moreover, the immune system is concomitantly activated upon combinatorial inhibitor treatment, hereby further augmenting the therapeutic efficiency.
Keywords: MLL-rearranged leukemia, MYC, MAT2A, CRISPR/Cas9, Immune defense
Introduction
Chromosomal translocations are the most common abnormalities observed in solid tumors and hematological malignancies and are responsible for tumorigenesis [1]. Likewise, the MLL/KMT2A gene can fuse to over 130 known partner genes like AF4 or AF9 leading to leukemias in both children and adults with a dismal prognosis [2]. Mostly, patients show early resistance to conventional chemotherapy leading to high mortality rates [3]. Moreover, chemotherapies induce severe side effects like infections due to cytopenia and disruption of efficient immune responses. Such immune responses are also required to prevent relapse once remission has been induced. Therefore, targeted therapies circumventing the damage of the immune system are urgently needed. For instance, the use of hypomethylating agents targeting the epigenetic regulation of acute myeloid leukemia (AML) blasts also results in infections and impaired immune responses [4]. Thus, targeting the pathogenic mechanisms that maintain MLLr leukemogenesis and determining the influence on the immune system are crucial steps in developing new targeted therapies.
By using CRISPR/Cas9, we developed a representative human MLLr model based on patient-specific sequences with complete translocation of the MLL and AF4 or AF9 gene modeling the consequences of endogenous oncogene activation and hereby mimicking the patient disease [5], [6], [7]. These MLLr cells with significantly increased in vitro growth potential are suitable for both uncovering pathways sustaining leukemogenesis and conducting pharmacological studies with high translational character.
In our model, we found MYC significantly overexpressed and most responsible for the genetic aberrations in our cells. It is known, that MLL fuses to important regulators, including the super elongation complex (SEC) leading to deregulated transcription like MYC overexpression being responsible for leukemogenesis. SEC consists of the Bromodomain and Extra Terminal domain (BET) protein family members BRD3 and BRD4 that are also components of the polymerase-associated factor complex (PAFc) and the interaction with MLL and MLL fusions is critical for the transcriptional activation [8]. Therefore, BET inhibitors, like JQ-1, lead to a reduction of MYC expression and have been successfully studied in MLLr cell lines and MLLr mouse models [9,10]. Although BET inhibition has an acceptable side effect profile (clinical trial NCT0230876111), it shows only modest clinical activity in adult leukemias, and further combination approaches are required to improve efficiency. Recently, we and others highlighted the important roles of protein arginine methyltransferase 5 (PRMT5) regulated by PAFc, and methionine adenosyltransferase 2A (MAT2A) synthesizing S-adenosylmethionine (SAM) as universal methyl donor in human cells, as important epigenetic regulators involved in promoting gene expression of critical oncogenes [11,12]. Inhibition of PRMT5 or MAT2A led to convincing anti-leukemic effects [5,6]. Therefore, we use our human MLLr model to unravel the potential synergistic anti-tumoral efficacy in a combinatorial treatment strategy by inhibition of MYC and MAT2A or PRMT5. In this context, we also want to shed light on the effect of these compounds on the immune system. Recently, it has been shown, that BRD4 and MAT2A inhibition can potentially improve T-cell function and prevent T-cell exhaustion [[13], [14], [15]]. Whereas few studies on PRMT5 in mouse models describe the opposite effect [16,17]. However, the effect of these inhibitors alone and in combinations on other human immune cells like natural killer (NK) or invariant natural killer T (iNKT) cells in the context of leukemia is not well studied and needs to be elucidated.
We administered inhibitors of BET, MAT2A and PRMT5 in our MLLr leukemia model resulting in anti-leukemic effects that were significantly enhanced by combinatorial drug delivery. Remarkably, these effects could be observed without any negative impact on healthy hematopoietic stem cells. Furthermore, all compounds had a positive effect on immune cells with activation of T, NK and iNKT cells, increase of immunostimulatory cytokines like interferon γ (IFNγ), downregulation of both programmed cell death-1 (PD-1) on the immune effector cells and the respective ligand (PD-L1) on the target cells leading to a more efficient killing of MLLr cells. We thereby establish the rationale to target BET, PRMT5 and MAT2A in the treatment of leukemia patients harboring MLL translocations by inducing both strong anti-leukemic effects and, moreover, positive stimulation of the immune system being essential to sustain remission.
Results
Human CRISPR/Cas9-MLLr model reveals MYC as target in MLLr leukemia
Recently, we established a reliable human CRISPR/Cas9-MLLr model by inducing translocations of the MLL and AF4 or AF9 gene in hematopoietic stem and progenitor cells (HSPCs) derived from both human cord blood (huCB) and bone marrow (huBM) [5], [6], [7]. Hereby, the fusion oncogene expression is under the endogenous promoter leading to unlimited growth potential in in vitro culture systems in contrast to patient samples undergoing apoptosis within a short time frame.
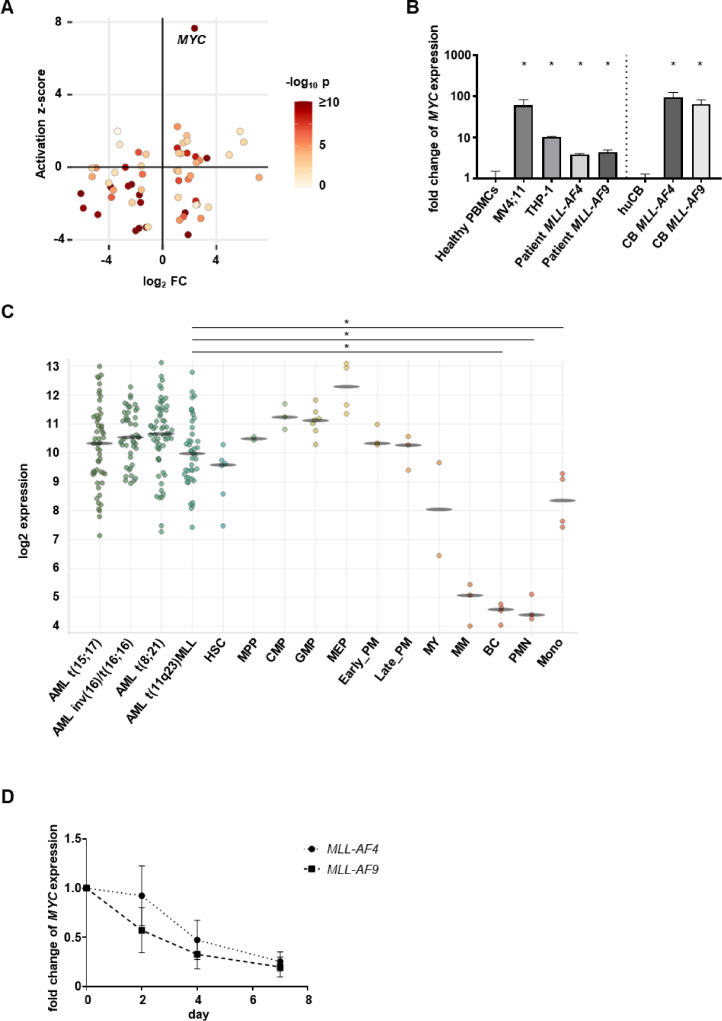
We performed comparative RNA sequencing (RNA-seq) analysis of the human CRISPR/Cas9-MLLr model with respective control cells (CD34+ huCB cells nucleofected with Cas9) and revealed MYC significantly overexpressed and with the highest activation z-score (Fig. 1A). The activation z-score identifies upstream regulators which are mostly responsible for the aberration of gene expression in cells. Therefore, these data indicate, that the MYC pathway plays an important role in MLLr leukemia. For further confirmation of the RNA-seq data, we performed RT-qPCR of our CRISPR/Cas9-MLLr model, cell lines and patient samples demonstrating again higher levels of MYC mRNA compared to control cells (Fig. 1B). Furthermore, we analyzed publicly available patient data (Gene Expression Omnibus (GEO) series accession numbers GSE42519 and GSE13159) supporting the findings of high MYC expression in leukemia and showing a downregulation in healthy cells upon myeloid differentiation (Fig. 1C, bloodspot.eu). Importantly, JQ-1 blocking BRD4 leads to efficient downregulation of MYC over time measured by RT-qPCR in our MLLr model (Fig. 1D). These data indicate that MYC plays a pivotal role in MLLr leukemogenesis that can be efficiently blocked by BRD4 inhibitor JQ-1 amenable for further pharmaceutical studies.
Fig. 1.
MYC is overexpressed in leukemia and can be inhibited by JQ-1. (A) RNA-seq of human CRISPR/Cas9-MLL-AF4 (two different donors, n=2) compared to the respective controls (ctrl, n=2, CD34+ huCB cells nucleofected with Cas9 alone and cultured for the same time) revealed MYC with the highest activation z-score. (B) MYC expression in MLL-AF4 and MLL-AF9 cell lines, patient cells and CRISPR/Cas9-MLLr model (CB MLL-AF4, CB MLL-AF9) determined with RT-qPCR in comparison to respective controls (peripheral blood mononuclear cells (PBMCs) or huCB). Experiment was performed in n=3 technical replicates for the MLL-AF4 patient and in n=3 biological replicates for all other cell types. Statistical analysis with Student's t test, *p < 0.05. (C) MYC expression in AML and normal hematopoiesis. Data from the BloodSpot database (GSE42519 and GSE13159). Statistical analysis with Student's t test, *p < 0.05 HSC: Hematopoietic stem cell, MPP: Multipotential progenitors, CMP: Common myeloid progenitor cell, GMP: Granulocyte monocyte progenitors, MEP: Megakaryocyte-erythroid progenitor cell, Early_PM: Early Promyelocyte, Late_PM: Late Promyelocyte, MY: Myelocyte, MM: Metamyelocytes, BC: Band cell, PMN: Polymorphonuclear cells, Mono: Monocytes. (D) MYC expression in MLL-AF4 and MLL-AF9 model cells following treatment with 100 nM JQ-1 for 2, 4 and 7 days determined by RT-qPCR. Fold change of gene expression was calculated in relation to DMSO-treated cells. Experiment was performed in n=3 biological replicates.
MYC inhibition induces apoptosis in MLLr cells and acts synergistically in combination with targeted therapies
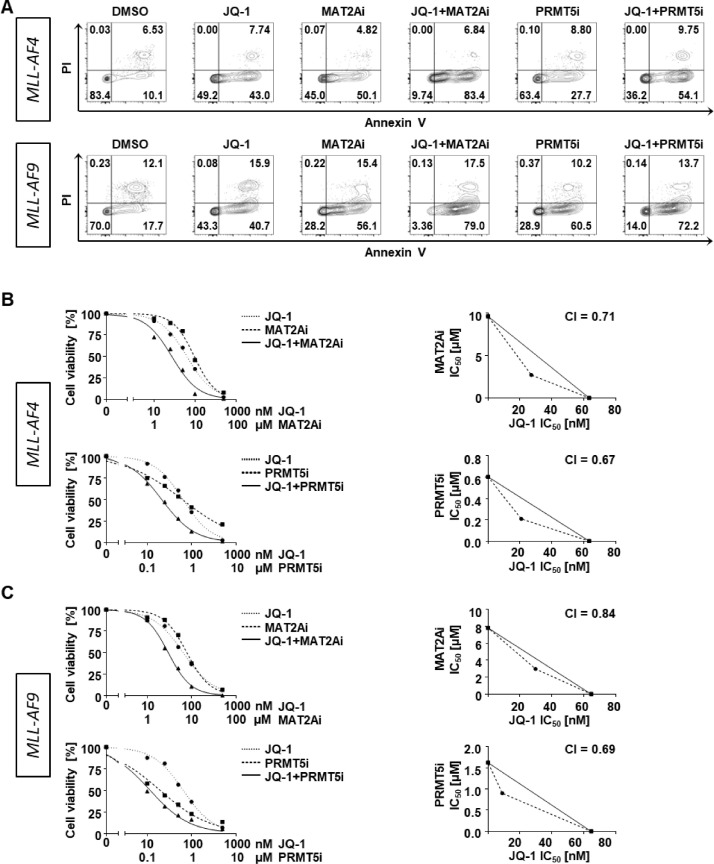
Although in clinical trials JQ-1 was well tolerated with only marginal side effects, the clinical activity was also limited. Therefore, we aimed to further improve the anti-leukemic capacity by combining JQ-1 with other drugs. Recently, MAT2A and PRMT5 have been identified as suitable epigenetic targets with only low side effects for the treatment of poor prognosis MLLr leukemia and we could demonstrate that the combination of both resulted in synergistic effects [5,6,11,18]. Therefore, we used our CRISPR/Cas9-MLL-AF4/-AF9 models to define the anti-leukemic effects of JQ-1 alone and in combination with inhibition of MAT2A (MAT2Ai) or PRMT5 (PRMT5i). Following a treatment period of 7 days, we measured apoptotic and dead cells by Annexin V and propidium iodide (PI) staining. We found an increase of apoptotic cells already by using the respective single agents that was further augmented by the combinatorial treatment (Fig. 2A). To assess if we achieved a synergistical effect, we determined dose response curves of single and combination treatments of both inhibitor variants (JQ-1+MAT2Ai and JQ-1+PRMT5i) in both CRISPR/Cas9-MLLr models at a constant ratio of equipotency of 1:10 or 1:100 using counting beads and flow cytometry. For synergy determination, IC50 values were interpolated at 50% effect level and isobolograms as well as the calculation of combination indices (CI<1) indicated synergism (Fig. 2 B, C).
Fig. 2.
Combinatorial treatments enhance cell apoptosis and promote synergistic effects. (A) MLL-AF4 and MLL-AF9 model cells were assessed for apoptotic and dead cells with Annexin V/PI staining after treatment with 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i for 7 days. Representative data are shown. Dose response curves and isobolograms of (B) MLL-AF4 and (C) MLL-AF9 model cells after treatment with increasing concentrations of JQ-1, MAT2Ai, PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i for 7 days are shown. Experiments were performed in n=3 biological replicates. CI = combination index.
These data provide evidence for the beneficial combinatorial treatment of JQ-1 with targeted therapies against MAT2A and PRMT5.
Combinatorial treatment inhibits cell growth and viability
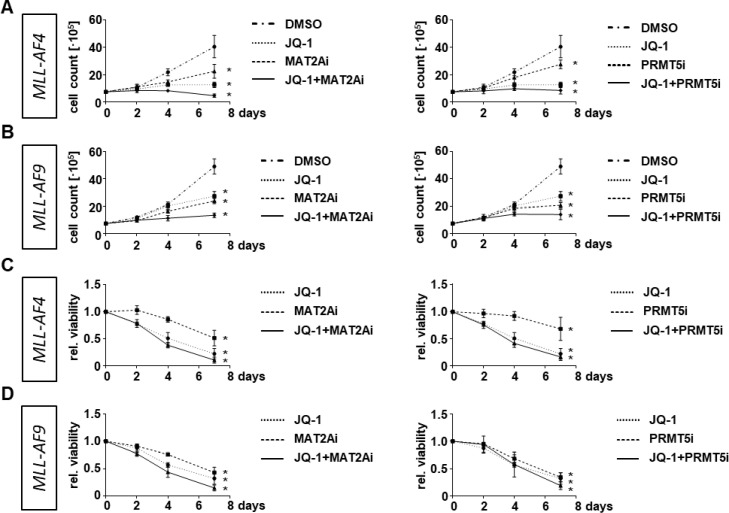
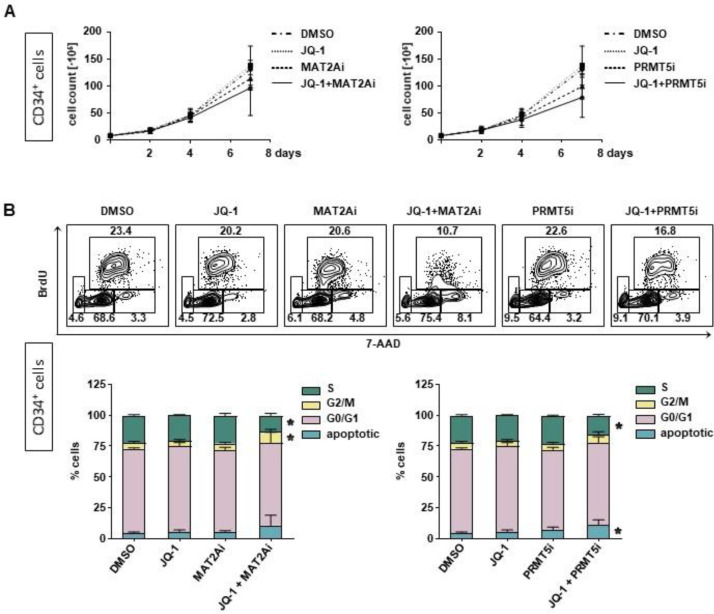
To further investigate the consequences of combinatorial treatment on MLLr cells we assessed the cell growth capacity and determined the metabolic activity upon single and combinatorial treatment. For proliferation, we treated the MLL-AF4 or MLL-AF9 CRISPR/Cas9 cells with JQ-1, MAT2Ai and PRMT5i alone and in combination and assessed the cell count by trypan blue staining and microscopy on day 2, 4 and 7. Cells treated with DMSO served as respective control. Whereas the single treatment led already to a significant reduction of cell proliferation, the combinatorial treatment further increased the anti-proliferative effect resulting in a complete proliferative stop (Fig. 3A, B). The decrease in viability upon inhibitor treatment could also be confirmed by performing AlamarBlue assays (Fig. 3C, D). To assess the potential side effects and toxicity especially of the combinatorial treatment, we performed the same experiments with freshly isolated CD34+ cells derived from huCB as the presumably most susceptible cell population towards a MYC inhibition since HSCs express a high basal level of MYC in publicly available datasets (Fig. 1C). Notably, we detected only a marginally effect on their proliferation capacity, viability and cell cycle distribution upon inhibitor treatment for 7 days (Fig. 4).
Fig. 3.
Targeted therapies and their combinations lead to decreased proliferation and viability in MLLr cells. Cell growth assay of (A) MLL-AF4 cells and (B) MLL-AF9 cells with assessment of cell count on days 2, 4 and 7 following treatments with 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i. Determination of cell viability with AlamarBlue assay of (C) MLL-AF4 and (D) MLL-AF9 cells normalized to DMSO-treated cells. Experiments were performed in n=3 biological replicates. Statistical analysis with Ordinary One-Way ANOVA in relation to DMSO-treated cells, *p < 0.05.
Fig. 4.
Combinatorial treatments spare healthy cells. Following treatment with 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i for 7 days HSPCs derived from huCB were assessed for their (A) cell growth capacity and (B) cell cycle analysis with BrdU labeling. Experiments were performed in n=3 biological replicates. Statistical analysis with Ordinary One-Way ANOVA in relation to DMSO-treated cells, *p < 0.05.
These data suggest that the inhibition of MYC results in a reduction of cell proliferation and viability in MLL fusion protein-driven leukemia without impact on control cells which was further improved by the combinatorial treatment.
The immune system is stimulated upon combinatorial treatment
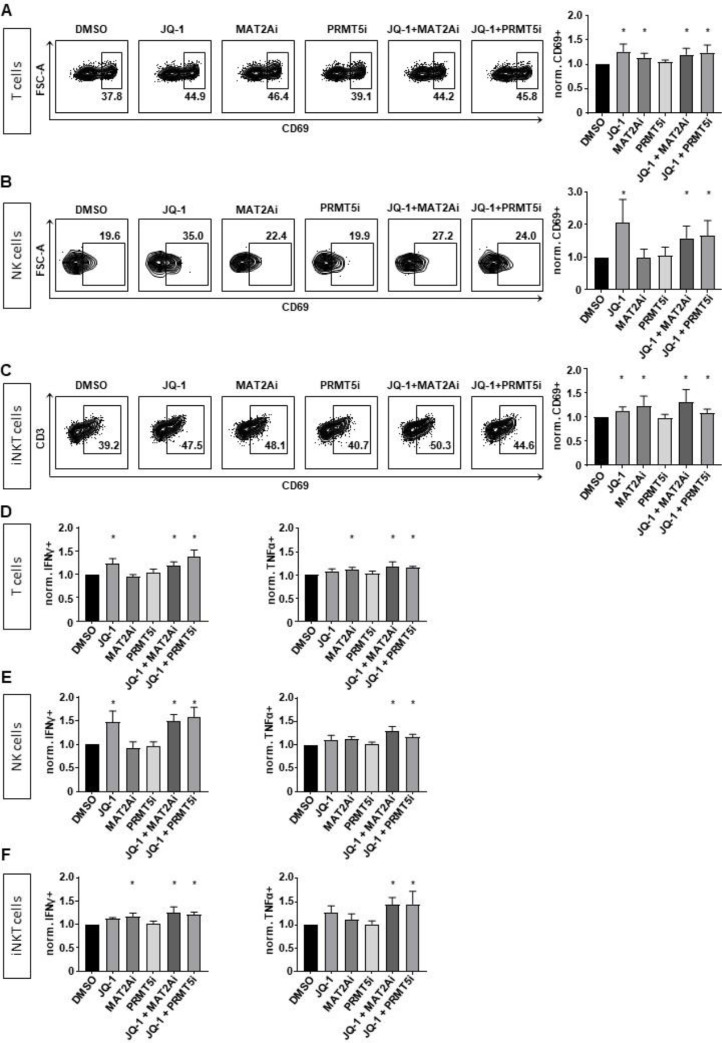
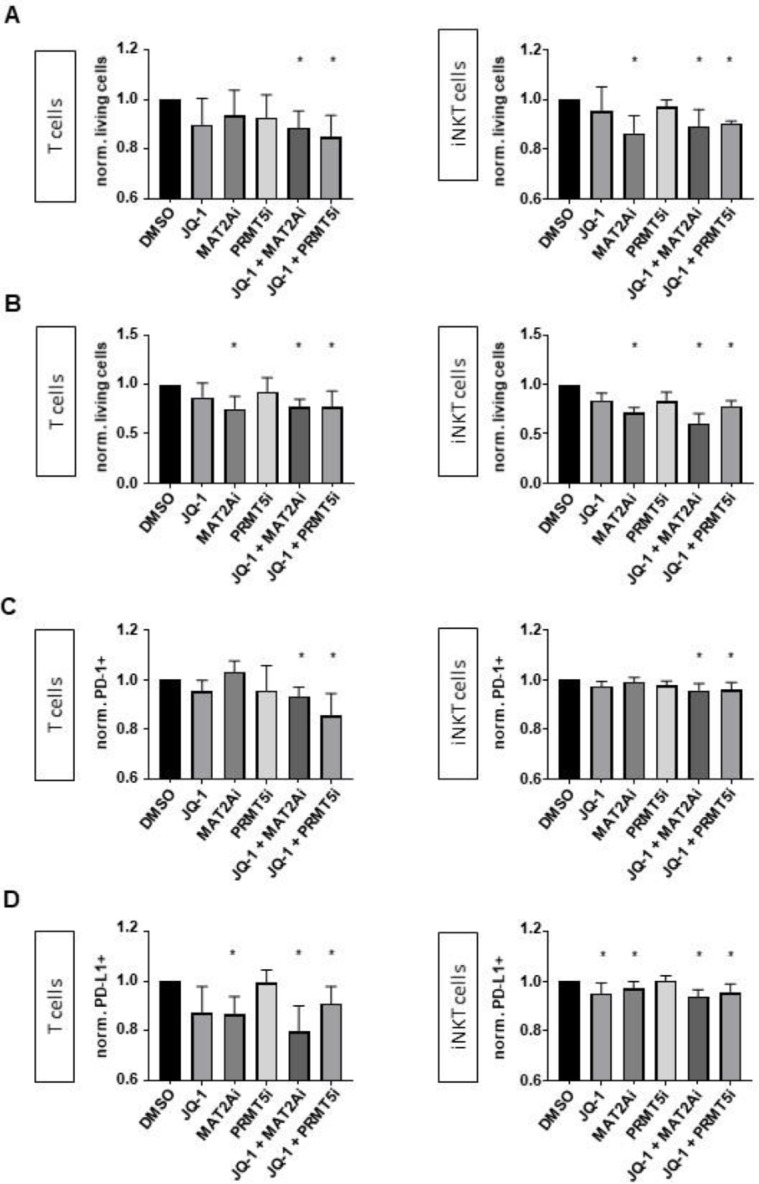
In our next experiments, we investigated the consequences of JQ-1, MAT2Ai and PRMT5i alone and in combinatorial treatment on cells of the immune system. We assessed the activation of T, NK and iNKT cells by measuring CD69 on the cell surface upon respective inhibitor treatment for 24 hours. Strikingly, mostly JQ-1 and MAT2Ai treatment led to a significant upregulation of CD69, whereas a pronounced effect was also achieved by both combinatorial treatments on all immune effector cells (Fig. 5A-C). We further tested if the activation resulted in an increased production of cytokines. Upon combinatorial inhibitor treatment, all immune cells showed a significant increase of the cytokines IFNγ and tumor necrosis factor alpha (TNFα) by intracellular staining and measurement with flow cytometry (Fig. 5D-F) [19]. We further performed killing assays against MLLr cells to determine their cytotoxicity with inhibitor treatment combinations. Regarding NK cells, increased activation did not result in improved killing of MLLr leukemia cells (data not shown, [20]). However, we found a significant increase of cytotoxicity when using the combinatorial pretreated iNKT and T cells (Fig. 6A). To translate the inhibitor treatment in a more clinical setting we pretreated the immune cells and the MLLr cells and determined the respective leukemia lysis following coincubation for 4 hours (Fig. 6B). Also in this setting, the most pronounced cytolytic activity was observed with the combinatorial treatment.
Fig. 5.
Combinatorial treatments activate the immune system. Surface expression of CD69 was assessed by flow cytometry on viable cells following inhibitor treatment alone or in combination for 24h on (A) T, (B) NK and (C) iNKT cells. Experiments were performed in n=5 (T cells), n=4 (NK cells) and n=6 (iNKT cells) biological replicates. Inhibitor concentrations: 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i. Statistical analysis with One-Sample t test. *p < 0.05. (D) T, (E) NK and (F) iNKT cells were treated for a total of 24 hours with the indicated inhibitors and after 20 hours PMA was added. IFNγ and TNFα were measured by intracellular staining and flow cytometry. Experiments were performed in n=4 (T and NK cells) and n=3 (iNKT cells) biological replicates. Inhibitor concentrations: 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i. Statistical analysis with Ordinary One-Way ANOVA in relation to DMSO-treated cells, *p < 0.05.
Fig. 6.
Combinatorial treatments enhance cytotoxicity. (A) Cytotoxicity assay of T and iNKT cells (both previously exposed to inhibitors for 24h) against MLLr target cells. Experiments were performed in n=4 biological replicates. (B) Cytotoxicity assay of T and iNKT cells (both previously exposed to inhibitors for 24h) against MLLr target cells (previously exposed to inhibitors for 48h); PD-1 (C) and PD-L1 (D) expression on effector and target cells was analyzed simultaneously. Experiments were performed in n=5 (T cells) and n=3 (iNKT cells) biological replicates. Inhibitor concentrations: 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i. Statistical analysis with One-Sample t test, *p < 0.05.
The PD-1/PD-L1 interaction is known to inhibit lymphocyte proliferation, survival and effector functions like cytotoxicity and cytokine release, induces apoptosis of tumor-specific T cells and most importantly leads to resistance of tumor cells against the attacking immune system [21,22]. As potential mechanistic explanation for the improved killing, we assessed PD-1 on T and iNKT cells and the respective ligand PD-L1 on the MLLr cells and found on both a significant downregulation of the surface expression upon combinatorial inhibitor treatment (Fig. 6C, D).
These data indicate that the inhibitor combinations not only unfold their anti-leukemic activity by direct cell lysis of target cells but also by improving the activity of the immune system.
Discussion
In this study, we demonstrated that inhibition of MYC in combination with inhibition of other epigenetic targets, MAT2A or PRMT5, serves as ideal approach for the treatment of poor prognosis MLLr leukemia. We used our recently developed human CRISPR/Cas9-MLLr model based on complete translocations of the MLL and AF4 or AF9 gene, respectively, to elucidate the treatment benefit.
MLL fusion proteins unfold their leukemogenic potential by the recruitment of transcriptional activation complexes, such as SEC leading to phosphorylation of RNA polymerase II and hereby promoting transcriptional activation of oncogenes like MYC [23]. The transcription factor BRD4 as part of the PAFc is a member of the BET protein family. An aberrant BRD4 activation has been shown to be important for MLLr leukemia [24]. Therefore, blocking MYC expression by BET inhibitor JQ-1 in MLLr leukemia has already been subjected to study and others were able to demonstrate encouraging preclinical activity of this therapeutic strategy [9,10]. Nevertheless, the final success is hampered by limited clinical activity as single agent showing no convincing survival benefit in adult leukemia patients in a clinical trial (NCT02308761 [25]). However, this trial is not specifically focusing on MLLr patients, and the real number of this leukemic subtype was rather low. Nevertheless, the involvement of the methyltransferases PRMT5 regulated by PAFc, and MAT2A producing SAM is necessary for all methylations in MLLr leukemia. This provides the rationale to further improve the anti-leukemic activity of JQ-1 by additionally blocking these methyltransferases [5,6].
We analyzed both publicly available datasets and our own generated data and retrieved MYC as major player in sustaining leukemogenesis. Our CRISPR/Cas9-MLLr model provided robust data about the inhibition of MYC leading to reduced viability and proliferation and finally apoptosis of the leukemic cells which was further improved by a combinatorial treatment against MAT2A or PRMT5. Strikingly, we could demonstrate synergistic anti-leukemic effects serving as basis for a reasonable therapeutic combinatorial strategy with minimal side effects.
With the immune system playing a key role in inducing and sustaining remissions, a particular focus is now set on maintaining and even promoting immune responses during or after induction therapy to prevent relapse. Especially, inhibition of MYC is known to improve T-cell function in a PD-1-mediated manner [13,14]. Recently, it has been described in hepatocellular carcinoma that MAT2A drives T-cell exhaustion as well [15]. For PRMT5 inhibition rather negative effects on T cells have been described [16,17,26]. However, little is known about the combinatorial effect on human T and even less on NK or iNKT cells which have been a particular focus of this study. NK and iNKT cells are part of the immune system and are known to play a major role in tumor control [27,28].
Upon combinatorial inhibitor treatment, we can observe an activation of these immune cells with an increase of pro-immunogenic cytokines resulting in a more pronounced cellular cytotoxicity. It has been described that PRMT5 is required for murine hematopoiesis and that inhibition can abolish T-cell development and proliferation [16,17]. However, we did not notice negative effects on human cells in our study. In contrast, the combinatorial treatment with JQ-1 turned out to be beneficial. However, all the above-mentioned studies were mainly done in mouse models which may explain the potential species-dependent differences to our human model. More detailed analyses of immune effector cell subsets in direct comparison to mouse cells are necessary to shed more light on the function of PRMT5 in human immune cells.
BET inhibition was recently described to rescue both PD-1-mediated T-cell and CAR T-cell exhaustion [13,29]. Likewise, we found a PD-1 downregulation on T cells and iNKT cells and a reduced expression of PD-L1 on leukemic cells upon combinatorial treatment. The PD-1/PD-L1 interaction has been shown to restrain T-cell activity [30]. Therefore, checkpoint inhibitors in the treatment of cancer patients have been successfully approved [31].
Taken together, in this study we demonstrate preclinical evidence that a combinatorial therapy against MYC and epigenetic key drivers in MLL leukemogenesis leads to both synergistic effects against leukemic cells and stimulation of the immune system by activation of immune cells, increase of immunomodulatory cytokines and downregulation of the PD-1/PD-L1 axis hereby augmenting cytotoxicity.
Our results provide novel insights into the myriad effects of BET, PRMT5 and MAT2A combinatorial inhibition on cancer therapy allowing to improve the poor prognosis of MLLr patients.
Materials and methods
Human CRISPR/Cas9-MLLr model
CD34+ HSPCs were isolated from huCB obtained from the Center for Women's Health (Department of Gynecology) of the University Hospital Tuebingen (IRB approval 751/2015BO2) and maintained in culture as previously described [4]. Written consent was obtained from all patients in compliance with the Declaration of Helsinki. CRISPR/Cas9 was used to target patient-specific MLL-AF4 and -AF9 breakpoints for MLLr model induction as previously described [5,[32], [33], [34]].
Magnetic-activated cell sorting (MACS) and cell culture
iNKT cells derived from PBMCs were expanded over 14-21 days as previously described [35]. Culture-expanded human iNKT cells were isolated using anti-iNKT MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). CD3+ T and NK cells were directly isolated from human PBMCs with CD3 MicroBeads or the NK Cell Isolation Kit (both Miltenyi Biotec, Bergisch Gladbach, Germany). MidiMACS Separator and LS Columns were used according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). Isolated T and NK cells were maintained in RPMI 1640 GlutaMAX Supplement Media (Gibco, Grand Island, USA) containing 10% FCS (Gibco, Grand Island, USA) and 100 IU/ml Penicillin/Streptomycin (Lonza, Basel, Switzerland).
RT-qPCR
RNA was isolated using the NucleoSpin RNA Kit (Macherey-Nagel, Dueren, Germany). RNA was transcribed to cDNA with RevertAid H Minus Reverse Transcriptase, RiboLock RNase Inhibitor, dNTP Mix and Random Hexamers (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's instructions. Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, USA) was used for the amplification of MYC with the following primers: fwd 5′ CCTGGTGCTCCATGAGGAGAC, rev. 5′ CAGACTCTGACCTTTTGCCAGG. Maxima Probe qPCR Master Mix (Thermo Fisher Scientific, Waltham, USA) was used for the amplification of the housekeeper 18S rRNA using the following primers: fwd 5′ CGGCTACCACATCCAAGGAA, rev 5′ GCTGGAATTACCGCGGCT and the 18S rRNA probe [JOE]TGCTGGCACCAGACTTGCCCTC[TAM]. The samples were analyzed on a LightCycler 480 Instrument II (Roche Life Science, Penzberg, Germany) and CT value calculations were performed with the LightCycler 480 Software version 1.5.1. Fold change of MYC expression was calculated as 2−ΔΔCT normalized to 18S rRNA and in relation to respective control cells.
Inhibitor treatment
MLLr cells were seeded at 0.75 * 106 cells/ml in culture media and treated with 100 nM JQ-1 (Adooq Bioscience, Irvine, USA), 10 µM PF-9366 (MAT2Ai, Selleckchem, Houston, USA), 1 µM EPZ01566 (PRMT5i, Sigma-Aldrich, Saint Louis, USA) or the combination of JQ-1 with MAT2Ai or PRMT5i for 7 days. On days 2, 4 and 7, cell counts were assessed using Trypan Blue solution (Sigma-Aldrich, Saint Louis, USA) and a Neubauer counting chamber. On days 2 and 4, cells were re-seeded and re-treated at the original concentrations.
T, NK and iNKT cells were seeded at 1 * 106 cells/ml in their respective culture media and treated with the same inhibitors and concentrations as MLLr cells for 24h.
Apoptosis analysis
Cells were analyzed for apoptosis with Annexin V/PI with the Annexin V-FITC Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions.
Cell viability assay with AlamarBlue
Cell viability was assessed by mixing 90 µl of cell suspension with 10 µl AlamarBlue Cell Viability Reagent (Invitrogen, Waltham, USA) and incubation at 37°C for 2h. Blue, cell-permeable, non-fluorescent resazurin is reduced to red, highly fluorescent resorufin upon entering living cells. The increase in fluorescent signal at 560 nm was measured on a Tecan Infinite 200 Pro M Plex Microplate Reader (Tecan, Maennedorf, Switzerland).
Determination of synergistic effects
An isobologram was generated with IC50 values calculated from dose response curves with increasing concentrations of JQ-1, MAT2Ai, PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i. The CI as an indicator of synergy was calculated according to the Chou-Talalay method [36]. CI=1 defines additivity, CI<1 defines synergism and CI>1 defines antagonism.
Cell cycle analysis
Cell cycle analysis was performed using the FITC BrdU Flow Kit (BD Biosciences, Franklin Lakes, USA) according to the manufacturer's instructions. Cells were incubated with 10 µM BrdU for 40 min, followed by fixation and permeabilization, DNase treatment and staining with anti-BrdU antibody and 7-AAD.
Flow cytometry
For flow cytometric analysis of surface markers, cells were stained with the following antibodies: CD69 (BV711, clone FN50, BioLegend, San Diego, USA), PD-1 (BV650, clone EH12.2H7, BioLegend, San Diego, USA), PD-L1 (BV785, clone 29E.2A3), CD107a (APC, clone H4A3, BioLegend, San Diego, USA). iNKT cells were detected with a PBS57-CD1d tetramer (PE, National Institutes of Health Tetramer Core Facility, Atlanta, USA).
For intracellular antigens the following antibodies were used: IFN-γ (BV421, clone 4S.B3, BioLegend, San Diego, USA), TNF-α (BV605, Mab11, BioLegend, San Diego, USA).
For measurement of cell viability, the following dyes were used: fixable viability dye eFluor506 (eBioscience, San Diego, USA), fixable viability dye eFluor780 (eBioscience, San Diego, USA) or 7-AAD (BD Biosciences, Franklin Lakes, USA).
All measurements were performed on a BD LSRFortessa flow cytometer (BD Biosciences, Franklin Lakes, USA) and analyzed with FlowJo Version 10.8 (BD Biosciences, Franklin Lakes, USA).
Intracellular cytokine staining
After 20h of inhibitor treatment with 100 nM JQ-1, 10 µM MAT2Ai, 1 µM PRMT5i or the combination of JQ-1 with MAT2Ai or PRMT5i, cells were incubated with protein transport inhibitor cocktail (Invitrogen, Waltham, USA) containing 10.6 µM Brefeldin A and 2 µM Monensin for 4h under continued inhibitor treatment. Additionally, Cell Stimulation Cocktail (Invitrogen, Waltham, USA) was added at a final concentration of 25 ng/ml phorbol-12-myristate-13-acetate (PMA) and 500 ng/ml ionomycin. Afterwards, cells were stained for extracellular antigens and live/dead for 20 min at 4°C and fixed in IC Fixation Buffer (eBioscience, San Diego, USA) for 20-60 min at room temperature. Cells were permeabilized with permeabilization buffer (eBioscience, San Diego, USA) and stained for intracellular antigens for another 20-60 min at room temperature, followed by flow cytometric analysis.
Cytotoxicity assay
Target cells were co-cultivated with effector cells for 4h, 16h or 24h at effector:target ratios ranging from 0.5:1 to 5:1 with or without inhibitor (pre-)treatment as indicated in the respective figure legends. Effector and target cells were distinguished by labeling of effector cells with 0.5 µM CellTrace Violet (Invitrogen, Waltham, USA) according to the manufacturer's instructions. Living tumor cell count was assessed by viability staining with 7-AAD (BD Biosciences, Franklin Lakes, USA) for flow cytometry and normalized to respective DMSO-treated controls using Latex Beads (polystyrene, 3.0 µm, Sigma-Aldrich, Saint Louis, USA).
RNA-seq
RNA-seq data have been analyzed as previously described and have been deposited in NCBI's Gene Expression Omnibus with the accession number GSE128342 [5].
Statistical analyses
Student's t test or One-Way ANOVA were used for statistical analysis as indicated in each figure legend after testing for normality with the Shapiro-Wilk test. A suitable post-hoc test was used for multiple comparisons (Dunnet or Holm-Šidák) and p<0.05 was considered statistically significant. Data were analyzed with GraphPad Prism version 9.4 (GraphPad Software, La Jolla, California, USA).
Data sharing statement
For original data, please contact corina.schneidawind@med.uni-tuebingen.de
Author Contributions
RF performed experiments, analyzed and interpreted data. KASG, HK, FK, RS and EE perfomed and analyzed data. DS contributed to the study design, interpreted data and supervised the study. CL provided important advice. TH and JMSH performed bioinformatic analyses. CS conceptualized the study, wrote the manuscript as lead author, interpreted data and supervised the study. All authors critically reviewed and edited the manuscript for content.
Funding
CS was supported by the Wuerttembergischer Krebspreis, José Carreras Leukaemia Foundation (DJCLS 04R/2019), German Research Foundation (DFG, grant number 442788847) and the Clinician Scientist Program (390-0-0) of the Faculty of Medicine Tuebingen. JMSH was supported by a Margarete-von-Wrangell fellowship through the Ministry of Science, Research and the Arts Baden-Wuerttemberg, and together with TH, received funding from the decipherPD transnational consortium on Epigenomics of Complex Diseases (BMBF grant number 01KU1503).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Flow Cytometry Core Facility Berg of the University Hospital Tuebingen for their excellent technical support. Likewise, we would like to thank the Department of Obstetrics and Gynecology of the University Hospital Tuebingen for providing human cord blood.
References
- 1.Nambiar M., Kari V., Raghavan S.C. Chromosomal translocations in cancer. Biochim. Biophys. Acta. 2008;1786(2):139–152. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Meyer C., Burmeister T., Groger D., et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32(2):273–284. doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaussmann A., Wenger T., Eberle I., et al. Combined effects of the two reciprocal t(4;11) fusion proteins MLL.AF4 and AF4.MLL confer resistance to apoptosis, cell cycling capacity and growth transformation. Oncogene. 2007;26(23):3352–3363. doi: 10.1038/sj.onc.1210125. [DOI] [PubMed] [Google Scholar]
- 4.Livio P., Alessandro B., Anna C., et al. Risk of infection in elderly patients with AML and MDS treated with hypomethylating agents. Acta Biomed. 2018;89(Suppl 11):5–39. [Google Scholar]
- 5.Secker KA, Keppeler H, Duerr-Stoerzer S, et al. Inhibition of DOT1L and PRMT5 promote synergistic anti-tumor activity in a human MLL leukemia model induced by CRISPR/Cas9. Oncogene. 2019;38(46):7181–7195. doi: 10.1038/s41388-019-0937-9. [DOI] [PubMed] [Google Scholar]
- 6.Secker KA, Bloechl B, Keppeler H, et al. MAT2A as Key Regulator and Therapeutic Target in MLLr Leukemogenesis. Cancers (Basel) 2020;12(5):1342. doi: 10.3390/cancers12051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Secker K.A., Bruns L., Keppeler H., et al. Only hematopoietic stem and progenitor cells from cord blood are susceptible to malignant transformation by MLL-AF4 translocations. Cancers (Basel) 2020;12(6):1487. doi: 10.3390/cancers12061487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson M.A., Prinjha R.K., Dittmann A., et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J., Shi J., Wang E., et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bill M., Goda C., Pepe F., et al. Targeting BRD4 in acute myeloid leukemia with partial tandem duplication of the MLL gene. Haematologica. 2021;106(9):2527–2532. doi: 10.3324/haematol.2020.271627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik S., Liu F., Veazey K.J., et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia. 2018;32(2):499–509. doi: 10.1038/leu.2017.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serio J., Ropa J., Chen W., et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene. 2018;37(4):450–460. doi: 10.1038/onc.2017.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong M., Gao R., Zhao R., et al. BET bromodomain inhibition rescues PD-1-mediated T-cell exhaustion in acute myeloid leukemia. Cell Death Dis. 2022;13(8):671. doi: 10.1038/s41419-022-05123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang K., Zhang Q., Fan Y., et al. MYC inhibition reprograms tumor immune microenvironment by recruiting T lymphocytes and activating the CD40/CD40L system in osteosarcoma. Cell Death Discov. 2022;8(1):117. doi: 10.1038/s41420-022-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung M.H., Lee J.S., Ma C., et al. Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma. Nat. Commun. 2021;12(1):1455. doi: 10.1038/s41467-021-21804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y., Nagai Y., Okumura M., Greene M.I., Kambayashi T. PRMT5 is required for T cell survival and proliferation by maintaining cytokine signaling. Front. Immunol. 2020;11:621. doi: 10.3389/fimmu.2020.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F., Cheng G., Hamard P.J., et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Investig. 2015;125(9):3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daigle SR, Olhava EJ, Therkelsen CA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evrard D., Dumont C., Gatineau M., et al. Targeting the tumor microenvironment through mTOR inhibition and chemotherapy as induction therapy for locally advanced head and neck squamous cell carcinoma: the CAPRA study. Cancers (Basel) 2022;14(18):4509. doi: 10.3390/cancers14184509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan W.K., Kung Sutherland M., Li Y., Zalevsky J., Schell S., Leung W. Antibody-dependent cell-mediated cytotoxicity overcomes NK cell resistance in MLL-rearranged leukemia expressing inhibitory KIR ligands but not activating ligands. Clin. Cancer Res. 2012;18(22):6296–6305. doi: 10.1158/1078-0432.CCR-12-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng S.Y., Otsuji M., Gorski K., et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001;193(7):839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson D.F., Ellies P., Pires R.T., Tseng S.C. Amniotic membrane transplantation for partial limbal stem cell deficiency. Br. J. Ophthalmol. 2001;85(5):567–575. doi: 10.1136/bjo.85.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amari K., Sasagawa S., Imayoshi N., et al. The CDK4/6-UCHL5-BRD4 axis confers resistance to BET inhibitors in MLL-rearranged leukemia cells by suppressing BRD4 protein degradation. Biochem. Biophys. Res. Commun. 2022;588:147–153. doi: 10.1016/j.bbrc.2021.12.063. [DOI] [PubMed] [Google Scholar]
- 24.Abedin S.M., Boddy C.S., Munshi H.G. BET inhibitors in the treatment of hematologic malignancies: current insights and future prospects. Onco Targets Ther. 2016;9:5943–5953. doi: 10.2147/OTT.S100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roboz G.J., Desai P., Lee S., et al. A dose escalation study of RO6870810/TEN-10 in patients with acute myeloid leukemia and myelodysplastic syndrome. Leukemia Lymphoma. 2021;62(7):1740–1748. doi: 10.1080/10428194.2021.1881509. [DOI] [PubMed] [Google Scholar]
- 26.Geoghegan V., Guo A., Trudgian D., Thomas B., Acuto O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat. Commun. 2015;6:6758. doi: 10.1038/ncomms7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid H., Ribeiro E.M., Secker K.A., et al. Human invariant natural killer T cells promote tolerance by preferential apoptosis induction of conventional dendritic cells. Haematologica. 2022;107(2):427–436. doi: 10.3324/haematol.2020.267583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buechele C., Baessler T., Wirths S., Schmohl J.U., Schmiedel B.J., Salih H.R. Glucocorticoid-induced TNFR-related protein (GITR) ligand modulates cytokine release and NK cell reactivity in chronic lymphocytic leukemia (CLL) Leukemia. 2012;26(5):991–1000. doi: 10.1038/leu.2011.313. [DOI] [PubMed] [Google Scholar]
- 29.Kong W., Dimitri A., Wang W., et al. BET bromodomain protein inhibition reverses chimeric antigen receptor extinction and reinvigorates exhausted T cells in chronic lymphocytic leukemia. J. Clin. Investig. 2021;131(16) doi: 10.1172/JCI145459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X., Gao R., Li Y., Zeng C. Regulation of PD-1 in T cells for cancer immunotherapy. Eur. J. Pharmacol. 2020;881 doi: 10.1016/j.ejphar.2020.173240. [DOI] [PubMed] [Google Scholar]
- 31.Wang D., Lin J., Yang X., et al. Combination regimens with PD-1/PD-L1 immune checkpoint inhibitors for gastrointestinal malignancies. J. Hematol. Oncol. 2019;12(1):42. doi: 10.1186/s13045-019-0730-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer C., Hofmann J., Burmeister T., et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27(11):2165–2176. doi: 10.1038/leu.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer T., Metzler M., Reinhardt D., et al. Analysis of t(9;11) chromosomal breakpoint sequences in childhood acute leukemia: almost identical MLL breakpoints in therapy-related AML after treatment without etoposides. Genes Chromosomes Cancer. 2003;36(4):393–401. doi: 10.1002/gcc.10167. [DOI] [PubMed] [Google Scholar]
- 34.Reichel M., Gillert E., Angermuller S., et al. Biased distribution of chromosomal breakpoints involving the MLL gene in infants versus children and adults with t(4;11) ALL. Oncogene. 2001;20(23):2900–2907. doi: 10.1038/sj.onc.1204401. [DOI] [PubMed] [Google Scholar]
- 35.Schmid H., Schneidawind C., Jahnke S., et al. Culture-expanded human invariant natural killer T cells suppress T-cell alloreactivity and eradicate leukemia. Front. Immunol. 2018;9:1817. doi: 10.3389/fimmu.2018.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]