Summary
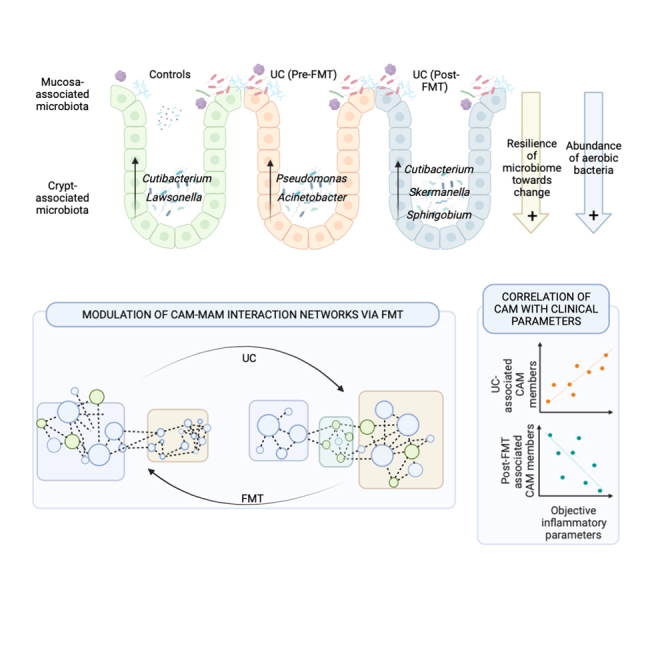
A crypt autochthonous microbial population called crypt-associated microbiota (CAM) is localized intimately with gut regenerative and immune machinery. The present report utilizes laser capture microdissection coupled with 16S amplicon sequencing to characterize the CAM in patients with ulcerative colitis (UC) before and after fecal microbiota transplantation with anti-inflammatory diet (FMT-AID). Compositional differences in CAM and its interactions with mucosa-associated microbiota (MAM) were compared between the non-IBD controls and in patients with UC pre- and post-FMT (n = 26). Distinct from the MAM, CAM is dominated by aerobic members of Actinobacteria and Proteobacteria and exhibits resilience of diversity. CAM underwent UC-associated dysbiosis and demonstrated restoration post-FMT-AID. These FMT-restored CAM taxa correlated negatively with disease activity in patients with UC. The positive effects of FMT-AID extended further in refurbishing CAM-MAM interactions, which were obliterated in UC. These results encourage investigation into host-microbiome interactions established by CAM, to understand their role in disease pathophysiology.
Subject areas: Gastroenterology, Microbiome
Graphical abstract
Highlights
-
•
CAM resides in the colonic crypts of patients with UC
-
•
CAM undergoes dysbiosis in UC and is refurbished by FMT
-
•
FMT-associated CAM members negatively correlate with disease activity in UC
-
•
CAM-MAM interactions undergo UC-associated distortion and are restored by FMT
Gastroenterology; Microbiome
Introduction
Intestinal mucosa accommodates distinct gut biogeographical niches, which host compositionally and functionally distinct microbial members. Crypts are one such pristine microhabitat possessing a unique nutrient profile and a high oxygen tension due to their intimacy with the underlying mucosal tissue. Moreover, crypts act as a senate of intestinal homeostasis, since they harbor the Lgr5+ intestinal stem cells and are seated just above the mucosal immune artillery.1 Once believed to be sterile, the crypts have been recently highlighted to house an autochthonous bacterial community called the “crypt-associated microbiota” (CAM).2 Initial reports provided fluorescence images that proved bacterial colonization of the crypt region and also went on to analyze quantitative variation in crypt colonization between healthy and inflamed mucosal tissue.3,4 The composition of the crypt-resident community, however, was still unexplored. This anticipated discovery was made by Pedron et al. who demonstrated the existence of bacterial communities in mice as well as human colonic crypts and described their core constituent members.2,5 Unsurprisingly, the majority of the colonic crypt inhabitants were either strict or facultative aerobes, in contrast to the luminal or mucosa-associated microbial (MAM) populations, and possessed strong tropism toward the crypt base. Given the strategic position of this microbial consortium in close proximity to the stem cells and the underlying immune system, it is important to understand if and how this population adapts itself in face of disease or microbiota-modulation therapies.
Inflammatory bowel diseases (IBD), comprising of ulcerative colitis (UC) and Crohn disease are debilitating, chronic intestinal inflammatory disorders, characterized by dysbiotic gut microbiota and distorted crypt architecture. Fecal microbiota transplantation (FMT) has been proven to be efficacious in the induction of remission in patients with UC, via modulation of the fecal and mucosal gut microbiota.6,7,8
The present report deciphers disease-associated alterations in CAM composition and tests if FMT combined with an anti-inflammatory diet (FMT-AID) can restore the microbial community in this preserved niche. The study also compares compositional differences between CAM and its close mucosal adherent neighbors - the MAM. Furthermore, we explored if the FMT-associated alterations in CAM correlated with the clinical, endoscopic, and biological inflammatory parameters of the patients with UC and whether CAM-MAM interaction networks undergo rewiring after FMT-AID.
Results
Subject recruitment
Nine patients with the mild-moderate activity of UC, receiving an anti-inflammatory diet along with a multi-donor FMT, administered colonoscopically once a week for a period of 7 weeks, who were recruited as a part of an open-labeled randomized control trial were included in this study.9 The mean age and BMI of the patient cohort were 33 ± 10.3 years and 23.15 ± 6.4 kg/m2, respectively, with a mean disease duration of 75.3 weeks. The median SCCAI and UCEIS scores at recruitment were 6 (IQR: 6–7) and 3 (IQR: 3–3), respectively (Table 1). In addition, 9 subjects with hemorrhoidal bleeds were recruited as non-IBD controls.
Table 1.
Demographic details of healthy controls and patients with ulcerative colitis
| Characteristics | Ulcerative Colitis (n = 9) |
|---|---|
| Age, years, mean ± SD | 33.22 ± 10.31 |
| Sex, Male, n (%) | 3 (33.33%) |
| BMI, kg/m2, mean ± SD | 23.15 ± 6.42 |
| Current alcohol intake, n (%) | 0 (0%) |
| Current smoker, n (%) | 1 (11.11%) |
| Diet (Veg:Non-veg) | 1:2 |
| Disease duration (weeks±SD) | 75.33 ± 37.36 |
| Disease extent | |
| E2 (left-sided [to the splenic flexure] | 6 (66.66%) |
| E3 (beyond the splenic flexure) | 3 (33.33%) |
| Controls (n = 9) | |
| Age, years, mean ± SD | 43.88 ± 11.99 |
| Gender, Male, n (%) | 5 (55.55%) |
| BMI, kg/m2, mean ± SD | 20.92 ± 3.26 |
| Current alcohol intake, n (%) | 1 (11.11%) |
| Current smoker, n (%) | 3 (33.33%) |
| Diet (Veg:Non-veg) | 1:8 |
UC, Ulcerative colitis; HC, Healthy controls, BMI, Body Mass Index; Veg, Vegetarian; Non-veg, Non-vegetarian.
A distinct microbial population inhabits colonic crypts in patients with UC and in controls
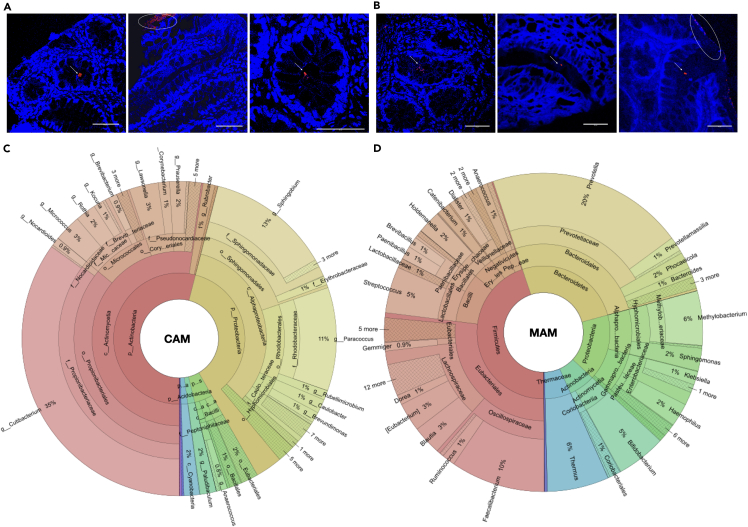
Fluorescence in situ hybridization performed using pan-bacterial Eub338 probes revealed the presence of sparse microbial clusters in colonic crypts, distinct from the overlying layer of MAM, in both controls as well as in patients with UC (Figures 1A and 1B). Microbiome composition analysis showed MAM being dominated by members of phyla Firmicutes (45%), followed by Bacteroidetes (26%), Proteobacteria (16%), and Actinobacteria (6%), while CAM comprised predominantly of aerobic members of Actinobacteria (54%) and Proteobacteria (38%), followed by minor proportions of Firmicutes (4%), Acidobacteria, and Cyanobacteria (2% each). The significant members of CAM included aerobic genera such as Cutibacterium (35%), Sphingobium (13%), Paracoccus (11%), Micrococcus (3%), Lawsonella (3%), Rothia (2%), Prauserella (2%), Kocuria, Corynebacterium, Rubrobacter, Caulobacter, Acinetobacter, and Brevundimonas (1% each) (Figures 1C and 1D).
Figure 1.
Colonic crypts harbor crypt-associated microbiota in distinct from the mucosa-association microbiota
(A and B) Confocal microscopy images of fluorescent in situ hybridization (FISH) of pan-bacterial probes (Alexa 488 labeled Eub338) performed on methacarn fixed tissue sections derived from the recto-sigmoid biopsies from non-UC controls and from patients with mild-moderate UC. Arrows indicate the crypt-associated bacteria, and ellipses indicate.
(C and D) Krona plots highlight the composition of crypt-associated microbiota (CAM) and mucosa-associated microbiota (MAM) in control samples. Scale bar for all FISH images is 50 μm.
Crypts harbor a relatively resilient and conserved microbial community
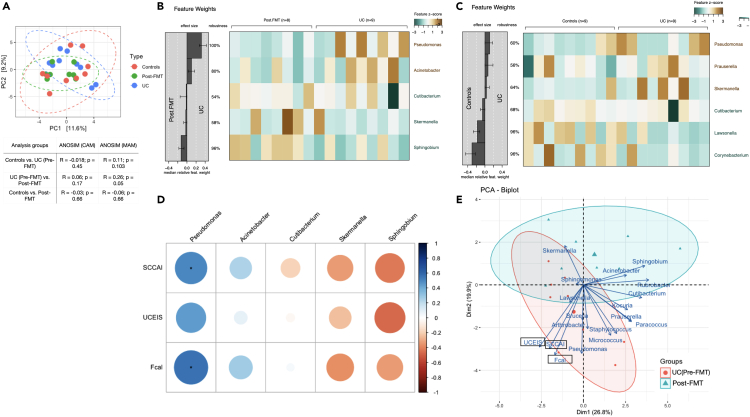
Analysis of CAM diversity in controls, and in patients with UC before and after FMT-AID, showed no significant alterations in the α-diversity indices of Pilou’s evenness, richness, and Shannon index. However, alterations in the β diversity measured through analysis of similarity (ANOSIM) statistic applied on the Aitchison distance showed the three CAM populations to have significantly similar β diversity (Figure 2A). ANOSIM statistic applied to the MAM population revealed dissimilarity in β diversity between control and UC samples (R = 0.11, p = 0.103) and between pre- and post-FMT samples (R = 0.26, p = 0.05), while controls and post-FMT samples showed high similarity (R = −0.06, p = 0.66). However, ANOSIM applied to the CAM populations showed significant similarity across the three groups (ANOSIM R = −0.018, p = 0.45, for controls vs. UC Pre-FMT groups; R = 0.06, p = 0.17 for UC Pre-FMT vs. Post-FMT groups and; R = −0.03; p = 0.66 for controls vs. Post-FMT groups) (Figure 2).
Figure 2.
CAM undergoes compositional shifts in response to FMT in patients with UC, and these shifts correlate with disease-associated clinical parameters
(A) PCA plot depicting the clustering of the three sample groups of CAM on the basis of the Bray Curtis diversity indices along with the table depicting the analysis of similarity (ANOSIM) values performed on the Aitchison Distances, for CAM and MAM samples.
(B and C) Analysis of differentially abundant bacterial genera between the UC pre-FMT and post-FMT samples and between Controls and UC samples performed through a generalized linear model fitted with Lasso regularization, constructed using the SIAMCAT R package.
(D) Heatmap depicting the Spearman correlation between the abundances of the differentially abundant CAM taxa (in the UC pre-FMT vs. Post-FMT analysis) and the UC-associated parameters – SCCAI, UCEIS, and FCal levels.
(E) Biplot ordination depicting clustering of CAM abundance data and UC-associated clinical parameters (SCCAI, UCEIS, and FCal levels). Constructed using “ade4” and “factoextra” R package.
Effect of FMT-AID on UC-associated dysbiosis of CAM
Analysis of differentially abundant bacterial genera in UC, when compared to controls, was carried out by constructing a generalized linear model fitted with Lasso regularization, performed using the SIAMCAT R package. Crypts in patients with UC were found to be enhanced with CAM genera, Pseudomonas, Prauserella, and Skermanella, while Cutibacterium, Lawsonella, and Corynebacterium showed a significant reduction when compared to the controls (Figure 2C). FMT-AID in UC reduced the abundances of CAM genera, Pseudomonas and Acinetobacter while enriching the crypts with Cutibacterium, Sphingobium, and Skermanella (Figure 2B).
Analysis of the correlation of the CAM genera found to be reduced post-FMT and clinical parameters of patients with UC (SCCAI, UCEIS, and FCal levels) showed a significant positive correlation between abundances of Pseudomonas and the SCCAI (R = 0.51, p = 0.04) and the FCal levels (R = 0.57, p = 0.02). The abundance of Pseudomonas also showed a trend of positive correlation with the UCEIS (R = 0.44, p = 0.07). Trends of positive correlation were also observed between the abundances of Acinetobacter and SCCAI (R = 0.24, p = 0.35) and the FCal levels (R = 0.28, p = 0.28). The abundance of CAM member Sphingobium, which was enhanced post-FMT, showed a trend of negative correlation with SCCAI (R = −0.43, p = 0.09), UCEIS (R = −0.47, p = 0.06), and FCal levels (R = −0.34, p = 0.18). Abundances of Skermanella, a CAM member enhanced in the post-FMT samples, showed trends of negative correlation with SCCAI (R = −0.33, p = 0.19), UCEIS (R = −0.25, p = 0.34), and FCal levels (R = −0.37, p = 0.14) (Figure 2D). Principal component analysis biplot of CAM abundance and FCal levels and the disease activity scores (UCEIS and SCCAI), built on the pre-and post-FMT dataset, revealed localization of Pseudomonas with the elevated FCal levels, and higher SCCAI and UCEIS. (Figure 2E).
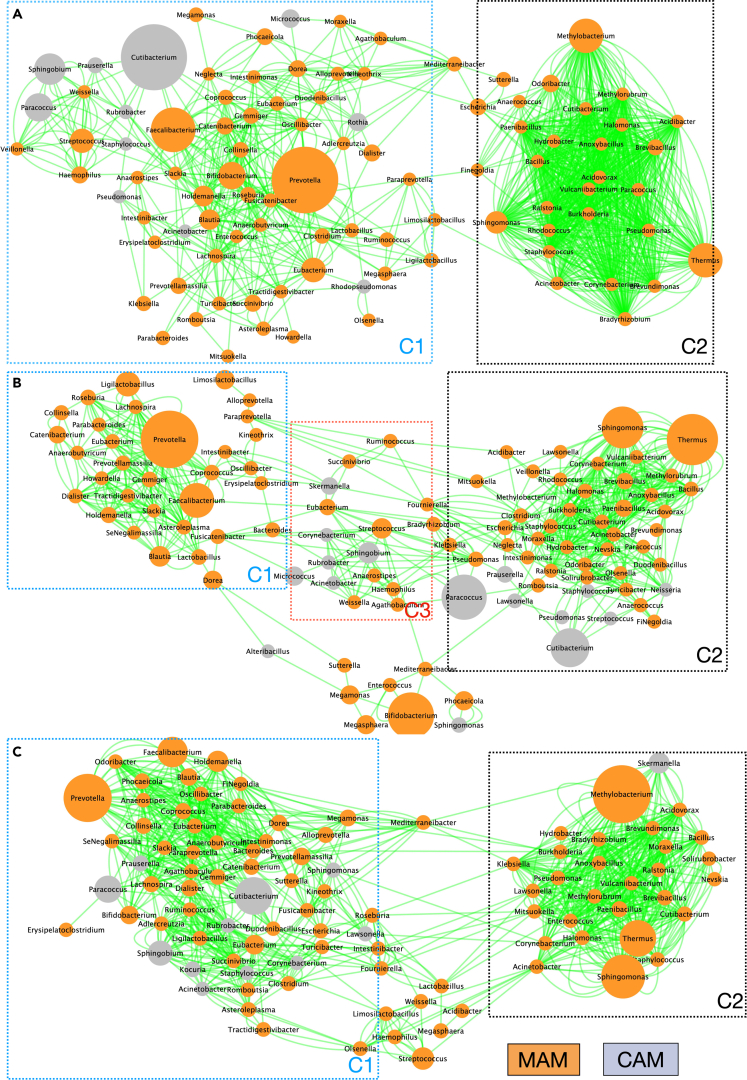
Impact of UC and FMT-AID on the inter-niche (CAM-MAM) interaction networks
To assess whether these evident compositional alterations in CAM and MAM, induced by colitis or FMT-AID, are accompanied by alterations in the inter-niche microbial communications, we constructed interaction networks between inhabitants from the two niches, based on their co-abundance data. The co-occurrence network across all three sample types (controls, UC, and Post-FMT) showed the bacterial genera formed two distinct clusters, with “Cluster-1” comprising majorly of beneficial MAM members – Faecalibacterium, Prevotella, Roseburia, Lachnospira, Bifidobacterium, Catenibacterium, Coprococcus, Gemmiger, Dialister, Eubacterium, Ruminococcus, Megasphaera, Ligilactobacillus, etc., while “Cluster-2” was composed of co-occurring MAM pathobionts such as Pseudomonas, Staphylococcus, Acinetobacter, Burkholderia, Corynebacterium, Brevundimonas, Thermus, Brevibacillus, Methylobacterium, Escherichia, Anoxybacillus, Halomonas, etc. The number of nodes comprising cluster-1 was reduced in UC and restored in post-FMT samples (66 nodes in controls, 32 nodes in UC and 57 nodes in Post-FMT samples) (Figure 3).
Figure 3.
Intern-niche co-occurrence network of the crypt- and mucosa-associated microbiota constructed using Cytoscape-based Co-Net application, for (A) controls, (B) UC, and (C) post-FMT samples
The node sizes vary as per the abundance of the taxa, Each edge depicts a co-occurrence, assessed using Pearson, Spearman, mutual information, Bray Curtis, and Kullback-Leibler dissimilarity, with bootstrap randomization routine. Orange nodes depict MAM members while gray nodes highlight CAM members.
The interaction network for controls showed all major CAM members (Cutibacterium, Prauserella, Sphingobium, Paracoccus, Rubrobacter, Staphylococcus, Acinetobacter, Rothia, and Micrococcus) co-occurring with cluster-1, with no evident interaction with cluster-2 (Figure 3A). While the UC network showed diminished interaction of CAM members with cluster-1, rather they formed their own cluster, overlapping with both major clusters. UC network also highlighted CAM members like Paracoccus, Prauserella, Lawsonella, Pseudomonas, Streptococcus, and Staphylococcus co-occurring with cluster-2 (Figure 3B). The post-FMT network showed restored interaction of all major CAM members with cluster-1 (Figure 3C).
Discussion
The composition and function of luminal and mucosal gut microbial communities and the implications of their dysbiosis have been thoroughly explored in IBD.10 The impact of microbiome restoration therapies such as FMT on IBD restitution and alleviation of gut microbial dysbiosis has also been recently highlighted.7,9,11 This study is the first report describing the composition of a unique and bio-geographically significant crypt-associated microbiota in the colonic tissue of patients with UC and how it is modulated via FMT-AID. In agreement with previous reports, the composition of CAM in health, as well as disease, was found to be dominated by aerobic members, which can be attributed to higher oxygen tension in the crypts.2,5 Even though the population was conserved as a whole, which was noted from the similarity of the diversity across the three sample groups, colitis and FMT-associated alterations of specific CAM members were evident. Interestingly, this FMT-associated enhancement and reduction in specific CAM genera showed their respective negative and positive correlations with FCal levels and disease activity scores (SCCAI and UCEIS), thus highlighting the potential link of CAM with the course of UC and its FMT-mediated restitution.
Delving into the inter-niche co-occurrence network of the crypt- and mucosa-associated bacterial genera, our results showed UC-mediated shrinkage of the cluster-1 (majorly composed of beneficial MAM members) and an expansion of cluster-2 (composed of detrimental pathobionts). This can be explained by inflammation-associated dysbiosis, reduced host antimicrobial defense, and loss of competitive inhibition against pathogenic bacteria in UC. Since the cluster of beneficial MAM bacteria is being curtailed in UC, their co-occurring CAM members are now seen to be adapting and forging newer interactions with the blooming pathobionts (members of cluster-2), as evident from the segregation of CAM members in a “transitional” cluster (cluster-3) between clusters-1 and 2. Apart from the UC-associated dysbiosis, the loss of architectural integrity of gut bio-geographical niches (such as loss of crypt structure) associated with inflammation and deterioration of the mucosal layer can also explain these results. Restoration of these interactions by FMT to a “control-like” network has further strengthened the claim of FMT as a potent microbiome-restorative therapy across distinct gut bio-geographical niches.
Appreciating the risk of contamination while isolating and profiling sparse microbial communities, the present study has adopted all necessary measures: experimental (maintenance of sterility during reagent preparation and sample processing) and analytical (prevalence-based decontamination using experimental controls), to highlight the true inhabitants of crypts.
Limitations of the study
The study is a preliminary work which shows compositional alterations in a relatively unexplored gut biogeographical niche - the crypts, during UC, before and after FMT-AID. Even though our sample counts gave significant insights into the structure of CAM, the study is exploratory in nature and likely underpowered, and the results must be validated in a larger cohort. In addition, the results rely on the 16S marker gene analysis, which limited our access to functional information about the inhabitants. In addition, the role of CAM in IBD pathophysiology can be further consolidated by correlating the abundance of these members with mucosal immunological markers.
Nevertheless, our study shows that CAM undergoes dysbiosis during UC along with a perturbation in its interaction with MAM, and a 7-week FMT-AID can not only restore these compositional shifts but can also recuperate the lost CAM-MAM interactions in the gut. These results encourage further investigation into the host-microbiome interactions established by CAM, to understand their role in disease pathophysiology.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| ProLong™ Gold Antifade Mountant | Invitrogen | Cat#P36930 |
| DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) | Roche | Cat#10236276001 |
| Mutanolysin | Sigma Aldrich | Cat#M9901 |
| Lysostaphin | Sigma Aldrich | Cat#L7386 |
| Lysozyme | Sigma Aldrich | Cat# L6876 |
| guanidine thiocyanate | Sigma Aldrich | Cat#G9277 |
| N-lauryl sarcosine | Sigma Aldrich | Cat#L5777 |
| Deposited data | ||
| Raw 16S rRNA gene sequencing data | This paper | NCBI Sequence Read Archive (SRA) : PRJNA918506 (http://www.ncbi.nlm.nih.gov/bioproject/918506) |
| Oligonucleotides | ||
| 5′- Alexa Fluor 488 labeled Pan-bacteria specific Probe: Eub338 (/5Alex488N/GCTGCCTCCCGTAGGAGT) | IDT (Amann et al.)12 | N/A |
| Critical commercial assays | ||
| Arcturus® PicoPure® DNA Extraction Kit | Thermo Fisher | Cat#KIT0103 |
| Software and algorithms | ||
| QIIME2-2022.11 | Bolyen et al.13 | N/A |
| R version 4.1.0 | CRAN | https://cran.r-project.org/bin/windows/base/old/4.1.0/ |
| QIIME2R version 0.99.6 | N/A | https://github.com/jbisanz/qiime2R.git |
| R Studio Version 1.4.1717 | CRAN | NA |
| decontam version 1.12.014 | Davis et al.14 | https://github.com/benjjneb/decontam |
| QIIME2R | N/A | https://github.com/jbisanz/qiime2R/ |
| SIAMCAT15 | Wirbel et al.15 | https://bioconductor.org/packages/SIAMCAT/ |
| Other | ||
| HistoBond®+S adhesive microscope slides | Paul Marienfeld | Cat#0810501 |
| PEN Membrane Glass Slides | Applied Biosystems | Cat#LCM0522 |
| Glass beads (2.3 mm) | Biospec, USA | Cat#11079125 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Vineet Ahuja (vineet.aiims@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patients (n = 35) with UC having mild-moderate disease activity (Simple Clinical Colitis Activity Index (SCCAI) of 3–9) following at the IBD clinic, All India Institute of Medical Sciences, New Delhi, recruited as a part of an open-labelled randomized control trial as per the inclusion and exclusion criteria were included in this study. The cohort and methods have previously been described.9 Briefly, thirty-five patients with UC were recruited for a seven-week multi-donor FMT, however, only sixteen patients could complete the seven weekly sessions of FMT along with the anti-inflammatory diet, as the trial was withheld due to the Covid lockdown. Out of the sixteen patients who completed the 7-week FMT, the crypt dissectates could be retrieved from the biopsies of only eleven patients. A subset of the above cohort (n = 9 pre-FMT samples and n = 8 post-FMT samples) including the samples which passed DNA quality control were used for the CAM and MAM analysis.
The mean age and BMI of the patients analyzed were 33 ± 10.3 years and 23.15 ± 6.4 kg/m2 respectively, with a mean disease duration of 75.3 weeks. The median SCCAI and UCEIS scores at recruitment were 6 (IQR: 6–7) and 3 (IQR: 3–3) respectively. 66% of the patients analyzed for CAM and MAM analysis in the study were females. Additionally, 9 subjects undergoing routine colonoscopic examination for suspected haemorrhoidal bleeding and having normal rectosigmoid mucosa were also enrolled in the study and their recto-sigmoidal biopsy specimens were also collected once, at the time of examination, which served as non-IBD controls for the present study. The demographic details of patients and controls have been described in Table 1. The study has been approved by the institutional ethics committee of All India Institute of Medical Sciences, New Delhi (IEC -147/06.03.2020).
Method details
Patient recruitment and sample collection
Patients (n = 35) with UC having mild-moderate disease activity (SCCAI of 3–9) following at the IBD clinic, AIIMS, New Delhi, recruited for a seven-week multi-donor FMT trial as per the inclusion and exclusion criteria were included in this study. Patients aged 18–65 years, with mild to moderate UC, UCEIS >1, along with a disease extent of left-sided colitis or pancolitis, who agreed to adhere to the diet schedule and provided written consent were included in the study. Permitted medications and their dosages included oral 5- ASA (stable dose for >4 weeks), azathioprine/6-MP (stable dose for <2 weeks), oral steroids (<20 mg prednisolone equivalent with a mandatory taper of 5 mg/week), and anti-TNF monoclonal antibody (if used more than 6 months back). Patients with severe disease activity or acute severe ulcerative colitis, patients requiring hospitalization or surgery, patients with a history of bowel surgery, concomitant GI infections, pregnancy, or other comorbid illnesses and patients administered topical steroids and/or antibiotics within 2 weeks and 4 weeks of recruitment respectively were excluded from the study. Out of the recruited cohort, only sixteen patients could complete the seven weekly sessions of FMT along with the anti-inflammatory diet, as the trial was withheld due to the Covid lockdown. The recruited patients underwent multi-donor FMT performed colonoscopically. All patients underwent a uniform baseline evaluation including clinical, laboratory and endoscopic assessments. The laboratory assessment included serum inflammatory markers (C-reactive protein). Stool samples were also collected in a sterile airtight container for fecal calprotectin (FCal) measurement and stored at −80°C. All patients underwent a baseline colonoscopy for assessment of endoscopic activity using UCEIS (at the area of maximum endoscopic inflammation).
A subset of the above cohort (n = 9) was used for the CAM and MAM analysis in the present study. Recto-sigmoidal biopsy specimens were collected from each patient at 0 and 8 weeks, for visualization and identification of crypt and mucosa-associated microbial populations. Additionally, 9 subjects undergoing routine colonoscopic examination for suspected haemorrhoidal bleeding and having normal rectosigmoid mucosa were also enrolled in the study and their recto-sigmoidal biopsy specimens were also collected once, at the time of examination, which served as normal controls for the present study. Five biopsies were collected from the recto-sigmoidal region of each enrolled patient and processed separately for FISH, laser capture microdissection and total mucosal DNA isolation, as per the protocols below.
Constitution and administration of anti-inflammatory diet (AID) in patients with UC
Patients with UC undergoing FMT were administered an anti-inflammatory diet, constituted to mediate expansion of T-regulatory cells, promotion of healthy gut microbiota, and improvement in intestinal barrier integrity, and lacked dietary components which have been implicated in mediating gut dysbiosis and/or breach of gut barrier permeability. The diet regimen involved avoidance of gluten-based grains,16 dairy products (except curd) and margarine, processed or canned red meat,17 oils rich in omega-six fatty acids, refined sugars, and food additives.18,19 AID recommended increased intake of fresh fruits and vegetables20, fermented foods, vegetables rich in AhR ligands (cruciferous vegetables)21 and flavonoids, and oils rich in omega-3 fatty acids (mustard, soybean, olive, canola oils etc.).22 Patients were prescribed a diet chart and were counseled to adhere to it, along with regular telephonic follow-ups.
Fluorescence in situ hybridization
One biopsy specimen from each subject was fixed in Methacarn, processed and embedded in paraffin blocks. These embedded tissues were sectioned at 4 μm thickness and sections were taken onto HistoBond+S adhesive microscope slides. The prepared slides were deparaffinized by heating at 60°C for 10 min and followed by treatment with pre-warmed xylene, twice for 2 min each. Slides were then treated with 99.5% ethanol for 5 min at room temperature and air dried. Eub338 probe was mixed with pre-warmed hybridization buffer (20 mM Tris-HCl, pH7.4, 0.9M NaCl, 0.1% SDS, 20% formamide in RNase free water) at a concentration of 1ug/ul and placed onto the tissue sections. Coverslips were placed onto the sections to spread the hybridization solution uniformly over the sections and the slides were then incubated overnight at 50°C in a hybridization chamber. Slides were then washed with FISH wash buffer (20 mM Tris-HCl, pH 7.4, 0.9M NaCl, in RNase-free water) with continuous shaking at 50°C for 20 min, followed by PBS washes for 10 min each. After air drying, a solution of 0.125ug/mL DAPI was applied to the sections and incubated for 5 min in dark. Slides were again washed thrice with PBS for 2 min each and allowed to air dry. Stained sections were mounted in Pro-Long Gold antifade reagent and covered with coverslips. These slides were visualized on a Leica confocal SP8 microscope with a 63x oil objective. Further processing of images was done using LAS-X software.
Laser capture microdissection and DNA isolation from crypt tissue
3 biopsies from each subject were fixed in Methacarn and embedded close together in a single paraffin block. These blocks were sectioned at 8 μm thickness and sections were taken onto Arcturus PEN membrane glass slides (Applied Biosystems) and left at 37°C for 10 min to dry. The slides were stored at 4°C until processed for laser capture microdissection. At the time of preparation, slides were heated at 60° for 2 min in a covered heating chamber and then dipped in pre-warmed xylene for 2 min. This was followed by 2 min in 100% ethanol before the slides were air dried. Sections were then stained with eosin for 20 s and subsequently washed with 100% ethanol twice, for 1 min each. Slides were then air-dried and used for isolating crypts from the mucosal tissue via laser capture microdissection on the Zeiss PALM Micro-Beam system. The dissected crypts were collected in the Zeiss adhesive caps. DNA was isolated from these crypt tissues using the Arcturus Pico Pure DNA extraction kit (Applied Biosystems) as per the manufacturer’s protocol.
DNA isolation for mucosa-associated microbiota
Single biopsy from each subject was snap frozen in liquid nitrogen and processed for whole biopsy DNA isolation as follows: biopsy specimens were thawed, precisely weighed to 2 mg and homogenised using glass beads-2.3 mm (Biospec, USA). This was followed by enzymatic cell lysis using lysozyme (10 mg/mL) (Sigma Aldrich), mutanolysin (25 KU/mL) (Sigma Aldrich) and lysostaphin (4 KU/mL) (Sigma Aldrich) at 37°C for 1 h. Post-incubation samples were subjected to treatment with 4M guanidine thiocyanate (Sigma-Aldrich) and 10% N-lauryl sarcosine (Sigma-Aldrich), before incubation at 37°C for 10 min and 70°C for 1 h. This was followed by mechanical lysis of cells by bead beating cycles before supernatants were transferred to fresh tubes and subjected to protein removal. Nucleic acids were pelleted using ice-cold ethanol (96%) and centrifugation at 14000 rcf for 10 min at 4°C. The final precipitation of DNA was achieved by adding 3M sodium phosphate and 1 mL of 96% ethanol and subjecting the pellet to centrifugation at 14000 rcf for 10 min at 4°C.23 Isolated DNA samples were subjected to quality control check by PCR amplification of 16s rRNA gene using universal primers, followed by library preparation and sequencing on the Illumina MiSeq platform.
Quantification and statistical analysis
Processing of raw reads and decontamination
High-quality raw reads were obtained by using Trimmomatic v0.38 to remove adapter sequences, ambiguous reads and low-quality reads (reads with more than 10% quality threshold (QV) < 20 Phred score). Raw paired-end reads from 26 subjects (9 controls, 9 pre-FMT UC and 8 post-FMT) were subjected to demultiplexing, denoising, and chimaera removal using the DADA2 pipeline of QIIME2-2022.2, which generated amplicon sequence variant (ASV) feature table, and representative sequences.13 The bacterial ASV feature table was then annotated via a taxonomy classifier built using reference sequence annotation and curation pipeline (RESCRIPt), based on the NCBI Bio projects 33175 and 33317 (NCBI 16s rRNA gene RefSeq database).24 The QIIME2 outputs and associated metadata were imported into R as a phyloseq object using QIIME2R (version 0.99.6). Considering the potential risk of contamination associated with the compositional analysis of sparse bacterial populations in low-biomass samples, we performed a “decontamination step” on the raw sequence reads. For the identification of contaminant sequences, the study used micro-dissected Polyethylene-napthalate (PEN) membrane sections obtained from the processed sides, at the sites devoid of tissue sections, as ‘experimental controls’ (ECs). The sequences found to be more prevalent in the ECs than in our samples were designated and discarded as contaminants using the prevalence-based contaminant identification of the ‘decontam’ R package. Decontaminated tables were then used for subsequent analysis through R packages.14
Microbiome data analysis
The β diversity analysis was performed using the ‘phyloseq’ R package using the plot_ordination function on the centred-log transformed data. Alterations in the β diversity were measured through analysis of similarity (ANOSIM) statistics applied to the Aitchison distance. To identify differentially abundant CAM members, the microbiome dataset filtered to include the features present in at least 20% of the samples was subjected to ‘compositional’ transformation and log.clr normalization, followed by 5-repeated 5-fold cross-validation Lasso logistic regression using the ‘SIAMCAT’ R package.15 Spearman correlation analysis between the clinical parameters and bacterial abundances was performed using the ‘psych’ R package and the heatmaps were plotted using the ‘corrplot’ package. PCA biplot of the clinical and CAM abundance datasets was constructed using the ‘ade4’ R package. The Krona charts were constructed using ‘KronaTools’ packages on R. CAM-MAM interaction networks were constructed using Cytoscape-based Co-Net application using Pearson, Spearman, mutual information, Bray Curtis and Kullback-Leibler dissimilarity, with bootstrap randomization routine.25
Acknowledgments
This work was supported by the Science and Engineering Research Board (SERB), Department of Science and Technology, under Grant CRG/2019/005292; and Scheme for Promotion of Academic and Research Collaboration (SPARC), Ministry of Human Resource Development, under Grant 1498. We would like to acknowledge Prof. Soumya Iyengar, National Brain Research Center, Manesar, Dr Divya Chandran, Regional Center for Biotechnology, Faridabad, and Central Instrumentation Facility, National Institute of Plant Genome Research, New Delhi, for their support and guidance in performing the LCM-related experimentation.
Author contributions
M.M., A.B., V.A.: study conception and design; A.B., M.M., V.A.: Sample processing, data curation, data analysis; A.B., M.M., V.A.: data interpretation and manuscript writing; V.A., S.K., G.M., C.V.S.: manuscript editing and review; P.G., M.M., C.V.S.: Imaging experimentations, acquisition and analysis; M.K.S., S.V., M.M., A.B., M.V.: Patient recruitment and sample collection; P.D., M.M., A.B.: Histological processing; V.A.: Study supervision and funding acquisition; All authors reviewed the results and approved the final version of the manuscript.
Declaration of interests
The authors report there are no competing interests to declare.
Inclusion and diversity
We worked to ensure that the study questionnaires were prepared in an inclusive way. We support inclusive, diverse, and equitable conduct of research.
Published: April 23, 2023
Data and code availability
-
•
Raw 16S rRNA gene sequencing data generated by next-generation sequencing platforms, with per-base quality scores, have been deposited at NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/bioproject/918506, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Markandey M., Bajaj A., Ilott N.E., Kedia S., Travis S., Powrie F., Ahuja V. Gut Microbiota: sculptors of the Intestinal stem cell niche in health and Inflammatory bowel disease. Gut Microb. 2021;13:1990827. doi: 10.1080/19490976.2021.1990827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saffarian A., Mulet C., Regnault B., Amiot A., Tran-Van-Nhieu J., Ravel J., Sobhani I., Sansonetti P.J., Pédron T. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio. 2019;10:e01315-19. doi: 10.1128/mBio.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swidsinski A., Weber J., Loening-Baucke V., Hale L.P., Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowan F., Docherty N.G., Murphy M., Murphy T.B., Coffey J.C., O’Connell P.R. Bacterial colonization of colonic crypt mucous gel and disease activity in ulcerative colitis. Ann. Surg. 2010;252:869–875. doi: 10.1097/SLA.0b013e3181fdc54c. [DOI] [PubMed] [Google Scholar]
- 5.Pédron T., Mulet C., Dauga C., Frangeul L., Chervaux C., Grompone G., Sansonettia P.J. A crypt-specific core microbiota resides in the mouse colon. mBio. 2012;3:00116-12. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moayyedi P., Surette M.G., Kim P.T., Libertucci J., Wolfe M., Onischi C., Armstrong D., Marshall J.K., Kassam Z., Reinisch W., et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109.e6. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Paramsothy S., Nielsen S., Kamm M.A., Deshpande N.P., Faith J.J., Clemente J.C., Paramsothy R., Walsh A.J., van den Bogaerde J., Samuel D., et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454.e2. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Narula N., Kassam Z., Yuan Y., Colombel J.F., Ponsioen C., Reinisch W., Moayyedi P. Systematic review and meta-analysis: fecal microbiota transplantation for treatment of active ulcerative colitis. Inflamm. Bowel Dis. 2017;23:1702–1709. doi: 10.1097/MIB.0000000000001228. [DOI] [PubMed] [Google Scholar]
- 9.Kedia S., Virmani S., Vuyyuru S.K., Kumar P., Kante B., Sahu P., Kaushal K., Farooqui M., Singh M., Verma M., et al. Faecal microbiota transplantation with anti-inflammatory diet (FMT-AID) followed by anti-inflammatory diet alone is effective in inducing and maintaining remission over 1 year in mild to moderate ulcerative colitis: a randomised controlled trial. Gut. 2022;71:2401–2413. doi: 10.1136/gutjnl-2022-327811. [DOI] [PubMed] [Google Scholar]
- 10.Caruso R., Lo B.C., Núñez G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020;20:411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 11.Lopetuso L.R., Ianiro G., Allegretti J.R., Bibbò S., Gasbarrini A., Scaldaferri F., Cammarota G. Fecal transplantation for ulcerative colitis: current evidence and future applications. Expert Opin. Biol. Ther. 2020;20:343–351. doi: 10.1080/14712598.2020.1733964. [DOI] [PubMed] [Google Scholar]
- 12.Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirbel J., Zych K., Essex M., Karcher N., Kartal E., Salazar G., Bork P., Sunagawa S., Zeller G. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021;22:93. doi: 10.1186/s13059-021-02306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menta P.L.R., Andrade M.E.R., Leocádio P.C.L., Fraga J.R., Dias M.T.S., Cara D.C., Cardoso V.N., Borges L.F., Capettini L.S.A., Aguilar E.C., et al. Wheat gluten intake increases the severity of experimental colitis and bacterial translocation by weakening of the proteins of the junctional complex. Br. J. Nutr. 2019;121:361–373. doi: 10.1017/S0007114518003422. [DOI] [PubMed] [Google Scholar]
- 17.Dong C., Chan S.S.M., Jantchou P., Racine A., Oldenburg B., Weiderpass E., Heath A.K., Tong T.Y.N., Tjønneland A., Kyrø C., et al. Meat intake is associated with a higher risk of ulcerative colitis in a large European prospective cohort study. J. Crohns Colitis. 2022;16:1187–1196. doi: 10.1093/ecco-jcc/jjac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chassaing B., Van De Wiele T., De Bodt J., Marzorati M., Gewirtz A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. 2017;66:1414–1427. doi: 10.1136/gutjnl-2016-313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laudisi F., Di Fusco D., Dinallo V., Stolfi C., Di Grazia A., Marafini I., Colantoni A., Ortenzi A., Alteri C., Guerrieri F., et al. The food additive maltodextrin promotes endoplasmic reticulum stress–driven mucus depletion and exacerbates intestinal inflammation. Cell Mol. Gastroenterol. Hepatol. 2019;7:457–473. doi: 10.1016/j.jcmgh.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J., McKenzie C., Potamitis M., Thorburn A.N., Mackay C.R., Macia L. Advances in Immunology. 2014. The role of short-chain fatty acids in health and disease; pp. 91–119. [DOI] [PubMed] [Google Scholar]
- 21.Hooper L.V. You AhR what you eat: linking diet and immunity. Cell. 2011;147:489–491. doi: 10.1016/j.cell.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang H.K., Yeh C.H., Iwamoto T., Satsu H., Shimizu M., Totsuka M. Dietary flavonoid naringenin induces regulatory T cells via an aryl hydrocarbon receptor mediated pathway. J. Agric. Food Chem. 2012;60:2171–2178. doi: 10.1021/jf204625y. [DOI] [PubMed] [Google Scholar]
- 23.Bag S., Saha B., Mehta O., Anbumani D., Kumar N., Dayal M., Pant A., Kumar P., Saxena S., Allin K.H., et al. An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci. Rep. 2016;6:26775. doi: 10.1038/srep26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robeson M.S., O’Rourke D.R., Kaehler B.D., Ziemski M., Dillon M.R., Foster J.T., Bokulich N.A. RESCRIPt: reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021;17:e1009581. doi: 10.1371/journal.pcbi.1009581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faust K., Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res. 2016;5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Raw 16S rRNA gene sequencing data generated by next-generation sequencing platforms, with per-base quality scores, have been deposited at NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/bioproject/918506, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.