Abstract
We discovered on the chromosome of Saccharomyces cerevisiae Σ1278b novel genes involved in l-proline analogue l-azetidine-2-carboxylic acid resistance which are not present in the standard laboratory strains. The 5.4 kb-DNA fragment was cloned from the genomic library of the l-azetidine-2-carboxylic acid-resistant mutant derived from a cross between S. cerevisiae strains S288C and Σ1278b. The nucleotide sequence of a 4.5-kb segment exhibited no identity with the sequence in the genome project involving strain S288C. Deletion analysis indicated that one open reading frame encoding a predicted protein of 229 amino acids is indispensable for l-azetidine-2-carboxylic acid resistance. The protein sequence was found to be a member of the N-acetyltransferase superfamily. Genomic Southern analysis and gene disruption showed that two copies of the novel gene with one amino acid change at position 85 required for l-azetidine-2-carboxylic acid resistance were present on chromosomes X and XIV of Σ1278b background strains. When this novel MPR1 or MPR2 gene (sigma 1278b gene for l-proline analogue resistance) was introduced into the other S. cerevisiae strains, all of the recombinants were resistant to l-azetidine-2-carboxylic acid, indicating that both MPR1 and MPR2 are expressed and have a global function in S. cerevisiae.
We previously investigated the cryoprotective effect of amino acids on freezing stress in the yeast Saccharomyces cerevisiae and found that proline, known as an osmoprotectant (6), has a cryoprotective activity nearly equal to that of glycerol or trehalose (37). In bacteria, it was found that feedback inhibition of glutamate kinase acted as the primary mechanism for the control of proline biosynthesis from glutamate (34). Proline-overproducing mutants of Escherichia coli (7), Salmonella enterica serovar Typhimurium (5), and Serratia marcescens (36) had mutations which resulted in desensitization of the feedback inhibition of glutamate kinase (25) and which did not lead to the production of proline oxidase (5). S. cerevisiae synthesizes proline from glutamate via the intermediates γ-glutamyl phosphate, γ-glutamyl semialdehyde, and Δ1-pyrroline-5-carboxylate by almost the same pathway as found in bacteria, but the rate-limiting step has not been determined (39). In general, the microorganisms that overproduce various amino acids have been obtained by isolating mutants resistant to analogues of corresponding amino acids (43). We therefore isolated l-proline analogue l-azetidine-2-carboxylic acid (AZC)-resistant mutants derived from an l-proline-nonutilizing strain of S. cerevisiae (37). Some of the AZC-resistant mutants were found to accumulate a larger amount of proline and showed a prominent increase in cell viability compared to the parent after freezing in the medium.
Recently, we showed that the strain with a disruption of the PUT1 gene, which encodes proline oxidase, accumulated higher levels of proline in the cells and conferred higher resistance to water stress conditions relative to wild-type strains (38). Our results indicated that the intracellular proline level and stress resistance of S. cerevisiae are directly correlated and that the increased flux in the metabolic pathway of proline is effective for constructing new freeze-tolerance yeasts. Therefore, it is of great interest to clarify the mechanism of the proline accumulation and the freeze tolerance in the AZC-resistant mutants.
In this work, we isolated the gene involved in AZC resistance from the genomic library of the mutant. We describe the unexpected discovery of an additional DNA fragment with novel genes MPR1 and MPR2 (sigma 1278b gene for l-proline analogue resistance) in S. cerevisiae Σ1278b and the partial characterization of the genes, which were present only in strains with the Σ1278b background.
MATERIALS AND METHODS
Strains and vectors.
The S. cerevisiae strains used in this study are described in Table 1. Strain MB329-17C was derived from a cross between S288C and Σ1278b (40). An AZC-resistant mutant strain, FH506, with higher levels of intracellular proline was isolated from strain MB329-17C after ethyl methanesulfonate mutagenesis (37). Strain CKY263 was used to induce expression of the MPR1 gene under control of the GAL1 gene promoter. Strain XU-1 is a haploid derived from sake yeast strain K-9 (16). E. coli strain JM109 [recA1 Δ(lac-proAB) endA1 gyrA96 thi-1 hsdR17 relA1 supE44/(F′ traD36 proAB+ lacIq ZΔM15)] was used for subcloning of the MPR1 gene.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Background and/or source |
|---|---|---|
| Σ1278b | α wild-type MPR1 MPR2 AZCr | M. Brandriss |
| MB329-17C | α ura3-52 trp1 put1-54 MPR1 MPR2 AZCr | S288C and Σ1278b, M. Brandriss |
| FH506 | α ura3-52 trp1 put1-54 MPR1 MPR2 AZCr | MB329-17C, this study |
| FH506D1 | α ura3-52 trp1 put1-54 mpr1::URA3 MPR2 AZCr | This study |
| FH506D2 | α ura3-52 trp1 put1-54 MPR1 mpr2::URA3 AZCr | This study |
| FH506D12 | α ura3-52 trp1 put1-54 mpr1::URA3 mpr2::TRP1 | This study |
| S288C | α wild-type mal1 gal2 | C. Kaiser |
| CKY2 | a ura3-52 his4-619 gal2 | S288C, C. Kaiser |
| CKY263 | a leu2-3,112 ura3-52 GAL | S288C, C. Kaiser |
| XU-1 | a ura3 | Japanese sake strain K-9, Y. Kubo |
Two S. cerevisiae-E. coli shuttle vectors, pYES2 (Invitrogen, San Diego, Calif.) and pRS406 (Stratagene, La Jolla, Calif.), both of which contain the bacterial ampicillin resistance gene and the S. cerevisiae URA3 gene, were used for the cloning and for chromosomal integration, respectively, of the MPR1 gene. Plasmids YEp24 (4) harboring the URA3 gene and pRS404 (Stratagene) (33) harboring the TRP1 gene were used for disruptions of the MPR1 and MPR2 genes.
Culture media.
The media used for growth of S. cerevisiae were SD (2% glucose, 0.67% Bacto Yeast Nitrogen Base without amino acids [Difco Laboratories, Detroit, Mich.]) and YPD (2% glucose, 1% Bacto Yeast Extract, 1% Bacto Peptone). SD medium contains ammonium sulfate (0.1%) as the nitrogen source. When appropriate, required supplements were added to the media for auxotrophic strains. Yeast strains were also cultured on SD agar plates containing AZC (Sigma Chemical Co., St. Louis, Mo.). The E. coli recombinant strains were grown in Luria-Bertani (LB) medium (31) containing ampicillin (50 μg/ml). If necessary, 2% agar was added to solidify the medium.
Cloning of the MPR1 and MPR2 genes.
The enzymes used for DNA manipulations were obtained from Takara Shuzo (Kyoto, Japan). Conventional techniques (29) were used for S. cerevisiae genomic DNA preparation and transformation. Genomic DNA was prepared from the AZC-resistant mutant FH506 and partially digested with Sau3AI. Sau3AI fragments larger than 5 kb were ligated into the unique BamHI site of pYES2. The genomic library containing over 10,000 independent E. coli clones was transformed into strain MB329-17C, and Ura+ colonies were replica plated onto SD agar plates containing 3 mg of AZC per ml. Two AZC-resistant colonies were isolated, and the AZC resistance was the plasmid-dependent phenotype. Two plasmids (pMH1 and pMH2) had different but overlapping 5.4-kb inserts based on restriction digestions and DNA sequence analysis. The nucleotide sequence of the cloned DNA fragment was confirmed with a model 377 DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.) by the dideoxy-chain termination method. Plasmid pMH3 was constructed by cloning the 3.7-kb SacI-SacI fragment from pMH1 into the SacI site of pRS406. The linearized pMH3 cut with StuI in the URA3 gene of pRS406 was introduced for integration of the MPR1 gene to the URA3 locus of the recipient strain. To place the open reading frame of the MPR1 gene under control of the GAL1 gene promoter, the 930-bp HindIII-MluI fragment from pMH1 was ligated to the large fragment of pYES2 digested with HindIII and MluI.
Disruptions of the MPR1 and MPR2 genes.
Plasmid pMPR1U or pMPR1T was constructed by deleting the 1.6-kb BglII-MluI fragment containing the MPR1 gene from plasmid pMH1 and inserting the 1.2-kb HindIII fragment containing the URA3 gene of plasmid YEp24 or the 1.6-kb AatII-NaeI fragment containing the TRP1 gene of plasmid pRS404, respectively, by blunt-end ligation. For MPR1 or MPR2 gene disruption, the 3.3-kb SacI-SacI fragment containing mpr1::URA3 of pMPR1U was integrated into the MPR1 or MPR2 locus in strain FH506 by transformation. The Ura+ phenotype was selected, and the gene disruption was verified either by Southern blotting or PCR. Subsequent disruption of the MPR2 or MPR1 gene was performed in a similar manner by using the 3.7-kb SacI-SacI fragment containing mpr1::TRP1 of pMPR1T. The nucleotide sequences of the MPR1 and MPR2 genes were confirmed by analyzing the genomic PCR products of the disruptant. The primers for DNA sequencing were 5′-TTGATATTTAGTGAAGGCGCA-3′, 5′-TTAGCTGAATCCGAGTTGATAGC-3′, 5′-GCTCGAGAAGCTTCGAATGC-3′, 5′-GCCAACCTTCTGACCTCTATG-3′, and 5′-CGACGCGTCGTTATTCGTTCTT-3′.
Southern blot analysis.
Southern blot analysis was carried out using ECL (enhanced chemiluminescence) direct nucleic acid labeling and detection systems (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). As a DNA probe, the DNA fragments of the PRO1 gene, which encodes γ-glutamyl kinase, and of MPR1 were prepared by PCR. For the PRO1 gene, the primers were designed based on the nucleotide sequence determined by Li and Brandriss (20). The forward primer was 5′-CGGAATTCGGCTCTTCATCGCTAGT-3′, and the reverse primer was 5′-CGGGATCCGGTCACTGTGCAAACCT-3′. For the MPR1 gene, the forward primer was 5′-TAGCTGAATCCGAGTTGATAGC-3′, and the reverse primer was 5′-GTGCAATGCATCAACCGGTTC-3′. Unique amplified bands of 1,161 bp for the PRO1 gene and 1,636 bp for the MPR1 gene were purified from agarose gel, and their nucleotide sequences were confirmed. The DNA fragments were then denatured and labeled with horseradish peroxidase according to the protocol recommended by the supplier.
Pulsed-field gel electrophoresis.
Stationary-phase cultures were obtained by growing cells at 30°C for 24 h in 10 ml of YPD medium. The harvested cells were resuspended in cold 50 mM EDTA (pH 8.0), and cell concentrations were determined. The agarose-embedded yeast DNA was prepared by using a CHEF Yeast Genomic DNA Plug kit (Bio-Rad, Hercules, Calif.). The plugs were washed in 0.5× TBE buffer (45 mM Tris-borate [pH 8.3], 1 mM EDTA) before loading. The genomic DNAs were separated in 1.0% low-melting-point preparative-grade agarose (Bio-Rad) on a CHEF-DR III apparatus (Bio-Rad). Pulsed-field gel electrophoresis was carried out for 40 h at 14°C in 0.5× TBE buffer with a switching interval of 75 s and a voltage gradient of 6 V/cm.
Computer analysis of DNA and amino acid sequences.
Sequence data for the 5.4-kb Sau3AI fragment were analyzed by a computer using the program DNASIS (version 3.6; Hitachi Software Engineering, Tokyo, Japan). Based on the DNA sequence, protein homology searches were performed via the World Wide Web by using the BLAST search engine at the National Center for Biotechnology Information (1).
Nucleotide sequence accession number.
The nucleotide sequence of the cloned 5.4-kb DNA fragment including the MPR1 gene found in plasmid pMH1 has been submitted to DDBJ/EMBL/GenBank databases under accession no. AB031349.
RESULTS
Identification of the gene involved in AZC resistance.
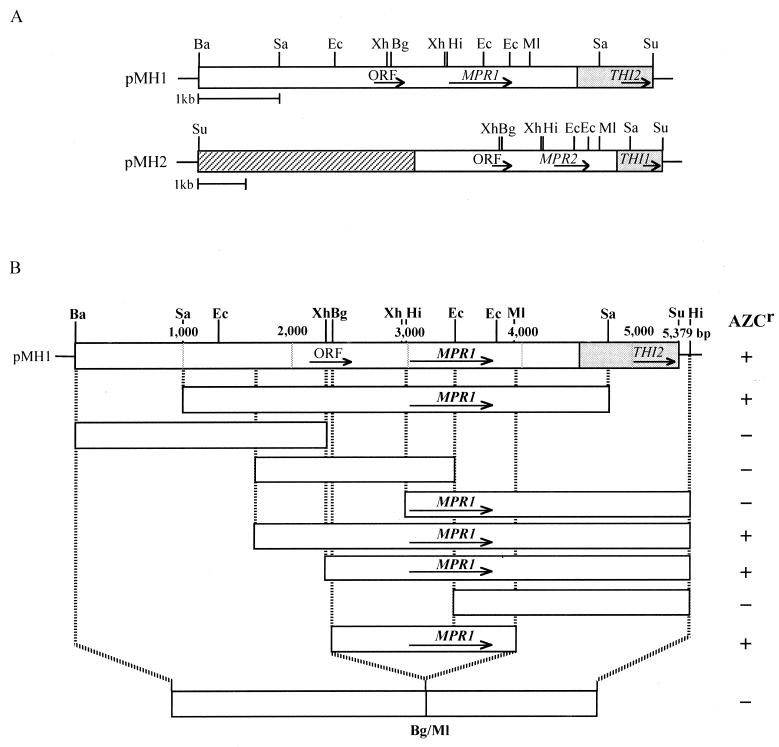
The genomic DNA library of the AZC-resistant mutant FH506 was constructed in a high-copy-number plasmid pYES2. In two Ura+ transformants which showed the AZC-resistant phenotype, two plasmids (pMH1 and pMH2) were isolated and had overlapping 5.4-kb inserts (Fig. 1A). It is worth noting that the nucleotide sequence of an approximately 4.5-kb fragment in the cloned DNA exhibited no sequence identity with the genome sequence of S. cerevisiae S288C, while the 0.9-kb fragment from the 3′ end completely matched those of chromosomes IV (accession no. SCYDL244W), VI (accession no. YSCCHRVIN), X (accession no. SCYJR156C), and XIV (accession no. SCDNANO). Plasmid pMH2 had an unknown 4.6-kb fragment, which is in part sequenced, in addition to the overlapping 5.4 kb (Fig. 1A). The novel 5.4-kb fragment had a G+C content of 35.9%, which is almost equivalent to that of the total DNA (39.5%) in S. cerevisiae S288C. Computer-assisted analysis of the sequenced region confirmed two possible open reading frames encoding a >10-kDa protein besides the THI2 gene, which encodes the transcriptional activator of thiamine biosynthetic genes (Fig. 1A). Deletion analysis indicated that the essential region for AZC resistance lay within the 1.6-kb BglII-MluI fragment containing the open reading frame (Fig. 1B). Thus, we concluded that the region is required for AZC resistance and named it the MPR1 gene.
FIG. 1.
Restriction map of the cloned DNA fragment and deletion analysis to identify the region required for AZC resistance. The predicted size and transcriptional orientation of each deduced open reading frame (ORF) is shown by an arrow. Restriction enzymes: Ba, BamHI; Bg, BglII; Ec, EcoRI; Hi, HindIII; Ml, MluI; Sa, SacI; Su, Sau3AI; Xh, XhoI. (A) Restriction map of the cloned DNA fragment in pMH1 and pMH2. Two plasmids had the overlapping 5.4-kb Sau3AI insert (open box). The region in each plasmid matching the sequence on chromosome XIV (pMH1) or X (pMH2) of S. cerevisiae S288C is indicated by a shaded box. The hatched box in pMH2 represents the unknown, partially sequenced 4.6-kb fragment. (B) Analysis of MPR1 deletion mutants. Each DNA fragment was subcloned into pYES2, and the resultant plasmids were introduced into strain CKY2. AZC resistance of the Ura+ transformants was examined on SD agar plates containing AZC (0.3 mg/ml) after incubation at 30°C for 3 days. +, growth; −, no growth.
Nucleotide sequence analysis of the MPR1 gene.
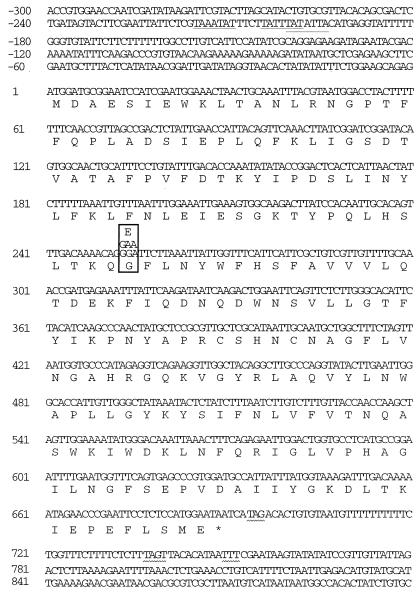
The sequence of 1,200 nucleotides containing the MPR1 gene and its 5′ and 3′ flanking regions is shown in Fig. 2. Sequence analysis revealed one open reading frame from positions +1 to +690, capable of encoding a polypeptide of 229 amino acids with a molecular mass of 26.2 kDa. The first ATG codon is surrounded by purines at positions −3 (G) and +4 (G), bases that have been proposed to play a role in translation (18). Three sequences with some relationship to the TATA box, the consensus sequence TATA(A/T)A(A/T) (3), were found in the upstream region: TAAATAT at −216, TATTTAT at −205, and TATATTA at −201 relative to the start site of the MPR1 open reading frame. All three were located upstream of the HindIII site at −67. When the open reading frame was placed under control of the galactose-inducible GAL1 promoter in pYES2, the recombinant strain CKY263 showed the AZC-resistant phenotype on SD agar plates containing 2% galactose instead of glucose as the source of carbon, indicating that the open reading frame would encode a polypeptide involved in AZC resistance (data not shown). In the 3′ untranslated sequence, a tripartite terminator, 5′-TAG....TAGT....TTT-3′ (44), was found in the region between nucleotides +694 and +752.
FIG. 2.
Nucleotide and predicted amino acid sequences of the MPR1 gene. +1 and the asterisk refer to the putative translational initiation site and the termination codon, respectively. One base change which leads to Gly and Glu at position 85 in MPR1 and MPR2, respectively, is boxed. Matches to known consensus sequences are marked as follows: TATA box (underline) and transcription termination (wavy underline).
Deduced amino acid sequence of the MPR1 gene product.
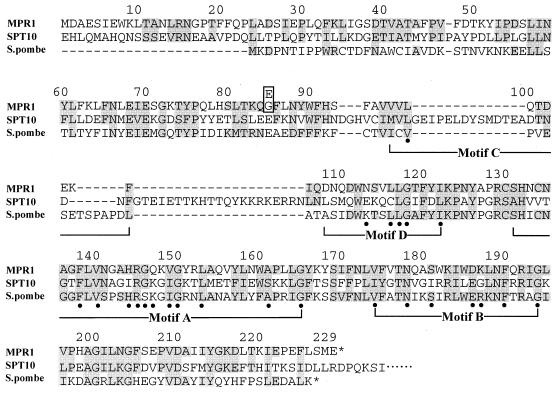
By comparison of the amino acid sequence of the predicted MPR1 protein to entries in the protein databases (SwissProt, PIR, and PRF), the protein sequence was found to be a member of the N-acetyltransferase superfamily (24). In particular, the sequence was homologous to the amino-terminal sequence of the S. cerevisiae SPT10 (SUD1)-encoded protein with 640 amino acids, a negative transcriptional regulator (23, 42). Within the overlapping region of 229 amino acids, 33% of the amino acids were identical, with 50% considered to be similar (Fig. 3). Also, the sequence showed 32% identity to the fission yeast Schizosaccharomyces pombe hypothetical 23.8-kDa protein (24) (Fig. 3).
FIG. 3.
Amino acid sequence deduced from the nucleotide sequence of the MPR1 gene (MPR1) and its alignment with S. cerevisiae Spt10p (SPT10) and the S. pombe hypothetical 23.8-kDa protein (S. pombe). Numbers above the sequences refer to the MPR1 gene. An amino acid residue at position 85 in the MPR1 gene product (Gly for the MPR1 gene product and Glu for the MPR2 gene product) is boxed. Amino acids with identity or similarity are shown in shaded boxes. A horizontal line indicates the absence of the corresponding amino acid residue at this position. Filled dots under the sequences indicate the highly conserved positions in consensus motifs (A to D) of the N-acetyltransferase superfamily (23). The GenBank accession numbers for S. cerevisiae Spt10p and the S. pombe hypothetical 23.8-kDa protein are L24435 and Z67999, respectively.
Chromosomal location of the MPR1 and MPR2 genes.
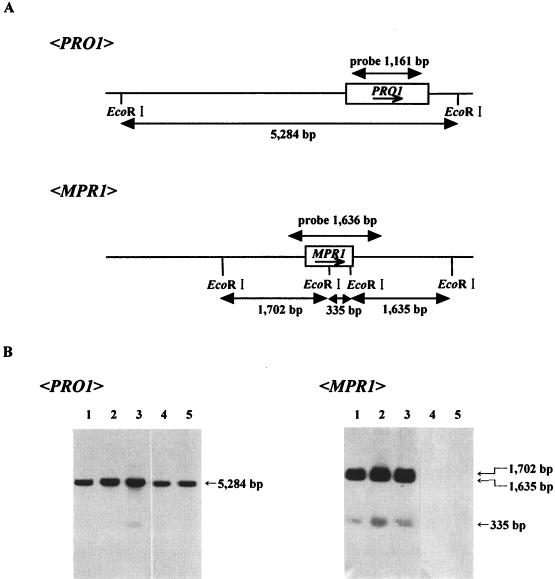
To confirm the origin of the MPR1 gene in Saccharomyces species, total DNA was isolated from each strain (S288C and Σ1278b background) and used for genomic Southern hybridization (Fig. 4A). When the 1,161-bp fragment within the PRO1 gene was used as a probe, a 5,284-bp fragment containing the entire PRO1 gene was detected in all strains tested (Fig. 4B). When the MPR1 gene was used as a probe, three bands corresponding to the 1,702-, 1,635-, and 335-bp fragments were observed only in the Σ1278b background strains (lanes 1 to 3).
FIG. 4.
Southern blot analysis of genomic DNAs from S. cerevisiae strains. (A) Construction used to identify the PRO1 or MPR1 gene. Locations of the EcoRI sites are marked. The PRO1 and MPR1 genes are indicated by shaded boxes; arrows show the direction of transcription. (B) Southern hybridization. Five micrograms of yeast genomic DNA from each strain was digested with EcoRI, electrophoresed on an 0.8% agarose gel, transferred onto a nylon membrane, and hybridized with a 1,161-bp fragment for the PRO1 gene (left) and 1,636 bp fragment for the MPR1 gene (right). Lane 1, Σ1278b; lane 2, FH506; lane 3, MB329-17C; lane 4, S288C; lane 5, XU-I. An EcoT14I-BglII digest of λDNA was used as the DNA size standard.
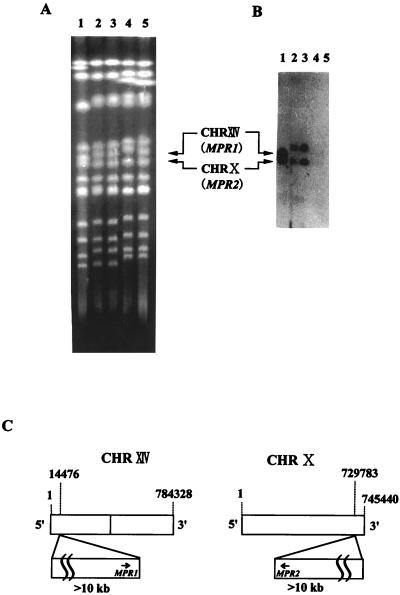
To further elucidate the location of the MPR1 gene, pulsed-field gel electrophoresis (Fig. 5A) and subsequent Southern analysis using the MPR1 gene (1.6-kb BglII-MluI fragment including the putative coding region and the 5′ and 3′ noncoding regions) (Fig. 5B) were carried out. The most striking result was that each of the Σ1278b strains had two copies of the gene, one on chromosome X and the other on chromosome XIV, although the electrophoretic karyotypes varied considerably between the strains tested. Therefore, the two genes at the different locations required for AZC resistance were given different names: MPR1 for the gene on chromosome XIV and MPR2 for the gene on chromosome X. Taken together with the DNA sequencing data, these findings suggested that the MPR1 and MPR2 genes were located on the left arm of chromosome XIV and the right arm of chromosome X, respectively, approximately 15 kb from the telomere in either case (Fig. 5C). These results indicate that the MPR1 and MPR2 genes are present only in strains with Σ1278b background.
FIG. 5.
Chromosomal locations of the MPR1 and MPR2 genes in S. cerevisiae strains. (A) Pulsed-field gel electrophoresis of genomic DNAs from S. cerevisiae strains. Electrophoresis was carried out as described in Materials and Methods, and the gel was stained with ethidium bromide. CHR, chromosome. (B) Southern hybridization. After electrophoresis, each genomic DNA was transferred onto a nylon membrane and hybridized with a 1,636-bp fragment for the MPR1 gene labeled using the ECL direct nucleic acid labeling system. Lane 1, Σ1278b; lane 2, FH506; lane 3, MB329-17C; lane 4, S288C; lane 5, XU-1. (C) Schematic map of the MPR1 gene on chromosome XIV (left) and the MPR2 gene on chromosome X (right) of strain Σ1278b. Predicted transcriptional orientation of MPR1 or MPR2 is shown by an arrow.
Structural comparison of the MPR1 and MPR2 genes.
We cloned and sequenced the 1.6-kb BglII-MluI fragments containing the MPR1 and MPR2 genes from each disruptant. Comparison of the MPR1 and MPR2 nucleotide sequences revealed that both sequences matched perfectly except for only one base at position 254, leading to Gly and Glu at position 85 in MPR1 and MPR2, respectively (Fig. 2). Therefore, the novel DNA fragments containing the MPR1 gene on chromosome XIV and the MPR2 gene on chromosome X were ascertained to be cloned into pYES2, resulting in pMH1 and pMH2, respectively (Fig. 1A).
In addition, the 1.6-kb BglII-MluI fragments containing the MPR1 and the MPR2 genes from each disruptant of the parent strain MB329-17C and the wild-type strain Σ1278b were sequenced. It should be noted that no mutations in the MPR1 and MPR2 genes were found in the three strains FH506, MB329-17C, and Σ1278b (data not shown).
Expression of the MPR1 and MPR2 genes in other S. cerevisiae strains.
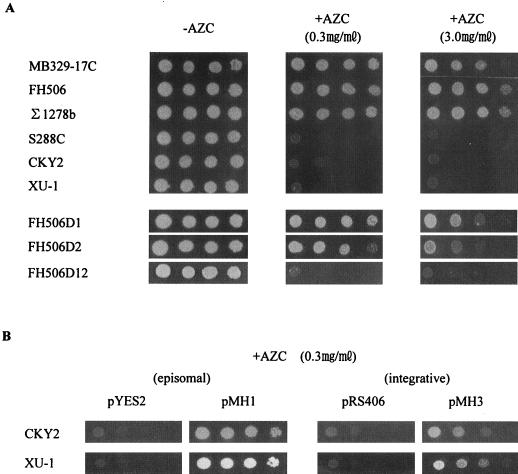
We examined the growth of various S. cerevisiae strains on SD agar plates containing AZC. The strains with Σ1278b background (Σ1278b, FH506, and MB329-17C) showed greater AZC resistance than the other strains (S288C, CKY2, and XU-1) (Fig. 6A). The sensitivities to heat shock and to osmotic stresses in Σ1278b background strains were similar to those of S288C strains (data not shown). MPR1 MPR2 double disruptants failed to grow on AZC-containing plates, whereas MPR1 or MPR2 single disruptants remained AZC resistant (Fig. 6A). These results demonstrate that the MPR1 or MPR2 gene is required for resistance of Σ1278b background strains to l-proline analogues.
FIG. 6.
Growth phenotype of S. cerevisiae strains on minimal medium containing AZC. Approximately 106 cells of each strain and serial dilutions of 10−1 to 10−3 (from left to right) were spotted onto SD plates with appropriate amino acids in the absence (−AZC) and presence (+AZC) of AZC. Plates were incubated at 30°C for 3 days. (A) Function of the MPR1 and MPR2 genes in Σ1278b background strains. The MPR1 and MPR2 disruptants derived from strain FH506 are represented by FH506D1 and FH506D2, respectively. The MPR1 MPR2 double disruptant is represented by FH506D12. (B) Function of the MPR1 gene in the other S. cerevisiae strains. Plasmids pMH1 and pMH3 were constructed from vectors pYES2 and pRS406, respectively.
The episomal plasmid pMH1 harboring the MPR1 gene was introduced into the other laboratory or sake yeast strains, and AZC resistance of the Ura+ transformants was examined. All of the recombinants were capable of growing on SD agar plates containing AZC, thereby acquiring the AZC-resistant phenotype (Fig. 6B), but did not show increases in proline content and cell viability after freezing to the host cells (data not shown). In the case of integration of the MPR1 gene to the URA3 locus of each strain by plasmid pMH3, a similar finding was obtained but the transformants showed AZC resistance slightly lower than that of pMH1 (Fig. 6B). Expression of the MPR2 gene found in plasmid pMH2 also conferred AZC resistance to the other strains (data not shown). These results indicate that both the MPR1 and MPR2 genes present in the Σ1278b strain are expressed in other S. cerevisiae strains, where they play global roles involved in l-proline analogue resistance.
DISCUSSION
In this paper, we describe novel genes involved in l-proline analogue resistance in the chromosome of S. cerevisiae Σ1278b. Previous works have reported only the observation of a point mutation or a deletion of a few bases in comparisons of some genes between Σ1278b and S288C strains. For example, the FLO8 gene encodes a nuclear protein required for diploid filamentous growth, haploid invasive growth, and flocculation (17). Strain S288C has a “naturally occurring mutation” consisting of a single base change and resulting in a stop codon in the coding sequence of the FLO8 gene; the mutation prevents both pseudohyphal development and haploid invasion (21). AQY2, encoding an aquaporin water channel protein, has an 11-bp deletion causing a frameshift in strain S288C (2). Further, the null mutant of the SEM1 gene is viable but is temperature sensitive in a Σ1278b background and not in an S288C background (15). Likewise, it is probable that the gene is essential in one background but not in the other. Some genes, especially those dealing with amino acid permeation, might exist only in strain Σ1278b because the strain seems to have unique genetic features for the specific transport systems of various amino acids (11). For instance, the general amino acid permease is largely inactive when cells are grown with ammonia or glutamate as the nitrogen source, but it is highly active when cells are grown with a poor nitrogen source such as proline or urea (41). To our knowledge, the present study is the first to report the discovery of a novel gene that is present in strain Σ1278b but not in other laboratory strains (S288C, etc.).
Unfortunately, our isolated clones did not cover the whole length of the unknown DNA fragment containing the MPR1 and MPR2 genes, which was not found in the genome project using strain S288C. The >10-kb fragments are inserted at the far right and left ends of chromosomes X and XIV, respectively (Fig. 5). In this study, only a junction at one end was determined, and accordingly we are now extending the sequence so that it will rejoin the complete genome sequence. In standard laboratory strains, chromosome length polymorphism is thought mainly to originate from movement of Ty elements in and out of chromosomes and from Ty-associated duplications or deletions (12). Moreover, the chromosome size variation observed in yeast strains suggests that more drastic chromosomal rearrangements might also occur. It is known that repeated sequences such as Ty elements or solo long terminal repeats are able to promote chromosomal translocations by ectopic recombination (26). If Ty-mediated rearrangements were responsible, one would consider that the novel fragment might reside at the ends of chromosomes, but no repeated sequence involved in Ty elements was found at the end of cloned DNA fragments, suggesting that these DNA fragments are in subtelomeric locations. Much more research on genome evolution is needed to elucidate the local genomic structure and origin of the gene.
When nucleotide sequences were compared, one amino acid change at position 85 was found between the MPR1 and MPR2 genes. This change might be regarded as significant to function, because Glu85 is conserved in Spt10p and the S. pombe hypothetical 23.8-kDa protein. The notable finding is that no mutation occurred in both MPR1 and MPR2 of strain FH506 compared to the parent strain MB329-17C and the wild-type strain Σ1278b, indicating that the MPR1 and MPR2 genes in the AZC-resistant mutant FH506 were both wild type, not mutant. The question arises as to why strain FH506 showed AZC resistance higher than that of strain MB329-17C. The higher AZC resistance of FH506 was dominant to the parent strain and the characteristic segregated 2:2 in tetrads, suggesting that the phenotype is due to a single nuclear mutation (data not shown). Expression of the MPR1 and MPR2 genes isolated from strain FH506 conferred AZC resistance to other S. cerevisiae strains but did not cause increases in proline content and cell viability after freezing of the host cells (data not shown). As we reported previously (37), strain FH506 showed a prominent increase in cellular proline content and cell viability after freezing in the medium compared to MB329-17C. Therefore, strain FH506 seems to have a mutation in the gene involved in proline biosynthesis or degradation, leading it to accumulate intracellular proline and to be more AZC resistant than strain MB329-17C, although the possibility that the mutation occurred in the cloned DNA fragment outside the 1.6-kb BglII-MluI fragment containing the MPR1 or MPR2 gene cannot be ruled out. The search for a mutated gene on FH506 is in progress.
At present, the detailed function of MPR1 or MPR2 is not clear yet. AZC, an l-proline analogue, is known to be taken up into cells and incorporated into proteins instead of proline (22) and to induce the synthesis of abnormal proteins, although probably without a prominent and immediate blockade of protein synthesis. Proline is transported into cells via two transporters, the general amino acid permease (encoded by GAP1) (11) and the proline-specific permease (encoded by PUT4) (19), which respond at the transcriptional and posttranscriptional levels to nitrogen repression (35). Recent reports have shown that the SEC13 gene, encoding an essential component for the secretory pathway, is responsible for targeting certain amino acid permeases to the plasma membrane, and the sec13-1 mutant was more resistant to toxic amino acid analogues such as AZC and 4-aza-dl-leucine (27). The mutant defects on amino acid uptake were specific for GAP1 and PUT4 gene products, and these permeases were unable to be exported to the cell surface (28).
Nucleotide sequencing of the MPR1 and MPR2 genes revealed that the putative protein sequence belongs to the N-acetyltransferase superfamily. Neuwald and Landsman (24) reported that GCN5-related histone acetyltransferases belong to a far more extensive superfamily of both known and putative N-acetyltransferases and that this superfamily is characterized by four conserved regions spanning over 100 residues. Several other members of this superfamily are also associated with gene regulation. These include the S. cerevisiae SPT10-encoded protein and two bacterial proteins, PaiA, which negatively controls sporulation and degradative enzymes in Bacillus subtilis (14), and FlaG, which regulates synthesis and assembly of flagellin proteins in Caulobacter crescentus (32). Spt10p, which has not been shown to contain histone acetyltransferase activity, is required for transcription of particular histone genes (8) and influences the transcription of a variety of other unlinked genes, including PUT1 and PUT2 (23, 42). We confirmed that the SPT10 gene is present in strain Σ1278b (data not shown), although the nucleotide sequence and the transcript have not yet been analyzed. Because we assume that the MPR1 or MPR2 gene functions as an Spt10p-like transcriptional regulator, we will identify the target gene(s) by using the AZC-sensitive mutant derived from the AZC-resistant S288C recombinant carrying the MPR1 gene on pYES2. Preliminary experiments revealed that GDH1 (encoding NADP-specific glutamate dehydrogenase) and RSP5 (encoding ubiquitin-protein ligase) might be the candidates (data not shown). Both gene products are known to repress GAP1 and PUT4 or inactivate Gap1p and Put4p in the presence of ammonia (11, 13). We are now investigating the MPR1 and MPR2 function based on the hypothesis that both genes repress or inactivate the GAP1 and PUT4 genes by regulating expression of the GDH1 and RSP5 genes, leading to the l-proline analogue resistance phenotype. Also, the amino acid sequence of the predicted MPR1 and MPR2 gene products is homologous with the S. pombe hypothetical 23.8-kDa protein. It should be of interest to investigate the S. pombe gene at both transcriptional and translational levels.
ACKNOWLEDGMENTS
We thank M. C. Brandriss, University of Medicine and Dentistry of New Jersey, Newark, for her gift of yeast strains and valuable discussions, and we thank C. Kaiser, Massachusetts Institute of Technology, Boston, and Y. Kubo, Fukui Food Processing Research Institute, Fukui, Japan, for providing yeast strains.
This work was supported by a grant from the Fukui Prefectural Scientific Research Foundation to H.T.
REFERENCES
- 1.Altschul S F, Boguski M S, Gish W, Wooten J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 2.Bonhivers M, Carbrey J M, Gould S J, Agre P. Aquaporins in Saccharomyces. Genetic and functional distinctions between laboratory and wild-type strains. J Biol Chem. 1998;273:27565–27572. doi: 10.1074/jbc.273.42.27565. [DOI] [PubMed] [Google Scholar]
- 3.Breathnach R, Chambon P. Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 4.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 5.Csonka L N. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet. 1981;182:82–86. doi: 10.1007/BF00422771. [DOI] [PubMed] [Google Scholar]
- 6.Csonka L N. Physical and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandekar A M, Uratsu S L. A single base pair change in proline biosynthesis genes causes osmotic stress tolerance. J Bacteriol. 1988;170:5943–5945. doi: 10.1128/jb.170.12.5943-5945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dollard C, Ricupero-Hovasse S L, Natsoulis G, Boeke J D, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenson I, Mousset M, Wiame J M, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- 10.Grenson M, Hou C, Grabeel M. Multiplicity of the amino acid permease in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol. 1970;103:770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grenson M, Hou C. Ammonia inhibition of the general amino acid permease and its suppression in NADPH-specific glutamate dehydrogenaseless mutants of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1972;48:749–756. doi: 10.1016/0006-291x(72)90670-5. [DOI] [PubMed] [Google Scholar]
- 12.Grivell L A, Planta R J. Yeast: the model “eukaryote.”. Trends Biotechnol. 1990;8:241–243. doi: 10.1016/0167-7799(90)90185-z. [DOI] [PubMed] [Google Scholar]
- 13.Hein C, Springael J Y, Volland G, Haguenauer-Tsapis R, Andre B. NP11, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 14.Honjo M, Nakayama A, Fukazawa K, Kawamura K, Ando K, Hori M, Furutani Y. A novel Bacillus subtilis gene involved in negative control of sporulation and degradative-enzyme production. J Bacteriol. 1990;172:1783–1790. doi: 10.1128/jb.172.4.1783-1790.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäntti J, Lahdenranta J, Olkkonen V M, Soderlund H, Keränen S. SEM1, a homologue of the split hand/split foot malformation candidate gene Dss1, regulates exocytosis and pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1999;96:909–914. doi: 10.1073/pnas.96.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitamoto K, Oda K, Gomi K, Takahashi K. Genetic engineering of a sake yeast producing no urea by successive disruption of arginase gene. Appl Environ Microbiol. 1991;57:301–306. doi: 10.1128/aem.57.1.301-306.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi O, Suda H, Ohtani T, Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Mol Gen Genet. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codons by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasko P F, Brandriss M C. Proline transport in Saccharomyces cerevisiae. J Bacteriol. 1981;148:241–247. doi: 10.1128/jb.148.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Brandriss M C. Proline biosynthesis in Saccharomyces cerevisiae: molecular analysis of PRO1 gene, which encodes γ-glutamyl kinase. J Bacteriol. 1992;174:4148–4156. doi: 10.1128/jb.174.12.4148-4156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Styles C A, Fink G R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizzen L A, Welch W J. Characterization of the thermotolerant cell. I. Effects on protein synthesis activity and the regulation of heat-shock protein to expression. J Cell Biol. 1988;106:1105–1116. doi: 10.1083/jcb.106.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natsoulis G, Winston F, Boeke J D. The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics. 1994;136:93–105. doi: 10.1093/genetics/136.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 25.Omori K, Suzuki S, Imai Y, Komatsubara S. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J Gen Microbiol. 1992;138:693–699. doi: 10.1099/00221287-138-4-693. [DOI] [PubMed] [Google Scholar]
- 26.Rachidi N, Barre P, Blondin B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:841–850. doi: 10.1007/s004380050028. [DOI] [PubMed] [Google Scholar]
- 27.Roberg K J, Rowley N, Kaiser C A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberg K J, Bickel S, Rowley N, Kaiser C A. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M D, Winston D F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schoenlein P V, Lui J, Gallman L, Ely B. The Caulobacter crescentus flaFG region regulates synthesis and assembly of flagellin proteins encoded by two genetically unlinked gene clusters. J Bacteriol. 1992;174:6046–6053. doi: 10.1128/jb.174.19.6046-6053.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith C J, Deuth A H, Rushlow K E. Purification and characteristics of a glutamate kinase involved in Escherichia coli proline biosynthesis. J Bacteriol. 1984;157:545–551. doi: 10.1128/jb.157.2.545-551.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanbrough M, Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugiura M, Kisumi M. Proline production by regulatory mutants of Serratia marcescens. Appl Environ Microbiol. 1985;49:782–786. doi: 10.1128/aem.49.4.782-786.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagi H, Iwamoto F, Nakamori S. Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl Microbiol Biotechnol. 1997;47:405–411. doi: 10.1007/s002530050948. [DOI] [PubMed] [Google Scholar]
- 38.Takagi H, Sakai K, Morida K, Nakamori S. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;184:103–108. doi: 10.1111/j.1574-6968.2000.tb08998.x. [DOI] [PubMed] [Google Scholar]
- 39.Tomenchok D M, Brandriss M C. Gene-enzyme relationship in the proline biosynthetic pathway of Saccharomyces cerevisiae. J Bacteriol. 1987;169:5364–5372. doi: 10.1128/jb.169.12.5364-5372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S-S, Brandriss M C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol. 1986;6:2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiame J-M, Grenson M, Arst H N. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv Microb Physiol. 1985;26:1–88. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita I. Isolation and characterization of the SUD1 gene, which encodes a global repressor of core promoter activity in Saccharomyces cerevisiae. Mol Gen Genet. 1993;241:616–626. doi: 10.1007/BF00279904. [DOI] [PubMed] [Google Scholar]
- 43.Yoshinaga F, Nakamori S. Production of amino acids. In: Herrmann K M, Sommerville R L, editors. Amino acids: biosynthesis and genetic regulation. London, England: Addison-Wesley Publishing Company; 1983. pp. 405–429. [Google Scholar]
- 44.Zaret K S, Sherman F. Mutationally altered 3′ ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. Cell. 1982;28:563–573. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]