Summary
Cancer and autism spectrum disorder/developmental delay (ASD/DD) are two common clinical phenotypes in individuals with germline PTEN variants (PTEN hamartoma tumor syndrome, PHTS). Burgeoning studies have shown that genomic and metabolomic factors may act as modifiers of ASD/DD versus cancer in PHTS. Recently, we showed copy number variations to be associated with ASD/DD versus cancer in these PHTS individuals. We also found that mitochondrial complex II variants occurring in 10% of PHTS individuals modify breast cancer risk and thyroid cancer histology. These studies suggest that mitochondrial pathways could act as important factors in PHTS phenotype development. However, the mitochondrial genome (mtDNA) has never been systematically studied in PHTS. We therefore investigated the mtDNA landscape extracted from whole-genome sequencing data from 498 PHTS individuals, including 164 with ASD/DD (PHTS-onlyASD/DD), 184 with cancer (PHTS-onlyCancer), 132 with neither ASD/DD nor cancer (PHTS-neither), and 18 with both ASD/DD and cancer (PHTS-ASDCancer). We demonstrate that PHTS-onlyASD/DD has significantly higher mtDNA copy number than PHTS-onlyCancer group (p = 9.2 × 10−3 in all samples; p = 4.2 × 10−3 in the H haplogroup). PHTS-neither group has significantly higher mtDNA variant burden than PHTS-ASDCancer group (p = 4.6 × 10−2); the PHTS-noCancer group (PHTS-onlyASD/DD and PHTS-neither groups) also shows higher variant burden than the PHTS-Cancer group (PHTS-onlyCancer and PHTS-ASD/Cancer groups; p = 3.3 × 10−2). Our study implicates the mtDNA as a modifier of ASD/DD versus cancer phenotype development in PHTS.
Keywords: PTEN, genomic modifiers, mitochondrial DNA (mtDNA), autism spectrum disorder, cancer
Cancer and autism spectrum disorder/developmental delay (ASD/DD) are seemingly disparate phenotypes in individuals with germline PTEN variants (PHTS). We investigated the mitochondrial DNA (mtDNA) landscape in PHTS and demonstrate differences in copy number and variant burden according to phenotype, thus serving as a modifier of ASD/DD versus cancer in PHTS.
Introduction
PTEN (MIM: 601728), a classical tumor suppressor gene, is a shared risk gene for cancer and autism spectrum disorder (ASD, MIM: 605309).1,2,3,4 PTEN dysfunction can cause a wide range of phenotypes including macrocephaly, benign overgrowths and malignant neoplasia, metabolic alterations, and neurodevelopmental disorders (NDD), such as ASD and developmental delay (DD).5 Two common, but seemingly disparate, clinical phenotypes of individuals with PTEN hamartoma tumor syndrome (PHTS, MIM: 158350), defined as having germline PTEN variants, are cancer and ASD/DD. Indeed, PTEN is one of the most prevalent and penetrant risk genes for ASD, with 2%–5% ASD individuals carrying germline PTEN variants.1,4,6,7,8,9,10 However, it is still impossible to accurately predict development of cancer and/or ASD/DD for any single PHTS individual (versus group), including in those having an identical PTEN genotype.
Burgeoning studies have shown that genomic and metabolic factors may act as modifiers of ASD/DD versus cancer in PHTS.11,12,13 We showed that variants in subunits of succinate dehydrogenase, also known as mitochondrial complex II (collectively referred to as SDHx), modify breast cancer risk and thyroid cancer histology in PHTS individuals though these occur only in 10%.14,15 Relatedly, we recently illustrated that metabolic profiles could serve as a biomarker and enable more accurate prediction of PHTS-ASD/DD versus PHTS-Cancer at the individual level.11,12 More recently, we showed that copy number variations in genomic DNA are associated with ASD/DD compared with cancer development in individuals with PHTS.13 These data support the hypothesis that genetic or genomic modifying factors exist in PHTS.
The crosstalk between SDHx and metabolism suggests that mitochondrial pathways may act as an important factor in PHTS phenotype development. Importantly, despite identifying several possible modifiers of phenotypic outcomes in PHTS,11,12,13 the vast majority of individuals with PHTS remain without known modifying factors. Mitochondria are not only the powerhouses of cells, but they also play a key role in regulating cellular metabolism.16 This semi-independent cellular organelle contains its own genome housed as mitochondrial DNA (mtDNA), which encodes both RNA and protein. The 13 mitochondrial gene-coding proteins are key components of enzyme complexes of the oxidative phosphorylation system/electron transport chain.16 Based on these observations and premises, we hypothesized that the mitochondrial genome (mtDNA) content and variation could also be modifiers of ASD/DD versus cancer phenotype development in PHTS.
Material and methods
Study participants, sample collection, processing, and sequencing
We performed whole-genome sequencing from germline genomic DNA derived from 599 peripheral blood samples from individuals with PHTS, followed at the Cleveland Clinic’s PTEN Multidisciplinary Clinic and Center of Excellence (Cleveland, Ohio, USA) (Table S3). Each individual had only one sequencing sample. We were interested in the dichotomy of cancer and ASD/DD phenotypes. We categorized samples into four groups by phenotype, including onlyASD/DD, onlyCancer, ASDCancer, and neither (no ASD/DD and no cancer). The ASD/DD group includes individuals diagnosed with ASD/DD regardless of cancer status; the Cancer group includes individuals diagnosed with cancer regardless of ASD/DD status. This study was approved by the Cleveland Clinic Institutional Review Board, and all participants gave written informed consent. To ensure uniform phenotyping, after informed consent was obtained, checklists to document the presence or absence of specific features were completed by specialist genetic counselors or physicians (who have served in the PTEN Multidisciplinary Clinic for years and led by C.E.) concurrently with withdrawal of blood specimen. DNA sequencing of germline genomic DNA was performed at the Broad Institute Genomic Services (Cambridge, Massachusetts, USA). Whole-genome sequencing was performed with Illumina HiSeq XTM Ten platform with over 30x coverage in 150-bp paired-end reads.
Alignment and extraction of chrM and chr22 reads
All sequencing reads were aligned to genome reference build hg38. Mapping of reads to chromosome 22 (chr22), positions 46,000,000 to 46,100,000, and to the mitochondrial chromosome (chrM) were extracted from the BAM files using SAMtools version 1.16.1.17 Mitochondrial reads were realigned to the revised Cambridge Reference Sequence using the Burrows-Wheeler Aligner (BWA) tool version 0.7.17.18
Copy number estimation of the mitochondrial genome
To assess mtDNA copy number (CN) for each sample, nuclear chromosome 22 positions 46,000,000 to 46,100,000 (genome build hg38) were chosen as a proxy representing two copies per cell due to its low level of copy number variants (CNVs) in the population.19 Using read depth as a proxy of DNA CN, the mtDNA CN was estimated as , where denotes the average read depth across the mitochondrial chromosome, and denotes the average read depth of the two-copy region, chr22: 46,000,000–46,100,000.
Variant calling and downstream quality control
We used the Genome Analysis Toolkit (GATK) version 4.2.0.0 and its mitochondrial mode to call mitochondrial DNA single-nucleotide variants (SNVs).20 We omitted all variants in the regions from mitochondrial base positions 302 to 316, 513 to 526, 566 to 573, 8,860, and 16,181 to 16,194, which are known to yield false positive variant calls.21 Because heteroplasmic variants generally impact disease phenotypes only at levels above 80%,22 all variants with heteroplasmy levels less than this threshold were excluded from further consideration. Population frequency of each variant was obtained from GenBank (data last accessed on Dec. 10, 2021).23 We used HaploGrep 2.1.21 to define the haplogroup for each sample.24
Statistical procedures
All statistical computations were performed using R version 4.2.0. Associations between mtDNA CN and PHTS clinical phenotypes were tested using the logistic regression models:
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
Where, α, β0, β1, and β2 are coefficients of the corresponding regression, and p values were extracted from the model fit with the anova() function, using the chi-squared test argument.
Rare SNV association analyses were performed using similar models:
| (Equation 5) |
| (Equation 6) |
| (Equation 7) |
| (Equation 8) |
Here, the “(rare) SNV” term represents either the numerical variable log(total number of mtDNA SNVs) (or) log(rare mtDNA SNVs). p values were computed as above.
Phenotype terms for all the above logistic regression models include PHTS-onlyASD/DD, PHTS-onlyCancer, PHTS-neither, PHTS-ASD, PHTS-Cancer, PHTS-ASD/Cancer, PHTS-noCancer, and PHTS-noASD.
We used the nonparametric Mann-Whitney test, Fisher’s exact test, and chi-squared test, which are the unadjusted tests of the above-described logistic regression models, to identify differences between the features of the groups clustered by PHTS clinical phenotypes. p values <0.05 were considered as statistically significant. Our participants belong to over 15 mitochondrial haplogroups, which makes it challenging to perform variant-level association analyses, knowing that mitochondrial variants are core determinants of the different haplogroups and may therefore lead to spurious associations. Since the H haplogroup is the largest one among all the haplogroups, we decided to perform targeted analyses limited to this haplogroup (n = 213).
Results
Mitochondrial haplogroup distribution and participant phenotypes
Our analytical series consisted of 498 samples after excluding samples from individuals who have maternal relationships, performing standard quality control, haplogroup calling, and excluding PTEN wild-type family members. Of the 498 samples, 213 (42.8%) belong to the H haplogroup, while the others mainly belong to the U, J, K T, L, and W haplogroups. In this series, 164 (32.93%) individuals have ASD/DD but not cancer (designated as onlyASD/DD), 184 (36.94%) have cancer but not ASD/DD (designated as onlyCancer), and 18 (3.61%) have both ASD/DD and cancer (designated as ASDCancer). The remaining 132 (26.51%) participants have neither ASD/DD nor cancer (designated as neither). When the samples are narrowed down within the H haplogroup, the estimated distribution of individuals within each of the phenotype groups parallels the distribution within the entire series.
Mitochondrial DNA copy number distribution in individuals with PHTS
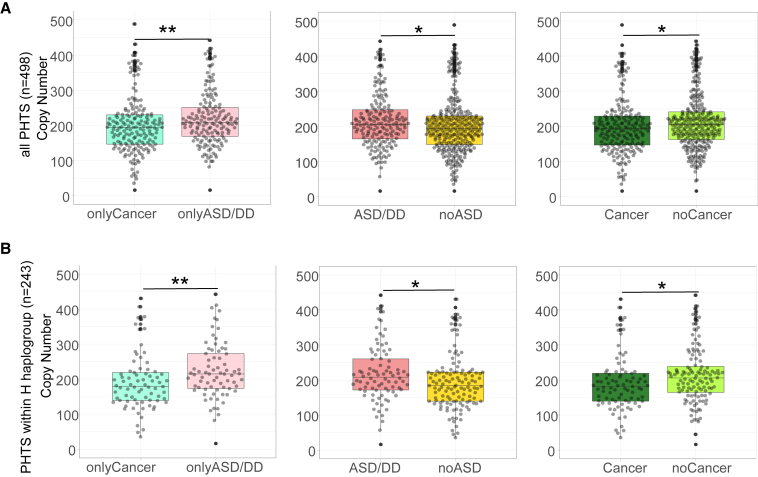
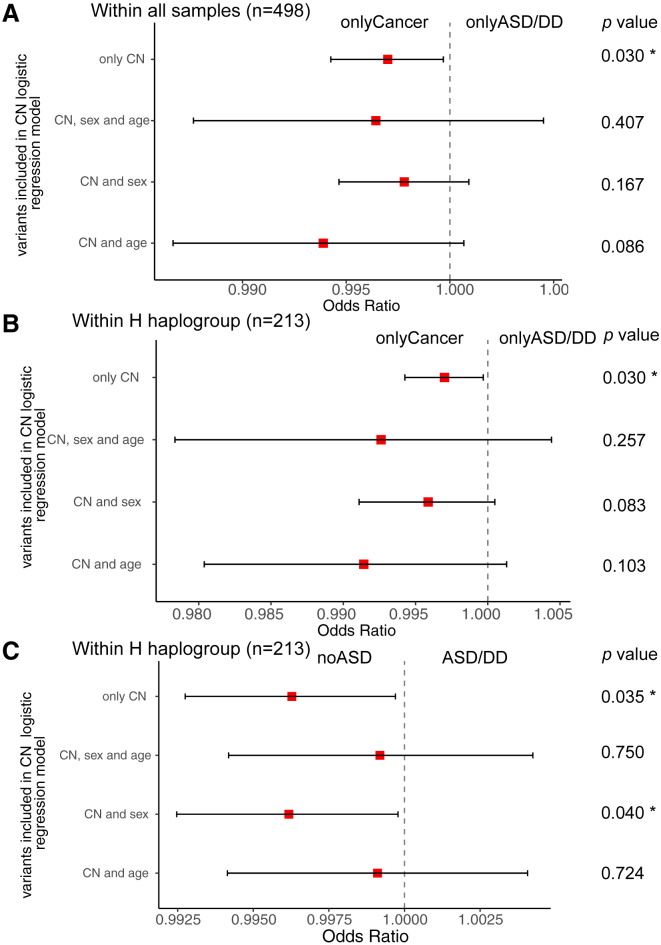
We previously showed that a higher genomic DNA autosomal CNV burden is associated with the PHTS-ASD/DD phenotype compared with PHTS individuals without ASD/DD.13 We therefore hypothesized that PHTS-ASD/DD individuals may also have a higher mtDNA CN. To address this hypothesis, we tested mtDNA CN differences among different PHTS-related phenotype groups (Figure S1). In addition to the onlyASD/DD, onlyCancer, ASDCancer, and neither groups, we also examined the ASD/DD group, which includes individuals diagnosed with ASD/DD regardless of cancer status, and the Cancer group, which includes individuals diagnosed with Cancer regardless of ASD/DD status. We observed that the onlyASD/DD group has a higher median mtDNA CN compared with the onlyCancer group (p = 9.2 × 10−3); the ASD/DD group has a higher mtDNA CN burden than the noASD group (p = 2.4 × 10−2); and the noCancer group has a higher mtDNA CN burden than the Cancer group (p = 3.2 × 10−2) (Figures 1A and S1). Since idiopathic ASD is more frequently diagnosed in males than in females, we performed statistical comparisons while controlling for biological sex. Additionally, because individuals with PHTS-ASD/DD are younger than PHTS individuals with cancer at the time of consent, we also performed statistical comparisons controlling for age alone and controlling for both age and sex. After controlling for age at consent, the onlyASD/DD group still showed a trend toward higher burden of mtDNA CN compared with the onlyCancer group (p = 8.6 × 10−2) (Figure 2A and Tables S1 and S2).
Figure 1.
Mitochondrial DNA copy number differences among different PHTS phenotype groups
(A) Beeswarm plots of copy number differences between different cancer/ASD phenotypes within all samples (n = 498). p values are calculated by the Mann-Whitney test.
(B) Beeswarm plots of copy number differences between different cancer/ASD phenotypes within the H haplogroup (n = 213). p values are calculated by the Mann-Whitney test.
The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). Each dot represents one sample’s mtDNA CN value. The upper whisker extends from the hinge to the largest value no further than 1.5 ∗ inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 ∗ IQR of the hinge. Data beyond the end of the whiskers are "outlying" points and are plotted individually.
∗∗0.001 < p value ≤ 0.01. ∗p value <0.05.
Figure 2.
Forest plots of mtDNA CN analysis among different PHTS clinical phenotype groups
(A) Logistic regression results of mtDNA CN between onlyCancer and onlyASD/DD group (all participants, n = 498) with unadjusted variants, adjusted sex and age at consent, sex only, and age at consent only, respectively.
(B) Logistic regression results of mtDNA CN between onlyCancer and onlyASD/DD group (limited to the H haplogroup, n = 213) with unadjusted variants, adjusted sex and age at consent, sex only, and age at consent only, respectively.
(C) Logistic regression results of mtDNA CN between noASD and ASD group (limited to the H haplogroup, n = 213) with unadjusted variants, adjusted sex and age at consent, sex only, and age at consent only, respectively.
The red squares indicate the odds ratios, and error bars indicate the 95% confidence intervals of the odds ratios. ∗p value < 0.05.
Mitochondrial DNA copy number distribution in PHTS individuals belonging to the H haplogroup
To test whether the mtDNA CN differences among the different phenotypes still held within the H haplogroup, we performed similar comparisons utilizing a more focused set of individuals. Within the H haplogroup, we similarly found that the onlyASD/DD group has a higher mtDNA CN compared with the onlyCancer group (p = 4.2 × 10−3) and with the neither group (p = 3.4 × 10−3 controlling for sex); the ASD/DD group has higher mtDNA CN than the noASD group (p = 1.4 × 10−2, p = 4.0 × 10−2 when controlling for age at consent); and the Cancer group has lower mtDNA CN than the noCancer group (p = 1.5 × 10−2) (Figures 1B, 2B, and S2).
Mitochondrial DNA single-nucleotide variant burden in PHTS individuals with the H haplogroup
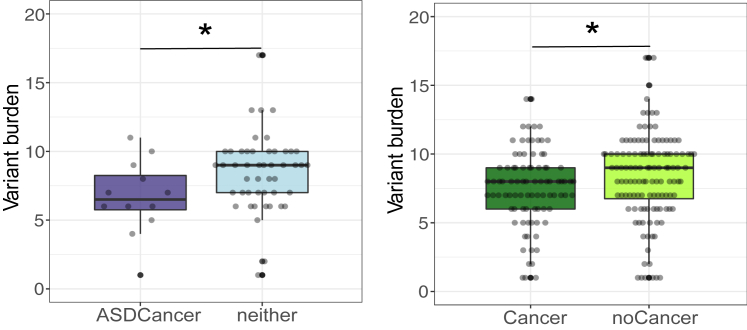
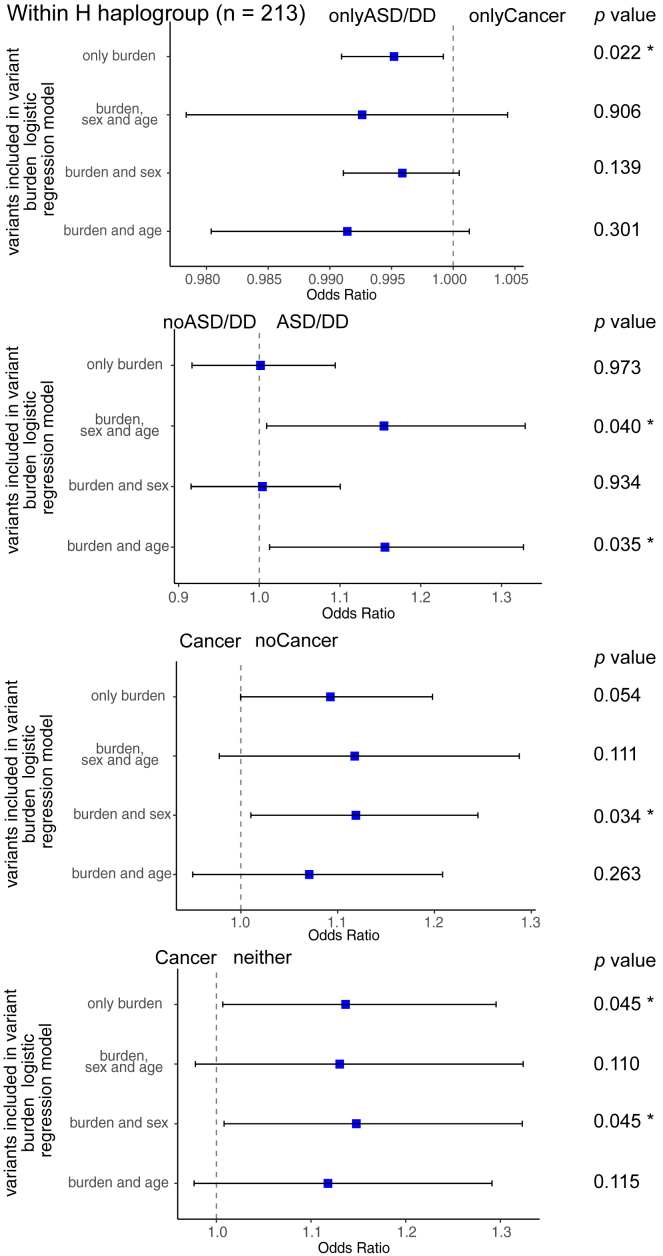
Next, we tested the association between mtDNA SNV burden and PHTS clinical phenotypes in individuals belonging to the H haplogroup. Haplogroups are defined by specific sets of SNVs.24 Therefore, we should either control for the haplogroups or analyze SNV burden within each haplogroup. However, with the limited sample size (n = 498), it became prudent to only focus on the largest haplogroup, the H haplogroup. Overall, the entire series included 8,156 non-reference SNVs (1,017 unique variants, Table S4). Within the H haplogroup, we identified 1,691 SNVs (369 unique variants). After performing all pairwise comparisons, the neither group showed a higher SNV burden compared with the ASDCancer group (p = 4.6 × 10−2) and with the Cancer group (p = 0.045 controlling for age at consent); the noASD group had a higher SNV burden than the ASD/DD group (p = 4.0 × 10−2, controlling for age and sex); and the noCancer group had a higher SNV burden than the Cancer group (p = 3.3 × 10−2, p = 3.4 × 10−2 controlling for sex) (Figures 3 and 4 and Tables S1 and S2).
Figure 3.
Beeswarm plots of mitochondrial DNA single-variant burden differences among different PHTS phenotypes within the H haplogroup (n = 213)
The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). Each dot represents one sample’s mtDNA CN value. The upper whisker extends from the hinge to the largest value no further than 1.5 ∗ inter-quartile range (IQR) from the hinge. The lower whisker extends from the hinge to the smallest value at most 1.5 ∗ IQR of the hinge. Data beyond the end of the whiskers are "outlying" points and are plotted individually.
p values are calculated by the Mann-Whitney test. ∗p value < 0.05.
Figure 4.
Forest plots of mtDNA variant burden analysis among different PHTS clinical phenotype groups within the H haplogroup (n = 213)
The blue squares indicate the odds ratios, and error bars indicate the 95% of confidence interval of the odds ratios. Clinical phenotype groups have been indicated at the top of each forest plot. ∗p value < 0.05.
Mitochondrial DNA single-nucleotide variant effects in PHTS individuals with the H haplogroup
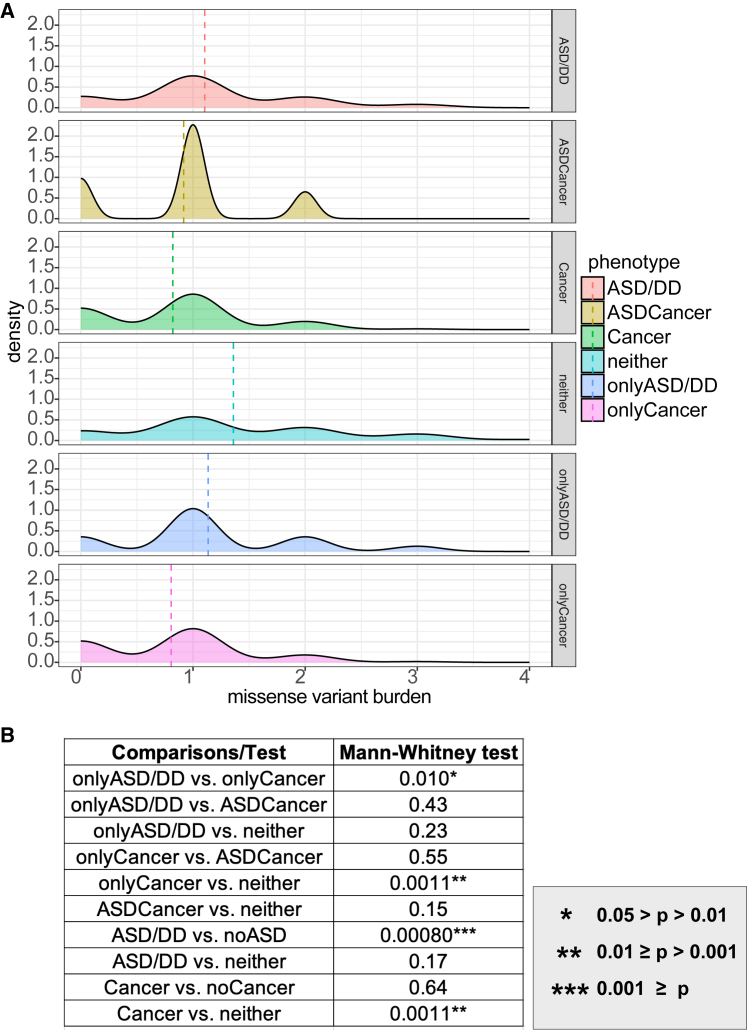
Within the H haplogroup, we identified 251 SNVs with predicted functional consequences (48 unique variants) based on the NCBI database.25 Among the 251 SNVs, 225 SNVs (35 unique variant) are missense variants, 25 non-coding transcript exon variants, and only 1 synonymous variant. Missense variants, non-coding transcript exon variants, and synonymous variants have moderate, modifier, and low impact, respectively. The missense variant histogram indicates that the neither group has more samples with higher missense variant burden than the onlyCancer group (Figure 5A). After performing all pairwise comparisons, the neither group showed a higher missense variant burden than the Cancer and onlyCancer groups (both p = 1.0 × 10−2); the noASD/DD group showed a higher missense variant burden than the ASD/DD group (p = 8.0 × 10−4); and the onlyASD/DD group showed a higher missense variant burden than the onlyCancer group (Figure 5B). We did not find any statistically significant comparisons after examining non-coding transcript exon variants and the synonymous variant.
Figure 5.
Missense variant burden among different PHTS phenotypes within the H haplogroup (n = 213)
(A) Density plot of missense variants among different phenotype groups. Dashed lines represent the mean missense variant burden.
(B) Mann-Whitney test results of the missense variant burden comparisons between different phenotype groups. ∗∗∗ p ≤ 0.001. ∗∗0.001 < p value ≤ 0.01. ∗0.01 < p value <0.05.
Discussion
Here, we present results investigating the relationship between PHTS clinical phenotypes and mtDNA content and variation. These observations begin to provide important insights into the clinical challenge of predicting ASD/DD and/or cancer development in PHTS.
ASD/DD has strong genetic underpinnings and metabolic comorbidities.26,27,28 Although previous studies have explored mtDNA within individuals with sporadic ASD/DD,29,30,31,32 the mtDNA landscape in PHTS-ASD/DD has never been investigated. On this PHTS background, we found that PHTS-ASD/DD has higher mtDNA CN than PHTS-Cancer or PHTS-neither. Notably, our observations remain consistent before and after controlling for age, which is highly associated with PHTS-ASD/DD and PHTS-Cancer. Intriguingly, the PHTS-ASD/DD group has lower mtDNA SNV (versus CN) burden than that of PHTS-neither. We attempted to annotate all the SNVs we identified from the whole-genome sequencing; however, only a small number has known annotations.33,34 This implies that most SNVs we found are novel, non-functional, or neutral. We interpret those SNVs as polymorphism-like and protective rather than pathogenic or likely pathogenic. Additionally and notably, we found that individuals with PHTS-ASD/DD have a higher burden of mitochondrial missense variants than individuals with PHTS-Cancer. Importantly, sporadic ASD/DD has been previously correlated with mitochondrial disorders.28 The mtDNA variant n.3397A>G (p. ND1:M31V) has been found in 25 individuals with sporadic non-PHTS-ASD/DD, but in our series, only one participant with ASD/DD harbored this variant.32 These observations may be explained by the fact that our series of individuals with PHTS represents syndromic ASD/DD rather than the sporadic form and the mtDNA variation likely being modifying instead of etiologic in this context.35
Our study does have certain limitations. Interrogating mtDNA in peripheral blood is, at best, an indirect measure of the mtDNA content in cancer tissues or in the brain, the primary tissues of interest based on phenotype. However, for such studies, it is impossible (perhaps unethical) to obtain brain tissues from living individuals with ASD/DD. Importantly, mtDNA from blood can serve as a representative tissue source for mtDNA.29,30 Although this may be a limitation when analyzing CN, our aim was to analyze germline mtDNA sequence content, which should largely be the same in all tissues. Owing to the rarity of PHTS, another limitation is sample size (n = 498), which left us underpowered to detect specific phenotype-associated SNVs, rare SNVs, and SNVs within specific genes or regions. The mtDNA gene-coding variants, n.8251G>A (p. COII:G222G), n.8269G>A (p. COII:∗228∗), n.8271A>G (non-coding variant in mt-NC7), and others, found by previous ASD studies, have not been found in our participant series.31,32 Because these SNVs were identified in a much limited sample size in previous studies, it is plausible that these SNVs are associated with sporadic versus PTEN-related syndromic ASD and are more etiologic versus modifying. A natural extension of our work would be to query the mitochondrial whole-genome sequence for large deletions. Together with the nuclear mitochondrial genome results, these data will help inform the association between ASD/DD and mtDNA within the PHTS background. Another caveat in our study is that although we follow these research participants prospectively for a median of 4 years (range = 1 month to 16 years; SD = 4 years), we cannot ascertain cancer status in individuals who did not have cancer at the time of consent nor during follow-up. However, we are more certain that individuals without ASD/DD indeed do not have these NDDs given the younger (typically pediatric) ages at diagnosis of NDD. Similarly, we cannot predict which ASD/DD individuals will develop cancer decades from consent. This is part of the motivation of our studies. It is also important to point out that the PHTS component cancers (except for thyroid cancer) do not arise until 30–35 years old and beyond, in the context that the median age of our ASD/DD persons is 11.35 years old at the time of consent.
Despite these limitations, our participant series comprises one of the largest PHTS cohorts worldwide, including comprehensive and uniform phenotyping by PHTS experts. The use of whole-genome sequencing yields extremely high-depth coverage of the mitochondrial genome, which enables comprehensive and sensitive detection of all mtDNA substitutions and avoids the error-prone imputation process. Because all samples were processed at the same center with the same sequencing protocols, batch effects are minimized, and meta-analysis is not necessary. This overall uniformity enabled us to uncover significant associations despite a limited sample size for each phenotype group.
In summary, we analyzed mtDNA content and variation in the peripheral blood of individuals with PHTS and found that individuals with ASD/DD have higher mtDNA CN on a PHTS background. We also found that the PHTS-ASD/DD group has lower mtDNA SNV (versus CN) burden than PHTS-neither, and PHTS-ASD/DD individuals have more mitochondrial missense variants than PHTS-Cancer, an observation that also held true within the H haplogroup. Thus, our data suggest that specific mtDNA content/variation could act as modifiers in the background of germline PTEN variants. However, future studies are needed to validate these findings in other PHTS cohorts and within the other mitochondrial haplogroups. Characterizing the mtDNA landscape is an important step for more precisely predicting ASD/DD versus cancer phenotypes in individuals with PHTS, a clinically heterogeneous syndrome.
Data and code availability
Data will be available on reasonable request to the corresponding author. All the bioinformatics and statistical tools are described in the material and methods. Scripts and code for generation of association statistics and additional statistical analyses are available at https://github.com/weiruipeng/EngLab_mtDNAproject.
Acknowledgments
We thank all our research participants who contributed to this study. We are grateful to the clinical research team at the PTEN Multidisciplinary Clinic and Center of Excellence at the Cleveland Clinic for administrative support. We also thank Dr. Thomas LaFramboise (Case Western Reserve University) for scientific advice and helpful discussions. This study was funded, in part, by the PTEN Research Foundation (CCF19-001), the Ambrose Monell Foundation (Switch grant), the Breast Cancer Research Foundation, and the NICHD/NIH (R01HD105049) (all to C.E.). L.Y. is an Ambrose Monell Cancer Genomic Medicine Fellow at the Cleveland Clinic Genomic Medicine Institute. C.E. is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic and an ACS Clinical Research Professor.
Declaration of interests
The authors have no relevant conflicts of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100199.
Web resources
OMIM: Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
Genome Analysis Toolkit (GATK), https://gatk.broadinstitute.org/hc/en-us.
MITOMAP, https://www.mitomap.org/MITOMAP.
Haplogrep2, https://haplogrep.i-med.ac.at/category/haplogrep2/
HmtDB, https://www.hmtdb.uniba.it/
Supplemental information
References
- 1.Butler M.G., Dasouki M.J., Zhou X.-P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawley J.N., Heyer W.-D., LaSalle J.M. Autism and cancer share risk genes, pathways and drug targets. Trends Genet. 2016;32:139–146. doi: 10.1016/j.tig.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forés-Martos J., Catalá-López F., Sánchez-Valle J., Ibáñez K., Tejero H., Palma-Gudiel H., Climent J., Pancaldi V., Fañanás L., Arango C., et al. Transcriptomic metaanalyses of autistic brains reveals shared gene expression and biological pathway abnormalities with cancer. Mol. Autism. 2019;10:17. doi: 10.1186/s13229-019-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trost B., Thiruvahindrapuram B., Chan A.J.S., Engchuan W., Higginbotham E.J., Howe J.L., Loureiro L.O., Reuter M.S., Roshandel D., Whitney J., et al. Genomic architecture of autism from comprehensive whole-genome sequence annotation. Cell. 2022;185:4409–4427.e18. doi: 10.1016/j.cell.2022.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yehia L., Keel E., Eng C. The clinical spectrum of PTEN mutations. Annu. Rev. Med. 2020;71:103–116. doi: 10.1146/annurev-med-052218-125823. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez B.A., Scherer S.W. Syndromic autism spectrum disorders: moving from a clinically defined to a molecularly defined approach. Dialogues Clin. Neurosci. 2017;19:353–371. doi: 10.31887/DCNS.2017.19.4/sscherer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein S., Sharifi-Hannauer P., Martinez-Agosto J.A. Macrocephaly as a clinical indicator of genetic subtypes in autism. Autism Res. 2013;6:51–56. doi: 10.1002/aur.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orrico A., Galli L., Buoni S., Orsi A., Vonella G., Sorrentino V. Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin. Genet. 2009;75:195–198. doi: 10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 9.Tilot A.K., Frazier T.W., Eng C. Balancing proliferation and connectivity in PTEN-associated autism spectrum disorder. Neurotherapeutics. 2015;12:609–619. doi: 10.1007/s13311-015-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varga E.A., Pastore M., Prior T., Herman G.E., McBride K.L. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet. Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 11.Yehia L., Ni Y., Feng F., Seyfi M., Sadler T., Frazier T.W., Eng C. Distinct alterations in tricarboxylic acid cycle metabolites associate with cancer and autism phenotypes in cowden syndrome and bannayan-riley-ruvalcaba syndrome. Am. J. Hum. Genet. 2019;105:813–821. doi: 10.1016/j.ajhg.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yehia L., Ni Y., Sadler T., Frazier T.W., Eng C. Distinct metabolic profiles associated with autism spectrum disorder versus cancer in individuals with germline PTEN mutations. NPJ Genom. Med. 2022;7:16–19. doi: 10.1038/s41525-022-00289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yehia L., Seyfi M., Niestroj L.-M., Padmanabhan R., Ni Y., Frazier T.W., Lal D., Eng C. Copy number variation and clinical outcomes in patients with germline PTEN mutations. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2019.20415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni Y., Zbuk K.M., Sadler T., Patocs A., Lobo G., Edelman E., Platzer P., Orloff M.S., Waite K.A., Eng C. Germline mutations and variants in the succinate dehydrogenase genes in cowden and cowden-like syndromes. Am. J. Hum. Genet. 2008;83:261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni Y., He X., Chen J., Moline J., Mester J., Orloff M.S., Ringel M.D., Eng C. Germline SDHx variants modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome via FAD/NAD-dependant destabilization of p53. Hum. Mol. Genet. 2012;21:300–310. doi: 10.1093/hmg/ddr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberts B. 6th ed. W.W. Norton & Company; 2017. Molecular Biology of the Cell. [DOI] [Google Scholar]
- 17.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup Genome project data processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon S., LaFramboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis. 2014;35:1046–1054. doi: 10.1093/carcin/bgu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M., Schröder R., Ni S., Madea B., Stoneking M. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl. Acad. Sci. USA. 2015;112:2491–2496. doi: 10.1073/pnas.1419651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart J.B., Chinnery P.F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 23.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geschwind D.H. Genetics of autism spectrum disorders. Trends Cognit. Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geschwind D.H., State M.W. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y., Vadodaria K.C., Lenkei Z., Kato T., Gage F.H., Marchetto M.C., Santos R. Mitochondria, metabolism, and redox mechanisms in psychiatric disorders. Antioxidants Redox Signal. 2019;31:275–317. doi: 10.1089/ars.2018.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patowary A., Nesbitt R., Archer M., Bernier R., Brkanac Z. Next generation sequencing mitochondrial DNA analysis in autism spectrum disorder. Autism Res. 2017;10:1338–1343. doi: 10.1002/aur.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalkia D., Singh L.N., Leipzig J., Lvova M., Derbeneva O., Lakatos A., Hadley D., Hakonarson H., Wallace D.C. Association between mitochondrial DNA haplogroup variation and autism spectrum disorders. JAMA Psychiatr. 2017;74:1161–1168. doi: 10.1001/jamapsychiatry.2017.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piryaei F., Houshmand M., Aryani O., Dadgar S., Soheili Z.-S. Investigation of the mitochondrial ATPase 6/8 and tRNA(lys) genes mutations in autism. Cell J. 2012;14:98–101. [PMC free article] [PubMed] [Google Scholar]
- 32.Weissman J.R., Kelley R.I., Bauman M.L., Cohen B.H., Murray K.F., Mitchell R.L., Kern R.L., Natowicz M.R. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS One. 2008;3:e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clima R., Preste R., Calabrese C., Diroma M.A., Santorsola M., Scioscia G., Simone D., Shen L., Gasparre G., Attimonelli M. HmtDB 2016: data update, a better performing query system and human mitochondrial DNA haplogroup predictor. Nucleic Acids Res. 2017;45:D698–D706. doi: 10.1093/nar/gkw1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grandhi S., Bosworth C., Maddox W., Sensiba C., Akhavanfard S., Ni Y., LaFramboise T. Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum. Mol. Genet. 2017;26:2912–2922. doi: 10.1093/hmg/ddx172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available on reasonable request to the corresponding author. All the bioinformatics and statistical tools are described in the material and methods. Scripts and code for generation of association statistics and additional statistical analyses are available at https://github.com/weiruipeng/EngLab_mtDNAproject.