Abstract
Red blood cells (RBCs) demonstrate immunomodulatory capabilities through the expression of nucleic acid sensors. However, little is known about bat RBCs, and no studies have examined the immune function of bat erythrocytes. Here we show that bat RBCs express the nucleic acid-sensing Toll-like receptors TLR7 and TLR9 and bind the nucleic acid ligands, single-stranded RNA, and CpG DNA. Collectively, these data suggest that, like human RBCs, bat erythrocytes possess immune function and may be reservoirs for nucleic acids. These findings provide unique insight into bat immunity and may uncover potential mechanisms by which virulent pathogens of humans are concealed in bats.
Introduction
A fundamental role of the immune system is nucleic acid-sensing, which is essential for pathogen detection and coordination of host defense. (1, 2) Toll-like receptors (TLRs) are critical for nucleic acid recognition and include TLR9, which detects DNA containing unmethylated CpG motifs present in bacterial, viral, and mitochondrial DNA (mt-DNA), and TLR7, which recognizes single-stranded RNA (ssRNA) present in viruses and host RNA. Most studies on TLR function have focused on their roles in classical immune cells, although some enucleated cells such as platelets also express TLRs.(3, 4) Because mammalian red blood cells (RBCs) are devoid of organelles, the prevailing assumption has been that they lack immune function. However, contrary to this dogma, a DNA-sensing role for RBCs through the expression of TLR9 has recently been discovered.(5) Whether bat RBCs bind nucleic acids or express TLRs is unknown.
Members of the order Chiroptera (bats) are exceptional in their ability to harbor deadly viruses without developing disease.(6) Although substantial evidence supports a bat origin for numerous viruses, how bats tolerate such viruses and other pathogens remains unknown.(7) Bats are hosts to diverse intraerythrocytic haemosporidian parasites and co-evolved with these parasites.(8) Additionally, as an adaption to flight, the number of erythrocytes per microliter of blood is two- to five-fold higher than in humans, increasing the total red cell surface area.(9) Despite these observations, little is known regarding bat erythrocytes and their interaction with the immune system. Because we identified TLRs on mammalian RBCs, we asked if bat RBCs similarly express TLRs and bind nucleic acids.
Methods
Animals and ethic statements
Animal studies were conducted using terminal blood samples obtained from bats in rehabilitation and provided by the Pennsylvania Game Commission. All studies were done in accordance with the Institutional Animal Care and Use Committee. Studies using human specimens were approved by the Institutional Review Board. Healthy volunteers provided informed consent prior to study participation.
Bat blood collection
Bats were anesthetized with isoflurane and then euthanized via cervical dislocation. Blood was collected either via cardiac puncture or from major vessels after decapitation and placed in EDTA microtainers. Samples were centrifuged for 15 min at 3,500 rpm (max RCF = 1,534g) and then transferred to the University of Pennsylvania for further analysis.
Human RBC Isolation
Healthy volunteers provided informed consent. Whole blood was collected in EDTA tubes prior to centrifugation at 3000g for 10 min. The plasma and buffy coat were aspirated. Red blood cells were purified from the remaining packed red cell fraction using magnetic-assisted cell sorting (MACS) as previously described.(10)
Blood processing and RBC isolation
Whole blood was centrifuged at room temperature for 5 min at 800g. Plasma and buffy coat were isolated and saved. The remaining packed red blood cells (RBCs) were resuspended in 1 mL PBS for washing. Then, the cells were centrifuged, and the supernatant was aspirated along with a small amount of the packed RBCs on the top. To determine the yield of RBCs, serially diluted packed RBCs were counted on a hemacytometer. Immediately following this, the RBCs were either used for functional assays, fixed, or frozen in aliquots. To freeze RBCs, 107 RBCs were added to DNA lo-bind tubes and pelleted by centrifugation at 10,000 rpm for 5 min. The supernatant was removed and the cell pellets were store at −80°C. Residual packed RBCs were stored in Adsol (Fenwal) at 3:1 volume ratio at 4°C.
RBC smears and staining
To generate smears for RBCs, 5 μL of PBS was added to a clean Superfrost Plus microscope slide (Fisherbrand), and 2 μL of washed packed RBCs were mixed with the PBS and spread along the slide. The slides were air-dried and stored at room temperature until usage. Air-dried smears were stained using DiffQuik.
Nucleic acid binding assay
250,000 RBCs in 100 μL sterile PBS were transferred to round-bottom polystyrene tubes (Falcon, 352052). Afterwards, oligonucleotides diluted in sterile PBS were added to the cells in a volume of 5 μL. For competitive binding assays, 1000 nM FITC-ODN2006 and various concentrations of unlabeled ODN2006 were used. The tubes were placed on a rack, sealed with parafilm, covered in aluminum foil, and rocked on a nutator at 37°C for 2 hours. To stop the binding reactions, the cells were washed twice in 1 mL PBS and pelleted at 800g for 5 min with slow deceleration. The cells were resuspended in 300 μL PBS and analyzed by flow cytometry (BD Fortessa). Immediately prior to data acquisition, the cells were resuspended by pipetting up and down repeatedly.
Sequences for the oligonucleotides are: RNA40-ATTO647: rG*rC*rC*rC*rG*rU*rC*rU*rG*rU*rU*rG*rU*rG*rU*rG*rA*rC*rU*rC (IDT, Integrated DNA Technology, Inc.)
ATTO647-ODN2006: T*C*G*T*C*G*T*T*T*T*G*T*C*G*T*T*T*T*G*T*C*G*T*T (IDT)
FITC-ODN2006 T*C*G*T*C*G*T*T*T*T*G*T*C*G*T*T*T*T*G*T*C*G*T*T (IDT)
APC-ODN1826: T*C*C*A*T*G*A*C*G*T*T*C*C*T*G*A*C*G*T*T (IDT)
FITC-ODN1826: T*C*C*A*T*G*A*C*G*T*T*C*C*T*G*A*C*G*T*T (Invivogen)
* denotes phosphothiorate bond; r denotes ribonucleotide.
Viral RNA binding assays
SARS-CoV-2 and zika virus (ZIKV) RNA were provided by Dr. Kellie Jurado, University of Pennsylvania. 107 RBCs in 100 μL sterile PBS were transferred to 2 mL DNA/RNA lo-bind tube (Eppendorf, 86-924) and mixed with indicated amount of viral RNA diluted in sterile PBS. The mix was incubated at 37°C for 2 hr on a nutator. At 1 hr after incubation, tubes were rotated to ensure cells remain resuspended. After the incubation, cells were washed with 1 mL sterile PBS three times and frozen at −80°C prior to RNA extraction.
Surface antibody staining
The cells were resuspended in FACS buffer (PBS + 2% FBS) to 107 cells/mL. For each staining reaction, 106 RBC were pelleted in round bottom polystyrene tubes and stained in 100 μL of an antibody mix diluted in FACS buffer. The cells were rocked at room temperature for 1 hr in the dark. The cells were washed with 1 mL PBS twice as described above and analyzed by flow cytometry.
RBC fixation
RBCs were resuspended to 107 cells/mL in PBS, and an equal volume of 0.1% glutaraldehyde diluted in PBS was added to the cells for a final concentration of 0.05%. The cells were fixed at room temperature in the dark for 10 min. Fixation was stopped by adding equal volume of FACS buffer to the cells. The cells were pelleted at 800 g for 5 min and washed a total of three times. For long-term storage, fixed cells were resuspended in FACS supplemented with 0.05% sodium azide and stored at 4°C.
Intracellular staining with flow cytometry
106 RBCs were used in each staining reaction. Fixed RBCs were permeabilized in 0.1% TritonX-100 diluted in PBS for 15 min at room temperature. The cells were washed and pelleted in FACS buffer, blocked in PBST (PBS + 0.05% tween 20) supplemented with 1% BSA for 1 hr and then pelleted and resuspended in antibody diluted in PBST. Information for the antibodies used is listed below. For flow cytometry staining, cells were incubated with either antibody or isotype controls diluted in FACS buffer for 1 hr in the dark at room temperature under gentle shaking conditions. Subsequently, the cells were washed with FACS buffer twice and stained with a secondary antibody diluted in PBST for 45 min at room temperature. The cells were then washed in FACS buffer three times prior to analysis.
Immunofluorescent staining
RBCs were fixed, permeabilized, and blocked as described above. The primary antibody was diluted in PBST with 1% BSA, and incubation was performed overnight at 4°C. Subsequently, cells were washed in PBST three times and resuspended in a secondary antibody diluted in PBST. After 1 hr of staining at room temperature, the cells were washed in PBST three times. Following the final wash, the buffer was aspirated, and the residual volume of the RBCs was further reduced by pipetting. The remaining cells were resuspended in no more than 10 μL PBS. To mount cells on a slide, no more than 5 μL of Fluoromount G was added to a microscope slide and 5 μL RBCs were mixed in it, and samples were sealed with a coverslip.
Antibodies used and their applications
For all antibodies the target, clone, dilution and vendor are listed, FC= flow cytometry, IF=immunofluorescence. TLR3, 40C1285.6, FC: 5μg (Novus Biological); TLR7-4G6, IF: 1:200, FC: 1:200(Novus Biological); TLR7-726606, IF: 1:200, FC: 5μg (R&D Systems); TLR9-5G5, FC: 5μg, TLR9-ab37154, IF: 1:200 (Abcam); TLR9-26C593.2, FC: 5μg (Novus Biological/Abcam).
Microscopy and quantification
All micrographs were taken on Nikon A2 microscope. Manual counting quantification was performed by at least two personnel, one of which is blinded.
Imaging Flow Cytometry
RBCs were washed in PBS three times. 7x105 RBCs were then incubated with the specified oligonucleotides at 100 nM or 1000 nM at a final volume of 200 μL for 2 hr at 37°C. Cells were washed in PBS twice and fixed in 0.05% glutaraldehyde for 10 min. Prior to Imagestream (Amnis) analysis, the cells were washed in FACS buffer twice. Analysis was performed using IDEAS software. The automated feature finder was used to discriminate between RBC populations of different morphology. The aspect ratio (the length to width ratio or elongation of the cell) and DNA binding differentiated populations that displayed distinct RBC morphology.(11)
DNA extraction
107 frozen RBCs were thawed on ice. DNA was extracted using DNeasy blood/tissue kit (Qiagen) according to the manufacturer’s protocol. DNA was eluted in 152 μL elution buffer at the final step.
RNA extraction and reverse transcription
1x107 frozen RBCs were thawed on ice. RNA was extracted using RNeasy Plus Kit (Qiagen) according to the manufacturer’s protocol. During the initial step, RBCs were lysed in 600 μL lysis buffer, and during the final step, RNA was eluted in 30 μL water. 8 μL of freshly isolated RNA was reverse transcribed to cDNA via the Superscript First Strand Synthesis system (ThermoFisher) following the manufacturer’s protocol.
qPCR
Nucleic acids were quantified via the QuantStudio7 Flex System. For 18S rRNA and ZIKV, cDNA were amplified in PowerUp SYBR Green master mix with the primers (18S rRNA F: AACCCGTTGAACCCCATT and 18S rRNA-R: CCATCCAATCGGTAGTAGCG; ZIKV-E: forward: TTGGTCATGATACTGCTGATTGC and reverse: CCTTCCACAAAGTCCCTATTGC; ZIKV-NS5 forward: GGCCACGAGTCTGTACCAAA and reverse: AGCTTCACTGCAGTCTTCC). For bacterial 16S rDNA, DNA was amplified in Taqman Fast Universal Mastermix with the primers (BSF8: AGA GTT GAT CCT GGC TCA G and BSR357: CTG CTG CCT YCC GTA) and probe (/56-FAM/TA A+CA +CAT G+CA +AGT +GGA /3BHQ_1/ where + denotes locked nucleic acid). SARS-CoV-2 was quantified using 2019-nCoV RUO Kit from IDT and Taqman Fast Universal Mastermix.
Statistical Analysis
Statistical analyses were performed with GraphPad Prism 9. Data normality was determined by Wilk-Shapiro test. Differences among groups were determined with t-test or one-way ANOVA wherever appropriate.
Results
Bat RBCs Display Diverse Morphologies
Big brown bats (Eptesicus fuscus) are a hibernating North American micro-bat species that commonly use anthropogenic structures for hibernation and maternal colonies, even to some degree in winter, and are frequently submitted to wildlife rehabilitation facilities throughout the year, often presenting with injuries. We obtained terminal blood samples from wild-caught big brown bats admitted to Pennsylvania wildlife rehabilitators that were determined to be non-releasable due to their conditions (Table 1). All bats were consistently in a euthermic state during their time at the facility. Samples were collected post-euthanasia following transfer to the Pennsylvania Game Commission. We first prepared peripheral blood smears to examine RBC morphology under basal conditions (Figure 1A). Consistent with previous reports, we noted crenated and poikilocytic cells (Figure 1B).(9) Unlike human RBCs where crenated RBCs were primarily observed following DNA binding and during acute inflammatory states, the number of non-biconcave/RBCs was equivalent to biconcave/discoid-appearing RBCs in bats that spent sufficient time in rehabilitation (Figure 1C).(5) We also noted a substantial amount of heterogeneity in the appearance of crenated cells. Two of the bats (J and K) demonstrated significant hemolysis and the presence of intra-erythrocytic inclusions, marked by erythrocyte ghosts and Howell-Jolly bodies, respectively (Figure 1D).
Table 1.
Big brown bat (Eptesicus fuscus) subjects
| Bat | Sex | Weight (g)* | Days in Rehabilitation | Clinical Presentation |
|---|---|---|---|---|
| A | M | 9.8 | 91 | Injured wing |
| B | F | 11.2 | 70 | Grounded due to unknown cause |
| C | F | 16.5 | 27 | Grounded and captured at remodeling work site |
| D | M | 11.7 | 33 | Multiple membrane holes and fracture to right wing, small distal fracture to left wing, possible frost bite |
| E | F | 13.5 | 98 | No visible injuries |
| F | M | 12.7 | 6 | No visible injuries |
| G | M | 9.4 | 4 | Shredded membrane and open fracture of left wing |
| H | F | 12.0 | 8 | Shredded membrane and exposed bone of right wing |
| I | M | 11.3 | 9 | Alopecia along left shoulder, unspecified injury to left wing, degloving injury to one toe of left foot |
| J | M | 20.9 | 193 | Injury and bruising to right leg, cuts along left leg, hip, and back |
| K | M | 16.4 | >240 | No visible injuries but unexplained significant weight loss despite healthy appetite |
Weight was measured at time of euthanasia
Figure 1. Bat RBCs bind nucleic acids.
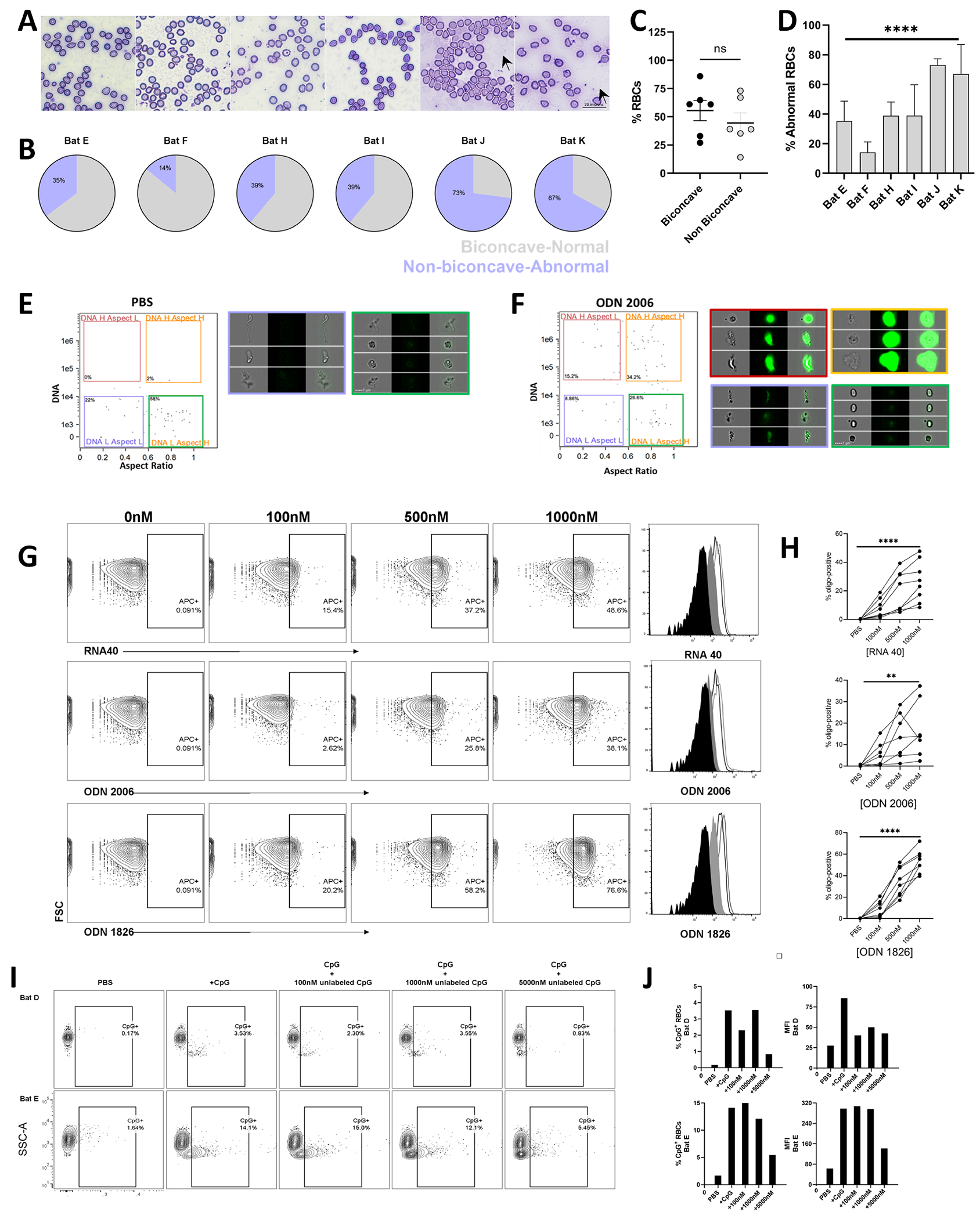
(A-D) Morphological Characteristics of Bat RBCs. (A) Peripheral smears of RBCs from six bats. (B) Percentage of normal and abnormal RBCs in individual bats. (C) Similar numbers of normally shaped and abnormally shaped RBCs are observed in bats. (D) Percentage of abnormal RBCs in bats. Arrows denote erythrocyte ghosts. (E and F) Imaging Flow Cytometry of CpG-treated Bat RBCs. (E) PBS- and (F) ODN2006-treated RBCs reveal four distinct populations of cells and acquisition of DNA by RBCs. (G and H) Dose-dependent binding of ssRNA (RNA40), CpG ODN 1826 or 2006; contour plots and histograms for one representative bat are shown in (G), and summary statistics for percent positive cells in (H). n=8 bats. (I and J) Competitive inhibition of CpG binding to bat RBCs; contour plots (I) and percent CpG-positive cells and MFI (J) are shown for two bats.
We next performed imaging flow cytometry to examine RBC morphology at the singlecell level. Concurring with earlier observations, naive untreated bat RBCs also displayed a range of morphologies (Figure 1E). Because we observed dramatic alterations in human RBC morphology following CpG-binding, we asked whether CpG would alter bat RBCs. Consistent with our observations of human RBCs, CpG acquisition by RBCs led to marked alteration of the bat RBCs (Figure 1F).(5)
Nucleic Acids Bind to Bat RBCs
Next, we measured the ability of bat RBCs to bind RNA and DNA. Fluorescently labeled GU-rich single-stranded RNA (ssRNA, RNA-40) and CpG oligodeoxynucleotides (ODNs) were incubated with bat RBCs before washing and analyzing by flow cytometry. Consistent with human RBCs, bat RBCs bound both RNA and CpG ODNs in a dose-dependent fashion (Figure 1G and H). We next performed competitive binding experiments by assessing the acquisition of fluorescently labeled CpG by bat RBCs in the presence of excess unlabeled CpG. Despite heterogeneity in RBC CpG acquisition, we observed attenuation in CpG binding in the presence of high doses of unlabeled CpG, indicative of specific binding to the oligonucleotide by RBCs. (Figure 1I and J).
Because microbial DNA binds to human RBCs, we assayed purified bat RBCs for the presence of RNA or DNA by performing qPCR for 18S rRNA and the 16S rDNA gene.(5) When compared with RBCs from healthy human donors, bat RBCs contained significantly less bacterial DNA but similar amounts of 18S rRNA (Figure 2A and B). Taken together, these data would suggest that bat RBCs exhibit differential nucleic acid-binding capability when compared with human RBCs. Becasue Zika virus (ZIKV) infection has recently been described in free ranging bats and persistent association of ZIKV to RBCs has been reported in the transfusion literature, we next asked whether RBCs bind RNA derived from ZIKV virus.(12, 13) We observed a dose-dependent binding of ZIKV RNA to RBCs (Figure 2C). Since a bat origin of SARS-CoV-2 has been proposed, we also asked whether bat RBCs would bind RNA from SARS-CoV-2. Although only one animal was available for studies, we observed robust binding of SARS-CoV-2 RNA to bat RBCs (Figure 2D). Collectively, these data demonstrate that bat RBCs bind RNA from pathogenic viruses.
Figure 2. Bat RBCs contain DNA and RNA and bind pathogen derived RNA.
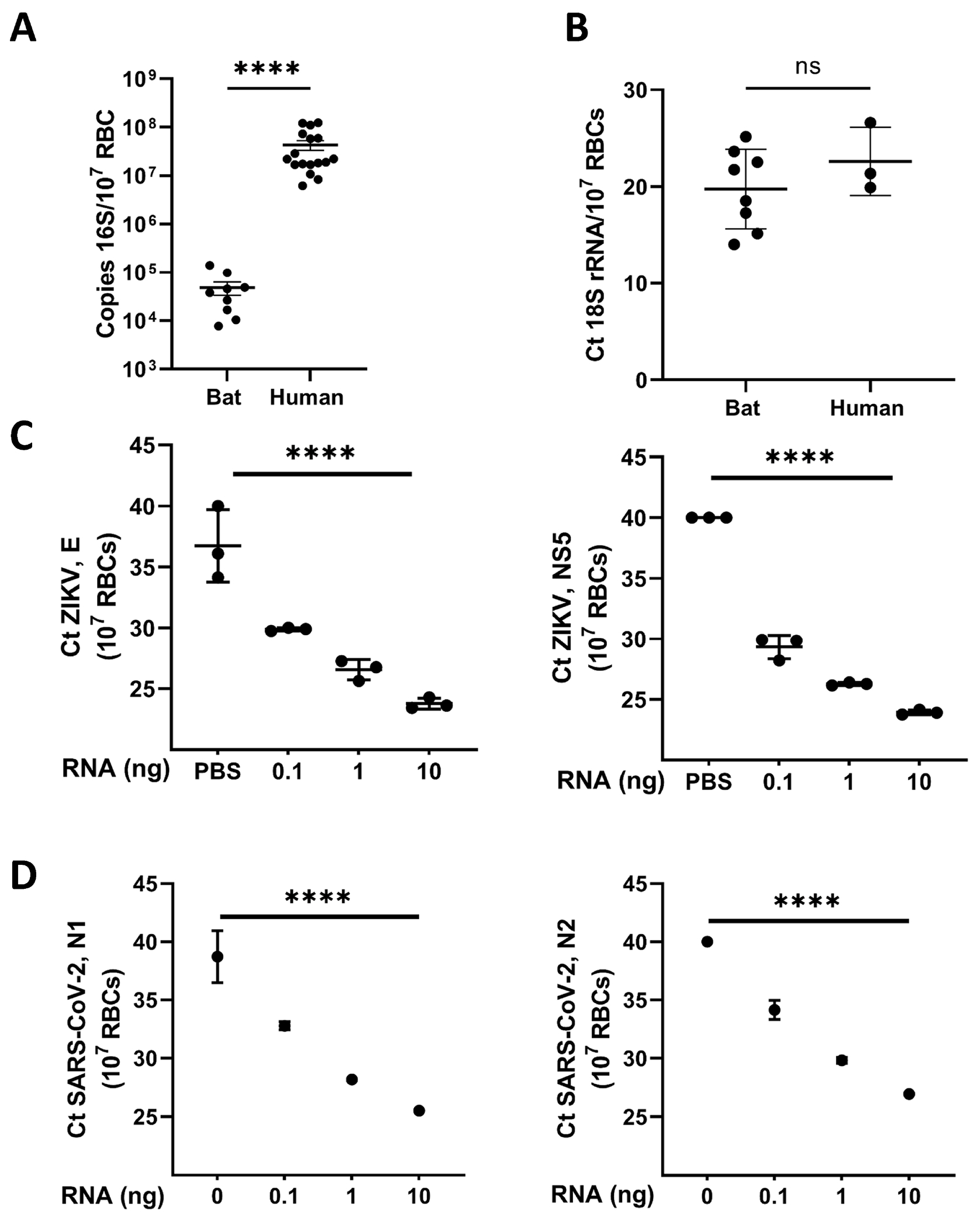
(A) 16S rDNA gene copies associated with RBCs derived from healthy human donors and bats. (B) 18S rRNA content of bat and human RBCs. For (A and B), each dot represents an individual bat or human donor sample,****p<0.0001. (C) qPCR quantitation for ZIKV RNA on RBCs incubated with varying concentrations of ZIKV RNA with PCR of the envelope (E) and non-structural protein 5 (NS5) genes. (D) qPCR for SARS-CoV-2 RNA on RBCs incubated with varying concentrations of SARS-CoV-2 RNA with PCR for two different amplicons for nucleocapsid gene (N1 and N2). n=1 bat. For (C and D), technical replicates are shown for each dose of RNA used. **P<0.01, ****p<0001.
Bat RBCs Express TLR7 and TLR9
Nucleic acid sensing in bats is a topic of intense investigation and the toll-like receptors (TLRs) have been extensively characterized in bats.(14, 15) We, therefore, asked if bat RBCs expressed TLRs. Flow cytometry of intact, unpermeablized bat RBCs did not identify TLRs on the RBC surface (data not shown). However, staining of permeabilized cells did reveal the presence of TLR9 and TLR7, although TLR3 was not detected (Figure 3A and B). We confirmed these findings with immunofluorescence (Figure 3C). However, only four of the five bats tested consistently demonstrated positive TLR7 and TLR9 staining, suggesting heterogeneity in erythroid TLR expression amongst mammals. Notably, the bats with the highest TLR7 staining were also the bats nearest to their injury before arrival to the rehabilitation facility (Table 1).
Figure 3. TLR expression on Bat RBCs.
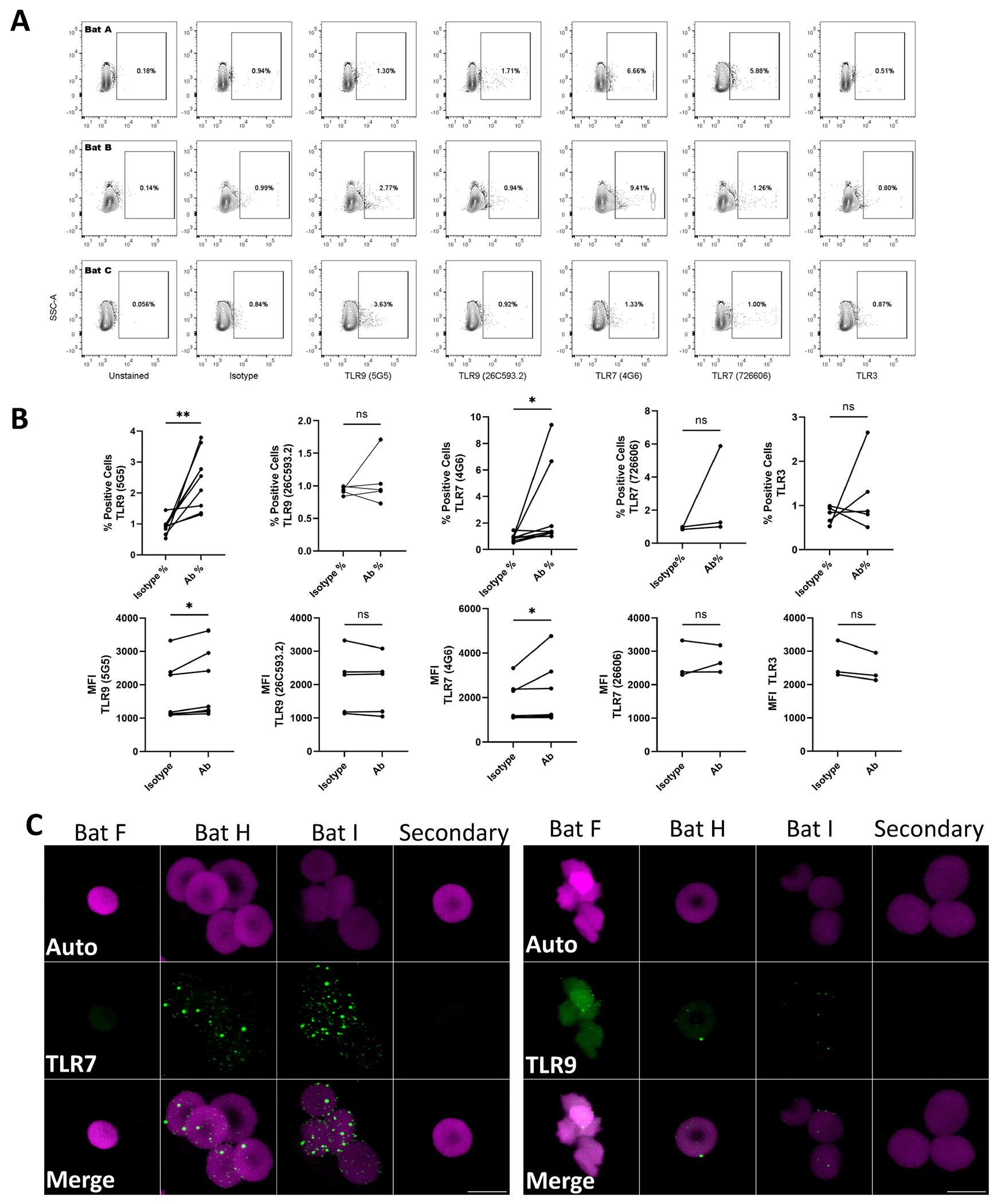
(A) Intracellular staining for TLR 9, 7, and 3 from three representative bats. (B) Summary statistics, percent positive cells, and MFI for TLRs, n=5, *P<0.05, **P<0.01. (C) Immunofluorescent staining for TLR7 and TLR9, magenta is autofluorescence, and green is TLR staining.
Discussion
Collectively our data demonstrate the presence of nucleic acid-sensing TLRs on bat RBCs and nucleic acid-binding capabilities of bat RBCs. Furthermore, bat RBCs contain RNA, suggesting that these RBCs may serve as reservoirs for RNA. As a result of the preponderance of bat-borne viral pathogens with pandemic potential, substantial interest in bat immunity has emerged. Recently, an immune function of RBCs through nucleic acid-sensing and delivery to classical immune cells has been identified.9 Given the large number of circulating RBCs present in Chiropteran species and the uniquely rich lipid composition of bat RBCs, we speculate that lipid-rich bat RBCs function to shelter RNA from the immune system, permitting host tolerance; however, further studies of bat RBCs including detailed analysis of surface proteins and lipid composition will be needed to further investigate this hypothesis.(16) Interestingly, in rehabilitated bats, RBCs exhibited a substantial number of abnormal cells and features, including excessive hemolysis, poikilocytes, Howell-Jolly bodies, and crenated cells, reminiscent of those found in hemolytic anemias, acute infection, and RBC membrane disorders, including pyropoikilocytosis and elliptocytosis. Additionally, bat TLRs display mutations in ligand binding sites that may alter their ability to bind nucleic acids, yet whether these adaptations affect erythrocyte TLRs and whether the bat RBC TLRs directly bind nucleic acids similar to human and mouse RBCs remains unknown as further characterization of the RBC membrane including the TLRs we report here is needed.(10, 14, 15, 17) Because bats undergo dramatic temperature changes from hypothermia during torpor to elevated temperature during flight, it is possible that the unique RBC features are not just a consequence of acclimatization to pathogens but also an adaption to rapid dynamic temperature changes, as has been observed in some congenital human red cell disorders, such as pyropoikilocytosis.(18) The extreme metabolic stress of flight has also driven red cell adaptations, including increased hematocrit and smaller RBCs, allowing for greater oxygen transport, and thus may also have influenced other RBC changes.(19) Although the driver of these unique RBC alterations in Chiroptera remain unclear, our findings of nucleic acid-binding by bat RBCs suggests an immune function for these RBCs.
The discovery of an immune role for enucleate, mature RBCs prompts the question of what evolutionary pressure led to such adaptations. Mounting evidence suggests that human nongas exchanging RBC adaptations arose in response to the dominant selective pressure of malaria-causing parasites of the genus Plasmodium, with chemokine binding through DARC (Duffy Antigen Receptor for Chemokines) and hemoglobin S (sickle cell) being the two most prominent examples. Mammals in the order Chiroptera co-exist with Plasmodium and other haemosporidians, and some have postulated a Chiropteran origin to primate malaria.(8) Future studies of mammalian erythrocyte immune function, including nucleic acid-binding and complement regulation in bats and other mammalian hosts, will be necessary to elucidate the complete picture of red cell immune function, potentially providing insight into mechanisms by which pathogens lethal to humans are tolerated in other mammals.
Acknowledgements:
We would like to thank Robyn Grabowski and Rebekah Jones at Centre Wildlife Care and Stephanie Stronsick at Pennsylvania Bat Rescue for their care of the animals and providing whole blood from the bats. We thank Dr. Kellie Jurado for providing SARS-CoV-2 and ZIKV RNA and Dr. Christopher A. Hunter for editorial assistance.
Funding:
Institute for Translational Medicine and Therapeutics of the Perelman School of Medicine and the School of Veterinary Medicine at the University of Pennsylvania Program in Comparative Biology to J.C.E and N.S.M. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing Interest Statement: The authors have no conflicts of interest to declare.
References
- 1.Diebold SS, Kaisho T, Hemmi H, Akira S, and Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531. [DOI] [PubMed] [Google Scholar]
- 2.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, and Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745. [DOI] [PubMed] [Google Scholar]
- 3.Aslam R, Speck ER, Kim M, Crow AR, Bang KW, Nestel FP, Ni H, Lazarus AH, Freedman J, and Semple JW. 2006. Platelet Toll-like receptor expression modulates lipopolysaccharide-induced thrombocytopenia and tumor necrosis factor-alpha production in vivo. Blood 107: 637–641. [DOI] [PubMed] [Google Scholar]
- 4.Semple JW, Italiano JE, and Freedman J. 2011. Platelets and the immune continuum. Nature Reviews Immunology 11: 264–274. [DOI] [PubMed] [Google Scholar]
- 5.Lam LKM, Murphy S, Kokkinaki D, Venosa A, Sherrill-Mix S, Casu C, Rivella S, Weiner A, Park J, Shin S, Vaughan AE, Hahn BH, Odom John AR, Meyer NJ, Hunter CA, Worthen GS, and Mangalmurti NS. 2021. DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Science Translational Medicine 13: eabj1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayman DTS 2019. Bat tolerance to viral infections. Nature Microbiology 4: 728–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schountz T, Baker ML, Butler J, and Munster V. 2017. Immunological Control of Viral Infections in Bats and the Emergence of Viruses Highly Pathogenic to Humans. Frontiers in immunology 8: 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaer J, Perkins SL, Decher J, Leendertz FH, Fahr J, Weber N, and Matuschewski K. 2013. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci U S A 110: 17415–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schinnerl M, Aydinonat D, Schwarzenberger F, and Voigt CC. 2011. Hematological survey of common neotropical bat species from Costa Rica. Journal of zoo and wildlife medicine : official publication of the American Association of Zoo Veterinarians 42: 382–391. [DOI] [PubMed] [Google Scholar]
- 10.Lam LKM, Murphy S, Kokkinaki D, Venosa A, Sherrill-Mix S, Casu C, Rivella S, Weiner A, Park J, Shin S, Vaughan AE, Hahn BH, John ARO, Meyer NJ, Hunter CA, Worthen GS, and Mangalmurti NS. 2021. DNA binding to TLR9 expressed by red blood cells promotes innate immune activation and anemia. Science Translational Medicine 13: eabj1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basiji DA, Ortyn WE, Liang L, Venkatachalam V, and Morrissey P. 2007. Cellular image analysis and imaging by flow cytometry. Clin Lab Med 27: 653–670, viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagre AC, Lewis J, Miller MR, Mossel EC, Lutwama JJ, Nyakarahuka L, Nakayiki T, Kityo R, Nalikka B, Towner JS, Amman BR, Sealy TK, Foy B, Schountz T, Anderson J, and Kading RC. 2021. Subgenomic flavivirus RNA (sfRNA) associated with Asian lineage Zika virus identified in three species of Ugandan bats (family Pteropodidae). Scientific reports 11: 8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone M, Bakkour S, Lanteri MC, Brambilla D, Simmons G, Bruhn R, Kaidarova Z, Lee T-H, Orlando Alsina J, Williamson PC, Galel SA, Pate LL, Linnen JM, Kleinman S, and Busch MP. 2020. Zika virus RNA and IgM persistence in blood compartments and body fluids: a prospective observational study. The Lancet Infectious Diseases 20: 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escalera-Zamudio M, Zepeda-Mendoza ML, Loza-Rubio E, Rojas-Anaya E, Méndez-Ojeda ML, Arias CF, and Greenwood AD. 2015. The evolution of bat nucleic acid-sensing Toll-like receptors. Molecular ecology 24: 5899–5909. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Li J, Li L, Zhang X, Yuan L, and Chen J. 2017. Selective evolution of Toll-like receptors 3, 7, 8, and 9 in bats. Immunogenetics 69: 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillett MP, and Wilson RB. 1985. Unusual lipid composition of erythrocytes from the insectivorous bat Molossus molossus. Comparative biochemistry and physiology. A, Comparative physiology 80: 149–150. [DOI] [PubMed] [Google Scholar]
- 17.Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, Rivella S, Worthen GS, and Mangalmurti NS. 2018. Red Blood Cells Homeostatically Bind Mitochondrial DNA through TLR9 to Maintain Quiescence and to Prevent Lung Injury. American Journal of Respiratory and Critical Care Medicine 197: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Costa L, Galimand J, Fenneteau O, and Mohandas N. 2013. Hereditary spherocytosis, elliptocytosis, and other red cell membrane disorders. Blood Rev 27: 167–178. [DOI] [PubMed] [Google Scholar]
- 19.Maina JN 2000. What it takes to fly: the structural and functional respiratory refinements in birds and bats. The Journal of experimental biology 203: 3045–3064. [DOI] [PubMed] [Google Scholar]


