Abstract
Ornithine racemase has been purified to homogeneity from Clostridium sticklandii, as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. This is the first racemase known to be highly specific to ornithine. This PLP-dependent enzyme has an Mr of 92,000, with a Km for l-ornithine of 0.77 ± 0.05 mM and a kcat of 980 ± 20 s−1.
A major finding from studies of amino acid fermentation in Clostridia is the Stickland reaction, where oxidation of one amino acid is coupled to reduction of another amino acid (1, 2). The catabolism of ornithine in Clostridium sticklandii is of great interest, as ornithine can be either reduced to 5-aminovaleric acid or oxidized to acetate, alanine, and ammonia. Enzymes involved in the l-ornithine oxidation pathway, including d-ornithine aminomutase, DAPA dehydrogenase, and 2-amino-4-ketopentanoic acid dehydrogenase, have been extensively studied (3, 7, 8). However, ornithine racemase, the enzyme responsible for the first step in this pathway, has never been purified or characterized from any Clostridia species. In order to understand more about the catabolism of ornithine in C. sticklandii on a molecular level, we have started to investigate this enzyme. This paper describes the purification of ornithine racemase to homogeneity, together with some properties of the enzyme.
Abbreviations.
The abbreviations used here are as follows: PLP, pyridoxal 5′-phosphate; AdoCbl, adenosylcobalamin; DAPA, 2,4-diaminopentanoic acid; HPLC, high-pressure liquid chromatography.
Bacterial strain and media.
Methods for culturing C. sticklandii (ATCC 12662) were modified from a previous report (6). Cells were grown in a medium consisting of 0.6% tryptone, 0.6% yeast extract, 10 μM CoCl2, 10 μM MnCl2, 10 μM Na2MoSO4, 100 mM MgCl2, 100 mM CaCl2, 1% l-arginine, and 40 mM potassium phosphate buffer (pH 7.5). Each batch of medium was inoculated with a 5% inoculum of an actively growing culture and was incubated at 37°C in the dark for 12 to 16 h. The cell paste was stored at −20°C until used.
Enzyme assay.
A spectrophotometric method was developed to assay ornithine racemase activity. The assay couples the formation of d-ornithine to reduction of NADP+ through the action of AdoCbl-dependent d-ornithine aminomutase and DAPA dehydrogenase. The ornithine racemase activity was assayed by monitoring, at 340 nm, the production of NADPH. The assay solution was buffered by 50 mM Tris-Cl, pH 8.5, and contained 10 mM l-ornithine, 25 μM AdoCbl, 40 μM PLP, and sufficient amounts of the coupling enzymes. This assay was carried out at room temperature in dim light to protect the coenzymes.
Protein purification and properties of the enzyme.
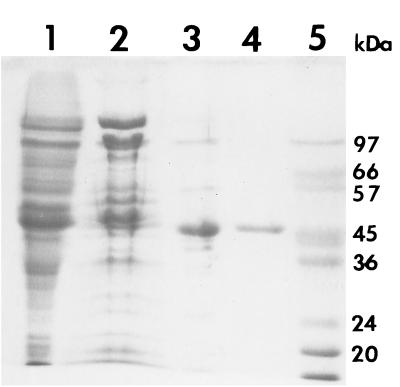
All steps were performed on ice or at 4°C. Cells (15 g) were resuspended in 60 ml of 50 mM potassium phosphate buffer, pH 7.0, and ruptured by sonication. Cell debris was removed by centrifugation, and the supernatant was brought to 25% saturation in ammonium sulfate. The precipitate was removed by centrifugation, and the supernatant was applied to a phenyl-Sepharose high-performance hydrophobic interaction column (2.6 by 25 cm) equilibrated in 50 mM potassium phosphate buffer, pH 7.0, containing 1 mM ditheothreitol, 1 M (NH4)2SO4, and 10% glycerol. After washing the column with 100 ml of the same buffer, the enzyme was eluted with a linear, descending gradient of ammonium sulfate in 1,000 ml of buffer at a flow rate of 1 ml/min. Active fractions were pooled and concentrated by ultrafiltration. After overnight dialysis against 1 liter of 10 mM potassium phosphate buffer, pH 6.2, the protein was loaded onto a 2.6- by 20-cm Q-Sepharose high-performance column equilibrated with 10 mM potassium phosphate buffer, pH 6.2. Protein was eluted with a 600-ml gradient from 0 to 0.5 M KCl at a flow rate of 1 ml/min. Active fractions were pooled and dialyzed overnight against 1 liter of 10 mM potassium phosphate buffer, pH 7.0. Protein was then applied to a Mono Q HR5/5 column equilibrated in 10 mM potassium phosphate, pH 7.0, and eluted with a 20-ml gradient from 0 to 0.5 M KCl at a flow rate of 1 ml/min. The purified enzyme showed a single band with an Mr of 46,800 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 1 and Table 1), whereas its apparent Mr was 92,000, as estimated by gel filtration on a calibrated Sephadex S 200 HR10/30 column. This result suggests that the enzyme retains a dimeric subunit structure. The optimum pH level was observed to be in the alkaline range; maximal rate was achieved at about pH 8.5. The kinetic properties of ornithine racemase were investigated in the direction of l- to d-ornithine. The apparent Km for l-ornithine and the apparent kcat were 0.77 ± 0.05 mM and 980 ± 20 s−1, respectively.
FIG. 1.
Purification of ornithine racemase. Shown are the results from sodium dodecyl sulfate-polyacrylamide gel electrophoresis of samples taken after each step of the purification (gel stained with Coomassie brilliant blue). Lane 1, crude cell extract; lane 2, pooled fractions after phenyl-Sepharose HP hydrophobic interaction chromatography; lane 3, pooled fractions after Q-Sepharose HP anion-exchange chromatography; lane 4, pooled fractions after Mono Q chromatography using the Pharmacia FPLC system; lane 5, marker proteins.
TABLE 1.
Purification of ornithine racemasea
| Column used for purification step | Vol (ml) | Amt of protein (mg) | Activity (U) | Specific activity (U/mg) |
|---|---|---|---|---|
| None | 50 | 699.8 | N/Ab | N/A |
| Phenyl-Sepharose HP | 77 | 89.8 | 6,594 | 73.4 |
| Q-Sepharose HP | 35 | 6.7 | 2,066 | 308.4 |
| Mono Q | 4.2 | 1.3 | 499 | 383.8 |
Enzyme activity was determined in the presence of 25 μM AdoCbl, 10 mM l-ornithine, 40 μM PLP, and NADP+. One unit of enzyme was that amount which catalyzed the formation of 1 μmol of d-ornithine per min in the assay.
N/A, not available. Due to the presence of strong activities of ornithine aminotransferase and cyclase in crude extracts, an accurate measurement of ornithine racemase activity was not available.
Substrate specificity.
Enantiomers of amino acids, except proline, were separated by HPLC on a CROWNPAK CR(+) chiral column. Compounds were eluted with 22% HClO4 at a flow rate of 0.25 ml/min and detected by monitoring at 210 nm. Another chiral column, CHIROBIOTIC T, was used to separate d- and l-proline. The eluent was 0.1% triethylamine (pH 4.1):methanol:water (5:15:80), and the flow rate was 0.25 ml/min. The conversion ratio of l-amino acid into d-amino acid was calculated by integration of the corresponding peak area. For each reaction, 62 ng of enzyme was incubated with 10 mM l-amino acid and 100 mM Tris-Cl, pH 8.5, in a total volume of 0.2 ml at 25°C for 20 min. The reactions were quenched by phenol-chloroform extraction. The chiral HPLC analysis showed that 19.5% of l-ornithine was converted into d-ornithine. However, no activity was detected when l-methionine, l-phenylalanine, l-leucine, l-lysine, l-tyrosine, l-arginine, l-alanine, l-glutamate, and l-proline were used as substrates, even after the addition of five times the amount of enzyme and extension of the incubation time to 60 min.
Cofactor requirement.
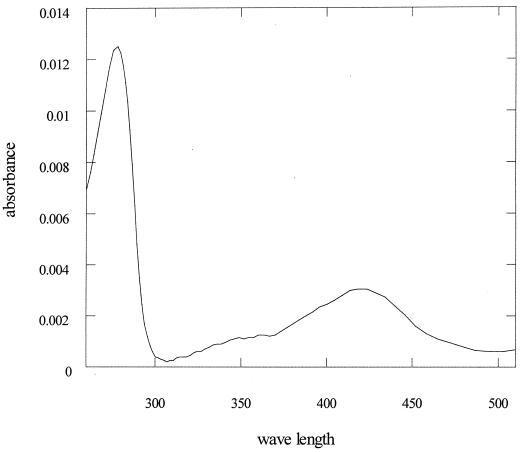
Purified enzyme was loaded onto a Superdex 200 HR10/30 column equilibrated in 50 mM potassium phosphate buffer, pH 7.0, containing 150 mM NaCl. The column was attached to an HPLC pump and a diode array detector. The absorbance of the well-resolved protein peak was recorded between 240 and 500 nm. The presence of an absorption maximum at 420 nm suggests that ornithine racemase binds PLP tightly (Fig. 2). In addition, the conversion rate of l-ornithine with exogenously added PLP (60 μM) was about three times that of l-ornithine with no additions.
FIG. 2.
UV visible spectrum of ornithine racemase. The maximal absorption at 420 nm indicated that PLP was bound to ornithine racemase.
In summary, ornithine racemase has been successfully purified from C. sticklandii and is the first racemase discovered which is highly selective for ornithine. In contrast, other amino acid racemases that are catalytically active to ornithine, such as arginine racemase from Pseudomonas graveolens and threonine racemase from Pseudomonas putida (4, 9), usually possess a very broad substrate specificity. Interestingly, another PLP-independent proline racemase with strict substrate specificity is also involved in the reduction pathway of ornithine in C. sticklandii (5). To investigate the mechanism of this specificity, further experiments are now in progress to clone and overexpress the ornithine racemase gene.
Acknowledgments
This work was supported by grant NSC-88-2314-B-039-035 from the National Scientific Council, Taiwan, Republic of China, and, in part, by grant CMC-89-M013 from China Medical College to H.-P. Chen.
REFERENCES
- 1.Andreesen J R, Bahl H, Gottschalk G. Introduction to the physiology and biochemistry of the genus Clostridium. In: Mintoz N P, Clarke D P, editors. Clostridia. New York, N.Y: Plenum Press; 1989. pp. 49–50. [Google Scholar]
- 2.Barker H A. Amino acid degradation by anaerobic bacteria. Ann Rev Biochem. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- 3.Jeng I M, Somack R, Barker H A. Ornithine degradation in Clostridium sticklandii; pyridoxal phosphate and coenzyme A dependent thiolytic cleavage of 2-amino-4-ketopentanoate to alanine and acetyl coenzyme A. Biochemistry. 1974;13:2898–2903. doi: 10.1021/bi00711a019. [DOI] [PubMed] [Google Scholar]
- 4.Lim Y G, Yokoigawa K, Easaki N, Soda K. A new amino acid racemase with threonine α-epimerase activity from Pseudomonas putida: purification and characterization. J Bacteriol. 1993;175:4213–4217. doi: 10.1128/jb.175.13.4213-4217.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudnick G, Abeles R H. Reaction mechanism and structure of the active site of proline racemase. Biochemistry. 1975;14:4515–4522. doi: 10.1021/bi00691a028. [DOI] [PubMed] [Google Scholar]
- 6.Somack R L, Bing D H, Costilow R N. Preparation and characterization of 2,4-diaminovaleric acid: an intermediate in the anaerobic oxidation of ornithine. Anal Biochem. 1971;41:132–137. doi: 10.1016/0003-2697(71)90198-9. [DOI] [PubMed] [Google Scholar]
- 7.Somack R, Costilow R N. 2,4-Diaminopentanoic acid C4 dehydrogenase. Purification and properties of the protein. J Biol Chem. 1973;247:385–388. [PubMed] [Google Scholar]
- 8.Somack R, Costilow R N. Purification and properties of a pyridoxal phosphate and coenzyme B12 dependent d-α-ornithine 5,4-aminomutase. Biochemistry. 1973;12:2597–2604. doi: 10.1021/bi00738a008. [DOI] [PubMed] [Google Scholar]
- 9.Yorifufi T, Misono H, Soda K. Arginine racemase of Pseudomonas graveolens. J Biol Chem. 1971;246:5085–5092. [PubMed] [Google Scholar]