Abstract
Mitochondria are dynamic membrane-bound organelles in eukaryotic cells. These are important for the generation of chemical energy needed to power various cellular functions and also support metabolic, energetic, and epigenetic regulation in various cells. These organelles are also important for communication with the nucleus and other cellular structures, to maintain developmental sequences and somatic homeostasis, and for cellular adaptation to stress. Increasing information shows mitochondrial defects as an important cause of inherited disorders in different organ systems. In this article, we provide an extensive review of ontogeny, ultrastructural morphology, biogenesis, functional dynamics, important clinical manifestations of mitochondrial dysfunction, and possibilities for clinical intervention. We present information from our own clinical and laboratory research in conjunction with information collected from an extensive search in the databases PubMed, EMBASE, and Scopus.
Keywords: Archezoan, Inner membrane, Intermembrane space, Matrix, Mitochondrial DNA, Mitophagy, Neonate, Ontogeny, Outer membrane, Parkin
INTRODUCTION
Mitochondria are membrane-bound organelles that orchestrate cellular energy production in almost all eukaryotic cells.1,2 These organelles generate adenosine triphosphate (ATP) through oxidative phosphorylation, the components of which are partially encoded in their own genome.3 Mitochondria are not only the cellular ‘powerhouses’, but also play critical roles in supplying intermediary metabolites, temperature maintenance, regulation of Ca2+ homeostasis, determination of cellular life-span, and integration of various signaling pathways.4–11 The physiological importance of mitochondria becomes evident early, and can be seen in the developing oocyte and the embryo, in the fetus, and throughout infancy.12–15 After birth, the increased energy requirements are associated with a significant increase in the mitochondrial number and function.16–19
In this review, we have summarized recent advances in our understanding of mitochondrial dynamics, the importance of the cytoskeleton, cellular signaling, cellular and organ differentiation, regulation of the function of other organelles, and cellular lifespan. The importance of mitochondria as mediators of epigenetic regulation and metabolic processes during development has been explored. The critical role of mitochondria in cellular homeostasis is evidenced by the wide range of clinical manifestations involving multiple organ systems seen in mitochondrial diseases. We present our own clinical and laboratory research, combined with an extensive search in the databases PubMed, EMBASE, and Scopus. To avoid bias, keywords were identified from discussions in our own group and from PubMed’s Medical Subject Heading (MeSH) thesaurus.20
Mitochondrial Ultrastructure
Mitochondria are dynamic intracellular organelles seen in all eukaryotic organisms; one exception might be the oxymonad monocercomonoides, which are obligate animal symbionts that live in the intestinal tracts of vertebrates.21 Human tissues contain mitochondria with a high degree of numerical heterogeneity; erythrocytes do not contain any, whereas hepatocytes and muscle cells may contain hundreds to thousands per cell.5,22,23 These numbers vary not only across various tissues, but also during development, cell cycle, and stress.24
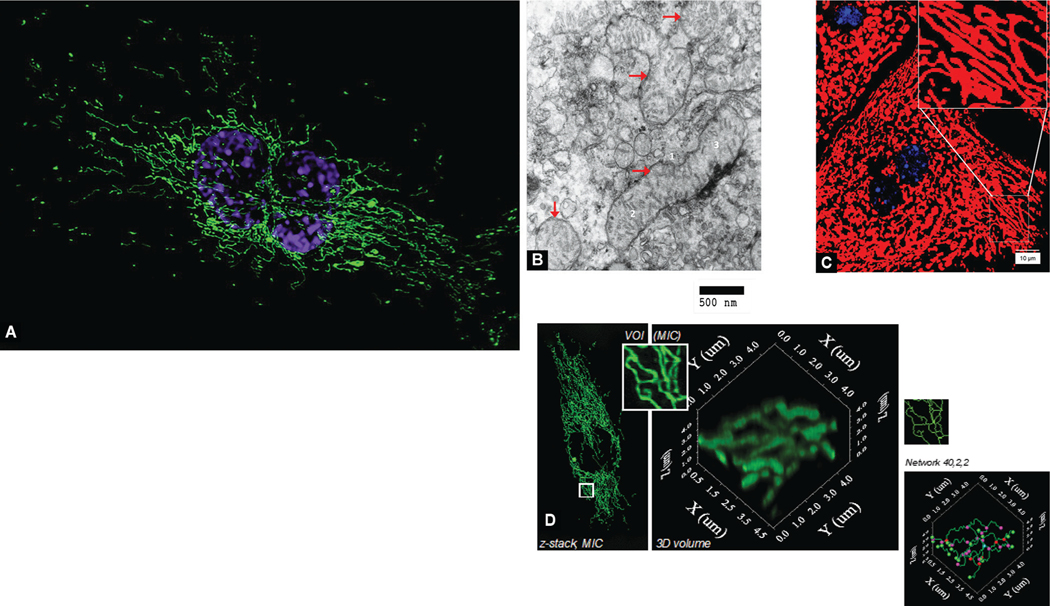
Mitochondria have been traditionally viewed as 0.5–1 μm ovoids, where the number per unit volume seems to be inversely related to size.1,25–27 However, the mitochondrial morphology varies between different cell types; cultured endothelial cells contain a mitochondrial reticulum around the nucleus (Fig. 1A). Similar to prokaryotes, mitochondria are uniquely covered in bilayered membranes. These organelles multiply by binary fission and consistently, electron micrographs show many mitochondria as a dumbbell or racket-shaped: Two larger halves with a narrow bridging tube prior to the separation of the daughter organelles.28–31 Electron micrographs show the mitochondrial membranes, cristae, and matrix (Fig. 1B).
Figs 1A to D:
Panoramic view of mitochondria. (A) Mitochondria in a cultured dividing endothelial cell. The image shows a cultured mitotic human vascular endothelial cell (HUVEC) expressing mitochondrial green fluorescent protein (GFP). Nuclei (blue/purple) were stained with DAPI. Mitochondrial morphology varies between different cell types; this dividing endothelial cell displays a mitochondrial reticulum around recently-duplicated nucleus; (B) Transmission electron microscopy of mouse liver cells show mitochondria (indicated with red arrow). Mitochondrial membranes, 1; cristae, 2; and the matrix, 3 can be seen; (C) Mouse primary hepatocytes stained with MitoTracker Red, a red-fluorescent dye that stains mitochondria in live cells and its accumulation is dependent upon membrane potential. The dye is well-retained after aldehyde fixation; (D) Quantitative image analysis of mitochondria in a human vascular endothelial cell. VOI, Volume-of-interest; MIC, 3-dimensional light microscopic image (described in Nikolaisen et al, DOI: 10.1371/journal.pone.0101365)
Advanced cellular imaging shows foci where mitochondria appear tubular and as forming a network (Fig. 1C) with active division and fusion.32,33 The mitochondrial content and morphology are altered by cellular stress. In living cells, the net mitochondrial content or the mitochondrial mass depends on the balance between mitochondrial biogenesis and degradation. Changes in spatiotemporal positioning, which have been described as the mitochondrial dynamics, and mitochondrial morphology are governed by mitochondrial fusion, fission, and motility.34–36 The dynamics can be seen in sub-nanometer resolution with cryo-electron tomography.32,33 There may be unique orientations and distribution in different types of cells, and a close association with microtubules in some regions.37,38 In sites where the energy requirements are relatively high, mitochondria may appear more static and provide ATP directly on-site.39 However, in other regions, there may be prominent motility either in surveillance for foci with high energy needs or in actual energy production once those are found.36 Fluorescence microscopy combined with quantitative image analysis is a useful method to study the amount and morphology of mitochondrial organelles within cells (Fig. 1D).40 Dynamin-related guanosine triphosphate hydrolases (GTPases) may play an important role in mitochondrial motility.41 Even if the mitochondrial network gets damaged during isolation from cells, some fragments may continue to show respiration and ATP synthesis.3 The mitochondrial membrane protects the structure and electrical potential of these organelles.42
Structural Models
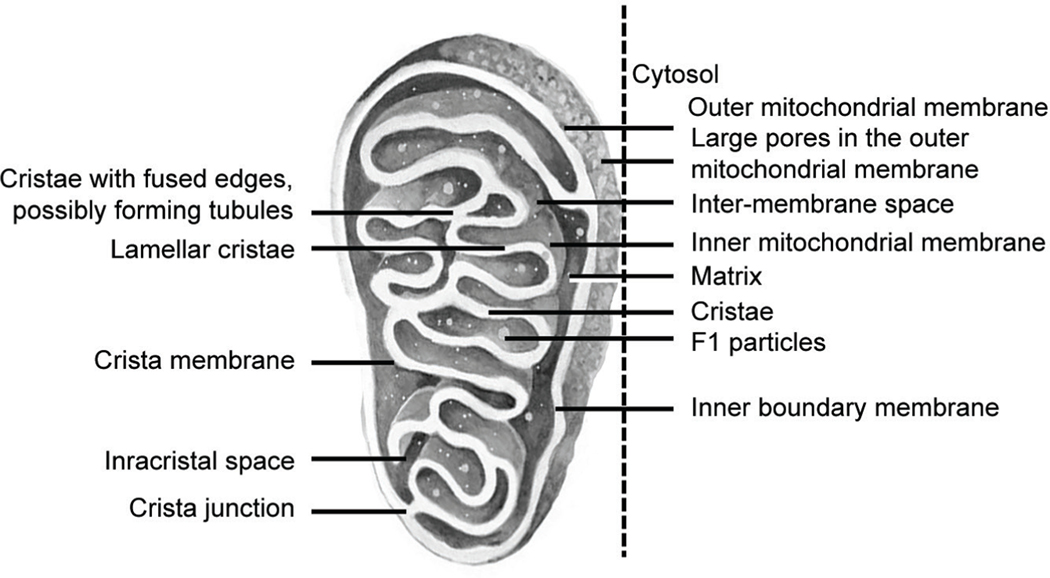
The following section summarizes current information on various compartments in mitochondria. The location of these key elements is shown in Figure 2:
Fig. 2:
Graphical depiction of mitochondrial infrastructure showing the location of the most important/better-known elements
- An outer mitochondrial membrane (OMM) that is freely traversed by ions and small molecules.3 It is highly porous, and hence no electrical potential difference is detectible across this membrane layer.3 There are nearly 200 proteins, but the following are some of the best characterized:43
- Mitochondrial import complex (MIM): it inserts α-helical proteins in the OMM independently of the TOM complex;47 and
- Endoplasmic reticulum-mitochondria encounter structure (ERMES).48
- Adenine nucleotide translocator (ANT), has been referred to by various other terms such as the ADP/ATP translocase, ADP/ATP carrier protein, or mitochondrial ADP/ATP carrier. It exchanges free ATP with free ADP across the inner mitochondrial membrane (IMM). Adenine nucleotide translocator is the most abundant carrier protein in the IMM;
-
An inter membrane space (IMS) between the inner and outer membranes contains about 5% of the mitochondrial proteins in an aqueous medium (3.8 μL/mg protein).3,53,54 The large physical size of the OMM pores makes the IMS largely continuous with the cytosol;55,56 Proteins synthesized on cytosolic ribosomes traverse these pores and bind carriers.57–61
The IMS may contain many pro-apoptotic factors such as the cytochrome c.62 Other proteins may display CX3C or CX9C motifs.63 The machinery for import and assembly of IMS proteins mitochondrial intermembrane space assembly (MIA) can bring in large proteins that may be up to 11 kDa in size.59,64
An IMM separates the IMS from the mitochondrial matrix. It is very selectively permeable to most molecules, and therefore, carries many specialized transporters.3,65,66 Anelectrochemical membrane potential of about 180 mV has been documented across the inner membrane.67 The membrane is also a site for oxidative phosphorylation, which is used to create electrochemical gradients for ATP synthesis.68 The IMM is extensively folded, where numerous invaginations called cristae increase its total surface area.3 These cristae are separated from inner boundary membranes by junctions, and the ends are partially closed by transmembrane proteins that bind opposing membranes.3,69–71 Cristae also affect the overall chemiosmotic function of mitochondria.65
The major role of the IMM is to facilitate molecular transport and signaling for oxidative phosphorylation and ATP synthesis.69,71 A junctional protein, the inner mitochondrial membrane translocase protein (IMMT), is expressed in the nucleus and is transported to the IMM, where it maintains the electrical potential and the structural invaginations seen in the inner membrane.72–74 On the matrix side, the crystal membranes are studded with small proteinaceous F1 particles, which promote proton-gradient-driven ATP synthesis.3 The electron transport chain on the cristae includes 5 complexes: complex I nicotinamide adenine dinucleotide, hydrogenated (NADH) (NADH: coenzyme Q oxidoreductase), complex II (succinate: coenzyme Q oxidoreductase), complex III (coenzyme Q: cytochrome c oxidoreductase), complex IV (cytochrome c oxidase) and ATP synthase.62,75,76 Overall, the cristae membranes are dynamic and can reshape in seconds. Cristae membrane remodeling is regulated by the mitochondrial contact site and cristae organizing system (MICOS) complex, optic atrophy-1 (OPA1), F1F0 ATP synthase, and the lipid microenvironment.77–81
The matrix in the core of these organelles is enclosed within the IMM. This gel-like material contains DNA, ribosomes, soluble enzymes, small organic molecules, nucleotide cofactors, and inorganic ions.3 The pH of 7.8 in the matrix is higher than the 7–7.4 seen in the IMS.82,83 The water content, about 0.8 μL/mg protein, is lower than that in the IMS.84 The restricted permeability of the IMM may regulate the osmotic balance.85 The aquaporin conduits in the membrane may also play a role.86 The matrix is the site for the tricarboxylic acid (TCA) cycle (citric acid cycle, Krebs cycle) metabolism for ATP production (Fig. 4).87 It contains the key regulators, including citrate synthase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, fumarase, and malate dehydrogenase; succinate dehydrogenase, located on the IMM, is an exception.88,89
Mitochondrial DNA (mtDNA) is comprised of one or more double-stranded, mainly circular DNA in the matrix. Mitochondrial DNA (mtDNA) encodes for 2 ribosomal ribonucleic acids (rRNAs), 22 transfer RNAs (tRNAs), and 13 proteins involved in the mitochondrial respiratory chain.90 It is rich in guanine and cytosine and contains 37 genes with about 17,000 base pairs.91
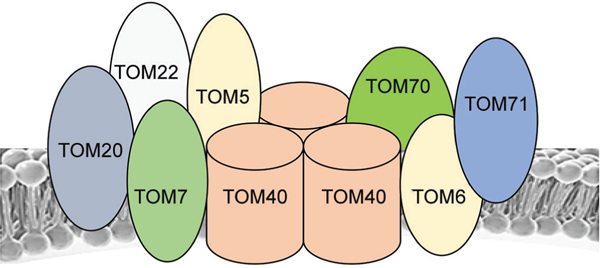
Fig. 3:
Translocases of the outer mitochondrial membrane (TOM): The TOM complex is comprised of many subunits; It recognizes, segregates, and translocates precursor proteins to various sites within the mitochondria
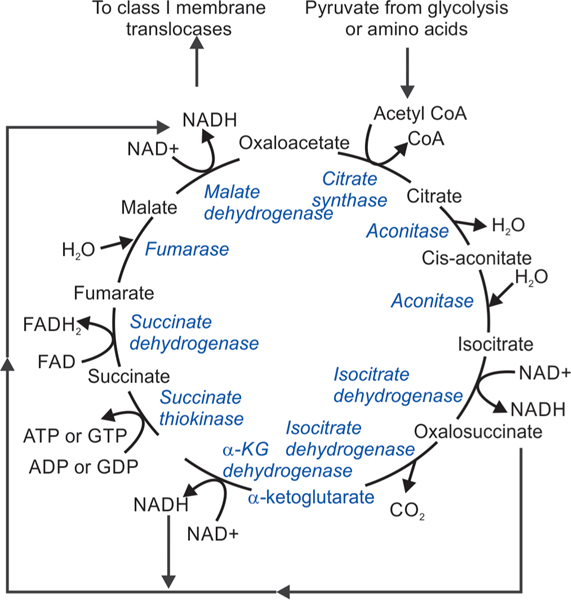
Fig. 4:
Graphical depiction of the TCA cycle. Metabolites enter the TCA cycle as acetyl-CoA, and progress to form α-ketoglutarate, succinyl-CoA, fumarate, and oxaloacetate. Nicotinamide adenine dinucleotide (NAD), a coenzyme central to metabolism is hydrogenated to form NADH; KG = ketoglutarate. Enzymes are depicted in blue font
The mtDNA accounts for about 1% of the total DNA in a cell. In humans, only 13 proteins are encoded in mtDNA; all are central, hydrophobic subunits of the respiratory chain complexes/ATP synthase.92 In total, there are 1,500 estimated different mitochondrial proteins; >99% of these proteins are likely encoded in the nucleus, synthesized in the cytosol, and imported into the mitochondria.93,94
Human mitochondria contain a unique protein translation machinery with ribosomes, transfer-RNAs (tRNAs), and associated protein factors that resemble those seen in bacteria.95,96 However, mitochondria make surprisingly little use of their specialized protein production machinery. Most of the mitochondrial proteins are synthesized in the cytoplasm and then imported into the organelle by protein translocases.57 Most of the mitochondrial proteins are transcribed in the nucleus, synthesized in the cytosol, and then imported back into the organelle.94 More than 3,000 mitochondrial proteins have been estimated in vertebrate animals.97 During evolution mitochondrial genes have relocated to the nucleus, whereas the translated proteins are imported back into the mitochondria to perform their function.97–99
Evolutionary Perspective
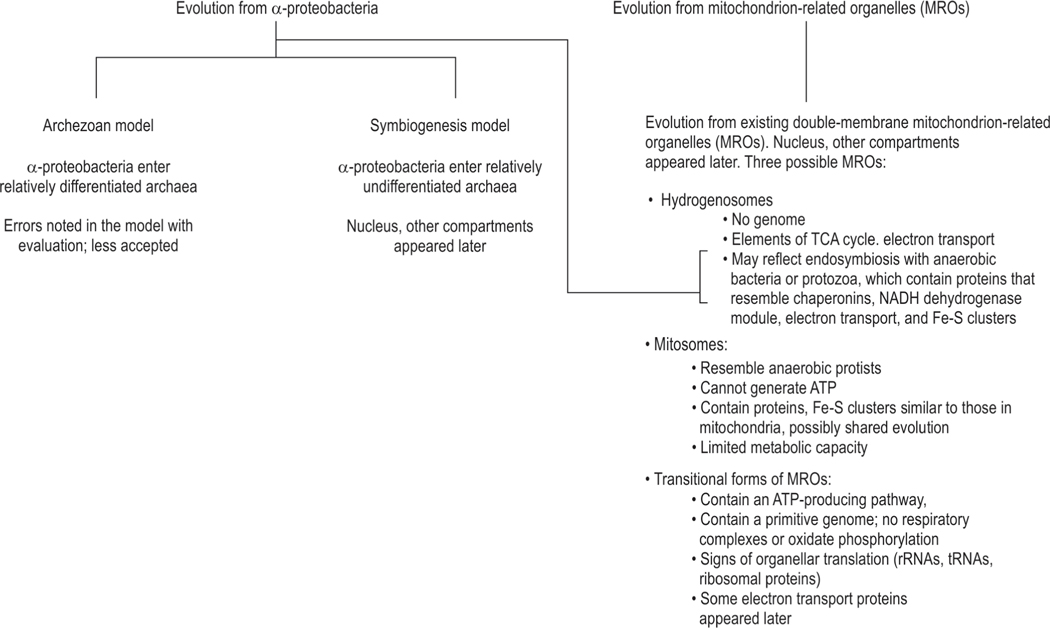
There are three models for mitochondrial development, where an existing cellular organism accepted proto-mitochondria.100 There are two endosymbiotic models, and a third where mitochondria could have evolved from related predecessor organelles (Fig. 5):
Fig. 5:
Existing models for mitochondrial development. Two endosymbiotic models suggest that proto-mitochondria entered an existing cellular organism. The third suggests that mitochondria evolved from related organelles that already existed in host cells
Archezoan scenario: A hypothetical primitive a mitochondrial eukaryote, termed archezoan, accepted a proto-mitochondrial endosymbiont.97,101,102 Rigorous studies have detected artifacts and raised doubts about the validity of these hypotheses.103
Symbiogenesis scenario: An archaeal cell underwent a single endosymbiotic event with an α-proteobacterium, which generated mitochondria.104 This event was followed by the evolution of the nucleus and compartmentalization of the eukaryotic cell.102
- Evolution from mitochondrion-related organelles (MROs).105 These models envisage three possible double-membrane mitochondrial precursors, which contained minimal or no DNA:
- Hydrogenosomes: These lack a genome but may have a few incomplete elements of the TCA cycle and the electron transport chain.106 The anaerobic metabolism seen in hydrogenosomes suggests that these might have originated through endosymbiosis with an anaerobic bacteria such as Clostridium.107 However, later studies showed that the hydrogenosomes in Trichomonas vaginalis may also contain several proteins resembling those in mitochondria, including chaperonins, the NADH dehydrogenase module of electron transport complex I, and components of the mitochondrial machinery for synthesis of iron-sulfur (Fe-S) clusters.108,109 Hydrogenosomes could very well be relict mitochondria;110
- Mitosomes: These have been seen in anaerobic, parasitic protists such as the amoebozoons Entamoeba histolytica and Mastigamoeba balamuthi. These organisms were initially considered to be amitochondriate and unable to generate ATP.105,111 However, these contain several proteins similar to those seen in mitochondria and hence could be evolutionarily related to conventional mitochondria.97,109 Compared to mitochondria, the metabolic capacity of mitosomes is relatively limited;112
- Transitional MROs: These are seen in anaerobic ciliates Nyctotherus ovalis, and Blastocystis spp., which are related to the brown algae, diatoms.97 There is a possibility of a shared origin between mitochondria, hydrogenosomes, and mitosomes.113 These MROs can generate H2 via an ATP-producing, hydrogenase-mediated pathway.114 The genome is relatively limited, and there are not many mtDNA-encoded genes similar to those seen in the respiratory complexes III, IV, and V.115 These MROs also lack the ability to generate ATP via coupled electron transport and oxidative phosphorylation.116 The MRO genome can contain organellar translation systems (rRNAs, tRNAs, ribosomal proteins) and a partial electron transport chain with subunits of electron transport complexes I and II.111
Evolutionary Precursors of Mitochondria
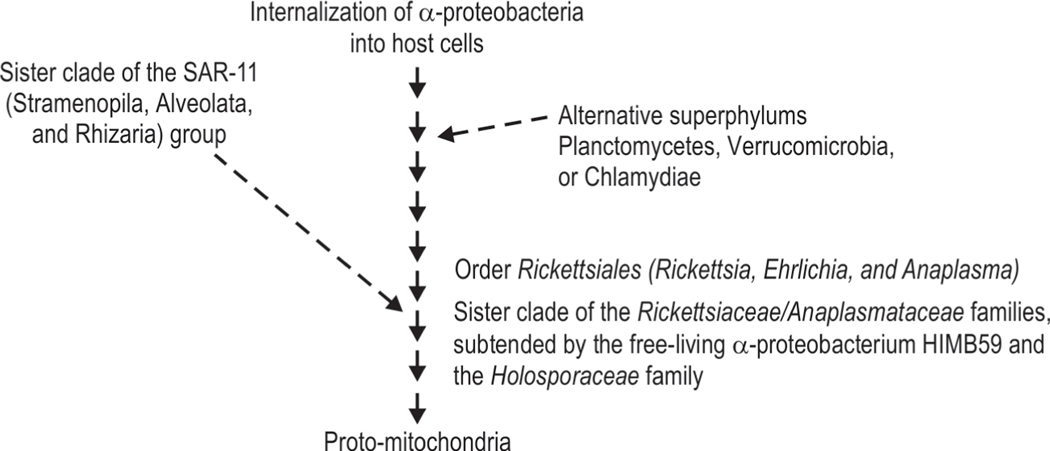
Mitochondria are generally believed to have evolved about 1.6–2 billion years ago from α-proteobacteria, a subgroup of the purple non-sulfur bacteria (Fig. 6).81–83 These bacteria were internalization into host cells and then differentiated through several transitional forms of proto-mitochondria.117–119 Increasing information places mitochondrial precursors in the order Rickettsiales, which is a subgroup of α-proteobacteria that include obligate intracellular bacterial parasites such as Rickettsia, Ehrlichia, and Anaplasma.120 Phylogenomic analysis based on the 32 genes shared by mitochondria and these bacteria show similarities with the order Rickettsiales.121 These similarities identify mitochondria to possibly be a sister clade of the Rickettsiaceae/Anaplasmataceae families, subtended by the free-living α-proteobacterium HIMB59 and the Holosporaceae family.122
Fig. 6:
Proteo-bacteria evolved into mitochondria following internalization into host cells
Mitochondria also seem to show a sister-clade relationship with a group of free-living bacteria known as the Stramenopila, Alveolata, and Rhizaria (SAR)-11 group.121 These are ubiquitous, free-living, small carbon-oxidizing bacteria with an estimated global population of 2.4 × 1028 cells, present in nearly 25% of all oceanic plankton.123,124 If confirmed, this relationship would suggest an alternative source of mitochondria in addition to those from Rickettsiales.125 However, these findings need confirmation through accurate reconstruction of genome trees and evolutionary models, better statistical support without stochastic noise, identification of composition biases in the sequence data, or systematic errors such as long-branch attraction.126–131
Unfortunately, the identification of these relationships has been difficult.130,132,133 There are many restrictions such as (a) weakness of the phylogenetic signal: Signals from small subunit rRNAs have weakened with time due to saturated mutations;134 (b) long-branch attraction: Mitochondria and the obligate intracellular α-proteobacteria have more rapid rates of evolution than the free-living bacteria, and therefore, there are more artifacts;135 and (c) sequence composition bias: AT-rich genome sequences in mitochondria can result in errors in phylogenetic reconstruction.136
Evolution of Prokaryotic Host Cells into Eukaryotic Ancestors
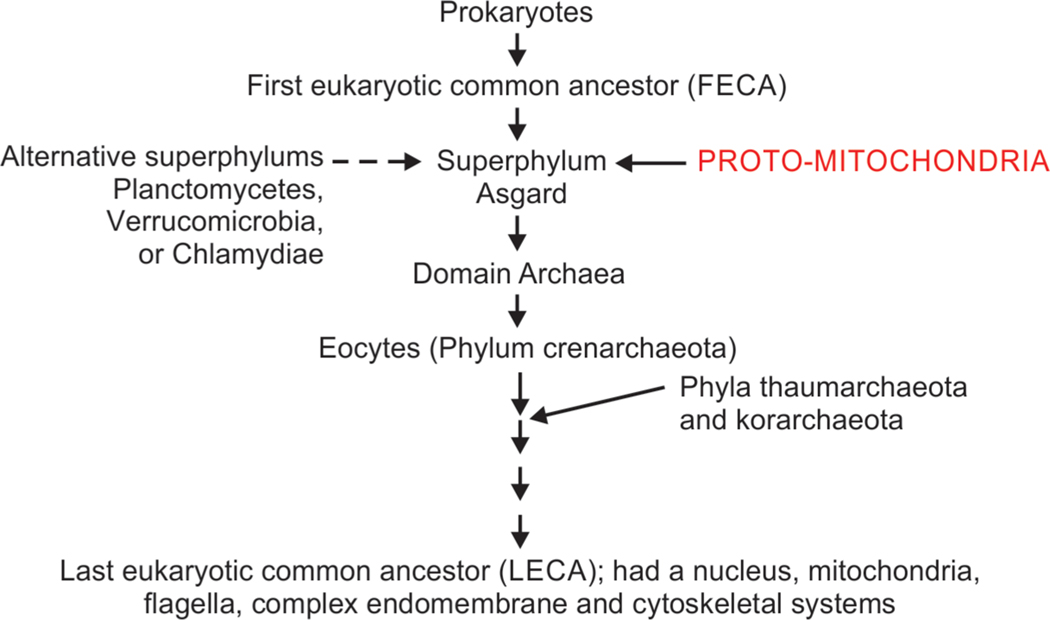
The evolutionary sequence in which the proto-mitochondrial bacteria entered an endosymbiotic relationship with prokaryotes is still uncertain. To place cellular evolution in perspective, prokaryotes are known to have acquired eukaryotic characteristics with differentiation.117 The first eukaryotic common ancestor (FECA) matured through several stages to be identified as the last eukaryotic common ancestor (LECA) about 1–1.9 million years ago (Fig. 7).113,137–139 These ancestor cells showed many features similar to modern eukaryotes.
Fig. 7:
Evolution of an endosymbiotic relationship between eukaryotic ancestors and proto-mitochondrial bacteria. The first eukaryotic common ancestors (FECAs) likely matured into the superphylum Asgard and internalized proto-mitochondrial bacterial ancestors. However, there is a possibility that the FECAs could have originated in other super-phyla. These cells matured over time, and the last eukaryotic common ancestors (LECAs) seen about 1–1.9 million years showed a nucleus, mitochondria, flagella, complex endomembrane, and cytoskeletal systems
Cells in the superphylum Asgard, which were the immediate descendants of the FECA, are considered to be the most likely hosts of the proto-mitochondria.113,140 Some alternative host lineages have also been considered in the superphylum Planctomycetes, Verrucomicrobia, or Chlamydiae.103,141 However, if the host cells were indeed proven to be Asgardian, these cells most likely evolved first to the domain archaea, and then to phyla such as Crenarchaeota, Thaumarchaeota, and Korarchaeota.142 With some capability of metabolizing oxygen, these cells have been viewed as evolutionarily closer to eukaryotes and termed eocytes.143–145
The eukaryotic ancestors continued to differentiate during this process. The LECA possessed most of the eponymous components of eukaryotic cells, including the nucleus with nuclear pores, associated complexes, and nuclear lamina.146 This nucleus is believed to have contained linear chromosomes with telomeres, encoding about 4,000 genes containing spliceosomal introns.147 It likely possessed complex gene regulatory mechanisms, including RNA interference systems and small non-coding RNAs, and histone packaging that affected the accessibility to chromatin.148 Transcription was uncoupled from translation and involved extensive RNA processing (including intron splicing, capping, and polyadenylation).149 This ancestor also had an elaborate protein regulation and recycling system composed of a proteasome and a ubiquitin signaling system.150
The cellular environment of the LECA was compartmentalized with endomembrane systems such as the endoplasmic reticulum, the golgi apparatus, endosomes, lysosomes, and peroxisomes.137 It displayed exocytic and various endocytic pathways such as phagocytosis.151 There was an actin-tubulin cytoskeleton that enabled intracellular trafficking, cell motility, and a complex cell cycle.152 The last eukaryotic common ancestor was likely able to synthesize phospholipids composed of glycerol 3-phosphate and fatty acids, as well as sterols and sphingolipids.153 These cells also show many genes of bacterial origin that were likely acquired when the mitochondrial ancestor was engulfed.154
The host cells are covered in a bilayered lipid membrane, a simple cell wall (S-layer) rich in N-glycosylated proteins, and a relatively well-developed cytoskeleton with homologs of actin and tubulin.155 During evolution into eukaryotes, the three most important changes were the acquisition of the nucleus, the endomembrane system, and the mitochondria.154 However, the sequence of these events remains unclear. There are at two possibilities:
Syntrophic consortium model: Simultaneous fusion of a symbiotic community that included the cytoplasm, nucleus, and mitochondria.156
Endospore model: The nucleus evolved when a cell engulfed a sister following cell division. This model resembled endospore development in Gram-positive bacteria. Mitochondria were acquired later.103 This model is so not well-supported by evidence.62
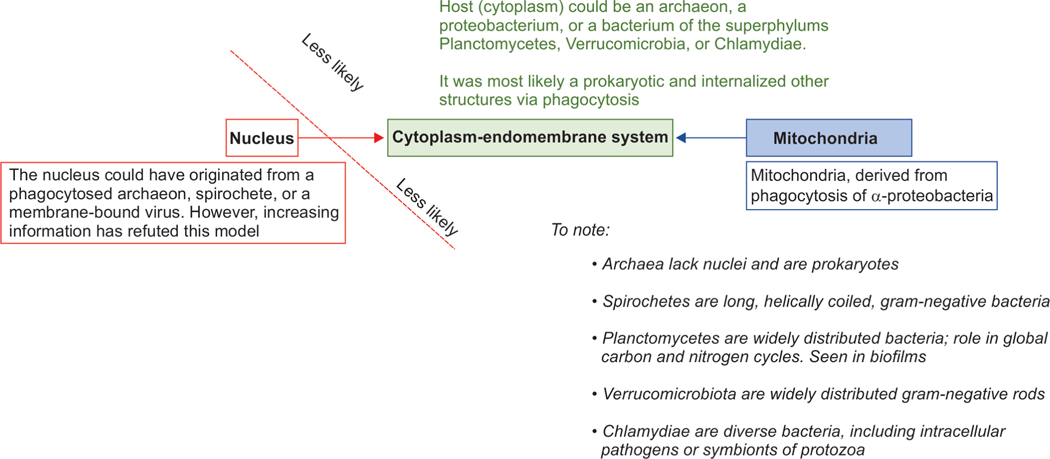
The nucleus was most likely not acquired from the internalization of another organism; phylogenomic analyses of the eukaryotic genome support the presence of an archaeal and a proteobacterial genome, but not the other genome donor(s) expected in nuclear endosymbiotic models (Fig. 8).155 Such endosymbiotic models would also require supplemental theories to explain the origin of the endomembrane system, the physical continuity of inner and outer nuclear membranes, and the formation of nuclear pores.62,104 Hence, the most compelling models suggest an autogenous origin of the nucleus. Infoldings/pinched-off sections of the plasma membrane formed the endoplasmic reticulum (ER) like internal compartments that later became organized around the chromatin to form the inner and outer nuclear envelope and enclosed a proto-nucleus.155
Fig. 8:
Differentiating host cells internalized proto-mitochondrial bacterial ancestors. There have been considerations that the nucleus could also have originated in a phagocytosed archaeon, spirochete, or a membrane-bound virus. However, increasing information has refuted these possibilities and favor an endogenous origin of the nucleus
Acquisition of Mitochondrial Precursors into Host Cells
Mechanistic Perspective
Phylogenetic data suggest that the proto-mitochondria were likely acquired through an intimate mutualistic association between the archaeal host cells with bacterial ancestors of mitochondria that lived on the surface of these cells.103 During evolution, these bacteria have exchanged genes to achieve lower GC contents.157 The strongest evidence of this evolutionary process is the close homology between bacterial and mitochondrial respiratory chain complexes.158,159 The mitochondrial endosymbiont gradually became less complex in its genome and proteome.97,113 It also adapted gradually to anaerobiosis.103 There are two possible mechanisms:
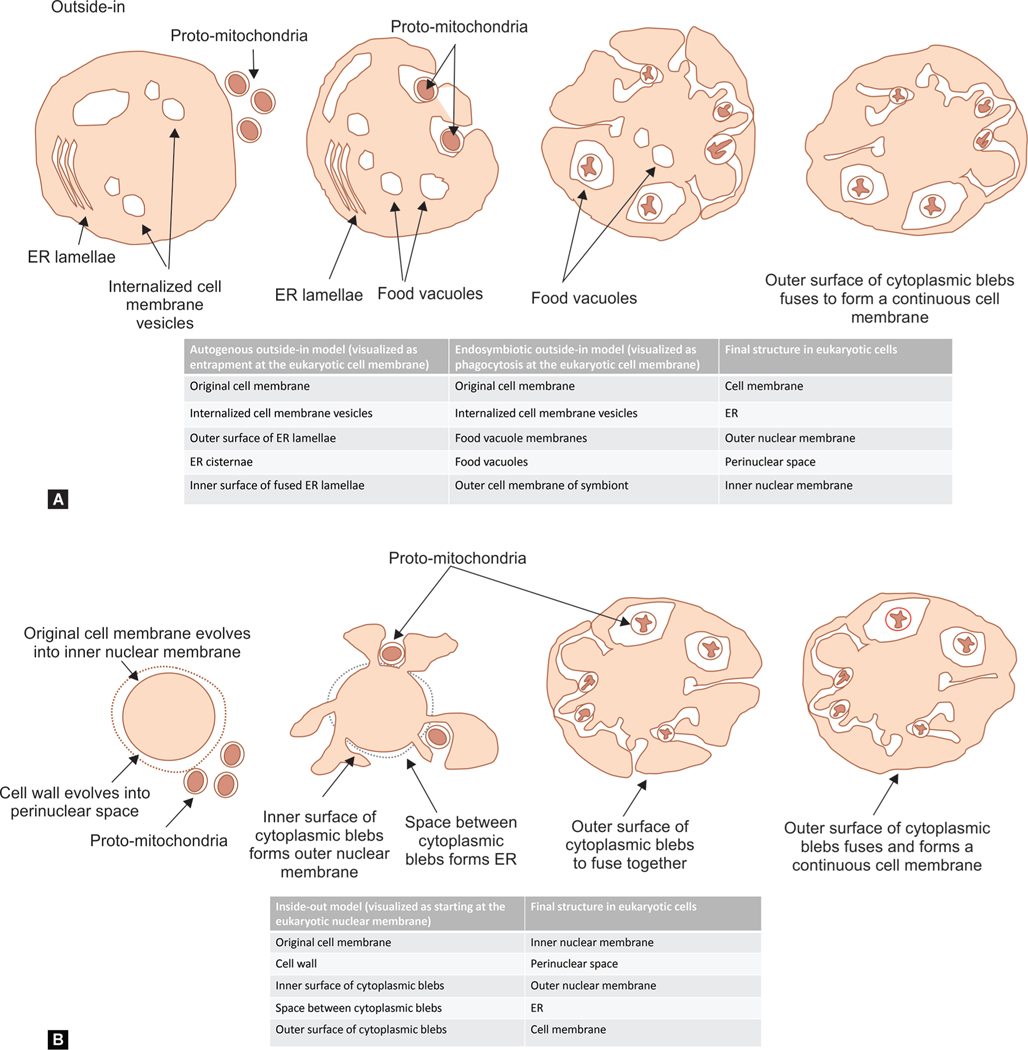
Outside-in models: The symbionts living on the host surface might have been internalized in cell membrane vesicles (Fig. 9A). The inner and outer nuclear membrane was formed by the ER lamellae and the perinuclear space by the ER cisternae.160 There is also a possibility that these bacteria could have been phagocytosed into food vacuoles, and then entered the cytoplasm by lysing the vacuolar membrane.161 The cell membrane covering these symbionts contributed to the formation of the nuclear membrane;155
Inside-out model: The host cells generated extracellular protrusions (blebs) to increase the total surface area155 (Fig. 9B). The ancestral prokaryotic cell body remained intact as the eventual nuclear compartment during this evolution into eukaryotes.155 The protrusions then fused and contained the cytoplasm and an endomembrane system, which evolved to make the outer nuclear and plasma membranes.155 The mitochondrial precursors were initially trapped within the ER but then penetrated the ER membrane to move into the cytoplasm.155 Finally, the formation of a continuous cell membrane closed off the ER from the exterior.162
Figs 9A and B:
Evolution of mitochondria. (A) Outside-in model: Symbionts could have been internalized in cellular vesicles. The inner and outer nuclear membrane may have originated in the endoplasmic reticulum lamellae, and the perinuclear space in the ER cisternae. There is also a possibility that these bacteria could have been phagocytosed into food vacuoles, and then entered the cytoplasm by lysing the vacuolar membrane; (B) Inside-out model: host cells generated extracellular protrusions (blebs). The ancestral prokaryotic cell body remained intact as the eventual nuclear compartment during this evolution into eukaryotes. The protrusions fused and formed the cytoplasm and an endomembrane system, which evolved to make the outer nuclear and the plasma membranes. The mitochondrial precursors moved from the ER into the cytoplasm. Finally, the formation of a continuous cell membrane closed off the ER from the exterior
The base of the cytoplasmic protrusions might have been stabilized by proteins homologous to the highly conserved coat protein II (COPII) in the outer ring of the nuclear pore.163 Exchange of materials such as hydrogen, sulfur, hydrogen sulfide, organic acids, and ATP may have expanded these protrusions.155 The blebs likely stabilized an outer ring of nucleoporins in the cell wall.155 Proteins such as the linker of nucleoskeleton and cytoskeleton (LINC), which physically connect the cytoskeleton with the nucleoskeleton, might have stabilized the nuclear envelope and promoted nuclear bleb formation.164
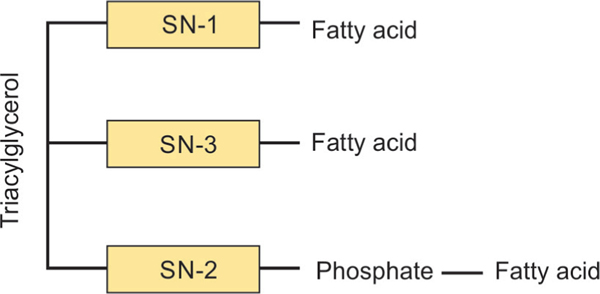
Most structural lipids in eukaryotic cell membranes resemble bacterial, not archaeal lipids. Bacterial and eukaryotic membrane lipids carry glycerol-3-phosphate lipids with ester-linked, straight-chain fatty acids, whereas archaea contain a glycerol-1-phosphate backbone and ether-linked fatty acids (Fig. 10).154,165 Additionally, eukaryotes and some bacteria, but not archaea, produce triterpenoids (for example, hopanoids and sterols) that modulate membrane fluidity.166 Eukaryotes may have acquired bacterium-like lipids from mitochondria.154 The genes for lipid biosynthesis from proto-mitochondria may have been transferred prior to the development of vesicle trafficking systems and phagocytosis.155
Fig. 10:
Bacterial and eukaryotic membranes carry glycerol-3-phosphate lipids with ester-linked, straight-chain fatty acids, unlike the glycerol-1-phosphate backbone seen in archaea; SN, Stereospecific numbering
The analysis of archaeal lipids has provided some support to the possibility that phagocytosis evolved after the acquisition of mitochondria. Archaeal membranes typically retain their physical properties across a wide range of temperatures, whereas bacterial and eukaryotic membranes retain structures best at a narrow range of physiological temperatures.167 These properties might be important for optimizing phagocytosis.
Phylogenetic Perspective
Mitochondria are seen in most eukaryotic host cells as the two may have evolved together.97 Consequent improvements in efficiency in metabolic processes and the encoding of interacting gene products have created an obligate codependence. However, the number of mitochondria per cell has changed through evolution and differs across phyla.168 Many unicellular eukaryotes contain only a few mitochondria, whereas others can contain up to 105.169 The number of mitochondria can vary in multicellular eukaryotes from 80 to 2,000 per cell.168
In proto-mitochondria, the electron transport chain and pathways for β-oxidation of fatty acids were likely present, indicating that the mitochondrial endosymbiont had an aerobic metabolism.170 The pathways for the synthesis of lipids, biotin, heme, and Fe-S clusters were also present.113 This proto-mitochondria might have been capable of facultative aerobic respiration.113
Unlike the mitochondrial genome, the proteome shows only limited similarity with that in α-proteobacteria.98 The mitochondrial ribosome also shows a high degree of evolutionary retailoring.171,172 In many eukaryotes, the mitochondrial large- and small subunits have become smaller than their bacterial counterparts, and many new ribosomal proteins have been added.97 The structure that was originally an RNA-rich complex has now become enriched in proteins.171
Mitochondrial Biogenesis
This process includes the growth and division of pre-existing mitochondria. Mitochondria are believed to have evolved from an α-protobacteria endosymbiont that became incorporated in a host cell.173 Due to this bacterial origin, mitochondria contain a characteristic genome and also show auto replication.174 Mitochondrial proteins are encoded by the mtDNA and specifically-encoded structures in the nuclear genome (described above).94
Major Molecular Components
A large number of mitochondrial proteins, nearly 1,000–1,500 are encoded in the nucleus.175 The mRNAs are transcribed in the nucleus and translated into the cytosol. Most precursor proteins, whether folded or not, pass through the mitochondrial membranes assisted by protein translocases.57 Many of these folded proteins are tagged with an N-terminal, positively-charged presequence.176,177 Once this presequence is cleaved off by a matrix protease, these proteins may get folded with the aid of molecular chaperones.176 Proteins moving to the other mitochondrial compartments may be transported by different protein-import pathways.178 Many precursor proteins that do not contain the N-terminal signals may carry the targeting information within the actual protein sequence.179 The mitochondrial membrane potential and the action of matrix Hsp70 (heat-shock protein 70) regulate this translocation.180
As mentioned above, the translocase of the outer membrane (TOM) protein is a universal entrygate for all proteins entering the mitochondria.44 Many different pathways diverge at this point:
translocase of the inner membrane (TIM), which sorts matrix-targeted precursors;181
presequence translocase-associated motor (PAM) regulates matrix Hsp70 action to drive precursors into the matrix;182
sorting and assembly machinery (SAM) on the outer membrane inserts β-barrel proteins into the outer membrane.183
These processes are an integral part of mitochondrial biogenesis, which may involve not only increased number but the size and mass. Mitochondrial biogenesis can also be altered by environmental stress as malnutrition, low temperature, oxidative stress, and cell division.184
An important regulator of mitochondrial biogenesis is peroxisome proliferator-activated receptor-gamma coactivator ([PGC]-1alpha) [PPAR (peroxisome proliferator-activated receptor)-γ coactivator-1α].185,186 Peroxisome proliferator-activated receptor-gamma coactivator (PGC)-1alpha stimulates the formation of new mitochondria by inducing UCP (uncoupling protein) 2, nuclear respiratory factor (NRF)-1, and NRF-2.175 Nuclear respiratory factor (NRF)-1 and NRF-2 then induce key mitochondrial enzymes.187 These also interact with Tfam, which drives transcription and replication of mtDNA and many nuclear-encoded mitochondrial components.184 Nuclear respiratory factor (NRF)-1 activates the transcription of δ-ALAS (δ-aminolevulinate synthase), and NRF-2 that of cyclo-oxygenase (COX) IV.188,189 Peroxisome proliferator-activated receptor-gamma coactivator (PGC)-1alpha also interacts with other modulators of transcription such as PPARs, thyroid hormones, glucocorticoids, estrogen and ERRs (estrogen-related receptors)-α and -γ.190,191 The ERRs are orphan nuclear receptors that target the gene networks involved in energy homoeostasis, and in mitochondrial biogenesis and function.192–194 Mitochondrial subdivision also involves the dynamin-like and the fis-type proteins.41 Regulation of the numerical density of mitochondria in various cells: Metabolic needs may regulate the mitochondrial mass via-a-vis the cellular size across the cell cycle and the total body weight according to a power law.168,195 In some tissues, the organelles appear as discrete units and the number and size may be related to the cell size. The cells resemble single-celled eukaryotes and may follow linear or sublinear scaling for mitochondria, not Kleiber’s power law (an animal’s metabolic rate scales to the 3/4 power of the animal’s mass).196,197 Some cells show context-dependent mitochondrial morphology, and sometimes a large filamentous organelle reticulum.34 The size of these organelles may not consistently scale strongly with cell size in single-celled eukaryotes, suggesting that the number of mitochondria per cell may be more important than the size of these organelles as a means of modulating cellular energetic requirements.168 There might be a possibility of having an optimal per-cell mitochondrial mass given the cell size and the nature of mitochondrial biogenesis.
In humans, the mitochondrial count varies across different tissues and organs.10,168 Mature erythrocytes do not contain any, whereas metabolically-active organs such as the liver, kidney, heart, and brain tissues contain large numbers.198 Mitochondria are important for metabolic functions and also for cellular maintenance.199,200 Many mitochondrial genes have been transferred to the nuclear genome during evolution. These relocations might have improved the numerical efficiency, proximity to up- and/or downstream genetic systems, or improved utilization of cytoskeletal space by preventing redundancy in transcription sites.201
Even though there might be some variance in the expression of a subset of genes expressed in the mitochondria, most of these genes are transcribed at a specific, constitutive level.91 One reason might be that the entire circular mitochondrial genome involves one strand at a time.202 The number of mitochondria may also be important because of the variations in the lability of these organelles and that of energetic constraints across tissue types.203
Intercellular transfer of mitochondria: Mitochondria and mtDNA can be transferred between cells.204 Transient focal cerebral ischemia can release mitochondria from astrocytes, which enter adjacent neurons via a calcium-dependent mechanism involving CD38 and cyclic ADP ribose signaling. This can amplify the survival signals.205,206 Horizontal transfers of mtDNA have also been noted in cancer cells; extracellular vesicles containing mtDNA can pass through tunneling nanotubes and connexin 43 gap junctions between cells.207,208
Epigenetic Changes Involved in Mitochondrial Biogenesis
The epigenetic landscape is extensively reprogrammed during embryonic and fetal development.209 Paternal genome also shows considerable DNA demethylation after fertilization.210 Depletion of mtDNA leads to alteration in the metabolism of amino acids including methionine, leading to increased DNA methylation.211 S-adenosylmethionine (SAM) acts as a co-factor in these methylation reactions; SAM is produced from methionine by methionine-adenosyl transferases (MATs).212,213 Interestingly, the development of the embryo prior to implantation requires appropriate histone demethylation mediated by the JMJ (jumonji, or the Jarid2) deamylase, which removes the methyl group from lysine residues. The JMJ demethylases catalyze the histone demethylation in an α-ketoglutarate-dependent manner.214,215 Thus, mitochondria regulate demethylation via α-ketoglutarate through the oxidation of glucose and glutamine in the mitochondrial citric acid cycle.216
Chromatin remodeling is also important in embryonic epigenetic programming.216 Histone acetylation relaxes the condensed chromatin and promotes gene transcription.217 Contrarily, deacetylation of histone condenses the chromatin and suppresses transcription.217 Histone acetylation by specific histone acyltransferases requires acetyl-CoA, which is the product of oxidative decarboxylation of pyruvate produced by glycolysis, β-oxidation of fatty acids, and amino acid metabolism, and then shuttled out of mitochondria in the form of citrate, acetyl-CoA precursor.218 In human embryonic stem cells, increasing acetylation suppresses differentiation, while inhibition of acetyl-CoA production from glucose results in the loss of pluripotency. The availability of nicotinamide adenine dinucleotide (NAD+) controls the activity of the conserved NAD+-dependent histone deacetylases, the sirtuins (SIRTs).219 SIRTs are involved in blastocyst development as the inhibition of SIRT activity decelerates blastocyst development. NAD+ can be synthesized de novo from the amino acid tryptophan or through the NAD+ salvage pathway from nicotinamide.220 However, cytoplasmic NAD+ levels are normally very low, and blastocyst development and placental and fetal growth can be maintained only when NAD+/NADH-reducing equivalents shuttle into mitochondria through either malate-aspartate or mitochondria glycerol 3-phosphate dehydrogenase.221 Histone acetylation during development deserves further study.
Mitochondrial Function
The following section summarizes currently available information on various aspects of mitochondrial function(s) (Table 1):
Table 1:
Genetic components of mitochondria and associated inherited disorders seen in young infants
| Composition of the gene/gene complex | Clinical features | ||
|---|---|---|---|
| Mitochondrial respiratory chain complex | Complex I | Complex I contains 33 genes that have been mapped to various autosomes, 1 to the X chromosome, and 7 to mtDNA | Lethal infantile mitochondrial disease, lactic acidosis, Leber’s Hereditary Optic Neuropathy, mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) syndrome |
| Complex II | Complex II (succinate: ubiquinone oxidoreductase) is the smallest complex in the respiratory chain and is composed of 4 subunits of the succinate dehydrogenase in the nucleus: SDHA, SDHB, SDHC, and SDHD | Leigh syndrome, mitochondrial leukoencephalopathy, Kearns-Sayre syndrome, cardiomyopathy, infantile leukodystrophy | |
| Complex III | Complex III contains 11 subunits, and only one, cytochrome b is of mitochondrial origin | Lactic acidosis, hypoglycemia, ketosis, hyperammonemia, cardiomyopathy, multisystemic dysfunction, encephalopathy; growth retardation, aminoaciduria, cholestasis, iron overload, lactic acidosis, and early death (GRACILE) syndrome | |
| Complex IV | Complex IV is comprised of 4 cytochrome c oxidase genes, which form a large transmembrane protein complex. It is the last enzyme in the respiratory electron transport chain located in the mitochondrial membrane. It converts molecular oxygen to water and helps establish a transmembrane electrochemical potential that the ATP synthase then uses to synthesize ATP | Steatosis, encephalopathy, myopathy, hypertrophic cardiomyopathy, hepatomegaly, liver dysfunction and hypotonia, delayed motor development, mental retardation | |
| Complex V | Complex V contains 24 genes. Synthesizes ATP using energy provided by the proton electrochemical gradient across the IMM. It is an F-type ATPase and consists of two structural domains: F1, a soluble portion situated in the mitochondrial matrix; and F0, which spans the IMM. Protons pass from the inter membrane space to the matrix through F0, which transfers the energy created by the proton-motive force to F1; ADP is phosphorylated to ATP | Severe neonatal encephalopathy, neonatal respiratory distress, lactic acidosis, peripheral neuropathy, dysmorphism, cataract, pulmonary arterial hypertension, bilateral cataracts, Reye-like syndrome | |
| Fatty acid metabolism | Carnitine palmitoyltransferase I | Hypoketotic hypoglycemia, hyperammonemia, elevated transaminases, and mild metabolic acidosis | |
| Carnitine-acylcarnitine translocase | Hypoglycemia, seizures, cardiomyopathy, cardiac arrhythmia, and apnea | ||
| Carnitine palmitoyltransferase II | Nonketotic hypoglycemia, hepatomegaly, encephalopathy, seizures, respiratory distress, and metabolic acidosis. cardiomyopathy and arrhythmia | ||
| Very long-chain acyl-CoA dehydrogenase (VLCAD) deficiency | Hypertrophic cardiomyopathy and fasting hypoketotic hypoglycemia | ||
| Short-chain acyl-CoA dehydrogenase (SCAD) deficiency | Hypotonia, muscle weakness, and seizure | ||
| Neonatal long-chain 3-ketoacyl CoA thiolase (LCKAT) deficiency | Lactic acidosis, pulmonary edema, and cardiomyopathy | ||
| Amino acid metabolism | Phenylalanine hydroxylase Cystathionine synthase Branched chain ketoacid dehydrogenase SLC6A19 solute carrier family 6, member 19 | Phenylketonuria Homocystinuria/Homocystinuria Maple syrup urine disease Hartnup disease | |
| Pyruvate metabolism | PDHA1 | Lactic acidosis, hypotonia, Seizure | |
| Pyruvate carboxylase | Hypercitrullinemia and hyperlysinemia | ||
| Krebs cycle metabolism | Dihydrolipoyl dehydrogenase | Severe persistent lactic acidosis, respiratory difficulties, seizures, dystonic movements, hypoglycemia, lethargy, hypotonia, vomiting, constipation, failure to thrive, and feeding difficulties | |
| α-ketoglutarate dehydrogenase | Choreoathetosis, opisthotonos, spasticity, hypertrophic cardiomyopathy, hepatomegaly, and sudden death | ||
| Fumarase | Lethargy, microcephaly, hypotonia, axial dystonia or opisthotonos, areflexia, or developmental delay | ||
| Pathological disorders caused by complex genetic alterations | Deletions with combinations of Class I, II, III | Steatosis, fibrosis/cirrhosis | |
| Syndromes due to deletions and duplications in the mitochondrial genome | Usually sporadic, heteroplasmic, and unique. Frequently occur between directly repeated sequences, suggesting that many occur because of de novo arrangements during oogenesis or early development | Syndromes described in text |
Energy production: Mitochondria play an important role in energy production and its storage as ATP.3 Glucose is broken down during glycolysis in the cytoplasm into two molecules of pyruvate, which are then translocated to the mitochondria by membrane-bound permeases.222 There, pyruvate dehydrogenase processes these molecules via oxidative decarboxylation to produce acetyl coenzyme A (acetyl-CoA), which triggers the TCA as described above.216 Metabolites enter the TCA cycle as acetyl-CoA, α-ketoglutarate, succinyl-CoA, fumarate, and oxaloacetate. Nicotinamide adenine dinucleotide (NAD), a coenzyme central to metabolism, is reduced to form hydrogenated NAD (NADH).223
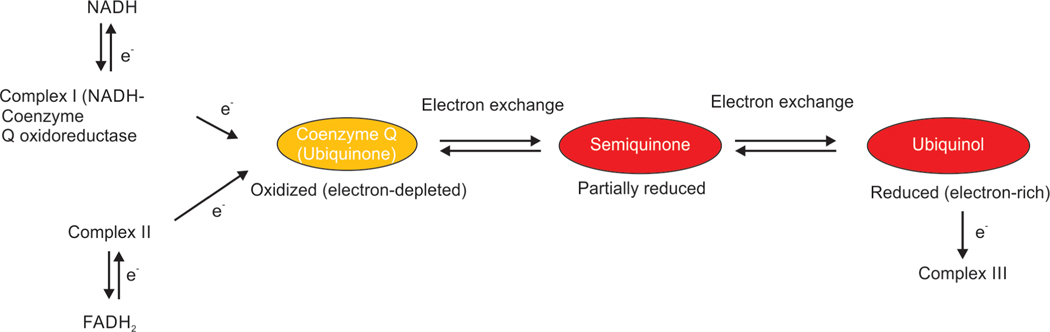
The mitochondrial respiratory chain has been conserved through evolution (as shown in Table 1 and details depicted in Fig. 11). Complex I and II oxidize NADH and Flavin adenine dinucleotide (FADH2), respectively, and transfer the resulting electrons to ubiquinol, which carries electrons to Complex III (Fig. 12).224 Complex III shunts the electrons across the intermembrane space to reduce cytochrome c (ubiquinone), which brings electrons to complex IV.225 Complex IV or cytochrome c oxidase (COX) is the last electron acceptor of the respiratory chain, involved in the reduction of O2 to H2O. This multimeric complex includes multiple structural subunits encoded in two different genomes, several heme groups (heme a and heme a3), and coordinated copper ions (CuA and CuB). About four electrons are removed from four molecules of cytochrome c and transferred to molecular oxygen (O2) and four protons, producing two molecules of water. About eight protons are removed from the mitochondrial matrix (although only four are translocated across the membrane), contributing to the proton gradient.226 Finally, mitochondrial ATP production is the main energy source for intracellular metabolic pathways. Complex V is a multi-subunit oxidative phosphorylation complex.227 There are two functional domains, including (a) F1, in the mitochondrial matrix; and (b) the F0, located in the IMM.228 This energy created in the proton electrochemical gradient is utilized to phosphorylate ADP to ATP.227
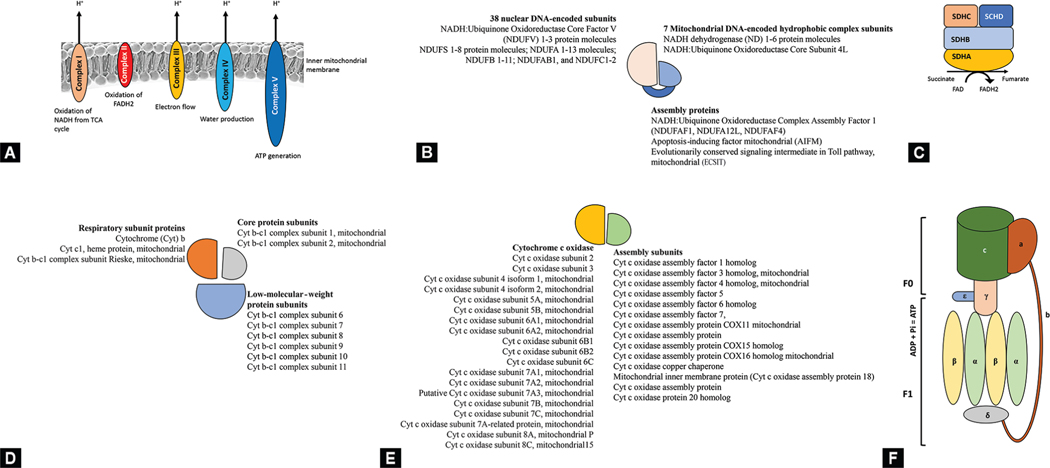
Figs 11A to F:
Mitochondrial genes. (A) Overall pattern of organization in the 5 mitochondrial gene complexes; (B) Complex I is comprised of 38 subunits that are encoded in the nuclear DNA, 7 in mitochondrial DNA, and several assembly units. This complex oxidizes NADH by transferring electrons to ubiquinol. NADH stands for “nicotinamide adenine dinucleotide (NAD) + hydrogen (H); (C) Complex II converts succinate to fumarate and reduces FAD to FADH2 during this process. The released electrons are transferred to ubiquinol. SDH = succinate dehydrogenase. The figure shows four components of the SDH complex, A-D; (D) Complex III (coenzyme Q: cytochrome c – oxidoreductase, or the cytochrome bc1 complex), contains 11 subunits: 3 respiratory subunits, 2 core proteins and 6 low-molecular-weight proteins; (E) Complex IV (cytochrome c oxidase or cytochrome AA3), contains two hemes, a cytochrome a and cytochrome a3, and two copper centers, the CuA and CuB centers; and (F) Complex V (mitochondrial ATP synthase) is a multisubunit oxidative phosphorylation complex. Complex V is composed of two functional domains: F1, which is situated in the mitochondrial matrix, and Fo, located in the inner mitochondrial membrane. Complex V uses the energy created by the proton electrochemical gradient to phosphorylate ADP to ATP. ADP, Adenosine diphosphate; ATP, Adenosine triphosphate; Pi, inorganic phosphate
Fig. 12:
Ubiquinol is an electron-rich (reduced) form of coenzyme Q (ubiquinone). The term most often refers to ubiquinol-10 that has a 10-unit tail; it exists in three redox states, the fully reduced (ubiquinol), partially reduced (semiquinone or ubisemiquinone), and fully oxidized (ubiquinone) forms. Ubiquinol can serve a redox function in cellular energy production and in antioxidant protection based on the ability to exchange two electrons in redox cycles. Complex I (NADH-Coenzyme Q oxidoreductase, or NADH dehydrogenase) can accept high energy electrons from NADH, and complex II interacts with FADH2.
Reduced NAD+ (NADH) is used by enzymes embedded in the mitochondrial cristae to produce ATP.229 Beta-oxidation of fatty acids also produces acetyl-CoA, NADH, and reduced flavin adenine dinucleotide (FADH2; FAD is a redox-active coenzyme involved in several metabolic pathways).230 Oxidative degradation of certain amino acids can also contribute to this process; the mitochondria are also the key regulators of the biosynthesis of amino acids, lipids, and gluconeogenesis.231 Under normal conditions, over 90% of ATP is made in mitochondria232 but most of the genetic machinery needed to produce ATP has been translocated to the nucleus during evolution.233 Only about 3% of the mitochondrial proteins are needed to synthesize ATP.229
Flavin adenine dinucleotide (FADH2) is another energy carrier that is produced in the mitochondrial matrix and is processed by oxidative phosphorylation in the electron transport chain to regenerate FAD.234 Protons are pulled into the intermembrane space by the energy of the electrons going through the electron transport chain. In the electron transport chain, 4 electrons are accepted by oxygen and the protons return to the mitochondrial matrix through the protein ATP synthase.224 The energy is used to activate ATP synthase, which then facilitates the passage of a proton to produce ATP.234 The difference in pH between the matrix and intermembrane space creates an electrochemical gradient by which ATP synthase can facilitate the passage of protons into the matrix.3 The oxidation of NADH and FADH2 also produces GTP from succinyl-CoA synthetase-mediated signaling.216
In oogonia, mitochondria are the prime source of energy.235 These mitochondria have a dense matrix and a few arch-like or transverse cristae, and are usually seen in the central cytoplasm.236 Mitochondria in metaphase I and II of oocytes still resemble those in the germinal vesicle, with an even distribution in the cytoplasm and aggregation around the smooth ER.236 At the pronuclear stage, mitochondria conglomerate around the pronuclei, and this persists up to syngamy.237 In the 8-cell embryo, the morula, and the blastocyst, mitochondria are relatively less electron-dense and show clear areas in the matrices. In expanding blastocysts, trophoblast, embryoblast, and endodermal cells, mitochondria look elongated with the IMM arranged into transverse cristae.3,236
Growing oocytes preferentially utilize pyruvate to make ATP, and early embryos also use pyruvate, lactate, and amino acids to support development. 238–240 Mature oocytes contain the highest mitochondrial DNA copy number and mass of any cell.241 Fertilized oocytes have even higher mtDNA copy numbers, Inhibition of mitochondrial metabolic activity blocks the maturation of oocytes and the subsequent embryonic development. 136,242–247 In humans, embryo development and implantation rates are closely correlated with ATP levels, and inhibiting mitochondrial activity suppresses embryonic stem cell differentiation. 247,248 After birth, there is a rapid increase in the density and activity of mitochondria in the heart and other metabolically active organs.249 The expression of mitochondrial respiratory chain genes is also increased.17
The density of mitochondria in cells does seem to be important. Fibroblasts and other cells from obese mothers have been shown to carry fewer mitochondria, which relates to higher levels of triglycerides, free fatty acids, and more lipids.250 The paucity of mitochondria alters the placental lipid metabolism and transfer of the lipids to the fetus, causing lipid-related diseases such as newborn adiposity.251 Likewise, the reduced mitochondrial function has been noted in brain injury in newborns. 252 Decreased mitochondrial proteins may lower neonatal pyruvate dehydrogenase and oxidative phosphorylation activity, and increase the risk of morbidity.253 Metabolic shifts are also important for the function of cardiomyocytes. Mutations in mtDNA and mitochondrial dysfunction were associated with dilated cardiomyopathy via transcription factor A, mitochondrial (TFAM).254
Changes in mitochondrial function based on maternal age: Mitochondria have important roles in oocyte maturation, fertilization, and early embryo development.255 In women of advanced reproductive age, aging oocytes often show less ATP and mtDNA copy number, mutations in mtDNA, and ultrastructural abnormalities.256
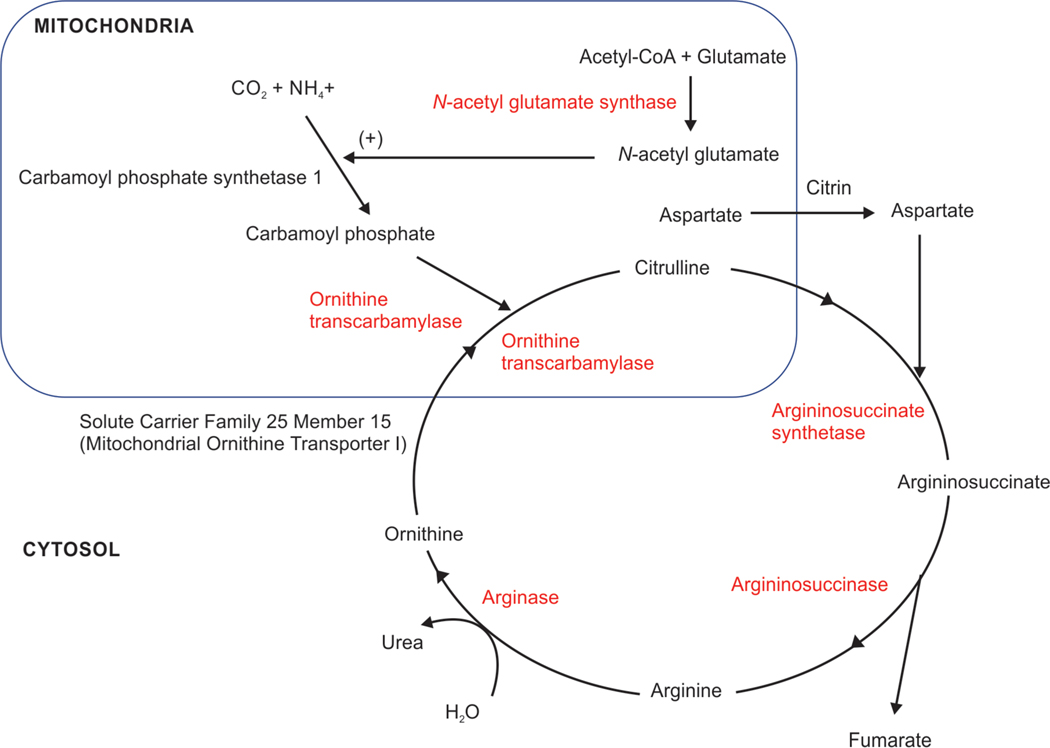
Urea Cycle
The initial steps of the urea cycle take place in the mitochondrial matrix, particularly in hepatocyte and renal epithelium (Fig. 13).257 In the first step, the carbamoyl phosphate synthetase I enzyme utilizes two ATP molecules to convert ammonia into carbamoyl phosphate.258 In the second, ornithine transcarbamylase converts carbamoyl phosphate and ornithine into citrulline.259 Subsequent steps continue in the cytoplasm until ornithine is re-transported into the matrix.257
Fig. 13:
The initial steps of the urea cycle take place in the mitochondrial matrix. Carbamoyl phosphate synthetase I (CPS I) combines ammonia with carbon dioxide to from carbamoyl phosphate and ornithine transcarbamylase promotes citrulline synthesis. N-acetyl glutamate (NAG) synthase increases the formation of NAG, which activates CPS I.
Transamination
Oxaloacetate can be transaminated to produce aspartate and asparagine in the matrix.260 Similarly, transamination of α-ketoglutarate produces glutamate, proline, and arginine.261
Metabolic Regulation
The concentrations of ions, various metabolites, and energy charge in mitochondria are closely regulated. Ca2+ ions regulate the TCA cycle (as shown in Fig. 4) by activating pyruvate dehydrogenase, isocitrate dehydrogenase, and α-ketoglutarate dehydrogenase.262 The concentration of intermediates and coenzymes in the matrix also influences the rate of ATP production.263 NADH can inhibit TCA enzymes α-ketoglutarate dehydrogenase, isocitrate dehydrogenase, citrate synthase, and pyruvate dehydrogenase.264 Adenosine triphosphate inhibits isocitrate dehydrogenase, pyruvate dehydrogenase, the electron transport chain, and ATP synthase.265 In contrast, ADP acts as an activator.266
Apoptosis
Mitochondrial apoptosis is the most common form of programmed cell death.267 It is mediated through proteins of the B-cell lymphoma-2 (bcl-2) family.268 There are two sub-classes of bcl-2 proteins: (a) the pro-apoptotic bcl-2 associated X-protein (bax) and bcl-2 antagonist/killer 1 (bak) proteins, which oligomerize to create “pores” on the OMM in response to apoptotic stimuli.269 Through these pores, proteins from the mitochondrial intermembrane space (IMS) reach the cytoplasm and activate the caspase cascade;51,270,271 (b) anti-apoptotic bcl-2 proteins such as bcl-2, bcl-xL or myeloid cell leukemia (MCL)-1, which inhibit bax and bak signaling.272
Apoptosis is induced when various conformations of activated bax accumulate on the mitochondrial surface.267 The ratio of mitochondrial/cytosolic levels of bak and bax determine the cellular response to apoptosis stimulation.273 Increasing information suggests that bax, either alone or complexed with other proteins such as cytochrome c, Smac/direct IAP binding protein with low pI (Diablo), HtrA Serine Peptidase 2 (HtrA2)/Omi, and apoptosis-inducing factor (AIF), forms pores in the OMM to release IMS proteins.274 Bcl-2 associated X-protein (Bax) can also modulate the function of permeability transition pore complexes formed with other mediators such as the voltage-dependent anion channels (VDAC–1, −2, −3; mitochondrial porin), the adenine nucleotide transporter cyclophilin D, and the F1Fo ATP synthase.275 These complexes promote the transport of nucleotides, phosphocreatine, Ca2+, and other small ions across the OMM.276 Some investigators have noted the resemblance between active bax/bak and the holin proteins that are involved in host cell membrane lysis by bacteriophages.277,278 The bax/bak oligomers form membrane lesions, which release endolysin, a muralytic enzyme, through these lesions to attack the cell wall.278,279
Ionic Content of Various Tissues
Mitochondria can absorb calcium ions and play an important role in the regulation of calcium content in various cells.11,280 Increased intracellular calcium can regulate many cellular processes. In neurons, these levels can alter neurotransmitter release.281 It can also alter other processes such as endocrine changes. muscle function, and coagulation.282 In infants, mitochondria can alter non-shivering thermogenesis in brown fat through proton leaks.283
Mitochondrial Genetic Defects in Neonates
Epidemiology and Clinical Features
Mitochondrial dysfunction affects one in 6, 000 – 8, 000 newborns.284–286 The diseases may occur in patterns consistent with autosomal recessive, autosomal dominant, mitochondrial, and random mutations.287,288 Young infants require mitochondrial energetic metabolism to support rapid growth. Key organs such as the muscle, heart, and brain require mitochondrial function/aerobic metabolism for adaption to extra-uterine life.289 Disorders of mitochondrial metabolism caused by defects in fatty acid oxidation; pyruvate metabolism; and those in the respiratory chain, including mitochondrial complex I, II, III, IV, and ATP synthase, can become symptomatic in the neonatal period.230 Mutations of both nuclear genes and mtDNA can cause mitochondrial dysfunction with adverse, outcomes in neonates.224–228,290
Neonatal-onset mitochondrial disease is associated with considerable, early mortality.291 In a recent study, Ebihara et al.285 reviewed the records of 281 patients with mitochondrial disease. The multisystem disease was noted in 194, Leigh syndrome in 26, cardiomyopathy in 38, and hepatopathy in 23 patients. Of the 321 with initial symptoms, 236 were recognized to have illness within two days of birth. The disorders were recognized by altered mitochondrial respiratory chain enzyme activity rate in 182, and abnormal oxygen consumption rate in 89. The remaining 10 patients were diagnosed using a genetic approach. Genetic analysis showed 69 to have nuclear DNA variants in 36 genes; 11 of 15 had mtDNA variants in 5 genes, and 4 had a single large deletion. Cyclo-oxygenase (Cox) proportional hazards regression showed significant differences in survival in those with Leigh syndrome [hazard ratio (HR) = 0.15, 95% confidence interval (CI) 0.04 to 0.63, p = 0.010] and in others with a molecular diagnosis (HR = 1.87, 95% CI 1.18 to 2.96, p = 0.008).
In the outpatient setting, mutations of mitochondrial complex I mutations can be seen with Leigh Syndrome, lethal infantile mitochondrial disease, lactic acidosis, MELAS syndrome, and Leber’s Hereditary Optic Neuropathy.292,293 Mutations in succinate dehydrogenase (SDH)-A, -B, and -AF1 genes in Complex II can cause mitochondrial leukoencephalopathy, cardiomyopathy, infantile leukodystrophy, and Kearns-Sayre syndrome.294 Mutations in Complex III can cause severe lactic acidosis with hypotonia, irritability, and muscle wasting.295 Complex III deficiency is mainly caused by mutations in maternally-transmitted mitochondrial chaperone BCS1 (BCS1L), Ubiquinol-Cytochrome C Reductase-Binding Protein (UQCRB), Ubiquinol-Cytochrome C Reductase Complex III Subunit VII (UQCRQ) and mitochondrially-encoded Cytochrome B (MTCYB) genes.296,297 Mutations in Complex IV are associated with neonatal hypertrophic cardiomyopathy, liver dysfunction, myopathy, hypotonia, developmental delay, and encephalopathy.294 The biogenesis and assembly of cyclooxygenase (COX) in Complex IV depends on numerous ancillary factors, including copper chaperones, all nuclear-encoded.298,299 Specifically, disease-causing mutations were found in the gene encoding the Surfeit locus protein 1 (SURF1), which is essential for the formation of early assembly intermediates.300,301 Mutations in Complex IV have been associated with neonatal encephalopathy, respiratory distress, pulmonary hypertension, lactic acidosis, peripheral neuropathy, dysmorphism, and cataracts.302 Defects in ATP synthase can also cause fatal encephalopathy in neonates.303,304
Pyruvate dehydrogenase complex (PDHc) catalyzes the oxidative decarboxylation of pyruvate to produce acetyl-CoA and initiates the TCA cycle.305 Pyruvate dehydrogenase complex (PDHc) deficiency is most often due to mutations in the first component of the enzyme complex, pyruvate dehydrogenase E1α (responsible for 70% of PDH deficiencies).306 There is a spectrum of clinical presentations in E1α mutations;307 the most severe mutations can manifest with lactic acidosis within a few hours of birth.308 Other infants may show hypotonia, lethargy, feeding, respiratory difficulties, and encephalopathy.309,310
Mitochondrial Disorders
Infections: The sepsis syndrome is a systemic host inflammatory response accompanied by organ dysfunction in response to invading microbial pathogens.311 The host recognizes both danger and pathogens through its pattern recognition receptors on immune cells.312 These receptors bind to pathogen associated molecular patterns (PAMP) and danger (DAMP) associated molecular patterns derived from microbes and host tissues, respectively.313 These DAMPs and PAMPs activate the formation of inflammasomes, which bind to the apoptosis-associated speck-like protein (ASC) containing a caspase recruitment domain (CARD).314–316 This forms a platform for the activation of caspase-1 and induction of interleukin (IL)-1β and IL-18.317,318 Caspase-1 triggers mitochondrial damage.319 It also inhibits mitophagy, a process that clears damaged mitochondria, leading to accumulation of defective mitochondria and damaged cells.319 Mitochondrial DNA (mtDNA) has also been detected in the extracellular traps formed by innate immune leukocytes in these infected lesions.320,321
Oxidative phosphorylation: Mitochondria are the site of oxidative phosphorylation in eukaryotes; the energy is produced by means of electron flow between four enzymes, of which three are proton pumps, in the inner mitochondrial membrane.322 NADH generated in the TCA cycle is oxidized and activates the electron transport chain, which is comprised of complexes I–IV, and ATP synthase.158,216 Acute inflammation may curtail these pathways. 323 Tumor necrosis factor (TNF) induces microRNAs that damage the mitochondrial complex-I, inhibit oxidative phosphorylation, and reduce ATP levels.324,325
Inflammation: Inflammatory stimuli can promote mitochondrial fragmentation by increased protein unfolding, ER stress, phosphorylation of pro-fission proteins, and decreased respiratory capacity.326–330 There is also increased oxidative stress; mitochondrial complex III generates superoxide during the ubiquinone (Q)-cycle.331 Some lesions may show mitochondrial fission, mitophagy, and decreased fusion.330,332,333 The intrinsic dynamicity of mitochondria also plays a role in proinflammatory signaling, identifying these organelles as a central platform for the control of innate immunity and the inflammatory response.334
During inflammation, cytokines such as TNF, interleukin (IL)-1, and IL-18 can promote necroptosis, a form of programmed necrosis mediated by various cytokines and pattern recognition receptors (PRRs).335–338 Cells dying by necroptosis show necrotic phenotypes, including swelling and membrane rupture, and release damage-associated molecular patterns (DAMPs), inflammatory cytokines, and chemokines, thereby mediating extreme inflammatory responses.339 Mitochondrial proteins such as the phosphoglycerate mutase (PGAM)-5 and dynamin-related protein (Drp)-1 have been identified as important activators of the receptor-interacting serine-threonine kinase (RIPK)-3 and consequent mitochondrial fission and necroptosis.340–342 Mitochondrial reactive oxygen species (ROS) may regulate TNF-mediated cell death in other diseases.343–345
Mitochondrial mediators such as the DNA polymerase γ (POLG) and the protein growth factor erv1-like can alter physiological mediators for cellular self-renewal and suppress signaling mediators such as the octamer-binding protein 4 (OCT4), nanog homeobox (NANOG), and the putative thiosulfate sulfurtransferase (SSEA).346–350
Mitophagy in birth asphyxia and neurological disorders: In asphyxiated infants with hypoxic-ischemic encephalopathy, neural energy failure is being increasingly documented.351 Currently, the options for timely diagnosis and treatment are limited. Mitochondrial dysfunction with increased permeability, altered dynamics with changes in fission and fusion, mitophagy, and biogenesis have been observed in many studies.352–355 Mitoprotective therapies may help prevent/treat brain injury and reduce the incidence of lifelong disabilities.355,356
In other neurological disorders, mitochondrial transmembrane potential loss with the involvement of PTEN (phosphatase and tensin homolog)-induced putative kinase 1 (PINK1), which then recruits Parkin, the E3 ubiquitin ligase, to the damaged mitochondria.357–360 The BCL2 (B-cell lymphoma 2 genes)-interacting protein 3-like (BNIP3L), a mitophagy receptor that recruits LC3 family proteins to the damaged mitochondria, can be altered. 361,362 The FUN14 domain-containing 1 (FUNDC1) is another mitophagy receptor located on the outer mitochondrial membrane.363,364
Mutations in mitochondrial genes: Many deletions and duplications in the mitochondrial genome can be seen sporadically. These may develop de novo or during early development (Table 1):365–367
Leber’s hereditary optic neuropathy (LHON) is the most common, maternally-inherited mitochondrial disorder in the respiratory chain, which causes degeneration of retinal ganglion cells, associated axons, and optic atrophy within a year of disease onset.368,369,319 An intriguing feature of LHON is that only 50% of males and 10% of the females with the mtDNA mutations actually become symptomatic.370 This incomplete penetrance and gender bias imply that additional mitochondrial and/or nuclear genetic factors must be modulating the phenotypic expression of LHON.371 It typically begins as a unilateral progressive optic neuropathy with the central visual loss with sequential involvement of the fellow eye months to years later.220 In about 90% of clinical cases, the disease is associated with three mutations in mtDNA complex I subunit genes, namely the G3460A, G11778A, and T14484C. These mutations are absent or very rare among normal controls.371–374
Pearson syndrome is a rare fatal mitochondrial disorder caused by single large-scale mitochondrial DNA deletions. Most patients present with sideroblastic anemia during infancy, followed by multi-organ dysfunction including lactic acidosis, pancreatic insufficiency, renal tubulopathy, failure to thrive, muscle hypotonia, and endocrine disorders.375,376 Bone marrow cytology shows vacuolization in erythroid and myeloid precursors and ring-sideroblasts; the diagnosis is established by the detection of mitochondrial DNA deletions.375,377,378 Most cases have a lethal outcome. Some survivors go on to develop Kearns-Sayre syndrome, a progressive cardio-encephalo-myopathy caused by a large deletion or rearrangement of mtDNA.379–382
Leigh syndrome, also termed subacute necrotizing encephalomyelopathy, is a rare, inherited progressive neurodegenerative disorder that usually manifests in infancy or early childhood.383,384 Many cases can be diagnosed in early infancy and present with developmental delay, pyramidal and extrapyramidal symptoms, leukodystrophy, and brainstem dysfunction.385,386 Neuroimaging shows focal, symmetrical, necrotic lesions in the thalamus, the brainstem, and the posterior columns of the spinal cord. Histopathology shows symmetric spongiform lesions with degeneration of basal ganglia, particularly in the corpus striatum; and demyelination, vascular proliferation, and astrocytosis in the brainstem.387–389 The lesions typically appear hyperintense on T2-weighted MRI.390 Pathogenic mutations are often seen in flavoprotein of complex II.391
Alpers-Huttenlocher syndrome manifests with seizures, developmental delay, hypotonia, and liver disease.326,327 It is a maternally-inherited disease with variable penetrance. It is usually caused by mutations in polymerase gamma (POLG).392,393 Mutations in mitochondrial tRNA synthetase genes, including the phenylalanyl-tRNA synthetase 2, mitochondrial (FARS2), asparaginyl-tRNA synthetase 2, mitochondrial (NARS2), and the prolyl-tRNA synthetase 2, mitochondrial (PARS2) have also been linked.394,395
A syndrome associated with MELAS syndrome is caused by mutations in mitochondrial transfer genes such as the mitochondrially-encoded tRNA leucine 1 (MT-TL1).396,397 It is characterized by mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes.398,399
OUR CURRENT UNDERSTANDING
Mitochondria provide energy and regulate key cellular events not only during the embryonic, fetal and neonatal period, but and throughout life.4 After birth, the newborn infant’s organs may show major changes in the number and function of mitochondria.400 There is some evidence that epigenetic changes in mitochondrial DNA during the perinatal period may be protective.401,402 These changes are most noticeable in the muscles, heart and brain.403 Disorders of mitochondrial metabolism related to nuclear/mtDNA defects in mitochondrial complexes I, II, III, IV, and ATP synthase may present in the neonatal period.404,405
There is a need for focused studies of mitochondrial function and its modulators in the fetal/neonatal period to ascertain the need for interventions.406 We still have major gaps in our understanding of the long-term effects of mitochondrial dysfunction in neonatal period and infancy.353 A framework is needed to focus future research on altered mitochondrial function as a mechanism of perinatal adaptation.
KEY POINTS.
Mitochondria are highly-dynamic, membrane-bound organelles that generate most of the chemical energy in eukaryotic cells.
These organelles most likely evolved about 2 billion years ago from α-proteobacteria, a subgroup of the purple non-sulfur bacteria. These precursors of mitochondria likely belong to the order Rickettsiales.
Besides the primary role in energy generation, mitochondria also perform numerous other cellular functions to support metabolism, epigenetic regulation, and cell cycle.
In this article, we have summarized the ontogeny, ultrastructure, structure-function correlation, biogenesis, and clinical manifestations of mitochondrial dysfunction.
Source of support:
This study was supported by DK120309, DK107641.
Footnotes
Conflict of interest: Dr. Akhil Maheshwari and Dr. Ling He are associated as the Editorial Board Members of this journal and this manuscript was subjected to this journal’s standard review procedures, with this peer review handled independently of these Editorial Board Members and their research group.
REFERENCES
- 1.Friedman JR, Nunnari J. Mitochondrial form and function. Nature 2014;505(7483):335–343. DOI: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picard M, Shirihai OS. Mitochondrial signal transduction. Cell Metab 2022;34(11):1620–1653. DOI: 10.1016/j.cmet.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Structure Kuhlbrandt W. and function of mitochondrial membrane protein complexes. BMC Biol 2015;13:89. DOI: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol 2006;16(14):R551–R560. DOI: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. Mitochondrial diseases in man and mouse. Science 1999;283(5407):1482–1488. DOI: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 6.Lopez J, Tait SW. Mitochondrial apoptosis: Killing cancer using the enemy within. Br J Cancer 2015;112(6):957–962. DOI: 10.1038/bjc.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munro D, Treberg JR. A radical shift in perspective: Mitochondria as regulators of reactive oxygen species. J Exp Biol 2017;220(Pt 7):1170–1180. DOI: 10.1242/jeb.132142. [DOI] [PubMed] [Google Scholar]
- 8.Bohovych I, Khalimonchuk O. Sending Out an SOS: Mitochondria as a Signaling Hub. Front Cell Dev Biol. 2016;4:109. DOI: 10.3389/fcell.2016.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill S, Van Remmen H. Mitochondrial stress signaling in longevity: A new role for mitochondrial function in aging. Redox Biol 2014;2: 936–944. DOI: 10.1016/j.redox.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia M, Zhang Y, Jin K, et al. Communication between mitochondria and other organelles: A brand-new perspective on mitochondria in cancer. Cell Biosci 2019;9:27. DOI: 10.1186/s13578-019-0289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kluge MA, Fetterman JL, Vita JA. Mitochondria and endothelial function. Circ Res 2013;112(8):1171–1188. DOI: 10.1161/CIRCRESAHA.111.300233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappel S. The role of mitochondria from mature oocyte to viable blastocyst. Obstet Gynecol Int 2013;2013:183024. DOI: 10.1155/2013/183024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med 2012;366(12):1132–1141. DOI: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 14.Xie JH, Li YY, Jin J. The essential functions of mitochondrial dynamics in immune cells. Cell Mol Immunol 2020;17(7):712–721. DOI: 10.1038/s41423-020-0480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulzebos CV, Sauer PJ. Energy requirements. Semin Fetal Neonatal Med 2007;12(1):2–10. DOI: 10.1016/j.siny.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Lai L, Leone TC, Zechner C, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev 2008;22(14):1948–1961. DOI: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Merhie N, Baumgart-Vogt E, Pilatz A, et al. Differential alterations of the mitochondrial morphology and respiratory chain complexes during postnatal development of the mouse Lung. Oxid Med Cell Longev 2017;2017:9169146. DOI: 10.1155/2017/9169146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton R, Pollak JK. Hormone-initiated maturation of rat liver mitochondria after birth. Biochem J 1980;186(1):361–367. DOI: 10.1042/bj1860361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastin J, Delaval E, Freund N, et al. Effects of birth on energy metabolism in the rat kidney. Biochem J 1988;252(2):337–41. DOI: 10.1042/bj2520337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shultz M. Mapping of medical acronyms and initialisms to Medical Subject Headings (MeSH) across selected systems. J Med Libr Assoc 2006;94(4):410–414. PMID: 17082832. [PMC free article] [PubMed] [Google Scholar]
- 21.Adl SM, Simpson AG, Lane CE, et al. The revised classification of eukaryotes. J Eukaryot Microbiol 2012;59(5):429–93. DOI: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZW, Cheng J, Xu F, et al. Red blood cell extrudes nucleus and mitochondria against oxidative stress. IUBMB Life 2011;63(7):560–565. DOI: 10.1002/iub.490. [DOI] [PubMed] [Google Scholar]
- 23.Aryaman J, Johnston IG, Jones NS. Mitochondrial Heterogeneity. Front Genet. 2018;9:718. DOI: 10.3389/fgene.2018.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 2014;15(10):634–646. DOI: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins HE, Kane MS, Litovsky SH, et al. Mitochondrial morphology and mitophagy in heart diseases: Qualitative and quantitative analyses using transmission electron microscopy. Front Aging 2021;2:670267. DOI: 10.3389/fragi.2021.670267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rube DA, van der Bliek AM. Mitochondrial morphology is dynamic and varied. Mol Cell Biochem 2004;256–257(1–2):331–339. DOI: 10.1023/b:mcbi.0000009879.01256.f6. [DOI] [PubMed] [Google Scholar]
- 27.Miettinen TP, Bjorklund M. Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev Cell 2016;39(3): 370–382. DOI: 10.1016/j.devcel.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen RE. Control of mitochondrial shape. Curr Opin Cell Biol 2005;17(4):384–388. DOI: 10.1016/j.ceb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012;337(6098):1062–1065. DOI: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaldakov GN, Kokosharov PN. An intracristal structure in rat liver dumbbell-shaped mitochondria. Preliminary communication. Acta Morphol Acad Sci Hung 1973;21(2):149–154. PMID: 4744686. [PubMed] [Google Scholar]
- 31.Cogliati S, Frezza C, Soriano ME, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 2013;155(1):160–171. DOI: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Englmeier R, Forster F. Cryo-electron tomography for the structural study of mitochondrial translation. Tissue Cell 2019;57:129–138. DOI: 10.1016/j.tice.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Rambold AS, Kostelecky B, Elia N, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA 2011;108(25):10190–10195. DOI: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tronstad KJ, Nooteboom M, Nilsson LI, et al. Regulation and quantification of cellular mitochondrial morphology and content. Curr Pharm Des 2014;20(35):5634–5652. DOI: 10.2174/1381612820666140305230546. [DOI] [PubMed] [Google Scholar]
- 35.Hu D, Liu Z, Qi X. Mitochondrial quality control strategies: Potential therapeutic targets for neurodegenerative diseases? Front Neurosci 2021;15:746873. DOI: 10.3389/fnins.2021.746873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roca-Portoles A, Tait SWG. Mitochondrial quality control: From molecule to organelle. Cell Mol Life Sci 2021;78(8):3853–3866. DOI: 10.1007/s00018-021-03775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frederick RL, Shaw JM. Moving mitochondria: Establishing distribution of an essential organelle. Traffic 2007;8(12):1668–1675. DOI: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, Kalyanasundaram A, Zhu J. Structural and biomechanical basis of mitochondrial movement in eukaryotic cells. Int J Nanomedicine 2013;8:4033–4042. DOI: 10.2147/IJN.S52132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonora M, Patergnani S, Rimessi A, et al. ATP synthesis and storage. Purinergic Signal 2012;8(3):343–357. DOI: 10.1007/s11302-012-9305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolaisen J, Nilsson LI, Pettersen IK, et al. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One 2014;9(7):e101365. DOI: 10.1371/journal.pone.0101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramachandran R. Mitochondrial dynamics: The dynamin superfamily and execution by collusion. Semin Cell Dev Biol 2018;76:201–212. DOI: 10.1016/j.semcdb.2017.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasan K, Clutter M, Fernandez Dunne S, et al. Genes involved in maintaining mitochondrial membrane potential upon electron transport chain disruption. Front Cell Dev Biol 2022;10:781558. DOI: 10.3389/fcell.2022.781558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazur M, Kmita H, Wojtkowska M. The diversity of the mitochondrial outer membrane protein import channels: Emerging targets for modulation. Molecules 2021;26(13):4087. DOI: 10.3390/molecules26134087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Model K, Prinz T, Ruiz T, et al. Protein translocase of the outer mitochondrial membrane: Role of import receptors in the structural organization of the TOM complex. J Mol Biol 2002;316(3):657–666. DOI: 10.1006/jmbi.2001.5365. [DOI] [PubMed] [Google Scholar]
- 45.Structure Zeth K. and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim Biophys Acta 2010;1797 (6–7):1292–1299. DOI: 10.1016/j.bbabio.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Meisinger C, Rissler M, Chacinska A, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell 2004;7(1):61–71. DOI: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Doan KN, Grevel A, Martensson CU, et al. The mitochondrial import complex MIM functions as main translocase for alpha-Helical outer membrane proteins. Cell Rep 2020;31(4):107567. DOI: 10.1016/j.celrep.2020.107567. [DOI] [PubMed] [Google Scholar]
- 48.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J Cell Sci 2010;123(Pt 9):1389–1393. DOI: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weeber EJ, Levy M, Sampson MJ, et al. The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J Biol Chem 2002;277(21):18891–18897. DOI: 10.1074/jbc.M201649200. [DOI] [PubMed] [Google Scholar]
- 50.Camara AKS, Zhou Y, Wen PC, et al. Mitochondrial VDAC1: A key gatekeeper as potential therapeutic target. Front Physiol 2017;8:460. DOI: 10.3389/fphys.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westphal D, Kluck RM, Dewson G. Building blocks of the apoptotic pore: How Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ 2014;21(2):196–205. DOI: 10.1038/cdd.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem J 1999;341 (Pt 2)(Pt 2):233–249. PMID: 10393078. [PMC free article] [PubMed] [Google Scholar]
- 53.Edwards R, Eaglesfield R, Tokatlidis K. The mitochondrial intermembrane space: The most constricted mitochondrial subcompartment with the largest variety of protein import pathways. Open Biol 2021;11(3):210002. DOI: 10.1098/rsob.210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Backes S, Herrmann JM. Protein translocation into the intermembrane space and matrix of mitochondria: Mechanisms and driving forces. Front Mol Biosci 2017;4:83. DOI: 10.3389/fmolb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vander Heiden MG, Chandel NS, Li XX, et al. Outer mitochondrial membrane permeability can regulate coupled respiration and cell survival. Proc Natl Acad Sci USA 2000;97(9):4666–4671. DOI: 10.1073/pnas.090082297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walther DM, Bos MP, Rapaport D, Tommassen J. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol Biol Evol 2010;27(4):887–895. DOI: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- 57.Fox TD. Mitochondrial protein synthesis, import, and assembly. Genetics 2012;192(4):1203–1234. DOI: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peleh V, Cordat E, Herrmann JM. Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. Elife 2016;5:e16177. DOI: 10.7554/eLife.16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chacinska A, Pfannschmidt S, Wiedemann N, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 2004;23(19):3735–3746. DOI: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fass D. The Erv family of sulfhydryl oxidases. Biochim Biophys Acta 2008;1783(4):557–566. DOI: 10.1016/j.bbamcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta 2008;1783(4):601–609. DOI: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 62.Zimorski V, Ku C, Martin WF, et al. Endosymbiotic theory for organelle origins. Curr Opin Microbiol 2014;22:38–48. DOI: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Khalimonchuk O, Winge DR. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim Biophys Acta 2008;1783(4):618–628. DOI: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stojanovski D, Muller JM, Milenkovic D, et al. The MIA system for protein import into the mitochondrial intermembrane space.Biochim Biophys Acta 2008;1783(4):610–617. DOI: 10.1016/j.bbamcr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Joubert F, Puff N. Mitochondrial cristae architecture and functions: Lessons from minimal model systems. Membranes (Basel) 2021;11(7):465. DOI: 10.3390/membranes11070465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frazier AE, Chacinska A, Truscott KN, et al. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol Cell Biol 2003;23(21): 7818–7828. DOI: 10.1128/MCB.23.21.7818-7828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamo N, Muratsugu M, Hongoh R, et al. Membrane potential of mitochondria measured with an electrode sensitive to tetraphenyl phosphonium and relationship between proton electrochemical potential and phosphorylation potential in steady state. J Membr Biol 1979;49(2):105–121. DOI: 10.1007/BF01868720. [DOI] [PubMed] [Google Scholar]
- 68.Klecker T, Westermann B. Pathways shaping the mitochondrial inner membrane. Open Biol 2021;11(12):210238. DOI: 10.1098/rsob.210238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel F, Bornhovd C, Neupert W, et al. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 2006;175(2):237–247. DOI: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf DM, Segawa M, Kondadi AK, et al. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J 2019;38(22):e101056. DOI: 10.15252/embj.2018101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mannella CA. Consequences of Folding the Mitochondrial Inner Membrane. Front Physiol 2020;11:536. DOI: 10.3389/fphys.2020.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darshi M, Mendiola VL, Mackey MR, et al. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 2011;286(4): 2918–2932. DOI: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie J, Marusich MF, Souda P, et al. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett 2007;581(18):3545–3549. DOI: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 74.Madungwe NB, Feng Y, Lie M, et al. Mitochondrial inner membrane protein (mitofilin) knockdown induces cell death by apoptosis via an AIF-PARP-dependent mechanism and cell cycle arrest. Am J Physiol Cell Physiol 2018;315(1):C28–C43. DOI: 10.1152/ajpcell.00230.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Enriquez JA, Lenaz G. Coenzyme q and the respiratory chain: Coenzyme Q pool and mitochondrial supercomplexes. Mol Syndromol 2014;5(3–4):119–40. DOI: 10.1159/000363364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao RZ, Jiang S, Zhang L, et al. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int J Mol Med 2019;44(1):3–15. DOI: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kondadi AK, Anand R, Reichert AS. Cristae Membrane Dynamics - A Paradigm Change. Trends Cell Biol 2020;30(12):923–936. DOI: 10.1016/j.tcb.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 78.Cadena LR, Gahura O, Panicucci B, et al. Mitochondrial contact site and cristae organization system and F(1)F(O)-ATP synthase crosstalk Is a fundamental property of Mitochondrial mcristae. mSphere 2021;6(3):e0032721. DOI: 10.1128/mSphere.00327-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramonet D, Perier C, Recasens A, et al. Optic atrophy 1 mediates mitochondria remodeling and dopaminergic neurodegeneration linked to complex I deficiency. Cell Death Differ 2013;20(1):77–85. DOI: 10.1038/cdd.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grover GJ, Marone PA, Koetzner L, et al. Energetic signalling in the control of mitochondrial F1F0 ATP synthase activity in health and disease. Int J Biochem Cell Biol 2008;40(12):2698–2701. DOI: 10.1016/j.biocel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 81.Field CS, Baixauli F, Kyle RL, et al. Mitochondrial integrity regulated by lipid metabolism is a cell-Intrinsic checkpoint for treg suppressive function. Cell Metab 2020;31(2):e5422–437 e5. DOI: 10.1016/j.cmet.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiederkehr A, Park KS, Dupont O, et al. Matrix alkalinization: A novel mitochondrial signal for sustained pancreatic beta-cell activation. EMBO J 2009;28(4):417–428. DOI: 10.1038/emboj.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selivanov VA, Zeak JA, Roca J, et al. The role of external and matrix pH in mitochondrial reactive oxygen species generation. J Biol Chem 2008;283(43):29292–29300. DOI: 10.1074/jbc.M801019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Halestrap AP. The regulation of the oxidation of fatty acids and other substrates in rat heart mitochondria by changes in the matrix volume induced by osmotic strength, valinomycin and Ca2+. Biochem J 1987;244(1):159–164. DOI: 10.1042/bj2440159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Makarov VI, Khmelinskii I, Javadov S. Computational modeling of in vitro swelling of mitochondria: A biophysical approach. Molecules 2018;23(4):783. DOI: 10.3390/molecules23040783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Calamita G, Ferri D, Gena P, et al. The inner mitochondrial membrane has aquaporin-8 water channels and is highly permeable to water. J Biol Chem 2005;280(17):17149–17153. DOI: 10.1074/jbc.C400595200. [DOI] [PubMed] [Google Scholar]
- 87.Smith AC, Robinson AJ. A metabolic model of the mitochondrion and its use in modelling diseases of the tricarboxylic acid cycle. BMC Syst Biol 2011;5:102. DOI: 10.1186/1752-0509-5-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase – Assembly, regulation and role in human disease. Mitochondrion 2010;10(4):393–401. DOI: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cavalcanti JH, Esteves-Ferreira AA, Quinhones CG, et al. Evolution and functional implications of the tricarboxylic acid cycle as revealed by phylogenetic analysis. Genome Biol Evol 2014;6(10):2830–2848. DOI: 10.1093/gbe/evu221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D’Souza AR, Minczuk M. Mitochondrial transcription and translation: overview. Essays Biochem 2018;62(3):309–320. DOI: 10.1042/EBC20170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1999;1410(2):103–123. DOI: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 92.Luo S, Valencia CA, Zhang J, et al. Biparental Inheritance of Mitochondrial DNA in Humans. Proc Natl Acad Sci USA 2018;115(51):13039–13044. DOI: 10.1073/pnas.1810946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pfanner N, Warscheid B, Wiedemann N. Mitochondrial proteins: From biogenesis to functional networks. Nat Rev Mol Cell Biol 2019;20(5):267–284. DOI: 10.1038/s41580-018-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 2010;11:25–44. DOI: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang F, Zhang D, Zhang D, et al. Mitochondrial protein translation: Emerging roles and clinical significance in disease. Front Cell Dev Biol 2021;9:675465. DOI: 10.3389/fcell.2021.675465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koripella RK, Sharma MR, Bhargava K, et al. Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation. Nat Commun 2020;11(1):3830. DOI: 10.1038/s41467-020-17715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. DOI: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gray MW. Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci USA 2015;112(33):10133–10138. DOI: 10.1073/pnas.1421379112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boussau B, Karlberg EO, Frank AC, et al. Computational inference of scenarios for alpha-proteobacterial genome evolution. Proc Natl Acad Sci USA 2004;101(26):9722–9727. DOI: 10.1073/pnas.0400975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gabaldon T. Relative timing of mitochondrial endosymbiosis and the “pre-mitochondrial symbioses” hypothesis. IUBMB Life. Dec 2018;70(12):1188–1196. DOI: 10.1002/iub.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koonin EV. Archaeal ancestors of eukaryotes: Not so elusive any more. BMC Biol 2015;13:84. DOI: 10.1186/s12915-015-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Archibald JM. Endosymbiosis and eukaryotic cell evolution. Curr Biol 2015;25(19):R911–R921. DOI: 10.1016/j.cub.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 103.Martin WF, Garg S, Zimorski V. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci 2015;370(1678):20140330. DOI: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aanen DK, Eggleton P. Symbiogenesis: Beyond the endosymbiosis theory? J Theor Biol 2017;434:99–103. DOI: 10.1016/j.jtbi.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 105.Shiflett AM, Johnson PJ. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol. 2010;64:409–29. DOI: 10.1146/annurev.micro.62.081307.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gawryluk RMR, Kamikawa R, Stairs CW, et al. The earliest stages of mitochondrial adaptation to low oxygen revealed in a novel nhizarian. Curr Biol 2016;26(20):2729–2738. DOI: 10.1016/j.cub.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 107.Muller M, Mentel M, van Hellemond JJ, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 2012;76(2):444–495. DOI: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hrdy I, Hirt RP, Dolezal P, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 2004;432(7017):618–622. DOI: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 109.Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci 2010;365(1541):799–817. DOI: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Embley TM, van der Giezen M, Horner DS, et al. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci 2003;358(1429):191–201; Discussion 201–202. DOI: 10.1098/rstb.2002.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Makiuchi T, Nozaki T. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 2014;100:3–17. DOI: 10.1016/j.biochi.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 112.Read AD, Bentley RE, Archer SL, et al. Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biol 2021;47:102164. DOI: 10.1016/j.redox.2021.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roger AJ, Munoz-Gomez SA, Kamikawa R. The Origin and Diversification of Mitochondria. Curr Biol 2017;27(21):R1177–R1192. DOI: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 114.van der Giezen M, Slotboom DJ, Horner DS, et al. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: A common origin for both organelles. EMBO J 2002;21(4):572–579. DOI: 10.1093/emboj/21.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reznik E, Wang Q, La K, et al. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 2017;6: e21592. DOI: 10.7554/eLife.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012;48(2):158–167. DOI: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zachar I, Boza G. Endosymbiosis before eukaryotes: Mitochondrial establishment in protoeukaryotes. Cell Mol Life Sci 2020;77(18): 3503–3523. DOI: 10.1007/s00018-020-03462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Degli Esposti M. Bioenergetic evolution in proteobacteria and mitochondria. Genome Biol Evol2014;6(12):3238–3251. DOI: 10.1093/gbe/evu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta RS, Mok A. Phylogenomics and signature proteins for the alpha proteobacteria and its main groups. BMC Microbiol 2007;7:106. DOI: 10.1186/1471-2180-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Wu M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep 2015;5:7949. DOI: 10.1038/srep07949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thrash JC, Boyd A, Huggett MJ, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 2011;1:13. DOI: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fan L, Wu D, Goremykin V, et al. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within alphaproteobacteria. Nat Ecol Evol 2020;4(9):1213–1219. DOI: 10.1038/s41559-020-1239-x. [DOI] [PubMed] [Google Scholar]
- 123.Morris RM, Rappe MS, Connon SA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 2002;420(6917): 806–810. DOI: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 124.Lopez-Perez M, Haro-Moreno JM, Coutinho FH, et al. The Evolutionary Success of the Marine Bacterium SAR11 Analyzed through a Metagenomic Perspective. mSystems 2020;5(5):e00605–e00620. DOI: 10.1128/mSystems.00605-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Munoz-Gomez SA, Hess S, Burger G, et al. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. Elife 2019;8:e42535. DOI: 10.7554/eLife.42535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodriguez-Ezpeleta N, Embley TM. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One 2012;7(1):e30520. DOI: 10.1371/journal.pone.0030520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grattepanche JD, Walker LM, Ott BM, et al. Microbial diversity in the eukaryotic SAR clade: Illuminating the darkness between morphology and molecular data. Bioessays 2018;40(4):e1700198. DOI: 10.1002/bies.201700198. [DOI] [PubMed] [Google Scholar]
- 128.Lio P, Goldman N. Models of molecular evolution and phylogeny. Genome Res 1998;8(12):1233–1244. DOI: 10.1101/gr.8.12.1233. [DOI] [PubMed] [Google Scholar]
- 129.McDonnell MD, Abbott D. What is stochastic resonance? Definitions, misconceptions, debates, and its relevance to biology. PLoS Comput Biol 2009;5(5):e1000348. DOI: 10.1371/journal.pcbi.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ross MG, Russ C, Costello M, et al. Characterizing and measuring bias in sequence data. Genome Biol 2013;14(5):R51. DOI: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Philippe H, Zhou Y, Brinkmann H, et al. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol 2005;5:50. DOI: 10.1186/1471-2148-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol 2011;9(3):e1000602. DOI: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bergsten J. A review of long-branch attraction. Cladistics 2005;21(2):163–193. DOI: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 134.Revell LJ, Harmon LJ, Collar DC. Phylogenetic s ignal, evolutionary process, and rate. Syst Biol 2008;57(4):591–601. DOI: 10.1080/10635150802302427. [DOI] [PubMed] [Google Scholar]
- 135.Susko E, Roger AJ. Long branch attraction biases in phylogenetics. Syst Biol 2021;70(4):838–843. DOI: 10.1093/sysbio/syab001. [DOI] [PubMed] [Google Scholar]
- 136.Foster PG, Hickey DA. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol 1999;48(3):284–290. DOI: 10.1007/pl00006471. [DOI] [PubMed] [Google Scholar]
- 137.Koumandou VL, Wickstead B, Ginger ML, et al. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol 2013;48(4):373–396. DOI: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Roger AJ, Susko E, Leger MM. Evolution: Reconstructing the timeline of eukaryogenesis. Curr Biol 2021;31(4):R193–R196. DOI: 10.1016/j.cub.2020.12.035. [DOI] [PubMed] [Google Scholar]
- 139.Eme L, Spang A, Lombard J, et al. Archaea and the origin of eukaryotes. Nat Rev Microbiol 2017;15(12):711–723. DOI: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 140.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017;541(7637):353–358. DOI: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 141.Martin WF. Physiology, anaerobes, and the origin of mitosing cells 50 years on. J Theor Biol 2017;434:2–10. DOI: 10.1016/j.jtbi.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 142.Nunoura T, Takaki Y, Kakuta J, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 2011;39(8): 3204–3223. DOI: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lopez-Garcia P, Eme L, Moreira D. Symbiosis in eukaryotic evolution. J Theor Biol 2017;434:20–33. DOI: 10.1016/j.jtbi.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lopez-Garcia P, Moreira D. Open Questions on the Origin of Eukaryotes. Trends Ecol Evol 2015;30(11):697–708. DOI: 10.1016/j.tree.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ryan DG, Frezza C, O’Neill LA. TCA cycle signalling and the evolution of eukaryotes. Curr Opin Biotechnol 2021;68:72–88. DOI: 10.1016/j.copbio.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koreny L, Field MC. Ancient eukaryotic origin and evolutionary plasticity of nuclear lamina. Genome Biol Evol 2016;8(9):2663–2671. DOI: 10.1093/gbe/evw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Energetics Lane N. and genetics across the prokaryote-eukaryote divide. Biol Direct 2011;6:35. DOI: 10.1186/1745-6150-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Torri A, Jaeger J, Pradeu T, et al. The origin of RNA interference: Adaptive or neutral evolution? PLoS Biol 2022;20(6):e3001715. DOI: 10.1371/journal.pbio.3001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Koonin EV. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early vs introns-late debate? Biol Direct 2006;1:22. DOI: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Grau-Bove X, Sebe-Pedros A, Ruiz-Trillo I. The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin. Mol Biol Evol 2015;32(3):726–739. DOI: 10.1093/molbev/msu334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mills DB. The origin of phagocytosis in Earth history. Interface Focus 2020;10(4):20200019. DOI: 10.1098/rsfs.2020.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol 2011;194(4):513–525. DOI: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guan XL, Souza CM, Pichler H, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 2009;20(7):2083–2095. DOI: 10.1091/mbc.e08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Poole AM, Gribaldo S. Eukaryotic origins: How and when was the mitochondrion acquired? Cold Spring Harb Perspect Biol 2014;6(12):a015990. DOI: 10.1101/cshperspect.a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol 2014;12:76. DOI: 10.1186/s12915-014-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lopez-Garcia P, Moreira D. The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol 2020;5(5):655–667. DOI: 10.1038/s41564-020-0710-4. [DOI] [PubMed] [Google Scholar]
- 157.Milner DS, Wideman JG, Stairs CW, et al. A functional bacteria-derived restriction modification system in the mitochondrion of a heterotrophic protist. PLoS Biol 2021;19(4):e3001126. DOI: 10.1371/journal.pbio.3001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sharma LK, Lu J, Bai Y. Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr Med Chem 2009;16(10):1266–1277. DOI: 10.2174/092986709787846578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moparthi VK, Hagerhall C. The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J Mol Evol 2011;72(5–6):484–497. DOI: 10.1007/s00239-011-9447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 2016;73(1):79–94. DOI: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol 2014;6(8): a016972. DOI: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta 2008;1778(10):1978–2021. DOI: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 163.Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol 2013;5(2):a013367. DOI: 10.1101/cshperspect.a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stroud MJ, Banerjee I, Veevers J, et al. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ Res 2014;114(3):538–548. DOI: 10.1161/circresaha.114.301236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jain S, Caforio A, Driessen AJ. Biosynthesis of archaeal membrane ether lipids. Front Microbiol. 2014; 5: 641. DOI: 10.3389/fmicb.2014.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Salvador-Castell M, Tourte M, Oger PM. In search for the membrane regulators of archaea. Int J Mol Sci2019;20(18):4434. DOI: 10.3390/ijms20184434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Siliakus MF, van der Oost J, Kengen SWM. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017;21(4):651–670. DOI: 10.1007/s00792-017-0939-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cole LW. The evolution of per-cell organelle number. Front Cell Dev Biol 2016;4:85. DOI: 10.3389/fcell.2016.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hjort K, Goldberg AV, Tsaousis AD, et al. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci 2010;365(1541):713–727. DOI: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y, Palmfeldt J, Gregersen N, et al. Mitochondrial fatty acid oxidation and the electron transport chain comprise a multifunctional mitochondrial protein complex. J Biol Chem 2019;294(33):12380–12391. DOI: 10.1074/jbc.RA119.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: Implications for human genetic disease. Gene 2002;286(1):73–79. DOI: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 172.Ferrari A, Del’Olio S, Barrientos A. The diseased mitoribosome. FEBS Lett 2021;595(8):1025–1061. DOI: 10.1002/1873-3468.14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cavalier-Smith T. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc Biol Sci 2006;273(1596):1943–1952. DOI: 10.1098/rspb.2006.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Falkenberg M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem 2018;62(3):287–296. DOI: 10.1042/EBC20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem 2010;47:69–84. DOI: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kunze M, Berger J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front Physiol 2015;6:259. DOI: 10.3389/fphys.2015.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Avendano-Monsalve MC, Mendoza-Martinez AE, Ponce-Rojas JC, et al. Positively charged amino acids at the N terminus of select mitochondrial proteins mediate early recognition by import proteins alphabeta’-NAC and Sam37. J Biol Chem 2022;298(6):101984. DOI: 10.1016/j.jbc.2022.101984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bolender N, Sickmann A, Wagner R, et al. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep 2008;9(1):42–49. DOI: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Diekert K, Kispal G, Guiard B, et al. An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA 1999;96(21):11752–11757. DOI: 10.1073/pnas.96.21.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Craig EA. Hsp70 at the membrane: Driving protein translocation. BMC Biol 2018;16(1):11. DOI: 10.1186/s12915-017-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ieva R, Heisswolf AK, Gebert M, et al. Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nat Commun 2013;4:2853. DOI: 10.1038/ncomms3853. [DOI] [PubMed] [Google Scholar]
- 182.Li Y, Dudek J, Guiard B, et al. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 2004;279(36):38047–38054. DOI: 10.1074/jbc.M404319200. [DOI] [PubMed] [Google Scholar]
- 183.Takeda H, Tsutsumi A, Nishizawa T, et al. Mitochondrial sorting and assembly machinery operates by beta-barrel switching. Nature 2021;590(7844):163–169. DOI: 10.1038/s41586-020-03113-7. [DOI] [PubMed] [Google Scholar]
- 184.Gureev AP, Shaforostova EA, Popov VN. Regulation of mitochondrial biogenesis as a way for active longevity: Interaction between the Nrf2 and PGC-1alpha signaling pathways. Front Genet 2019;10:435. DOI: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): Transcriptional coactivator and metabolic regulator. Endocr Rev 2003;24(1):78–90. DOI: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 186.Liang H, Ward WF. PGC-1alpha: A key regulator of energy metabolism. Adv Physiol Educ 2006;30(4):145–151. DOI: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 187.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol 2005;25(4):1354–1366. DOI: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev 1993;7(12A):2431–2445. DOI: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 189.Saha S, Buttari B, Panieri E, et al. An overview of Nrf2 signaling pathway and its role in inflammation. Molecules. 2020;25(22):5474. DOI: 10.3390/molecules25225474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol 2009;20(2):98–105. DOI: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fan W, Evans R. PPARs and ERRs: Molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol 2015;33:49–54. DOI: 10.1016/j.ceb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Audet-Walsh E, Giguere V. The multiple universes of estrogen-related receptor alpha and gamma in metabolic control and related diseases. Acta Pharmacol Sin. Jan 2015;36(1):51–61. DOI: 10.1038/aps.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev2008;29(6):677–696. DOI: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 194.Deblois G, Giguere V. Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim Biophys Acta 2011;1812(8):1032–1040. DOI: 10.1016/j.bbadis.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 195.Melser S, Lavie J, Benard G. Mitochondrial degradation and energy metabolism. Biochim Biophys Acta 2015;1853(10 Pt B):2812–2821. DOI: 10.1016/j.bbamcr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 196.Okie JG, Smith VH, Martin-Cereceda M. Major evolutionary transitions of life, metabolic scaling and the number and size of mitochondria and chloroplasts. Proc Biol Sci 2016;283(1831). DOI: 10.1098/rspb.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Thommen A, Werner S, Frank O, et al. Body size-dependent energy storage causes Kleiber’s law scaling of the metabolic rate in planarians. Elife 2019;8:e38187. DOI: 10.7554/eLife.38187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol 1990;143(1):160–164. DOI: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- 199.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun 2017;482(3):426–431. DOI: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 200.Pickles S, Vigie P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 2018;28(4):R170–R185. DOI: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab 2012;23(9):459–466. DOI: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Byrnes J, Garcia-Diaz M. Mitochondrial transcription: how does it end? Transcription 2011;2(1):32–36. DOI: 10.4161/trns.2.1.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Picard M, McEwen BS, Epel ES, et al. An energetic view of stress: Focus on mitochondria. Front Neuroendocrinol 2018;49:72–85. DOI: 10.1016/j.yfrne.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Torralba D, Baixauli F, Sanchez-Madrid F. Mitochondria know no boundaries: Mechanisms and functions of intercellular mitochondrial transfer. Front Cell Dev Biol 2016;4:107. DOI: 10.3389/fcell.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hayakawa K, Esposito E, Wang X, et al. Corrigendum: Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016;539(7627):123. DOI: 10.1038/nature19805. [DOI] [PubMed] [Google Scholar]
- 206.Hayakawa K, Esposito E, Wang X, et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016;535(7613):551–555. DOI: 10.1038/nature18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dong LF, Kovarova J, Bajzikova M, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife 2017;6:e22187. DOI: 10.7554/eLife.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Takenaga K, Koshikawa N, Nagase H. Intercellular transfer of mitochondrial DNA carrying metastasis-enhancing pathogenic mutations from high- to low-metastatic tumor cells and stromal cells via extracellular vesicles. BMC Mol Cell Biol 2021;22(1):52. DOI: 10.1186/s12860-021-00391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Burton A, Torres-Padilla ME. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics 2010;9(5–6):444–454. DOI: 10.1093/bfgp/elq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000;10(8):475–478. DOI: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 211.Zhang N. Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Anim Nutr 2018;4(1): 11–16. DOI: 10.1016/j.aninu.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lin H. S-Adenosylmethionine-dependent alkylation reactions: When are radical reactions used? Bioorg Chem. Dec 2011;39(5–6):161–170. DOI: 10.1016/j.bioorg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Struck AW, Thompson ML, Wong LS, et al. S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 2012;13(18):2642–2655. DOI: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 214.Tollervey JR, Lunyak VV. Epigenetics: Judge, jury and executioner of stem cell fate. Epigenetics 2012;7(8):823–840. DOI: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Johansson C, Tumber A, Che K, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics 2014;6(1):89–120. DOI: 10.2217/epi.13.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Martinez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020;11(1):102. DOI: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog 2015;20(1–2):35–47. DOI: 10.1615/critrevoncog.2015012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J Biol Chem 2012;287(28):23865–23876. DOI: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 2004;5(5):224. DOI: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yen MY, Wang AG, Wei YH. Leber’s hereditary optic neuropathy: A multifactorial disease. Prog Retin Eye Res 2006;25(4):381–396. DOI: 10.1016/j.preteyeres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 221.Peng J, Ramatchandirin B, Pearah A, et al. Development and functions of mitochondria in early life. Newborn 2022;1(1):131–141. DOI: 10.5005/jp-journals-11002-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zangari J, Petrelli F, Maillot B, Martinou JC. The multifaceted pyruvate metabolism: Role of the mitochondrial pyruvate carrier. Biomolecules 2020;10(7):1068. DOI: 10.3390/biom10071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lozoya OA, Martinez-Reyes I, Wang T, et al. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol 2018;16(4):e2005707. DOI: 10.1371/journal.pbio.2005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol 2020;37:101674. DOI: 10.1016/j.redox.2020.101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chandel NS. Mitochondrial complex III: An essential component of universal oxygen sensing machinery? Respir Physiol Neurobiol 2010;174(3):175–181. DOI: 10.1016/j.resp.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li Y, Park JS, Deng JH, et al. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembr 2006;38(5–6):283–291. DOI: 10.1007/s10863-006-9052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: Architecture, function and pathology. J Inherit Metab Dis 2012;35(2):211–225. DOI: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Faccenda D, Campanella M. Molecular regulation of the mitochondrial F(1)F(o)-ATPsynthase: Physiological and pathological significance of the inhibitory factor 1 (IF(1)). Int J Cell Biol 2012;2012:367934. DOI: 10.1155/2012/367934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Paumard P, Vaillier J, Coulary B, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 2002;21(3):221–230. DOI: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nsiah-Sefaa A, McKenzie M. Combined defects in oxidative phosphorylation and fatty acid beta-oxidation in mitochondrial disease. Biosci Rep 2016;36(2):e00313. DOI: 10.1042/BSR20150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 2018;20(7):745–754. DOI: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bhatti JS, Bhatti GK, Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders - A step towards mitochondria based therapeutic strategies. Biochim Biophys Acta Mol Basis Dis 2017;1863(5):1066–1077. DOI: 10.1016/j.bbadis.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull 2013;106(1):135–159. DOI: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Le NQ, Ou YY. Prediction of FAD binding sites in electron transport proteins according to efficient radial basis function networks and significant amino acid pairs. BMC Bioinformatics 2016;17:298. DOI: 10.1186/s12859-016-1163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kirillova A, Smitz JEJ, Sukhikh GT, et al. The Role of Mitochondria in Oocyte Maturation. Cells 19 2021;10(9):2484. DOI: 10.3390/cells10092484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod 2000;15(Suppl 2):148–59. DOI: 10.1093/humrep/15.suppl_2.148. [DOI] [PubMed] [Google Scholar]
- 237.Au HK, Yeh TS, Kao SH, et al. Abnormal mitochondrial structure in human unfertilized oocytes and arrested embryos. Ann N Y Acad Sci 2005;1042:177–185. DOI: 10.1196/annals.1338.020. [DOI] [PubMed] [Google Scholar]
- 238.Dumesic DA, Meldrum DR, Katz-Jaffe MG, et al. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril 2015;103(2):303–316. DOI: 10.1016/j.fertnstert.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 239.Houghton FD, Thompson JG, Kennedy CJ, et al. Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev 1996;44(4):476–485. DOI: . [DOI] [PubMed] [Google Scholar]
- 240.Gardner DK, Wale PL. Analysis of metabolism to select viable human embryos for transfer. Fertil Steril 2013;99(4):1062–1072. DOI: 10.1016/j.fertnstert.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 241.May-Panloup P, Chretien MF, Jacques C, et al. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod 2005;20(3):593–597. DOI: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
- 242.Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006;85(3):584–591. DOI: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 243.Giles RE, Blanc H, Cann HM, et al. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA 1980;77(11):6715–6719. DOI: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Reynier P, May-Panloup P, Chretien MF, et al. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod 2001;7(5):425–429. DOI: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- 245.Murakoshi Y, Sueoka K, Takahashi K, et al. Embryo developmental capability and pregnancy outcome are related to the mitochondrial DNA copy number and ooplasmic volume. J Assist Reprod Genet 2013;30(10):1367–1375. DOI: 10.1007/s10815-013-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ge H, Tollner TL, Hu Z, et al. The importance of mitochondrial metabolic activity and mitochondrial DNA replication during oocyte maturation in vitro on oocyte quality and subsequent embryo developmental competence. Mol Reprod Dev 2012;79(6):392–401. DOI: 10.1002/mrd.22042. [DOI] [PubMed] [Google Scholar]
- 247.Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod 1995;10(2):415–424. DOI: 10.1093/oxfordjournals.humrep.a135954. [DOI] [PubMed] [Google Scholar]
- 248.Mandal S, Lindgren AG, Srivastava AS, et al. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 2011;29(3):486–495. DOI: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Piquereau J, Ventura-Clapier R. Maturation of cardiac energy metabolism during perinatal development. Front Physiol 2018;9:959. DOI: 10.3389/fphys.2018.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Abraham M, Collins CA, Flewelling S, et al. Mitochondrial inefficiency in infants born to overweight African-American mothers. Int J Obes (Lond) 2018;42(7):1306–1316. DOI: 10.1038/s41366-018-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Delhaes F, Giza SA, Koreman T, et al. Altered maternal and placental lipid metabolism and fetal fat development in obesity: Current knowledge and advances in non-invasive assessment. Placenta 2018;69:118–124. DOI: 10.1016/j.placenta.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 252.Ten VS, Stepanova AA, Ratner V, et al. Mitochondrial dysfunction and permeability transition in neonatal brain and lung injuries. Cells 2021;10(3):569.DOI: 10.3390/cells10030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci 2014;71(14):2577–2604. DOI: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sommakia S, Houlihan PR, Deane SS, et al. Mitochondrial cardiomyopathies feature increased uptake and diminished efflux of mitochondrial calcium. J Mol Cell Cardiol 2017;113:22–32. DOI: 10.1016/j.yjmcc.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wang LY, Wang DH, Zou XY, et al. Mitochondrial functions on oocytes and preimplantation embryos. J Zhejiang Univ Sci B 2009;10(7):483–492. DOI: 10.1631/jzus.B0820379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zhang D, Keilty D, Zhang ZF, et al. Mitochondria in oocyte aging: current understanding. Facts Views Vis Obgyn 2017;9(1):29–38. PMID: 28721182. [PMC free article] [PubMed] [Google Scholar]
- 257.Haskins N, Bhuvanendran S, Anselmi C, et al. Mitochondrial enzymes of the urea cycle cluster at the inner mitochondrial membrane. Front Physiol 2020;11:542950. DOI: 10.3389/fphys.2020.542950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.de Cima S, Polo LM, Diez-Fernandez C, et al. Structure of human carbamoyl phosphate synthetase: Deciphering the on/off switch of human ureagenesis. Sci Rep 2015;5:16950. DOI: 10.1038/srep16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Couchet M, Breuillard C, Corne C, et al. Ornithine transcarbamylase - From structure to metabolism: An update. Front Physiol 2021;12:748249. DOI: 10.3389/fphys.2021.748249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Borst P. The malate-aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life 2020;72(11):2241–2259. DOI: 10.1002/iub.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Cooper AJ, Kuhara T. Alpha-Ketoglutaramate: An overlooked metabolite of glutamine and a biomarker for hepatic encephalopathy and inborn errors of the urea cycle. Metab Brain Dis 2014;29(4):991–1006. DOI: 10.1007/s11011-013-9444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Traaseth N, Elfering S, Solien J, et al. Role of calcium signaling in the activation of mitochondrial nitric oxide synthase and citric acid cycle. Biochim Biophys Acta 2004;1658(1–2):64–71. DOI: 10.1016/j.bbabio.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 263.Heikal AA. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 2010;4(2):241–263. DOI: 10.2217/bmm.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: A target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci 29 2005;360(1464):2335–2345. DOI: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Williams GS, Boyman L, Lederer WJ. Mitochondrial calcium and the regulation of metabolism in the heart. J Mol Cell Cardiol 2015;78:35–45. DOI: 10.1016/j.yjmcc.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Chance B. Reaction of oxygen with the respiratory chain in cells and tissues. J Gen Physiol 1965;49(1):Suppl:163–195. DOI: 10.1085/jgp.49.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol 2007;35(4):495–516. DOI: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008;9(1):47–59. DOI: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 269.Luna-Vargas MP, Chipuk JE. The deadly landscape of pro-apoptotic BCL-2 proteins in the outer mitochondrial membrane. FEBS J 2016;283(14):2676–2689. DOI: 10.1111/febs.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wei MC, Zong WX, Cheng EH, et al. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science 2001;292(5517):727–730. DOI: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Dewson G, Kluck RM. Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J Cell Sci 2009;122(Pt 16):2801–2808. DOI: 10.1242/jcs.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Carrington EM, Zhan Y, Brady JL, et al. Anti-apoptotic proteins BCL-2, MCL-1 and A1 summate collectively to maintain survival of immune cell populations both in vitro and in vivo. Cell Death Differ 2017;24(5):878–888. DOI: 10.1038/cdd.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Karch J, Kwong JQ, Burr AR, et al. Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. Elife 2013;2:e00772. DOI: 10.7554/eLife.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet 2009;43:95–118. DOI: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Baines CP, Kaiser RA, Sheiko T, et al. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol 2007;9(5):550–555. DOI: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Mnatsakanyan N, Jonas EA. The new role of F(1)F(o) ATP synthase in mitochondria-mediated neurodegeneration and neuroprotection. Exp Neurol 2020;332:113400. DOI: 10.1016/j.expneurol.2020.113400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Saier MH Jr, Reddy BL Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol 2015;197(1):7–17. DOI: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pang X, Moussa SH, Targy NM, et al. Active Bax and Bak are functional holins. Genes Dev 2011;25(21):2278–2290. DOI: 10.1101/gad.171645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Catalao MJ, Gil F, Moniz-Pereira J, et al. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol Rev 2013;37(4):554–571. DOI: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 280.Xu Z, Zhang D, He X, et al. Transport of Calcium Ions into Mitochondria. Curr Genomics 2016;17(3):215–219. DOI: 10.2174/1389202917666160202215748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sudhof TC. Calcium control of neurotransmitter release. Cold Spring Harb Perspect Biol 2012;4(1):a011353. DOI: 10.1101/cshperspect.a011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Brini M, Cali T, Ottolini D, et al. Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci 2014;71(15):2787–2814. DOI: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lettieri-Barbato D. Redox control of non-shivering thermogenesis. Mol Metab 2019;25:11–19. DOI: 10.1016/j.molmet.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Pompei M, Pompei F. Overcoming bioethical, legal, and hereditary barriers to mitochondrial replacement therapy in the USA. J Assist Reprod Genet 2019;36(3):383–393. DOI: 10.1007/s10815-018-1370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ebihara T, Nagatomo T, Sugiyama Y, et al. Neonatal-onset mitochondrial disease: Clinical features, molecular diagnosis and prognosis. Arch Dis Child Fetal Neonatal Ed 2022;107(3):329–334. DOI: 10.1136/archdischild-2021-321633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ahuja AS. Understanding mitochondrial myopathies: A review. PeerJ 2018;6:e4790. DOI: 10.7717/peerj.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ng YS, Turnbull DM. Mitochondrial disease: Genetics and management. J Neurol2016;263(1):179–191. DOI: 10.1007/s00415-015-7884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Orsucci D, Caldarazzo Ienco E, Rossi A, et al. Mitochondrial Syndromes Revisited. J Clin Med 2021;10(6):1249. DOI: 10.3390/jcm10061249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol 2010;5:297–348. DOI: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Saneto RP, Sedensky MM. Mitochondrial disease in childhood: mtDNA encoded. Neurotherapeutics 2013;10(2):199–211. DOI: 10.1007/s13311-012-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Muraresku CC, McCormick EM, Falk MJ. Mitochondrial Disease: Advances in clinical diagnosis, management, therapeutic development, and preventative strategies. Curr Genet Med Rep 2018;6(2):62–72. DOI: 10.1007/s40142-018-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Rodenburg RJ. Mitochondrial complex I-linked disease. Biochim Biophys Acta 2016;1857(7):938–945. DOI: 10.1016/j.bbabio.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 293.Koene S, Rodenburg RJ, van der Knaap MS, et al. Natural disease course and genotype-phenotype correlations in Complex I deficiency caused by nuclear gene defects: What we learned from 130 cases. J Inherit Metab Dis 2012;35(5):737–747. DOI: 10.1007/s10545-012-9492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Goldstein AC, Bhatia P, Vento JM. Mitochondrial disease in childhood: Nuclear encoded. Neurotherapeutics 2013;10(2):212–226. DOI: 10.1007/s13311-013-0185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Mori M, Goldstein J, Young SP, et al. Complex III deficiency due to an in-frame MT-CYB deletion presenting as ketotic hypoglycemia and lactic acidosis. Mol Genet Metab Rep 2015;4:39–41. DOI: 10.1016/j.ymgmr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Meunier B, Fisher N, Ransac S, et al. Respiratory complex III dysfunction in humans and the use of yeast as a model organism to study mitochondrial myopathy and associated diseases. Biochim Biophys Acta 2013;1827(11–12):1346–1361. DOI: 10.1016/j.bbabio.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 297.Tucker EJ, Wanschers BF, Szklarczyk R, et al. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet 2013;9(12):e1004034. DOI: 10.1371/journal.pgen.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hamza I, Gitlin JD. Copper chaperones for cytochrome c oxidase and human disease. J Bioenerg Biomembr 2002;34(5):381–388. DOI: 10.1023/a:1021254104012. [DOI] [PubMed] [Google Scholar]
- 299.Soto IC, Fontanesi F, Liu J, et al. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta 2012;1817(6):883–897. DOI: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kose M, Canda E, Kagnici M, et al. SURF1 related Leigh syndrome: Clinical and molecular findings of 16 patients from Turkey. Mol Genet Metab Rep 2020;25:100657. DOI: 10.1016/j.ymgmr.2020.100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Danis D, Brennerova K, Skopkova M, et al. Mutations in SURF1 are important genetic causes of Leigh syndrome in Slovak patients. Endocr Regul 2018;52(2):110–118. DOI: 10.2478/enr-2018-0013. [DOI] [PubMed] [Google Scholar]
- 302.Abdulhag UN, Soiferman D, Schueler-Furman O, et al. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur J Hum Genet 2015;23(2):159–164. DOI: 10.1038/ejhg.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Emmanuel IA, Olotu FA, Agoni C, et al. In Silico Repurposing of J147 for neonatal encephalopathy treatment: Exploring molecular mechanisms of mutant mitochondrial ATP synthase. Curr Pharm Biotechnol 2020;21(14):1551–1566. DOI: 10.2174/1389201021666200628152246. [DOI] [PubMed] [Google Scholar]
- 304.Dautant A, Meier T, Hahn A, et al. ATP synthase diseases of mitochondrial genetic origin. Front Physiol 2018;9:329. DOI: 10.3389/fphys.2018.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Martin E, Rosenthal RE, Fiskum G. Pyruvate dehydrogenase complex: Metabolic link to ischemic brain injury and target of oxidative stress. J Neurosci Res 2005;79(1–2):240–247. DOI: 10.1002/jnr.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Patel KP, O’Brien TW, Subramony SH, et al. The spectrum of pyruvate dehydrogenase complex deficiency: Clinical, biochemical and genetic features in 371 patients. Mol Genet Metab 2012;106(3): 385–394. DOI: 10.1016/j.ymgme.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Singhi P, De Meirleir L, Lissens W, et al. Pyruvate dehydrogenase-e1alpha deficiency presenting as recurrent demyelination: an unusual presentation and a novel mutation. JIMD Rep 2013;10:107–111. DOI: 10.1007/8904_2012_211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Gupta N, Rutledge C. Pyruvate dehydrogenase complex deficiency: An unusual cause of recurrent lactic acidosis in a paediatric critical care unit. J Crit Care Med (Targu Mures) 2019;5(2):71–75. DOI: 10.2478/jccm-2019-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Bravo-Alonso I, Navarrete R, Vega AI, et al. Genes and variants underlying human congenital lactic acidosis-from genetics to personalized treatment. J Clin Med 2019;8(11):1811. DOI: 10.3390/jcm8111811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Toyoshima M, Oka A, Egi Y, et al. Thiamine-responsive congenital lactic acidosis: clinical and biochemical studies. Pediatr Neurol 2005;33(2):98–104. DOI: 10.1016/j.pediatrneurol.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 311.Hotchkiss RS, Moldawer LL, Opal SM, et al. Sepsis and septic shock. Nat Rev Dis Primers 2016;2:16045. DOI: 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Amarante-Mendes GP, Adjemian S, Branco LM, et al. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol 2018;9:2379. DOI: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Gentile LF, Moldawer LL. DAMPs, PAMPs, and the origins of SIRS in bacterial sepsis. Shock 2013;39(1):113–114. DOI: 10.1097/SHK.0b013e318277109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Santoni G, Cardinali C, Morelli MB, et al. Danger- and pathogen-associated molecular patterns recognition by pattern-recognition receptors and ion channels of the transient receptor potential family triggers the inflammasome activation in immune cells and sensory neurons. J Neuroinflammation 2015;12:21. DOI: 10.1186/s12974-015-0239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Demento SL, Siefert AL, Bandyopadhyay A, et al. Pathogen-associated molecular patterns on biomaterials: A paradigm for engineering new vaccines. Trends Biotechnol 2011;29(6):294–306. DOI: 10.1016/j.tibtech.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Hruz P, Eckmann L. Caspase recruitment domain-containing sensors and adaptors in intestinal innate immunity. Curr Opin Gastroenterol 2008;24(2):108–114. DOI: 10.1097/MOG.0b013e3282f50fdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kaneko N, Kurata M, Yamamoto T, et al. The role of interleukin-1 in general pathology. Inflamm Regen 2019;39:12. DOI: 10.1186/s41232-019-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zheng D, Liwinski T, Elinav E. Inflammasome activation and regulation: Toward a better understanding of complex mechanisms. Cell Discov 2020;6:36. DOI: 10.1038/s41421-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Yu J, Nagasu H, Murakami T, et al. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci USA 2014;111(43):15514–15519. DOI: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Santoni K, Pericat D, Gorse L, et al. Caspase-1-driven neutrophil pyroptosis and its role in host susceptibility to Pseudomonas aeruginosa. PLoS Pathog 2022;18(7):e1010305. DOI: 10.1371/journal.ppat.1010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.McIlroy DJ, Jarnicki AG, Au GG, et al. Mitochondrial DNA neutrophil extracellular traps are formed after trauma and subsequent surgery. J Crit Care 2014;29(6):1133 e1–e5. DOI: 10.1016/j.jcrc.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 322.Wilson DF. Oxidative phosphorylation: Regulation and role in cellular and tissue metabolism. J Physiol 2017;595(23):7023–7038. DOI: 10.1113/JP273839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Lee SR, Kwon KS, Kim SR, et al. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem 1998;273(25):15366–15372. DOI: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 324.Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ, et al. Biochemical and molecular basis for mitochondrial cardiomyopathy in neonates and children. J Inherit Metab Dis 2000;23(6):625–633. DOI: 10.1023/a:1005638231195. [DOI] [PubMed] [Google Scholar]
- 325.Russell AE, Doll DN, Sarkar SN, et al. TNF-alpha and beyond: Rapid mitochondrial dysfunction mediates TNF-alpha-Induced neurotoxicity. J Clin Cell Immunol 2016;7(6):467. DOI: 10.4172/2155-9899.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Altara R, Zouein FA, Booz GW. Untangling the interplay between mitochondrial fission and NF-kappa B signaling in endothelial inflammation. Hypertension 2020;76(1):23–25. DOI: 10.1161/HYPERTENSIONAHA.120.14854. [DOI] [PubMed] [Google Scholar]
- 327.Kapetanovic R, Afroz SF, Ramnath D, et al. Lipopolysaccharide promotes Drp1-dependent mitochondrial fission and associated inflammatory responses in macrophages. Immunol Cell Biol 2020;98(7):528–539. DOI: 10.1111/imcb.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Dasgupta D, Delmotte P, Sieck GC. Inflammation-Induced protein unfolding in airway smooth muscle triggers a homeostatic response in mitochondria. Int J Mol Sci 2020;22(1):363. DOI: 10.3390/ijms22010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Delmotte P, Sieck GC. Endoplasmic reticulum stress and mitochondrial function in airway smooth muscle. Front Cell Dev Biol 2019;7:374. DOI: 10.3389/fcell.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Geto Z, Molla MD, Challa F, et al. Mitochondrial dynamic dysfunction as a main triggering factor for inflammation associated chronic non-communicable diseases. J Inflamm Res 2020;13:97–107. DOI: 10.2147/JIR.S232009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sun J, Trumpower BL. Superoxide anion generation by the cytochrome bc1 complex. Arch Biochem Biophys 2003;419(2): 198–206. DOI: 10.1016/j.abb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 332.Ma K, Chen G, Li W, et al. Mitophagy, mitochondrial homeostasis, and cell fate. Front Cell Dev Biol 2020;8:467. DOI: 10.3389/fcell.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Gkikas I, Palikaras K, Tavernarakis N. The role of mitophagy in innate immunity. Front Immunol 2018;9:1283. DOI: 10.3389/fimmu.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Missiroli S, Genovese I, Perrone M, et al. The role of mitochondria in inflammation: From cancer to neurodegenerative disorders. J Clin Med 2020;9(3):740. DOI: 10.3390/jcm9030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Tanzer MC. A proteomic perspective on TNF-mediated signalling and cell death. Biochem Soc Trans 2022;50(1):13–20. DOI: 10.1042/BST20211114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kearney CJ, Cullen SP, Tynan GA, et al. Necroptosis suppresses inflammation via termination of TNF- or LPS-induced cytokine and chemokine production. Cell Death Differ 2015;22(8):1313–1327. DOI: 10.1038/cdd.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Place DE, Kanneganti TD. Cell death-mediated cytokine release and its therapeutic implications. J Exp Med 2019;216(7):1474–1486. DOI: 10.1084/jem.20181892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Dhuriya YK, Sharma D. Necroptosis: A regulated inflammatory mode of cell death. J Neuroinflammation 2018;15(1):199. DOI: 10.1186/s12974-018-1235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Seo J, Nam YW, Kim S, et al. Necroptosis molecular mechanisms: Recent findings regarding novel necroptosis regulators. Exp Mol Med 2021;53(6):1007–1017. DOI: 10.1038/s12276-021-00634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Lu W, Sun J, Yoon JS, et al. Mitochondrial protein PGAM5 regulates mitophagic protection against cell necroptosis. PLoS One 2016;11(1):e0147792. DOI: 10.1371/journal.pone.0147792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Zhang S, Che L, He C, et al. Drp1 and RB interaction to mediate mitochondria-dependent necroptosis induced by cadmium in hepatocytes. Cell Death Dis 2019;10(7):523. DOI: 10.1038/s41419-019-1730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Rayamajhi M, Miao EA. The RIP1-RIP3 complex initiates mitochondrial fission to fuel NLRP3. Nat Immunol 2014;15(12):1100–1102. DOI: 10.1038/ni.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Marshall KD, Baines CP. Necroptosis: Is there a role for mitochondria? Front Physiol 2014;5:323. DOI: 10.3389/fphys.2014.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Xue C, Gu X, Li G, et al. Mitochondrial mechanisms of necroptosis in liver diseases. Int J Mol Sci 23 2020;22(1):66. DOI: 10.3390/ijms22010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Khoshnan A, Tindell C, Laux I, et al. The NF-kappa B cascade is important in Bcl-xL expression and for the anti-apoptotic effects of the CD28 receptor in primary human CD4+ lymphocytes. J Immunol 2000;165(4):1743–1754. DOI: 10.4049/jimmunol.165.4.1743. [DOI] [PubMed] [Google Scholar]
- 346.Davis AF, Ropp PA, Clayton DA, et al. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res 1996;24(14):2753–2759. DOI: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Graziewicz MA, Day BJ, Copeland WC. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res 2002;30(13):2817–2824. DOI: 10.1093/nar/gkf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Sedlic F, Seiwerth F, Sepac A, et al. Mitochondrial ROS induce partial dedifferentiation of human mesothelioma via upregulation of NANOG. Antioxidants (Basel) 2020;9(7):606. DOI: 10.3390/antiox9070606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim Biophys Acta 2009;1787(5):312–319. DOI: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006;38(4):431–440. DOI: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 351.Allen KA, Brandon DH. Hypoxic ischemic encephalopathy: Pathophysiology and experimental treatments. Newborn Infant Nurs Rev 2011;11(3):125–133. DOI: 10.1053/j.nainr.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Thornton C, Jones A, Nair S, et al. Mitochondrial dynamics, mitophagy and biogenesis in neonatal hypoxic-ischaemic brain injury. FEBS Lett 2018;592(5):812–830. DOI: 10.1002/1873-3468.12943. [DOI] [PubMed] [Google Scholar]
- 353.Ten VS. Mitochondrial dysfunction in alveolar and white matter developmental failure in premature infants. Pediatr Res 2017;81(2):286–292. DOI: 10.1038/pr.2016.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Jones A, Thornton C. Mitochondrial dynamics in the neonatal brain – A potential target following injury? Biosci Rep 2022;42(3):BSR20211696. DOI: 10.1042/BSR20211696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Leaw B, Nair S, Lim R, et al. Mitochondria, bioenergetics and excitotoxicity: New therapeutic targets in perinatal brain injury. Front Cell Neurosci 2017;11:199. DOI: 10.3389/fncel.2017.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Samaiya PK, Krishnamurthy S, Kumar A. Mitochondrial dysfunction in perinatal asphyxia: Role in pathogenesis and potential therapeutic interventions. Mol Cell Biochem 2021;476(12):4421–4434. DOI: 10.1007/s11010-021-04253-8. [DOI] [PubMed] [Google Scholar]
- 357.Wang Z, Jiang H, Chen S, et al. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012;148(1–2):228–243. DOI: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 358.Morais VA, Haddad D, Craessaerts K, et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 2014;344(6180):203–207. DOI: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 359.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008;183(5):795–803. DOI: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Yoo SM, Jung YK. A molecular approach to mitophagy and mitochondrial dynamics. i 2018;41(1):18–26. DOI: 10.14348/molcells.2018.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kim YJ, Choo OS, Lee JS, et al. BCL2 interacting protein 3-like/NIX-mediated mitophagy plays an important role in the process of age-related hearing loss. Neuroscience 2021;455:39–51. DOI: 10.1016/j.neuroscience.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 362.Iriondo MN, Etxaniz A, Varela YR, et al. LC3 subfamily in cardiolipin-mediated mitophagy: A comparison of the LC3A, LC3B and LC3C homologs. Autophagy 2022;18(12):2985–3003. DOI: 10.1080/15548627.2022.2062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Zhang W. The mitophagy receptor FUN14 domain-containing 1 (FUNDC1): A promising biomarker and potential therapeutic target of human diseases. Genes Dis 2021;8(5):640–654. DOI: 10.1016/j.gendis.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Chen M, Chen Z, Wang Y, et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 2016;12(4): 689–702. DOI: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Diaz F, Kotarsky H, Fellman V, et al. Mitochondrial disorders caused by mutations in respiratory chain assembly factors. Semin Fetal Neonatal Med 2011;16(4):197–204. DOI: 10.1016/j.siny.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res 2009;19(7):802–815. DOI: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Szczepanowska J, Malinska D, Wieckowski MR, et al. Effect of mtDNA point mutations on cellular bioenergetics. Biochim Biophys Acta 2012;1817(10):1740–1746. DOI: 10.1016/j.bbabio.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 368.Hage R, Vignal-Clermont C. Leber hereditary optic neuropathy: Review of treatment and management. Front Neurol 2021;12:651639. DOI: 10.3389/fneur.2021.651639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Graham EC, You Y, Yiannikas C, et al. Progressive loss of retinal ganglion cells and axons in nonoptic neuritis eyes in multiple sclerosis: A longitudinal optical coherence tomography study. Invest Ophthalmol Vis Sci 2016;57(4):2311–2317. DOI: 10.1167/iovs.15-19047. [DOI] [PubMed] [Google Scholar]
- 370.Meyerson C, Van Stavern G, McClelland C. Leber hereditary optic neuropathy: current perspectives. Clin Ophthalmol. 2015;9:1165–1176. DOI: 10.2147/OPTH.S62021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Yu-Wai-Man P, Turnbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet2002;39(3):162–169. DOI: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Liang M, Guan M, Zhao F, et al. Leber’s hereditary optic neuropathy is associated with mitochondrial ND1 T3394C mutation. Biochem Biophys Res Commun 2009;383(3):286–292. DOI: 10.1016/j.bbrc.2009.03.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Guo DY, Wang XW, Hong N, et al. A Meta-analysis of the association between different genotypes (G11778A, T14484C and G3460A) of Leber hereditary optic neuropathy and visual prognosis. Int J Ophthalmol 2016;9(10):1493–1498. DOI: 10.18240/ijo.2016.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Shafa Shariat Panahi M, Houshmand M, Tabassi AR. Mitochondrial D-loop variation in leber hereditary neuropathy patients harboring primary G11778A, G3460A, T14484C mutations: J and W haplogroups as high-risk factors. Arch Med Res 2006;37(8):1028–1033. DOI: 10.1016/j.arcmed.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 375.Yoshimi A, Ishikawa K, Niemeyer C, et al. Pearson syndrome: A multisystem mitochondrial disease with bone marrow failure. Orphanet J Rare Dis 2022;17(1):379. DOI: 10.1186/s13023-022-02538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Broomfield A, Sweeney MG, Woodward CE, et al. Paediatric single mitochondrial DNA deletion disorders: An overlapping spectrum of disease. J Inherit Metab Dis 2015;38(3):445–457. DOI: 10.1007/s10545-014-9778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica 2011;96(6):789–792. DOI: 10.3324/haematol.2011.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Wild KT, Goldstein AC, Muraresku C, et al. Broadening the phenotypic spectrum of Pearson syndrome: Five new cases and a review of the literature. Am J Med Genet A 2020;182(2):365–373. DOI: 10.1002/ajmg.a.61433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Khambatta S, Nguyen DL, Beckman TJ, et al. Kearns-Sayre syndrome: A case series of 35 adults and children. Int J Gen Med 2014;7:325–332. DOI: 10.2147/IJGM.S65560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Channer KS, Channer JL, Campbell MJ, et al. Cardiomyopathy in the Kearns-Sayre syndrome. Br Heart J 1988;59(4):486–490. DOI: 10.1136/hrt.59.4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Sabella-Jimenez V, Otero-Herrera C, Silvera-Redondo C, et al. Mitochondrial DNA deletion and duplication in Kearns-Sayre Syndrome (KSS) with initial presentation as Pearson Marrow-Pancreas Syndrome (PMPS): Two case reports in Barranquilla, Colombia. Mol Genet Genomic Med 2020;8(11):e1509. DOI: 10.1002/mgg3.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zhu Q, Chen C, Yao J. Kearns-Sayre syndrome with a novel large-scale deletion: A case report. BMC Ophthalmol 2022;22(1):35. DOI: 10.1186/s12886-021-02224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Paltiel HJ, O’Gorman AM, Meagher-Villemure K, et al. Subacute necrotizing encephalomyelopathy (Leigh disease): CT study. Radiology 1987;162(1 Pt 1):115–118. DOI: 10.1148/radiology.162.1.3786750. [DOI] [PubMed] [Google Scholar]
- 384.Salama M, El-Desouky S, Alsayed A, et al. FOXRED1 silencing in mice: A possible animal model for Leigh syndrome. Metab Brain Dis 2019;34(1):367–372. DOI: 10.1007/s11011-018-0334-z. [DOI] [PubMed] [Google Scholar]
- 385.Takada R, Tozawa T, Kondo H, et al. Early infantile-onset Leigh syndrome complicated with infantile spasms associated with the m.9185 T > C variant in the MT-ATP6 gene: Expanding the clinical spectrum. Brain Dev 2020;42(1):69–72. DOI: 10.1016/j.braindev.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 386.Baertling F, Rodenburg RJ, Schaper J, et al. A guide to diagnosis and treatment of Leigh syndrome. J Neurol Neurosurg Psychiatry 2014;85(3):257–265. DOI: 10.1136/jnnp-2012-304426. [DOI] [PubMed] [Google Scholar]
- 387.Walker MA, Miranda M, Allred A, Mootha VK. On the dynamic and even reversible nature of Leigh syndrome: Lessons from human imaging and mouse models. Curr Opin Neurobiol 2022;72:80–90. DOI: 10.1016/j.conb.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Arii J, Tanabe Y. Leigh syndrome: Serial MR imaging and clinical follow-up. AJNR Am J Neuroradiol 2000;21(8):1502–1509. PMID: 11003287. [PMC free article] [PubMed] [Google Scholar]
- 389.Lai LM, Gropman AL, Whitehead MT. MR Neuroimaging in pediatric inborn errors of metabolism. Diagnostics (Basel) 2022;12(4):861. DOI: 10.3390/diagnostics12040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ardissone A, Bruno C, Diodato D, et al. Clinical, imaging, biochemical and molecular features in Leigh syndrome: A study from the Italian network of mitochondrial diseases. Orphanet J Rare Dis 2021;16(1):413. DOI: 10.1186/s13023-021-02029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Parfait B, Chretien D, Rotig A, et al. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum Genet2000;106(2):236–243. DOI: 10.1007/s004390051033. [DOI] [PubMed] [Google Scholar]
- 392.Saneto RP. Alpers-Huttenlocher syndrome: The role of a multidisciplinary health care team. J Multidiscip Healthc 2016;9: 323–333. DOI: 10.2147/JMDH.S84900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Saneto RP, Cohen BH, Copeland WC, et al. Alpers-Huttenlocher syndrome. Pediatr Neurol 2013;48(3):167–178. DOI: 10.1016/j.pediatrneurol.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sofou K, Kollberg G, Holmstrom M, et al. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genomic Med 2015;3(1):59–68. DOI: 10.1002/mgg3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Fine AS, Nemeth CL, Kaufman ML, et al. Mitochondrial aminoacyl-tRNA synthetase disorders: An emerging group of developmental disorders of myelination. J Neurodev Disord 2019;11(1):29. DOI: 10.1186/s11689-019-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Henry C, Patel N, Shaffer W, et al. Mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes-MELAS syndrome. Ochsner J. Fall 2017;17(3):296–301. PMID: 29026367. [PMC free article] [PubMed] [Google Scholar]
- 397.Schild C, Hahn D, Schaller A, et al. Mitochondrial leucine tRNA level and PTCD1 are regulated in response to leucine starvation. Amino Acids 2014;46(7):1775–1783. DOI: 10.1007/s00726-014-1730-2. [DOI] [PubMed] [Google Scholar]
- 398.Mustafa MF, Fakurazi S, Abdullah MA, et al. Pathogenic mitochondria DNA mutations: Current detection tools and interventions. Genes (Basel) 2020;11(2):192. DOI: 10.3390/genes11020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.El-Hattab AW, Adesina AM, Jones J, et al. MELAS syndrome: Clinical manifestations, pathogenesis, and treatment options. Mol Genet Metab 2015;116(1–2):4–12. DOI: 10.1016/j.ymgme.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 400.Gyllenhammer LE, Entringer S, Buss C, et al. Developmental programming of mitochondrial biology: A conceptual framework and review. Proc Biol Sci 2020;287(1926):20192713. DOI: 10.1098/rspb.2019.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Pizzorno J. Mitochondria-fundamental to life and health. Integr Med (Encinitas) 2014;13(2):8–15. PMID: 26770084. [PMC free article] [PubMed] [Google Scholar]
- 402.Yue Y, Ren L, Zhang C, et al. Mitochondrial genome undergoes de novo DNA methylation that protects mtDNA against oxidative damage during the peri-implantation window. Proc Natl Acad Sci USA 2022;119(30):e2201168119. DOI: 10.1073/pnas.2201168119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Hanley B, Dijane J, Fewtrell M, et al. Metabolic imprinting, programming and epigenetics – A review of present priorities and future opportunities. Br J Nutr 2010;104(Suppl 1):S1–S25. DOI: 10.1017/S0007114510003338. [DOI] [PubMed] [Google Scholar]
- 404.Kanungo S, Morton J, Neelakantan M, et al. Mitochondrial disorders. Ann Transl Med 2018;6(24):475. DOI: 10.21037/atm.2018.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Angelini C, Bello L, Spinazzi M, et al. Mitochondrial disorders of the nuclear genome. Acta Myol 2009;28(1):16–23. PMID: 19772191. [PMC free article] [PubMed] [Google Scholar]
- 406.Apostolova N, Victor VM. Molecular strategies for targeting antioxidants to mitochondria: Therapeutic implications. Antioxid Redox Signal 2015;22(8):686–729. DOI: 10.1089/ars.2014.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]