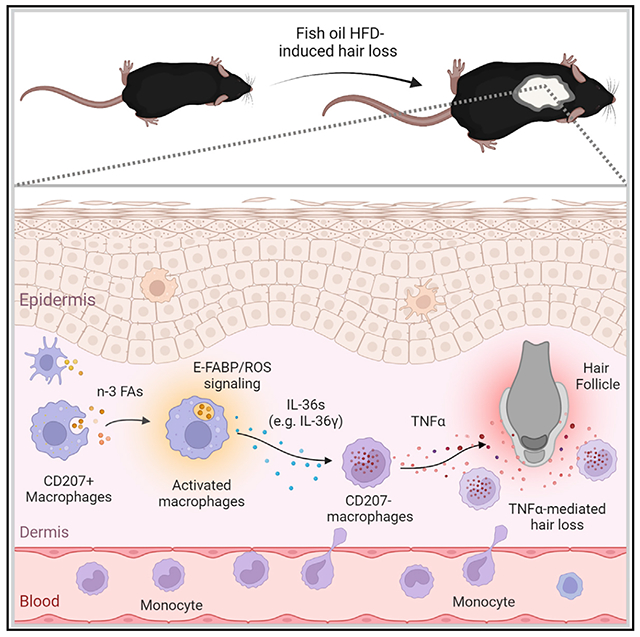
SUMMARY
Fats are essential in healthy diets, but how dietary fats affect immune cell function and overall health is not well understood. Mimicking human high-fat diets (HFDs), which are rich in different fatty acid (FA) components, we fed mice various HFDs from different fat sources, including fish oil and cocoa butter. Mice consuming the fish oil HFD exhibit a hair-loss phenotype. Further studies show that omega-3 (n-3) FAs in fish oil promote atypical infiltration of CD207− (langerin−) myeloid macrophages in skin dermis, which induce hair loss through elevated TNF-α signaling. Mechanistically, epidermal fatty acid binding protein (E-FABP) is demonstrated to play an essential role in inducing TNF-α-mediated hair loss by activating the n-3 FA/ROS/IL-36 signaling pathway in dermal resident macrophages. Absence of E-FABP abrogates fish oil HFD-induced murine hair loss. Altogether, these findings support a role for E-FABP as a lipid sensor mediating n-3 FA-regulated macrophage function and skin health.
Graphical abstract
In brief
Dietary fats regulate immune cell function and skin health. Hao et al. show that high-fat diets rich in fish oil, but not cocoa butter, induce hair loss in mouse models. They identify that E-FABP expressed in skin macrophages plays an essential role in mediating fish oil-induced effects.
INTRODUCTION
Fats are essential for human health.1,2 Dietary fatty acids (FAs) are saturated (e.g., 16:0 palmitic acid, PA), monounsaturated (e.g., 18:1 oleic acid, OA), or polyunsaturated (e.g., 18:2 linoleic acid, LA). Depending on the double-bond position counting from the methyl end of the FA carbon chain, polyunsaturated FAs (PUFAs) include omega-6 (n-6) and omega-3 (n-3) FAs. It is generally believed that intake of n-3 FAs (e.g., docosapentaenoic acid, DPA, or eicosapentaenoic acid, EPA) inhibits inflammation and prevents inflammatory diseases,3,4 whereas saturated fat ingestion increases chronic inflammation and promotes obesity-associated diseases, including cardiovascular disease, type II diabetes, and multiple types of cancer.5,6 However, mounting evidence from recent randomized clinical trials does not support the favorable benefits of dietary n-3 FA supplements,7,8 and the controversy of saturated fat on adverse health effects also appears far from conclusive.9 These conflicting results highlight the mechanistic gap in how metabolism of different fats, in particular n-3 and saturated FAs, regulates cellular function and inflammatory responses in vivo.
Fats are insoluble in aqueous environments. FA binding proteins (FABPs) are a group of evolutionarily conserved proteins evolved to facilitate FA transport and responses across species.10 The FABP family consists of at least nine members, which were named according to their distinct pattern of tissue distribution. For example, epidermal FABP (E-FABP, also known as FABP5) is mainly expressed in the skin, while adipose FABP (A-FABP, also known as FABP4) is predominantly expressed in adipose tissue. Recent studies from our group and others have demonstrated that, in addition to the tissues where they were originally identified, some FABP members are also expressed in different subsets of immune cells, regulating immune cell lipid metabolism and function.11–13 In the skin, E-FABP expression in skin macrophages promotes saturated FA-induced IL-1β production and instigates chronic inflammatory skin lesions. In the tumor stroma, A-FABP expression in tumor-associated macrophages enhances oncogenic IL-6 signaling and promotes mammary tumor growth and progression.14 Thus, FABPs have been recognized to play a critical role in immunoregulation via integration of lipid metabolic and inflammatory signals, especially in obesity-associated diseases.10
Characterized by phagocytosis, macrophages are known to perform an array of accessory functions, including antigen presentation, inflammatory cytokine production, and recruitment and activation of other immune cells.15 There is a growing realization that macrophages are critical in lipid metabolism, especially in exogenous dietary lipid uptake and processing in the setting of obesity.16,17 While lipid-laden macrophage-derived foam cells are believed to be essential for the occurrence and development of atherosclerosis, the detailed mechanisms of lipid uptake and processing in macrophages are still unclear.18 Depending on the tissue, some macrophages (e.g., skin, liver) are believed to be more specialized in lipid metabolism than others,19,20 but how these tissue macrophages metabolize different dietary fats and whether FABPs are essential in mediating individual FA metabolism and responses remain to be further determined.
In our recent studies, we fed mice either a fish oil high-fat diet (HFD) or a cocoa butter HFD to determine whether n-3 or saturated FAs exhibited different effects on diet-induced murine obesity and obesity-associated mammary tumor growth.17 Serendipitously, we observed that mice fed the fish oil HFD exhibited significant hair loss, while littermates fed the cocoa butter HFD or control low-fat diet (LFD) did not. Further analysis demonstrated that n-3 FAs specifically activated reactive oxygen species (ROS)/IL-36 signaling in dermal macrophages, further leading to TNF-α-mediated hair loss. More importantly, we identified E-FABP as an essential lipid sensor in mediating n-3 FA effects and fish oil HFD-induced hair loss.
RESULTS
Consumption of fish oil HFD induces mouse hair loss
To determine the effect of consumption of n-3 or saturated FAs on diet-induced obesity and immune cell function, we custom-made two HFDs (45% fat), with the fat source being either fish oil or cocoa butter, and one LFD (5% fat). All these diets contained identical base ingredients except for the fat components (see food components in Table S1). The cocoa butter HFD was rich in saturated FAs, whereas the fish oil HFD contained unique n-3 FAs (Figure 1A). When female C57BL/6 littermates were randomly weaned on these diets, both cocoa butter and fish oil HFDs induced significant increases in mouse body weight compared with the LFD after 3 months (Figure 1B). Serendipitously, we observed that a majority of mice on the fish oil HFD exhibited severe hair loss, whereas mice on the cocoa butter HFD or LFD did not show such phenomenon (Figure 1C). Mice fed the fish oil HFD exhibited patches of non-scarring, complete loss of hair shafts accounting for over 20% of the skin area (Figures 1D and 1E). To further verify this finding, we fed male mice the same fish oil HFD, cocoa butter HFD, or LFD. Similar observations were made in that only the male mice on the fish oil HFD developed hair loss, although at a reduced rate compared with female mice (Figures S1A and S1B). These results suggested a hair loss phenotype specifically induced by the fish oil diet. As diet-associated gut dysbiosis has been linked to hair loss,21 we treated female mice on the fish oil HFD with or without antibiotics. Antibiotic treatment had no effect on the increase in body weight (Figure S1C) or the diet-induced hair loss (Figures S1D and S1E). This striking phenotype of hair loss induced by the fish oil HFD, but not by the cocoa butter HFD or LFD, indicated that FA components in dietary fats were critical in determining skin hair health independent of the gut microbiome.
Figure 1. Consumption of fish oil HFD induces hair loss in mice.
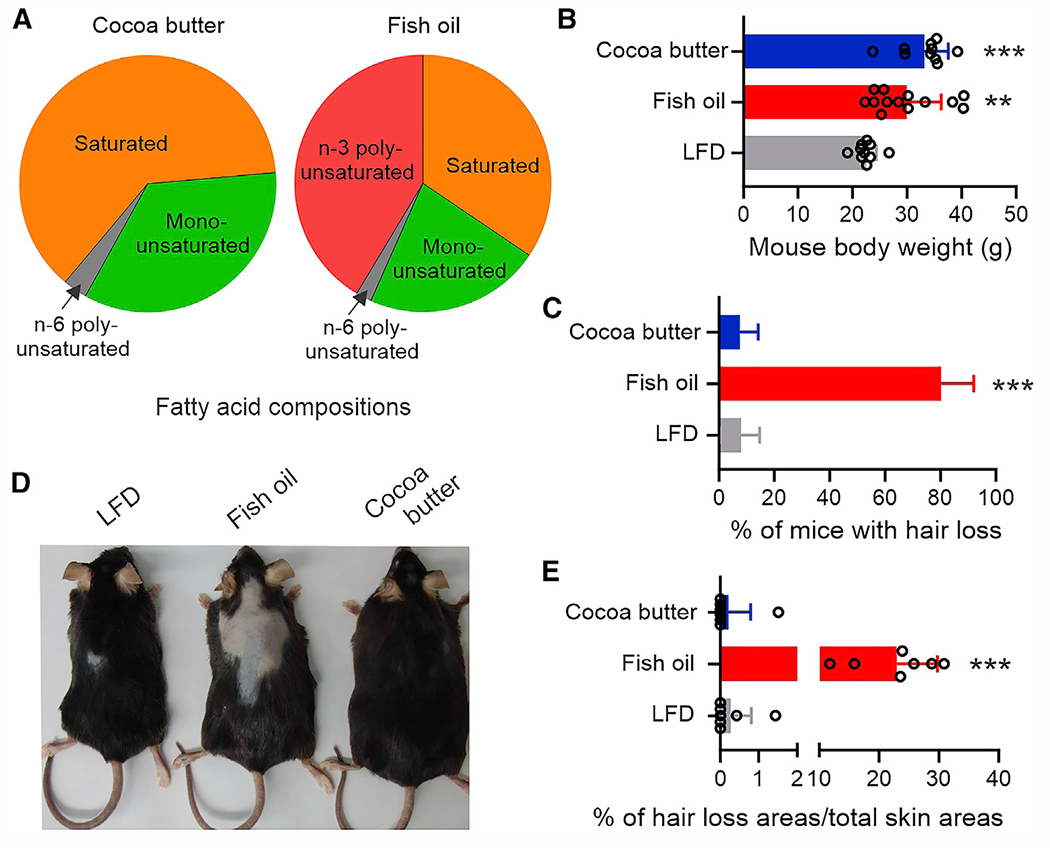
(A) Percentage of FA composition (g/100 g fat) in HFDs from different fat sources.
(B) Body weight of C57BL/6 female littermates fed the cocoa butter HFD (n = 13), fish oil HFD (n = 10), or LFD (n = 10) for 3 months.
(C) Hair loss percentage of female mice after 3 months on LFD (n = 25), fish oil HFD (n = 19), or cocoa butter HFD (n = 28).
(D) Representative pictures of mouse hair loss on LFD or fish oil or cocoa butter diet for 3 months.
(E) Quantification of hair loss areas in comparison with the total skin areas (n = 7/group). Data are shown as mean ± SD in (B), (C), and (E) (**p = 0.0018, ***p < 0.001, compared with LFD group; unpaired Student’s t test). See also Figure S1.
Fish oil HFD induces langerin-negative macrophage accumulation in skin dermis
To determine if consumption of different diets induced alterations in the immune environment in the skin, we first compared the skin histology of mice fed the different diets. We found that mice fed the fish oil, but not cocoa butter or LFD, had a significant accumulation of mononuclear inflammatory infiltrates surrounding hair follicles in the dermis (Figure 2A), suggesting a possible inflammation-mediated hair follicle inhibition with the fish oil diet. Consistent with the histological data, we observed that a cell population with non-specific autofluorescent staining had accumulated in the skin dermis of fish oil HFD-fed mice, but not in cocoa butter-or LFD-fed mice, by flow cytometric analysis (Figure 2B). The autofluorescent characteristic of these cells was mainly detected by lasers (UV, violet, and blue) with shorter light wavelengths (e.g., antibodies labeled with FITC, PE, Pacific blue, BV605, etc.), but not by the red laser with longer light wavelengths (e.g., antibodies labeled with APC or Alexa Fluor 700) (Figures 2C and S2A). To further characterize the phenotype of the autofluorescent cells, we purified them with a flow sorter and analyzed them using high-dimensional time-of-flight mass cytometry (CyTOF), which eliminated the interference of autofluorescence by detecting antibody-coupled metal isotopes. Compared with unpurified dermal populations (Figure 2D), purified autofluorescent dermal cells were mainly enriched for a langerin (CD207)-negative macrophage phenotype (CD207−CD45+CD11b+F4/80+MHCII+CD11C−CD3−TCRβ−TCRγδ−) (Figures 2E and 2F). Fish oil HFD significantly induced accumulation of this population compared with the LFD or cocoa butter HFD (Figure S2B). Immunohistochemistry (IHC) staining further confirmed that macrophages (Figure 2G), but not CD8+ or CD4+ T lymphocytes (Figures 2H and S2C), extensively accumulated around hair follicles in the skin of fish oil HFD-fed mice. To determine whether these fish oil HFD-induced macrophages were enriched in other tissues, we analyzed their presence in the skin epidermis (Figures S2D and S2E), spleen (Figure S2F), lymph nodes (Figure S2G), and peripheral blood (Figure S2H). Although trending higher in the spleen, the prevalence of these macrophages was not significantly increased in these tissues, suggesting a dermis-specific accumulation of this subset. These results demonstrated that consumption of fish oil HFD specifically induced accumulation of langerin-negative non-skin-resident macrophages in skin dermis.
Figure 2. Fish oil HFD induces accumulation of langerin− macrophages in the skin dermis.
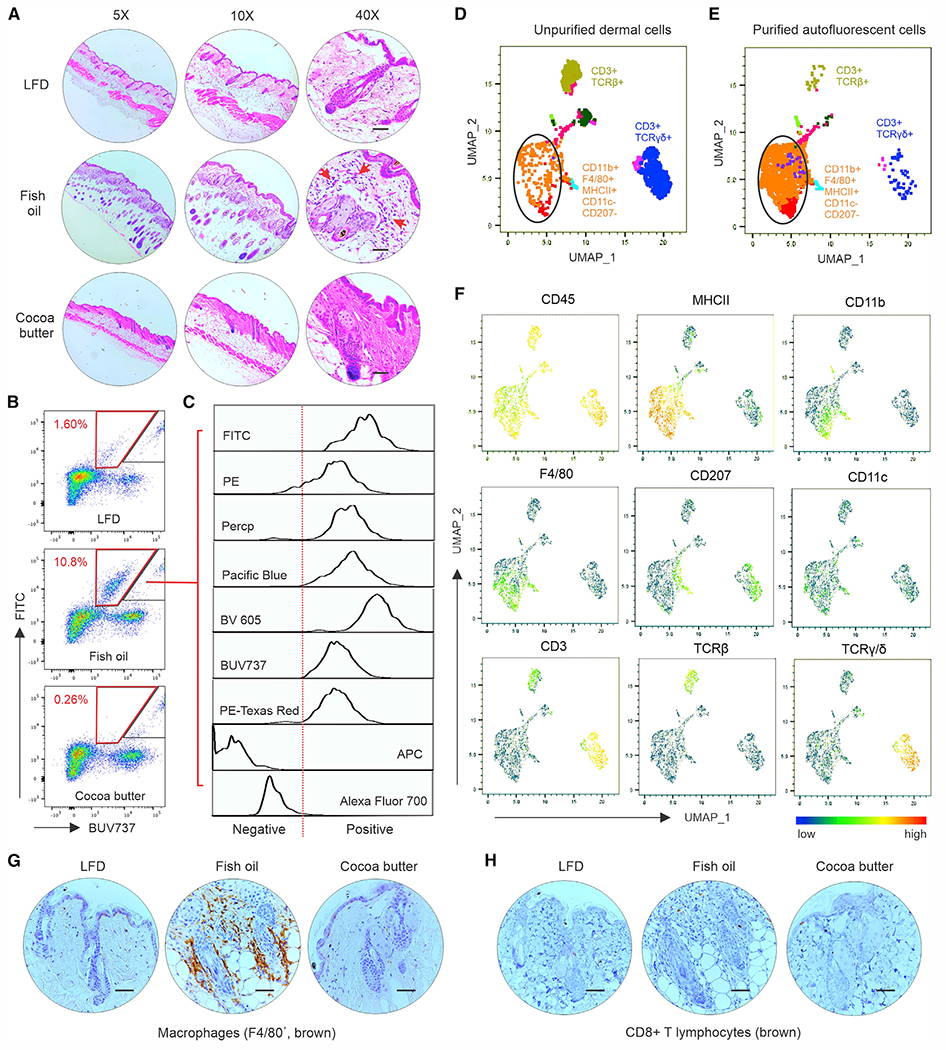
(A) Representative hematoxylin and eosin staining of skin tissues from mice fed the LFD, fish oil HFD, or cocoa butter diet at different magnifications (red arrows: infiltrating inflammatory cells). Scale bars, 100 μm.
(B and C) Flow cytometric analysis of immune cell phenotype in the dermis of mice on different diets for 3 months. A cell population with strong autofluorescence (red trapezoid gate) was specifically accumulated in the dermis of mice fed the fish oil HFD (B). Multichannel signals of the autofluorescent cells were analyzed using a BD Fortessa flow cytometer (C).
(D–F) Autofluorescent cells in the dermis of fish oil HFD-fed mice were purified using a BD FACSAria II flow sorter and stained with a panel of metal-tagged CyTOF antibodies. Uniform manifold approximation and projection (UMAP) was used to visualize and identify immune cell populations in unsorted dermal cells (D) and sorted autofluorescent dermal cells (E). Individual surface marker signatures in the CyTOF panel are shown in (F).
(G and H) Representative IHC images of F4/80+ macrophages (brown staining, G) and CD8+ T cells (brown staining, H) in the skin of mice fed the LFD, fish oil HFD, or cocoa butter HFD. Scale bars, 100 μm. See also Figure S2. These in vitro experiments were repeated with at least three biological replicates.
Fish oil-induced dermal macrophages mediate hair loss by TNF-α signaling
We next evaluated the function of the fish oil HFD-induced dermal macrophages by CyTOF surface and intracellular staining. As shown in Figures 3A and 3B, there were several main immune populations in the skin of mice fed the fish oil HFD, including CD207+CD11c+ macrophages (pop 13), CD207−CD11c− macrophages (pop 18), TCRγδ+ T cells (pops 0, 1, and 3), and TCRβ+ T cells (pop 4). Interestingly, fish oil HFD-induced macrophages (pop 18) mainly expressed TNF-α, whereas CD3+CD69+ TCRγδdim (pop 3) and CD3+TCRβ+CX3CR1dim (pop 4) mainly expressed IL-17 and IFNγ, respectively. Compared with LFD- or cocoa butter HFD-fed mice, fish oil-fed mice exhibited similar frequencies of IFNγ- and IL-17-producing dermal T cells (Figures 3C and 3D), but significantly higher frequencies of TNF-α-producing dermal CD207− macrophages (Figure 3E). IHC staining further confirmed that TNF-α production was upregulated in fish oil HFD-fed mice, but not in LFD-or cocoa butter HFD-fed mice (Figure 3F). We speculated that the TNF-α produced by fish oil-induced macrophages might drive the hair loss. As hair follicle stem cells (HFSCs) are essential to hair growth,22 we first treated mouse HFSCs with TNF-α and measured their proliferation, stemness, and cell apoptosis. Interestingly, TNF-α treatment significantly inhibited expression of cyclin B1, Ki67, and ALDH1 in mouse HFSCs (Figures S3A–S3C). Moreover, TNF-α (Figures 3G and 3H), but not IFNγ (Figures S3D and S3E) or IL-17 (Figures S3F and S3G), directly induced HFSC apoptosis. Second, we evaluated the direct cytotoxic effect of macrophages by coculturing them with HFSCs in the presence or absence of anti-TNF-α blocking antibody. Macrophage-mediated HFSC cytotoxicity was significantly inhibited by anti-TNF-α antibody (Figure 3I), suggesting a specific role for macrophage-derived TNF-α in mediating HFSC cytotoxicity in vitro. By analyzing CD34+ dermal HFSCs in vivo, we found that mice fed the fish oil HFD exhibited a significant increase in apoptotic CD34+ HFSCs compared with mice on LFD or cocoa butter HFD (Figure 3J). To further confirm the role of TNF-α in fish oil HFD-induced hair loss, we treated these hair-loss mice with anti-TNF-α antibody in vivo. Strikingly, compared with mice treated with vehicle PBS control, mice treated with anti-TNF-α antibody successfully recovered from fish oil HFD-induced hair loss (Figures 3K and 3L). IHC staining further demonstrated that perifollicular macrophage cells (brown) were abrogated by anti-TNF-α antibody treatment (Figure S3H). As n-3 FAs (e.g., DPA or EPA) in the fish oil diet did not directly affect HFSC viability (Figures S3I and S3J), our results indicated that TNF-α signaling generated by dermal CD207− macrophages represented a main molecular mechanism underlying fish oil HFD-induced murine hair loss.
Figure 3. Fish oil HFD induces murine hair loss by activating TNF-α signaling.
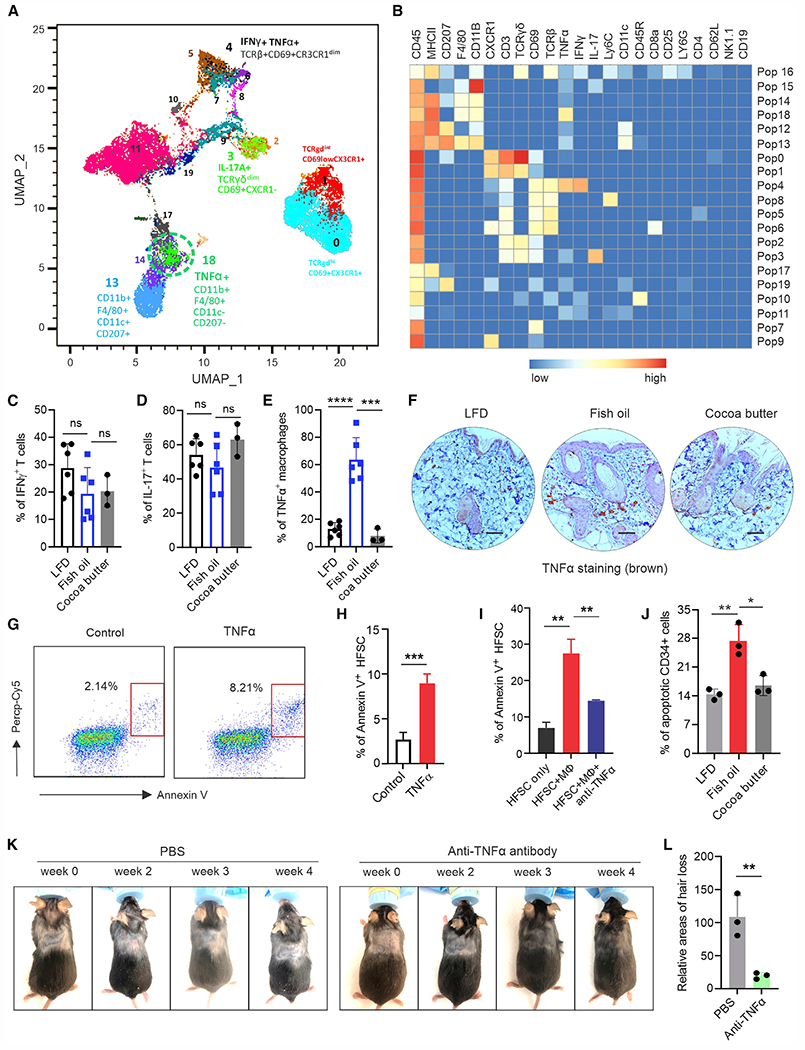
(A and B) Surface and intracellular staining of skin immune cell signatures using CyTOF-based analysis. (A) Representative plot of immune cell populations from fish oil HFD-fed mice. (B) Heatmap of the indicated markers across the 20 cell populations in the skin.
(C and D) Intracellular staining of IFNγ (C) and IL-17 (D) in skin T cells from mice fed the LFD (n = 5), fish oil HFD (n = 5), or cocoa butter HFD (n = 3).
(E) Intracellular staining of TNF-α production in CD207− dermal macrophages from mice fed the LFD (n = 5), fish oil HFD (n = 5), or cocoa butter HFD (n = 3).
(F) Representative IHC images of TNF-α production (brown) in the skin of mice fed the LFD, fish oil HFD, or cocoa butter HFD. Scale bars, 100 μm.
(G and H) Flow cytometric analysis of TNF-α-induced apoptosis of HFSCs using Annexin V staining (G). Average percentage of apoptotic HFSCs in three replicates is shown in (H).
(I) Analysis of bone marrow-derived macrophage-mediated cytotoxicity to HFSCs in the presence or absence of anti-TNF-α blocking antibody (1 μg/mL) in vitro (n = 3/group).
(J) Analysis of in vivo apoptosis of dermal CD34+ HFSCs in mice fed the LFD, fish oil HFD, or cocoa butter HFD for 3 months (n = 3/group).
(K and L) Mice with fish oil HFD-induced hair loss (n = 6) were subcutaneously treated with Ultra-LEAF purified anti-TNF blocking antibody (10 mg/kg) or PBS for 4 weeks (twice/week). Representative pictures of hair regrowth in each group are shown in (K). Quantification of hair loss areas is shown in (L). Data are shown as mean ± SD in (C), (D), (E), (H), (I), (J), and (L) (****p < 0.0001, ***p < 0.001, **p < 0.01, *p = 0.0159; ns, non-significant, unpaired Student’s t test). See also Figure S3.
TNF-α signaling in dermal macrophages is activated by the n-3 FA/IL-36 axis
To dissect how consumption of fish-oil HFD activated TNF-α signaling in the accumulated dermal macrophages, we speculated that the unique n-3 FAs in the fish oil diet (Figure 1A) might cause the effect. As cocoa butter HFD, which was rich in saturated FAs, did not cause hair loss, we first fluorescently labeled n-3 FAs (e.g., DPA) and saturated FAs (e.g., PA) and tracked their distribution in vivo. Near-infrared fluorescence imaging showed that DPA and PA exhibited similar distributions in metabolic organs (e.g., liver, kidney), but not in the fat tissue. Specifically, PA was predominantly accumulated in epididymal fat, whereas DPA was mainly enriched in subcutaneous fat (Figure 4A), suggesting a specific skin distribution pattern of n-3 FAs.
Figure 4. n-3 FAs activate IL-36/TNF-α signaling in skin macrophages.
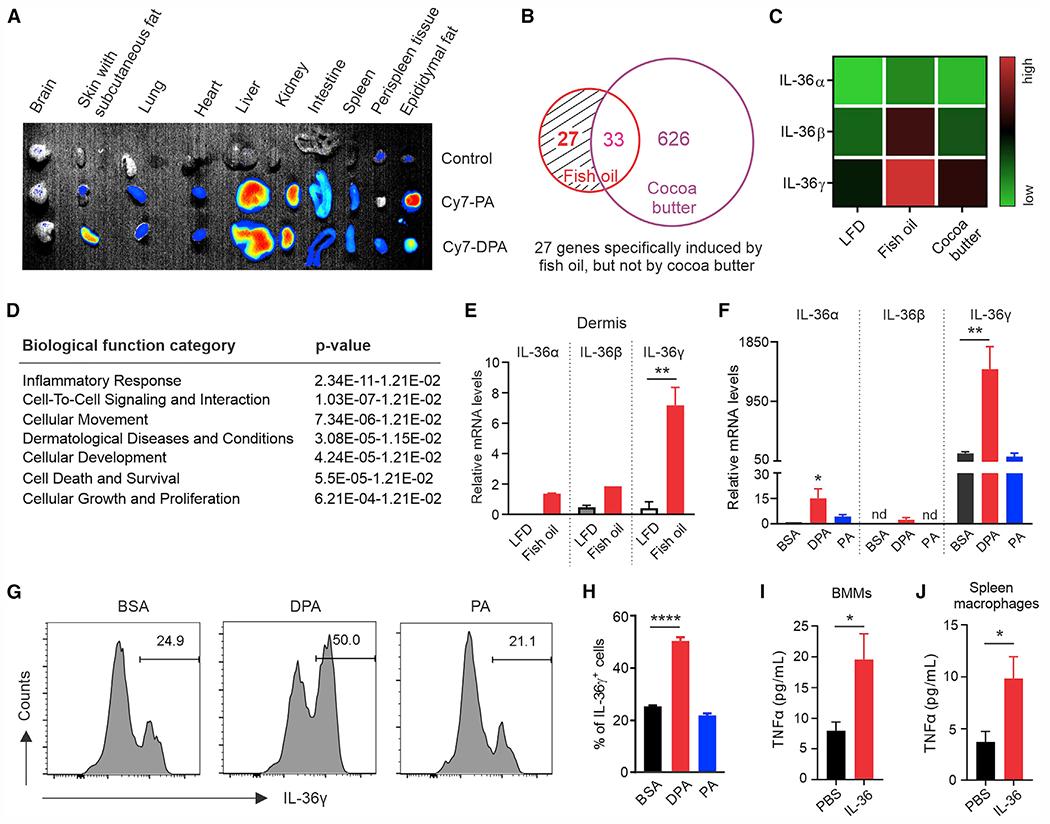
(A) In vivo trafficking: the distribution of fluorescence-labeled dietary FAs (PA and DPA) in different tissues of mice.
(B) RNA-sequencing analysis of differentially expressed genes upregulated in the skin of mice fed the fish oil HFD or cocoa butter HFD compared with the LFD-fed mice (n = 3/group).
(C) Heatmap of IL-36 genes in skin of mice fed the LFD, fish oil HFD, or cocoa butter HFD, respectively.
(D) Analysis of IL-36-involved biological functions by Ingenuity pathway analysis (IPA).
(E) Real-time PCR analysis of IL-36 gene expression in the dermis of mice fed the LFD or fish oil HFD (n = 3/group).
(F) Analysis of expression levels of IL-36 genes in CD11c+ macrophages treated with DPA (200 μM), PA (200 μM), or BSA control for 24 h by real-time PCR (n = 3/group).
(G and H) Flow intracellular staining of IL-36γ expression in CD11c+ macrophages treated with DPA (200 μM), PA (200 μM), or BSA control for 24 h. Average percentage of IL-36γ+ macrophages is shown in (H).
(I and J) Measurement of TNF-α production by bone marrow-derived CD11c− macrophages (BMMs) (I) or splenic macrophages (J) treated with or without IL-36 (100 ng/mL) for 3 h by ELISA (n = 3/group). All in vitro experiments were repeated with at least three biological replicates. Data are shown as mean ± SD in (E), (F), (H), (I), and (J) (****p < 0.0001, **p < 0.01, *p < 0.05, unpaired Student’s t test). See also Figure S4.
To further investigate the underlying molecular mechanisms of the n-3 FA-induced effect, we performed RNA-sequencing analysis using skin tissues from mice fed the LFD or fish oil or cocoa butter HFD. Compared with LFD mice, there were 686 differentially expressed genes induced by HFDs, among which 27 genes were specifically upregulated in the fish oil HFD-fed mice (Figures 4B and S4A). Of the 27 genes, three members of the IL-36 gene family, IL-36α, IL-36β, and IL-36γ (Figure 4C), attracted our attention, due to the extensive involvement in the top-ranked pathways (24 of 30 top pathways) in the Ingenuity Canonical Pathways analysis (Table S2). Biological function analyses further supported that IL-36 genes were mainly involved in the inflammatory response, cell-to-cell signaling and interaction, dermatological disease, and cellular proliferation (Figure 4D, Table S3). Using quantitative PCR, we identified that the fish oil-upregulated IL-36 genes in the skin (Figure S4B) were mainly from the dermis (Figure 4E), and not from the epidermis (Figure S4C).
As CD207+CD11c+ macrophages (pop 13 in Figures 3A and 3B) represented a main resident population in the skin regardless of fish oil HFD, we observed that this population also surrounded hair follicles in fish oil-fed, but not in LFD- or cocoa butter diet-fed mice (Figure S4D), suggesting a potential role for these skin-resident macrophages as the first line of defense when exposed to enriched n-3 FAs in the skin. Indeed, n-3 DPA, but not saturated PA, induced significant expression of IL-36 genes, especially IL-36γ, in CD11c+ macrophages (Figure 4F). Higher levels of IL-36γ protein were also detected in CD11c+ macrophages when treated with DPA, but not with PA (Figures 4G and 4H), indicating dermal-resident CD11c+ macrophages as a potent source of IL-36 in response to n-3 FA skin accumulation. Given that IL-36 signaling is well known to promote myeloid macrophage infiltration and activation in skin inflammation,23,24 we treated macrophages from different myeloid sources (e.g., spleen, bone marrow derived) with IL-36 and demonstrated that IL-36 treatment significantly induced TNF-α production in these myeloid-derived macrophages (Figures 4I and 4J). These data supported a critical role for the n-3 FA/IL-36 axis in activating TNF-α signaling in CD207− myeloid macrophages, thus leading to hair loss in fish oil HFD-fed mice.
n-3 FAs induce IL-36 production via ROS
To further determine how n-3 FAs specifically induced IL-36γ expression in CD11c+ macrophages, we treated them with DPA, PA, or BSA control and compared the expression pattern of multiple FA metabolism-related cytokines (TNF-α, IL-6, IL-1β), enzymes (ALOX5, COX2, iNOS, Arg1), chemokines (CXCL1, CXCL2, CCL2, CCL5), and immune checkpoint molecules (PD-1, PD-L1, PD-L2) (Figure S5A). However, we did not observe any significant differences among these factors that could explain the upregulation of IL-36 genes in response to n-3 FA treatment. Instead, we noticed that DPA-treated CD11c+ macrophages exhibited dramatic lipid accumulation compared with PA- or BSA-treated cells (Figure 5A). Moreover, DPA-induced lipid accumulation significantly induced the generation of ROS (Figures 5B and 5C), which could be abrogated by N-acetyl-L-cysteine (NAC), a commonly used ROS inhibitor, and diphenyleneiodonium (DPI), a specific inhibitor of NADPH oxidase (Figure 5D). Similar observations were confirmed when other n-3 FAs, such as EPA, were used to treat CD11c+ macrophages (Figures S5B–S5F), suggesting that n-3 FAs, but not saturated FAs, induced a specific NADPH oxidase/ROS response in skin macrophages. In line with these observations, n-3 FA-induced IL-36γ production in macrophages was blocked at both the mRNA (Figure 5E) and the protein (Figure 5F) levels when ROS were inhibited by DPI. These data demonstrated a pivotal role for n-3 FA-induced ROS signaling in IL-36 production in macrophages.
Figure 5. ROS mediates n-3 FA-induced IL-36 signaling in skin macrophages.
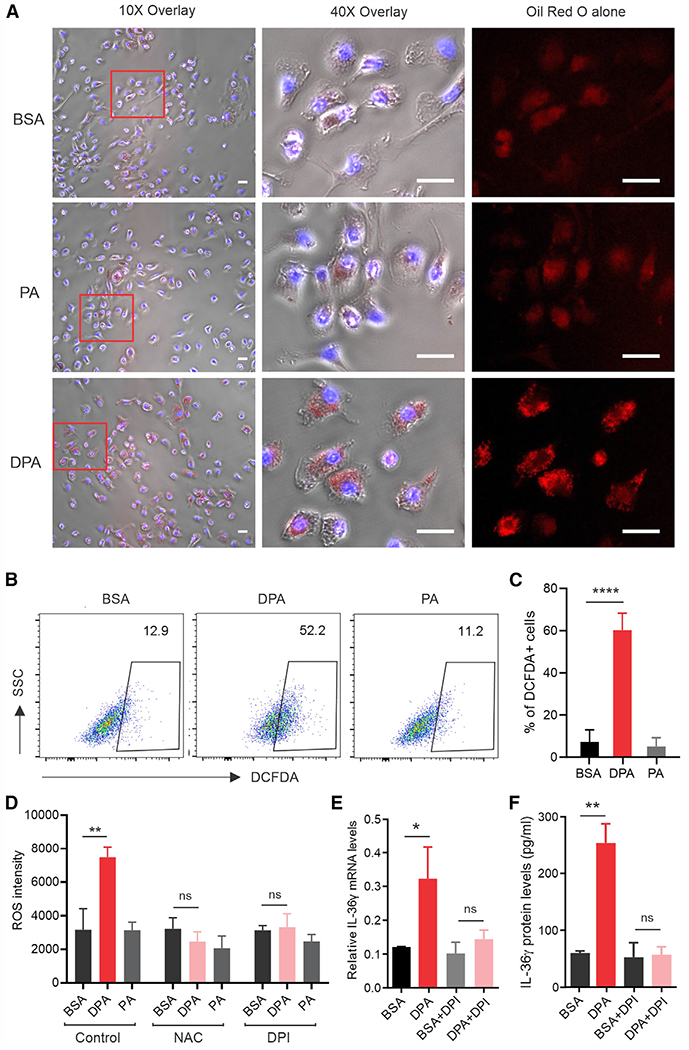
(A) CD11c+ macrophages were treated with 200 μM DPA, PA, or BSA. Oil red O staining was performed to observe lipid accumulation in these cells. DAPI was used for nucleus staining. Scale bars, 20 μm.
(B and C) Flow cytometric analysis of ROS production in CD11c+ macrophages treated with 200 μM DPA, PA, or BSA. Average percentage of DCFDA+ macrophages is shown in (C) (n = 3/group).
(D) Measurement of ROS intensity in CD11c+ macrophages treated with 200 μM DPA, PA, or BSA in the presence of absence of ROS inhibitor NAC or DPI (n = 3/group).
(E and F) Analysis of IL-36 gene expression (E) and protein levels (F) in CD11c+ macrophages treated with DPA (200 μM) in the presence or absence of the ROS inhibitor DPI (n = 3/group). All in vitro experiments were repeated with at least three biological replicates. Data are shown as mean ± SD in (C), (D), (E), and (F) (****p < 0.0001, **p < 0.01, *p < 0.05; ns, non-significant, unpaired Student’s t test). See also Figure S5.
E-FABP is essential in n-3 FA-induced ROS/IL-36 signaling
Dietary long-chain FAs are insoluble in an aqueous host environment. As such, FABPs evolved to facilitate hydrophobic FA absorption and transport inside cells.10,25 Our previous studies demonstrated that skin CD11c+ macrophages predominantly expressed E-FABP, but not other FABP members.11,26 We thus speculated that E-FABP was essential in mediating n-3 FA-induced effects in skin macrophages. Indeed, when CD11c+ macrophages were treated with DPA or PA in vitro, we noticed that DPA treatment strongly upregulated the expression of E-FABP, but not A-FABP or other FA metabolism-related molecules, such as Cpt1, PPAR, Nrf2, etc. (Figures 6A and S6A). Dermal skin from fish oil HFD-fed mice also showed a significant upregulation of E-FABP, but not A-FABP (Figures 6B and 6C), confirming E-FABP as a major lipid sensor in the skin. More importantly, genetic ablation of E-FABP dramatically impaired DPA-induced ROS production in macrophages as shown by fluorescent DCFDA staining (Figure 6D). Dynamic measurement of ROS production in saturated FA (e.g., PA, SA)- or n-3 FA (e.g., DPA, EPA)-treated macrophages further supported that E-FABP played a pivotal role in n-3 FA-induced ROS production (Figures 6E and S6B). To ascertain why E-FABP was able to mediate n-3 FA-induced ROS responses, we demonstrated that purified recombinant E-FABP (Figure S6C) was able to bind both PA and DPA (Figures S6D and S6E). However, DPA binding increased E-FABP thermal stability, while PA binding did the opposite, suggesting that DPA-induced ROS signaling is dependent on DPA/E-FABP structural stability. Consistent with these data, E-FABP−/− macrophages exhibited significant reduction of DPA-induced IL-36³ gene transcription (Figure 6F) and protein production (Figure 6G) compared with wild-type (WT) macrophages. ROS inhibition eliminated the difference of IL-36γ between WT and E-FABP−/− macrophages (Figure 6G), further supporting an essential role for E-FABP in facilitating n-3 FA-induced ROS/IL-36 signaling.
Figure 6. E-FABP is essential in n-3 FA-induced ROS production.
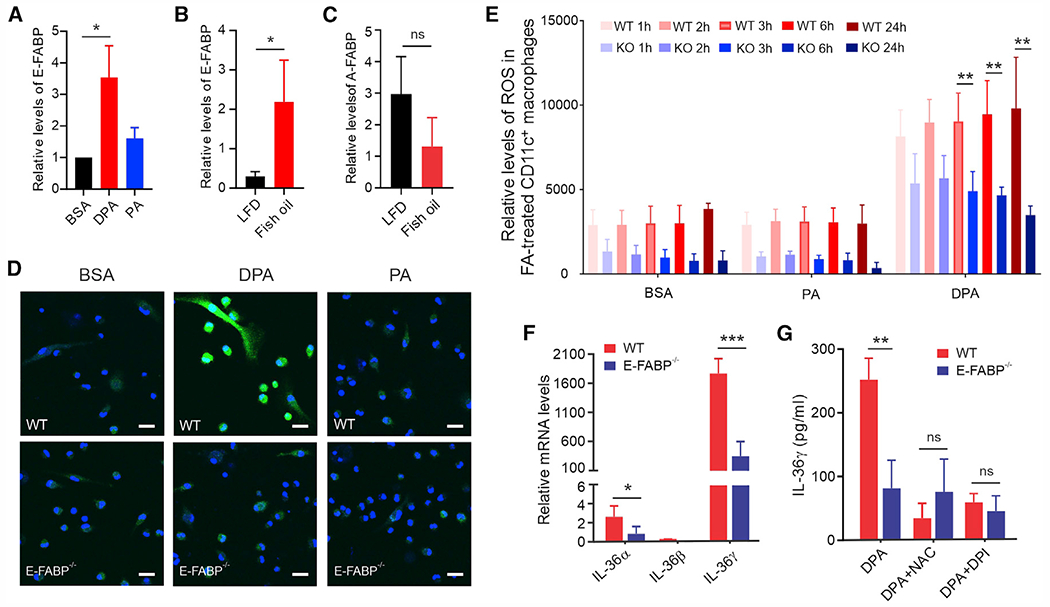
(A) Real-time PCR analysis of E-FABP expression in CD11c+ macrophages treated with 200 μM DPA, PA, or BSA for 24 h (n = 3/group).
(B and C) Measurement of expression levels of E-FABP (B) and A-FABP (C) in the skin tissue of mice fed the LFD or fish oil HFD for 3 months by real-time PCR analysis (n = 3/group).
(D) Confocal microscopy analysis of ROS (green) production in wild-type (WT) and E-FABP-deficient CD11c+ macrophages treated with 200 μM DPA, PA, or BSA control for 1 h (DAPI, blue). Scale bars, 20 μm.
(E) ROS production in WT and E-FABP−/− CD11c+ macrophages treated with 200 μM PA, DPA, or BSA control at the indicated time points (n = 3/group).
(F) Real-time PCR measurement of IL-36 gene expression in WT and E-FABP−/− CD11c+ macrophages treated with 200 μM DPA for 24 h (n = 3/group).
(G) Measurement of IL-36γ levels in supernatants of cultured WT and E-FABP−/− CD11c+ macrophages treated with 200 μM DPA or BSA for 24 h in the presence of ROS inhibitor DPI or NAC by ELISA (n = 3/group). Data are shown as mean ± SD in all except (D) (***p < 0.001, **p < 0.01, *p < 0.05; ns, non-significant, unpaired Student’s t test). See also Figure S6.
E-FABP deficiency abrogates fish oil HFD-induced murine hair loss
Given the findings that E-FABP was essential in mediating n-3 FA-indued ROS/IL-36 responses in macrophages, we reasoned that fish oil HFD-induced hair loss in WT mice should be compromised in E-FABP-deficient mice. To this end, we fed WT and E-FABP−/− mice with fish oil HFD, cocoa butter HFD, or LFD. Strikingly, fish oil HFD-induced hair loss in WT mice was completely abrogated in E-FABP−/− mice (Figures 7A and 7B). Skin histological analysis demonstrated that fish oil-induced accumulation of perifollicular inflammatory cells in the dermis of WT mice was obviously reduced in the E-FABP−/− mice (Figures 7C, S7A, and S7B). CyTOF analysis of immune cell components in the skin of these mice identified at least 14 immune cell populations in the epidermis and dermis, mainly consisting of TCRγδ+ T cells (pops 0 and 1), TCRβ+ T cells (pop 5), CD207+ macrophages (pops 12 and 13), and CD207− macrophages (pop 11) (Figures S7C and S7D). In contrast with epidermal skin (Figures 7D and 7E), CD207− macrophages (pop 11) were specifically distributed in the dermis (Figure 7F), and E-FABP deficiency significantly lowered the infiltration of this population (Figure 7G). IHC staining further confirmed that these perifollicular macrophages in the skin of WT mice were absent when E-FABP was deficient (Figure 7H). Consistent with in vitro functional studies showing that E-FABP deficiency significantly impaired IL-36-induced TNF-α production in macrophages (Figures S7E and S7F), TNF-α was mainly expressed by these CD207− dermal macrophages (Figure 7I), and TNF-α+ macrophages were significantly reduced when E-FABP was deficient (Figure 7J). Of note, consumption of fish oil HFD did not seem to affect circulating levels of E-FABP (Figure S7G) and TNF-α (Figure S7H) in mice, corroborating a local effect mediated by E-FABP expression in the skin. Collectively, our data uncovered an essential role for E-FABP in mediating fish oil HFD-induced hair loss in mice.
Figure 7. E-FABP deficiency abrogates fish oil HFD-induced murine hair loss.
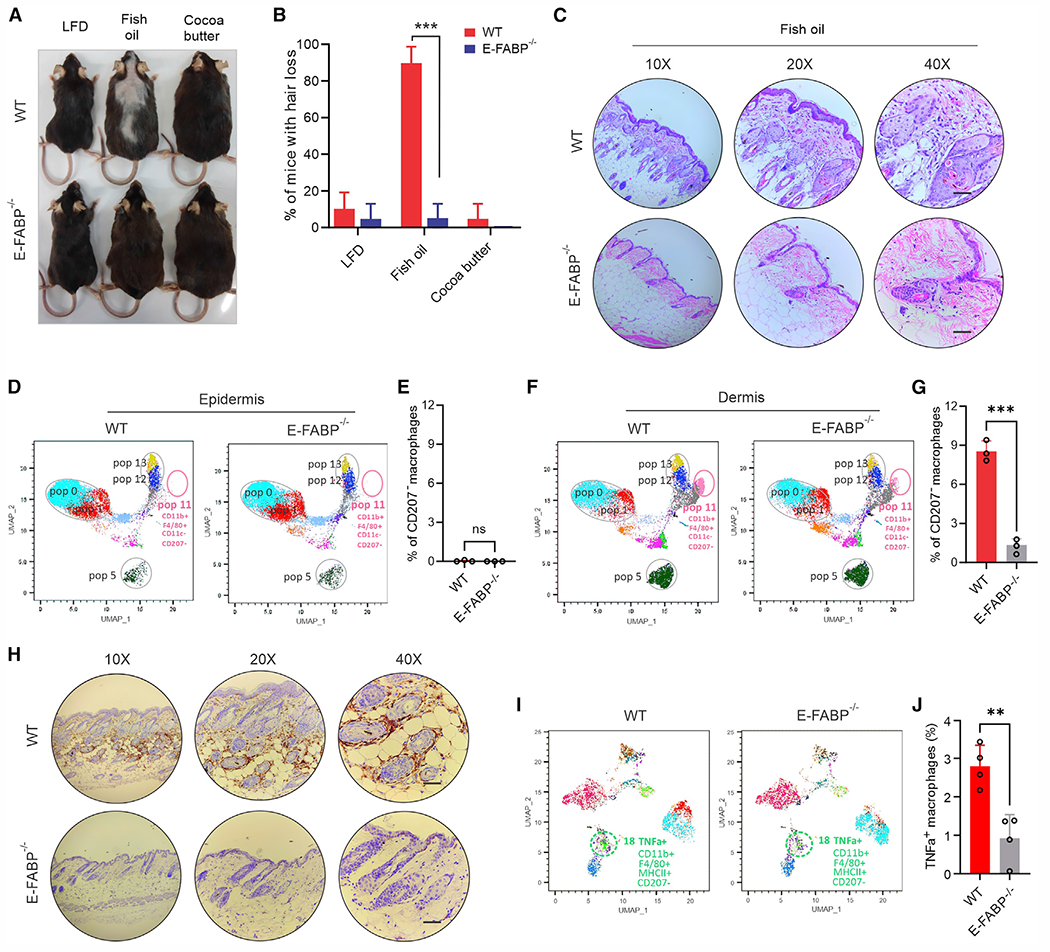
(A) Representative hair-loss pictures of WT and E-FABP−/− mice fed the LFD, fish oil HFD, or cocoa butter HFD for 3 months.
(B) Average percentage of hair loss in WT or E-FABP−/− mice after feeding the LFD, fish oil HFD, or cocoa butter HFD for 3 months (n = 20/group).
(C) Representative hematoxylin and eosin staining of skin tissues from WT and E-FABP−/− mice fed the fish oil HFD for 3 months. Scale bars, 100 μm.
(D and E) CyTOF-based surface staining of epidermal immune cell signatures in WT and E-FABP−/− mice fed the fish oil HFD for 3 months. Average percentage of CD207− macrophages in the epidermis is shown in (E) (n = 3/group).
(F and G) CyTOF-based surface staining of dermal immune cell signatures in WT and E-FABP−/− mice fed the fish oil HFD for 3 months. Average percentage of CD207− macrophages in the dermis is shown in (G) (n = 3/group).
(H) Representative IHC images of F4/80+ macrophages in the skin of WT and E-FABP−/− mice fed the fish oil HFD for 3 months. Scale bars, 100 μm.
(I and J) CyTOF-based intracellular staining of dermal immune cell signatures in WT and E-FABP−/− mice fed the fish oil HFD for 3 months. Average percentage of TNF-α+ CD207− macrophages in the dermis is shown in (J) (n = 3/group). Data are shown as mean ± SD in (B), (E), (G), and (J) (***p < 0.001, **p < 0.01; ns, non-significant, unpaired Student’s t test). See also Figure S7.
DISCUSSION
n-3 FAs in fish oil are generally believed to have multiple health benefits, such as promoting brain and skin health, reversing/preventing inflammation, lowering blood pressure, and reducing cardiovascular diseases.27 Our own studies also demonstrated that consumption of fish oil HFD exhibited an anti-tumor effect when compared with cocoa butter HFD.17 However, fish oil diets have many downstream effects. The possibility exists that not all may be considered favorable. In the current studies, we reported a serendipitous but consistent observation that consumption of fish oil HFD induced substantial hair loss in murine models. Further analysis uncovered a molecular mechanism underlying this intriguing phenotype.
Dietary fats mainly composed of long-chain FAs are absorbed and transported in the form of chylomicrons via lymphatic vessels. After draining into large veins, chylomicrons are delivered throughout the body and delipidated through the action of lipoprotein lipases (LPLs) on the endothelial surface of the capillaries in peripheral tissues.28 Depending on the distribution and organ-specific LPL actions, different tissues exhibit different capabilities and preferences in lipid uptake.29 When we tracked the distribution of n-3 FAs and saturated FAs in vivo, we observed that saturated FAs were predominantly distributed in the epididymal visceral fat, while n-3 FAs were accumulated more in skin subcutaneous fat. Given that visceral fat is more metabolically active than subcutaneous fat,30 it is not surprising to see that consumption of cocoa butter HFD was associated with elevated systemic inflammation and accelerated tumor growth when compared with fish oil HFD.17 However, when mice were fed fish oil HFD over 3 months, 80% of the mice exhibited obvious patch-like hair loss, whereas randomized littermates on the LFD or cocoa butter HFD did not, indicating a specific skin phenotype induced by the fish oil diet.
To determine the mechanisms of fish oil diet-induced hair loss, we excluded the possibility of diet-related behavioral alterations, such as grooming or fighting, as singly housed mice also exhibited hair loss. Gut dysbiosis was also unlikely to be involved in this diet-induced alopecia, as mice treated with or without antibiotics developed similar degrees of hair loss. C57BL/6 mice usually exhibit highly coordinated hair follicle cycling consisting of anagen (1–3 weeks), catagen (~2 days), and telogen (~2 weeks) phases early in life.31 When littermates (3–4 weeks of age) were randomly weaned on different diets, we noticed that fish oil HFD-induced hair loss started on the back of the mice around 2 months on the diet, which corresponded to the telogen phase at this stage. HFSCs are essential in cyclic growth of hair follicles and regeneration.22,32 Notably, n-3 FAs did not directly impact HFSC apoptosis in vitro, but mice consuming fish oil HFD exhibited extensive perifollicular macrophage infiltration and apoptosis of CD34+ HFSCs in vivo, suggesting that fish oil HFD-induced immune environmental alterations in the skin impaired HFSC activity.
Emerging studies suggest that human skin macrophages are engaged in hair growth control.33 Using the CyTOF technology, we identified a subset of dermal macrophages (CD207−CD11b+F4/80+MHCII+CD11c−) that were specifically upregulated in the fish oil HFD-fed mice. Interestingly, this macrophage subset highly expressed TNF-α, a paradoxical cytokine involved in hair follicle biology. On one hand, appropriate TNF-α signaling can stimulate hair follicle (HF) regeneration in homeostatic and wound-induced conditions,34 which was supported by random case reports that hair loss occurred during clinical therapy with TNF-α inhibitors.35 On the other hand, superfluous levels of TNF-α have been associated with forms of human alopecia.36 Given elevated levels of TNF-α in the skin of mice with fish oil-induced hair loss, we treated these mice with anti-TNF-α neutralizing antibody to see if blocking TNF-α signaling would revive hair growth. Indeed, fish oil-induced hair loss was reversed following treatment with anti-TNF-α antibody. These data indicate a mechanism by which the fish oil diet induced activation of dermal CD207− macrophages for excessive TNF-α signaling, which acts as an immune checkpoint inhibiting hair regeneration.
CD207 (also known as langerin) is a well-known surface receptor for skin-resident Langerhans cells,37 which function as the main lipid-processing population and provide the first immunological barrier to foreign substances in the skin.16,38 Our flow cytometric and CyTOF data confirmed that CD207+ cells were distributed in both epidermis and dermis regardless of diet. However, CD207− myeloid macrophages were accumulated only in the dermis of fish oil HFD-fed mice. To determine how the fish oil diet induced the infiltration of CD207− myeloid macrophages, we found that n-3 FAs were preferentially enriched in the skin and induced ROS/IL-36 signaling in skin-resident macrophages. In line with the well-characterized effect of IL-36 in promoting infiltration and activation of myeloid macrophages in skin,23,24 our data support the critical role of IL-36 in myeloid macrophage skin accumulation and identified n-3 FAs as upstream activators of IL-36 signaling.
To further dissect how n-3 FAs activated ROS/IL-36 signaling in the skin, we found that E-FABP, a skin-specific FA chaperone, was essential in mediating n-3 FA responses. E-FABP was originally identified in the skin as psoriasis-associated FABP with unknown functions. Studies from our group and others demonstrated that E-FABP binds an array of hydrophobic ligands, including n-3 FAs, retinoic acid (RA), and saturated FAs, and facilitates different ligand-mediated lipid responses.11,12,39–41 For example, E-FABP can channel RA to activate PPARβ/δ signaling for cell survival, whereas E-FABP binds saturated FAs to promote inflammasome activation. Herein, our data uncovered a function of E-FABP, which facilities n-3 FA-mediated ROS/IL-36 signaling in skin macrophages. Specifically, E-FABP functions at at least two checkpoints: (1) facilitation of n-3 FA-induced ROS production and (2) promotion of IL-36-mediated TNF-α production. Moreover, genetic deletion of E-FABP expression completely abrogated fish oil HFD-induced hair loss, further highlighting the essential role of E-FABP in n-3 FA-mediated unfavorable skin responses.
Fish oil diet is not typically associated with hair loss disorders in humans. Instead, although lacking solid scientific evidence, it is generally thought that n-3 FAs benefit hair follicles and scalp by improving blood circulation. It is worth noting that common American diets contain very little n-3 FA, and the FDA-approved dose of n-3 FA supplements/drugs accounts for only 1.2%–5% of the total daily fat intake.8 Thus, the adverse effects of n-3 FAs on hair growth may be underestimated at these low consumption levels. Interestingly, we noticed that nations that rely heavily on fatty fish as a food source (such as Japan) exhibit the most hair loss when compared with other Asian countries.42 Case reports that a diet high in tuna induced hair loss43 also support our observations in animal studies. While hair loss takes on a wide variety of forms and may be due to multiple inborn properties and environmental triggers,44 our well-controlled animal studies have provided solid evidence to support that overconsumption of n-3 FAs confers a detrimental effect by causing diet-induced hair loss.
In summary, we demonstrated that consumption of fish oil HFD induces hair loss via E-FABP-dependent immunoregulatory effects. Following lipid absorption, n-3 FAs were preferably enriched in the skin, where E-FABP expression in CD207+ resident skin macrophages plays an essential role in mediating n-3 FA/ROS/IL-36 signaling, which further promotes infiltration/activation of CD207− myeloid macrophages, therefore leading to TNF-α-mediated immune inhibition on HFSCs. Altogether, these studies not only provide a cellular and molecular mechanism by which consumption of n-3 FAs activates skin macrophages through E-FABP/ROS/IL-36/TNF-α signaling, thus promoting fish oil-induced hair loss, but also have clinical implications for understanding the etiology of dietary fat-associated hair health.
Limitations of the study
There are several limitations to the current studies: (1) although we noticed that consumption of fish oil HFD induced skin macrophages to generate ROS-dependent production of IL-36, it is possible that other skin cells (e.g., keratinocytes) might also produce IL-36 in response to fish oil HFD; (2) we observed that macrophages infiltrating the skin dermis of fish oil-fed mice disappeared upon anti-TNF-α neutralizing antibody treatment, but the mechanisms by which the macrophages either died or left upon treatment remain unclear; moreover, including mouse models with a genetic loss of IL-36 or TNF-α in future studies may render the identified molecular mechanism more compelling; (3) unlike murine diets, there are many confounding factors in human diets, which are hard control well. Of note, Western diets contain little n-3 fat, which gives this biological pathway less relevance to people who consume little fish. While consumption of fish oils is generally thought to be beneficial, overconsumption may lead to unexpected health problems, as is currently reported.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact, Bing Li (bing-li@uiowa.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
RNA-seq data have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. The accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rat monoclonal anti-mouse CD8α - PE | Biolegend | Cat#100708; RRID:AB_312747 |
| Hamster monoclonal anti-mouse TCR γ/δ - FITC | Biolegend | Cat#118106; RRID:AB_313830 |
| Rat monoclonal anti-mouse F4/80 - Pacific Blue | Biolegend | Cat#123124; RRID:AB_893475 |
| Rat monoclonal anti-mouse F4/80 - Alexa Fluor 488 | Biolegend | Cat#123120; RRID:AB_893479 |
| Hamster monoclonal anti-mouse CD11c - APC-Cy7 | Biolegend | Cat#117324; RRID:AB_830649 |
| Hamster monoclonal anti-mouse CD11c - PerCP | Biolegend | Cat#117325; RRID:AB_893236 |
| Mouse monoclonal anti-mouse NK1.1 - APC | Biolegend | Cat#108710; RRID:AB_313397 |
| Rat monoclonal anti-mouse/human CD45R/B220 - Alexa Fluor 700 | Biolegend | Cat#103232; RRID:AB_493717 |
| Rat monoclonal anti-mouse I-A/I-E - Brilliant Violet 605 | Biolegend | Cat#107639; RRID:AB_2565894 |
| Rat monoclonal anti-mouse CD16/32 | Biolegend | Cat#101302; RRID:AB_312801 |
| Rat monoclonal anti-mouse CD4 - APC | Biolegend | Cat#100412; RRID:AB_312697 |
| Rat monoclonal anti-mouse TNFα - Brilliant Violet 605 | Biolegend | Cat#506329; RRID:AB_11123912 |
| Rat monoclonal anti-mouse IL-17A - Pacific Blue | Biolegend | Cat#506918; RRID:AB_893544 |
| Rat monoclonal anti-mouse IFNγ - APC | Biolegend | Cat#505810; RRID:AB_315404 |
| Rat monoclonal anti-mouse TNFα (Ultra-LEAF purified for antibody treatment) | Biolegend | Cat#506347; RRID:AB_2616671 |
| Rat monoclonal anti-mouse CD11b - BUV737 | BD Biosciences | Cat#612800; RRID:AB_2870127 |
| Hamster monoclonal anti-mouse CD3ε - BUV737 | BD Biosciences | Cat#612771; RRID:AB_2870100 |
| Rat monoclonal anti-mouse CD34 - FITC | ThermoFisher Scientific | Cat#11-0341-082; RRID:AB_465021 |
| Goat polyclonal anti-Rabbit IgG(H + L) - APC | Jackson ImmunoResearch Laboratories INC. | Cat#111-136-144; RRID:AB_2337987 |
| Rabbit polyclonal anti-mouse IL-36α - APC | Assaypro | Cat#32103-05161; RRID:AB_2925172 |
| Rabbit polyclonal anti-mouse IL-36γ | Cloud-clone corp. | Cat#PAL621Mu01; RRID:AB_2925173 |
| Rat monoclonal anti-mouse CD45 - 89Y | Fluidigm | Cat#3089005B; RRID:AB_2651152 |
| Hamster monoclonal anti-mouse CD11c - 142Nd | Fluidigm | Cat#3142003B; RRID:AB_2815737 |
| Hamster monoclonal anti-mouse CD69 - 143Nd | Fluidigm | Cat#3143004B; RRID:AB_2827881 |
| Rat monoclonal anti-mouse CD4 - 145Nd | Fluidigm | Cat#3145002B; RRID:AB_2687832 |
| Rat monoclonal anti-mouse F4/80 - 146Nd | Fluidigm | Cat#3146008B; RRID:AB_2895117 |
| Rat monoclonal anti-mouse CD19 - 149Sm | Fluidigm | Cat#3149002B; RRID:AB_2814679 |
| Rat monoclonal anti-mouse Ly6C - 150Nd | Fluidigm | Cat#3150010B; RRID:AB_2895118 |
| Hamster monoclonal anti-mouse CD3ε - 152Sm | Fluidigm | Cat#3152004B; RRID:AB_2687836 |
| Rat monoclonal anti-mouse CD62L - 162Gd | Fluidigm | Cat#3160008B; RRID:AB_2687840 |
| Rat monoclonal anti-mouse CD44 - 162Dy | Fluidigm | Cat#3162030B; RRID:AB_2814898 |
| Rat monoclonal anti-mouse CD8a - 168Er | Fluidigm | Cat#3168003B; RRID:AB_2811241 |
| Mouse monoclonal anti-mouse NK1.1 - 170Er | Fluidigm | Cat#3170002B; RRID:AB_2885023 |
| Rat monoclonal anti-mouse CD11b - 172Yb | Fluidigm | Cat#3172012B; RRID:AB_2661809 |
| Rat monoclonal anti-mouse I-A/I-E - 209Bi | Fluidigm | Cat#3209006B; RRID:AB_2885025 |
| Hamster monoclonal anti-mouse TCR γ/δ - 159Tb | Fluidigm | Cat#3159012B; RRID:AB_2922919 |
| Hamster monoclonal anti-mouse TCRβ - 169Tm | Fluidigm | Cat#3169002B; RRID:AB_2827883 |
| Mouse monoclonal anti-mouse CX3CR1 - 164Dy | Fluidigm | Cat#3164023B; RRID:AB_2832247 |
| Mouse monoclonal anti-mouse/human CD207 - 175Lu | Fluidigm | Cat#3175016B; RRID:AB_2861414 |
| Rat monoclonal anti-mouse IL-17A - 174Yb | Fluidigm | Cat#3174002B; RRID:AB_2925175 |
| Rat monoclonal anti-mouse IFNγ - 165Ho | Fluidigm | Cat#3165003B; RRID:AB_2925174 |
| Rat monoclonal anti-mouse TNFα - 162Dy | Fluidigm | Cat#3162002B; RRID:AB_2801437 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Endotoxin-free BSA | Akronbiotech | Cat#AK8917-0100 |
| Palmitate acid | Nu-Chek prep | Cat#S-1109-JA30-X |
| Stearic acid | Nu-Chek prep | Cat#S-1111 |
| Linoleic acid | Nu-Chek prep | Cat#S-1127 |
| Oleate acid | Nu-Chek prep | Cat#S-1120-S19-W |
| Docosapentaenoate | Nu-Chek prep | Cat#S-1145 |
| Eicosapentaenoate | Nu-Chek prep | Cat#S-1144 |
| Collagenase type 2 | Worthington Biochemical | Cat#LS004177 |
| Hyaluronidase | MP Biomedicals | Cat#100740 |
| DNase I | ZYMO research | Cat#E1009-A |
| Gentamycin | VWR | Cat#0304-10G |
| Thermo Scientific Oil Red O | Fisher Scientific | Cat#A12989.14 |
| 2-Propanol | Millipore Sigma | Cat#190764 |
| Diphenyleneiodonium chloride(DPI) | Millipore Sigma | Cat#D2926 |
| N-acetyl-L-cysteine(NAC) | Millipore Sigma | Cat#A7250 |
| Tissue-Tek O.C.T. compound | Sakura Finetek | Cat#4583 |
| GM-CSF | Biolegend | Cat#576306 |
| M-CSF | Biolegend | Cat#576404 |
| Recombinant mouse TNFα | Biolegend | Cat#575202 |
| Recombinant mouse IL-17A | Biolegend | Cat#576002 |
| Recombinant mouse IFNγ | Biolegend | Cat#575302 |
| Recombinant mouse IL-36α | Biolegend | Cat#555902 |
| Recombinant mouse IL-36β | Biolegend | Cat#554502 |
| Recombinant mouse IL-36γ | Biolegend | Cat#552802 |
| Annexin V (APC) | Biolegend | Cat#640941 |
| Annexin V (PE) | Biolegend | Cat#640947 |
| Cell activation cocktail with Brefeldin A | Biolegend | Cat#423304 |
| Low fat diet (5% fat) | Research Diets. Inc | Cat#D20071505 |
| Fish oil high fat diet (45% fat) | Research Diets. Inc | Cat#D20071502 |
| Cocoa butter high fat diet (45% fat) | Research Diets. Inc | Cat#D20071501 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat#4368708 |
| SYPRO Orange dye | ThermoFisher Scientific | Cat#S6650 |
| PureLink RNA mini kit | Life technologies | Cat#12183025 |
| QuantiTect reverse transcription kit | Qiagen | Cat#205314 |
| ROS detection assay kit | Abcam | Cat#ab113851 |
| Mouse IIL-36γ ELISA kit | Mybiosource | Cat#MBS288173 |
| TruSeq Stranded mRNA Library Prep kit | Illumina | Cat#20020594 |
| TruSeq RNA Single Indexes Set A | Illumina | Cat#20020492 |
| TruSeq RNA Single Indexes Set B | Illumina | Cat#20020493 |
| MiSeq Reagent Nano Kit | Illumina | Cat#MS-103-1001 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Cat#Q32851 |
| DNA High Sensitivity Kit | Agilent Technologies | Cat#5067-4626 |
| EasySep mouse PE positive selection kit | Stemcell technologies | Cat#17666 |
|
| ||
| Deposited data | ||
|
| ||
| Skin tissue RNA-seq data | This paper | GEO: GSE161296 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse hair follicle stem cells(mHFSC) | Celprogen | Cat#66007-08 |
| Bone marrow-derived macrophages | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: E-FABP KO | This paper | N/A |
| Mouse: E-FABP WT | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| PCR primer sequences | This paper | Table S4 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 9 | Graph Pad Software | https://www.graphpad.com/ |
| CorelDRAW X7 | CorelDRAW | https://www.coreldraw.com/en/?link=wm |
| ImageJ 1.0 | National Institutes of Health(NIH) | https://imagej.nih.gov/ij/index.html |
| Flowjo v10.8.0 | BD | https://www.flowjo.com/solutions/flowjo/downloads |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and diet-induced hair loss mouse models
The protocol of mouse studies was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Iowa. Wild type (WT) and E-FABP−/− mice (all C57BL/6 background) were bred and housed in the animal facility at the University of Iowa. Both female and male mice were used in our experiments. Mice were weaned at 3–4 weeks old and randomly divided into 3 groups, fed a LFD (5% fat), fish oil HFD (45% fat), or cocoa butter HFD (45% fat), respectively (see detailed ingredient components of these custom diets in Table S1). Mice were euthanized for calculating hair loss area, analyzing immune cell phenotype in various tissues and organs after 3 months on diets. Mouse body weight was monitored biweekly. Images that were captured from posterior view of mice were processed using Image J software. The quantification of hair loss area vs total skin area was calculated with the equation: percentage of hair loss (%) = (value of hair loss areas/value of total skin) × 100.
Culture of bone marrow-derived macrophages (BMDMs) and mHFSC
For culture of macrophages in vitro, bone marrow cells from WT and E-FABP−/− female mice were plated in 10 mL RPMI 1640 supplemented with 5% heat inactivated FBS, 10 μg/mL gentamicin, 2 mM glutamax (referred to as R5) at 37°C. Two hours after plating, non-adherent cells were collected and cultured at the concentration of 4×106/15 mL in the R5 medium supplemented with either 20 ng/mL GM-CSF for CD11c+ macrophages or 40 ng/ml M-CSF for CD11c− macrophages. On days 2 and 5, half of the medium was replaced with fresh medium supplemented with GM-CSF or M-CSF. On day 7, cells from WT and E-FABP−/− mice were collected for various treatments.
Mouse hair follicle stem cells (mHFSC) (Cat# 66007-08, CELPROGEN) were grown to confluence in a 24-well plate in a HFSC culture Complete Growth Medium (M66007-08S, CELPROGEN) at 37°C before using for experiments. Cells were negative for Mycoplasma test using both PCR and mycoplasma agar methods.
METHOD DETAILS
Isolation of primary skin cells
Primary keratinocytes were isolated according to Dr. Morris’ protocol.45 Briefly, mice were euthanized, and hair was removed with an electric trimmer. Mice were disinfected in 70% ethanol for 3 min, and dorsal skin was removed using thumb forceps and scissors. Skin tissue was transferred into a 100 mm sterile dish containing 1 × PBS (17-516F, LONZA) with a final concentration of 50 μg/mL gentamycin (0304-10G, VWR). After subcutaneous fat tissue was scraped, the skin was cut into 1 cm × 1 cm pieces and floated in 20 mL 0.25% trypsin solution (25-053-CI, CORNING) on a gentle shaker placed in the 30°C incubator for 1.5 h. Hairy layer was removed from skin tissue and pipetted thoroughly at least 10 min in 1 × PBS with gentamycin. The suspension with isolated keratinocytes was filtrated through a 70 μm cell strainer and centrifuged at 1000 rpm for 7 min at 4°C. Epidermal cells were resuspended in epithelial cell culture medium (CnT-PR, CELLNTEC) by pipetting 20–25 times before experiments. For preparation of single cells from dermis, dermis was cut into small pieces and incubated in a 60 mm dish containing of digestion solution of RPMI medium (15-040-CV, CORNING) with 5% FBS, 0.5 mg/mL collagenase type 2 (S004177, Worthington Biochemical), 0.2 mg/mL hyaluronidase (0215127500, MP Biomedicals), and 0.02 mg/mL DNase I (E1009-A, ZYMO Research) in 37°C incubator for 40 min. The suspension was filtered through a 70 μm cell strainer, and then centrifuged at 1000 rpm for 7 min at 4°C. Cells were resuspended with 1 × PBS and kept on ice before experiments.
FA preparation and treatment
Endotoxin-free BSA (AK8917, Akron) was prepared in PBS with a stocking concentration of 2 mM. Different FAs, including PA (S-1109, Nu-Chek Prep), SA (S-1111, Nu-Chek Prep), EPA (S-1144, Nu-Chek Prep) and DPA (DPA n-3, S-1145, Nu-Chek Prep) were sonicated until dissolved at a stocking concentration of 5 mM with the BSA.17,46 For in vitro treatment of epidermal or dermal cells, isolated single cells from epidermis or dermis were cultured in epithelial cell culture medium with 100 or 200 μM FAs, such as DPA, PA or control BSA for 48 h. Cells were then collected for flow cytometric analysis.
Treatment of BMDMs
For Oil Red O (A12989.14 Thermo Fisher Scientific) staining, CD11c+ or CD11c− macrophages were fixed in 4% paraformaldehyde for 15 min, washed twice with dH2O, permeabilized with 60% isopropanol (190764, Sigma for 5 min, stained with an Oil Red O (Stock solution 75 mg in 25 mL 100% isopropanol and working solution 3 parts Oil Red O to 2 parts dH2O 0.2 μm filtered) for 20 min, images were collected using an Echo RVL2-K microscope.
For FA treatment effects, CD11c+ or CD11c− macrophages were treated with 200 μM PA, DPA or control BSA in the presence or absence of 1 mM N-acetyl-L-cysteine (NAC, A7250, Sigma-Aldrich) or 5μM Diphenyleneiodonium chloride (DPI, Sigma-Aldrich) for 24 h for gene expression analysis by real-time PCR. For IL-36 intracellular staining, cells were treated with 200 μM PA, DPA or control BSA for 48 h. After surface staining with anti-mouse CD11c and anti-mouse F4/80, cells were permeabilized for intracellular staining of IL-36α (32103-05161, Assaypro) or IL-36γ (PAL621Mu01, Cloud-clone Corp). For measurement of IL-36γ concentration in cultural supernatants, CD11c+ macrophages were treated with 200 μM PA, DPA or control BSA for 48 h. Supernatants were collected and detected using mouse Interleukin-36γ ELISA Kit (MBS288173, Mybiosource) according to the manufacturer’s protocol.
CyTOF mass cytometry sample preparation
The metal-tagged antibodies and reagents were all purchased from Fluidigm, and staining panels were designed using Maxpar Panel Designer. For samples preparation, cells from epidermis and dermis were suspended and washed in serum free RPMI 1640 (11875-093, Gibco), and single cell solution was harvested through a 70 μm cell strainer. All samples were checked for viability by staining with 5 uM cisplatin (Fluidigm) in serum-free RPMI 1640 for 10 min, then washed and centrifuged at 300 × g for 5 min at room temperature (RT). For surface antibodies staining, samples were stained at RT for 20 min and washed with Maxpar Cell staining buffer (Fluidigm). For checking on intracellular cytokines, cells were cultured with or without Cell Activation Cocktail with Brefeldin A for 4 h, then collected cells were stained with surface antibodies and washed twice. Cells were fixed with 1 mL of 1 × Maxpar Fix I buffer at RT for 30 min and then washed twice with 2 mL of 1 × Maxpar Perm-S buffer at room temperature for 5 min at 500 × g, cells were stained with intracellular staining antibodies in 1 × Maxpar Perm-S buffer at room temperature for 30 min. Finally, cells were fixed at RT for 10 min using 1.6% formaldehyde and washed at 800 × g for 10 min, then removed the fix buffer and incubated cell pellet overnight in 125 nM of IntercalatorIridium (Fluidigm) at 4°C.
Tissue HE & IHC staining
To observe if different diets affected hair follicle and skin structure, fresh skin samples from mice on different diets were immediately fixed in 10% neutral buffered formalin for at least 24 h. The fixed samples were cut into 5 μm sections and stained with hematoxylin and eosin (H&E). Sections for IHC staining were treated with Proteinase K for antigen retrieval and incubated with F4/80 primary antibody (Cat#MCA497, 1:200, Bio-Rad) at room temperature for 2 h, then stained with Rat-on-Mouse HRP-Polymer Kit (Cat#RT517, Biocare Medical). For other marker staining, sections were treated with Diva Decloaker (citrate buffer, pH6.0, Cat#DV2004, Biocare Medical), and incubated with anti-CD4 (Cat#14-9766-37, 1:200, Invitrogen), anti-CD8 (Cat#14-0808-82, at 1:500, Invitrogen), anti-CD207 (Cat# PA1-41053, at 1:500, ThermoFisher) or anti-TNF (Cat# AF-410-NA, at 1:1000, R&D) primary antibody, separately, at room temperature for 1 h, then stained with biotinylated secondary antibody (1:500) using VECTASTAIN Elite ABC-HRP Kits (Cat#PK-6100, Vector Laboratories). All methods were used with DAB (Cat#K346811-2, Agilent) for 5 min and DAB Enhancer (Cat#S196131-2, Agilent) for 3 min. Sections were subsequently counterstained with hematoxylin and mounted in mounting medium. IHC images were taken with ECHO Revolve Microscope (RVL2-K) in brightfield mode using 4X, 10x, 20x, and 40x objective lenses, respectively.
Analysis of ROS production
ROS generation was detected using the DCFDA (2′,7′-dichlorofluorescin diacetate) cellular ROS detection assay kit (ab113851, Abcam). For ROS measurement using flow cytometry, macrophages were seeded (1×105 cells/well) overnight in a 24-well plate. Cells were pretreated with 5 μM diphenyleneiodonium chloride (DPI, Sigma-Aldrich) for 15 min, and then treated with 200 μM PA, EPA, DPA or control BSA for 30 min. Cells were then collected and stained with 10 μM DCFDA for 45 min at 37°C. Samples were acquired using FACS LSR Fortessa, and data were analyzed using FlowJo software. For ROS detection by microplate assay, macrophages were seeded (5 × 104 cells/well) overnight in a 96-well plate. Cells were treated with 200 μM different FAs, including PA, SA, EPA and DPA, or BSA for the indicated time points (e.g. 1 h, 2 h, 3 h, 6 h, 24 h). After FA stimulation, cells were washed once with 1 × buffer and stained with 10 μM DCFDA (ab113851, Abcam) for 45 min at 37°C. After washing, cells were measured for the fluorescent signal at Ex/Em 485/535 nm using a Biotek plate reader. Data are shown as mean ± SD after background subtraction.
Macrophage-mediated cytotoxicity assay with antibody blocking
To evaluate macrophage-mediated cytotoxicity to mHFSCs, BMDMs were harvested on day 7 for cytotoxicity assay. mHFSCs were seeded in 48-well plates (1 × 105/well) with a final volume of 500 μL Complete Growth Medium for HFSCs. The effector macrophages were added to the target cells (mHFSCs) at a ratio of 1:2, and stimulated with Cell Activation Cocktail (423304, Biolegend) for 48 h in the presence of 1 μg/ml anti-mouse TNFα neutralizing antibody (510801, Biolegend), or control mIgG, respectively. All cells were stained with Annexin V-APC (640941, Biolegend) with Binding Buffer (422201, Biolegend) for 30 min and acquired with a BD LSR Fortessa. Data were analyzed by Flowjo software.
RNA-seq analysis
RNA sequence data was analyzed by the KBRIN Bioinformatics Core at the University of Louisville. Quality control (QC) of the raw sequence data was performed using FastQC (version 0.10.1). The quality value remained above 30 (99.9% base call accuracy) across the reads except the last base of all samples. Therefore, trimming was not deemed necessary. The sequences were directly aligned to the Mouse mm10 reference genome assembly using tophat2 (version 2.0.13),47 generating alignment files in bam format. The alignment rate for each sample was above 97.5%. Differential expression analysis between two conditions was performed using the Tuxedo Suite program cuffdiff2 (version 2.2.1)48,49 with Ensembl v84 genes. A p-value cutoff ≤0.05, q-value cutoff ≤0.05 with |FC| ≥ 1 was used to determine differential expression. RNA-seq data are available at GEO accession number (GSE161296).
In vivo trafficking of dietary FAs
Dietary FAs, such as DPA and PA, were first conjugated with Cy7-amine as described below. DPA or PA (1 eq), Cy7-amine (1 eq, 250C0, Lumiprobe), N-(3-dimethyl-aminopropyl)-N′-ethylcarbodiimide hydrochloride (1.1 eq), 1-Hydroxybenzotriazole hydrate (1.1 eq) in 0.5 mL DMF were mixed together at 0°C. N,N-Diisopropylethylamine (0.6 eq) was added and stirred at R.T. overnight. Afterwards, ice-cold water was added, and the mixture was extracted with ethyl acetate. The organic layer was dried with Na2SO4 and concentrated in vacuo to provide the product as a blue solid (61% yield). Typically, 100 nmole of Cy7-DPA or PA was reconstituted with 2 mL solvents (polyoxyethylene castor oil, absolute ethanol and saline at 1:1:2).50 100 μL of Cy-7-DPA or Cy-7-PA was intraperitoneally injected into each mouse for optical imaging (Photo imager optima, Biospace). Near infrared fluorescence imaging of mice was acquired at different time post-injection using photon acquisition software.
Flow cytometry surface and intracellular staining
For surface staining, immune cells in skin tissues were first blocked with anti-mouse CD16/32 (101302, Biolegend) for 30 min at 4°C in PBS containing 1% FBS. After washing, cells were stained with following antibodies: anti-mouse TCRβ-AF700(109224, Biolegend), anti-mouse TCR γ/δ-FITC antibody (118106, Biolegend), anti-mouse CD8α-PE (100708, Biolegend), anti-mouse CD11c-PE (117308, Biolegend), anti-mouse F4/80-AF488 (123120, Biolegend), anti-mouse NK1.1-APC(108710, Biolegend), anti-mouse B220-Alexa Fluor 700 (103232, Biolegend), anit-mouse Ly6C (128026, Biolegend), anti-mouse I-A/I-E-BV605 (107639, Biolegend), anti-mouse CD3ε-BUV737 (612771, BD Bioscience), anti-mouse CD11b-BUV737 (612800, BD Bioscience; For intracellular staining, cells were washed with PBS, fixed and permeabilized with Intracellular Staining Buffer Set (00-5523-00, Invitrogen) according to the manufacturer’s protocol. Cells were stained for 1 h at 4°C with different antibodies: anti-mouse TNFα-BV605 (506329, Biolegend), anti-mouse IL-17A-Pacific blue (506918, Biolegend), anti-mouse IL-36α-APC (32103-05161, Assaypro), anti-mouse IL-36γ (PAL621Mu01, Cloud-clone Corp), or control antibody (goat anti-rabbit IgG, 111-136-144, Jackson ImmunoResearch). Cells were acquired either by BD FACS Fortessa or BD FACS Aria II Cell Sorter. All the data were analyzed using FlowJo 10.1.
CyTOF data acquisition and analysis
Samples from overnight incubation were washed twice with Cells Staining Buffer (Fluidigm) and centrifuged at 4°C for 5 min at 500×g, then the pellet was suspended within a 1:9 solution of Cell Acquisition Solution: EQ 4 element calibration beads (Fluidigm). Samples were harvested using the Helios CyTOF system at speed of 500 events/second. CyTOF data was analyzed using the CytoBank software package, the CyTOF workflow which includes a suite of packages available in R, and Flowjo. Raw data were normalized in CyTOF software and analyzed on target populations through optimizing special gates to remove dead cells, debris, and doublets.
Confocal microscopy
Confocal microscopy was used in multiple settings. For ROS production analysis, CD11c+ macrophage from WT or E-FABP−/− mice were seeded into 35 mm Glass Bottom Dish (D35-14-1.5-N, Cellvis) for overnight culture. Cells were treated with 200 μM DPA, PA or control BSA for 30 min, and stained with DCFDA.12 For assessment of inflammatory immune cell infiltration in the skin, skin samples obtained from mice fed the different diets were snap-frozen with cryo-embedding media OCT (4583, Sakura Finetek USA) in −80°C. Frozen samples cut into 8 μm sections were mounted on gelatin-coated slides, and immediately incubated with blocking buffer (5% BSA) for 30 min on ice. Samples were stained with different immune cell surface markers, such as anti-mouse TCRγδ-FITC (118106, Biolegend), anti-mouse CD8α-PE (100708, Biolegend), anti-mouse CD11c-PE (117308, Biolegend), etc., for 30 min on ice, respectively, followed by nucleus staining with 200 nM DAPI (4038S, Cell Signaling Technology) for 15 min. All samples were analyzed with a Nikon A1 laser scanning confocal microscope, and images were captured by Olympus confocal microscopy and analyzed by FV10-ASW Viewer software (Ver 4.2b).
Analysis of cytokines and FA-metabolism related molecules with real-time PCR
RNA was extracted from macrophages treated with different FAs using a PureLink RNA Mini Kit (12183025, Life technologies). Complementary DNA synthesis was performed with a QuantiTect Reverse Transcription Kit (205314, Qiagen). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (4367659, Applied Biosystems) using a Step One Plus™ Real-time PCR System (Applied Biosystems). Relative mRNA levels were determined using the housekeeping HPRT1 gene as a reference. Detailed primers are shown in Table S4.
Cell viability analysis of mHFSCs
mHFSC were grown to confluence in a 24-well plate at least 12 h before experiments. For cytokine-induced apoptosis assay, mHFSCs were added with IL-17, IFNγ, TNFα at a range of physio- pathological levels (from 0 to 200 ng/mL) into the HFSC culture Complete Growth Medium for 24 h. To monitor FA-induced apoptosis, mHFSCs were added with 200 μM DPA, EPA, or control BSA into the medium for 24 h. Cells were gently harvested using Cell Detachment Solution (Accutase, AT-104, Innovative Cell Technologies), washed with Annexin V Binding Buffer (422201, Biolegend) twice and stained with Annexin-V (640941 for cytokine-induced apoptosis and 640947 for FA-induced apoptosis from Biolegend, respectively) in Binding Buffer for 20 min. For in vivo HFSC staining, dermal cells isolated from mice fed the LFD, fish oil HFD, or cocoa butter HFD were stained with anti-CD34-FITC (11-0341-082, eBioscience) and Zombie blue. All samples acquired using FACS LSR Fortessa flow cytometer. Data were analyzed using FlowJo software.
E-FABP protein purification and thermal shift assay
Recombinant E-FABP was purified from BL21 (DE3) and endogenous lipids of purified E-FABP were removed by Lipidex-1000 column.51,52 A fluorescent based thermal shift assay53 was performed to measure the binding of E-FABP with DPA and PA. Reactions were set-up in PCR tubes in a 20 μL volume containing 10 μM E-FABP and 10× SYPRO Orange dye (Invitrogen) in 20 mM HEPES pH 7 and 150 mM NaCl, containing either test FAs or solvent (ethanol) only controls. PCR tubes were sealed, centrifuged and heated from 25 to 95° at a rate of 1°/min on 7500 Real-Time PCR machine (Applied Biosystems). Tm values were calculated based on raw data analysis and curve fitting.53
RNA-seq library preparation
RNA was extracted from the skin of mice fed the LFD, fish oil diet, or cocoa butter diet for 3 months using mRNA extraction kit (Qiagen) (n = 3/group). RNA-seq Libraries were prepared using the TruSeq Stranded mRNA Library Prep kit (Illumina Cat# 20020594), TruSeq RNA Single Indexes Set A (Illumina Cat# 20020492) and Set B (Illumina Cat# 20020493). 200 ng of samples (in a volume of 50 μL) were treated with RNA Purification Beads and denatured for 5 min at 65°C. Then the supernatant was discarded, and the beads were washed with bead wash buffer. Captured polyadenylated RNA was eluted using Elution buffer at 80°C for 2 min. mRNA was further purified in a second bead clean-up, as well as fragmented and primed during elution by adding 19.5 μL of Elute, Prime, Fragment High Mix to the beads and incubating the samples for 8 min at 94°C. After fragmentation, 17ul of supernatant was removed from the beads and proceeded immediately to synthesize first strand cDNA. Following the protocol, 8 μL of First Strand Synthesis Mix Act D and SuperScript II mix were added to each sample and heated on a thermocycler using preprogrammed thermal conditions. Once the reaction finished and reached 4°C, we immediately proceeded for second strand cDNA synthesis. Second Strand Marking Mix was added, mixed well and incubated in a pre-heated thermocycler at 16°C for one hour. Then the DNA was purified using Agencourt AMPure XP Beads. Finally, samples were eluted with Resuspension Buffer and 15 μL of elute was collected and stored at −20°C. A-Tailing Mix was added to the purified samples and the samples were incubated on the preprogrammed thermal cycler. Once the incubation completed, we proceeded immediately to adapter ligation. Ligation Mix and barcodes were added and incubated in a pre-heated thermocycler at 30°C for 10 min. Stop Ligation Buffer was immediately added to each sample and mixed well. Then the ligated samples were purified using Agencourt AMPure XP Beads. We eluted with 50 μL of Resuspension Buffer and the elute was again purified for a second time using Agencourt AMPure XP Beads. Afterwards, the final elution, consisting of 20 μL of the elute was collected and used for DNA enrichment. Samples were barcoded with Illumina TruSeq Adapters. PCR Primer Cocktail Mix and PCR Master Mix were added to the samples and incubated on a preprogrammed thermal cycler. Then the samples were purified using Agencourt AMPure XP Beads. Finally, 20 ul of eluted libraries were collected and stored at −20°C. The concentration of libraries was measured by Qubit dsDNA HS Assay Kit (Invitrogen Q32851). Libraries were diluted and normalized to the optimal range for Agilent Bioanalyzer analysis using the DNA High Sensitivity Kit (Agilent Technologies, Cat# 5067-4626). The final fragment size for all the samples was approximately 250-270 bp which is expected according to the protocol. The same amount of libraries was pooled based on the molar concentration from Bioanalyzer. Pooled library was run on MiSeq to test quantity and quality, using the MiSeq Reagent Nano Kit V2 300 cycles (Illumina, Cat. No. MS-103-1001). Library and PhIX control (Illumina, Cat. No. FC-110-3001) were denatured and diluted using the standard normalization method following manufacturer’s directions, to a final concentration of 6 pM. 300 ul of library and 300 ul of PhIX were combined and sequenced on Illumina MiSeq. Library and PhIX were denatured and diluted using the standard normalization method following manufacturer’s directions. The total volume of library was 1.3 mL at 1.8 pM, with 1% PhIX spike in. Sequencing was performed on the University of Louisville Center for Genetics and Molecular Medicine’s (CGeMM) Illumina NextSeq 500 using the NextSeq 500/550 75 cycle High Output Kit v2 (FC-404-2002), resulting in approximately 35 million single-end reads per sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
All quantitative data were shown as the mean ± SD as indicated by at least three independent experiments or technical replicates. For both in vitro and in vivo experiments, a two-tailed, unpaired Student t test provided by GraphPad Prism 9 was performed for the comparison of differences between groups. A p value of <0.05 is regarded as statistically significant.
Supplementary Material
Highlights.
Consumption of fish oil HFD, but not cocoa butter HFD, induces murine hair loss
TNF-α signaling in CD207− macrophages mediates fish oil HFD-induced hair loss
E-FABP is essential in the n-3 fatty acid-induced ROS/IL-36/TNF-α axis in skin macrophages
E-FABP deficiency protects mice from fish oil HFD-induced hair loss
ACKNOWLEDGMENTS
We thank Dr. Rebecca Morris for assistance with the skin cell separation protocol and helpful discussion regarding skin histology. We also thank the Comparative Pathology Laboratory at the University of Iowa for skin histological and immunohistochemistry staining. B.L. is grateful for the funding support from NIH grants R01AI137324, R01CA180986, and U01CA272424. Part of this work was performed with the assistance of the University of Louisville Functional Immunomics Core (supported by NIH P20GM135004) and Genomics Facility (supported by NIH P20GM103436, NIH P30GM106396, and the J.G. Brown Foundation).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111804.
REFERENCES
- 1.Diekman C, and Malcolm K (2009). Consumer perception and insights on fats and fatty acids: knowledge on the quality of diet fat. Ann. Nutr. Metab 54, 25–32. [DOI] [PubMed] [Google Scholar]
- 2.Liu AG, Ford NA, Hu FB, Zelman KM, Mozaffarian D, and Kris-Etherton PM (2017). A healthy approach to dietary fats: understanding the science and taking action to reduce consumer confusion. Nutr. J 16, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordeleau L, Yakubovich N, Dagenais GR, Rosenstock J, Probstfield J, Chang Yu P, Ryden LE, Pirags V, Spinas GA, Birkeland KI, et al. ORIGIN Trial Investigators (2014). The association of basal insulin glargine and/or n-3 fatty acids with incident cancers in patients with dysglycemia. Diabetes Care 37, 1360–1366. [DOI] [PubMed] [Google Scholar]
- 4.Wu JHY, and Mozaffarian D (2014). omega-3 fatty acids, atherosclerosis progression and cardiovascular outcomes in recent trials: new pieces in a complex puzzle. Heart 100, 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louie SM, Roberts LS, and Nomura DK (2013). Mechanisms linking obesity and cancer. Biochim. Biophys. Acta 1831, 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper L, Martin N, Jimoh OF, Kirk C, Foster E, and Abdelhamid AS (2020). Reduction in saturated fat intake for cardiovascular disease. Cochrane Database Syst. Rev 8, CD011737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keaney JF Jr., and Rosen CJ (2019). VITAL signs for dietary supplementation to prevent cancer and heart disease. N. Engl. J. Med 380, 91–93. [DOI] [PubMed] [Google Scholar]
- 8.Manson JE, Cook NR, Lee IM, Christen W, Bassuk SS, Mora S, Gibson H, Albert CM, Gordon D, Copeland T, et al. VITAL Research Group (2019). Marine n-3 fatty acids and prevention of cardiovascular disease and cancer. N. Engl. J. Med 380, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siri-Tarino PW, Sun Q, Hu FB, and Krauss RM (2010). Meta-analysis of prospective cohort studies evaluating the association of saturated fat with cardiovascular disease. Am. J. Clin. Nutr 91, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Hao J, Zeng J, and Sauter ER (2020). SnapShot: FABP functions. Cell 182, 1066–1066.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Li Q, Rao E, Sun Y, Grossmann ME, Morris RJ, Cleary MP, and Li B (2015). Epidermal Fatty Acid binding protein promotes skin inflammation induced by high-fat diet. Immunity 42, 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng J, Zhang Y, Hao J, Sun Y, Liu S, Bernlohr DA, Sauter ER, Cleary MP, Suttles J, and Li B (2018). Stearic acid induces CD11c expression in proinflammatory macrophages via epidermal fatty acid binding protein. J. Immunol 200, 3407–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. (2017). Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao J, Yan F, Zhang Y, Triplett A, Zhang Y, Schultz DA, Sun Y, Zeng J, Silverstein KAT, Zheng Q, et al. (2018). Expression of adipocyte/macrophage fatty acid-binding protein in tumor-associated macrophages promotes breast cancer progression. Cancer Res. 78, 2343–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okabe Y, and Medzhitov R (2016). Tissue biology perspective on macrophages. Nat. Immunol 17, 9–17. [DOI] [PubMed] [Google Scholar]
- 16.Remmerie A, and Scott CL (2018). Macrophages and lipid metabolism. Cell. Immunol 330, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Jin R, Hao J, Zeng J, Yin D, Yi Y, Zhu M, Mandal A, Hua Y, Ng CK, et al. (2020). Consumption of the fish oil high-fat diet uncouples obesity and mammary tumor growth through induction of reactive oxygen species in protumor macrophages. Cancer Res. 80, 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin R, Hao J, Yi Y, Sauter E, and Li B (2021). Regulation of macrophage functions by FABP-mediated inflammatory and metabolic pathways. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1866, 158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Immunological Genome Consortium (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovászi M, Mattii M, Eyerich K, Gácsi A, Csányi E, Kovács D, Rühl R, Szegedi A, Kemény L, Ståhle M, et al. (2017). Sebum lipids influence macrophage polarization and activation. Br. J. Dermatol 177, 1671–1682. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi A, Mikami Y, Miyamoto K, Kamada N, Sato T, Mizuno S, Naganuma M, Teratani T, Aoki R, Fukuda S, et al. (2017). Intestinal dysbiosis and biotin deprivation induce alopecia through overgrowth of lactobacillus murinus in mice. Cell Rep. 20, 1513–1524. [DOI] [PubMed] [Google Scholar]
- 22.Chen CL, Huang WY, Wang EHC, Tai KY, and Lin SJ (2020). Functional complexity of hair follicle stem cell niche and therapeutic targeting of niche dysfunction for hair regeneration. J. Biomed. Sci 27, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito K, Iwata Y, Fukushima H, Watanabe S, Tanaka Y, Hasegawa Y, Akiyama M, and Sugiura K (2020). IL-36 receptor antagonist deficiency resulted in delayed wound healing due to excessive recruitment of immune cells. Sci. Rep 10, 14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster AM, Baliwag J, Chen CS, Guzman AM, Stoll SW, Gudjonsson JE, Ward NL, and Johnston A (2014). IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J. Immunol 192, 6053–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hotamisligil GS, and Bernlohr DA (2015). Metabolic functions of FABPs–mechanisms and therapeutic implications. Nat. Rev. Endocrinol 11, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Sun Y, Rao E, Yan F, Li Q, Zhang Y, Silverstein KAT, Liu S, Sauter E, Cleary MP, and Li B (2014). Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-beta responses in tumor-associated macrophages. Cancer Res. 74, 2986–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siriwardhana N, Kalupahana NS, and Moustaid-Moussa N (2012). Health benefits of n-3 polyunsaturated fatty acids: eicosapentaenoic acid and docosahexaenoic acid. Adv. Food Nutr. Res 65, 211–222. [DOI] [PubMed] [Google Scholar]
- 28.Acquistapace S, Patel L, Patin A, Forbes-Blom E, Cuenoud B, and Wooster TJ (2019). Effects of interesterified lipid design on the short/medium chain fatty acid hydrolysis rate and extent (in vitro). Food Funct. 10, 4166–4176. [DOI] [PubMed] [Google Scholar]
- 29.Merkel M, Eckel RH, and Goldberg IJ (2002). Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res 43, 1997–2006. [DOI] [PubMed] [Google Scholar]
- 30.Ellulu MS, Patimah I, Khaza’ai H, Rahmat A, and Abed Y (2017). Obesity and inflammation: the linking mechanism and the complications. Arch. Med. Sci 13, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller-Röver S, Handjiski B, van der Veen C, Eichmüller S, Foitzik K, McKay IA, Stenn KS, and Paus R (2001). A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol 117, 3–15. [DOI] [PubMed] [Google Scholar]
- 32.Owczarczyk-Saczonek A, Krajewska-Włodarczyk M, Kruszewska A, Banasiak Ł, Placek W, Maksymowicz W, and Wojtkiewicz J (2018). Therapeutic potential of stem cells in follicle regeneration. Stem Cell. Int 2018, 1049641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardman JA, Muneeb F, Pople J, Bhogal R, Shahmalak A, and Paus R (2019). Human perifollicular macrophages undergo apoptosis, express Wnt ligands, and switch their polarization during catagen. J. Invest. Dermatol 139, 2543–2546.e9. [DOI] [PubMed] [Google Scholar]
- 34.Rahmani W, Sinha S, and Biernaskie J (2020). Immune modulation of hair follicle regeneration. NPJ Regen. Med 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Béné J, Moulis G, Auffret M, Lefevre G, Coquerelle P, Coupe P, Péré P, and Gautier S (2014). Alopecia induced by tumour necrosis factor-alpha antagonists: description of 52 cases and disproportionality analysis in a nationwide pharmacovigilance database. Rheumatology 53, 1465–1469. [DOI] [PubMed] [Google Scholar]
- 36.Gohary YM, and Abdel Fattah DS (2017). Detection of tumor necrosis factor-alpha in nonlesional tissues of alopecia are at a patients: a prove for a systemic disease. Int. J. Trichol 9, 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merad M, Ginhoux F, and Collin M (2008). Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol 8, 935–947. [DOI] [PubMed] [Google Scholar]
- 38.Tay SS, Roediger B,Tong PL,Tikoo S, and Weninger W (2014). The skin-resident immune network. Curr. Dermatol. Rep 3, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Figueroa JD, Serrano-Illan M, Licero J, Cordero K, Miranda JD, and De Leon M (2016). Fatty acid binding protein 5 modulates docosahexaenoic acid-induced recovery in rats undergoing spinal cord injury. J. Neurotrauma 33, 1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levi L, Wang Z, Doud MK, Hazen SL, and Noy N (2015). Saturated fatty acids regulate retinoic acid signalling and suppress tumorigenesis by targeting fatty acid-binding protein 5. Nat. Commun 6, 8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schug TT, Berry DC, Shaw NS, Travis SN, and Noy N (2007). Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagino T, Okazaki S, Serizawa N, Suzuki K, Kaga M, Otsuka Y, Mikami E, Hoashi T, Saeki H, Matsuda H, et al. (2021). Dietary habits in Japanese patients with alopecia areata. Clin. Cosmet. Invest. Dermatol 14, 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters JB, and Warren MP (2019). Reversible alopecia associated with high blood mercury levels and early menopause: a report of two cases. Menopause 26, 915–918. [DOI] [PubMed] [Google Scholar]
- 44.McElwee KJ, Gilhar A, Tobin DJ, Ramot Y, Sundberg JP, Nakamura M, Bertolini M, Inui S, Tokura Y, King LE Jr., et al. (2013). What causes alopecia areata? Exp. Dermatol 22, 609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park H, Lad S, Boland K, Johnson K, Readio N, Jin G, Asfaha S, Patterson KS, Singh A, Yang X, et al. (2018). Bone marrow-derived epithelial cells and hair follicle stem cells contribute to development of chronic cutaneous neoplasms. Nat. Commun 9, 5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Hao J, Sun Y, and Li B (2017). Saturated fatty acids induce ceramide-associated macrophage cell death. J. Vis. Exp 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, and Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S, Qin J, Tian C, Cao J, Fida G, Wang Z, Chen H, Qian Z, Chen WR, and Gu Y (2014). The targeting mechanism of DHA ligand and its conjugate with Gemcitabine for the enhanced tumor therapy. Oncotarget 5, 3622–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao E, Singh P, Li Y, Zhang Y, Chi YI, Suttles J, and Li B (2015). Targeting epidermal fatty acid binding protein for treatment of experimental autoimmune encephalomyelitis. BMC Immunol. 16, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao E, Singh P, Zhai X, Li Y, Zhu G, Zhang Y, Hao J, Chi YI, Brown RE, Cleary MP, and Li B (2015). Inhibition of tumor growth by a newly-identified activator for epidermal fatty acid binding protein. Oncotarget 6, 7815–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niesen FH, Berglund H, and Vedadi M (2007). The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc 2, 2212–2221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. The accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rat monoclonal anti-mouse CD8α - PE | Biolegend | Cat#100708; RRID:AB_312747 |
| Hamster monoclonal anti-mouse TCR γ/δ - FITC | Biolegend | Cat#118106; RRID:AB_313830 |
| Rat monoclonal anti-mouse F4/80 - Pacific Blue | Biolegend | Cat#123124; RRID:AB_893475 |
| Rat monoclonal anti-mouse F4/80 - Alexa Fluor 488 | Biolegend | Cat#123120; RRID:AB_893479 |
| Hamster monoclonal anti-mouse CD11c - APC-Cy7 | Biolegend | Cat#117324; RRID:AB_830649 |
| Hamster monoclonal anti-mouse CD11c - PerCP | Biolegend | Cat#117325; RRID:AB_893236 |
| Mouse monoclonal anti-mouse NK1.1 - APC | Biolegend | Cat#108710; RRID:AB_313397 |
| Rat monoclonal anti-mouse/human CD45R/B220 - Alexa Fluor 700 | Biolegend | Cat#103232; RRID:AB_493717 |
| Rat monoclonal anti-mouse I-A/I-E - Brilliant Violet 605 | Biolegend | Cat#107639; RRID:AB_2565894 |
| Rat monoclonal anti-mouse CD16/32 | Biolegend | Cat#101302; RRID:AB_312801 |
| Rat monoclonal anti-mouse CD4 - APC | Biolegend | Cat#100412; RRID:AB_312697 |
| Rat monoclonal anti-mouse TNFα - Brilliant Violet 605 | Biolegend | Cat#506329; RRID:AB_11123912 |
| Rat monoclonal anti-mouse IL-17A - Pacific Blue | Biolegend | Cat#506918; RRID:AB_893544 |
| Rat monoclonal anti-mouse IFNγ - APC | Biolegend | Cat#505810; RRID:AB_315404 |
| Rat monoclonal anti-mouse TNFα (Ultra-LEAF purified for antibody treatment) | Biolegend | Cat#506347; RRID:AB_2616671 |
| Rat monoclonal anti-mouse CD11b - BUV737 | BD Biosciences | Cat#612800; RRID:AB_2870127 |
| Hamster monoclonal anti-mouse CD3ε - BUV737 | BD Biosciences | Cat#612771; RRID:AB_2870100 |
| Rat monoclonal anti-mouse CD34 - FITC | ThermoFisher Scientific | Cat#11-0341-082; RRID:AB_465021 |
| Goat polyclonal anti-Rabbit IgG(H + L) - APC | Jackson ImmunoResearch Laboratories INC. | Cat#111-136-144; RRID:AB_2337987 |
| Rabbit polyclonal anti-mouse IL-36α - APC | Assaypro | Cat#32103-05161; RRID:AB_2925172 |
| Rabbit polyclonal anti-mouse IL-36γ | Cloud-clone corp. | Cat#PAL621Mu01; RRID:AB_2925173 |
| Rat monoclonal anti-mouse CD45 - 89Y | Fluidigm | Cat#3089005B; RRID:AB_2651152 |
| Hamster monoclonal anti-mouse CD11c - 142Nd | Fluidigm | Cat#3142003B; RRID:AB_2815737 |
| Hamster monoclonal anti-mouse CD69 - 143Nd | Fluidigm | Cat#3143004B; RRID:AB_2827881 |
| Rat monoclonal anti-mouse CD4 - 145Nd | Fluidigm | Cat#3145002B; RRID:AB_2687832 |
| Rat monoclonal anti-mouse F4/80 - 146Nd | Fluidigm | Cat#3146008B; RRID:AB_2895117 |
| Rat monoclonal anti-mouse CD19 - 149Sm | Fluidigm | Cat#3149002B; RRID:AB_2814679 |
| Rat monoclonal anti-mouse Ly6C - 150Nd | Fluidigm | Cat#3150010B; RRID:AB_2895118 |
| Hamster monoclonal anti-mouse CD3ε - 152Sm | Fluidigm | Cat#3152004B; RRID:AB_2687836 |
| Rat monoclonal anti-mouse CD62L - 162Gd | Fluidigm | Cat#3160008B; RRID:AB_2687840 |
| Rat monoclonal anti-mouse CD44 - 162Dy | Fluidigm | Cat#3162030B; RRID:AB_2814898 |
| Rat monoclonal anti-mouse CD8a - 168Er | Fluidigm | Cat#3168003B; RRID:AB_2811241 |
| Mouse monoclonal anti-mouse NK1.1 - 170Er | Fluidigm | Cat#3170002B; RRID:AB_2885023 |
| Rat monoclonal anti-mouse CD11b - 172Yb | Fluidigm | Cat#3172012B; RRID:AB_2661809 |
| Rat monoclonal anti-mouse I-A/I-E - 209Bi | Fluidigm | Cat#3209006B; RRID:AB_2885025 |
| Hamster monoclonal anti-mouse TCR γ/δ - 159Tb | Fluidigm | Cat#3159012B; RRID:AB_2922919 |
| Hamster monoclonal anti-mouse TCRβ - 169Tm | Fluidigm | Cat#3169002B; RRID:AB_2827883 |
| Mouse monoclonal anti-mouse CX3CR1 - 164Dy | Fluidigm | Cat#3164023B; RRID:AB_2832247 |
| Mouse monoclonal anti-mouse/human CD207 - 175Lu | Fluidigm | Cat#3175016B; RRID:AB_2861414 |
| Rat monoclonal anti-mouse IL-17A - 174Yb | Fluidigm | Cat#3174002B; RRID:AB_2925175 |
| Rat monoclonal anti-mouse IFNγ - 165Ho | Fluidigm | Cat#3165003B; RRID:AB_2925174 |
| Rat monoclonal anti-mouse TNFα - 162Dy | Fluidigm | Cat#3162002B; RRID:AB_2801437 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Endotoxin-free BSA | Akronbiotech | Cat#AK8917-0100 |
| Palmitate acid | Nu-Chek prep | Cat#S-1109-JA30-X |
| Stearic acid | Nu-Chek prep | Cat#S-1111 |
| Linoleic acid | Nu-Chek prep | Cat#S-1127 |
| Oleate acid | Nu-Chek prep | Cat#S-1120-S19-W |
| Docosapentaenoate | Nu-Chek prep | Cat#S-1145 |
| Eicosapentaenoate | Nu-Chek prep | Cat#S-1144 |
| Collagenase type 2 | Worthington Biochemical | Cat#LS004177 |
| Hyaluronidase | MP Biomedicals | Cat#100740 |
| DNase I | ZYMO research | Cat#E1009-A |
| Gentamycin | VWR | Cat#0304-10G |
| Thermo Scientific Oil Red O | Fisher Scientific | Cat#A12989.14 |
| 2-Propanol | Millipore Sigma | Cat#190764 |
| Diphenyleneiodonium chloride(DPI) | Millipore Sigma | Cat#D2926 |
| N-acetyl-L-cysteine(NAC) | Millipore Sigma | Cat#A7250 |
| Tissue-Tek O.C.T. compound | Sakura Finetek | Cat#4583 |
| GM-CSF | Biolegend | Cat#576306 |
| M-CSF | Biolegend | Cat#576404 |
| Recombinant mouse TNFα | Biolegend | Cat#575202 |
| Recombinant mouse IL-17A | Biolegend | Cat#576002 |
| Recombinant mouse IFNγ | Biolegend | Cat#575302 |
| Recombinant mouse IL-36α | Biolegend | Cat#555902 |
| Recombinant mouse IL-36β | Biolegend | Cat#554502 |
| Recombinant mouse IL-36γ | Biolegend | Cat#552802 |
| Annexin V (APC) | Biolegend | Cat#640941 |
| Annexin V (PE) | Biolegend | Cat#640947 |
| Cell activation cocktail with Brefeldin A | Biolegend | Cat#423304 |
| Low fat diet (5% fat) | Research Diets. Inc | Cat#D20071505 |
| Fish oil high fat diet (45% fat) | Research Diets. Inc | Cat#D20071502 |
| Cocoa butter high fat diet (45% fat) | Research Diets. Inc | Cat#D20071501 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat#4368708 |
| SYPRO Orange dye | ThermoFisher Scientific | Cat#S6650 |
| PureLink RNA mini kit | Life technologies | Cat#12183025 |
| QuantiTect reverse transcription kit | Qiagen | Cat#205314 |
| ROS detection assay kit | Abcam | Cat#ab113851 |
| Mouse IIL-36γ ELISA kit | Mybiosource | Cat#MBS288173 |
| TruSeq Stranded mRNA Library Prep kit | Illumina | Cat#20020594 |
| TruSeq RNA Single Indexes Set A | Illumina | Cat#20020492 |
| TruSeq RNA Single Indexes Set B | Illumina | Cat#20020493 |
| MiSeq Reagent Nano Kit | Illumina | Cat#MS-103-1001 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Cat#Q32851 |
| DNA High Sensitivity Kit | Agilent Technologies | Cat#5067-4626 |
| EasySep mouse PE positive selection kit | Stemcell technologies | Cat#17666 |
|
| ||
| Deposited data | ||
|
| ||
| Skin tissue RNA-seq data | This paper | GEO: GSE161296 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse hair follicle stem cells(mHFSC) | Celprogen | Cat#66007-08 |
| Bone marrow-derived macrophages | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: E-FABP KO | This paper | N/A |
| Mouse: E-FABP WT | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| PCR primer sequences | This paper | Table S4 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 9 | Graph Pad Software | https://www.graphpad.com/ |
| CorelDRAW X7 | CorelDRAW | https://www.coreldraw.com/en/?link=wm |
| ImageJ 1.0 | National Institutes of Health(NIH) | https://imagej.nih.gov/ij/index.html |
| Flowjo v10.8.0 | BD | https://www.flowjo.com/solutions/flowjo/downloads |


