Abstract
The CcrM DNA methyltransferase of the α-proteobacteria catalyzes the methylation of the adenine in the sequence GAnTC. Like Dam in the enterobacteria, CcrM plays a regulatory role in Caulobacter crescentus and Rhizobium meliloti. CcrM is essential for viability in both of these organisms, and we show here that it is also essential in Brucella abortus. Further, increased copy number of the ccrM gene results in striking changes in B. abortus morphology, DNA replication, and growth in murine macrophages. We generated strains that carry ccrM either on a low-copy-number plasmid (strain GR131) or on a moderate-copy-number plasmid (strain GR132). Strain GR131 has wild-type morphology and chromosome number, as assessed by flow cytometry. In contrast, strain GR132 has abnormal branched morphology, suggesting aberrant cell division, and increased chromosome number. Although these strains exhibit different morphologies and DNA content, the replication of both strains in macrophages is attenuated. These data imply that the reduction in survival in host cells is not due solely to a cell division defect but is due to additional functions of CcrM. Because CcrM is essential in B. abortus and increased ccrM copy number attenuates survival in host cells, we propose that CcrM is an appropriate target for new antibiotics.
Changes in DNA methylation patterns signal changes in cellular physiology in both prokaryotes and eukaryotes. In bacteria, DNA methyltransferases not only participate in restriction-modification systems (5) but also play regulatory roles in the cell. For example, methylation of the origin of replication by the Dam methyltransferase governs the timing of the initiation of DNA replication in Escherichia coli (6, 29). Dam methylation also contributes to strand discrimination in methyl-directed DNA mismatch repair (19) and plays a role in pathogenesis (10, 12). For instance, changes in Dam methylation patterns control pyelonephritis-associated pilus (pap) transcription in uropathogenic E. coli by altering the binding of Lrp (leucine-responsive protein) to the pap promoter (21, 38). In addition, Dam methylation either directly or indirectly regulates the transcription of a number of genes in Salmonella enterica serovar Typhimurium that are induced following infection of the host (12), suggesting that DNA methylation plays a role in the virulence of this organism.
The Dam methyltransferases of E. coli and other γ-proteobacteria have a counterpart in α-proteobacteria, a DNA adenine methyltransferase called CcrM (for cell cycle-regulated methyltransferase). This enzyme was originally described in Caulobacter crescentus, a bacterium that is easily synchronized and therefore amenable to cell cycle studies (42). Both Dam and CcrM catalyze the transfer of the methyl group from S-adenosylmethionine to the adenine of their target DNA sequences. Both enzymes apparently lack cognate restriction enzymes and instead regulate cell cycle events. However, unlike Dam, CcrM is required for cell viability, and its activity is tightly regulated during the cell cycle (36). In Caulobacter, CcrM is present only in late predivisional cells, just prior to cell division (36). This pattern of CcrM expression is achieved by strict temporal regulation of ccrM transcription (37) and rapid turnover by Lon-mediated degradation (41). The CtrA response regulator, which controls multiple cell cycle events, activates ccrM transcription in late predivisional cells (23, 27). Constitutive transcription of ccrM throughout the cell cycle, resulting in chromosomes that remain fully methylated at all times, yields elongated cells that divide abnormally and exhibit relaxed control of DNA replication initiation (41, 42). Although CcrM-catalyzed DNA methylation appears to play a role in the initiation of DNA replication, little is known about the other essential functions of this enzyme.
CcrM homologs are widely distributed among the α-proteobacteria, including C. crescentus, the nitrogen-fixing soil bacterium Rhizobium meliloti, and the animal pathogen Brucella abortus (36, 40). CcrM functions and its essential nature are conserved in at least two of these bacteria, C. crescentus and R. meliloti (40). Alignment of the amino acid sequences of the Brucella and Caulobacter CcrM homologs reveals extensive conservation (78% similarity) throughout the entire proteins (40). Although the B. abortus homolog has an N-terminal extension of 15 amino acids, the putative catalytic and AdoMet binding sites are highly conserved in both proteins (18). Their similar structures suggest that the two enzymes share a common function.
Unlike the free-living bacterium C. crescentus, B. abortus and other members of the α-proteobacteria live in close association with eukaryotic cells. The brucellae are small, nonmotile bacteria that infect many mammalian species, including humans. Because the brucellae are intracellular pathogens, the ability of these organisms to survive and replicate in macrophages is critical to their ability to cause disease in the host (7). The cellular mechanisms used by the brucellae to survive and multiply within host cells, however, are poorly understood (33).
In this report, we show that the B. abortus ccrM promoter contains a binding site for the CtrA response regulator, suggesting that the regulation of ccrM transcription may be similar to that observed in C. crescentus. Moreover, B. abortus CcrM is required for cell viability, and increased copy number of the ccrM gene results in changes in cell morphology and chromosome replication. Aberrant ccrM expression also alters the ability of B. abortus to replicate in murine macrophages. Our results indicate that CcrM functions play a role in allowing the brucellae to appropriately adapt to their intracellular lifestyle. This role may be directly related to the role of CcrM in maintaining normal cellular physiology; alternatively, it may be related to other cellular functions of this enzyme, such as controlling the expression of as-yet-undescribed virulence factors.
MATERIALS AND METHODS
Bacterial strains and media.
B. abortus 2308 is a laboratory strain which is virulent in both natural and experimental hosts. Brucellae were grown at 37°C in brucella broth or on Trypticase soy agar (Difco Laboratories) supplemented with 5% defibrinated bovine blood (BA) and antibiotics as appropriate (kanamycin, 30 μg/ml; chloramphenicol, 5 to 15 μg/ml; and ampicillin, 50 μg/ml).
Primer extension analysis.
RNA was isolated using freeze-thaw extraction with acidified phenol (11). Primer extension was conducted as described elsewhere (30) using Moloney murine leukemia virus reverse transcriptase (New England Biolabs). The primer for this reaction was 5′-GCGCGGAAACGCAATCACCTTTGAT-3′. The corresponding nucleotide sequence was determined using dideoxy chain termination (31) with the same primer.
DNase I protection experiments.
DNase I footprinting experiments were performed with purified C. crescentus His6-CtrA that was phosphorylated using a maltose-binding protein–EnvZ fusion protein as described previously (27). The template DNA, a 490-bp B. abortus ccrM promoter fragment (PccrM −89 to +402), was generated by PCR with the oligonucleotides 5′-CGGGCTTTCCCTGTGATATT-3′ and 5′-AAGCCCGGATCCTGCAACT-3′ and end labeled with [γ-32P]ATP by use of T4 DNA polynucleotide kinase. The 3′ end of the labeled template was removed by digestion at an introduced BamHI site.
Construction of a B. abortus ccrM null mutant.
The B. abortus ccrM gene has been previously cloned and characterized (40). Brucella ccrM was disrupted by removing a 0.4-kb HindIII-XmnI fragment from the ccrM coding region of pRW378 and inserting the chloramphenicol resistance gene (cat) from pBlue-Cm2 to generate p378::Cm. The resultant ccrM::cat cassette was then removed by digestion with HincII and EcoRV and ligated to the SmaI site of the pUC-based vector pEX100T, containing the sacB and bla genes (32), to generate plasmid pGR17. pGR17 was introduced by electroporation into B. abortus 2308, and two chloramphenicol-resistant (Cmr) and ampicillin-resistant (Apr) integrants, GR121 and GR122, were selected for further analysis. In strains GR121 and GR122, pGR17 was integrated via homologous recombination 5′ of the native ccrM gene (see Fig. 2). Cells were grown in brucella broth in the absence of antibiotic selection to promote recombination, and sucrose-resistant clones were isolated by plating cells on modified Luria-Bertani sucrose plates (containing, per liter: 10 g of tryptone, 5 g of yeast extract, 50 g of sucrose, and 16 g of agar).
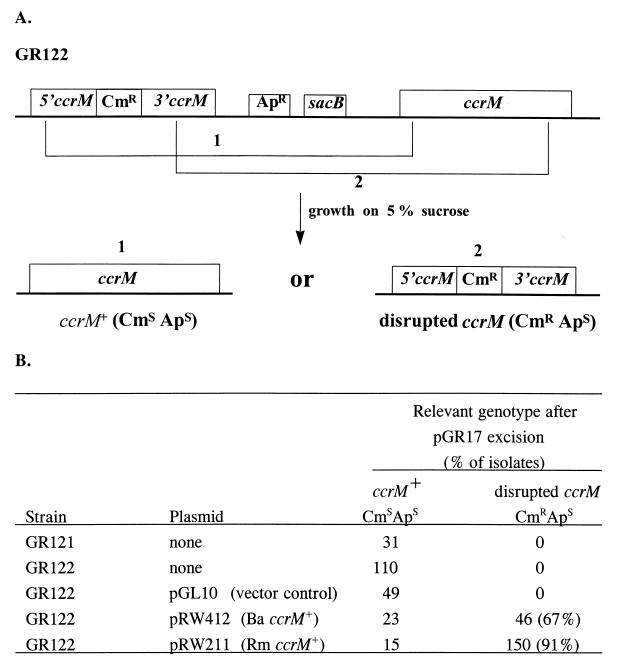
FIG. 2.
The B. abortus ccrM gene is essential for viability. (A) Strategy for disrupting the B. abortus ccrM gene. A schematic of the ccrM locus in GR122 is shown. Plasmid pGR17 was integrated into the B. abortus ccrM chromosomal locus, yielding strain GR122. Growth on 5% sucrose selects for a second recombination event that generates two types of isolates (labeled 1 and 2). (i) If excision occurs 5′ of cat, the gene encoding Cmr, then wild-type ccrM is retained and sucrose-resistant colonies will be Cms and Aps. (ii) If excision occurs on the opposite side of cat, then disrupted ccrM is retained and sucrose-resistant colonies will be Cmr and Aps. (B) Genotype of the ccrM chromosomal locus after growth on sucrose to select for excision of plasmid pGR17. The percentage of isolates that retained disrupted ccrM is shown in parentheses. Ba, B. abortus; Rm, R. meliloti.
Fixation procedures and microscopy.
Bacteria were harvested from BA plates, placed in brucella broth, pelleted by centrifugation, and either fixed in buffered neutral formalin (3.7% formaldehyde, 145 mM NaCl, 30 mM KH2PO4, 45 mM Na2HPO4) for phase microscopy or harvested in 2.5% glutaraldehyde–50 mM sodium cacodylate buffer (pH 7.4) for evaluation by transmission electron microscopy as previously described (4).
For phase microscopy, formalin-fixed cells were immobilized by placing them on a microscope slide with a thin pad of 1% agarose. The cells were observed with a Nikon Eclipse E800 microscope with a Nikon Plan Apo 100×/1.40 objective. The images were acquired with a cooled charge-coupled device camera and processed with Metamorph 4.01 (Universal Imaging Corp.). To ensure that fixation procedures did not influence cell morphology, live cells were also examined with an Olympus BH2 phase-contrast microscope under Biosafety Level 3 conditions.
Flow cytometry.
For flow cytometry analysis, the B. abortus strains GR129, GR130, GR131, and GR132 were subcultured in brucella broth at 37°C for 6 h (mid-exponential-phase growth). The cells were harvested and fixed in 70% ethanol at 4°C for 4 days. For fluorescence-activated cell sorter analysis, an aliquot of cells was centrifuged at 4,000 × g, washed with 1 ml of PBS (10 mM phosphate buffer [pH 7.4], 150 mM NaCl), stained in PBS containing Hoechst dye no. 33342 (10 μg/ml) for 30 min at 37°C, washed again, and resuspended in 1 ml of PBS. For each flow cytometry experiment, DNA content in a population of 10,000 cells was measured with a Becton Dickinson FACStar Plus machine with excitation at 358 nm and emission at 440 nm. The data were collected and analyzed using FlowJo software (Tree Star Inc., San Carlos, Calif.).
Isolation and infection of resident peritoneal macrophages.
Macrophages were harvested from the peritoneal cavities of euthanatized 9-week-old BALB/c mice by lavage with Dulbecco's modified Eagle's medium (DMEM)–5% fetal calf serum (FCS) supplemented with 5 U of heparin per ml. Pooled macrophages in 200 μl of DMEM–5% FCS were cultivated in 96-well plates at a concentration of 1.6 × 105 per well at 37°C with 5% CO2. Cell cultures were enriched for macrophages by washing away nonadherent cells after overnight incubation. B. abortus cells were opsonized for 30 min with a subagglutinating dilution (1:2,000) of hyperimmune BALB/c mouse serum in DMEM–5% FCS. Opsonized cells were added to macrophages at a ratio of approximately 40 bacteria per macrophage, and the mixture was incubated at 37°C for 1.5 h to allow time for phagocytosis. At this point, the culture medium was replaced with 200 μl of DMEM–5% FCS containing 50 μg of gentamicin per ml for 1 h to kill extracellular B. abortus. Macrophages were then washed three times in warm PBS–0.5% FCS and maintained in DMEM–5% FCS with 12.5 μg of gentamicin per ml. At various times after the addition of 12.5 μg of gentamicin per ml, individual cultures were lysed with 0.1% deoxycholate. The CFU were determined by serial dilution in PBS and duplicate plating on BA and on BA with 15 μg of chloramphenicol or 30 μg of kanamycin per ml. Statistical comparisons were made using Student's t test (34). P values of less than 0.05 were considered significant.
Biological containment and animal use.
All procedures involving live Brucella were performed in a Biosafety Level 3 containment facility following Centers for Disease Control and Prevention-National Institutes of Health guidelines (7a). In conducting experiments with animals, the investigators adhered to the Guide for the Care and Use of Laboratory Animals (7b).
RESULTS
The CtrA response regulator binds the B. abortus ccrM promoter.
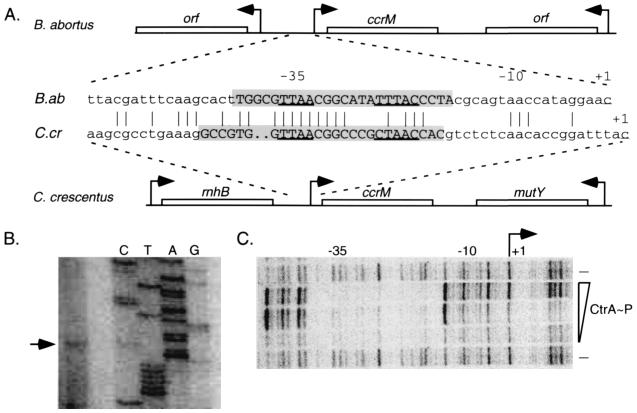
The transcriptional start site of the B. abortus ccrM gene was determined by primer extension assays using RNA isolated from wild-type cells. A single transcript initiating 34 bp upstream of the ccrM translational start site was detected (Fig. 1B). A comparison of the 5′ untranslated sequences upstream of ccrM in B. abortus and C. crescentus revealed extensive sequence homology and a putative CtrA binding site overlapping the −35 region of the B. abortus ccrM promoter (Fig. 1A). In C. crescentus, ccrM transcription is activated by the phosphorylated CtrA response regulator (CtrA-P) which binds to this region of the promoter (23, 27). Because a CtrA homolog has been identified in B. abortus that is 80% identical to the Caulobacter protein (J. J. Letesson, personal communication), we investigated the possibility that CtrA binds to the promoter of the B. abortus ccrM gene. Based on the extensive amino acid identity shared by the Brucella and Caulobacter CtrA proteins and because a purified version of the Brucella CtrA was not available, we used purified Caulobacter CtrA for these studies.
FIG. 1.
B. abortus ccrM promoter region. (A) Comparison of the genes adjacent to ccrM and the ccrM promoter regions in B. abortus (B.ab) and C. crescentus (C.cr). The CtrA recognition sequence is underlined, and the bases protected by His6–CtrA-P are in uppercase letters and shaded; identical bases are indicated by vertical lines. (B) Primer extension mapping of the B. abortus ccrM transcriptional start site. The first lane shows the primer extension product; the last four lanes show a sequencing ladder generated using the same primer. The lanes are labeled to reflect the sense strand. (C) DNase I protection of the ccrM promoter with purified C. crescentus His6–CtrA-P. His6–CtrA-P concentrations in the reactions were 50, 25, and 10 μg/ml. His6–CtrA-P was omitted from the lanes marked with a minus sign. The transcriptional start site (+1) and the −10 and −35 elements are indicated.
We assessed the binding of the Caulobacter His6–CtrA-P fusion protein to the Brucella ccrM promoter by DNase I footprinting analysis (Fig. 1C). Template DNA was generated by PCR and labeled at the 5′ end as described in Materials and Methods. Purified His6-CtrA was phosphorylated in vitro by use of the E. coli EnvZ histidine kinase (27) and was used in the footprinting reactions. As shown in Fig. 1, CtrA-P specifically protected a single 25-bp region from −18 to −42 relative to the transcriptional start site in the Brucella ccrM promoter. In both the Brucella and the Caulobacter ccrM promoters, the protected sequence overlaps the −35 region. These in vitro data suggest that CtrA-P regulates ccrM transcription in Brucella. The Caulobacter ccrM promoter contains two inverted repeat structures that are not found in the Brucella ccrM promoter; one encompasses the CtrA binding site, and the other is immediately downstream of the transcriptional start site and includes a pair of CcrM methylation sites (37). These differences in promoter structure suggest that although ccrM transcription is likely to be regulated by CtrA-P in Brucella, this bacterium may have additional modes of controlling ccrM expression.
B. abortus ccrM is essential for viability.
To determine if CcrM is essential for viability in B. abortus, we attempted to inactivate ccrM using a sacB counterselection technique. First, a plasmid-borne ccrM gene was disrupted by inserting a cat (Cmr) selectable marker; then, the resulting plasmid, pGR17, was integrated into the B. abortus ccrM chromosomal locus, generating strains GR121 and GR122 (Table 1 and Fig. 2A). This recombination event resulted in the separation of the complete wild-type chromosomal copy of ccrM and the disrupted ccrM gene by a plasmid sequence containing the sacB and bla (Apr) genes (Fig. 2A). Subsequent growth of these strains on 5% sucrose selected for excision of the sacB gene and a second recombination event between the homologous regions of the two ccrM genes. Cells retaining the wild-type ccrM gene and cells with the disrupted gene were distinguished by sensitivity to chloramphenicol. As shown in Fig. 2B, it was not possible to obtain Cmr Aps strains containing only the disrupted ccrM gene. To confirm that ccrM is essential, we showed that the chromosomal ccrM locus could be inactivated when a functional copy of B. abortus ccrM was provided on a plasmid. The chromosomal copy of ccrM could be disrupted in the presence of pRW412 but not in the presence of the pGL10 vector alone. These data demonstrate that ccrM is required for B. abortus viability under normal growth conditions. The ccrM gene from the closely related species R. meliloti also complemented the B. abortus ccrM null strain.
TABLE 1.
Bacterial strains and vectors used in this study
| B. abortus strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 2308 | Wild type, virulent, smooth lipopolysaccharide | Laboratory stock |
| GR121 | 2308 ccrM/ccrM::cat sacB bla, integration of pGR17 occurred on the 5′ side of cat | This study |
| GR122 | 2308 ccrM/ccrM::cat sacB bla, integration of pGR17 occurred on the 5′ side of cat | This study |
| GR129 | 2308 containing plasmid pBBR1MCS | This study |
| GR130 | 2308 containing plasmid pGL10 | This study |
| GR131 | 2308 containing plasmid pRW412 | This study |
| GR132 | 2308 containing plasmid pRW414 | This study |
| Plasmids | ||
| pBBR1MCS | Chloramphenicol-resistant broad-host-range cloning vector (moderate copy number [10 to 12 copies per genome equivalent]) (9) | 15 |
| pGL10 | Stable kanamycin-resistant RK2 derivative (low copy number [2 to 4 copies per genome equivalent]) (this study) | K. Andersen |
| pBlue-Cm2 | cat amplified by PCR from pBC and cloned into the EcoRV site of pBluescript KS(+) | M. E. Kovach |
| pEX100T | Ampicillin-resistant cloning vector containing the B. subtilis sacB gene | 32 |
| pRW378 | pBBR1MCS with a 2.0-kb SalI-EcoRV insert containing the B. abortus ccrM gene and promoter region | This study |
| p378::Cm | pRW378 with an internal 0.4-kb HindIII-XmnI fragment removed and replaced with cat excised from pBlue-Cm2 as a 1.0-kb HindIII-SmaI fragment | This study |
| pGR17 | pEX100T with a 3.1-kb HincII-EcoRV fragment containing ccrM::cat from p378::Cm in the SmaI site | This study |
| pRW211 | pGL10 with a 1.8-kb PstI-HindIII fragment containing the R. meliloti ccrM gene and promoter | This study |
| pRW412 | pGL10 with a 1.6-kb XbaI-KpnI fragment containing the B. abortus ccrM gene and promoter region | This study |
| pRW414 | pBBR1MCS with a 1.6-kb XbaI-KpnI fragment containing the B. abortus ccrM gene and promoter region | This study |
Increased ccrM copy number alters cell morphology and DNA replication.
Constitutive expression of ccrM throughout the C. crescentus cell cycle and overexpression of ccrM in R. meliloti cause aberrant morphology and relaxation of the control of chromosome replication (40). To test the effects of increased copy number of the ccrM gene on B. abortus, the ccrM coding sequence and 315 bp of the 5′ region were cloned into the low- and moderate-copy-number vectors pGL10 (2 to 4 copies/cell) and pBBR1MCS (10 to 12 copies/cell) (9). The resulting plasmids, pRW412 and pRW414, were introduced into wild-type B. abortus 2308 to generate strains GR131 and GR132, respectively (Table 1). Strain GR132 exhibited a strikingly aberrant morphology. Electron microscopy (Fig. 3) and light microscopy (Fig. 4) showed that the majority of GR132 cells were enlarged and branched. The branched GR132 cells represented single bacteria with atypical morphology and not several attached daughter cells (Fig. 3). A similar branched phenotype has been observed for R. meliloti cells that overexpress ccrM (40). In this bacterium, the branched phenotype correlates with blocked cell division (16). In both strains GR129 (control) and GR131, the cells were short rods typical of wild-type cells (Fig. 3 and 4).
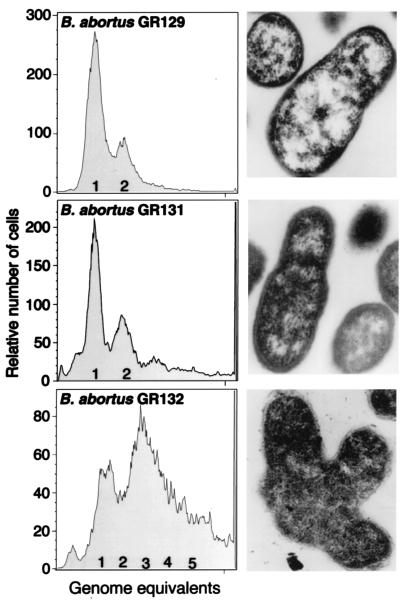
FIG. 3.
Effect of overexpressing B. abortus ccrM on DNA replication and cell morphology. Shown are flow cytometry analysis and electron micrographs (magnification, ×27,500) of strains GR129 (B. abortus 2308 with vector alone), GR131 (B. abortus 2308 expressing ccrM from a low-copy-number vector), and GR132 (B. abortus 2308 expressing ccrM from a moderate-copy-number vector). For the flow cytometry analysis, the vertical axis shows the relative number of cells measured by fluorescence intensity, and the horizontal axis shows the DNA content (presented as genome equivalents).
FIG. 4.
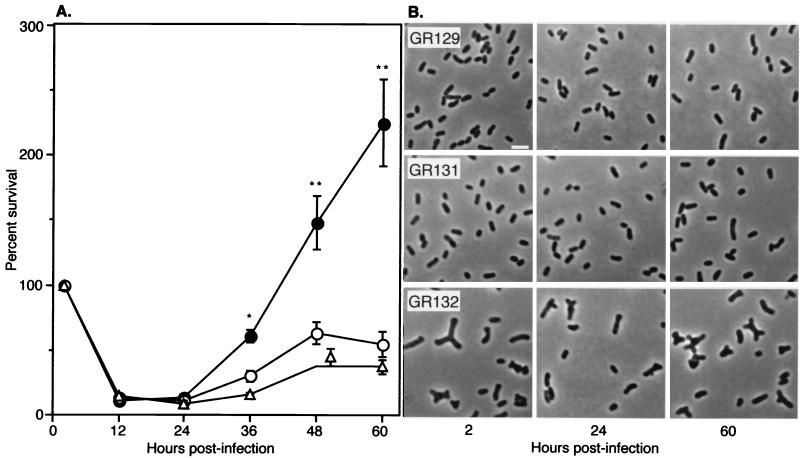
Survival of B. abortus in cultured murine macrophages. (A) Murine peritoneal macrophages were infected with B. abortus GR129 (closed circles), GR131 (open circles), and GR132 (open triangles) as described in Materials and Methods. At each time point after phagocytosis, the number of surviving intracellular bacteria in four replicate wells was determined. Percent survival represents the number of intracellular bacteria present at each sampling time divided by the number present at time zero and multiplied by 100. Data are mean ± standard error. Symbols: ∗, survival of GR132 is significantly different from that of the GR129 control (P < 0.05) (Student's t test); ∗∗, survival of both GR131 and GR132 is significantly different from that of the GR129 control (P < 0.05). (B) Morphology of B. abortus isolated from macrophages. B. abortus strains GR129, GR131, and GR132 were isolated from macrophages 2, 24, and 60 h after infection, fixed in buffered neutral formalin, and examined by phase microscopy. Bar, 2 μm.
In R. meliloti and C. crescentus, multiple copies of ccrM result not only in aberrant cell division and cell morphology but also in relaxation of the control of the initiation of DNA replication (40, 41, 42). To determine if the overexpression of ccrM alters the control of DNA replication in B. abortus, we used flow cytometry to compare the DNA contents of control strain GR129 and the two strains that have multiple copies of ccrM (Fig. 3). In control strain GR129, a single peak representing the two chromosomes in the B. abortus genome (13) was observed for 70% of the cells (Fig. 3). The remaining 30% of the cells contained two genome equivalents. Similar results were obtained with control strain GR130 (B. abortus containing the pGL10 vector; data not shown). In strain GR131, which carries ccrM on a low-copy-number vector, about 40% of cells contained two or more genome equivalents (Fig. 3). Therefore, a small increase in ccrM copy number had little effect on DNA content or cell morphology. In contrast, over 70% of GR132 cells contained more than one genome equivalent, and many contained three to five genome equivalents. Many of the GR132 cells were also branched, suggesting a defect in the control of DNA replication or the coupling of DNA replication to cell division in this strain.
Increased ccrM copy number attenuates the intracellular replication of B. abortus in macrophages.
To evaluate the intracellular replication of B. abortus strains carrying multiple copies of ccrM in cultured murine peritoneal macrophages, macrophages were infected with immunoglobulin G-opsonized B. abortus control strain GR129, GR131 (low ccrM copy number), or GR132 (moderate ccrM copy number). Each strain remained viable during the opsonization and internalization steps. Equivalent numbers of cells were internalized by phagocytosis, as judged by the numbers of intracellular bacteria present after gentamicin treatment (data not shown). As expected, the number of intracellular bacteria decreased dramatically during the first 24 h after infection in all groups (Fig. 4A). However, 36 to 60 h after infection the number of viable intracellular brucellae increased significantly in macrophages infected with the control strain. For both B. abortus strains containing multiple copies of ccrM, the number of intracellular bacteria increased slightly but not to control levels. These data indicate that the overexpression of ccrM did not affect uptake of the bacteria by macrophages but attenuated the intracellular replication of B. abortus in these phagocytes. Despite the absence of antibiotic selection in these experiments, no significant loss of vector-encoded antibiotic resistance was detected, indicating that the plasmids were retained throughout the study (data not shown). The difference in antibiotic resistance imparted by pRW414 (kanamycin resistance) and pRW414 (chloramphenicol resistance) also allowed us to verify the identities of GR131 and GR132 following isolation from cultured macrophages. In liquid cultures, strains GR131 and GR132 grew at the same rate as the vector-only controls (data not shown). This result suggests that gross alterations in cell growth rate are not responsible for attenuation of the replication of these strains in macrophages.
Bacteria isolated from macrophages 2, 24, and 60 h after phagocytosis were examined for morphologic abnormalities by light microscopy. As shown in Fig. 4B, cells of the B. abortus control strain GR129 and strain GR131 retained wild-type morphology at all times, while the majority of GR132 cells remained enlarged and branched throughout the study. Because the intracellular replication of both GR131 and GR132 was attenuated (Fig. 4A), the aberrant morphology and relaxation of the control of DNA replication observed for strain GR132 (Fig. 3) were not solely responsible for the failure of these strains to replicate within macrophages. Thus, the impaired intracellular replication of the B. abortus strains bearing multiple copies of ccrM is likely to be due to additional cellular functions of CcrM. Although the data presented in Fig. 4 are the results of a single representative experiment, equivalent results were obtained from multiple experiments.
DISCUSSION
The phylogenetic relationship between B. abortus and α-proteobacteria has been well established (20). We have further investigated this relationship by examining the role of the CcrM DNA methyltransferase in the maintenance of normal physiology in B. abortus. As in C. crescentus and the plant symbiont R. meliloti, ccrM is required for the viability of B. abortus. This requirement is not believed to be due to the restriction of self DNA, as CcrM does not appear to have an associated restriction endonuclease (36). Therefore, CcrM methylation is required for essential cellular processes that appear to be common to at least three members of the α-proteobacteria. The physiologic basis for the essential nature of this enzymatic reaction is not known, although CcrM has been linked to normal cell cycle progression in C. crescentus (41, 42), and it seems likely that deregulation of this process could be detrimental to the maintenance of normal cellular physiology. Our studies suggest a similar role for CcrM in B. abortus, as strains bearing multiple copies of ccrM exhibit aberrant cell morphology and relaxation of the control of DNA replication. These processes do not appear to be significantly altered in strains bearing fewer copies of ccrM, suggesting that a threshold level of the enzyme must be reached to obtain the gross morphological changes observed with strains bearing higher-copy-number plasmids. In C. crescentus, the Lon protease is responsible for clearing CcrM from the cells (41). Perhaps proteolysis can effectively clear CcrM from B. abortus cells when it is overproduced at low levels but fails to do so at higher levels.
In addition to the proposed role of CcrM in cell cycle progression in C. crescentus (26), there is growing evidence that CcrM-mediated methylation also regulates gene expression. Methylation sites are present in the promoter regions of several Caulobacter genes, including the ctrA response regulator, the ftsZ cell division gene, and the flagellar genes fliL and fliQ (26). Moreover, expression of the Caulobacter ccrM gene appears to be autoregulated by the methylation of its own promoter, as mutation of the GAnTC methylation sites in the mRNA leader region results in prolonged ccrM expression (37). These findings support the contention that CcrM-mediated DNA methylation controls the transcription of certain genes and may provide a means to link gene expression to cell cycle progression in this bacterium. Brucella and Caulobacter are closely related genera with at least two homologous regulatory proteins, CcrM and CtrA. The CtrA response regulator orchestrates cell cycle events in C. crescentus. It controls the transcription of both ccrM and ftsZ (14, 23), initiates biogenesis of the flagellum (23), and inhibits DNA replication initiation (24). The fact that the Caulobacter CtrA protein binds to the Brucella ccrM promoter strengthens the premise that the Caulobacter and Brucella CtrA proteins are functionally interchangeable. Our findings also suggest that the transcription of ccrM in B. abortus is likely to be controlled by CtrA.
In enteric bacteria, Dam methylation plays critical roles in the timing and control of basic physiologic processes, such as the initiation of DNA replication, mismatch repair, and transcription (3). Precise levels of Dam also appear to be required for the virulence of Salmonella enterica serovar Typhimurium (10, 12). Specifically, deletion or overproduction of Dam results in severe attenuation of this organism in experimentally infected mice (12). In uropathogenic E. coli, Dam methylation also regulates the expression of the pap operon (21, 38) and consequently the production of pili, structures known to be significant virulence determinants (8).
Brucellae are intracellular pathogens and, arguably, the true environmental niche for these organisms is within phagosomes of host macrophages (2). Within these cells, B. abortus exhibits major changes in gene expression (17), resulting in either the induction or the repression of at least 73 proteins (25). This alteration in gene expression cannot be mimicked in the presence of in vitro stress conditions, such as heat, acid, and nutritional and oxidative stress. These findings suggest that within macrophages, the brucellae are subject to complex global regulation of gene expression. Despite extensive studies to identify dedicated virulence factors for this organism, however, only a select subset of genetically defined Brucella strains demonstrating significant and stable attenuation has been constructed. This subset includes strains with mutations resulting in the loss of normal lipopolysaccharide O-side-chain biosynthesis (1) and the inactivation of genes encoding a type IV secretion apparatus (22), the BvrR-BvrS two-component regulatory system (35), and a putative RNA binding protein (encoded by the hfq gene) that appears to be required for the maintenance of stationary phase (28). Intriguingly, the Brucella hfq also contains a putative CtrA binding sequence overlapping the −10 region of its promoter and therefore may be under the negative control of this transcriptional regulator. This finding provides indirect evidence that CtrA may regulate the expression not only of ccrM but also of at least one additional gene required for survival in macrophages and virulence in BALB/c mice.
The defective intracellular replication in macrophages of the B. abortus strains with ccrM on low-copy-number plasmids strongly suggests that the physiologic basis for this defect is independent of that which produces aberrant cell morphology. For example, CcrM methylation may control the expression of genes which are specifically required for successful adaptation to the intracellular environment. This activity could be dependent on some function that is directly regulated by DNA methylation status. Alternatively, this activity could be mediated by a second regulator which is itself controlled by DNA methylation. A prime candidate for the latter is the recently identified Brucella CtrA response regulator, which has six CcrM methylation sites within the 240 bases immediately upstream of the ATG (G. T. Robertson and R. M. Roop II, unpublished observation). CtrA likely contributes to the regulation of a wide array of cellular functions, including the regulation of ccrM and hfq transcription (28). Understanding the basis for the essential nature of CcrM in B. abortus and the cellular functions that are controlled by this enzyme should provide insights into the mechanisms used by brucellae to adapt to and survive within host macrophages.
It has been proposed that the Dam methyltransferase of the enteric bacteria may represent an ideal target for the design of new antimicrobial agents and the development of vaccines (12). In this same vein, our data suggest that the CcrM methyltransferase is an appropriate target for new antibiotics. CcrM homologs are widely distributed among the α-proteobacteria (40), and this group contains a number of important animal and plant pathogens, including, in addition to B. abortus, Agrobacterium, Bartonella, and Ochrobactrum (39). Given that CcrM is essential for the viability of at least three members of the α-proteobacteria, it seems likely that its essential functions may be conserved throughout this group of bacteria.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant GM51426 and by a contract from the U.S. Army Defense Advanced Research Projects Agency (MDA972-97-1-0008-0002).
REFERENCES
- 1.Allen C A, Adams L G, Ficht T A. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect Immun. 1998;66:1008–1016. doi: 10.1128/iai.66.3.1008-1016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin C L, Roop R M., II . Brucella infections and immunity. In: Paradise L J, Friedman H, Bendinelli M, editors. Opportunistic intracellular bacteria and immunity. New York, N.Y: Plenum Press; 1998. pp. 255–279. [Google Scholar]
- 3.Barras F, Marinus M G. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 4.Beveridge T J, Popkin T J, Cole R M. Electron microscopy. In: Gerhardt P, et al., editors. Methods for general and molecular biology. Washington, D.C.: American Society for Microbiology; 1994. pp. 42–71. [Google Scholar]
- 5.Bickle T A, Kruger D H. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boye E, Løbner-Olesen A. The role of Dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 7.Canning P C. Phagocyte function in resistance to brucellosis. In: Adams L G, editor. Advances in brucellosis research. College Station: Texas A & M University Press; 1990. pp. 151–163. [Google Scholar]
- 7a.Centers for Disease Control and Prevention. Biosafety in microbiological and biomedical laboratories. CDC publication no. 93-8395. Atlanta, Ga: Centers for Disease Control and Prevention; 1993. [Google Scholar]
- 7b.Committee on Care and Use of Laboratory Animals, Institute of Laboratory Animal Resources, National Research Council. 1985. Guide for the care and use of laboratory animals. NIH publication no. 86-23.
- 8.Domingue G J, Roberts J A, Laucirica R, Ratner M H, Bell D P, Suarez G M, Kallenius G, Svenson S. Pathogenic significance of P-fimbriated Escherichia coli in urinary tract infections. J Urol. 1985;133:983–989. doi: 10.1016/s0022-5347(17)49341-4. [DOI] [PubMed] [Google Scholar]
- 9.Elzer P H, Phillips R W, Kovach M E, Peterson K M, Roop R M., II Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect Immun. 1994;62:4135–4139. doi: 10.1128/iai.62.10.4135-4139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Del Portillo F, Pucciarelli M G, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 13.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 16.Latch J N, Margolin W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol. 1997;179:2373–2381. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Ficht T A. Protein synthesis in Brucella abortus induced during macrophage infection. Infect Immun. 1995;63:1409–1414. doi: 10.1128/iai.63.4.1409-1414.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 19.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 20.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nou X, Skinner B, Braaten B, Blyn L, Hirsch D, Low D. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol Microbiol. 1993;7:545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 22.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz Y M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 23.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 24.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafie-Kolpin M, Essenberg R C, Wyckoff J H., III Identification and comparison of macrophage-induced proteins and proteins induced under various stress conditions in Brucella abortus. Infect Immun. 1996;64:5274–5283. doi: 10.1128/iai.64.12.5274-5283.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reisenauer A, Kahng L S, McCollum S, Shapiro L. Bacterial DNA methylation: a cell cycle regulator? J Bacteriol. 1999;181:5135–5139. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson G T, Roop R M., II The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- 29.Russell D W, Zinder N D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987;50:1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 33.Smith L D, Ficht T A. Pathogenesis of Brucella. Crit Rev Microbiol. 1990;17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 34.Snedecor G W, Cochran W G. Statistical methods. 8th ed. Ames: Iowa State University Press; 1989. [Google Scholar]
- 35.Sola-Landa A, Pizarro-Cerda J, Grillo M J, Moreno E, Moriyon I, Blasco J M, Gorvel J P, Lopez-Goni I. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29:125–138. doi: 10.1046/j.1365-2958.1998.00913.x. [DOI] [PubMed] [Google Scholar]
- 36.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens C M, Zweiger G, Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 39.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 42.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]