ABSTRACT
Previous study has demonstrated the high expression of circular RNA 3-oxoacid CoA-transferase 1 (circ-OXCT1) in lung adenocarcinoma tumor tissues. However, the role and possible mechanism of circ-OXCT1 in non-small cell lung cancer (NSCLC) progression was unclear.Quantitative real-time PCR (qRT-PCR), western blotting and immunohistochemistry (IHC) staining assay were performed to detect the expression of circ-OXCT1, microRNA-516b-5p (miR-516b-5p), solute carrier family 1 member 5 (SLC1A5) and other indicated protein markers. Cell proliferation was measured by Cell counting kit 8 (CCK8), colony formation and 5-Ethynyl-2’-deoxyuridine (EdU) assays. Flow cytometry was employed to detect the rate of apoptotic cells. Cell migration and invasion were measured using transwell assay. The relative glutamine uptake and α-ketoglutarate (α-KG) production was determined using commercial kits. Interaction between miR-516b-5p and circ-OXCT1 or SLC1A5 was predicted by bioinformatics analysis and confirmed via luciferase reporter and RNA immunoprecipitation (RIP) assays. In vivo assay was implemented to demonstrate the effect of circ-OXCT1 in tumor growth.Circ-OXCT1 and SLC1A5 were upregulated and miR-516b-5p was downregulated in NSCLC tissues and cells. Functional experiments revealed that circ-OXCT1 silencing suppressed cell proliferation, migration and invasion, but promoted cell apoptosis in vitro. Circ-OXCT1 knockdown repressed tumor formation in vivo. Besides, miR-516b-5p was a target of circ-OXCT1, and miR-516b-5p inhibitor could relieve circ-OXCT1 absence-mediated effects in NSCLC cells. SLC1A5 was identified as a target of miR-516b-5p. Circ-OXCT1 promoted SLC1A5 expression by target binding with miR-516b-5p.Circ-OXCT1 facilitated NSCLC progression via miR-516b-5p-dependent regulation of SLC1A5, which provided a possible circRNA-targeted therapy for NSCLC.
KEYWORDS: NSCLC, circ-OXCT1, MiR-516b-5p
Introduction
Lung cancer is a common malignant tumor with high morbidity and mortality characteristics [1]. Non-small cell lung cancer (NSCLC) is one common type of lung cancer and account for about 85% of lung cancer cases [2]. Surgical resection combined with radiotherapy and chemotherapy is the main treatment strategy for NSCLC [3,4]. With the development and application of molecular targeted therapy and immunotherapy, the current status of NSCLC treatment has improved [5,6]. However, due to the late diagnosis, imperfect development of targeted therapy drugs and high recurrence of NSCLC, current treatment strategies only have a good effect on partly patients, and the overall clinical results of the advanced patients are still poor [5,7]. Therefore, the exact mechanism behind the progression of NSCLC needs to be explored urgently to identify potential diagnostic and therapeutic biomarkers.
Circular RNA (circRNA), a non-coding RNA, is formed by the back-splicing of linear pre-mRNA [8]. And it is widely expressed in multiple species and has a conserved sequence [9]. Compared with linear RNA, the circular structure of circRNA makes it resistant to degradation induced by RNase R and has better stability [10]. In addition, the results of RNA sequencing showed that multiple circRNAs were dysregulated in cancer patient samples, playing a cancer-promoting or anti-tumor effect [8,11]. Therefore, circRNA is an ideal biomarker for cancer due to the stability and differential expression. With the deepening of research, it has been confirmed many times that circRNA acts as miRNA sponge to regulate the mRNA expression of target gene, thereby regulating the cell biological functions. For example, hsa-circRNA-G004213 contributed to PRPF39 expression and suppressed the cisplatin resistance by targeting miR-513b-5p in liver cancer [12]. CircRHOT1 facilitated pathogenesis of NSCLC by regulating miR-330-5p-dependent YY1 expression [13]. Circ_RPPH1 promoted breast cancer cell growth and motility while boosted apoptosis via miR-146b-3p/E2F2 axis [14]. Therefore, research on the role and mechanism of circular RNA may provide new support for molecular targeted therapy of NSCLC.
CircRNA 3-oxoacid CoA-transferase 1 (circ-OXCT1, ID: hsa_circ_0004873) is a newly discovered circRNA formed by the cyclization of exons 8–13 of the OXCT1 gene. In previous study, circ-OXCT1 expression was highly expressed in NSCLC tumor samples and cells [15]. However, the role of circ-OXCT1 in the pathogenesis of NSCLC and the underlying molecular mechanism remain unclear. The starBase and circInteractome databases were used for bioinformatics analysis, and the results revealed that circ-OXCT1 and miR-516b-5p had the binding sites in their sequences. In addition, Song et al. have confirmed that miR-516b-5p played an active role in the process of NSCLC [16]. Solute carrier family 1 member 5 (SLC1A5) is a glutamine transporter, which is mainly responsible for transporting glutamine from the outside to the inside of the cell, and plays an important role in the glutamine metabolism process [17]. The prediction results of the starBase online software revealed that there were binding sites between SLC1A5 3ʹUTR and miR-516b-5p. However, it was not clear whether there was a circ-OXCT1/miR-2516b-5p/SLC1A5 regulatory axis during the development of NSCLC.
Therefore, we first investigated the roles of circ-OXCT1 in NSCLC, and then further explored the involvement of miR-516b-5p/SLC1A5 axis in the regulation of circ-OXCT1 in NSCLC.
Materials and methods
Clinical tissues and cell lines
NSCLC tissues (including lung adenocarcinoma and lung squamous carcinoma tissues) and paired adjacent normal tissues were obtained from 66 identified NSCLC patients in Changsha Central Hospital between June 2019 to January 2021. Before the surgery, patients had not received chemotherapy or radiation therapy. After operation, tissue specimens were immediately frozen at −80°C. This study was approved by Changsha Central Hospital, and all subjects provided the written informed consents.
NSCLC cell lines (A549, H1975 and H1650) and control cell line (16HBE) were bought from Tongpai Biotechnology (Shanghai, China). 16HBE, H1975 and H1650 were growth in RPMI-1640 (Gibco, Thermo Fisher Scientific, Rockville, MD, USA). A549 cells were cultivated in Ham’s F12 K (Procell, Wuhan, China) at 37°C with 5% CO2. And 10% fetal calf serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco) were added into all mediums. We (all authors) were aware of the group allocation at the different stages of the experiment.
Quantitative real-time PCR (qRT-PCR)
TRIzol Reagent (Beyotime, Shanghai, China) was employed to obtain total RNA from NSCLC tissues and cells. After testing RNA concentration by NanoDrop One/OneC (Thermo Scientific, Shanghai, China), Synthesis Kit (Invitrogen, Carlsbad, CA, USA) was used to obtain cDNA. Then, specific primers and SYBR Green (Solarbio, Beijing, China) were mixed and incubated with the cDNA to conduct qRT-PCR. Primer sequences were shown in Table 1. Relative expression was calculated by 2−ΔΔCT method and normalized to β-actin or U6.
Table 1.
Primers sequences used for PCR.
| Name | Primers for PCR (5’-3’) | |
|---|---|---|
| Circ-OXCT1 | Forward | ACACGTCGATCTGACAATGCT |
| Reverse | TGGATGTCTTCTGGAGCAAATG | |
| OXCT1 | Forward | GGCACACTTGCAGAGAGGAT |
| Reverse | GGCTTACTGGCAATGGCAAC | |
| miR-136-5p | Forward | GCCGAGACTCCATTTGTTTTG |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| miR-145-5p | Forward | GCCGAGGTCCAGTTTTCCC |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| miR-665 | Forward | GTATGAGACCAGGAGGCTGAGGC |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| miR-516b-5p | Forward | GCCGAGATCTGGAGGTAAGA |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| SLC1A5 | Forward | GAGACTCCAAGGGGCTCGC |
| Reverse | CACAAGCAGGTTGGCTCGAAG | |
| β-actin | Forward | TGGATCAGCAAGCAGGAGTA |
| Reverse | TCGGCCACATTGTGAACTTT | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
RNase R assay
One portion of RNA obtained from A549 and H1650 cells was treated with 3 U/μg RNase R (Geneseed, Guangzhou, China). Then circ-OXCT1 expression and linear OXCT1 expression were measured by qRT-PCR.
Cell transfection
Lipofectamine 2000 (Invitrogen) was used for cell transfection. The short hairpin RNA (shRNA) of circ-OXCT1 (sh-circ-OXCT1), miR-516b-5p mimic (miR-516b-5p), miR-516b-5p inhibitor (in- miR-516b-5p), SLC1A5 overexpression plasmid (SLC1A5) and corresponding controls (sh-NC, miR-NC, in-miR-NC and pcDNA) were purchased from Ribobio (Guangzhou, China).
Cell counting kit 8 (CCK8) assay
Transfected NSCLC cells were cultured until the predetermined time points. Then, CCK-8 reagent (Beyotime) was added into medium and mixed well. 4 hours later, the absorbance at 450 nm was detected by an enzyme immunoassay analyzer (Bio-Tek, Winooski, VT, USA).
Colony formation assay
Transfected A549 and H1650 cells were cultured for 2 weeks. After removing the culture medium, the paraformaldehyde and crystal violet bought from Phygene (Fuzhou, China) were used to fix and stain cells. And the number of colonies were counted under microscope.
5-Ethynyl-2’-deoxyuridine (EdU) assay
To measure the cell proliferative ability, the DNA synthesis was monitored using BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime). Briefly, transfected NSCLC cells were co-incubated with click reaction buffer, CuSO4, Azide 488, click additive solution and Hoechst 33,342 solution. Finally, EdU positive cell rate were calculated via fluorescence images.
Flow cytometry
After transfection, NSCLC cells were harvested by centrifugation. Afterward, cells were stained with Annexin V-FITC and 10 μL of PI (MultiSciences, Hangzhou, China) in the dark for 15 min. At last, flow cytometer was utilized to quantify apoptotic cells.
Transwell assay
Transwell chambers (BD Bioscience, San Jose, CA, USA) were employed to measure NSCLC cell migration ability. Transfected NSCLC cells suspended with medium without FBS was seeded into the upper of chambers, and medium containing 10% FBS (Gibco) was added into the bottom chambers. For cell invasion detection, transwell chambers were pre-coated with matrigel (BD Bioscience), and cells need to invade the matrigel-coated polycarbonate transwell filter. After 24-hour of incubation, paraformaldehyde (Phygene) and 0.1% crystal violet (Phygene) were used to fix and dye migrated or invaded cells respectively. And the migrated and invaded cells was photographed and counted.
Western blotting
The NSCLC tissues and cells were lysed by RIPA buffer (Beyotime) to obtain total protein, which was separated by SDS-PAGE gel and transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA). Then, the membranes with protein signals were blocked using 5% defatted milk for 1 h. After being co-incubated with primary and secondary antibodies that presented in Table 2 at 4°C overnight, BeyoECL Plus kit (Beyotime) was used to visualize the protein signals of the membranes. Image Lab software was used to analyze the gray value of proteins.
Table 2.
The antibodies in Western blotting and IHC.
| Antibody | Cat. | Dilution ratio | Source |
|---|---|---|---|
| PCNA | ab92552 | 1:10000 | Abcam |
| Bax | ab32503 | 1:10000 | Abcam |
| E-cadherin | 20874-1-AP | 1:10000 | Proteintech |
| SLC1A5 | ab237704 | 1:1000 | Abcam |
| β-actin | ab8227 | 1:5000 | Abcam |
| ki-67 | ab15580 | 1:200 | Abcam |
| Goat Anti-Rabbit IgG H&L (HRP) | ab6721 | 1:20000 | Abcam |
Detection of glutamine uptake and α-ketoglutarate (α-KG) production
The transfected A549 and H1650 cells were grown in medium containing 2 mM glutamine (Beyotime) for 24 h. According to the manufacturers’ instructions, the glutamine uptake and α-KG production were detected using Glutamine Colorimetric Assay Kit (Biovision, Milpitas, CA, USA) and α-KG Colorime-tric Assay Kit (Biovision) respectively.
Bioinformatics analysis and dual-luciferase reporter system
The miRNAs in circ-OXCT1 was retrieved by starBase and CircInteractome online software, and the retrieval results were showed in Venn diagram. Besides, starBase online software was also used to predict miR-516b-5p-binding sites in SLC1A5. Afterward, fragments of wild or mutant type (WT or MUT) circ-OXCT1 and 3ʹUTR of SLC1A5 mRNA were cloned into the upstream of pmirGLO luciferase reporter to generate luciferase reporter for WT or MUT circ-OXCT1 and SLC1A5. The above reporter vectors and miR-516b-5p mimic were co-transfected into A549 and H1650 cells, the luciferase activity was determined by Dual-Lumi™ II Luciferase Assay Kit (Beyotime).
RNA immunoprecipitation (RIP) assay
RIP Kit (Millipore, Billerica, MA, USA) was employed to perform Magna RIP assay. Briefly, RIP lysis buffer was used to obtain NSCLC cell lysates, which were mixed with magnetic beads pre-coated with antibodies against Ago2 or IgG. The enrichment of circ-OXCT1 and miR-516b-5p was detected by qRT-PCR.
Xenograft model
Ten healthy BALB/c nude mice (5-week-old, female, 16–20 g) from the same animal cage were purchased from Vital River Laboratory (Beijing, China), and animal study obtained the approval of the Animal Ethics Committee of Changsha Central Hospital. Animals were adaptive feeding for three days under standard laboratory conditions (specific-pathogen-free, 23°C ± 2°C, 45%~65% humidity) and randomly divided into two group (n = 5/group). H1650 cells transfected with sh-NC or sh-circ-OXCT1 were subcutaneously inoculated into the dorsal side of the mice after anesthesia with 2% isoflurane. Tumor volume was monitored via the method of length×width2 × 0.5 every 7 d for 35 d. On the 35th day after cells injection, after mice were euthanized by overdose CO2 for 10 min, tumors were taken out and weighed. To measured RNA and protein expression in tumor tissues of nude mice, qRT-PCR and western blotting were performed. In addition, immunohistochemistry (IHC) assay was conducted to detect the expression of SLC1A5 and ki-67. Antibodies used in IHC were listed in Table 2. In this work, the potential confounders were not controlled. Dead and ailing animals were excluded from the study. There were no experimental animals were excluded in this study because there were no dead or ailing animals. Each experimental group, report any animals, experimental units or data analysis were no exclusions. The data distribution of all animal experimental results was normal, and comparison was analyzed by Student’s t-test.
Statistical analysis
Data were presented as the means ± standard deviations. All experiments were repeated at least 3 times. The significant differences between the two groups were compared with unpaired t test (two-tailed). 1/2-way analysis of variance (ANOVA) with Tukey’s test was employed to calculate the P value among three or more groups. Mann-Whitney U test was accessed to analyze the differences between two group clinical specimens. Pearson correlation coefficient was adopted to test intermolecular linear relations. All data met the assumptions of the statistical approach. Statistical analysis was carried out using GraphPad Prism 8.0 software, and P < 0.05 (confidence level: 95%) indicated statistical significance.
Results
Circ-OXCT1 expression was upregulated in NSCLC tissues and cells
Circ-OXCT1 was formed by the back-splicing of exon 8–13 of OXCT1 gene, and the chromosomal location of circ-OXCT1 was shown in Figure 1a. The expression of circ-OXCT1 in NSCLC tumor tissues and was upregulated compared to adjacent normal tissues (Figure 1(b)). Also, qRT-PCR data presented the high expression of circ-OXCT1 in cell lines (A549, H1975, and H1650) in comparison with 16HBE cells (Figure 1(c)). Additionally, we proved that circ-OXCT1 could resistant to the digestion of RNase R, while linear OXCT1 could be digested by RNase R (Figure 1(d-e)). These data demonstrated that circ-OXCT1 might be implicated in the development of NSCLC.
Figure 1.
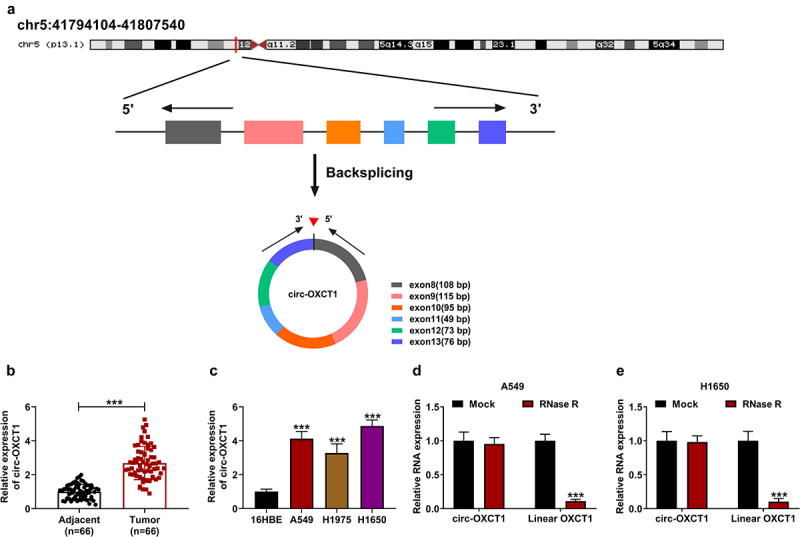
Circ-OXCT1 was highly expressed in NSCLC tissues and cell lines. (a) The basic information of circ-OXCT1 was shown. (b-c) Circ-OXCT1 expression was measured by qRT-PCR in clinical NSCLC tumor (n = 66), adjacent normal tissues (n = 66) as well as in 16HBE, A549, H1975 and H1650 cells. (d-e) RNase R assay was performed to verify the circular characteristic of circ-OXCT1. n = 3 independent biological replicates. Data were showed as mean ± SD. ***P < 0.001 by Mann-Whitney U test, one-ANOVA followed by Tukey’s post hoc test or unpaired t test (two-tailed).
Circ-OXCT1 depletion inhibited cell proliferation, motility and glutamine metabolism, and induced cell apoptosis in NSCLC cells
To explore the role of circ-OXCT1 in NSCLC cells, sh-circ-OXCT1 was transfected into A549 and H1650 cells, and the transfection efficiency results revealed that circ-OXCT1 knockdown downregulated circ-OXCT1 expression (Figure 2a). Subsequently, CCK-8 assay, colony formation assay and EdU staining were performed to measure cell proliferation of NSCLC cells. As data showed in Figure 2b-e, circ-OXCT1 depletion suppressed the cell viability, the colony numbers and the EdU positive cells of A549 and H1650 cells. On the contrary, the apoptosis rate of A549 and H1650 cells was increased after circ-OXCT1 silencing (figure 2f). Moreover, the effects of circ-OXCT1 silencing on the migration and invasion of A549 and H1650 cells were determined by transwell assay. The number of migrated and invaded NSCLC cells was restrained by circ-OXCT1 knockdown (Figure 2g-h). In support, we found that circ-OXCT1 silencing repressed the protein expression of PCNA, whereas facilitated Bax and E-cadherin protein expression in A549 and H1650 cells (Figure 2i-j). Besides, the influence of circ-OXCT1 in NSCLC cells glutamine metabolism was investigated, and results showed that the relative glutamine uptake and α-KG production were reduced by circ-OXCT1 silencing (Figure 2k-l). These findings illuminated that circ-OXCT1 might facilitate NSCLC progression.
Figure 2.
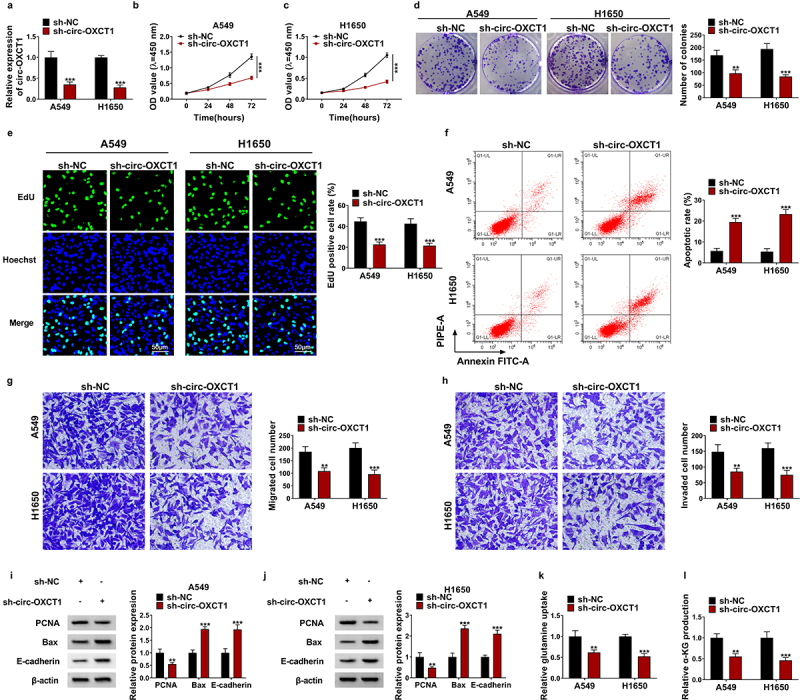
Circ-OXCT1 knockdown suppressed the malignant behaviors of NSCLC cells. (a-l) Both A549 and H1650 cells were transfected with sh-NC or sh-circ-OXCT1. (a) Circ-OXCT1 expression was determined by qRT-PCR to identify the transfection efficiency. (b-e) CCK-8 assay (b-c), colony formation assay (d) and EdU assay (e) were used to investigate the proliferation ability of NSCLC cells. (f) The cell apoptosis rate of A549 and H1650 cells was detected by flow cytometry. (g-h) The number of migrated and invaded cells was measured by transwell assay. (i-j) The protein levels of PCNA, Bax and E-cadherin were detected by western blotting. (k-l) The relative glutamine uptake and α-KG production were determined using commercial kits. n = 3 independent biological replicates. Data were showed as mean ± SD. **P < 0.01 and ***P < 0.001 by one-ANOVA followed by Tukey’s post hoc test or unpaired t test (two-tailed).
Circ-OXCT1 acted as a molecular sponge for miR-516b-5p
To explore circ-OXCT1-related miRNAs, we found that four miRNAs (miR-136-5p, miR-145-5p, miR-665 and miR-516b-5p) were predicted as potential targets of circ-OXCT1 both in starBase and CircInteractome databases (Figure 3a). After silencing circ-OXCT1 in A549 and H1650 cells, the expression levels of miR-136-5p, miR-145-5p and miR-516b-5p were significantly increased (Figure 3b-c). Considering that miR-516b-5p had the highest response to the change of circ-OXCT1 expression, we chosen miR-516b-5p for follow-up research. According to the binding sites between circ-OXCT1 and miR-516b-5p, the circ-OXCT1 WT/MUT vectors were constructed (Figure 3d). Transfection of miR-516b-5p mimic increased miR-516b-5p expression A549 and H1650 cells (Figure 3e), indicating a high transfection efficiency. MiR-516b-5p mimic and the circ-OXCT1 WT/MUT vectors were co-transfected into A549 and H1650 cells. Dual-luciferase reporter assay results revealed that miR-516b-5p overexpression only decreased the luciferase activity of circ-OXCT1 WT vector whereas had little effect on the luciferase activity of the MUT vector (figure 3f-g). Both circ-OXCT1 and miR-516b-5p were enriched in beads pre-coated with Ago2 antibody instead of IgG antibody (Figure 3h-i). Besides, the expression of miR-516b-5p was downregulated in NSCLC tissues and cells when compared with the controls (Figure 3j-k). And there was a negative correlation between the levels of circ-OXCT1 and miR-516b-5p in NSCLC tumor tissues (Figure 3l). The above data verified that circ-OXCT1 could interact with miR-516b-5p.
Figure 3.
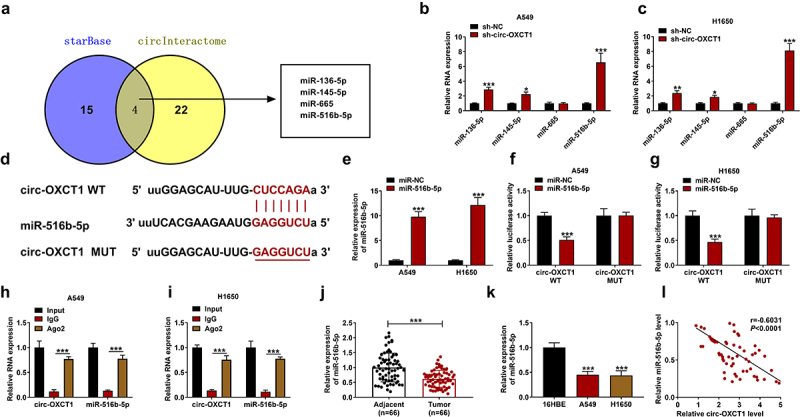
Circ-OXCT1 interacted with miR-516b-5p in NSCLC cells. (a) Venn diagram showed the number of miRNAs that were predicted as targets for circ-OXCT1 in starBase and circInteractome online softwares. (b-c) The expression of miR-136-5p, miR-145-5p, miR-665 and miR-516b-5p in A549 and H1650 cells was measured by qRT-PCR. (d) The supposed binding sites between circ-OXCT1 and miR-516b-5p were predicted by online starBase. (e) MiR-516b-5p expression was detected by qRT-PCR to identify the transfection efficiency. (f-g) Dual-luciferase reporter assay was performed to determine the relative luciferase activity of circ-OXCT1 WT and MUT reporter vectors. (h-i) The relative RNA enrichment of circ-OXCT1 and miR-516b-5p was detected by RIP assay. (j-k) The expression of miR-516b-5p was determined by qRT-PCR in clinical NSCLC tumor (n = 66), adjacent normal tissues (n = 66) as well as 16HBE, A549 and H1650 cells. (l) Pearson correlation analysis was performed between relative circ-OXCT1 and miR-516b-5p levels in NSCLC tumor tissues. n = 3 independent biological replicates. Data were showed as mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 by Mann-Whitney U test, one-ANOVA followed by Tukey’s post hoc test or unpaired t test (two-tailed).
Circ-OXCT1 regulated NSCLC progression by sponging miR-516b-5p
The rescue experiments were performed to explore the involvement of miR-516b-5p in the circ-OXCT1-medieted regulation effect in NSCLC cells. Transfection of miR-516b-5p inhibitor repressed miR-516b-5p expression (Figure 4a). Next, sh-OXCT1 and in-miR-516b-5p were co-incubated into A549 and H1650 cells, and results displayed that OXCT1 depletion upregulated the expression of miR-516b-5p, miR-516b-5p inhibitor restored the promotion of OXCT1 depletion (Figure 4a). Through CCK-8 assay, colony formation assay and EdU staining, we found that the repressive effect of circ-OXCT1 silencing on cell proliferation was reverted by miR-516b-5p inhibitor (Figure 4c-g). Circ-OXCT1 knockdown-induced apoptosis of NSCLC cells was rescued by miR-516b-5p inhibitor (Figure 4h). The transfection of miR-516b-5p inhibitor also restored the suppressive effects of circ-OXCT1 knockdown on migration and invasion abilities of NSCLC cells (Figure 4i-j). In support, miR-516b-5p silencing relieved circ-OXCT1 knockdown-mediated repressive effect of PCNA expression and the promotion of Bax and E-cadherin expression (Figure 4k-l). In addition, the suppressive effects of circ-OXCT1 silencing on the relative glutamine uptake and α-KG production of NSCLC cells were abolished by miR-516b-5p inhibition (Figure 4m-n). These results demonstrated that circ-OXCT1 regulated biological behaviors of NSCLC cells by targeting miR-516b-5p.
Figure 4.
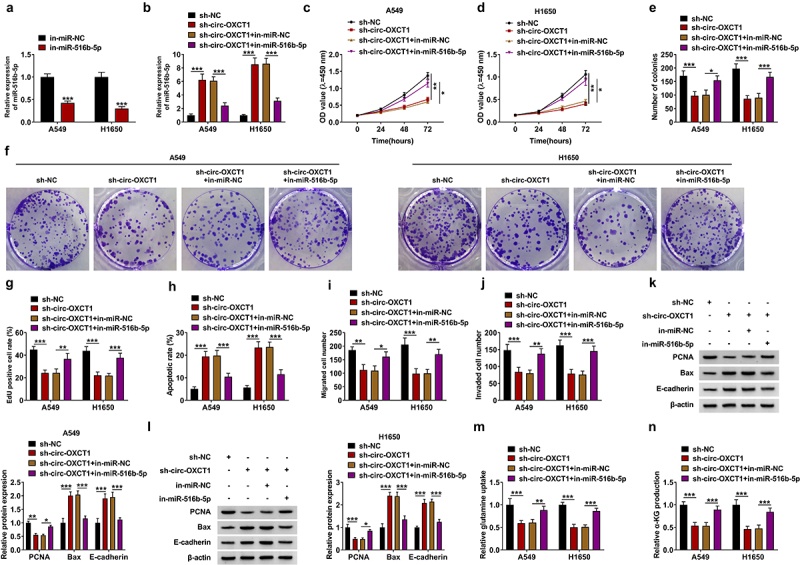
MiR-516b-5p inhibitor restored circ-OXCT1 depletion-mediated effects in NSCLC cells. (a) In-miR-NC or in-miR-516b-5p was transfected into A549 and H1650 cells, and the transfection efficiency was confirmed by qRT-PCR. (b-l) A549 and H1650 cells were transfected with sh-NC or sh-circ-OXCT1 and in-miR-NC or in-miR-516b-5p. (b) The expression of miR-516b-5p was tested by qRT-PCR. (c-g) CCK-8 assay (c-d), colony formation assay (e-f) and EdU assay (g) were performed to investigate the proliferation ability of NSCLC cells. (h) The cell apoptosis rate of A549 and H1650 cells was detected by flow cytometry. (i-j) The number of migrated and invaded cells was measured by transwell assay. (k-l) The protein levels of PCNA, Bax and E-cadherin were detected by western blotting. (m-n) The relative glutamine uptake and α-KG production were determined using commercial kits. n = 3 independent biological replicates. Data were showed as mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 by one-ANOVA followed by Tukey’s post hoc test.
SLC1A5 was the target gene of miR-516b-5p
Based on the starBase results, we found that there were many genes targeted by miR-516b-5p. We searched for genes that were highly expressed in NSCLC and promoted tumor growth. Next, qRT-PCR was used to detect the effect of miR-516b-5p on the expression of these genes. We found that overexpression of miR-516b-5p downregulated NOVA2, EIF4G2, RGS17, SLC1A5, and SRSF7; moreover, SLC1A5 was the most downregulated gene by upregulating miR-516b-5p (Supplementary Fig 1). So, we selected SLC1A5 for further study. The binding sites between miR-516b-5p and SLC1A5 were presented in Figure 5a. After co-transfecting with the miR-516b-5p mimic and SLC1A5 3ʹUTR WT/MUT vectors into A549 and H1650 cells, we discovered that the luciferase activity of SLC1A5 3ʹUTR WT vector, but not SLC1A5 3ʹUTR MUT vector was dramatically reduced (Figure 5(b-c)), indicating that SLC1A5 was a target gene of miR-516b-5p. The results of qRT-PCR showed that the mRNA expression of SLC1A5 was upregulated in NSCLC tumor tissues in comparison with adjacent normal tissues (Figure 5(d)). In addition, IHC staining and western blotting assays data further verified the high expression of SLC1A5 in NSCLC tumor tissues relative to that in adjacent normal tissues (Figure 5(e-f)). The data in Figure 5(g-h) exhibited that SLC1A5 expression was also upregulated in NSCLC cell lines (A549 and H1650) compared with 16HBE cells. Circ-OXCT1 silencing reduced the protein level of SLC1A5, and this effect was relieved by the addition of in-miR-516b-5p (Figure 5(i-k)). Correlation analysis presented that SLC1A5 mRNA level was negatively correlated with miR-516b-5p level and positively correlated with circ-OXCT1 level in NSCLC tumor tissues (Figure 5(l-m)). Taken together, we confirmed that circ-OXCT1 upregulated SLC1A5 expression by sponging miR-516b-5p.
Figure 5.
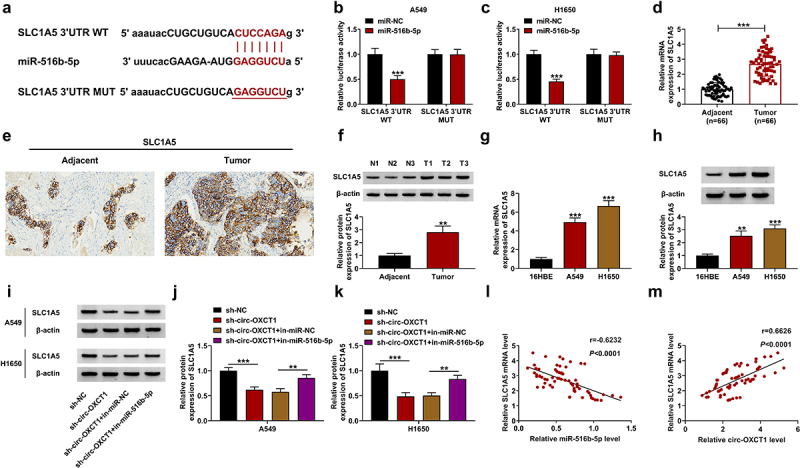
MiR-516b-5p interacted with SLC1A5 in NSCLC cells. (a) The binding sites between miR-516b-5p and SLC1A5 3ʹUTR WT or MUT was showed. (b-c) The relative luciferase activity of SLC1A5 3ʹUTR WT and MUT reporter vectors was determined by dual-luciferase reporter assay. (d-f) qRT-PCR (n = 66) (d), IHC staining assay (e) and western blotting (f) were used to detect SLC1A5 expression in clinical NSCLC tumor and adjacent normal tissues. (g-h) SLC1A5 expression was measured by qRT-PCR (g) and western blotting (h) in 16HBE, A549 and H1650 cells. (i-k) SLC1A5 protein level was detected by western blotting in A549 and H1650 cells-transfected with sh-NC or sh-circ-OXCT1 and in-miR-NC or in-miR-516b-5p. (l-m) Pearson correlation analysis was performed between relative SLC1A5 level and miR-516b-5p or circ-OXCT1 levels in NSCLC tumor tissues. n = 3 independent biological replicates. Data were showed as mean ± SD. **P < 0.01 and ***P < 0.001 by Mann-Whitney U test, one-ANOVA followed by Tukey’s post hoc test or unpaired t test (two-tailed).
Overexpression of miR-516b-5p inhibited cell proliferation, motility and glutamine metabolism, and induced cell apoptosis by targeting SLC1A5
To confirm the effect of miR-516b-5p and SLC1A5 in NSCLC cell malignancy, SLC1A5 overexpression plasmid was transfected into A549 and H1650 cells, and results revealed that overexpressed SLC1A5 remarkably upregulated the protein level of SLC1A5 (Figure 6(a)). Then, miR-516b-5p mimic and SLC1A5 overexpression plasmid were co-transfected into A549 and H1650 cells, and results displayed that miR-516b-5p mimic notably repressed SLC1A5 protein expression, which was relieved by overexpression of SLC1A5 (Figure 6b-c). In functional experiments, we found that miR-516b-5p mimic suppressed cell proliferation, but facilitated cell apoptosis; however, these effects were almost reverted by SLC1A5 overexpression (Figure 6d-h). Additionally, overexpressed SLC1A5 reverted the influence of miR-516b-5p mimic in cell migration and invasion (Figure 6i-j). In support, reduced expression of PCNA and increased expression of Bax and E-cadherin caused by miR-516b-5p mimic were rescued after transfecting SLC1A5 overexpression plasmid (Figure 6k-l). In addition, miR-516b-5p mimic-mediated the suppressive effects on the relative glutamine uptake and α-KG production were relieved by overexpressed SLC1A5 (Figure 6m-n). These data explained that miR-516b-5p overexpression suppressed malignant behaviors of NSCLC cells by targeting SLC1A5.
Figure 6.
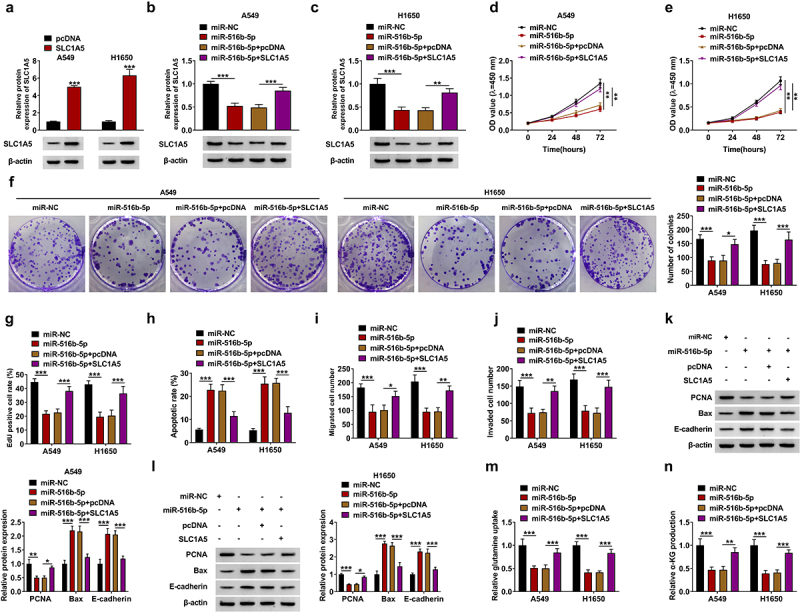
MiR-516b-5p mimic-mediated effects were reverted by SLC1A5 overexpression in NSCLC cells. (a) A549 and H1650 cells was transfected with pcDNA or SLC1A5, and the transfection efficiency was confirmed by western blotting. (b-n) MiR-NC or miR-516b-5p and pcDNA or SLC1A5 were transfected into A549 and H1650 cells. (b-c) The protein expression of SLC1A5 was detected by western blotting. (d-g) CCK-8 assay (d-e), colony formation assay (f) and EdU assay (g) were performed to investigate the proliferation ability of NSCLC cells. (h) The cell apoptosis rate of A549 and H1650 cells was detected by flow cytometry. (i-j) The number of migrated and invaded cells was measured by transwell assay. (k-l) The protein levels of PCNA, Bax and E-cadherin were detected by western blotting. (m-n) The relative glutamine uptake and α-KG production were determined using commercial kits. n = 3 independent biological replicates. Data were showed as mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 by one-ANOVA followed by Tukey’s post hoc test.
Circ-OXCT1 absence repressed tumor growth in vivo
The xenograft mice model was established to further confirm the role of circ-OXCT1 in NSCLC tumorigenesis in vivo. As showed in Figure 7a-c, circ-OXCT1 knockdown notably restrained the tumor volume and weight in comparison with the vector group. Comparing to the sh-NC group, the expression of circ-OXCT1 and SLC1A5 protein was reduced whereas miR-516b-5p expression was upregulated in tumor tissues (Figure 7d). Besides, western blotting and IHC staining assays data further confirmed the circ-OXCT1 silencing remarkably repressed the expression of SLC1A5 in tumor tissues (Figure 7e-f). Through IHC staining assay, we also found that the protein expression of proliferation marker ki-67 was reduced by circ-OXCT1 silencing (Figure 7g). These findings displayed that circ-OXCT1 knockdown restrained xenograft tumor growth in vivo.
Figure 7.
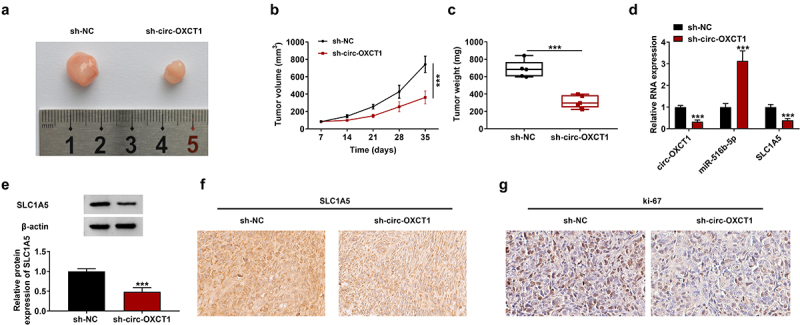
Silencing of circ-OXCT1 repressed NSCLC tumor growth in vivo. After stably transfecting with sh-NC or sh-circ-OXCT1, H1650 cells were injected into nude mice (n = 5/group) to induce xenograft tumors. (a) The image of most representative tumors were presented. (b) Tumor volume was monitored every 7 days for 35 days. (c) Tumor weight was recorded on the 35th day. (d) Circ-OXCT1, miR-516b-5p and SLC1A5 expression were measured by qRT-PCR analysis. (e) SLC1A5 protein expression was detected by western blotting. (f-g) The SLC1A5 (f) and ki-67 (g) positive cells were stained by IHC assay. Data were showed as mean ± SD. ***P < 0.001 by unpaired t test (two-tailed).
Discussion
NSCLC is one of the leading causes of cancer-related deaths worldwide, posing a fatal threat to human health [18]. The research for safe and effective biological treatment targets will positively promote the development of NSCLC treatment methods [19]. Here, we proved that cir-OXCT1 was highly expressed in NSCLC tissues and cell lines through in vivo and in vitro experiments. Knockdown of cir-OXCT1 restrained the malignant behaviors of NSCLC cells and glutamine metabolism. In addition, we discovered and confirmed the mechanism that cir-OXCT1 promoted the NSCLC process via the miR-516b-5p/SLC1A5 axis.
CircRNA could regulate a variety of biological behaviors, such as cell growth, metastasis and apoptosis [20,21]. With the development and application of RNA sequencing technology, many abnormally expressed circRNAs in NSCLC have been discovered [22]. Further studies verified that some differentially expressed circRNAs were related to NSCLC process and used as disease diagnosis and treatment markers [22–24]. For instance, Jiang et al. confirmed that hsa_circ_0007385 was upregulated in NSCLC tumor samples and cells, and hsa_circ_0007385 silencing repressed NSCLC cell growth and motility in vivo and in vitro [24]. Previous studies have shown that circ-OXCT1 was upregulated in NSCLC [15]. However, the effects of circ-OXCT1 in NSCLC progression were unclear. Consistent with previous studies, the results revealed that circ-OXCT1 expression was higher in NSCLC samples and cells than the controls. And the results of functional experiments confirmed that circ-OXCT1 knockdown suppressed the growth, metastasis and glutamine metabolism and induced apoptosis in NSCLC cells. In support, animal experiments verified that circ-OXCT1 silencing inhibited xenograft tumor growth.
CircRNA has been reported to regulate cell biological functions by sponging miRNA [25]. For instance, circPVT1 facilitated cell growth, invasion and radioresistance via absorbing miR-1208 [26]. To further explored the mechanism by which circ-OXCT1 promoted the progression of NSCLC, bioinformatics analysis, dual luciferase report and RIP assays were performed, and results showed that circ-OXCT1 acted as a sponge for miR-516b-5p. It has been reported that miR-516b-5p, a cancer-related miRNA, acts as a tumor suppressor in esophageal squamous cell carcinoma [27], osteosarcoma [28], bladder cancer [29] and NSCLC [16]. Our data showed that miR-516b-5p was downregulated in NSCLC tissues and cell lines when compared to the control group. In addition, we also confirmed that overexpression of miR-516b-5p restrained the malignant behaviors of NSCLC cells. And miR-516b-5p silencing weakened the inhibition of circ-OXCT1 knockdown on the malignancy of NSCLC cells.
Glutamine metabolism is a metabolic process that converts glutamine into α-KG thereby entering the tricarboxylic acid cycle to release energy [30]. Glutamine metabolism provides energy for rapidly proliferating cancer cells and is involved in the occurrence and metastasis of tumors [31]. SLC1A5 is an important glutamine transmembrane transporter, which exists as a cancer-promoting factor in NSSLC and Ovarian Cancer [32,33]. In addition, studies have confirmed that targeted interference glutamine metabolism notably inhibited the progression of NSCLC [34]. These findings indicated that targeting SLC1A5 to influence glutamine metabolism might regulate the progression of NSCLC. In current study, we confirmed that SLC1A5 was a target of miR-516b-5p. Functional experiments further displayed that overexpression of SLC1A5 almost reversed the suppressive impact of miR-516b-5p on the malignant behaviors of NSCLC cells. Since circ-OXCT1 was rarely researched, there was no more information accessed to further verify its prognostic role in more NSCLC samples. Besides, how circ-OXCT1 was dysregulated in NSCLC was unknown, which await further research.
In summary, our study proved that circ-OXCT1 silencing hindered the proliferation, motility and glutamine metabolism and facilitated the apoptosis of NSCLC cells by regulating the miR-516b-5p/SLC1A5 signaling pathway, which provided new basis for NSCLC molecular targeted therapy.
Supplementary Material
Funding Statement
This study was supported by:Fund of Hunan Provincial Health Commission (202104022248)
Highlights
(1) Circ-OXCT1 expression was upregulated in NSCLC tissues and cells.
(2) Circ-OXCT1 silencing repressed NSCLC cell malignancy.
(3) Circ-OXCT1 promoted SLC1A5 expression by interacting with miR-516b-5p.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Protocol registration
A protocol (including the research question, key design features, and analysis plan) was prepared before the study and it was not registered.
Ethics approval and consent to participate
Written informed consents were obtained from all participants and this study was permitted by the Ethics Committee of Changsha Central Hospital.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2071565
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Lee HYJ, Meng M, Liu Y, et al. Medicinal herbs and bioactive compounds overcome the drug resistance to epidermal growth factor receptor inhibitors in non-small cell lung cancer. Oncol Lett. 2021;22(3):646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Duma N, Santana-Davila R, Molina JR.. Non-Small Cell Lung Cancer: epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc. 2019;94(8):1623–1640. [DOI] [PubMed] [Google Scholar]
- [4].Van Meerbeeck JP, De Pooter C, Raskin J, et al. Local treatment of stage IIIA-N2 nonsmall cell lung cancer: surgery and/or radiotherapy. Curr Opin Oncol. 2020;32(1):54–62. [DOI] [PubMed] [Google Scholar]
- [5].Friedlaender A, Addeo A, Russo A, et al. Targeted Therapies in Early Stage NSCLC: hype or Hope? Int J Mol Sci. 2020;21(17):6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang M, Herbst RS, Boshoff C. Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. 2021;27(8):1345–1356. [DOI] [PubMed] [Google Scholar]
- [7].Berzenji L, Debaenst S, Hendriks JMH, et al. The role of the surgeon in the management of oligometastatic non-small cell lung cancer: a literature review. Transl Lung Cancer Res. 2021;10(7):3409–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2020;12(12):911–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Feng J, Xiang Y, Xia S, et al. CircView: a visualization and exploration tool for circular RNAs. Brief Bioinform. 2018;19(6):1310–1316. [DOI] [PubMed] [Google Scholar]
- [10].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Haddad G, Lorenzen JM. Biogenesis and Function of Circular RNAs in Health and in Disease. Front Pharmacol. 2019;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qin L, Zhan Z, Wei C, et al. HsacircRNAG004213 promotes cisplatin sensitivity by regulating miR513b5p/PRPF39 in liver cancer. Mol Med Rep. 2021;23(6). DOI: 10.3892/mmr.2021.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bing ZX, Zhang JQ, Wang GG, et al. Silencing of circ_0000517 suppresses proliferation, glycolysis, and glutamine decomposition of non-small cell lung cancer by modulating miR-330-5p/YY1 signal pathway. Kaohsiung J Med Sci. 2021;37(12):1027–1037. [DOI] [PubMed] [Google Scholar]
- [14].Feng H, Sun SZ, Cheng F, et al. Mediation of circ_RPPH1 on miR-146b-3p/E2F2 pathway to hinder the growth and metastasis of breast carcinoma cells. Aging (Albany NY). 2021;13(16):20552–20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mo WL, Deng LJ, Cheng Y, et al. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging (Albany NY). 2021;13(8):11629–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Song H, Li H, Ding X, et al. Long noncoding RNA FEZF1AS1 facilitates nonsmall cell lung cancer progression via the ITGA11/miR516b5p axis. Int J Oncol. 2020;57(6):1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cormerais Y, Massard PA, Vucetic M, et al. The glutamine transporter ASCT2 (SLC1A5) promotes tumor growth independently of the amino acid transporter LAT1 (SLC7A5). J Biol Chem. 2018;293(8):2877–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou H, Feng B, Abudoureyimu M, et al. The functional role of long non-coding RNAs and their underlying mechanisms in drug resistance of non-small cell lung cancer. Life Sci. 2020;261:118362. [DOI] [PubMed] [Google Scholar]
- [19].Malapelle U, Leprieur EG, Kamga PT, et al.Editorial: emerging Biomarkers for NSCLC: recent Advances in Diagnosis and Therapy. Front Oncol. 2021;11:694578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao B, Li Z, Qin C, et al. Mobius strip in pancreatic cancer: biogenesis, function and clinical significance of circular RNAs. Cell Mol Life Sci. 2021;78(17–18):6201–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Soghli N, Qujeq D, Yousefi T, et al. The regulatory functions of circular RNAs in osteosarcoma. Genomics. 2020;112(4):2845–2856. [DOI] [PubMed] [Google Scholar]
- [22].Wang C, Tan S, Liu WR, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wan J, Hao L, Zheng X, et al. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515(2):303–309. [DOI] [PubMed] [Google Scholar]
- [24].Jiang MM, Mai ZT, Wan SZ, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang M, Li T, Wang Q, et al. Silencing circPVT1 enhances radiosensitivity in non-small cell lung cancer by sponging microRNA-1208. Cancer Biomark. 2021;31(3):263–279. [DOI] [PubMed] [Google Scholar]
- [27].Huang Y, Jiang L, Wei G. Circ_0006168 Promotes the Migration, Invasion and Proliferation of Esophageal Squamous Cell Carcinoma Cells via miR-516b-5p-Dependent Regulation of XBP1. Onco Targets Ther. 2021;14:2475–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pan X, Tan J, Tao T, et al. LINC01123 enhances osteosarcoma cell growth by activating the Hedgehog pathway via the miR-516b-5p/Gli1 axis. Cancer Sci. 2021;112(6):2260–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang C, Mou Z, Wu S, et al. High-throughput sequencing identified circular RNA circUBE2K mediating RhoA associated bladder cancer phenotype via regulation of miR-516b-5p/ARHGAP5 axis. Cell Death Dis. 2021;12(8):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16(11):749. [DOI] [PubMed] [Google Scholar]
- [31].Xia M, Li X, Diao Y, et al. Targeted inhibition of glutamine metabolism enhances the antitumor effect of selumetinib in KRAS-mutant NSCLC. Transl Oncol. 2021;14(1):100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang X, Luo Y, Li X. Circ_0072995 Promotes Ovarian Cancer Progression Through Regulating miR-122-5p/SLC1A5 Axis. Biochem Genet. 2021;60(1):153–172. [DOI] [PubMed] [Google Scholar]
- [33].Xue M, Hong W, Jiang J, et al. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol. 2020;17(12):1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Teixeira E, Silva C, Martel F. The role of the glutamine transporter ASCT2 in antineoplastic therapy. Cancer Chemother Pharmacol. 2021;87(4):447–464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.


