Significance
Earth scientists rely on chemical fossils like the carbon isotope record to derive ancient atmospheric CO2concentrations, but interpretation of this record is calibrated using modern organisms. We tested this assumption by measuring the carbon isotope fractionation of a reconstructed ancestral rubisco enzyme (>1 billion years old) in vivo and in vitro. Our results contradicted prevailing models of carbon flow in Cyanobacteria, but our data could be rationalized if light-driven uptake of CO2is considered. Our study suggests that the carbon isotope record tracks both the evolution of photosynthetic physiology as well as changes in atmospheric CO2, highlighting the importance of considering both evolution and physiology for comparative biological approaches to understanding Earth’s history.
Keywords: evolution, carbon isotopes, rubisco, cyanobacteria, Precambrian
Abstract
The history of Earth’s carbon cycle reflects trends in atmospheric composition convolved with the evolution of photosynthesis. Fortunately, key parts of the carbon cycle have been recorded in the carbon isotope ratios of sedimentary rocks. The dominant model used to interpret this record as a proxy for ancient atmospheric CO2 is based on carbon isotope fractionations of modern photoautotrophs, and longstanding questions remain about how their evolution might have impacted the record. Therefore, we measured both biomass (εp) and enzymatic (εRubisco) carbon isotope fractionations of a cyanobacterial strain (Synechococcus elongatus PCC 7942) solely expressing a putative ancestral Form 1B rubisco dating to ≫1 Ga. This strain, nicknamed ANC, grows in ambient pCO2 and displays larger εp values than WT, despite having a much smaller εRubisco (17.23 ± 0.61‰ vs. 25.18 ± 0.31‰, respectively). Surprisingly, ANC εp exceeded ANC εRubisco in all conditions tested, contradicting prevailing models of cyanobacterial carbon isotope fractionation. Such models can be rectified by introducing additional isotopic fractionation associated with powered inorganic carbon uptake mechanisms present in Cyanobacteria, but this amendment hinders the ability to accurately estimate historical pCO2 from geological data. Understanding the evolution of rubisco and the CO2 concentrating mechanism is therefore critical for interpreting the carbon isotope record, and fluctuations in the record may reflect the evolving efficiency of carbon fixing metabolisms in addition to changes in atmospheric CO2.
Photoautotrophs have evolved over geologic time to harness energy from the sun in order to “fix” external, inorganic carbon (Ci) into reduced, organic carbon (Co), thereby creating biomass for growth and energy storage. Today, and likely for much of Earth’s history (1), the most widespread strategy for carbon fixation is the Calvin–Benson–Bassham (CBB) cycle, where the key carbon fixation step is catalyzed by ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (rubisco) (2, 3). But rubisco’s central role in the CBB cycle and oxygenic photosynthesis poses a conundrum because it is usually considered to be a nonspecific and slow enzyme. The first issue concerns rubisco’s dual carboxylase and oxygenase activities: The RuBP intermediate (enediolate) is susceptible to both O2 and CO2 attacks (4). Consequently, instead of fixing a CO2 molecule during photosynthesis, rubisco can instead assimilate O2 to yield 2-phosphoglycolate (2-PG), which is not part of the CBB cycle and therefore must be salvaged through photorespiratory pathways that consume adenosine triphosphate (ATP), reducing power, and carbon (5). The second issue concerns rubisco’s maximum carboxylation rate (VC), which is ≈7 to 10 times slower than other central metabolic enzymes (6), and displays very limited variation across large phylogenetic distances (7).
Both issues—its dual carboxylase/oxygenase activity and limited maximum carboxylation rate—are typically rationalized by considering its evolutionary history in the context of long-term changes in environmental CO2 and O2 concentrations. Rubisco is thought to have been the primary carboxylating enzyme of global photosynthesis since the Great Oxygenation Event and potentially far prior (1). It is also thought to have evolved when there was trace O2 and much higher CO2 concentrations in the atmosphere, in contrast to the modern atmosphere where O2 is roughly 20% while CO2 is only about 0.04% by partial pressure (1).
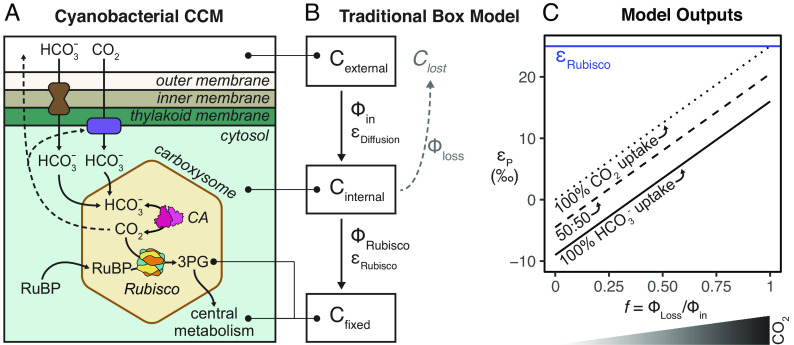
Likely in response to these changing environmental concentrations, many aquatic photoautotrophs evolved CO2 concentrating mechanisms (CCMs) that enhance carboxylation and suppress oxygenation by immersing rubisco in a high-CO2 environment. Even with CCMs, the effective in vivo rates of extant rubiscos are estimated to be lower (≈1% for terrestrial and ≈15% for marine rubiscos) than the maximal catalytic rates measured at 25 °C (2). Today, all known Cyanobacteria have CCMs, as do many bacterial chemolithoautotrophs, many aquatic algae and some plants (8). The bacterial CCM has two main components: i) Ci pumps producing high cytosolic HCO3− concentrations, and ii) coencapsulation of carbonic anhydrase (CA) and rubisco inside proteinaceous organelles known as carboxysomes (Fig. 1A) (9–11). These Ci pumps include BCT1 (ATP-dependent powered HCO3− transporter), SbtA (Na+/HCO3− symporters), BicA (Na-dependent HCO3− transporter), NDH-1MS, and NDH-1MS’ (NADPH-dependent powered CO2 uptake; see ref. 12 for review). It is unclear exactly when the bacterial CCM arose, with proposals ranging from the Proterozoic to the Phanerozoic Eon (8, 13). Therefore, for up to half of Earth’s history, cyanobacterial rubiscos have functioned in concert with a system that pumps Ci into and around the cell.
Fig. 1.
Comparing the cyanobacterial CO2 concentrating mechanism (CCM) to the traditional box model of photosynthetic C isotope discrimination. (A) Cyanobacterial CCMs rely on i) active Ci uptake into the cell, and ii) coencapsulation of carbonic anhydrase (CA) and rubisco within the carboxysome. Independent, powered transporters for HCO3− and CO2 are shown in brown and purple; both work to increase cytosolic concentrations of HCO3− (see ref. 12 for review). All CCM components work to produce a high carboxysomal CO2 concentration that enhances CO2 fixation by rubisco and suppresses oxygenation. Limited CO2 escapes from the carboxysome—some is scavenged by CO2 pumps while the rest leaves the cell. (B) Architecture of the traditional box model based on (14–17); see SI Appendix for full discussion of this model. Boxes denote carbon pools of interest, and fluxes between boxes are denoted by Φ. Each flux has its own isotopic fractionation denoted by ε; no fractionation is assumed for Φloss. Model assumes an infinitely large external carbon pool, that carbon not fixed by rubisco (Clost) returns to this pool, and that fluxes are at steady state. Note that this architecture does not include a box for the carboxysome. (C) Model solution for the traditional model is εP = a*εequil + f*εRubisco (Eq. 2), where εP is defined as the difference in δ13C of Cexternal and Cfixed, f is defined as the fraction of Ci lost (Φloss/Φin), and a is the fractional contribution of HCO3− to total Ci uptake. When a = 0, all Ci uptake is as CO2 (dotted line); when a = 1, all Ci uptake is as HCO3− (solid line). This model is presented in ref. 18, which is a generalization of (19) that accounts for the fact that Ci uptake (Φin in Panel B) ranges in composition between CO2 and HCO3− based on which Ci uptake system is used. Values of εRubisco = 25‰ and εequil = −9‰ were used for this illustration (20). Model outputs indicate that at high external CO2 concentrations (dark wedge under graph), there is greater net Ci leakage (larger f values) from the cell.
Rubisco displays a kinetic isotope effect (KIE) where it preferentially fixes 12CO2 over 13CO2 due in part to the VC being slightly faster for 12CO2 than 13CO2 (21), leading the reaction product, 3-phosphoglycerate (3-PG), to be relatively depleted in 13C by several percent (tens of ‰) relative to the isotopic composition of the CO2 substrate. This effect is typically reported in delta (δ13C) and epsilon (ε) notation in units of per mille (‰), where δ13C = [13Rsa/13Rref - 1]*1000 and 13R is the ratio of 13C/12C in the sample or reference, respectively; see Materials and Methods. The difference in δ13C of the CO2 substrate and the 3-PG product is reported as εRubisco and varies between 18 and 30‰ for extant rubiscos (22, 23), with the exception of the coccolithophore E. huxleyi with εRubisco ≈ 11‰ (24). Because autotrophs utilizing the CBB cycle synthesize biomass from 3-PG, biomass is 13C-depleted compared to external Ci pools—the magnitude of this difference is called εp.
The KIE of rubisco, along with other and more minor processes that affect carbon isotope ratios, is recorded in the carbon isotope record, which is comprised of measurements of the relative ratios of 13C to 12C isotopes in C-bearing phases of sedimentary rocks over time (25). Carbon isotope data have been assembled globally from myriad of ancient environments to cover ≈3.8 billion years (Ga) of Earth’s 4.5 Ga history (26). Contemporaneous Ci pools are preserved as carbonate salts (e.g., limestones and dolomites), while contemporaneous biomass and Co pools are preserved in the organic-rich components (e.g., kerogen) of many different lithologies and are measured as rock total organic carbon (TOC) (25). There are additional fractionations associated with the preservation of biomass and Ci as rocks, so the magnitude of fractionation between rock Ci and Co is termed εTOC and differs slightly from εp (27). Therefore, if one can derive εp from the rock record (εTOC) and pair it with a model relating εp to pCO2, in principle one can infer the history of atmospheric pCO2 from the carbon isotope record.
The carbon isotope record is particularly important for constraining ancient atmospheric pCO2 (28, 29) because direct observations of the past atmosphere from trapped gas in ice cores only extends back ≈1 million years (30). One notable feature of the record from ≈3.8 Ga to the present is that rock Co is depleted in 13C by ≈25‰ compared to Ci (23, 25, 26), and this offset roughly matches the KIE of extant rubiscos (25). The dominant model used to derive ancient atmospheric CO2 from the geological record (referred to as the “C Isotope Record Model” here; SI Appendix, Fig. S7 and Eq. 1) reflects this observation by fixing the maximum possible fractionation of biomass to be that of rubisco:
| [1] |
where εf is the maximum isotopic fractionation for carbon fixation and is typically set to equal εRubisco, [CO2(aq)] is the concentration of dissolved CO2 in solution around the cells, and b is a fitted parameter derived from experiments (31). This physiological factor, b (‰ kg μM−1), is fit from pure culture experiments of eukaryotic and bacterial algae, and encompasses all physiological effects that may affect cellular isotopic fractionation including the CCM, growth rate, cell size and geometry, membrane permeability, growth medium composition (e.g., pH, salinity, limiting nutrient), strain genetics, and physiological state (31–35). In the limit of high [CO2(aq)], the term b/[CO2(aq)] goes to zero and εP = εf, which is assumed to equal εRubisco. Therefore, with this model framework the maximum value of εP is εRubisco, and the term b sets how quickly εP approaches the limit of εRubisco.
The term b and the assumption that εRubisco sets the upper limit of εP directly follows from the “traditional model” (Fig. 1 B and C and Eq. 2) that was developed by measuring εP of plants and algae while parameters like pCO2 were varied:
| [2] |
where f is a ratio describing how much Ci exits vs. enters the organism (f = 1 is all Ci that enters is lost), εequil is the equilibrium isotope effect, and a is the fraction of Ci entering the cell as CO2 (a = 0) or HCO3− (a = 1) (18, 19). The diffusion isotope effect (εDiffusion) is considered negligible. This model (Fig. 1 B and C and Eq. 2) is therefore the physiological underpinning Eq. 1 and subsequent interpretations of the C isotope record; both show a limit where the maximum εP is εRubisco.
This traditional model was originally developed from studies of C isotope fractionation in plants (dotted line in Fig. 1C; all Ci uptake is as CO2 for plants) and was later adapted to eukaryotic and bacterial algae. The primary architecture of the traditional model stems from a seminal study by Park and Epstein (16) who proposed a “two step model” to explain εP of tomato plants grown in varied CO2 concentrations and light levels. In this model, carbon can be viewed as residing in one of three pools or “boxes” (Fig. 1B)—Ci outside the cell (Cext), Ci inside the cell (Cinternal), or Co as biomass (Cfixed). A “leakiness” term, f, is defined as the ratio of fluxes (Φ) of Ci exiting or entering the plant, where all of the Ci that entered the cell is lost when f = 1. In this simplified model, εp is determined by the isotopic effect of two distinct steps: i) the diffusion of CO2 into the plant [εDiffusion; <1‰ across a diaphragm cell in water at 25 °C (36)]; and ii) the carbon fixation step catalyzed by rubisco (εRubisco; ≈18 to 30‰). Notably, Park and Epstein proposed that the isotopic fractionations of these two steps are not additive in vivo (i.e., εp ≠ εDiffusion + εRubisco) but instead reflects the process by which photosynthesis is limited, either entry of CO2 into the cell (εp = εDiffusion) or CO2 fixation by rubisco (εp = εRubisco) (16).
Solving the traditional model at steady state results in a linear relationship between εp and f where the minimum and maximum εp values are εDiffusion and εRubisco, respectively (Fig. 1C). This allows experimentally measured values of εp to be used to solve for CO2 leakage (f, Fig. 1C). When εp ≈ εDiffusion, nearly all carbon entering the cell is used (f ≈ 0) and rubisco’s 12C preference is not “expressed”; conversely, when εp ≈ εRubisco, very little of the carbon entering the cell is fixed (f ≈ 1, nearly all carbon leaks from the cell) and rubisco can “choose” between 12C and 13C substrates so that rubisco’s KIE is fully expressed. Farquhar et al. (17) later derived a relationship between εp and the ratio of external vs. intracellular CO2 partial pressures, allowing CO2 concentrations at the site of rubisco to be roughly estimated from εp. Therefore, given the assumption that Ci is taken up passively, it is possible to derive an increasing relationship between Cext and εP from this model, where large εP indicates that high external CO2 concentrations generate excess CO2 at rubisco and ultimately cause more CO2 to leak out of the cell than can be fixed [see SI Appendix and (15)].
This model was later adapted to algae to account for CCMs—mainly active uptake of Ci as HCO3− and/or CO2—and physiological parameters including growth rate and cell geometry (19, 32, 33, 37, 38). These studies grew eukaryotic and bacterial algae in a range of pCO2 and culturing conditions to test if the linear relationship between εp and pCO2 observed in plants still held. Interestingly, cyanobacterial εp was found to be roughly constant independent of environmental pCO2 and growth rate (32). Because cyanobacterial εp values were less than known corresponding εRubisco values, additional isotopic fractionation factors were not needed to explain εp, even though some active Ci transport processes, which may fractionate carbon isotopes, were known in cyanobacteria at the time (39–41). Therefore, though different versions of this “traditional model” exist, all variations essentially modified the plant model by shifting the y-intercept of Fig. 1C to account for uptake of HCO3− in addition to CO2. If Ci entering the cell is primarily CO2, the model effectively represents plants (dotted line in Fig. 1C). If Ci is taken up primarily as HCO3−, as in many algae, εp is shifted to lower values (solid line in Fig. 1C) because of the equilibrium isotopic effect (εequil) between CO2 and HCO3− [≈ −9‰ (20)]. In Fig. 1C, we plot the traditional model as derived in Eichner et al. (18), which is an adaptation of (19).
The C Isotope Record Model (Eq. 1 and SI Appendix, Fig. S7) and the traditional model (Eq. 2 and Fig. 1C) have a limit where εp cannot exceed εRubisco. Yet, the largest εp values observed in the Archaean Eon exceed 30‰ (25, 26) and also exceed all current measurements of εRubisco (23). In addition, recent studies in dinoflagellates have shown that εp can regularly exceed εRubisco under certain growth conditions (22), and detailed studies of Cyanobacteria imply that leakage estimates derived from εp are not physiologically possible (18). These studies motivated updated models of algal carbon isotope fractionation that account for the isotopic fractionations associated with different Ci uptake mechanisms in order to rationalize anomalous εp values (18, 22).
These experiments made clear that the physiology of algae and Cyanobacteria—e.g., how they take up Ci as CO2 or HCO3− and by which mechanism—affects the C isotopic content of biomass, εp. Further, these Ci transporters and other integral components of modern CCMs were once absent from ancient autotrophs, who used various forms of rubisco alone to grow in Archaean or Proterozoic atmospheres (42). Efforts to draw inferences about the ancient Earth from the C isotope record must, therefore, include some understanding of the physiology and evolution of CCMs in Cyanobacteria and eukaryotic algae (22, 42, 43). Recent studies have attempted to address this issue by characterizing model organisms that may better resemble an ancestral counterpart, including a cyanobacterial strain lacking a CCM (43), a strain that overexpresses rubisco (44), and a strain expressing an inferred ancestral rubisco dating from ≈1 to 3 Ga (45, 46).
Here, we measured the εp of a control strain of Synechococcus elongatus PCC 7942 expressing the wild-type rubisco (NS2-KanR, referred to as “WT” for "wild-type", see Materials and Methods), as well as a strain, nicknamed “ANC” for "ancestral", engineered to express an inferred ancestral Form 1B enzyme (dating to >1 Ga) as its sole rubisco (47) in varied CO2 and light conditions. This putative ancestral rubisco was previously purified and its kinetics were characterized in vitro. Its sequence was then inserted into the genome of a modern cyanobacterium, though the genome of the strain in that study contained both extant and ancestral rubisco sequences (47). Here we study a strain where the extant rubisco was fully removed and replaced with the reconstructed ancestor. In contrast to (46), we also measured εRubisco of the present-day and ancestral rubiscos in vitro. We observed that: i) biomass εp is greater for ANC than WT for all conditions tested, even though ANC εRubisco (17.23 ± 0.61‰) is considerably smaller than WT εRubisco (25.18 ± 0.31‰); ii) ANC εp increases with light levels while WT εp increases with CO2; iii) ANC displays a growth defect at ambient pCO2 that is rescued at high pCO2; and iv) ANC growth is severely inhibited in high light. Consistent with recent studies of eukaryotic algae (18, 22), ANC εp exceeding εRubisco in all conditions implies that the traditional box model is incomplete and additional isotope fractionations are needed to rationalize measured εp. In addition, modulation of ANC εp with light suggests that some light-powered component of the CCM is responsible for excess fractionation beyond εRubisco. We posit that fractionation due to Ci uptake might explain isotopic measurements that deviate from traditional model predictions in both extant and ancient organisms.
Results and Discussion
Ancestral Rubisco Enzyme Fractionates Less Than the Modern Rubisco.
We measured the carbon isotope fractionations of WT and ANC rubiscos in vitro using the substrate depletion method (48–51). Note that there exists experimental variation in εRubisco measurements, both within and across studies, and its cause remains uncertain at present [see SI Appendix, section 4a and (52)]; so we employed the same general approach as others (the substrate depletion method) to be consistent with prior literature. Previous work on rubisco isotope discrimination predicted that εRubsico should correlate positively with specificity (SC/O), a unitless measure of the relative preference for CO2 over O2 (53). We therefore expected ANC and WT εRubisco values to be the same within uncertainty because of their similar SC/O values (previously measured in ref. 47), but found that the fractionation factor (εRubisco) of the ancestral rubisco (17.23 ± 0.61‰) was about 8‰ lower than that of the extant rubisco (25.18 ± 0.31‰, Table 1).
Table 1.
Rubisco characteristics
| Rubisco | εRubsico (‰) | VC (s−1) | KCAir (μM) | VC/KCAir (s−1mM−1) | SC/O |
|---|---|---|---|---|---|
| Ancestral form IB | 17.23 ± 0.61 | 4.72 ± 0.14 | 168.7 | 28 | 49.6 ± 1.8 |
| Modern form IB | 25.18 ± 0.31* | 9.78 ± 0.48* | 184.1* | 53.1* | 50.3 ± 2.0* |
Starred values (*) for the modern Form 1B were measured in rubiscos purified from Synechococcus sp. PCC 6301, which has the same small and large subunit (RbcS RbcL) sequences as our working WT strain, Synechococcus sp. PCC 7942 (47). Kinetic isotope effect (εRubisco, avg. ± SE) was measured in this study using the substrate depletion method (48–51). Carboxylation turnover under substrate-saturated conditions (VC); Michaelis constant for CO2 in ambient levels of O2 (KCAir); the catalytic efficiency toward CO2 in ambient air (VC/KCAir); and specificity, a unitless measure of the relative preference for CO2 over O2; (SC/O) are from ref. 47.
Ancestral Rubisco Strain Grows at Ambient CO2 Concentrations.
Working in S.elongatus PCC 7942, we produced a mutant strain lacking the native Form 1B rubisco and expressing instead an ancestral Form 1B rubisco produced by computational ancestral sequence reconstruction (47) as its sole rubisco enzyme. We then grew this strain, termed ANC, and a control strain, termed wild-type or “WT” (Materials and Methods), in a variety of light and CO2 levels: i) a reference condition (ambient pCO2 of 0.04% v/v, standard light flux (120 µE)); ii) high CO2 (5% pCO2, 120 µE); and iii) high light (0.04% pCO2, 500 µE). The CO2 gas at ambient and high CO2 conditions had δ13C values of −12.46‰ and −36.84‰, respectively.
Remarkably, as in ref. 46, the ANC strain managed to grow in ambient pCO2 and standard light conditions (Fig. 2), even though the ancestral rubisco has a VC roughly half that of WT (Table 1). This implies that its rubisco enzyme is properly encapsulated in the carboxysome, since improper carboxysome formation prohibits growth in ambient air (54, 55). Additional characterization of the physiology of the ANC could be valuable, but our inference of proper carboxysome encapsulation is supported by several experiments and analyses as follows. First, electron micrographs of WT and ANC cells grown in ambient CO2 and light conditions (Materials and Methods) showed multiple carboxysomes per cell in both strains (Fig. 3 and SI Appendix, Fig. S13). Rubisco density can be seen within some of the carboxysomes (Fig. 3C). Second, the rubisco amino acid residues necessary for protein interactions mediating β-carboxysome encapsulation were recently identified (56), and the ANC sequence retains fourteen of the sixteen residues involved (SI Appendix, Tables S8 and S9 and Fig. S14). In addition, WT and ANC strains harvested during exponential growth in the reference condition exhibit similar photosystem stoichiometry, as indicated by absorbance spectra (SI Appendix, Fig. S15). Taken together, these data indicated that carboxysomes form in ANC and the ancestral rubisco is encapsulated within these structures.
Fig. 2.
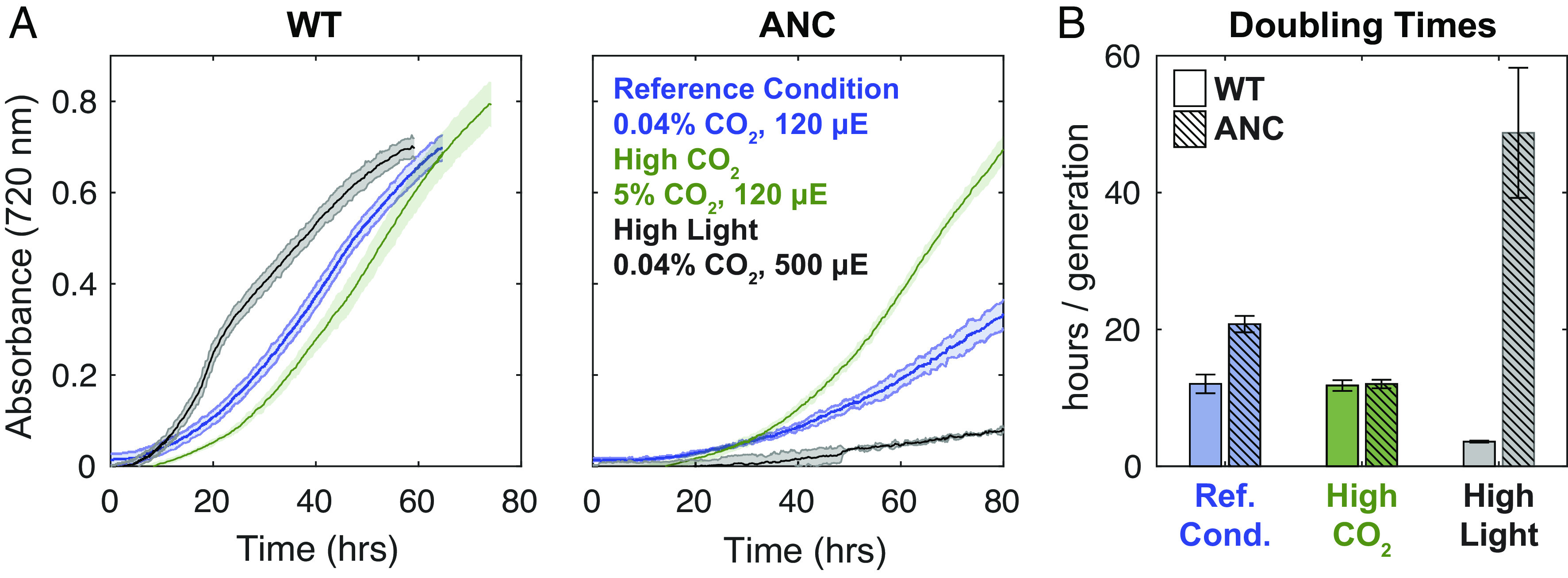
Growth curves for WT and ANC strains across experimental conditions. (A) Averaged growth curves shown for WT and ANC strains to 80 h, colored by growth condition as indicated in figure. Data were smoothed with a rolling median (Materials and Methods); see full ANC growth curves in SI Appendix, Fig. S12. (B) Average doubling times with SDs. See SI Appendix for details of doubling time calculation. ANC displayed a growth defect relative to the WT at the reference condition, which was rescued by high CO2. ANC grew slowest in high light, while WT grew fastest in that condition.
Fig. 3.
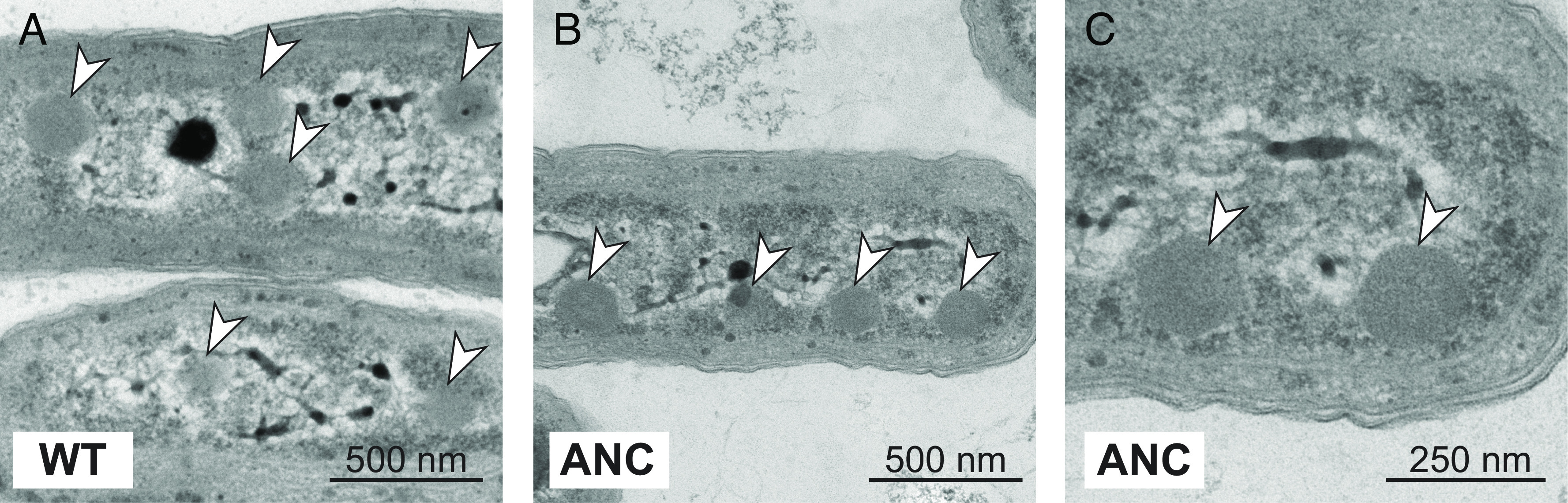
WT and ANC strains both produce carboxysomes at ambient pCO2. Transmission electron micrographs of WT (A) and ANC (B and C) strains that were harvested during exponential growth in the reference condition (ambient pCO2, standard light flux). Both strains show multiple carboxysomes per cell, as indicated by white arrows, and carboxysomes exhibit the typical hexagonal shape (55). (C) is the same image as in (B) but enlarged to show that rubisco density seen can be within the carboxysomes of ANC. The dark internal body in (A) is likely a polyphosphate body (57). See SI Appendix, Fig. S13 for additional images.
In addition, the difference in VC between the ancestral and modern rubiscos was mirrored in the doubling times of WT and ANC strains (Fig. 2B and SI Appendix, Table S2), where ANC doubling times were roughly twice that of WT in the reference condition (20.8 ± 1.2 vs. 12.0 ± 1.4 h, respectively). This suggested that ANC’s growth was limited by its ability to fix CO2 from ambient air. This growth defect was ameliorated by high pCO2, where doubling times for both strains were the same within uncertainty (WT 11.8 ± 0.8 h; ANC 12.0 ± 0.6 h). In contrast to WT, elevated CO2 greatly accelerated the growth of ANC, reducing its doubling time from ≈21 to ≈12 h (Fig. 2B), supporting our inference that CO2 availability limits the growth of ANC in ambient air, implicating the CCM in its growth defect. Similar results were found in ref. 46.
We observed the greatest differences in doubling times between ANC and WT when the strains were grown in high light (500 µmol photons m−2 s−1, Fig. 2 and SI Appendix, Table S2). In these conditions, WT cultures were a dark, blue-green color typical of healthy cyanobacterial cells while ANC cultures were yellow-green (SI Appendix, Fig. S11), suggesting degradation of phycobilisomes via a known starvation pathway to reduce the cell’s capacity for light harvesting and photochemical electron transport (58, 59). Note that this is a very high light intensity for Cyanobacteria and may induce a severe photoinhibitory response (60). We therefore inferred that ANC could not fix CO2 at a rate matching its light harvesting capability, and hence expressed this regulatory pathway to decrease light harvesting capacity. WT, in contrast, grew rapidly in high light.
The ANC Strain Fractionates More than WT.
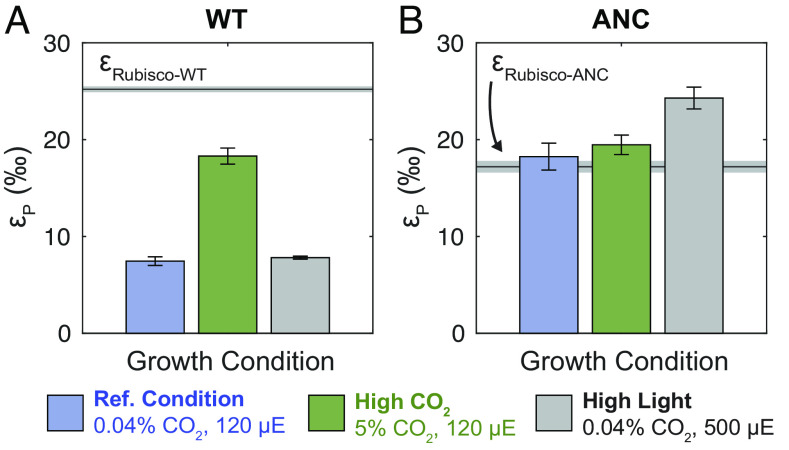
Counter to expectations based on εRubisco (Table 1), ANC εp was as large or larger than WT εp in all conditions tested (Fig. 4). This was consistent with recent results from a similar ancestral mutant, where that mutant’s εp values exceeded WT in ambient and elevated CO2 levels (46). In this study, the highest ANC εp values were observed for cultures grown in high light, where growth was significantly slower than the WT (doubling time ≈ 50 vs. 4 h, respectively, Fig. 3 and SI Appendix, Table S3). ANC εp values were also modulated differently by light and CO2 compared to WT. Compared to the reference condition, WT εp values were indifferent to high light and only increased in high CO2 (Fig. 4A). In contrast, ANC εp values did not increase in high CO2 and only increased in high light (Fig. 4B). This result contrasted with the ancestral mutant in ref. 46 where εp values increased by ≈10‰ at 2% CO2.
Fig. 4.
Whole-cell carbon isotope fractionation by WT and ANC strains. εp (‰) values (avg. ± SE) for (A) WT and (B) ANC strains across growth conditions. For each strain, the maximum εp possible based on the traditional model (εp = εRubisco) is shown as a gray line (avg. ± SE). Most measured ANC εP values exceed the theoretical limit (εp > εRubisco-ANC + SE), while WT εp values do not (εp < εRubisco-WT + SE). WT εp values increase in response to elevated CO2 concentrations, while ANC εp values increase in response to elevated light flux. See SI Appendix, Table S3 for full results.
As discussed above, the traditional box model cannot accommodate εp values in excess of εRubisco (Fig. 1C). However, average ANC εp values exceeded ANC εRubisco in all growth conditions (Fig. 4), particularly under high-light conditions where the largest difference was seen (εp = 24.30 ± 0.12‰ vs. εRubisco = 17.23 ± 0.61‰). The traditional box model also states that εp values are solely modulated by changing external pCO2 concentrations, which is plainly contradicted by Fig. 4B.
Proposed Influence of a Light-Powered, Vectoral Carbonic-Anhydrase.
Recent studies in extant bacterial and eukaryotic algae have shown that εp can regularly exceed εRubisco under certain growth conditions (22), motivating updated models of carbon isotope fractionation in both eukaryotic and bacterial algae (18, 22, 61). Taken together, these studies indicated that observed εp values could only be rationalized if an additional fractionation factor was present. Several studies argued that this factor is an energy-coupled CA catalyzing the vectoral hydration of intracellular CO2 to HCO3−, as this reaction is calculated to have a large isotopic effect and would allow εp to exceed εRubisco (18, 22, 61). Energy-coupled CAs can facilitate CO2 uptake by converting extracellular CO2 that passively translocates the membrane to intracellular HCO3− (Fig. 1A), which is advantageous in acidic conditions where CO2 is the dominant form of extracellular Ci (10, 62, 63). Vectoral CAs are also thought to potentially “recycle” CO2 that leaks from the carboxysome by converting it to HCO3− (12).
Cyanobacteria and eukaryotic algae have two general modes of active Ci uptake: uptake of hydrated Ci (predominantly H2CO3 and HCO3−) and of CO2 (63). In order for the CCM to function, either mode must produce a high, nonequilibrium concentration of HCO3− in the cytoplasm (8, 10). This is thought to be achieved by coupling CA to an energy source (e.g., light or an ion gradient) that drives the vectoral hydration of CO2 to HCO3− in the cytoplasm (64). There is now excellent data supporting this hypothesis in Cyanobacteria, where accessory proteins that bind to the NDH complex, the cyanobacterial homolog of the respiratory Complex I NADH-dehydrogenase, are known to mediate CO2 uptake specifically (65–67). Additionally, one of these accessory proteins, CupA/B, is reminiscent of a CA and contains a telltale zinc active site situated near a proton channel in a membrane subunit (68). The prevailing understanding of these data is, therefore, that these complexes couple Ci uptake to energy supplied by photochemical electron transport (68, 69). Moreover, a similar protein complex has been described in proteobacterial chemoautotrophs, suggesting that energy-coupled CO2 hydration is widespread (62).
A vectoral CA would affect εp for two reasons. First, CO2 and HCO3− are isotopically distinct. At equilibrium in standard conditions, HCO3− is ≈9‰ more enriched in 13C than CO2 (20, 70, 71). Therefore, if a cyanobacterium is predominantly taking up CO2, the internal Ci pool from which biomass is formed would be isotopically lighter (13C-depleted) than if HCO3− is the dominant source of Ci. We focused only on Ci uptake as CO2 because we were interested in a modification to the traditional model that could achieve large εp values (indicating 13C-depleted biomass) to account for at least an additional ~8‰ of fractionation in εp (maximum of ~25‰ in the high-light condition) greater than εRubisco (~17‰) in ANC. Though HCO3- uptake through bicarbonate transporters (e.g., SbtA) was likely occurring under our experimental conditions (65), isotopically it would not help us achieve the measured large εp values because it would shift all εp values to be maximally 9‰ more negative (i.e., 13C-enriched biomass, Fig. 1C) when we seek to explain values that are ~8‰ more positive. Second, unidirectional CO2 hydration (CO2 + H2O → HCO3– + H+) is expected to impart a substantial KIE, with calculated values ranging from ≈19 to 32‰ (70, 72–75). Therefore, there are two mechanistic reasons (CO2 vs. HCO3− uptake; unidirectional CO2 hydration) that εp could exceed εRubisco in conditions where energized CO2 uptake and hydration is active. Indeed, a recent model of C-isotope fractionation in Cyanobacteria specifically invoked the NDH complex to rationalize εp values that exceed εRubisco (18).
Because energy-coupled CO2 uptake and hydration by the NDH complex is driven by light energy, e.g., via cyclic electron flow around photosystem I (68), and because the vectoral hydration of CO2 to HCO3− is thought to have a large carbon isotope fractionation (70, 72–75), εp should increase with light intensity. Indeed, we observed the largest ANC εp values in the high-light condition and found that ANC εp varies primarily with light and not CO2 (Fig. 4). This observation is counter to the traditional model, which proposes εp as a direct correlate of external pCO2 (14, 15). Furthermore, on short timescales (≈minutes) cyanobacterial Ci uptake can be modulated by light intensity alone, fully independent of external Ci concentrations (76), and CO2 uptake can occur in the absence of carbon fixation (77, 78). Based on these physiological and isotopic observations, our study also supports the hypothesis that an energy-coupled vectoral CA like the NDH complex permits εp > εRubisco, as observed here for ANC in all growth conditions.
Conceptual Model for Carbon Isotope Fractionation in Cyanobacteria.
As discussed above, the traditional box model cannot produce εp > εRubisco (Fig. 1C). In this model, the Ci leakage term (f ) is fit from measured εp values and f = 1 implies that all carbon uptake leaks out of the cell. Though the traditional box model can accommodate both CO2 and HCO3- uptake, which differ in their equilibrium isotopic composition, it does not account for the isotopic effect of vectoral CO2 hydration. As such, even modeling 100% CO2 uptake gave physiologically infeasible values of f > 1 for ANC in all conditions (Fig. 5A and SI Appendix, Fig. S8), yet ANC grew reproducibly in all conditions tested (Fig. 2). We also encountered challenges using the traditional model to rationalize WT data: fitting the model gave f < 1 in ambient pCO2 conditions, but high-CO2 conditions yielded f > 1 unless all Ci uptake was assumed to be as HCO3− (see SI Appendix, Fig. S8 for discussion). Therefore, to rationalize our results, we developed a simple modified box model that permits εp > εRubisco by including fractionation due to Ci uptake through vectoral CAs.
Fig. 5.
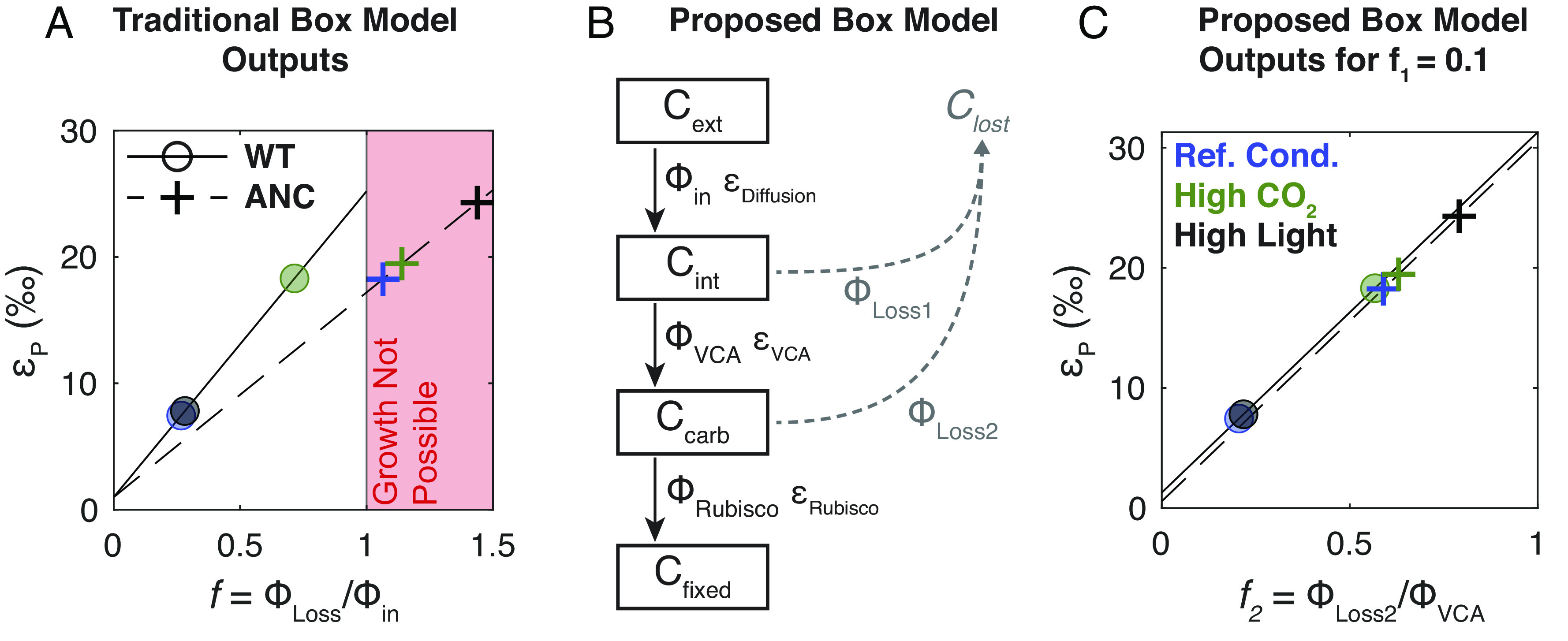
Proposed box model based on experimental results. (A) Experimental results (circles and crosses) plotted onto traditional box model outputs (solid and dashed lines) for WT and ANC, respectively, if Ci uptake is all CO2. See SI Appendix, Table S11 for quantification of uncertainty. Colors indicate growth conditions as in Fig. 2. The red shaded region demarcates the physiologically infeasible region where f > 1. (B) Our proposed box model architecture. Subscripts indicate external (ext), internal (int), carboxysome (carb), and fixed (fixed) carbon pools. Fluxes are denoted by Φ where subscripts indicate fluxes into the cell (in), out of the cell (Loss1, Loss2), into the carboxysome (VCA for Vectoral Carbonic Anhydrase), and into fixed biomass (Rubisco), each with a corresponding isotopic fractionation denoted with ε. Loss fluxes were assumed to have no isotopic fractionation. In this model, f1 is defined as ΦLoss1/Φin, and f2 is defined as ΦLoss2/ΦVCA. (C) Experimental results plotted onto proposed box model outputs for f1 = 0.1; colors and symbols are the same as Panel A; see SI Appendix, Table S11 for quantification of uncertainty. εp is defined as the difference in δ13C between Cext and Cfixed. Here only results for f1 = 0.1 are shown; see SI Appendix, Supplementary Text for full description of model assumptions and results.
In this modified model, we explicitly represent the CCM by distinguishing between carbon in the cytosol (Cint) and carbon in the carboxysome (Ccarb), allowing carbon to be lost from the carboxysome (ΦLoss2, Fig. 5B). Therefore, external Ci enters the cell (flux Φin) where it can either leak out (flux ΦLoss1) or undergo active hydration (flux ΦVCA, where VCA denotes Vectoral CA). Intracellular Ci can then enter the carboxysome, where it is either fixed (flux ΦRubisco) or ultimately leaks out of the cell (flux ΦLoss2).
We made similar simplifying assumptions as the traditional box model: i) an infinite supply of external carbon, ii) no isotopic fractionation for carbon lost from the cell, iii) Φin has the isotopic fractionation associated with εDiffusion, and iv) the system is at steady state. We did not add an explicit term for light energy used to power Ci uptake. Instead, the model included an energized CA (denoted VCA) and its associated isotopic fractionation as free parameters. In modeling each strain, we used the appropriate εRubisco measurements (Table 1). We do not know the true value for εVCA, but used a value of 30‰ similar to a recent model that explicitly invoked the NDH complex in Cyanobacteria (18). For comparison with the traditional model, we plotted Fig. 5C with f1 = 0.1 so that it could be represented in two dimensions; see SI Appendix, Fig. S10 for full model outputs. In this updated model, each value of εp corresponds to a set of feasible f1 and f2 values that fall along a line (SI Appendix, Fig. S10). Therefore, our model constrains but does not uniquely determine f1 and f2, nor does it allow for estimation of external Ci levels because many pairs of f1 and f2 values can produce the same εp.
The modified model was able to rationalize our experimental data of εp > εRubisco with leakage values compatible with cell growth (f2 < 1, Fig. 5C). It may also explain why ANC and WT responded so differently to high light. Our model results implied that ANC lost more carbon than WT at the branch point before rubisco (ΦLoss2); i.e., even though carbon was present in the cell, it could not be fixed by the ancestral rubisco, perhaps due to its lower VC (Table 1). Excess CO2 allowed rubisco’s KIE (εRubisco) to be expressed in εp. These results indicated that, in high light, the vectoral CA was delivering high amounts of CO2 to both the WT and ANC rubiscos. The faster WT rubisco was able to match this flux, which was reflected in its fast growth rate (Fig. 2) and no change in εp vs. the reference condition (Fig. 4). However, the slower ANC rubisco was not, which led to its slowest growth rate (Fig. 2), and highest εp values across all conditions (Fig. 4). Conditions where εp exceeded εRubisco in ANC suggested that, in addition to ΦLoss2 being large (allowing εRubisco to be expressed), ΦLoss1 was high as well, which allows εVCA to be expressed. However, since we could not independently determine ΦLoss1 and ΦLoss2 —i.e., what proportion of εp reflects the contribution of εRubisco vs. εVCA —we could only conclude that overall the slower ANC rubisco created a “backup” where leakage increased all along the CO2 fixation pathway and that this effect was exaggerated at high light.
We also note that our use of the term “vectoral” CO2 hydration connotes a net flux that is dominantly in the direction of CO2 hydration (CO2 + H2O → HCO3− + H+), rather than implying that the flux of HCO3− dehydration (HCO3− + H+ → CO2 + H2O) is zero. As such, there is likely some bidirectional activity (CO2 + H2O ⇌ HCO3− + H+) of the NDH complex. It is difficult to experimentally measure the isotope effect associated with the CO2 hydration reaction, but transition state theory and quantum chemical modeling (70, 71, 74) suggest that the value is large (roughly 25‰, see ref. 22 for review). HCO3− dehydration, and equilibration in general, would tend to reduce the isotopic fractionation (70). Our results here do not require a larger isotopic effect, however. Rather, a smaller value of εVCA = 10‰ (SI Appendix, Fig. S10) would have allowed us to rationalize our measurements, as we need only account for an additional ≈8‰ of fractionation in εp (maximum of ≈25‰) above εRubsico (≈17‰) in ANC. See SI Appendix, Fig. S10 for further discussion.
In addition, ours is not the only model structure that can permit εp > εRubisco. We tested other models by fitting our data to them (SI Appendix, Fig. S9). Models that incorporated an explicitly one-way, “CA-like” enzyme (61) or the NDH complex specifically (18) were mostly able to rationalize our data as well. The poorest fits are when Ci uptake was mostly as HCO3− (SI Appendix, Fig. S9) which is not surprising since we need more positive εp values and HCO3− uptake would shift all εp values to be 9‰ more negative (Fig. 1C). Altogether, model fitting indicates adding an additional carbon isotope fractionation step produces a model capable of rationalizing our data by enabling εp > εRubisco with plausible leakage values f < 1.
Consequences for Understanding the Evolution of Carbon-Fixing Metabolisms.
Our goal was to test if prevailing models of carbon fixation and isotopic fractionation apply to an ancestral analogue strain that may be relevant to understanding the carbon cycle over geologic time. We did so by measuring the isotopic fractionation of a reconstructed ancestral rubisco both inside and outside a living cyanobacterium. We emphasize that ANC is not a true ancestral Cyanobacteria; rather it is a chimeric construct—a modern strain saddled with a predicted Precambrian enzyme. This reconstructed ancestral rubisco is characterized by slower carboxylation kinetics (47) and a much lower εrubisco than the modern strain’s native enzyme (17.23 ± 0.61‰ vs. 25.18 ± 0.31‰, Table 1).
Recent studies in extant bacterial (18) and eukaryotic algae (22) have motivated updated models of C isotope fractionation in cells; these models address observations that: i) εp can exceed εRubisco in certain conditions; ii) factors other than pCO2 can modulate εp. Our results emphasize that similar caveats apply to Cyanobacteria, where ANC εp exceeded εRubisco in all conditions tested. Inference of Archaean and early Proterozoic pCO2 from the C isotopic record relies intimately on models of cyanobacterial physiology due to their distinction as the oldest oxygenic photoautotrophs (1). Yet, our results show that the traditional form of these models is not generally reliable.
To date, such anomalous εp values have been observed during relatively slow growth; in ref. 61 εp > εRubisco occurred early in the growth curve as cells were acclimating to fresh culture media, in ref. 22 εp > εRubisco occurred during nitrogen and phosphorus limitation, and in this study εp > εRubisco was observed in a mutant strain growing slowly while expressing a reconstructed ancestral rubisco. These observations indicated that growth physiology affects isotopic fractionation by photosynthetic algae and, in all cases, motivated a rethinking of the traditional box model (Fig. 1 B and C) to include more physiological detail relating to the presence of a CCM.
As high light consistently slowed growth of ANC, induced chlorosis (yellowing of cultures, SI Appendix, Fig. S11), and increased εp, we were motivated to consider the effects of light-related physiology on εp. The yellowing of ANC cultures in high light was consistent with starvation and taken to indicate that light levels exceeded the downstream capacity for CO2 fixation (79, 80). We interpreted these observations as indicating that the replacement of the native rubisco with a slower enzyme decreased capacity for CO2 fixation (Table 1).
Low-CO2 fixation capacity would not, on its own, explain anomalously high εp values, however. An additional fractionating process is required to explain εp values in excess of εRubisco, which we assume is due to light-coupled vectoral hydration of CO2, which has a large calculated isotope effect (70, 72–75). It is well established that modern Cyanobacteria have light-coupled CO2 uptake systems (65, 68) and in model Cyanobacteria, this activity is due to the Cup proteins (CupAS/B, also known as Chp proteins), which bind the NDH complex (68, 81). In order for CO2 uptake to drive the CCM and promote CO2 fixation, it would need to produce a high, nonequilibrium HCO3− concentration in the cytoplasm (8, 10). We and others therefore assumed that the complex of NDH-1 and CupAS/B couples light energy to the vectoral hydration of CO2 to HCO3− at a CA-like active site (68). Disruption of a Cup protein by point mutation was also shown to largely affect cell growth (69), suggesting that the energy-induced directionality is important for Cyanobacteria.
It is apparent from our experiments that εRubisco does not set an upper bound on εp, nor does it predict which strains will have larger εp values (Fig. 4). This was only apparent because we measured the isotopic fractionation due to the ancestral rubisco enzyme (εRubisco) and compared it to ANC strain biomass (εp), in contrast with (46), which measured εp but not εRubisco. While our ANC εp values (≈18 to 24‰) fell within the range of εp values derived from the carbon isotope record (43), they exceeded ANC εRubisco (Fig. 2). Attention has been paid to outliers in the carbon isotope record where εp exceeds εRubisco precisely because they violate the assumptions underlying the dominant models Eqs. 1 and 2) used to interpret the record (22). In addition, ANC εRubsico (17.23 ± 0.61‰) is anomalously low; not only is it ≈8‰ less than WT εRubsico (25.18 ± 0.31‰) but it is among the lowest measured rubisco KIEs. However, only thirteen unique rubisco KIEs have been measured thus far (23) while ≈300 distinct rubiscos have been kinetically characterized (7, 82), suggesting that measuring the isotopic effects of several well-chosen rubisco variants is worthwhile.
Turning to trends in carbon isotope data from the geological record, our results suggested there are at least two nonunique ways to achieve the large εp values observed earlier in Earth history: i) High external concentrations of Ci, or ii) Active CO2 uptake driven by photochemical electron transport. Our proposed model (an idealized extension of the traditional model, Eq. 2) cannot be applied readily to the C Isotope Record Model (Eq. 1). Doing so currently gives nonsensical values of b because ANC εp > εRubisco (see SI Appendix, section 6 and Fig. S16 for further discussion), and because we cannot independently constrain the extra degree of freedom introduced (two loss fluxes, ΦLoss1 and ΦLoss2, instead of one, f ). In addition, these parameters could vary over evolutionary history as the CCM and the efficiency of carbon fixation evolves. Additional measurements that constrain these parameters (i.e., ΦLoss1 and ΦLoss2) could enable pCO2 to be back-calculated from εp, but further work must be done to then adapt those observations to the C Isotope Record Model (Eq. 2). Importantly, the modified model framework proposed here is not the only approach to producing εp > εRubisco with physiologically feasible leakage fluxes. Rather than advocating for our specific model, we offer it as an example form of a solution – showing that εp can only exceed εRubisco if additional fractionating process is considered. As shown in SI Appendix, Fig. S9, several approaches to extending the traditional box model can accommodate εp > εRubisco (18, 61), yet all of these models represent substantial simplifications of bacterial and algal CCMs. Overall, our study supports the conclusion of prior studies (18, 22) that a modified traditional model that engages more fully with photosynthetic physiology, like the CCM, is required to more accurately constrain environmental Ci concentrations from εp.
In addition, this study and other recent work (43, 46) have raised a greater question for the Earth Sciences: What is uniformitarianism for biology? Earth scientists often apply uniformitarian assumptions—assuming that physical and chemical processes behave the same now as they did billions of years ago—in order to reason about the past. This approach is powerful, but these assumptions are challenged by biological processes that undergo substantial evolution on geologic timescales. Ongoing discoveries of novel metabolisms have supported some principles like “the principle of microbial infallibility”—that microbes will always find a way to take advantage of available energy sources (83)—but it is not clear what principles apply to the details of metabolism. Take rubisco, for example – most extant autotrophs use rubisco to fix carbon, but rubisco sits within a variety of physiologies—e.g., C3, C4, CAM in plants—that temper the effect of εRubisco on εp (23). We are far from having a clear answer to this question, but recent work at the interface of molecular biology and isotope geochemistry show that these ideas can be tested in the lab. Here and in other recent papers (42, 43, 46), we used synthetic biology to construct organisms with ancestral components so that specific aspects of ancient organisms can be isolated and tested. These “ancestral-like” organisms helped sharpen our understanding of the physiological and environmental factors determining growth (42) and isotopic fractionation (this work) in both ancient and modern autotrophs, showing that models rigidly based on modern taxa are likely not universally applicable across geologic time.
Overall, carbon fixation was a fundamental challenge that autotrophs overcame early in the history of Earth’s biosphere (1). These early processes were recorded in some fashion in the carbon isotope record, but robust interpretation of this record must grapple with the fact that the carbon cycle is an amalgam of both environmental changes and evolutionary processes, mediated by physiology. We now have synthetic biological approaches that offer a way to probe these long timescale coevolutionary problems by producing ancient process analogs of carbon fixation in the laboratory. Utilizing these tools will enable us to better understand how the evolution of key metabolisms have shaped Earth’s chemistry over time.
Materials and Methods
Ancestral Enzyme Reconstruction.
Ancestral Rubisco enzyme sequences were previously reported and characterized by Shih et al. (47). Briefly, for both the large subunit and small subunit of Rubisco, encoded by rbcL and rbcS, respectively, the most recent common ancestor (MRCA) for Form 1A (α), 1B (β), and 1A/B (α/ β) clades were predicted from independently derived phylogenetic trees for RbcL and RbcS containing a broad diversity of Form 1A and 1B Rubisco (>100 sequences). Maximum-likelihood algorithms were used to reconstruct the most probable ancestral sequence for each clade. Ancestral sequences were then expressed in Escherichia coli and purified, and enzyme kinetics were measured.
ANC Strain Generation.
The “ANC” strain studied here was generated by replacing the native large and small rubisco subunits (cbbL and cbbS, respectively) of the parent strain (Synechococcus elongatus PCC 7942) with the reconstructed β ancestral cbbL and cbbS sequences. The NS2-KanR (“WT” strain) was generated by inserting a KanR cassette into neutral site 2 (NS2) (GenBank: U44761.1). This was done as a control for having the KanR in the neutral site. Synechococcus elongatus PCC 7942 were transformed from the WT strain using the approach of Golden and Sherman (84). Briefly, cultures were grown to OD750 nm = 0.5. Cultures were centrifuged at 18,000 x g for 2 min. Pellets were washed with 100 mM CaCl2 and spun again at 18,000 x g for 2 min. Pellets were resuspended in BG-11 media followed by addition of plasmid and grown for 16 h in the dark at 30 °C. Transformants were then plated onto BG-11 + KAN100 agar plates and placed under 100 µE of light at 30 °C. Single colonies were selected in media with antibiotic until segregation and then genotyped by PCR amplification of the rubisco locus followed by sequencing to confirm homoplasmic ANC strain rubisco sequence. SI Appendix, Table S1 lists plasmids and primers used in this study.
Growth Conditions.
For ambient CO2 growth, NS2-KanR (“WT”) and β Ancestral Rubisco-KanR (“ANC”) strains were grown in quadruplicate in a photobioreactor (Photon Systems Instruments–MC 1000) at the University of California, Berkeley (UC Berkeley) for four biological replicates total. Cultures were grown in buffered BG-11 media with 50mM HEPES at pH 8. Cultures were inoculated at a starting OD720 nm = 0.015 and cultivated at 120 µmol photons m−2 s−1, 30 °C, and bubbled with ambient air. High CO2 growth was performed using the same conditions as ambient growth with the exception of placing the photobioreactor in a 5% CO2 chamber (Percival AR22L) and bubbling in air from the chamber. High-light growth was performed using the ambient conditions above with the exception of using 500 µmol photons m−2 s−1 for light intensity. Cells were harvested by centrifugation at 6000 x g for 20 min at 4 °C. Decanted pellets were then flash frozen with liquid N2 and lyophilized overnight with the Millrock Technology Model BT85A freeze dryer. Doubling time was calculated by fitting the exponential phase of growth (k) using a Markov Chain Monte Carlo (MCMC) approach, using the generic model y = a*EXP(k*x)+b. Growth curves displayed in Fig. 3 were smoothed with a rolling median (n = 12) to remove errant readings caused by bubbles advected in front of the detector. See SI Appendix for more information.
Carbon Isotope Analysis.
Carbon isotope data are reported using delta notation (δ13C) in units of per mille (‰) where δ13C = [(13C/12C)sa/(13C/12C)ref-1]*1000, where the subscripts “sa” and “ref” denote sample and reference respectively. The reference used is the Vienna Pee Dee Belemnite (VPDB). δ13C values of cyanobacterial cells were measured on an EA-IRMS (Elemental Analyzer Isotope Ratio Mass Spectrometer; Costech Thermo Delta-V) at the California Institute of Technology (Caltech) in Pasadena, CA. Each biological replicate was run four times with two different isotope standards—urea (−27.8‰) and sucrose (−10.45‰). A suite of urea and sucrose standards were run at the beginning, middle, and end of run for sample bracketing and to assess drift throughout the run. An average δ13C and SE were calculated and reported for each biological replicate (see SI Appendix for more information). The δ13C of the starting CO2 gas was measured on the Thermo Mat 253 Ultra at Caltech; the CALT-2049C standard was used, which has a δ13CVPDB value of −3.62‰. CO2 gas from high-pCO2 experiments was sourced from a CO2 tank, while the CO2 gas in ambient pCO2 experiments was distilled from ambient lab air through cryogenic distillation at Caltech. In addition, we labored to keep gas pressures approximately constant during our experiments (i.e., equilibrating to ambient pressure by bubbling) because of potential unwanted isotopic pressure effects. εp, the carbon isotope fractionation between CO2 gas and bulk cyanobacterial cells, was calculated as (αCO2/bio - 1)*1000, where αCO2/bio = 13RCO2/13Rbio, where 13R is the ratio of 13C to 12C in the analyte. We note this in contrast to other isotope literature where εp is calculated as αbio/CO2 - 1)*1000, which would cause the positive values in this study to be negative. In this study, more positive εp values indicate more 13C-depleted; see SI Appendix for more detail.
Rubisco KIE Assay.
Syn6301 and β-MRCA rubisco were purified according to previous methodologies (85, 86) at University of California, Davis and then shipped on dry ice to Caltech. Clarified lysate from a BL21 DE3 Star E. coli culture expressing rubisco was subjected to ammonium sulfate precipitation, at the 30 to 40% cut for Syn6301 and at the 40 to 50% cut for β-MRCA, followed by anion exchange chromatography and size exclusion chromatography. We then used the substrate depletion method to measure the KIE of the Syn6301 and β-MRCA rubiscos (εRubisco), as used previously in similar studies (48–51). Briefly, an assay mix of HCO3−, bovine CA, rubisco, ribulose 1,5-bisphosphate (RuBP), MgCl2, bicine, and dithiothreitol (DTT) was prepared. As the reaction progressed to completion, aliquots of that assay mix were injected into prefilled exetainers containing phosphoric acid that both stopped the reaction and converted all inorganic carbon species to gaseous CO2. The δ13C of these CO2 aliquots was then measured on a Delta-V Advantage with Gas Bench and Costech elemental analyzer at Caltech. Here, instead of RuBP being given in excess, CO2 was given in excess. In addition, instead of determining the fraction of CO2 (f ) consumed independently to create a Rayleigh plot, we fit the curvature of the δ13C results to find f before converting to a Rayleigh plot to calculate εRubisco, similar to previous studies (49). See SI Appendix for more information.
Transmission Electron Microscopy Imaging of Whole Cells.
WT and ANC strains were grown in the reference condition—buffered BG-11 media, shaking at 250 rpm, with white cool fluorescent light at 120 µE, 30 °C, ambient air (0.04% CO2 (v/v). WT and ANC cells were collected at mid-log (40 and 80 h, respectively) at OD730 nm = 0.4 and pelleted by centrifugation (10,000 x g for 10 min). Pelleted cells were then resuspended in 1 mL cold solution 2.5% Glutaraldehyde in 0.1M Sodium Cacodylate Buffer, pH 7.4 (Electron Microscopy Sciences) and stored in the fixative solution at 4 °C until imaging. Sample preparation and sectioning were performed in the Electron Microscope Laboratory core facility at the University of California Berkeley. Briefly, samples were stabilized in 1% low melting-point agarose, cut into small cubes, and then washed at room temperature with 0.1 M sodium cacodylate buffer, pH 7. Samples were then mixed with 1% osmium tetroxide, 1.6% potassium ferricyanide and 0.1 M cacodylate buffer pH 7.2 for an hour in the dark with rotation. These were washed again with a cacodylate buffer pH 7.2, then DI water, and subjected to a 1-h incubation with uranyl acetate 0.5% solution. After a new wash with DI water, samples were dehydrated by an ascending series of acetone concentration (35%, 50%, 75%, 80%, 90%, 100%, 100%). Later, samples were progressively infiltrated in resin (Epon solution: Eponate 12, DDSA NMA and BDMA (Electron Microscopy Sciences) with rotation, followed by a final step at 60˚C until polymerized. Thin sections (70 nm) were cut using a Reichert Ultracut E (Leica Microsystems) and collected on 100 mesh formvar-coated copper grids. Sections were poststained using 2% uranyl acetate in 70% methanol and followed with Reynold’s lead citrate. The sections were imaged using a FEI Tecnai 12 transmission electron microscope operated at 120 kV (FEI). Images were collected using UltraScan 1000 digital micrograph software (Gatan Inc).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Newton Nguyen for valuable guidance in the MCMC model used to calculate doubling times from growth curve data. We thank Victoria Orphan and Alex Sessions for access to lab space and analytical instruments, as well as lab managers Stephanie A. Connon, Fenfang Wu, and Nami Kitchen for assistance. This research was supported by the David and Lucille Packard Foundation (12540178), Simons Foundation (554187), NASA Exobiology (00010652), and the Schwartz-Reisman Collaborative Science Program (12520057). R.Z.W. was supported by a NSF Graduate Research Fellowship. Work in the lab of D.F.S. was supported by the US Department of Energy (DE-SC00016240). Work in the lab of P.M.S. was supported by a Society in Science-Branco Weiss fellowship from ETH Zürich and a Packard Fellowship from the David Lucile Packard Foundation. We thank Danielle Jorgens and Reena Zalpuri at the University of California Berkeley Electron Microscope Laboratory for advice and assistance in electron microscopy sample preparation and data collection.
Author contributions
R.Z.W., R.J.N., A.K.L., D.F.S., J.M.E., P.M.S., and W.W.F. designed research; R.Z.W., R.J.N., A.K.L., J.A., and D.M.B. performed research; R.Z.W., R.J.N., A.K.L., A.I.F., and J.A. analyzed data; D.F.S., J.M.E., P.M.S., and W.W.F. advised on project; and R.Z.W. and A.I.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Fischer W. W., Hemp J., Johnson J. E., Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci. 44, 647–683 (2016). [Google Scholar]
- 2.Bar-On Y. M., Milo R., The global mass and average rate of rubisco. Proc. Natl. Acad. Sci. U.S.A. 116, 4738–4743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wildman S. G., Along the trail from Fraction I protein to Rubisco (ribulose bisphosphate carboxylase-oxygenase). Photosyn. Res. 73, 243–250 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Lorimer G. H., Andrews T. J., Plant photorespiration—an inevitable consequence of the existence of atmospheric oxygen. Nature 243, 359–360 (1973). [Google Scholar]
- 5.Andrews T. J., Lorimer G. H., The Biochemistry of Plants: A Comprehensive Treatise, Vol. 10, Photosynthesis, Hatch M. D., Boardman N. K., Eds. (Academic Press, 1987). [Google Scholar]
- 6.Bar-Even A., et al. , The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Flamholz A. I., et al. , Revisiting trade-offs between rubisco kinetic parameters. Biochemistry 58, 3365–3376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flamholz A., Shih P. M., Cell biology of photosynthesis over geologic time. Curr. Biol. 30, R490–R494 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Rae B. D., Long B. M., Badger M. R., Price G. D., Functions, compositions, and evolution of the two types of carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangan N. M., Flamholz A., Hood R. D., Milo R., Savage D. F., pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, E5354–E5362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raven J. A., Beardall J., CO 2 concentrating mechanisms and environmental change. Aquatic Botany 118, 24–37 (2014). [Google Scholar]
- 12.Price G. D., et al. , The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64, 753–768 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Riding R., Cyanobacterial calcification, carbon dioxide concentrating mechanisms, and Proterozoic?Cambrian changes in atmospheric composition Geobiology 4, 299–316 (2006). [Google Scholar]
- 14.Hayes J. M., Factors controlling 13C contents of sedimentary organic compounds: Principles and evidence. Mar. Geol. 113, 111–125 (1993). [Google Scholar]
- 15.Francois R., et al. , Changes in the δ13 C of surface water particulate organic matter across the subtropical convergence in the SW Indian Ocean. Global Biogeochem. Cycles 7, 627–644 (1993). [Google Scholar]
- 16.Park R., Epstein S., Carbon isotope fractionation during photosynthesis. Geochim. Cosmochim. Acta 21, 110–126 (1960). [Google Scholar]
- 17.Farquhar G. D., O’Leary M. H., Berry J. A., On the relationship between carbon isotope discrimination and the intercellular carbon dioxide concentration in leaves. Aust. J. Plant Physiol. 9, 121 (1982). [Google Scholar]
- 18.Eichner M., Thoms S., Kranz S. A., Rost B., Cellular inorganic carbon fluxes in Trichodesmium: A combined approach using measurements and modelling. J. Exp. Bot. 66, 749–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharkey T. D., Berry J. A., “Carbon isotope fractionation of algae as influenced by an inducible CO2 concentrating mechanism” in Inorganic Carbon Uptake by Aquatic Photosynthetic Organisms, Lucas W. J., Berry J. A., Eds. (The American Society of Plant Physiologists, 1985), pp. 389–401. [Google Scholar]
- 20.Mook W. G., Bommerson J. C., Staverman W. H., Carbon isotope fractionation between dissolved bicarbonate and gaseous carbon dioxide. Earth Planet. Sci. Lett. 22, 169–176 (1974). [Google Scholar]
- 21.Farquhar G. D., Ehleringer J. R., Hubick K. T., Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 503–537 (1989). [Google Scholar]
- 22.Wilkes E. B., Pearson A., A general model for carbon isotopes in red-lineage phytoplankton: Interplay between unidirectional processes and fractionation by RubisCO. Geochim. Cosmochim. Acta 265, 163–181 (2019), 10.1016/j.gca.2019.08.043. [DOI] [Google Scholar]
- 23.Garcia A. K., Cavanaugh C. M., Kacar B., The curious consistency of carbon biosignatures over billions of years of Earth-life coevolution. ISME J. 15, 2183–2194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boller A. J., Thomas P. J., Cavanaugh C. M., Scott K. M., Low stable carbon isotope fractionation by coccolithophore RubisCO. Geochim. Cosmochim. Acta 75, 7200–7207 (2011). [Google Scholar]
- 25.Schidlowski M., A 3,800-million-year isotopic record of life from carbon in sedimentary rocks. Nature 333, 313–318 (1988). [Google Scholar]
- 26.Krissansen-Totton J., Buick R., Catling D. C., A statistical analysis of the carbon isotope record from the Archean to Phanerozoic and implications for the rise of oxygen. Am. J. Sci. 315, 275–316 (2015). [Google Scholar]
- 27.Hayes J. M., Strauss H., Kaufman A. J., The abundance of 13C in marine organic matter and isotopic fractionation in the global biogeochemical cycle of carbon during the past 800 Ma. Chem. Geol. 161, 103–125 (1999). [Google Scholar]
- 28.Jasper J. P., Hayes J. M., A carbon isotope record of CO2 levels during the late quaternary. Nature 347, 462–464 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Pagani M., et al. , The role of carbon dioxide during the onset of Antarctic glaciation. Science 334, 1261–1264 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Higgins J. A., et al. , Atmospheric composition 1 million years ago from blue ice in the Allan Hills, Antarctica. 112, 6887–6891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bidigare R. R., et al. , Consistent fractionation of13 C in nature and in the laboratory: Growth-rate effects in some haptophyte algae. Global Biogeochem. Cycles 11, 279–292 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Popp B. N., et al. , Effect of phytoplankton cell geometry on carbon isotopic fractionation. Geochim. Cosmochim. Acta 62, 69–77 (1998). [Google Scholar]
- 33.Laws E. A., Popp B. N., Bidigare R. R., Kennicutt M. C., Macko S. A., Dependence of phytoplankton carbon isotopic composition on growth rate and [CO2)aq: Theoretical considerations and experimental results. Geochim. Cosmochim. Acta 59, 1131–1138 (1995). [Google Scholar]
- 34.Zhang Y. G., Henderiks J., Liu X., Refining the alkenone-pCO2 method II: Towards resolving the physiological parameter ‘b’. Geochim. Cosmochim. Acta 281, 118–134 (2020). [Google Scholar]
- 35.Rau G. H., Riebesell U., Wolf-Gladrow D., A model of photosynthetic 13C fractionation by marine phytoplankton based on diffusive molecular CO2 uptake. Mar. Ecol. Prog. Ser. 133, 275–285 (1996). [Google Scholar]
- 36.O’Leary M. H., Measurement of the isotope fractionation associated with diffusion of carbon dioxide in aqueous solution. J. Phys. Chem. 88, 823–825 (1984). [Google Scholar]
- 37.Cassar N., Laws E. A., Popp B. N., Carbon isotopic fractionation by the marine diatom Phaeodactylum tricornutum under nutrient- and light-limited growth conditions. Geochim. Cosmochim. Acta 70, 5323–5335 (2006). [Google Scholar]
- 38.Berry J. A., “Studies of mechanisms affecting the fractionation of carbon isotopes in photosynthesis” in Stable Isotopes in Ecological Research, Ecological Studies, Rundel P. W., Ehleringer J. R., Nagy K. A., Eds. (Springer, New York, 1989), pp. 82–94. [Google Scholar]
- 39.Gimmler H., Weiss C., Baier M., Hartung W., The conductance of the plasmalemma for co2. J. Exp. Bot. 41, 785–795 (1990). [Google Scholar]
- 40.Rotatore C., Lew R. R., Colman B., Active uptake of CO2 during photosynthesis in the green alga Eremosphaera viridis is mediated by a CO2-ATPase. Planta 188, 539–545 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Sultemeyer D., Biehler K., Fock H. P., Evidence for the contribution of pseudocyclic photophosphorylation to the energy requirement of the mechanism for concentrating inorganic carbon in Chlamydomonas. Planta 189, 235–242 (1993). [Google Scholar]
- 42.Flamholz A. I., et al. , Trajectories for the evolution of bacterial CO2-concentrating mechanisms. Proc. Natl. Acad. Sci. U.S.A. 119, e2210539119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurley S. J., Wing B. A., Jasper C. E., Hill N. C., Cameron J. C., Carbon isotope evidence for the global physiology of Proterozoic cyanobacteria. Sci. Adv. 7, eabc8998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia A. K., et al. , Effects of RuBisCO and CO2 concentration on cyanobacterial growth and carbon isotope fractionation. Geobiology 21, 390–403 (2023), 10.1111/gbi.12543. [DOI] [PubMed] [Google Scholar]
- 45.Kacar B., Hanson-Smith V., Adam Z. R., Boekelheide N., Constraining the timing of the Great Oxidation Event within the Rubisco phylogenetic tree. Geobiology 15, 628–640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kędzior M., et al. , Resurrected Rubisco suggests uniform carbon isotope signatures over geologic time. Cell Rep. 39, 110726 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Shih P. M., et al. , Biochemical characterization of predicted Precambrian RuBisCO. Nat. Commun. 7, 10382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guy R. D., Fogel M. L., Berry J. A., Photosynthetic fractionation of the stable isotopes of oxygen and carbon. Plant Physiol. 101, 37–47 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNevin D. B., Badger M. R., Kane H. J., Farquhar G. D., Measurement of (carbon) kinetic isotope effect by Rayleigh fractionation using membrane inlet mass spectrometry for CO2-consuming reactions. Funct. Plant Biol. 33, 1115 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Scott K. M., Schwedock J., Schrag D. P., Cavanaugh C. M., Influence of form IA RubisCO and environmental dissolved inorganic carbon on the delta13C of the clam-chemoautotroph symbiosis Solemya velum. Environ. Microbiol. 6, 1210–1219 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Thomas P. J., et al. , Isotope discrimination by form IC RubisCO from Ralstonia eutropha and Rhodobacter sphaeroides, metabolically versatile members of “Proteobacteria” from aquatic and soil habitats. Environ. Microbiol. 21, 72–80 (2019), 10.1111/1462-2920.14423. [DOI] [PubMed] [Google Scholar]
- 52.Wang R. Z., Liu A. K., Banda D. M., Fischer W. W., Shih P. M., A bacterial form I’ Rubisco has a smaller carbon isotope fractionation than its form I counterpart. Biomolecules 13, 596 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tcherkez G. G. B., Farquhar G. D., Andrews T. J., Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 103, 7246–7251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price G. D., Badger M. R., Isolation and characterization of high CO(2)-requiring-mutants of the cyanobacterium synechococcus PCC7942: Two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO(2) within the carboxysome. Plant Physiol. 91, 514–525 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H., et al. , Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566, 131–135 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Jensen T. E., Electron microscopy of polyphosphate bodies in a blue-green alga, Nostoc pruniforme. Archiv. Mikrobiol. 62, 144–152 (1968). [Google Scholar]
- 58.Śliwińska-Wilczewska S., Konarzewska Z., Wiśniewska K., Konik M., Photosynthetic pigments changes of three phenotypes of picocyanobacteria Synechococcus sp. under different light and temperature conditions. Cells 9, 2030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grébert T., et al. , Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E2010–E2019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson K., Beardall J., Raven J. A., Adaptation of unicellular algae to irradiance: An analysis of strategies. New Phytol. 93, 157–191 (1983). [Google Scholar]
- 61.Erez J., Bouevitch A., Kaplan A., Carbon isotope fractionation by photosynthetic aquatic microorganisms: Experiments withSynechococcus PCC7942, and a simple carbon flux model. Can. J. Bot. 76, 1109–1118 (1998). [Google Scholar]
- 62.Desmarais J. J., et al. , DABs are inorganic carbon pumps found throughout prokaryotic phyla. Nat. Microbiol. 4, 2204–2215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogawa T., Kaplan A., Inorganic carbon acquisition systems in cyanobacteria. Photosyn. Res. 77, 105–115 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Volokita M., Zenvirth D., Kaplan A., Reinhold L., Nature of the inorganic carbon species actively taken up by the Cyanobacterium Anabaena variabilis. Plant Physiol. 76, 599–602 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price G. D., Maeda S., Omata T., Badger M. R., Modes of active inorganic carbon uptake in the cyanobacterium, Synechococcus sp. PCC7942. Funct. Plant Biol. 29, 131 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Maeda S., Badger M. R., Price G. D., Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 43, 425–435 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Klughammer B., Sültemeyer D., Badger M. R., Price G. D., The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol. Microbiol. 32, 1305–1315 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Schuller J. M., et al. , Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 11, 494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Artier J., et al. , Modeling and mutagenesis of amino acid residues critical for CO2 hydration by specialized NDH-1 complexes in cyanobacteria. Biochim. Biophys. Acta Bioenerg. 1863, 148503 (2022). [DOI] [PubMed] [Google Scholar]
- 70.Sade Z., Halevy I., New constraints on kinetic isotope effects during CO 2(aq) hydration and hydroxylation: Revisiting theoretical and experimental data. Geochim. Cosmochim. Acta 214, 246–265 (2017). [Google Scholar]
- 71.Zeebe R. E., Wolf-Gladrow D., CO2 in Seawater: Equilibrium, Kinetics, Isotopes (Elsevier, 2001). [Google Scholar]
- 72.Clark I. D., Lauriol B., Kinetic enrichment of stable isotopes in cryogenic calcites. Chem. Geol. 102, 217–228 (1992). [Google Scholar]
- 73.Guo W., “Carbonate clumped isotope thermometry: Applications to carbonaceous chondrites and effects of kinetic isotope fractionation”, Dissertation (Ph.D.), California Institute of Technology; (2009). [Google Scholar]
- 74.Zeebe R. E., Kinetic fractionation of carbon and oxygen isotopes during hydration of carbon dioxide. Geochim. Cosmochim. Acta 139, 540–552 (2014). [Google Scholar]
- 75.Boettger J. D., Kubicki J. D., Equilibrium and kinetic isotopic fractionation in the CO2 hydration and hydroxylation reactions: Analysis of the role of hydrogen-bonding via quantum mechanical calculations. Geochim. Cosmochim. Acta 292, 37–63 (2021). [Google Scholar]
- 76.Tchernov D., et al. , Passive entry of CO2 and its energy-dependent intracellular conversion to HCO3- in cyanobacteria are driven by a photosystem I-generated deltamuH+. J. Biol. Chem. 276, 23450–23455 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Espie G. S., Miller A. G., Canvin D. T., High affinity transport of CO(2) in the cyanobacterium synechococcus UTEX 625. Plant Physiol. 97, 943–953 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaplan A., Reinhold L., Co2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 539–570 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Collier J. L., Grossman A. R., Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J. Bacteriol. 174, 4718–4726 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adir N., Dines M., Klartag M., McGregor A., Melamed-Frank M., “Assembly and disassembly of phycobilisomes” in Complex Intracellular Structures in Prokaryotes, Microbiology Monographs, Shively J. M., Ed. (Springer, Berlin Heidelberg, 2006), pp. 47–77. [Google Scholar]
- 81.Battchikova N., Eisenhut M., Aro E.-M., Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochim. Biophys. Acta 1807, 935–944 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Iñiguez C., et al. , Evolutionary trends in RuBisCO kinetics and their co-evolution with CO2 concentrating mechanisms. Plant J. 101, 897–918 (2020). [DOI] [PubMed] [Google Scholar]
- 83.O’Malley M. A., Walsh D. A., Rethinking microbial infallibility in the metagenomics era. FEMS Microbiol. Ecol. 97, fiab092 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Golden S. S., Sherman L. A., Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 158, 36–42 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saschenbrecker S., et al. , Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129, 1189–1200 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Banda D. M., et al. , Novel bacterial clade reveals origin of form I Rubisco. Nat. Plants 6, 1158–1166 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.