Significance
Gastric cancer is a clinically challenging disease with poor prognosis. Heparan sulfate proteoglycans (HSPGs) have emerged as key molecules in tumor progression and communication. However, the functional relevance of HSPGs during gastric cancer development remains poorly known. Here, we demonstrate that Syndecan 4 (SDC4) is highly expressed in intestinal subtype gastric tumors and associates with higher invasion and patient poor survival. We provide evidence that glycosylated-SDC4 is packed on extracellular vesicles (EVs) and impacts EV protein cargo, uptake by recipient cells and, importantly, defines the tropism of cancer EVs to the common metastatic sites of gastric cancer. Thus, our data uncover SDC4 potential as a therapeutic target in precision oncology for limiting gastric cancer invasion and communication routes.
Keywords: syndecan-4, heparan sulfate proteoglycans, gastric cancer, extracellular vesicles, cancer cell invasion
Abstract
Gastric cancer is a dominating cause of cancer-associated mortality with limited therapeutic options. Here, we show that syndecan-4 (SDC4), a transmembrane proteoglycan, is highly expressed in intestinal subtype gastric tumors and that this signature associates with patient poor survival. Further, we mechanistically demonstrate that SDC4 is a master regulator of gastric cancer cell motility and invasion. We also find that SDC4 decorated with heparan sulfate is efficiently sorted in extracellular vesicles (EVs). Interestingly, SDC4 in EVs regulates gastric cancer cell-derived EV organ distribution, uptake, and functional effects in recipient cells. Specifically, we show that SDC4 knockout disrupts the tropism of EVs for the common gastric cancer metastatic sites. Our findings set the basis for the molecular implications of SDC4 expression in gastric cancer cells and provide broader perspectives on the development of therapeutic strategies targeting the glycan-EV axis to limit tumor progression.
Gastric cancer persists as a global health challenge, as it is the fifth most frequent malignancy with over one million new cases annually, and the fourth leading cause of cancer-related death worldwide (1). The clinical aggressiveness of gastric cancer, which is translated into poor survival of patients, is mainly attributed to the late diagnosis due to silent disease progression and the lack of efficient therapeutic options (2, 3).
Paramount evidence shows that alterations in the cellular glycosylation pathways are drivers of tumor progression (4, 5), as glycans and glycoconjugates are key regulators of the cross talk between cancer cells and the different components of the tumor microenvironment (5, 6). Particularly in gastric cancer, aberrant glycan signatures have been shown to govern important oncogenic processes, including proliferation, cell–cell and cell-extracellular matrix (ECM) adhesion, receptor tyrosine kinase (RTK) signaling, migration, invasion, angiogenesis, and metastatic dissemination (7–9).
Syndecans (SDCs) are a family of four transmembrane heparan sulfate proteoglycans (HSPGs). SDCs act in cooperation with key transmembrane receptors and ECM molecules mediating pleiotropic functions, such as signaling, adhesion, proliferation, migration, apoptosis, and differentiation (10). HSPG expression patterns, and particularly SDC signatures, have been described as aberrant in different cancer models, including tumors of the gastrointestinal tract (11, 12). However, there is a considerable gap of knowledge on the functional mechanisms and relevance of HSPGs and their ECM interactomes during gastric cancer development.
Syndecan-4 (SDC4) is one of the carriers of HS glycosaminoglycan chains in epithelial cells with key roles in the control of focal adhesion formation, migration, invasion, and endocytosis (13–16). SDC4 expression was shown to be up-regulated in response to oncogenic stimulus during the early stages of the gastric carcinogenesis pathway (17, 18).
Remarkably, SDCs are also key regulators of extracellular vesicle (EV) biogenesis (19). EVs are a heterogeneous group of secreted vesicles, which include exosomes and microvesicles, found in all body fluids. EVs are secreted by all cell types, including cancer cells, and hold major roles in tumor cell communication due to their capacity of reprograming recipient cells, and ultimately defining metastasis organ tropism (20, 21). SDCs together with their small intracellular adaptor syntenin and their associated regulators Alix (an accessory component of the endosomal-sorting complex required for transport), the small GTPase ARF6, the lipid modifying enzyme PLD2, and the HS glycosidase heparanase, have been shown to regulate the formation of EVs of endosomal origin and to impact their cargo composition (19, 22, 23). Moreover, the interaction of tetraspanin 6 (TSPN6) with SDC4 was shown to functionally control EV degradation and release (24). On the recipient cell side, HS chains, including those carried by SDCs, act as critical internalizing receptors of cancer cell-derived EVs with impact on their functional activity (25). Taking together the key roles played by HSPGs in both axes of cancer cell communication, EVs and their recipient cells, it is of utmost relevance to understand how SDC4, a master HSPG, impacts EV-mediated signaling and cancer-derived EV organ tropism.
In this study, we show that SDC4 is highly expressed in intestinal subtype gastric tumors and lymph node (LN) metastasis and associates with poor patient survival. Moreover, we mechanistically demonstrate, using SDC4 knockout (KO) gastric cancer cells, that SDC4 is a key regulator of gastric cancer aggressive features, such as migration and invasion. On top of modulating gastric cancer cell motility, we reveal that SDC4 conjugated with HS glycosaminoglycans is present in the EV cargo and defines EV organ distribution, uptake, and oncogenic functional effects on recipient cell populations.
Results
SDC4 Expression Is Up-Regulated in Intestinal Subtype Gastric Tumors and Is a Marker of Poor Patient Survival.
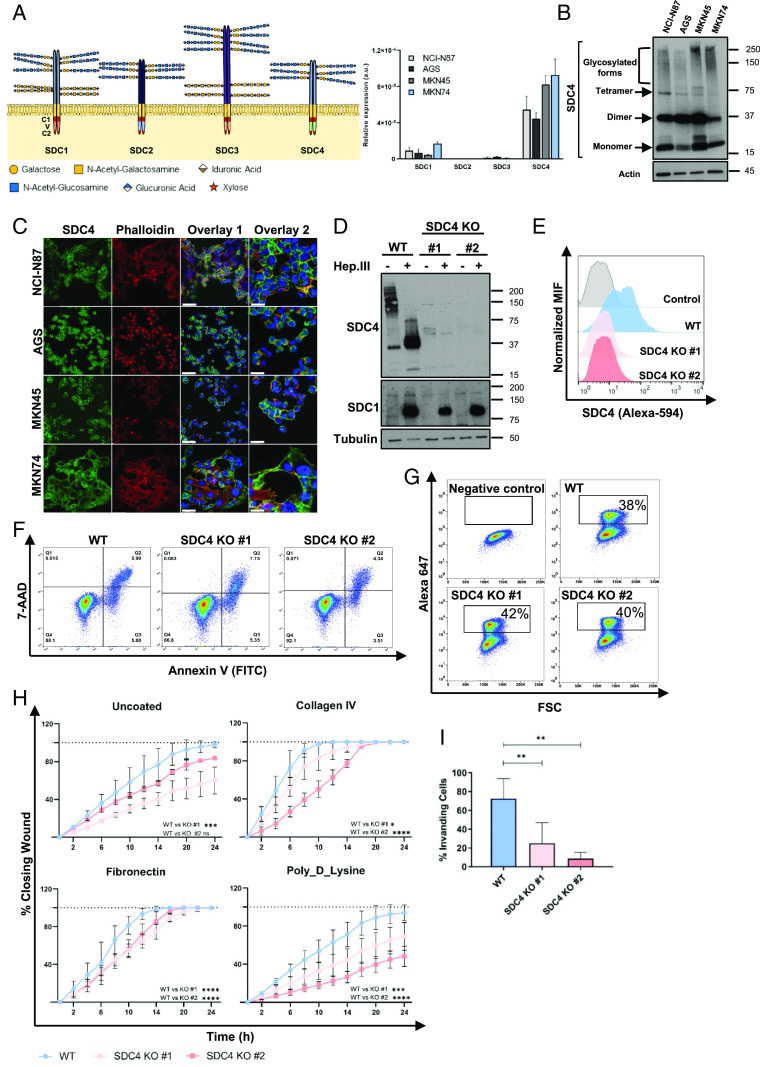
Aberrant expression of SDC4 has been reported in several cancer models (26–34). We have previously demonstrated that SDC4 transcription in gastric cells is up-regulated in response to inflammatory stimulus, such as cytokines or bacterial infection. In addition, we showed that SDC4 expression on Helicobacter pylori-inflamed gastric mucosa is associated with the virulence of the infecting strain (17, 18). However, the expression profile of SDC4 in gastric tumors remained to be elucidated. Thus, in this study we evaluated SDC4 expression in a retrospective series of 152 stage I–IV gastric carcinomas. SI Appendix, Table S1 summarizes the clinicopathological information of this cohort of gastric cancer patients. We observed that SDC4 tissue staining, using the 8G3 antibody which binds to the extracellular domain (ECD) of SDC4 (35), was significantly associated with tumor histological classification (P = 0.031), with 71.4% of the intestinal subtype tumors showing SDC4 positivity (Fig. 1 A and C and SI Appendix, Table S1), whereas among diffuse subtype tumors, SDC4 staining was lower in intensity and frequency and near half of the evaluated cases showed no staining (47.6% of positive cases) (Fig. 1 B and C and SI Appendix, Table S1). Interestingly, the SDC4 staining profile was also preserved in the LN metastases, with the majority of cases of intestinal subtype tumors showing SDC4-positive LN metastases (Fig. 1 A and D and SI Appendix, Fig. S1 and Table S1).
Fig. 1.
SDC4 is highly expressed in intestinal subtype gastric cancer and associates with patient poor survival. (A and B) Representative images of SDC4 immunohistochemistry (IHC) analysis of (A) intestinal and (B) diffuse subtypes of primary gastric tumor tissue samples paired with respective LN metastases. Three regions (R1-3) were selected: R1-adjacent mucosa; R2-primary tumor; R3-LN metastasis. (C and D) Bar graphs show the percentage of (C) primary tumor cases or (D) LN metastasis positive (red bar) or negative (black bar) for SDC4, for intestinal and diffuse gastric cancer subtypes. The chi-square test was performed and * represents P < 0.005. (E–G) Kaplan–Meier (KM) analysis of SDC4 tissue expression levels and gastric cancer patients overall survival (OS) for (E) all gastric cancer, (F) intestinal subtype and (G) diffuse subtype patients, the categorization of these clinical samples was assigned into SDC4 IHC positive (red lines) or SDC4 IHC negative (black lines). Comparison between curves in KM plots was performed using the log-rank test. (H) SDC4 expression signature in gastric cancer (STAD-TCGA cohort) was clustered using the heatmap CbioPortal algorithm (1, 2) and correlated with clinicopathological data, including metastasis, recurrence, Lauren subtype, TCGA molecular subtype [chromosomal instability (CIN); Epstein–Barr virus (EBV); microsatellite instability (MSI); genomically stable (GS)], MSI and aneuploid scores. (I and J) Distribution of SDC4 mRNA expression levels in the STAD-TCGA cohort by (I) Lauren classification and by (J) tumor grade. Samples classified as undetermined gastric cancer NOS (Not otherwise specified) in this dataset were excluded from the analysis. RNA sequencing of SDC4 expression is represented as fragments per kilobase million. (K and L) KM analysis of SDC4 mRNA levels and gastric cancer (K) OS or (L) disease-free survival (DFS) (STAD-TCGA cohort). The categorization of patient samples was divided into low (lowest 25%, black lines) and high (highest 25%, red lines) subgroups according to the levels of SDC4 mRNA expression. Comparison between curves in KM plots was performed using the log-rank test. References: 1. E. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-4. 2. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
Kaplan–Meier analysis of patient prognosis, showed that SDC4 expression was significantly associated with overall poor survival of patients (OS) (80.4 vs. 59.7 mo, P = 0.025) (Fig. 1E and SI Appendix, Table S2). When stratified by clinicopathological variables, this association was also observed in younger (<66 y) (P = 0.005), male (P = 0.028), patients without metastatic disease, pN0 (P = 0.016) and pM0 (P = 0.044) and exclusively treated with surgery (P = 0.026). Considering the histological classification, the OS association with SDC4 expression was only significant in patients within the intestinal gastric cancer subtype group (88.8 vs. 60.6 mo, P = 0.018) (Fig. 1F and SI Appendix, Table S2), while for diffuse subtype there was no association between SDC4-positivity and survival (Fig. 1G and SI Appendix, Table S2).
The independent prognostic value of SDC4 expression was assessed via stepwise multivariate Cox regression with backward elimination. The analysis was performed in all patients and then stratified by histological (Lauren) classification, since SDC4 expression was associated with poor prognosis especially in patients with intestinal subtype. Variables such as gender, age, disease stage, and treatment were added to the model, and disease stage was the only covariable retained. In patients with intestinal subtype tumors, the association of SDC4 expression remained significant when adjusted to disease stage (HR = 1.893; 95% CI: [1.033–3.471]; P = 0.039), highlighting its value as an independent prognostic factor.
To further validate our observations in an independent gastric cancer patient cohort, we analyzed the SDC4 transcriptomic profile of gastric cancer samples from The Cancer Genome Atlas Stomach Adenocarcinoma (STAD-TCGA). We observed that SDC4 high-transcript levels clustered with cancer aggressive features, such as metastasis (M1) and disease recurrence (Fig. 1H). Again, SDC4 expression was clustered with the intestinal subtype (Fig. 1 H and I). Noteworthy, high SDC4 transcription levels also clustered with the CIN molecular subtype and with low MSI/high aneuploidy scores (Fig. 1H). In this STAD-TCGA cohort, SDC4 expression showed a trend toward higher expression in higher disease grades (Fig. 1J). Kaplan–Meier analysis of SDC4 mRNA levels and gastric cancer patient OS and DFS was performed in two additional independent cohorts: the STAD-TCGA cohort (Fig. 1 K and L and SI Appendix, Fig. S2 A and B) and in the cohort from the Kaplan–Meier Plotter (KM plotter) database (SI Appendix, Fig. S2 C and D). These analyses validated a significant association between SDC4 expression and patient worse prognosis (DFS) for all patients and in intestinal subtype in STAD-TCGA dataset (Fig. 1L and SI Appendix, Fig. S2B). A significant association between SDC4 and OS and DFS was also confirmed in KM plotter cohort for intestinal subtype (SI Appendix, Fig. S2 C and D).
A multivariate Cox regression with backward elimination was performed to assess the independent prognostic value of SDC4 expression in all gastric cancer patients and then in the intestinal subtype patients, within the STAD-TCGA database. The clinicopathological parameters gender, Lauren classification, age, and disease stage were used as covariables for OS and DFS. Of note, the expression data from all patients from this dataset were collected before treatment. We found that the covariables disease stage and age (≥67 y) were associated with OS. However, SDC4 was not associated with OS (HR = 1.782; 95%CI: [0.747 – 4.258]; P = 0.187). When the multivariate analysis was performed for DFS, we found that the clinical parameter grade was the only covariable retained only for the intestinal subtype. Importantly, this analysis validated the independent prognostic value of SDC4 for DFS, when adjusted to disease stage in all gastric cancer patients (HR = 4.004; 95%CI: [1.080 – 14.837]; P = 0.038) and when subclassified by intestinal subtype (HR = 3.912; 95%CI: [0.109–12.872]; P = 0.017).
SDC4 Promotes Promigratory and Invasive Phenotypes in Gastric Cancer Cells.
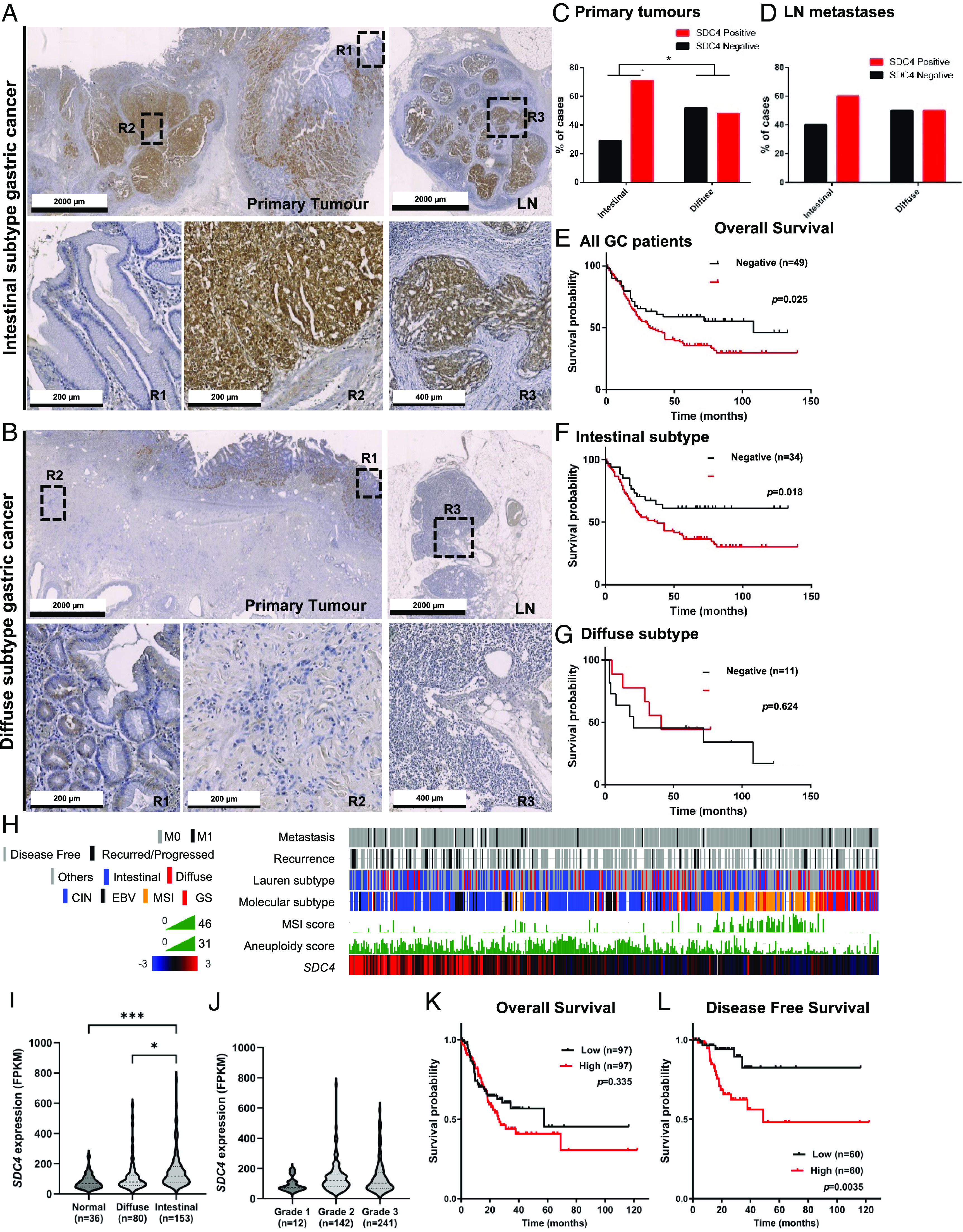
Taking into consideration the high expression of SDC4 in gastric tumors and its clinical association with patient poor outcome, we sought to determine the functional role of SDC4 in cancer cell features. SDC transcription analysis in four gastric cancer cell lines showed that SDC4 is highly expressed, SDC1 and SDC3 were detected in lower levels, whereas SDC2 was not detected in all cells (Fig. 2A). Next, we evaluated the SDC4 protein expression and cellular distribution in gastric cancer cells (Fig. 2 B and C). The WB analysis revealed a high MW smear corresponding to the fully glycosylated forms of SDC4, tetrameric (70 kDa), dimeric (35 kDa), and monomeric (17 kDa) forms. Confocal microscopy showed that SDC4 was predominantly expressed in the plasma membrane of most gastric cancer cells, with AGS cells displaying also punctuated cytoplasmic staining (Fig. 2C).
Fig. 2.
SDC4 KO impairs gastric cancer cell motility features. (A) Schematic representation of SDC family, showing the cytoplasmic domain with two highly conserved regions (C1 and C2) and the SDC-specific variable (V) region, and the ECD that may be decorated with heparan sulfate (HS) and chondroitin sulfate (CS) chains; Bar graph displays the qRT-PCR analysis of SDC1-4 mRNA levels in four gastric cancer cell lines (NCI-N87, AGS, MKN45, MKN74). (B) WB analysis of SDC4 protein expression showing high-MW heterogeneous bands corresponding to the glycosylated forms of SDC4, tetramer, dimer, and monomer; Actin was used as loading control. (C) Confocal imaging of SDC4 (8G3) (green), phalloidin (red), and DAPI (blue) staining in gastric cancer cell lines. Scale bar corresponds to 50 µm (Overlay 1) and 20 µm (Overlay 2). (D) WB analysis of SDC4 and SDC1 in SDC4 KO models and parental cells with (+) and without (−) Heparinase III (Hep. III) digestion. Tubulin was used as loading control. (E) FACS analysis of SDC4 (8G3 clone) cell surface expression on KO models and parental cells. MFI-mean fluorescence intensity. (F) Cell viability analysis by 7-AAD/ Annexin V-FITC cell labeling. Dot plots show the percentages of (Q1) cells in necrosis, (Q2) cells in late apoptosis, (Q3) cells in early apoptosis, and (Q4) viable cells. Representative dot plot of two independent biological replicates. (G) Evaluation of cell proliferation by EdU incorporation assessed by flow cytometry. Dot plots show the percentage of proliferating cells as EdU-Alexa647-positive cells, highlighted in the black box. A representative dot plot of a negative control (without EdU) is shown. Data from one representative assay of two independent biological replicates. (H) Migration capacity of parental and KO cells by wound healing migration assays in different coatings. Graphs represent the percentage of closing rate of the wound. Uncoated and poly-D-Lysine data from two independent biological replicates (each n = 6); Collagen IV data from three independent biological replicates (each n = 6), Fibronectin data from six independent biological replicates (each n = 6). Statistical significance was calculated by two-way ANOVA, with error bars representing SD. (I) Invasion capacity of parental and KO cells was evaluated by Matrigel invasion assays. Bar graphs show the mean of the percentage of invading cells ± SD. A total of at least n = 6 from four independent experiments is shown. Statistical significance was determined using Student’s t test with Welch’s correction. ns-nonsignificant; *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine the functional impact of SDC4, its expression was KO in the MKN74 intestinal subtype gastric cancer cell line using the CRISPR/Cas9 system. Two independent clones (SDC4 KO #1 and SDC4 KO #2) were selected (SI Appendix, Fig. S3). We validated the complete abrogation of SDC4 expression at the protein level by WB (Fig. 2D) and flow cytometry (Fig. 2E). Concerning WB analysis, the hindrance effect of HS chains in antibody binding was discarded by treating the protein extracts with the Heparinase III (Hep.III) enzyme that digests HS chains and confirmed the abrogation of SDC4 (Fig. 2D). As a control of Hep.III-digestion efficiency, we evaluated SDC1 expression using the SDC1 B-A38 clone antibody that only binds to the SDC1 core protein upon HS chains removal.
We then evaluated the impact of SDC4 KO in cellular features. We did not find any statistically significant difference either in cellular viability (Fig. 2F ) or proliferation (Fig. 2G). However, we could show that WT gastric cancer cells presented a higher migration rate comparing to SDC4 KO models in all coatings tested (Fig. 2H). Furthermore, we found that the lack of SDC4 expression remarkably decreased the invasion capacity of gastric cancer cells (Fig. 2I ). The impact of SDC4 in cancer cell invasion was also validated by siRNA strategy in MKN74 and AGS cell models (SI Appendix, Fig. S4). It was confirmed that SDC4 was specifically targeted and silenced in more than 75% (SI Appendix, Fig. S4).
In order to identify molecular cues underlying the deficient motility of SDC4 KO cells, we performed a screening of RTK activation status. This analysis showed that the total phosphorylated forms of ephrin type A receptor 4 (EphA4), fibroblast growth factor receptor 3 (FGFR3), and epidermal growth factor receptor (EGFR) were up-regulated in SDC4 KO cells (SI Appendix, Fig. S5 A and B). However, WB analysis of specific p-Tyr sites in EphA4 (Tyr602) and EGFR (Tyr1068) failed to confirm this increase (SI Appendix, Fig. S5C). In addition, based on the key role of SDC4 in focal adhesion processes (36–38), we evaluated the effect of SDC4 KO on the expression of several focal adhesion components. Protein expression levels were evaluated by WB for pFAK (Tyr397)/FAK levels, and total forms of PKCα, paxillin and β1-integrin, in cells cultured in complete (FBS+) or FBS-depleted medium. No major differences were observed regarding FAK activation, PKCα and β1-integrin expression (SI Appendix, Fig. S5 D and E). However, paxillin showed a trend toward lower expression when SDC4 was depleted, which was only significant in one of the KO clones for FBS+ condition (SI Appendix, Fig. S5E).
Glycosylated SDC4 Is Sorted in EVs and Impacts Their Features.
As the evaluated cellular partners of SDC4 did not reveal any major alteration and taking into account the SDCs’ role in EV-mediated extracellular communication (19, 22–25), we addressed the impact of SDC4 KO on gastric cancer cells’ EV secretion and biological activity. WB analysis showed that SDC4 was detected in EVs isolated from WT cells (Fig. 3A). Ponceau staining (SI Appendix, Fig. S6A) is shown as a control of sample loading. Remarkably, when SDC4 abundance in the WT EVs was compared with the abundance in the WT cells, there was an enrichment of high MW glycosylated forms, as well as of the monomeric form (17 kDa), while the dimeric conformation was not detected in the EVs (Fig. 3A). Gold-immunolabeling methodology validated the presence of SDC4 in WT EVs (Fig. 3B). Additionally, MS-based proteomic analysis identified five SDC4 peptides in WT EVs: three located in the ECD, demonstrating that SDC4 ECD is part of EV cargo, and two in the intracellular domain (ICD), one of these in the variable region and the other in the conserved portion (Fig. 3C and SI Appendix, Table S3). MS analysis further confirmed the enrichment of SDC4 abundance in EVs when compared with cells (SI Appendix, Table S3).
Fig. 3.
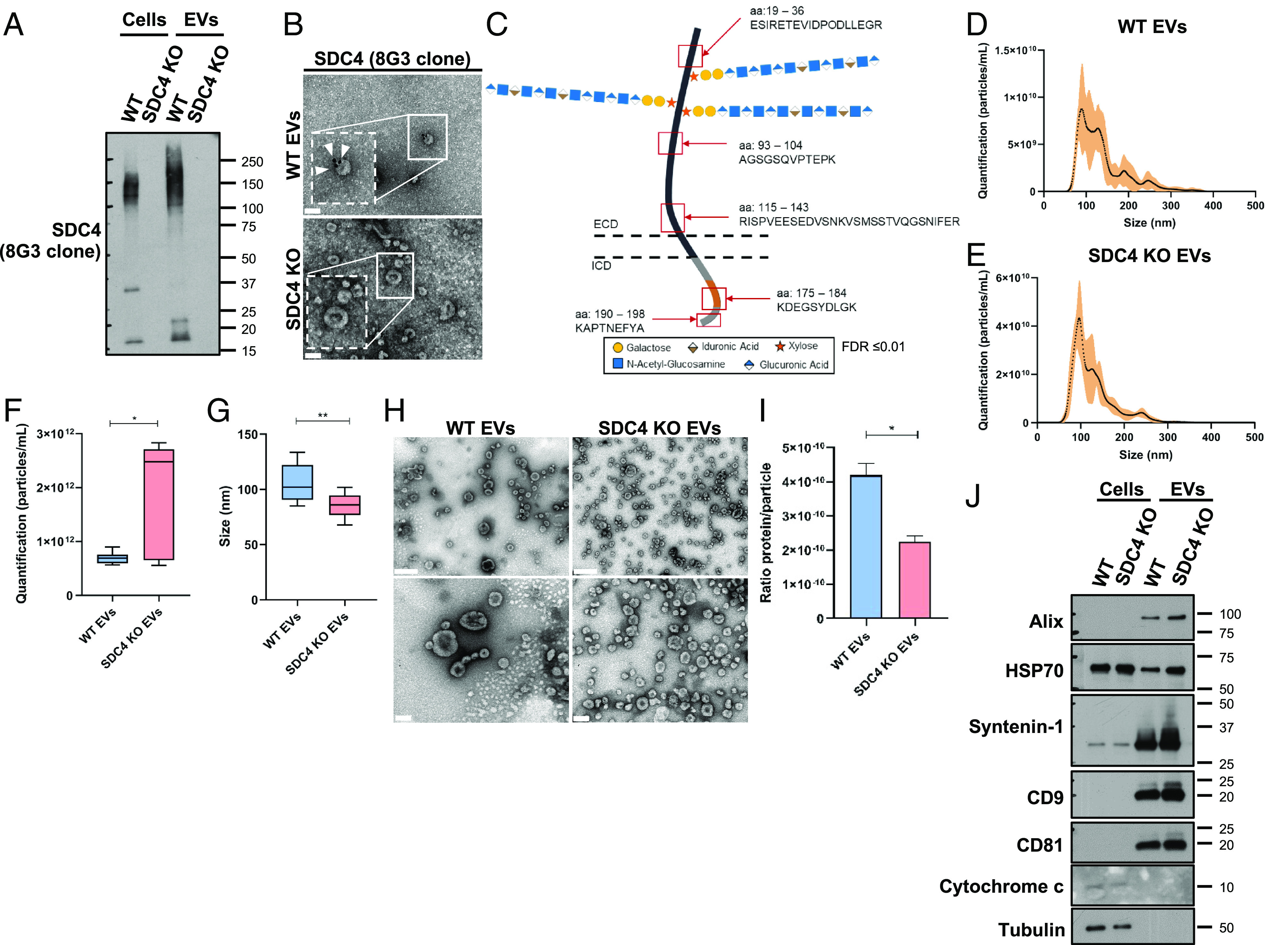
SDC4 is carried by gastric cancer cell EVs and affects EV secretion and size. (A) WB of SDC4 (8G3) in WT and KO cells and EVs. (B) Gold-immunolabeling of SDC4 (8G3) in EVs isolated from WT and KO cells. Scale bar corresponds to 100 nm and white arrowheads point the labeling. (C) Schematic representation of SDC4 showing the sequenced peptides (red squares) in ECD and ICD (variable region in orange and conserved region in grey) by mass spectrometry (MS) analysis. (D and E) Concentration and size distribution analysis using NTA of EVs isolated from (D) WT and (E) KO cells. (F and G) Quantification of the (F) concentration and (G) size of the WT and KO EVs. Bar graphs show the median of the concentration and the median of the size of EVs ± SD. Data from two independent biological replicates. (H) Representative images of negative staining in TEM of the WT and KO EVs. On the Top panel, the scale bar corresponds to 200 nm and below to 100 nm. (I) Bar graphs show the average of the ratio protein per particle ± SD. Data from two independent biological replicates. (J) WB analysis of EV-markers [Programmed cell death 6-interacting protein (Alix), Heat shock 70 (HSP70), syntenin-1, CD9 antigen (CD9), CD81 antigen (CD81)] using WT and KO, cells and EVs. Cytochrome c was used as control for EV fraction isolation and α-tubulin as loading control. Statistical significance was assessed using Student’s t test with Welch’s correction. *P ≤ 0.05, **P < 0.01.
Regarding EV size analysis, both WT and SDC4 KO populations presented size between 50 and 300 nm (Fig. 3 D and E) with the peak fraction between 70 and 150 nm. Analysis of overall EV abundance, measured by NTA, revealed that the quantity of EVs secreted by SDC4 KO cells was higher (median 2.4 × 1012 particles/mL) and displayed smaller size (86 nm), when compared to EVs from WT cells (median concentration: 0.7 × 1012 particles/mL and size: 102.1 nm) (Fig. 3 F and G). These results were confirmed by negative staining analysis by TEM (Fig. 3H). Interestingly, the ratio protein/particle was significantly higher in WT EVs compared to SDC4 KO EVs (Fig. 3I ). Importantly, WB confirmed the expression of a panel of EV-markers, including Alix, HSP70, syntenin-1, CD9, and CD81 in both WT and SDC4 KO EVs (Fig. 3J ). Cytochrome-c and α-tubulin were used as cell lysate controls. Ponceau staining (SI Appendix, Fig. S6B) is shown as a control of samples loading.
SDC4 KO Leads to a Different Proteomic Profile in the EVs’ Cargo.
In view of the differences observed between WT and SDC4 KO EVs, we proceeded to evaluate the protein cargo of the two EV populations (Fig. 4 A and B). A total of 2,431 proteins were identified by MS analysis of gastric cancer cells-derived EVs. The identified proteins were compared with the ones present in the open EV-database Vesiclepedia (Fig. 4A). Of note, 2,289 of the identified proteins were also present in this database, and 142 proteins were identified only in this study (Fig. 4A). Next, the comparison of the identified proteins with the top 100 EV proteins reported in Vesiclepedia showed that most of the proteins detected in our study overlap with the top 100 proteins from Vesiclepedia (Fig. 4B).
Fig. 4.
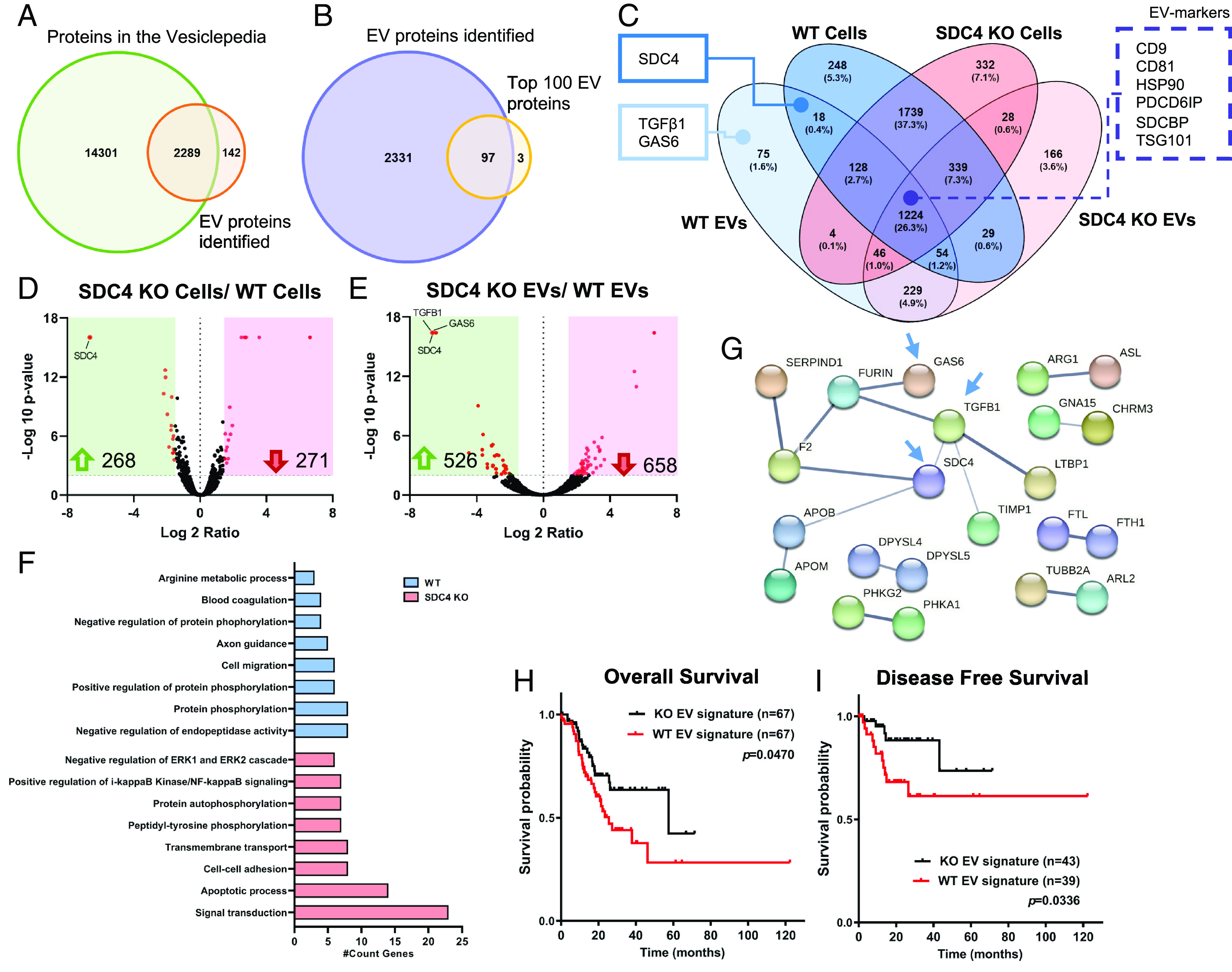
SDC4 impacts EVs proteomic signature. Protein identification was evaluated by LC-MS/MS proteomic analysis. (A) Venn diagram displays the number of unique and overlapping proteins identified in this study for WT and KO EVs, comparing to all proteins deposited in the Vesiclepedia database (1). (B) Venn diagram shows comparative analysis between the EV proteins detected in this study and the Top100 EV proteins described in the Vesiclepedia. (C) Venn diagram illustrates the number (and percentage) of the unique and overlapping proteins from WT and KO cell and EV populations. Two key cancer-related proteins and SDC4 are highlighted in full line box, as well, EV-markers are highlighted in dashed line box. (D and E) Volcano plot analysis displaying differentially expressed proteins (red dots) when comparing the protein cargo of (D) KO vs. WT cells and (E) KO vs. WT EVs. Each protein is represented as a dot and is mapped according to its fold change on the x axis, and the P-value on the y axis. Red dots indicate proteins exhibiting significantly altered expression [−Log10 (p-value) > 2 and Log 2 ratio < −1.5 or >1.5], and black dots indicate proteins whose expression did not vary significantly. The number of proteins up-regulated (green arrow) and down-regulated (red arrow) are highlighted. (F) Functionally grouped network of gene ontology (GO) biological process terms for the exclusive proteome in WT and KO EVs (Benjamini P < 0.01). (G) Protein–protein interaction for the SDC4-specific proteome in WT EVs. Blue arrows point to SDC4, GAS6 and TGFβ1. (H and I) In silico analysis of the prognostic value of WT and KO EV signatures. Analysis of (H) OS and (I) DFS was assessed using the STAD-TCGA database. Reference: 1. Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLOS Biology. 2012;10(12):e1001450.
The combined MS analysis of WT and SDC4 KO EVs and cells identified 4,659 proteins (Fig. 4C). The segregation of the different biological conditions was confirmed by the principal component analysis of the MS data (SI Appendix, Fig. S7A). Gastric cancer cells showed a higher amount of identified proteins (4,189 proteins) when compared with EVs (2,431 proteins). Considering only the cell populations (WT and KO), the Venn diagram revealed that most of the identified proteins were common (1,739 proteins, 37.3%) (Fig. 4C). Moreover, we observed that 26.3% of the proteins overlapped in all populations examined (cells and EVs) (Fig. 4C). Noteworthy, EV-markers (Alix, syntenin-1, CD9 and CD81), that were previously identified by WB (Fig. 3J), were present in this group of overlapping proteins (Fig. 4C and SI Appendix, Table S4). Syntenin-1 abundance was higher in WT cells comparing with KO. Both Syntenin-1 and Alix were highly enriched in the EV populations (SI Appendix, Table S4).
Interestingly, SDC4 KO population (cells and EVs) presented a higher amount of unique identified proteins (4,318 proteins), compared to the WT population (4,133 proteins).
In addition to SDC4, the cancer-associated molecules transforming growth factor beta-1 protein (TGFβ1) and growth arrest-specific protein 6 (GAS6) were exclusively present in the cargo of EVs secreted from WT cells (Fig. 4C). This observation was validated in two independent EV preparations. Furthermore, volcano plots (Fig. 4 D and E), highlighting the significantly different protein content of each population, show that SDC4 KO significantly changed the proteome of both cells and secreted EVs. The EV proteomic data demonstrated that SDC1 abundance is higher in KO EVs. Interestingly, this effect is specific of EV cargo and not observed in the parental cell lines (SI Appendix, Table S4). Network analysis for GO biological process terms for SDC4-exclusive proteins in WT EVs showed an enrichment in several relevant categories, including protein phosphorylation, cell migration, and axon guidance (Fig. 4F). In contrast, the exclusive proteome of KO EVs displayed enriched signal transduction, apoptotic process, and cell–cell adhesion biological processes (Fig. 4F). Analysis of GO molecular function revealed enrichment in categories such as calcium ion binding on WT and ubiquitin protein ligase binding in KO EVs (SI Appendix, Fig. S7B). Protein–protein interaction analysis revealed a high degree of connectivity between of SDC4, TGFβ1, and GAS6 in WT EVs (Fig. 4G). Analysis of KO EVs interconnected several proteins involved in the Wnt pathway, such as Frizzled (FZD) receptors and E3 Ubiquitin-Protein Ligase (ZNRF3) (SI Appendix, Fig. S7C). In addition, we evaluated whether the exclusive proteomic signature of WT and KO EVs would associate with patient clinical prognosis. Remarkably, the WT EV signature significantly associated with patient poor OS and DFS, based on the STAD-TCGA database (Fig. 4 H and I).
SDC4 in EVs Is Critical for EV Uptake and Functional Effects on Recipient Cells.
Since EVs are crucial regulators of intercellular communication, we next sought to investigate the impact of SDC4 on EV uptake and functional signaling on recipient cells. We evaluated WT and SDC4 KO EVs uptake by two macrophage cell lines (RAW and Kupffer) and MKN74 gastric cancer cells. The results showed a significant decrease in the uptake of SDC4 KO EVs by all recipient cells, as illustrated in Fig. 5 (Fig. 5 A–C). Moreover, treatment of the WT EVs with Hep. III resulted in more than 70% uptake inhibition (Fig. 5C). EV uptake was reduced by 17% when performed in the presence of heparin and by 72% when EVs where preincubated with an HS-blocking antibody (Fig. 5C). The efficient Hep. III digestion of HS chains in EVs was confirmed by HS detection in WB (SI Appendix, Fig. S8).
Fig. 5.
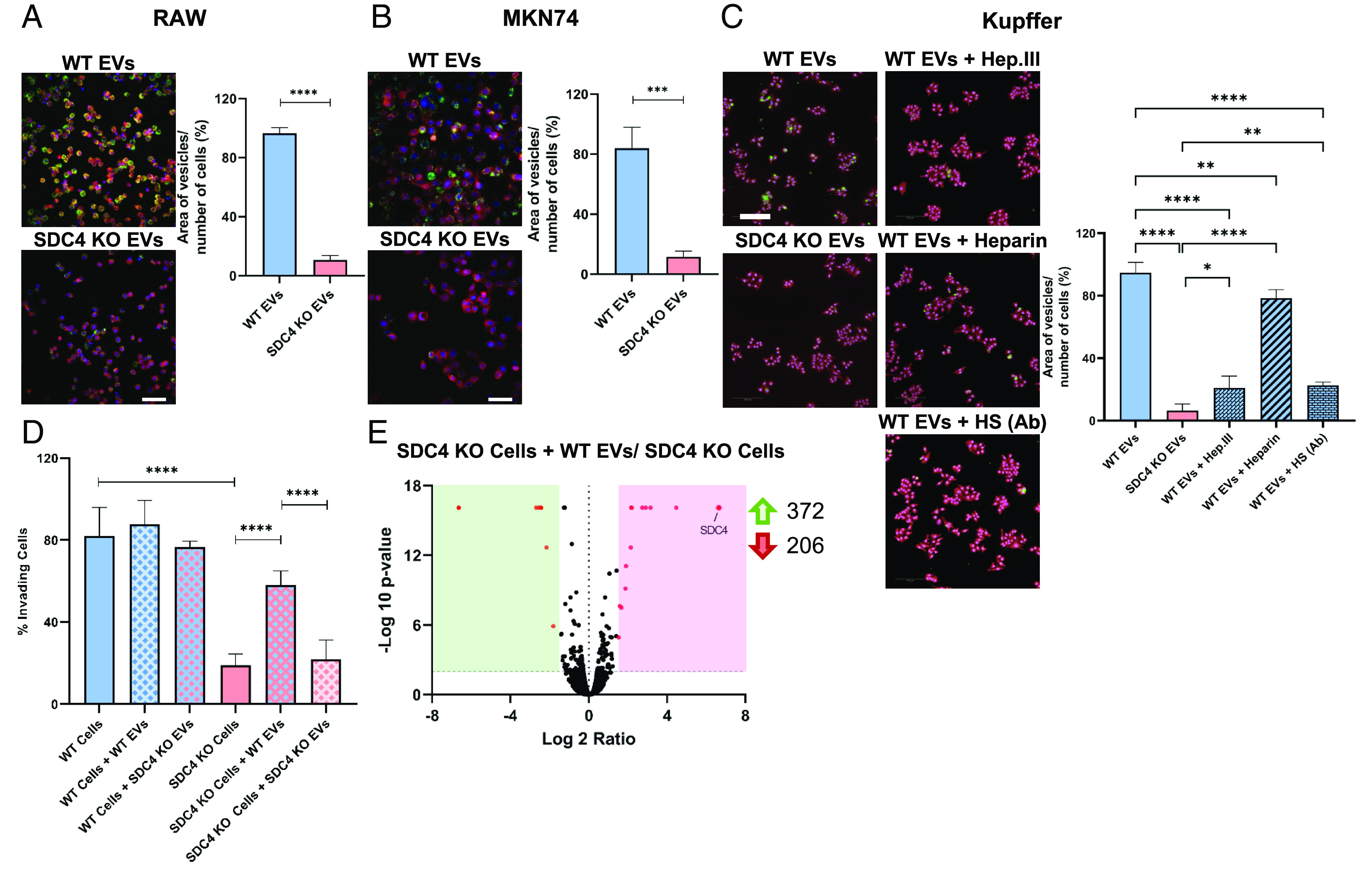
EVs depend on SDC4 for their internalization and functional activity. (A and B) WT and KO EV uptake was evaluated in (A) RAW and (B) MKN74. (C) EV uptake on Kupffer cells was evaluated including WT and KO EVs, WT EVs pretreated with Heparinase III (WT EVs + Hep. III), EVs blocked with heparan sulfate (HS)-antibody (WT EVs + HS Ab), and coincubation with WT EVs and heparin (WT EVs + Heparin). EVs were labeled with PKH67 (green), cells with HCS CellMask (red) and nuclei with Hoechst (blue). Bar graphs show the average of EV area per number of cells (%) ± SD. A representative image of each condition is shown. Data from 3 (RAW), 1 (MKN74) and 2 (Kupffer) independent biological replicates (each n = 2 and 25 field captures). (Scale bar corresponds to 20 µm). (D) Invasion assay of WT and KO cells upon treatment with WT and KO EVs. Bar graphs show the mean of the invading cells percentage ± SD. Data from at least two independent biological replicates (each n = 3). (E) Volcano plot displaying the differentially expressed proteins (red dots) when comparing the protein cargo of KO cells treated with WT EVs vs. KO cells without treatment. Statistical significance was assessed using Student’s t test for two conditions or one-way ANOVA when comparing more than two conditions. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further assess whether EV internalization would modulate cancer cells’ malignant features, we treated the WT and SDC4 KO gastric cancer cells with both WT and SDC4 KO EVs and evaluated cell invasion (Fig. 5D). Remarkably, when SDC4 KO cells were treated with WT EVs, their invasion capacity was restored to values similar to the WT. On the other hand, when KO cells were treated with KO EVs, the invasion capacity was not affected (Fig. 5D). Comparative analysis of the cellular proteome of KO cells treated with WT EVs with the KO cells (without EV treatment) revealed that SDC4 was present in the EV-treated cell population (Fig. 5E).
The Gastric Cancer Cell EV Tropism to the Liver and Lung Is Disrupted by SDC4 KO.
Following the in vitro functional characterization, we tested whether the presence of SDC4 in EVs influences their organ distribution in NOD SCID mice (Fig. 6A). First, we confirmed that WT and KO EVs were equally labeled with the near-infrared fluorescent dye NIR815 (SI Appendix, Fig. S9A). Organ NIR fluorescence analysis revealed that the three organs with higher intensity signal upon EV treatment were the liver, spleen, and lung (Fig. 6 B–E and SI Appendix, Fig. S9 B–E). Among these, the liver was the organ that presented the highest intensity, followed by the spleen (Fig. 6 B–D). The comparative analysis of WT and KO EVs showed a significantly lower uptake of KO EVs in the liver (Fig. 6 B and C), spleen (Fig. 6 B and D), and lung (Fig. 6 B and E), while no differences were observed between both EV populations when analyzing brain, bone, kidney, and pancreas (SI Appendix, Fig. S9 B–E). Flow cytometry analysis further confirmed the lower uptake of KO EVs by the liver cells (Fig. 6F and SI Appendix, Fig. S10A), and a tendency for decreased uptake of KO EVs was observed in spleen and lung (SI Appendix, Fig. S11 A–C). The use of complementary labeling and gating strategies (SI Appendix, Fig. S10B) showed that a decreased uptake of KO EVs was occurring specifically by the liver Kupffer macrophages (CD45+, CD11b+, F4/80+ /Tim4+) (Fig. 6G). No significant differences were noticed on EV uptake by Tim4- macrophages (CD45+, CD11b+, F4/80+) in the bone marrow, spleen, and lung (SI Appendix, Fig. S12).
Fig. 6.
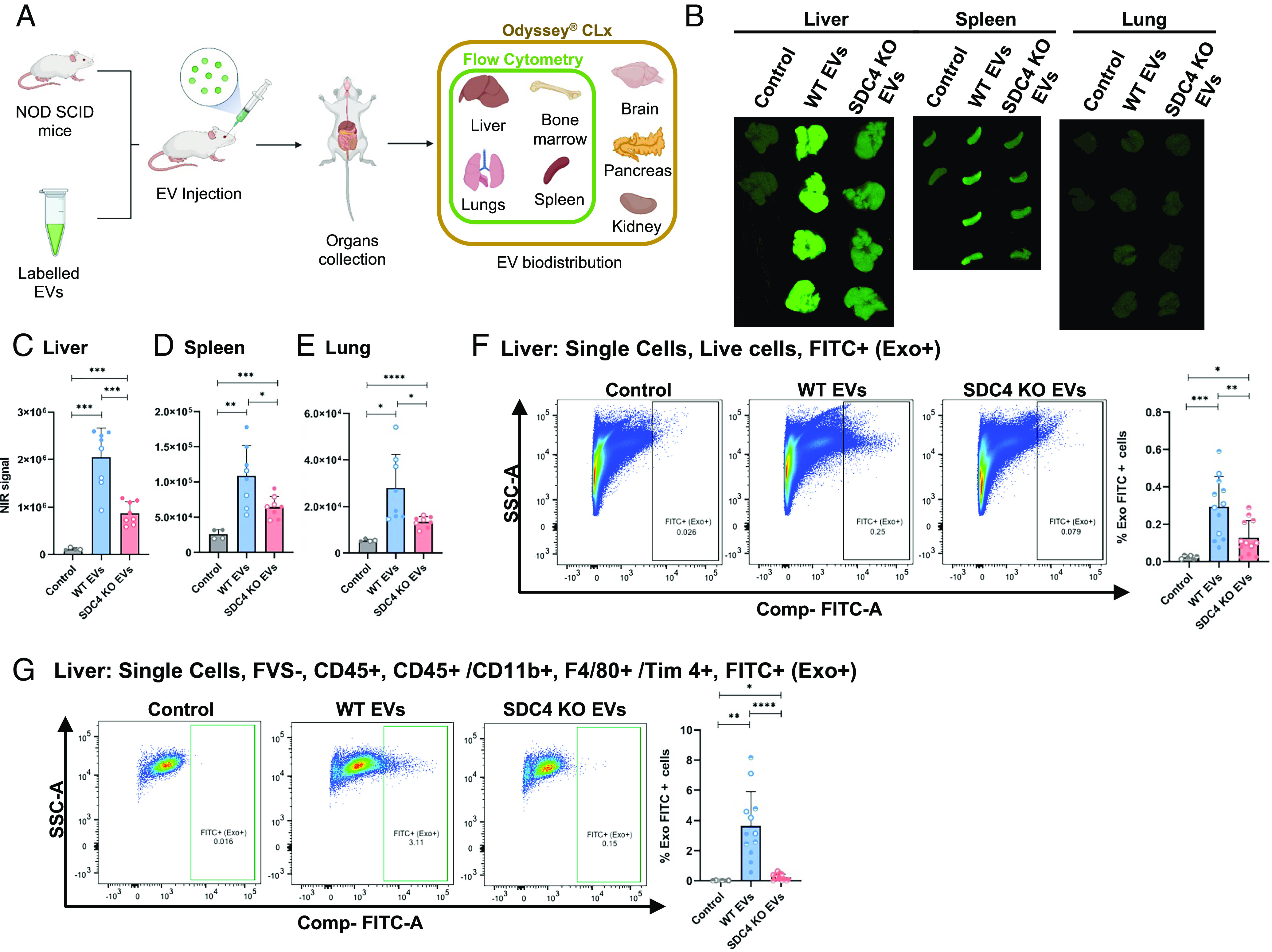
SDC4 impacts EVs organ distribution. (A) Schematic representation of the experimental design. (B) EV organ distribution in NOD SCID mice by measurement of NIR fluorescence. Quantification of NIR signaling in (C) liver, (D) spleen and (E) lung. Bar graphs represent the average of fluorescence quantification ± SD. Data from two independent biological replicates for WT and KO EVs (each n = 4, per condition group), and for control (n = 2). (F and G) FACS analysis of liver cells collected from mice injected with PKH67-labeled WT and KO EVs: (F) all live FITC+ (Exo+) cells and (G) all CD45+/CD11b+, F4/80+ Tim4+, FITC+ (Exo+) cells. Bar graphs represent the average of the percentage of cells that incorporated EVs. Data from three independent biological replicates for WT and KO EVs (each n = 4, per condition group), and for control (each n = 2). Statistical significance was assessed using Student’s t test. *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
In the present era of precision oncology, glycans and glycoconjugates are emerging as major players in cancer initiation and progression with strong potential both as biomarkers and as therapeutic targets (4, 5, 7, 12). In this study, we reveal that the HS proteoglycan SDC4 is highly expressed in intestinal subtype gastric tumors and that this glycosylation profile correlates with cancer aggressiveness features and poorer prognosis. We addressed SDC4 functional implications in gastric cancer cell biology and demonstrated that SDC4 promotes a promigratory phenotype with higher invasion capacity, which was abrogated by SDC4 KO. Interestingly, we further demonstrate that glycosylated SDC4 is sorted in EVs secreted by gastric cancer cells and showed that SDC4 KO impacted EV secretion and proteomic profile. Moreover, our results disclose that SDC4 KO impacts EV uptake by recipient cells and disrupts the tropism of gastric cancer cell-derived EVs.
The aberrant expression of HS proteoglycans has been described in different tumor models (12). In particular, SDC4 was previously shown to be up-regulated in glioma, melanoma, osteosarcoma, papillary thyroid carcinoma, breast, liver, kidney, and bladder carcinomas (27–34). In our series of gastric cancer patients, we demonstrate that SDC4 protein expression is associated with intestinal subtype and patient poor OS. Of note, we observed a significant worse OS in male patients with SDC4 expression, which was not observed in women. This observation aligns with previous reports on hormonal regulation of HSPGs during cancer progression (39). Among patients presenting metastatic disease, SDC4 expression was not associated with OS, yet it should be noticed that the number of individuals in this category, in the IPO-Porto cohort, is relatively small. Moreover, in STAD-TCGA dataset, SDC4 high-transcript levels clustered with metastasis and disease recurrence. When considering the stratification of patients by treatment regiment, SDC4 expression only associated with poor OS in the subgroup that exclusively received surgical treatment. Multivariate analysis demonstrated the independent prognostic value of SDC4 expression in intestinal subtype gastric cancer.
In breast and liver carcinoma cells, SDC4 has been associated with higher migration and invasion (40, 41). In line with these observations, SDC4 KO has been shown to result in decreased invasion capacity of glioblastoma cells (42). Moreover, shedding of the N-terminal portion of the ectodomain of SDC4 by members of the ADAMTS protease family was shown to enhance the migration of cancer cells (43). In this work, we show that SDC4 has a role in intestinal subtype gastric cancer cell migration and invasion, independently of the presence of different matrix components. Noteworthy, it was possible to restore the invasive capacity of SDC4 KO cells by coculturing them with EVs secreted by WT cells with proinvasive phenotype. MS analysis of KO cells with restored invasion capacity demonstrated de novo incorporation of SDC4, further supporting the important role of EVs cargo in the SDC4-associated malignant phenotype.
Several seminal studies have comprehensively addressed the EV cargo, including nucleic acids, proteins, lipids, and metabolites (44–46). However, the glycans and glycoconjugates carried by EVs remain poorly characterized. Of note, HS-rich proteoglycans, namely SDCs, together with syntenin and Alix have been shown to play important roles in EV biogenesis (19, 24, 47). Furthermore, heparanase, the enzyme responsible for HS remodeling, can stimulate intraluminal budding of SDC–syntenin–Alix complex promoting EV secretion (23, 48, 49). Our data revealed an enrichment of highly glycosylated SDC4 forms in EVs when compared to the parental cells. Specifically, MS analysis identified three peptides, corresponding to the ECD of SDC4, that were exclusively sequenced in EVs. Of notice, SDC4 KO impacted EV biogenesis with KO cells producing higher amounts of vesicles with smaller size and decreased protein/particle ratio. SDC4 KO EVs presented higher SDC1 levels, supporting a compensatory effect for the lack of SDC4. The HSPG SDC1 has been previously reported to control the biogenesis of intraluminal vesicles and exosomes (EVs) (19).
Interestingly, proteomic analysis revealed that TGFβ1 and GAS6 were exclusively identified on WT EVs. In addition, network analysis of SDC4-exclusive proteins in WT EVs, clustered together SDC4, GAS6, and TGFβ1, showing an enrichment in biological processes involved in protein phosphorylation, cell migration, and axon guidance. This proteomic signature is associated with the proinvasive phenotype induced by these EVs. TGFβ1 is a key regulator of migration, invasion, and epithelial–mesenchymal transition in gastric carcinogenesis (50, 51) and GAS6 was described as involved in the recruitment and polarization of macrophages (52, 53), therefore supporting EV contribution for cancer cell invasion and tumor progression. Protein network analysis of the KO EV proteome revealed several proteins of the Wnt pathway, including ZNRF3 that acts as a negative feedback regulator of Wnt signaling (54), therefore, corroborating the less aggressive phenotype of KO EVs. In line with these observations, the SDC4-specific signature of WT EVs was significantly associated with gastric cancer patient poor overall and DFS.
The functional effects of EVs are dependent on an efficient uptake by recipient cells, and the different pattern of surface components on EVs, e.g., integrins, cytokines, and glycoconjugates, has been shown to dictate EV uptake (21, 55, 56). Furthermore, from the recipient cell side HSPGs have been described as regulators of EV internalization and functional activity (57). The glycosaminoglycan chains that decorate HSPGs, particularly the 2-O- and N-sulfation domains, were demonstrated to be key for exosome uptake (25). Additionally, it was shown that enzymatic treatment with heparanase, that trims HS chains, and xylosides, which hinder HS biosynthesis, also resulted in EV uptake inhibition (25). Here, we provide evidence that SDC4 plays a crucial role on EV uptake, as KO of this specific proteoglycan impacts the EV cellular internalization and organ distribution. Our data further support that HS glycosaminoglycans in EVs are mechanistically involved in the EV internalization process, since enzymatic digestion of EV HS-chains or EV blocking with HS-antibody significantly impaired EV uptake. However, we cannot exclude the possibility of SDC4-independent alternative uptake pathways since SDC4 KO did not completely abolished EV uptake. Interestingly, we observed a significant decrease in SDC4 KO EV uptake by the liver, lung and spleen, which are common metastatic sites of gastric primary tumor (58). These results highlight the specific tropism of the gastric cancer EVs to these organs. Furthermore, our data provide evidence that Kupffer cells are major players in gastric cancer-derived EV uptake in the liver. The uptake of gastric cancer-derived EVs by macrophages, with consequences on macrophage differentiation and tumor dissemination, has been previously reported (59). Moreover, Kupffer cells have been demonstrated as a key cell subpopulation in the uptake of pancreatic cancer exosomes, having a functional contribution for the establishment of a liver premetastatic niche (60, 61).
The understanding of the functional implications of proteoglycans glycosylation in EV cellular recognition and uptake holds great potential for the therapeutic inhibition of EV-mediated tumor progression (62). This knowledge is particularly relevant in gastric cancer where the identification of molecular targets to design personalized therapeutic strategies is of utmost need. Moreover, proteoglycans carried by EVs have been already recognized as biomarkers for minimally invasive cancer diagnosis: Glypican-1 in pancreatic cancer (63) and SDC1 in glioma (62). Therefore, the identification of SDC4 in gastric cancer cell-derived EVs holds biomarker potential.
The role of SDC4 in the definition of EV tropism to common metastatic sites of gastric cancer is of utmost relevance within the emerging concept that EVs prime a premetastatic niche. This knowledge prompts future research on the impact of glycosylated-SDC4 in EVs for gastric cancer dissemination. However, the current challenge is the lack of good experimental models that mimic the human gastric cancer metastasis features. Therefore, the foreseeable study of EV education using gastric cancer orthotopic patient-derived xenograft models has particular pertinence. Moreover, it would be important to decipher the TGFβ1, GAS6, and SDC4 molecular interplay and the role of these partners in EV-mediated immune regulation, namely on macrophage polarization, within the tumor microenvironment.
Collectively, our findings reveal a previously unappreciated role of SDC4 in determining gastric cancer cell aggressiveness with implications in patient clinical outcome and further uncover the potential of this HSPG as a therapeutic target. Furthermore, our data contribute to decipher the glycan code of cancer-derived EVs and for the identification of key molecular cues that determine gastric cancer communication routes.
Materials and Methods
Detailed description of reagents, immunohistochemistry, in silico analyses, cell culture, CRISPR/Cas9 KO methodology, qPCR, western blot, flow cytometry, immunofluorescence, enzymatic digestion, viability and proliferation assays, RTK phosphorylation analysis, EV isolation, NTA, EV protein quantification, TEM, gold-immunolabeling microscopy, LC-MS/MS analysis, EV uptake, and biodistribution experiments are provided in SI Appendix.
Ethic Statements.
All procedures concerning the inclusion of patients were approved by the institutional Ethics Committee (Comissão de ética para a saúde—CES-IPOFG-EPE 87/2017) after patients’ informed consent. Additional details regarding this cohort are provided in SI Appendix, Materials and Methods.
All animal procedures were conducted in accordance with National Animal Experimentation Guidelines (DGAV) on the use and care of experimental animals, and approved by the Champalimaud Foundation Ethics Committee (0421/000/000/2018).
In Silico Analysis of SDC4 Expression and Prognostic Impact.
SDC4 transcript levels were investigated in STAD-TCGA and the KM plotter database (64), as described in SI Appendix, Materials and Methods.
SDC4 KO by CRISPR/Cas9.
The SDC4 KO was performed in MKN74 cell line using CRISPR/Cas9 genome editing and validated, as previously described (65, 66). Additional information is provided in SI Appendix, Materials and Methods.
Isolation of EVs.
The EV isolation and characterization were performed according to the International Society for Extracellular Vesicles (ISEV) (67) guidelines and are described in SI Appendix, Materials and Methods.
Statistical Analysis.
Prism software (GraphPad, v8.0) and IBM® SPSS® Statistics v.22 were used for statistical analysis, as detailed in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank all patients and clinicians for their participation in this study. we would like to acknowledge Guido David for providing the 8G3 antibody, Henrik Clausen for providing GFP-Cas9 and David Lyden for the RAW and Kupffer cells. We would like to thank Henrique O. Duarte for manuscript proof reading. We acknowledge the support of the i3S Scientific Platforms: Proteomics, Bioinformatics, Translation Cytometry, Advanced Light Microscopy, BioSciences Screening, Bioimaging, and Histology and Electron Microscopy, members of the PPBI (PPBI-POCI-01-0145-FEDER-022122). This work was funded by FEDER funds through the Operational Programme for Competitiveness Factors-COMPETE (POCI-01-0145-FEDER-028489; POCI-01-0145-FEDER-029780) and National funds through the Foundation for Science and Technology (FCT), under the projects PTDC/MED-ONC/28489/2017 (to A.M.); PTDC/MED-QUI/29780/2017 and PTDC/MED-QUI/2335/2021 (C.G.), EXPL/BTM-ORG/1450/2021 (F.P.) and PTDC/MEDOUT/2512/2021 (J.A.F.); and the project Norte-01-0145-FEDER-000051-“Cancer Research on Therapy Resistance: From Basic Mechanisms to Novel Targets,” and “P.CCC.: Porto Comprehensive Cancer Center Raquel Seruca“ - NORTE-01-0145-FEDER-072678 both supported by NORTE 2020, under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund; and COST Action CA18103 INNOGLY: INNOvation with Glycans: new frontiers from synthesis to new biological targets. This work was also financed by the Portuguese Mass Spectrometry Network, integrated in the National Roadmap of Research Infrastructures of Strategic Relevance (ROTEIRO/0028/2013; LISBOA-01-0145-FEDER-022125). J.P. and C.M. acknowledge FCT PhD scholarships SFRH/BD/137319/2018 and 2020.06412.BD, respectively. F.P. acknowledges the SFRH/BPD/115730/2016 FCT fellowship. A.H.O. acknowledges the FAPESP 19/00740-0 fellowship. B.C. is supported by a JUNIOR_RESEARCHER/CEEC_INST21/IBMC/2802/2022. J.A.F. acknowledges FCT position CEECIND/03186/2017.
Author contributions
J.P., B.C.-S., and A.M. designed research; J.P., C.M., C.G., A.H.O., F.P., M.F., T.S., I.F.-R., R.M., A.R.R., E.S., S.B., J.M., J.A.M., L.P.A., A.P., and H.O. performed research; F.P. and B.C. performed bioinformatic analysis; C.G., C.A.R., B.C.-S., and A.M. contributed new reagents/analytic tools; J.P., A.R.R., E.S., B.C., L.L., J.A.F., L.L.S., A.P., H.O., M.B., C.A.R., B.C.-S., and A.M. analyzed data; and J.P. and A.M. wrote the paper.
Competing interests
J.P., C.M., C.G., C.A.R., and A.M. have submitted a provisional patent application (reference: 20221000003006) related to the exploitation of SDC4 as a biomarker of poor survival and organ tropism. The remaining authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All relevant data from EV experiments were submitted to the EV-TRACK knowledgebase (EV-TRACK ID: EV210302) (68). The MS-based proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (69) partner repository with the dataset identifier PXD035679 and 10.6019/PXD035679 (70). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Sung H., et al. , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Duarte H. O., Gomes J., Machado J. C., Reis C. A., Gastric cancer: Basic aspects. Helicobacter 23, e12523 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H., Gastric cancer. Lancet 388, 2654–2664 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C. A., Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell 36, 6–16 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Pinho S. S., Reis C. A., Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Magalhães A., Duarte H. O., Reis C. A., The role of O-glycosylation in human disease. Mol. Aspects Med. 79, 100964 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Ferreira J. A., et al. , Protein glycosylation in gastric and colorectal cancers: Toward cancer detection and targeted therapeutics. Cancer Lett. 387, 32–45 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Gomes C., et al. , Expression of ST3GAL4 leads to SLe(x) expression and induces c-Met activation and an invasive phenotype in gastric carcinoma cells. PLoS One 8, e66737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitas D., et al. , O-glycans truncation modulates gastric cancer cell signaling and transcription leading to a more aggressive phenotype. EBioMedicine 40, 349–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couchman J. R., Multhaupt H., Sanderson R. D., Recent insights into cell surface heparan sulphate proteoglycans and cancer. F1000 Res. 5, 1541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faria-Ramos I., et al. , Heparan sulfate glycosaminoglycans: (Un)expected allies in cancer clinical management. Biomolecules 11, 136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bratulic S., et al. , Noninvasive detection of any-stage cancer using free glycosaminoglycans. Proc. Natl. Acad. Sci. U.S.A. 119, e2115328119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal S., et al. , Heparan sulfate chain valency controls syndecan-4 function in cell adhesion. J. Biol. Chem. 285, 14247–14258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass M. D., et al. , Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 177, 527–538 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echtermeyer F., et al. , Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J. Clin. Invest. 107, R9–R14 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longley R. L., et al. , Control of morphology, cytoskeleton and migration by syndecan-4. J. Cell Sci. 112, 3421–3431 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Magalhaes A., et al. , Helicobacter pylori cag pathogenicity island-positive strains induce syndecan-4 expression in gastric epithelial cells. FEMS Immunol. Med. Microbiol. 56, 223–232 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Marcos N. T., et al. , Helicobacter pylori induces β3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl–Lewis x. The J. Clin. Invest. 118, 2325–2336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baietti M. F., et al. , Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Mathieu M., Martin-Jaular L., Lavieu G., Théry C., Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Peinado H., et al. , Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Friand V., David G., Zimmermann P., Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 107, 331–341 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Roucourt B., Meeussen S., Bao J., Zimmermann P., David G., Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 25, 412–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghossoub R., et al. , Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. U.S.A. 117, 5913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christianson H. C., Svensson K. J., van Kuppevelt T. H., Li J.-P., Belting M., Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U.S.A. 110, 17380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onyeisi J. O. S., Lopes C. C., Götte M., Syndecan-4 as a pathogenesis factor and therapeutic target in cancer. Biomolecules 11, 503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe A., et al. , Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: Participation of nuclear factor-κB in upregulation of syndecan-1 expression. J. Neuro. Oncol. 77, 25–32 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Baba F., et al. , Syndecan-1 and syndecan-4 are overexpressed in an estrogen receptor-negative, highly proliferative breast carcinoma subtype. Breast Cancer Res. Treat. 98, 91–98 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Roskams T., De Vos R., David G., Van Damme B., Desmet V., Heparan sulphate proteoglycan expression in human primary liver tumours. J. Pathol. 185, 290–297 (1998). [DOI] [PubMed] [Google Scholar]
- 30.O’Connell M. P., et al. , Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J. Biol. Chem. 284, 28704–28712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Na K. Y., Bacchini P., Bertoni F., Kim Y. W., Park Y. K., Syndecan-4 and fibronectin in osteosarcoma. Pathology 44, 325–330 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Erdem M., et al. , Up-regulation of TGM2 with ITGB1 and SDC4 is important in the development and metastasis of renal cell carcinoma. Urol. Oncol. 32, e13-20 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Chen L. L., et al. , SDC4 gene silencing favors human papillary thyroid carcinoma cell apoptosis and inhibits epithelial mesenchymal transition via Wnt/β-catenin pathway. Mol. Cells 41, 853–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marzioni D., et al. , Expression of basic fibroblast growth factor, its receptors and syndecans in bladder cancer. Int. J. Immunopathol. Pharmacol. 22, 627–638 (2009). [DOI] [PubMed] [Google Scholar]
- 35.David G., van der Schueren B., Marynen P., Cassiman J. J., van den Berghe H., Molecular cloning of amphiglycan, a novel integral membrane heparan sulfate proteoglycan expressed by epithelial and fibroblastic cells. J. Cell Biol. 118, 961–969 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gopal S., Multhaupt H. A. B., Pocock R., Couchman J. R., Cell-extracellular matrix and cell-cell adhesion are linked by syndecan-4. Matrix Biol. 60–61, 57–69 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Lim H. C., Multhaupt H. A., Couchman J. R., Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Mol. Cancer 14, 15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denhez F., et al. , Syndesmos, a syndecan-4 cytoplasmic domain interactor, binds to the focal adhesion adaptor proteins paxillin and Hic-5. J. Biol. Chem. 277, 12270–12274 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Tsonis A. I., et al. , Evaluation of the coordinated actions of estrogen receptors with epidermal growth factor receptor and insulin-like growth factor receptor in the expression of cell surface heparan sulfate proteoglycans and cell motility in breast cancer cells. FEBS J. 280, 2248–2259 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Habes C., Weber G., Goupille C., Sulfated glycoaminoglycans and proteoglycan syndecan-4 are involved in membrane fixation of LL-37 and its pro-migratory effect in breast cancer cells. Biomolecules 9, 481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charni F., et al. , Syndecan-1 and syndecan-4 are involved in RANTES/CCL5-induced migration and invasion of human hepatoma cells. Biochim. Biophys. Acta 1790, 1314–1326 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Ochieng J., et al. , Extracellular histones are the ligands for the uptake of exosomes and hydroxyapatite-nanoparticles by tumor cells via syndecan-4. FEBS Lett. 592, 3274–3285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez-Manzaneque J. C., et al. , Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. Int. J. Biochem. Cell Biol. 41, 800–810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolte-’t Hoen E. N. M., et al. , Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272–9285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Jaular L., et al. , Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection. EMbo J. 40, e105492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haraszti R. A., et al. , High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 5, 32570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kashyap R., et al. , Syntenin-knock out reduces exosome turnover and viral transduction. Sci. Rep. 11, 4083 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanderson R. D., Elkin M., Rapraeger A. C., Ilan N., Vlodavsky I., Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 284, 42–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David G., Zimmermann P., Heparanase involvement in exosome formation. Adv. Exp. Med. Biol. 1221, 285–307 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Li T., et al. , TGF-β1-SOX9 axis-inducible COL10A1 promotes invasion and metastasis in gastric cancer via epithelial-to-mesenchymal transition. Cell Death Dis. 9, 849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Peng K. W., Wang B., Yang X. F., Zhang Z. M., EDIL3 regulates gastric cancer cell migration, invasion and epithelial-mesenchymal transition via TGF-β1/XIST/miR-137 feedback loop. Transl. Cancer Res. 9, 6313–6330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F., et al. , Interaction with CD68 and regulation of GAS6 expression by endosialin in fibroblasts drives recruitment and polarization of macrophages in hepatocellular carcinoma. Cancer Res. 80, 3892–3905 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Zdżalik-Bielecka D., et al. , The GAS6-AXL signaling pathway triggers actin remodeling that drives membrane ruffling, macropinocytosis, and cancer-cell invasion. Proc. Natl. Acad. Sci. U.S.A. 118, e2024596118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Z. A., Michalski M. N., Stevens P. D., Sall E. A., Williams B. O., Regulation of Wnt receptor activity: Implications for therapeutic development in colon cancer. J. Biol. Chem. 296, 100782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lima L. G., et al. , Tumor microenvironmental cytokines bound to cancer exosomes determine uptake by cytokine receptor-expressing cells and biodistribution. Nat. Commun. 12, 3543 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams C., et al. , Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 9, 11920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerezo-Magaña M., Bång-Rudenstam A., Belting M., The pleiotropic role of proteoglycans in extracellular vesicle mediated communication in the tumor microenvironment. Semin. Cancer Biol. 62, 99–107 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Li W., Ng J. M., Wong C. C., Ng E. K. W., Yu J., Molecular alterations of cancer cell and tumour microenvironment in metastatic gastric cancer. Oncogene 37, 4903–4920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito A., et al. , Extracellular vesicles shed from gastric cancer mediate protumor macrophage differentiation. BMC Cancer 21, 102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoshino A., et al. , Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa-Silva B., et al. , Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Indira Chandran V., et al. , Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin. Cancer Res. 25, 3115–3127 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Melo S. A., et al. , Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szász A. M., et al. , Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonowski L. A., et al. , Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protocols 12, 581–603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y.-H., et al. , The GAGOme: A cell-based library of displayed glycosaminoglycans. Nat. Methods 15, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Théry C., et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Deun J., et al. , EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 14, 228–232 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poças J., Osório H., Magalhães A., Identification and quantification of proteins from MKN74 WT and SDC4 KO cells and EVs. The PRoteomics IDEntifications (PRIDE) database. www.ebi.ac.uk/pride/archive/projects/PXD035679. Deposited 1 August 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All relevant data from EV experiments were submitted to the EV-TRACK knowledgebase (EV-TRACK ID: EV210302) (68). The MS-based proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (69) partner repository with the dataset identifier PXD035679 and 10.6019/PXD035679 (70). All study data are included in the article and/or SI Appendix.