Abstract
We report an analysis of a sample of the SOS response of Salmonella enterica serovar Typhimurium using the differential display of RNA fingerprinting gels of arbitrarily primed PCR products. The SOS response was induced by the addition of mitomycin C to an exponentially growing culture of serovar Typhimurium, and the RNA population was sampled during the following 2 h. These experiments revealed 21 differentially expressed PCR fragments representing mRNA transcripts. These 21 fragments correspond to 20 distinct genes. All of these transcripts were positively regulated, with the observed induction starting 10 to 120 min after addition of mitomycin C. Fifteen of the 21 transcripts have no homologue in the public sequence data banks and are therefore classified as novel. The remaining six transcripts corresponded to the recE, stpA, sulA, and umuC genes, and to a gene encoding a hypothetical protein in the Escherichia coli lysU-cadA intergenic region; the recE gene was represented twice by nonoverlapping fragments. In order to determine if the induction of these 20 transcripts constitutes part of a classical SOS regulon, we assessed the induction of these genes in a recA mutant. With one exception, the increased expression of these genes in response to mitomycin C was dependent on the presence of a functional recA allele. The exception was fivefold induced in the absence of a functional RecA protein, suggesting another layer of regulation in response to mitomycin C, in addition to the RecA-LexA pathway of SOS induction. Our data reveal several genes belonging to operons known to be directly involved in pathogenesis. In addition, we have found several phage-like sequences, some of which may be landmarks of pathogenicity determinants. On the basis of these observations, we propose that the general use of DNA-damaging agents coupled with differential gene expression analysis may be a useful and easy method for identifying pathogenicity determinants in diverse organisms.
Maintaining the integrity of genetic information is essential to all organisms. Consequently, all species have evolved diverse ways of maintaining the fidelity of their genetic material, particularly in response to a harsh environment (10, 30). Perhaps the most comprehensively studied system for the repair of DNA damage is the SOS regulon of Escherichia coli.
The SOS regulon of E. coli is composed of at least 20 genes scattered throughout the chromosome (10, 12). These genes all share the property that their transcription is induced or elevated by DNA damage. The mechanism whereby E. coli's SOS regulon is induced has been intensively studied with a variety of DNA-damaging agents over the past 30 years. These studies have provided a detailed picture of the molecular mechanism of SOS activation.
A variety of proteins that recognize damaged bases and effect their repair exist in the cell. Nucleotide excision repair is a common mechanism for DNA repair shared by many species (22). In E. coli and Salmonella, this method of repair does not require SOS induction (although its component enzymes are under SOS regulation) and acts on double-stranded DNA.
When the nature or level of DNA damage is such that excision repair is overwhelmed, DNA replication enzymes will encounter the damaged bases. In this event the replication enzymes can bypass the damage and reinitiate replication downstream, leaving a single-stranded gap of approximately 1,500 nucleotides called a postreplication daughter strand gap (11, 22, 23). This gap harbors the damaged bases, which must be repaired using the complementary strand on the sister chromosome (31). The formation of a postreplication daughter strand gap can lead to a double-strand break (14). This double-strand break can be repaired using the sister chromosome as a template (27).
Both the gap and the double-strand break are substrates for recombination enzymes (10). These enzymes create single-stranded DNA (ssDNA), a necessary intermediate, leading to the recombinational repair of the damaged DNA. This ssDNA is also one of the key effectors of SOS induction (2).
Once ssDNA is generated, the RecA protein polymerizes on the single strands, forming a nucleoprotein filament (13). This RecA-ssDNA filament acts as a catalyst for the cleavage of the LexA repressor protein. The LexA protein negatively regulates the transcription of the SOS regulon by binding to sites found in the 5′ ends of these genes. Thus, the generation of ssDNA leads to the RecA-mediated cleavage of the LexA repressor and the consequential derepression of the SOS operon (2, 21).
The derepression of the SOS operon results in the induction of DNA repair and recombination enzymes, which effect the recombination and repair of the damaged DNA. Once recombination and repair have ceased, the concentration of ssDNA returns to normal levels, cleavage of LexA is terminated (the lexA gene is itself regulated by LexA), and homeostatic levels of LexA are reached. Thus, the SOS regulon is repressed and the cell returns to its “wild-type” physiological state.
While the SOS response in E. coli is well documented, relatively little is known about the regulon in Salmonella. It is certain that Salmonella enterica serovar Typhimurium exhibits an SOS response, since one can isolate mitomycin C-inducible LacZ fusions (J. Roth, unpublished data) and Salmonella serovar Typhimurium has homologues of E. coli SOS genes including the lexA, recA, sulA, recN, uvrABD, and umuDC genes (10, 25).
In this work, we investigate the SOS response to DNA damage using a classical inducer of SOS, mitomycin C. We present an analysis of differential gene expression using the technique of RNA fingerprinting by arbitrarily primed PCR (RAP-PCR). The results suggest that, in the 120 million years since E. coli and Salmonella serovar Typhimurium diverged from a common ancestor, their SOS regulons—while retaining similar regulatory and enzymatic features—have otherwise significantly diverged. In addition, we reveal that Salmonella serovar Typhimurium possesses an mitomycin C-induced, RecA-independent pathway of gene activation.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
Our wild-type strain of Salmonella serovar Typhimurium is DB7000 [originally from David Botstein; leuA414(Am) Fels2−] and was provided by Mimi Susskind. Our recA mutant strain, TT18642 [leuA414(Am) Fels2− recA1] is described in reference 3. Standard growth medium is Luria-Bertani (LB) broth supplemented with 1× E salts and 0.2% glucose (7). The pCR2.1 cloning vector, transformation strains, methods, and supplies for cloning differentially expressed genes were provided in the Original TA Cloning Kit obtained from Invitrogen Corp. and used according to the manufacturer's protocols. DNase I (RNase free) and RNase inhibitor (human placenta) were purchased from Boehringer Mannheim. Moloney murine leukemia virus (MMLV) reverse transcriptase was purchased from Promega. The Stoffel fragment of Taq DNA polymerase was purchased from Perkin-Elmer.
Synthetic oligonucleotides.
All oligonucleotides were purchased, desalted, from Genosys and resuspended in 10 mM Tris, pH 8. The sequences of the oligonucleotides used to identify regulated genes in RAP-PCR experiments are listed in Table 1. Additional oligonucleotides were used to confirm differential expression and were based on the sequence of the particular fragment. Due to the length of the latter list, we have chosen not to present it here but will gladly provide it to interested individuals.
TABLE 1.
Oligonucleotide primers used for the discovery of mitomycin C-induced genes
| Sequence | Oligomer designation |
|---|---|
| GCCATCCGGC | REP |
| TTCAGCCAGCG | EA10 |
| GCACCAGGGG | OPN28 |
| CAGGGGCACC | OPN23 |
| CCAGGGGCAC | OPN22 |
| GGCTGCAGAA | N1 |
| GAGGATCCCT | N2 |
| TGCTCATCCT | U7 |
| TAAGGAGGTAG | Shine |
| AGTGGCACGGTTCCTGGAATAC | A94 |
| TTTCTGCGGTGCTTACTGTCCC | J43B |
| GTAATGGCGGCGTTGTTGAC | J43F |
| TAACGACGGCGAGATTCTGG | 85B |
| GCTTGTAGTTCCAGCTCATC | 85F |
| TGACCTGTAGTACATGCCATCCG | 84B |
| GATTATCGCCCTGAAAGCCG | 84F |
| GGAGGCACGTATTGCGGC | 91B |
| TGCAAACAACGTCGGTCTG | 91F |
| CCAAAAGGCAATCACG | 94B |
| ACACAATGAGACAGAGGAG | 94F |
| CCAATATGCCTGCACTAAACCAG | G4B |
| CGTGGATACTGATTCAGGAAAACC | G4F |
| GCCAGCTCCATAGAAATCAGCAG | X2B |
| CCAGAGAAGCGTGAATTTTGGG | X2F |
Mitomycin C induction protocol.
A fresh overnight culture of a wild-type or recA mutant strain was diluted 4,000-fold into LB broth supplemented with 1× E salts and 0.2% glucose. This culture (50 ml) was grown to a density of ∼2 × 107 cells/ml. The culture was then split into 2 equal volumes, to which an equal volume of LB broth at 37°C (supplemented as above) was added. The two cultures were allowed to shake at 37°C until the concentration of cells was between 3 × 107 and 4 × 107 cells/ml. At this point 6 ml of each culture was withdrawn, distributed into 4 × 1.5-ml microcentrifuge tubes, and microcentrifuged for 45 s; then the supernatant was aspirated off and the tube was immersed in liquid nitrogen. These samples were the time zero samples. While the time zero samples were being processed, mitomycin C was added to one of the cultures (final concentration, 0.5 or 2.0 μg/ml; the remaining culture serves as the uninduced control) and both flasks were returned to the shaking water bath. Ten minutes (t = 10) after time zero, another 6 ml was withdrawn from each flask and treated like the time zero samples. The same procedure was used for the 30-min samples. The same procedure was also used for the 60-, 90-, and 120-min samples except that only 3-ml samples were withdrawn. The frozen cell pellets were stored at −80°C until RNA extraction.
Extraction and purification of RNA.
Frozen cell pellets were treated in groups of no more than six tubes. Each tube received 0.4 ml of 30 mM sodium acetate (pH 5.2) and 20 μl of 20% sodium dodecyl sulfate (SDS) (both preheated to 70°C). Each tube was vortexed vigorously and incubated in a 70°C water bath for 30 to 60 s. Each tube then received 0.4 ml of phenol (preheated to 70°C; equilibrated with 30 mM sodium acetate, pH 5.2) and was vortexed vigorously. Each tube was incubated at 70°C for an additional 10 min with occasional vortexing, then microcentrifuged for 15 min at high speed. The aqueous top layer was removed, extracted with 0.5 ml of phenol and microcentrifuged for 5 min. The aqueous layer was removed, extracted with 0.5 ml of chloroform, and microcentrifuged for 2 min. The aqueous layer was removed, brought up to a total volume of 0.4 ml with water, and combined with 45 μl of 3 M sodium acetate (pH 5.2). RNA was precipitated by addition of 0.9 ml of 100% ethanol, and the precipitated RNA was pelleted by microcentrifuging for 20 min at high speed. The nucleic acid pellet was briefly dried under a vacuum and resuspended in 10 mM Tris, pH 8, such that each time point sample was resuspended in a total of 80 μl. This preparation was then treated with DNase I.
Treatment of RNA preparations with DNase I.
Each 80-μl preparation of crude RNA was treated with DNase I by combination with 20 μl of a 5× cocktail (100 mM Tris [pH 8], 50 mM MgCl2, 1.2 U of RNase inhibitor/ml, 0.4 U of DNase I/ml) and incubation at 37°C for 40 min. The RNA was purified using RNeasy spin columns (Qiagen) according to the manufacturer's specifications. Purified RNA was eluted in water and adjusted to a final concentration of 100 ng/μl in water. RNA preparations were stored at −80°C.
General methods for the identification of differentially expressed fragments.
In this work, we have used two different methods of identifying differentially expressed fragments. These methods differ in the method of synthesizing cDNA. In our original procedure, we used a 9- or 10-nucleotide primer of defined sequence to prime the reverse transcription (RT) reaction. The cDNA made from this “specific-RT” reaction was amplified (under arbitrary conditions) by the addition of a second primer. In this procedure, the cDNA amplification is accomplished by a “primer pair”: the RT primer and the cDNA primer. Our alternate method (the random-hexamer RT method) uses random hexamers to prime cDNA synthesis; the cDNA is then amplified (under arbitrary conditions) by adding a gene-specific primer pair. The use of random hexamers to prime cDNA synthesis is particularly useful when one wishes to compare many different primer pair combinations (i.e., different genes) against one RT reaction.
RT.
Final reaction conditions for the RT reaction were 4 U of MMLV reverse transcriptase/ml, 2 μM arbitrary primer (or random hexamers), 50 mM Tris (pH 8.3; at 37°C), 50 mM KCl, and 4 mM MgCl2. Total amounts of RNA in a 10-μl reaction volume ranged from 1 to 0.2 μg. The reaction mixtures were assembled by spotting a 5-μl aliquot of a 2× cocktail containing everything except RNA and 5 μl of RNA on the sides of a 0.2-ml PCR tube. The reaction was initiated by briefly centrifuging the tubes; the reaction tubes were incubated at 37°C for 1 h, heated to 95°C for 10 min, diluted with a 3× volume of water, and stored at −20°C. This mixture was used directly in the cDNA amplification reaction.
cDNA amplification.
The cDNA amplification procedure has been described elsewhere (16, 17).
Identification, isolation, purification, cloning, and sequencing of candidate differentially expressed fragments.
The identification, isolation, and purification of differentially expressed genes have been described in detail previously (15, 16). Briefly, 3 μl of a RAP-PCR mixture was diluted with 12 μl formamide of loading dye (95% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol) and denatured at 94°C for 5 min, and 2 μl was loaded onto a 7 M urea–5% polyacrylamide gel (Accugel; National Diagnostics) and run for 2 h at a constant 50 W. The gel was dried and exposed to film, and differentially expressed fragments were identified. If a differentially expressed fragment was observed, the remainder of the reaction mix was run on a preparative gel; the band was excised from the gel and the DNA was eluted by soaking the gel slice in 10 mM Tris (pH 8) for 5 to 24 h at 50°C. The eluted nucleic acid was precipitated with sodium acetate and ethanol, pelleted, dried, and resuspended in 10 μl of SSCP running buffer (10 mM NaOH, 95% formamide, 0.1% bromophenol blue, 0.1% xylene cyanol). The sample was denatured for 3 min at 95°C, cooled on ice, loaded onto a SSCP gel (FMC Bioproducts) (15), and run overnight at 8 W (constant). The gel was dried, the fragment was visualized by autoradiography, and the band was excised and eluted by soaking (as before) in 150 μl of 10 mM Tris (pH 8). An aliquot of this mixture was amplified using the appropriate primers and PCR was run on a preparative 2% agarose (SeaPlaque; FMC Bioproducts) gel. The amplified fragment was visualized with ethidium bromide and UV lighting, excised, and purified with the QIAquick Gel Extraction Kit (Qiagen). This nucleic acid was directly cloned into the vector pCR2.1 supplied with the Original TA Cloning Kit (Invitrogen). Clones harboring inserts were identified and sequenced on a Licor 4200 automated sequencing apparatus. Fragments were sequenced on both strands using the vector's M13 priming sites. All sequence data have been deposited with GenBank. Accession numbers are listed in Table 2.
TABLE 2.
Salmonella serovar Typhimurium genes induced by mitomycin C treatment and their identities
| Designation | Size (nt)a | Primer pairb | Description (accession no.) (source or reference)c | Scored | Ed |
|---|---|---|---|---|---|
| 2p257 | 372 | 84F–84B | BLASTX: Head-tail preconnector protein GP5, bacteriophage 21 (NCBI P36273; PID g549296) (26) | 82.7 | 3 × e−31 |
| 5p257 | 349 | 85F–85B | BLASTX: Host specificity protein J, bacteriophage lambda (NCBI P03749; PID g138412) (24) | 111 | 3 × e−24 |
| 4p90 | 176 | OPN23–U7 | BLASTX: Bacteriophage N15, gp11 (NCBI AFO64539) | 42 | 7 × e−07 |
| 7p257 | 306 | G4F–G4B | BLASTN: Serovar Typhimurium recE gene (NCBI AF001386) (N. Figueroa-Bossi and L. Bossi, unpublished data) | 599 | e−169 |
| X1p203 | 87 | A94–91F | BLASTN: Serovar Typhimurium recE gene (32) | 435 | 4.2 × e−27 |
| Ap88 | 169 | EA10–N2 | BLASTX: Bacteriophage ES18, gp19, serovar | 42 | 7 × e−4 |
| 10-1p257 | 111 | X2F–X12B | BLASTN: H-NS homologue, serovar Typhimurium stpA gene (NCBI AF009363) | 218 | 2 × e−55 |
| Jp94 | 189 | N2–OPN22 | Novel | ||
| 1p85 | 191 | REP–OPN22 | Novel | ||
| 6p84 | 249 | OPN22–Shine | BLASTN: sulA gene of serovar Typhimurium | 0.0 | |
| 6p257 | 232 | 85F–85B | Novel | ||
| 4p85 | 174 | EA10–OPN22 | Novel | ||
| Fp94G1 | 61 | N2–OPN28 | Novel | ||
| 1p257 | 346 | J43B–J43F | Novel | ||
| RW1 | 127 | 91F–91B | Novel | ||
| 8p275 | 320 | 94F–94B | BLASTX: Lipase, Pseudomonas sp. strain B11-1 (NCBI AA38151; PID g2853612) (6) | 46 | 9 × e−05 |
| 1p90 | 241 | N1–OPN22 | Novel | ||
| 12p257 | 630 | 91F–91B | BLASTN: umuC gene of serovar Typhimurium (GenBank M57431) | 1,246 | 0.0 |
| Fp94G2 | 69 | N2-OPN28 | Novel | ||
| LP94 | 111 | N2–OPN22 | BLASTX: Tail fiber protein Gp37 of E. coli (NCBI BAA14966; PID g1742233) | 60 | 3 × e−09 |
| Ep43 | 215 | EA10-U7 | BLASTX: Hypothetical 53.1-kDa protein encoded in lysU-cadA intergenic region (SwissProt P39276) (5) | 122 | 3 × e−28 |
Does not include the primer sequences.
Differentially expressed fragments were isolated in two different experiments, as described in Materials and Methods. Primer pairs are identified in a “primer 1– primer 2” format.
Descriptions are derived from BLASTN or BLASTX as noted in the field. Searches were performed and gene assignments were made as described in Materials and Methods. Descriptions are taken from the National Center for Biotechnology Information (NCBI).
Scores and expectation values are provided by the Blast programs (http://www/ncbi.nih.gov/blast/blast.cgi).
Verification of differential expression.
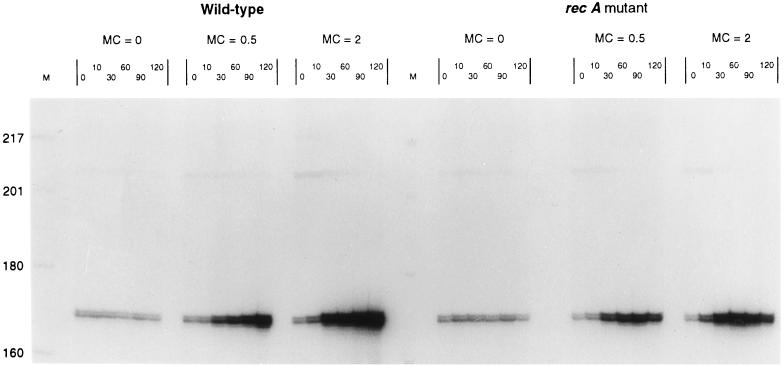
Once a candidate differentially expressed fragment was sequenced, complementary primers 18 to 25 nucleotides in length were designed which specifically amplified a portion of the cloned sequence. Differential expression of the sequence in question was then confirmed with those primer pairs by the following procedure. RAP-PCR was performed on cDNA (primed from random hexamers) using a gene-specific primer pair under the following conditions: 25 cycles of 30 s at 95°C, 45 s at 35°C, and 60 s at 72°C, followed by 10 min at 72°C. Synthesized DNA was quantified by 32P incorporation. If the appropriately sized band showed the expected differential expression, this was considered confirmation of differential expression. Each fragment identified in Table 2 was shown to be differentially expressed at two different concentrations of mitomycin C (0.5 and 2.0 μg/ml), although we present data only for a mitomycin C concentration of 0.5 μg/ml (see Fig. 2 and Table 3). In addition, differential expression at each concentration of mitomycin C was confirmed on RNA derived from minimum of three independent induction experiments.
FIG. 2.
Confirmation of differential expression using a standard quantitative RAP-PCR gel. In this experiment, RNA was reverse transcribed by priming with random hexamers, and the subsequent PCR amplification was accomplished with a gene-specific primer pair. Molecular weight markers are on the extreme left and in the middle of the gel. The experiment for which results are shown on the left (with respect to the central molecular weight marker) was done on wild-type Salmonella serovar Typhimurium at 0, 0.5, and 2.0 μg of mitomycin C (MC)/ml. Samples were withdrawn and analyzed at the indicated time points. The right side of the figure shows the results of the same protocol carried out on a recA mutant strain. The data clearly indicate that the gene is not induced in the recA mutant, while other background bands remain unaffected.
TABLE 3.
Quantitative analysis of gene induction by 0.5 μg of mitomycin C/ml on wild-type and recA mutant Salmonella serovar Typhimurium
| Fragment | Fold induction at the indicated time (min)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type
|
recA mutant
|
|||||||||||
| 0 | 10 | 30 | 60 | 90 | 120 | 0 | 10 | 30 | 60 | 90 | 120 | |
| 2p257 | 1.0 | 0.9 | 1.2 | 2.2 | 45.2 | 80.8 | 1.0 | 0.9 | 1.0 | 0.9 | 0.8 | 0.8 |
| 5p257 | 1.0 | 1.0 | 1.0 | 1.4 | 14.8 | 76.1 | 1.0 | 0.9 | 0.8 | 0.8 | 0.8 | 0.9 |
| 4p90 | 1.0 | 0.9 | 1.0 | 4.1 | 18.5 | 54.2 | 1.0 | 0.8 | 0.8 | 0.7 | 0.6 | 0.7 |
| 7p257(recE) | 1.0 | 1.0 | 1.4 | 20.3 | 35.8 | 52.6 | 1.0 | 1.0 | 1.0 | 1.0 | 0.9 | 1.0 |
| X1p203 | 1.0 | 1.0 | 1.3 | 22.7 | 31.8 | 46.3 | 1.0 | 1.0 | 1.0 | 0.8 | 0.8 | 0.8 |
| Ap88 | 1.0 | 1.1 | 1.0 | 1.7 | 23.5 | 41.4 | 1.0 | 0.8 | 0.7 | 0.6 | 0.5 | 0.4 |
| 10-1p257(StpA) | 1.0 | 1.2 | 0.9 | 1.3 | 27.0 | 39.6 | 1.0 | 0.8 | 0.8 | 0.7 | 0.4 | 0.4 |
| Jp94 | 1.0 | 1.0 | 1.3 | 10.0 | 21.9 | 37.2 | 1.0 | 0.9 | 0.8 | 0.8 | 0.7 | 0.7 |
| 1p85 | 1.0 | 1.0 | 0.9 | 1.2 | 9.7 | 31.5 | 1.0 | 0.9 | 0.9 | 0.8 | 0.7 | 0.6 |
| 6p84(sulA) | 1.0 | 3.5 | 7.4 | 13.3 | 21.4 | 24.6 | 1.0 | 0.7 | 0.9 | 0.8 | 0.6 | 0.5 |
| 6p257 | 1.0 | 1.1 | 1.0 | 1.5 | 12.2 | 24.6 | 1.0 | 0.8 | 0.8 | 0.6 | 0.5 | 0.4 |
| 4p85 | 1.0 | 1.0 | 1.1 | 2.0 | 8.0 | 23.5 | 1.0 | 0.9 | 1.0 | 0.9 | 0.9 | 0.9 |
| Fp94G1 | 1.0 | 1.0 | 1.1 | 1.2 | 3.7 | 18.8 | 1.0 | 1.0 | 0.8 | 0.8 | 0.6 | 0.6 |
| 1p257 | 1.0 | 1.0 | 0.9 | 1.6 | 6.4 | 16.8 | 1.0 | 0.9 | 1.0 | 0.9 | 0.6 | 0.6 |
| RW1 | 1.0 | 1.2 | 1.6 | 3.1 | 6.3 | 16.3 | 1.0 | 0.9 | 2.0 | 3.9 | 4.4 | 4.8 |
| 8p257 | 1.0 | 1.7 | 3.9 | 6.7 | 13.8 | 14.4 | 1.0 | 0.9 | 1.0 | 0.9 | 0.7 | 0.6 |
| 1p90 | 1.0 | 1.0 | 1.5 | 1.5 | 16.7 | 12.7 | 1.0 | 0.9 | 1.2 | 1.1 | 0.9 | 0.4 |
| 12p257(umuC) | 1.0 | 1.9 | 5.1 | 7.0 | 8.8 | 12.5 | 1.0 | 1.0 | 1.0 | 1.0 | 1.2 | 1.4 |
| FP94G2 | 1.0 | 1.0 | 0.8 | 1.8 | 4.8 | 6.7 | 1.0 | 0.8 | 0.7 | 1.7 | 0.6 | 0.5 |
| Lp94 | 1.0 | 0.9 | 1.2 | 1.5 | 1.0 | 2.3 | 1.0 | 1.1 | 1.0 | 0.8 | 0.8 | 0.8 |
| Ep43 | 1.0 | 1.0 | 0.9 | 0.9 | 1.8 | 1.6 | 1.0 | 1.0 | 1.1 | 1.0 | 0.9 | 0.8 |
Quantification of differential expression.
The differential expression of the gene fragments listed in Table 2 was quantified using a Molecular Dynamics Storm PhosphorImager. For each time point, the differentially expressed band was normalized to a constant band or region within each lane. The same background band or region is used across all time points for a particular fragment. Each of these “internally” normalized time points is then normalized to the time zero point. The data presented in Table 3 are the results of quantifying a typical induction experiment for a mitomycin C concentration of 0.5 μg/ml. Data for a mitomycin C concentration of 2.0 μg/ml were also generated with similar results (data not shown).
Identification of fragment sequences.
The sequence of each differentially expressed fragment was compared to the National Institutes of Health (NIH) nonredundant (nr) sequence database using the BLASTN and BLASTX programs (1). The results of the BLASTN search were used to classify the sequence as a novel or known sequence. A sequence was classified as known if our sequence had a greater than 98% match with the database sequence with an expectation value (E) of ≤10−10. This criterion allowed an unambiguous classification of the data. If there was no significant nucleotide homology with the database, a BLASTX search was performed. The BLASTX program translates the query sequence in all six reading frames and compares this to a protein database. The highest-scoring result(s) of the BLASTX search is reported in Table 2 if the expectation value is ≤10−3.
Nucleotide sequence accession numbers.
The sequences of the following differentially expressed fragments have been deposited in GenBank under the accession numbers given in parentheses: 10-1p257 (AF206676), 12p257 (AF206677), 1p257 (AF206678), 1p85 (AF206679), 1p90 (AF206680), 2p257 (AF206681), 4p85 (AF206682), 4p90 (AF206683), 5p257 (AF206684), 6p257 (AF206685), 7p257 (AF206686), 8p257 (AF206687), ap88 (AF206688), ep43 (AF206689), fp94g1 (AF206690), fp94g2 (AF206691), jp94 (AF206692), lp94 (AF206693), rw1 (AF206694), and x1p203 (AF206695).
RESULTS
Identification of genes induced by treatment of Salmonella serovar Typhimurium with mitomycin C.
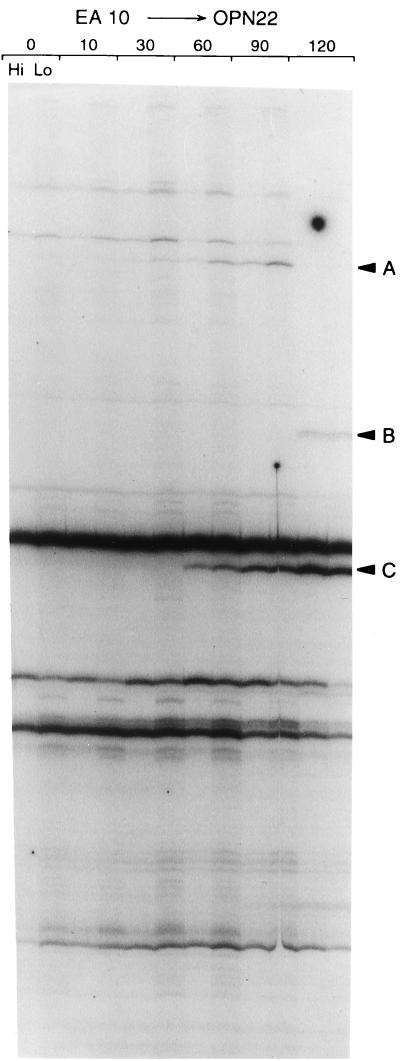
A fresh culture of Salmonella serovar Typhimurium was treated with two different concentrations of mitomycin C as described in Materials and Methods. An untreated control for each time point was also taken, although time points other than time zero were not included in the initial experiments. RNA fingerprinting reactions were run on the RNA isolated from treated and untreated cells. A typical RNA fingerprinting gel is shown in Fig. 1. This figure shows an RNA fingerprint derived from the EA10–OPN22 primer pair.
FIG. 1.
Standard RAP gel for a mitomycin C treatment of 2.0 μg/ml. In this experiment, the RT primer is EA10 (Table 1) and the cDNA amplification primer is OPN22. The time zero point is used as the untreated control. At each time point, two RNA samples were analyzed, using a total of 500 μg (Hi) or 250 μg (Lo) for each time point. In this particular experiment, bands A, B, and C were extracted and isolated as described in Materials and Methods.
Using 36 primer pairs, 42 candidates for differentially expressed genes were identified in gels such as that shown in Fig. 1. Of these 42 candidates, the 21 listed in Table 2 were ultimately shown to be differentially expressed, as described below (see also Materials and Methods). Of these 21 candidates, 2 were independent isolates from different regions of the recE gene; thus, these 21 transcripts represent 20 distinct genes.
Confirmation and quantification of differential expression.
Once a candidate gene fragment such as that shown in Fig. 1 was identified, it was extracted from the gel, purified, cloned, and sequenced. From the sequence data, we then designed a specific, homologous PCR primer pair that would amplify a portion of the fragment. This specific PCR primer pair was employed to amplify first-strand cDNA made using random hexamers. These specific PCRs were used to independently confirm the observation of differential expression. Figure 2 shows the results of such a confirmation experiment for two different concentrations of mitomycin C.
The amplification conditions used to confirm differential expression were low-temperature annealing conditions (i.e., an annealing temperature of 35°C). Under such conditions not only is the specific band corresponding to the target RNA generated, but a fainter, background fingerprint of bands is also generated by arbitrary priming events. This background fingerprint is useful because the constant background bands serve as internal controls for each lane (18). Thus, by normalizing the band of interest to a common, unchanging background band within each lane, samples taken at different time points can be accurately compared.
Table 3 shows the results of quantifying gels such as that shown in Fig. 2 for a concentration of mitomycin C equal to 0.5 μg/ml. Inspection of the data in Table 3 shows a range of induction from 1.8-fold (fragment Ep43) to 80-fold (fragment 2p257) with respect to the time zero. Similar results have been found for a mitomycin C concentration of 2.0 μg/ml (data not shown). The data also show that induction can be observed as soon as 10 min (fragment 6p84 [the sulA gene]) or as long as 120 min (fragment Lp94) after mitomycin C exposure.
The genetic regulation of mitomycin C-induced operons in Salmonella serovar Typhimurium.
In E. coli, the SOS response to DNA damage is under the direct control of the LexA protein. The RecA protein is the effector that passes on the signal for SOS induction by stimulating cleavage of the LexA repressor. In E. coli, recA null mutants are unable to stimulate the LexA cleavage reaction and are unable to establish the SOS response. If the mitomycin C-inducible regulon we have revealed in Salmonella serovar Typhimurium is analogous to the E. coli SOS regulon, we would predict the Salmonella regulon to be under LexA-RecA control.
In Fig. 2 and Table 3 we show the results of treating wild-type Salmonella serovar Typhimurium versus a recA mutant strain with mitomycin C. Figure 2 demonstrates that in the recA mutant strain, the transcript is not induced by mitomycin C treatment. Table 3 shows quantitative data for the regulation of all 20 transcripts in the wild-type and recA mutant backgrounds. Clearly, 19 of the 20 transcripts fail to be induced in a recA mutant strain. We conclude that the mitomycin C-inducible regulon characterized in Table 3 is a Salmonella serovar Typhimurium analogue of the E. coli SOS regulon.
The one exception is fragment RW1, which is induced by mitomycin C in the absence of a functional RecA protein. The RecA-independent induction of RW1 is illustrated in Fig. 3 and quantified in Table 3. The data in Table 3 demonstrate that, in the recA mutant strain, the RW1 transcript is induced fivefold by mitomycin C treatment. The substantial regulation of RW1, in the absence of functional RecA protein, suggests the existence of an unknown regulatory component for the mitomycin C-induced pathway in Salmonella serovar Typhimurium.
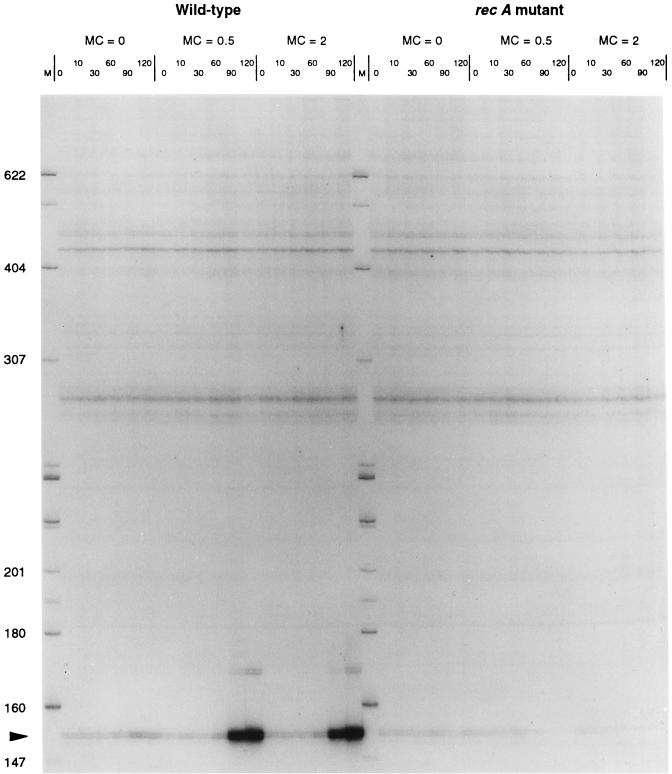
FIG. 3.
An experiment similar to that described for Fig. 2 was performed for the RW1 sequence. The transcript shows induction by mitomycin C (MC) (at 0.5 and 2.0 μg/ml) in the presence and absence of a functional RecA protein. The gene is clearly induced in the recA mutant strain and therefore is the first example of an SOS-induced gene with a regulatory structure different from that of the standard MC-induced regulon. Quantified induction levels are shown in Table 3.
DISCUSSION
We have conducted a survey of genes induced by the DNA-damaging agent mitomycin C in Salmonella serovar Typhimurium. The technique we have used, RAP-PCR, is capable of identifying both increased and decreased mRNA expression against a background of unchanging expression (17). We have described 20 differentially expressed genes, all of which are activated by mitomycin C. We did not find any transcripts which were repressed by mitomycin C, although these experiments revealed at least five genes whose transcripts were repressed by entry into stationary phase (data not shown).
The fact that only induced genes were found may be a bias of the RAP-PCR system. Repressed genes will be observed only among those genes that generate PCR products in the controls. In contrast, induced genes can be sampled from the PCR fragments that are not visible in the control but become visible upon induction. The class of genes not visible on the gel is obviously larger than the visible class, so the class of observed up-regulated genes will usually be larger than the class of observed down-regulated genes.
The regulation of the 20 mitomycin C-induced genes was investigated in a recA mutant background. We found that 19 of the 20 genes required the presence of an active RecA protein for induction by mitomycin C. The requirement for an active RecA protein, and the identity of some of the genes revealed, affirms that Salmonella has an SOS operon analogous to that in E. coli, as other investigators have shown (20, 25). The discovery that the RW1 gene is induced in the absence of a functional RecA protein reveals the presence of a novel, RecA-independent, SOS transcriptional activation mechanism.
These data represent the application of an approach to gene expression different from that used in E. coli. The SOS system in E. coli has been largely analyzed with genetic methods. Most of the genes found in the E. coli SOS regulon were revealed by use of Mu dlac operon fusions (reviewed in references 10 and 30). While the utility of such fusions is widely known, their potential as a gene discovery tool can be limited by the effect of the Mu dlac insertion on the viability and growth rate of the cell. In contrast, RAP-PCR is a biochemical approach whose sampling method has no genetic consequences on the cell. In principle, if a large enough spectrum of PCR primers is employed, RAP-PCR could detect every regulated gene regardless of the role that gene plays in the cell's health.
Our data show that a variety of genes are induced in Salmonella serovar Typhimurium, with a spread of induction time points and with variable induction ratios. These data, shown in Table 3, are all within the parameters for components of the SOS regulon described for E. coli (10).
Identities of gene fragments.
We have classified the gene fragments found in this study into two broad categories—genes of known function and novel genes, including those distantly related to known genes. This information is summarized in Table 2 for each gene fragment. Those fragments whose sequence (or a homologous sequence) has been previously reported in the public databases are described here as “known” genes (5 of 20 genes). Those gene fragments for which no close match or homologue could be found are described here as novel genes (15 of 20 genes). The criterion used as the basis for these designations is discussed in Materials and Methods.
Known genes.
The sequences characterized here as known Salmonella genes are sulA, stpA (h-ns), umuC, and recE. A fifth sequence corresponds to a hypothetical gene in the lysU-cadA intergenic region of E. coli and is presumed to be the Salmonella homologue. The results of BLAST searches for these sequences are shown in Table 2.
The E. coli regulon is composed of more than 20 genes, of which at least half have been identified by sequence analysis in Salmonella serovar Typhimurium. Two of the genes we found, the sulA and umuC genes, are known to be components of the E. coli SOS regulon. That we have found only 2 of the more than 10 shared genes is likely attributable to the arbitrary component of the RAP-PCR technique. It is likely that the combination of the sequences of the primers used in this work, the specific sequences of genes which have been missed, and the induction level of those genes has simply masked the regulation of those genes.
It is interesting to note that the Salmonella recE operon, reported here for the first time as mitomycin C inducible, has been previously reported as a suppressor of recBC mutations in E. coli and is induced by hydrogen peroxide treatment of Salmonella (9, 32). The Salmonella recE gene is part of a larger, peroxide-inducible operon, which includes the Gifsy-1 prophage and has been implicated in the pathogenicity of Salmonella serovar Typhimurium (9). In E. coli, the recE gene is part of a cryptic operon and has not been associated with the SOS response.
Novel genes.
The majority of fragments (15 of 20) revealed in this work have poor nucleotide homology with the public sequence databases. The range of BLASTN expectation values for this group was e−10 to 2.8, with two sequences having no hit whatsoever.
In order to analyze this group further, the sequences were subjected to BLASTX analysis. The BLASTX program translates the query nucleic acid in all six reading frames and compares the translations with public protein databases. This analysis allowed the group to be divided into two subcategories—entirely novel sequences and sequences distantly related to previously reported proteins. The results of the BLASTX analyses for these sequences are shown in Table 2.
A fragment was classified as novel (9 of 15 sequences) if BLASTX analysis could not find a match (E > 1 × e−3) in any public protein database. A fragment was classified as of unknown function if the BLASTX expectation value was less than or equal to 1 × e−3. The designation “unknown” is meant to imply that a sequence might be a new member of a gene family, the identity of that family being suggested by the best hit with the protein database (Table 2).
Of the six sequences classified as novel, five had their best BLASTX hits with bacteriophage proteins. The remaining novel sequence has a best hit with a Pseudomonas sp. lipase as well as several lesser hits with lipases from other organisms (data not shown).
The proportion of Salmonella genomes with close homology to the E. coli K-12 genome is 60 to 70% (19, 33). Thus, the fact that 15 of 20 of the identified sequences are encoded in regions not shared with E. coli indicates a bias for mitomycin C induction in areas of the genome that carry “loops” that are not shared by related enterobacteria. It is these genomic differences between species which account for differences in pathogenicity, host range, etc.
Of the 15 unknown or novel genes described here, 6 have protein similarity to phage gene products (Table 2). In addition, one of these genes (Lp94) has 22 bases, at one end, perfectly matched with the macrophage-induced Salmonella mig-3 gene (29). The significance of this homology is unclear (BLASTN score, 44; E = 0.006); it could represent either a rearrangement of the mig-3 locus or a repeated sequence.
We have presented data showing the induction of 20 genes of the Salmonella SOS regulon. It is not yet clear if these 20 genes represent 20 different operons. The cloned sequences were all searched against the almost completed sequences of Salmonella serovar Typhimurium, Salmonella enterica serovar Paratypi A, and Klebsiella pneumoniae (http://genome.wustl.edu/gsc/bacterial/Salmonella.shtml), Salmonella enterica serovar Typhi and Yersinia pestis (http://www.sanger.ac.uk/Projects/), and other partly completed genomes (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html). All the fragments in Table 2, with the exception of 8p257 and RW1, mapped to regions that were found only in Salmonella serovar Typhimurium and not in the other genomes (at a P value of <e−10), indicating that they are very recent additions to the Salmonella serovar Typhimurium genome. Furthermore, five products, Ap88, 2p257, 4p90, 1p85, and Fp94G, mapped to the same 15-kb unique region, and two other pairs of products mapped within a few kilobases of each other elsewhere in the genome. All these regions had patches of high homology with bacteriophage genes.
The general subjects of phage remnants and pathogenicity determinants are relevant because it appears that many, if not all, pathogenicity determinants are peppered with phage remnants. It appears that these remnants may be landmarks of pathogenicity islands (8, 9, 29). Because our protocol has revealed a high percentage of novel phage remnants, this may be an indication that the use of differential gene expression analysis to analyze the effects of DNA-damaging agents is a simple and efficient method for revealing pathogenicity determinants in any pathogen that can be cultured in vitro.
We have recently shown that RAP-PCR products can be used to probe cDNA arrays and that these probes detect RNAs that are not visible using total labeled cDNA (reference 28 and references therein). The experiments performed here have been archived, and we are constructing arrays of open reading frames for Salmonella in order to further increase the throughput of the method.
In conclusion, we note that the components of the Salmonella SOS operon revealed here are significantly different from the components of the E. coli SOS operon. Although both operons share common genes, it appears that the regulons have significantly diverged. In addition, this work has revealed the existence of a mitomycin C-inducible gene that has a regulatory structure different from that of the standard SOS regulon. Apparently, this gene does not have an E. coli homologue.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI 34829-09, AI 43283-02, and CA81667-02.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D G, Kowalczykowski S C. Reconstitution of an SOS response pathway: derepression of transcription in response to DNA breaks. Cell. 1998;95:975–979. doi: 10.1016/s0092-8674(00)81721-3. [DOI] [PubMed] [Google Scholar]
- 3.Benson N R, Roth J. A Salmonella phage-P22 mutant defective in abortive transduction. Genetics. 1997;145:17–27. doi: 10.1093/genetics/145.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Burland V, Plunkett III G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo D W, Kurihara T, Suzuki T, Soda K, Esaki N. A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol. 1998;64:486–491. doi: 10.1128/aem.64.2.486-491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 8.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 11.Hanawalt P C, Cooper P K, Ganesan A K, Smith C A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- 12.Humayun M Z. SOS and Mayday: multiple inducible mutagenic pathways in Escherichia coli. Mol Microbiol. 1998;30:905–910. doi: 10.1046/j.1365-2958.1998.01120.x. [DOI] [PubMed] [Google Scholar]
- 13.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 15.Mathieu-Daude F, Benson N, Kullmann F, Honeycutt R, McClelland M, Welsh J. Screening differentially displayed PCR products by single-strand conformation polymorphism gels. In: Innis M A, Gelfand D H, Sninsky J J, editors. PCR applications. San Diego, Calif: Academic Press; 1999. pp. 341–354. [Google Scholar]
- 16.Mathieu-Daude F, Trenkle T, Welsh J, Jung B, Vogt T, McClelland M. Identification of differentially expressed genes using RNA fingerprinting by arbitrarily primed polymerase chain reaction. Methods Enzymol. 1999;303:309–324. doi: 10.1016/s0076-6879(99)03020-7. [DOI] [PubMed] [Google Scholar]
- 17.Mathieu-Daude F, Welsh J, Davis C, McClelland M. Differentially expressed genes in the Trypanosoma brucei life cycle identified by RNA fingerprinting. Mol Biochem Parasitol. 1998;92:15–28. doi: 10.1016/s0166-6851(97)00221-1. [DOI] [PubMed] [Google Scholar]
- 18.Mathieu-Daude F, Welsh J, Vogt T, McClelland M. DNA rehybridization during PCR: the ‘Cot effect’ and its consequences. Nucleic Acids Res. 1996;24:2080–2086. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClelland M, Wilson R K. Comparison of sample sequences of the Salmonella typhi genome to the sequence of the complete Escherichia coli K-12 genome. Infect Immun. 1998;66:4305–4312. doi: 10.1128/iai.66.9.4305-4312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang P P, Walker G C. The Salmonella typhimurium LT2 uvrD gene is regulated by the lexA gene product. J Bacteriol. 1983;154:1502–1504. doi: 10.1128/jb.154.3.1502-1504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehrauer W M, Lavery P E, Palmer E L, Singh R N, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. I. LexA repressor cleavage is competitive with binding of a secondary DNA molecule. J Biol Chem. 1996;271:23865–23873. [PubMed] [Google Scholar]
- 22.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. . (Erratum, 66:VII, 1997.) [DOI] [PubMed] [Google Scholar]
- 23.Sancar A, Sancar G B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- 24.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 25.Smith C M, Arany Z, Orrego C, Eisenstadt E. DNA damage-inducible loci in Salmonella typhimurium. J Bacteriol. 1991;173:3587–3590. doi: 10.1128/jb.173.11.3587-3590.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith M P, Feiss M. Sequence analysis of the phage 21 genes for prohead assembly and head completion. Gene. 1993;126:1–7. doi: 10.1016/0378-1119(93)90583-o. [DOI] [PubMed] [Google Scholar]
- 27.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 28.Trenkle T, Welsh J, McClelland M. Differential display probes for cDNA arrays. BioTechniques. 1999;27:554–564. doi: 10.2144/99273rr03. [DOI] [PubMed] [Google Scholar]
- 29.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 30.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 31.Wang T C, Smith K C. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol. 1983;156:1093–1098. doi: 10.1128/jb.156.3.1093-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong R M, Wong K K, Benson N R, McClelland M. Sample sequencing of a Salmonella typhimurium LT2 lambda library: comparison to the Escherichia coli K-12 genome. FEMS Microbiol Lett. 1999;173:411–423. doi: 10.1111/j.1574-6968.1999.tb13533.x. [DOI] [PubMed] [Google Scholar]