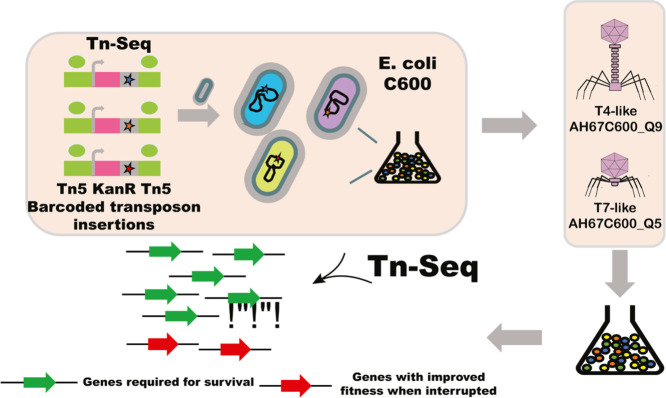
Highlights
-
•
Successfully characterized a high lytic T7-like phage infecting E. coli C600.
-
•
Replication and transcription regions show high genetic variability compared to T7.
-
•
TraDIS strategy is applied in uncovering host determinants involved in fitness.
-
•
wbbL probably play an important role in two kinds of lytic phage susceptibility.
-
•
Two-component system susceptible gene QseE and RstB are also co-selected.
Keywords: Lytic T4-like phage; Lytic T7-like phage; Antimicrobial resistance (AMR); Tn5 transposons, Phage therapy
Abstract
As antimicrobial resistance (AMR) continues to increase, the therapeutic use of phages has re-emerged as an attractive alternative. However, knowledge of phage resistance development and bacterium-phage interaction complexity are still not fully interpreted. In this study, two lytic T4-like and T7-like phage infecting model Escherichia coli strain C600 are selected, and host genetic determinants involved in phage susceptibility and resistance are also identified using TraDIS strategy. Isolation and identification of the lytic T7-like show that though it belongs to the phage T7 family, genes encoding replication and transcription protein exhibit high differences. The TraDIS results identify a huge number of previously unidentified genes involved in phage infection, and a subset (six in susceptibility and nine in resistance) are shared under pressure of the two kinds of lytic phage. Susceptible gene wbbL has the highest value and implies the important role in phage susceptibility. Importantly, two susceptible genes QseE (QseE/QseF) and RstB (RstB/RstA), encoding the similar two-component system sensor histidine kinase (HKs), also identified. Conversely and strangely, outer membrane protein gene ompW, unlike the gene ompC encoding receptor protein of T4 phage, was shown to provide phage resistance. Overall, this study exploited a genome-wide fitness assay to uncover susceptibility and resistant genes, even the shared genes, important for the E. coli strain of both most popular high lytic T4-like and T7-like phages. This knowledge of the genetic determinants can be further used to analysis the behind function signatures to screen the potential agents to aid phage killing of MDR pathogens, which will greatly be valuable in improving the phage therapy outcome in fighting with microbial resistance.
Graphical abstract
1. Introduction
Antimicrobials have been widely used in important clinical procedures and have saved millions of lives since their discovery in 1920s. Unfortunately, antimicrobial resistance among clinically pathogens is increasing, and the wide and inappropriately use of antimicrobials has exacerbated the development of resistance (Murray et al., 2022). Thus, alternative treatments that are successful and efficient in eliminating resistant bacteria are desperately needed (Butler and Paterson, 2020; Theuretzbacher et al., 2020). Nowadays, this growing antimicrobial resistance has rekindled the interest in bacteriophage therapy (BT) (Schooley and Strathdee, 2020; Watts, 2017). BT has been used successfully in former Soviet republics like Georgia and Russia for decades (Pirnay et al., 2021), whereas was largely abandoned later in the West following the discovery of antibiotics and the lacking of basic understanding of phage biology (Lavigne and Loessner, 2021).
Bacteriophages (phages), bacterial viruses, are obligate bacterial parasites that infect specific strains and represent the most abundant biological entities on earth and body system, showing a key ecological status in driving microbial community dynamics, activity, and adaptation (Dion et al., 2020; Liang and Bushman, 2021). During the treatment in multidrug-resistant (MDR) bacteria, the success of BT largely be limited by the development of phage resistance by bacteria. Also, the huge number of phage applications could lead to the uncertain result of phage-bacterial interaction and probably induced the occurrence of multi-phage resistance bacteria (Egido et al., 2022). Despite multiple mechanisms by which wide type bacteria defend against diversity phages have been uncovered (Gao et al., 2020), unbiased and overall genetic screens that are involved phage susceptibility and resistance are still not fully interpreted. Comprehensive genome-wide screens for studying host factors essential in phage infection would be important for obtaining a deeper understanding of phage resistance phenotypes (Qimron et al., 2006).
Considering E. coli strain C600 has been widely used in laboratories worldwide for molecular microbiology studies, such as transformation and conjugation assay for gene function analysis (Allue-Guardia et al., 2019). Historically, the Caudovirales traditionally were morphologically classified into the Myoviridae, Siphoviridae and Podoviridae. The Myophage (T4-like) have long and contractile tails while the Siphophage (lambda-like) have long and non-contractile tails and Podophage (T7-like) have short and non-contractile tails(Nobrega et al., 2018). However, the existing nomenclature has not kept pace with modern phage research and the latest viral taxonomic database (https://ictv.global/taxonomy) has now elevated the order Caudovirales to class Caudoviricetes (Turner et al., 2021). Nevertheless, the T4-like and T7-like phages still represent the most commonly encountered phage in many environments and possess broad host ranges(Dion et al., 2020). Since the phage lambda usually leads to lysogenic cycle, resulting in integration of phage DNA with bacterial chromosome (Casjens and Hendrix, 2015), its value for BT is greatly reduced. To this end, method that more accurately characterize T4-like and T7-like phages susceptibility or resistance determinants for E. coli strain C600 will provide a glimpse of phage fitness landscape.
Since phenotypic methods (Castledine et al., 2022; Wang et al., 2021b) and whole-genome sequencing (Brown et al., 2021; Markwitz et al., 2021) of phage-resistant mutants displayed difficulties, such as cumbersome and time consuming. (Hesse et al., 2020), we employed transposon-directed insertion site sequencing (TraDIS), a method combining phenotypic fitness selection with genomics to simultaneously assay all genes for their involvement in survival under selective conditions (Zhang et al., 2019), which are suitable for discovering host genes critical for phages susceptibility or resistance. In this study, E. coli strain C600 is selected as the model strain and two kinds of lytic phages (a T4-like phage and a T7-like phage) are also selected. The comparison of two lineages of dsDNA phage allowed us to examine how phage-level divergence of genotype lead to differential susceptibility and resistance under phage pressure, these datasets can provide a foundation for deeper understanding phage-host interactions and engineering phage for future therapeutic applications.
2. Material and methods
2.1. Strain and phages collection
E. coli strain C600 (NZ_CP031214) is selected as the model strain. The T4-like and T7-like lytic phage were isolated from a pig farm wastewater sample through additional pretreatment steps. T4-like phage, named Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Straboviridae; Tevenvirinae; Tequatrovirus; Tequatrovirus AH67C600_Q9 (Accession number: MZ681930.1), was documented in our previous study (Zhang et al., 2021). In this study, a T7-like phage, named Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Autographiviridae; Studiervirinae; Teseptimavirus; Teseptimavirus AH67C600_Q5, is further isolated and characterized.
Briefly, the sewage samples were concentrated by an ultrafiltration device (Millipore Lab-scale tangential flow filtration System, Merck, Germany), and then filtered through a 0.22 µm membrane (Millex-GP, Millipore, MA, USA). The filtrate was incubated with logarithmic phase cells of E. coli strain C600 overnight at 37 °C with shaking at 200 rpm. The mixture was centrifuged and filtered to enrich the primary phage preparation. The final filtrates were plaque-purified using the double-layer soft agar plate method. Single plaques were selected and diluted in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris–HCl pH 7.5) and stored at 4 °C. Single phage was plaque-purified three times by E. coli strain C600 before.
2.2. Transmission electron microscopy of T7-like phage
Transmission Electron Microscopy of T7-like phage AH67C600_Q5 were conducted. High titer phage stocks, about 108 PFU/ml, were concentrated 50-fold using 100 kDa Amicon Ultra centrifugal filter units and about 15 μL of phage concentrate was dropped on carbon-coated formvar covered grids and let stand for 15 min. The preparations were then stained with 2% w/v phosphotungstic acid pH 7.0 and air-dried. The T7-like phage AH67C600_Q5 was examined using a FEI transmission electron microscope (Thermo Fisher, Hillsboro, USA) at an acceleration voltage of 80 kV.
2.3. One step growth curve of T7-like phage
One-step growth experiments were carried out. Briefly, T7-like phage AH67C600_Q5 was added at the MOI of 10 and allowed to absorb to E. coli strain C600 cells for 10 min at room temperature. The mixture was centrifuged at 2700 × g for 10 min to remove unabsorbed phages and infected cells were resuspended with LB to 5 mL. The culture samples were harvested every 10 min and phage numbers were quantified as per above. This experiment was performed in triplicate.
2.4. Optimal multiplicity of infection (OMOI)
Bacterial colony forming units (CFU) per mL and phage titers (PFU per mL) were determined as previously described (Kropinski et al., 2009). Optimal multiplicity of infection (OMOI) was examined by using appropriate susceptible host cultured to early log phase growth and incubated with phage for 2 h in the following ratios: T7-like phage AH67C600_Q5: E. coli strain C600 = 10:1, 1:1, 0.1:1, 0.01:1, 0.001:1 and 0.0001:1. Then phage titers were calculated again in each group using standard protocols in triplicate.
2.5. Thermostability and pH tolerance of T7-like phage
Phage thermostability was examined by incubating phage stocks (≥108 PFU/mL) at 4 °C, 37 °C, 50 °C, 55 °C, 60 °C and 70 °C for 1 h and pH tolerance was examined by incubating over a pH range of 1.0 to 12.0 for 1 h at 37 °C. Phage titers were determined by double-layer agar plate as above. Both experiments were performed three duplications.
2.6. DNA extraction of T7-like phage
The purified phage lysates were concentrated by polyethylene glycol 8000 (PEG8000) and treated with DNase I (Sangon Biotech, Shanghai, China) at 100 U per mL prior to DNA extraction. Phage DNA extraction was conducted. An aliquot of a phage lysates was amplified for bacterial 16S rDNA determinations using conventional polymerase chain reaction (PCR) and only negative samples were used for the subsequent sequencing (Wang et al., 2021a, 2018). DNA was suspended in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5) and DNA concentrations were determined by UV spectroscopy (NanoDrop, Wilmington, USA).
2.7. Genome sequencing and assembly of T7-like phage
Phage DNA samples were sent for de novo sequencing at GENEWIZ Biological Technology (Suzhou, China) and a library of these DNA samples was constructed for sequencing. Trimmomatic-0.36 was used to trim and remove adaptors and low-quality nucleotides to obtain clean data (Bolger et al., 2014). Spades (version 3.8.1) was used to assemble the post-processed readings into contig sequences (Bankevich et al., 2012). SSPACE (version 3.0) was used to further assemble contig sequences into scaffold sequences and GapFiller (version 1.10) was used to complement and extend the scaffold sequence to obtain the final scaffold sequence of the whole phage genome.
An online website (www.ncbi.nlm.nih.gov/orffinder/), RASTtk and GeneMarks were jointly used to predict all genes in the genome. After comparisons with protein sequences of known function using BLASTP with an E-value of ≤1 × 10−5 and an identity of ≥ 70%(Brettin et al., 2015), the best hit was selected for the annotation of the gene products. CGview Server (http://cgview.ca/) was used to draw the whole genome map of the T7-like phage AH67C600_Q5. The evolutionary relationship between T7-like phage AH67C600_Q5 and other download phages were analyzed using phylogenetic trees based on the amino acid sequences of the terminase large subunits (Black, 2015). Phylogenetic analysis was performed with MEGA7 by the neighbor-joining method (Bootstrap: 1000 with the Poisson Model) (Yang et al., 2020). Alignment analysis was based on the ClustalW alignment of amino acid sequences by MEGA7. (Parameter setting: Gap Opening Penalty: 10, Gap Extension Penalty: 0.2) (Lu et al., 2017).
2.8. Competent cells producing of library E. coli strain C600
E. coli strain C600 was used to produce the competent cells library according to the previous study (Recacha et al., 2017). Single colonies of E. coli strain C600 was inoculated into 3 ml volumes of LB broths and left to propagate overnight at 37 °C. The 3 ml overnight culture was added to 300 ml of LB broth and incubated at 37 °C until a mid-log phase (OD600 = 0.5) was achieved. The cultures were then recentrifuged at 5000 rpm/min for 10 min at 4 °C in 6 × 50 ml aliquots. The supernatant was decanted, and the cells were suspended in about 20 ml of ice-cold 10% glycerol and repeated twice. Subsequently, the supernatant was centrifuged at 5000 rpm/min for 10 min at 4 °C and then discarded the supernatant, and the cells were suspended in 6.25 ml ice-cold 10% glycerol. Again, centrifuged at 9000 rpm for 10 min and removed supernatant, and the cells were suspended in 0.5 ml of ice-cold 10% glycerol. Finally, the cells were suspended in 50 µl of ice-cold 10% glycerol and kept on ice before electroporation.
2.9. Generation of the transposon mutant library
A 2-µl volume of pUTmini-Tn5km2 plasmid (Ampr Kmr) was added to each of the 100-µl competent cell on ice. The mixture of Tn5 transposome and competent cells was transferred into electroporation cuvettes and electroporated (with settings of 1.4 kV, 25 µF, and 200 Ω) in a Gene Pulser Xcell electroporation system (Bio-Rad). Cells were immediately recovered in 1 ml of Super Optimal Broth (SOB) after electroporation. The recovered cells were then incubated for 2 h at 37 °C in a shaking incubator.
To construct high saturation transposon mutant library of E. coli strain C600, the recovered cells were harvested and washed three times in PBS. After discarding the supernatant, the cells were resuspended in 100 μl PBS, plated onto LB agar plates, and incubated at 37 °C for 4 h. Colonies were collected, resuspended in PBS, and plated onto LB agar containing Kan and Rfp. After incubation overnight at 37 °C, the total number of colonies was estimated by counting a proportion from each of multiple plates. Kan- and Rfp-resistant colonies were resuspended in sterilized LB broth using a bacteriological spreader before adding sterile PBS. Each batch contained an estimated 30,000 mutants, and a total of over 100,000 transconjugants was generated by pooling 4 mutant batches. The entire mini-Tn5 mutant collection was grown three times in LB for 12 h at 37 °C, and the resulting pool was used as the inoculum for experimental infections.
2.10. T-4 like and T7-like phage selections on library
The library was interrogated with T4-like phage AH67C600_Q9 (Zhang et al., 2021) and a T7-like phage AH67C600_Q5 (isolated in this study), which was fully lytic when exposed to E. coli strain C600. A 100-µl volume of the log phase culture was inoculated into three 10-ml aliquots of LB broth. Then, T4-like or T7-like phage (OMOI) of were inoculated into two of the broths and into one used as a control that was prepared without phage selection but cultured in parallel under the same conditions. DNA is then extracted by using a TIANamp bacterial DNA kit (TianGen, Beijing, China) following the manufacturer's instruction. TraDIS-specific sequencing was performed on a HiSeq 2500 machine using 50-bp single-end reads to produce transposon-directed reads as described previously (Zhang et al., 2019).
2.11. TraDIS sequencing and bioinformatics analysis
To determine that insertion sites of each transposon that had randomly inserted into the genome of each mutant, a TraDIS library was prepared using specifically designed TraDIS adapters for the Tn5 transposon. This method increased the enrichment of genuine transposon-chromosome junctions by preventing hybridization of the reverse primer (Barquist et al., 2016). This ensured that the first 10 bp of every read consisted of transposon sequence and that the remaining sequence was downstream of where the transposon was inserted. These reads were then mapped using SMALT (WTSI). The reference strain was annotated with Prokka (Seemann, 2014) to identify the genes in the insertions were found.
A change in the number of reads that mapped to each gene between the control and the selections was measured using LogFC (log2 fold change), calculated from log counts per million using in-house TraDIS analysis scripts (https://github.com/sanger-pathogens/Bio-Tradis) (Barquist et al., 2016). LogFC was used as a measure of comparison of fold changes in read numbers compared to the LB control; ≥2 and ≤−2 were used as cutoff values for genes with different numbers of insertions. Only those that had a q value of <0.01 and a P value of <0.05 were included.
2.12. Recombinant DNA techniques
Deletion mutants were constructed using the bacteriophage lambda-red recombinase system. All oligonucleotide primers used in this study were purchased from GENEWIZ (Nanjing, China). All mutants were created using the E. coli C600 strain, and homologous recombination constructions were generated using PCR-purified products harboring a selectable antibiotic resistance gene (ARG) and 60-nucleotide homology extensions. The ARG was removed by transforming the pCP20 plasmid carrying a flippase. Mutants were confirmed by PCR and sequencing. On the other hand, the C600 and mutants (adjust the OD600 to 1) were challenged to decimal dilutions of phage AH67C600_Q5 and AH67C600_Q9 ranging from 109 to 102, and finally perform phage plaque reading.
2.13. Accession number(s)
The sequences of T7-like phage AH67C600_Q5 (MZ833439.1) and TraDIS sequencing (SAMN30170428, SAMN30170428 and SAMN30170428) have been submitted to the NCBI.
3. Results
3.1. Isolation of T7-like lytic phage AH67C600_Q5
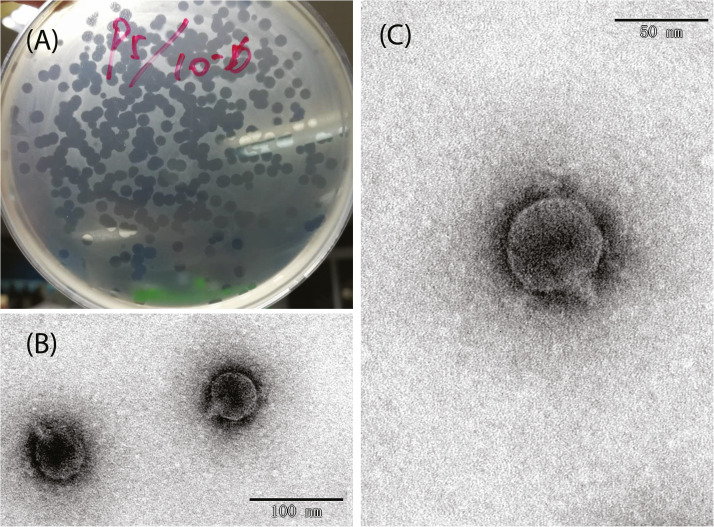
A lytic phage was isolated from a pig farm sewage treatment pond, China, named Enterobacteria phage AH67C600_Q5 that was able to lyse E. coli strain C600. Phage AH67C600_Q9 formed transparent, large, and round plaques of approximately 3.0 mm in diameter on a lawn of E. coli strain C600 (Fig. 1A). TEM observations of the phage revealed an icosahedral capsid with a cross diameter of about 50 nm, the longitudinal diameter of capsid was also about 50 nm, and the central tail spike length was about 4 nm (Fig. 1B and 1C). Phage AH67C600_Q5 showed a high similar to phage T7 and could be classified morphologically T7-like phage belonged to the genus Teseptimavirus; subfamily Studiervirinae; family Autographiviridae; class Caudoviricetes; phylum Uroviricota; kingdom Heunggongvirae; realm Duplodnaviria.
Fig. 1.
Morphological characteristics of lytic phage AH67C600_Q5. (A) Culture map of phage AH67C600_Q5 plaques in double-layer agar. (B) TEM images of phage AH67C600_Q5 in 100 nm vision. (C) TEM images of phage AH67C600_Q5 in 50 nm vision.
3.2. Characterization of T7-like lytic phage AH67C600_Q5
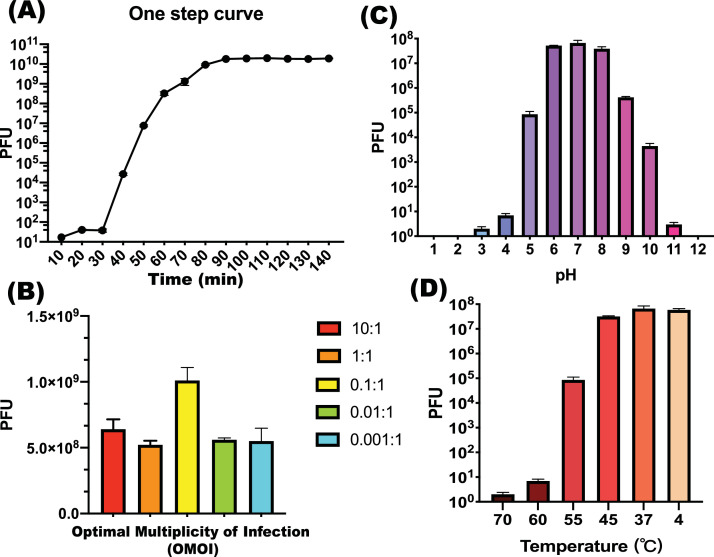
The one-step growth curve for the phage AH67C600_Q5 was analyzed by infecting the host during the exponential growth phase at a MOI of 10 to ensure complete culture lysis without bacterial regrowth. The phage AH67C600_Q5 had a latent period of about 30 min and a burst period of about 50 min (Fig. 2A). MOI ranging from 10 to 0.001 was tested to determine the optimal values for producing maximal phage titers. When the ratio of the phage AH67C600_Q5 and host E. coli strain C600 was 0.1, the titer was maximal at 9.6 × 108 PFU per mL and this OMOI was used to guide follow-up experiments (Fig. 2B). The pH and thermostability tolerance of the phage were analyzed. The phage titer was stable from pH 6 to 8 after 60 min incubation, whereas the titers decreased by 2–3 logs at pH 5 or 9 and by 4 logs at pH 10 for 60 min. The phage had little activity in the pH ranges of 1 to 4 and 11 to 12 (Fig. 3C). Incubations at 4 and 37 °C for 60 min did not alter the titers, whereas the titer gradually decreased with incubations at 45, 55 and 60 °C and at 70 °C the phage AH67C600_Q5 was inactive (Fig. 3D).
Fig. 2.
Physiological characteristics of T7-like lytic phage AH67C600_Q5. (A) One step curve of phage AH67C600_Q5. 10 to 30 min is the latent time and 30 to 80 min is the burst time. (B) Optimal MOI test. The phage AH67C600_Q5 achieved the highest titer after cultured while the MOI is 0.1:1 (0.96 × 109 PFU per mL) (C) pH stability tests. The phage AH67C600_Q5 is stable at the pH of 6 to 8, then sharply decreased with the pH increase and decrease. (D) Thermal stability tests. The phage AH67C600_Q5 had stable biological activity at temperatures of 4 to 45 °C, then sharply decreased with the temperatures increase.
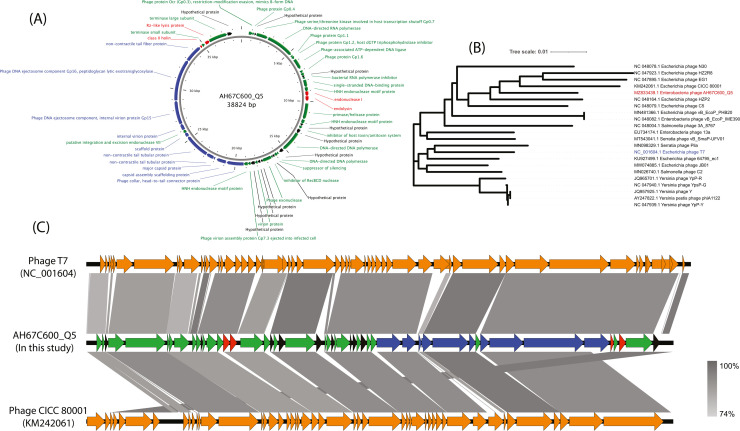
Fig. 3.
Bioinformatics analysis of T7-like lytic Phage AH67C600_Q5. (A) Annotation of the predicted genes of phage AH67C600_Q5. The genes located at upper right are replication and transcription -related and the structural proteins are concentrated in the lower left corner. (B) Phylogenetic analysis based on the large subunit of terminase for Enterobacteriaceae phage, demonstrating an independent branch for phage AH67C600_Q5. (C) Genome sequence alignment of phage T7, phage AH67C600_Q5, and phage CICC 80,001. The functional modules indicated by color, and similarities were shown in gray according to the scale on the bottom right corner, with green representing replication and transcription -related, red representing lytic-related gene, blue representing structure-related genes and black representing hypothetical genes.
3.3. Bioinformatics analysis of T7-like lytic phage AH67C600_Q5
Sequence for AH67C600_Q5 had been submitted to GenBank under accession number MZ833439.1. Genome of phage AH67C600_Q5 was 38,824 bp with a GC content of 48.38% with 50 predicted genes (Fig. 3A). Based on the amino acid sequences of the terminase large subunits, phylogenetic tree showed that phage AH67C600_Q5 had an independent branch and full taxonomy of this phage is Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Autographiviridae; Studiervirinae; Tseptimavirus (Fig. 3B). On the other hand, it differed from the prototype phage T7 but had a closer evolutionary relationship with phage CICC80001 (KM242061). In addition, the reference phage T7 (NC_001604) of this group and phage CICC80001, coupled with AH67C600_Q5, were further used to analysis the modular arrangement in structure (Fig. 3C). Results showed that the three genomes were almost the same but with a little difference that genes encoding structural related protein had a higher homology compared to the genes encoding replication and transcription protein.
In the structural related regions, a significant difference is genes 10A and 10B encoding phage major capsid protein of phage T7 is replaced by a gene encoding a small low-related major capsid protein (site: 21,977–22,168) of AH67C600_Q5. In the replication and transcription regions, we can find that phage AH67C600_Q5 acquired several genes, such as genes encoding HNH endonuclease motif protein (site: 11,759–12,187), putative integration and excision endonuclease VII (site: 25,748–26,062) and other hypothetical protein. It is worth noting that phage AH67C600_Q5 also lost several genes compared to phage T7.
3.4. Genes involved in phage susceptibility
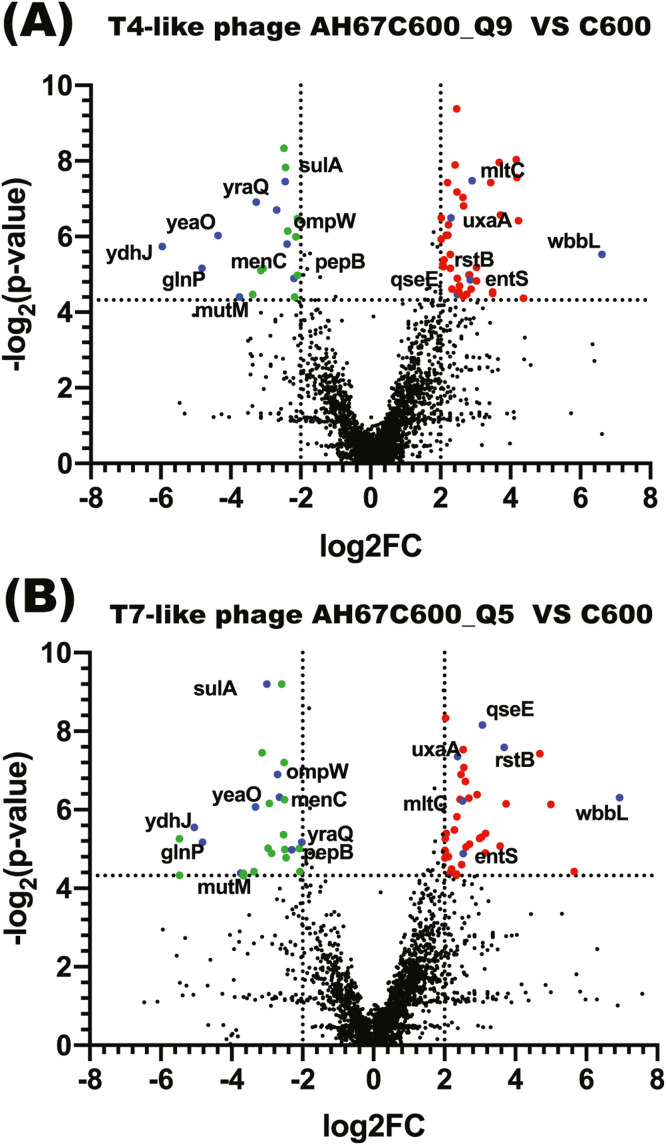
Under prey pressure of T4-like phage AH67C600_Q9, a total of 44 susceptibility genes that led to the mutants significantly outcompeting and expanding in the population of the E. coli strain C600 (≥ 4-fold increase or log fold change Log2FC ≥ 2 in the number of insertions) were identified (Fig. 4A). Under prey pressure of T7-like phage AH67C600_Q5, a total of 42 susceptibility genes were identified (Fig. 4B).
Fig. 4.
Volcano plot showing changes in prevalence of mutants in the mutant pool compared to the control during the addition of T4-like phage AH67C600_Q9 (A) and T7-like phage AH67C600_Q5 (B) selections, as shown by the relationship of log2 fold change in selection condition compared to the control (x axis), with the q value (y axis) indicating the false-discovery rate. Colored points show those shared genes in T4-like phage and T7-like phage group that a log2FC of greater than 2 (red) or less than 2 (green). Those shared genes are labeled with annotated gene name.
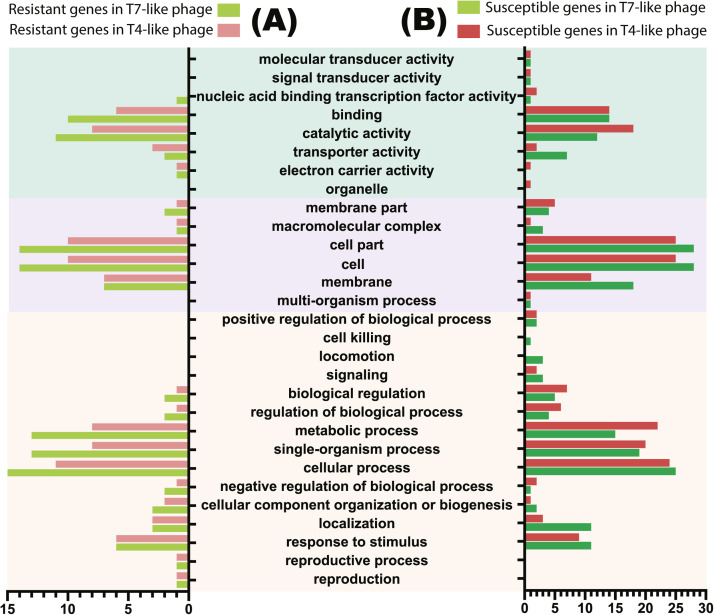
Gene ontology (Go) enrichment analysis of these genes showed that total of 27 GO terms were identified, among which 14 GO terms turn out to describe biological processes, 7 turn out to describe molecular functions and 6 turn out to describe cellular component (Fig. 5B). Furthermore, cell and cell part in cellular component and cellular process in biological processes were the most frequently annotated process. On the other hand, though GO terms had the similar trend under two kinds of phage pressure, there were still several differences. Genes were more likely to involve in metabolic process and catalytic activity in T4-like phage group than T7-like phage groups, whereas genes involved in localization, membrane and transporter activity had the opposite trend.
Fig. 5.
Gene ontology (Go) enrichment analysis of genes involved in phage resistance(A) and susceptibility (B) during the addition of T4-like phage AH67C600_Q9 (A) and T7-like phage AH67C600_Q5 (B) selections. The X-axis represents the number of genes involved in a GO terms, and the Y-axis represents the GO terms. Colored column shows genes number in different groups, with light green and green representing resistant and susceptible gene in T7-like group respectively, and light red and red representing resistant and susceptible gene in T4-like group respectively. Background color shows the GO terms involved in different process, with cyan representing cellular component, purple representing molecular functions and light yellow representing biological processes.
3.5. Genes involved in phage resistance
Under prey pressure of T4-like phage AH67C600_Q9, results showed that 19 genes had a significant reduction in the number of insertions compared to the control (log fold change Log2FC less than −2) (Fig. 4A). Under prey pressure of T7-like phage AH67C600_Q5, we showed that 28 genes had a significant reduction (Fig. 4B). These genes are likely to play a functional role in phage resistance, as their inactivation results in cells that are easier to be killed in the presence of T4-like or T7-like phage.
Gene ontology (Go) enrichment analysis of these genes showed that total of 21 GO terms were identified, among which 11 GO terms turn out to describe biological processes, 5 turn out to describe molecular functions and 5 turn out to describe cellular component (Fig. 5A). Similar to the results of genes involved in phage susceptibility, cell and cell part in cellular component and cellular process in biological processes were also the most frequently annotated process under the two kinds of phage pressure. Interestingly, we also observed the similar trend of GO terms but with a slight difference that genes in T7-like phage groups were more incline to be enriched in several cellular and metabolic process than T4-like phage group.
3.6. Shared genes associated with phage susceptibility and resistance
There were six genes involved in susceptibility were co-selected under two kinds of phage pressure (Fig. 4). Two genes QseE and RstB encoding the similar two-component system sensor histidine kinase (HKs), QseE/QseF system and RstB/RstA system, showed an increase in the number of insertions. Gene wbbL encoding a N-acetylglucosaminyl-diphospho-decaprenol l-rhamnosyltransferase also showed an increase in the number of insertions (more than 6 Log2FC). Gene mltC encoding membrane-bound lytic murein transglycosylase, gene uxaA encoding altronate dehydratase and gene entS encoding enterobactin transporter also showed an increase in the number of insertions.
There were nine genes involved in resistance were co-selected under two kinds of phage pressure. Gene ydhJ encoding a secretion protein showed a dramatic decrease in the number of insertions (less than 5 Log2FC). Strangely, gene ompW encoding outer membrane protein, unlike the gene ompC encoding popular outer membrane protein exhibiting important role in susceptibility to T4 phage, showed a resistance to the two kinds of phage. The other seven genes with the reduction in the number of insertions were involved in different functions, including the glnP, mutM, yeaO, menC, pepB, yraQ and sulA.
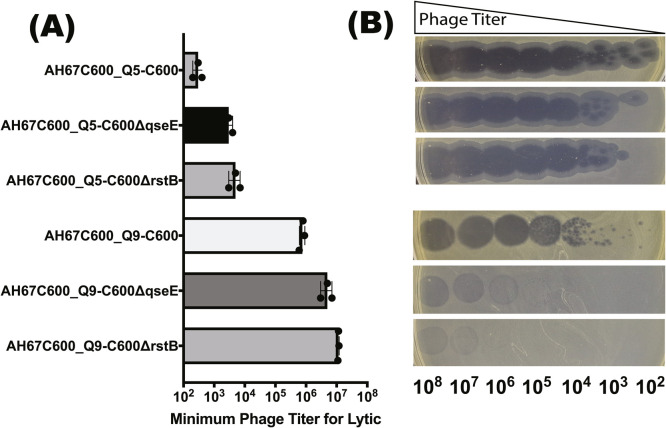
3.7. Confirmation of genes associated with phage susceptibility and resistance
The shared genes (QseE and RstB) associated with phage susceptibility were selected to confirm the results of TraDIS strategy. Results showed that the mutant strains showed resistance to phage AH67C600_Q5 and phage AH67C600_Q9, but with some differences (Fig. 6). Under pressure of phage AH67C600_Q5, though mutant strains (C600ΔqseE and C600ΔrstB) showed a resistance, this resistance is very weak with a decrease of about 1 log (10). On the contrary, under pressure of phage AH67C600_Q9, we found that mutant strains (C600ΔqseE and C600ΔrstB) showed a huge resistance with a decrease of about 3–4 log (10).
Fig. 6.
Changes of phage host range and efficiency of plating. (A) Minimum phage titer of three stains (E. coli strain C600, E. coli strain C600ΔqseE and E. coli strain C600ΔrstB) for two test phages (AH67C600_Q5 and AH67C600_Q9). (B) Spot assay phenotypes.
4. Discussion
With alarming rise in infections caused by multidrug-resistant (MDR) bacteria, the therapeutic use of phages has started to come back to the modern medicine. Regrettably, the success of phage therapy is largely limited by the development of phage resistance by its host bacteria. Also, the complexity of phage-bacteria interactions is still not fully interpreted, and these could be a barrier in the development of successful phage-based therapies. Fortunately, a huge number of studies have stated the safety of phage therapy in clinical studies and trials (Pirnay et al., 2021), thereby suggesting a promising outcome underlying the different phage application. Considering that addressing and tackling phage resistance can lead to improved treatment outcomes (Egido et al., 2022), high-throughput screening and characterizing phage-host interaction determinants involved in phage susceptibility and resistance will provide new insights into the coevolution of phage and their hosts, and ultimately be of great value in designing better phage therapeutic treatments and tools for precision phage engineering.
TraDIS is a genome-wide mutagenesis methodology combining phenotypic fitness selection that has proven useful in identifying genes for their involvement in survival under phage pressure (Pickard et al., 2013). In this study, TraDIS was used to investigate phage-host interactions of two high lytic T4-like and T7-like phages and an E. coli C600 strain, thereby uncovering novel genes involved in phage susceptibility and resistance. The lytic T4-like phage, named Enterobacteria phage AH67C600_Q9, has been documented in previous study (Zhang et al., 2021). In this study, a lytic T7-like phage, named Enterobacteria phage AH67C600_Q5, is further isolated and characterized. Thoroughly understanding of the molecular interactions as well as uncovering determinants governing the two kinds of phage infection of E. coli might aid the development of phage therapy in the future. Under phages prey pressure, a total of 44 susceptibility genes in T4-like group and 42 susceptibility genes in T7-like group were identified.
It worth noting that the go enrichment analysis showed that these susceptible genes were located at the similar pathways, and they were inclined to be enriched in several fixed process, such as cellular and metabolic process in biological processes. Usually, the phage lytic life cycle can be accomplished throughout six steps: adsorption, DNA injection, replication, transcription-translation, assembly, and lysis (Dy et al., 2014). The susceptible genes enriched in the biological processes probably imply that these genes played an important role in phage replication, transcription, and translation. Interestingly, go enrichment analysis of these resistant genes showed that they were also inclined to be enriched in cellular and metabolic process in biological processes. In the structural related regions, apart from the difference of phage major capsid protein, we can find that the replication and transcription regions of phage AH67C600_Q5 showed a significant difference, such as acquiring genes encoding HNH endonuclease motif protein. We all know that HNH proteins are a very common proteins generally associated with nuclease activity. Also, evidence had documented that HNH proteins were required for the specific endonuclease activity of phage HK97 terminase (Kala et al., 2014). Thus, whether the additional acquisition of HNH endonuclease motif protein implies that the assembly ability of AH67C600_Q5 is improved, thus stimulating the host reaction.
Though the application of phage cocktail greatly improves the killing effect of MDR pathogens (Cieplak et al., 2018), the huge number of phage applications could lead to the uncertain result of phage-bacterial interaction. Also, these diversity phages could induce the occurrence of multi-resistance bacteria that immune to different phages (Rohde et al., 2018; Yuan et al., 2019). Nowadays, there is a growing body of literature showing that phage agent (e.g., antibiotic) synergistic showed a promising future in facilitating the kill and eradication of MDR pathogens (Duarte et al., 2021; Liu et al., 2021). Possibility, the shared genes function in signatures would be of great value as potential adjuvant in facilitating phage-agent synergistic therapy (Oyejobi et al., 2022).
Two shared genes QseE and RstB associated with phage susceptibility were detected and confirmed by the gene knocking-out experiment. They all encoded the similar two-component system sensor HKs, QseE/QseF system and RstB/RstA system. Usually, bacterial pathogens sense external cues through HKs to activate expression of related genes. These systems comprise the main sensory mechanism in bacteria, in which QseE is a second bacterial adrenergic receptor that gauges the stress signals Epi, sulfate, and phosphate (Reading et al., 2009). On the other hand, study had showed that silencing of RstB/RstA could lead to damage in adhesion, biofilm production, motility, hemolysis, and virulence (Huang et al., 2018). Though no researches presently had stated the relationship among QseE/QseF, RstB/RstA and phages, our results showed that disruption of QseE and RstB resulting in a decreasing of infection ability of phages. With this knowledge, it may be possible to use phages to target these genes, then based on the fitness-cost, finally making them more tractable to be eliminated when infect the human and animal host (Mangalea and Duerkop, 2020).
Gene wbbL encoding a N-acetylglucosaminyl-diphospho-decaprenol l-rhamnosyltransferase had a large fold increase to the control (more than 6 Log2FC), indicating that it clearly plays an important role in susceptibility to the T4-like or T7-like phage. Evidence had documented that deletion of a 209 kb genetic fragment of E. coli genome, genetic regions from ompC to wbbL, conferred resistance to an assortment of infectious phages. Meanwhile, restoration of the OmpC protein by plasmid-mediated complementation did not completely restore the susceptibility to cognate phages, implying gene wbbL was essential for successful infection (Tanji et al., 2008). When screening the genes content similarity of phage AH67C600_Q5 and phage T7, results showed that, though genes encoding structural related protein were almost the same, genes 10A and 10B encoding phage major capsid protein (MCP) of phage T7 is replaced by a small low-related MCP of AH67C600_Q5. Evidence had shown that MCP sequences is an important implication of capsid morphology of different subgroups of the T4 or T7 superfamily (Comeau and Krisch, 2008). Meanwhile, the MCP PCR assay was also shown to be a simple method for identifying new lytic phages to a certain group and have the potential to become the biocontrol candidates (Born et al., 2019). This difference may imply that the host surface receptor genes, such as wbbL, probably play a vital role under pressure of phage AH67C600_Q5 and AH67C600_Q9 with an evolutionary difference to the traditional phage T4 and T7. Gene mltC encoding membrane-bound lytic murein transglycosylase function in catalyzing the nonhydrolytic cleavage of the glycan strands of cell wall in Gram-negative bacterial (Artola-Recolons et al., 2014). A study had shown that lytic transglycosylases MltC can assist in separating of daughter cell in Vibrio cholerae (Weaver et al., 2019), showing that our lytic T4-like and T7-like phage might hijack and use this gene for their own needs while inhibit cell segregation, whereas the detailed mechanism needs to be further clarified in the future.
Although only 19 genes in T4-like group and 28 genes in T7-like group involved in the resistance to phage, there were 9 genes were co-selected, showing a broad spectrum of resistance when confronted with diverse phages. Interestingly, unlike the gene ompC encoding popular outer membrane protein exhibiting important role in susceptibility to T4 phage (Mutalik et al., 2020), gene ompW encoding outer membrane protein showed a resistance to the two kinds of phage. As a β-barrel protein, previous studies had demonstrated that ompW is involved in protection of bacteria against various environmental stress, such as osmosis, antibiotics, oxidation, and the unavailability of nutrients and oxygen(Morales et al., 2012). Also, deletion of ompW exhibited a significantly increased phagocytosis rate (Wu et al., 2013). The significant reduction of ompW in the insertion number might imply that this gene is also play an important role in resistance to our T4- and T7-like phages. Gene ydhJ encoding a HlyD family secretion protein had the largest fold decrease to the control (less than 5 Log2FC), whereas there was almost no literature further investigate the detailed function under phage pressure. Impressively, gene mutM showed a high decrease in the number of insertions. Protein MutM is a DNA repair glycosylase, which function in removing DNA damage generated from oxidative stress and then preventing mutations and genomic instability (Landova and Silhan, 2020). The host greatly decreased after Tn5 insertion of this gene implied that gene mutM increased the resistance ability throughout stabilize the genome to prevent mutations when facing diversity phages. More direct evidences were still needed to verify our conjecture
5. Conclusions
In summary, we used a TraDIS strategy to successfully identify susceptibility and resistant genes important for the E. coli strain of both high lytic T4-like and T7-like phage infection, of which 6 shared genes important for the susceptibility, and 9 shared genes important for the resistance. Nowadays, MDR pathogens invoked the renewed interest in phage therapy. However, the phage applications could lead to the uncertain result of phage-bacterial interaction and inevitably induced the occurrence of multi-resistance bacteria to diversity phages. In this study, application of TraDIS sequencing could provide a glimpse of phage susceptibility and resistance landscape. Further analysis of shared genes, we could focus on the shared these genes encoding function signatures to different kind of lytic phages identify potential agents to aid phage killing of MDR pathogens, which will greatly improve the therapeutic effect. Additionally, although the candidate genes were identified in this study, their underlying mechanisms remain unknown. Future studies will, it is hoped, provide a more comprehensive understanding of the mechanisms involved in susceptibility and resistant. If these findings could be proven, it would be great valuable in improving the phage therapy outcome and provide a foundation in fighting with microbial resistance.
CRediT authorship contribution statement
Mianzhi Wang: Conceptualization, Methodology, Software, Investigation, Formal analysis, Writing – original draft. Heng Zhu: Data curation. Jingyi Wei: Visualization, Investigation. Li Jiang: Resources, Supervision. Lei Jiang: Software, Validation. Ziyi Liu: Visualization, Software. Ruichao Li: Visualization, Software. Zhiqiang Wang: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (32102717), Natural Science Foundation of the Higher Education Institutions of Jiangsu Province, China (21KJB230010), Jiangsu Agricultural Science and Technology Innovation Fund (CX(21)2010) and Postgraduate Reasearch & Practice Innovation Program of Jiangsu Province (KYCX22_3550).
Contributor Information
Mianzhi Wang, Email: wangmz@yzu.edu.cn.
Zhiqiang Wang, Email: zqwang@yzu.edu.cn.
Data availability
Data will be made available on request.
References
- Allue-Guardia A., Nyong E.C., Koenig S.S.K., Vargas S.M., Bono J.L., Eppinger M. Closed genome sequence of Escherichia coli K-12 group strain C600. Microbiol Resour Announc. 2019;8(2) doi: 10.1128/MRA.01052-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artola-Recolons C., Lee M., Bernardo-García N., Blázquez B., Hesek D., Bartual S.G., Mahasenan K.V., Lastochkin E., Pi H., Boggess B., Meindl K., Usón I., Fisher J.F., Mobashery S., Hermoso J.A. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem. Biol. 2014;9(9):2058–2066. doi: 10.1021/cb500439c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquist L., Mayho M., Cummins C., Cain A.K., Boinett C.J., Page A.J., Langridge G.C., Quail M.A., Keane J.A., Parkhill J. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics. 2016;32(7):1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black L.W. Old, new, and widely true: the bacteriophage T4 DNA packaging mechanism. Virology. 2015;479-480:650–656. doi: 10.1016/j.virol.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born Y., Knecht L.E., Eigenmann M., Bolliger M., Klumpp J., Fieseler L. A major-capsid-protein-based multiplex PCR assay for rapid identification of selected virulent bacteriophage types. Arch. Virol. 2019;164(3):819–830. doi: 10.1007/s00705-019-04148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomason J.A., 3rd, Stevens R., Vonstein V., Wattam A.R., Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P., Chen Y., Parsons C., Brown E., Loessner M.J., Shen Y., Kathariou S. Whole genome sequence analysis of phage-resistant listeria monocytogenes serotype 1/2a strains from turkey processing plants. Pathogens. 2021;10(2) doi: 10.3390/pathogens10020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.S., Paterson D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020;73(6):329–364. doi: 10.1038/s41429-020-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S.R., Hendrix R.W. Bacteriophage lambda: early pioneer and still relevant. Virology. 2015;479-480:310–330. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castledine M., Padfield D., Sierocinski P., Soria Pascual J., Hughes A., Makinen L., Friman V.P., Pirnay J.P., Merabishvili M., de Vos D., Buckling A. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. Elife. 2022:11. doi: 10.7554/eLife.73679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplak T., Soffer N., Sulakvelidze A., Nielsen D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes. 2018:1–19. doi: 10.1080/19490976.2018.1447291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau A.M., Krisch H.M. The capsid of the T4 phage superfamily: the evolution, diversity, and structure of some of the most prevalent proteins in the biosphere. Mol. Biol. Evol. 2008;25(7):1321–1332. doi: 10.1093/molbev/msn080. [DOI] [PubMed] [Google Scholar]
- Dion M.B., Oechslin F., Moineau S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020;18(3):125–138. doi: 10.1038/s41579-019-0311-5. [DOI] [PubMed] [Google Scholar]
- Duarte A.C., Fernandez L., De Maesschalck V., Gutierrez D., Campelo A.B., Briers Y., Lavigne R., Rodriguez A., Garcia P. Synergistic action of phage phiIPLA-RODI and lytic protein CHAPSH3b: a combination strategy to target Staphylococcus aureus biofilms. NPJ Biofilms Microbiomes. 2021;7(1):39. doi: 10.1038/s41522-021-00208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy R.L., Richter C., Salmond G.P., Fineran P.C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 2014;1(1):307–331. doi: 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- Egido J.E., Costa A.R., Aparicio-Maldonado C., Haas P.J., Brouns S.J.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022;46(1) doi: 10.1093/femsre/fuab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Altae-Tran H., Bohning F., Makarova K.S., Segel M., Schmid-Burgk J.L., Koob J., Wolf Y.I., Koonin E.V., Zhang F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science. 2020;369(6507):1077–1084. doi: 10.1126/science.aba0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse S., Rajaure M., Wall E., Johnson J., Bliskovsky V., Gottesman S., Adhya S. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. MBio. 2020;11(1) doi: 10.1128/mBio.02530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Xu W., Su Y., Zhao L., Yan Q. Regulatory role of the RstB-RstA system in adhesion, biofilm production, motility, and hemolysis. Microbiologyopen. 2018;7(5):e00599. doi: 10.1002/mbo3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kala S., Cumby N., Sadowski P.D., Hyder B.Z., Kanelis V., Davidson A.R., Maxwell K.L. HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. U.S.A. 2014;111(16):6022–6027. doi: 10.1073/pnas.1320952111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A.M., Waddell T.E., Lingohr E., Johnson R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009;501:69–76. doi: 10.1007/978-1-60327-164-6_7. M.A. [DOI] [PubMed] [Google Scholar]
- Landova B., Silhan J. Conformational changes of DNA repair glycosylase MutM triggered by DNA binding. FEBS Lett. 2020;594(18):3032–3044. doi: 10.1002/1873-3468.13876. [DOI] [PubMed] [Google Scholar]
- Lavigne, R., Loessner, M.J., 2021. Editorial overview: phage therapy in the 21st century - inspired by biotechnology! Current opinion in biotechnology. [DOI] [PubMed]
- Liang G., Bushman F.D. The human virome: assembly, composition and host interactions. Nat. Rev. Microbiol. 2021;19(8):514–527. doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhao Y., Hayes A., Hon K., Zhang G., Bennett C., Hu H., Finnie J., Morales S., Shearwin L., Psaltis A.J., Shearwin K., Wormald P.J., Vreugde S. Overcoming bacteriophage insensitivity in Staphylococcus aureus using clindamycin and azithromycinat subinhibitory concentrations. Allergy. 2021 doi: 10.1111/all.14883. [DOI] [PubMed] [Google Scholar]
- Lu L., Cai L., Jiao N., Zhang R. Isolation and characterization of the first phage infecting ecologically important marine bacteria Erythrobacter. Virol J. 2017;14(1):104. doi: 10.1186/s12985-017-0773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalea M.R., Duerkop B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020;88(7) doi: 10.1128/IAI.00926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwitz P., Lood C., Olszak T., van Noort V., Lavigne R., Drulis-Kawa Z. Genome-driven elucidation of phage-host interplay and impact of phage resistance evolution on bacterial fitness. ISME J. 2021 doi: 10.1038/s41396-021-01096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales E.H., Calderón I.L., Collao B., Gil F., Porwollik S., McClelland M., Saavedra C.P. Hypochlorous acid and hydrogen peroxide-induced negative regulation of Salmonella enterica serovar Typhimurium ompW by the response regulator ArcA. BMC Microbiol. 2012;12(1):63. doi: 10.1186/1471-2180-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.J.L., Ikuta K.S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., Han C., Bisignano C., Rao P., Wool E., Johnson S.C., Browne A.J., Chipeta M.G., Fell F., Hackett S., Haines-Woodhouse G., Kashef Hamadani B.H., Kumaran E.A.P., McManigal B., Agarwal R., Akech S., Albertson S., Amuasi J., Andrews J., Aravkin A., Ashley E., Bailey F., Baker S., Basnyat B., Bekker A., Bender R., Bethou A., Bielicki J., Boonkasidecha S., Bukosia J., Carvalheiro C., Castañeda-Orjuela C., Chansamouth V., Chaurasia S., Chiurchiù S., Chowdhury F., Cook A.J., Cooper B., Cressey T.R., Criollo-Mora E., Cunningham M., Darboe S., Day N.P.J., De Luca M., Dokova K., Dramowski A., Dunachie S.J., Eckmanns T., Eibach D., Emami A., Feasey N., Fisher-Pearson N., Forrest K., Garrett D., Gastmeier P., Giref A.Z., Greer R.C., Gupta V., Haller S., Haselbeck A., Hay S.I., Holm M., Hopkins S., Iregbu K.C., Jacobs J., Jarovsky D., Javanmardi F., Khorana M., Kissoon N., Kobeissi E., Kostyanev T., Krapp F., Krumkamp R., Kumar A., Kyu H.H., Lim C., Limmathurotsakul D., Loftus M.J., Lunn M., Ma J., Mturi N., Munera-Huertas T., Musicha P., Mussi-Pinhata M.M., Nakamura T., Nanavati R., Nangia S., Newton P., Ngoun C., Novotney A., Nwakanma D., Obiero C.W., Olivas-Martinez A., Olliaro P., Ooko E., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik V.K., Adler B.A., Rishi H.S., Piya D., Zhong C., Koskella B., Kutter E.M., Calendar R., Novichkov P.S., Price M.N., Deutschbauer A.M., Arkin A.P. High-throughput mapping of the phage resistance landscape in E. coli. PLoS Biol. 2020;18(10) doi: 10.1371/journal.pbio.3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega F.L., Vlot M., de Jonge P.A., Dreesens L.L., Beaumont H.J.E., Lavigne R., Dutilh B.E., Brouns S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018;16(12):760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- Oyejobi G.K., Xiong D., Shi M., Zhang X., Yang H., Xue H., Ogolla F., Wei H. Genetic signatures from adaptation of bacteria to lytic phage identify potential agents to aid phage killing of multidrug-resistant acinetobacter baumannii. J. Bacteriol. 2022;204(3) doi: 10.1128/jb.00593-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D., Kingsley R.A., Hale C., Turner K., Sivaraman K., Wetter M., Langridge G., Dougan G. A genomewide mutagenesis screen identifies multiple genes contributing to Vi capsular expression in Salmonella enterica serovar Typhi. J. Bacteriol. 2013;195(6):1320–1326. doi: 10.1128/JB.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirnay J.P., Ferry T., Resch G. Recent progress towards the implementation of phage therapy in Western medicine. FEMS Microbiol. Rev. 2021 doi: 10.1093/femsre/fuab040. [DOI] [PubMed] [Google Scholar]
- Qimron U., Marintcheva B., Tabor S., Richardson C.C. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2006;103(50):19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading N.C., Rasko D.A., Torres A.G., Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106(14):5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recacha E., Machuca J., Alba P.D.d., Ramos-Güelfo M., Docobo-Pérez F., Rodriguez-Beltrán J., Blázquez J., Pascual A., Rodríguez-Martínez J.M. Quinolone resistance reversion by targeting the SOS response. MBio. 2017;8(5):e00971. doi: 10.1128/mBio.00971-17. -00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde C., Resch G., Pirnay J.-.P., Blasdel B., Debarbieux L., Gelman D., Górski A., Hazan R., Huys I., Kakabadze E., Łobocka M., Maestri A., Almeida G., Makalatia K., Malik D., Mašlaňová I., Merabishvili M., Pantucek R., Rose T., Štveráková D., Van Raemdonck H., Verbeken G., Chanishvili N. Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses. 2018;10(4):178. doi: 10.3390/v10040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T., Strathdee S. Treat phage like living antibiotics. Nature Microbiol. 2020;5(3):391–392. doi: 10.1038/s41564-019-0666-4. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Tanji Y., Hattori K., Suzuki K., Miyanaga K. Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl. Environ. Microbiol. 2008;74(14):4256–4263. doi: 10.1128/AEM.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuretzbacher U., Outterson K., Engel A., Karlen A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020;18(5):275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D., Kropinski A.M., Adriaenssens E.M. A roadmap for genome-based phage taxonomy. Viruses. 2021;13(3) doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Sun Y., Zeng Z., Wang Z. Metagenomics of wastewater phageome identifies an extensively cored antibiotic resistome in a swine feedlot water treatment environment. Ecotoxicol. Environ. Saf. 2021;222 doi: 10.1016/j.ecoenv.2021.112552. [DOI] [PubMed] [Google Scholar]
- Wang M., Xiong W., Liu P., Xie X., Zeng J., Sun Y., Zeng Z. Metagenomic insights into the contribution of phages to antibiotic resistance in water samples related to swine feedlot wastewater treatment. Front. Microbiol. 2018;9:2474. doi: 10.3389/fmicb.2018.02474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wan M., Huang R., Zhang Y., Xie Y., Wei Y., Ahmad M., Wu D., Hong Y., Deng Z., Chen S., Li Z., Wang L. SspABCD-SspFGH constitutes a new type of DNA phosphorothioate-based bacterial defense system. MBio. 2021;12(2) doi: 10.1128/mBio.00613-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts G. Phage therapy: revival of the bygone antimicrobial. The Lancet. 2017;390(10112):2539–2540. doi: 10.1016/S0140-6736(17)33249-X. [DOI] [PubMed] [Google Scholar]
- Weaver A.I., Jimenez-Ruiz V., Tallavajhala S.R., Ransegnola B.P., Wong K.Q., Dorr T. Lytic transglycosylases RlpA and MltC assist in Vibrio cholerae daughter cell separation. Mol. Microbiol. 2019;112(4):1100–1115. doi: 10.1111/mmi.14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.B., Tian L.H., Zou H.J., Wang C.Y., Yu Z.Q., Tang C.H., Zhao F.K., Pan J.Y. Outer membrane protein OmpW of Escherichia coli is required for resistance to phagocytosis. Res. Microbiol. 2013;164(8):848–855. doi: 10.1016/j.resmic.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Yang M., Liang Y., Huang S., Zhang J., Wang J., Chen H., Ye Y., Gao X., Wu Q., Tan Z. Isolation and Characterization of the Novel Phages vB_VpS_BA3 and vB_VpS_CA8 for Lysing Vibrio parahaemolyticus. Front. Microbiol. 2020;11:259. doi: 10.3389/fmicb.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wang L., Li X., Tan D., Cong C., Xu Y. Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii. Virus Res. 2019;272 doi: 10.1016/j.virusres.2019.197734. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chen X., Nolan L.K., Zhang W., Li G. Identification of host adaptation genes in extraintestinal pathogenic escherichia coli during infection in different hosts. Infect. Immun. 2019;87(12) doi: 10.1128/IAI.00666-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., He X., Shen S., Shi M., Zhou Q., Liu J., Wang M., Sun Y. Effects of the newly isolated T4-like phage on transmission of plasmid-borne antibiotic resistance genes via generalized transduction. Viruses. 2021;13(10) doi: 10.3390/v13102070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.